Abstract
Characterizing the structure of membrane proteins (MPs) generally requires extraction from their native environment, most commonly with detergents. Yet, the physicochemical properties of detergent micelles and lipid bilayers differ markedly and could alter the structural organization of MPs, albeit without general rules. Dodecylphosphocholine (DPC) is the most widely used detergent for MP structure determination by NMR, but the physiological relevance of several prominent structures has been questioned, though indirectly, by other biophysical techniques, e.g., functional/thermostability assay (TSA) and molecular dynamics (MD) simulations. Here, we resolve unambiguously this controversy by probing the functional relevance of three different mitochondrial carriers (MCs) in DPC at the atomic level, using an exhaustive set of solution-NMR experiments, complemented by functional/TSA and MD data. Our results provide atomic-level insight into the structure, substrate interaction and dynamics of the detergent–membrane protein complexes and demonstrates cogently that, while high-resolution NMR signals can be obtained for MCs in DPC, they systematically correspond to non-functional states.
Keywords: CPMG NMR, protein stability, transport activity, detergent-protein interactions, MD simulation
Mitochondrial carriers (MCs) constitute a large family of transport proteins of ca. 30-35 kDa molecular weight that play important roles in the intracellular translocation of metabolites, nucleotides, and coenzymes1. The transport mechanism involves the alternating exposure of a central cavity to the cytosolic side (c-state) or the mitochondrial matrix (m-state)2. Crystallographic structures of the most prominent member, the ADP/ATP carrier (AAC), extracted from the the native mitochondrial membrane3,4 could only be obtained in the presence of the strong inhibitor carboxyatractyloside (CATR) which locks the carrier in an aborted c-state. These structures revealed a central cavity, in which CATR is bound, surrounded by six transmembrane helices. No other atomic-resolution structures of MCs are available, underscoring the difficulty of obtaining crystals of these dynamic proteins. Solution-NMR may overcome the difficulties that crystallography faces for dynamic MPs. Substrate, lipid and inhibitor binding5–10 as well as dynamics8,11, detected by NMR have been reported and related to functional mechanisms of this class of membrane proteins. Other biophysical techniques, e.g. functional/thermostability assays and molecular-dynamics simulations, have challenged some of these interpretations12–14. Solving this apparent controversy has been hampered by the lack of direct experimental atomic-resolution data and stringent control experiments. Here, we resort to an extensive set of NMR experiments to address the structure substrate interactions and dynamics of MCs in DPC alongside with functional/thermostability assays and molecular-dynamics simulations and conclude on their physiological significance.
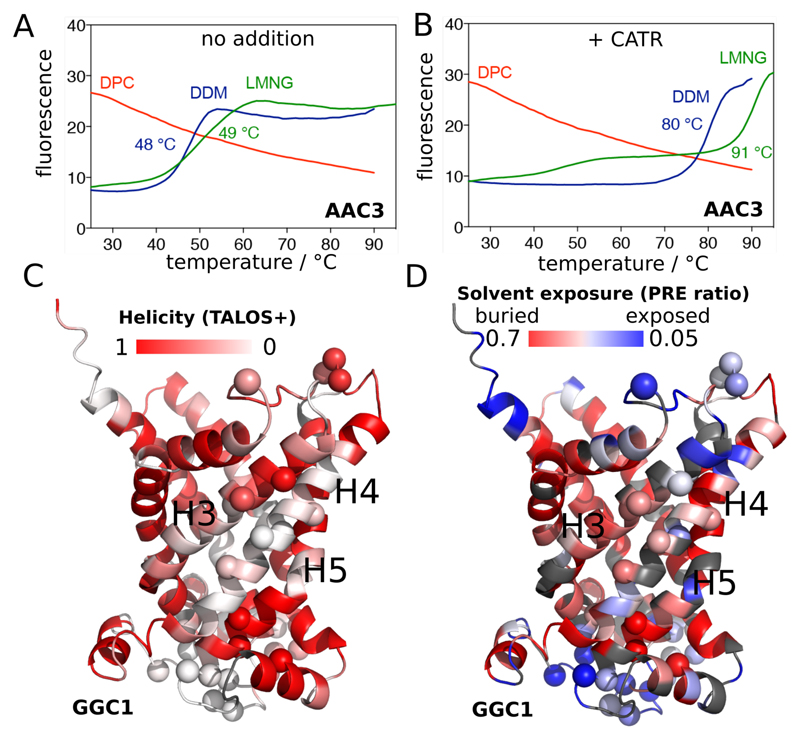
To probe the structural integrity of the yeast ADP/ATP carrier 3 (AAC3) in different detergents, we first investigated its thermal stability, consistently with our previous investigations13,14. We produced AAC3 in the yeast mitochondrial membrane, extracted it with the mild detergent dodecylmaltoside (DDM), diluted it into solutions of either DDM, lauryl maltoside neopentyl glycol (LMNG) or DPC, and performed thermostability shift assay (TSA) experiments14,15. In DDM and LMNG, a typical protein melting curve reveals a cooperative thermal denaturation event with an apparent melting temperature of ca. 49°C (Figure 1, blue/green lines). In the CATR-bound state, the melting temperature is dramatically increased to 80-90°C. In contrast, the same protein preparation diluted into DPC already