Abstract
Evolution depends on mutations. For an individual genotype, the rate at which mutations arise is known to increase with various stressors (stress-induced mutagenesis – SIM) and decrease at high final population density (density-associated mutation-rate plasticity – DAMP). We hypothesised that these two forms of mutation rate plasticity would have opposing effects across a nutrient gradient. Here we test this hypothesis, culturing Escherichia coli in increasingly rich media. We distinguish an increase in mutation rate with added nutrients through SIM (dependent on error-prone polymerases Pol IV and Pol V) and an opposing effect of DAMP (dependent on MutT, which removes oxidised G nucleotides). The combination of DAMP and SIM result in a mutation rate minimum at intermediate nutrient levels (which can support 7×108 cells ml-1). These findings demonstrate a strikingly close and nuanced relationship of ecological factors – stress and population density – with mutation, the fuel of all evolution.
Keywords: fluctuation test, antibiotic resistance, fitness, rifampicin, nalidixic acid, mutagenesis
Introduction
How and why the rate of spontaneous genetic mutation varies is a fundamental and enduring biological issue (Lynch et al 2016). Mutation rate can vary both among species (Sung et al 2012) and within a genotype (Maharjan and Ferenci 2017b). Intra-genotypic variation can depend upon stressful environmental conditions such as nutrient limitation, growth-rate reduction, high osmotic pressure, low pH, extreme shifts in temperature or various DNA damaging agents (Foster 2007). In these environments double stranded breaks can induce stress responses that in turn increase mutation rates via DNA polymerases with different error rates (Rosenberg et al 2012) – a phenomenon known as stress-induced mutagenesis (SIM).
Recently, we found that across microbes, the mutation rate of a particular genotype is closely associated with the final density to which the population grows (that is, the carrying capacity of the environment divided by its volume) (Krašovec et al 2017). In this so-called Density-Associated Mutation-rate Plasticity (DAMP), bacterial and yeast populations show a power law (log-log linear) reduction in mutation rate with D when grown in a defined minimal medium with glucose as the sole carbon source (Krašovec et al 2014, Krašovec et al 2017). DAMP and SIM modify mutation rates in Escherichia coli via different genetic pathways. DAMP requires a Nudix hydrolase protein, whose primary role is degrading highly mutagenic 8-oxo-dGTP (Michaels and Miller 1992), while error-prone polymerase Pol IV is not involved in DAMP (Krašovec et al 2017). Differences in the underlying mechanism and the fact that the densest populations, experiencing the highest stress, show the lowest mutation rates, suggest that DAMP is not obviously associated with stress.
Growth in minimal medium on a single carbon source does not, however, reflect the environmental complexity or range of population densities experienced by many species. E. coli population density in host environments varies over 5 orders of magnitude among host species, and can be higher than 109 colony forming units per gram of faeces (reviewed in (Tenaillon et al 2010)). As the highest population densities, with the greatest competition, rely on high nutrient availability, we reasoned that the addition of nutrients to minimal nutrient environments could indirectly increase both population densities and the level of stress. We therefore hypothesised that effects of density and stress on mutation rates, DAMP and SIM respectively, will act in opposition to one another across such a nutrient gradient – DAMP decreasing mutation rate and SIM increasing it as nutrients and final population density increase.
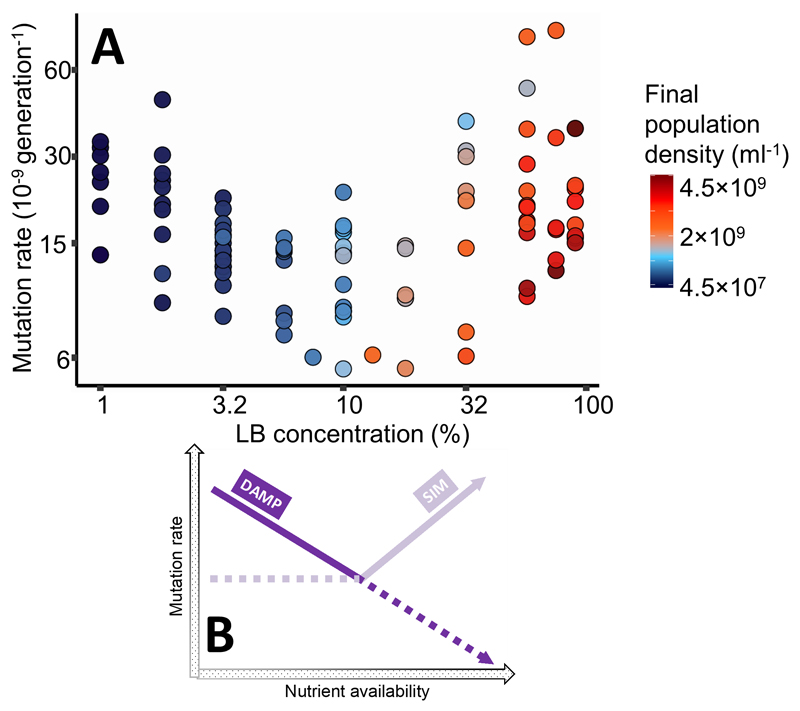
Here we test this hypothesis by determining E. coli mutation rates across a nutrient gradient, while genetically manipulating DAMP and SIM independently. As hypothesised, we identify genetically separable and opposed associations of mutation rate with nutrient availability – a negative association requiring mutT (DAMP) and a positive association requiring polymerases IV and V (dinB and umuC respectively; SIM). We find that these associations combine to minimise average mutation rates in environments with intermediate nutrient availability and final population density (Fig. 1B).
