Abstract
Motivation
Genome-wide association studies have identified thousands of loci associated with human disease, but identifying the causal genes at these loci is often difficult. Several methods prioritise genes most likely to be disease causing through the integration of biological data, including protein-protein interaction and phenotypic data. Data availability is not the same for all genes however, potentially influencing the performance of these methods.
Results
We demonstrate that whilst disease genes tend to be associated with greater numbers of data, this may be at least partially a result of them being better studied. With this observation we develop PhenoRank, which prioritises disease genes whilst avoiding being biased towards genes with more available data. Bias is avoided by comparing gene scores generated for the query disease against gene scores generated using simulated sets of phenotype terms, which ensures that differences in data availability do not affect the ranking of genes. We demonstrate that whilst existing prioritisation methods are biased by data availability, PhenoRank is not similarly biased. Avoiding this bias allows PhenoRank to effectively prioritise genes with fewer available data and improves its overall performance. PhenoRank outperforms three available prioritisation methods in cross-validation (PhenoRank area under receiver operating characteristic curve [AUC]=0.89, DADA AUC=0.87, EXOMISER AUC=0.71, PRINCE AUC=0.83, P < 2.2 × 10-16).
Availability
PhenoRank is freely available for download at https://github.com/alexjcornish/PhenoRank.
1. Introduction
Genome-wide association studies (GWAS) have identified thousands of genomic variants associated with a range of human traits, including susceptibilities to many diseases. Disease-associated variants are themselves rarely causal and instead ‘tag’ regions of the genome containing variants in linkage disequilibrium, any one of which may be the causal variant. These causal variants may be located in the coding region of a gene, or in a regulatory region and disrupt the expression of a gene through cis or trans-acting regulatory mechanisms (Jäger et al., 2015), making the identification of causal genes often difficult. This has led to the development of methods that integrate biological data to prioritise likely causal genes (Köhler et al., 2008; Vanunu et al., 2010; Erten et al., 2011; Yates et al., 2014; Smedley et al., 2015).
Network-based methods have been demonstrated to be effective at prioritising disease-causing genes (Köhler et al., 2008; Vanunu et al., 2010; Erten et al., 2011; Yates et al., 2014; Smedley et al., 2015; Cowen et al., 2017). These approaches often score genes more highly if they, or their protein products, interact with genes known to be associated with the query disease, or genes associated with diseases that are phenotypically similar to the query disease (Vanunu et al., 2010; Erten et al., 2011; Smedley et al., 2015). Yates et al. found that disease proteins tend to occupy more central positions in PPI networks than non-disease proteins (Yates and Sternberg, 2013), whilst Das et al. found this to be true, but only for PPI networks generated through literature-curation (Das and Yu, 2012). Some network-based methods therefore score genes and gene variants whose protein products are more central in PPI networks higher than those that are less central (Fu et al., 2014; Yates et al., 2014). The centrality measures used by these methods are correlated with the number of interactions a protein is involved in (Valente et al., 2008). It has been suggested however that some proteins may be involved in more interactions in literature-curated PPI networks as a result of them being better studied (Das and Yu, 2012; Gillis and Pavlidis, 2012). If this is true, then network-based methods that score genes central in a network more highly may be less effective at prioritising genes that are less well studied, as these genes may be more peripheral in a network, as a result of them having fewer available data.
Databases such as ClinVar (Landrum et al., 2016), OMIM (Amberger et al., 2015) and UniProtKB (The UniProt Consortium, 2014) collate data on the relationships between genetic variation and human disease. Databases that associate genetic variation with phenotypic abnormalities have also been established for model organisms (Bult et al., 2016) and used to study human disease (Chen et al., 2012; Smedley et al., 2015). It has been demonstrated that disease genes can be prioritised by identifying genes implicated in phenotypically similar diseases (Vanunu et al., 2010; Smedley et al., 2015). For example, novel causal genes for prostate cancer may be inferred by identifying genes implicated in other cancers (Vanunu et al., 2010). Similarly, candidate disease genes in humans can be prioritised by identifying orthologous mouse genes whose mutation causes similar phenotypes in mice (Chen et al., 2012; Smedley et al., 2015).
Multiple approaches have been proposed to quantify the phenotypic similarity of human diseases. Phenotype ontologies, such as the Human Phenotype Ontology (HPO) (Köhler et al., 2014) and the Mammalian Phenotype Ontology (MP) (Smith, Goldsmith and Eppig, 2005), provide standardized and structured vocabularies of observed phenotypic abnormalities. Multiple phenotype ontology terms can be mapped to a human disease to describe the phenotypic features of the disease. These features can include abnormalities associated with the disease (for example ‘Abnormality of the outer ear’), its mode of inheritance (for example ‘Autosomal dominant inheritance’) and clinical features (for example ‘Childhood onset’). The structured nature of ontologies allows the similarity of terms to be quantified. For example, the HPO terms ‘IgM deficiency’ and ‘IgE deficiency’ are both subclasses of ‘Decreased antibody level in blood’ and may therefore be considered similar. The HPO terms ‘IgM deficiency’ and ‘Dementia’ are less well connected in the ontology and may therefore be considered less similar. Semantic similarity methods such as simGIC measure the similarity of sets of terms in ontologies (Pesquita et al., 2008) and can therefore quantify the similarity of sets of phenotype terms annotating human diseases and mouse mutants, thereby providing a measure of their phenotypic similarity. It has been demonstrated however that semantic similarity methods can be biased by data availability. For example, the simGIC method tends to identify larger sets of terms, sets of terms that are more similar in size, and sets of terms from deeper ontology levels, as being more similar (Kulmanov and Hoehndorf, 2017). Gene prioritisation methods that use semantic similarity to quantify phenotypic similarity may therefore be biased by the numbers of phenotype terms annotating human diseases and model organism mutants, which may reflect how well studied these entities are.
