Abstract
Oaks are an important part of our natural and cultural heritage. Not only are they ubiquitous in our most common landscapes1, they have also supplied human societies with invaluable services, including food and shelter, since prehistoric times2. With 450 species spread throughout Asia, Europe and America3, oaks constitute a critical global renewable resource. The longevity of oaks (several hundred years) probably underlies their emblematic cultural and historical importance. Such long-lived sessile organisms must persist in the face of a wide range of abiotic and biotic threats over their lifespans. We investigated the genomic features associated with such a long lifespan, by sequencing, assembling, and annotating the oak genome. We then used the growing number of whole-genome sequences for plants (including tree and herbaceous species) to investigate the parallel evolution of genomic characteristics potentially underpinning tree longevity. A further consequence of the long lifespan of trees is their accumulation of somatic mutations (SMs) during mitotic divisions of the stem cells present in the shoot apical meristems. Empirical4 and modeling5 approaches have shown that intra-organismal genetic heterogeneity can be selected6 and provides direct fitness benefits in the arms race with short-lived pests and pathogens, through a patchwork of intra-organismal phenotypes7. However, there is no clear proof that large-statured trees consist of a genetic mosaic of clonally distinct cell lineages within and between branches. Through this case study of oak, we demonstrate the accumulation-transmission of SMs and the expansion of R-gene families in trees.
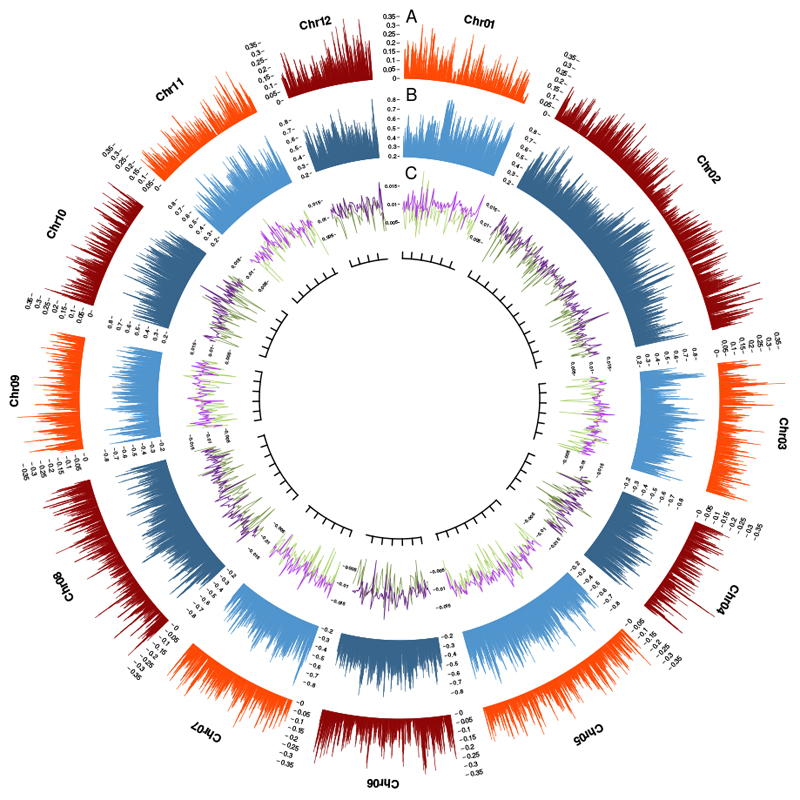
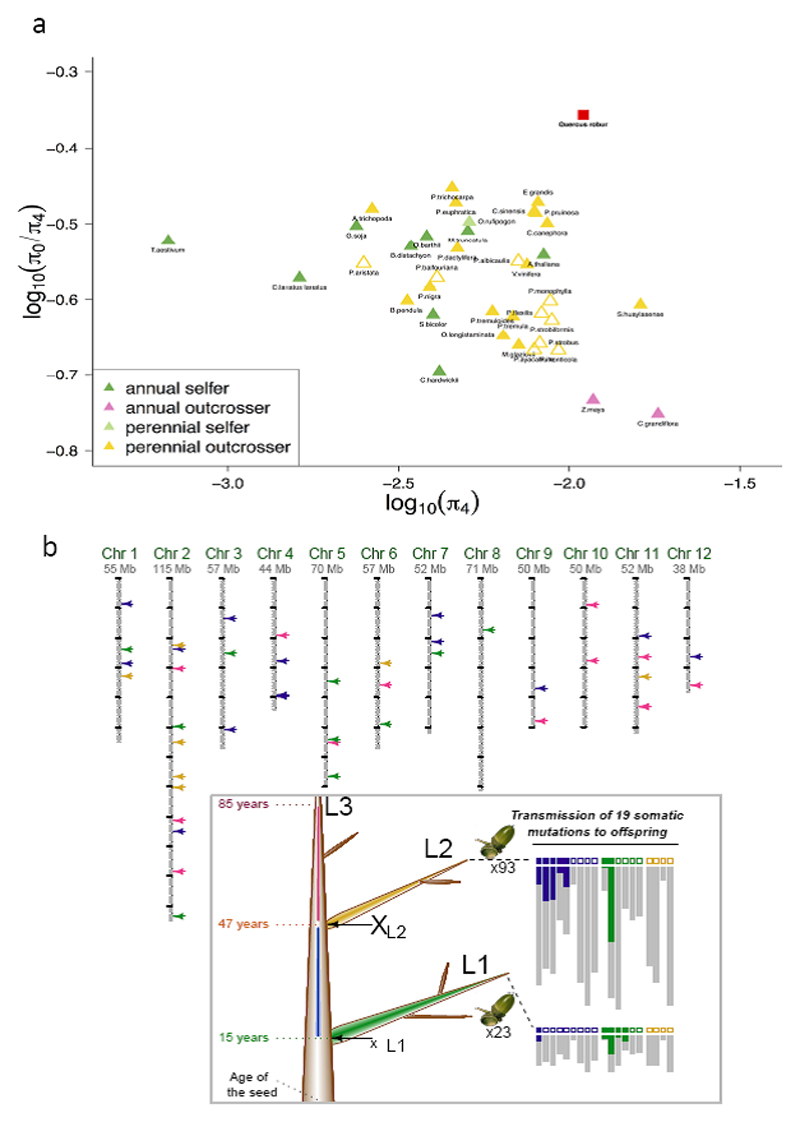
We sequenced the highly heterozygous genome of pedunculate oak (Quercus robur L., Supplementary Notes 1 and 2), using a combination of long and short sequence reads (Supplementary Table 1). We generated a highly contiguous haploid genome sequence of a heterozygous tree comprising 1,409 nuclear scaffolds, with an N50 of 1.35 Mb (Supplementary Note 2.2, Supplementary Table 2, Supplementary Fig. 1). Comparison with existing tree genomes is shown in Supplementary Table 3. In total, 871 scaffolds, covering 96% (716.6 Mb) of the estimated physical size of the oak genome and containing 90% of the 25,808 predicted protein-coding genes (Supplementary Data Set 1, Supplementary Note 3.3), were anchored to the 12 oak chromosomes. To this end, we used the existing high-density oak gene-based linkage map8 combined with a synteny-driven approach using Prunus persica as a pivotal genome. Non anchored scaffolds harbouring genes syntenic to peach were placed on the pseudomolecules considering the local microsynteny identified between oak and peach (Fig. 1, Supplementary Note 2.3, Supplementary Fig. 2, Supplementary Data Set 2). Overall, 52% of the genome was found to consist of diverse transposable elements (TEs), dominated by class I retrotransposons (70%) (Supplementary Table 4, Supplementary Fig. 3, Supplementary Notes 3.1 and 3.4). Genome-wide genetic diversity, as assessed by an analysis of single-nucleotide polymorphisms at the individual level (heterozygosity rate) and using a population of 20 genotypes (π), amounted to ~1%, with significant variation within and between chromosomes (Fig. 1, Supplementary Fig. 4). Nucleotide diversity in protein-coding genes was 0.011 for four-fold degenerate sites, and 0.005 for non-degenerate sites, with a non-synonymous-to-synonymous nucleotide diversity ratio (π0/π4) of 0.44. A comparison of these values with those obtained in a recent survey of plant and animal species9 indicated that oak was remarkable in terms of both its high nucleotide diversity (π4) and the high rate at which it accumulates deleterious mutations (Fig. 2a, Supplementary Note 4.1). Indeed, the value of oak shows the largest deviation from the regression line with the largest residual (0.25) compared to the other 37 plant species (ranging from -0.13 to 0.12).
Figure 1. Genomic landscape of the 12 assembled oak chromosomes.
Gene (A) and TE (B) density, percentage heterozygosity (purple in C) and genetic diversity (green in C). These four metrics are calculated in 1 Mb sliding windows, moved in 250-kb steps. A ruler is drawn on each chromosome, with tick marks every 10 Mb.
Figure 2. Genetic diversity and somatic mutations.
(A) Distribution of π0/π4 plotted against π4 among plants (modified from Chen et al.9) including oak (red square). (B) Genomic location of somatic mutations along the 12 chromosomes of a 100-year-old oak tree. Mutations are represented as colored arrows, according to where they took place during tree growth (see inset). Location and age (left of the trunk) of the three levels (L1, L2 and L3) sampled for somatic mutation detection in the reference pedunculate oak genotype “3P”. L1, L2 and L3 = end of selected branches. For each branch, the recovery or non-recovery of mutations in acorns is indicated by closed and open squares, respectively. Below each square, the numbers of copies of the alternative (colored) and reference (gray) alleles are shown.
In addition to the spontaneous meiotic mutations in each generation, long-lived plants are expected to accumulate somatic mutations throughout their lifetime. These mutations occur during the mitotic divisions of stem cells in the shoot apical meristems4. In trees, unlike animals, these mutations can be passed from the soma to the reproductive tissue and on to the offspring. SMs may therefore increase genetic diversity in long-lived trees such as oaks. Oak has weak apical control (i.e. an inability to control the flushing and growth of lateral buds from the previous year10), resulting in a multi-stemmed morphology. As such, oaks constitute a particularly appropriate model for studies of the somatic generation of diversity. We sampled buds at the extremities of branches initiated at the ages 15, 47 and 85 years on the reference tree sequenced in this study (Fig. 2b, Supplementary Fig. 5). Using a frequency-dependent methods for detecting somatic point mutations in genomic DNA11, we identified 46 reliable SMs (Supplementary Note 4.2, Supplementary Table 5) most of which (44) were located on scaffolds anchored to the 12 chromosomes (Fig. 2b). Compared to a recent report using also the pedunculate oak as a model system12, we detected 2.7 times more SMs on a three times younger tree. This difference is probably due to our superior ability to detect SMs on a higher fraction of the genome (owing to the quality of our genome assembly) and to detect smaller changes in allele frequency by applying a frequency-explicit method, which was developed for cancer research and in our case accounts for the mosaic of mutated and non-mutated stem cells in shoot apical meristems (SAM). Given that most SMs have at low allele frequency (1/2N stem cells) during growth13, most SMs are expected to remain at frequencies too low to be unambiguously detected. Thus, while this work provides clear evidence that SMs exist in trees, it still remains particularly challenging to determine the actual rate of SMs. Consequently, we consider that we only reported the tip of the iceberg of the total number of SMs in the studied genotype. The hypothesis of a protection of stem cell mutagenesis against UV damage in the SAM formulated by Schmid-Siegert et al.12 and based on the discrepancy between theoretical expectations and the low number of empirically identified SMs is an interesting working hypothesis. However, considering the detection bias for low allele frequency variants, it remains unsupported by the best genomic data available to date. We then investigated the transmission of mutations to the offspring, by evaluating a subset of 19 SMs (Supplementary Table 6) in 116 acorns collected from the extremities of lateral branches (Fig. 2b). Despite the limited number of seeds collected, we recovered 47% (9/19) of the SMs in the embryonic tissues of the acorns, confirming intergenerational transmission (Fig. 2b). Our work demonstrates that SMs exist in oak and are passed on to the next generation. However, our results do not allow conclusions to be drawn on the contribution of SMs to the high genetic diversity level and large-scale evolution of oaks.
