Abstract
Background
The genetic etiology of primary immunodeficiency disease (PID) carries prognostic information.
Objective
We conducted a whole-genome sequencing study assessing a large proportion of the NIHR-BioResource – Rare Disease cohort.
Methods
In the predominantly European study population of principally sporadic unrelated PID cases (n=846), a novel Bayesian method identified NFKB1 as one most strongly associated with PID, and the association was explained by 16 novel heterozygous truncating, missense and gene deletion variants. This accounted for 4% of common variable immunodeficiency (CVID) cases (n=390) in the cohort. Amino-acid substitutions predicted to be pathogenic were assessed by analysis of structural protein data. Immunophenotyping, immunoblotting and ex vivo stimulation of lymphocytes determined the functional effects of these variants. Detailed clinical and pedigree information was collected for genotype-phenotype co-segregation analyses.
Results
Both sporadic and familial cases demonstrated evidence of the non-infective complications of CVID, including massive lymphadenopathy (24%), unexplained splenomegaly (48%) and autoimmune disease (48%), features prior studies correlate with worse clinical prognosis. Although partial penetrance of clinical symptoms was noted in certain pedigrees, all carriers have a deficiency in B lymphocyte differentiation. Detailed assessment of B lymphocyte numbers, phenotype and function identifies the presence of a raised CD21low B cell population: combined with identification of the disease-causing variant, this distinguishes between healthy individuals, asymptomatic carriers and clinically affected cases.
Conclusion
We show that heterozygous loss-of-function variants in NFKB1 are the most common known monogenic cause of CVID that results in a temporally progressive defect in the formation of immunoglobulin-producing B cells.
Keywords: B cells, Common Variable Immunodeficiency, NF-κB1
Introduction
Common variable immunodeficiency (CVID [MIM 607594]), which occurs in approximately 1:25,000 people1–3, is a clinically and genetically heterogeneous disorder characterized by susceptibility to sinopulmonary infections, hypogammaglobulinemia and poor vaccine responses. CVID is the most common primary immune deficiency requiring life-long clinical follow up and the clinical course is highly variable with substantial excess mortality. Affected individuals frequently present with recurrent respiratory infections as well as immune dysregulatory features. The antibody deficiency is often not as marked as the agammaglobulinemia seen in the genetically defined conditions leading to B lymphocyte aplasia4,5. Conversely, while patients with B lymphocyte aplasia have a favorable prognosis on adequate replacement immunoglobulin treatment, the response of CVID patients is highly variable.
Past studies focused on familial cases with CVID and used techniques ranging from traditional linkage analysis to more recent exome sequencing to characterize the genetic etiology. This has revealed that monogenic gene dysfunction accounts for 10% of cases4,5. Several of the variants in these genes have been characterized as partially penetrant; it remains unclear whether genetic or environmental factors determine disease onset. Multiple recent studies identified variants in NFKB1 as a monogenic cause of CVID and reported on the clinical features of these cases6–11.
As part of this NIHR-BioResource – Rare Diseases study, we sequenced the genomes of 846 unrelated individuals with predominantly sporadic primary immunodeficiency (PID) who were recruited from across the UK and by European collaborators. Application of a recently developed statistical method BeviMed12 to the 846 PID cases and over 5,000 control genomes, identified NFKB1 as the gene with the highest probability of association with the disease, and with the largest number of cases explained by variants in that gene. Further investigations revealed a series of 16 heterozygous loss-of-function (LOF) variants in NFKB1 as the most common genetic cause of CVID.
Mutations in genes that affect NF-κB-dependent signaling are associated with a number of immunodeficiencies13–26. NF-κB is a ubiquitous transcription factor member of the Rel proto-oncogene family. NF-κB regulates the expression of several genes involved in inflammatory and immune responses. The classical activated form of NF-κB consists of a heterodimer of the p50/p65 protein subunits. The NF-κB family of transcription factors comprises five related proteins, c-Rel, p65 (RelA), RelB, p50 (NF-κB1) and p52 (NF-κB2) that interact to form homodimers and heterodimers with distinct gene regulatory functions13,27–29. Each Rel NF-κB protein has a conserved 300-amino-acid N-terminal Rel homology domain (RHD) that encompasses sequences needed by NF-κB proteins to bind DNA motifs (κB elements), to form dimers, to interact with regulatory inhibitor IκB proteins and to enter the nucleus. The ten different NF-κB dimers identified have distinct transcriptional properties28. In most cells, NF-κB is retained in the cytosol in a latent state through interaction with IκB proteins (such as IκBα, IκBβ and IκBε), a family of proteins with ankyrin repeats, that mediate IκB interaction with the RHD of NF-κB, masking the nuclear localization sequence and DNA-binding domains. Signal-dependent activation of an IκB kinase (IKK) complex comprising catalytic (α and β) and regulatory (NEMO) subunits, induces the phosphorylation and degradation of IκB29, which permits NF-κB factors to enter the nucleus and regulate gene expression.
We show that variants in NFKB1 culminate in a progressive humoral immunodeficiency indistinguishable from CVID, with a highly variable penetrance. We demonstrate the utility of an in silico protein prediction model for validating the predicted disease-causing substitutions, and we report on the clinical spectrum, immunological phenotype and functional consequences of heterozygous NFKB1 variants.
Methods
Cohort
NIHR-BioResource – Rare Disease (NIHRBR-RD) study was established in the UK to further the clinical management of patients with rare diseases, by providing a national resource of whole genome sequence data. All participants provided written informed consent and the study was approved by the East of England Cambridge South national institutional review board (13/EE/0325). At the time of our analysis, the NIHRBR-RD study included whole genome sequence data from 8,066 individuals, of which 1,299 were part of the PID cohort. These were predominantly singleton cases, but additional affected and/or unaffected family members of some of the patients were also sequenced; in total there were 846 unrelated index cases.
The PID patients were recruited by specialists in clinical immunology (either trained in pediatrics or internal medicine) from 26 hospitals in the UK, and a smaller number came from the Netherlands, France and Italy. The recruitment criteria included: clinical diagnosis of CVID according to the ESID criteria (ESID Registry – Working Definitions for Clinical Diagnosis of PID, 2014, latest version: April, 25, 2017); extreme autoimmunity; or recurrent (and/or unusual) infections suggestive of severely defective innate or cell-mediated immunity. Exclusion of known causes of PID was encouraged, and some of the patients were screened for one or more PID genes prior to enrollment into the PID cohort. The ethnic make-up of the study cohort represented that of the general UK population: 82% were European, 6% Asian, 2% African, and 10% of mixed ethnicity, based on the patients’ whole genome data.
Given that PID is a heterogeneous disease, with overlap in phenotypes and genetic etiologies across different diagnostic categories, we decided to perform an unbiased genetic analysis of all of 846 unrelated index cases. Whole genome sequence data was additionally available for 63 affected and 345 unaffected relatives. Within a broad range of phenotypes, CVID is the most common disease category, comprising 46% of the NIHRBR-RD PID cohort (n=390 index cases, range 0-93 years of age).
Sequencing and variant filtering
Whole genome sequencing of paired-end reads was performed by Illumina on their HiSeq X Ten system. Reads of 100, 125 or 150 base pairs in length were aligned to the GRCh37 genome build using Isaac aligner, variants across the samples were jointly called with AGG tool, and large deletions were identified using Canvas and Manta algorithms (all software by Illumina), as described previously30. Average read depth was 35, with 95% of the genome covered by at least 20 reads.
Single nucleotide variants (SNVs) and small insertions/deletions (InDels) were filtered based on the following criteria: passing standard Illumina quality filters in >80% of the genomes sequenced by the NIHRBR-RD study; having a Variant Effect Predictor (VEP)31 impact of either MODERATE or HIGH and having a minor allele frequency (MAF) <0.001 in the Exome Aggregation Consortium (ExAC) dataset, and MAF <0.01 in the NIHRBR-RD cohort. Large deletions called by both Canvas and Manta algorithms, passing standard Illumina quality filters, overlapping at least one exon, absent from control datasets32, and having frequency of <0.01 in the NIRBR-RD genomes were included in the analysis.
All variants reported as disease-causing in this study were confirmed by Sanger sequencing using standard protocols. Large deletions were inspected in IGV plot (Figure E1) and breakpoints were confirmed by sequencing the PCR products spanning each deletion.
Gene and variant pathogenicity estimation
In order to evaluate genes for their association with PID, we applied the BeviMed inference procedure12 to the NIHRBR-RD whole genome dataset. BeviMed (https://CRAN.R-project.org/package=BeviMed) evaluates the evidence for association between case/control status of unrelated individuals and allele counts at rare variant sites in a given locus. The method infers the posterior probabilities of association under dominant and recessive inheritance and, conditional on such an association, the posterior probability of pathogenicity of each considered variant in the locus. BeviMed was applied to rare variants and large rare deletions in each gene, treating the 846 unrelated PID index cases as ‘cases’ and 5,097 unrelated individuals from the rest of the NIHRBR-RD cohort as ‘controls’. All genes were assigned the same prior probability of association with the disease of 0.01, regardless of their previously published associations with an immune deficiency phenotype. Variants which had a VEP impact labelled HIGH were assigned higher prior probabilities of pathogenicity than variants with a MODERATE impact, as described previously12.
