Abstract
The cleavage of a protecting group from a protein or drug under bioorthogonal conditions enables accurate spatiotemporal control over protein or drug activity. Despite recent advances in the decaging of proteins and small molecules, the ability to release alcohol-containing molecules has been elusive. Herein, we disclose vinyl ethers as protecting groups for alcohol-containing molecules and as reagents for bioorthogonal bond cleavage reactions. As a proof-of-concept we installed the vinyl ether moiety into a range of molecules including amino acids, a monosaccharide, a fluorophore and an analogue of the cytotoxic drug duocarmycin. Tetrazine-mediated decaging proceeded under biocompatible conditions with good yields and reasonable kinetics. Importantly, the non-toxic, vinyl ether duocarmycin double prodrug was successfully decaged in live cells, to reinstate cytotoxicity. This bioorthogonal reaction presents broad applicability and may be suitable for in vivo applications.
Keywords: tetrazine, bioorthogonal, decaging, fluorogenic probes, drug-delivery
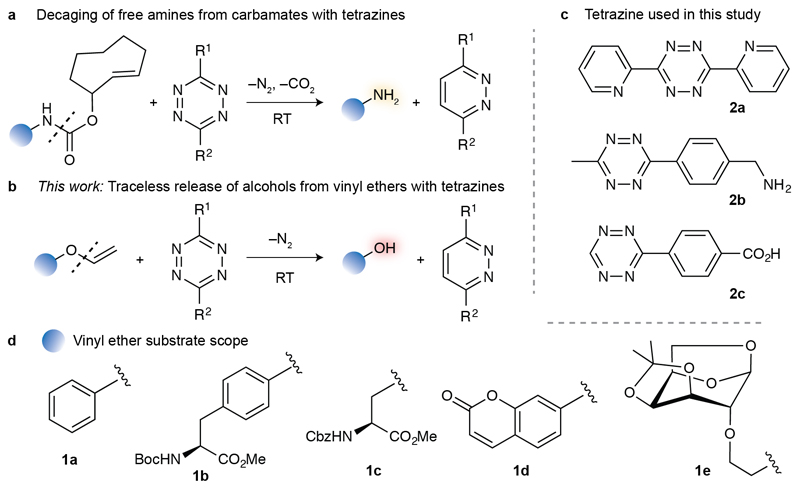
Bioorthogonal chemistry to covalently conjugate synthetic molecules at a predefined protein residue has been a major focus of research in the past two decades.[1] Just very recently, focus has been placed on reactions that can instead cleave specific bonds under bioorthogonal conditions.[2] This strategy holds great potential for the precise spatiotemporal control of protein function in vivo.[1c, 2] For example, photodeprotection of a genetically encoded caged cysteine could be used to reveal the active native protein in live cells.[3] Similarly, palladium-mediated depropargylation,[4] phosphine-mediated Staudinger reduction,[5] and tetrazine-triggered inverse electron-demand Diels–Alder (IEDDA) elimination reactions[6] were successfully employed to restore the activity of proteins bearing a caged lysine residue in the active site. Bond cleavage reactions are also attractive for drug-delivery applications. Palladium-catalysed deprotection of a 5-fluoroacil prodrug was shown as a method for controlled drug release in vivo.[7] The IEDDA reaction between a tetrazine and a caged doxorubicin derivative efficiently releases the cytotoxic drug.[8] Strategies based on IEDDA elimination reactions with tetrazines are particularly attractive for decaging relevant molecules in cells and interrogating biology, due to the favourable kinetics and the abiotic nature of tetrazines when compared to photo- and metal-catalyzed reactions. One limitation, however, has been the breadth of protecting groups available for stable, yet conditionally reversible linkages. Typically, IEDDA elimination reactions have been used with strained alkene protecting groups connected through a carbamate, resulting in a cascade release of a primary amine (Figure 1a).[2, 9] Furthermore, the reduced metabolic stability of strained alkenes constitutes a major caveat for its utility. For instance, cis-cyclooctene easily isomerises to the non-reactive trans-cyclooctene, thus limiting the efficiency of the decaging process in cells.[10]
Figure 1.
a) Tetrazine-mediated decaging of amines from strained alkenes connected through a carbamate linker. b) Decaging of alcohols from vinyl ethers triggered by c) tetrazines. d) Vinyl ether-caged alcohols studied.