In this study, we demonstrate that whilst disease genes are involved in greater numbers of PPIs than non-disease genes in some PPI databases, this may be at least partly a result of them being better studied. Scoring genes with more available data more highly may reduce the ability of a method to prioritise less-well-studied genes, for which fewer data are likely to be available. We therefore develop PhenoRank, which uses PPI and phenotype data from multiple species to prioritise disease genes, whilst avoiding being biased by the number of data associated with each gene. Bias is avoided by comparing gene scores generated for the query disease against gene scores generated using simulated sets of phenotype terms. Using this simulation-based approach ensures PhenoRank is not biased towards genes with more available data and improves its performance.
2. Methods
2.1. PPI data
PPI data were downloaded from four databases: BioGRID (version 3.4.131) (Chatr-Aryamontri et al., 2015), HI-II-14 (on 27 November 2015) (Rolland et al., 2014), HPRD (on 30 March 2015) (Keshava Prasad et al., 2009) and IntAct (on 4 January 2016) (Orchard et al., 2014). Only direct interactions, associations and physical associations were obtained from BioGRID and IntAct. Duplicate interactions, looping interactions and interactions that did not occur between two H. sapiens proteins were excluded. Some PPI resources do not record interactions between different protein isoforms and we therefore considered all interactions at the gene level. Combined data from the four resources, containing 210,914 unique interactions spanning 16,184 genes, were used in PhenoRank (Supplementary Table 1).
2.2. Human disease variant data
Data downloaded from ClinVar (on 22 October 2016), OMIM (on 1 November 2016) and UniProtKB (on 22 October 2016) were used to define the disease-gene associations used in PhenoRank. ClinVar variants not marked as pathogenic or likely pathogenic, or whose review status was less than two stars were excluded. Non-disease variants from UniProtKB were excluded. Disease-gene associations from OMIM were not considered if the molecular basis of the disease is unknown. Diseases reported using vocabularies other than OMIM were mapped to OMIM terms using the cross-referencing provided by the Disease Ontology (DO) (Kibbe et al., 2015). Using these data, we define a disease as being associated with a gene if a gene variant is reported as being disease causing. The combined data set contains 5,685 unique associations between 4,729 diseases and 3,713 genes (Supplementary Table 2).
2.3. Mouse phenotype data
Genotypes and phenotype term annotations for 24,834 mouse mutants and human-mouse gene orthology data were downloaded from the Mouse Genomics Database (MGD, on 13 October 2016). Human orthologs of the mutated gene in 21,143 mouse mutants were identified using the orthology data.
2.4. Annotating diseases with phenotype terms
Mappings between disease terms (from OMIM) and phenotype terms (from HPO and MP) from HPO (Köhler et al., 2014) and Hoehndorf et al. (Hoehndorf et al., 2015) are used in PhenoRank to measure the phenotypic similarity of the query disease and diseases in OMIM. Hoehndorf et al. mapped phenotype terms (from HPO and MP) to disease terms (from DO) through automated text mining. Hoehndorf et al. determined that the 21 phenotype terms most strongly associated with each disease were most informative when quantifying phenotypic similarity and we therefore include these phenotype term mappings in PhenoRank. The cross-referencing provided by the DO was used to transfer the mapped phenotype terms to the corresponding OMIM diseases. The combined data set contains 128,695 unique mappings between 7,042 OMIM diseases and 8,313 unique HPO and MP phenotype terms (Supplementary Table 3).
2.5. Measuring phenotypic similarity
PhenoRank uses the simGIC similarity measure to compute the phenotypic similarity of human diseases and mouse mutants (Supplementary Figure 1). Let Wi and Wj be two sets of phenotype ontology terms. In our case, these phenotype terms are terms from the HPO or MP that either annotate a disease or describe abnormalities observed in a mouse mutant (Supplementary Figure 2). Uberpheno, a cross-species ontology generated by integrating multiple phenotype ontologies, including the HPO and MP, is used to compare sets of terms (Köhler et al., 2013). The true path rule states that association with an ontology term implies association with all ancestors of the term (Pesquita et al., 2008), and we therefore add the ancestors of each term in Wi and Wj to the respective set using Uberpheno. Each term in Uberpheno is weighted by its information content (IC), defined as the negative logarithm of the probability that a given disease or mouse mutant is annotated with the term. Using simGIC, the similarity S of Wi and Wj is the Jaccard similarity coefficient weighted by the IC of each term:
2.6. Disease gene prioritisation using PhenoRank
Let D be the diseases represented in ClinVar, OMIM and UniProtKB, M be the mouse mutants reported in the MGD, q be the query disease so that q∈D, and Wi be the set of phenotype terms mapped to phenotype data source i, which can be either a human disease or mouse mutant. In PhenoRank, all diseases in D and mouse mutants in M are first scored by their phenotypic similarity to query disease q (Figure 1A). Phenotypic similarity is measured by comparing the ontological similarity of the set of phenotype terms mapped to q, to the sets of phenotype terms mapped to each disease in D and mouse mutant in M, using the simGIC method. Diseases and mouse mutants are therefore scored as being phenotypically similar to q if they are mapped to phenotype terms that are closely related in Uberpheno.
Figure 1.
PhenoRank overview. (A) Phenotypic similarity of the query disease (q) to each disease in OMIM (D) and each mouse mutant in MGD (M) is quantified. Thicker lines represent stronger phenotypic similarities. (B) Phenotypic similarity scores are applied to genes in a PPI network, using known disease-gene associations and mouse-human gene orthology data. Darker red nodes represent genes with greater relevance to the disease of interest. (C) Phenotypic relevance scores are propagated across a PPI network, so that genes that interact with many high scoring genes are also scored highly. (D) Gene scores generated for the query disease are compared against gene scores generated using simulated sets of phenotype terms (S). By comparing the score the gene receives for the query disease against the distribution of scores the gene receives for the simulated sets of phenotype terms, a P value for each gene is generated. In this illustrative example, the computation of a P value for the gene marked X is shown. While only two simulated sets of disease phenotypes are shown, PhenoRank is run with 1,000 simulated sets by default.
