Abstract
Sorting endosomes (SEs) are the regulatory hubs for sorting cargo to multiple organelles, including lysosome-related organelles such as melanosomes in melanocytes. In parallel, melanosome biogenesis is initiated from SEs with the processing and sequential transport of melanocyte-specific proteins toward maturing melanosomes. However, the mechanism of cargo segregation on SEs is largely unknown. RNAi screening in melanocytes revealed that knockdown of Rab4A results in defective melanosome maturation. Rab4A-depletion increases vacuolar endosomes and disturbs the cargo sorting, which in turn lead to the mislocalization of melanosomal proteins to lysosomes, cell surface and exosomes. Rab4A localizes to the SEs and forms endosomal complex with AP-3 adaptor, Rabenosyn-5 effector and KIF3 motor, which possibly coordinates cargo segregation on SEs. Consistently, inactivation of Rabenosyn-5 or KIF3A/B phenocopied the defects observed in Rab4A-knockdown melanocytes. Further, Rabenosyn-5 associates with Rabaptin-5 or Rabip4/4’ and differentially regulate cargo sorting from SEs. Thus, Rab4A acts a key regulator of cargo segregation on SEs.
Keywords: Rab4A, AP-3, KIF3, Rabenosyn-5, Rabaptin-5, RabIP4’, sorting endosome and melanosome biogenesis
Introduction
Organelles of the endocytic system constantly mature into terminal organelles such as lysosomes in all cells and lysosome-related organelles (LROs) in specialized cells (Luzio et al., 2014; Raposo et al., 2007). Melanosomes are the LROs of melanocytes present in the skin and eye, which provide color and photoprotection. These organelles are derived by post-sequential trafficking of multiple melanosomal cargo from sorting/recycling endosomes (SEs/REs) to maturing melanosomes (Marks et al., 2013; Sitaram and Marks, 2012). For example, pre-melanosomal protein (PMEL) is segregated into intraluminal vesicles (ILVs) of SEs (also called stage I melanosomes) for fibril formation (matures to stage II) (van Niel et al., 2011); tyrosinase-related protein-1 (TYRP1), copper transporter (ATP7A) and other cargo are sorted into RE tubular structures by a BLOC-1-dependent transport mechanism (Setty et al., 2008; Setty et al., 2007); and tyrosinase (TYR) is sorted into endosomal vesicles by AP-3-dependent transport pathway (Theos et al., 2005) on SEs, which are then targeted to stage II for melanin synthesis (matures to stage III and IV) (Marks et al., 2013). This process is essential for the step-wise maturation of melanosomes from stage I to IV to avoid the pigment formation in SEs. However, the mechanism of melanocytic cargo sorting on SEs is poorly understood.
Domain organization and cargo sorting on endocytic membranes are predicted to be mediated by Rab GTPases (Rabs) and adaptor proteins (APs) (Bonifacino and Lippincott-Schwartz, 2003; Stenmark, 2009; Zerial and McBride, 2001). In general, Rabs recruit effector proteins, including kinesin motors and SNAREs, during vesicle budding/transport and membrane fusion, respectively (Bonifacino and Glick, 2004; Ohbayashi and Fukuda, 2012; Pfeffer, 2013). In contrast, APs such as AP-3 and AP-1 complexes have been shown to segregate both melanocytic and non-melanocytic cargoes on endosomes by binding to unique amino acid motifs in the cargo tails (Bonifacino and Traub, 2003; Park and Guo, 2014). Studies using live-cell imaging have reported the existence of AP-1- and AP-3-independent domains on SEs (D'Souza et al., 2014). Moreover, tetraspanin-like protein CD63 in melanocytes or Cos proteins in yeast are shown to regulate sorting of multiple cargoes on SEs (MacDonald et al., 2015; van Niel et al., 2011). Nevertheless, the mechanism by which melanosomal cargoes are segregated into subdomains on SEs is poorly studied. Additionally, several Rabs, such as Rab4, 5, 7, 9, 11 and 22 (Grant and Donaldson, 2009; Pfeffer, 2013; Stenmark, 2009; Zerial and McBride, 2001), and their multiple effectors or tethering proteins, like Rabenosyn-5, Rabaptin-5, Rabip4’, EEA1 and the HOPS complex, have been shown to localize to these domains (de Renzis et al., 2002; Deneka et al., 2003; Fouraux et al., 2004; Ivan et al., 2012; Kalin et al., 2015; Zhu et al., 2009) for the regulation of different cargo transport processes. Furthermore, Rab7 and 9 have been shown to control different transport steps during melanosome biogenesis by functioning on late endosomes (LEs) or melanosomes (Hida et al., 2011; Mahanty et al., 2016). Nonetheless, function of Rab4A in regulating the trafficking of melanocytic cargo during melanosome maturation has not been studied.
Rab4A has been shown to be involved in many cellular processes, including fast recycling of cargo to the cell surface (Mohrmann et al., 2002; van der Sluijs et al., 1992) and conversion of Rab5-postive early endosomes (EEs) to Rab11-positive REs (de Renzis et al., 2002; Sonnichsen et al., 2000). Interestingly, Rab4A independently binds to the Rab5A effectors such as Rabenosyn-5, Rabaptin-5 and Rabip4’ as well as to the AP-1 and AP-3 complexes (D'Souza et al., 2014; de Renzis et al., 2002; Deneka et al., 2003; Fouraux et al., 2004; Ivan et al., 2012; Vitale et al., 1998). However, the specific step of cargo trafficking/sorting in which Rab4A interacts with these multiple molecules is unclear. Moreover, Rab4A co-fractionates with the kinesin-2 motor protein, KIF3A/B and regulates endosomal positioning/distribution (Bananis et al., 2004). Recently, Rab4 has been shown to associate with either the KIF3A or KIF13A motors on anterograde transport vesicles in Drosophila and regulate synapse organization (Dey et al., 2017). Nevertheless, the importance of Rab4A-KIF3 interaction in endosomal organization or its role in organelle biogenesis is not well studied. Rab4A has also been shown to modulate autophagy directly (Talaber et al., 2014) or in response to mechanical membrane stretch (Yao et al., 2016) and has a role in exocytosis of phagosomes containing pathogenic bacteria (Takeuchi et al., 2015). Together these studies suggest that Rab4A either participates in multiple pathways by interacting with different effectors or forms an unique protein complex assembled on the endosomal membrane that regulates different transport steps.
In this study, we aimed to dissect the role of Rab4A in melanosome biogenesis by taking advantage of well-known melanocytic cargo transport steps between SEs/REs and maturing melanosomes. Our studies provide evidence that Rab4A acts as a key regulator in sorting multiple cargoes on SEs by forming an unique protein complex with AP-3, Rabenosyn-5 and KIF3A/B. Moreover, this complex associate with Rabaptin-5 to sort PMEL to stage II melanosomes and with Rabip4/4’ to sort TYRP1/TYR to REs in melanocytes. Importantly, our study showed that the absence of Rab4A expression blocks melanosome maturation at stage II and up-regulates melanophagosome formation, and alters the cargo sorting into exosomes. Thus, Rab4A is essentially required for cargo segregation on SEs, which possibly occurs by creating different endosomal domains using its multiple effector molecules.
Results
Rab4A is required for cargo sorting on SEs and melanocyte pigmentation
SEs act as the key intermediary organelles during biogenesis of melanosomes in melanocytes (Bissig et al., 2016; Jani et al., 2016; Marks et al., 2013) in addition to their role in cargo transport to the cell surface, Golgi apparatus or lysosomes as similar to other mammalian cells (Grant and Donaldson, 2009; Klumperman and Raposo, 2014). On the SE membrane, multiple melanocyte-specific cargoes must be segregated and transported through different routes to the melanosomes for sequential maturation from stage I to IV. However, the mechanism of cargo segregation on SEs is poorly understood. We hypothesized a role for small Rab GTPases and performed an RNAi screen using shRNAs (transfected transiently) against endo/late endosomal Rab proteins, Rab3A, 4A, 4B, 5A, 5B, 5C, 7A and 11A in wild-type mouse melanocytes (WT, melan-Ink) (Fig. S1A-B). We confirmed the gene knockdown (obtained 30 - 40% of gene depletion except in Rab5B sh, Fig. S1C) and analyzed the cells for following cellular phenotype. We predict that the reduced Rab expression mislocalizes melanocytic cargoes to the lysosomes for degradation following hypopigmentation of melanocytes. Visual quantification of pigmentation loss by bright-field microscopy (BFM) showed that more than 40% of Rab3A-, 4A-, 5A-, 7A- and 11A-depleted melanocytes have a hypopigmentation phenotype when compared to control cells (grey bars, Fig. S1A). Quantitative immunofluorescence microscopy (IFM) showed reduced TYRP1 and TYR intensities (indicative of their lysosomal degradation) in Rab3A, 4A, 5A and 11A-depleted melanocytes (Fig. S1A). Among these, Rab4A and 11A (but not 5A)-knockdown melanocytes displayed reduced levels of melanin content compared to control cells and their respective protein levels were also reduced in these cells (Fig. S1D). In contrast, the other melanosomal protein, PMEL was mislocalized to lysosomes in Rab4A-depleted melanocytes compared to control or other Rab-inactivated cells (Fig. S1B). Thus, we wanted to evaluate Rab4A’s role in the cargo transport pathways to melanosome.
Retroviral transduction of WT melanocytes with two different shRNAs (sh-1 and sh-2) specific to mouse Rab4A showed a severe pigmentation defect (arrows) compared to control shRNA-transduced melanocytes (Fig. 1A). Additionally, a large number of melanosome clusters (MC) that resembled the melanophagosomes (Boissy et al., 1987) (arrowheads, Fig. 1A) were also observed in Rab4A-depleted melanocytes (see below). Estimation of melanin pigments in Rab4A-knockdown cells showed a moderate reduction in melanin content compared to control melanocytes (Fig. S1E). However, the visual quantification of pigmented melanocytes during four constitutive experiments displayed 75-80% hypopigmented cells (similar to Fig. 1A) in Rab4A-inactivated compared to control condition (Fig. S1F). IFM and biochemical analyses showed that Rab4 staining (Fig. 1B), transcript (Fig. S1G) and protein levels (see Fig. 1E) were dramatically reduced in Rab4A-knockdown compared to control cells. Consistently, the corrected total cell fluorescence (CTCF) of Rab4 staining in Rab4A-depleted cells was notably reduced compared to control melanocytes (CTCF=1.6±0.2x106 AU, p=ns in Rab4A sh-1, 1.4±0.2x106 AU, p≤0.05 in Rab4A sh-2 and 2.1±0.2x106 AU in control cells) (Fig. 1B).
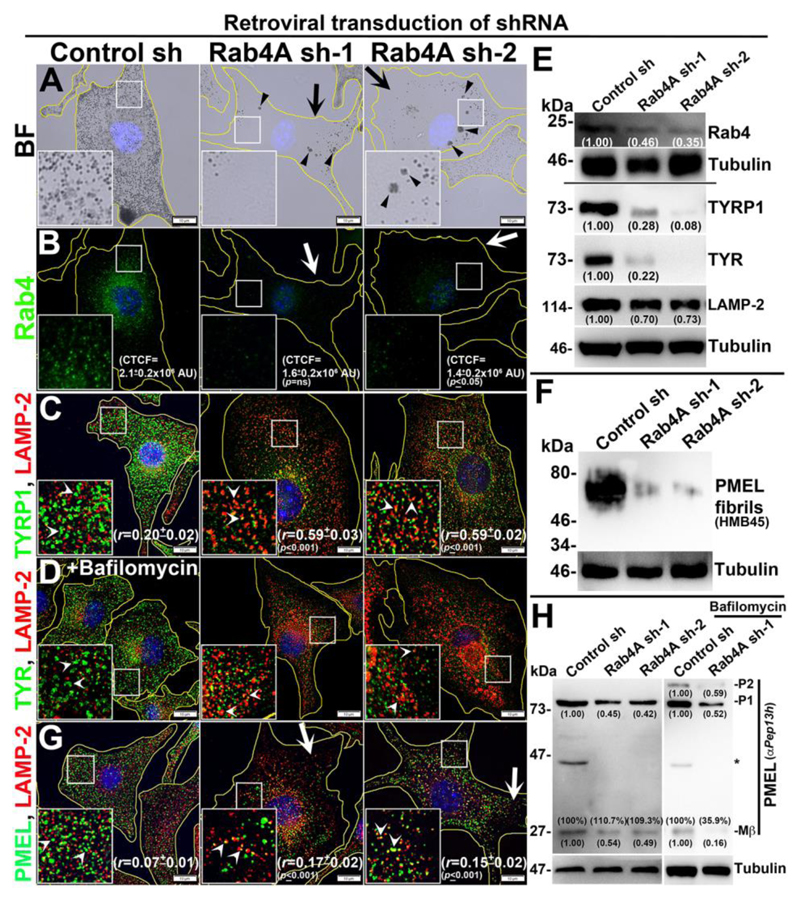
Fig. 1. Rab4A-knockdown affects melanocyte pigmentation and cargo transport to melanosomes.