Immunophenotyping and B and T cell functional assays
Peripheral blood mononuclear cells (PBMCs) were isolated using standard density gradient centrifugation techniques using Lymphoprep (Nycomed, Oslo, Norway). Absolute numbers of lymphocytes, T cells, B cells and NK cells were determined with Multitest six-color reagents (BD Biosciences, San Jose, USA), according to manufacturer’s instructions. For the immunophenotyping of the PBMCs we refer to the Supplemental Methods of the Online Repository.
To analyze the ex vivo activation of T and B cells, PBMCs were resuspended in PBS at a concentration of 5–10×106 cells/ml and labeled with 0.5μM CFSE (Molecular Probes) as described previously33 and in the Supplemental Methods of the Online Repository. Proliferation of B and T cells was assessed by measuring CFSE dilution in combination with the same mAbs used for immunophenotyping. Analysis of cells was performed using a FACSCanto-II flowcytometer and FlowJo software. Patient samples were analyzed simultaneously with PBMCs from healthy controls.
ELISA
The secretion of immunoglobulins by mature B cells was assessed by testing supernatants for secreted IgM, IgG and IgA with an in-house ELISA using polyclonal rabbit anti-human IgM, IgG and IgA reagents and a serum protein calibrator all from Dako (Glostrup, Denmark), as described previously33.
SDS PAGE and Western Blot analysis
Blood was separated into neutrophils and PBMCs. Neutrophils (5×106) were used for protein lysates, separated by SDS polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. Individual proteins were detected with antibodies against NF-κB p50 (mouse monoclonal antibody E-10, Santa Cruz Biotechnology, Texas, USA), against IκBα (rabbit polyclonal antiserum C-21, Santa Cruz Biotechnology) and against human glyceraldehyde-3-phosphate dehydrogenase, GAPDH (mouse monoclonal antibody, Merck Millipore, Darmstadt, Germany).
Secondary antibodies were either goat anti-mouse-IgG IRDye 800CW, goat anti-rabbit-IgG IRDye 680CW or goat anti-mouse-IgG IRDye 680LT (LI-COR Biosciences, Lincoln, NE, USA). Relative fluorescence quantification of bound secondary antibodies was performed on an Odyssey Infrared Imaging system (LI-COR Biosciences, Nebraska, USA), and normalized to GAPDH.
NF-κB1 protein structure
To gain structural information on the NF-κB1 RHD, a previously resolved crystal structure of the p50 homodimer (A43-K353) bound to DNA was used34. Ankyrin repeats of NF-κB1 (Q498-D802) were modelled employing comparative homology modelling (Modeller 9v16) using the Ankyrin repeats crystal structure of NF-κB2 as template35,36. There is no structural information on the region between the 6th and 7th Ankyrin repeat (F751-V771)36 and was therefore omitted in the model. The death domain (G807-S894) structure has been resolved by Saito and co-workers using NMR (pdb: 2bdf).
Statistical analysis of lymphocyte data
Differences between groups with one variable were calculated with a non-paired Student’s t-test or one-way ANOVA with Bonferroni post-hoc test, differences between groups with two or more variables were calculated with two-way ANOVA with Bonferroni post-hoc test using GraphPad Prism 6. A P-value less than 0.05 was considered significant.
Results
Pathogenic variants in NFKB1 are the most common monogenic cause of CVID
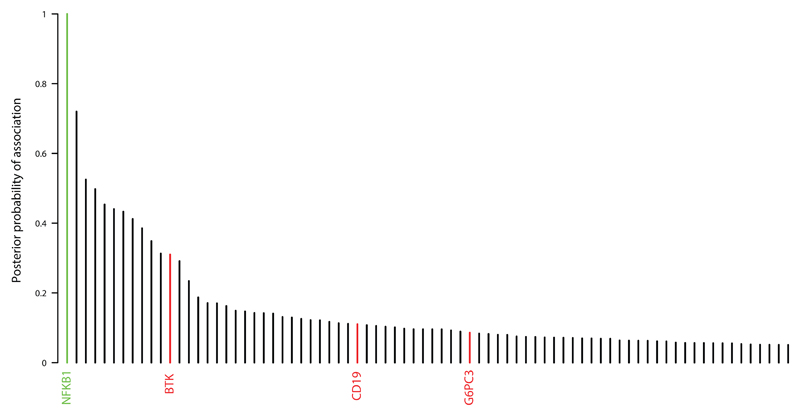
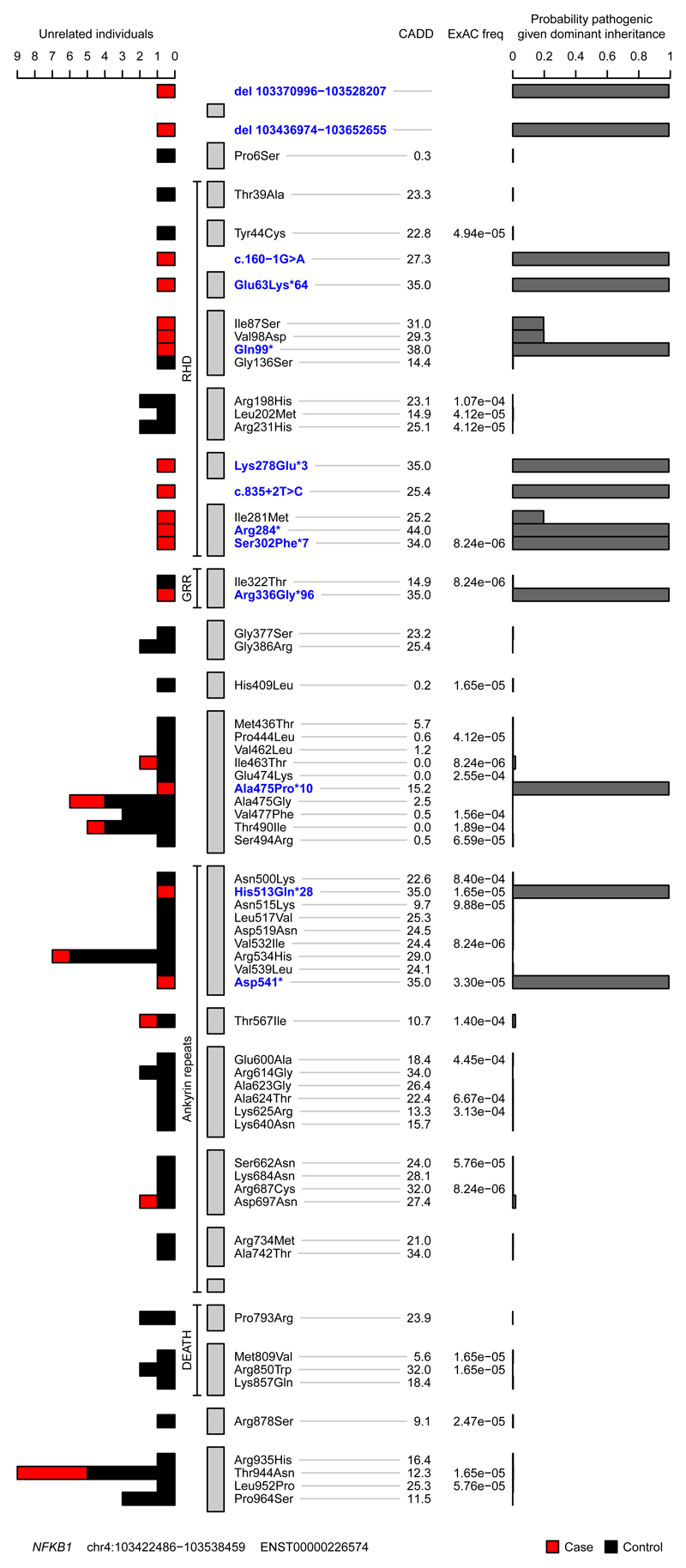
In an unbiased approach to analysis, we obtained BeviMed posterior probabilities of association with PID for every individual gene in all 848 unrelated PID patients in the NIHR-BioResource – Rare Disease (NIHRBR-RD) study. Genes with posterior probabilities greater than 0.05 are shown in Figure 1, showing that NFKB1 has the strongest prediction of association with disease status (1.000). All 13 HIGH impact variants (large deletion, nonsense, frameshift and splice site) in NFKB1 were observed in cases only, resulting in the very high posterior probabilities of pathogenicity (mean 0.99) for this class of variants (Figure 2). On the other hand, MODERATE impact variants (missense substitutions) were observed both in cases and controls. The majority had near zero probability of pathogenicity, but three substitutions were observed in the PID cases only, and had posterior probabilities greater than 0.15 (Figure 2), suggesting their potential involvement in the disease. Genomic variants with a high Combined Annotation Dependent Depletion (CADD) score were found within both the PID and control cohorts, suggesting that this commonly used metric of variant deleteriousness cannot reliably distinguish disease-causing from benign variants in NFKB1. All 16 predicted likely pathogenic variants were private to the PID cohort, and further investigation revealed that all 16 individuals were within the diagnostic criteria of CVID (Table 1).