shows a high fluorescence signal at low temperature, and no transition in the course of the temperature scan, irrespective of the presence of CATR. These observations indicate that AAC3 in DPC does not form a stable tertiary structure, nor binds CATR, in line with previous findings13. To understand this behavior at the atomic level, we turned to solution-NMR of three members of the MC family, namely AAC3, GDP/GTP carrier (GGC1) and ornithine carrier (ORC1). Given their low sequence identity (<32%, Figure S1), common traits observed in DPC are likely shared by the entire MC family. Samples of AAC3, GGC1 and ORC1, prepared through refolding from inclusion bodies, just like in previous NMR studies of MCs5,7,8,11, are all homogeneous (as shown by size-exclusion chromatograms and analytical ultra-centrifugation; Figure S2). Consistently with previous NMR studies5–9,11, all these samples yield high-quality NMR spectra (Figure S3), a criterion that is commonly employed to assess the functional relevance of MP samples and motivate structural studies. To reconcile these data with TSA experiments and to characterize the structural organization of MCs that is responsible for high-quality NMR spectra, we performed an NMR-observed temperature series up to ca. 70°C (Figure S3). One expects a discontinuity in the NMR parameters if collective unfolding arises. In contrast, we observe linear chemical-shift changes up to high temperatures, mirroring the TSA data and suggesting that these MCs undergo a gradual change of the structural ensemble without defined denaturation transition.
Figure 1. Stability of secondary and tertiary structures in MCs.
(A, B) Thermostability of AAC3, extracted from yeast mitochondria and purified in DDM, diluted twentyfold into 0.1% DDM (blue line), 0.1% LMNG (green line) or 0.1% DPC (red line) in the absence (A) or presence (B) of CATR, measured with TSA experiments (see Supplementary methods). (C) Residue-wise helix propensity in GGC1, determined from NMR chemical shifts using the program TALOS+ 16, plotted onto a structural model of GGC1. Residues in white color are not in a helical conformation. (D) Residue-wise solvent accessibility in GGC1, as probed with the paramagnetic agent gadodiamide. Residues shown in white-to-blue colors are accessible to solvent. In (C) and (D), amide sites undergoing μs-ms dynamics (discussed further below) are indicated by spheres.
We probed the structural and dynamical properties of MCs in DPC further with three different NMR observables. First, the residue-specific chemical-shift assignments were used with the TALOS+ software16 to derive local backbone geometry at the level of each residue (Figures 1C and S4). While there is an overall good agreement between the residues that, according to their chemical shifts, have α-helical conformation and the location of helices in the crystal structures of AAC3, several stretches of residues in the TM helices appear disordered in DPC, particularly in helices H4 (GGC1), and H2/H3 (AAC3). Independent support for the flexible nature of these parts comes from 15N transverse relaxation rate constants (R2), which reveal enhanced flexibility in GGC1's H4 (Figure S5). Second, we performed solvent-paramagnetic relaxation-enhancement (sPRE) experiments with the soluble hydrophilic paramagnetic complex Gd-DTPA-BMA (gadodiamide), which probe the solvent-accessibility of each amide site (Figure 1D). As expected, the most exposed parts are located in loops of GGC1 and AAC3. Unexpectedly, however, sizable sPRE effects are also found in TM helices, in particular those for which TALOS+ finds a loop conformation. Third, we measured amide hydrogen/deuterium exchange (HDX) by dilution of GGC1 from H2O- into D2O-buffer. The observed very fast HDX kinetics (< 1 minute) indicates that backbone hydrogen bonds are marginally stable (Fig. S6).