BF (A) and IFM (B-D, G) images of Rab4A-depleted (sh-1 and sh-2) and control melanocytes. Black arrows and arrowheads indicate the pigmentation loss and melanosome clusters respectively (A). White arrows indicate the loss in fluorescence staining of Rab4 (B) or PMEL (G) in knockdown cells. White arrowheads (C, D, G) point to the cargo localization to lysosomes. In Panel D, cells were treated with bafilomycin. Nuclei are stained with Hoechst33258. The insets are a magnified view of the white boxed areas. The Pearson’s coefficient (r) between the two markers and CTCF values were indicated separately (mean±s.e.m.). Scale bars, 10 μm. (E, F, H) Immunoblotting analysis of Rab4, melanosomal and lysosomal proteins, and PMEL fibrils in Rab4A-knockdown cells and the tubulin was used as a loading control. P1/P2 and Mβ, full length/glycosylated ER-form and processed PMEL bands. *, non-specific band. Protein band intensities were quantified and indicated on the gels.
However, the gross localization of Rab5 was unaffected but its fluorescence intensity was slightly reduced in the peripheral cytosol of Rab4A-depleted melanocytes (arrows, Fig. S1H). Further, rescuing of Rab4A-depleted cells with GFP-Rab4A (susceptible to Rab4A shRNA) marginally restored the melanocyte pigmentation and cargo (TYRP1) stability (Fig. S1Ia). In line, transfection of Rab4A sh-2-knockdown melanocytes with GFP-Rab4Ash2R (resistant to shRNA and localizes as similar to Rab4AWT – data not shown) rescued melanocyte pigmentation and restored the TYRP1 protein levels and its localization to melanosomes (Fig. S1Ib, data not shown for cargo levels). These results indicate that the phenotypes observed in Rab4A sh cells are specific to Rab4A. IFM analysis further revealed that fluorescence intensities of both melanosome-localized TYRP1 and TYR were dramatically reduced in Rab4A-depleted compared to control melanocytes (Figs. 1C and S1J). In addition, localization of the remaining TYRP1 appeared as punctate structures and colocalized significantly with LAMP-2-positive compartments in Rab4A-knockdown melanocytes (r=0.59±0.03, p≤0.001 in Rab4A sh-1; 0.59±0.02, p≤0.001 in Rab4A sh-2 and 0.20±0.02 in control cells) (Fig. 1C). Moreover, the localization of TYRP1 to LAMP-2-positive structures was further enhanced with the treatment of bafilomycin A1 (vacuolar ATPase inhibitor) in Rab4A sh cells compared to control sh cells (see Fig. S1L), indicating that TYRP1 is targeted for lysosomal degradation upon Rab4A depletion in melanocytes. In contrast, TYR appeared as diffused cytosolic signal and its localization to LAMP-2-positive compartments was slightly restored upon treatment with bafilomycin in Rab4A-depleted cells (Figs. S1J and 1D). Consistent with these results, the activity of TYR measured through DOPA (3,4-dihydroxyphenylalanine) assay was completely reduced in Rab4A shRNA-transduced cells compared to control cells (Fig. S1K). As expected, both TYRP1 and TYR were localized as ring-like structures (Figs. 1C and S1J) and positive for BF melanosomes (not shown) in control cells. Immunoblotting analysis showed both TYRP1 and TYR protein levels were dramatically reduced and nearly restored to that of control cells upon lysosomal inhibition with bafilomycin in Rab4A-depleted melanocytes (Figs. 1E and S1M). These studies indicate that Rab4A-inactivation in melanocytes results in hypopigmentation due to mistargeting of both TYRP1 and TYR to lysosomes.
Next, we examined whether Rab4A-knockdown affects formation of PMEL fibrils and maturation of stage II melanosomes in melanocytes. IFM staining of PMEL (using the HMB45 antibody) in Rab4A-depleted melanocytes was dramatically reduced (arrows) and remaining PMEL partially colocalized with LAMP-2-positive lysosomes (arrowheads) compared to control cells (r=0.17±0.02, p≤0.001 in Rab4A sh-1, 0.15±0.02, p≤0.001 in Rab4A sh2 and 0.07±0.01 in control cells) (Fig. 1G). Consistent with these results, the total PMEL fibrils isolated from Rab4A-knockdown cells was drastically reduced compared to control cells (Fig. 1F). Previous studies have shown that PMEL (P1 form) undergo endosomal processing into Mα, Mβ and CTF (C-terminal fragment) (can be detected with αPep13h antibody as shown in (Rochin et al., 2013), and Mα further matures into fibrils in SEs/stage I (Bissig et al., 2016). Immunoblotting of Rab4A-knockdown cells showed reduced levels of both the P1 and Mβ forms compared to control cells (Fig. 1H). Surprisingly, the percentage of Mβ generated from the total PMEL was not affected in Rab4A-depleted compared to control cells (Fig. 1H), suggesting that proteolytic processing of PMEL is not affected upon Rab4A inactivation.
Further, the decreased levels of P1 and Mβ in Rab4A-depleted cells were not restored in the presence of bafilomycin (Fig. 1H) or protease inhibitors (not shown), indicating that PMEL is not targeted for lysosomal degradation; however, a cohort of PMEL was mislocalized onto lysosomal membranes (Figs. 1G and 1H). These results encouraged us to investigate the mislocalization of PMEL into other organelles. It is most likely that unprocessed PMEL would be segregated into the ILVs of LEs and then secreted as exosomes upon Rab4A-depletion in melanocytes. As predicted, the P1 form of PMEL was notably increased in the exosomes derived from Rab4A-knockdown melanocytes compared to control cells (Fig. S1N). Moreover, a small cohort of Rab4 was associated with the exosomes released from the control cells (Fig. S1N), similar as reported previously (Vidal and Stahl, 1993). These studies suggest that Rab4A-depletion in melanocytes causes mislocalization of full-length PMEL to the lysosomes and exosomes that possibly reduces the total cellular fibrils.
The sorting of proteolytically processed PMEL into ILVs is partially dependent on the non-melanocytic cargo CD63 (van Niel et al., 2011). We then examined whether Rab4A-knockdown affects trafficking of other endocytic cargoes such as CD63, LAMP-1/-2 and transferrin receptor (TfR) from SEs in melanocytes. IFM analysis showed that localization of GFP-CD63 to lysosomes was moderately increased in Rab4A-depleted melanocytes compared to control cells (r=0.40±0.04, p=ns in Rab4A sh-1 and 0.30±0.05 in control cells) (Fig. S1O). As expected, the colocalization between PMEL and GFP-CD63 was also increased in these cells (r=0.49±0.03, p≤0.001 in Rab4A sh-1 and 0.23±0.04 in control cells) (Fig. S1O). Further, quantitative IFM colocalization experiments showed that a pool of lysosomal proteins such as LAMP-2 was accumulated in the endosomal compartments upon Rab4A-knockdown compared to control melanocytes (Fig. S1P). Consistently, the protein levels of LAMP-2 were slightly reduced in Rab4A-knockdown cells (Fig. 1E). In contrast, the cell surface expression of LAMP-1, but not TfR, was moderately increased as similar to melanocytic cargo PMEL and TYRP1 (despite a slight increase in the total protein levels of LAMP-1 and TfR) in Rab4A shRNA cells compared to control cells (Fig. S1Q). These studies indicate a slight defect in the trafficking of lysosomal proteins but not fast recycling cargo upon Rab4A-depletion in wild-type melanocytes. Overall, these studies illustrate that Rab4A controls trafficking of structural melanosome cargoes from SEs to melanosomes and thus form a critical regulator of melanosome biogenesis.
Rab4A-depletion in melanocytes increases accumulation of enlarged endosomes and alters the stages of melanosome biogenesis
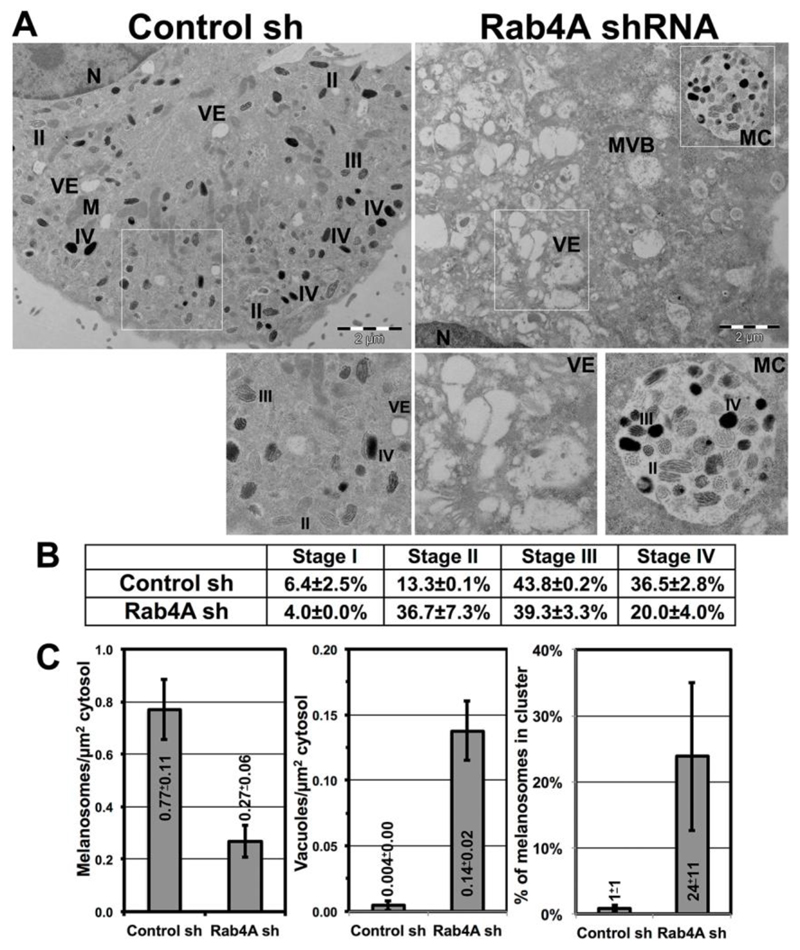
Rab4A-depletion possibly alters endosomal morphology/dynamics in wild-type melanocytes and thus mislocalizes the melanocytic cargo. Electron microscopy (EM) analysis showed enrichment of enlarged vacuolar structures in Rab4A-knockdown melanocytes (Fig. 2A right panel and inset VE); whereas, in control melanocytes, all melanosome biogenesis stages (II to IV) were observed (Fig. 2A left panel and inset). Quantification of the data demonstrated that percentage of stage II melanosomes was dramatically increased (mostly present in the phagosomes, see below) in Rab4A-depleted compared to control melanocytes (13.3±0.1% in control shRNA and 36.7±7.3% in Rab4A shRNA cells) (Fig. 2B). Concurrently, the percentage of Stage IV melanosomes was notably reduced (also present in phagosomes, see below) in Rab4A-knockdown compared to control cells (Fig. 2B). As expected, the number of melanosomes or vacuoles per μm2 of cytosol was reciprocal in Rab4A shRNA compared to control shRNA cells (Fig. 2C). Surprisingly, melanosomes in Rab4A-knockdown cells formed clusters (referred to here as melanosomes clusters, MCs) resembling melanophagosome structures (Fig. 2A inset MC) (Boissy et al., 1987), similar to the clusters observed in cells using BFM (arrowheads, Fig. 1A). Moreover, IFM analysis showed a cohort of LC3 puncta (but not LAMP-2) were associated with these MCs (Fig. S1R). Notably, the percentage of melanosomes in MCs was increased in Rab4A-depleted cells compared to control melanocytes (Fig. 2C). Consistent with these results, the cultured medium turned black during the initial stages (2 to 3 days) of Rab4A-knockdown compared to control melanocytes (data not shown), suggesting that these MCs possibly undergo exocytosis. However, the mechanism by which Rab4A-depletion increases the formation of these MCs is unknown. We hypothesized that Rab4A-deficiency possibly alters the autophagy in these cells (Talaber et al., 2014; Yao et al., 2016). These results indicate that Rab4A-knockdown results in enlarged endosomes that likely alter the cargo segregation on SEs and melanosomes intermediates.
Fig. 2. Rab4A-depletion increases vacuolar endosomes and inhibits melanosome maturation in melanocytes.
(A-C) Electron microscopy analysis of control and Rab4A-knockdown melanocytes. M, mitochondria; MVB, multi-vesicular bodies; MC, melanosome cluster; N, nucleus; VE, vacuolar endosomes; and II/III/IV, stages of melanosomes. Scale bars, 2 μm. Table represents the percent quantification of melanosome stages (mean±s.e.m., detailed in materials and methods) in both conditions (B). Significant changes in the values were highlighted as bold. Melanosomes or vacuoles per μm2 of cytosol, or % of melanosomes in a cluster was plotted (mean±s.e.m.) for both the conditions (C).