Here, we report the development of a vinyl ether–tetrazine system as IEDDA reaction partners for the traceless decaging of alcohol-containing molecules. We demonstrate the broad applicability of this reaction on several chemotypes including the protected amino acids serine and tyrosine, an 1,6-anhydro sugar, a fluorophore and a drug. Importantly, the reaction proceeds under physiological conditions (aqueous buffer pH 7.4 and 37 °C) and was applied to activate a potent toxic derivative of the drug duocarmycin in cancer cells.
To harness the current chemical biology toolbox and develop a broadly applicable technology for the controlled release of alcohol-containing chemotypes, we set out to develop bioorthogonal bond cleavage reactions. In particular, we envisaged that vinyl ethers could efficiently mask both aliphatic and aromatic hydroxyl groups, and be used for traceless release of alcohols through a tetrazine IEDDA bond cleavage reaction. In fact, the reactivity of the vinyl group with tetrazines has been detailed in organic synthesis[11] and such a reactive pair was very recently employed to visualize and detect RNA under bioorthogonal conditions.[12] We first used the commercial phenyl vinyl ether 1a as a model compound, and tetrazine 2a to challenge our decaging hypothesis (Figure 1b–d). After reaction with tetrazine 2a, phenol 3a and 3,6-di(pyridin-2-yl)pyridazine 4a were obtained in 49% and 61% yield, respectively (Table 1). The reaction was performed in dichloromethane to ensure that all the reagents were soluble and at room temperature with only two molar equivalents of tetrazine. Increasing the amount of tetrazine showed no significant improvement in reaction yield. To assess the scope of the reaction we then synthetized vinyl ether derivatives 1b–e.[13] Tyrosine and serine residues play a paramount role in the building of binding pocket architecture and controlling catalytic cycles, e.g. as in tyrosine kinases and serine proteases. Indeed, bioorthogonal decaging of catalytically crucial residues is emerging as a disruptive technology in chemical biology.[2] Furthermore chromone-based fluorophores and sugars can efficiently be used to interrogate biological systems.[14] Remarkably, decaging vinyl ether derivatives of such molecules 1b–e with 2a gave the corresponding free hydroxyl derivatives in good yields (50–68% isolated yields after purification by column chromatography, Table 1). Of note, the vinyl ether reagents were generally stable under biocompatible conditions (PBS pH 7.4 at 37 °C) over 8 hours, as assessed by HPLC/UV traces (Table 1, Supporting Information (SI)). While some degree of unstability was observed for 1c, for instance, the free hydroxyl-containing molecule was not detected.
Table 1.
Stability of vinyl ethers 1a–e and their decaging with tetrazine 2a.
| Compound[a] | Alcohol[c] | Pyridazine[c] | Conversion | Stability[d] |
|---|---|---|---|---|
| 1a[b] | 61% | 49% | 100% | 100%[e] |
| 1b | 68% | 65% | 73% | 100% |
| 1c | 57% | 72% | 100% | 77% |
| 1d | 50% | 47% | 56% | 100% |
| 1e | 65% | 50% | 100% | n.d.[f] |
The reactions were performed in dichloromethane at room temperature with 1 equiv. of vinyl ether (100 mM) 1a–e and 2 equiv. of tetrazine 2a (200 mM) for 72 hours.
The reaction was complete after 40 hours.
Yield of isolated product.
The stability (as % of remaining starting compound) was assessed by HPLC in PBS pH 7.4 at 37 °C with a concentration of vinyl ether of 200 μM using acetophenone as an internal standard.
10% H2O in DMF.
1e does not absorb in the UV. n.d., not determined.