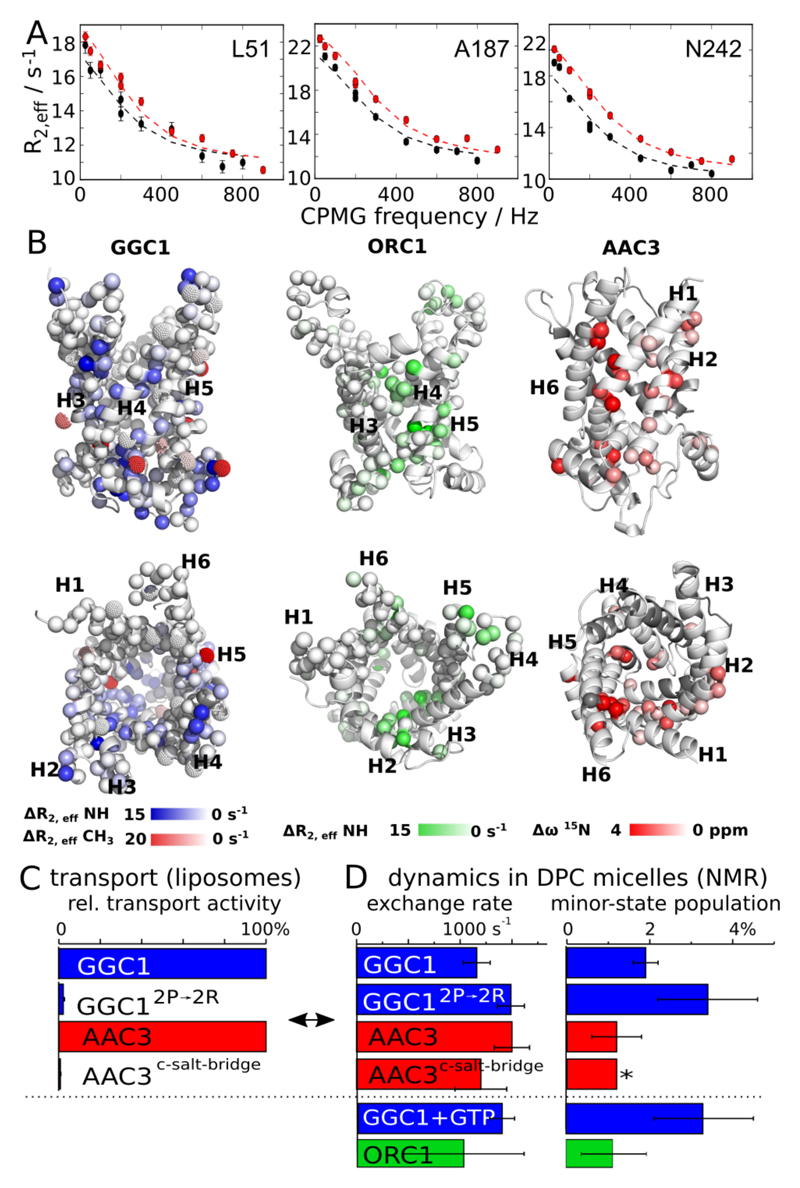
To gain further insight into the properties and possibly functional relevance of MCs in DPC, we investigated the details of their dynamics. Functional turnover of MCs in the membrane occurs at a rate of ca. 500 s-1.17 Functional MCs are, thus, expected to undergo motions on this time scale, and these motions are expected to be sensitive to substrates or mutations. Carr-Purcell-Meiboom-Gill (CPMG) relaxation-dispersion (RD) NMR experiments18 are ideally suited to probe µs-ms motions at the level of individual residues. In this study we performed extensive CPMG RD measurements on a total of ten different samples, including wild-type (WT) and mutant proteins, as well as samples with substrates and lipids, and on up to three magnetic field strengths per sample (see Table S1 and Figures S7-S17); in addition, previous work reported CPMG data for WT AAC38. In AAC3, GGC1 and ORC1, we find extensive µs-ms motions for about 20% of the 15N backbone amide sites, and in all cases the exchanging residues are clustered on one side of the molecule (Figure 2). We have performed additional 13C CPMG experiments on methyl groups of Ala, Val and Leu residues in GGC1 (Table S1 and Figure S8), independently confirming the presence of µs-ms dynamics in the region revealed by 15N CPMG experiments (red in Figure 2B, left). In order to obtain more insight into this process, we fitted a two-state exchange model to the CPMG RD data. Statistical analyses, shown in Figure S18, indicate that this simple model describes the data satisfyingly. The fitted parameters are similar for all three proteins, with an exchange rate constant, kex=kAB+kBA, on the order of 1000-2000 s-1, and populations of the minor state, pB, of ca. 1-3%. (Figure 2D and Table S1).
Figure 2. Millisecond dynamics in MCs in DPC detergent micelles.
(A) Representative 3D-HNCO-detected CPMG RD profiles of 15N amide sites in GGC1 (WT, no substrate). All non-flat RD profiles of this study are shown in Figures S7 to S17. (B) Location of residues with non-flat CPMG RD curves in GGC1, ORC1 and AAC3 plotted onto structural models based on the crystal structure of AAC3. For GGC1 and ORC1, the color code represents the extent of relaxation-dispersion, as determined from the difference in R2,eff of the first and last points of the RD profiles; for AAC3, the color code represents Δω values reported in reference 8. A qualitatively similar picture is obtained for two mutants studied here, GGC12P→2R and AAC3-c-saltbridge, see main text. (C) Transport activity and (D) NMR-derived dynamics parameters in MCs. The pB value denoted with an asterisk was fixed to the value found for AAC3, which resulted in satisfactory fits. All transport activity and dynamics data reported in this study are provided in Tables S1-S3.