Rab4A associates with Rab5A-shared effectors and provides the specificity to cargo segregation on SE membranes
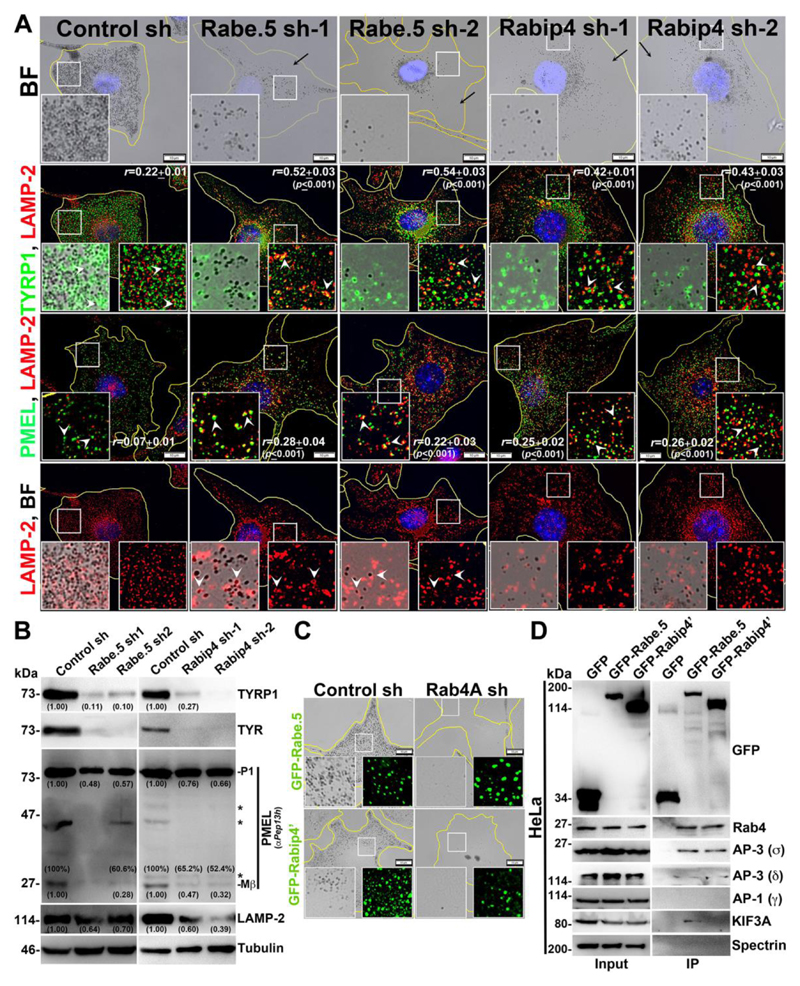
As observed, Rab4A regulates trafficking of different melanosomal and lysosomal cargoes; however, Rab4A alone may not be sufficient to segregate the cargo into subdomains on SEs. We hypothesized that Rab4A-Rab5A shared effectors possibly play a role during this process. Studies have shown that Rab4A associates with many endosomal effectors in which Rabenosyn-5, Rabip4/4’ (4’ is the longer isoform of 4) and Rabaptin-5 are either recruited by or associate with Rab5A on endosomal membranes (Fig. S2A, referred to here as Rab4A-Rab5A shared effectors) (de Renzis et al., 2002; Fouraux et al., 2004; Mattera et al., 2003). Additionally, Rabenosyn-5 and Rabip4/4’ possess a FYVE domain (conserved in Fab1, YOTB, Vac1 and EEA1 proteins) that binds to PI3P (phosphatidylinositol 3-phosphate) lipids on the endosomal membranes (Mari et al., 2001; Nielsen et al., 2000). Nevertheless, the role of these shared effectors in cargo transport to melanosomes is unknown. Sequential knockdown of individual effectors resulted in severe hypopigmentation defect similar to Rab4A-depleted melanocytes (Figs. 3A and S2B). Further, IFM analysis of Rabenosyn-5- or Rabip4 (depletes both Rabip4 and 4’ isoforms)-knockdown melanocytes showed a dramatic loss in peripheral staining of TYRP1 and a pool was targeted to lysosomes for degradation (Fig. 3A) (r=0.52±0.03, p≤0.001 in Rabenosyn-5 sh-1; 0.54±0.03, p≤0.001 in Rabenosyn-5 sh-2; 0.42±0.01, p≤0.001 in Rabip4 sh-1; 0.43±0.03, p≤0.001 in Rabip4 sh-2 and 0.22±0.01 in control cells). In contrast, TYRP1 in Rabaptin-5-depleted melanocytes was moderately affected and a small population was targeted to lysosomes (Fig. S2B) (r=0.47±0.02, p≤0.001 in Rabaptin-5 sh-1; 0.44±0.03, p≤0.001 in Rabaptin-5 sh-2 and 0.22±0.01 in control cells). In line with these data, immunoblot analysis showed that TYRP1 levels were dramatically reduced in Rabenosyn-5- or Rabip4-knockdown cells but moderately decreased in Rabaptin-5-knockdown cells (Figs. 3B and S2C) compared to respective control melanocytes. Similarly, the protein levels of TYR (AP-3-dependent cargo) phenocopied TYRP1 levels in the respective effector-knockdown cells (Figs. 3B and S2C). This data suggests that both Rabenosyn-5 and Rabip4/4’ either separately or cooperatively regulate the initial segregation of these cargoes on SEs.
Fig. 3. Rabenosyn-5 and Rabip4 regulate the cargo trafficking to melanosomes and Rabenosyn-5 forms a complex with Rab4A-AP-3-KIF3.
(A, C) BF and IFM analysis of Rabenosyn-5 or Rabip4 -knockdown cells (sh-1 and sh-2) or GFP-Rabenosyn-5/GFP-Rabip4’ expression in Rab4A-depleted melanocytes. Black arrows indicate the loss in pigmentation and arrowheads point to the cargo localization to lysosomes or melanosomes. The colocalization efficiency (r) between the proteins was indicated separately. Nuclei are stained with Hoechst33258. The insets are a magnified view of the white boxed areas. Scale bars, 10 μm. (B) Immunoblotting analysis of melanosomal and lysosomal proteins in knockdown cells and the tubulin was used as a loading control. P1 and Mβ, full length and processed PMEL bands. *, non-specific bands. Protein band intensities were quantified and indicated on the gels. (D) Immunoprecipitation of GFP-Rabenosyn-5, GFP-Rabip4’ and GFP (control) in HeLa cells. Both cell lysate (input) and IP blots were probed as indicated. Spectrin was used as positive control for IP.
Interestingly, PMEL fluorescence intensity was dramatically reduced and a pool was targeted to lysosomes in all effectors-depleted melanocytes (Figs. 3A and S2B), similar to Rab4A-knockdown melanocytes (Fig. 1G). Consistent with this result, the P1 form of PMEL was drastically reduced in Rabenosyn-5 shRNA (Fig. 3B), similar to Rab4A shRNA cells (Fig. 1H); and was moderately decreased in Rabip4 shRNA or Rabaptin-5 shRNA cells (Figs. 3B and S2C). In contrast to Rab4A-depleted melanocytes, the percentage of Mβ generated after proteolytic processing was drastically affected in Rabaptin-5-knockdown cells and moderately affected in both Rabenosyn-5 and Rabip4-depleted melanocytes (Figs. 3B and S2C), suggesting that Rabaptin-5 likely regulate either PMEL segregation on SEs or its maturation in melanocytes. In line with these results, fibrils isolated from the Rabenosyn-5-depleted or Rabaptin-5 knockdown (data not shown) melanocytes were notably reduced compared to control cells, which was similar to Rab4A-depleted melanocytes (Fig. 1F). Similarly, the fluorescence intensity and protein level of lysosomal membrane protein LAMP-2 were also dramatically reduced in both Rabenosyn-5- and Rabip4-, but not Rabaptin-5-, depleted melanocytes (Figs. 3A, 3B, S2B and S2C). Moreover, LAMP-2 was partially mislocalized to melanosomes in Rabenosyn-5- and Rabaptin-5-knockdown cells, indicating that Rabenosyn-5 and Rabaptin-5 either individually or jointly regulate the trafficking of lysosomal proteins and PMEL. Overall, these studies illustrate that Rabenosyn-5-depleted cells share the phenotypes of both Rabip4- and Rabaptin-5-knockdown melanocytes. Based on these data, we hypothesized (see below) that Rabip4/4’ in association with Rabenosyn-5, possibly regulate the segregation/sorting of TYRP1 and TYR cargo; whereas Rabaptin-5 in association with Rabenosyn-5, likely control the processing or maturation of PMEL fibrils in melanocytes.
Rab4A-Rab5A-shared effectors interact with each other and are independently recruited to the endosomal membranes upstream of Rab4A
We further examined whether any of the Rab4A-Rab5A shared effectors can rescue the hypopigmentation defect of Rab4A-depleted melanocytes. Unexpectedly, none of these effectors (GFP-Rabenosyn-5, GFP-Rabip4’ or mCherry-Rabaptin-5) individually improved the pigmentation defect of Rab4A-inactivated melanocytes (Figs. 3C and S2D) upon overexpression. This result suggests that Rab4A possibly co-ordinates these effectors by functioning downstream in the trafficking pathway (Kalin et al., 2015). Further, we analyzed the stability and localization of these effector proteins in different knockdown melanocytes to understand their molecular regulation. Upon Rab4A-depletion, Rabip4 level, but not Rabenosyn-5 or Rabaptin-5, were moderately reduced in melanocytes (Fig. S3A). Similarly, Rabenosyn-5-depleted melanocytes displayed reduced levels of Rab4 and Rabip4, but not Rabapatin-5, compared to control cells (Fig. S3A). In contrast, knockdown of either Rabip4 or Rabaptin-5 did not change the protein levels of other effectors, including Rab4 levels (Fig. S3A). These studies indicate that Rabenosyn-5 possibly mediates the molecular interaction between these effectors. Similar to fibroblasts, GFP-tagged shared effectors localized as punctate structures (de Renzis et al., 2002; Fouraux et al., 2004; Mattera et al., 2003); the pattern resembled EEA1-localized early endosomes in wild-type melanocytes (Setty et al., 2007) and colocalized with Rab5-positive endosomes (Fig. S3B inset for GFP-Rabenosyn-5 and data not shown for others). IFM studies showed that GFP-Rabenosyn-5 (but not GFP-Rabip4’ or GFP-Rabaptin-5)-positive punctate structures were dispersed throughout the cell upon Rab4A-depletion in wild-type melanocytes (Fig. S3B). In contrast, reduced GFP-Rabip4’ punctate structures (in the periphery, arrows) or increased cytosolic signal of GFP-Rabaptin-5 (arrows) was observed in Rabenosyn-5-depleted melanocytes. In line with these results, the distribution of GFP-Rabenosyn-5-positive punctate structures was moderately affected in either Rabip4- or Rabaptin-5-depelted melanocytes (Fig. S3B). Taken together, these studies indicate that Rabenosyn-5 possibly regulate the localization of Rabip4/4’ and Rabaptin-5 in melanocytes. Consistent with this conclusion, subcellular fractionation revealed that distribution of Rabip4/4’ to multiple membrane fractions and the localization of Rabaptin-5 to cytosol was increased in Rabenosyn-5-knockdown melanocytes (Fig. S3C). Thus, Rabenosyn-5 plays a key role in regulating the localization of Rabip4/4’ and Rabaptin-5 to the specific membranes. Based on this regulation, we predicted that Rabenosyn-5 possibly interacts with Rabip4/4’ or Rabaptin-5 either in a complex or independently. Due to low plasmid transfection efficiency of melanocytes, we immunoprecipitated Rabip4’ from HeLa cells expressing GFP-Rabip4’, which showed an interaction with Rabenosyn-5 (Fig. S3D), but not with Rabaptin-5 (data not shown). This may be due to the low endogenous expression of Rabaptin-5 in HeLa cells (Fig. S3E). However, the expression of Rabaptin-5 in melanocytes was considerably higher than in HeLa cells (Fig. S3E). Immunoprecipitation of endogenous Rabaptin-5 showed no interaction with either Rabenosyn-5 or Rabip4, but strong binding to Rab4 in melanocyte lysates (Fig. S3F) (Kalin et al., 2015; Mattera et al., 2003), indicating that interaction between Rabaptin-5 and other effectors is likely to be very transient in nature. Overall, these results suggest that the recruitment of Rab4A-Rab5A shared effectors to endosomal membranes is independent of Rab4A and possibly function upstream of Rab4A. Moreover, Rabenosyn-5 regulates recruitment of Rabaptin-5, stability/membrane distribution of Rabip4/4’ and cargo transport to melanosomes.
