Abstract
Ubiquitin specific protease 15 (USP15) is a widely expressed deubiquitylase that has been implicated in diverse cellular processes in cancer. Here we identify topoisomerase II (TOP2A) as a novel protein that is regulated by USP15. TOP2A accumulates during G2 and functions to decatentate intertwined sister chromatids at prophase, ensuring the replicated genome can be accurately divided into daughter cells at anaphase. We show that USP15 is required for TOP2A accumulation, and that USP15 depletion leads to the formation of anaphase chromosome bridges. These bridges fail to decatenate, and at mitotic exit form micronuclei that are indicative of genome instability. We also describe the cell cycle-dependent behaviour for two major isoforms of USP15, which differ by a short serine-rich insertion that is retained in isoform-1 but not in isoform-2. Whilst USP15 is predominantly cytoplasmic in interphase, we show that both isoforms move into the nucleus at prophase, but that isoform-1 is phosphorylated on its unique S229 residue at mitotic entry. The micronuclei phenotype we observe on USP15 depletion can be rescued by either USP15 isoform and requires USP15 catalytic activity. Importantly however, an S229D phospho-mimetic mutant of USP15 isoform-1 cannot rescue either the micronuclei phenotype, or accumulation of TOP2A. Thus, S229 phosphorylation selectively abrogates this role of USP15 in maintaining genome integrity in an isoform-specific manner. Lastly, we show that USP15 isoform-1 is preferentially upregulated in a panel of non-small cell lung cancer cell lines, and propose that isoform imbalance may contribute to genome instability in cancer. Our data provide the first example of isoform-specific deubiquitylase phospho-regulation and reveal a novel role for USP15 in guarding genome integrity.
Keywords: TOP2A, micronuclei, ubiquitin specific protease 15, isoform, phosphorylation, lung cancer
Introduction
Ubiquitylation is a reversible post-translational modification that can target proteins for degradation or regulate their activity or cellular localisation1. Monoubiquitin or polyubiquitin chains are appended to substrates by E1/E2/E3 ligases, and may subsequently be removed by a family of almost 100 deubiquitylases (DUBs) to reverse signals or stabilise proteins2,3. As specific substrates are gradually assigned to each DUB4–6, it is becoming apparent that many play roles in cell cycle progression and maintenance of genome integrity7–10. DUBs can be regulated by conformational changes, adaptor proteins or post-translational modifications, which control their activity or recruitment to specific complexes11,12. In particular, phosphorylation may regulate the localisation, stability, or substrate interactions of DUBs12,13. For example, during the cell cycle, periodic phosphorylation activates USP16 and USP3714,15 but inactivates USP8 through recruitment of 14-3-3 proteins16. The regulated expression of DUBs may also control their cellular availability, and alternative splicing can generate DUB isoforms that are targeted to distinct subcellular compartments, as described for USP3317, or exhibit different substrate specificity, as recently suggested for USP1518.
USP15 is a widely expressed DUB19 that regulates diverse cellular processes. Importantly, USP15 copy number gains have been reported in glioblastoma, breast and ovarian cancers20 and copy number losses identified in pancreatic cancer21. The proposed targets for USP15 include numerous cancer-associated proteins and signalling pathways, such as the HPV E6 oncoprotein22, APC tumour suppressor23, NF-κB inhibitor IκBα24, pro-apoptotic caspase-325, the TGFβ receptor20 and its R-SMAD effectors26, p5327, MDM228 and the ubiquitin E3 ligase BRAP associated with the Ras-MAPK signalling cascade29. USP15 substrates include both polyubiquitylated and monoubiquitylated proteins. In the case of BRAP, USP15 reverses polyubiquitin promoting its stability29, whilst USP15 removes monoubiquitin from R-SMADs enhancing their transcriptional activity26. A systematic interaction study revealed prominent association of USP15 with RNA-binding proteins and splicing factors30, and USP15 depletion affects CRAF transcript levels29.
These diverse targets and modes of action for USP15 suggest that its activity must be tightly regulated and directed within cells. Although USP15 predominantly localises to the cytoplasm31, it performs specific functions in the nucleus32, and at mitochondria33 or polysomes34. Mechanisms to control USP15 activity within cells are suggested by evidence that USP15 is alternatively spliced18,35 and can be ubiquitylated or phosphorylated29,34,36–39. Despite these insights, it remains unclear how the multifarious cellular functions of USP15 are directed and regulated.
We recently discovered that USP15 controls stability of the transcription factor REST, a context-dependent tumour suppressor or oncogene, which is acutely degraded at mitosis40 before being rapidly replenished in early G1 in a USP15-dependent manner34. We also observed that USP15 is regulated during the cell cycle. USP15 increases in abundance and becomes phosphorylated at the start of mitosis, coinciding with mitotic degradation of REST. Subsequently, USP15 is dephosphorylated in early G1 as REST re-accumulates34. Thus, USP15 can exhibit temporal activity during the cell cycle to promote accumulation of a specific protein. As pervasive mitotic phosphorylation often inactivates protein functions37, USP15 phosphorylation may abrogate its role in stabilising REST at mitosis, allowing REST to degrade.
Here we set out to investigate the importance of alternative splicing and phosphorylation in regulating the functions of USP15 during the cell cycle. We identify topoisomerase II (TOP2A) as a novel protein that is regulated by USP15. Depletion of USP15 leads to a failure of TOP2A to accumulate as cells approach mitosis, and the subsequent formation of anaphase chromosome bridges, which fail to resolve leading to micronuclei. We find that both USP15 isoforms localise to the nucleus at prophase, and that USP15 isoform-1 is selectively phosphorylated on S229 at mitotic entry. Intriguingly, the role of USP15 in maintaining genomic stability through TOP2A is abrogated by S229 isoform-1 phosphorylation. As USP15 isoform-1 can be selectively upregulated in lung cancer cell lines, we propose this isoform imbalance may contribute to genome instability in cancer.
Results
USP15 promotes accumulation of TOP2A during G2
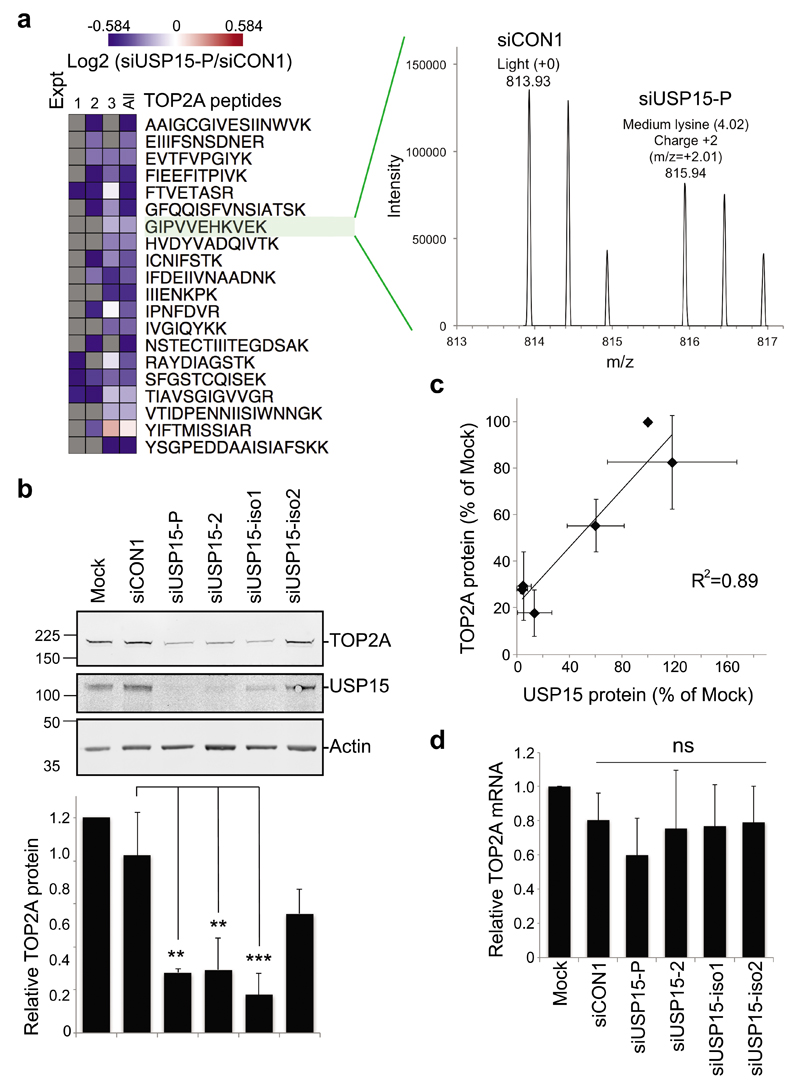
Two ~110kDa isoforms of USP15 have until recently been interchangeably studied in the literature. These isoforms arise through alternative splicing that skips (isoform-2) or includes (isoform-1) exon 7, coding for a serine-rich cassette of 28 amino acids (Supplementary Figure S1a). We found that A549 lung adenocarcinoma cells express both USP15 isoforms, with higher levels of isoform-1, that can be distinguished by isoform-specific siRNAs and RT-PCR primers (Supplementary Figure S1). We wished to identify new candidates that may respond to dynamic USP15 regulation during the cell cycle. To this end, we interrogated a quantitative mass spectrometry dataset of protein abundance in response to USP15 depletion in A549 cells (Concannon et al, unpublished data) for likely candidates. We identified topoisomerase II (TOP2A) as a novel protein that was consistently downregulated by a pool of USP15 siRNA (Figure 1a). We confirmed the relationship between USP15 and TOP2A expression levels by immunoblotting; TOP2A decreased significantly on depletion of total USP15, or of the most abundant isoform-1 (Figure 1b). Overall, there was a strong correlation between residual USP15 and TOP2A proteins across all samples (Figure 1c), suggesting both USP15 isoforms can function to maintain TOP2A levels. Of note, USP15 depletion did not significantly alter TOP2A mRNA expression (Figure 1d).