These phenotypic similarity scores are used to score each gene in a PPI network. Let G=(V,E) be this network, with V being nodes representing genes and E being edges representing physical interactions between the protein products of the genes. Gene scores are computed using disease-gene associations from ClinVar, OMIM and UniProtKB, and human-mouse orthology data from the MGD (Figure 1B). The score of gene i is defined as the sum of the phenotypic similarity of each associated disease and each mutant of an orthologous mouse gene, to q, divided by the numbers of associated diseases and mouse mutants:
where Yi is the set of diseases associated with gene i, Zi is the set of mutants of mouse genes orthologous to gene i and Wq is the set of phenotype terms annotating q. Genes are therefore scored highly if they are associated with a disease that is phenotypically similar to q, or if they are orthologous to a mouse gene whose mutation produces phenotypic abnormalities similar to q.
These gene scores are next propagated across G using the random walk with restart (RWR) method (Figure 1C), as this approach has been shown to be effective when prioritising disease genes and variants using network data (Köhler et al., 2008; Vanunu et al., 2010). Propagation of gene scores ensures that genes that interact with many genes that are phenotypically relevant to the query disease are also scored highly.
To account for the differing availability of data between genes, gene scores generated for the query disease are compared against gene scores generated using simulated sets of phenotype terms (Figure 1D). Simulated sets of phenotype terms are generated by sampling from phenotype terms mapped to the same diseases by HPO and Hoehndorf et al. (Supplementary Figure 3B), to ensure that the simulated sets of terms closely resemble the sets of phenotype terms mapped to real diseases. Sets of phenotype terms equal in size to the set of terms mapped to q are simulated. To simulate a set of terms of size |Wq|, a single seed term is first sampled from HPO or MP. All phenotype terms in HPO and MP are then ranked by the number of times they are mapped to the same disease as the seed term, with ties ordered randomly. The seed term is itself included in this ranking. If fewer than |Wq| terms are mapped to the same disease as the seed term, then a new seed term is sampled. The top |Wq| ranked terms are used as the simulated set of phenotype terms, ensuring that the simulated sets contain terms that are frequently mapped to the same disease.
For each simulated set of phenotype terms, all genes are rescored using the simulated set of terms in place of the query disease, and these scores again propagated across the PPI network. When rescoring genes all data are unchanged, ensuring that the effect of data availability on each gene score is the same for the query disease and each simulated set of phenotype terms. Comparing the gene scores generated using the query disease against the gene scores generated using each simulated set of phenotype terms therefore allows differences in data availability to be negated. An empirical P value is computed for each gene by taking the proportion of simulated sets of phenotype terms in which the gene is scored higher than when q is considered. These P values represent the probability of observing a gene score at least as great as that observed, given that the gene is not associated with the query disease. A minimum P value of 1/u, where u is the number of simulated sets of phenotype terms used, is applied to ensure that no P values equal zero. We run PhenoRank using 1,000 simulated sets of phenotype terms and use these P values to prioritise candidate genes. Through the application of PhenoRank to 100 randomly selected diseases, we demonstrate that PhenoRank correctly controls the type-1 error rate (Supplementary Figure 4). PhenoRank data is available to download (https://github.com/alexjcornish/PhenoRank_Data).
2.7. Propagating scores across PPI networks
PhenoRank propagates gene scores across network G using the RWR method (Supplementary Figure 5) (Köhler et al., 2008). Let n be the number of vertices in G, A be the column-normalised adjacency matrix of G and Qt be a vector of length n and the distribution of scores across vertices at time t. Q0 is the initial distribution of gene scores. The distribution of scores across time points is computed iteratively:
where r is the restart probability. To make these scores comparable across the query disease and the simulated sets of phenotype terms, a fixed number of iterations are completed and scores ranked before PhenoRank computes P values.
2.8. Evaluating method bias
We measured the correlation between the gene scores computed by PhenoRank and three published gene prioritisation methods (DADA (Erten et al., 2011), EXOMISER (Smedley et al., 2015) and PRINCE (Vanunu et al., 2010)) and the numbers of data associated with each gene, to determine whether the methods are biased towards genes with more available data. PhenoRank was run with and without simulated sets of phenotype terms (we refer to these method versions as PhenoRank-Simulation and PhenoRank-NoSimulation), to establish whether the use of these simulated sets of terms affects how biased PhenoRank is. EXOMISER prioritises disease genes and variants by combining a gene-based scoring method, which uses PPI data and phenotype data from multiple species, with variant-based pathogenicity prediction. We use the gene-level scores produced by EXOMISER when evaluating performance and therefore refer to the method as EXOMISER-Walker for clarity. Gene scores were generated by applying each method to 200 diseases, randomly selected from those diseases than can be considered by all methods. We considered how five features of the data used by each method correlate with the computed gene scores:
Network degree of each gene.
Number of sources of phenotype data. This is defined as the number of human diseases and (if used by the method) model organism mutants associated with each gene.
Number of annotating phenotype terms. This is defined as the median number of phenotype terms annotating the sources of phenotype data associated with each gene.
Difference in the number of annotating phenotype terms. This is defined as the median absolute difference between the number of phenotype terms annotating the query disease, and the numbers of phenotype terms annotating each source of phenotype data associated with each gene.
Ontology depth of annotating phenotype terms. This is defined as the median of the maximum ontology depth of the phenotype terms annotating the sources of phenotype data associated with each gene.
PhenoRank and EXOMISER-Walker both use phenotype terms annotating each source of phenotype data to measure phenotypic similarity, whilst DADA and PRINCE measure phenotypic similarity using text mining. We therefore test all five data features when considering PhenoRank and EXOMISER-Walker, but restrict our analysis to data features (i) and (ii) when considering DADA and PRINCE.