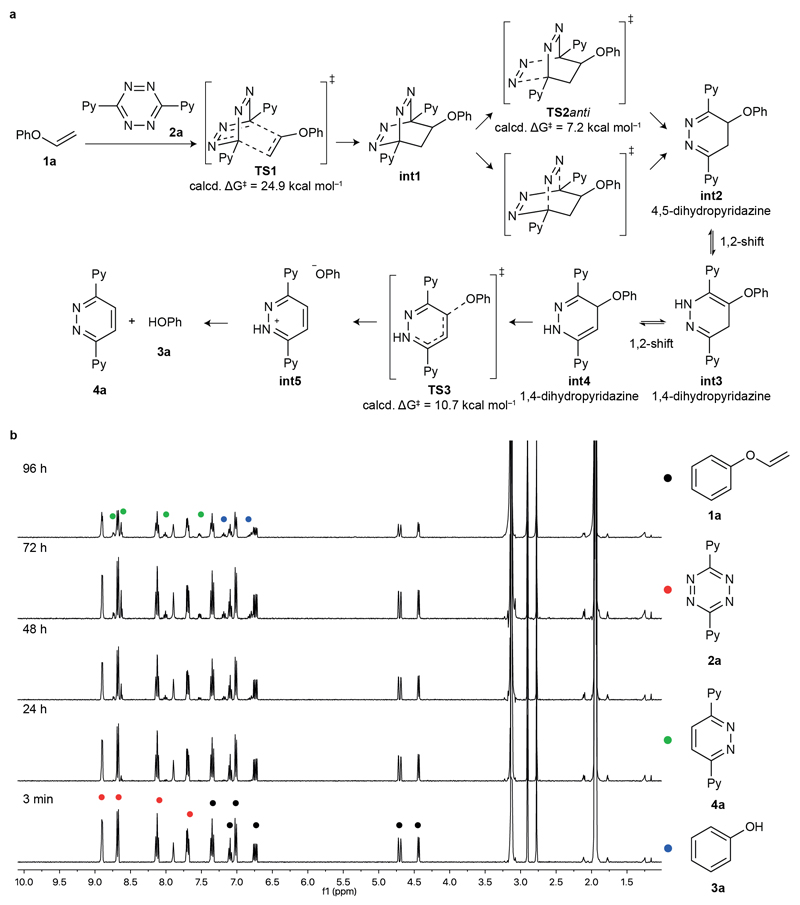
After confirming the stability of the vinyl ether molecules in pH 7.4 buffer and their successful tetrazine-mediated decaging, we proceeded to study the reaction mechanism in detail through quantum mechanics at the M06-2X/6-31+G(d,p) level of theory (Figure 2a, SI). Unlike tetrazine reactions with highly reactive strained alkenes,[6] our data indicate that the first step, i.e. the IEDDA cycloaddition, is the rate-limiting step of the reaction (TS1, ΔG‡ ~ 25 kcal mol−1, reaction time ~3 days) followed by very fast retro-Diels Alder (TS2anti, ΔG‡ ~ 7 kcal mol−1) and phenoxy group cleavage (TS3, ΔG‡ ~ 11 kcal mol−1). Once the 4-phenoxy-4,5-dihydropyridazine (int2) is obtained, it readily tautomerizes into 4-phenoxy-1,4-dihydropyridazine (int4), which can swiftly decage through an elimination reaction. Of note, other dihydropyridazine tautomers previously reported to undergo decaging (int3),[6] are unreactive in this reaction. To support the theoretical data, we mixed 1a and tetrazine 2a in 10% D2O in CD3CN, and recorded the 1H NMR spectra at selected times to gain insight into the reaction mechanism (Figure 2b). Peaks in the aromatic region assigned to the final products could be identified after 24 hours. Conversely, no peaks in the δ 2.5–6 ppm region that could be attributed to the 4,5-dihydropyridazine int2 and 1,4-dihydropyridazine int3,4 intermediate species were observed over the course of the experiment. Given that no intermediate species were identified through 1H NMR, unlike amines decaging from carbamates,[8] our data clearly supports that the Diels-Alder cycloaddition is rate-limiting in this case. A fast and irreversible decaging step after IEDDA cycloaddition is thus responsible for the experimental observations, and fully in line with the predicted reaction coordinate diagram (SI, Figure S3). Furthermore, our spectral data also confirm that no reaction intermediates are trapped, and that all of them evolve to a common tautomer before the decaging step (Figure 2b).
Figure 2.
a) Proposed mechanism based on quantum mechanics for the IEDDA cycloaddition of vinyl ether 1a and tetrazine 2a, followed by in situ alcohol release. Only the relevant activation free energies (ΔG‡) are shown. The initial cycloaddition is the rate-limiting step. After very fast nitrogen cleavage, different dihidropyridazine tautomers int2–4 equilibrate before irreversibly decaging to the experimentally obtained products (pyridazine 4a and phenol 3a). See Figure S3 for the whole calculated minimum energy pathway. b) 1H NMR release studies of 1a upon reaction with tetrazine 2a. The reaction was performed at 3 mM of 1a and 2a in 10% D2O/CD3CN. Ph, phenyl; Py, pyridine. The reaction was followed for 96 hours; while the reaction was not always complete at 96 hours, the results obtained were consistent with the mechanism supported by the theoretical calculations.