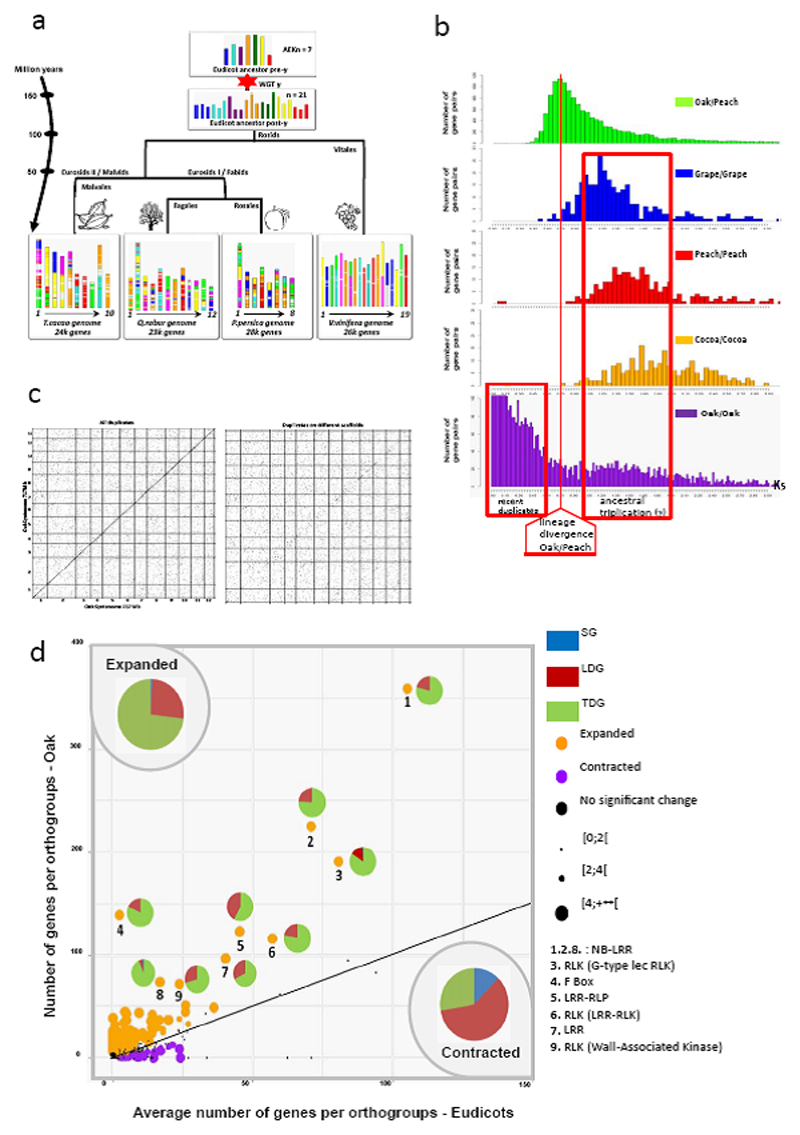
We searched for genomic features specific to oak that might contribute to its longevity, by first reconstructing its paleohistory within the rosid clade. We compared the Ancestral Eudicot karyotype (AEK14) reconstructed from the comparison of Vitales (grape15), Rosales (peach16) and Malvales (cocoa17) major subfamilies to reveal that that oak experienced five fissions and fourteen fusions from 21 AEK18 chromosomes to reach the modern 12 chromosomes (Fig. 3a). The Ks distribution of paralogs (Fig. 3b) indicated that oak did not experience lineage-specific whole-genome duplication beside the ancestral triplication shared among the Eudicots (γ19). We also found that oak experienced a recent burst of local gene duplications (accounting for 35.6% of the oak gene repertoire) after oak-peach lineage divergence (Fig. 3b). The eucalyptus genome is the only other plant genome shown to date to display such high levels of tandem duplication20 (34%), contrasting strongly with the other four genomes investigated (< 25% tandem duplicates). We validated that recent TDGs were true duplicates rather than different alleles or duplication artifacts generated during haplome construction (i.e. during the scaffolding or merging steps of our hierarchical assembly pipeline) by applying two checking procedures based on the comparison with polymorphism of allelic gene pairs (Supplementary Fig. 22), and sequence coverage analysis (Supplementary Fig. 23).
Figure 3. Evolutionary history of the oak genome.
(A) Evolutionary scenario of oak from the Eudicot Ancestral Karyotypes (AEK) of 21 (post-γ) and 7 (pre-γ) protochromosomes, reconstructed from comparison of Vitales (grape), Rosales (peach) and Malvales (cocoa) major subfamilies. The modern genomes (bottom) are illustrated with different colours reflecting the seven ancestral chromosomes of AEK origin (top). WGT refers to the whole-genome triplication (γ), shared among the eudicots. (B) Ks (synonymous substitution rate) distribution of gene pairs for oak/peach orthologs (green) as well as the shared γ triplication in grape (blue), peach (red), cocoa (brown), and oak (purple). Ks distribution of all gene pairs in oak (purple) illuminate gene pairs from the γ triplication as well as recent duplicates. (C) Dot plot representation of the oak genome against itself for the complete set of paralogous pairs (left) and without tandemly duplicated genes (right) representing the disappearance of the diagonal (tandemly duplicated genes) when low Ks values are removed. (D) Expansion (524 orthogroups, orange dots) and contraction (72 orthogroups, purple dots) in oak relative to 15 other eudicot species. The pie chart reflects the contribution of tandemly duplicated genes (TDGs, green), long distance-duplicated genes (LDGs, red) and singleton genes (SGs, blue) to the significantly expanded/contracted orthogroups and to outstanding outliers, labeled from #1-9.
A comparison of gene families (for 36,844 orthogroups, including 435,095 genes from sixteen plant species (Supplementary Table 7) provided further clues to the functional significance of tandem duplications. Of the 524 orthogroups found to have undergone expansion in oak relative to the other 15 species (Supplementary Data Set 3), 73% of the genes concerned were tandem duplicates (Supplementary Data Set 4). Such a tight relationship between tandemly duplicated genes and lineage-specific selection is not a novel observation21, and it seems to be particularly common for disease resistance (R) genes22. However, the higher frequency of such relationships in long-lived plants, such as oak and eucalyptus, suggests that there may be a convergent mechanism in trees, towards an expansion of these families of genes in long lived species.
The orthogroups expanded in oaks are clearly enriched in Gene Ontology (GO) terms relating to biotic interactions. They included 95% of the 1,091 NB-LRR (nucleotide-binding site leucine-rich repeat)-related protein genes and 55% of the 1,247 RLK (receptor-like kinase)-encoding genes (Supplementary Data Sets 5 and 6, Supplementary Table 8, Supplementary Notes 3.5.6 and 3.5.7). We detected a particularly strong expansion of two major clades of TIR-NB-LRRs in orthogroup #1 (shaded areas in Fig. 4a and Supplementary Fig. 6). In addition, three of the nine orthogroups displaying the strongest expansions (Fig. 3d, Supplementary Data Set 3) corresponded to intracellular receptors (NB-LRR: #1, #2, #8) and four corresponded to cell surface receptors of the innate immune response (RLK: #3, #6, #9 and LRR-RLP receptor-like protein: #5). The entire complement of NB-LRR and RLK genes accounts for 9% of all oak genes, a proportion about twice that reported for other plants22,24, and 75% and 65% of the NB-LRR and RLK expansions, respectively, can be accounted for by tandem duplications. The distribution of the LRR-RLK genes between the established subgroups (SGs) based on the analysis of 31 angiosperms25 also revealed remarkable expansions, with SG-XIIa (#6 in Fig. 3d) and SG-XIIb harboring the highest global expansion rates in oak: 102 copies for SG-XIIa and 50 copies for SG-XIIb, corresponding to an expansion rate of 11.3 and 12.5-fold, respectively. SG-XIIa (with FLS2: Flagellin-sensitive 2; EFR: EF-TU receptor and Xa21) and SG-XIIb (with XIK1: Xoo-induced kinase 1) included receptors known to play a role in the response to bacterial infections26. The orthogroups expanded in oaks also presented a significantly (P-value < 2×10-16) higher π0/π4 ratio than contracted or stable orthogroups (Supplementary Table 9), and the efficacy of purifying selection was remarkably low for the NB-LRR and RLK gene families, with mean π0/π4 ratios of 0.68 and 0.58, respectively (Supplementary Note 4.1).
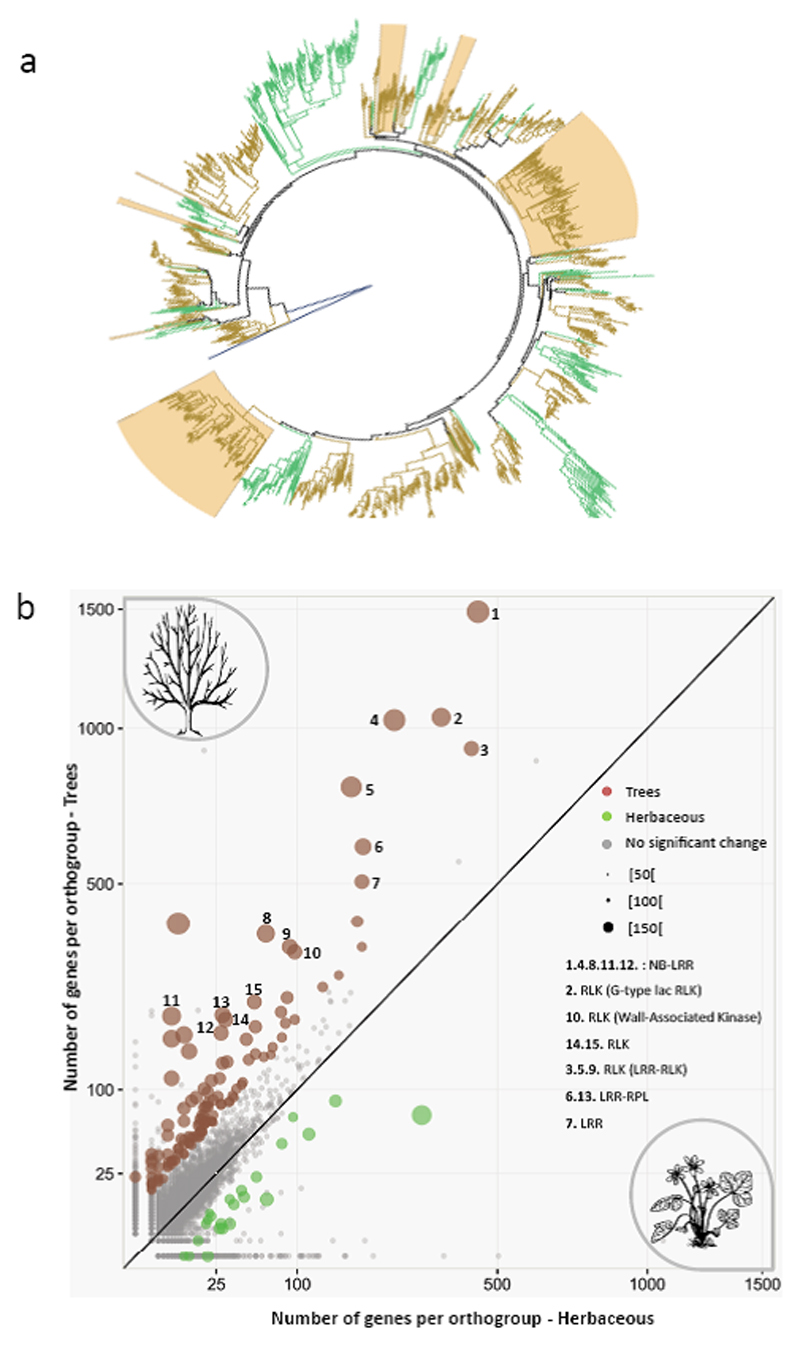
Figure 4. Expanded gene families in trees.
