Abstract
To identify novel coding association signals and facilitate characterization of mechanisms influencing glycemic traits and type 2 diabetes risk, we analyzed 109,215 variants derived from exome array genotyping together with an additional 390,225 variants from exome sequence in up to 39,339 normoglycemic individuals from five ancestry groups. We identified a novel association between the coding variant (p.Pro50Thr) in AKT2 and fasting insulin, a gene in which rare fully penetrant mutations are causal for monogenic glycemic disorders. The low-frequency allele is associated with a 12% increase in fasting plasma insulin (FI) levels. This variant is present at 1.1% frequency in Finns but virtually absent in individuals from other ancestries. Carriers of the FI-increasing allele had increased 2-hour insulin values, decreased insulin sensitivity, and increased risk of type 2 diabetes (odds ratio=1.05). In cellular studies, the AKT2-Thr50 protein exhibited a partial loss of function. We extend the allelic spectrum for coding variants in AKT2 associated with disorders of glucose homeostasis and demonstrate bidirectional effects of variants within the pleckstrin homology domain of AKT2.
The increasing prevalence of type 2 diabetes is a global health crisis, making it critical to promote development of more efficient strategies for prevention and treatment. Individuals with type 2 diabetes display both pancreatic beta-cell dysfunction and insulin resistance. Genetic studies of surrogate measures of these glycemic traits can identify variants that influence these central features of type 2 diabetes (2) highlighting potential pathways for therapeutic manipulation. Comprehensive surveys of the influence of common genetic variants on fasting plasma glucose (FG) and fasting plasma insulin (FI) have highlighted defects in pathways involved in glucose metabolism, and insulin processing, secretion, and action (3). Recent studies have identified type 2 diabetes-associated alleles that are common in one population but rare or absent in others (4–6). These associations were observed either due to an increase in frequency of older alleles based on population dynamics and demography (5), or the emergence of population-specific alleles (4; 6).
We set out to identify and characterize low-frequency allele (minor allele frequency [MAF]<5%) glycemic trait associations by meta-analysis of exome sequence and exome array genotype data in a multi-ancestry sample. We also performed in vitro functional studies of protein expression, localization and activity to understand the consequences of our novel findings.
Methods
Genetic association studies
Study Samples
The Genetics of Type 2 Diabetes (GoT2D) study and Type 2 Diabetes Genetic Exploration by Next-generation sequencing in multi-Ethnic Samples (T2D-GENES) study were initially designed to evaluate the contribution of coding variants to type 2 diabetes risk (7). We performed a discovery association analysis to find novel coding variants associated with fasting glycemic traits in 14 studies from GoT2D that contributed exome array information on 33,231 non-diabetic individuals of European ancestry. Further discovery analysis was performed with GoT2D and T2D-GENES studies with exome sequence data (average 80x coverage) in five ancestral groups comprised of 12,940 individuals (6,504 with type 2 diabetes, 6,436 without) with measured FG or FI levels available in 2,144 European, 508 South Asian, 1,104 East Asian, 844 Hispanic, and 508 African American non-diabetic individuals. We performed a replication analysis and an assessment of allele frequency distributions in 5,747 individuals from four Finnish cohorts: Cardiovascular Risk in Young Finns Study (YFS) (8), Helsinki Birth Cohort (HBCS) (9), Health 2000 GenMets Study (GenMets) (10), and National FINRISK Study 1997 and 2002 (FR) (11). We also assessed the allele frequencies of novel findings in 46,658 individuals from CHARGE studies with available exome array data (12), although none of the studies passed our QC filter of a minor allele count greater than 5 for inclusion in our replication analysis. See Supplementary Table 1 for study details, sample characteristics, ascertainment criteria, and detailed genotype calling and quality control procedures for each cohort. The relevant institutional review boards, conducted according to the Declaration of Helsinki, approved all human research and all participants provided written informed consent. A detailed description of ethical permissions is provided in the Supplementary Materials.
Phenotypes
For the discovery and replication analysis, we excluded individuals from the analysis if they had a diagnosis of type 2 diabetes, were currently receiving oral or injected diabetes treatment, had FG measures ≥ 7mmol/L, had 2-hour post-load glucose (2hrG) measures ≥ 11.1mmol/L, or had HbA1c measures ≥ 6.5% (48mmol/mol). Additional exclusions occurring at the study level included pregnancy, non-fasting at time of exam, type 1 diabetes, or impaired glucose tolerance. See Supplementary Table 1A for details. Within each study, we adjusted FG and log transformed FI levels for age, sex, body mass index (BMI), and additional study specific covariates. We applied rank-based inverse-normal transformations to study- or ancestry-specific residuals to obtain satisfactory asymptotic properties of the exome-wide association tests.
We tested for genetic associations with type 2 diabetes, hypertension, and other related quantitative traits in the Finnish discovery and replication cohorts. We analyzed lipid levels (total cholesterol, high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), and triglycerides (TG)), blood pressure (systolic (SBP) and diastolic (DBP) blood pressures and hypertension (HTN)), height, BMI, central adiposity measures (waist-to-hip ratio (WHR), waist circumference, hip circumference), adiponectin level, 2-hour insulin level, and Matsuda index, which is known to correlate with whole-body insulin sensitivity as measured by the hyperinsulinemic euglycemic clamp (r=0.7, P<1.0×10-4) (13). For quantitative traits and HTN, we adjusted for age, sex, BMI (for glycemic, blood pressure, and central adiposity traits), stratified by type 2 diabetes status and sex (for central adiposity measures) within study. We adjusted LDL and total cholesterol for use of lipid-lowering medication, by dividing total cholesterol by 0.8 if on lipid-lowering medication, prior to calculating LDL using the Friedewald equation (14). SBP and DBP were adjusted for use of blood pressure-lowering medication by adding 15 mmHg to SBP and 10 mmHg to DBP measurements if an individual reported taking blood pressure-lowering medication (15). The Matsuda Index was log transformed and analyzed in non-diabetic individuals only. After adjusting for covariates, traits were inverse-normalized within strata. In addition to studying these metabolic outcomes, we used international classification of diseases (ICD) codes to query electronic medical records in the METSIM and FINRISK 1997 and 2002 cohorts (in all individuals regardless of type 2 diabetes status) and categorized affection status for lipodystrophy, polycystic ovary disease, and ovarian or breast cancer.
Statistical Analysis
Discovery Analysis
We performed association analyses within each study for the exome array data sets and within ancestry for the exome sequence data sets. We used linear mixed models implemented in EMMAX (16) to account for relatedness. Within each study/ancestry, we required variants to have a minor allele count (MAC) greater than or equal to five alleles for single variant association tests. We meta-analyzed the single variant results from the (European-ancestry) exome array studies using the inverse variance meta-analysis approach implemented in METAL (17) and combined these with the European ancestry exome sequence results. Then, we meta-analyzed summary statistics across ancestries. We used P<5×10-7 as exome-wide statistical significance thresholds for the single variant tests (18). We used the binomial distribution to assess enrichment of previously reported associations with FG or FI by calculating a P-value for the number of non-significant variants with consistent direction of effects.
Gene based association analysis
We performed gene-based association tests using variants with MAF <1% (including rare variants with MAC≤5), annotating and aggregating variants based on predicted deleteriousness using previously described methods (7). Briefly, we defined four different variant groupings: “PTV-only”, containing only variants predicted to severely impair protein function, “PTV+missense”, containing PTV and NS variants with MAF <1%, “PTV+NSstrict” composed of PTV and NS variants predicted damaging by five algorithms (SIFT, LRT, MutationTaster, polyphen2 HDIV, and polyphen2 HVAR), and “PTV+NSbroad” composed of PTV and NS variants with MAF<1% and predicted damaging by at least one prediction algorithm above. We used the sequence kernel association test (SKAT) (19) and a frequency-weighted burden test to conduct exome array meta-analyses in an unrelated subset of individuals using RareMETAL (20). We conducted exome sequence gene-based analyses within ancestry using a linear mixed model to account for relatedness and combined results across ancestries with MetaSKAT (21), which accounts for heterogeneous effects. We further combined gene-based results from exome array and exome sequences using Stouffer’s method with equal weights. For gene-based tests, we considered P<2.5×10-6 as exome-wide significant, corresponding to Bonferroni correction for 20,000 genes in the genome (18).
