Abstract
Eco-evolutionary dynamics of the gut microbiota at macroscale level, i.e. in across species comparisons, are largely driven by ecological variables and host genotype. The repeated explosive radiations of African cichlid fishes in distinct lakes, following a dietary diversification in a context of reduced genetic diversity, provide a natural setup to explore convergence, divergence and repeatability in patterns of microbiota dynamics as a function of the host diet, phylogeny and environment. Here we characterized by 16S rRNA amplicon sequencing the gut microbiota of 29 cichlid species from two distinct lakes/radiations (Tanganyika and Barombi Mbo) and across a broad dietary and phylogenetic range. Within each lake, a significant deviation between a carnivorous and herbivorous lifestyle was found. Herbivore species were characterized by an increased bacterial taxonomic and functional diversity and converged in key compositional and functional community aspects. Despite a significant lake effect on the microbiota structure, this process has occurred with remarkable parallels in the two lakes. A metabolic signature most likely explains this trend, as indicated by a significant enrichment in herbivores/omnivores of bacterial taxa and functions associated to fiber degradation and detoxification of plant chemical compounds. Overall, compositional and functional aspects of the gut microbiota individually and altogether validate and predict main cichlid dietary habits, suggesting a fundamental role of gut bacteria in cichlid niche expansion and adaptation.
Introduction
Trophic adaptation has driven most of animal phenotypic diversity, ranging from morphology to behavior, and has favored the occupation of novel or unoccupied niches, enhancing the speciation process (e.g. adaptive radiations) (Muschick et al 2012). Besides efficient food harvesting and intake, optimization of nutrient extraction and absorption, with limited waste (digestive efficiency), has been equally crucial to animal trophic adaptation.
In both vertebrates and invertebrates, nutrient metabolism and assimilation largely occur in the intestine as a cooperative process between host cells and the gut microbiota (Karasov and Martinez del Rio 2007). Gut bacteria, in particular, can increase energy transformation from diet and facilitate nutrient absorption (Ayayee et al 2016, Larsen et al 2016). Evidence from humans and mice clearly indicates that breaking this symbiotic equilibrium results in several host nutritional deficiencies and metabolic disorders (Baothman et al 2016, Ridaura et al 2013). While bacterial contribution to host biology goes far beyond processing of nutrients, their metabolic function has certainly played a key role in shaping this symbiosis in the first steps (Hansen and Moran 2014, Russell et al 2009).
Carbon dietary needs in animals are met through either an animal-rich diet (proteins and simple polysaccharides), a plant-rich diet (fibers or structural polysaccharides), or then a mix (Edward Stevens and Hume 2004). Adaptation of gut morphology best exemplifies such metabolic differences, with gut length and complexity typically increasing with increased fiber consumption (Davis et al 2013, Karasov and Martinez del Rio 2007, Karasov et al 2011, Wagner et al 2009). As eukaryotic cells cannot break down complex polysaccharides, animals heavily rely on bacterial fermentation for this task (Flint et al 2012). Moreover, as a plant-based diet is nutritionally unbalanced and typically comes with noxious chemicals, gut microbes are also known to supply nutrients (Karasov and Martinez del Rio 2007) and degrade toxins produced by plants in response to herbivory, as recently documented in insects and few vertebrates (Hammer and Bowers 2015, Kohl and Dearing 2012, Kohl et al 2014, Kohl et al 2016b, Pennisi 2017). The increased gut microbial diversity observed in terrestrial herbivores compared to carnivores clearly underlines these metabolic differences (Kohl and Dearing 2012, Muegge et al 2011).
At an evolutionary timescale, quantifying how much of the extant gut microbiota variation seen across species is attributable to ecological and environmental factors, rather than to the host genotype, remains a challenge. Unclear is also whether this variability is largely adaptive. At present, the gut microbiota literature on vertebrates covers both examples of host phylogeny and ecology-driven microbial features (Delsuc et al 2014, Hird et al 2015, Muegge et al 2011, Soverini et al 2016). Still, the large interdependency between phylogeny and ecology currently represents a major challenge for assessing microbial community adaptation in vertebrate evolution (Spor et al 2011). In a few case studies where the two factors uncouple, such as in pandas and whales (Sanders et al 2015, Xue et al 2015), their individual strength emerged, with clear examples of microbial phylogenetic inertia (i.e. the resemblance of microbial communities according to close ancestry in contradiction with the trophic niche), indicating a tight link between gut morphology/physiology and microbiota.
Vertebrate adaptive radiations provide an especially interesting system to explore the dynamics of the gut microbiota resulting from the trade-off between host constraints and feeding ecology at a macroscale level, i.e. in across-species comparisons. In such systems divergence of trophic niches during host speciation occurred rapidly, often within an ecological timescale, and thus largely uncoupled from the host genetic adaptation (Brawand et al 2014).
The great radiations of African cichlid fishes offer a particularly powerful study system in this respect. The majority of cichlid species are found in the three great lakes from East Africa (Victoria, Malawi and Tanganyika), but they also occur in numerous smaller lakes and rivers, providing several examples of adaptive radiations at different evolutionary time-scales (Day et al 2008, Genner et al 2007). Cichlids have occupied most available trophic niches, some species as highly specialized feeders, others as opportunist feeders (Muschick et al 2012, Salzburger 2009). Their outstanding trophic morphology (including jaws, teeth, body and gut) largely correlates with the feeding ecology, regardless of species phylogenetic relatedness, hence supporting a process of “ecological speciation” (Muschick et al 2012). Similar ecomorphs have repeatedly evolved through convergent evolution, due to the discrete number of trophic niches available and/or to a putative ontogenetic-driven canalization of shape variance.
Given the high genetic similarity among cichlid species (Brawand et al 2014), examples of microbiota phylogenetic inertia are to be expected. Yet, as is the case for morphological traits, we can also imagine that optimization of the gut microbiota composition in function of diet can increase the host fitness and contribute to its niche specialization, by enhancing the efficiency in extracting energy from food. If indeed the gut microbiota acts predominantly as a trophic trait then we might state a few predictions: 1) the gut microbiota should largely mirror the adaptation of the other eco-morphological traits of cichlids, and examples of divergence and convergence of this trait should occur across species as a function of diet rather than phylogeny; 2) herbivores should present a larger taxonomic and functional bacterial diversity than strict carnivores, consistently with their additional needs for fiber degradation, potential nutrient supplementation and chemical detoxification; 3) functional profiles of herbivores should reflect enrichment in the above bacterial functions; 4) if these patterns are deterministic and therefore predictable, then they should replicate across independent radiations.
Here we set to test the above predictions using wild specimens from two cichlid radiations: the oldest radiation from lake Tanganyika (Zambia, 9-12 Myr), and the recent monophyletic radiation from lake Barombi Mbo (Cameroon, 0.5–1 Myr) (Genner et al 2007). The two radiations diverged around 55.14 Mya from the last common ancestor (Matschiner et al 2016). Lake Tanganyika harbors the most phenotypically and genetically diverse african species assemblage, comprising around 250 species currently subdivided into 12-16 tribes (Meyer et al 2014). Lake Barombi Mbo is a volcanic crater lake, approximately 2-km in diameter, hosting a small sympatric radiation of 11 cichlid species (Schliewen et al 1994, Trewavas et al 1972).
Both lakes offer multiple species representatives of main diet categories, thus providing a natural setup to explore convergence, divergence and repeatability of patterns in microbiota eco-evolutionary diversification.
Materials and Methods
Sampling and diet
We sampled 19 cichlid species from lake Tanganyika (Zambia) encompassing major tribes and diets, and 10 of the 11 cichlid species from lake Barombi Mbo (Cameroon) (115 specimens, Supplementary Table 1).
Given some inconsistencies in calling dietary categories across literature, here we used a standard approach based on food preferences, as inferred by published gut content data from (Muschick et al 2012) for all Tanganyika species and from (Trewavas et al 1972) and unpublished data for Barombi Mbo. We discriminated among carnivores (C), herbivores (H), omnivores (O), planktivores (P) and specialized insectivores (I) and scale-eaters (SC) (Supplementary Table 1). Specifically, carnivory indicates a restricted feeding on animal sources (including mollusks, crustaceans, fishes and insects) to the virtual exclusion of vegetable matter; omnivory, the regular inclusion of both animal and vegetable matter (including algae and plants), with no overfeeding on the latest; herbivory, a disproportional feeding on algae and plants. In this respect, the definition of herbivory used in this study is not strict. For Barombi Mbo, which lacks specialized herbivores, we approximated this category to the two debris/algae feeding species within the genus Sarotherodon. Diet assignment was validated by stable isotopes, trophic morphologies (including gut lengths) and behavior (Hata et al 2015, Hata et al 2014, Muschick et al 2012, Muschick et al 2014, Sturmbauer et al 1992, Wagner et al 2009, Trewavas et al 1972). Species were further classified according to whether they regularly include or not vegetable material in their diet (fiber and protein-associated categories F and Pr, respectively), thus largely grouping herbivores, omnivores and phytoplanktivores against carnivores. For diet details see Supplementary Table 1 and Information.