Figure 1.
Effect of nutrient availability on mutation rate A) to rifampicin resistance in wild-type E. coli K-12 MG1655 (N=97). Cells were grown in Davis minimal medium mixed with 1% to 90% of lysogeny broth (LB) medium. Colours represent final population density measured by colony forming units (see Fig. S2 for details). See Figure S4 for a plot of mutation rate directly against final population density (measured by ATP-based assay) and S10 for mutation rates co-estimated with the relative fitness of resistant mutants and model S-I in Supplementary Information for analysis. Note the non-linear axes. B) Schematic of mechanisms involved in mutation rate plasticity (density-associated mutation rate plasticity, DAMP, and stress-induced mutagenesis, SIM). At low nutrient availabilities, DAMP is present and SIM is absent. At higher nutrient availabilities, SIM becomes dominant. Solid lines correspond to the mutation rate plasticity measured in Fig. 1A. Genetically removing one or the other mechanism (Figs. 2-3) reveals mutation rate plasticity indicated by the dotted lines.
Results
We assayed mutation rates to rifampicin resistance using fluctuation tests in E. coli K-12 MG1655 grown across a gradient of nutrient availability: a range of concentrations (1-90% v/v) of lysogeny broth (LB) mixed with Davis minimal (DM) medium (LB/DM). We find that the relationship of mutation rate to LB concentration is non-linear (Fig. 1A, likelihood ratio test of a quadratic effect of log nutrient availability on log mutation rate: N=97, LR8,7=105, P=1.2×10-24, model S-I in Supplementary Information). Repeating this experiment using a different marker of mutation (nalidixic acid resistance) gives a similar non-linear relationship (Fig. S1). Mutation rate to both rifampicin and nalidixic acid resistance decreases as LB/DM is increased from 1% to 10% LB (increasing final population density, D, from 4.5×107 to 1×109 cells ml-1 Figs. S3-S5). This is comparable to DAMP in DM with glucose (Krašovec et al 2014, Krašovec et al 2017). However, mutation rate increases again in richer media, with 90% LB reaching similar or higher mutation rates than in 1% LB.
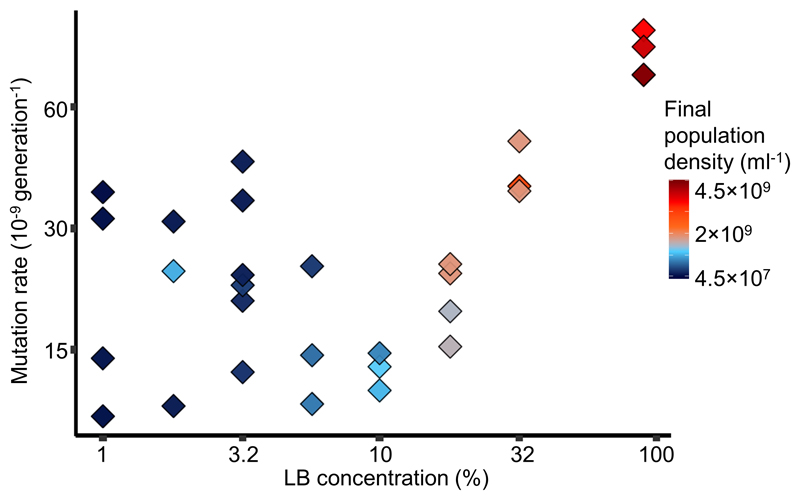
We next asked whether the increase in mutation rate at higher nutrient availability is genetically separable from the decrease in mutation rate due to DAMP. DAMP in E. coli requires the 8-oxo-dGTP diphosphatase MutT protein, meaning that, in minimal medium with glucose, the mutation rate in a ΔmutT mutant does not decrease with increased nutrient concentration (Krašovec et al 2017). We therefore performed fluctuation tests to nalidixic acid resistance in LB/DM with a ΔmutT mutant. We find that in LB/DM, as in DM with glucose, mutation rate in ΔmutT shows no relationship with increased nutrients or final population density below 10% LB (Fig. 2). However, even more clearly than in the wild-types (both MG1655 [Fig. 1A] and the immediate parent of the ΔmutT mutant [Fig. S6]), mutation rate of the ΔmutT mutant increases with the nutrient availability above 10% LB (final population density of ~1×109 cells ml-1). The only other E. coli mutant reported not to exhibit DAMP is E. coli K-12 MG1655 ΔluxS (Krašovec et al 2014). However, the deficiency in DAMP of this mutant is functionally complemented by added aspartate (Krašovec et al 2014), and LB is a medium rich in amino acids (Sezonov et al 2007). If variation in mutation rate at 10% LB/DM and below is the same phenomenon as DAMP, we expect this mutant strain to behave more similarly to wild-type than the ΔmutT mutant. We find that the ΔluxS mutant’s mutation rate is indistinguishable from the wild-type MG1655 across LB/DM environments (Fig. S7, N=167, likelihood ratio tests of the luxS deletion on the interaction between quadratic response of mutation rate to LB concentration and genotype [N = 167, LR11,12=2, P=0.16], and on the fixed effect of genotype [N = 167, LR11,10=1.4, P=0.24]).