Replication Analysis
The AKT2 p.Pro50Thr variant was observed at sufficient frequency in the independent Finnish cohorts to perform single-variant association test of association with FI. We tested association in SNPTEST (22) (v.2.4.0) in each study with the same additive linear model used in the discovery analysis. Covariate adjustments for FI levels were sex, age, and ten principal components (PCs), and models were run with and without adjustment for BMI.
Estimate of effect on raw FI level and variance explained
To characterize the association between AKT2 p.Pro50Thr and FI, we examined full regression models with raw FI in three studies (FUSION, METSIM, and YFS). We estimated the raw effect on log-transformed FI levels with a fixed-effects meta-analysis. The variance in log-transformed FI explained by AKT2 p.Pro50Thr was estimated by a weighted average of the narrow-sense heritability of AKT2 p.Pro50Thr seen in these three studies.
Population genetics and constraint
We used the Exome Aggregation Consortium (ExAC) for constraint metrics and allele frequencies (23). We obtained sequence alignments for AKT proteins and mRNAs in 100 vertebrates from the UCSC Genome Browser (24), used Shannon’s entropy (normalized K=21) as a conservation score (25) and plotted the sequence logos in R using the RWebLogo library (26).
Associations with other traits
We conducted association tests for traits other than FI and FG within studies for both discovery studies as well as the independent Finnish studies used for replication. P-values for type 2 diabetes and HTN came from EMMAX (16) or the Wald test from logistic regression (Finnish replication data sets) and meta-analyzed using an N weighted meta-analysis (17). Odds ratios (OR) were obtained from logistic regression adjusting for age, sex, with and without BMI, and PCs and meta-analyzed using an inverse variance meta-analysis.
Trait distributions and phenotype clustering
We examined distributions of traits among AKT2 missense allele carriers (p.Pro50Thr, p.Arg208Lys, and p.Arg467Trp) in the T2D-GENES exome sequencing data set. We used non-parametric rank based methods (kruskal.wallis and permKS functions in R) on both the inverse-normalized covariate-adjusted traits used in the genetic association studies and normalized raw trait values (scale function in R). We clustered AKT2 missense allele carriers on scaled trait values (pheatmap function in R).
In vitro functional studies
Plasmids and cell lines
The generation of the AKT2 allelic series was initiated by the production of pDONR223-AKT2 through PCR of the human AKT2 open reading frame with the integration of terminal attR sites using primers (see below). HeLa, HuH7, and 293T cells were obtained at The Broad Institute and maintained in 10% FBS DMEM, 100U/ml penicillin and 100µg/ml streptomycin, and documented mycoplasma-free. HeLa and HuH7 cells were starved for 18 hours and stimulated for 15 minutes with 100nM insulin for activation analyses.
Primers for functional work
The generation of the AKT2 allelic series was initiated by the production of pDONR223-AKT2 through PCR of the human AKT2 open reading frame with the integration of terminal attR sites using primers FWD: 5 ’- GGGGACAAGTTTGTACAAAAAAGTTGGCACCATGAATGAGGTGTCTGTCATC -3’ REV: 5’- GGGGACCACTTTGTACAAGAAAGTTGGCAACTCGCGGATGCTG -3’, and subsequent Gateway BP reaction into pDONR223 obtained from The Broad Institute Genetics Perturbation Platform. Site-directed mutagenesis was then performed to generate AKT2.E17K (AKT2.Lys17), AKT2.P50T (AKT2.Thr50), AKT2.R208K (AKT2.Lys208), AKT2.R274H (AKT2.His274), AKT2.R467W (AKT2.Trp467) with the following primers: AKT2.E17K: FWD: 5'- GGCTCCACAAGCGTGGTAAATACATCAAGACCTGG -3' REV: 5'- CCAGGTCTTGATGTATTTACCACGCTTGTGGAGCC -3'; AKT2.P50T: FWD: 5'- AGGCCCCTGATCAGACTCTAACCCCCTTAAAC -3' REV: 5'- GTTTAAGGGGGTTAGAGTCTGATCAGGGGCCT -3'; AKT2.R208K: FWD: 5'- GTCCTCCAGAACACCAAGCACCCGTTCC -3' REV: 5'- GGAACGGGTGCTTGGTGTTCTGGAGGAC -3'; AKT2.R274H: FWD: 5'- GGGACGTGGTATACCACGACATCAAGCTGGA -3'REV3'REV: 5'- TCCAGCTTGATGTCGTGGTATACCACGTCCC -3'; AKT2.R467W: FWD: 5'- GGAGCTGGACCAGTGGACCCACTTCCC -3' REV: 5'- GGGAAGTGGGTCCACTGGTCCAGCTCC -3'. C-terminal, V5-tagged lentiviral pLX304-AKT2.E17K, pLX304-AKT2.P50T, pLX304- AKT2.R208K, pLX304-AKT2.R274H, and pLX304- AKT2.R467W were each generated by subsequent Gateway LR reactions with pDONR223-AKT2.E17K, pDONR223-AKT2.P50T, pDONR223-AKT2.R208K, pDONR223-AKT2.R274H, and pDONR223-AKT2.R467W, respectively, and pLX304 obtained from The Broad Institute Genetics Perturbation Platform. Control plasmid pLX304- empty vector was additionally acquired from The Broad Institute Genetics Perturbation Platform.
Antibodies
Anti-Akt (#4685), anti-phospho-Akt S473 (#4060), anti-phospho-Akt T308 (#9275), anti-β Actin (#4970), anti-GSK3β (#9315), anti-phospho-GSK3β (#9336), anti-GST (#2625), and anti-V5 (#13202) were purchased from Cell Signaling Technologies (product numbers listed for each). Horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin G (IgG) antibodies were purchased from Millipore.
3D modeling
The 3D structure of AKT2 with the full allelic series was predicted using IntFOLD (27) and visualized in PyMOL (28).
In vitro kinase assays
We isolated V5-AKT2, V5-AKT2.Lys17, V5-AKT2.Thr50, V5-AKT2.Lys208, V5-AKT2.His274, and V5-AKT2.Trp467 variants from lentivirally infected and 5μg/mL blasticidin selected HeLa cell lysate with V5 agarose beads (SIGMA) and incubated with 150ng GST-GSK3β substrate peptide (Cell Signaling Technologies) and 250mM cold ATP in kinase assay buffer (Cell Signaling Technologies) for 35 minutes at 30°C.
Proliferation assay
We cultured lentiviral pLX304 V5-AKT2 variants and control empty vector infected and 5μg/mL blasticidin selected HuH7 cells in 24 well plate for 72 hours in 10% FBS /phenol red-free DMEM for 72 hours. We added WST-1 (Takara Clontech) to each well at the manufacture recommended 1:10 ratio and incubated for 4 hours at 37°C prior to absorbance measurement at 450nm with BioTek Synergy H4 plate reader.
Immunoblots
We washed cells with phosphate buffered saline and lysed in EBC buffer (120mM NaCl, 50mM TRIS-HCl (pH7.4), 50nM calyculin, cOmplete protease inhibitor cocktail (Roche), 20mM sodium fluoride, 1mM sodium pyrophosphate, 2mM ethylene glycol tetraacetic acid, 2mM ethylenediaminetetraacetic acid, and 0.5% NP-40) for 20 minutes on ice. To preclear cell lysates, we centrifuged at 12,700 rmp at 4°C for 15 minutes. We measured protein concentration with Pierce BCA protein assay kit using a BioTek Synergy H4 plate reader. We resolved lysates on BioRad any kD mini-PROTEAN TGX polyacrylamide gels by SDS-PAGE and transferred by electrophoresis to nitrocellulose membrane (Life Technologies) at 100V for 70 minutes. We blocked membranes in 5% nonfat dry milk/ TBST (10mM Tris-HCl, 150mM NaCl, 0.2% Tween 20) buffer pH 7.6 for 30 minutes. We incubated blots with indicated antibody overnight at 4°C. The membrane was then washed in TBST, three times at 15 minute intervals, before 1 hour secondary horseradish peroxidase-conjugated antibody incubation at room temperature. We again washed nitrocellulose membranes in TBST, three times for 15 minutes, prior to enhanced chemiluminescent substrate detection (Pierce).