2.9. Evaluating method performance
The performance of each gene prioritisation method was evaluated using leave-one-out cross-validation. To ensure that any observed performance differences were a result of methodology, rather than the data releases used, we ran DADA, EXOMISER-Walker and PRINCE using the same disease-gene association data used by PhenoRank. Gene-phenotype associations used by EXOMISER-Walker were also updated and the IC of each phenotype term recalculated. HPO terms mapped to OMIM diseases by the HPO were used as input when running EXOMISER-Walker. We used our own implementation of the PRINCE algorithm, which is available in the PhenoRank package.
Leave-one-out cross-validation was completed using a set of 2,708 associations between diseases that can be input into all four methods, and genes that can be scored by all four methods. In each cross-validation trial, an association from this set (between disease Di and gene gj ∈ Si) was masked (i.e. removed from the data used by each method). Each method was then run using disease Di as input. All other genes associated with disease Di (gk ∈ Si, k ≠ j) and genes not scored by all four methods were excluded from the results. The score of gene gj relative to the scores of the other genes in the results was used to evaluate the performance of each method. This process was repeated for each of the 2,708 disease-gene associations. Receiver operating characteristic (ROC) curves and the areas under these curves (AUCs) were computed using the pROC R package (Robin et al., 2011).
2.10. Selecting method parameters
Scores propagated across a network using the RWR algorithm converge on a steady-state distribution (Cowen et al., 2017). To determine the number of RWR algorithm iterations required by PhenoRank for convergence, we ran PhenoRank using 200 randomly selected OMIM terms and calculated the mean absolute difference between the gene scores computed using between 1 and 29 iterations, and the gene scores computed using 30 iterations, demonstrating that scores converge and that the mean absolute change in gene score after 20 iterations is <10-5 for all tested parameters (Supplementary Figure 6A). We next conducted leave-one-out cross-validation to select an optimal value for restart probability r, using only the 2,977 disease-gene associations reported by ClinVar, OMIM or UniProtKB that were not in the set of 2,708 associations used in performance evaluation. This ensured that parameter selection and performance evaluation were independent, therefore avoiding circularity. PhenoRank performs optimally when r=0.1 (Supplementary Table 4) and we therefore ran PhenoRank using 20 iterations and r=0.1. We used the same approach to determine the number of RWR algorithm iterations required by PRINCE and select optimal values for the two PRINCE parameters (α and c). Convergence is achieved by 20 iterations (Supplementary Figure 6B) and performance is optimal when α=0.5 and c=-15 (Supplementary Table 4), and we therefore ran PRINCE using these values.
2.11. Disease classes
A disease class was identified for each OMIM disease using the ontological structure of the DO (Supplementary Table 5). Each OMIM disease was first mapped to a DO term using the cross references provided by the DO. Ancestors of these DO terms at the third level of the DO were then identified and these broader disease definitions used as disease classes. If an OMIM disease mapped to multiple third-level DO terms, then the third-level DO term mapped to the greatest number of OMIM diseases was used to classify the disease, to reduce the number of classes considered.
3. Results
3.1. Study bias in PPI databases
We analysed the numbers of PPIs involving disease and non-disease genes to determine whether disease genes are involved in greater numbers of PPIs than non-disease genes, and whether study bias is likely to contribute to any differences. PPI data were downloaded from BioGRID, HI-II-14, HPRD and IntAct and disease-gene association data were obtained from ClinVar, OMIM and UniProtKB. If a gene is reported as being disease associated by at least one resource, then it is defined as a disease gene. Otherwise it is defined as a non-disease gene.
BioGRID, HPRD and IntAct contain PPIs curated from the literature. The proteins screened in the studies contributing to these resources depend on the aims of the studies and the generation of these data was therefore hypothesis-driven (HD). Conversely, HI-II-14 contains interactions identified in a single unbiased screen of 14,000 proteins (Rolland et al., 2014) and the generation of these data was therefore hypothesis-free (HF). If disease genes are truly involved in greater numbers of PPIs than non-disease genes, we would expect disease genes to be involved in greater numbers of PPIs than non-disease genes in data sets generated using both HD and HF approaches. However, whilst disease genes are involved in greater numbers of PPIs than non-disease genes in each of the HD data sets, disease genes are involved in similar numbers of PPIs as non-disease genes in the HF data set (Table 1).
Table 1.
Numbers of PPIs disease and non-disease genes are involved in. Differences tested using a two-sided Mann-Whitney U test.
| Database | Disease genes | Non-disease genes | Difference | ||
|---|---|---|---|---|---|
| Mean n. PPIs |
Median n. PPIs |
Mean n. PPIs |
Median n. PPIs |
||
| BioGRID | 26.3 | 10.0 | 18.5 | 8.0 | P<2.2×10-16 |
| HI-II-14 | 5.7 | 2.0 | 6.7 | 2.0 | P=0.685 |
| HPRD | 10.7 | 5.0 | 7.2 | 3.0 | P<2.2×10-16 |
| IntAct | 18.8 | 7.0 | 14.3 | 6.0 | P<2.2×10-16 |
If disease genes are better studied, and better studied genes are involved in greater numbers of PPIs in each of the HD data sets, then study bias may at least partially explain why disease genes are involved in more interactions than non-disease genes in the HD data sets, but not the HF data set. To determine whether disease genes are better studied, we used the number of PubMed-indexed publications related to each gene in gene2pubmed (NCBI Resource Coordinators, 2016) (downloaded 11 August 2017) as a measure of how well studied each gene is. Disease genes tend to be related to greater numbers of publications (median 59 publications) than non-disease genes (median 13 publications, P < 2.2 × 10-16, Wilcoxon rank sum test) indicating that they are better studied. Better-studied genes are involved in more PPIs than less-well-studied genes in each of the PPI data sets (Table 2), although this difference is much greater in the HD data sets than in the HF data set. The fact that disease genes tend to be better studied than non-disease genes, and that better-studied genes are involved in more PPIs than less-well-studied genes in the HD data sets, may partly explain why disease genes are observed as being involved in more PPIs than non-disease genes in the HD data sets. Study bias may therefore at least partially account for the differences in the numbers of PPIs that disease and non-disease genes are involved in in the HD data sets.