A major challenge in the interpretation of relaxation dispersion data is to understand the underlying motional process structurally. An intriguing observation is that the timescale of motion observed in all three MCs – ca. 1 ms – is similar to the kinetics of transmembrane transport17, suggesting a possible role in functional turnover. To test this hypothesis, we investigated whether the introduction of function-abolishing mutations or the presence of substrates and lipids alter the dynamics. We designed two mutants that address different structural/functional aspects of the mitochondrial carriers, reasoning that if they inactivate the proteins, they would do so for different reasons. In a first mutant, termed GGC12P→2R, two highly conserved Pro residues of the MC signature motifs (PX[ED]XX[KR]) in H3 and H5 were mutated to Arg. Given the importance of these Pro kinks in the structure and mechanism2, and the unfavorable energetics of placing Arg residues in the core of TM helices19, we expected significant changes in the activity and dynamics. A second mutant targets Asp, Lys and Gln residues on the cytosolic side of the TM helices of AAC3, which have been proposed to form a salt bridge network in the elusive m-state4. In the AAC3-c-saltbridge mutant, we inverted charges such that the putative salt-bridge network cannot form any more. Liposome transport assays show that the residual transport activity in GGC12P→2R and AAC3-c-saltbridge is below 3%, i.e. that the mutations render the proteins essentially non-functional (Figure 2C and Table S3).
Strikingly, however, the characteristics of the mutants in DPC micelles are hardly different from those of the WT protein samples. NMR spectra of these mutants are highly similar to the respective WT proteins. Chemical-shift differences are small, and restricted to residues in the immediate vicinity of the mutation sites (Figure S19). If the helices were in close contact to each other, as in the crystal structure, one would expect pronounced chemical-shift changes in the neighboring helices in these mutants. Another surprising observation is that the μs-ms dynamics in these non-functional mutants is very similar to the one of WT proteins, even though the mutants are non-functional, likely suggesting that the µs-ms motions are not directly related to function (Figures 2D, S12-S14 and Table S1). Along the same lines, we find that the presence of substrate (GTP) does not lead to a detectable difference in the dynamics compared to apo-GGC1 (Figure 2D), indicating that substrate cannot influence this dynamic process. Put together, the observed dynamics does not appear to be related to function. We reasoned that the functionally important2 lipid cardiolipin (CL) might resurrect the native properties of the protein. However, CPMG experiments with CL reveal an identical behavior for WT GGC1 and GGC12P→2R (see Supplementary Discussion and Figures S9, S11 and S20), showing that these MCs in DPC are non-functional, irrespective of added lipids.
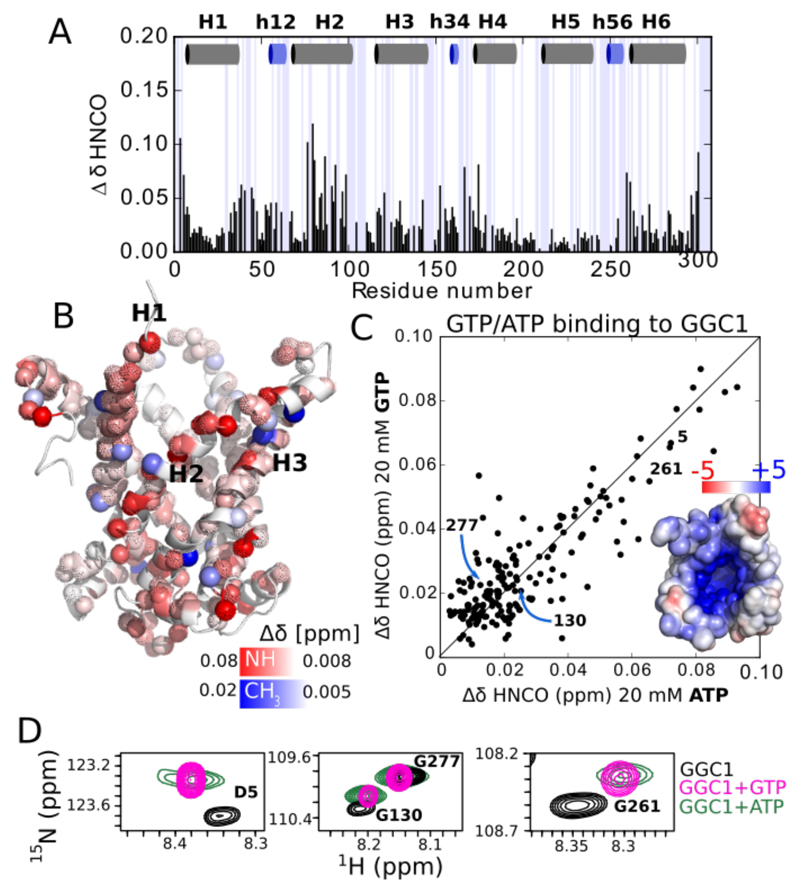