Figure 1. Overall BeviMed results showing that NFKB1 has the highest posterior probability of association with disease in the NIHRBR-RD PID cohort.
Genes with variants previously reported to cause PID are highlighted in red. Genes with posterior probabilities > 0.05 are shown.
Figure 2. Plot of rare missense, truncating and gene deletion NFKB1 variants identified in the NIHRBR-RD genomes of unrelated individuals, and their location relative to NFKB1 domains.
The tracks from left to right show: number of unrelated case (red) and control (black) individuals in whom each variant was observed; the four major NFKB1 domains; gray bars representing each exon in transcript ENST00000226574; variant annotation relative to transcript ENST00000226574 and genomic location of large deletions, with VEP HIGH impact variants and large deletions highlighted in blue; CADD scores of all nonsense, frameshift, splice and missense variants; ExAC allele frequencies; conditional probability of variant pathogenicity inferred using BeviMed. Only variants labelled as MODERATE or HIGH impact relative to the canonical transcript ENST00000226574 are shown. The initial inference that formed part of the genome-wide analysis included variant chr4:103423325G>A, which was observed in one control sample. This variant is intronic (LOW impact) relative to ENST00000226574 but is a splice variant (HIGH impact) relative to the minor transcript ENST00000505458. As variants were filtered based on the highest impact variant annotation against any Ensembl transcript, this variant was originally included in the inference. For this plot, the inference was re-run including only missense, truncating and gene deletion variants relative to the canonical transcript.
Table 1.
Summary of the CVID patients’ clinical presentation and their NFKB1 variants.
| Case | Sporadic/ Familial |
Infections | Auto-immunity | Malignancy | chr4 position (GRCh37) |
Nucleotide change |
Type of variant |
cDNA(NM_00 3998.3);Amin oAcid |
|---|---|---|---|---|---|---|---|---|
| A | Familial | ● | 103504037 | C>T | nonsense | c.850C>T;Arg284* | ||
| B | Familial | ● | ● | ○ | 103518717 | delCATGC | frameshift | c.1539_1543del;His513Glnfs*28 |
| C | Familial | ● | ● | ●○ | 103459014 | G>A | splice-acceptor | c.160-1G>A;? |
| D | Familial | ● | ● | 103518801 | delGA | nonsense | c.1621_1622del; Asp541* | |
| E | Sporadic | ● | ● | ○ | 103504030 | C>G | missense | c.843C>G;Ile281Met |
| F | Sporadic | ● | ● | 103488178 | T>A | missense | c.293T>A;Val98Asp | |
| G | Sporadic | ● | 103488145 | T>G | missense | c.260T>G;Ile87Ser | ||
| H | Familial | ● | ● | 103501798 | T>C | splice-donor | c.835+2T>C;? | |
| I | Sporadic | ● | 103370996-103528207 | - | large deletion | - | ||
| J | Familial | ● | 103517415 | delG | frameshift | c.1423del;Ala475Profs*10 | ||
| K | Sporadic | ● | ● | ● | 103436974-103652655 | - | large deletion | - |
| L | Familial | ● | ● | 103459041 | delG | frameshift | c.187del;Glu63Lysfs*64 | |
| M | Sporadic | ● | ● | 103501790 | insA | frameshift | c.830dup;Lys278Glufs*3 | |
| N | Sporadic | ● | ● | ● | 103504086 | insT | frameshift | c.904dup;Ser302Phefs*7 |
| O | Sporadic | ● | 103488180 | C>T | nonsense | c.295C>T;Gln99* | ||
| P | Sporadic | ● | ● | 103505914 | delG | frameshift | c.1005del;Arg336Glyfs*96 | |
Closed dots, presence of symptoms in index patient
Open dots, presence of symptoms in family member of index patient
Assessment of all 390 CVID cases in our cohort for pathogenic variants showed that the next most commonly implicated genes are NKFB2 and BTK, with three explained cases each (Figure E2). Importantly, based on the gnomAD dataset of 135,000 predominantly healthy individuals, none of the NFKB1 variants reported here are observed in a single gnomAD individual, even though 90% of our CVID cohort, and all of the NFKB1-positive cases, had European ancestry. Therefore, our results suggest that LOF variants in NFKB1 are the most commonly identified monogenic cause of CVID in the European population, with 16 out of 390 CVID cases explaining up to 4.1% of our cohort. None of the variants identified here had been reported in the previously described NFKB1 cases6–11.
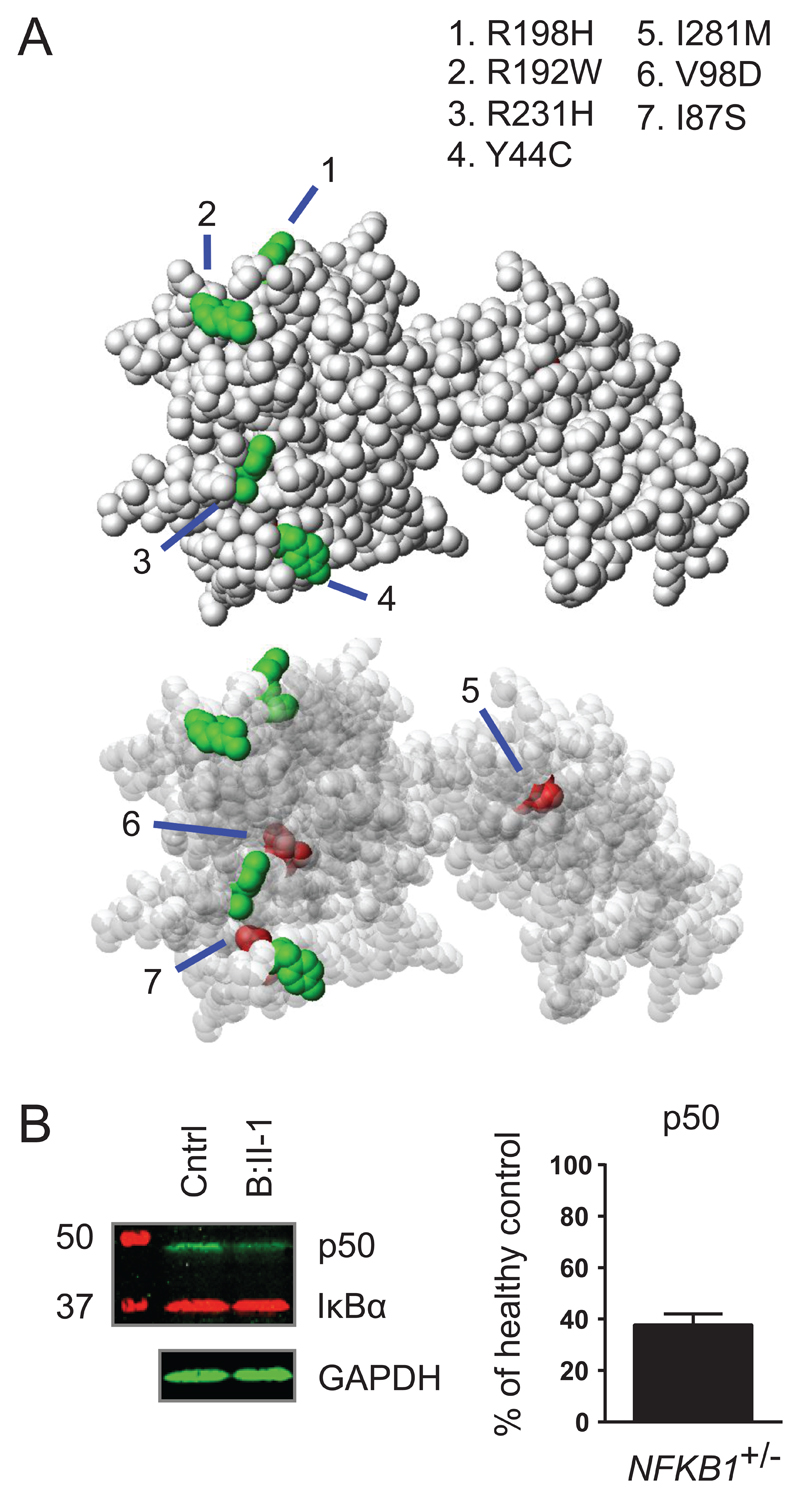
The NFKB1 gene encodes the p105 protein that is processed to produce the active DNA-binding p50 subunit13. The 16 potentially pathogenic variants we identified were all located in the N-terminal p50 part of the protein (Figure 2). The effects of the three rare substitutions on NF-κB1 structure were less clear than those of the truncating and gene deletion variants, so we assessed their position in the crystal structure of the p50 protein. Their location in the inner core of the RHD (Figure 3A) suggested a potential impact on protein stability, whereas other rare substitutions in the NIHRBR-RD cohort were found in locations less likely to affect this (Figure 2 and 3A, Figure E3).