With a detailed assessment of the reaction mechanism, we then performed kinetics studies by following the decrease of tetrazine absorbance, i.e. the rate-limiting step, in the visible region. We used compound 1a and screened tetrazines 2a–c, entailing different substituents (Figure 1 and Table 2). Stability studies of compound 1a in the system solvent used (10% H2O in DMF) showed no significant degradation after 8 hours. As expected, tetrazines bearing electron withdrawing groups (2a and 2c) led to faster reactions compared to the one bearing electron donating ones (2b). Finally, tetrazine 2c proved to have the fastest kinetics probably due to the less steric hindrance induced by hydrogen substituted tetrazines.[15] We also compared the kinetics of the vinyl ether/tetrazine decaging with a very reactive, strained alkene as reference, 5-norbornen-2-ol. The second order rate constant determined (k2 = 0.189 M−1s−1) in 10% H2O in DMF, although lower, compares well with literature values for the same reaction (k2 = 1.3 M−1s−1 in H2O at 20 °C).[16] The differences observed are attributed to the faster IEDDA reactions in polar protic solvents.
Table 2.
Kinetics of the reaction of vinyl ether 1a with tetrazines 2a–c.
| Tetrazine[a] | Dienophile | k2 x 10–4 M–1s–1 |
|---|---|---|
| 2a | 1a | 3.92 ± 0.11 |
| 2b | 1a | 0.063 ± 0.013 |
| 2c | 1a | 5.37 ± 0.13 |
| 2a | 5-norbornen-2-ol | 1890 ± 40 |
The reactions were performed in 10% H2O in DMF and were followed by UV through the decay of UV absorption of the tetrazines. An excess of 150–350 fold of 1a was used. In the case of 5-norbornen-2-ol the kinetic rate was determined using the same solvent system with tetrazine 2a with a 20–100 fold excess of 5-norbornen-2-ol.
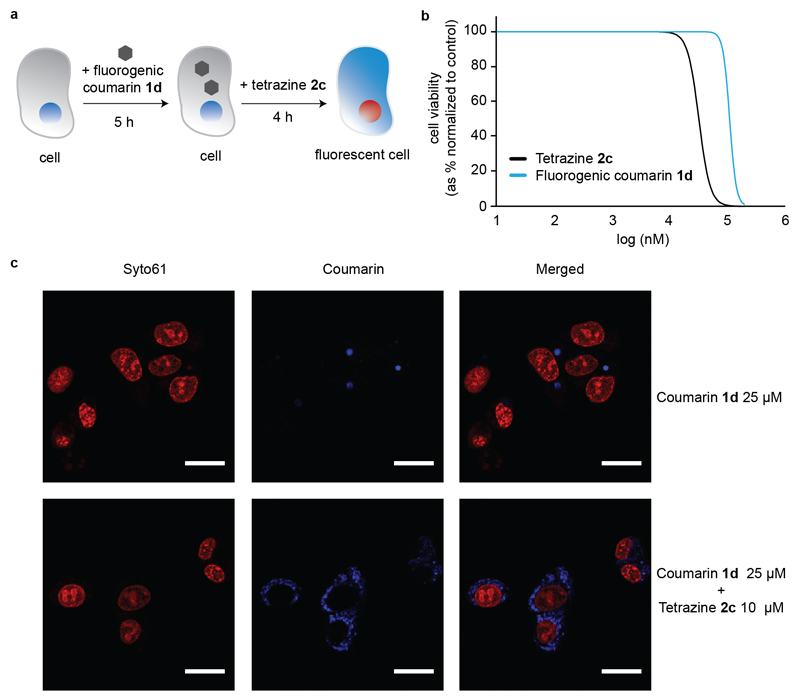
In order to show the potential bioorthogonality of this bond cleavage reaction, we next studied its use for the decaging of a vinyl ether fluorogenic probe in live cells (Figure 3a). To this end, we used a vinyl ether non-fluorescent coumarin derivative 1d and tetrazine 2c, which were both shown to be non-toxic to HepG2 cells at the concentrations used (Figure 3b, and SI for A549 cells). In short, upon incubation of 25 μM of 1d for 5 h, tetrazine 2c (10 μM) was subsequently added to the cells for 4 h. At this time, cells were imaged using confocal microscopy and the turn-on fluorescence of the released coumarin was recorded as a result of the successful tetrazine decaging of the vinyl ether protecting group installed in 1d (Figure 3c). Importantly, this study highlights the biocompatibility of this approach for turn-on live cell imaging applications.