An important criterion to assess functionality of a MP is its ability to interact with substrates. MCs are highly substrate-specific transporters. In liposome transport assays, we find that recombinantly expressed and reconstituted AAC3 and GGC1 bind and transport specifically their substrate (ATP/GTP, respectively), while they do not bind the other nucleotide (GTP/ATP; see Table S3). Furthermore, CATR inhibits AAC3, but not GGC1 (see Supplementary discussion). Testing the binding specificity is, thus, a convenient way to investigate the structural integrity of MCs in DPC. Addition of the substrate GTP to GGC1 leads to significant CSPs for many amides and Ala, Leu and Val methyls (Figure 3A). The largest CSPs are observed for residues lining H2, and on the cytoplasmic side of H1 and H6. Weaker yet significant effects are observed in the three amphipathic matrix helices (Figure 3B); these CSPs are in good agreement with previously reported data11. However, addition of ATP – which does not interact with GGC1 in membranes – unexpectedly produces essentially the same CSPs as GTP (Figure 3C,D). Likewise, addition of GTP or ATP to AAC3 produces CSPs that are nearly identical to each other (Figure S21). Thus, while AAC3 and GGC1 in their lipid-bilayer environment can discriminate between GTP and ATP, this ability is lost in DPC micelles. We repeated GTP-titration experiments with the non-functional GGC12P→2R mutant. Even though this protein is non-functional and likely distorted through the Arg in the TM part, the GTP-induced CSPs are very similar to those observed in WT protein (Figure S22). This observation suggests that the observed GTP interaction is not dependent on structural integrity, and is thus non-specific.
Figure 3. GGC1 in DPC lacks the expected binding specificity.
(A) Chemical-shift perturbations in GGC1 upon addition of GTP. (B) Plot of amide (H/N/CO) and methyl (H/C) CSPs onto a structural model of GGC1. (C, D) GTP and ATP produces very similar CSPs in GGC1, as exemplified with extracts from HNCO spectra (D) and shown as a correlation plot in (C). The inset shows the electrostatic surface of a AAC-derived structural model of GGC1. Equivalent data for AAC3 are shown in Figure S21.
We then titrated WT GGC1 with CATR, a doubly negatively charged inhibitor of AAC, but not of GGC120. Despite the absence of GGC1/CATR interaction in membranes, the addition of CATR to GGC1 in DPC produces significant CSPs. The affected set of residues partly coincides with the sites that are modulated by the presence of GTP and ATP, and the effects are similar to those observed in AAC3 upon addition of CATR (Figure S23). These observations lead us to propose that the reported interactions of CATR with GGC1 or AAC3 in DPC are non-specific and not reminiscent of binding in biological membranes. Indeed, the affinity of CATR to AAC3 in DPC (Kd = 20-150 μM8) is ca. 3 orders of magnitude weaker than the AAC/CATR interaction in lipid bilayers (Kd = 10-20 nM21) or of AAC extracted from mitochondria with LAPAO detergent (Kd = 310 nM21). A plausible driving force for all these non-specific low-affinity interactions is the electrostatic attraction between the negatively charged solutes (nucleotides, CATR) and the positively charged cavity of GGC1 and AAC3 (Figures 3C and S21D). In agreement with this view, ORC1, which has a significantly lower electrostatic charge, does not show significant CSPs upon addition of L-histidine (Fig. S24).