Figure 2.
Effect of nutrient availability on mutation rate to nalidixic acid resistance in cells without DAMP (ΔmutT, N=30). Cells were grown in Davis minimal medium mixed with 1% to 90% of lysogeny broth (LB) medium. Colours represent final population density measured by colony forming units (see Fig. S2 for details). See Fig. S6 for a plot of mutation rate directly against final population density and S11 for mutation rates co-estimated with the relative fitness of resistant mutants. Nalidixic acid resistance is used as the marker here rather than rifampicin resistance (as in Fig. 1) because the mutation rate of this ΔmutT strain is too high to assay with rifampicin (for which at least 69 different resistance mutations are known (Garibyan et al 2003), the ‘target size’ for nalidixic acid resistance is much smaller). Directly comparable data for wildtype MG1655 mutation rate to nalidixic acid resistance is shown in Fig. S1. Note the non-linear axes.
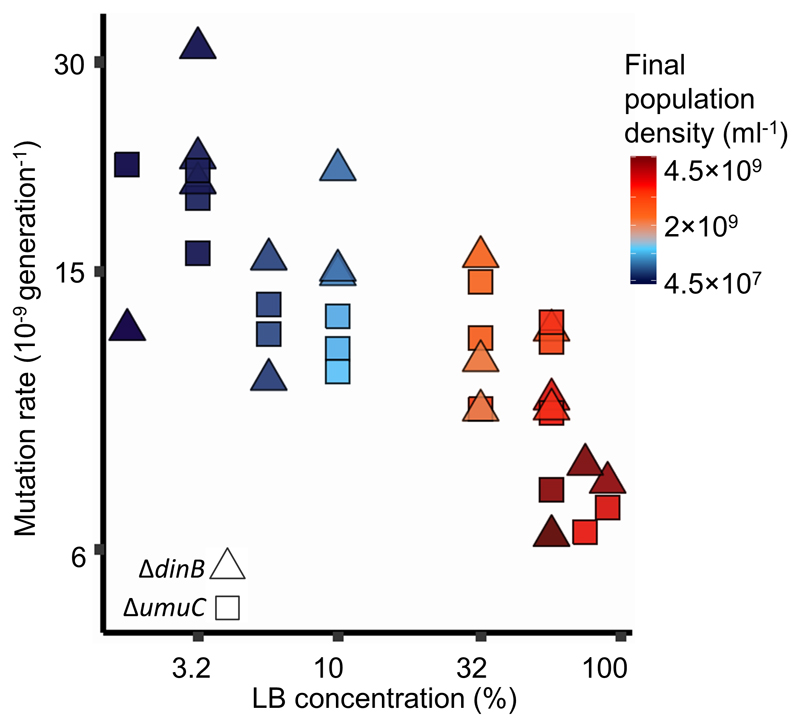
The fact that mutation rate increases at high LB concentrations in a ΔmutT mutant (Fig. 2), where DAMP is absent, suggests that high LB concentrations increase mutation rate via a DAMP-independent mechanism. We hypothesised that higher LB concentrations increase the level of stress (e.g. by promoting competition), thereby causing error-prone polymerases Pol IV and Pol V (coded by dinB and umuCD respectively) to increase the mutation rate at very high LB concentrations. We tested this hypothesis by estimating mutation rates to rifampicin resistance in E. coli ΔdinB and ΔumuC mutants growing in LB/DM. We find that, unlike E. coli MG1655 (Fig. 1A) and ΔmutT (Fig. 2), mutation rates of the ΔdinB and ΔumuC deletants (Fig. 3, Model S-III in Supplementary Information) decrease with increasing nutrients above 10% LB (above final population densities of 7×108 cells ml-1, Fig. S8). This continued decrease indicates that these polymerases are required for the rise in mutation rates as nutrients increase, and that DAMP continues to affect mutation rates at high nutrient levels.
Figure 3.