Figure 3. NFKB1 LOF variants lead to haploinsufficiency of the p50 protein.
(A) Localization of RHD substitutions with a high CADD score (>20) within the structure of the NF-κB p50 monomer. Shown is a solid (top panel) and a transparent (bottom panel) sphere representation of the NF-κB p50 monomer. Perturbed residues indicated in green were observed in a control dataset and are located on the outside of the structure, while the residues shown in red were perturbed exclusively in the PID cohort and are buried inside the structure. (B) Western blot analysis targeting p50, IκBα and GAPDH of NFKB1 variant carriers. Left, representative blot of a healthy control and patient B-II:1; Right, summary of 16 NFKB1 variant carriers showing haploinsufficiency, expressed as percentage of healthy controls on the same blot corrected for GAPDH, mean ± SEM.
NF-κB1 LOF as the disease mechanism
Twelve patients with truncating variants (Arg284*, His513Glnfs*28, c.160-1G>A and Asp451*), one patient with gene deletion (del 103370996-103528207) and three patients with putative protein destabilizing missense variants (Ile281Met, Val98Asp and Ile87Ser) were investigated for evidence of reduced protein level. Assessment of the NF-κB1 protein level in PBMCs or neutrophils in 9 index cases and 7 NFKB1 variant carrying relatives demonstrated a reduction in all individuals (Figure 3B, Figure E4). Relative fluorescence quantification of the bands confirmed this and demonstrated a protein level of 38 ± 4.3% (mean ± SEM) compared to healthy controls. There was no difference between clinically affected and clinically unaffected individuals (36 ± 4.4% versus 42 ± 10.1%, n=11 versus n=5, P=0.50). Our observations indicate that the pathogenic NFKB1 variants result in LOF of the NF-κB1 p50 subunit, as reduction in protein levels was seen in all carriers regardless of their clinical phenotype, and was absent in family members that were non-carriers.
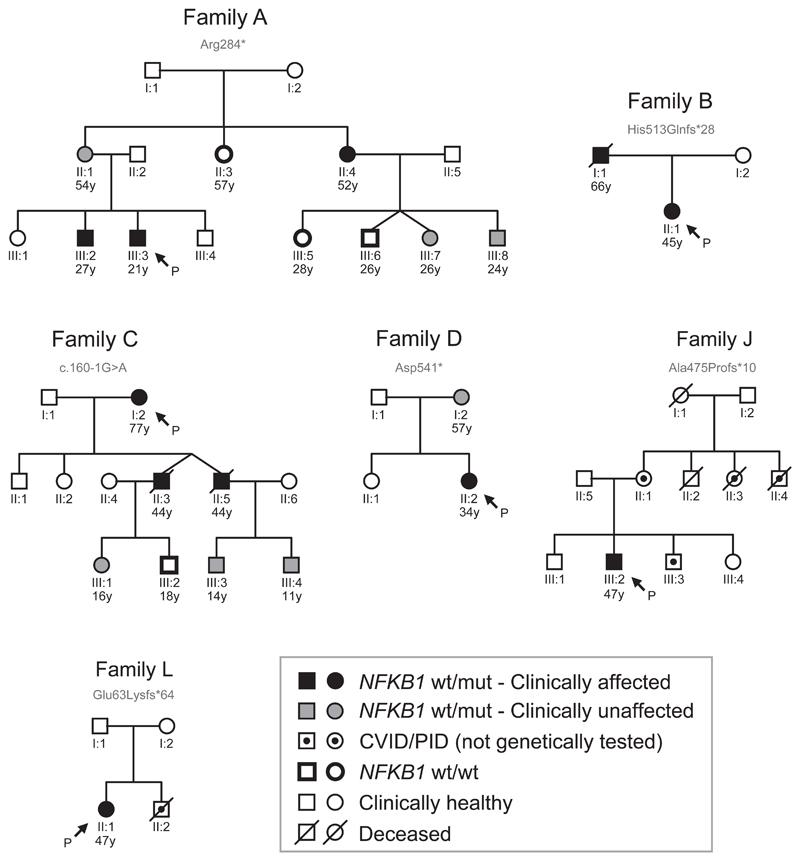
Variable disease manifestations in NFKB1 LOF individuals
Seven individuals had evidence of familial disease (Table 1), prompting us to investigate genotype-phenotype co-segregation and disease penetrance in cases for which pedigree information and additional family members were available (Figure 4, Tables E1-3). The age at which hypogammaglobulinemia becomes clinically overt is highly variable (Figure E5), as illustrated by pedigree C in which the grandchildren carrying the c.160-1G>A splice-site variant had IgG subclass deficiency (C:III-3 and C-III-4), in one case combined with an IgA deficiency (C:III-3). Although not yet overtly immunodeficient, the clinical course of their fathers (C:II-3 and C:II-5) and grandmother (C:I-2) predicts this potential outcome, and warrants long-term clinical follow up of these children.
Figure 4. Pedigrees of familial NFKB1 cases.
Six affected families for which pedigree information and additional family members were available. Proband/index cases indicated with P.
We also observed variants in individuals who were clinically asymptomatic. Pedigree A highlights variable disease penetrance: the healthy mother (A:II-1) carries the same Arg284* variant as two of her clinically affected children (A:III-2 and A:III-3). The identification of this nonsense variant prompted clinical assessment of the extended kindred and demonstrated that her sister (A:II-4) suffered from recurrent sinopulmonary disease and nasal polyps with serum hypogammaglobulinemia consistent with a CVID diagnosis. Overall, based on the clinical symptoms observed at the time of this study across six pedigrees, the penetrance of NFKB1 variants with respect to the clinical manifestation of CVID is incomplete (about 60% in our cohort, 11 affected individuals among 18 variant carriers), with varied expressivity not only of age at disease onset, but of specific disease manifestations even within the same pedigree.
The clinical disease observed among the NFKB1 variant carriers is characteristic of progressive antibody deficiency associated with recurrent sinopulmonary infections (100% of clinically affected individuals), with encapsulated microbes such as Streptococcus pneumoniae and Haemophilus influenzae species (Table E1). The clinical spectrum of NFKB1 LOF includes massive lymphadenopathy (24%), unexplained splenomegaly (48%) and autoimmune disease (48%) – either organ-specific and/or hematological of nature (mainly autoimmune hemolytic anemia and idiopathic thrombocytopenic purpura, Tables E1 and E2). The percentage of autoimmune complications is based on the presence of autoimmune cytopenias (autoimmune hemolytic anemia, idiopathic thrombocytopenic purpura (<50-75x106/mL), autoimmune neutropenia, Evans syndrome), alopecia areata/totalis, vitiligo and Hashimoto thyroiditis among the clinically affected cases. Granulomatous-lymphocytic interstitial lung disease and splenomegaly were considered lymphoproliferation. Enteropathy, liver disease, colitis and a mild decrease in platelet count (>100x106/mL) were neither included in those calculations nor scored separately. Histological assessment of liver disease found in three patients showed no evidence of autoimmune or granulomatous liver disease, though fibrosis and cirrhosis was observed, in these male patients. Finally, the number of oncological manifestations, predominantly hematological, was noticeable. There were two cases with solid tumors (parathyroid adenoma, breast cancer) and four cases with hematological malignancies (B-cell non-Hodgkin lymphoma, diffuse large B-cell lymphoma, follicular lymphoma, peripheral T-cell lymphoma), which add up to 6/21 cases, 28.6%.