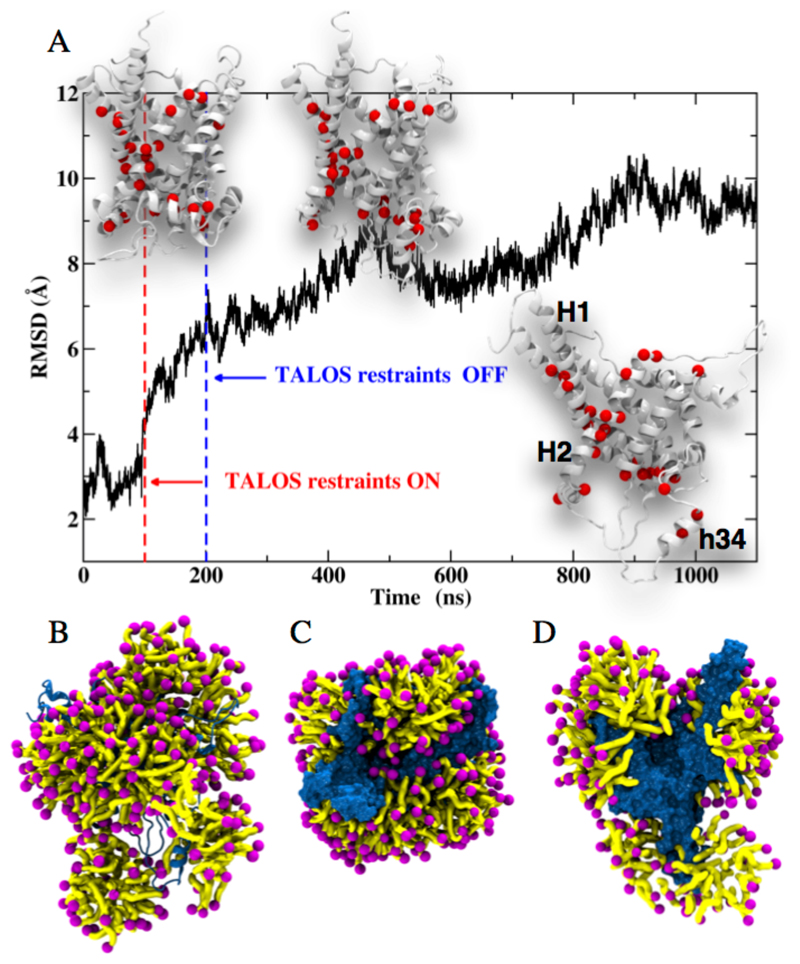
To obtain a mechanistic understanding of the structural organization of the protein and detergent, we turned to all-atom molecular dynamics (MD) simulations in explicit DPC. MD simulations have been applied previously to MCs in a membrane environment22–24 and in DPC12. We specifically sought a description of the structural organization of AAC3 in DPC, which would reflect our experimental data. In initial unrestrained all-atom simulations over 0.7 μs, AAC3 remains close to the conformation adopted in crystals4, which is in disagreement with the TALOS-derived secondary structure in DPC. Presumably, the time scale accessible to simulation is too short to allow the transition of the compact conformation in crystals to the one in DPC micelles (see Supplementary Discussion and Figures S25, S26). To better reflect the experimental data, we, thus, used chemical-shift derived backbone torsional angles (limited to the ϕ,ψ angles for which the TALOS+ result was unambiguous; see Supplementary Methods), and introduced a soft harmonic potential to these residues over a simulation period of 0.1 μs. This strategy, which rests on the use of NMR restraints to guide conformational analyses of proteins, is similar in spirit to that proposed by others25,26. When the torsional angles coincide with those inferred from the NMR data, the overall structure of AAC3 is markedly distorted, corresponding to a shift from ca. 3 to 6 Å of the root-mean-square deviation (RMSD) of atomic positions (Figure 4A). The evolution of the tertiary structure upon introduction of the TALOS+ restraints is asymmetrical and chiefly involves the interfacial helix h34 (connecting H3 and H4) and the cytoplasmic side of H1, H2 and H3 (Figure 4 and S27). In a last step, the restraints were removed and the simulation was extended to 1.1 μs. Over this timescale AAC3 remains distorted. The cytoplasmic side of the TM helices H1, H2 and the interfacial helix h34 undergo relatively slow dynamics on the simulation timescale (see Movie M1). Interestingly, this region coincides with the segments that were found experimentally to undergo μs-ms motions (red spheres in Figure 4). We speculate that the motions detected experimentally might correspond to the kind of slow rearrangements seen by MD. Most importantly, the simulations provide a view of how the detergent may organize around the protein. Figures 4B-D show that DPC molecules do not only form a corona around the hydrophobic core of the protein, as one might expect, but also form micelles interacting with both sides of the protein (cf. Movie M2). DPC molecules protrude between the TM helices and penetrate deep within the protein cavity (Figure 4C,D and Movie 2).
Figure 4. All-atom MD simulations of AAC3 in DPC micelles.
(A) Time evolution of the root-mean-square displacement (RMSD) of yAAC3 backbone atomic positions with respect to those in the crystal structure. From 0 to 100 ns, the protein embedded in DPC is free to move. From 100 to 200 ns, a subset of dihedral angles is restrained to TALOS+ values inferred from NMR chemical shifts (see Methods and Table S4). At 200 ns, restraints are turned off. The secondary structures of the protein before and after applying the TALOS+ restraints and at the end of the trajectory are depicted chronologically in the insets. Amide sites for which μs-ms motion is found in CPMG experiments are highlighted with red spheres. (B-D) Snapshots at 1 μs simulation time showing the DPC (purple/yellow) organization around yAAC3 from side view (B), top view (C) and cut-open side view (D). Supplementary Movies 1 and 2 show the time evolution of AAC3 and its surrounding micelle.