Statistical analysis
The quantified results of the in vitro kinase and proliferation assays were normalized to internal control values for each replicate. We used generalized linear models of the quantified assay results to assess effects of variants within and across replicate rounds, allowing for interaction by replicate. The graphical representation was produced using functions in the effects (v 3.0-3) package in R.
Gene Expression Studies
Study samples
GTEx: We compared the expression pattern of AKT2 to the two other members of the AKT gene family, AKT1 and AKT3, using multi-tissue RNA sequencing (RNA-seq) data from the pilot phase of the GTEx project (dbGaP accession number: phs000424.v3.p1) in 44 tissues with data from more than one individual. Detailed procedures for sample collection, RNA extraction, RNA-seq, and gene and transcript quantifications have been previously described (29). EuroBATs: Samples from photo protected subcutaneous adipose tissue from 766 twins were extracted (130 unrelated individuals, 131 monozygotic and 187 dizygotic twin pairs) and processed as previously described (30; 31). METSIM: Subcutaneous fat biopsy samples were obtained from a sample of 770 participants from the METSIM study and processed as previously described (32).
Phenotypes
We studied the association of age, body mass index (BMI) and fasting insulin levels with gene expression levels and with expression-associated SNPs (eQTLs) in the AKT2 region. Age and sex were available for the GTEx study samples. In additional to age and BMI, fasting insulin level was measured at the same time point as the fat biopsies in the EuroBATs sample data, following a previously described protocol (33). Baseline age, BMI and fasting insulin levels were used for the METSIM study participants (34)
Statistical analysis
The comparison of expression levels of AKT2 versus AKT1, and AKT2 versus AKT3 was performed using log2-transformed reads per kilobase per million mapped reads (RPKMs). The percent increase in AKT2 expression was calculated with the following formula: 2^log-fold-change (AKT2 vs AKT1). We studied BMI, age, and fasting insulin (not available in GTEx data) associations with AKT2 expression using linear mixed models as implemented in the lme4 package in R. The gene expression RPKM values were inverse variance rank normalized for these analyses. Covariates included study-specific fixed and random effects (see Supplementary Note 4 for additional details on each cohort), using sex, BMI and age as additional fixed effects as appropriate. The expression quantitative trait loci (eQTL) analysis was performed on single nucleotide polymorphisms (SNPs) within a 1 Mb of AKT2 using linear mixed models to assess the association of the SNPs with the inverse normalized RPKM expression values.
Results
Genetic association studies
We tested the association of FI and FG with 390,225 variants from exome sequence data (GoT2D and T2D-GENES studies) and 109,215 variants derived from exome array genotyping (GoT2D studies) (7) (individual study λGC<1.06; Supplementary Figure S1). We examined variants that had been previously associated with FG and FI (3; 18). Of 28 FG and 14 FI loci with the reported SNPs or close proxies in our data set, 13 FG and four FI showed directionally consistent significant associations. Among the remaining GWAS loci not significant in our data, we observed directionally consistent associations in 14/15 FG and 9/10 FI loci (Penrichment =5×10-4 for FG and 0.01 for FI) (Supplementary Note 1; Supplementary Table 2).
In addition, we identified a novel significant single variant association between rs184042322 and FI (MAF=1.2%, P=1.2×10-7), a coding variant in AKT2 (V-AKT Murine Thymoma Viral Oncogene Homolog 2) where amino acid Pro50 is substituted with a threonine (NP_001617.1:p.Pro50Thr) (Figure 1; Supplementary Figure S1). The same allele drove a significant FI signal for AKT2 in gene-based analysis (P=6.1×10-7), in which we discovered two additional significant gene-based associations between GIMAP8 and FG (PPTV=2.3×10-6), and between NDUFAF1 and FI (PPTV+NSBroad=9.2×10-7) (Supplementary Figure S2; Supplementary Table 2D).
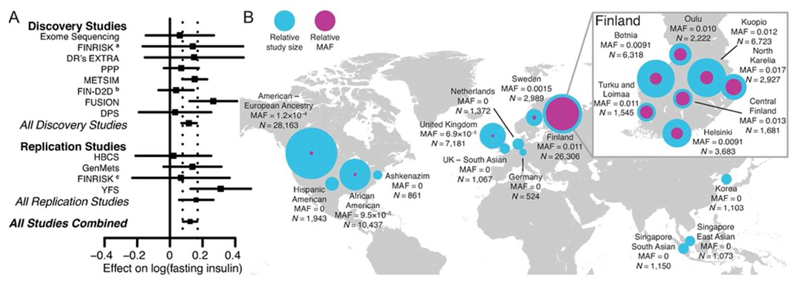
Figure 1. AKT2 Pro50Thr association with fasting insulin levels.
(a) For each study, the square represents the estimate of the additive genetic effect for the association of the AKT2 Pro50Thr allele with log-transformed fasting insulin (FI) levels and the horizontal line gives the corresponding 95% confidence interval of the estimate. Inverse-variance meta-analyses were performed for All Discovery Studies, All Replication Studies, and All Studies Combined. The vertical dashed lines indicate the 95% confidence interval for the estimate obtained in the meta-analysis of All Studies Combined. (b) Minor allele frequency for each available region and ancestry. Across countries the world, the MAF ranges from 0% to 1.1%. The relative sample sizes (N) for each region/ancestry are displayed with the blue circles and the relative minor allele frequencies of AKT2 Pro50Thr are displayed with the purple circles, with the size of the circles showing comparative differences. Within Finland (inset), where the MAF ranges from 0.9% to 1.7%, birthplace and study center data were used to show the allele distribution across the country. a FINRISK 2007; b FIN-D2D 2007; c FINRISK 1997 and 2002
In an effort to replicate the single variant association of AKT2 Pro50Thr with FI, we aggregated the allele frequency estimates of AKT2 Pro50Thr in our data with data from the CHARGE consortium and the four Finnish studies. In ExAC, rs184042322 is multi-allelic (p.Pro50Thr and p.Pro50Ala) but Pro50Ala is observed only twice in the Latino population sample and not seen in our exome sequencing data, which includes 1,021 individuals of Hispanic ancestry. AKT2 Pro50Thr was observed at a much higher frequency in Finnish individuals (MAF=1.1%) than other European (MAF=0.2%), African American (MAF=0.01%), Asian (MAF<0.01%), or Hispanic (MAF<0.01%) individuals (Figure 1). We replicated the association between FI and AKT2 Pro50Thr by meta-analysis of the association in the four Finnish studies (P=5.4×10-4, N=5,747) with the discovery studies (Pcombined=9.98×10-10, N=25,316). We observed no evidence of effect-size heterogeneity between studies (PHeterogeneity=0.76). The minor T allele was associated with a 12% (95% CI=7%-18%) increase in FI levels in the discovery and replication studies, a per allele effect of 10.4pmol/L (95% CI=6.6-14.3pmol/L).
The serine/threonine protein kinases AKT1, AKT2, and AKT3 are conserved across all vertebrates (Figure 2). Pro50 and the seven preceding residues in the pleckstrin homology (PH) domain appear to be specific for the AKT2 isoform. Population genetic studies show a strong intolerance to missense and loss of function variation in AKT2 (Supplementary Note 2; Supplementary Figure S3; Supplementary Figure S4; Supplementary Table 3). Notably, in ExAC data, AKT2 contains fewer missense variants than expected (the missense constraint metric, Z=3.5, is in the 94th percentile of all genes) and extreme constraint against loss-of-function (LoF) variation (estimated probability of being LoF intolerant (pLI)=1).