(a) Phylogeny of orthogroup #1 from Fig. 3d and Fig. 4b, established from the nucleotide-binding domains of 1,641 NB-LRR genes. Branches for trees and herbaceous species are shown in brown and green, respectively. Branches expanded in oak are shaded. For a higher resolution see Supplementary Figure 6. (B) Scatter plot showing orthogroups expanded in trees (dots in brown) and herbaceous plants (green) (images from https://openclipart.org/).
The enrichment of gene families relating to receptor-mediated signaling in oak led us to investigate whether similar enrichment had occurred in other trees by comparing trees and herbaceous species among the sixteen plant genomes investigated. In eudicots, each distinct tree lineage provides an independent evolutionary experiment for investigating the genomic features relating to the tree habit27. We found that 126 of the 36,844 orthogroups had undergone tree-specific expansion (Fig. 4b, Supplementary Data Set 7). These orthogroups were enriched in 61 GO terms, largely (63%) related to plant immunity (Supplementary Data Set 8, Supplementary Fig. 7). Ten of the 15 gene families displaying striking expansion in tree genomes (Fig. 4b) corresponded to NB-LRRs (orthogroup #1, #4, #8, #11, #12), LRR-RLKs (#3 SG-XIIb, #5 SG-XIIa, #9) or LRR-RLPs (#6, #13). Phylogenetic analysis of the orthogroup most strongly expanded in trees (#1 in Figs. 3d and 4b) clearly highlighted the expansion of TIR-NB-LRRs in woody perennials relative to herbaceous species (Fig. 4a, Supplementary Fig. 6). Several TIR-NB-LRR genes from this cluster are involved in the perception of bacterial or oomycete pathogens in Arabidopsis (e.g. Rps4 or Rpp528,29). We also investigated the adaptive value of R-gene within expanded orthogroups, making use of a recent meta-analysis of these membrane-bound receptor genes in 31 angiosperm genomes25. We isolated 24 groups of oak lineage-specific expanded LRR-RLK paralogs and explored footprints of positive selection (Supplementary Data Set 9) based on the divergence between paralogous copies. In total, 19 groups (80%) had a significant signature of positive selection, with similar proportions reported only for two other tree species (Malus, 73% and Populus, 87%). We identified 260 sites subject to positive selection after the manual curation of protein sequence alignments in oak. More than 78% of these sites were located in LRR domains. As reported in a previous study25, positive selection mostly targeted four amino acids of the hypervariable region of the characteristic LXXLXLXX β-sheet/β-turn structure of LRRs (Supplementary Fig. 8), which has been implicated in protein-protein interactions30. The high proportion of sites under positive selection in this domain thus confirms the amino acid sequence diversification of these genes through fixation of amino acid changes.
In their opinion article, Tobias and Guest31 suggested three non-exclusive mechanisms that could allow plants “to grow old without antibobies”: i/ have numerous and highly diversified defense genes, ii/ favor R-gene families expansion, and iii/ accumulate somatic mutations which can be transmitted to the next generation. Our study tackles all three genomic features that may contribute to the success of long-lived trees and finds support for all three suggested mechanisms.
In conclusion, we sequenced the oak genome and revealed its considerable genetic diversity, to which heritable somatic mutations may contribute. This work poses new research questions about the contribution of this mutational load in adaptation, in particular with regard to defenses against new pests and pathogens. We also showed that the genome of this iconic tree went through a single paleohexaploidization event (γ, shared among the eudicots), followed by a massive burst of recent local gene duplication. These duplications have amplified families of genes involved in defense against pathogens. We observed a parallel expansion of R-gene related gene families across multiple tree species, suggesting that the immune system makes an essential contribution to the survival of long-lived plants over several centuries. The remarkable relaxation of purifying selection observed in oaks may facilitate the evolution of a richer and more diverse set of R-genes, thereby conferring an advantage on these trees in their continuous arm race with pathogens32. This dynamic is likely to apply, in particular, to oaks, with their remarkably long lifespan. However, the maintenance of such a diversity of R-genes may be costly, and future studies should look at how trees control the expression of these immune receptors, through micro-RNA control, for example22.
Methods
Tree material
Pedunculate oak (Quercus robur L, 2n=2x=24) is an outcrossing, highly heterozygous diploid species. Flow cytometry analysis has shown this species to have a genome of 740 Mb/C33. The “3P” accession selected for establishment of the reference genome sequence for pedunculate oak is a tree of about 100 years of age located at the INRA Pierroton forestry research station (Aquitaine, France; 44°44′N, 00°46′W). It had already been characterized at the genetic34,35 and genomic36,37 levels. This tree (used as a female parent) has also been crossed with accession A4 (used as a male parent) to generate a full-sib progeny for studies of the genetic architecture of quantitative traits38–47. A graft copy of “3P” was placed in darkness in July 2009, to trigger the release of as much starch as possible from second-flush leaves, in an in-house procedure that has been shown to improve the quality of DNA extraction from oak leaves. We harvested 140 g of etiolated leaves and stored them at -80°C before DNA extraction.
DNA sample preparation for reference genome sequencing
The Invisorb Spin Plant Mini Kit (Stratec Molecular, GmbH,Berlin, Germany) was used to isolate genomic DNA and prepare short-read libraries for the Roche-454 and Illumina sequencing platforms. DNA concentrations were determined with the Quant-iT dsDNA Assay Kit (Life Technologies, Carlsbad, California, USA) and a Qubit Fluorometer (Invitrogen, Carlsbad, CA, USA). We checked the integrity of the genomic DNA by agarose gel electrophoresis and pulsed-field gel electrophoresis. Agarose-embedded high-molecular weight (HMW) DNA was prepared as described by Peterson et al.48, and modified as described by Zhang et al.49, to construct Illumina TruSeq Synthetic Long Read (TSLR) libraries. Agarose gel plugs were washed three times in TE buffer, and subjected to digestion with 8 U of β-agarase (New England Biolabs, Ipswich, Massachusetts, USA) for 12-16 hours at 42°C. HMW DNA was then drop-dialyzed for 2.5 hours. DNA concentrations were quantified with the Quant-iT dsDNA Assay Kit (Life Technologies). DNA quality was then checked with Argus™ Qcard kit (OpGen, Maryland, USA) and was estimated at 20 to 100 kb.
Sequencing
We prepared 454 single-end read libraries according to the standard Roche procedure, with RL adaptors (GS FLX Titanium Rapid Library Preparation Kit, Roche Diagnostic, Indianapolis, USA). The libraries were sequenced with titanium chemistry, on a 1/2 Pico Titer Plate on a 454 GS FlX instrument (Roche Diagnostic).
Illumina overlapping and tightly sized paired-end libraries were prepared with a semi-automated protocol. Briefly, 250 ng of genomic DNA was sheared with the Covaris E210 instrument (Covaris, Inc., Woburn, Massachusetts, USA), to generate fragments of 150-400 bp or 200-800 bp in size for the overlapping and tightly sized libraries, respectively. End repair, A-tailing and ligation with Illumina compatible adaptors (Bioo Scientific Austin, Texas, USA) were performed with the SPRIWorks Library Preparation System and a SPRI TE instrument (Beckmann Coulter, Danvers, Massachusetts, USA), according to the manufacturer’s protocol. We selected fragments of 200-400 bp or 300-600 bp in size for the overlapping and tightly sized libraries, respectively. DNA fragments were then amplified by 12 cycles of PCR with the Pfx Platinum Taq polymerase (ThermoFisher, Waltham, Massachusetts, USA) and Illumina adapter-specific primers. We selected amplified library fragments of about 300 bp in size by electrophoresis in a 3% agarose gel for the overlapping libraries, and fragments of about 600 bp in size by electrophoresis in a 2% agarose gel for tightly sized libraries.
The 3 kb mate-pair (MP) library was prepared according to the initial Illumina protocol (Illumina Mate Pair library kit, Illumina, San Diego, CA), with about 10 µg of genomic DNA subjected to Covaris fragmentation in the first step. The other MP libraries were prepared with the Nextera Mate Pair Sample Preparation Kit (Illumina). Briefly, genomic DNA (4 µg) was simultaneously fragmented by enzymatic treatment and tagged with a biotinylated adaptor. The resulting fragmented and tagged (tagmented) DNA was subjected to size selection (3-5, 5-8 and 8-11 kb) by regular gel electrophoresis, and circularized by overnight incubation with a ligase. Linear, non-circularized fragments were digested and circularized DNA was fragmented to generate fragments of 300-1000 bp in size with the Covaris E210 system. Biotinylated DNA was immobilized on streptavidin beads, end-repaired, then 3’-adenylated, and Illumina adapters were added. DNA fragments were amplified by PCR with Illumina adapter-specific primers and purified.
All Illumina library traces were evaluated with an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) and quantified by qPCR with the KAPA Library Quantification Kit (KapaBiosystems Inc., Woburn, MA, USA) on a MxPro instrument (Agilent Technologies). Libraries were then sequenced as described in Supplementary Table 1.
Finally, 39,092 BACs (corresponding to a physical coverage of 3.5x, Supplementary Note 2.1) were end-sequenced with dye terminator chemistry an ABI 3730 sequencer (Applied Biosystems, Foster City, CA, USA), as described by Plomion et al.50. The sequences can be obtained from Genbank, accession numbers: HN154083-HN174138, JS673272-JS676292, JS676293-JS684825, FO926004-FO981373.
We prepared 14 libraries (Supplementary Table 1) from five different extracted HMW DNA samples, with TSLR technology (previously known as Moleculo), according to the Illumina protocol. Briefly, 500 ng of gDNA was sheared into fragments of approximately 10 kb in size with g-Tube (Covaris). The fragments were subjected to end repair, A-tailing and adaptor ligation, and the ligated products were size-selected by gel electrophoresis to obtained fragments of 8-10 kb in size, which were quantified by qPCR. The long-insert library was then diluted such that each well of a 384-well plate contained 3 fg of the library. The diluted products were subjected to long-range PCR, tagmentation and barcoding with 384 different barcoding PCR primers. The 384 barcoded libraries were pooled, purified and subjected to size selection. Each library was sequenced by 100 or 150 base-length read chemistry, in a paired-end flow cell on an Illumina HiSeq2500 machine (Illumina).