DPC has been employed for solving about 40% of all known NMR structures, but less than 1% of all crystal structures (Figure S28). Our data provide possible clues about the origin of this intriguing track record. In our hands, DPC is the only detergent able to maintain MCs in solution for extended time periods – a prerequisite for solution-NMR –, and the embedded proteins exhibit well-resolved spectra and solute interactions that may be interpreted as signs of a functionally relevant sample. Our study reveals that these observations do not necessarily point to a functionally relevant protein sample, and stresses the importance of stringent control experiments.
Supplementary Material
Supporting Information. A PDF file containing detailed experimental materials and methods, tables summarizing the results of transport activity measurements, CPMG fits and assigned chemical shifts and figures with supporting experimental data is available free of charge.
Acknowledgment
We thank Klaus Zangger for providing gadodiamide, and Astrid C. Sivertsen, Guillaume Bouvignies, Eva Pebay-Peyroula, Stéphanie Ravaud, Ewen Lescop, Cécile Breyton, Sven Brüschweiler, Christine Ebel and Aline LeRoy for insightful discussions, Jason R. Schnell for help in preparing Figure S28 and Michael Kohlhaas for inspiring thoughts. This work was supported by the European Research Council (ERC-Stg-311318) to P. S. and the UK Medical Research Council (U105663139) to E.R.S.K. This work used the platforms of the Grenoble Instruct Center (ISBG; UMS 3518 CNRS-CEA- UJF-EMBL) with support from FRISBI (ANR-10-INSB-05- 02) and GRAL (ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology (PSB).
Footnotes
Author Information
The authors declare no competing financial interests.
References
- (1).Palmieri F, Monné M. Discoveries, Metabolic Roles and Diseases of Mitochondrial Carriers: A Review. Biochim Biophys Acta - Mol Cell Res. 2016;1863(10):2362–2378. doi: 10.1016/j.bbamcr.2016.03.007. [DOI] [PubMed] [Google Scholar]
- (2).Kunji ERS, Aleksandrova A, King MS, Majd H, Ashton VL, Cerson E, Springett R, Kibalchenko M, Tavoulari S, Crichton PG, et al. The Transport Mechanism of the Mitochondrial ADP/ATP Carrier. Biochim Biophys Acta. 2016;1863(10):2379–2393. doi: 10.1016/j.bbamcr.2016.03.015. [DOI] [PubMed] [Google Scholar]
- (3).Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trezeguet V, Lauquin G, Brandolin G. Structure of Mitochondrial ADP/ATP Carrier in Complex with Carboxyatractyloside. Nature. 2003;426(6962):39–44. doi: 10.1038/nature02056. [DOI] [PubMed] [Google Scholar]
- (4).Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ERS. Structures of Yeast Mitochondrial ADP/ATP Carriers Support a Domain-Based Alternating-Access Transport Mechanism. Proc Natl Acad Sci USA. 2014;111(4):E426–34. doi: 10.1073/pnas.1320692111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Run C, Yang Q, Liu Z, OuYang B, Chou JJ. Molecular Basis of MgATP Selectivity of the Mitochondrial SCaMC Carrier. Structure. 2015;23(8):1–10. doi: 10.1016/j.str.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Zhao L, Wang S, Zhu Q, Wu B, Liu Z, OuYang B, Chou JJ. Specific Interaction of the Human Mitochondrial Uncoupling Protein 1 with Free Long-Chain Fatty Acid. Structure. 2017;25(9):1371–1379.e3. doi: 10.1016/j.str.2017.07.005. [DOI] [PubMed] [Google Scholar]
- (7).Zhao L, Wang S, Run C, OuYang B, Chou JJ. Specific Lipid Binding of Membrane Proteins in Detergent Micelles Characterized by NMR and Molecular Dynamics. Biochemistry. 2016;55(38):5317–5320. doi: 10.1021/acs.biochem.6b00836. [DOI] [PubMed] [Google Scholar]