Figure 1. USP15 regulates the cellular level of TOP2A.
a, TOP2A was identified from a dataset of proteins that differed in abundance on USP15 depletion. SILAC-labelled A549 cells were transfected with pooled siRNA targeting USP15 or a non-targeting control (siCON1) for mass spectrometry analysis. Multiple TOP2A peptides were downregulated in USP15 depleted cells across 3 independent experiments (left); spectra for an example peptide shown (right). b-c, TOP2A protein expression correlates with total USP15 levels. A549 cells were transfected with siRNAs as indicated and whole cell lysates analysed 72hr later. b, Representative immunoblot, with quantification of TOP2A protein expression relative to actin below (mean of 3 independent experiments, error bars SD, one-way ANOVA with Tukey's multiple comparison test **P≤0.01, ***P≤0.005). c, Correlation of TOP2A protein and total USP15 protein in these experiments. d, TOP2A mRNA level was not affected by USP15 depletion. qRT-PCR analysis of TOP2A relative to actin (mean of 4 independent experiments, error bars SD, ns=not significant by one-way ANOVA with Tukey's multiple comparison test).
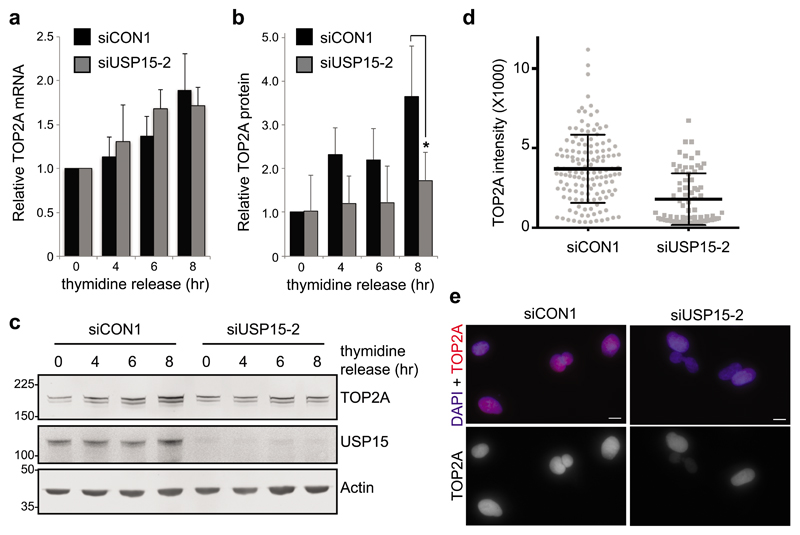
As TOP2A transcription increases during G241, anticipating the requirement for TOP2A in decatenation checkpoint signaling at late G242,43, we wondered whether USP15 may play a role in this temporal accumulation. In A549 cells released from G1/S thymidine arrest, TOP2A transcript and protein levels both increased during G2 as the cells approached mitosis (Figure 2). USP15 depletion does not affect cell cycle phase distribution within an asynchronous population (Supplementary Figure S2) and did not prevent TOP2A transcription during G2 (Figure 2a), it did however impede accumulation of TOP2A protein (Figure 2b and 2c). Quantification of TOP2A within individual cells 8hr after release from G1/S arrest confirmed that, despite heterogeneity within the population as synchronization diminished, cells with USP15 knockdown failed to accumulate TOP2A to the same degree as control cells (Figure 2d and 2e). This role in supporting temporal accumulation of TOP2A protein during G2 is analogous to the role of USP15 in accumulation of newly synthesized REST during early G134.
Figure 2. USP15 supports TOP2A accumulation in G2 as cells approach mitosis.
A549 cells were transfected with either control or USP15 siRNA, arrested at G1/S by double thymidine block and then released for the indicated times (0hr=G1/S, 4hr=S to G2, 6hr=G2, 8hr=G2 to M); RNA and protein lysates were made in parallel. a, qRT-PCR analysis of TOP2A mRNA expression relative to actin (mean of 3 independent experiments, error bars SD). b-c, Immunoblotting for TOP2A, USP15 and actin: (b) quantification of TOP2A protein levels (mean 3 independent experiments, error bars SD, paired T-Test *P≤0.05), and (c) a representative gel. d-e, Immunofluorescence for TOP2A expression levels 8hr after release from double-thymidine block. Quantification of the intensity of staining for >75 cells per condition (d), and representative images from control and USP15 depleted cells; scale bars 10μm (e).
USP15 is required for correct mitotic chromosome segregation
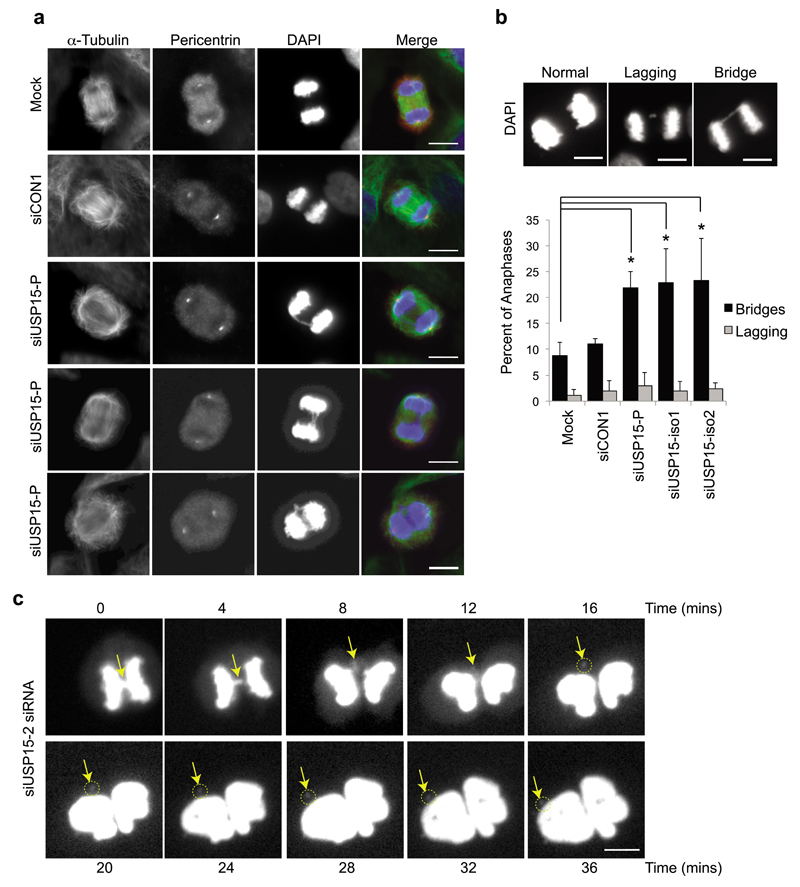
TOP2A catalyzes transient breaking and rejoining of two strands of duplex DNA, allowing the strands to pass through one another. It accumulates at centromeres during prophase, where it is required for chromatin condensation and to decatenate intertwined sister chromatids to ensure their correct separation during anaphase44,45. TOP2A depletion leads to the formation of bulky DAPI-positive anaphase bridges and ultrafine bridges, due to the persistence of catenations between sister chromatids46–49. We found that USP15 depletion similarly caused frequent defects in chromosome segregation during anaphase (Figure 3a). Defects were categorized as lagging chromosomes, which fail to attach to kinetochores, or as anaphase bridges, where sister chromatids are attached to opposite spindle poles but fail to separate due to catenation (Figure 3b). The frequency of lagging chromosomes was low in A549 cells and did not increase on USP15 depletion. However, anaphase bridges were evident in ~10% of cells exiting mitosis, and their frequency was significantly increased in cells depleted of total USP15 or of either USP15 isoform (Figure 3b). To explore the consequence of these anaphase bridges, we followed USP15 depleted cells by time-lapse microscopy. Figure 3c shows an example of an anaphase bridge, which resolves to form a distinct micronucleus that persists after the nuclei of the daughter cells reform. Taken together with the requirement of USP15 for TOP2A accumulation as cells approach mitosis (Figure 2), this suggests that USP15-dependent mis-segregation of chromosomes during mitosis most likely arises through defective decatenation of sister chromatids by TOP2A.
Figure 3. USP15 depletion causes anaphase bridges that can lead to micronuclei formation.
a-b, USP15 depletion in A549 cells significantly increases the number of anaphase chromosome bridges but not the number of lagging chromosomes. (a) Representative examples of anaphases observed in A549 cells, showing chromosome bridges in USP15 depleted cells. (b) Examples of each phenotype (top) with quantification of their prevalence below, data show mean of 3 independent experiments with >50 anaphases counted per condition per experiment (error bars SD, one-way ANOVA with Dunnett's multiple comparison test *P≤0.05). c, Live cell imaging demonstrates that anaphase chromosome bridges (arrow) can resolve into micronuclei (circled) in USP15 depleted cells; the timecourse begins at metaphase and extends beyond cytokinesis. All scale bars are 10μm.
USP15 depletion leads to CENP-A positive micronuclei
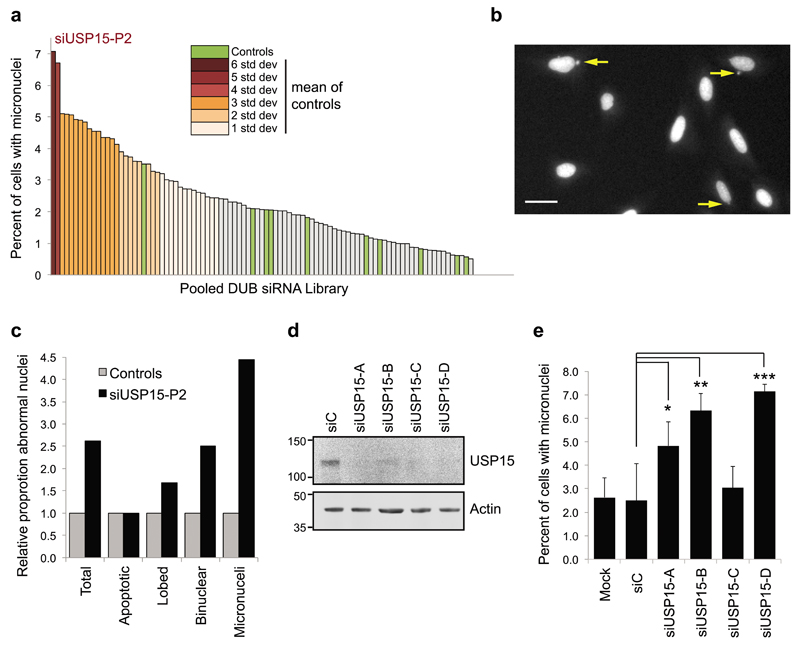
Independently of these experiments, we performed an unbiased screen in U2OS cells for the phenotypic effects of depleting each of the DUBs using an siRNA library, which we scored for nuclear abnormalities. Compared to the controls, depletion of USP15 led to the highest increase in formation of micronuclei amongst the 92 DUBs tested (Figure 4a – 4c). The siRNA library comprises pools of four oligonucleotides targeting each DUB, which for USP15 were distinct from the siRNAs employed in our earlier experiments. Each of the USP15 siRNAs from the library pool effectively depleted USP15 (Figure 4d) and three siRNAs individually recapitulated the micronuclei phenotype (Figure 4e).