DNA extractions and Illumina Sequencing
DNA was extracted from individual guts following the protocol described in (Baldo et al 2015). The region V3-V4 (~464 bp) of 16S rRNA was amplified with non-barcoded primers with TruthSeq adapters. Amplicons from individual specimens were barcoded and pooled (Supplementary information). The final library was sequenced on a MiSeq v3 instrument (600 cycle cartridge, 300 paired-end) with 10% PhiX, at the Centre for Genomic Regulation (CRG) in Barcelona (Spain).
Sequence processing
Sequences were quality-filtered to remove ambiguities and chimeras and input into the Qiime pipeline (Caporaso et al 2010). Operational taxonomic units (OTUs), defined at 97% similarity, were taxonomically assigned with uclust against the Greengenes 13_8 database, filtered to exclude singletons, retaining only Bacteria and excluding Archaea and Cyanobacteria (Supplementary information). The OTU abundance table was rarefied to 13730 reads and used for all analyses, unless otherwise specified. Tables of taxa relative abundances were created by summarizing the rarefied OTU table.
Functional metagenomes were predicted using PICRUSt (Langille et al 2013). After removal of de-novo OTUs from the original table and rarefaction to 8000 reads, the table was input into the PICRUSt pipeline in Galaxy (Afgan et al 2016). Functions were predicted according to KEGG orthology.
Correlation analyses between taxonomic and functional diversity were performed with the vegan R package (Oksanen et al 2015).
Alpha diversity was calculated as Shannon entropy, converted to effective number of species (Jost 2006) and Faith’s phylogenetic diversity (PD whole tree).
Deconstructing the microbiota phylogenetic and ecological signals
To specifically separate the confounding phylogenetic effect from the main dietary one in affecting the microbiota clustering, we used the Tanganyika dataset for which host phylogeny and stable isotopes (15N and 13C) are publicly available (Muschick et al 2012).
Starting from the rarefied OTU table we retained an OTU if present in at least 50% of the conspecifics of at least 10/19 species, summing ≥20 total reads, and using the median across conspecifics as a proxy per species. Reads were log-transformed and dimensionally reduced with Principal Components (PCs). PCs were considered as a multivariate traits and tested with the Kmult statistics (Adams 2014), an n-dimension generalization of the Blomberg’s K test (Blomberg et al 2003). Both approaches compare the quantitative values of the traits on the tips of a given phylogeny with the values resulting from a Brownian motion null evolution model, here iterated by 999 simulations.
A redundancy analysis (RDA) was applied on a second table of average stable isotopic signatures of each cichlid species, as proxies for their trophic position in the lake food web. The first two bacterial axes were constrained to be dependent on the isotopic matrix (RDAs), while the remaining 16 axes were unconstrained by the ecological matrix (PCs). To compare both PC2 obtained from the two different PCAs performed herein, two separate Euclidian distance matrices were built among the tips of both virtually equivalent components and compared through a Mantel test.
Discriminatory taxa and functions
Discriminatory taxa/OTUs were identified through group_significance.py script with Kruskal-Wallis (kw) test, after removing low-abundance taxa/OTUs (<50 reads total). Individual results from each lake were merged, retaining all taxa/OTUs with at least 10 differences between means of categories compared and significance according to false discovery rate (FDR, p<0.05). Indicator functions were similarly estimated, after filtering for sum of gene counts above 100 and retaining functions with at least 50 differences between means of categories compared.
Results
Taxonomic composition of wild cichlids gut bacterial communities
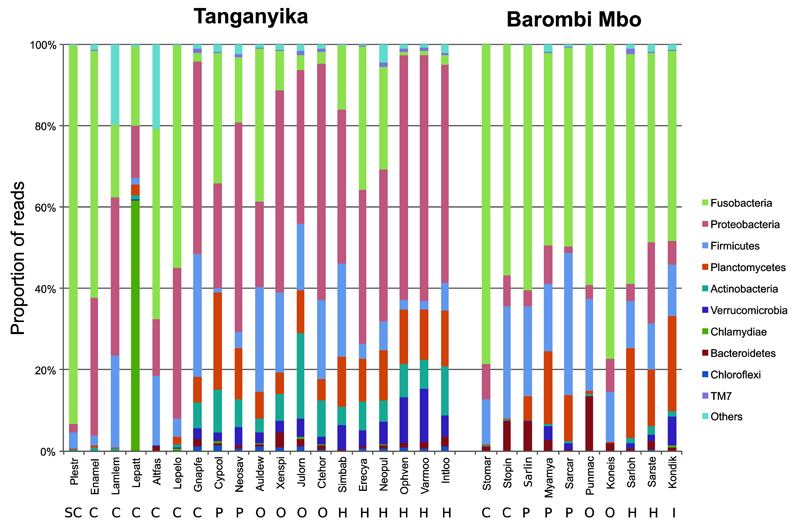
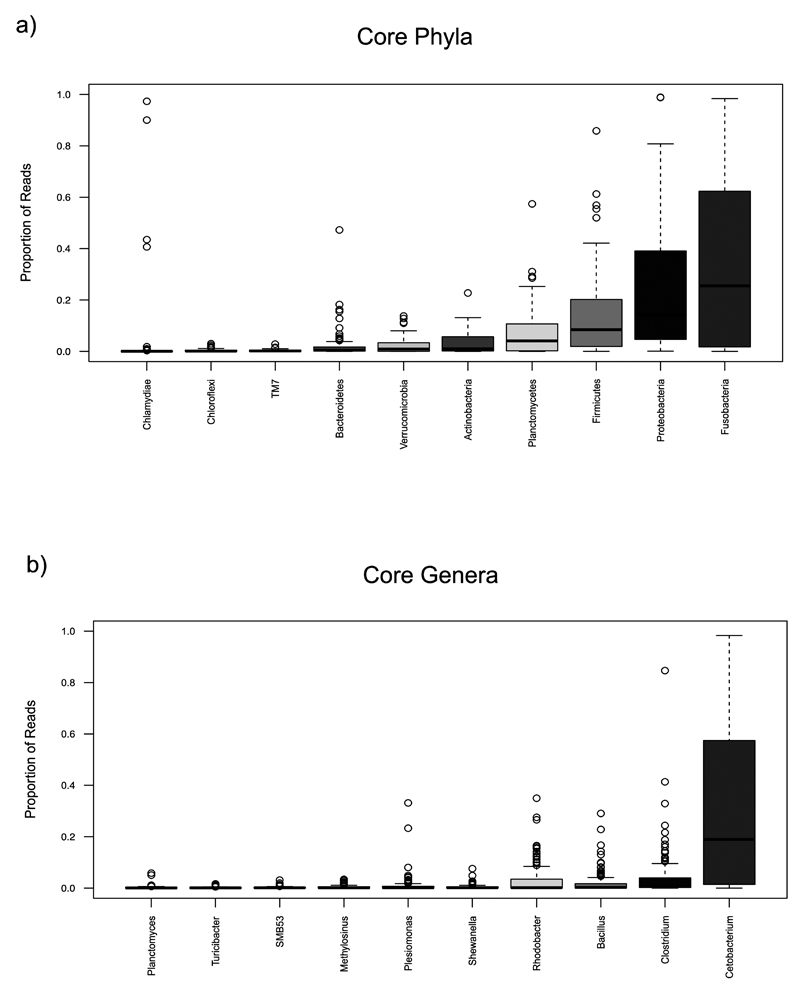
Ten phyla represented the core signature of the cichlid gut microbiota in both Tanganyika and Barombi Mbo, accounting on average for 89% of the total reads/specimen (Figure 1 and 2a). Fusobacteria (average 36%), Proteobacteria (22%), and Firmicutes (13%) represented the dominant components (Figure 2a). The most prevalent orders were Fusobacteriales (Fusobacteria) and Clostridiales (Firmicutes), together accounting for 47% of the reads, followed by Rhodobacterales, Vibrionales and Rhizobiales (Proteobacteria). Within the small fraction of OTUs classified to genus level (23%), 11 genera were systematically recovered (Figure 2b), the most prevalent being Cetobacterium (average 32% of reads), with a single most abundant OTU (828162), classified as C. somerae, being universally found in all specimens, also in our previous work (Baldo et al 2015).