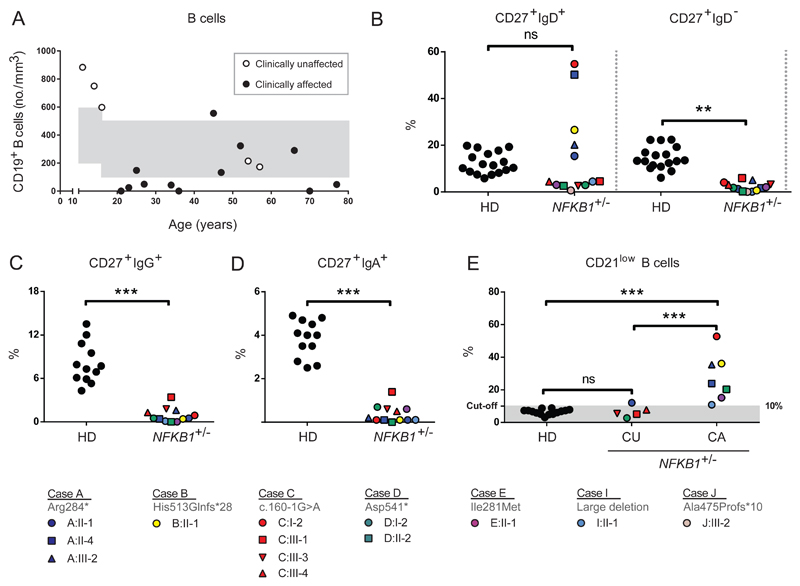
B cell phenotype in NFKB1 LOF individuals and immune cell activation
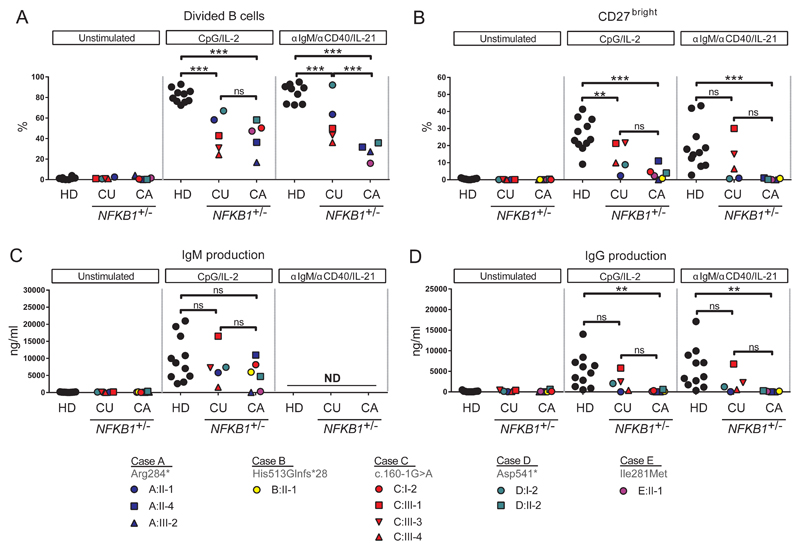
Index cases and family members carrying NFKB1 variants were approached for repeat venipuncture for further functional assessment. In clinically affected individuals the B cell numbers and phenotype were indistinguishable from that described for CVID37 (Figure 5, Figure E6). However, in clinically unaffected individuals, the absolute B cell count was often normal or raised (Figure 5A). In all individuals with NFKB1 LOF variants the numbers of switched memory B cells were reduced (Figure 5B-5D), while a broad range of non-switched memory B cells was observed. This demonstrates that while the clinical phenotype of NFKB1 LOF variants is partially penetrant, all carriers have a deficiency in class-switched memory B cell generation. The presence of raised numbers of the CD21low population described in CVID discriminates between clinically affected and unaffected individuals with NFKB1 LOF variants (Figure 5E). The B cells from individuals with NFKB1 LOF variants demonstrated impaired proliferative responses to anti-IgM/anti-CD40/IL-21 and CpG/IL-2 (Figure 6A); this corresponded with the inability to generate plasmablasts (CD38+/CD27++), most pronounced in the more extreme phenotypes, i.e. clinically affected cases (Figure 6B, Figure E7B). Similarly, ex vivo IgG production was reduced in individuals with LOF variants, whereas the level of IgM in the supernatants was normal (Figure 6C and 6D, Figure E7C), compatible with hypogammaglobulinemia.
Figure 5. Decreased class-switched memory B cells and increased CD21low B cells in NFKB1 LOF variant carriers.
(A) Absolute numbers of CD19+ B cells; each dot represents a single individual and their age. In grey are age-dependent reference values. (B-E) Percentages within CD19+CD20+ B lymphocytes of (B) CD27+IgD+ (non-switched memory or marginal-zone B cells) and CD27+IgD- (switched memory B cells), or (C) CD27+IgG+, or (D) CD27+IgA+, or (E) CD21lowCD38low/dim. (HD healthy donor; CU clinically unaffected or CA clinically affected individuals with LOF variant in NFKB1.) Gating strategy is shown in Figure E6A. Only individuals with sufficient B cells could be analyzed. P-values were determined by one-way (Figure 5E) or two-way (Figure 5B) ANOVA with Bonferroni post-hoc test or unpaired Student’s t-test (Figure 5C,D), ns not significant, **P≤0.01, ***P≤0.001.
Figure 6. The ex vivo class switch recombination defect of individuals carrying NFKB1 LOF variants is linked to the more extreme phenotype.
6 day culture of CFSE-labeled lymphocytes normalized for B cell number unstimulated, CpG/IL-2 (T cell independent activation) or anti-IgM/anti-CD40/IL-21 (T cell dependent activation). (A) Percentage of divided B cells as measured by CFSE dilution (B) Percentage of CD27++ plasmablasts. Gating strategy is shown in Figure E7A. (C, D) IgM and IgG production in supernatant of 6 day culture. (HD healthy donor; CU clinically unaffected or CA clinically affected individuals with LOF variant in NFKB1.) Only individuals with sufficient B cells could be analyzed. P-values were determined by two-way ANOVA with Bonferroni post-hoc test,ns not significant, **P≤0.01, ***P≤0.001.
T cell phenotype in NFKB1 LOF individuals
The T cell phenotype was largely normal in the subset distribution (Figure E8 and E9). Similar to the knockout mouse model38, we found an aberrant number of invariant Natural Killer T (iNKT) cells in the clinically affected individuals (Figure E8). T cell proliferation was intact upon anti-CD3/anti-CD28 or IL-15 activation (Figure E10). Since iNKT cells have been implicated in diverse immune reactions39, this deficiency may contribute to the residual disease burden in replacement immunoglobulin treated patients, some of whom had acute or chronic relapsing infection with herpes virus and, in one case, JC-virus.
Discussion
In our study we show that LOF variants in NFKB1 are present in 4% of our cohort of CVID cases, being the most commonly identified genetic cause of CVID. Furthermore, we highlight specific features of these patients that distinguish them within the diagnostic category of "CVID", which otherwise applies to an indiscrete phenotype, acquired over time, that is termed ‘common’ and ‘variable’. The majority of the genetic variants we report here truncate or delete one copy of the gene; together with pedigree co-segregation analyses, protein expression and the B cell functional data, we conclude that NFKB1 LOF variants are causing autosomal dominant haploinsufficiency. This has now been recognized as the genetic mode of inheritance for at least 17 known PIDs, including that associated with previously reported variants in NFKB16,40–42. In monogenic causes of PID, incomplete penetrance has been more frequently described in haploinsufficient, relative to dominant-negative, PID disease, having been reported in more than half of the monogenic autosomal dominant haploinsufficient immunological conditions described40. This may be because dominant-negative gain-of-function mutations cause disease by expression of an abnormal protein at any level while, as seen in this study, haploinsufficiency is predicted to lead to 50% residual function of the gene product. Incomplete penetrance of a genetic illness by definition will be associated with substantial variation in the clinical spectrum of disease, and the spectrum seen in this study is consistent with prior reports; in three pedigrees with 20 individuals6, harboring heterozygous mutant NFKB1 alleles, the age of onset varied from 2-64 years, with a high variety of disease severity, including two mutation carriers that were completely healthy at the age of 2 and 43 years.
It is important to temper skepticism of partial penetrance of immune genetic lesions with our knowledge that individual immune genes may have evolved in response to selection pressure for host protection against specific pathogens43. Consequently, within the relatively pathogen-free environment of developed countries, the relevant pathogen for triggering disease may be scarce and reports documenting partial penetrance of the clinical phenotype will increase. This makes the traditional approaches of genetics for determining causality difficult. The BeviMed algorithm used in this study prioritized both the gene NFKB1 and individual variants within NFKB1 for contribution to causality; the power of methods like this will increase with greater data availability. The identification of a number of rare NFKB1 variants with high CADD scores both in PID and control datasets highlights the potential for false attribution of disease causality when the genetics of an individual case is considered outside the context of relevant control data.
Currently healthy family members carrying the same NFKB1 LOF variant demonstrated similar reductions in p50 expression and low numbers of switched memory B cells as their relatives suffering from CVID. The longitudinal research investigation of these individuals could help identify the additional modifiers, including epigenetic or environmental factors, which influence the clinical penetrance of these genetic lesions. The similarity of results seen in patients with large heterozygous gene deletions and in those with more discrete substitutions is consistent with haploinsufficiency as the shared disease mechanism.