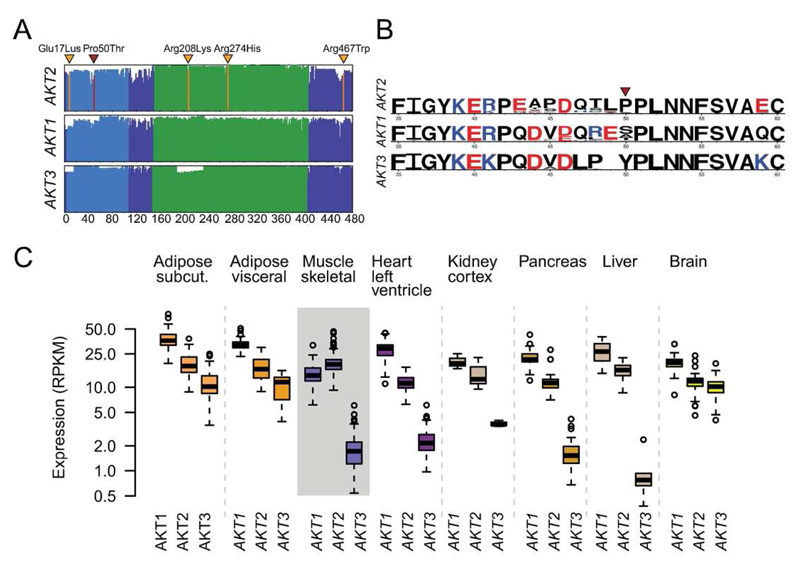
Figure 2. Expression and conservation properties.
(a) Amino acid alignment and conservation of the three AKT proteins in vertebrates. The x axis gives the amino acid position and the height of the lines shows the conservation score across 100 vertebrate genome alignments. The functional domains are the pleckstrin homology (PH) domain (blue) and the kinase domain (green). The position of AKT2 Pro50Thr is shown in red while the locations of the other AKT2 disease-causing mutations (37–40) are shown in orange: Glu17Lys, Arg208Lys, Arg274His, and Arg467Trp. (b) WebLogo plots of amino acids 35-60 are shown for AKT2, AKT1, and AKT3 contrasting the homology of the three isoforms. The height of letters gives the relative frequency of different amino acids across the 100 vertebrate species, with the colors showing amino acids with similar charge. (c) Expression of AKT1, AKT2, and AKT3 in eight insulin-sensitive tissues using RNA sequencing data from the GTEx consortium.
AKT2 is a primary transducer of phosphoinositide 3-kinase (PI3K) signaling downstream of the insulin receptor and is responsible for mediating the physiological effects of insulin in tissues including liver, skeletal muscle, and adipose. Akt2 null mice are characterized by hyperglycemia and hyperinsulinemia, and some develop diabetes (35; 36). In humans, highly penetrant rare alleles in AKT2 cause familial partial lipodystrophy and hypoinsulinemic hypoglycemia with hemihypertrophy (Glu17Lys) (37; 38) and a syndrome featuring severe insulin resistance, hyperinsulinemia, and diabetes mellitus (Arg274His) (39). Additional rare alleles have been observed in individuals with severe insulin resistance (Arg208Lys and Arg467Trp) but no variant has been associated with glycemic traits at the population level (40).
Given the spectrum of diseases and traits associated with AKT2 (41), we hypothesized that AKT2 Pro50Thr would be associated with features of metabolic syndrome or lipodystrophy. In quantitative trait analysis in the initial discovery and replication cohorts, we did observe a constellation of features indicative of a milder ‘lipodystrophy-like phenotype’ associated with the rare allele: associations with increased 2-hour insulin values (effect=0.2 SD of log-transformed 2-hour insulin, 95% CI=0.1-0.4; P=7.9×10-8, N=14,150), lower insulin sensitivity (effect=-0.3 SD of the log-transformed Matsuda index, 95% CI=-0.5 to -0.2, P=1.2×10-6, N=8,566), and increased risk of type 2 diabetes (odds ratio (OR)=1.05 95% CI=1.0-1.1, P=8.1×10-5 ; 9,783 type 2 diabetes cases; 22,662 controls), with no effects on fasting glucose, postprandial glucose, or fasting lipid levels (P≥0.01; Supplementary Table 4). In the T2D-GENES exome sequencing data where FG and FI levels were available in diabetic individuals, we observed one individual who was homozygous for the P50T allele with FI and FG levels in the 99.8th and 98.8th percentiles, respectively. There was a significant difference in trait distributions by P50T genotype (FI P=0.002; FG P=0.02; Supplementary Figure S5; Supplementary Table 4). Next, we used electronic health records available in the Finnish METSIM and FINRISK cohorts to characterize the impact of AKT2 Pro50Thr on disease risk. We found no evidence for association with any cancer, polycystic ovary disease, or acanthosis nigricans (Supplementary Table 5); however, these tests are underpowered due to the low number of cases and potential for misclassification. Nor did we find evidence for enrichment of low-frequency associations in any AKT2 related pathways or genes implicated in monogenic forms of glycemic disease (Supplementary Note 3; Supplementary Table 6; Supplementary Table 7; Supplementary Figure S6; Supplementary Figure S7).
In vitro functional studies
To understand the functional consequences of the AKT2 Pro50Thr variant on the protein, we investigated protein expression, activation, kinase activity, and downstream effector phosphorylation.
First, we used in silico classifiers that predict potential functional consequences of alleles on protein function. Two of the five classifiers predicted AKT2 Pro50Thr to be deleterious (Supplementary Table 3). Second, we used 3D models of AKT2 viewed in the PyMol software, which predicted that the Pro50Thr variant causes a change in the conformations of the lipid binding PH domain (Figure 3, Supplementary Figure S8). We hypothesized that the variant protein is inefficiently recruited to the plasma membrane thereby impacting AKT2 phosphorylation and downstream activity.
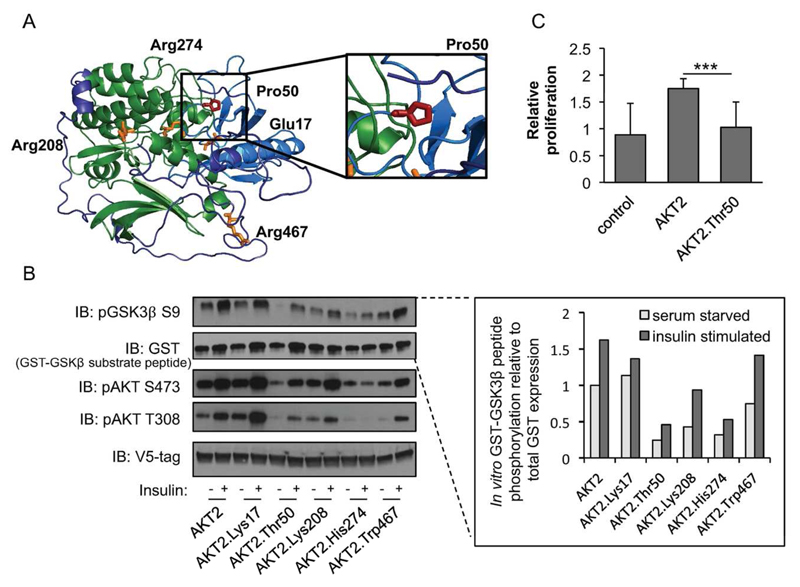
Figure 3. Functional properties of AKT2-Thr50.
(a) Predicted protein structure of AKT2. Domain and variants are highlighted as in Figure 2. The relative spatial positioning of the AKT2-Pro50 residue is magnified within the inset. (b) HeLa cells were infected with lentiviral V5-AKT2, V5-AKT2-Lys17, V5-AKT2-Thr50, V5-AKT2-Lys208, V5-AKT2-His274, V5-AKT2-Trp467, starved for 18 hours (white bar), and stimulated for 20 minutes with 100nm insulin (grey bar). V5-tagged AKT2 was isolated from cell lysates with anti-V5 agarose beads and incubated with GSK3β-GST peptide in an in vitro kinase (IVK) assay. Quantification of phosphorylated substrate peptide (pGSK3β) relative to total peptide (GST-GSK3β) is shown at the inset. Immunoblots and quantification shown are representative of three independent replicates. Linear model (LM) statistical analyses across all three independent replicates are available in Supplementary Figure S9. The IVK was immunoblotted (IB) with the indicated antibodies. (c) HuH7 cells were infected with lentiviral V5-AKT2, V5-AKT2-Thr50, or control pLX304. At 72 hours relative cellular proliferation was determined with WST-1 assay of HuH7 cells. Error bars represent the standard deviation (SD). *** P=4.5×10-5.