Figure 4. In a DUB family screen USP15 depletion induces the most micronuclei.
a-c, An unbiased DUB siRNA library screen in FKHRL1-U2OS cells. The screen was performed in triplicate and >100 cells were scored for nuclear defects for each control (siC or mock-transfected) or DUB siRNA condition. a, USP15 depletion causes most micronuclei formation. Data for the DUB siRNA library are ranked according to the percentage of cells exhibiting micronuclei, SD from the mean of the controls (green) is shown as a color gradient. b, Representative examples of micronuclei in USP15 depleted cells; scale bar 10μm. c, Formation of micronuclei is the most common nuclear defect in USP15 depleted cells; scores for the indicated categories were collated for siUSP15-P2 transfected cells in the screen. Data are shown as fold-change relative to the mean score for control conditions. d-e, Multiple USP15 siRNAs cause micronuclei formation in U2OS cells. (d), A representative immunoblot showing USP15 knockdown with the individual siRNAs that comprise the library pool. (e) Quantification for the percent of cells with micronuclei, mean data from 3 independent experiments with >270 cells scored per condition across experiments (error bars SD, one-way ANOVA with Dunnett's multiple comparison test *P≤0.05, **P≤0.01, ***P≤0.005).
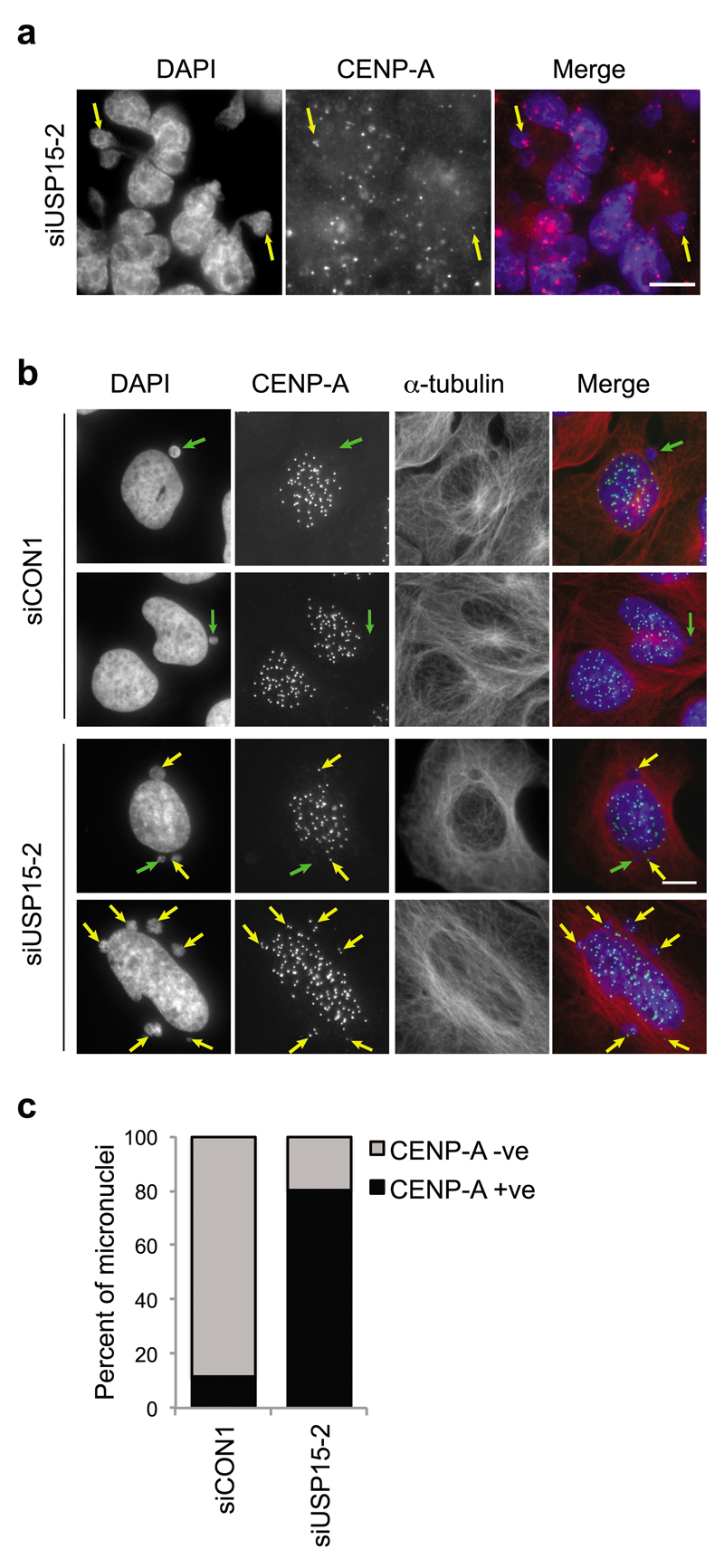
Micronuclei may arise because defective DNA replication or repair results in chromosome fragments or because mitotic defects such as sister chromatid catenation lead to mis-segregation of chromosomes during mitosis49,50. Micronuclei arising from DNA replication or repair errors are likely to be acentric50, whereas catenated chromosome bridges that break to from micronuclei often contain centromeres51. Therefore, to further characterize the observed micronuclei, we stained cells for the centromere protein CENP-A. Micronuclei positive for CENP-A were evident in USP15-depleted A549 cells (Figure 5a) and U2OS cells (Figure 5b). In U2OS cells transfected with non-targeting siRNA only ~20% of micronuclei were CENP-A positive. However, in cells depleted of total USP15 with an independent siRNA, the number of micronuclei increased (Figure 5b) and ~80% were CENP-A positive (Figure 5c). These data further support the idea that USP15 is required to ensure correct mitotic chromosome segregation.
Figure 5. Additional micronuclei in USP15 depleted cells are CENP-A positive.
Representative images of micronuclei in USP15 depleted A549 cells (a) or U2OS cells (b), arrows indicate micronuclei that are CENP-A negative (green) or CENP-A positive (yellow). The percentage of micronuclei that are CENP-A positive were scored for U2OS for >80 micronuclei in each condition (c).
Mitotic regulation of USP15 isoforms
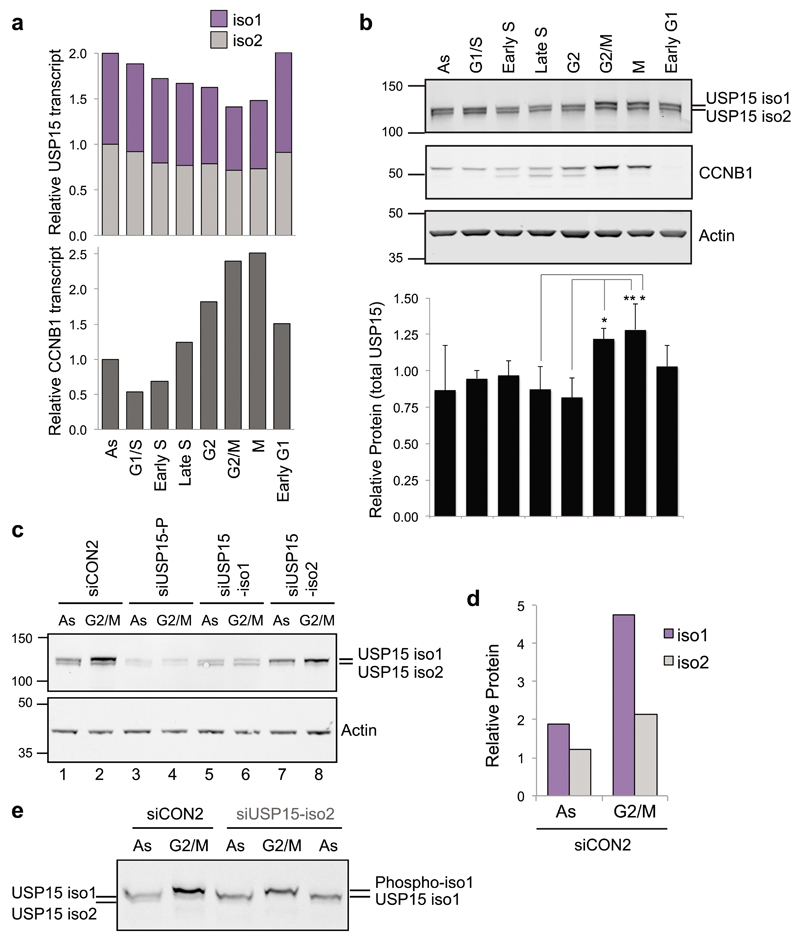
As we have previously shown that, like TOP2A, total USP15 expression increased during G234, we next asked whether both USP15 isoforms behave in the same manner. A549 cells were synchronised to enrich for populations in specific cell cycle phases to investigate how the USP15 isoforms oscillate during the cell cycle. Although their transcript levels and splice variant ratios remain relatively stable (Figure 6a), USP15 protein levels were significantly increased by the time cells reached mitosis compared to S-phase, most noticeably for isoform-1 (Figure 6b – 6e). However, depletion of either isoform had no effect on distribution between cell cycle phases in a population of cells (Supplementary Figure S2).