Rab4A associates with Rabenosyn-5 and coordinates cargo sorting by forming a Rabenosyn5-KIF3-AP-3 complex
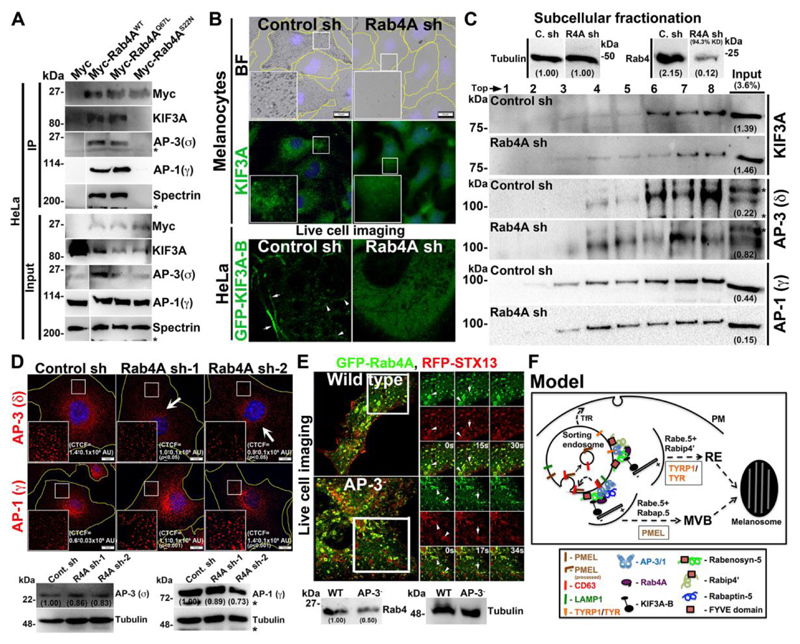
To understand the molecular regulation between Rab4A and the shared effectors in controlling cargo sorting on SEs, we studied large-scale interactome of Rabenosyn-5 and Rabip4’ in HeLa cells due to its high plasmid transfection efficiency compared to melanocytes (data not shown). Interestingly, the shared effectors showed an interaction with endosomal adaptor proteins (AP-3/AP-1) and kinesin-2 family motor KIF3. To validate these interactions, we performed immunoprecipitation (IP) of GFP-Rabenosyn-5, GFP-Rabip4’ or GFP (as a control) using HeLa cell lysates. GFP-Rabenosyn-5 showed a strong interaction with the Rab4, AP-3 and KIF3A, but not with AP-1 (Fig. 3D). However, GFP-Rabip4’ interacted only with the Rab4 and AP-3 (Ivan et al., 2012), but not with KIF3A or AP-1 (Fig. 3D). We predicted that Rabip4’-Rab4-AP-3 interaction is possibly mediated through Rabenosyn-5, since Rabip4’ and Rabenosyn-5 associate with each other (Fig. S3D). Moreover, we hypothesize a similar molecular interactions exist in melanocytes. To validate whether Rabenosyn5-Rab4A-AP-3-KIF3A complex localize to the endosomal membranes in melanocytes, we carried out subcellular fractionation. Upon membrane fractionation of wild-type melanocytes Rab4, KIF3A, Rabenosyn-5, and AP-3 (δ and σ) molecules were segregated into the same membrane fractions 5 to 7 (Fig. S4A, quantified and plotted as graph in S4B). As observed, these fractions were positive for several organelle-specific markers such as Rab5 (EEs), STX13 (REs) and LIMPII (late endosomes and Golgi), but not with LAMP-2 (lysosomes), suggesting that Rab4A-Rabenosyn-5-associated complex possibly localize to the intermediate organelles of early/sorting-recycling-late endosomal membranes (Fig. S4A). Interestingly, the membrane association of this complex was independent of AP-3, since the Rab4A-Rabenosyn-5-associated complex fractionated to the same membranes (5 to 7 fractions) even in the absence of AP-3 expression in melanocytes (Fig. S4A AP-3-, melan-mh cells-deficient for delta subunit) (Jani et al., 2015), suggesting that these molecules associate to the membranes independent of AP-3 recruitment. This is possibly due to the interaction of the Rabenosyn-5 FYVE domain with PI3P on endosomal membranes (Nielsen et al., 2000). Thus, these results identified a new endosomal complex Rab4A-Rabenosyn-5-KIF3A-AP-3 that possibly controls the segregation and trafficking of multiple cargoes on SEs. Moreover, coimmunoprecipitation of endogenous Rab4A showed an interaction with AP-3 (also with AP-1) and KIF3A/B but not with Rab4A-Rab5A shared effectors in melanocytes (Fig. S4C), indicating that the association of these dual effectors with the complex is very transient in nature. However, the role of KIF3A, but not AP-3 (Theos et al., 2005), in cargo sorting at SEs or in melanosome biogenesis is unknown.
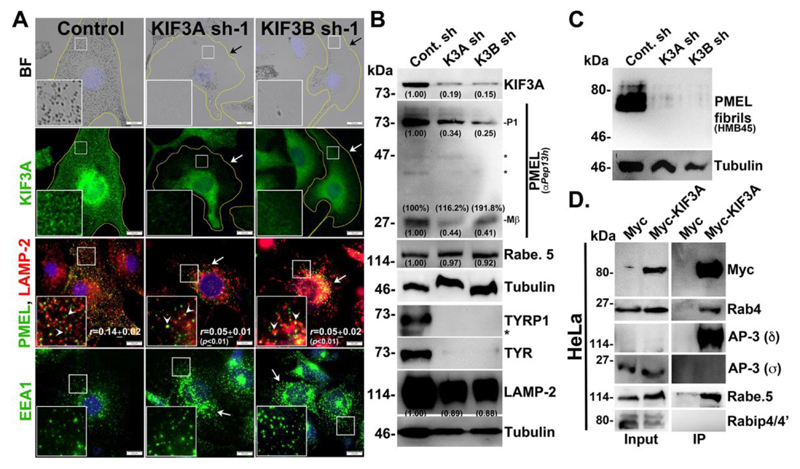
Previous studies have shown that KIF3A (Kinesin-2 family) and 5B (Kinesin-1 family) localize to the Rab4A-positive early endosomal membrane fractions (Bananis et al., 2004). Studies have also shown that KIF3A interacts with AP-3β1 for the release of HIV-1 Gag protein (Azevedo et al., 2009) and form a heterotrimeric complex with KIF3B (Yamazaki et al., 1995) and KAP3 (Yamazaki et al., 1996). We tested whether KIF3A or 3B has any role in cargo trafficking to melanosomes. Depletion of either KIF3A or 3B in wild-type melanocytes resulted in severe loss in pigmentation (Fig. 4A). As expected, KIF3A fluorescence staining and protein levels were reduced in KIF3A/3B-depleted melanocytes (Fig. 4A and 4B). Similar to Rab4A-depleted melanocytes, KIF3A- or 3B-knockdown melanocytes showed reduced protein levels of TYRP1, TYR, PMEL and LAMP-2, and decreased fibril formation (Fig. 4A, 4B and 4C). In line with these results, the fluorescence staining of PMEL was dramatically reduced in the peripheral cytosol (arrows) and a pool was mislocalized to lysosomes (arrowheads) (r=0.05±0.01, p≤0.01 in KIF3A sh-1; 0.05±0.02, p≤0.01 in KIF3B sh-1 and 0.14±0.02 in control cells) (Fig. 4A). Moreover, the formation of Mβ from total PMEL was not affected in KIF3A/3B shRNA cells (Fig. 4B), similar to Rab4A-depleted melanocytes (Fig. 1H). Additionally, LAMP-2- or EEA1-positive structures were clustered near the perinuclear region (arrows) upon KIF3A/B-knockdown in melanocytes, indicating that KIF3 regulates the positioning of these organelles (Fig. 4A) and melanosome biogenesis similar to Rab4A.
Fig. 4. KIF3 regulates melanocyte pigmentation and forms a complex with Rab4A-Rabenosyn-5-AP-3.
(A) BF and IFM analysis of KIF3A or 3B-knockdown melanocytes. Black arrows indicate the loss of pigmentation. White arrows show the loss in KIF3A or PMEL staining; or clustering of EEA1 in KIF3A or 3B sh cells. White arrowheads point to the colocalization of PMEL with LAMP-2 and their colocalization efficiency (r) was indicated separately. Nuclei are stained with Hoechst33258. The insets are a magnified view of the white boxed areas. Scale bars, 10 μm. (B, C) Immunoblotting analysis of melanosomal and lysosomal proteins, and PMEL fibrils in KIF3A/B-knockdown cells and the tubulin was used as a loading control. P1 and Mβ, full length and processed PMEL bands. *, non-specific bands. Protein band intensities were quantified and indicated on the gels. (D) Immunoprecipitation of Myc-KIF3A in HeLa cells. Both input and IP blots were probed as indicated.
Next, we examine the KIF3 interaction with Rab4A, AP-3, Rabenosyn-5 or any other effectors in HeLa cells due to low plasmid transfection efficiency of melanocytes. Surprisingly, Myc-KIF3A showed strong interaction with the Rab4A, AP-3 (δ subunit) and Rabenosyn-5, but not with Rabip4/4’ (Fig. 4D). We predict that these molecular interactions also exist in melanocytes. In line with this hypothesis, localization of AP-3 near the perinuclear region was drastically reduced in KIF3A- or 3B-depleted melanocytes (data not shown). In addition, the total Rabenosyn-5 level was unaffected upon KIF3A- or 3B-depletion in wild-type melanocytes (Fig. 4B). These results suggest that KIF3 associates with Rab4A-Rabenosyn-5-AP-3, in which Rab4A possibly mediates the assembly of this complex.
Rab4A regulates endosomal localization of KIF3 and AP-3 and controls cargo segregation on SEs
We examined whether the interaction of Rab4A with KIF3 and AP-3 is dependent on the nucleotide status of Rab4A. Immunoprecipitation of ectopically expressed Myc-tagged Rab4AWT and Rab4AQ67L (constitutively active mutant), but not Rab4AS22N (dominant negative mutant), showed a strong interaction with both KIF3A and AP-3 (σ), indicating that GTPase cycle of Rab4A is required for their interaction. Interestingly, we also observed an interaction between Rab4A and AP-1 (used as negative control) in the HeLa cell lysates (Fig. 5A), which suggests that Rab4A independently interacts with these adaptor complexes (see below). Additionally, we predict that these molecular interactions exist in melanocytes. Next, we studied whether the recruitment of KIF3 and AP-3 on endosomal membranes is dependent on Rab4A. IFM analysis showed that KIF3A localized as punctate structures and appeared as diffused cytosolic staining in wild-type and Rab4A-depleted melanocytes, respectively (Fig. 5B). Due to the difficulty in transfecting KIF3A/B constructs into melanocytes, we tested the motor localization in HeLa cells. Live-cell imaging analysis showed that GFP-KIF3A-B localized as both punctate structures resembling endosomes and long tubular structures, which is a hallmark of recycling endosomes in HeLa cells (Delevoye et al., 2014) (Fig. 5B, shown one of the movie frame, video is not shown). As predicted, GFP-KIF3A-B completely localized to the cytosol in Rab4A-knockdown HeLa cells (Fig. 4B). Consistent with this result, subcellular fractionation showed that localization of KIF3A to endosomal membranes (fractions 6-8) was notably reduced upon Rab4A-knockdown compared to wild-type melanocytes (Fig. 5C). These studies indicate that Rab4A recruits KIF3 onto endosomal membranes.
Fig. 5. Rab4A regulates the recruitment and association of KIF3 and AP-3 to endosomal membranes and the model illustrating Rab4A function in sorting cargo on SEs.
(A) Immunoprecipitation of Myc-Rab4A (WT, Q67L and S22N mutants) in HeLa cells. Both input and IP blots were probed as indicated. Spectrin was used as positive control for IP. (B, D) BF, IFM and live cell imaging of Rab4A-knockdown cells. Arrowheads point to the KIF3 localization in HeLa cells. Arrows show the loss in AP-3 staining. Nuclei are stained with Hoechst33258. The insets are a magnified view of the white boxed areas. The CTCF values were indicated separately (mean±s.e.m.). Scale bars, 10 μm. (C) Subcellular fractionation of control and Rab4A sh melanocytes and probed the fractions for localization of KIF3A, AP-3 or AP-1. (D, E) Immunoblotting analysis of adaptor subunits and Rab4 in respective cell types as indicated and the tubulin was used as a loading control. *, non-specific bands. Protein band intensities were quantified and indicated on the gels. (E) Super-resolution live cell imaging of GFP-Rab4A with respect to RFP-STX13 in wild-type and AP-3- melanocytes. Arrows and arrowheads point to the localization of proteins to the REs and vesicles arising from vacuolar/SEs respectively. The insets are a magnified view of the white boxed areas at indicated time points (Movies S1 and S2). (F) Proposed model wherein TYRP1 and TYR are segregated at the Rabenosyn-5-Rabip4’ domains, and PMEL, CD63 and LAMP-1 are segregated at the Rabenosyn-5-Rabapin-5 domains. In both these domains, cargo tails bind to the AP-3, which associates with Rab4A and KIF3 motor for positioning the domains. Post segregation, TYRP1 and TYR enters into the REs for targeting toward maturing melanosomes; whereas PMEL is proteolytically cleaved, internalized into ILVs of MVB along with CD63 (but not LAMP-1) for the biogenesis of stage II melanosomes.