Table 2.
Numbers of PPIs better and less-well-studied genes are involved in. Better and less-well-studied genes are defined as those in the top and bottom thirds of genes ranked by the number of related publications in gene2pubmed. Differences tested using a two-sided Mann-Whitney U test.
| Better-studied genes | Less-well-studied genes | ||||
|---|---|---|---|---|---|
| Database | Mean n. PPIs |
Median n. PPIs |
Mean n. PPIs |
Median n. PPIs |
Difference |
| BioGRID | 37.8 | 18.0 | 7.2 | 3.0 | P<2.2×10-16 |
| HI-II-14 | 6.8 | 2.0 | 6.2 | 2.0 | P=0.013 |
| HPRD | 15.2 | 8.0 | 3.4 | 2.0 | P<2.2×10-16 |
| IntAct | 26.4 | 12.0 | 6.5 | 3.0 | P<2.2×10-16 |
3.2. Bias in network-based gene prioritisation methods
We measured the correlations between gene scores computed by PhenoRank and three other prioritisation methods, and features of the data used by each method, to determine whether the methods are biased by the numbers of data associated with each gene. Gene scores computed by DADA, EXOMISER and PRINCE are positively correlated with the network degree of each gene and the numbers of associated sources of phenotype data (i.e. the number of human diseases and model organism mutants associated with each gene), suggesting that these methods score genes more highly if they are associated with more data (Table 3). The scores computed by PhenoRank-NoSimulation are similarly correlated with network degree and the number of associated sources of phenotype data, whilst the gene scores computed by PhenoRank-Simulation are less strongly correlated with these data features, indicating that the use of simulated sets of phenotype terms ensures that PhenoRank is less biased by data availability.
Table 3.
Correlations between the gene scores computed by each method and features of the data used by each method. DADA and PRINCE do not quantify phenotypic similarity using terms from phenotype ontologies, and therefore correlations involving phenotype terms were not measured for these methods. Correlations measured using Spearman’s rank correlation coefficient.
| Data feature | PhenoRank-Simulation | PhenoRank-NoSimulation | DADA | EXOMISER | PRINCE |
|---|---|---|---|---|---|
| Network degree of each gene | -0.04 | 0.91 | 0.64 | 0.46 | 0.47 |
| Number of sources of phenotype data | -0.03 | 0.21 | 0.32 | 0.18 | 0.26 |
| Number of annotating phenotype terms | -0.04 | 0.13 | NA | 0.01 | NA |
| Difference in the number of annotating phenotype terms | -0.01 | -0.03 | NA | -0.05 | NA |
| Ontology depth of annotating phenotype terms | -0.03 | 0.18 | NA | 0.02 | NA |
Gene scores computed by PhenoRank-NoSimulation also correlate with the number of phenotype terms annotating the sources of phenotype data associated with each gene, and the ontology depth of these phenotype terms (Table 3). This suggests that PhenoRank-NoSimulation scores genes more highly if the human diseases and mouse mutants associated with the gene are annotated with greater numbers of terms from phenotype ontologies. Differences between the number of phenotype terms annotating the query disease, and the numbers of phenotype terms annotating gene-associated human diseases and mouse mutants, do not correlate with computed gene scores, suggesting that this data feature is not a major source of bias. Gene scores computed by PhenoRank-Simulation are less strongly correlated with the number of annotating phenotype terms and the ontology depth of these terms, indicating that the use of simulated sets of phenotype terms reduces bias introduced by how well human diseases and mouse mutants are annotated with phenotype terms. Despite EXOMISER-Walker also using phenotype terms to score genes, the gene scores computed by EXOMISER-Walker are not strongly correlated with the numbers of annotating phenotype terms, or the ontology depths of these terms, possibly reflecting differences in the EXOMISER-Walker and PhenoRank-NoSimulation methodologies.
3.3. Evaluation of method performance
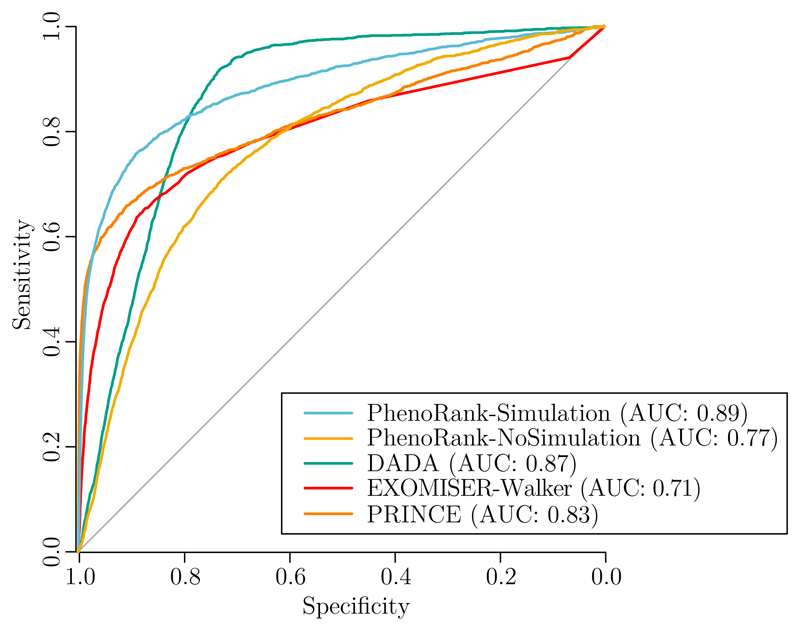
We used leave-one-out cross-validation to evaluate the performances of PhenoRank-Simulation, PhenoRank-NoSimulation, DADA, EXOMISER-Walker and PRINCE (Figure 2). Using simulated sets of phenotype terms improves the performance of PhenoRank (PhenoRank-Simulation AUC=0.89, PhenoRank No-Simulation AUC=0.77, P < 2.2 × 10-16, two-sided DeLong’s method). Reducing the bias of PhenoRank towards genes with more available data therefore also improve its performance. PhenoRank-Simulation outperforms DADA (AUC=0.87, P < 2.2 × 10-16), EXOMISER-Walker (AUC=0.71, P < 2.2 × 10-16) and PRINCE (AUC=0.83, P < 2.2 × 10-16). Whilst DADA is the method with overall performance most similar to PhenoRank-Simulation, it performs much worse than PhenoRank at higher specificities, with PhenoRank-Simulation and DADA achieving sensitivities of 87% and 47% at 90% specificity respectively. DADA however outperforms PhenoRank-Simulation at lower specificities, with PhenoRank-Simulation and DADA achieving sensitivities of 92% and 98% at 50% specificity respectively.