Figure 3.
a) General protocol for tetrazine 2c-mediated intracellular decaging of fluorogenic coumarin 1d. b) Cytotoxicity dose-response curves of tetrazine 2c and coumarin analogue 1d in HepG2 cells, obtained after 48 hours of exposure. c) Detection of fluorescent coumarin (blue) upon tetrazine decaging inside HepG2 cells by confocal microscopy. Cells were incubated for 5 hours with 25 μM 1d and then for 4 hours with 10 μM of tetrazine 2c (bottom panel) or equivalent vehicle control (top panel). Before image acquisition, nuclei were stained with Syto61 (red). Scale bar represents 20 μm.
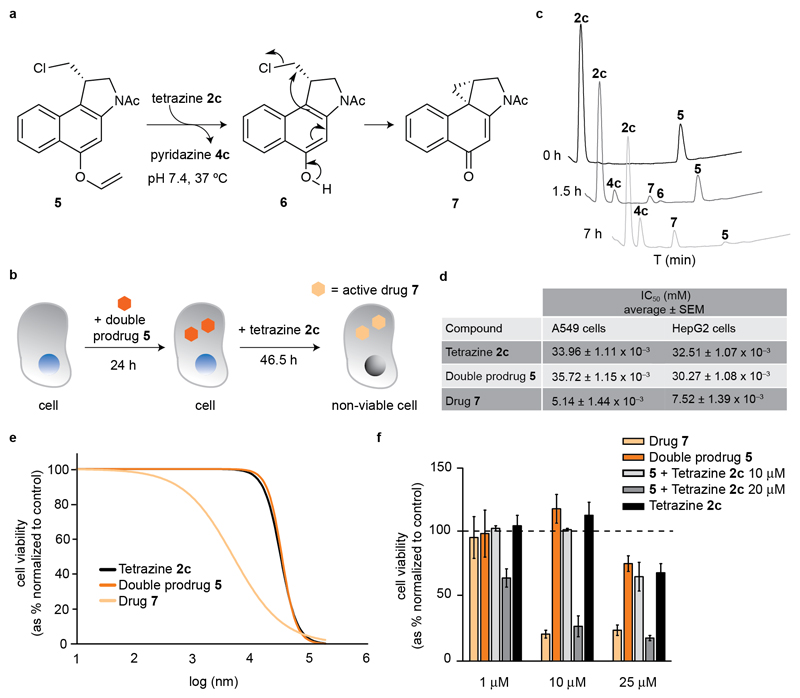
The targeted delivery of drugs to diseased tissues remains a topic of intensive study, and an unsolved issue in modern drug discovery. Currently, alcohol-containing drugs account for ca. 50% of all small FDA-approved chemical entities (cf. DrugBank v5.0, SI), providing ample opportunity for the design of innovative drug delivery constructs. As a proof-of-concept for our technology we assessed the spatiotemporal delivery of a duocarmycin-like natural product (5, Figure 4a). Duorcamycins are isolated from Streptomyces spp. bacteria[17] and have attracted considerable attention as payloads in antibody-drug conjugates, given their potent cytotoxic activity.[18] Interestingly, halogen-bearing duorcamycin cytotoxics undergo a Winstein spirocyclization reaction to afford the bioactive cyclopropanyl, DNA-alkylating species. This feature has been explored in antibody-directed enzyme prodrug therapy, where a non-toxic glycosydic derivative of duocarmycin is activated by a conjugate of an enzyme and a tumor-specific antibody.[19] To demonstrate that our tetrazine mediated IEDDA cleavage of vinyl ethers could be applied for the traceless release of an alcohol-containing drug, we synthetized the N-Ac-double prodrug 5 in three steps from the N-Boc protected 1,2,9,9a-tetrahydrocyclopropa[1,2-c]benz[1,2-e]-indol-4-one (CBI) starting material (SI for details).[20] We chose this simple duocarmycin analogue featuring only an acetyl group attached to the DNA-alkylating CBI core because it has been shown to be very toxic to rapidly replicating cells.[21] We envisioned that upon tetrazine IEDDA deprotection of the vinyl ether, the halogen prodrug 6 would be readily formed and undergo a rapid Winstein spirocyclization reaction to afford active drug 7. This tetrazine-triggered cascade formation of an active drug through an intermediate prodrug is known as the double prodrug concept.[22]
Figure 4.
