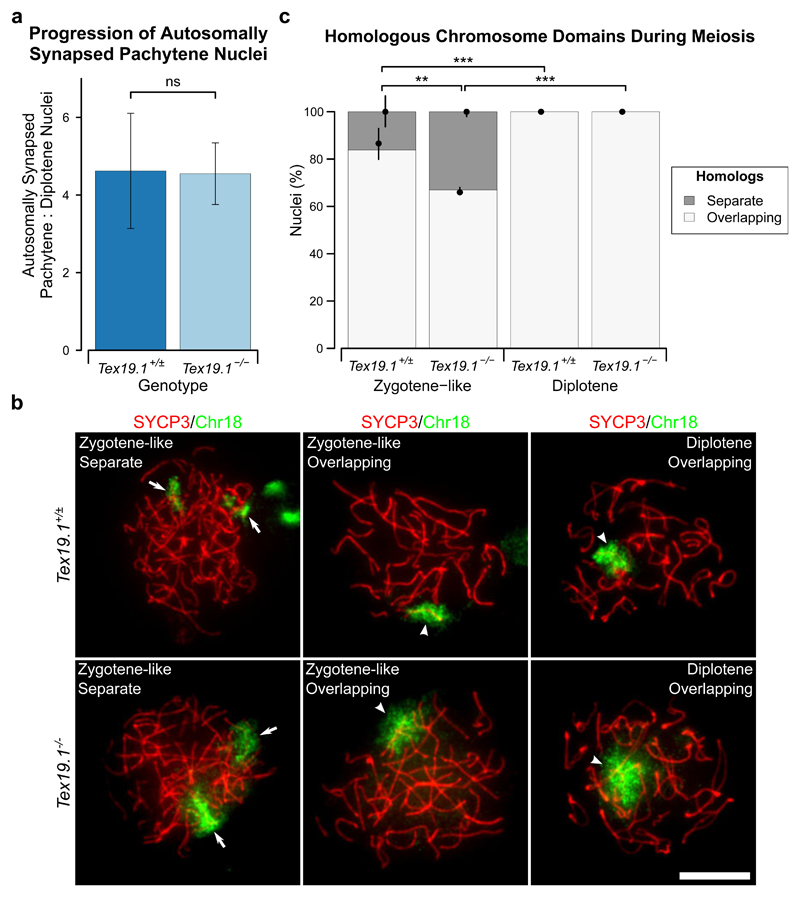
Fig. 4. Autosomally Synapsed, but Not Asynapsed, Tex19.1-/- Spermatocytes Progress to Diplotene.
a Ratio of autosomally synapsed pachytene to diplotene nuclei in meiotic chromosome spreads from Tex19.1+/± and Tex19.1-/- testes. Ratios are derived from the meiotic progression data shown in Online Resource 1 and were obtained from a total of 462 Tex19.1+/± and 459 Tex19.1-/- nuclei from three animals for each genotype. ns indicates no significant difference (4.6±1.5, 4.5±0.8; p = 0.94, Student’s t-test). b ImmunoFISH for chromosome 18 in zygotene-like and diplotene Tex19.1+/± and Tex19.1-/- spermatocytes. SYCP3 (red) marks the lateral element of the synaptonemal complex, the chromosome 18 paint is shown in green. The two homologs were scored as having overlapping (arrowheads) or separate (arrows) chromatin domains. Chromosome 18 FISH signals from an adjacent SYCP3-negative nucleus are visible in the top left panel. Scale bar 10 μm. c Quantification of chromosome domain configurations in Tex19.1+/± and Tex19.1-/- spermatocytes. 273 Tex19.1+/± and 335 Tex19.1-/- spermatocyte nuclei at zygotene-like or diplotene stage of meiosis I were categorised. FISH data for chromosome 18 and chromosome 19 were similar between genotypes and meiotic stage and were combined to provide more statistical power. 16.2% of 102 Tex19.1+/± zygotene-like nuclei and 33.1% of 121 Tex19.1-/- zygotene-like nuclei had separate chromosome domains, all diplotene nuclei had overlapping domains. Asterisks denote significance (Fisher’s exact test, ** p < 0.01, *** p < 0.001). Nuclei were obtained from three animals for each genotype, and circles and vertical lines represent the means and interquartile distances of animals within each genotype.