Figure 2.
Performances of PhenoRank-Simulation, PhenoRank-NoSimulation, DADA, EXOMISER-Walker and PRINCE in leave-one-out cross-validation.
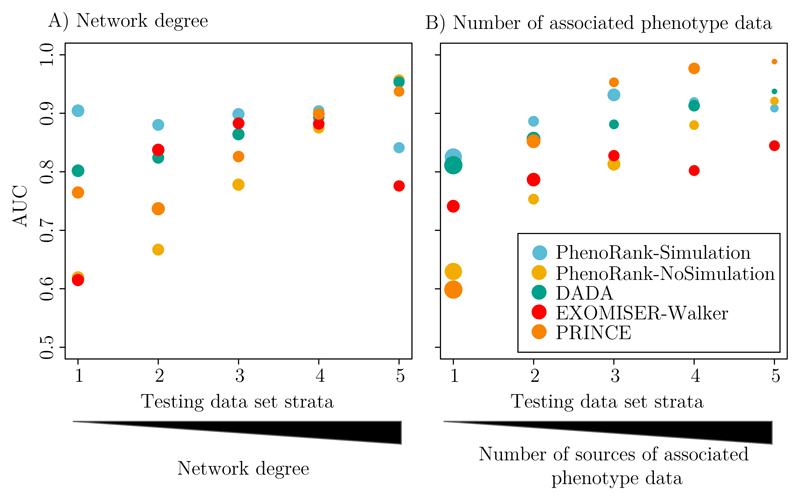
The bias of DADA, EXOMISER-Walker and PRINCE towards genes with more available data may affect their ability to effectively prioritise genes associated with fewer available data. To test this, we stratified the cross-validation procedure by the PPI network degree of each gene and the number of phenotype data associated with each gene. The set of 2,708 disease-gene associations used in method performance evaluation was split into five strata based on the degree of the gene in each disease-gene association, and five strata based on the number of phenotype data associated with the gene in each disease-gene association. Cross-validation was then run using each of the strata (Figure 3, Supplementary Table 6). Each method uses different data and how the disease-gene associations were stratified was therefore not the same for each method. DADA, EXOMISER-Walker and PRINCE perform better when applied to genes with greater degrees and associated with more phenotype data, reflecting their biases towards genes associated with greater numbers of data. Whilst the performance of PhenoRank-NoSimulation is similarly affected by the numbers of associated data, PhenoRank-Simulation performs more consistently across the strata, demonstrating that the use of simulated sets of phenotype terms reduces the effect of data availability on method performance. PhenoRank may therefore be especially useful when prioritising genes for which fewer data are available.
Figure 3.
Method performance when applied to genes with different numbers of associated data. For each method, the testing data set of 2,708 disease-gene associations was stratified based on (A) the network degree of each gene and (B) the number of sources of phenotype data associated with each gene, and leave-one-out cross-validation completed. The size of each circle represents the numbers of disease-gene associations in the testing data set strata.
Genes associated with diseases of different classes are associated with different numbers of data (Supplementary Table 7) possibly reflecting how well studied different disease classes are. To determine whether the performances of PhenoRank-Simulation, DADA, EXOMISER-Walker and PRINCE vary between disease classes, we ran the cross-validation procedure using disease-gene associations stratified by disease class (Supplementary Figure 7, Supplementary Table 8). PhenoRank-Simulation was the best performing method in 20 of the 30 disease classes, DADA in 3 disease classes, EXOMISER-Walker in 2 disease classes and PRINCE in 6 disease classes. These performance differences may be influenced by the biases exhibited by the methods. PhenoRank-Simulation outperforms DADA, EXOMISER-Walker and PRINCE in the Monogenic Disease and Integumentary System Disease classes (P<0.05), but PRINCE outperforms PhenoRank in the Cancer class (P<0.05). This may reflect the fact that the mean degree of genes in the PPI network used by PRINCE is higher for genes associated with diseases in the Cancer class (69.8), than genes associated with diseases in the Monogenic Disease (42.8) and Integumentary System Disease (29.6) classes.
In the data sets used by PhenoRank-Simulation, 3,713 human protein-coding genes are associated with at least one human disease, 8,607 with mouse phenotype data and 9,618 with either. To determine whether the performance of PhenoRank-Simulation is improved by using data from both species, we ran the cross-validation procedure using only human disease data and only mouse phenotype data. PhenoRank-Simulation performs better when using both human and mouse data (AUC=0.89), than when using only human data (AUC=0.85, P < 2.2 × 10-16) and only mouse data (AUC=0.80, P < 2.2 × 10-16) demonstrating that PhenoRank successfully integrates data from the two species.