a) N-Ac CBI double prodrug 5 reaction with tetrazine 2c leads to the formation of intermediate 6 that undergoes a Winstein spirocyclization to afford the bioactive cyclopropanyl 7. b) General protocol for tetrazine 2c-mediated intracellular decaging of 5. c) HPLC time-course of the reaction between 5 and 2c. d) Cytotoxicity fitted dose-response curves of tetrazine 2c, double prodrug 5 and drug 7 in A549 cells, obtained after 46 hours 30 min of exposure. e) Half maximal Inhibitory concentration (IC50) of tetrazine 2c, double prodrug 5 and drug 7, in A549 and HepG2 cells. f) Cytotoxic effects of intracellular activation of the prodrug 5 by tetrazine 2c inside A549 cells. For data on HepG2 cells, see the SI.
Next we studied the stability of the double prodrug 5 in PBS pH 7.4 at 37 °C using HPLC. While species 6 and the active drug 7 were not formed, we detected some degree of degradation of 5 overtime (of note, no formation of either 6 or 7 was observed). Importantly, the double prodrug 5 was found to be less toxic when compared with the active drug 7 in both HepG2 and A549 cells (Figure 4c,d, SI). Having a suitable masked vinyl ether double prodrug in hand, we performed a decaging reaction under physiological conditions (PBS pH 7.4 at 37 °C) with tetrazine 2c. Remarkably, close to complete formation of 7 was achieved after 7 h at 37 °C in PBS pH 7.4, with the short-lived species 6 as intermediate (Figure 4b). After successful demonstration of decaging of 5 under physiologically relevant conditions, we next proceeded to evaluate the feasibility of this approach for the tetrazine-mediated drug-delivery. A549 Cells were first incubated with double prodrug 5 for 24 h, after which time tetrazine 2c was added for an additional 48 h period (twice the doubling time of these cells and comparable to our cytotoxicity studies) (Figure 4c). Satisfyingly, we observed that at 10 μM, the product formed upon tetrazine-decaging of 5 is as toxic as the drug 7 alone (Figure 4e, see SI for identical study on HepG2 cells), suggesting complete drug activation in cells. Hence, this data advocates that tetrazine-mediated bond cleavage of vinyl ethers may be used for the traceless release of alcohol-containing drugs.
In summary, we described a vinyl ether–tetrazine pair as IEDDA reaction partners for the efficient traceless decaging of alcohol-containing molecules in live cells. Considering the wealth of hydroxyl groups in chemical probes and drugs, coupled to the need of circumventing adverse drug reactions, the spatiotemporal delivery method disclosed herein may find broad applicability in chemical biology and molecular medicine by unraveling new biology and leveraging the controlled modulation of (patho)physiological events. Additionally, and in combination with strategies for the genetic encoding of vinyl ether protected tyrosine and serine derivatives, this tetrazine IEDDA decaging reaction is likely to find use for precise control of protein function in vivo.
Experimental Section
Decaging of vinyl ether duocarmycin prodrug in vitro: The N-Ac CBI prodrug 5 was diluted in PBS pH 7.4 to a final concentration of 100 μM from a 10 mM stock in acetonitrile. Then the benzoic acid tetrazine 2c was added to a final concentration of 500 μM from a 50 mM stock in DMSO. The reaction was performed at 37 °C and was monitored by HPLC/UV at different times until completion.
Decaging of vinyl ether duocarmycin double prodrug 5 in cells: Cells were incubated with increasing concentrations of 5 or equivalent vehicle controls for 24 h. The culture medium was then exchanged to complete medium supplemented with increasing concentrations of tetrazine 2c, drug or equivalent vehicle controls. Cells were incubated for another 48 h until proceeding with the CellTiter-Blue Cell Viability Assay (Promega). Relative fluorescence units (R.L.U.) were normalized to the values obtained for the appropriate vehicle controls. Bars represent the average of 3 independent experiments and error bars represent standard error of the mean (SEM).