- (8).Brüschweiler S, Yang Q, Run C, Chou JJ. Substrate-Modulated ADP/ATP-Transporter Dynamics Revealed by NMR Relaxation Dispersion. Nat Struct Mol Biol. 2015;22(8):636–641. doi: 10.1038/nsmb.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Berardi MJ, Shih WM, Harrison SC, Chou JJ. Mitochondrial Uncoupling Protein 2 Structure Determined by NMR Molecular Fragment Searching. Nature. 2011;476(7358):109–113. doi: 10.1038/nature10257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Berardi MJ, Chou JJ. Fatty Acid Flippase Activity of UCP2 Is Essential for Its Proton Transport in Mitochondria. Cell Metab. 2014;20(3):541–552. doi: 10.1016/j.cmet.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Sounier R, Bellot G, Chou JJ. Mapping Conformational Heterogeneity of Mitochondrial Nucleotide Transporter in Uninhibited States. Angew Chemie Int Ed. 2015;54(8):2436–2441. doi: 10.1002/anie.201408417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Zoonens M, Comer J, Masscheleyn S, Pebay-Peyroula E, Chipot C, Miroux B, Dehez F. Dangerous Liaisons between Detergents and Membrane Proteins. The Case of Mitochondrial Uncoupling Protein 2. J Am Chem Soc. 2013;135(40):15174–15182. doi: 10.1021/ja407424v. [DOI] [PubMed] [Google Scholar]
- (13).Dehez F, Schanda P, King MS, Kunji ERS, Chipot C. Mitochondrial ADP/ATP Carrier in Dodecylphosphocholine Binds Cardiolipins with Non-Native Affinity. Biophys J. 2017;113(11):2311–2315. doi: 10.1016/j.bpj.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Crichton PG, Lee Y, Ruprecht JJ, Cerson E, Thangaratnarajah C, King MS, Kunji ERS. Trends in Thermostability Provide Information on the Nature of Substrate, Inhibitor, and Lipid Interactions with Mitochondrial Carriers. J Biol Chem. 2015;290(13):8206–8217. doi: 10.1074/jbc.M114.616607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Alexandrov AI, Mileni M, Chien EYT, Hanson MA, Stevens RC. Microscale Fluorescent Thermal Stability Assay for Membrane Proteins. Structure. 2008;16(3):351–359. doi: 10.1016/j.str.2008.02.004. [DOI] [PubMed] [Google Scholar]
- (16).Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: A Hybrid Method for Predicting Protein Backbone Torsion Angles from NMR Chemical Shifts. J Biomol NMR. 2009;44(4):213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gropp T, Brustovetsky N, Klingenberg M, Muller V, Fendler K, Bamberg E. Kinetics of Electrogenic Transport by the ADP/ATP Carrier. Biophys J. 1999;77(2):714–726. doi: 10.1016/S0006-3495(99)76926-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Mittermaier AK, Kay LE. Observing Biological Dynamics at Atomic Resolution Using NMR. Trends Biochem Sci. 2009;34(12):601–611. doi: 10.1016/j.tibs.2009.07.004. [DOI] [PubMed] [Google Scholar]
- (19).Zhou H-X, Cross TA. Influences of Membrane Mimetic Environments on Membrane Protein Structures. Ann Rev Biophys. 2013;42(1):361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Vozza A, Blanco E, Palmieri L, Palmieri F. Identification of the Mitochondrial GTP/GDP Transporter in Saccharomyces Cerevisiae. J Biol Chem. 2004;279(20):20850–20857. doi: 10.1074/jbc.M313610200. [DOI] [PubMed] [Google Scholar]
- (21).Krämer R, Klingenberg M. Reconstitution of Inhibitor Binding Properties of the Isolated Adenosine 5’-diphosphate, Adenosine 5’-triphosphate Carrier-Linked Binding Protein. Biochemistry. 1977;16(23):4954–49561. doi: 10.1021/bi00642a002. [DOI] [PubMed] [Google Scholar]
- (22).Wang Y, Tajkhorshid E. Electrostatic Funneling of Substrate in Mitochondrial Inner Membrane Carriers. Proc Natl Acad Sci USA. 2008;105(28):9598–9603. doi: 10.1073/pnas.0801786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Dehez F, Pebay-Peyroula E, Chipot C. Binding of ADP in the Mitochondrial ADP/ATP Carrier Is Driven by an Electrostatic Funnel. J Am Chem Soc. 2008;130(38):12725–12733. doi: 10.1021/ja8033087. [DOI] [PubMed] [Google Scholar]
- (24).Hedger G, Rouse SL, Domanski J, Chavent M, Koldso H, Sansom MSP. Lipid-Loving ANTs: Molecular Simulations of Cardiolipin Interactions and the Organization of the Adenine Nucleotide Translocase in Model Mitochondrial Membranes. Biochemistry. 2016;55(45):6238–6249. doi: 10.1021/acs.biochem.6b00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Vallurupalli P, Hansen DF, Kay LE. Structures of Invisible, Excited Protein States by Relaxation Dispersion NMR Spectroscopy. Proc Natl Acad Sci USA. 2008;105(33):11766–11771. doi: 10.1073/pnas.0804221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Robustelli P, Kohlhoff K, Cavalli A, Vendruscolo M. Using NMR Chemical Shifts as Structural Restraints in Molecular Dynamics Simulations of Proteins. Structure. 2010;18(8):923–933. doi: 10.1016/j.str.2010.04.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.