In contrast to KIF3, the IFM intensity of AP-3 (stained for δ subunit), but not AP-1 (stained for γ subunit), was significantly reduced in Rab4A-depleted compared to control melanocytes (CTCF for AP-3=1.0±0.1x106 AU, p≤0.05 in Rab4A sh-1, 0.9±0.1x106 AU, p≤0.05 in Rab4A sh-2 and 1.4±0.1x106 AU in control cells; CTCF for AP-1=1.1±0.1x106 AU, p≤0.001 in Rab4A sh-1, 1.4±0.1x106 AU, p≤0.001 in Rab4A sh-2 and 0.6±0.03x106 AU in control cells) (Fig. 5D). However, the total AP-3 or AP-1 subunit protein levels were not affected in Rab4A-knockdown melanocytes (Fig. 5D), suggesting that Rab4A either regulates the recruitment of AP-3, but not AP-1, onto endosomal membranes or is required for AP-3 stability onto those membranes. Consistent with the earlier hypothesis, subcellular fractionation analysis showed that membrane bound AP-3 levels, but not AP-1, were reduced and the localized AP-3 was distributed to the multiple organelle membranes (not shown) in Rab4A-depleted compared to control melanocytes (Fig. 5C). Similarly, Rabenosyn-5-knockdown in melanocytes also reduced the perinuclear distribution of AP-3 (data not shown). These studies suggest that Rab4A-Rabenosyn-5 partly regulates the recruitment/association of AP-3 onto selective endosomal membranes.
We examined whether the AP-3 plays a role in regulating the localization or activity of Rab4A in melanocytes. Previous studies have shown that Rab4A localizes to early/recycling tubular structures in fibroblasts (D'Souza et al., 2014; Sonnichsen et al., 2000; Van Der Sluijs et al., 1991). Super-resolution live-cell imaging of wild-type melanocytes showed that GFP-Rab4A localized as enlarged vacuolar ring-like structures with emanating vesicles positive for RFP-STX13 (arrowheads, syntaxin 13) (Movie S1 and Fig. 5E), possibly representing EEs/SEs (Jani et al., 2015; Setty et al., 2007). Additionally, a large population of GFP-Rab4A also appeared as punctate structures positive for STX13 and emanating only STX13 tubular structures (arrows) (Movie S1 and Fig. 5E), likely corresponding to SEs/REs. Consistently, Rab4A fractionated into multiple subcellular fractions that correspond to EEs/REs (Fig. S4A). Whereas GFP-Rab4A in AP-3- melanocytes localized to both STX13-positive enlarged vacuolar structures (arrowheads), which was similar to its localization in WT cells, and to longer tubular structures (arrows) (Movie S2 and Fig. 5E), representing the early/sorting and REs respectively (Dennis et al., 2015). Although, Rab4 level was slightly reduced, but its localization to the recycling tubular endosomes was moderately increased in AP-3-deficient compared to WT melanocytes (Fig. 5E). Thus, these studies in melanocytes demonstrate that Rab4A predominantly localizes to SEs and partly to EEs/REs, and its recruitment is independent of AP-3.
We further tested whether Rab4A-regulated cargoes are the sorting substrates of AP-3 and/or AP-1 complexes. Amino acid sequence analysis of the cargoes showed that PMEL and TYR contain a (D/E)XXXL(L/I) motif and that CD63 and LAMP-1 contain a YXXϕ motif in the C-terminus (Fig. S4D) (Bonifacino and Traub, 2003). Studies have shown that AP-3 regulates the sorting of TYR (Theos et al., 2005), CD63 (Rous et al., 2002) and LAMP-1 (Dell'Angelica et al., 1999; Peden et al., 2004) proteins on endosomal membranes. However, the role of AP-3 and/or AP-1 in regulating the trafficking of PMEL remains unclear, although C-terminus PMEL contains a putative acidic dileucine motif. In contrast, studies have shown that dileucine motif of PMEL interacts with AP-2 during its internalization from the cell surface (Valencia et al., 2006). Here, we examined the interaction of Rab4A-dependent cargo tails with AP-3 or AP-1 subunits by yeast two-hybrid (Y2H) assay (Fig. S4D). Similar to earlier studies, dileucine motif of PMEL (a longer C-terminal tail than previously studied) did not interact with either AP-3 or AP-1 subunits (Fig. S4D). However, the tyrosine-based motif of CD63 showed strong interaction with AP-3 (μ) but not with AP-1 subunits. We predict that PMEL trafficking in melanocytes possibly regulated through CD63 (van Niel et al., 2011). Consistently, AP-3-deficient melanocytes mislocalize both PMEL and GFP-CD63 to LAMP-2-positive compartments (Fig. S4E) as similar to Rab4A-knockdown cells (Fig. S1O). In line with these results, AP-3-deficient cells showed defective PMEL processing and fibril formation (Fig. S4E). However, PMEL was not segregated into exosomes (Fig. S4E) in AP-3-deficient cells in contrast to Rab4A-depleted melanocytes (Fig. S1N), suggesting that PMEL is directly targeted to lysosomes for degradation upon loss in expression of AP-3 subunits. Thus, this data indicates that both AP-3 and Rab4A are required for sorting of PMEL as well as CD63 on SEs. As expected, LAMP-1 showed strong interaction with the μ subunit of both AP-3 and AP-1 (Fig. S4D). Overall, these studies suggest that majority of Rab4A-dependent cargoes is sorted by the AP-3 on endosomal membranes.
Discussion
SEs are the central regulatory hubs for targeting cargo either to the cell surface through recycling or to lysosomes for degradation. These organelles originate from EEs and mature into recycling/late endosomes on post cargo sorting. Interestingly, in melanocytes, few of these pathways are diverted towards the biogenesis of pigment granules. Within these pathways, the structural and enzymatic proteins of melanosome follow three independent transport routes: BLOC-1-mediated TYRP1 and ATP7A transport, AP-3-dependent TYR transport, and CD63-dependent PMEL transport to melanosomes. However, the specific cargo segregation mechanisms on SEs are unknown. Moreover, these processes are essential for both proper trafficking of cargo to the target organelle and organelle homeostasis.
In general, Rab GTPases function in membrane identity, cargo sorting and membrane fusion processes (Ohbayashi and Fukuda, 2012; Stenmark, 2009; Zerial and McBride, 2001). These GTPases recruit specific effector proteins onto the membranes and form transient local subdomains, which are involved in cargo segregation, packaging, vesicle/tubule generation and then delivered towards the target membranes (Gruenberg, 2001). We hypothesized that similar subdomains exist on SEs for the segregation of melanosome-specific and general cargo in melanocytes. In search for a Rab regulator of melanosome cargo segregation, our RNAi screen identified Rab4A, which mislocalized all primary melanocytic cargoes to lysosomes upon its depletion. In contrast, Rab4A in fibroblasts has been shown to regulate the fast recycling of TfR (van der Sluijs et al., 1992) from EEs or GLUT4 vesicles (Aledo et al., 1995) to the plasma membrane. Furthermore, Rab4A has been shown to localize to the intermediates of early and recycling compartments (Sonnichsen et al., 2000; Van Der Sluijs et al., 1991) and orchestrates the endosomes by interacting with AP-1 or AP-3 adaptors via Arf1-ARL1-dependent GTPase cascade (D'Souza et al., 2014). Nevertheless, the function of Rab4A in cargo segregation/ transport pathways during melanosome biogenesis remains elusive. In this study, we extensively characterized the function of Rab4A in segregating and organizing the cargo into different subdomains, followed by trafficking to the premelanosomes. During this process, Rab4A associates with Rabenosyn-5 on endosomes and interacts with AP-3 on one side and KIF3 motor on other side (Fig. 5F). These interactions form a novel endosomal complex that further associate with either Rabip4/4’ or Rabaptin-5 molecules, which possibly facilitate the melanosomal and lysosomal cargo segregation on SEs. This segregation was found to be essential for targeting cargo to REs and for the maturation of late endosomes or premelanosomes in case of melanocytes. Thus, Rab4A acts as a key organizer of endosomal domains during segregation and trafficking of multiple cargoes on SEs. This endosomal domain organization possibly either generate cargo-specific membrane domains on SEs or regulate positioning/distribution of specific cargo containing endosomes. We favor the earlier model due to the following: (1) Rab4A-depletion increases accumulation of vacuolar endosomes compared to the control cells; (2) Rab4A-knockdown increases the secretion of proteolytically unprocessed PMEL into exosomes and mislocalizes both TYRP1 and TYR to lysosomes; (3) Rab4A-inactivation alters the trafficking of non-melanocytic cargoes CD63 and LAMP1/2; (4) Rab4A associates with AP-3, Rabenosyn-5 and KIF3 (referred to here as Rab4A-complex) on endosomes, as evident by subcellular fractionation; (5) Rab4A-, Rabenosyn-5- and KIF3-specific depletions in melanocytes phenocopy the hypopigmentation and defective fibril formation phenotypes; (6) Rabenosyn-5, Rabaptin-5 and Rabip4’ independently get recruited and moderately regulate the expression/endosomal localization of each other, however, the association of Rabenosyn-5 with Rabip4/4’ or Rabaptin-5 distinguishes the cargo specificity during the segregation; and (7) finally, Rab4A-knockdown phenotypes are specific and not attributed to any change in either the transcription profile or Rab5A-localization/recruitment to the membranes. Thus, these results strongly support that Rab4A acts as master regulator in segregating melanosomal and lysosomal cargo on SEs following individual transport to premelanosomes or lysosomes.
Our studies demonstrate that Rab4A regulates the formation of subdomains on SEs by interacting with Rab5A-effectors and the cargo sorting adaptor AP-3 (Fig. 3D). Moreover, we predicted that Rab4A coordinates these molecules through its transient interactions on the endosomal membranes, where Rabenosyn-5 and AP-3 (but not KIF3) are recruited independently of Rab4A. Additionally, Rabenosyn-5 further recruits or associates with either Rabip4/4’ or Rabaptin-5 and stabilizes Rab4A-complex (Fig. S3). This model is concurrent with our results that depletion of either Rabip4 or Rabaptin-5 partially mimics the cargo trafficking defects observed in Rabenosyn-5- or Rab4A-knockdown melanocytes. Thus, the Rab4A-Rab5A shared effectors are very likely to act as adaptors on endosomal membranes to assemble the complex, which is mediated through Rab4A. Consistent with this proposal, Rab4A-depletion reduced the association of AP-3 on the endosomal membranes, dissociated the KIF3 motor and dispersed the enlarged Rabenosyn-5 or Rabip4’-positive endosomes to the periphery (Figs. 5 and S3). Several individual studies support these interactions: (1) Rabip4 has been shown to interact with AP-3 (β subunit) and regulate endosomal cargo recycling and the distribution of lysosomes (Fouraux et al., 2004; Ivan et al., 2012) in fibroblasts; (2) Rabaptin-5 has been shown to interact with AP-1 (γ subunit) and regulate Tf recycling (Deneka et al., 2003); similarly, Rabenosyn-5 regulates TfR recycling (Navaroli et al., 2012); (3) Rab4A has been shown to associate with AP-1 or AP-3 localized domains on endosomes (D'Souza et al., 2014); (4) Rab4A has been shown to localize to the KIF3A-enriched membrane fractions (Bananis et al., 2004); and (5) Rab4-KIF3 has been shown to mediate the insulin-induced GLUT4 exocytosis (Imamura et al., 2003). Although, these interactions were observed primarily in fibroblasts, none of these studies integrated their role in other cargo transport pathways. To our knowledge, this study is the first to show the specificity of these interactions in selective cargo transport to melanosomes. However, our study did not illustrate the role of following known interactions in melanosome biogenesis, which need to be evaluated in future: (1) Rabenosyn-5 has been shown to interact with VPS45 and EEA1 (Nielsen et al., 2000); (2) Rabaptin-5 has been shown to interact with Rab4 and AP-1 (γ) (Deneka et al., 2003; Pagano et al., 2004); (3) Rab4A-GTP has been shown to recruit various effectors such as GRASP1 (Hoogenraad et al., 2010), D-AKAP2 (Eggers et al., 2009), Gadkin (Schmidt et al., 2009) and RCP (Lindsay et al., 2002); and (4) KIF3A/B has been shown to interact with KAP3 (Yamazaki et al., 1996). Here, we predicted that Rab4A or Rab4A-Rab5A shared effectors might have additional roles in regulating trafficking steps other than the melanosomal and lysosomal cargoes.
Our studies show that Rab4A act as a key regulator in the cargo trafficking to melanosomes. We demonstrated that Rab4A regulates sorting of PMEL (through CD63) and LAMP-1 through its interaction with AP-3. Here, we hypothesized that PMEL/CD63 and LAMP-1 bound AP-3, associate with Rab4A-Rabenosyn-5-Rabaptin-5-KIF3 molecules on SEs and generate a distinct subdomain that directs cargo towards late endosomes (Fig. 5F). Inactivation of Rab4A reduces membrane association of AP-3, which results in the internalization of unproteolyzed PMEL/CD63 into the ILVs and then secreted as exosomes, resulting in reduced number of stage II melanosomes (Figs. S1N and 2). In contrast, Rab4A possibly segregate both TYRP1 and TYR on SEs by interacting with AP-3, which further associates with Rabenosyn-5-Rabip4/4’-KIF3 and generates a different subdomain that guide the cargo towards REs (Fig. 5F). Upon depletion of Rab4A, both TYRP1 and TYR enter into the classical ubiquitin-dependent lysosomal degradation pathway and a pool recycles back to the cell surface (Figs. 1, S1M and S1Q). Moreover, this model further supports the BLOC-1-dependent TYRP1 trafficking to the melanosome, which occurs on REs (Delevoye et al., 2009). Thus, Rab4A acts a master regulator of cargo segregation by generating different subdomains through its association with the combination of Rab4A-Rab5A shared effectors on SE membranes.