Figure 6. USP15 isoforms are dynamically expressed and differentially phosphorylated during the cell cycle.
a & b, A549 cells were synchronised using standard thymidine/nocodazole protocols to enrich for the indicated phases. a, USP15 transcript levels remain stable during the cell cycle. Expression of USP15 splice variants and cyclin B1 (CCNB1) were quantified by qRT-PCR, mean data from 3 independent experiments are expressed relative to actin and normalised to the expression in asynchronous cells (As) for each splice variant. b, USP15 protein expression levels oscillate through the cell cycle. USP15 was evaluated by immunoblotting; a representative gel is shown with quantification of total USP15 expression relative to actin below (mean of 3 independent experiments, error bars SD, one-way ANOVA with Tukey's multiple comparison test *P≤0.05, **P≤0.01). c-e, USP15 isoform-1 increases in abundance by G2/M and becomes phosphorylated. A549 cells were depleted of USP15 isoforms as indicated and protein extracts were compared by immunoblotting for asynchronous cells (As) or cells arrested at G2/M. Separation by 4-12% gradient SDS-PAGE (c) showing quantification of the siCON2 samples (d), and analysis by Phostag gel electrophoresis (e).
We previously showed that a proportion of USP15 is phosphorylated at mitosis34, so we selectively depleted each isoform to establish whether they are differentially phosphorylated. In G2/M arrested cells, a gel mobility shift indicative of phosphorylation was seen for isoform-1 but not isoform-2 (Figure 6b). This was confirmed by depletion of the individual USP15 isoforms (Figure 6c) and analysis by Phos-tag electrophoresis (Figure 6e). To determine which residues are subject to mitotic phosphorylation, we immunoprecipitated the GFP-tagged USP15 isoforms from A549 cells. Mass spectrometry identified two peptides that span the region encoded by exon 7 in isoform-1. One of these was phosphorylated only in mitotic extracts, mapping with high confidence to S229, the first serine in the peptide (Supplementary Figure S3). Data for the HeLa mitotic phosphoproteome also report S229 phosphorylation37,52. Thus USP15 isoform-1 is regulated independently of isoform-2, accumulating in G2 as cells approach mitosis, and subsequently undergoing S229 mitotic phosphorylation.
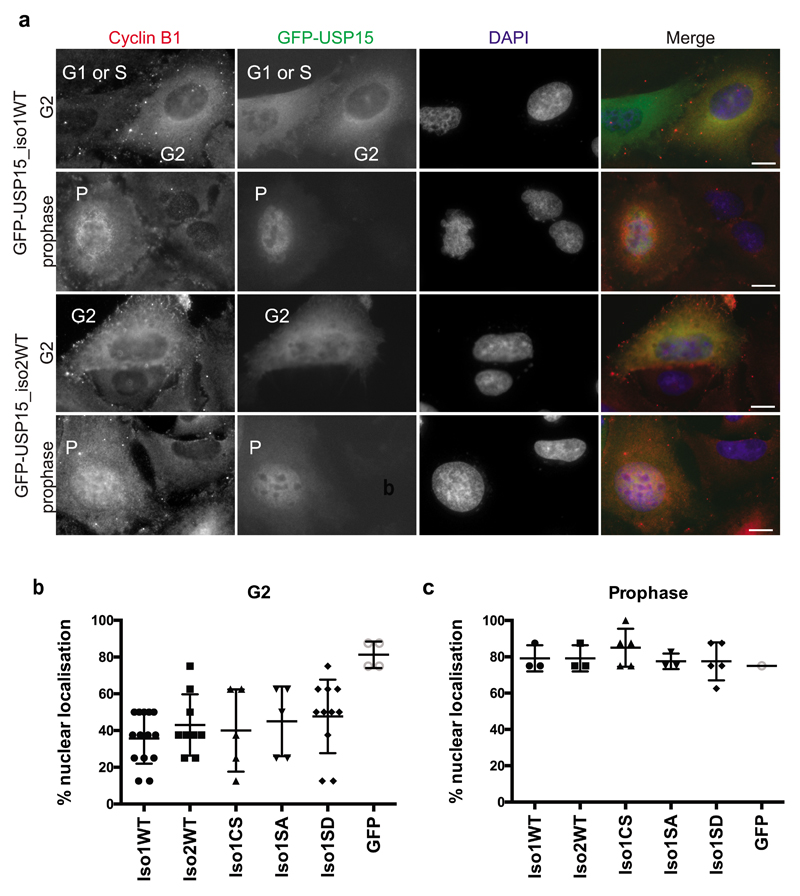
USP15 localises predominantly to the cytosol, in contrast to its closest paralogues USP4 and USP11, which respectively show uniform distribution and tight nuclear localisation31. We used GFP-tagged expression constructs to establish whether the two USP15 isoforms localise differentially, or relocalise during the cell cycle. Cells were co-stained with cyclin B1, which accumulates in the cytoplasm during G2, before translocating into the nucleus at prophase as mitosis initiates53. We found that GFP-USP15 localisation mirrored that of cyclin B1, being predominantly cytoplasmic in G2 cells, but localising to the nucleus at prophase (Figure 7a). This was observed for both USP15 isoforms, was independent of catalytic activity, and was unaffected by phospho-null or phospho-mimetic mutation of S229 (Figure 7b and 7c, Supplementary Figure S4). Likewise, the USP15 isoforms and phospho-mutants showed no gross difference in reactivity towards the ubiquitin active site directed probe Ub-VME (Supplementary Figure S5). Thus the two USP15 isoforms are similarly localised and catalytically active within cells, but both exhibit redistribution at the onset of mitosis that is independent of S229 phosphorylation. The movement of USP15 into the nucleus at prophase occurs at a time when TOP2A accumulates at centromeres to aid chromatin condensation and decatenation of sister chromatids44,45.
Figure 7. Both USP15 isoforms are predominantly cytosolic but localise to the nucleus at prophase.
A549 cells were transfected with the indicated GFP-USP15 constructs or GFP only as a control. Cells were fixed and co-stained for the cell cycle phase marker cyclin B1 and DAPI, to classify as G1 or S-phase (low cytoplasmic cyclin B1), G2 (high cytoplasmic cyclin B1) or prophase (nuclear cyclin B1, uncondensed chromatin). a, Representative immunofluorescence images of cells expressing the two wild-type (WT) USP15 isoforms in G2 cells and prophase cells; scale bars 10μm. b-c, Transfected cells were scored for relative intensity of GFP-USP15 variant expression in the nucleus compared to the cytoplasm, for cells in G2 (b) or prophase (c); counts for >100 cells per GFP-USP15 construct were pooled from 3 independent experiments. Additional images are shown in Supplementary Figure S4.
Protection of genomic integrity and TOP2A accumulation are abrogated by S229 phosphorylation of USP15 isoform-1
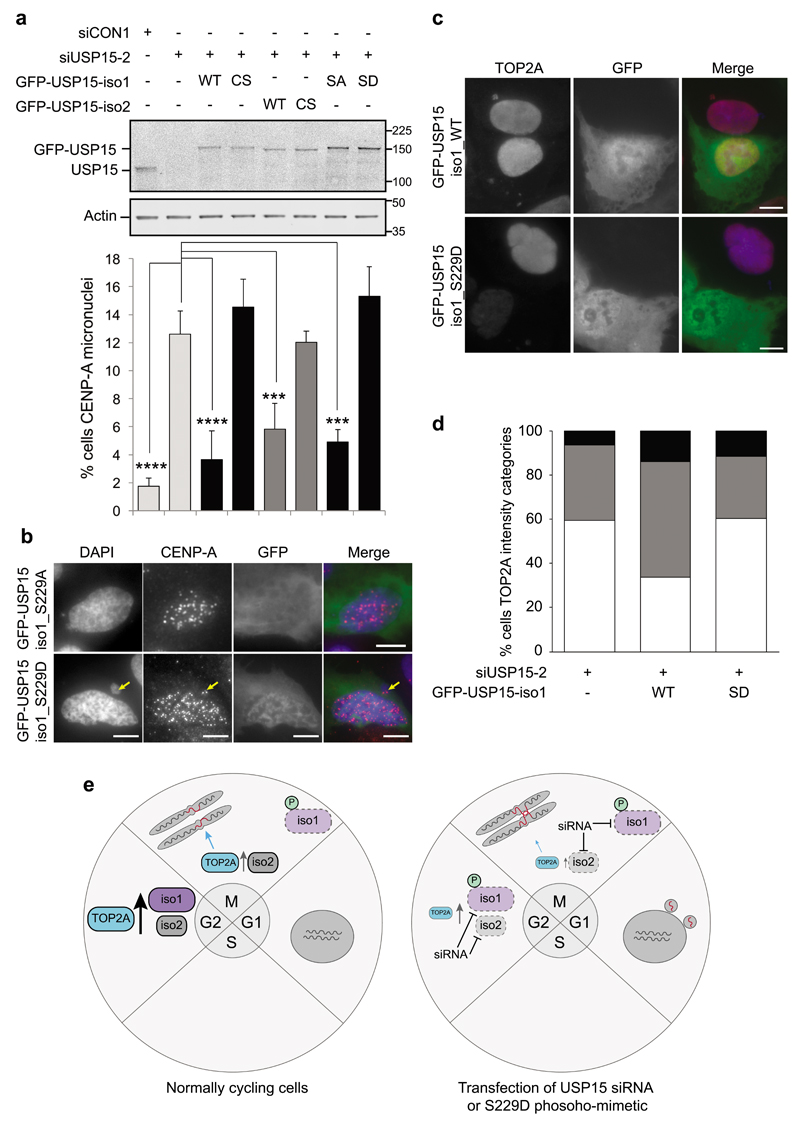
Given the premise that pervasive mitotic phosphorylation often inactivates protein function37, we explored whether mitotic S229 phosphorylation of USP15 isoform-1 might impede its roles in supporting TOP2A accumulation, and in preventing micronuclei forming from unresolved anaphase bridges. To this end, we performed rescue experiments in U2OS cells depleted of total USP15. The appearance of CENP-A positive micronuclei in USP15 depleted cells was reversed by exogenous expression of either USP15 isoform (Figure 8a). This is consistent with our data showing that depletion of either USP15 isoform reduces TOP2A accumulation during G2 (Figure 2). The catalytically inactive USP15 (C298S/C269S) isoforms could not rescue micronuclei, confirming that this role of USP15 in genome maintenance requires its deubiquitylase activity. Importantly, in contrast to the phospho-null mutant (S229A), expression of phospho-mimetic (S229D) USP15-isoform-1 did not reduce the number of CENP-A positive micronuclei (Figure 8a and 8b). Thus S229 phosphorylation impedes the ability of USP15 isoform-1 to protect genome integrity. In contrast to the S229 phospho-mimetic, wild-type USP15 isoform-1 can rescue the phenotype, presumably as it remains unphosphorylated in G2, and can support TOP2A accumulation during this phase. Consistent with our hypothesis that USP15 affects genome stability through TOP2A, wild-type USP15 isoform-1, but not S229D phospho-mimetic USP15, increased expression levels of TOP2A in USP15 depleted U2OS cells (Figure 8c and 8d).