Figure 1. Taxonomic composition at phylum-level of wild cichlid gut microbiotas.
Bars show proportions of taxa per cichlid species as average across conspecifics, estimated from the rarefied OTU table (13730 reads). “Others” groups all phyla with relative abundance below 1% over the total number of reads. Approximate diet categories are shown below species names: SC: scale-eaters; C: carnivores: P: planktivores; O: omnivores; H: herbivores; I: insect-eaters (see Supplementary Table 1 and Information for diet details). Within each lake, sampling was done in close areas and in a few-days time (less than a month) to minimize the effect of geographical and seasonal variables. Species names abbreviations: Plestr: Plecodus straeleni; Lamlem: Lamprologus lemairii; Lepelo: Lepidiolamprologus eolongatus; Lepatt: Lepidiolamprologus attenuatus; Altfas: Altolamprologus fasciatus; Enamel: Enantiopus melanogenys; Gnapfe: Gnathochromis pfefferi; Cypcol: Cyprichromis coloratus; Neosav: Neolamprologus savoryi; Audelw: Aulonochranus dewindtii; Ctehor: Ctenochormis horei; Julorn: Julidochromis ornatus; Xenspi: Xenotilapia spilotera; Varmoo: Variabilichromis moorii; Neopul: Neolamprologus pulcher; Intloo: Interochromis loockii; Ophven: Ophthalmotilapia ventralis; Erecya: Eretmodus cyanostictus; Simbab: Simochromis babaulti; Kondik: Konia dikume; Punmac: Pungu maclareni; Stomar: Stomatepia mariae; Stopin: Stomatepia pindu; Myamya: Myaka myaka; Sarcar: Sarotherodon caroli; Sarlin: Sarotherodon linnellii; Koneis: Konia eisentrauti; Sarloh: Sarotherodon lohbergeri; Sarste: Sarotherodon steinbachi.
Figure 2. Relative abundance of core phyla (a) and genera (b) across all samples.
Core taxa were defined by presence in all species and at least 80% of the specimens. Interquartile ranges (25th and 75th percentiles) and whiskers show data dispersion across specimens. Medians are shown as central horizontal lines.
After excluding species-specific differences, cichlids from the two lakes shared remarkably similar bacterial qualitative profiles at all taxonomic levels, suggesting important common host constraints on taxa retention. Only two taxa were lake-specific (i.e. shared by 50% of the specimens in one lake but absent in the other): the family Pseudoalteromonadaceae and the genus Verrucomicrobium from Tanganyika (kw, Bonferroni p<0.05). The main plastic component of the cichlid microbiota as a function of geography/large phylogenetic distance is seen at OTU level (see also below) and in terms of relative abundance of common taxa. At phylum level, Barombi Mbo cichlids were significantly enriched in Fusobacteria and depleted in Proteobacteria, Tenericutes and Actinobacteria (p<0.001) (Figure 1). Among all other shared taxa, 18% of classes and orders, 19% of families, 9% of genera and 1.3% of OTUs were differentially abundant between lakes (p<0.05).
Bacterial taxonomic and functional diversity as a function of diet
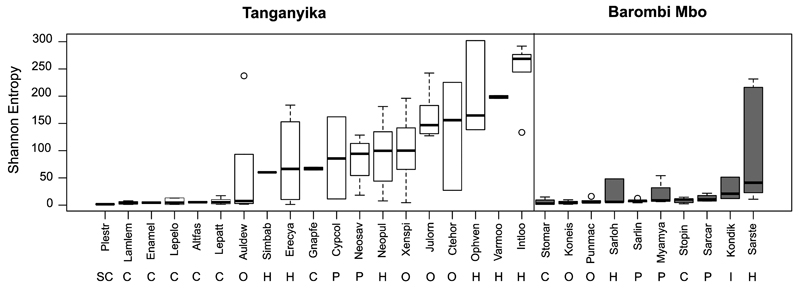
Cichlid species encompass a wide spectrum of bacterial diversity (Figure 3). Barombi Mbo carries less diverse communities than Tanganyika at common dietary categories, while no significant differences were observed among Tanganyika tribes (two-sample t-test, p>0.05), thus excluding a major phylogenetic effect. A main trend is clearly seen as a function of diet: herbivores showed on average the highest bacterial diversity at both lakes, while carnivores and scale-eaters were the most depleted (Figure 3 and 4). Omnivores and planktivores shared a comparable diversity within each lake, which was slightly reduced compared to herbivores (Figure 4b, p<0.05 PD whole tree only for Barombi Mbo).
Figure 3. Alpha diversity by species as effective number of OTUs (Shannon Entropy).
Boxplots summarize values of alpha diversity per species calculated as Shannon entropy on collated results after multiple rarefactions at a maximum depth of 13730 reads (1000 reads step, 10 iterations). Boxplots are centered at and ordered by the median, while whiskers show data dispersion across conspecifics.
Diversity can be substantially high at intraspecific level for some species, while remarkably low in others (mostly carnivores). A clear trend is seen as a function of diet at both lakes, with carnivores showing a highly depleted diversity compared to herbivores. Removal of either poorly represented OTUs (<10 reads total) or selective inclusion only of the three main phyla (Firmicutes, Fusobacteria and Proteobacteria) provided a comparable pattern of alpha-diversity across species.
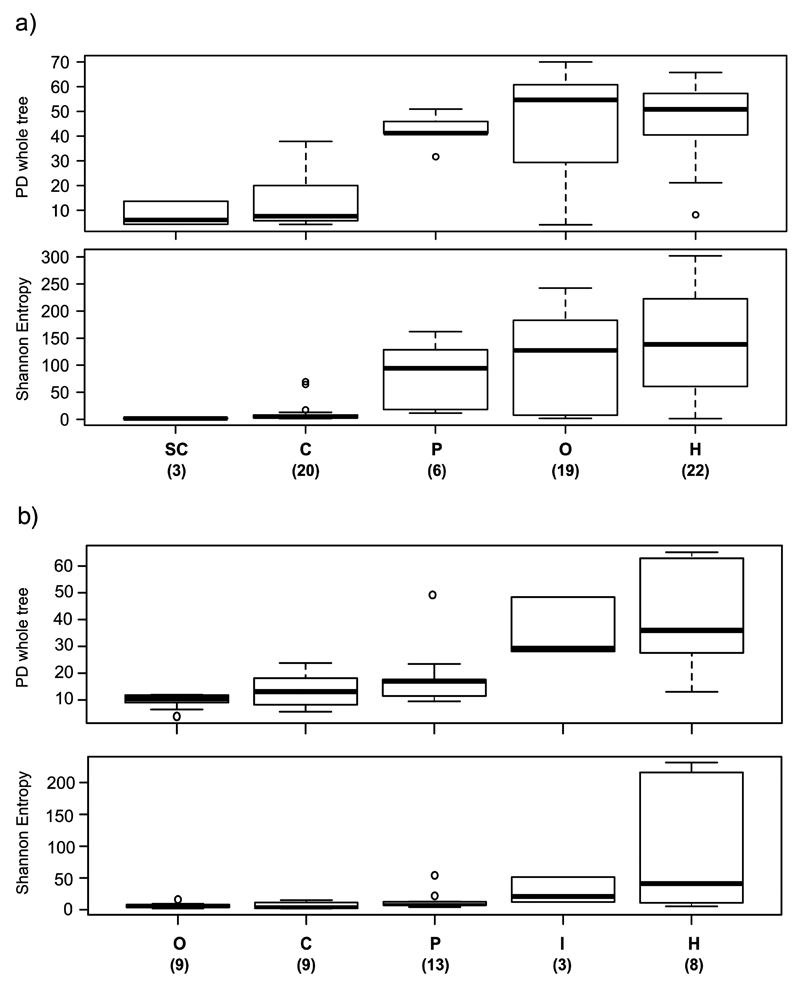
Figure 4. Alpha diversity (Shannon entropy and PD whole tree) by diet category in (a) Tanganyika and (b) Barombi Mbo.