Materials and methods
Reagents and antibodies
All chemicals and reagents were purchased either from Sigma-Aldrich (Merck) or ThermoFisher Scientific (Invitrogen). Puromycin from Calbiochem and Matrigel from BD Biosciences were purchased. Commercial antibodies with their specific use (IB, immunoblotting; IFM, immunofluorescence microscopy; IP, immunoprecipitation and FACS, fluorescence-activated cell sorting) and the catalogue numbers were indicated. Antibodies against KIF3A (IF, IB; ab11259), LIMPII (IB; ab16522) and PMEL (HMB45; IF, IB; represent pre-melanosomes in IF and detects fibrils on IB; ab787) were from Abcam; TYRP1 (TA99; IF, FACS; HB-8704) was from American Type Culture Collection; γ-adaptin (AP-1; IF, IB; 610385), LAMP1 (IB; 553792), Rab4 (IB, IF, IP; 610889; specific to Rab4A and low affinity against mouse Rab4), Rabaptin-5 (IB, IP; 610676) and TfR (CD71; FACS; 553264) were from BD Biosciences; alpha II-spectrin (SPTAN1; IB; A301-249A) was from Bethyl; EEA1 (IF; 3288), HSP90 (IB; 4877), LC3A/B (IF; 4108), Rab5 (IF, IB; 3547) and Rab11 (IB, 5589) were from Cell Signaling Technology; δ-adaptin (AP-3, IF; SA4), LAMP-1 (IB, FACS; 1D4B) and LAMP-2 (IF, IB; GL2A7) were from Developmental Studies Hybridoma Bank; GFP (IB; A11122) was from Invitrogen; γ-tubulin (GTU88; IB; T6557) was from Sigma-Aldrich; σ3-adaptin (AP-3; IB; sc-136338), GAPDH (IB; sc-25778), c-Myc (IB; sc-789), TfR (CD71; IB; sc-7087) and TYRP1 (IB; sc-25543) were from Santa Cruz Biotechnology. All secondary antibodies were either from Invitrogen or Jackson Immunoresearch. Antisera to Rabip4/4’ (IB)(Ivan et al., 2012), STX13 (IF, IB) (Prekeris et al., 1998) and TYR (PEP7h; IF, IB)(Theos et al., 2005) have been described previously. Other antisera such as δ-adaptin (dh2, AP-3; IB) (Andrew Peden, University of Sheffield, Sheffield, UK); αPmelN (N-terminus to PMEL; IB) and αPep13h (PMEL-C, C-terminus to PMEL; IB; used for PMEL processing) (Michael S. Marks, University of Pennsylvania, Philadelphia, USA) and Rabenosyn-5 (Silvia Corvera, UMADD Medical School, Worcester, USA) were obtained as gift from respective laboratories mentioned in the parenthesis.
Plasmids and shRNAs
Expression constructs: Myc-Rab4AWT– human full-length Rab4A was PCR amplified with N-terminal Myc epitope sequence from human cDNA and subcloned into the BamH1 and XhoI sites of pCDNA3.1(+) (Invitrogen). GFP-Rab4AWT - PCR amplified full-length human Rab4A (without Myc tag) was digested with EcoRI and XhoI enzymes and subcloned into the EcoRI and SalI sites of pEGFP-C2 (Clontech). GFP-Rab4Ash2R – mutagenesis of Rab4AWT DNA sequence at 436 – 459 bases (changed at wobble base of amino acid sequence QENELMFL; 5’-CAAGAAAATGAGCTGATGTTTTTG converted to 5’-CAGGAAAACGAATTAATGTTTTTG) was carried out using QuickChange multi site-directed mutagenesis kit (Agilent Technology). Note, this plasmid is resistant against the Rab4A shRNA-2. Similarly, Myc-Rab4AQ67L and Myc-Rab4AS22N – mutagenesis of amino acids glutamine (Q) at 67th position to leucine (L) and serine (S) at 22nd position to asparagine (N) separately in Myc-Rab4AWT was carried out using QuickChange multi site-directed mutagenesis kit. Empty vector, pCMV-Myc was from Clontech. GFP-CD63 (62964) and GFP-Rabenosyn-5 (37538) were obtained from Addgene. GFP-STX13 and RFP-STX13 (Jani et al., 2015); Myc-KIF3A (Azevedo et al., 2009); pCI-Pmel117 (referred to here as PMEL) (Berson et al., 2001) have been described or kind gift from their respective laboratories. GFP-KIF3A and GFP-KIF3B constructs were obtained from Alistair Hume (with the permission from Tetsu Akiyama, Japan), University of Nottingham Medical School, Nottingham, UK (Haraguchi et al., 2006). GFP-Rabip4’ has been described (Ivan et al., 2012), and mCherry-Rabaptin-5 and CFP-Rabaptin-5 were subcloned from pCDNA3-Rabaptin-5 (Nagelkerken et al., 2000). TRC shRNA vectors: We have selected the human shRNA plasmids encoding target sequence against the multiple Rab proteins that are highly conserved for mouse Rab GTPases. These shRNAs were purchased from TRC Genome-wide shRNA library (Sigma-Aldrich). The target sequence and their % homology with mouse proteins are listed in Table S1. Retroviral shRNA vectors: Oligodeoxyribonucleotide duplexes containing the target sequences (listed in Table S2) were cloned into the BamH1 and HindIII sites of pRS shRNA vector (OriGene Technologies). Empty pRS shRNA plasmid was used as a control in all shRNA knockdown experiments. Rabip4 shRNAs also target the longer Rabip4’ isoform in the wild-type melanocytes. Y2H vectors: Empty vectors and the plasmids containing different subunits of AP-3 (δ, μ3, β3A, β3A-hinge and σ3) or AP-1 (γ, μ1 and σ1) have been described previously (Jani et al., 2015). Oligodeoxyribonucleotide duplexes corresponds to C-terminal tails of hPMEL623-668, hCD63227-238, hLAMP-1406-417 and mTYR502-533 were cloned into EcoRI and SalI sites of pGBKT7. All plasmid inserts were verified by DNA sequencing.
Yeast two-hybrid assay
The detailed protocol of yeast two-hybrid (Y2H) assay has been described in (Jani et al., 2015). Briefly, the Y2HGold yeast strain (Clontech) was transformed with different bait and prey plasmids as indicated in the figure (Table S4) by lithium acetate transformation protocol. The yeast transformants were selected on minimal medium plates supplemented with complete amino acid mix (Y0750 - Sigma-Aldrich), lacking leucine and tryptophan (referred to here as +His medium). Further, transformants were grown to exponential phase, serial diluted and then spotted on +His, -His (Y2146 –Sigma-Aldrich) and –His (+2 or 10 mM 3AT [3-Amino-1,2,4-triazole]) plates. Plates were incubated for 3-5 days at 30°C and then imaged under white light in a Bio-Rad Molecular Imager. The yeast transformants that were grown on –His (+3AT) considered as positive interaction between bait and prey proteins.
Cell culture, transfection and retroviral transduction
Immortal mouse melanocyte cell lines used in this study – Wild-type melan-Ink4a: derived from C57BL/6J a/a Ink4a-Arf-/- mice, formerly called melan-Ink4a-1 and referred to here as WT or melan-Ink4a (Ha et al., 2007). AP-3- melan-mh: derived from C57BL/6J Ap3dmh/mh mice and referred to here as AP-3- or melan-mh (Jani et al., 2015). Cells were maintained as described previously (Jani et al., 2015). DNA vectors were transfected into the melanocytes or PLAT-E cells (Cell Biolabs) or HeLa cells (ATCC) by Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For gene knockdown in wild-type melanocytes - cells were transduced with retroviruses encoding different target sequences in pRS shRNA plasmids, isolated from PLAT-E cells (Morita et al., 2000). Melanocytes were selected twice with puromycin (2 μg/ml) on the 2nd and 5th day of retrovirus transduction. In some experiments, control/shRNA knockdown cells or AP-3- melanocytes were transfected with GFP-CD63, GFP-Rabenosyn-5, GFP-Rabip4’ or mCherry-Rabaptin-5 separately or mixture of GFP-KIF3A and KIF3B or GFP-Rab4A and RFP-STX13. In one experiment, HeLa cells were directly transfected with Rab4A shRNA (listed in Table S1) using Lipofectamine 2000 and selected the cells twice with puromycin (2 μg/ml) on 2nd and 5th day.
Transcript analysis by semiquantitative PCR
Melanocytes grown in a 60 mm dish was treated with Trizol reagent (Sigma-Aldrich) and then extracted with chloroform at room temperature. Further, the aqueous layer was precipitated with isopropanol followed by a wash with 70% ethanol. Finally, the isolated RNA pellet was air dried and suspended in 0.01% DEPC treated water (Sigma-Aldrich). The cDNA was prepared by using a cDNA synthesis kit (Fermentas) after estimating the RNA concentration using NanoDrop 2000C spectrophotometer (Thermo Scientific). Gene transcript levels were analyzed by semiquantitative PCR (Bio-Rad S1000 Thermal Cycler) using gene specific primers (listed in Table 3) and an equal amount of cDNA from each sample. In all PCRs, GAPDH was used as a loading control. Band intensities were measured, normalized with GAPDH, quantified fold change with respect to the control and then listed in the figure.
Melanin estimation
Intracellular melanin content of melanocytes was measured using a protocol as described previously (Mahanty et al., 2016). Cells were transfected with respective shRNAs or transduced with virus encoding control or different Rab4A shRNAs. After puromycin selection, cells were harvested and lysed in lysis buffer [50 mM Tris-Cl pH 7.4, 2 mM EDTA, 150 mM NaCl, 1 mM DTT and 1X protease inhibitor cocktail (Sigma-Aldrich)] and then centrifuged for 15 min at 20,000 g, 4 °C. Supernatants were subjected to protein estimation using Bradford protein estimation kit (Bio-Rad). The melanin pellet fractions were washed with ethanol:diethyl ether (1:1 ratio), air dried and resuspended in a buffer containing 2 M NaOH and 20% DMSO followed by incubation at 60 °C for 30 min. Optical density (O.D) of melanin pigments were measured at 492 nm using Tecan multi-well plate reader (Tecan) and then normalized with respective protein concentration.
Immunoblotting
Cell lysates were prepared using a protocol described previously (Setty et al., 2007) and γ-tubulin used as a loading control in all experiments. Immunoblots were developed with the Clarity Western ECL substrate (Bio-Rad) and imaged in a Bio-Rad Molecular Imager ChemiDoc XRS+ imaging system equipped with Supercooled (-30°C) CCD camera (Bio-Rad) using Image Lab 4.1 software. Protein band densities were measured, normalized with γ-tubulin, quantified the fold change with respect to control and then indicated in the figure. % Mβ formation was calculated from the total PMEL (sum of P1, P2 and Mβ band densities) after γ-tubulin normalization. In certain experiments, % knockdown was also quantified after γ-tubulin normalization.
Exosome preparation
Condition media from subconfluent mouse melanocytes was collected every 48 h and stored at 4°C before use. Initially, medium was cleared for cell debris by centrifuging consecutively at 2000 g and 4000 g for 15 min (4°C). The supernatant was further centrifuged at 10000 g for 30 min (4°C) followed by 100000 g spin for 60 min (4°C) in a Ultracentrifuge (Beckman L-80) using 80 TI rotor. The exosome pellet was washed once with 1XPBS (pH 7.4), lysed in Urea lysis buffer (8 mM Urea, 50 mM Tris-Cl pH 7.4, 50 mM Na2HPO4, 300 mM NaCl, 0.5% NP40 and protease inhibitor cocktail) and then analysed by immunoblotting.
Extraction of melanosomal fibrils
Control and knockdown melanocytes were washed with 1XPBS and suspended in lysis buffer (20 mM Tris-Cl pH 7.4, 150 mM NaCl, 1mM EDTA, 1% TX-100 and protease inhibitor cocktail) followed by lysing the cells using G25 syringe. Further, lysates were incubated on ice for 1 h and then centrifuged for 10 min at 12000 g (4°C). The pellets were suspended in 8M Urea containing lysis buffer and then boiled for 30 min at 60°C. The suspensions were centrifuged and collected the supernatants followed by analyzing with immunoblotting using HMB45 antibody.
In vitro tyrosinase activity and protease inhibitor assays
Cells on the Matrigel-coated coverslips were fixed and assayed for the tyrosinase activity using the substrate L-DOPA as described previously (Atul Jani et al., 2016). Similarly, cells were treated with or without 50 nM bafilomycin A1 for 4 h at 37°C before fixation and then stained with respective antibodies followed by IFM (Jani et al., 2015). In some experiments, control and bafilomycin treated cells were subjected to immunoblotting.