Effect of nutrient availability on mutation rates to rifampicin resistance in cells without error-prone polymerases Pol IV (ΔdinB, N=18, triangles) and Pol V (ΔumuC, N=18, squares). Cells were grown in Davis minimal medium mixed with 1% to 90% of lysogeny broth (LB) medium. The mutation rates of the two strains are not distinguishable (likelihood ratio test of the effect of genotype [N=36, LR9,8=2.6, P=0.11]). There is no evidence of a non-linear relationship of log mutation rate with nutrient availability (likelihood ratio test of a quadratic effect [N=36, LR8,7=0.15, P=0.70]), but there is a highly significant linear effect of nutrient availability (likelihood ratio test of linear effect [N=36, LR7,6=45, P=1.6×10-11]), see model S-III in Supplementary Information. Colours represent final population density measured by colony forming units (see Fig. S2 for details). See Figure S8 for an equivalent plot using final population density and S12 for mutation rates co-estimated with the relative fitness of resistant mutants. Note the non-linear axes.
The fitness effects of resistance mutations are known to be variable among non-selective environments, particularly for rifampicin (Maharjan and Ferenci 2017a). This variation has the potential to give artefactual differences in mutation rates among environments. We therefore estimated the fitness effects of resistance mutations in the fluctuation tests reported in Figs. 1-3 (Fig. S9). As expected, resistance mutations were, on average, somewhat deleterious across nutrient environments for both rifamipcin and nalidixic acid. Surprisingly, the average effect was least deleterious in intermediate nutrient environments. This suggests that, if anything, mutation rates in intermediate nutrient environments are over-estimated, relative to high and low nutrient environments. Thus, the results reported in Figs. 1-3 are robust to environmentally-dependent fitness effects of resistance mutations (Fig. S10-12).
Discussion
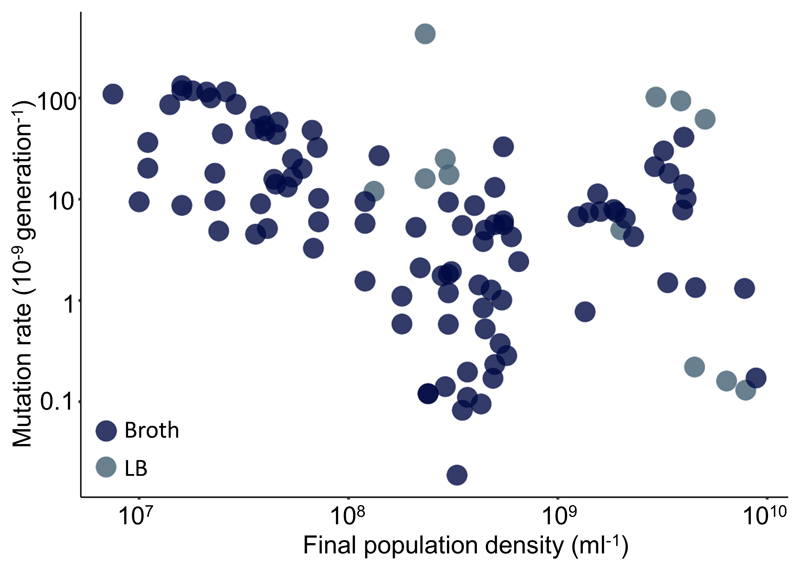
Our previous work on density associated mutation rate plasticity (DAMP,(Krašovec et al 2017) contained a paradox. In the laboratory, E. coli displays a substantial and highly significant decrease in mutation rate associated with final population density, much more so than the related bacterium Pseudomonas aeruginosa PAO1. Yet, in the published literature, of all 56 species with appropriate data (including P. aeruginosa), the one with least negative association was E. coli. Here we have resolved that paradox. We have shown how two mechanistically independent plastic processes act on the mutation rate: DAMP – apparent at lower final population densities (< 7×108 cells ml-1) – causing mutation rate to decrease with increasing nutrient concentration; SIM – apparent at higher final population densities (> 1×109 cells ml-1) – causing mutation rate to increase with nutrient concentration. E. coli is the organism whose mutation rate has, across the last 75 years of literature, been measured across the broadest range of final population densities (7.5×106 to 8.9×109 cells ml-1) (Krašovec et al 2017). This means that, like Fig. 1A (and Fig. S4), the published literature includes a range of rich media and shows a minimum in E. coli’s mutation rate at around 7×108 cells ml-1 (Fig. 4). This explains why attempting to fit a linear trend to this data does not yield a steep negative relationship (Krašovec et al 2017). It is also consistent with DAMP acting at low final population densities and SIM at high densities across diverse published studies (111 individual estimates across 12 studies), as we find here in a single, controlled study. We cannot currently say what particular aspect(s) or component(s) of the rich medium used here is/are most important in effecting the observed mutation rate changes. However, the fact that a similar pattern is seen in published data using different ‘broth’ media suggests that, like DAMP itself (Krašovec et al 2017), the interplay of opposing pressures on mutation rate that we have dissected here (Fig. 1B) is not unique to the particular media we used.
Figure 4.
Distribution of E. coli mutation rates estimated in rich media and published from 1943 to 2016 (re-plotted from reference (Krašovec et al 2017), N=111). Broth – rich nutrient medium, LB – rich lysogeny broth medium. These data also include a range of strains and phenotypic markers (see Supplementary Data File). Note the non-linear axes.