For each diet category, boxplots are centered at the median and whiskers show data dispersion across specimens (number within brackets) and ordered according to Shannon median values. Herbivores are significantly more diverse than carnivores at both lakes (p<0.05, two-sample t-test with Bonferroni correction). Statistically significant pairs are: for Tanganyika (both indexes), H versus SC, H vs C, O vs SC, O vs C, P vs C; for Barombi Mbo (PD whole tree only), H vs C, H vs O, H vs P, I vs C, and I vs O (p<0.05).
The two categories F and Pr (Supplementary Table 1) were significantly distinct within Tanganyika (p=0.001, both indexes), but not within Barombi Mbo (p=0.008), suggesting that at least for Tanganyika main metabolic differences related to the consumption of vegetable matter can alone explain the trend in alpha diversity observed, beyond range and type of food.
Taxonomic diversity couples with functional diversity
Following we addressed whether an increased OTU diversity confer the host with a higher functional diversity. From the predicted metagenomes we recovered a total of 6909 Kos, corresponding to 328 KEGG L3 pathways. The pattern of functional diversity largely followed the trend in taxonomic diversity (Supplementary Figure 1 and 2). After approximately the first 200 OTUs the curve starts leveling off, with diminishing increases in functionality (Supplementary Figure 1). As expected, while the compositional space (i.e. number of distinct OTUs) is potentially unlimited, the functional bacterial capacity rapidly reaches saturation. The same trend is observed for OTU phylogenetic diversity (p< 2.2e-16, rho 0.828), which removes the potential inflating effect of closely related OTUs on the compositional diversity. Overall, herbivores showed a greater bacterial taxonomic and functional evenness than carnivores (Supplementary Figure 3).
Diet drives microbiota taxonomic and functional clustering within lakes
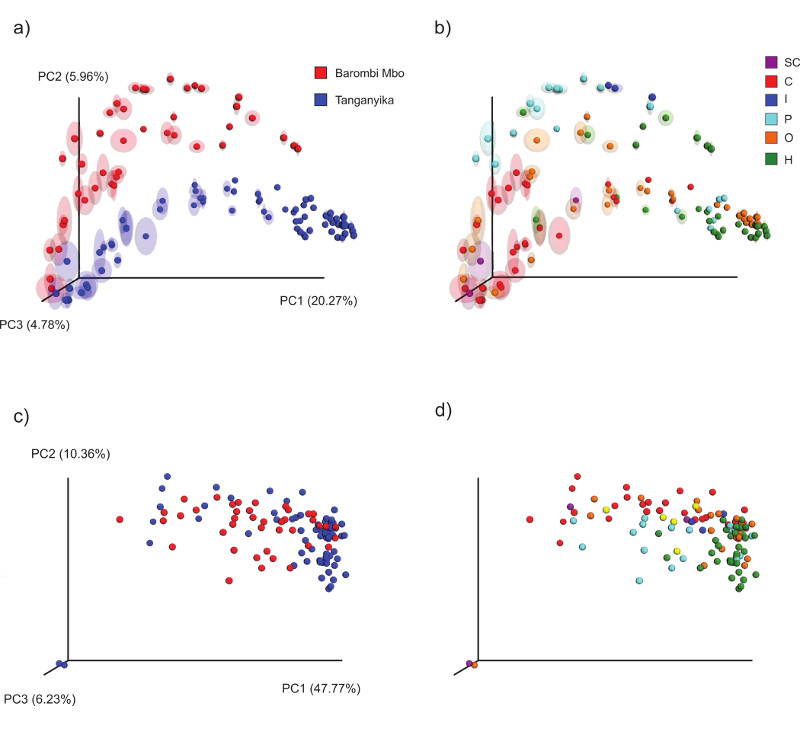
The two lakes significantly separated in their microbial communities clustering at both OTU and KO level (Figure 5a-c, Unweighted unifrac distances, adonis, p=0.001), indicating a strong environmental/deep phylogenetic effect. Within each radiation, a dietary gradient can be observed, with a strong parallel trend at both lakes (Figure 5b): distance typically increased from carnivores to planktivores, omnivores and herbivores. This trend alone explained most of the diversity among bacterial communities (20% of the total variance along PC1) and significantly discriminated between carnivores and herbivores at the two extremes of PC1 (p<0.01, both lakes). The functional profile supported a similar trend (Figure 5d), with carnivores and herbivores discriminating along PC1 (48% of variance along PC1, p<0.01, both lakes). Notably, within Tanganyika in particular, while the carnivores were sparser in their clustering at both taxonomic and functional profiles, most herbivores, omnivores and planktivores strongly converged, beyond tribe boundaries (see also below). At functional level, the convergence among herbivores even spanned across lakes (Figure 5d).
Figure 5. Principal coordinate analysis (PCoA) of cichlid gut bacterial communities according to lake (a, c) and diet (b, d).
(a-b) taxonomic (OTU) clustering based on Unweighted unifrac distances. Circles represent individual specimens with ellipses showing range of variation after multiple rarefactions to an even depth (13000 reads). (c-d) functional (KO) composition clustering based on Binary Jaccard, after rarefaction to 1835713 gene counts. Barombi Mbo (red) and Tanganyika (blue) specimens significantly separate at both taxonomic (along PC2) and functional level (along PC3). Given the time of divergence between the two radiations, deep phylogenetic and geographic effects are here intrinsically linked. Within lakes, diet explains most taxonomic and functional bacterial variance, with a clear divergence between carnivores (C) and herbivores (H).
Contribution of the host phylogenetic signal is significant but small
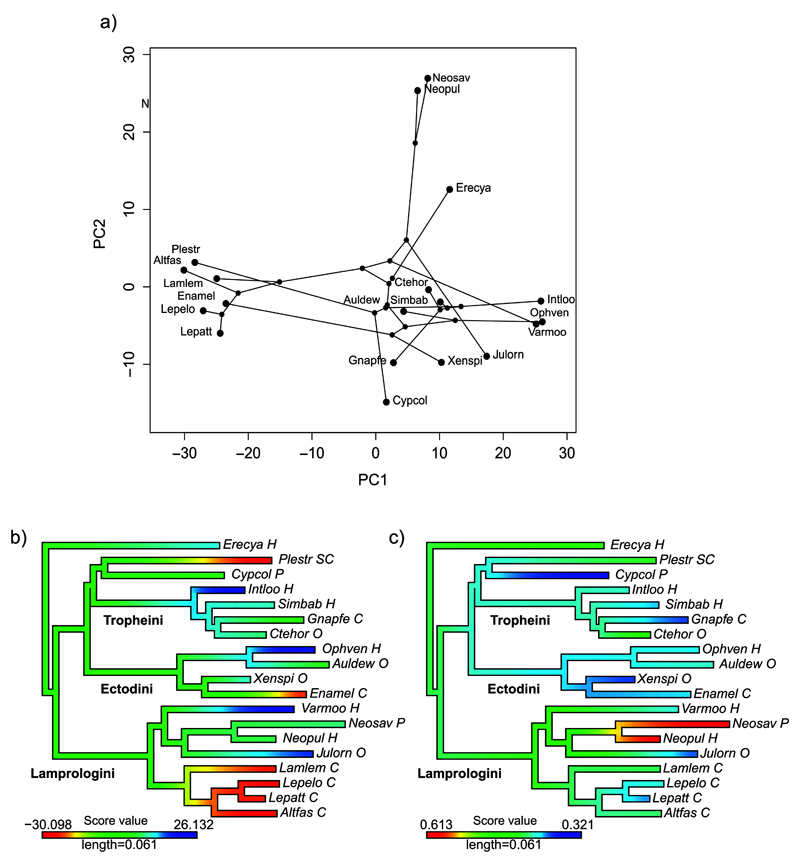
According to multivariate analyses on the Tanganyika dataset (see Methods), a clear phylogenetic signal was detectable at the level of the whole microbiota composition (Kmult p=0.005). Of the 12 significant PCs obtained with the broken stick as a null model, only the first two retained a phylogenetic signal (Kmult p=0.005) (Figure 6). PC1 (42% of variance, Figure 6b) is heavily dependent on both cichlid phylogeny and trophic niche given that the majority of carnivores belong to the same tribe Lamprologini, whereas herbivores spread in several clades. Interestingly, it also indicates strong cases of microbiota convergence for phylogenetically distant species, confirming previous observations (Figure 5). Conversely, PC2 (12% of variance, Figure 6c) is affected only by a phylogenetic signal, largely shaped by the coordinates of two closely related species, Neolamprologus savoryi (Neosav) and N. pulcher (Neopul).