Supplementary Material
Acknowledgements
We thank the European Commission (Marie Skłodowska-Curie ITN ProteinConjugates; Marie Skłodowska-Curie IEF to E.J.M. and B.L.O.; Marie Curie IEF to O.B.), China Scholarship Council (PhD studentship to Z.G.), FCT Portugal (FCT Investigator to G.J.L.B.), MINECO (CTQ2015-70524-R and RYC-2013-14706 to G.J.O.) and the EPSRC for financial support. We also thank BiFi (Memento cluster) for computer support. G.J.L.B. is a Royal Society University Research Fellow and the recipient of a European Research Council Starting Grant (TagIt).
Contributor Information
Ester Jiménez-Moreno, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK.
Zijian Guo, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK.
Bruno L. Oliveira, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK
Inês S. Albuquerque, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028, Lisboa, Portugal
Annabel Kitowski, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028, Lisboa, Portugal.
Ana Guerreiro, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028, Lisboa, Portugal.
Omar Boutureira, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK.
Tiago Rodrigues, Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028, Lisboa, Portugal.
Gonzalo Jiménez-Osés, Departamento de Química, Universidad de La Rioja, Centro de Investigación en Síntesis Química, 26006 Logroño, Spain; Institute of Biocomputation and Physics of Complex Systems (BIFI), University of Zaragoza, BIFI-IQFR (CSIC), Zaragoza, Spain.
Gonçalo J. L. Bernardes, Department of Chemistry, University of Cambridge, Lensfield Road, CB2 1EW Cambridge, UK; Instituto de Medicina Molecular, Faculdade de Medicina, Universidade de Lisboa, Avenida Professor Egas Moniz, 1649-028, Lisboa, Portugal
References
- [1].a) Sletten EM, Bertozzi CR. Angew Chem. 2009;121:7108–7133. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lang K, Chin JW. Chem Rev. 2014;114:4764–4806. doi: 10.1021/cr400355w. [DOI] [PubMed] [Google Scholar]; c) Krall N, da Cruz FP, Boutureira O, Bernardes GJL. Nat Chem. 2016;8:103–113. doi: 10.1038/nchem.2393. [DOI] [PubMed] [Google Scholar]
- [2].Li J, Chen PR. Nat Chem Biol. 2016;12:129–137. doi: 10.1038/nchembio.2024. and references therein. [DOI] [PubMed] [Google Scholar]
- [3].Nguyen DP, Mahesh M, Elsässer SJ, Hancock SM, Uttamapinant C, Chin JW. J Am Chem Soc. 2014;136:2240–2243. doi: 10.1021/ja412191m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Li J, Yu J, Zhao J, Wang J, Zheng S, Lin S, Chen L, Yang M, Jia S, Zhang X, Chen PR. Nat Chem. 2014;6:352–361. doi: 10.1038/nchem.1887. [DOI] [PubMed] [Google Scholar]
- [5].Luo J, Liu Q, Morihiro K, Deiters A. Nat Chem. 2016;8:1027–1034. doi: 10.1038/nchem.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li J, Jia S, Chen PR. Nat Chem Biol. 2014;10:1003–1005. doi: 10.1038/nchembio.1656. [DOI] [PubMed] [Google Scholar]
- [7].Weiss JT, Dawson JC, Macleod KG, Rybski W, Fraser C, Torres-Sánchez C, Patton EE, Bradley M, Carragher NO, Unciti-Broceta A. Nat Commun. 2014;5:3277. doi: 10.1038/ncomms4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Versteegen RM, Rossin R, ten Hoeve W, Janssen HM, Robillard MS. Angew Chem. 2013;125:14362–14366. doi: 10.1002/anie.201305969. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2013;52:14112–14116. doi: 10.1002/anie.201305969. [DOI] [PubMed] [Google Scholar]