Figure 8. Phospho-mimetic S229D USP15 isoform-1 cannot rescue TOP2A expression or the CENP-A micronuclei phenotype.
a-b, Catalytic activity and non-phosphorylated S229 are required for USP15 to rescue the CENP-A micronuclei phenotype. U2OS cells were transfected with siUSP15-2 (targeting total USP15) or a non-targeting control siRNA for 72hr, the indicated siUSP15-2 resistant GFP-USP15 expression constructs were transfected for the final 24hr. (a) A representative immunoblot (top) and scoring of CENPA positive micronuclei from 3 independent experiments (below); >50 cells, with low to moderate GFP expression, were scored per condition per experiment (error bars SD, one-way ANOVA with Dunnett's multiple comparison test *** P≤0.005, **** P≤0.001). (b) Representative images for cells in the phospho-null and phospho-mimetic rescue conditions. All scale bars are 10μm. c-d, Non-phosphorylated S229 is required for USP15 to rescue TOP2A expression. Experiments were performed as above in U2OS cells, and immunofluorescence intensity for TOP2A assessed following rescue with WT or S229D USP15 isoform-1, (c) representative images, and (d) quantification for >100 cells per condition. e, schematic illustrating our working model for USP15 regulation of TOP2A, anaphase bridges and micronuclei.
Taken together, our data suggest that G2 accumulation of TOP2A, and its function in mitotic decatenation, are sensitive to the combined amount of the two USP15 isoforms in cells. When available USP15 is reduced, by siRNA depletion of either isoform, or by S229-phosphorylation of isoform-1, this promotes genome instability (Figure 8e).
USP15 isoform-1 is selectively upregulated in NSCLC
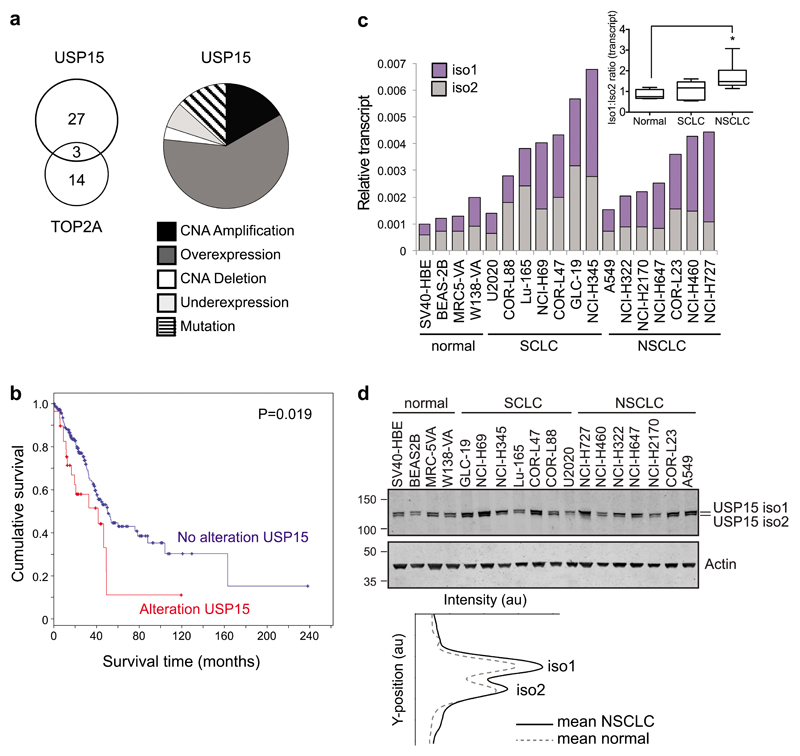
Lastly, we asked whether USP15 may be dysregulated in lung cancer in a way that might promote genome instability. Copy number alterations for USP15 are reported in glioblastoma, breast, ovarian and pancreatic cancers20,21. In relation to our data for A549 cells, we examined the provisional TCGA dataset for lung adenocarcinoma and found that USP15 is altered in 30 of 230 cases (13%), most commonly being amplified and/or overexpressed (Figure 9a). Interestingly, whilst TOP2A is modified in 17 cases (7%), there is little overlap with cases harbouring USP15 alterations (Figure 9a). Of note, alterations in USP15 are associated with worse prognosis within this cohort of lung adenocarcinoma patients (Figure 9b).
Figure 9. USP15 is amplified and USP15 isoform-1 is overexpressed in NSCLC.
a-b, Data from the TCGA provisional cohort of 230 lung adenocarcinoma patients. a, Alterations in USP15 (n=30), highlighting the frequency of mutations, copy number alterations (CNA) or transcript expression changes (left) and overlap with cases exhibiting TOP2A alterations (right). b, Kaplan–Meier overall survival curves stratified according to alterations in USP15; log rank test P=0.019 for patients with (n=30) compared to those without (n=200) alterations. c-e, USP15 isoform-1 is selectively upregulated in NSCLC. Endogenous expression of USP15 isoform-1 and isoform-2 in a panel of 18 cell lines representing normal lung and lung cancer sub-types. c, qRT-PCR for splice variants; relative expression is shown normalised to actin. Inset boxplot shows the ratio of USP15 isoform 1 to 2 splice variants for each cell type: normal (n=4), SCLC (n=7) and NSCLC (n=7); one-way ANOVA with Dunnett’s multiple comparison test *P≤0.05. d, Immunoblotting of whole cell protein extracts for USP15 isoforms, with mean quantification of the two isoforms in normal or NSCLC cells below.
To explore the relative cellular abundance of the two USP15 isoforms in lung cancer, we used qRT-PCR primers that specifically detect the splice variants encoding USP15 isoform-1 or isoform-2 (Supplementary Figure S1). We quantified transcript expression across a panel of cell lines derived from normal lung, small cell lung cancer (SCLC) or non-small cell lung cancer (NSCLC), the latter being predominantly adenocarcinoma (Figure 9c). Both splice variants were endogenously expressed in all the cell lines examined, and they accounted for equivalent proportions of the total USP15 in normal lung and SCLC cell lines. However, the overall abundance of USP15 mRNA was increased in both SCLC and NSCLC, and in the latter was mainly attributable to the transcript variant encoding isoform-1 (Figure 9c, inset). Broadly speaking, these transcript expression profiles concur with the relative abundance of the two USP15 isoforms on immunoblotting, with isoform-1 more abundant in NSCLC than the normal lung cell lines (Figure 9d). Together these data suggest that increased expression of USP15 in lung adenocarcinoma is biased towards higher expression of isoform-1, which once S229-phosphorylated is less able to protect against micronuclei formation through mitotic mis-segregation.
Discussion
Here we report TOP2A as a novel USP15 regulated protein, revealing a new role for USP15 in guarding genome integrity. In addition, we describe the first example of isoform-specific deubiquitylase phospho-regulation, as we show that mitotic phosphorylation of S229 selectively abrogates this role for USP15 isoform-1. USP15 and its most closely related DUBs, USP4 and USP11, have been implicated in other aspects of genome integrity at the level of DNA repair54–56, but here we identify a distinct role for USP15 in G2/M decatenation by TOP2A.
TOP2A expression increases during G2 and decreases when cells complete mitosis. It is essential for the decatenation checkpoint in late G2, and for resolving intertwined sister chromatids and facilitating chromatin condensation at the onset of mitosis, thus ensuring accurate division of the genome during anaphase42–45. We show that G2 accumulation of TOP2A and its function in mitotic decatenation are sensitive to the total amount of cellular USP15, which if limited by siRNA depletion or S229-phosphorylation, results in genome instability (Figure 8e).
Our data also shed new light on temporal regulation for the two major USP15 isoforms during the cell cycle. The alternative splicing mechanism that generates these isoforms remains unclear, although VEZF1-dependent RNA pol II pausing is reported to promote exon inclusion favouring isoform-1, whilst the polypyrimidine tract binding protein PTB promotes exon skipping to favour isoform-257. USP4, the closest homologue of USP15, is also alternatively spliced in a similar fashion, although the skipped amino acid cassette differs in sequence to that in USP15, and the two USP4 isoforms are proposed to act in different cellular compartments58.
The USP15 isoforms were historically studied interchangeably, although an example of isoform-dependent substrate-specificity was recently reported18. In contrast, the USP15 role that we describe here, can be performed by either isoform. Importantly though, we show for the first time that the USP15 isoforms are differentially regulated during the cell cycle, with S229-phosphorylation specifically abrogating the role of USP15 isoform-1 in regulating TOP2A expression and preventing micronuclei formation. In cycling cells, endogenous USP15 isoform-1 remains unphosphorylated during G2 when it is required for TOP2A accumulation. S229-phosphorylation occurs as cells enter mitosis, at around the time USP15 relocalises to the nucleus and TOP2A localizes to centromeres to perform its role in decatenation. In our experiments (Figure 8), exogenously expressed USP15 isoform-1 can support G2 accumulation of TOP2A and this appears sufficient to rescue the micronuclei phenotype. In contrast, activity of the S229D phospho-mimetic towards TOP2A is lacking not only during mitosis, but also in G2, rendering it unable to rescue either the TOP2A or micronuclei phenotypes.