In patients with mild antibody deficiency it is often difficult to decide when to initiate replacement immunoglobulin therapy; this may be the case for individuals and their family members identified with LOF NFKB1 variants. Two measures seem to correlate well with clinical disease: first, the class-switch defect and lower IgG and IgA production ex vivo. Immunoglobulin class-switching is known to be regulated by NF-κB. Mutations in the NF-κB essential modulator (NEMO) cause a class-switch defective hyper-IgM syndrome in humans20 as well as in the p50 knockout mice13,44,45. Haploinsufficiency of NF-κB may result in defective class-switch recombination due to poor expression of activation-induced cytidine deaminase (AID), a gene regulated by NF-κB, that, when absent, is also associated with immunodeficiency46. Secondly, the ability to measure the CD21low B cell population is widespread in diagnostic immunology laboratories and our study identifies this marker to correlate with NF-κB-disease activity. Although the function of these cells remain to be fully elucidated47, this laboratory test may be useful for the longitudinal assessment of clinically unaffected individuals identified with LOF NFKB1 variants.
Apart from suffering from recurrent and severe infections (including viral disease) for which these patients had been diagnosed with PID in the first place, autoimmunity and unexplained splenomegaly are very common manifestations in our patient cohort, similar to the other heterozygous NFKB1 cases described6–11. Although autoimmunity has been subject to variable percentages per cohort study3,48,49, it seems that these complications occur more frequently in NFKB1-haploinsufficient patients compared to unselected CVID cohorts. In contrast to IKAROS defects, but similar to CTLA4 haploinsufficiency, we observed that NFKB1-haploinsufficiency may also result in chronic and severe viral disease, as noted for CMV and JC virus infections in three of our patients. In the study of Maffucci et al.11, one of the NFKB1-affected cases also suffered from Pneumocystis jirovecii and progressive multifocal leukoencephalopathy (PML), which is suggestive for JC virus infection. Whether the B cell defect in NFKB1-haploinsufficiency is responsible for these non-bacterial infections is unclear50,51. PML is most often discovered in the context of an immune reconstitution inflammatory syndrome, as seen in HIV patients on antiretroviral therapy, and in multiple sclerosis patients after natalizumab discontinuation52. Although the exact contribution of B cell depletion in PML pathogenesis is unknown, the increased PML risk in rituximab-treated patients53 suggests a protective role for B cells.
Three individuals in this cohort suffered from liver failure and an additional three of transaminitis. Although autoimmunity is suspected, a non-hematopoietic origin of liver disease cannot be excluded in the absence of autoantibodies and nodular regenerative disease. Mouse models have suggested a non-immune role for NF-κB signaling in liver failure13,54–56.
In the cohort of NFKB1 patients we identified a number of malignancies. Malignancies in PID patients have been cited as the second-leading cause of death after infection57,58, and murine-models have demonstrated that haploinsufficiency of NF-κB1 is a risk factor for hematological malignancy59. In a large CVID registry study on 2,212 patients, 9% had malignancies, with one-third being lymphomas, some presenting prior to their CVID diagnosis49. Despite the fact that our cohort is relatively small, we found oncological manifestations in 29% of our cases (two-third being lymphoma), suggesting that malignancies in NFKB1-haploinsufficiency may occur more often than in unselected CVID patients. In a study in 176 CVID patients, among the 626 relatives of patients with CVID, no increase in cancer risk was observed60, suggesting that when this does occur, as in this study (three out of seven), it may be due to a shared genetic lesion. Therefore, in a pedigree with a LOF variant in NFKB1, any relatives with cancer should be suspected of sharing the same pathogenic variant.
In conclusion, previous publications61,62 have suggested that CVID is largely a polygenic disease. Our results provide further evidence that LOF variants in NFKB1 are the most common monogenic cause of disease to date, even in seemingly sporadic cases. In these patients there is a clear association with complications such as malignancy, autoimmunity and severe non-immune liver disease; this is important since the excess mortality seen in CVID occurs in this group48. The screening for defined pathogenic NFKB1 variants accompanied by B cell phenotype assessment, has prognostic management and is effective in stratifying these patients.
Supplementary Material
Key Messages.
Pathogenic variants in NFKB1 are currently the most common known monogenic cause of CVID.
There is a clear association with complications such as autoimmunity and malignancy, features associated with worse prognosis.
These patients can be stratified by NFKB1 protein level and the B cell phenotype.
Capsule Summary.
Whole genome sequencing revealed 16 novel pathogenic truncating, missense and gene deletion variants in NFKB1. Most mutations were linked to reduced protein expression, perturbed immunophenotyping and ex vivo stimulation assays of patients and relatives.
Acknowledgments
We thank the patients and their family members for participation in our study.
Funding:
This study was supported by The National Institute for Health Research England (grant number RG65966), and by the Center of Immunodeficiencies Amsterdam (CIDA). JET is supported by an MRC Clinician Scientist Fellowship (MR/L006197/1). AJT is supported by both the Wellcome Trust (104807/Z/14/Z) and by the National Institute for Health Research Biomedical Research Centre at Great Ormond Street Hospital for Children NHS Foundation Trust and University College London. EO receives personal fees from CSL Behring and MSD.
Abbreviations used
- CADD
Combined Annotation Dependent Depletion
- CVID
Common variable immunodeficiency
- ExAC
Exome Aggregation Consortium
- IKK
IκB kinase
- IMDM
Iscove’s Modified Dulbecco’s medium
- InDels
Insertions/deletions
- iNKT cells
Invariant Natural Killer T cells
- LOF
Loss-of-function
- MAF
Minor allele frequency
- PBMCs
Peripheral blood mononuclear cells
- PID
Primary immunodeficiency disease
- PML
Progressive multifocal leukoencephalopathy
- RHD
Rel homology domain
- SNVs
Single nucleotide variants
- VEP
Variant Effect Predictor
Footnotes
Conflicts of Interest
The authors do not have any conflicts of interest to declare.
Consortia
The members of the NIHR-BioResource – Rare Diseases PID Consortium are: Zoe Adhya, Hana Alachkar, Ariharan Anantharachagan, Richard Antrobus, Gururaj Arumugakani, Chiara Bacchelli, Helen Baxendale, Claire Bethune, Shahnaz Bibi, Barbara Boardman, Claire Booth, Michael Browning, Mary Brownlie, Siobhan Burns, Anita Chandra, Hayley Clifford, Nichola Cooper, Sophie Davies, John Dempster, Lisa Devlin, Rainer Doffinger, Elizabeth Drewe, David Edgar, William Egner, Tariq El-Shanawany, Bobby Gaspar, Rohit Ghurye, Kimberley Gilmour, Sarah Goddard, Pavel Gordins, Sofia Grigoriadou, Scott Hackett, Rosie Hague, Lorraine Harper, Grant Hayman, Archana Herwadkar, Stephen Hughes, Aarnoud Huissoon, Stephen Jolles, Julie Jones, Peter Kelleher, Nigel Klein, Taco Kuijpers (PI), Dinakantha Kumararatne, James Laffan, Hana Lango Allen, Sara Lear, Hilary Longhurst, Lorena Lorenzo, Jesmeen Maimaris, Ania Manson, Elizabeth McDermott, Hazel Millar, Anoop Mistry, Valerie Morrisson, Sai Murng, Iman Nasir, Sergey Nejentsev, Sadia Noorani, Eric Oksenhendler, Mark Ponsford, Waseem Qasim, Ellen Quinn, Isabella Quinti, Alex Richter, Crina Samarghitean, Ravishankar Sargur, Sinisa Savic, Suranjith Seneviratne, Carrock Sewall, Fiona Shackley, Ilenia Simeoni, Kenneth G.C. Smith (PI), Emily Staples, Hans Stauss, Cathal Steele, James Thaventhiran, Moira Thomas, Adrian Thrasher (PI), Steve Welch, Lisa Willcocks, Sarita Workman, Austen Worth, Nigel Yeatman, Patrick Yong.
The members of the NIHR-BioResource – Rare Diseases Management Team are: Sofie Ashford, John Bradley, Debra Fletcher, Tracey Hammerton, Roger James, Nathalie Kingston, Willem Ouwehand, Christopher Penkett, F Lucy Raymond, Kathleen Stirrups, Marijke Veltman, Tim Young.
The members of the NIHR-BioResource – Rare Diseases Enrolment and Ethics Team are: Sofie Ashford, Matthew Brown, Naomi Clements-Brod, John Davis, Eleanor Dewhurst, Marie Erwood, Amy Frary, Rachel Linger, Jennifer Martin, Sofia Papadia, Karola Rehnstrom.