3.4. Application of PhenoRank to genes in loci associated with rheumatoid arthritis
We prioritised likely causal genes in loci identified in a GWAS of rheumatoid arthritis (Okada et al., 2014) using PhenoRank. Candidate genes in the loci were identified by first selecting single nucleotide polymorphisms (SNPs) in linkage disequilibrium with the lead SNP (r2>0.05) in European populations (Machiela and Chanock, 2014). Regions spanning these SNPs were then defined and all genes whose protein-coding regions at least partially overlap these regions were considered candidates. Loci containing a gene already known to be associated with rheumatoid arthritis were not considered. PhenoRank was then run using rheumatoid arthritis as the input disease and the scores of the genes in these loci extracted from the generated results file. Four genes (PADI2, SYT7, LGALS1 and PLCL2) in the identified loci were implicated by PhenoRank as being involved in rheumatoid arthritis development (P<0.05, after within-locus correction for multiple testing, Supplementary Table 9). None of these four genes are associated with any human disease in the data used by PhenoRank, although inflammatory and autoimmune phenotypes have been observed in mutants of their mouse orthologs, including “increased susceptibility to experimental autoimmune encephalomyelitis” and “abnormal adaptive immunity” (Supplementary Table 9). These genes also interact with genes with immune system functions, including CD4 and CD8A (Zhu, Yamane and Paul, 2010). It is for these reasons that PhenoRank identifies them as being potential candidates. Some of these genes have been previously implicated in autoimmune disease: PADI2 expression has been demonstrated to correlate with arthritis severity in mice (Johnsen et al., 2011), LGALS1 damages cartilage via inflammation in osteoarthritis (Toegel et al., 2016) and PLCL2 has been associated with systemic sclerosis (Arismendi et al., 2015).
4. Conclusions
It has been suggested that the proteins involved in greater numbers of interactions may be less able to tolerate mutations, as a greater proportion of their sequence may be required to facilitate the interactions, thereby increasing the likelihood that they are disease-associated (Yates and Sternberg, 2013). In this study, we show that whilst the protein products of disease genes tend to be involved in greater numbers of PPIs in HD data sets, this may be at least partly a result of them being better studied. PPI networks generated through high-throughput approaches, which are less susceptible to study bias, currently cover only a small proportion of the PPIs thought to occur in cells (Rolland et al., 2014). It may not be possible to determine whether the number of interactions a protein is involved in affects the likelihood of it being disease-associated until we have a more comprehensive, accurate and unbiased map of the interactome.
Genes associated with diseases of difference classes tend to be involved in different numbers of PPIs (Supplementary Table 7). In the PPI network used by PhenoRank, genes involved in cancer have a mean degree of 99.5, whilst genes involved in inherited metabolic disorders have a mean degree of only 18.2. The better characterisation of pathways involved in cancer may at least partly explain this difference. The differing performances of PhenoRank, DADA, EXOMISER-Walker and PRINCE across disease classes suggests that a user should consider how well studied a disease is when selecting a prioritisation method. A method that performs well when more data are available, such as DADA, EXOMISER-Walker or PRINCE, may be more suitable for studying diseases with more available data, whilst methods that are less biased by data availability, such as PhenoRank, may be more suitable for studying diseases with few available data.
Gillis and Pavlidis (2012) describe study bias in PPI networks in relation to predicting gene function. They suggest that genes involved in more interactions may represent highly studied genes, and that these genes may be more open to the accumulation of false positive interactions as a result of this. If highly studied genes are involved in more false positive interactions, then this may partially explain why the use of simulated sets of disease phenotype terms improves PhenoRank performance, as using this simulation-based approach reduces the score of high-degree genes, thereby reducing the influence of interactions that are more likely to be false positives.
The gene scores computed by PhenoRank-NoSimulation, DADA, EXOMISER-Walker and PRINCE most strongly correlate with the degree of the genes in the networks used by the methods. This suggests that differences in PPI network degree is a major source of bias affecting these methods. Whilst DADA adjusts for the degree of candidate genes in PPI networks in order to better prioritise genes of low degree, it is still biased towards genes of high degree. This is likely because DADA employs a “uniform scoring strategy”, in which raw and degree-adjusted gene scores are combined to produce the final gene ranking.
While PhenoRank was developed to prioritise genes in disease-associated loci, the method could be extended to prioritise disease variants. Methods such as EXOMISER and eXtasy (Sifrim et al., 2013) have demonstrated that the integration of a variant effect predictor with gene-level prioritisation can aid in pathogenic variant identification. The use of a method that is less biased towards genes for which more data are available, such as PhenoRank, alongside a variant effect predictor may allow for the more effective prioritisation of variants in genes that are less well studied.
Whilst existing network-based gene prioritisation methods are biased toward genes for which more data are available, the use of simulated sets of phenotype terms ensures that PhenoRank is not similarly biased. Although high-throughput phenotypic screens are being completed (Brown and Moore, 2012), many data sources are still likely to be influenced by study bias. Approaches similar to the simulated sets of phenotype terms used by PhenoRank could be incorporated into existing prioritisation methods, such as DADA, PRINCE and EXOMISER, to reduce the influence of study bias.
Supplementary Material
Supplementary information: Supplementary data are available at Bioinformatics online.
Funding
This work is supported by a British Heart Foundation PhD studentship (AJC) and The Wellcome Trust ref WT/104955/Z/14/Z (AD).
Footnotes
Conflict of Interest: MJES is a Director and shareholder in Equinox Pharma Ltd., which uses bioinformatics and chemoinformatics in drug discovery research and services.