It is perhaps surprising that both error-prone polymerases Pol IV and Pol V are required for the increase in mutation rate observed here at high LB concentrations (Fig. 3) since, in some cases, only pol IV is required for SIM (Galhardo et al 2009). Nonetheless, in SIM, increases of base substitutions (as opposed to insertion or deletion events), which are the primary form of mutation underlying the rifampicin resistance assayed here (Garibyan et al 2003), do frequently depend on both of these polymerases (Al Mamun et al 2012, Petrosino et al 2009, Shee et al 2011). Such different molecular processes leave different signatures in the precise spectrum of mutations observed. However, the dependence of such mutational spectra on nutrient environment has recently been shown to be very complex (Maharjan and Ferenci 2017b). It is thus unlikely that such spectra will explain the mechanisms of DAMP, SIM and potentially other environmental plasticity in mutation rates, which vary simultaneously, via different molecular mechanisms, even across a simple nutrient gradient. However, the sort of genetic separation of environmental effects reported here may in future help understand the complexities of those spectra, which have the potential to affect the course of evolution (Couce et al 2013).
It seems likely a priori that the dynamics of population growth and cell division, which differ among nutrient environments, are involved in the mutation rate changes observed here. For instance, environmental differences that affect growth rate will in turn affect ploidy (Pecoraro et al 2011), which can affect mutation rate estimates (Sun et al 2018). Therefore, we cannot exclude the possibility that the effects of either DAMP or SIM on mutation rate considered here are mediated by some aspect(s) of the culture cycle that differ across different nutrient environments. Such dynamics are largely inaccessible to fluctuation tests, as used here, or indeed other standard methods of assaying mutation rate (Foster 2006) that consider at least one full population growth cycle. Continuous culture, specifically continuous culture at fixed population density, a turbidostat (Myers and Clark 1944), may be a useful tool with which to factor out some of these dynamics. However, better still may be to examine the mechanistic detail of such dynamics directly using single-cell mutation monitoring approaches (Elez et al 2010, Uphoff et al 2016).
Variation in mutation rate among members of a population can itself provide evolutionary advantages (Alexander et al 2017), and modulating mutation rate in response to the environment could hypothetically allow organisms to optimise their rate of adaptation (Belavkin et al 2016). Such ‘optimal’ variation involves minimising mutation rates at high fitness, but allowing them to increase away from fitness peaks. However, environmental cues do not give direct information about an individual’s fitness. An individual simply receives information about the levels of particular molecules in the environment. These could give information, for instance, about availability of a particular nutrient or about population density, and that information may be linked to mechanisms involved in the plastic control of mutation rate, e.g. (Krašovec et al 2014). But a cue indicative of high population density could be an indicator of an individual having high fitness (if it is part of a successful clone), or high competition and therefore low individual fitness. Similarly, un-utilised nutrients may be indicative of a benign environment and therefore high fitness, or of a clone that has been unable to utilise resources and therefore low individual fitness. Organisms receive many environmental cues and may therefore be able to parse them to give a clear picture of the competitive environment, enabling appropriate responses (Cornforth and Foster 2013). How far this occurs in terms of mutation rate is unclear. And adaptive explanations are probably unnecessary to explain the existence of SIM and perhaps DAMP, given more direct and/or non-adaptive explanations (Lynch et al 2016, MacLean et al 2013). Nonetheless, it is reasonable to speculate about the evolutionary effects of these plastic mutation rate traits, whatever their origins, either evolutionarily or mechanistically in terms of environmental cues. The effect of minimising mutation rate in intermediate nutrient (and final population density) environments (Figs. 1A, S1 and S4) may be to minimise evolutionary change for organisms that are doing well in a relatively benign nutrient environment, but without excessive competition, which is potentially advantageous (Belavkin et al 2016).
Without clearer evidence around the evolution of these traits and their effects on evolution beyond fluctuation tests, any reasoning about their adaptive effects remains speculative. Nonetheless, final population density and nutrient availability are focal points of microbial ecological competition (Hoek et al 2016, Oliveira et al 2015). Microbes have evolved numerous strategies to sense and increase acquisition of resources (Hibbing et al 2010), and they possess efficient ways of sensing population density (Xavier and Bassler 2005). A threshold population density (known as quorum) is often required to regulate a diverse array of physiological activities (Walters and Sperandio 2006), many of which promote stress tolerance (Williams et al 2007). SIM is not known to depend on quorum-sensing. Although DAMP requires LuxS, central to autoinducer 2 quorum sensing, in minimal media (Krašovec et al 2014), we do not find any role for LuxS in the conditions studied here (Fig. S7). This is consistent with a metabolic rather than quorum sensing effect on DAMP (Krašovec et al 2014). Nonetheless, mutation rate can respond to the luxS genotype of a co-cultured strain (Krašovec et al 2014) indicating that, in addition to the nutrient environment, studied here, biotic environment also has a role in determining an organism’s mutation rate. The fact that mutation rate responds in such complex ways to these diverse environmental factors indicates that, for the de novo evolution of traits such as antibiotic resistance, ecological circumstances and evolutionary outcomes are tightly linked.