The members of the NIHR-BioResource – Rare Diseases Bioinformatics Team are: William Astle, Antony Attwood, Marta Bleda, Keren Carss, Louise Daugherty, Sri Deevi, Stefan Graf, Daniel Greene, Csaba Halmagyi, Matthias Haimel, Fengyuan Hu, Roger James, Hana Lango Allen, Vera Matser, Stuart Meacham, Karyn Megy, Christopher Penkett, Olga Shamardina, Kathleen Stirrups, Catherine Titterton, Salih Tuna, Ernest Turro, Ping Yu, Julie von Ziegenweldt.
The members of the Cambridge Translational GenOmics Laboratory are: Abigail Furnell, Rutendo Mapeta, Ilenia Simeoni, Simon Staines, Jonathan Stephens, Kathleen Stirrups, Deborah Whitehorn, Paula Rayner-Matthews, Christopher Watt.
References
- 1.Primary immunodeficiency diseases. Report of a WHO scientific group. Clin Exp Immunol. 1997;109(Suppl 1):1–28. [PubMed] [Google Scholar]
- 2.Oksenhendler E, Gerard L, Fieschi C, Malphettes M, Mouillot G, Jaussaud R, et al. Infections in 252 patients with common variable immunodeficiency. Clin Infect Dis. 2008;46(10):1547–54. doi: 10.1086/587669. [DOI] [PubMed] [Google Scholar]
- 3.Cunningham-Rundles C, Bodian C. Common variable immunodeficiency: clinical and immunological features of 248 patients. Clin Immunol. 1999;92(1):34–48. doi: 10.1006/clim.1999.4725. [DOI] [PubMed] [Google Scholar]
- 4.Bousfiha A, Jeddane L, Al-Herz W, Ailal F, Casanova JL, Chatila T, et al. The 2015 IUIS Phenotypic Classification for Primary Immunodeficiencies. J Clin Immunol. 2015;35(8):727–38. doi: 10.1007/s10875-015-0198-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Picard C, Al-Herz W, Bousfiha A, Casanova JL, Chatila T, Conley ME, et al. Primary Immunodeficiency Diseases: an Update on the Classification from the International Union of Immunological Societies Expert Committee for Primary Immunodeficiency 2015. J Clin Immunol. 2015;35(8):696–726. doi: 10.1007/s10875-015-0201-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fliegauf M, Bryant VL, Frede N, Slade C, Woon ST, Lehnert K, et al. Haploinsufficiency of the NF-kappaB1 Subunit p50 in Common Variable Immunodeficiency. Am J Hum Genet. 2015;97(3):389–403. doi: 10.1016/j.ajhg.2015.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boztug H, Hirschmugl T, Holter W, Lakatos K, Kager L, Trapin D, et al. NF-kappaB1 Haploinsufficiency Causing Immunodeficiency and EBV-Driven Lymphoproliferation. J Clin Immunol. 2016;36(6):533–40. doi: 10.1007/s10875-016-0306-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lougaris V, Moratto D, Baronio M, Tampella G, van der Meer JW, Badolato R, et al. Early and late B-cell developmental impairment in nuclear factor kappa B, subunit 1-mutated common variable immunodeficiency disease. J Allergy Clin Immunol. 2017;139(1):349–52 e1. doi: 10.1016/j.jaci.2016.05.045. [DOI] [PubMed] [Google Scholar]
- 9.Kaustio M, Haapaniemi E, Goos H, Hautala T, Park G, Syrjanen J, et al. Damaging heterozygous mutations in NFKB1 lead to diverse immunological phenotypes. J Allergy Clin Immunol. 2017 doi: 10.1016/j.jaci.2016.10.054. [DOI] [PubMed] [Google Scholar]
- 10.Schipp C, Nabhani S, Bienemann K, Simanovsky N, Kfir-Erenfeld S, Assayag-Asherie N, et al. Specific antibody deficiency and autoinflammatory disease extend the clinical and immunological spectrum of heterozygous NFKB1 loss-of-function mutations in humans. Haematologica. 2016;101(10):e392–e6. doi: 10.3324/haematol.2016.145136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maffucci P, Filion CA, Boisson B, Itan Y, Shang L, Casanova JL, et al. Genetic Diagnosis Using Whole Exome Sequencing in Common Variable Immunodeficiency. Front Immunol. 2016;7:220. doi: 10.3389/fimmu.2016.00220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene D, BioResource N, Richardson S, Turro E. A Fast Association Test for Identifying Pathogenic Variants Involved in Rare Diseases. Am J Hum Genet. 2017;101(1):104–14. doi: 10.1016/j.ajhg.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Q, Lenardo MJ, Baltimore D. 30 Years of NF-kappaB: A Blossoming of Relevance to Human Pathobiology. Cell. 2017;168(1–2):37–57. doi: 10.1016/j.cell.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshioka T, Nishikomori R, Hara J, Okada K, Hashii Y, Okafuji I, et al. Autosomal dominant anhidrotic ectodermal dysplasia with immunodeficiency caused by a novel NFKBIA mutation, p.Ser36Tyr, presents with mild ectodermal dysplasia and non-infectious systemic inflammation. J Clin Immunol. 2013;33(7):1165–74. doi: 10.1007/s10875-013-9924-z. [DOI] [PubMed] [Google Scholar]
- 15.Courtois G, Smahi A, Reichenbach J, Doffinger R, Cancrini C, Bonnet M, et al. A hypermorphic IkappaBalpha mutation is associated with autosomal dominant anhidrotic ectodermal dysplasia and T cell immunodeficiency. J Clin Invest. 2003;112(7):1108–15. doi: 10.1172/JCI18714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burns SO, Plagnol V, Gutierrez BM, Al Zahrani D, Curtis J, Gaspar M, et al. Immunodeficiency and disseminated mycobacterial infection associated with homozygous nonsense mutation of IKKbeta. J Allergy Clin Immunol. 2014;134(1):215–8. doi: 10.1016/j.jaci.2013.12.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mousallem T, Yang J, Urban TJ, Wang H, Adeli M, Parrott RE, et al. A nonsense mutation in IKBKB causes combined immunodeficiency. Blood. 2014;124(13):2046–50. doi: 10.1182/blood-2014-04-571265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pannicke U, Baumann B, Fuchs S, Henneke P, Rensing-Ehl A, Rizzi M, et al. Deficiency of innate and acquired immunity caused by an IKBKB mutation. N Engl J Med. 2013;369(26):2504–14. doi: 10.1056/NEJMoa1309199. [DOI] [PubMed] [Google Scholar]
- 19.Doffinger R, Smahi A, Bessia C, Geissmann F, Feinberg J, Durandy A, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-kappaB signaling. Nat Genet. 2001;27(3):277–85. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 20.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–8. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 21.Greil J, Rausch T, Giese T, Bandapalli OR, Daniel V, Bekeredjian-Ding I, et al. Whole-exome sequencing links caspase recruitment domain 11 (CARD11) inactivation to severe combined immunodeficiency. J Allergy Clin Immunol. 2013;131(5):1376–83 e3. doi: 10.1016/j.jaci.2013.02.012. [DOI] [PubMed] [Google Scholar]
- 22.Stepensky P, Keller B, Buchta M, Kienzler AK, Elpeleg O, Somech R, et al. Deficiency of caspase recruitment domain family, member 11 (CARD11), causes profound combined immunodeficiency in human subjects. J Allergy Clin Immunol. 2013;131(2):477–85 e1. doi: 10.1016/j.jaci.2012.11.050. [DOI] [PubMed] [Google Scholar]
- 23.Torres JM, Martinez-Barricarte R, Garcia-Gomez S, Mazariegos MS, Itan Y, Boisson B, et al. Inherited BCL10 deficiency impairs hematopoietic and nonhematopoietic immunity. J Clin Invest. 2014;124(12):5239–48. doi: 10.1172/JCI77493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jabara HH, Ohsumi T, Chou J, Massaad MJ, Benson H, Megarbane A, et al. A homozygous mucosa-associated lymphoid tissue 1 (MALT1) mutation in a family with combined immunodeficiency. J Allergy Clin Immunol. 2013;132(1):151–8. doi: 10.1016/j.jaci.2013.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKinnon ML, Rozmus J, Fung SY, Hirschfeld AF, Del Bel KL, Thomas L, et al. Combined immunodeficiency associated with homozygous MALT1 mutations. J Allergy Clin Immunol. 2014;133(5):1458–62. doi: 10.1016/j.jaci.2013.10.045. 62 e1-7. [DOI] [PubMed] [Google Scholar]
- 26.Punwani D, Wang H, Chan AY, Cowan MJ, Mallott J, Sunderam U, et al. Combined immunodeficiency due to MALT1 mutations, treated by hematopoietic cell transplantation. J Clin Immunol. 2015;35(2):135–46. doi: 10.1007/s10875-014-0125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25(51):6706–16. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 28.Oeckinghaus A, Hayden MS, Ghosh S. Crosstalk in NF-kappaB signaling pathways. Nat Immunol. 2011;12(8):695–708. doi: 10.1038/ni.2065. [DOI] [PubMed] [Google Scholar]