The only kinases described to date for USP15 are ATM, which phosphorylates the two isoforms at S678/S649 respectively 39, and CK2 that targets undetermined residues 36. There is no strong kinase consensus around S229 and the various prediction algorithms suggest a number of candidates, including CK1, GS3K and ERK. The kinase which phosphorylates USP15 at S229 remains to be experimentally determined.
Post-translational modifications including ubiquitylation are important regulators of TOP2A function59 and E3 ligases for TOP2A include BRCA160, SCFFBXW761 and RNF16862. Likewise, in addition to our data for USP15, USP1062 and OTUD363 were recently reported as DUBs that regulate TOP2A. Such apparent redundancy may be essential to regulate critical checkpoint proteins. A notable example is P53, which is regulated by multiple DUBs acting at different cell cycle stages, or in different cellular compartments, to co-ordinate fine-tuned, temporal control over P53 expression (reviewed in10). OTUD3, a predominantly cytosolic DUB31, is proposed to regulate TOP2A specifically when complexed with the tumour suppressor PTEN63. In contrast USP10, also a cytosolic DUB31, deubiquitylates TOP2A in complex with the E3 ligase RNF16862, that is also important in DNA damage response signalling for double-strand break repair64. Thus a suite of E3 ligases and DUBs may regulate TOP2A in specific contexts to fine-tune TOP2A function in response to cellular needs.
Importantly, we also demonstrate that both isoforms of USP15 are commonly endogenously expressed in cells, whilst isoform-1 expression is preferentially upregulated in NSCLC cell lines. Interestingly, USP15 amplification is reported in certain cancers20, and here we show this is also the case for lung cancer, where elevated USP15 is associated with worse prognosis in clinical lung adenocarcinoma (Figure 9). However, in non-small cell lung cancer cell lines, we find the USP15 isoform-1 is preferentially upregulated. As USP15 has multiple verified and candidate substrates, it is difficult to assess the relative importance for regulation of TOP2A in USP15 amplified and/or USP15 isoform-1 overexpressing cancer cells. However, we postulate that such an imbalance, in favour of isoform-1 that can be mitotically inactivated by phosphorylation, may dampen the temporal role of USP15 in promoting genome stability through TOP2A, whilst not impacting on the other roles for USP15 in regulating alternative oncogenic pathways.
Materials and Methods
Cell culture
A549 and U2OS cells (ECACC) were cultured in high glucose DMEM supplemented with 10% fetal bovine serum (FBS) and 1% non-essential amino acids. All other lung cancer cell lines (sourced as previously described65) were cultured in RPMI supplemented with 10% FBS. FKHRL1-U2OS cells (Thermo Scientific, Bioimage Products, Lafayette, USA) were maintained in high glucose DMEM supplemented with 10% FBS, 100units/ml Penicillin/Streptomycin and 0.5mg/ml Geneticin. All cells were cultured in a humidified incubator at 37°C and 5% CO2. Cell lines were authenticated by STR profiling, verified as mycoplasma free and cultured for limited passage numbers.
Cell synchronisation
To obtain cell populations synchronised at G1/S or in early S, late S or G2, A549 were subject to double thymidine block (2mM thymidine (Sigma) for 18hr, released into fresh media for 8hr, arrested with thymidine for 17hr) then lysed directly or released into full medium for 2.5hr, 5.5hr or 7.5hr. To obtain cell populations synchronised at G2/M or in M, or early G1, A549 were subject to a thymidine/nocodazole block (2mM thymidine for 24hr, released into full medium containing 100ng/ml nocodazole (Sigma) for 14hr) then mitotic cells were collected by knocking off the dish, and either lysed directly or replated into full medium for 0.5hr or 4hr. Propidium iodide staining with flow cytometry analysis, and immunoblotting for a panel of stage-specific cell cycle markers, are routinely used to confirm enrichment of cell cycle phases. For analysis of each timepoint, adherent cells were trypsinised and pooled with non-adherent cells from the medium, and then processed for RNA or protein extraction.
RNA interference
A549 or U2OS cells were seeded at 6x104 cells per well in 6-well plates and transfected the following day with 40nM siRNA using Oligofectamine (Invitrogen); cells were analyzed 72hr later by immunoblotting or imaging. For rescue experiments, 48hr after siRNA transfection, U2OS cells were transfected with 1μg of plasmid using GeneJuice (Novagen) and processed for immunofluorescence 24hr later. siGenome siRNAs (Dharmacon) that target both of the USP15 isoforms were used as a pool named siUSP15-P [siUSP15-1 (D-006066-01), siUSP15-2 (D-006066-02) and siUSP15-17 (D-006066-17)]. siGenome non-targeting control siRNAs were siCON1 (D-001210-01) and siCON2 (D-001210-02). siGenome custom siRNAs specific for each USP15 isoform were: siUSP15-iso1 (5’-ACAACAUGAACAACAGAAAUU-3’) and siUSP15-iso2 (5’-CUUUCUACUCCUAAUGUGAUU-3’).
A custom designed DUB siRNA library consisting of four pooled oligos for each of 92 human DUBs (Qiagen) was used for the RNAi screen. Individual Qiagen siRNAs targeting both USP15 isoforms were used for follow-up studies [siUSP15-A (SI00087353), siUSP15-B (SI00087360), siUSP15-C (SI00087367), siUSP15-D (SI03072909)], either alone or as a pool (USP15-P2), together with the All-Stars negative control siRNA (1027281, siC). For the siRNA library screen, FKHRL1-U20S cells were seeded at 3000 cells per well in 96-well plates and reverse transfected with 20nM siRNA using Lipofectamine RNAiMax (Life Technologies). 48hr after transfection, media was replaced with HBSS (Gibco) containing 2.5µM DRAQ5 (Biostatus) and the cells then imaged.
Plasmid DNA Constructs
USP15 isoform-1 (NM_001252078.1) and USP15 isoform-2 (NM_006313.2) were cloned using the Gateway system into pDONR233 entry constructs. siRNA-resistant (to siUSP15-2), catalytically inactive and phospho-mutant forms of USP15 were generated by Quickchange site-directed mutagenesis in pDONR233 using complementary primer pairs: USP15-siRes2, 5’-CCATGAAAAAAGAACGCACTTTAGAGGTATACTTAGTTAGAATG-3’, USP15 isoform-1 (C298S) or USP15 isoform-2 (C269S), 5’-GTAACTTGGGAAATACGAGTTTCATGAACTCAGC-3’, USP15 isoform-1 phospho-null (S229A) 5’-CTTCTACTCCTAAGGCCCCAGGTGCATCC-3’ and phospho-mimetic (S229D) 5’-CTTCTACTCCTAAGGACCCAGGTGCATCC-3’. Each construct was sequence verified and shuttled into expression vectors. H2B-Cherry was a gift from Stephen Royle, University of Warwick, UK.
RNA extraction and real-time PCR
Total RNA was extracted using RNeasy columns (Qiagen) and cDNA was reverse transcribed from 1μg RNA with RevertAid H-minus M-MuLV reverse transcriptase (Fermentas) using an oligo-dT primer (Promega). Quantitative real-time RT-PCR (qRT-PCR) was performed in triplicate using SYBR Green supermix and a CFX real-time PCR detection system (Bio-Rad). Primer sequences were: ACTB (For: 5’-CACCTTCTACAATGAGCTGCGTGTG-3’, Rev: 5’-ATAGCACAGCCTGGATAGCAACGTAC-3’), total USP15 (For: 5’-CAGACAGCACCATTCAGGATGC-3’, Rev: 5’-GAGTTTTTCACATTAGGAGTAG-3’), USP15 isoform-1 (For: 5’-CAGACAGCACCATTCAGGATGC-3’, Rev: 5’-AAAATTGGATGCACCTGGGGAC-3’), USP15 isoform-2 (For: 5’-CAGACAGCACCATTCAGGATGC-3’, Rev: 5’-GAGTTTTTCACATTAGGAGTAG-3’), cyclin B1 (For: 5’-GCTCTTCTCGGCGTGCTGC-3’, Rev: 5’-CCTGCCATGTTGATCTTCG-3’), TOP2A (For: 5’-TGAAAACCCAACCTTTGACTC-3’, Rev: 5’-GCTTTCTACAATACCACAGCC-3’). Samples underwent 2-step amplification at 94°C (30 s) and 60°C (60 s); melt curves were analyzed after 40 cycles. The Ct values for test genes were normalized to ACTB and relative expression represented as 2-[ΔCt] for the cell panel or 2-[ΔΔCt] relative to a comparator sample in experiments.
Antibodies
Antibodies used were mouse anti-USP15 (H00009958-M01, Abnova), anti-TOP2A (sc-165986, Santa Cruz), anti-cyclinB1 (05-373, Millipore), anti-β-actin (ab6276, Abcam), anti-α-tubulin (DM1A, Sigma) and anti-CENPA (ab13939, Abcam); rabbit anti-actin (A2066, Sigma), anti-pericentrin (ab4448, Abcam). Polyclonal affinity-purified sheep anti-GFP was generated in house (IAP).
Immunofluorescence and microscopy
Cells were fixed in 4% paraformaldehyde, quenched with ammonium chloride and permeabilised with 0.1% triton prior to blocking and primary antibody incubation. Alexa-Fluor 488 and Alexa-Fluor 594 coupled secondary antibodies (Molecular probes) were used. Coverslips were mounted on Moviol supplemented with 4′,6-diamidino-2-phenylindole (DAPI) at 1:10 000. Cells were imaged using a Nikon Eclipse Ti (CFI Plan Apochromat 40× N.A. 0.95, W.D. 0.14mm) microscope. For TOP2A quantitation, nd2 files were opened in FIJI and the DAPI channel used to create regions of interest, which were overlaid on the red channel to measure the mean TOP2A intensity per cell. Micronuclei rescue experiment slides were scored blind, with the identity of samples anonymized until after scoring was complete. For live cell imaging, an Okolab incubator maintained cells at 37 °C, 5% CO2 and CFI Plan-Fluor 10X N.A.0.3 or CFI Super-Plan-Fluor 20X N.A.0.45 objectives were used.