Materials and Methods
Strains used in this study
Escherichia coli K-12 strain KX1228 (ΔluxS) was derived from the wild-type K-12 MG1655 (luxS+) (Xavier and Bassler 2005). E. coli ΔmutT mutant is part of Keio collection (Baba et al 2006) designated as JW0097-1 (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔmutT790::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514). E. coli ΔdinB mutant is part of Keio collection designated as JW0221-1 (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔdinB749::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514. E. coli ΔumuC mutant is part of Keio collection designated as JW1173-1 (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, ΔumuC773::kan, rph-1, Δ(rhaD-rhaB)568, hsdR514). Parent of Keio collection is an E. coli strain BW25113 (F-, Δ(araD-araB)567, ΔlacZ4787(::rrnB-3), λ-, rph-1, Δ(rhaD-rhaB)568, hsdR514).
Media
We used MilliQ water for all media. Strains were grown with shaking (250 rpm) at 37°C in lysogeny broth (LB) medium (10g NaCl, 5g yeast extract and 10g tryptone per litre [l-1]) mixed with Davis minimal (DM) medium (0.5g C6H5Na3O7×2H20, 1g (NH4)2SO4, 2g H2KO4P and 7g HK2O4P×3H2O l-1). 100mg L-1 MgSO4×7H20 (406 µmol) and 4μg L-1 thiamine hydrochloride were added to DM after autoclaving. We used 1% to 90%LB with a content of Mg2+ ranging from 40 to 35.5 µmol respectively, assuming that LB contains on average 35 µmol l-1 of Mg2+ (Papp-Wallace and Maguire 2008). Selective tetrazolium arabinose agar (TA) medium (10g tryptone, 1g yeast extract, 5g NaCl, 3g arabinose and 0.05g 2,3,5-Triphenyl-tetrazolium chloride l-1) was supplemented with freshly prepared rifampicin (50µg ml-1) or nalidixic acid (30µg ml-1). For all cell dilutions sterile saline (8.5 g l-1 NaCl) was used. All media were solidified as necessary with 15 g l-1 of agar (Difco).
Fluctuation tests
We did fluctuation tests with Escherichia coli as already explained (Krašovec et al 2014, Krašovec et al 2017). In short, strains were first inoculated from frozen stock and grown in liquid LB medium at 37°C and then transferred to non-selective liquid media (LB or DM with glucose) and allowed to grow overnight shaking at 37°C. E. coli cells were again diluted into fresh LB/DM, giving a mean initial population size (N0) of 2,373 (range 1.5×102 – 1.3×104). Various volumes (0.35–1 ml) of parallel cultures were grown to saturation for 24 hours at 37°C in 96 deep-well plates. The position of each culture on a 96-deep-well polypropylene plate was chosen randomly. Final population size (Nt) was determined by colony forming units (CFU) where appropriate dilution was plated on solid non-selective TA medium. Final population density (D) was estimated was determined by two independent techniques using CFU and an ATP based assay: luminescence (LUM) was measured using a Promega GloMax luminometer and the Promega Bac-Titer Glo kit, according to manufacturer's instructions. We measured luminescence of each culture 0.5 and 510 seconds after adding the Bac-Titer Glo reagent and calculated net luminescence as LUM = luminescence510s – luminescence0.5s. Each estimate of D and Nt was averaged across 3 independent cultures. Evaporation (routinely monitored by weighing plate before and after 24h incubation) was accounted for in the Nt value determined by CFU and was also used in statistical modelling as a variance covariate. We obtained the observed number of mutants resistant to rifampicin or nalidixic acid, r, by plating the entirety of remaining cultures onto solid selective TA medium (4.5cm plates in Figs. 1-2 and 9cm plates in Fig. 3) that allows spontaneous mutants to form colonies. Plates were incubated at 37°C and mutants were counted at the earliest possible time after plating. For rifampicin plates this was 44–48 hours, when nalidixic acid was used the incubation time was 68–72 hours.
For figures 1A, 2 and 3 we used 13, 3 and 4 independent experimental blocks, respectively, carried out on different days. Within an experimental block multiple 96-well plates were used. Any individual mutation rate estimate requires multiple parallel cultures, which were all carried out on a particular plate. For figures 1A, 2 and 3 the median (with interquartile range) of parallel cultures used per mutation rate estimate was 16 (21–16), 16 (16–16) and 16 (16–16), respectively.