- 29.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 30.Carss KJ, Arno G, Erwood M, Stephens J, Sanchis-Juan A, Hull S, et al. Comprehensive Rare Variant Analysis via Whole-Genome Sequencing to Determine the Molecular Pathology of Inherited Retinal Disease. Am J Hum Genet. 2017;100(1):75–90. doi: 10.1016/j.ajhg.2016.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17(1):122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16(3):172–83. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 33.aan de Kerk DJ, Jansen MH, ten Berge IJ, van Leeuwen EM, Kuijpers TW. Identification of B cell defects using age-defined reference ranges for in vivo and in vitro B cell differentiation. J Immunol. 2013;190(10):5012–9. doi: 10.4049/jimmunol.1201807. [DOI] [PubMed] [Google Scholar]
- 34.Muller CW, Rey FA, Sodeoka M, Verdine GL, Harrison SC. Structure of the NF-kappa B p50 homodimer bound to DNA. Nature. 1995;373(6512):311–7. doi: 10.1038/373311a0. [DOI] [PubMed] [Google Scholar]
- 35.Webb B, Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr Protoc Bioinformatics. 2014;47:5 6 1–32. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- 36.Tao Z, Fusco A, Huang DB, Gupta K, Young Kim D, Ware CF, et al. p100/IkappaBdelta sequesters and inhibits NF-kappaB through kappaBsome formation. Proc Natl Acad Sci U S A. 2014;111(45):15946–51. doi: 10.1073/pnas.1408552111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wehr C, Kivioja T, Schmitt C, Ferry B, Witte T, Eren E, et al. The EUROclass trial: defining subgroups in common variable immunodeficiency. Blood. 2008;111(1):77–85. doi: 10.1182/blood-2007-06-091744. [DOI] [PubMed] [Google Scholar]
- 38.Sivakumar V, Hammond KJ, Howells N, Pfeffer K, Weih F. Differential requirement for Rel/nuclear factor kappa B family members in natural killer T cell development. J Exp Med. 2003;197(12):1613–21. doi: 10.1084/jem.20022234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brennan PJ, Brigl M, Brenner MB. Invariant natural killer T cells: an innate activation scheme linked to diverse effector functions. Nat Rev Immunol. 2013;13(2):101–17. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 40.Rieux-Laucat F, Casanova JL. Immunology. Autoimmunity by haploinsufficiency. Science. 2014;345(6204):1560–1. doi: 10.1126/science.1260791. [DOI] [PubMed] [Google Scholar]
- 41.Kuehn HS, Boisson B, Cunningham-Rundles C, Reichenbach J, Stray-Pedersen A, Gelfand EW, et al. Loss of B Cells in Patients with Heterozygous Mutations in IKAROS. N Engl J Med. 2016;374(11):1032–43. doi: 10.1056/NEJMoa1512234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen K, Coonrod EM, Kumanovics A, Franks ZF, Durtschi JD, Margraf RL, et al. Germline mutations in NFKB2 implicate the noncanonical NF-kappaB pathway in the pathogenesis of common variable immunodeficiency. Am J Hum Genet. 2013;93(5):812–24. doi: 10.1016/j.ajhg.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(51):E7128–37. doi: 10.1073/pnas.1521651112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sha WC, Liou HC, Tuomanen EI, Baltimore D. Targeted disruption of the p50 subunit of NF-kappa B leads to multifocal defects in immune responses. Cell. 1995;80(2):321–30. doi: 10.1016/0092-8674(95)90415-8. [DOI] [PubMed] [Google Scholar]
- 45.Snapper CM, Zelazowski P, Rosas FR, Kehry MR, Tian M, Baltimore D, et al. B cells from p50/NF-kappa B knockout mice have selective defects in proliferation, differentiation, germ-line CH transcription, and Ig class switching. J Immunol. 1996;156(1):183–91. [PubMed] [Google Scholar]
- 46.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102(5):565–75. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 47.Rakhmanov M, Keller B, Gutenberger S, Foerster C, Hoenig M, Driessen G, et al. Circulating CD21low B cells in common variable immunodeficiency resemble tissue homing, innate-like B cells. Proc Natl Acad Sci U S A. 2009;106(32):13451–6. doi: 10.1073/pnas.0901984106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Resnick ES, Moshier EL, Godbold JH, Cunningham-Rundles C. Morbidity and mortality in common variable immune deficiency over 4 decades. Blood. 2012;119(7):1650–7. doi: 10.1182/blood-2011-09-377945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gathmann B, Mahlaoui N, Ceredih. Gerard L, Oksenhendler E, Warnatz K, et al. Clinical picture and treatment of 2212 patients with common variable immunodeficiency. J Allergy Clin Immunol. 2014;134(1):116–26. doi: 10.1016/j.jaci.2013.12.1077. [DOI] [PubMed] [Google Scholar]
- 50.Durali D, de Goer de Herve MG, Gasnault J, Taoufik Y. B cells and progressive multifocal leukoencephalopathy: search for the missing link. Front Immunol. 2015;6:241. doi: 10.3389/fimmu.2015.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tan CS, Koralnik IJ. Progressive multifocal leukoencephalopathy and other disorders caused by JC virus: clinical features and pathogenesis. Lancet Neurol. 2010;9(4):425–37. doi: 10.1016/S1474-4422(10)70040-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palazzo E, Yahia SA. Progressive multifocal leukoencephalopathy in autoimmune diseases. Joint Bone Spine. 2012;79(4):351–5. doi: 10.1016/j.jbspin.2011.11.002. [DOI] [PubMed] [Google Scholar]
- 53.Carson KR, Evens AM, Richey EA, Habermann TM, Focosi D, Seymour JF, et al. Progressive multifocal leukoencephalopathy after rituximab therapy in HIV-negative patients: a report of 57 cases from the Research on Adverse Drug Events and Reports project. Blood. 2009;113(20):4834–40. doi: 10.1182/blood-2008-10-186999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanaka M, Fuentes ME, Yamaguchi K, Durnin MH, Dalrymple SA, Hardy KL, et al. Embryonic lethality, liver degeneration, and impaired NF-kappa B activation in IKK-beta-deficient mice. Immunity. 1999;10(4):421–9. doi: 10.1016/s1074-7613(00)80042-4. [DOI] [PubMed] [Google Scholar]
- 55.Pasparakis M, Luedde T, Schmidt-Supprian M. Dissection of the NF-kappaB signalling cascade in transgenic and knockout mice. Cell Death Differ. 2006;13(5):861–72. doi: 10.1038/sj.cdd.4401870. [DOI] [PubMed] [Google Scholar]
- 56.Geisler F, Algul H, Paxian S, Schmid RM. Genetic inactivation of RelA/p65 sensitizes adult mouse hepatocytes to TNF-induced apoptosis in vivo and in vitro. Gastroenterology. 2007;132(7):2489–503. doi: 10.1053/j.gastro.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 57.Mueller BU, Pizzo PA. Cancer in children with primary or secondary immunodeficiencies. J Pediatr. 1995;126(1):1–10. [PubMed] [Google Scholar]
- 58.Vajdic CM, Mao L, van Leeuwen MT, Kirkpatrick P, Grulich AE, Riminton S. Are antibody deficiency disorders associated with a narrower range of cancers than other forms of immunodeficiency? Blood. 2010;116(8):1228–34. doi: 10.1182/blood-2010-03-272351. [DOI] [PubMed] [Google Scholar]
- 59.Voce DJ, Schmitt AM, Uppal A, McNerney ME, Bernal GM, Cahill KE, et al. Nfkb1 is a haploinsufficient DNA damage-specific tumor suppressor. Oncogene. 2015;34(21):2807–13. doi: 10.1038/onc.2014.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mellemkjaer L, Hammarstrom L, Andersen V, Yuen J, Heilmann C, Barington T, et al. Cancer risk among patients with IgA deficiency or common variable immunodeficiency and their relatives: a combined Danish and Swedish study. Clin Exp Immunol. 2002;130(3):495–500. doi: 10.1046/j.1365-2249.2002.02004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Orange JS, Glessner JT, Resnick E, Sullivan KE, Lucas M, Ferry B, et al. Genome-wide association identifies diverse causes of common variable immunodeficiency. J Allergy Clin Immunol. 2011;127(6):1360–7 e6. doi: 10.1016/j.jaci.2011.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Schouwenburg PA, Davenport EE, Kienzler AK, Marwah I, Wright B, Lucas M, et al. Application of whole genome and RNA sequencing to investigate the genomic landscape of common variable immunodeficiency disorders. Clin Immunol. 2015;160(2):301–14. doi: 10.1016/j.clim.2015.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.