Sequence processing
Raw Roche/454 reads were used for subsequent analyses without processing. Illumina paired-end and mate-pair reads were cleaned in a three-step procedure: i) sequencing adapters and low-quality nucleotides (quality value < 20) were removed, ii) sequences between the second unknown nucleotide (N) and the end of the read were removed, iii) reads shorter than 30 nucleotides after trimming were discarded, together with reads and their mates mapping onto run quality control sequences (PhiX genome). The TSLRs were generated with the BaseSpace workflow; the primary sequencing data were uploaded without modification to the BaseSpace cloud and then processed with the standard Illumina workflow to generate long synthetic reads.
Genome size estimation by K-mer analysis
Before assembly, we analyzed the k-mer distribution of Illumina 100 bp paired-end reads (two lanes representing 95-fold coverage of the haploid genome), to obtain an independent estimate of the haploid size of the oak genome. The 31-mer distribution was generated with Jellyfish51(with the following parameters: -m 31 -s 2048M -C) and was uploaded to the GenomeScope website (http://qb.cshl.edu/genomescope/). We obtained an estimated haploid genome size of 736 Mb (Supplementary Fig. 25), a value very close to the 740 Mb estimated by flow cytometry33.
Genome assembly
We first assembled the longest reads together (obtained from 454 and Moleculo libraries) to maximize the separation of the two haplotypes of accession “3P” and to overcome the high level of heterozygosity. We used two OLC asssemblers: Newbler and Celera52. We used Newbler software (version MapAsmResearch-04/19/2010-patch-08/17/2010) with default parameters, with the addition of the -large and -sio options. As Newbler does not accept reads longer than 2 kb, we split Moleculo reads into overlapping 1,999 bp fragments (with overlaps of 1,499 bp) and retained the origin of each fragment for further analysis (see next section). We obtained an assembly (named A1 in Supplementary Table 10) of 300,113 contigs with an N50 of 9.3 kb and a cumulative size of 1.31 Gb, corresponding roughly to the size of the two haplotypes. We ran Celera with the following parameters: unitigger=bogart; merSize=31; merThreshold=auto*2; ovlMinLen=800; obtErrorRate=0.03; obtErrorLimit=4.5; ovlErrorRate=0.03; utgErrorRate=0.015; utgGraphErrorRate=0.015; utgGraphErrorLimit=0; utgMergeErrorRate=0.03; batThreads=20; utgMergeErrorLimit=0. It produced an assembly (named C1 in Supplementary Table 11) composed of 29,255 contigs with an N50 of 9.5 kb and a cumulative size of 1.31 Gb. The Celera assembler allows the direct input of raw Moleculo reads and we performed the scaffolding (i.e. ordering and orienting of contigs) step directly on the Celera contigs of the C1 assembly.
Use of long reads to simplify the contig graph
Once the initial Newbler assembly had been obtained, we used long-range information from Moleculo reads to simplify the contig graph. The Newbler output file “454ContigGraph.txt” describes the contig graph, in which the nodes are contigs and the edges are links between two contigs spanned by a read. Contigs were generally fragmented due to the presence of repeat or heterozygous regions. We extracted links between the contigs created from different parts of a single long reads. Finally, a file containing all the links was generated (in DE format) and used as input for the SGA scaffolding module53. We obtained an assembly (named A2 in Supplementary Table 10) composed of 198,695 contigs with a N50 of 16.2 kb and a cumulative size of 1.33 Gb.
Scaffolding step
We used Illumina paired-end and mate-pair libraries to organize contigs and produce scaffolds. We ran three iterations of the SSPACE scaffolder54 with the parameters -k 5 and -a 0.7, using the following libraries, ranked by increasing fragment size: 400 bp paired-end (PE), 3 kb mate-pairs (MP), 5 kb MP and 8 kb MP. We then ran SSPACE again, with -k 2 and -a 0.7, using the Sanger BAC-ends and the previously scaffolded assembly. Sanger reads were transformed into Illumina-like reads by selecting the 100 bp window with the highest quality according to Sickle software55. We obtained two assemblies (A3 and C2 in Supplementary Tables 10 and 11, respectively). The most contiguous of these assemblies (A3) consisted of 9,025 scaffolds with an N50 of 818 kb and a cumulative size of 1.45 Gb (including 11.19% ambiguous bases).
Choice of the final assembly
The choice of the final assembly was based on the metrics of the two assemblies obtained with Celera and Newbler (assemblies C2 and A3) and comparisons with high-quality BACs (see Supplementary Note 2.1.3 and examples in Supplementary Fig. 9). We chose the Newbler assembly because it gave better discrimination between the two haplotypes.
Gap filling
The scaffold gaps of the A3 assembly were closed with GapCloser software56 and Illumina paired-end reads. As input, we used 95x coverage (of the haploid genome) of overlapping paired-end reads and 95x coverage (of the haploid genome) of a standard paired-end library (400-600 bp fragments). We obtained an assembly (named A4 in Supplementary Table 10) consisting of 9,025 scaffolds with an N50 of 821 kb and a cumulative size of 1.46 Gb (including 4.63% ambiguous bases).
Bacterial decontamination
SNAP gene finder57 was applied to the whole assembly for draft gene prediction. We used an optimized calibration of SNAP based on the genewise alignment of Prunus persica coding sequences with the oak genome assembly. Predicted genes were then aligned against NCBI NR database with BLAST-p. We kept the best match for each predicted protein and used the corresponding taxon. The 198 scaffolds containing more than 50% bacterial genes for the assigned proteins were considered to be putative contaminants and were removed from the assembly file (assembly A5 in Supplementary Table 10).
Single-haplotype assembly
We used the Haplomerger v1 pipeline58 to reconstruct allelic relationships in the released polymorphic diploid assembly and to reconstruct a reference haploid assembly. The diploid genome was first soft-masked with i) TRF59 to mask tandem repeats, ii) RepeatMasker60 to mask simple repeats, low-complexity and viridiplantae-specific TEs, iii) DUST61 to mask low-complexity sequences, and iv) RepeatScout62 to mask unknown TEs. We then inferred a scoring matrix specific to the oak genome sequence, using 5% of the diploid assembly. The haploid genome was obtained from the soft-masked assembly and the specific scoring matrix, with Haplomerger. We used the ‘selectLongHaplotype=1’ parameter to maximize gene content as recommended in the Haplomerger documentation, as we knew this would generate frequent switches between haplotypes (Supplementary Fig. 11). We also prevented Haplomerger from creating false joins between scaffolds by using external information. We used the genetic linkage map (see Supplementary Note 2.3) and prevented Haplomerger from joining scaffolds from different linkage groups by modifying the “hm.new_scaffolds” file. We obtained an assembly (named H1, Supplementary Table 2) composed of 1,409 scaffolds with an N50 of 1,343 kb and a cumulative size of 814 Mb (including 2.94% ambiguous bases). We halved the size of the assembly, whilst retaining a completeness of gene content (evaluated with BUSCO,63 similar to that of the diploid assembly, see Supplementary Table 2). The haploid scaffolds were aligned with BACs, for visual inspection to determine the correctness of this final release (see Supplementary Figs. 11, 12 and 13). Comparison with existing heterozygous plant genome shows that our assembly ranks among the best for a series of metrics (number of contigs and scaffolds, scaffold N50 size, Supplementary Table 3). As introduced in Supplementary note 2.3, a chromosome-scale genome was finally established using a high-density linkage map based on SNP markers8. We assessed the linear association between the genetic and physical positions of the SNPs using Spearman rank correlation.
Detection and annotation of transposable elements
The REPET pipeline (http://urgi.versailles.inra.fr/Tools/REPET) was used for the detection, classification (TEdenovo64,65 and annotation (TEannot66) of TEs. The TEdenovo pipeline detects TE copies, groups them into families and defines the consensus sequence for each family containing at least five copies. The TEannot pipeline then annotates TEs using the library of consensus sequences.
The TEdenovo pipeline was used to search for repeats in contigs longer than 29,034 bp (50% of the genome) from the first diploid version (V1) of the Quecus robur reference genome sequence 50. The first step used Blaster with the following parameters [identity > 90 %, HSP (high-scoring segment pairs) length >100 bp and <20 kb, e-value ≤ 1e-300]. The HSPs detected were clustered by three different methods: Piler67, Grouper66 and Recon68. Multiple alignments (with MAP69) of the 20 longest members of each cluster (n clusters) containing at least 5 members were used to derive a consensus. Consensus sequences were then classified on the basis of their structure and similarities relative to Repbase Update (v17.11)70 and PFAM domain library v26.071, before the removal of redundancy (with Blaster + Matcher as in the TEdenovo pipeline). Consensus sequences with no known structure or similarity were classified as “unknown”.
The library of 4,552 classified consensus sequences provided by the TEdenovo pipeline was used to annotate TE copies throughout the genome with the TEannot pipeline. Three methods were used for annotation (Blaster, Censor, RepeatMasker). The resulting HSPs were filtered and combined. Three methods (TRF, Mreps and RepeatMasker) were also used to annotate simple sequence repeats (SSRs). TE annotation covered only by SSRs were then removed. Finally a “long join procedure”72 was used to address the problem of nested TEs. This procedure finds and connects fragments of TEs interrupted by other more recently inserted TEs, to build a TE copy. The nesting patterns of such insertions must respect three constraints: fragments must be collinear (both in the genome and with the same reference TE consensus sequence), of the same age and separated by more recent TE insertion. The percentage identity to the reference consensus sequence was used to estimate the age of the copy. Using the results of this first TEannot pipeline, we filtered out 2,047 consensus sequences with no full-length copy in the genome. A copy may be built from one or more fragments joined by the TEannot long join procedure. We then used manual curation to improve TE annotation. We removed TE copies with consensus sequences identified as part of the host gene. These consensus sequences were built from a family of repeats containing at least five members and were classified as unknown by the TEdenovo pipeline. They were predicted to be host genes from multigene families. We also filtered out consensus sequences identified as chimeric. We obtained a final library of 1,750 consensus sequences, which together captured 52% of the oak genome, a value in the upper range of the values previously reported for plants.
Gene prediction and functional annotation of protein-encoding genes
We used EuGene version 4.073 to predict gene structure. EuGene predicts gene models from combination of several lines of in silico evidence (ab initio and similarity). The EuGene pipeline was trained on a set of 342 genomic/full-coding cDNA pairs for which coding sequences were confirmed by protein evidence. One third of the dataset was used for training the ab initio gene structure prediction software: (i) Eugene_IMM74 based on probabilistic models for discriminating between coding and non-coding sequences, (ii) SpliceMachine75 was used to predict CDS start and intron splicing sites, and (iii) FGENESH, an ab initio gene finder (http://linux1.softberry.com/berry.phtml) was used with Populus trichocarpa parameters. A second third of the dataset was used to optimize EuGene parameters. The final third of the training dataset was used to calculate EuGene accuracy. We estimated sensitivity at 85.8% and 75.2%, and specificity at 87.7% and 74.6%, for exons and genes, respectively.