Estimation of mutation rates
To estimate number of mutational events, m, from the observed number of mutants we employed the Ma-Sandri-Sarkar maximum-likelihood method implemented by the FALCOR web tool (Hall et al 2009). The mutation rate per cell per generation is calculated as m divided by the final population size, Nt, determined by CFU. This approach does not account for potentially important issues that may affect mutation rate estimates. Crucially, if there is a cost to carrying a resistance allele in the fluctuation test environment, this can result in an under-estimation of the mutation rate. This issue can be corrected for by co-estimating the average fitness effect of resistance mutations with the mutation number of mutational events (Zheng 2005). In addition, variation in Nt may also affect estimates and may also be accounted for (Ycart and Veziris 2014). We therefore co-estimated mutation rates and fitness effects, accounting for variability in Nt using the flan package in R (Adrien et al 2017), also setting the Winsorization parameter to remove the effects of ‘jackpots’ with un-countably large numbers of mutants (greater than 150 on 4.5cm plates and greater than 1000 on 9cm plates). Since the estimated fitnesses (Fig. S9) tend to reinforce the patterns seen in Fig. 1-3, we report the results of the simpler and more widely used calculations in the main text as being more conservative.
Statistical analysis
All statistical analysis was executed in R v3.3.1 (R Core Team 2016) and nlme v3.1 packages for linear mixed effects modelling (Pinheiro and Bates 2000). This enabled the inclusion within the same model of experimental factors (fixed effects), blocking effects (random effects) and factors affecting variance (giving heteroscedasticity) as described in the Supplementary Information. In all cases log2 mutation rates were used. Details of models and their fitting are given in the Supplementary Information: diagnostic plots in Supplementary figures S12-S14, ANOVA tables for each model are given in Supplementary Tables S3-S5. Code and data to reproduce the main text figures are given in the accompanying R script, and Supplementary Data File respectively. The content of the Supplementary Data File is explained in Supplementary Table S2.
Supplementary Material
Supplementary Information is available at ISME Journal’s website.
Acknowledgments
We thank Karina B. Xavier for E. coli MG1655 strains. We thank César Aguilar for providing data. We thank Mark Foster and John Parfitt for technical assistance.
Funding: The BBSRC supported this work through grant BB/L009579/1, HR was supported by BB/M011208/1 and DG by BB/M020975/1. Additional support was provided to RK by EMBO fellowship ASTF 642 – 2014, Wellcome Trust Institutional Strategic Support Fund 105610/Z/14/Z and the University of Manchester School of Biological Sciences.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
References
- Adrien M, Rémy D, Stéphane D, Bernard Y. flan: An R package for inference on mutation models. The R Journal. 2017;9:334. [Google Scholar]
- Al Mamun AAM, Lombardo M-J, Shee C, Lisewski AM, Gonzalez C, Lin D, et al. Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science. 2012;338:1344–1348. doi: 10.1126/science.1226683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander HK, Mayer SI, Bonhoeffer S. Population heterogeneity in mutation rate increases the frequency of higher-order mutants and reduces long-term mutational load. Mol Biol Evol. 2017;34:419–436. doi: 10.1093/molbev/msw244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belavkin RV, Channon A, Aston E, Aston J, Krašovec R, Knight CG. Monotonicity of fitness landscapes and mutation rate control. J Math Biol. 2016;73:1–34. doi: 10.1007/s00285-016-0995-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornforth DM, Foster KR. Competition sensing: the social side of bacterial stress responses. Nat Rev Microbiol. 2013;11:285–293. doi: 10.1038/nrmicro2977. [DOI] [PubMed] [Google Scholar]
- Couce A, Guelfo JR, Blazquez J. Mutational spectrum drives the rise of mutator bacteria. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elez M, Murray AW, Bi LJ, Zhang XE, Matic I, Radman M. Seeing mutations in living cells. Curr Biol. 2010;20:1432–1437. doi: 10.1016/j.cub.2010.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Methods for determining spontaneous mutation rates. Methods Enzymol. 2006;409:195–213. doi: 10.1016/S0076-6879(05)09012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Stress-induced mutagenesis in bacteria. Crit Rev Biochem Mol Biol. 2007;42:373–397. doi: 10.1080/10409230701648494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galhardo RS, Do R, Yamada M, Friedberg EC, Hastings PJ, Nohmi T, et al. Dinb upregulation is the sole role of the SOS response in stress-induced mutagenesis in Escherichia coli. Genetics. 2009;182:55–68. doi: 10.1534/genetics.109.100735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garibyan L, Huang T, Kim M, Wolff E, Nguyen A, Nguyen T, et al. Use of the rpoB gene to determine the specificity of base substitution mutations on the Escherichia coli chromosome. DNA Repair. 2003;2:593–608. doi: 10.1016/s1568-7864(03)00024-7. [DOI] [PubMed] [Google Scholar]
- Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbrück fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek TA, Axelrod K, Biancalani T, Yurtsev EA, Liu J, Gore J. Resource availability modulates the cooperative and competitive nature of a microbial cross-feeding mutualism. PLoS Biol. 2016;14:e1002540. doi: 10.1371/journal.pbio.1002540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krašovec R, Belavkin RV, Aston JA, Channon A, Aston E, Rash BM, et al. Mutation rate plasticity in rifampicin resistance depends on Escherichia coli cell-cell interactions. Nat Commun. 2014;5:3742. doi: 10.1038/ncomms4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krašovec R, Richards H, Gifford DR, Hatcher C, Faulkner KJ, Belavkin RV, et al. Spontaneous mutation rate is a plastic trait associated with population density across domains of life. PLoS Biol. 2017;15:e2002731. doi: 10.1371/journal.pbio.2002731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, et al. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet. 2016;17:704–714. doi: 10.1038/nrg.2016.104. [DOI] [PubMed] [Google Scholar]
- MacLean RC, Torres-Barcelo C, Moxon R. Evaluating evolutionary models of stress-induced mutagenesis in bacteria. Nat Rev Gen. 2013;14:221–227. doi: 10.1038/nrg3415. [DOI] [PubMed] [Google Scholar]
- Maharjan R, Ferenci T. The fitness costs and benefits of antibiotic resistance in drug-free microenvironments encountered in the human body. Environ Microbiol Rep. 2017a;9:635–641. doi: 10.1111/1758-2229.12564. [DOI] [PubMed] [Google Scholar]
- Maharjan RP, Ferenci T. A shifting mutational landscape in 6 nutritional states: Stress-induced mutagenesis as a series of distinct stress input–mutation output relationships. PLoS Biol. 2017b;15:e2001477. doi: 10.1371/journal.pbio.2001477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels ML, Miller JH. The GO system protects organisms from the mutagenic effect of the spontaneous lesion 8-hydroxyguanine (7,8-dihydro-8-oxoguanine) J Bacteriol. 1992;174:6321–6325. doi: 10.1128/jb.174.20.6321-6325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers J, Clark LB. Culture conditions and the development of the photosynthetic mechanism. J Gen Physiol. 1944;28:103–112. doi: 10.1085/jgp.28.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, et al. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp-Wallace K, Maguire M. Magnesium transport and magnesium homeostasis. EcoSal Plus. 2008 doi: 10.1128/ecosalplus.5.4.4.2. [DOI] [PubMed] [Google Scholar]
- Pecoraro V, Zerulla K, Lange C, Soppa J. Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS One. 2011;6:e16392. doi: 10.1371/journal.pone.0016392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrosino JF, Galhardo RS, Morales LD, Rosenberg SM. Stress-induced β-lactam antibiotic resistance mutation and sequences of stationary-phase mutations in the Escherichia coli chromosome. J Bacteriol. 2009;191:5881–5889. doi: 10.1128/JB.00732-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinheiro JC, Bates DM. Mixed effects models in S and S-Plus. Springer; New York: 2000. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2016. [Google Scholar]
- Rosenberg SM, Shee C, Frisch RL, Hastings PJ. Stress-induced mutation via DNA breaks in Escherichia coli: A molecular mechanism with implications for evolution and medicine. Bioessays. 2012;34:885–892. doi: 10.1002/bies.201200050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sezonov G, Joseleau-Petit D, D'Ari R. Escherichia coli physiology in Luria-Bertani broth. J Bacteriol. 2007;189:8746–8749. doi: 10.1128/JB.01368-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shee C, Gibson JL, Darrow MC, Gonzalez C, Rosenberg SM. Impact of a stress-inducible switch to mutagenic repair of DNA breaks on mutation in Escherichia coli. Proc Natl Acad Sci. 2011;108:13659–13664. doi: 10.1073/pnas.1104681108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Alexander HK, Bogos B, Kiviet DJ, Ackermann M, Bonhoeffer S. Effective polyploidy causes phenotypic delay and influences bacterial evolvability. PLoS Biol. 2018;16:e2004644. doi: 10.1371/journal.pbio.2004644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W, Ackerman MS, Miller SF, Doak TG, Lynch M. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci USA. 2012;109:18488–18492. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- Uphoff S, Lord ND, Okumus B, Potvin-Trottier L, Sherratt DJ, Paulsson J. Stochastic activation of a DNA damage response causes cell-to-cell mutation rate variation. Science. 2016;351:1094–1097. doi: 10.1126/science.aac9786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters M, Sperandio V. Quorum sensing in Escherichia coli and Salmonella. Int J Med Microbiol. 2006;296:125–131. doi: 10.1016/j.ijmm.2006.01.041. [DOI] [PubMed] [Google Scholar]
- Williams P, Winzer K, Chan WC, Cámara M. Look who's talking: communication and quorum sensing in the bacterial world. Philos Trans R Soc B-Biol Sci. 2007;362:1119–1134. doi: 10.1098/rstb.2007.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier KB, Bassler BL. Regulation of uptake and processing of the quorum-sensing autoinducer AI-2 in Escherichia coli. J Bacteriol. 2005;187:238–248. doi: 10.1128/JB.187.1.238-248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ycart B, Veziris N. Unbiased estimation of mutation rates under fluctuating final counts. PLoS One. 2014;9:e101434. doi: 10.1371/journal.pone.0101434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q. New algorithms for Luria-Delbrück fluctuation analysis. Math Biosci. 2005;196:198–214. doi: 10.1016/j.mbs.2005.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.