Figure 6. Phylo-ecospace of the Tanganyika microbiota dataset shown by the first two significant PCs.
a) PC1 and PC2 correlation scores; b-c) color-plot of (b) PC1 and (c) PC2 scores on the cichlid phylogeny, with similar colors indicating comparable PC scores (i.e. similar microbiotas). (b) PC1 carries a mix of ecological and phylogenetic signals, with clear examples of microbiota convergence among the herbivores Ophven, Varmoo and Intloo, belonging to three distinct tribes, and the scale eater Plestr (tribe Perissodini) and the carnivorous Enamel (tribe Ectodini) with the main carnivorous Lamprologini clade. (c) PC2 shows a strong phylogenetic signal largely driven by the two Neolamprologus species (Neosav and Neopul) and partly by the two species Erecya (Eretmodini) and Cypcol (Cyprichromini), unique representatives of two distant tribes. Stable isotopes and phylogenetic data for Cyprichromis coloratus (Cypcol) were approximated by the sister species C. leptosoma (sharing a similar trophic niche).
According to the RDA analysis using isotopic data as independent ecological variables, as expected, the first two constrained RDAs had no phylogenetic signal (Kmult, P=0.123), while they strongly correlated with both 15N and 13C trophic signatures (Supplementary Figure 4). Of the two unconstrained PCs selected, only PC2 carries a significant phylogenetic signal, again mainly for the two Neolamprologus species (Blomberg’s K, p=0.034). The tree that emerged from this procedure is equivalent to that obtained with the second axis of the former PC analysis (Figure 6c) (Mantel test, P=0.001), indicating the robustness of the phylogenetic signal extracted.
Conserved discriminatory taxa in the cichlid trophic adaptation
Which taxa and functions do drive the difference in bacterial communities observed across diets? Here we limited our analyses to species at the extremes of diet range within each lake, carnivores and herbivores, usually more restricted in their diet range, while keeping in mind that the definition of herbivory in Barombi Mbo is less stringent (see Supplementary Information).
Patterns of taxonomic enrichment between carnivores and herbivores were strikingly concordant across lakes, at all levels (Supplementary Figure 5). At taxa level above OTU (Supplementary Figure 5), three main observations can be made: 1) discriminatory taxa in Barombi Mbo represented a subset of those recovered from Tanganyika; 2) shared taxa between lakes showed a fully concordant pattern of enrichment; and 3) most taxa were enriched in herbivores. Down to OTU level, of the 243 discriminatory OTUs obtained (kw, FDR p<0.05), the large majority was unique to either Tanganyika or Barombi Mbo (<20 OTUs overlap, see Supplementary Table 2 for full data). Nonetheless, again most OTUs were enriched in herbivores (>83%) and those overlapping between lakes had a concordant pattern of enrichment, with the majority of OTUs belonging to Proteobacteria (particularly the family Rhodobacteraceae), followed by Planctomycetes. Carnivores in Tanganyika showed an interesting enrichment in Chlamydia OTUs of the genus Candidatus Rhabdochlamydia, while in Barombi Mbo they were largely enriched in Clostridiales (Supplementary Table 2).
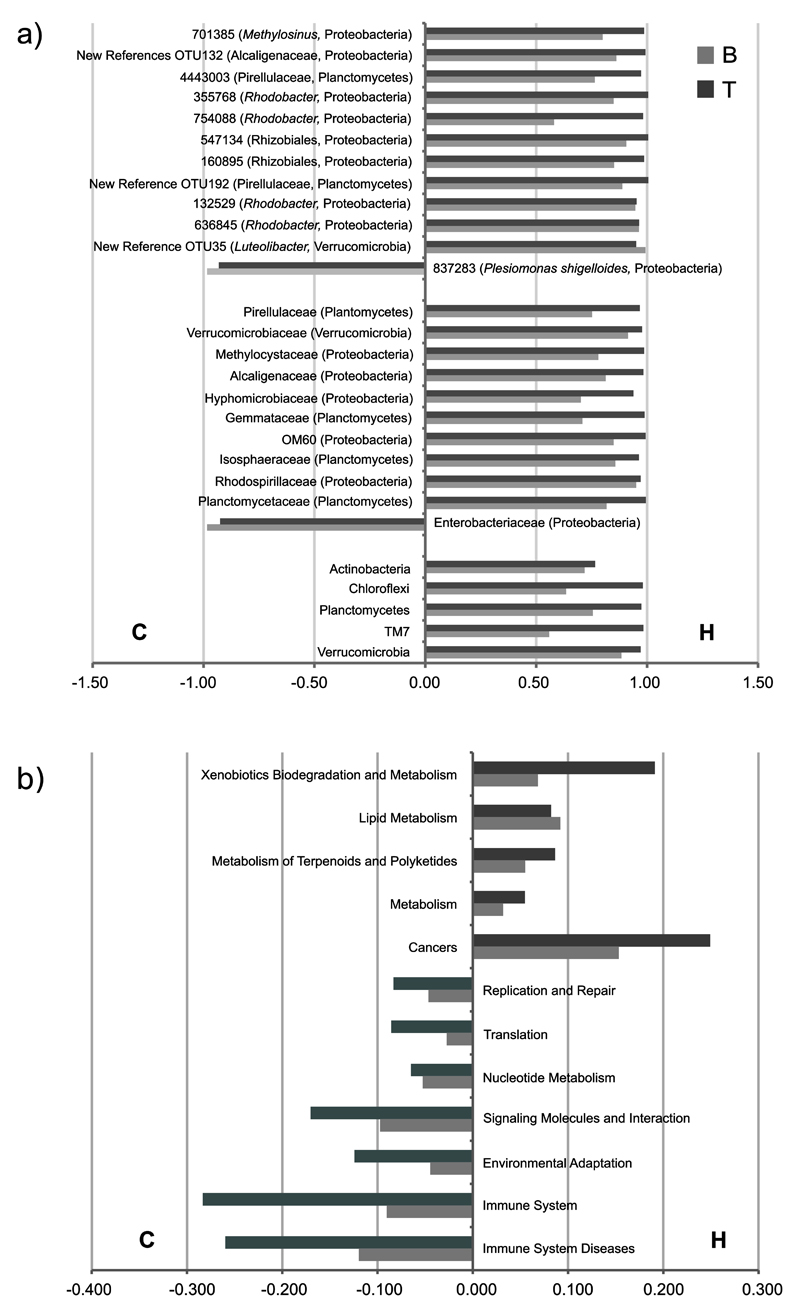
In terms of shared discriminatory taxa between two lakes (five phyla, 11 families and 12 OTUs), all had concordant patterns of enrichment. Only a single family (Enterobacteriaceae) and OTU (Pseudomonas shigelloides) were significantly enriched in carnivores of both lakes (Figure 7a).
Figure 7. Discriminatory (a) OTUs, families and phyla and (b) functional pathways (KEGG L2) between herbivores and carnivores, according to both lakes.
Bars show differences between average reads for diet category, normalized to the sum of means (right-side bars: H-enriched; left-side bars: C-enriched) (kw, p<0.05, Bonferroni corrected for Tanganyika and FDR corrected for Barombi Mbo). OTUs were classified to their highest level of taxonomic resolution followed by phylum. The two lakes show a consistent trend of shared taxa and functional pathways enrichment according to diet categories. The core OTU-837283 (present in 85% of all specimens), corresponding to P. shigelloides, was detected also in our previous work (corresponding to former OTU-137, (Baldo et al 2015) and was here significantly enriched in carnivores of both lakes (a). See complete list of discriminatory OTUs at Supplementary Table 2 and functions at Supplementary Table 4.
A highly similar pattern of taxa enrichment was also found for comparisons between Pr and F categories (Supplementary Table 3), indicating that most discriminatory taxa found are not an exclusive signature of herbivory, but rather correlated with general consumption of vegetable matter.
Bacterial functional signature of trophic adaptation
Lake Tanganyika and Barombi Mbo provided a virtually identical pattern of enrichment at shared functions and KEGG pathways between carnivores and herbivores, reflecting previous findings for taxa (Supplementary Table 4). Of the 1147 shared discriminatory KOs, only 13 showed discordant enrichment between lakes. Herbivores were consistently enriched in a larger number of KOs, despite further normalization, likely reflecting their higher bacterial taxonomic diversity, which translated into partial redundant functional information (Supplementary Figure 1).