- [9].a) Rossin R, van Duijnhoven SMJ, ten Hoeve W, Janssen HM, Kleijn LHJ, Hoeben FJM, Versteegen RM, Robillard MS. Bioconjugate Chem. 2016;27:1697–1706. doi: 10.1021/acs.bioconjchem.6b00231. [DOI] [PubMed] [Google Scholar]; b) Zhang G, Li J, Xie R, Fan X, Liu Y, Zheng S, Ge Y, Chen PR. ACS Cent Sci. 2016;2:325–331. doi: 10.1021/acscentsci.6b00024. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Mejia Oneto JM, Khan I, Seebald L, Royzen M. ACS Cent Sci. 2016;2:476–482. doi: 10.1021/acscentsci.6b00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Rossin R, van den Bosch SM, ten Hoeve W, Carvelli M, Versteegen RM, Lub J, Robillard MS. Bioconjugate Chem. 2013;24:1210–1217. doi: 10.1021/bc400153y. [DOI] [PubMed] [Google Scholar]
- [11].a) Sueur S, Lagrenee M, Abraham F, Bremard C. J Heterocycl Chem. 1987;24:1285–1289. [Google Scholar]; b) Boger DL, Sakya SM. J Org Chem. 1988;53:1415–1423. [Google Scholar]; c) Sauer J, Heldmann DK, Hetzenegger J, Krauthan J, Sichert H, Schuster J. Eur J Org Chem. 1998;1998:2885–2896. [Google Scholar]; d) Che D, Wegge T, Stubbs MT, Seitz G, Meier H, Methfessel C. J Med Chem. 2001;44:47–57. doi: 10.1021/jm000949w. [DOI] [PubMed] [Google Scholar]; e) Soenen DR, Zimpleman JM, Boger DL. J Org Chem. 2003;68:3593–3598. doi: 10.1021/jo020713v. [DOI] [PubMed] [Google Scholar]; f) Hamasaki A, Ducray R, Boger DL. J Org Chem. 2006;71:185–193. doi: 10.1021/jo051832o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu H, Alexander SC, Jin S, Devaraj NK. J Am Chem Soc. 2016;138:11429–11432. doi: 10.1021/jacs.6b01625. [DOI] [PubMed] [Google Scholar]
- [13].a) Okimoto Y, Sakaguchi S, Ishii Y. J Am Chem Soc. 2002;124:1590–1591. doi: 10.1021/ja0173932. [DOI] [PubMed] [Google Scholar]; b) Bosch M, Schlaf M. J Org Chem. 2003;68:5225–5227. doi: 10.1021/jo034376h. [DOI] [PubMed] [Google Scholar]
- [14].a) Laughlin ST, Bertozzi CR. Proc Natl Acad Sci U S A. 2009;106:12–17. doi: 10.1073/pnas.0811481106. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Xue L, Karpenko IA, Hiblot J, Johnsson K. Nat Chem Biol. 2015;11:917–923. doi: 10.1038/nchembio.1959. [DOI] [PubMed] [Google Scholar]
- [15].Karver MR, Weissleder R, Hilderbrand SA. Bioconjugate Chem. 2011;22:2263–2270. doi: 10.1021/bc200295y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lang K, Davis L, Torres-Kolbus J, Chou C, Deiters A, Chin JW. Nat Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Takahashi I, Takahashi K, Ichimura M, Morimoto M, Asano K, Kawamoto I, Tomita F, Nakano H. J Antibiot. 1988;41:1915–1917. doi: 10.7164/antibiotics.41.1915. [DOI] [PubMed] [Google Scholar]
- [18].Elgersma RC, Coumans RGE, Huijbregts T, Menge WMPB, Joosten JAF, Spijker HJ, de Groot FMH, van der Lee MMC, Ubink R, van den Dobbelsteen DJ, Egging DF, et al. Mol Pharm. 2015;12:1813–1835. doi: 10.1021/mp500781a. [DOI] [PubMed] [Google Scholar]
- [19].Tietze LF, von Hof JM, Müller M, Krewer B, Schuberth I. Angew Chem. 2010;122:7494–7497. doi: 10.1002/anie.201002502. [DOI] [PubMed] [Google Scholar]; Angew Chem Int Ed. 2010;49:7336–7339. doi: 10.1002/anie.201002502. [DOI] [PubMed] [Google Scholar]
- [20].Boger DL, Ishizaki T, Kitos PA, Suntornwat O. J Org Chem. 1990;55:5823–5832. [Google Scholar]
- [21].a) Aristoff PA, Johnson PD, Sun D, Hurley LH. J Med Chem. 1993;36:1956–1963. doi: 10.1021/jm00066a004. [DOI] [PubMed] [Google Scholar]; b) Tercel M, Stribbling SM, Sheppard H, Siim BG, Wu K, Pullen SM, Botting KJ, Wilson WR, Denny WA. J Med Chem. 2003;46:2132–2151. doi: 10.1021/jm020526p. [DOI] [PubMed] [Google Scholar]
- [22].Bundgaard H. Adv Drug Deliv Rev. 1989;3:39–65. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.