Immunoblotting
Whole cell extracts were prepared by direct addition of hot Laemmli buffer and incubation at 110 C for 10min with intermittent vortexing. Following Bicinchoninic Acid (BCA) assay (Thermo Scientific) equivalent amounts of proteins were resolved by SDS-PAGE and transferred to BiotraceNT membrane (VWR) for incubation with primary antibodies. To accentuate band shifts caused by phosphorylation, samples were run on 6% polyacrylamide gels with 20µM Phos-Tag Acrylamide (NARD institute, AAL-107) and 40µM MnCl2. Proteins were visualized using donkey anti-mouse, anti-rabbit or anti-sheep secondary antibodies conjugated to the IRDyes IR680-LT, or IR800 (LI-COR) and a LI-COR Odyssey 2.1 system, with 16-bit images quantified in ImageStudio.
Mass spectrometry
SILAC-labelled A549 cells were transfected with siUSP15-P or siCON1 for 72hr prior to analysis. Whole cell protein lysates were separated on NuPAGE Novex 4–12% Bis-Tris Gels (Invitrogen). Gel slices were chopped into cubes <1mm3 and de-stained with 50% acetonitrile/50% 100mM ammonium bicarbonate. Samples were reduced (10mM DTT, 56°C, 1hr) and alkylated (50mM iodoacetamide, 30min, room temp). After dehydration with acetonitrile, gel pieces were incubated with mass spectrometry grade Trypsin Gold (Promega) at 10ng/μl (18hr, 37°C). Peptides were extracted by incubation in acetonitrile, followed by two 1% formic acid incubations, and a final acetonitrile extraction. Samples were dried by rotary evaporation in a Speedvac and peptides re-suspended in 1% formic acid.
Peptides were separated using a nanoACQUITY UPLC system (Waters), coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Scientific) with a Proxeon nano-electrospray source. 5μl of the digest was injected into a 180μm x 20mm, 5μm C18 symmetry trapping column (Waters) in 0.1% formic acid at 10μl/min before being resolved on a 25cm × 75μm BEH-C18 column (Waters), in an acetonitrile gradient in 0.1% formic acid, with a flow rate of 400 nl/min. Full scan MS spectra (m/z 350–2000) were generated at 30,000 resolution, ions fragmented by collision-induced dissociation (collision energy 35%, 30ms) and subjected to MS/MS in the linear quadrapole ion trap. All spectra were acquired using Xcalibur software (version 2.0.7; Thermo Fisher Scientific). RAW files were analysed using Maxquant version 1.4.1.2.
Bioinformatics and statistical analysis
The provisional TCGA dataset for lung adenocarcinoma was accessed and analysed using cBioportal66,67. Biochemical measurements represent several thousand cells; these data are represented as the mean value from at least three independent experiments, with error bars showing standard deviation (as indicated in Figure legends). No statistical method was used to predetermine sample size. For expression levels or phenotypes of individual cells, where feasible, we aimed to count/score at least 100 cells per condition, except where the experiment precluded this. All statistical tests for experimental data were performed using GraphPad Prism version 6.00 for Mac; P values less than 0.05 were considered to be significant. Data were analysed by T-test or ANOVA as appropriate (indicated in Figure legends). These parametric tests are suitable for continuous data sets without off-scale measurements, and assume Gaussian distribution, which was confirmed by the D'Agostino & Pearson omnibus normality test in Prism, and equivalent variance between samples confirmed by a variance homogeneity test.
Supplementary Material
Supplementary Information accompanies the paper
Acknowledgements
The authors are supported by a North West Cancer Research project grant CR942 (A.B.F.) and Wellcome Trust PhD studentships 099793/Z/12/Z (M.C.) and 096567/Z/11/Z (S.D.). We acknowledge the contribution of Ruth Owen and Bethan Gay towards experiments using the lung cell panel.
Footnotes
Author contributions
A.F. performed the majority of experiments. M.C., S.D. and J.J.S. contributed additional experiments, E.R.-J. assisted with mass spectrometry. J.M.C. directed the work and wrote the manuscript with input from A.F. All authors discussed the work, analyzed the data and commented on the manuscript.
Conflict of interest
M.J.C. and S.U. are part of the DUB Alliance that includes Cancer Research Technology and FORMA Therapeutics.
References
- 1.Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12:295–307. doi: 10.1038/nrm3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S. Deubiquitylases from genes to organism. Physiological reviews. 2013;93:1289–1315. doi: 10.1152/physrev.00002.2013. [DOI] [PubMed] [Google Scholar]
- 3.Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, et al. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63:146–155. doi: 10.1016/j.molcel.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sacco JJ, Coulson JM, Clague MJ, Urbe S. Emerging roles of deubiquitinases in cancer-associated pathways. IUBMB Life. 2010;62:140–157. doi: 10.1002/iub.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clague MJ, Coulson JM, Urbé S. Cellular functions of the DUBs. J Cell Sci. 2012;125:277–286. doi: 10.1242/jcs.090985. [DOI] [PubMed] [Google Scholar]
- 6.Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. Biochem J. 2015;467:191. doi: 10.1042/bj4670191. [DOI] [PubMed] [Google Scholar]
- 7.Fournane S, Krupina K, Kleiss C, Sumara I. Decoding ubiquitin for mitosis. Genes Cancer. 2012;3:697–711. doi: 10.1177/1947601912473477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pinto-Fernandez A, Kessler BM. DUBbing Cancer: Deubiquitylating Enzymes Involved in Epigenetics, DNA Damage and the Cell Cycle As Therapeutic Targets. Front Genet. 2016;7:133. doi: 10.3389/fgene.2016.00133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lim KH, Song MH, Baek KH. Decision for cell fate: deubiquitinating enzymes in cell cycle checkpoint. Cell Mol Life Sci. 2016;73:1439–1455. doi: 10.1007/s00018-015-2129-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darling S, Fielding AB, S-P D, Prior IA, Coulson JM. Regulation of the cell cycle and centrosome biology by deubiquitylases. Biochem Soc Trans. 2017 doi: 10.1042/BST20170087. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes-Turcu FE, Ventii KH, Wilkinson KD. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu Rev Biochem. 2009;78:363–397. doi: 10.1146/annurev.biochem.78.082307.091526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahtoe DD, Sixma TK. Layers of DUB regulation. Trends in biochemical sciences. 2015;40:456–467. doi: 10.1016/j.tibs.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 13.Kessler BM, Edelmann MJ. PTMs in Conversation: Activity and Function of Deubiquitinating Enzymes Regulated via Post-Translational Modifications. Cell Biochem Biophys. 2011;60:21–38. doi: 10.1007/s12013-011-9176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang X, Summers MK, Pham V, Lill JR, Liu J, Lee G, et al. Deubiquitinase USP37 Is Activated by CDK2 to Antagonize APC(CDH1) and Promote S Phase Entry. Mol Cell. 2011;42:511–523. doi: 10.1016/j.molcel.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 15.Joo HY, Zhai L, Yang C, Nie S, Erdjument-Bromage H, Tempst P, et al. Regulation of cell cycle progression and gene expression by H2A deubiquitination. Nature. 2007;449:1068–1072. doi: 10.1038/nature06256. [DOI] [PubMed] [Google Scholar]
- 16.Mizuno E, Kitamura N, Komada M. 14-3-3-dependent inhibition of the deubiquitinating activity of UBPY and its cancellation in the M phase. Exp Cell Res. 2007;313:3624–3634. doi: 10.1016/j.yexcr.2007.07.028. [DOI] [PubMed] [Google Scholar]
- 17.Thorne C, Eccles RL, Coulson JM, Urbé S, Clague MJ. Isoform-Specific Localization of the Deubiquitinase USP33 to the Golgi Apparatus. Traffic. 2011;12:1563–1574. doi: 10.1111/j.1600-0854.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 18.Kotani Y, Morito D, Sakata K, Ainuki S, Sugihara M, Hatta T, et al. Alternative exon skipping biases substrate preference of the deubiquitylase USP15 for mysterin/RNF213, the moyamoya disease susceptibility factor. Sci Rep. 2017;7:44293. doi: 10.1038/srep44293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baker RT, Wang XW, Woollatt E, White JA, Sutherland GR. Identification, functional characterization, and chromosomal localization of USP15, a novel human ubiquitin-specific protease related to the UNP oncoprotein, and a systematic nomenclature for human ubiquitin-specific proteases. Genomics. 1999;59:264–274. doi: 10.1006/geno.1999.5879. [DOI] [PubMed] [Google Scholar]
- 20.Eichhorn PJ, Rodon L, Gonzalez-Junca A, Dirac A, Gili M, Martinez-Saez E, et al. USP15 stabilizes TGF-beta receptor I and promotes oncogenesis through the activation of TGF-beta signaling in glioblastoma. Nat Med. 2012;18:429–435. doi: 10.1038/nm.2619. [DOI] [PubMed] [Google Scholar]
- 21.Srihari S, Ragan MA. Systematic tracking of dysregulated modules identifies novel genes in cancer. Bioinformatics. 2013;29:1553–1561. doi: 10.1093/bioinformatics/btt191. [DOI] [PubMed] [Google Scholar]
- 22.Vos RM, Altreuter J, White EA, Howley PM. The ubiquitin-specific peptidase USP15 regulates human papillomavirus type 16 E6 protein stability. J Virol. 2009;83:8885–8892. doi: 10.1128/JVI.00605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, Langelotz C, Hetfeld-Pechoc BK, Schwenk W, Dubiel W. The COP9 signalosome mediates beta-catenin degradation by deneddylation and blocks adenomatous polyposis coli destruction via USP15. J Mol Biol. 2009;391:691–702. doi: 10.1016/j.jmb.2009.06.066. [DOI] [PubMed] [Google Scholar]