To assess the molecular and cellular consequence of the AKT2 Thr50 variant on protein function, we performed a comparative analysis of AKT2-Thr50 with inactivating and activating alleles implicated in monogenic disorders of insulin signaling. Analysis of AKT2-Thr50 expression showed that while AKT2 protein levels remained unchanged, there was a partial loss of AKT2-Thr50 phosphorylation at its activation sites (Thr308 and Ser473) in HeLa cells, suggesting impaired AKT2 signaling (Figure 3; Supplementary Figure S9). Similar effects were observed in human liver derived HuH7 cells (Supplementary Figure S10). AKT2-Thr50 also showed a reduced ability to phosphorylate its downstream target glycogen synthase kinase 3 beta (GSK3β). These defects in AKT2-Thr50 activity were confirmed through an in vitro kinase assay (P<0.01) (Figure 3). AKT2-Thr50 showed a similar decrease in kinase function to the lipodystrophy-causing AKT2-His274 variant. Using a four-hour time course analysis of AKT2 activity, we verified a reduction in both maximally phosphorylated Thr308 and Ser473 in AKT2-Thr50 (Supplementary Figure S11). To understand how this loss of activity could manifest as a defect in a known cellular function of AKT2 (42), we determined the impact of AKT2-Thr50 on cell proliferation in HuH7 cells. While the addition of AKT2 stimulated hepatocyte proliferation, the response to AKT2-Thr50 was reduced (effect=-1.2, P<1.0×10-3) (Figure 3C; Supplementary Figure S12).
Gene expression studies
We queried RNA sequencing data from the Genotype Tissue Expression (GTEx) Project and found that, in agreement with previous studies (43), AKT2 is highly and ubiquitously expressed across all tissues (44 tissue types, 3-156 individuals/tissue). Notably the AKT2 Pro50Thr containing exon is expressed in all tissues and individuals (Supplementary Figure S13), suggesting that the PH domain is important to AKT2 function (44). Of the three AKT homologs, AKT2 had 1.4-fold higher expression in skeletal muscle than AKT1 (P=1.5×10-19) and 11-fold higher expression than AKT3 (P=7.8×10-91). Skeletal muscle was the only tested tissue displaying such pronounced AKT2 enrichment (Figure 2; Supplementary Note 4; Supplementary Figure S14; Supplementary Table 8).
Motivated by the age-related loss of adipose tissue in Akt2 null mice (35; 36) and the growth and lipodystrophy phenotypes in carriers of fully-penetrant alleles (37–40), we examined associations of expression levels of AKT2 with BMI, FI, and age in the three adipose tissue data sets (Supplementary Table 9). We found an association between lower BMI levels and higher AKT2 expression in two cohorts (EuroBATS effect=-0.07 SD, P=6.1×10-28; METSIM effect=-0.06 SD, P=8.1×10-8) and also observed that higher AKT2 expression was associated with lower log-transformed FI (EuroBATS, effect=-0.04 SD, P=1.1×10-3, METSIM, effect=-0.4 SD, P=3.3×10-11). We next tested for gene expression quantitative trait loci (eQTL) and found an eQTL in the 5’UTR of AKT2 (rs11880261; MAF=35%; r2=0.002, D’=0.47 in the Finnish 1000 Genomes samples) with the common allele associated with lower AKT2 expression levels (METSIM P=6.9×10-14; EuroBATS P=2.3×10-8; GTEx P=0.08) (Supplementary Figure S15). No association was detected between rs11880261 and FI levels, suggesting that the common variant eQTL does not drive the initial FI association (Supplementary Note 4; Supplementary Table 10).
Discussion
Meta-analyses of exome sequence and array genotyping data in up to 38,339 normoglycemic individuals enabled the discovery, characterization, and functional validation of a FI association with a low-frequency AKT2 coding variant. Rare, penetrant variants in genes encoding components of the insulin signaling pathway, including AKT2, cause monogenic but heterogeneous glycemic disorders (45). In parallel, common alleles in or near many of these genes impact FI levels —the AKT2 Pro50Thr association shows an effect 5 to 10 times larger than those of these previous published associations (3). This discovery expands both the known genetic architecture of glucose homeostasis and the allelic spectrum for AKT2 coding variants associated with glucose homeostasis into the low-frequency range, and highlights the effects of both locus and allelic heterogeneity (Figure 4).
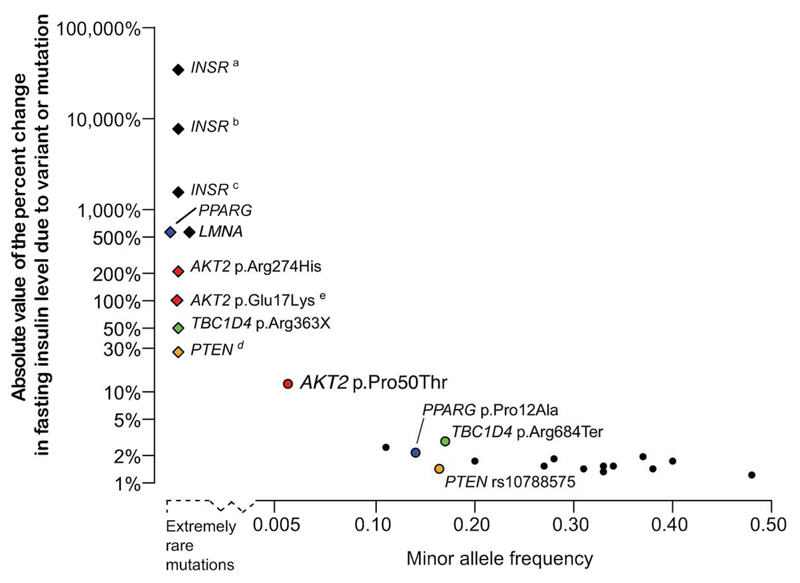
Figure 4. Genetic architecture of rare, low frequency, and common variants associated with FI levels.
In this plot, the absolute values of the percent change in fasting insulin level due to rare monogenic mutations (diamonds) and common genetic variants (circles) are plotted against the minor allele frequency of the variant. The extremely rare monogenic mutations (above the dashed line to the left of the x axis) were observed in 2 to 18 individuals (3; 37–40; 47; 53; 54) with the height of the point indicating the percent change in fasting insulin levels of mutation carriers from 40 pmol/L, an estimate of population mean fasting insulin level. Mutations in INSR and AKT2 p.Arg274His cause compensatory hyperinsulinemia, individuals with TBC1D4 p.Arg363Ter show normal fasting insulin levels but postprandial hyperinsulinemia, and mutations in PTEN cause enhanced insulin sensitivity providing protection against type 2 diabetes. For common variants, the percent change in fasting insulin levels per insulin-increasing allele is plotted above the solid horizontal axis. These observations are from sequencing (6) and array-based GWAS (3). For several genes, the effects from rare mutations can be compared to the effects of common variants in or near the gene: PPARG (blue), TBC1D4 (green), PTEN (orange), and AKT2 (red). a Donohue syndrome: Biallelic loss-of-function mutations in INSR (54). b Rabson-Mendenhall syndrome: Biallelic loss-of-function mutations in INSR (54). c Post-pubertal severe IR: Heterozygous or homozygous loss-of-function mutations in INSR (54). d Loss of function PTEN mutations cause Cowden Syndrome in which carriers exhibit a lowered fasting insulin level (mean=29 pmol/l) compared to matched controls (3). e Carriers with the AKT2 p.Glu17Lys mutation were described with hypoinsulinemic hypoketotic hypoglycemia and hemihypertrophy with undetectable serum insulin (37; 38).