References
- Amberger JS, et al. OMIM.org: Online Mendelian Inheritance in Man (OMIM), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:789–798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arismendi M, et al. Identification of NF-κB and PLCL2 as new susceptibility genes and highlights on a potential role of IRF8 through interferon signature modulation in systemic sclerosis. Arthritis Res Ther. 2015;17(1):71. doi: 10.1186/s13075-015-0572-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SDM, Moore MW. The International Mouse Phenotyping Consortium: Past and future perspectives on mouse phenotyping. Mamm Genome. 2012;23(9):632–640. doi: 10.1007/s00335-012-9427-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, et al. Mouse genome database 2016. Nucleic Acids Res. 2016;44:D840–D847. doi: 10.1093/nar/gkv1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, et al. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470–D478. doi: 10.1093/nar/gku1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, et al. Mousefinder: Candidate disease genes from mouse phenotype data. Hum Mutat. 2012;33(5):858–866. doi: 10.1002/humu.22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowen L, et al. Network propagation: a universal amplifier of genetic associations. Nat Rev Genet. 2017;18(9):551–562. doi: 10.1038/nrg.2017.38. [DOI] [PubMed] [Google Scholar]
- Das J, Yu H. HINT: High-quality protein interactomes and their applications in understanding human disease. BMC Syst Biol. 2012;6(1):92. doi: 10.1186/1752-0509-6-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erten S, et al. DADA: Degree-Aware Algorithms for Network-Based Disease Gene Prioritization. BioData Min. 2011;4(1):19. doi: 10.1186/1756-0381-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, et al. FunSeq2: A framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15(10):480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillis J, Pavlidis P. ‘Guilt by association’ is the exception rather than the rule in gene networks. PLOS Comput Biol. 2012;8(3):e1002444. doi: 10.1371/journal.pcbi.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoehndorf R, et al. Analysis of the human diseasome reveals phenotype modules across common, genetic, and infectious diseases. Sci Rep. 2015;5:10888. doi: 10.1038/srep10888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäger R, et al. Capture Hi-C identifies the chromatin interactome of colorectal cancer risk loci. Nat Commun. 2015;6:6178. doi: 10.1038/ncomms7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen AK, et al. Genome-wide and species-wide dissection of the genetics of arthritis severity in heterogeneous stock mice. Arthritis Rheum. 2011;63(9):2630–2640. doi: 10.1002/art.30425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshava Prasad TS, et al. Human Protein Reference Database-2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbe WA, et al. Disease Ontology 2015 update: an expanded and updated database of human diseases for linking biomedical knowledge through disease data. Nucleic Acids Res. 2015;43:D1071–D1078. doi: 10.1093/nar/gku1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, et al. Walking the interactome for prioritization of candidate disease genes. Am J Hum Genet. 2008;82:949–958. doi: 10.1016/j.ajhg.2008.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, et al. Construction and accessibility of a cross-species phenotype ontology along with gene annotations for biomedical research. F1000Research. 2013;2:30. doi: 10.12688/f1000research.2-30.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler S, et al. The Human Phenotype Ontology project: Linking molecular biology and disease through phenotype data. Nucleic Acids Res. 2014;42:D966–D974. doi: 10.1093/nar/gkt1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulmanov M, Hoehndorf R. Evaluating the effect of annotation size on measures of semantic similarity. J Biomed Semantics. 2017;8(1):7. doi: 10.1186/s13326-017-0119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landrum MJ, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela MJ, Chanock SJ. LDlink: A web-based application for exploring population-specific haplotype structure and linking correlated alleles of possible functional variants. Bioinformatics. 2014;31(21):3555–3557. doi: 10.1093/bioinformatics/btv402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016;44:D7–D19. doi: 10.1093/nar/gkv1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, et al. Genetics of rheumatoid arthritis contributes to biology and drug discovery. Nature. 2014;506(7488):376–381. doi: 10.1038/nature12873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard S, et al. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesquita C, et al. Metrics for GO based protein semantic similarity: a systematic evaluation. BMC Bioinformatics. 2008;9:S4. doi: 10.1186/1471-2105-9-S5-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland T, et al. A Proteome-Scale Map of the Human Interactome Network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sifrim A, et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10(11):1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- Smedley D, et al. Next-generation diagnostics and disease-gene discovery with the Exomiser. Nat Protoc. 2015;10(12):2004–2015. doi: 10.1038/nprot.2015.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CL, Goldsmith C-AW, Eppig JT. The Mammalian Phenotype Ontology as a tool for annotating, analyzing and comparing phenotypic information. Genome Biol. 2005;6:R7. doi: 10.1186/gb-2004-6-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium. Activities at the Universal Protein Resource (UniProt) Nucleic Acids Res. 2014;42:D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toegel S, et al. Galectin-1 Couples Glycobiology to Inflammation in Osteoarthritis through the Activation of an NF-κB-Regulated Gene Network. J Immunol. 2016;196(4):1910–1921. doi: 10.4049/jimmunol.1501165. [DOI] [PubMed] [Google Scholar]
- Valente TW, et al. How Correlated Are Network Centrality Measures? Connections. 2008;28(1):16–26. [PMC free article] [PubMed] [Google Scholar]
- Vanunu O, et al. Associating genes and protein complexes with disease via network propagation. PLOS Comput Biol. 2010;6(1):e1000641. doi: 10.1371/journal.pcbi.1000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates CM, Sternberg MJE. Proteins and domains vary in their tolerance of non-synonymous single nucleotide polymorphisms (nsSNPs) J Mol Biol. 2013;425(8):1274–1286. doi: 10.1016/j.jmb.2013.01.026. [DOI] [PubMed] [Google Scholar]
- Yates CM, et al. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol. 2014;426(14):2692–2701. doi: 10.1016/j.jmb.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Yamane H, Paul WE. Differentiation of Effector CD4 T Cell Populations. Annu Rev Immunol. 2010;28(1):445–489. doi: 10.1146/annurev-immunol-030409-101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.