Immunofluorescence microscopy and image analysis
For steady state localization studies, cells on coverslips were fixed with 2% formaldehyde (in PBS) and then stained with primary antibodies followed by the respective secondary antibodies as described previously (Setty et al., 2007). Bright field (BF) and immunofluorescence (IF) microscopy of cells was performed on an Olympus IX81 motorized inverted fluorescence microscope equipped with a CoolSNAP HQ2 (Photometrics) CCD camera using 60X (oil) U Plan super apochromat objective. Acquired images were deconvolved and analyzed using cellSens Dimension software (Olympus). Pigmentation (normal or hypopigmentation) in cells was quantified from BF images visually by counting ~100 cells in each experiment. Average pigmentation in cells was calculated and then plotted. Similarly, reduced fluorescence staining of TYRP1 or TYR in shRNA-depleted cells was quantified visually and then plotted as percentage of cells lost the staining of respective proteins (Fig. S1A). In Fig. S1F, melanocytes with approximately 50 or lesser melanosomes/cell were considered as hypopigmented cells, counted visually from the randomly taken BF images of each condition and then plotted. The colocalization between two colors was measured by selecting equal square areas in entire cell excluding the perinuclear area and then estimated the Pearson’s correlation coefficient (r) value using cellSens Dimension software. The average r value per each cell was calculated and then plotted or represented as mean value along with s.e.m. Note that maximum intensity projection of undeconvolved Z-stack images were used for estimating the r values. The mean fluorescence intensity (MFI) of immunostained melanocytes was measured using Image J software and corrected total cell fluorescence (CTCF) was calculated using below formula. CTCF (in arbitory units, A.U. or AU) =area of the cell (MFI of cell-MFI of background). The averaged CTCF values from 10-25 cells/condition were calculated and indicating in the figure. The analyzed images were assembled using Adobe Photoshop.
Live cell imaging
Cells were plated on 2-cm glass-bottomed dishes (Mat Tek Corporation) and then transfected with respective constructs. Post 24 h, cells were visualized under Olympus IX81 fluorescence microscope equipped with an environmental chamber maintained at 37°C with 5% CO2 and analyzed by cellSens Dimension software. Time-lapse microscopy of both GFP and RFP was performed by capturing image streams over 3-5 minutes using a CoolSNAP HQ2 (Photometrics) CCD camera. Similarly, the time lapse imaging was performed on a Zeiss LSM880 laser scanning microscope with Airyscan mode using Zen lite 2.0 software to obtain the videos equivalent to the super resolution. Images were analyzed and converted into either TIFF or avi format for visualization.
Electron microscopy
Control and Rab4A-knockdown melanocytes were seeded on Matrigel coated glass coverslips. Post 24 h, cells were fixed initially with 0.5% Karnovsky’s fixative (4% paraformaldehyde, 72 mM sodium cacodylate pH 7.4, 4 mM CaCl2, 0.5% glutaraldehyde) for 2 h followed by overnight fixation with 2% Karnovsky’s fixative (contains 2% glutaraldehyde). Cells were processed for Epon embedding as described (Raposo et al., 2001). Ultrathin sections of cell monolayers were prepared with a Reichert UltracutS ultramicrotome (Leica Microsystems) and contrasted with uranyl acetate and lead citrate as described (Raposo et al., 2001). Samples were examined with a FEI Tecnai Spirit electron microscope (FEI Company), and digital acquisitions were made with a numeric camera (Quemesa; Soft Imaging System). For quantification, melanosome stages were defined by morphology (Raposo et al., 2001) and vacuoles were defined as empty organelles. Melanosomes and vacuoles per µm2 cytosol were counted using ImageJ software. We have counted 10 cells each from control sh and Rab4A sh condition. Further, we have estimated the melanosome stages from 883 total melanosomes of control sh and 300 total melanosomes of Rab4A sh cells.
Cell surface expression using Flow cytometry
Cells were harvested, washed with 1XPBS and then suspended in growth medium (supplemented with 25 mM HEPES pH 7.4) containing saturating concentrations of unconjugated primary antibodies on ice for 30-45 min. Cells were washed and incubated with respective Alexa 488-conjugated secondary antibody for 30-45 min on ice. Finally, cells were washed, suspended in ice-cold FACS buffer (5% FBS, 1 mM EDTA and 0.02% sodium azide in PBS) and measured the fluorescence intensity using FACS Canto (BD biosciences). Data was analyzed using FlowJo (Tree Star) software and plotted the mean fluorescence intensity (MFI) as described previously (Setty et al., 2007).
Subcellular fractionation
Subcellular fractionation was carried out using a protocol described earlier (Mahanty et al., 2016). Briefly, melanocytes were harvested, washed with 1XPBS and suspended in 0.25 M sucrose buffer (0.25 M sucrose, 1 mM EDTA, 25 mM HEPES pH 7.4, 0.02% sodium azide and protease inhibitor cocktail). Cells were homogenized on ice using Dounce homogenizer and then clarified by centrifugation at 600 g for 10 min at 4°C. The cell lysate was fractionated on a sucrose step gradient (2.0 M, 1.6 M, 1.4 M and 1.2 M sucrose buffers manually layered from bottom to top in ultracentrifuge tube) using SW55TI rotor by spinning at 160000 g at 4°C for 4 - 6 h in a Beckman L-80 ultracentrifuge. Fractions were manually separated and subjected to immunoblotting. In Fig. S4, percent enrichment of each protein in the fraction was calculated from the protein band densities, normalized with 1st fraction and then plotted as a graph.
Immunoprecipitation
HeLa cells expressing GFP alone (as a control) or GFP- tagged expression constructs were subjected to immunoprecipitation using GFP-Trap_A beads (Chromotek). Briefly, cells were lysed in lysis buffer (20 mM HEPES pH7.4, 100 mM KCl, 10 mM MgCl2, 5 mM EDTA, 1% Triton-100, 100 μM GTPγS, and protease inhibitor cocktail) on ice for 30 min. The lysates were centrifuged at 20000 g for 10 min at 4°C and then incubated with equilibrated GFP-Trap_A beads for 4 – 5 h under constant mixing at 4°C. The beads were washed twice with wash buffer (20 mM HEPES pH7.4, 100 mM KCl, 10 mM MgCl2, 5 mM EDTA and 0.1% Triton-100), suspended in 2X SDS-sample buffer and then analyzed by the immunoblotting. Similarly, immunoprecipitation of Myc-tagged proteins expressing in HeLa cells or endogenous Rabaptin-5 or Rab4 in melan-Ink cells was carried out using anti-Myc or anti-Rabaptin-5 or anti-Rab4 antibodies respectively. These lysates were incubated with Protein G-Sepharose 4B beads (Invitrogen) for overnight under constant mixing at 4°C. Finally, beads were washed with wash buffer, suspended in sample buffer and analyzed by immunoblotting.
Statistical analysis
Statistical significance was determined by the unpaired Student’s t-test and variance analysis using the GraphPad software. All values are described as the mean±s.e.m. Ns, not significant (p=ns); *, p≤0.05; **, p≤0.01 and ***, p≤0.001.
Supplementary Material
Summary.
Sorting endosomes segregate the cargo to recycling/late endosomes and melanosomes in melanocytes. Nag et al. show that Rab4A controls cargo segregation by forming a complex with Rabenosyn-5, KIF3 and AP-3 on sorting endosomes to divert the cargo toward melanosomes.
Acknowledgements
We thank M.S. Marks, J.S. Bonifacino and A. Peden for generous gifts of reagents; E.V. Sviderskaya and D.C. Bennett for mouse melanocytes. We also thank D. Dey, R. Keerthana, C. Praneeth and A.B. Sneha for their technical help. We acknowledge the shRNA resource center, Bioimaging, flow cytometry and PICT-IBiSA EM facilities. This work was supported by a Indo-French Centre for the Promotion of Advanced Research (CEFIPRA) Project (4903-1 to S.R.G.S. and G.R.); Wellcome Trust-DBT India Alliance Senior Fellowship (500122/Z/09/Z to S.R.G.S.); Department of Biotechnology (DBT)-RNAi task-force (BT/PR4982/AGR/36/718/2012 to S.R.G.S.); IISc-DBT partnership program (to S.R.G.S.); Institut Curie, CNRS-Foundation ARC pour la Re-cherche sur le Cancer (SL220100601359 to G.R.); Labex CelTisPhyBio. Post-doctoral fellowship (to C.B.); DBT-RA program (to P.A.) and Medical Research Council (MRC) core support to the MRC/UCL Laboratory for Molecular Cell Biology University Unit (MC_UU_1201814 to AS and CA).
Abbreviations
- EE
early endosomes
- SE
sorting endosome
- RE
recycling endosome
- AP
adaptor protein
- BF
bright-field
- HPS
Hermansky-Pudlak syndrome
- IFM
immunofluorescence microscopy
- TYRP1
tyrosinase-related protein
Footnotes
Author contributions. S.N. designed and performed majority of the experiments in this study. S.R. performed all Y2H assays and the few experiments relating to subcellular fractionation, IP, IFM. S.M. carried out experiments required for manuscript revision. C.B. performed electron microscopy. P.A. standardized the exosome experiments. C.A and A.S. provided several DNA constructs and technical help in few experiments. P.v.d.S., C.D., G.v.N. and G.R. provide the scientific support throughout the project period and shared the crucial reagents for several experiments. S.R.G.S. oversaw the entire project, coordinated and discussed the work with coauthors, and wrote the manuscript.