Individuals of Finnish ancestry drove the AKT2 Pro50Thr association signal. This demonstrates the value of association studies in different ancestries where frequencies of rare alleles may increase due to selective pressure or stochastic changes from population bottlenecks and genetic drift. The allele associated with increased FI most likely rose to a higher frequency due to genetic drift and exists within the spectrum of rare and low-frequency variation observed in Finland, the excess of which facilitates the study of complex trait associations (46).
While the AKT2 Pro50Thr allele shows a strong effect on all of the insulin measures and modest increased type 2 diabetes risk (OR=1.05) we see no effect on any of the glucose measures in individuals without diabetes. Due to the effects of both type 2 diabetes and its treatment on glucose homeostasis, we have not tested genetic associations of FG and FI in individuals with type 2 diabetes, although we observed a diabetic individual homozygous for P50T with extreme FI and FG levels. The mechanism for such heterogeneous effects is unclear and detailed in vivo physiological studies are needed.
We leveraged similar findings to generate hypotheses for future work on AKT2 and downstream targets to further illuminate tissue-specific mechanisms. All reported carriers of the lipodystrophy causing AKT2 Arg274His allele are hyperinsulinemic, and three of the four carriers have diabetes mellitus (39). These observations are similar to the ones made for TBC1D4 (which encodes a protein that acts as a substrate immediately downstream of AKT2 in the PI3K pathway). In TBC1D4 a population specific, protein-truncating variant (Arg684Ter) is associated with increased type 2 diabetes risk (OR = 10.3), increased postprandial glucose and insulin levels, and a modest decrease in FI and FG levels (6) (Figure 4). Another stop codon allele in TBC1D4, Arg363Ter that is rare (not observed in ExAC) has been reported with a modest elevation in FI levels but extreme postprandial hyperinsulinemia and acanthosis nigricans (47). siRNA-mediated gene knock-down of AKT2 in human primary myotubes completely abolishes insulin action on glucose uptake and glycogen synthesis (48), which highlights the importance of an intact AKT2-TBC1D4 signaling pathway in the regulation of insulin sensitivity in humans. TBC1D4 is ubiquitously expressed with adipose and skeletal muscle tissue ranking among the tissues with highest expression in GTEx. TBC1D4 Arg363Ter seems to have an effect in adipocytes (47), while Arg684Ter falls in an exon that is exclusively expressed in skeletal and heart muscle (6; 49). This is a likely cause of the TBC1D4 Arg684Ter tissue specificity, which appears to differ from the other TBC1D4 Arg363Ter variant as well as the AKT2 variants.
The phenotypes exhibited by carriers of rare, penetrant AKT2 alleles reflect differential AKT2 activation with kinetically inactivating variants resulting in hyperinsulinemia and lipodystrophy while kinetically activating variants lead to hypoglycemia (37–39). The decrease of cellular proliferation we observe demonstrates that the downstream signaling changes caused by AKT2-Thr50 are sufficient in hepatocytes to impair AKT2 function at the cellular level while maintaining varying portions of regulatory capacity. Along with the observed association with increased fasting insulin levels in human populations, these results support AKT2 Pro50Thr as a partial loss-of-function variant. The inactivating AKT2 Pro50Thr variant contrasts with the known activating AKT2 Glu17Lys mutation and showcases bidirectional effects within the PH domain of AKT2. While the Pro50 residue is conserved in AKT2 throughout all vertebrates, the variant lies within the PH domain that is not conserved between AKT isoforms (Figure 2). These residues, harboring the Pro50 variant, may functionally distinguish AKT2 from AKT1 and AKT3. Although AKT isoforms are activated in the same mechanism within the PI3K pathway downstream of insulin, the Akt2-/- mouse is the only knockout of the gene family to be characterized by insulin resistance and diabetes (35; 50–52). A deeper understanding of what makes the AKT2 isoform distinct could offer potential sites for therapeutic intervention and enable more targeted approaches to disease prevention.
Supplementary Material
Acknowledgements
C.M.L. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
We thank the more than 44,412 volunteers who participated in this study. We acknowledge the following funding sources: Academy of Finland (129293, 128315, 129330, 131593, 139635, 139635, 121584, 126925, 124282, 129378, 258753); Action on Hearing Loss (G51); Ahokas Foundation; American Diabetes Association (#7-12-MN-02); Atlantic Canada Opportunities Agency; Augustinus foundation; Becket foundation; Benzon Foundation; Biomedical Research Council; British Heart Foundation (SP/04/002); Canada Foundation for Innovation; Commission of the European Communities, Directorate C-Public Health (2004310); Copenhagen County; Danish Centre for Evaluation and Health Technology Assessment; Danish Council for Independent Research; Danish Heart Foundation (07-10-R61-A1754-B838-22392F); Danish Medical Research Council; Danish Pharmaceutical Association; Emil Aaltonen Foundation; European Research Council Advanced Research Grant; European Union FP7 (EpiMigrant, 279143; FP7/2007-2013; 259749); Finland's Slottery Machine Association; Finnish Cultural Foundation; Finnish Diabetes Research Foundation; Finnish Foundation for Cardiovascular Research; Finnish Foundation of Cardiovascular Research; Finnish Medical Society; Finnish National Public Health Institute; Finska Läkaresällskapet; Folkhälsan Research Foundation; Foundation for Life and Health in Finland; German Center for Diabetes Research (DZD) ; German Federal Ministry of Education and Research; Health Care Centers in Vasa, Närpes and Korsholm; Health Insurance Foundation (2012B233) ; Helsinki University Central Hospital Research Foundation; Hospital districts of Pirkanmaa, Southern Ostrobothnia, North Ostrobothnia, Central Finland, and Northern Savo; Ib Henriksen foundation; Juho Vainio Foundation; Korea Centers for Disease Control and Prevention (4845–301); Korea National Institute of Health (2012-N73002-00); Li Ka Shing Foundation; Liv och Hälsa; Lundbeck Foundation; Marie-Curie Fellowship (PIEF-GA-2012-329156); Medical Research Council (G0601261, G0900747-91070, G0601966, G0700931); Ministry of Education in Finland; Ministry of Social Affairs and Health in Finland; MRC-PHE Centre for Environment and Health;Municipal Heath Care Center and Hospital in Jakobstad; Närpes Health Care Foundation; National Institute for Health Research (RP-PG-0407-10371); National Institutes of Health (U01 DK085526, U01 DK085501, U01 DK085524, U01 DK085545, U01 DK085584, U01 DK088389, RC2-DK088389, DK085545, DK098032, HHSN268201300046C, HHSN268201300047C, HHSN268201300048C, HHSN268201300049C, HHSN, R01MH107666 and K12CA139160268201300050C, U01 DK062370, R01 DK066358, U01DK085501, R01HL102830, R01DK073541, PO1AG027734, R01AG046949, 1R01AG042188, P30AG038072, R01 MH101820, R01MH090937, P30DK020595, R01 DK078616, NIDDK K24 DK080140, 1RC2DK088389, T32GM007753); National Medical Research Council; National Research Foundation of Korea (NRF-2012R1A2A1A03006155); Nordic Center of Excellence in Disease Genetics; Novo Nordisk; Ollqvist Foundation; Orion-Farmos Research Foundation; Paavo Nurmi Foundation; Perklén Foundation; Samfundet Folkhälsan; Signe and Ane Gyllenberg Foundation; Sigrid Juselius Foundation; Social Insurance Institution of Finland; South East Norway Health Authority (2011060); Swedish Cultural Foundation in Finland; Swedish Heart-Lung Foundation; Swedish Research Council; Swedish Research Council (Linné and Strategic Research Grant); The American Federation for Aging Research; The Einstein Glenn Center; The European Commission (HEALTH-F4-2007-201413); The Finnish Diabetes Association; The Folkhälsan Research Foundation; The Påhlssons Foundation; The provinces of Newfoundland and Labrador, Nova Scotia, and New Brunswick; The Sigrid Juselius Foundation; The Skåne Regional Health Authority; The Swedish Heart-Lung Foundation; Timber Merchant Vilhelm Bang’s Foundation; Turku University Foundation; Uppsala University; Wellcome Trust (064890, 083948, 085475, 086596, 090367, 090532, 092447, 095101/Z/10/Z, 200837/Z/16/Z, 095552, 098017, 098381, 098051, 084723, 072960/2/03/2, 086113/Z/08/Z, WT098017, WT064890, WT090532, WT098017, 098051, WT086596/Z/08/A and 086596/Z/08/Z). Detailed acknowledgment of funding sources is provided in the Additional Acknowledgements section of the Supplementary Materials.