- 24.Schweitzer K, Bozko PM, Dubiel W, Naumann M. CSN controls NF-kappaB by deubiquitinylation of IkappaBalpha. Embo J. 2007;26:1532–1541. doi: 10.1038/sj.emboj.7601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu M, Takanashi M, Oikawa K, Tanaka M, Nishi H, Isaka K, et al. USP15 plays an essential role for caspase-3 activation during Paclitaxel-induced apoptosis. Biochem Biophys Res Commun. 2009;388:366–371. doi: 10.1016/j.bbrc.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 26.Inui M, Manfrin A, Mamidi A, Martello G, Morsut L, Soligo S, et al. USP15 is a deubiquitylating enzyme for receptor-activated SMADs. Nat Cell Biol. 2011;13:1368–1375. doi: 10.1038/ncb2346. [DOI] [PubMed] [Google Scholar]
- 27.Liu WT, Huang KY, Lu MC, Huang HL, Chen CY, Cheng YL, et al. TGF-beta upregulates the translation of USP15 via the PI3K/AKT pathway to promote p53 stability. Oncogene. 2017;36:2715–2723. doi: 10.1038/onc.2016.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou Q, Jin J, Hu H, Li HS, Romano S, Xiao Y, et al. USP15 stabilizes MDM2 to mediate cancer-cell survival and inhibit antitumor T cell responses. Nature immunology. 2014;15:562–570. doi: 10.1038/ni.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes SD, Liu H, MacDonald E, Sanderson CM, Coulson JM, Clague MJ, et al. Direct and indirect control of mitogen-activated protein kinase pathway-associated components, BRAP/IMP E3 ubiquitin ligase and CRAF/RAF1 kinase, by the deubiquitylating enzyme USP15. J Biol Chem. 2012;287:43007–43018. doi: 10.1074/jbc.M112.386938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;138:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Urbé S, Liu H, Hayes SD, Heride C, Rigden DJ, Clague MJ. Systematic survey of deubiquitinase localization identifies USP21 as a regulator of centrosome- and microtubule-associated functions. Mol Biol Cell. 2012;23:1095–1103. doi: 10.1091/mbc.E11-08-0668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Long L, Thelen JP, Furgason M, Haj-Yahya M, Brik A, Cheng D, et al. The U4/U6 recycling factor SART3 has histone chaperone activity and associates with USP15 to regulate H2B deubiquitination. J Biol Chem. 2014;289:8916–8930. doi: 10.1074/jbc.M114.551754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cornelissen T, Haddad D, Wauters F, Van Humbeeck C, Mandemakers W, Koentjoro B, et al. The deubiquitinase USP15 antagonizes Parkin-mediated mitochondrial ubiquitination and mitophagy. Hum Mol Genet. 2014;23:5227–5242. doi: 10.1093/hmg/ddu244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Faronato M, Patel V, Darling S, Dearden L, Clague MJ, Urbe S, et al. The deubiquitylase USP15 stabilizes newly synthesized REST and rescues its expression at mitotic exit. Cell cycle. 2013;12:1964–1977. doi: 10.4161/cc.25035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Faronato M, Urbé S, Coulson JM. USP15 (ubiquitin specific peptidase 15) Atlas Genet Cytogenet Oncol Haematol. 2011;15:645–651. [Google Scholar]
- 36.Hetfeld BK, Helfrich A, Kapelari B, Scheel H, Hofmann K, Guterman A, et al. The zinc finger of the CSN-associated deubiquitinating enzyme USP15 is essential to rescue the E3 ligase Rbx1. Curr Biol. 2005;15:1217–1221. doi: 10.1016/j.cub.2005.05.059. [DOI] [PubMed] [Google Scholar]
- 37.Olsen JV, Vermeulen M, Santamaria A, Kumar C, Miller ML, Jensen LJ, et al. Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci Signal. 2010;3:ra3. doi: 10.1126/scisignal.2000475. [DOI] [PubMed] [Google Scholar]
- 38.Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A Proteome-wide, Quantitative Survey of In Vivo Ubiquitylation Sites Reveals Widespread Regulatory Roles. Mol Cell Proteomics. 2011;10 doi: 10.1074/mcp.M111.013284. M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mu JJ, Wang Y, Luo H, Leng M, Zhang J, Yang T, et al. A proteomic analysis of ataxia telangiectasia-mutated (ATM)/ATM-Rad3-related (ATR) substrates identifies the ubiquitin-proteasome system as a regulator for DNA damage checkpoints. J Biol Chem. 2007;282:17330–17334. doi: 10.1074/jbc.C700079200. [DOI] [PubMed] [Google Scholar]
- 40.Guardavaccaro D, Frescas D, Dorrello NV, Peschiaroli A, Multani AS, Cardozo T, et al. Control of chromosome stability by the beta-TrCP-REST-Mad2 axis. Nature. 2008;452:365–369. doi: 10.1038/nature06641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gauthier NP, Larsen ME, Wernersson R, de Lichtenberg U, Jensen LJ, Brunak S, et al. Cyclebase.org--a comprehensive multi-organism online database of cell-cycle experiments. Nucleic Acids Res. 2008;36:D854–859. doi: 10.1093/nar/gkm729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luo K, Yuan J, Chen J, Lou Z. Topoisomerase IIalpha controls the decatenation checkpoint. Nat Cell Biol. 2009;11:204–210. doi: 10.1038/ncb1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bower JJ, Karaca GF, Zhou Y, Simpson DA, Cordeiro-Stone M, Kaufmann WK. Topoisomerase IIalpha maintains genomic stability through decatenation G(2) checkpoint signaling. Oncogene. 2010;29:4787–4799. doi: 10.1038/onc.2010.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nagasaka K, Hossain MJ, Roberti MJ, Ellenberg J, Hirota T. Sister chromatid resolution is an intrinsic part of chromosome organization in prophase. Nat Cell Biol. 2016;18:692–699. doi: 10.1038/ncb3353. [DOI] [PubMed] [Google Scholar]
- 45.Samejima K, Samejima I, Vagnarelli P, Ogawa H, Vargiu G, Kelly DA, et al. Mitotic chromosomes are compacted laterally by KIF4 and condensin and axially by topoisomerase IIalpha. J Cell Biol. 2012;199:755–770. doi: 10.1083/jcb.201202155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gorbsky GJ. Cell cycle progression and chromosome segregation in mammalian cells cultured in the presence of the topoisomerase II inhibitors ICRF-187 [(+)-1,2-bis(3,5-dioxopiperazinyl-1-yl)propane; ADR-529] and ICRF-159 (Razoxane) Cancer Res. 1994;54:1042–1048. [PubMed] [Google Scholar]
- 47.Downes CS, Mullinger AM, Johnson RT. Inhibitors of DNA topoisomerase II prevent chromatid separation in mammalian cells but do not prevent exit from mitosis. Proc Natl Acad Sci U S A. 1991;88:8895–8899. doi: 10.1073/pnas.88.20.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Broderick R, Nieminuszczy J, Blackford AN, Winczura A, Niedzwiedz W. TOPBP1 recruits TOP2A to ultra-fine anaphase bridges to aid in their resolution. Nat Commun. 2015;6:6572. doi: 10.1038/ncomms7572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, et al. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun. 2015;6:8962. doi: 10.1038/ncomms9962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, et al. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis. 2011;26:125–132. doi: 10.1093/mutage/geq052. [DOI] [PubMed] [Google Scholar]
- 51.Naim V, Rosselli F. The FANC pathway and BLM collaborate during mitosis to prevent micro-nucleation and chromosome abnormalities. Nat Cell Biol. 2009;11:761–768. doi: 10.1038/ncb1883. [DOI] [PubMed] [Google Scholar]
- 52.Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr Biol. 1999;9:680–689. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- 54.Nishi R, Wijnhoven P, le Sage C, Tjeertes J, Galanty Y, Forment JV, et al. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat Cell Biol. 2014;16:1016–1026. doi: 10.1038/ncb3028. 1011-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, Zhang H, Wang X, Tian Q, Hu Z, Peng C, et al. The Deubiquitylating Enzyme USP4 Cooperates with CtIP in DNA Double-Strand Break End Resection. Cell Rep. 2015;13:93–107. doi: 10.1016/j.celrep.2015.08.056. [DOI] [PubMed] [Google Scholar]
- 56.Wiltshire TD, Lovejoy CA, Wang T, Xia F, O'Connor MJ, Cortez D. Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J Biol Chem. 2010;285:14565–14571. doi: 10.1074/jbc.M110.104745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gowher H, Brick K, Camerini-Otero RD, Felsenfeld G. Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II. Proc Natl Acad Sci U S A. 2012;109:2370–2375. doi: 10.1073/pnas.1121538109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vlasschaert C, Xia X, Gray DA. Selection preserves Ubiquitin Specific Protease 4 alternative exon skipping in therian mammals. Sci Rep. 2016;6:20039. doi: 10.1038/srep20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen T, Sun Y, Ji P, Kopetz S, Zhang W. Topoisomerase IIalpha in chromosome instability and personalized cancer therapy. Oncogene. 2015;34:4019–4031. doi: 10.1038/onc.2014.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shinagawa H, Miki Y, Yoshida K. BRCA1-mediated ubiquitination inhibits topoisomerase II alpha activity in response to oxidative stress. Antioxid Redox Signal. 2008;10:939–949. doi: 10.1089/ars.2007.1851. [DOI] [PubMed] [Google Scholar]
- 61.Chen MC, Chen CH, Chuang HC, Kulp SK, Teng CM, Chen CS. Novel mechanism by which histone deacetylase inhibitors facilitate topoisomerase IIalpha degradation in hepatocellular carcinoma cells. Hepatology. 2011;53:148–159. doi: 10.1002/hep.23964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guturi KK, Bohgaki M, Bohgaki T, Srikumar T, Ng D, Kumareswaran R, et al. RNF168 and USP10 regulate topoisomerase IIalpha function via opposing effects on its ubiquitylation. Nat Commun. 2016;7:12638. doi: 10.1038/ncomms12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kang X, Song C, Du X, Zhang C, Liu Y, Liang L, et al. PTEN stabilizes TOP2A and regulates the DNA decatenation. Sci Rep. 2015;5:17873. doi: 10.1038/srep17873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mattiroli F, Vissers JH, van Dijk WJ, Ikpa P, Citterio E, Vermeulen W, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 65.Moss AC, Jacobson GM, Walker LE, Blake NW, Marshall E, Coulson JM. SCG3 transcript in peripheral blood is a prognostic biomarker for REST-deficient small cell lung cancer. Clin Cancer Res. 2009;15:274–283. doi: 10.1158/1078-0432.CCR-08-1163. [DOI] [PubMed] [Google Scholar]
- 66.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.