We refined alignments with nucleotide similarity-based methods (Blat and Sim4), using transcript contigs from Quercus robur and Quercus petraea76. We ensured that alignment quality was high, by respecting the following criteria: (i) 100% coverage and 98% identity for alignments with contigs shorter than 300 bp; (ii) < 98% coverage and 98% identity for alignments with contig lengths between 300 and 500 bp, (iii) < 95% coverage and an identity of 98% for alignments with contigs longer than 500 bp, and (iv) < 95% identity for all other cases. We also used BLAST-x 2.2.29+ to match protein sequences with sequences in protein databases, such as SwissProt, and databases built for species phylogenetically related to oak, such as Prunus persica V1.39, Vitis vinifera V1.45, Populus trichocarpa V 2.10, Eucalyptus grandis V 2.01, and Arabidopsis thaliana V1.67. We filtered out predicted genes overlapping TEs identified with the REPET package (see previous section), but retained TEs in introns and UTRs. The results of the various analyses were combined in EuGene, to predict the final gene models. Predicted genes of less than 100 nucleotides in length were automatically filtered out by EuGene.
We initially predicted 77,043 protein-coding genes from the diploid version (V2) of the Quecus robur genome sequence. In total, 2,067 genes from different gene families were manually curated by experts (see Supplementary Note 3.5). From the 77,043 predicted genes, 43,240 were entirely recovered in the haplome, including 1,176 of the manually curated genes. Genes were tagged as “unreliable” if their coding sequences were less than 500 bp long (corresponding to 166 aa), transcript coverage was less than 90%, or the genes were not curated manually. Based on these criteria, 13,575 genes were tagged as “unreliable”, and the remaining genes were tagged as “regular” (28,484 genes) or “manual” (1,176 genes).
We then performed a manual analysis of the 43,240 candidate gene models, guided by a first orthoMCL run of the 16 genome sequences used in the evolutionary analysis (see section entitled “Oak karyotype evolution and genome organization” further down), in which we filtered out genes from orthoMCL clusters associated with: (i) domains identified as plant mobile element domains (PMD domain) or TE domains (e.g. transposases or GAG, a structural protein for virus-like particles within which reverse transcription takes place), and (ii) similarity to TE proteins, based on BLAST analyses against KEGG library results. We also checked that the orthoMCL clusters contained more than 90% Q. robur genes (i.e. with only a minor contribution from other species): (i) we filtered out “potential pseudogenes” or small gene fragments predicted in regions of dubious assembly due to a high repeat content (i.e. presence of TEs or repeated motifs in genes, such as NBS-LRR), (ii) we also filtered out unreliable and regular singletons (single genes not clustered with orthoMCL) with a CDS < 500 bp. Some small genes were classified as “regular”, as they were sufficiently covered by mRNA contigs, but they could be mapped to multiple sites within the genome and could not therefore be considered specific for the gene tagged.
Automated functional annotation was performed on the 25,808 predicted proteins (listed in Supplementary Data Set 1), with an in-house pipeline (FunAnnotPipe), mostly largely on the InterProScan v5.13-52.077 webservice for domain/motif searches. This included all the manually curated genes, 78% of the “regular” set and 17% of the “unreliable” set. Subcellular targeting signals and transmembrane domains were predicted with SignalP, TargetP and TMHMM78 and InterProScan. We also carried out similarity searches with BLAST-x V2.2.29+ against PDB, Swissprot and KEGG79, and rpsBLAST (Jun-14-2009) searches for conserved domains against CDD database80 and KOG81. We also used the BLASTKoala webservice [http://www.kegg.jp/blastkoala/, January 2016] to associate Kegg orthology groups, and E2P2 to identify the associated enzyme codes when relevant (https://dpb.carnegiescience.edu/labs/rhee-lab/software, version3.0).
We assigned “definitions” to the predicted proteins as proposed by Phytozome82 and DM Goodstein personal communication). We used the annotation from the most accurate analysis as input: EC number (E2P2), KEGG orthology group (KO; KEGGKOALA), PANTHER (InterProScan), KOG (conserved domain database for eukaryotic organisms), and PFAM (InterProScan). We then calculated the multiplicity (M) of annotations across the entire genome, both as single (e.g., KOG0157, PF0064, PF0005) and same-type compound keys (e.g. PF0064//PF0005). Mixed compound keys were not considered (e.g. KOG0157//PF0064). Weighting (W) factors were applied to protein definitions to give priority to the most informative annotations, as follows: EC=1, KO=1.1, PANTHER=2, KOG=3, PFAM=4. The final protein definition corresponds to the least frequent description (minimum M*W value) from this analysis. The key advantage of this approach is that it makes it possible to assign a protein definition without over-representing a single type of annotation found at multiple locations. As a result, a protein definition was assigned to 87% of the predicted oak proteins (Supplementary Data Set 1).
Estimation of heterozygosity of the reference genotype “3P”
All the short Illumina paired-end reads used to produce the “3P” oak reference genome were mapped against the haplome assembly with bowtie283, using standard parameters for the “fast end-to-end” mode. Duplicated mapped reads were removed with Picard (http://broadinstitute.github.io/picard/). SAMtools/bcftools84 were used to call variants. We then used a combination of custom-made scripts (available at http://www.oakgenome.fr) to calculate coverage and estimated allele frequency from the “DP4” tag of the VCF file. We discarded all SNPs with a MAF<0.25 and all INDELs; the proportion of heterozygous sites on the chromosomes was then calculated with a sliding window approach. For each window, this proportion was weighted by the N% and the fraction covered, defined here as the proportion of bases within a window satisfying the same sequence depth criteria as used for SNP calling.
Pool-seq-based estimator of oak genetic diversity
Branches from 38 pedunculate oak trees were sampled in spring 2011, from oak stands within the maritime pine forest (Supplementary Table 17, Erreur ! Source du renvoi introuvable.53) of the Landes (Southwest France). Branches were harvested with a telescopic pole pruner and placed in darkness for three days to trigger the release of starch from chloroplasts. Etiolated leaves were then harvested and their DNA was extracted using the DNeasy plant minikit, according to the manufacturer’s instructions (Qiagen, Hilden, Germany). The amount of DNA was assessed with a NanoDrop spectrometer (Technologies, Montchanin, DE, USA) and DNA quality was assessed visually, by electrophoresis in a 1.2% agarose gel. The 38 genotypes were genotyped with a 12-plex of EST-SSRs and an 8-plex of genomic SSRs85. We estimated genetic relatedness between genotypes with COANCESTRY86, as described by87, and the degree of introgression of sequences from sessile oak (Q. petreae) was assessed with STRUCTURE88, as described by Guichoux et al.85. Following this analysis, we excluded three samples identified as possibly related and eight samples displaying a large degree of introgression from sessile oak. We then randomly selected 20 of the remaining 27 trees (Supplementary Table 18) for whole-genome sequencing by pool-seq techniques89.
DNA from these 20 oaks was re-extracted from individual samples with the Invisorb Spin Plant Mini Kit (Stratec Molecular, GmbH, Berlin, Germany). We visually checked DNA quality by gel electrophoresis (1.5% agarose) and estimated concentration and purity with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies). We then pooled DNA from individual samples, to obtain an equimolar solution with a final concentration of 570 ng/µL. We used this pool of DNA to prepare a paired-end genomic library with the Paired-End DNA Sample Preparation Kit (Illumina). This library was sequenced on 10 lanes of a HiSeq2000 sequencer (Illumina) (2 x 100 bp paired-end reads), generating 1,732,899,595 paired-end reads (331 Gb, i.e. ~400x haploid genome coverage).
Raw reads were trimmed to remove low-quality bases, as described in the “sequence processing” section. All reads were then mapped against the oak haplome assembly with bowtie283, using standard parameters for the “sensitive end-to-end” mode. Potential PCR duplicates were removed with Picard (http://broadinstitute.github.io/picard/). Samtools84 and Popoolation290 were then used to call SNPs with counts of at least 10 for the alternate allele and a depth between 50 and 1000x at the position concerned. All SNPs with a minor allele frequency (MAF) below 0.05 were discarded. After subsampling the pileup at all retained positions to a uniform coverage of 30x (“subsample-pileup.pl”, Popoolation suite91, we used the “variance-sliding.pl” script (Popoolation91) to calculate π along chromosomes by a sliding window approach (1 Mb sliding windows, 250-kb steps, Supplementary Figs. 4 and 15).
Estimate of genetic diversity and π0/π4 ratio
We estimated genetic diversity as pairwise nucleotide diversity (π) at 0-fold and 4-fold sites for each protein-coding gene, as described by Chen et al.9. We then defined the π0/π4 ratio as the ratio of mean π0 to mean π4 over all genes. We also computed these metrics on manually curated genes only to show that gene model quality did not compromise our findings. We compared estimates between genes from expanded, contracted, and unchanged gene families (orthogroups) in oak. We accounted for the different gene family sizes, by randomly sampling 1000 genes from each of these three categories and repeating the operation 100 times.
Detection of somatic mutations
Our objective was to show that somatic mutations (in terms of single nucleotide polymorphisms) exist in a long-lived plant and transmitted to the next generation. Because we did not intent to provide a comprehensive estimate of the number of somatic mutations in the studied 100-year old tree, it is meaningless to compare our result to an expected number of somatic mutations juts because: i) the substitution rate per site and per generation, ii) the number and pattern of mitotic divisions from zygote and axillary buds, and iiii) cell death and bud abortion rates, are unknown.
We investigated somatic mutations, by resampling the “3P” genotype used to sequence and assemble the reference genome, as described below.
Vegetative buds were collected from the extremities of three second-order branches of the 2011 increment in February 2012: two lateral branches (L1 and L2) and the tree apex (L3). We used dendrochronology (tree-ring dating) to date the time of initiation of the L1 and L2 branches (Supplementary Fig. 5). To this end, we collected 5 mm diameter wood cores from the insertion point of the selected branches with an increment borer. We also dated the age of the tree by taking a core just above ground level and counting the number of rings under a microscope. We estimated that the L1 and L2 branches had been initiated 15 and 47 years earlier, respectively, and that the terminal branch was at least 85 years old.
DNA was extracted from three sets of vegetative buds sampled at location L1, L2, and L3, with the Invisorb Spin Plant Mini Kit (Stratec Molecular). For each sample, six independent DNA extractions were carried out on a pool of buds. DNA quality was checked by electrophoresis in a 1.5% agarose gel. DNA concentration and purity were assessed with a NanoDrop 1000 spectrophotometer (NanoDrop Technologies Inc). Individual DNA samples from the same branch were the pooled in an equimolar solution with a final concentration of 769-1,388 ng/µL. We prepared tightly sized paired-end libraries (600bp in size) as described in the “sequencing” section and sequenced each of these libraries on one to four lanes of a HiSeq2000 or HiSeq2500 sequencer (Illumina) (Supplementary Table 19, 100 bp or 250 bp paired-end reads). We obtained 284- (L1), 250.5- (L2) and 264.9-fold (L3) haploid genome coverage for these samples. For each of the three branches (L1, L2 and L3), reads were mapped against the reference genome sequence with BWA-MEM92 using the default parameters, except for minimum seed length (k=79). After sorting, PCR duplicates were removed with Picard (http://broadinstitute.github.io/picard/). We searched for somatic mutations, using MuTect (a program developed for the detection of somatic point mutations in heterogeneous cancer samples,11) to compare the three libraries (6 pairwise combinations, Supplementary Table 20). This frequency-dependent detection approach was considered as particularly well suited to identify somatic mutations in plants.