Footnotes
Author Contributions:
Sample Collection and Phenotyping: NG, A Mahajan, NPB, C Ladenvall, JB-J, NRR, NWR, RAS, APG, AUJ, CJG, CB, D Buck, GB, GJ, HMS, JRH, J Murphy, JMJ, J Trakalo, KSS, MM, MN, M Hollensted, RO, SG, ARW, ATH, HEA, AC, RAD, A Stančáková, AHR, A Metspalu, AJF, A-CS, A Käräjämäki, YAK, RA, A Swift, TA, BL, BG, BIF, B-GH, C Meisinger, CG, C Langenberg, D Pasko, D Aguilar, D Bowden, DH, EST, EC, C-YC, WYL, EM, SPF, FBH, G Atzmon, GWSr, DEH, HG, HK, HO, HATJr, TI, JSK, J Sehmi, J Lindstrom, J Kravic, JEC, CPJ, JEB, J Kriebel, JH, J Li, J Fadista, JCC, JCL, KRO, KSC, C-CK, LLB, J-YL, LK, DML, LH, L Milani, J Liu, L Liang, M Loh, MO-M, MW, MM-N, TM, MG, MR, MCYN, NDP, NN, LQ, NJW, NB, OM, OR, PJH, PWF, PN, A Peters, QQ, RM, S-TT, S Kumar, SKM, SPO'R, S Puppala, K Strauch, TMF, TK, TE, FT, BT, TVV, TYW, TAL, T Lauritzen, T Forsén, TIP, UA, VSF, WRS, YSC, ADM, ASFD, AL, BI, CNAP, FSC, CC, EI, FK, GLS, I Brandslund, J Tuomilehto, J Kuusisto, L Lannfelt, L Lind, LG, MEJ, MU, OP, RR, RNB, T Tuomi, TDS, TH, TJ, VS, GIB, JGW, JB, NJC, RD, KLM, M Laakso, CLH, APM, MB, D Altshuler, MIM
Replication and Expression Studies: T Tukiainen, AV, AAB, YW, A Palotie, AJ, JGE, OTR, S Koskinen, T Lehtimäki, JW, AYC, RAS, MOG, VS, JD, SR, JCF, JBM, ML, KLM
Data Production (Sequencing and Genotyping): XS, NG, A Mahajan, CF, NPB, C Hartl, C Ladenvall, JB-J, NRR, NWR, APG, AUJ, CJG, CB, D Buck, GB, GJ, HMS, JRH, J Murphy, JMJ, J Trakalo, KSS, MM, MN, M Hollensted, RO, PSC, SG, MOC, MD, EB, YF, MHdA, K Shakir, RP, T Fennell, TS, TW, TMS, K Stirrups, TM, PD, MB, MIM
Variant Calling & Panel Generation: MAR, KJG, HMK, GJ, BMN, GG, J Maguire, J Carey, JDS, JIG, S Purcell
Statistical Analysis: A Manning, HMH, JG, XS, T Tukiainen, P Fontanillas, NG, MAR, A Mahajan, AEL, P Cingolani, T Pers, J Flannick, CF, ERG, KJG, HKI, TMT, A Kumar, NPB, C Hartl, C Ladenvall, HMK, JB-J, YC, JRBP, LJS, C Ma, MvdB, L Moutsianas, NRR, RDP, TWB, TG, NWR, APG, AUJ, CJG, CB, D Buck, GB, GJ, HMS, JRH, J Murphy, JMJ, J Trakalo, KSS, MM, MN, M Hollensted, RO, SG, JBM, APM, MB, MIM, CML
Functional Studies: JG, SJ, ALG
Wrote the paper: A Manning, HMH, JG, XS, T Tukiainen, P Fontanillas, JCF, MB, MIM, ALG, CML
Study Design: XS, LJS, ATH, HEA, RAD, BG, EST, G Atzmon, JSK, CPJ, JCC, KSC, J-YL, DML, TM, TMF, TIP, YSC, C Hu, GRC, D Bharadwaj, PJD, D Prabhakaran, EZ, I Barroso, J Scott, J Chan, GM, MJD, M Sandhu, NT, PE, P Froguel, RCWM, RS, SBE, YYT, T Park, T Fingerlin, WJ, RMW, J Tuomilehto, LG, GIB, G Abecasis, JGW, JB, M Seielstad, NJC, RD, JD, IP, JCF, KLM, M Laakso, JBM, CLH, APM, MB, D Altshuler, MIM, ALG, CML
Study Supervision: GIB, G Abecasis, JGW, JB, M Seielstad, NJC, RD, JCF, KLM, JBM, CLH, APM, MB, D Altshuler, MIM, ALG, CML
The authors have no relevant conflicts of interest to disclose.
References
- 1.Kahn SE, Hull RL, Utzschneider KM. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature. 2006;444:840–846. doi: 10.1038/nature05482. [DOI] [PubMed] [Google Scholar]
- 2.Phillips DI, Clark PM, Hales CN, Osmond C. Understanding oral glucose tolerance: comparison of glucose or insulin measurements during the oral glucose tolerance test with specific measurements of insulin resistance and insulin secretion. Diabet Med. 1994;11:286–292. doi: 10.1111/j.1464-5491.1994.tb00273.x. [DOI] [PubMed] [Google Scholar]
- 3.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I, Amin N, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.SIGMA Type 2 Diabetes Consortium. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.SIGMA Type 2 Diabetes Consortium. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT, Gjesing AP, et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512:190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 7.Fuchsberger C, Flannick J, Teslovich TM, Mahajan A, Agarwala V, Gaulton KJ, Ma C, Fontanillas P, Moutsianas L, McCarthy DJ, Rivas MA, et al. The genetic architecture of type 2 diabetes. Nature. 2016 [Google Scholar]
- 8.Raitakari OT, Juonala M, Ronnemaa T, Keltikangas-Jarvinen L, Rasanen L, Pietikainen M, Hutri-Kahonen N, Taittonen L, Jokinen E, Marniemi J, Jula A, et al. Cohort profile: the cardiovascular risk in Young Finns Study. Int J Epidemiol. 2008;37:1220–1226. doi: 10.1093/ije/dym225. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson JG. Epidemiology, genes and the environment: lessons learned from the Helsinki Birth Cohort Study. J Intern Med. 2007;261:418–425. doi: 10.1111/j.1365-2796.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- 10.Perttila J, Merikanto K, Naukkarinen J, Surakka I, Martin NW, Tanhuanpaa K, Grimard V, Taskinen MR, Thiele C, Salomaa V, Jula A, et al. OSBPL10, a novel candidate gene for high triglyceride trait in dyslipidemic Finnish subjects, regulates cellular lipid metabolism. J Mol Med (Berl) 2009;87:825–835. doi: 10.1007/s00109-009-0490-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Mannisto S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty-five-year trends in cardiovascular risk factors in Finland. Int J Epidemiol. 2010;39:504–518. doi: 10.1093/ije/dyp330. [DOI] [PubMed] [Google Scholar]
- 12.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, Gudnason V, et al. Best practices and joint calling of the HumanExome BeadChip: the CHARGE Consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 14.Peloso GM, Auer PL, Bis JC, Voorman A, Morrison AC, Stitziel NO, Brody JA, Khetarpal SA, Crosby JR, Fornage M, Isaacs A, et al. Association of low-frequency and rare coding-sequence variants with blood lipids and coronary heart disease in 56,000 whites and blacks. Am J Hum Genet. 2014;94:223–232. doi: 10.1016/j.ajhg.2014.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Statistics in medicine. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 16.Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E. Variance component model to account for sample structure in genome-wide association studies. Nat Genet. 2010;42:348–354. doi: 10.1038/ng.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM, Locke AE, Grarup N, Im HK, Cingolani P, Flannick J, et al. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 2015;11:e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu MC, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am J Hum Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu DJ, Peloso GM, Zhan X, Holmen OL, Zawistowski M, Feng S, Nikpay M, Auer PL, Goel A, Zhang H, Peters U, et al. Meta-analysis of gene-level tests for rare variant association. Nat Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee S, Teslovich TM, Boehnke M, Lin X. General framework for meta-analysis of rare variants in sequencing association studies. Am J Hum Genet. 2013;93:42–53. doi: 10.1016/j.ajhg.2013.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 23.Exome Aggregation Consortium (ExAC) Cambridge, MA: [Accessed 02/2015]. (URL: http://exac.broadinstitute.org/) [Google Scholar]