Competing financial interest: Authors have no competing financial interest
References
- Aledo JC, Darakhshan F, Hundal HS. Rab4, but not the transferrin receptor, is colocalized with GLUT4 in an insulin-sensitive intracellular compartment in rat skeletal muscle. Biochem Biophys Res Commun. 1995;215:321–8. doi: 10.1006/bbrc.1995.2469. [DOI] [PubMed] [Google Scholar]
- Atul Jani R, Nag S, Setty SR. Visualization of Intracellular Tyrosinase Activity in vitro. Bio Protoc. 2016;6:e1794. doi: 10.21769/bioprotoc.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Burton A, Ruiz-Mateos E, Marsh M, Saiardi A. Inositol pyrophosphate mediated pyrophosphorylation of AP3B1 regulates HIV-1 Gag release. Proc Natl Acad Sci U S A. 2009;106:21161–6. doi: 10.1073/pnas.0909176106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bananis E, Nath S, Gordon K, Satir P, Stockert RJ, Murray JW, Wolkoff AW. Microtubule-dependent movement of late endocytic vesicles in vitro: requirements for Dynein and Kinesin. Mol Biol Cell. 2004;15:3688–97. doi: 10.1091/mbc.E04-04-0278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berson JF, Harper DC, Tenza D, Raposo G, Marks MS. Pmel17 initiates premelanosome morphogenesis within multivesicular bodies. Mol Biol Cell. 2001;12:3451–64. doi: 10.1091/mbc.12.11.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bissig C, Rochin L, van Niel G. PMEL Amyloid Fibril Formation: The Bright Steps of Pigmentation. Int J Mol Sci. 2016;17 doi: 10.3390/ijms17091438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissy RE, Moellmann GE, Lerner AB. Morphology of melanocytes in hair bulbs and eyes of vitiligo mice. Am J Pathol. 1987;127:380–8. [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–66. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nat Rev Mol Cell Biol. 2003;4:409–14. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- D'Souza RS, Semus R, Billings EA, Meyer CB, Conger K, Casanova JE. Rab4 orchestrates a small GTPase cascade for recruitment of adaptor proteins to early endosomes. Curr Biol. 2014;24:1187–98. doi: 10.1016/j.cub.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Renzis S, Sonnichsen B, Zerial M. Divalent Rab effectors regulate the sub-compartmental organization and sorting of early endosomes. Nat Cell Biol. 2002;4:124–33. doi: 10.1038/ncb744. [DOI] [PubMed] [Google Scholar]
- Delevoye C, Hurbain I, Tenza D, Sibarita JB, Uzan-Gafsou S, Ohno H, Geerts WJ, Verkleij AJ, Salamero J, Marks MS, et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J Cell Biol. 2009;187:247–64. doi: 10.1083/jcb.200907122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delevoye C, Miserey-Lenkei S, Montagnac G, Gilles-Marsens F, Paul-Gilloteaux P, Giordano F, Waharte F, Marks MS, Goud B, Raposo G. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 2014;6:445–54. doi: 10.1016/j.celrep.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Deneka M, Neeft M, Popa I, van Oort M, Sprong H, Oorschot V, Klumperman J, Schu P, van der Sluijs P. Rabaptin-5alpha/rabaptin-4 serves as a linker between rab4 and gamma(1)-adaptin in membrane recycling from endosomes. EMBO J. 2003;22:2645–57. doi: 10.1093/emboj/cdg257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis MK, Mantegazza AR, Snir OL, Tenza D, Acosta-Ruiz A, Delevoye C, Zorger R, Sitaram A, de Jesus-Rojas W, Ravichandran K, et al. BLOC-2 targets recycling endosomal tubules to melanosomes for cargo delivery. J Cell Biol. 2015;209:563–577. doi: 10.1083/jcb.201410026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey S, Banker G, Ray K. Anterograde Transport of Rab4-Associated Vesicles Regulates Synapse Organization in Drosophila. Cell Rep. 2017;18:2452–2463. doi: 10.1016/j.celrep.2017.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggers CT, Schafer JC, Goldenring JR, Taylor SS. D-AKAP2 interacts with Rab4 and Rab11 through its RGS domains and regulates transferrin receptor recycling. J Biol Chem. 2009;284:32869–80. doi: 10.1074/jbc.M109.022582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouraux MA, Deneka M, Ivan V, van der Heijden A, Raymackers J, van Suylekom D, van Venrooij WJ, van der Sluijs P, Pruijn GJ. Rabip4' is an effector of rab5 and rab4 and regulates transport through early endosomes. Mol Biol Cell. 2004;15:611–24. doi: 10.1091/mbc.E03-05-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg J. The endocytic pathway: a mosaic of domains. Nat Rev Mol Cell Biol. 2001;2:721–30. doi: 10.1038/35096054. [DOI] [PubMed] [Google Scholar]
- Ha L, Ichikawa T, Anver M, Dickins R, Lowe S, Sharpless NE, Krimpenfort P, Depinho RA, Bennett DC, Sviderskaya EV, et al. ARF functions as a melanoma tumor suppressor by inducing p53-independent senescence. Proc Natl Acad Sci USA. 2007;104:10968–10973. doi: 10.1073/pnas.0611638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi K, Hayashi T, Jimbo T, Yamamoto T, Akiyama T. Role of the kinesin-2 family protein, KIF3, during mitosis. J Biol Chem. 2006;281:4094–9. doi: 10.1074/jbc.M507028200. [DOI] [PubMed] [Google Scholar]
- Hida T, Sohma H, Kokai Y, Kawakami A, Hirosaki K, Okura M, Tosa N, Yamashita T, Jimbow K. Rab7 is a critical mediator in vesicular transport of tyrosinase-related protein 1 in melanocytes. J Dermatol. 2011;38:432–41. doi: 10.1111/j.1346-8138.2010.01004.x. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Popa I, Futai K, Martinez-Sanchez E, Wulf PS, van Vlijmen T, Dortland BR, Oorschot V, Govers R, Monti M, et al. Neuron specific Rab4 effector GRASP-1 coordinates membrane specialization and maturation of recycling endosomes. PLoS Biol. 2010;8:e1000283. doi: 10.1371/journal.pbio.1000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura T, Huang J, Usui I, Satoh H, Bever J, Olefsky JM. Insulin-Induced GLUT4 Translocation Involves Protein Kinase C- -Mediated Functional Coupling between Rab4 and the Motor Protein Kinesin. Molecular and Cellular Biology. 2003;23:4892–4900. doi: 10.1128/MCB.23.14.4892-4900.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan V, Martinez-Sanchez E, Sima LE, Oorschot V, Klumperman J, Petrescu SM, van der Sluijs P. AP-3 and Rabip4' coordinately regulate spatial distribution of lysosomes. PLoS One. 2012;7:e48142. doi: 10.1371/journal.pone.0048142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani RA, Mahanty S, Setty SR. SNAREs in the maturation and function of LROs. Bioarchitecture. 2016;6:1–11. doi: 10.1080/19490992.2015.1131890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jani RA, Purushothaman LK, Rani S, Bergam P, Setty SR. STX13 regulates cargo delivery from recycling endosomes during melanosome biogenesis. J Cell Sci. 2015;128:3263–76. doi: 10.1242/jcs.171165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalin S, Hirschmann DT, Buser DP, Spiess M. Rabaptin5 is recruited to endosomes by Rab4 and Rabex5 to regulate endosome maturation. J Cell Sci. 2015;128:4126–37. doi: 10.1242/jcs.174664. [DOI] [PubMed] [Google Scholar]
- Klumperman J, Raposo G. The complex ultrastructure of the endolysosomal system. Cold Spring Harb Perspect Biol. 2014;6:a016857. doi: 10.1101/cshperspect.a016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay AJ, Hendrick AG, Cantalupo G, Senic-Matuglia F, Goud B, Bucci C, McCaffrey MW. Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J Biol Chem. 2002;277:12190–9. doi: 10.1074/jbc.M108665200. [DOI] [PubMed] [Google Scholar]
- Luzio JP, Hackmann Y, Dieckmann NM, Griffiths GM. The biogenesis of lysosomes and lysosome-related organelles. Cold Spring Harb Perspect Biol. 2014;6:a016840. doi: 10.1101/cshperspect.a016840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald C, Payne JA, Aboian M, Smith W, Katzmann DJ, Piper RC. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev Cell. 2015;33:328–42. doi: 10.1016/j.devcel.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahanty S, Ravichandran K, Chitirala P, Prabha J, Jani RA, Setty SR. Rab9A is required for delivery of cargo from recycling endosomes to melanosomes. Pigment Cell Melanoma Res. 2016;29:43–59. doi: 10.1111/pcmr.12434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mari M, Macia E, Le Marchand-Brustel Y, Cormont M. Role of the FYVE finger and the RUN domain for the subcellular localization of Rabip4. J Biol Chem. 2001;276:42501–8. doi: 10.1074/jbc.M104885200. [DOI] [PubMed] [Google Scholar]
- Marks MS, Heijnen HF, Raposo G. Lysosome-related organelles: unusual compartments become mainstream. Curr Opin Cell Biol. 2013;25:495–505. doi: 10.1016/j.ceb.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattera R, Arighi CN, Lodge R, Zerial M, Bonifacino JS. Divalent interaction of the GGAs with the Rabaptin-5-Rabex-5 complex. EMBO J. 2003;22:78–88. doi: 10.1093/emboj/cdg015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohrmann K, Gerez L, Oorschot V, Klumperman J, van der Sluijs P. Rab4 function in membrane recycling from early endosomes depends on a membrane to cytoplasm cycle. J Biol Chem. 2002;277:32029–35. doi: 10.1074/jbc.M203064200. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–6. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Nagelkerken B, Van Anken E, Van Raak M, Gerez L, Mohrmann K, Van Uden N, Holthuizen J, Pelkmans L, Van Der Sluijs P. Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem J. 2000;346(Pt 3):593–601. [PMC free article] [PubMed] [Google Scholar]
- Navaroli DM, Bellve KD, Standley C, Lifshitz LM, Cardia J, Lambright D, Leonard D, Fogarty KE, Corvera S. Rabenosyn-5 defines the fate of the transferrin receptor following clathrin-mediated endocytosis. Proc Natl Acad Sci U S A. 2012;109:E471–80. doi: 10.1073/pnas.1115495109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen E, Christoforidis S, Uttenweiler-Joseph S, Miaczynska M, Dewitte F, Wilm M, Hoflack B, Zerial M. Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J Cell Biol. 2000;151:601–12. doi: 10.1083/jcb.151.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohbayashi N, Fukuda M. Role of Rab family GTPases and their effectors in melanosomal logistics. J Biochem. 2012;151:343–51. doi: 10.1093/jb/mvs009. [DOI] [PubMed] [Google Scholar]
- Pagano A, Crottet P, Prescianotto-Baschong C, Spiess M. In vitro formation of recycling vesicles from endosomes requires adaptor protein-1/clathrin and is regulated by rab4 and the connector rabaptin-5. Mol Biol Cell. 2004;15:4990–5000. doi: 10.1091/mbc.E04-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Guo X. Adaptor protein complexes and intracellular transport. Biosci Rep. 2014;34 doi: 10.1042/BSR20140069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J. Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol. 2004;164:1065–76. doi: 10.1083/jcb.200311064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer SR. Rab GTPase regulation of membrane identity. Curr Opin Cell Biol. 2013;25:414–9. doi: 10.1016/j.ceb.2013.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris R, Klumperman J, Chen YA, Scheller RH. Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J Cell Biol. 1998;143:957–971. doi: 10.1083/jcb.143.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Marks MS, Cutler DF. Lysosome-related organelles: driving post-Golgi compartments into specialisation. Curr Opin Cell Biol. 2007;19:394–401. doi: 10.1016/j.ceb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G, Tenza D, Murphy DM, Berson JF, Marks MS. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J Cell Biol. 2001;152:809–24. doi: 10.1083/jcb.152.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochin L, Hurbain I, Serneels L, Fort C, Watt B, Leblanc P, Marks MS, De Strooper B, Raposo G, van Niel G. BACE2 processes PMEL to form the melanosome amyloid matrix in pigment cells. Proc Natl Acad Sci U S A. 2013;110:10658–63. doi: 10.1073/pnas.1220748110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rous BA, Reaves BJ, Ihrke G, Briggs JA, Gray SR, Stephens DJ, Banting G, Luzio JP. Role of adaptor complex AP-3 in targeting wild-type and mutated CD63 to lysosomes. Mol Biol Cell. 2002;13:1071–82. doi: 10.1091/mbc.01-08-0409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MR, Maritzen T, Kukhtina V, Higman VA, Doglio L, Barak NN, Strauss H, Oschkinat H, Dotti CG, Haucke V. Regulation of endosomal membrane traffic by a Gadkin/AP-1/kinesin KIF5 complex. Proc Natl Acad Sci U S A. 2009;106:15344–9. doi: 10.1073/pnas.0904268106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Sviderskaya EV, Bennett DC, Raposo G, Marks MS. Cell-specific ATP7A transport sustains copper-dependent tyrosinase activity in melanosomes. Nature. 2008;454:1142–6. doi: 10.1038/nature07163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setty SR, Tenza D, Truschel ST, Chou E, Sviderskaya EV, Theos AC, Lamoreux ML, Di Pietro SM, Starcevic M, Bennett DC, et al. BLOC-1 is required for cargo-specific sorting from vacuolar early endosomes toward lysosome-related organelles. Mol Biol Cell. 2007;18:768–80. doi: 10.1091/mbc.E06-12-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaram A, Marks MS. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology. 2012;27:85–99. doi: 10.1152/physiol.00043.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnichsen B, De Renzis S, Nielsen E, Rietdorf J, Zerial M. Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J Cell Biol. 2000;149:901–14. doi: 10.1083/jcb.149.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–25. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Takeuchi H, Takada A, Kuboniwa M, Amano A. Intracellular periodontal pathogen exploits recycling pathway to exit from infected cells. Cell Microbiol. 2015 doi: 10.1111/cmi.12551. [DOI] [PubMed] [Google Scholar]
- Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, Banki K, Perl A. HRES-1/Rab4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy. PLoS One. 2014;9:e84392. doi: 10.1371/journal.pone.0084392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, Tenza D, Martina JA, Hurbain I, Peden AA, Sviderskaya EV, Stewart A, Robinson MS, Bennett DC, Cutler DF, et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell. 2005;16:5356–5372. doi: 10.1091/mbc.E05-07-0626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia JC, Watabe H, Chi A, Rouzaud F, Chen KG, Vieira WD, Takahashi K, Yamaguchi Y, Berens W, Nagashima K, et al. Sorting of Pmel17 to melanosomes through the plasma membrane by AP1 and AP2: evidence for the polarized nature of melanocytes. J Cell Sci. 2006;119:1080–91. doi: 10.1242/jcs.02804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Webster P, Male P, Goud B, Mellman I. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell. 1992;70:729–40. doi: 10.1016/0092-8674(92)90307-x. [DOI] [PubMed] [Google Scholar]
- Van Der Sluijs P, Hull M, Zahraoui A, Tavitian A, Goud B, Mellman I. The small GTP-binding protein rab4 is associated with early endosomes. Proc Natl Acad Sci U S A. 1991;88:6313–7. doi: 10.1073/pnas.88.14.6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel G, Charrin S, Simoes S, Romao M, Rochin L, Saftig P, Marks MS, Rubinstein E, Raposo G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev Cell. 2011;21:708–21. doi: 10.1016/j.devcel.2011.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal MJ, Stahl PD. The small GTP-binding proteins Rab4 and ARF are associated with released exosomes during reticulocyte maturation. Eur J Cell Biol. 1993;60:261–7. [PubMed] [Google Scholar]
- Vitale G, Rybin V, Christoforidis S, Thornqvist P, McCaffrey M, Stenmark H, Zerial M. Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 1998;17:1941–51. doi: 10.1093/emboj/17.7.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. KIF3A/B: a heterodimeric kinesin superfamily protein that works as a microtubule plus end-directed motor for membrane organelle transport. J Cell Biol. 1995;130:1387–99. doi: 10.1083/jcb.130.6.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Nakata T, Okada Y, Hirokawa N. Cloning and characterization of KAP3: a novel kinesin superfamily-associated protein of KIF3A/3B. Proc Natl Acad Sci U S A. 1996;93:8443–8. doi: 10.1073/pnas.93.16.8443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao P, Zhao H, Mo W, He P. Laminar Shear Stress Promotes Vascular Endothelial Cell Autophagy Through Upregulation with Rab4. DNA Cell Biol. 2016;35:118–23. doi: 10.1089/dna.2015.3041. [DOI] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–17. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhu GD, Salazar G, Zlatic SA, Fiza B, Doucette MM, Heilman CJ, Levey AI, Faundez V, L'Hernault SW. SPE-39 family proteins interact with the HOPS complex and function in lysosomal delivery. Mol Biol Cell. 2009;20:1223–40. doi: 10.1091/mbc.E08-07-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.