Because considering sequencing error (i.e. false positives) is essential for mutation detection and vital to draw valid conclusions, especially with respect to the detection of somatic mutations within a single individual, we addressed this concern and took all possible actions to minimize it. Thus, the accuracy of somatic point mutations was ensured, by considering as reliable somatic mutations only those sites with the following characteristics: (i) a minimum depth of 50x in both the reference and potentially mutated libraries, (ii) no mutant (i.e. alternative) allele in the reference library, and (iii) a minimum frequency of 20% for the mutant allele in the potentially mutated library, i.e. each somatic mutation was supported by 10 alternative alleles or more. We then filtered out candidate SMs by using a cross validation procedure. Across all pairwise comparisons, we only kept somatic mutations with a temporal pattern coherent with the chronology of branch development (see Supplementary Table 20 for details). These multiple comparisons made it possible both to validate the detected mutations and to reconstruct their mutational history along the trunk or the two branches. Finally, we discarded 15 additional candidate mutations among the set of 61 reliable SMs. Indeed, for this 15 SMs, we recovered the same alternate allele in the pool of 20 pedunculate individuals (see section “Pool-seq-based estimator of oak genetic diversity”) at a frequency greater than 0.005. Note that f(alt)<0.005 remains a stringent criterion considering Illumina sequencing error calls (0.024). As a consequence, we cannot rule out that some true positives were excluded at this step. However, our objective was to be as conservative as possible in order to study the transmission of these SMs to the next generation. (Supplementary Table 5).
We studied the transmission of somatically acquired mutations to the offspring, by extracting DNA with the DNAeasy 96-Plant Kit (Qiagen), from 116 acorns sampled from the extremities of the L1 and L2 branches (see Fig. 2b). DNA was extracted after the dissection of embryonic tissues (radicle and plumule) from the acorn. We used 15 ng DNA to genotype the offspring with the MassArray iPLEX assay (Agena Bioscience, Hamburg, Germany) according to the manufacturer's instructions. Primers were designed and 33 SNPs were multiplexed in the Assay Design Suite (Agena Bioscience). Allele calling was processed in Typer Viewer v 4.0.26.75 (Agena Bioscience). This 39-plex assay contained 12 control SNPs and 21 candidate somatic mutations (Supplementary Table 5). Control SNPs were used to provide an estimate of the selfing rate likely to impair interpretation of the segregation of somatic mutations in the offspring. The control SNPs were loci homozygous in the reference genotype “3P” and found at very low frequency in the pool of 20 pedunculate oaks, i.e. with MAFs ranging from 0.02 to 0.05. Embryos resulting from the self-pollination of “3P” were expected to be homozygous for the reference allele and most outcrossed embryos were expected to be heterozygous. We observed a mean heterozygosity (Ho) of 0.54 over the 12 control loci. In the absence of selfing and based on allele frequencies estimated in the pool of 20 individuals, mean Ho would have been close to 0.96, thus suggesting a relatively high rate of selfing (44%). Unamplified loci (2/21 SNPs, Supplementary Table 6) were excluded from the analysis. The overall rate of missing data was quite high (39% for missing SMs and 54% for control SNPs), so all polar plots from Typer Viewer software of the MassArray iPLEX assay were inspected visually to check that genotyping calls were accurate.
Oak karyotype evolution and genome organization
We used the two parameters defined by Salse93 to increase the stringency and significance of BLAST sequence alignment, by either parsing BLAST results and rebuilding HSPs (high-scoring pairs) or using pairwise sequence alignments to identify accurate paralogous relationships within oak (25,808 gene models, Supplementary Data Set 1), and orthologous relationships between oak and grape (26,346 genes on 19 chromosomes,15), peach (28,086 genes on 8 chromosomes,16) and cocoa (23,529 genes on 10 chromosomes,17). We estimated the sequence divergence of paralogs and orthologs from the rate of synonymous substitutions per synonymous site (Ks) calculated with the PAML 4 package94 for oak/peach, grape/grape, peach/peach, cocoa/cocoa and oak/oak gene pairs. Dotplot representations of synteny and paralogy were obtained with the R package ggplot2 (http://ggplot2.org/, Supplementary Fig. 16).
Gene family expansion/contraction in oak
A classification of groups of orthologous sequences (orthogroups, as also referred to here as gene families or clusters) was developed for 16 eudicot plant species: all the predicted oak proteins (corresponding to 25,808 gene models) and the proteins catalogued from 15 other eudicot species (Supplementary Table 21, Supplementary Note 5.1.2): Arabidopsis lyrata, Arabidopsis thaliana, Citrus clementina, Carica papaya, Eucalyptus grandis, Fragaria vesca, Glycine max, Malus domestica, Prunus persica, Populus trichocarpa, Ricinus communis, Solanum tuberosum, Theobroma cacao, Vitis vinifera (genomes available from https://phytozome.jgi.doe.gov) and Citrullus lanatus (genome available from http://www.icugi.org/cgi-bin/ICuGI/index.cgi). These 15 plant genomes were selected on the basis of the following criteria: i) availability of genome sequences and gene models from public databases, ii) assembly quality (N50 length of assembled fragments) and the number of predicted genes, iii) classification (order, family, and genus), the main goal being to cover the entire range of eudicots. The classification was based on a BLAST-p all-against-all comparison of the complete proteomes (E-value < 10-5) of these species, followed by clustering with OrthoMCL 2.0.995, using default parameters. GO terms for the 15 of the plant proteomes were retrieved from Phytozome. For watermelon, the CDS were downloaded from the following web site (http://cucumber.genomics.org.cn/page/cucumber/index.jsp) and we used Interproscan96 to assign GO terms. GO term enrichment analysis was then carried out on the expanded orthogroups in oak (see Supplementary Note 5.4).
We then used CAFE version 3.197,98 with phylogenetic tree information (drawn from http://etetoolkit.org/treeview/) derived from previous studies (see in Salse99, Supplementary Fig. 17) to identify the orthogroups displaying expansion and contraction in oak, using a p-value threshold of 0.01.
Identification and validation of tandemly duplicated genes in oak
Duplicated genes (DG) in oak were identified from the Ks paralog distribution (see purple Ks distribution in Supplementary Fig. 20) and are illustrated in the dot blot Supplementary Fig. 21 (see Supplementary Note 5.2). We extracted DGs from the complete repertoire of paralogs, and generated pairwise alignments of protein sequences with BLAST-p and filters based on alignment identity and length (CIP (for cumulative identity percentage)/CALP (for cumulative alignment length percentage) = 50%/50%). Then we sorted protein sequences by their coordinates on each of the 12 oak chromosomes. We defined TDGs as duplicates separated by up to three genes and LDGs as duplicates separated by more than three genes. The remaining genes were classified as singletons (SGs).
We checked that these recent TDGs in oak were true duplicates rather than different alleles or duplication artifacts arising during haplome construction (during the scaffolding or merging steps of our hierarchical assembly pipeline), by applying two verification procedures based on sequence variation and sequence coverage. First, we obtained pairwise nucleotide sequence alignments, using MUSCLE with standard parameters100, for all 9,189 putative TDGs. For each alignment, summary statistics were calculated with AMAS101. We found that 15 pairs of genes involved in local duplications presented no gaps or polymorphisms and could be considered to be putative assembly artifacts. This corresponds to only a minor fraction (0.13%) of the 11,695 pairwise alignments. In contrast, we found that 8,115 pairs of TDGs (69.4%) displayed substantial sequence divergence (gap length > 10% and a proportion of variable sites > 2%), greater than that between pairs of alleles (Supplementary Fig. 22). Indeed, from the 12,603 allelic pairs obtained by comparing the diploid and haploid versions of the oak genome sequence available for this comparison (indicated as 2:1 relationships in the last column of Supplementary Data Set 1), 1,278 (i.e. 10.1%) had a gap length > 10% and a proportion of variable sites > 2%. Second, a per-base coverage analysis based on reads from the genes classified as TDGs, LDGs and SGs indicated that TDGs did not present half the coverage of the other two categories (illustrated for the longest scaffold in Supplementary Fig. 23), ruling out the alternative hypothesis that TDGs are allelic regions or artifactual duplications due to errors in the assembly process.
Detection of significant expansion/contraction in woody perennials
Particular outcomes of gene family expansion/contraction may be associated with a tree lifestyle, but no study of differential gene gains and losses has ever been performed at the genomic scale in Eudicots (Supplementary Note 5.3). We therefore applied an additional criterion when selecting the 15 plant species for comparative genomic analyses: growth habit (woody perennial versus herbaceous). The genomes of nine woody perennials and seven herbaceous species were available for the investigation of orthogroup expansion in woody species (trees). These two categories were homogeneous in terms of OrthoMCL orthogroups. For a range of variable, including the number of genes per orthogroup (Supplementary Fig. 24), the mean number of genes per orthogroup, the percentage of orthogroups with no gene, and the number of species-specific orthogroups (Supplementary Table 22) no statistical difference was found between the two categories.
We investigated whether a given orthogroup showed significant expansion or contraction in trees by comparing the total number of genes per orthogroup between the two types of growth habit. Given the relatively small number of species per category, we performed a binomial test with a probability of success of p(W) = 9/16. From the initial set of 36,844 orthogroups, we retained orthogroups displaying a statistically significant outcome in terms of gene counts (FDR-adjusted p-value <0.05,102). The minimal contribution to each category was set to five for trees and four for herbaceous species, to minimize bias due to the number of species analyzed. We found that 126 orthogroups were expanded (corresponding to 23,321 genes, i.e. 155.1 genes per orthogroup on average) and 23 were contracted in woody perennials relative to herbaceous species. Functional identities and orthogroup sizes are presented for all significantly expanded or contracted orthogroups in Supplementary Data Set 7 (sheet#2 and #4). GO term enrichment analysis was carried out on the 126 expanded and 23 contracted orthogroups (see next section). We also identified a set of remarkable orthogroups (outliers in Fig. 3d), differing between trees and herbaceous species and including at least five genes in five different species.
Gene ontology (GO) enrichment analysis
All GO term enrichment analyses were performed with R 3.3.1 software103 and the topGO 2.22.0 package104. The weight01 algorithm104 and Fisher’s exact test were used to detect significantly enrichment in GO terms in the various test sets. As stated by the authors of the topGO, the P-value of a GO term is conditioned on the neighbouring terms. The tests are therefore not independent and the multiple testing theory does not directly apply. P-values should therefore be interpreted as corrected or not affected by multiple testing.
Fold-enrichment was defined as illustrated below:
-
(i)
At the gene level, if 52/9,189 (i.e. 0.56%) of input genes are involved in "chitinase activity" and the background level is 60/25,808 genes (i.e. 0.23%) associated with "chitinase activity", the fold-enrichment is approximately 0.56% / 0.23% = 2.43 for this molecular function.
-
(ii)
At the orthogroup level, if 6/126 (i.e. 4.76%) input orthogroups are involved in "protein serine/threonine kinase activity" and the background level is 50/36,844 orthogroups (i.e. 0.136%) associated with "protein serine/threonine kinase activity", the fold-enrichment is approximately 4.76% / 0.136% = 35 for this molecular function.
The first example corresponds to the fold-enrichment calculations performed for TDGs, LDGs, SGs and orthogroups expanded in oak. The second corresponds to the fold-enrichment calculation for orthogroups expanded in woody perennials.