In terms of KEGG pathways (L2) (Figure 7b), herbivores of both lakes were particularly enriched in “Xenobiotics Biodegradation and Metabolism”, and “Metabolism of Terpenoids and Polyketides”, and “Biosynthesis of Other Secondary Metabolites” (significance only for Tanganyika). Metabolism and particularly lipid metabolism were also a hallmark of herbivores at both lakes. On the other hand, carnivores were enriched in more conservative, housekeeping functions, such as “replication and repair”, “translation” and “nucleotide metabolism”.
Discussion
We built our study on previous findings for a sample of cichlids within a single tribe, suggesting a role of diet in shaping the gut microbiota (Baldo et al 2015). Using a much larger sample of 29 species and two lakes, here we showed a clear pattern of gut microbiota eco-evolutionary dynamics following dietary habits and largely independent from the host phylogeny and geography. The gut microbiota in carnivores was dominated by few and abundant taxa, while in herbivores (and partly omnivores) it distinguished for an increased bacterial richness while converging in key compositional and functional features across species, beyond host phylogenetic boundaries.
Parallel diversification of bacterial community structure following dietary shifts
Lake Tanganyika and Barombi Mbo are inherently different in geography, ecology and timing of species divergence, with dietary preferences, particularly herbivory, that are less marked within the Barombi young radiation where they have not yet transformed into exclusive specialization. Nonetheless, major differences in microbiota compositional structure between general carnivores and herbivores (i.e. alpha and beta diversity, taxa and functional enrichment) were largely conserved across lakes. These remarkable parallelisms suggest that the gut nutritional environment is largely responsible for structuring the gut microbiota, as also seen for higher vertebrates (Ley et al 2008). In mammals in particular, fiber consumption (amount and type) is known to have dramatic effects on the composition of the intestinal microbiota (De Filippo et al 2010, Kohl and Dearing 2012, Ley et al 2008, Schnorr et al 2014), including a typical increase in bacterial richness associated with a more complex metabolic cascade (Flint et al 2012, Sonnenburg and Backhed 2016). Fish literature does not provide a clear pattern in this respect, mostly owing to limited studies targeting such comparison and/or presence of confounding multivariate factors (e.g. phylogeny and diet) (Liu et al 2016, Sullam et al 2012). Our findings for cichlids indicated a comparable pattern to higher vertebrates, with herbivores of both lakes showing a significantly increase in bacterial taxonomic and functional diversities compared to carnivores. While the actual bacterial functional capacity and expression should be further explored through shotgun metagenomics and transcriptomics, currently a challenge for wild specimens from remote field regions, the relative functional diversity across main diet categories should nonetheless hold true for samples that were equally processed.
Not only herbivores hosted more diverse bacterial communities, they also largely converged at compositional and functional levels across species. This is especially relevant within Tanganyika, where herbivory has evolved independently in multiple tribes (our study includes members of four tribes), and reached high specialization (Hata et al 2014, Hata et al 2015, Muschick et al 2012). Such convergence was largely independent from the phylogenetic context as no clear evidence for microbiota phylogenetic inertia was found, except for the two Neolamprologus species. This is unlike what seen for some terrestrial mammals (Delsuc et al 2014, Xue et al 2015) and could be partly explained by the high plasticity of the cichlid gut/trophic morphology in response to diet changes (Muschick et al 2012, Muschick et al 2014, Wagner et al 2009), potentially favoring a rapid adaptation of microbial communities.
In Barombi Mbo, ecology and phylogeny follows a strict parallel path (Trewavas et al 1972) and unpublished observations), which currently prevents a clear disentangling of the contribution of each factor on the microbiota dynamics. Moreover, the recent cichlid ecological diversification within this lake is likely responsible for the less discrete pattern of microbiota clustering observed. Nevertheless, the several parallelisms detected with Tanganyika suggest a minor impact of species-specific genotypes on major microbial features.
Bacterial community signature of metabolic differences
The large OTU convergence among most herbivore species (Figure 5 and 6) might be partly explained by host retention of a limited pool of bacterial taxa able to process vegetable matter, as shown in humans (De Filippo et al 2010). Indeed, herbivores were mostly enriched in Verrucomicrobia, Actinobacteria and Planctomycetes, and among families, in methanotrophic bacteria (Methylocistaceae) (Graham et al 2002) and a major nitrogen-fixing group (Hyphomicrobaceae) (Clark et al 2009). The three above phyla have been repeatedly linked to herbivory in terrestrial vertebrates (Frey et al 2006, Garcia-Amado et al 2012, Nelson et al 2003); Verrucomicrobia, in particular, are known as fermentative bacteria with potential to degrade cellulose (Wertz et al 2012). Up to date, herbivory in freshwater fish species has been correlated with an increase in few bacterial taxa, including Clostridiales and Bacteriodetes (Sullam et al 2012), as well as the genera Citrobacter, Leptotrichia and Clostridium (Liu et al 2016). We could not corroborate the above results for cichlids; on the contrary, we found that Clostridium was particularly enriched among carnivores of both lakes (significantly in Tanganyika). Although most discriminatory taxa here identified appear associated to fiber consumption, we nonetheless suggest caution in linking their enrichment to specific functions in the gut. We also note that the extent of intestinal fermentation in freshwater fishes is still poorly understood (Clements et al 2014) and quantification of microbial fermentation products (i.e. short-chain fatty acids) is necessary.
On the other hand, lack of strong compositional convergence among carnivores, as also shown by the fewer discriminatory taxa detected for this category, suggests that a more basal protein metabolism and faster transit times could be responsible for a reduced stability and diversity of microbial communities. The Enterobacteriacae, and particularly P. shigelloides, were the only significantly enriched taxa in carnivores reported by both lakes; the species has been previously described in humans with a low-fiber diet (De Filippo et al 2010), suggesting it could represent a signature species of carnivores in vertebrates.
Alternative explanations exist for the observed trends in microbiota structure, particularly when considering the unknown contribution of the allochtonous component to the actual resident gut microbiota. Indeed, several discriminatory taxa in herbivores corresponded to common environmental Proteobacteria (e.g. Rhodobacteraceae) and Planctomycetes. Whereas the contribution of bacteria present in the water might be negligible to explain the differences and convergence observed, given that all our species were sampled in a close area, common food-associated bacteria could be driving part of the observed pattern, either as spurious contamination or as selectively retained by the gut. Undoubtedly, sharing the same environment and, within this, the same trophic niche increases accessibility to a common bacterial pool by restricted dispersal. For instance, the herbivores Interochromis loockii (Intloo) and Variabilichromis moorii (Varmoo) coexist in the same rocky habitat in a few meters range (Hata et al 2014) and despite belonging to distinct tribes, show strikingly similar microbiotas (84% of shared OTUs). These syntopic species, however, like other herbivore species in Tanganyika, show strong niche specialization, with significant differences in their algal farms, partly due to habitat-depth segregation (Hata et al 2014). Food contamination alone might, therefore, not fully explain the observed convergence, nor the similarities in taxa enrichment detected across lakes, especially when considering that such transient component is not under host-selection and its distribution across species should follow a more stochastic process (mostly reflecting the type of ingesta at the time of capture). This is also in accordance with recent studies in fishes, pointing to a minor role of the allochtonous component in shaping the abundant gut communities (Bolnick et al 2014, Giatsis et al 2015). An intriguing question remains on whether the fish gut could provide a transient/stable niche for environmental-associated bacteria, as recently found for Planctomycetes in the human gut (Cayrou et al 2013). We note, for instance, that Planctomyces and Rhodobacter, two typical environmental bacteria, were found as core genera in cichlids. Rhodobacter has been previously detected in the tilapia gut (Giatsis et al 2015), where it shows gastrointestinal transit tolerance (Zhou et al 2007). Collectively, the major microbial differences detected between carnivores and herbivores most likely reflect a differential selective retention by the gut. Nonetheless, an ecosystem-level study of microbial communities is needed to explore forces in bacterial metacommunity dynamics (particularly dispersal and niche selection), beyond the gut (Costello et al 2012).