- 24.Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, Harte RA, et al. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 2014;42:D764–770. doi: 10.1093/nar/gkt1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 26.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roche DB, Buenavista MT, Tetchner SJ, McGuffin LJ. The IntFOLD server: an integrated web resource for protein fold recognition, 3D model quality assessment, intrinsic disorder prediction, domain prediction and ligand binding site prediction. Nucleic Acids Res. 2011;39:W171–176. doi: 10.1093/nar/gkr184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 29.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buil A, Brown AA, Lappalainen T, Vinuela A, Davies MN, Zheng H-F, Richards JB, Glass D, Small KS, Durbin R, Spector TD, et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat Genet. 2015;47:88–91. doi: 10.1038/ng.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brown AA, Buil A, Viñuela A, Lappalainen T, Zheng H-F, Richards JB, Small KS, Spector TD, Dermitzakis ET, Durbin R. Genetic interactions affecting human gene expression identified by variance association mapping. Elife. 2014;3 doi: 10.7554/eLife.01381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Civelek M, Wu Y, Pan C, Raulerson C, Ko A, He A, Tilford C, Saleem N, Stancakova A, Scott L, Fuchsberger C, et al. Genetic regulation of adipose gene expression and integration with GWAS loci and cardio-metabolic traits. doi: 10.1016/j.ajhg.2017.01.027. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falchi M, Wilson SG, Paximadas D, Swaminathan R, Spector TD. Quantitative Linkage Analysis for Pancreatic B-cell Function and Insulin Resistance in a Large Twin Cohort. Diabetes. 2008;57:1120–1124. doi: 10.2337/db07-0708. [DOI] [PubMed] [Google Scholar]
- 34.Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58:1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKB beta) Science. 2001;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- 36.Garofalo RS, Orena SJ, Rafidi K, Torchia AJ, Stock JL, Hildebrandt AL, Coskran T, Black SC, Brees DJ, Wicks JR, McNeish JD, et al. Severe diabetes, age-dependent loss of adipose tissue, and mild growth deficiency in mice lacking Akt2/PKB beta. J Clin Invest. 2003;112:197–208. doi: 10.1172/JCI16885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hussain K, Challis B, Rocha N, Payne F, Minic M, Thompson A, Daly A, Scott C, Harris J, Smillie BJ, Savage DB, et al. An activating mutation of AKT2 and human hypoglycemia. Science. 2011;334:474. doi: 10.1126/science.1210878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arya VB, Flanagan SE, Schober E, Rami-Merhar B, Ellard S, Hussain K. Activating AKT2 mutation: hypoinsulinemic hypoketotic hypoglycemia. J Clin Endocrinol Metab. 2014;99:391–394. doi: 10.1210/jc.2013-3228. [DOI] [PubMed] [Google Scholar]
- 39.George S, Rochford JJ, Wolfrum C, Gray SL, Schinner S, Wilson JC, Soos MA, Murgatroyd PR, Williams RM, Acerini CL, Dunger DB, et al. A family with severe insulin resistance and diabetes due to a mutation in AKT2. Science. 2004;304:1325–1328. doi: 10.1126/science.1096706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan K, Kimber WA, Luan J, Soos MA, Semple RK, Wareham NJ, O'Rahilly S, Barroso I. Analysis of genetic variation in Akt2/PKB-beta in severe insulin resistance, lipodystrophy, type 2 diabetes, and related metabolic phenotypes. Diabetes. 2007;56:714–719. doi: 10.2337/db06-0921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parikh C, Janakiraman V, Wu WI, Foo CK, Kljavin NM, Chaudhuri S, Stawiski E, Lee B, Lin J, Li H, Lorenzo MN, et al. Disruption of PH-kinase domain interactions leads to oncogenic activation of AKT in human cancers. Proc Natl Acad Sci U S A. 2012;109:19368–19373. doi: 10.1073/pnas.1204384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawlor MA, Alessi DR. PKB/Akt: a key mediator of cell proliferation, survival and insulin responses? J Cell Sci. 2001;114:2903–2910. doi: 10.1242/jcs.114.16.2903. [DOI] [PubMed] [Google Scholar]
- 43.Zinda MJ, Johnson MA, Paul JD, Horn C, Konicek BW, Lu ZH, Sandusky G, Thomas JE, Neubauer BL, Lai MT, Graff JR. AKT-1, -2, and -3 are expressed in both normal and tumor tissues of the lung, breast, prostate, and colon. Clin Cancer Res. 2001;7:2475–2479. [PubMed] [Google Scholar]
- 44.Peng XD, Xu PZ, Chen ML, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Rahilly S. Human genetics illuminates the paths to metabolic disease. Nature. 2009;462:307–314. doi: 10.1038/nature08532. [DOI] [PubMed] [Google Scholar]
- 46.Lim ET, Wurtz P, Havulinna AS, Palta P, Tukiainen T, Rehnstrom K, Esko T, Magi R, Inouye M, Lappalainen T, Chan Y, et al. Distribution and medical impact of loss-of-function variants in the Finnish founder population. PLoS Genet. 2014;10:e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dash S, Sano H, Rochford JJ, Semple RK, Yeo G, Hyden CS, Soos MA, Clark J, Rodin A, Langenberg C, Druet C, et al. A truncation mutation in TBC1D4 in a family with acanthosis nigricans and postprandial hyperinsulinemia. Proc Natl Acad Sci U S A. 2009;106:9350–9355. doi: 10.1073/pnas.0900909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouzakri K, Zachrisson A, Al-Khalili L, Zhang BB, Koistinen HA, Krook A, Zierath JR. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006;4:89–96. doi: 10.1016/j.cmet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 49.Baus D, Heermeier K, De Hoop M, Metz-Weidmann C, Gassenhuber J, Dittrich W, Welte S, Tennagels N. Identification of a novel AS160 splice variant that regulates GLUT4 translocation and glucose-uptake in rat muscle cells. Cell Signal. 2008;20:2237–2246. doi: 10.1016/j.cellsig.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- 51.Toker A, Marmiroli S. Signaling specificity in the Akt pathway in biology and disease. Adv Biol Regul. 2014;55:28–38. doi: 10.1016/j.jbior.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tschopp O, Yang ZZ, Brodbeck D, Dummler BA, Hemmings-Mieszczak M, Watanabe T, Michaelis T, Frahm J, Hemmings BA. Essential role of protein kinase B gamma (PKB gamma/Akt3) in postnatal brain development but not in glucose homeostasis. Development. 2005;132:2943–2954. doi: 10.1242/dev.01864. [DOI] [PubMed] [Google Scholar]
- 53.Savage DB, Tan GD, Acerini CL, Jebb SA, Agostini M, Gurnell M, Williams RL, Umpleby AM, Thomas EL, Bell JD, Dixon AK, et al. Human metabolic syndrome resulting from dominant-negative mutations in the nuclear receptor peroxisome proliferator-activated receptor-gamma. Diabetes. 2003;52:910–917. doi: 10.2337/diabetes.52.4.910. [DOI] [PubMed] [Google Scholar]
- 54.Semple RK, Sleigh A, Murgatroyd PR, Adams CA, Bluck L, Jackson S, Vottero A, Kanabar D, Charlton-Menys V, Durrington P, Soos MA, et al. Postreceptor insulin resistance contributes to human dyslipidemia and hepatic steatosis. J Clin Invest. 2009;119:315–322. doi: 10.1172/JCI37432. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.