Web resources
We set up several tools and a browser based on the international open source project Generic Model Organism Database (GMOD: http://www.gmod.org) to provide us with access to both structural information and functional annotation (Supplementary Note 6). WebApollo/JBrowse105 was set up (https://urgi.versailles.inra.fr/WebApollo_oak_PM1N/jbrowse/) and populated with the oak reference genome sequence (i.e. 12 chromosomes comprising 876 scaffolds and 533 unassigned scaffolds) and 34 BAC sequences. Several “tracks” were superimposed on these sequences, including predicted genes, predicted TEs, predicted non-coding RNAs, proteins from several species, oak unigenes, RNA-Seq data and QTLs. The “chunk” track represents virtual contig sequences separated by “N” stretches of no more than 11 consecutive bases. Intermine (V1.3.9)106 was used to gather and make available all the information (structural and functional) produced for each protein-coding gene (https://urgi.versailles.inra.fr/OakMine_PM1N/begin.do). All the details about data sources are available from the application in the datasource panel. For JBrowse, tracks have been generated from the reference genome, using data generated for EuGene prediction.
Data availability
The oak haploid genome assembly and corresponding annotation have been deposited in the European Nucleotide Archive under project accession code PRJEB19898. Other sequence release data are indicated in Supplementary Tables 1, 13, 14 and 19, and Supplementary Data Set 10. Data (including intermediate genome assemblies, vcf files used to detect somatic mutations and estimate heterozygosity) are available at the oak genome web site hosted as a permanent resource by Inra (http://www.oakgenome.fr/).
Code availability
The source code for the prediction of miRNA is available as a workflow at https://forgemia.inra.fr/genotoul-bioinfo/ngspipelines/tree/master/workflows/srnaseq. Custom-made scripts for estimation of heterozygosity of the reference genotype “3P” are available at the oak genome web site (http://www.oakgenome.fr/?page_id=587).
Supplementary Material
Acknowledgements
The main sources of funding were the ANR (GENOAK 2022-BSV6-009-02), INRA and CEA. T.L. was supported by fellowships from the ANR and a European Research Council Advanced Grant (ERC TREEPEACE FP7-339728). I.Le. and T.A. were supported by fellowships from the ANR. N.F. received funding from the ANR (ARBRE ANR-22-LABX-0002-02) and the ERC. J.C. was supported by the Swedish Foundation for Strategic Research. We thank the four anonymous reviewers for their careful reading of our manuscript and their many insightful comments.
Footnotes
Author Contributions
C.P., JM.A. and J.A. are the lead investigators. They are joint first authors. T.L. and F.M. are joint authors and contributed equally to the work. C.P. conceived and coordinated the project, supervised the research, organized the main text of the manuscript and the supplementary materials, with contributions from all authors. T.L. estimated heterozygosity from the reference genome sequence and genetic diversity from pool seq-data, and performed somatic mutation analyses together with E.C. G.L.P. participated in the annotation jamboree, coordinated tissue sampling from the reference genotype, and organized the collection of the genotypes used for genetic diversity and somatic mutation analyses. C.B. established the composite linkage map of oak and participated in the annotation jamboree. I.Le. identified the allelic gene pairs and tandem duplicated genes. She prepared the RNAseq tracks for JBrowse, concatenated the functional annotations for the orthogroups and participated in the annotation jamboree. F.H. was involved in database development and prepared the QTL tracks of JBrowse. E.G., C.L. and F.S. were involved in genomic DNA and RNA extraction, genotyping before the pooling of pedunculate oak genotypes and the genotyping of somatic mutations in the offspring. ML.DL analyzed the MLO gene family. A.Kr. contributed to the writing and critical review of the manuscript. B.B. provided critical comments on earlier versions of the manuscript, help to reorganize the final version and reviewed the supplementary materials. J.B. performed the GO term enrichment and gene family expansion analysis comparing trees and herbaceous species. JM.A. directed the sequencing and assembly parts of the project. S.F., A.C., C.D.S., C.D., MA.M and J.M. were involved in the bioinformatic analyses (BAC, genome, transcriptome). K.L., V.B., C.B., A.L. and S.M. prepared the libraries and undertook most of the sequencing activities. P.W. supervised the sequencing activity. J.A. led the genome annotation and transposable elements (TEs) analysis, organized the gene annotation jamboree, and directed the setting up of genome browsers and OakMine interfaces. N.F. performed genome annotation and set up the web interfaces. I.Lu. and T.A. participated in TE annotation. F.M. annotated the endovirus. C.M. was responsible for the insertion of genetic data into the GnpIS database. T.A. helped to improve the REPET pipeline used to annotate TEs and participated in TE annotation. H.Q. supervised the TE analysis. F.M. and J.S. performed the macroevolutionary analyses. S.D., C.M. and E.M. analyzed the NB-LRR gene family. A.Ko. and F.M. annotated genes involved in biotic interactions with ectomycorrhizal fungi and jointly organized the gene annotation jamboree. I.H., D.C. and MB.BT analyzed the aquaporin gene family. A.H. and N.R. analyzed the thioredoxin, glutaredoxin and glutathione transferase gene families. P.H., C.R. and A.V. were involved in the manual curation of candidate genes for gallotannin production. JC.L annotated the laccase gene family. P.FR annotated the BAC clone sequences. C.G., C.K., and O.R. analyzed non-coding RNAs and carried out orthoMCL analysis. H.B. provided high-molecular weight DNA for sequencing. M.S. and J.GP analyzed the MYB gene family. N.C. and AD analyzed the LRR-RLK and LRR-RLP gene families. ML.B, S.H. and M.T. analyzed the SWEET gene family and expression levels, and annotated genes involved in ectomycorrhizal interaction. A.Z. carried out the prospective analysis to identify genes related to the tree habit and provided critical comments on earlier version of the manuscript. J.C. and M.L. estimated the efficacy of purifying selection in oak. O.P., E.L., and N.P.S were involved in the analysis of TEs and genome dynamics.
Competing interests
The authors declare no competing financial interests.
References
- 1.Camus A. Les Chênes: monographie du genre Quercus et Monographie du genre Lithocarpus. P. Lechevalier; 1954. [Google Scholar]
- 2.Logan WB. Oak: the frame of civilization. WW Norton & Company; 2005. [Google Scholar]
- 3.Manos PS, Stanford AM. The historical biogeography of Fagaceae: tracking the tertiary history of temperate and subtropical forests of the Northern Hemisphere. Int J Plant Sci. 2001;162:S77–S93. [Google Scholar]
- 4.Whitham TG, Slobodchikoff CN. Evolution by individuals, plant-herbivore interactions, and mosaics of genetic variability: The adaptive significance of somatic mutations in plants. Oecologia. 1981;49:287–292. doi: 10.1007/BF00347587. [DOI] [PubMed] [Google Scholar]
- 5.Folse HJ, Roughgarden J. Direct benefits of genetic mosaicism and intraorganismal selection: Modeling coevolution between a long-lived tree and a short-lived herbivore. Evolution (N. Y) 2012;66:1091–1113. doi: 10.1111/j.1558-5646.2011.01500.x. [DOI] [PubMed] [Google Scholar]
- 6.Pineda-Krch M, Fagerström T. On the potential for evolutionary change in meristematic cell lineages through intraorganismal selection. J Evol Biol. 1999;12:681–688. [Google Scholar]
- 7.Padovan A, et al. Transcriptome sequencing of two phenotypic mosaic Eucalyptus trees reveals large scale transcriptome re-modelling. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodénès C, et al. High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Res. 2016;23:115–124. doi: 10.1093/dnares/dsw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, Gl S, Lascoux M. Genetic diversity and the efficacy of purifying selection across plant and animal species. Mol Biol Evol. 2017;34:1417–1428. doi: 10.1093/molbev/msx088. [DOI] [PubMed] [Google Scholar]
- 10.Brown CL, Mcalpine RG, Kormanik PP. Apical dominance and form in woody plants : a reappraisal. Am J Bot. 1967;54:153–162. [Google Scholar]
- 11.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid-Siegert E, et al. Low number of fixed somatic mutations in a long-lived oak tree. Nat Plants. 2017;12:926–929. doi: 10.1038/s41477-017-0066-9. [DOI] [PubMed] [Google Scholar]
- 13.Gill DE, Chao L, Perkins SL, Wolj JB. Genetic mosaicism in plants and clonal animals. Ann~ Rev Ecol Syst. 1995;26:423–44. [Google Scholar]
- 14.Murat F, Armero A, Pont C, Klopp C, Salse J. Reconstructing the genome of the most recent common ancestor of flowering plants. Nat Genet. 2017;49:490–496. doi: 10.1038/ng.3813. [DOI] [PubMed] [Google Scholar]
- 15.Jaillon O, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–7. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 16.The International Peach Genome Initiative et al. The high-quality draft genome of peach (Prunus persica) identifies unique patterns of genetic diversity, domestication and genome evolution. Nat Genet. 2013;45:487–494. doi: 10.1038/ng.2586. [DOI] [PubMed] [Google Scholar]
- 17.Argout X, et al. The genome of Theobroma cacao. Nat Genet. 2011;43:101–8. doi: 10.1038/ng.736. [DOI] [PubMed] [Google Scholar]
- 18.Salse J. Ancestors of modern plant crops. Curr Opin Plant Biol. 2016;30:134–42. doi: 10.1016/j.pbi.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 19.Murat F, Zhang R, Guizard S, Gavranović H, Flores R, Steinbach D, Quesneville H, Tannier E, Salse J. Karyotype and gene order evolution from reconstructed extinct ancestors highlight contrasts in genome plasticity of modern rosid crops. Genome Biol Evol. 2015;7(3):735–49. doi: 10.1093/gbe/evv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Q, et al. Explosive tandem and segmental duplications of multigenic families in Eucalyptus grandis. Genome Biol Evol. 2015;7:1068–1081. doi: 10.1093/gbe/evv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu S-H. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, Xia R, Kuang H, Meyers BC. The diversification of plant NBS-LRR defense genes directs the evolution of microRNAs that target them. Mol Biol Evol. 2016;33:2692–2705. doi: 10.1093/molbev/msw154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mun JH, Yu HJ, Park S, Park BS. Genome-wide identification of NBS-encoding resistance genes in Brassica rapa. Mol Genet Genomics. 2009;282:617–631. doi: 10.1007/s00438-009-0492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jupe F, et al. Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics. 2012;13:75. doi: 10.1186/1471-2164-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer I, Diévart A, Droc G, Dufayard J-F, Chantret N. Evolutionary dynamics of the leucine-rich repeat receptor-like kinase (LRR-RLK) subfamily in angiosperms. Plant Physiol. 2016;170:1595–1610. doi: 10.1104/pp.15.01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greeff C, Roux M, Mundy J, Petersen M. Receptor-like kinase complexes in plant innate immunity. Front Plant Sci. 2012;3:1–7. doi: 10.3389/fpls.2012.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitzjohn RG, et al. How much of the world is woody? J Ecol. 2014;102:1266–1272. [Google Scholar]
- 28.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 29.Parker JE, et al. The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell. 1997;9:879–894. doi: 10.1105/tpc.9.6.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins Struct Funct Genet. 2004;54:394–403. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 31.Tobias PA, Guest DI. Tree immunity: growing old without antibodies. Trends Plant Sci. 2014;19:367–370. doi: 10.1016/j.tplants.2014.01.011. [DOI] [PubMed] [Google Scholar]