Potential detoxifying role of the gut microbiota
At functional level, an important link was found between herbivory and the enrichment in Xenobiotics biodegradation and Terpenoids and Polyketides metabolism, consistently in both lakes. Interestingly, such enrichment indicates a putative role of gut bacteria in the processing of plant/algae secondary compounds (including toxins), which, together with indigestible fibers, impose strong physiological constraints to animals, affecting their foraging behavior and limiting diet breadth (Kohl et al 2016a). Such bacterial detoxifying ability has been virtually neglected in vertebrates until recent studies (Kohl and Dearing 2012, Kohl et al 2014, Kohl et al 2016a, Kohl et al 2016b), and remains, to our knowledge, unstudied in fishes. Exposure to plant toxins has also been shown to enhance the gut microbial diversity in woodrats (Kohl and Dearing 2012), which could partly explain also the pattern observed in herbivore cichlids. Eco-evolutionary implications are large as it is apparent that, beyond nutrient metabolism, the detoxifying effect of gut bacteria could be crucial in the animal-plant dietary shift, favoring trophic niche expansion. This is an intriguing hypothesis for the evolution of herbivory in cichlids, as recently proposed for other animal systems (Pennisi 2017), which should be tested with ad-hoc experimental manipulation of diet and/or microbiota.
Overall, while demonstration of causality is far from being reached, the strong correlations found between cichlid diet/metabolism and gut microbial features set the bases for more targeted studies on the eco-evolutionary dynamics of this symbiotic association.
Additionally, the gut microbiota emerges here as a powerful tool to validate and/or predict dietary habits in cichlids. Without a prior clustering, we found that compositional and functional aspects of the gut microbiota largely support the main dietary categories previously assigned according to extensive trophic data (e.g. morphology and stable isotopes). In principle, such microbiota features could individually and altogether be used to predict or revise diets for species with unknown/uncertain trophic niches. As examples, according to richness and beta taxonomic and functional diversity, G. pfefferi, here classified as carnivore (Supplementary Table 1), might be largely omnivore (Figure 3 and 6, thus putatively feeding also on vegetable material) while S. steinbachi displays the strongest herbivore-like profile within Barombi Mbo.
Extending the analyses to other cichlid adaptive radiations from Africa and Central America (currently undergoing), together with a finer characterization of their ecological niches and metabolic differences, coupled with a more mechanistic approach, will help corroborating these findings and provide important insights into the role of these communities in cichlid trophic adaptation and potential niche expansion.
Supplementary Material
Supplementary information is available at ISME’s website http://www.nature.com/ismej/
Acknowledgments
We especially thank Fyodor Kondrashov from the Centre for Genomics Regulation (CRG) in Barcelona, for providing laboratory support and facilities; Adrian Indermaur, Hugo Gante, both members of the Salzburger’s lab, for the sampling effort; prof. Joaquim Gutiérrez i Fruitós, from the University of Barcelona, for use of equipments; and Jochen Hecht, head of the Genomics Unit at CRG, for technical assistance. This work was supported by the European Research Council (ERC; Starting Grant "INTERGENADAPT") to WS, funding from the Spanish Ministerio de Economía y Competitividad to LB, and Basler Stiftung für Biologische Forschung and Czech Science Foundation (16-09784Y) to ZM.
Footnotes
Conflict of Interest
The authors declare no conflict of interest
Availability of data and materials
OTU and KO original abundance tables are available as Dataset 1 and 2, respectively.
Raw sequences are available in the Sequence Read Archive (SRA) database at NCBI under BioProject ID PRJNA341982.
The alignment of representative OTU sequences generated and analyzed during the current study is available in the Dryad repository doi:10.5061/dryad.q6s1m.
Additional datasets and scripts used for the current study are available from the corresponding author on reasonable request.
References
- Adams DC. A generalized K statistic for estimating phylogenetic signal from shape and other high-dimensional multivariate data. Syst Biol. 2014;63:685–697. doi: 10.1093/sysbio/syu030. [DOI] [PubMed] [Google Scholar]
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Cech M, et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayayee PA, Larsen T, Rosa C, Felton GW, Ferry JG, Hoover K. Essential Amino Acid Supplementation by Gut Microbes of a Wood-Feeding Cerambycid. Environ Entomol. 2016;45:66–73. doi: 10.1093/ee/nvv153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, Riera JL, Tooming-Klunderud A, Alba MM, Salzburger W. Gut Microbiota Dynamics during Dietary Shift in Eastern African Cichlid Fishes. PloS one. 2015;10:e0127462. doi: 10.1371/journal.pone.0127462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baothman OA, Zamzami MA, Taher I, Abubaker J, Abu-Farha M. The role of Gut Microbiota in the development of obesity and Diabetes. Lipids Health Dis. 2016;15:108. doi: 10.1186/s12944-016-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg SP, Garland T, Jr, Ives AR. Testing for phylogenetic signal in comparative data: behavioral traits are more labile. Evolution. 2003;57:717–745. doi: 10.1111/j.0014-3820.2003.tb00285.x. [DOI] [PubMed] [Google Scholar]
- Bolnick DI, Snowberg LK, Hirsch PE, Lauber CL, Knight R, Caporaso JG, et al. Individuals' diet diversity influences gut microbial diversity in two freshwater fish (threespine stickleback and Eurasian perch) Ecology letters. 2014 doi: 10.1111/ele.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nature methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C, Sambe B, Armougom F, Raoult D, Drancourt M. Molecular diversity of the Planctomycetes in the human gut microbiota in France and Senegal. APMIS : acta pathologica, microbiologica, et immunologica Scandinavica. 2013;121:1082–1090. doi: 10.1111/apm.12087. [DOI] [PubMed] [Google Scholar]
- Clark DP, Dunlap P, Madigan M, Martinko J. Brock Biology of Microorganisms. Scientific Publisher; Beijing: 2009. [Google Scholar]
- Clements KD, Angert ER, Montgomery WL, Choat JH. Intestinal microbiota in fishes: what's known and what's not. Molecular ecology. 2014;23:1891–1898. doi: 10.1111/mec.12699. [DOI] [PubMed] [Google Scholar]
- Costello EK, Stagaman K, Dethlefsen L, Bohannan BJ, Relman DA. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AM, Unmack PJ, Pusey BJ, Pearson RG, Morgan DL. Ontogenetic development of intestinal length and relationships to diet in an Australasian fish family (Terapontidae) BMC Evol Biol. 2013;13:53. doi: 10.1186/1471-2148-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day JJ, Cotton JA, Barraclough TG. Tempo and mode of diversification of lake Tanganyika cichlid fishes. PloS one. 2008;3:e1730. doi: 10.1371/journal.pone.0001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, et al. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Metcalf JL, Wegener Parfrey L, Song SJ, Gonzalez A, Knight R. Convergence of gut microbiomes in myrmecophagous mammals. Molecular ecology. 2014;23:1301–1317. doi: 10.1111/mec.12501. [DOI] [PubMed] [Google Scholar]
- Edward Stevens C, Hume ID. Comparative Physiology of the Vertebrate Digestive System. 2nd edn. Cambridge University Press; 2004. [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey JC, Rothman JM, Pell AN, Nizeyi JB, Cranfield MR, Angert ER. Fecal bacterial diversity in a wild gorilla. Applied and environmental microbiology. 2006;72:3788–3792. doi: 10.1128/AEM.72.5.3788-3792.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Amado MA, Godoy-Vitorino F, Piceno YM, Tom LM, Andersen GL, Herrera EA, et al. Bacterial diversity in the cecum of the world's largest living rodent (Hydrochoerus hydrochaeris) Microbial ecology. 2012;63:719–725. doi: 10.1007/s00248-011-9963-z. [DOI] [PubMed] [Google Scholar]
- Genner MJ, Seehausen O, Lunt DH, Joyce DA, Shaw PW, Carvalho GR, et al. Age of cichlids: new dates for ancient lake fish radiations. Molecular biology and evolution. 2007;24:1269–1282. doi: 10.1093/molbev/msm050. [DOI] [PubMed] [Google Scholar]
- Giatsis C, Sipkema D, Smidt H, Heilig H, Benvenuti G, Verreth J, et al. The impact of rearing environment on the development of gut microbiota in tilapia larvae. Scientific reports. 2015;5:18206. doi: 10.1038/srep18206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham D, Kim H, Lindner A. Methanotrophic bacteria. In: B G, editor. Encyclopedia of environmental microbiology. Wiley; New York: 2002. pp. 1923–1936. [Google Scholar]
- Hammer TJ, Bowers MD. Gut microbes may facilitate insect herbivory of chemically defended plants. Oecologia. 2015;179:1–14. doi: 10.1007/s00442-015-3327-1. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Moran NA. The impact of microbial symbionts on host plant utilization by herbivorous insects. Molecular ecology. 2014;23:1473–1496. doi: 10.1111/mec.12421. [DOI] [PubMed] [Google Scholar]
- Hata H, Tanabe AS, Yamamoto S, Toju H, Kohda M, Hori M. Diet disparity among sympatric herbivorous cichlids in the same ecomorphs in Lake Tanganyika: amplicon pyrosequences on algal farms and stomach contents. BMC Biol. 2014;12:90. doi: 10.1186/s12915-014-0090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata H, Shibata J, Omori K, Kohda M, Hori M. Depth segregation and diet disparity revealed by stable isotope analyses in sympatric herbivorous cichlids in Lake Tanganyika. Zoological Lett. 2015;1:15. doi: 10.1186/s40851-015-0016-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SM, Sanchez C, Carstens BC, Brumfield RT. Comparative Gut Microbiota of 59 Neotropical Bird Species. Frontiers in microbiology. 2015;6:1403. doi: 10.3389/fmicb.2015.01403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. Entropy and diversity. Oikos. 2006;113:363–375. [Google Scholar]
- Karasov WH, Martinez del Rio C. Physiological Ecology: How Animals Process Energy, Nutrients, and Toxins. Princeton University Press; Princeton, New Jersey: 2007. [Google Scholar]
- Karasov WH, Martinez del Rio C, Caviedes-Vidal E. Ecological physiology of diet and digestive systems. Annu Rev Physiol. 2011;73:69–93. doi: 10.1146/annurev-physiol-012110-142152. [DOI] [PubMed] [Google Scholar]
- Kohl KD, Dearing MD. Experience matters: prior exposure to plant toxins enhances diversity of gut microbes in herbivores. Ecology letters. 2012;15:1008–1015. doi: 10.1111/j.1461-0248.2012.01822.x. [DOI] [PubMed] [Google Scholar]
- Kohl KD, Weiss RB, Cox J, Dale C, Dearing MD. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecology letters. 2014;17:1238–1246. doi: 10.1111/ele.12329. [DOI] [PubMed] [Google Scholar]
- Kohl KD, Brun A, Magallanes M, Brinkerhoff J, Laspiur A, Acosta JC, et al. Physiological and microbial adjustments to diet quality permit facultative herbivory in an omnivorous lizard. J Exp Biol. 2016a;219:1903–1912. doi: 10.1242/jeb.138370. [DOI] [PubMed] [Google Scholar]
- Kohl KD, Connelly JW, Dearing MD, Forbey JS. Microbial detoxification in the gut of a specialist avian herbivore, the Greater Sage-Grouse. FEMS Microbiol Lett. 2016b:363. doi: 10.1093/femsle/fnw144. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen T, Ventura M, Maraldo K, Triado-Margarit X, Casamayor EO, Wang YV, et al. The dominant detritus-feeding invertebrate in arctic peat soils derives its essential amino acids from gut symbionts. J Anim Ecol. 2016 doi: 10.1111/1365-2656.12563. [DOI] [PubMed] [Google Scholar]
- Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. Worlds within worlds: evolution of the vertebrate gut microbiota. Nature reviews Microbiology. 2008;6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Guo X, Gooneratne R, Lai R, Zeng C, Zhan F, et al. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Scientific reports. 2016;6:24340. doi: 10.1038/srep24340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matschiner M, Musilova Z, Barth JM, Starostova Z, Salzburger W, Steel M, et al. Bayesian Phylogenetic Estimation of Clade Ages Supports Trans-Atlantic Dispersal of Cichlid Fishes. Syst Biol. 2016 doi: 10.1093/sysbio/syw076. [DOI] [PubMed] [Google Scholar]
- Meyer BS, Matschiner M, Salzburger W. A tribal level phylogeny of Lake Tanganyika cichlid fishes based on a genomic multi-marker approach. Molecular phylogenetics and evolution. 2014;83:56–71. doi: 10.1016/j.ympev.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, Gonzalez A, Fontana L, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muschick M, Indermaur A, Salzburger W. Convergent evolution within an adaptive radiation of cichlid fishes. Current biology : CB. 2012;22:2362–2368. doi: 10.1016/j.cub.2012.10.048. [DOI] [PubMed] [Google Scholar]
- Muschick M, Nosil P, Roesti M, Dittmann MT, Harmon L, Salzburger W. Testing the stages model in the adaptive radiation of cichlid fishes in East African Lake Tanganyika. Proceedings. Biological sciences / The Royal Society. 2014;281 doi: 10.1098/rspb.2014.0605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KE, Zinder SH, Hance I, Burr P, Odongo D, Wasawo D, et al. Phylogenetic analysis of the microbial populations in the wild herbivore gastrointestinal tract: insights into an unexplored niche. Environmental microbiology. 2003;5:1212–1220. doi: 10.1046/j.1462-2920.2003.00526.x. [DOI] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, et al. vegan: Community Ecology Package. R package version 2.3-0. 2015 http://CRAN.R-project.org/package=vegan.
- Pennisi E. How do gut microbes help herbivores? Counting the ways. Science. 2017;355:236–236. doi: 10.1126/science.355.6322.236. [DOI] [PubMed] [Google Scholar]
- Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, Kau AL, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JA, Moreau CS, Goldman-Huertas B, Fujiwara M, Lohman DJ, Pierce NE. Bacterial gut symbionts are tightly linked with the evolution of herbivory in ants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21236–21241. doi: 10.1073/pnas.0907926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Molecular ecology. 2009;18:169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- Sanders JG, Beichman AC, Roman J, Scott JJ, Emerson D, McCarthy JJ, et al. Baleen whales host a unique gut microbiome with similarities to both carnivores and herbivores. Nat Commun. 2015;6:8285. doi: 10.1038/ncomms9285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliewen UK, Tautz D, Paabo S. Sympatric speciation suggested by monophyly of crater lake cichlids. Nature. 1994;368:629–632. doi: 10.1038/368629a0. [DOI] [PubMed] [Google Scholar]
- Schnorr SL, Candela M, Rampelli S, Centanni M, Consolandi C, Basaglia G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. 2014;5:3654. doi: 10.1038/ncomms4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnenburg JL, Backhed F. Diet-microbiota interactions as moderators of human metabolism. Nature. 2016;535:56–64. doi: 10.1038/nature18846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soverini M, Quercia S, Biancani B, Furlati S, Turroni S, Biagi E, et al. The bottlenose dolphin (Tursiops truncatus) faecal microbiota. FEMS microbiology ecology. 2016;92:fiw055. doi: 10.1093/femsec/fiw055. [DOI] [PubMed] [Google Scholar]
- Spor A, Koren O, Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nature reviews Microbiology. 2011;9:279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- Sturmbauer C, Mark W, Dallinger R. Ecophysiology of Aufwuchs-eating cichlids in Lake Tanganyika: niche separation by trophic specialization. Environmental Biology of Fishes. 1992;35:1573–5133. [Google Scholar]
- Sullam KE, Essinger SD, Lozupone CA, O'Connor MP, Rosen GL, Knight R, et al. Environmental and ecological factors that shape the gut bacterial communities of fish: a meta-analysis. Molecular ecology. 2012;21:3363–3378. doi: 10.1111/j.1365-294X.2012.05552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewavas E, Green J, Corbe SA. Ecological studies on crater lakes in West Cameroon Fishes of Barombi Mbo. Journal of Zoology. 1972;167:41–95. [Google Scholar]
- Wagner CE, McIntyre PB, Buels KS, Gilbert DM, Michel E. Diet predicts intestine length in Lake Tanganyika’s cichlid fishes. Functional Ecology. 2009;23:1122–1131. [Google Scholar]
- Wertz JT, Kim E, Breznak JA, Schmidt TM, Rodrigues JL. Genomic and physiological characterization of the Verrucomicrobia isolate Diplosphaera colitermitum gen. nov., sp. nov., reveals microaerophily and nitrogen fixation genes. Applied and environmental microbiology. 2012;78:1544–1555. doi: 10.1128/AEM.06466-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z, Zhang W, Wang L, Hou R, Zhang M, Fei L, et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio. 2015;6:e00022–00015. doi: 10.1128/mBio.00022-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou XX, Pan YJ, Wang YB, Li WF. In vitro assessment of gastrointestinal viability of two photosynthetic bacteria, Rhodopseudomonas palustris and Rhodobacter sphaeroides. J Zhejiang Univ Sci B. 2007;8:686–92. doi: 10.1631/jzus.2007.B0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.