- 32.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–9. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 33.Kremer A. In: Genome Mapping and Molecular Breeding in Plants, Vol 7 Forest Trees. Kole CR, editor. Springer; 2007. pp. 165–187. [Google Scholar]
- 34.Bodénès C, et al. Comparative mapping in the Fagaceae and beyond with EST-SSRs. BMC Plant Biol. 2012;12 doi: 10.1186/1471-2229-12-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodénès C, Chancerel E, Ehrenmann F, Kremer A, Plomion C. High-density linkage mapping and distribution of segregation distortion regions in the oak genome. DNA Res. 2016;23:115–124. doi: 10.1093/dnares/dsw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faivre Rampant P, et al. Analysis of BAC end sequences in oak, a keystone forest tree species, providing insight into the composition of its genome. BMC Genomics. 2011;12:292. doi: 10.1186/1471-2164-12-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lesur I, et al. A sample view of the pedunculate oak (Quercus robur) genome from the sequencing of hypomethylated and random genomic libraries. Tree Genet Genomes. 2011;7:1277–1285. [Google Scholar]
- 38.Saintagne C, et al. Distribution of genomic regions differentiating oak species assessed by QTL detection. Heredity (Edinb) 2004;92:20–30. doi: 10.1038/sj.hdy.6800358. [DOI] [PubMed] [Google Scholar]
- 39.Scotti-Saintagne C, et al. Detection of quantitative trait loci controlling bud burst and height growth in Quercus robur L. Theor Appl Genet. 2004;109:1648–59. doi: 10.1007/s00122-004-1789-3. [DOI] [PubMed] [Google Scholar]
- 40.Scotti-Saintagne C, Bertocchi E, Barreneche T, Kremer A, Plomion C. Quantitative trait loci mapping for vegetative propagation in pedunculate oak. Ann For Sci. 2005;62:369–374. [Google Scholar]
- 41.Gailing O. QTL analysis of leaf morphological characters in a Quercus robur full-sib family (Q. robur x Q. robur ssp. slavonica) Plant Biol. 2008;10:624–634. doi: 10.1111/j.1438-8677.2008.00063.x. [DOI] [PubMed] [Google Scholar]
- 42.Gailing O, Langenfeld-Heyser R, Polle A, Finkeldey R. Quantitative trait loci affecting stomatal density and growth in a Quercus robur progeny: implications for the adaptation to changing environments. Glob Chang Biol. 2008;14:1934–1946. [Google Scholar]
- 43.Casasoli M, et al. Comparison of quantitative trait loci for adaptive traits between oak and chestnut based on an expressed sequence tag consensus map. Genetics. 2006;172:533–546. doi: 10.1534/genetics.105.048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parelle J, et al. Quantitative trait loci of tolerance to waterlogging in a European oak (Quercus robur L.): physiological relevance and temporal effect patterns. Plant, Cell Environ. 2007;30:422–434. doi: 10.1111/j.1365-3040.2006.01629.x. [DOI] [PubMed] [Google Scholar]
- 45.Brendel O, et al. Quantitative trait loci controlling water use efficiency and related traits in Quercus robur L. Tree Genet Genomes. 2008;4:263–278. [Google Scholar]
- 46.Derory J, et al. Contrasting relationships between the diversity of candidate genes and variation of bud burst in natural and segregating populations of European oaks. Heredity (Edinb) 2010;104:438–48. doi: 10.1038/hdy.2009.134. [DOI] [PubMed] [Google Scholar]
- 47.Song J, et al. X-ray computed tomography to decipher the genetic architecture of tree branching traits: oak as a case study. Tree Genet Genomes. 2017;13:5. [Google Scholar]
- 48.Rani J, Chauhan P, Tripathi R. Li-Fi (Light Fidelity)-the future technology in wireless communication. Int J Appl Eng Res. 2012;7:1517–1520. [Google Scholar]
- 49.Zhang H-B, et al. Construction of BIBAC and BAC libraries from a variety of organisms for advanced genomics research. Nat Protoc. 2012;7:479–99. doi: 10.1038/nprot.2011.456. [DOI] [PubMed] [Google Scholar]
- 50.Plomion C, et al. Decoding the oak genome: public release of sequence data, assembly, annotation and publication strategies. Mol Ecol Resour. 2016;16:254–265. doi: 10.1111/1755-0998.12425. [DOI] [PubMed] [Google Scholar]
- 51.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 53.Simpson JT, Durbin R. Efficient construction of an assembly string graph using the FM-index. Bioinformatics. 2010;26:367–373. doi: 10.1093/bioinformatics/btq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15:211. doi: 10.1186/1471-2105-15-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Joshi N, Fass J. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files. 2011 [Google Scholar]
- 56.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang S, et al. HaploMerger: reconstructing allelic relationships for polymorphic diploid genome assemblies. Genome Res. 2012;22:1581–1588. doi: 10.1101/gr.133652.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Benson G. Tandem Repeats Finder: a program to analyse DNA sequences. Nucleic Acids Res. 1999;27:573–578. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smit AFA, Hubley R, Green P. RepeatMasker Open-3.0. 1996.
- 61.Morgulis A, Gertz EM, Schäffer AA, Agarwala R. A fast and symmetric DUST implementation to mask low-complexity DNA sequences. J Comput Biol. 2006;13:1028–40. doi: 10.1089/cmb.2006.13.1028. [DOI] [PubMed] [Google Scholar]
- 62.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21:351–358. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 63.Simão FA, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;19:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 64.Flutre T, Duprat E, Feuillet C, Quesneville H. Considering transposable element diversification in de novo annotation approaches. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hoede C, et al. PASTEC: an automatic transposable element classification tool. PLoS One. 2014;9:e91929. doi: 10.1371/journal.pone.0091929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Quesneville H, et al. Combined evidence annotation of transposable elements in genome sequences. PLoS Comput Biol. 2005;1:166–75. doi: 10.1371/journal.pcbi.0010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edgar R, Myers E. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(Suppl 1):i152–8. doi: 10.1093/bioinformatics/bti1003. [DOI] [PubMed] [Google Scholar]
- 68.Bao Z, Eddy S. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–76. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang X. On global sequence alignment. Bioinformatics. 1994;10:227–235. doi: 10.1093/bioinformatics/10.3.227. [DOI] [PubMed] [Google Scholar]
- 70.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 71.Finn RD, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ahmed I, Sarazin A, Bowler C, Colot V, Quesneville H. Genome-wide evidence for local DNA methylation spreading from small RNA-targeted sequences in Arabidopsis. Nucleic Acids Res. 2011;39:6919–6931. doi: 10.1093/nar/gkr324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Foissac S, et al. Genome annotation in plants and fungi: EuGene as a model platform. Curr Bioinform. 2008;3:87–97. [Google Scholar]
- 74.Schiex T, Moisan A, Rouzé P. EuGene: an eukaryotic gene finder that combines several sources of evidence. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2001;2066:111–125. [Google Scholar]
- 75.Degroeve S, Saeys Y, De Baets B, Rouzé P, Van de Peer Y. SpliceMachine: predicting splice sites from high-dimensional local context representations. Bioinformatics. 2005;21:1332–1338. doi: 10.1093/bioinformatics/bti166. [DOI] [PubMed] [Google Scholar]
- 76.Lesur I, et al. The oak gene expression atlas: insights into Fagaceae genome evolution and the discovery of genes regulated during bud dormancy release. BMC Genomics. 2015;16:112. doi: 10.1186/s12864-015-1331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Quevillon E, et al. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–20. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Krogh A, Larsson B, von Heijne G, Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 79.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Marchler-Bauer A, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tatusov RL, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Guichoux E, Lagache L, Wagner S, Léger P, Petit RJ. Two highly validated multiplexes (12-plex and 8-plex) for species delimitation and parentage analysis in oaks (Quercus spp.) Mol Ecol Resour. 2011;11:578–585. doi: 10.1111/j.1755-0998.2011.02983.x. [DOI] [PubMed] [Google Scholar]
- 86.Wang J. Coancestry: a program for simulating, estimating and analysing relatedness and inbreeding coefficients. Mol Ecol Resour. 2011;11:141–145. doi: 10.1111/j.1755-0998.2010.02885.x. [DOI] [PubMed] [Google Scholar]
- 87.Lagache L, Leger JB, Daudin JJ, Petit RJ, Vacher C. Putting the biological species concept to the test: using mating networks to delimit species. PLoS One. 2013;8:1–11. doi: 10.1371/journal.pone.0068267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: dominant markers and null alleles. Mol Ecol Notes. 2007;7:574–578. doi: 10.1111/j.1471-8286.2007.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Futschik A, Schlötterer C. The next generation of molecular markers from massively parallel sequencing of pooled DNA samples. Genetics. 2010;186:207–218. doi: 10.1534/genetics.110.114397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kofler R, Pandey RV, Schlötterer C. PoPoolation2: identifying differentiation between populations using sequencing of pooled DNA samples (Pool-Seq) Bioinformatics. 2011;27:3435–3436. doi: 10.1093/bioinformatics/btr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kofler R, et al. Popoolation: a toolbox for population genetic analysis of next generation sequencing data from pooled individuals. PLoS One. 2011;6 doi: 10.1371/journal.pone.0015925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv Prepr arXiv. 2013;0:3. [Google Scholar]
- 93.Salse J, Abrouk M, Murat F, Quraishi UM, Feuillet C. Improved criteria and comparative genomics tool provide new insights into grass paleogenomics. Brief Bioinform. 2009;10:619–630. doi: 10.1093/bib/bbp037. [DOI] [PubMed] [Google Scholar]
- 94.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 95.Li L, Stoeckert CJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 97.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 98.Han MV, Thomas GWC, Lugo-Martinez J, Hahn MW. Estimating gene gain and loss rates in the presence of error in genome assembly and annotation using CAFE 3. Mol Biol Evol. 2013;30:1987–1997. doi: 10.1093/molbev/mst100. [DOI] [PubMed] [Google Scholar]
- 99.Salse J. Ancestors of modern plant crops. Curr Opin Plant Biol. 2016;30:134–142. doi: 10.1016/j.pbi.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 100.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Borowiec ML. AMAS: a fast tool for alignment manipulation and computing of summary statistics. PeerJ. 2016;4:e1660. doi: 10.7717/peerj.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hochberg Y, Benjamini Y. More powerful procedures for multiple statistical significance testing. Stat Med. 1990;9:811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
- 103.Sasaki T, Massaki N, Kubo T. Wolbachia variant that induces two distinct reproductive phenotypes in different hosts. Heredity (Edinb) 2005;95:389–393. doi: 10.1038/sj.hdy.6800737. [DOI] [PubMed] [Google Scholar]
- 104.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 105.Lee E, et al. Web Apollo: a web-based genomic annotation editing platform. Genome Biol. 2013;14:R93. doi: 10.1186/gb-2013-14-8-r93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kalderimis A, et al. InterMine: extensive web services for modern biology. Nucleic Acids Res. 2014;42:W468–72. doi: 10.1093/nar/gku301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The oak haploid genome assembly and corresponding annotation have been deposited in the European Nucleotide Archive under project accession code PRJEB19898. Other sequence release data are indicated in Supplementary Tables 1, 13, 14 and 19, and Supplementary Data Set 10. Data (including intermediate genome assemblies, vcf files used to detect somatic mutations and estimate heterozygosity) are available at the oak genome web site hosted as a permanent resource by Inra (http://www.oakgenome.fr/).