Abstract
Polyphenols have shown promising bioactivity in experimental in vitro and in vivo models for cancer chemoprevention. However, consumed orally, they are often transformed by gut microbes into new active principles with so far incompletely deciphered molecular mechanisms. Here, enterolacton, S-equol and urolithin A as representatives of metabolites of lignans, isoflavones and ellagitannins, respectively, were examined for their impact on HCT116 colon cancer cell growth, cooperativity with oxaliplatin and p53 dependency in vitro. Whereas enterolacton and S-equol (≤60 μM) did not elicit growth inhibition or positive cooperativity with oxaliplatin, urolithin A showed an IC50 value of 19 μM (72 h) and synergism with oxaliplatin. Urolithin A induced p53 stabilization and p53 target gene expression, and absence of p53 significantly dampened the antiproliferative effect of urolithin A (IC50(p53−/−) = 38 μM). P53 was dispensable for the G2/M arrest in HCT116 cells but required for induction of a senescence-like phenotype upon long-term exposure and for the observed synergism with oxaliplatin. Moreover, extracellular flux analyses and knockdown approaches uncovered a reduced glycolytic potential via the p53/TIGAR axis which was linked to the higher susceptibility of wildtype cells to urolithin A. Overall, the p53 status turned out to be an important determinant for the potential benefit of dietary ellagitannins in cancer chemoprevention or use in adjuvant therapy.
Introduction
Polyphenols are readily consumed by diet or herbal supplements and claimed to exert multiple beneficial bioactivities including cancer chemoprevention (1). Statements on bioactivity are mainly based on observational studies in animals or epidemiological studies in humans and/or in vitro tests with the isolated compounds/mixtures often at (high) micromolar concentrations. However, the relevance of the latter may be questioned as oral consumption of those compounds is in most cases accompanied with low systemic bioavailability and bacterial metabolization in the gut. The arising new chemical entities usually possess higher bioavailability and/or altered activity profile. Thus, in order to understand the effect of orally consumed phenolic constituents and give valid recommendations for use, one needs to capture among others (i) the activity profile of the bioavailable metabolite and its possible interaction with drugs and (ii) the individual prerequisites on the patients’ side to allow optimal efficacy.
In this line, we focused on the antiproliferative activity of the selected microbial metabolites from polyphenolic lignans, isoflavones and ellagitannins toward cancer cells from colon, i.e. the site of their formation and presumably high achievable local concentrations after dietary consumption. The main source of dietary lignans, such as secoisolariciresinol (glycoside), is flaxseed, and together with their bacterial metabolites, enterolacton and enterodiol, they are considered to reduce risk factors of cancer, cardiovascular disease and diabetes (2). After dietary intervention with flaxseed for several weeks, plasma concentrations of enterolactone increased significantly in the range of nM, with age as a determinant for bioavailability. Higher tissue concentrations were found in various organs including intestine, kidney and uterus (3,4). Enterolignans may hold promise as antioxidative, anti-inflammatory, antiproliferative as well as weak estrogenic or antiestrogenic entities (4,5). Isoflavones occur in a variety of leguminous plants such as soybeans containing the glycoside forms daidzin and daidzein. Bacterial β-glucosidases are capable of releasing the unconjugated isoflavones which enter the circulation. After mainly glucuronidation and sulfation, they are excreted via bile to the intestine, where they are microbially converted to equol. Due to a chiral center, gut bacteria only synthesize the S-(−) equol enantiomer. Notably, of the adult population only 25–30% in western countries, but 50–60% in Asian countries, harbor the required colonic bacteria and are thus equol producers (6–8). After the intake of soy-based formulations, maximum plasma concentrations of equol occurred after ~16 h at around 130 ng/ml (~0.5 μM) (9,10). S-(−) equol shows both estrogenic properties as a selective estrogen-receptor β modulator and antiproliferative effects on prostatic epithelial cells (7,11). Orally consumed ellagitannins are hydrolyzed in the gut to release ellagic acid, which is further processed by certain gut bacteria into a series of urolithins with distinct hydroxylation pattern and subjected to phase 2 metabolism. Strongly depending on the composition of the gut microbiome, prevalent metabolites in humans are conjugates with glucuronic acid of urolithin A, isourolithin A and urolithin B. These circulate in human plasma with huge interindividual variability in the range of 0.01–70 μM. Under a dietary approach, it is unlikely that substantial amounts of free urolithin aglycones reach the systemic circulation. However, a local tissue distribution of 4.8–507.3 ng/g for several aglycones was found in colon (12,13). With respect to their bioactivity, urolithins were already shown to inhibit proliferation of different cancer cells and to exert anti-inflammatory or lifespan prolonging properties (14–18).
In this study, we examined growth inhibition in HCT116 colon cancer cells by enterolacton, S-equol and urolithin A and their interaction with the standard chemotherapeutic drug oxaliplatin. Moreover, we assessed the importance of the tumor suppressor p53, commonly mutated in (colon) cancer and a known determinant of drug efficacy, and its downstream signaling events for an observed growth inhibition.
Materials and methods
Chemicals, siRNA and antibodies
Urolithin A and S-equol (purity ≥98%) were obtained from Santa Cruz (Germany), siRNA targeting human TIGAR (ON-Targetplus; LQ-020597-01), and scrambled control siRNA were purchased from Dharmacon (via THP, Austria), Oligofectamine came from Invitrogen (via Life Tech, Austria) and all other chemicals, including oxaliplatin and enterolacton, were obtained from Sigma–Aldrich (Austria). The primary antibody against p21 (#ab109520) was obtained from Abcam (UK), the anti-p53 (#9282), and anti-TIGAR (#14751), anti-tubulin (#2144) and secondary antibodies were from Cell Signaling Technology (Germany) and the anti-actin (Clone C4; #08691001) antibody was from MP Biomedicals (Germany).
Cell cultivation
The human colon carcinoma HCT116 (WT, p53−/− and p21−/−) cell lines were kind gifts from Bert Vogelstein, Johns Hopkins University, USA. The cell lines were authenticated and declared free from other cell contaminations (22 June 2018) by short tandem repeat profiling (Microsynth, Switzerland), and respective knockouts were confirmed on protein and mRNA level by qPCR and immunoblot. Cells were maintained in DMEM medium (phenol-red free; Lonza, Switzerland) supplemented with 10% fetal calf serum (Gibco, Germany), 2 mM glutamine (Lonza), 100 U/ml benzylpenicillin (Lonza), 100 μg/ml streptomycin (Lonza) at 37°C and 5% CO2 in a humidified atmosphere. For subcultivation, cells at 75–90% confluency were detached from the cell culture dish, and an appropriate aliquot was transferred to a new dish and medium: 5–8 × 103 cells/well were seeded in 96-well plates (48–72 h incubation), 0.5–1 × 106 cells/well in 6-well plates (24-h incubation) and 3 × 106 cells/well in 6 cm dishes (24-h incubation). All test compounds were applied as a stock solution in DMSO. Solvent concentration was even throughout all samples of one experiment and never exceeded 0.3%.
Determination of biomass and metabolic activity after single and combinatorial treatments
Cells were seeded in 96-well plates and treated with test compounds in quadruplicates as indicated. After the desired incubation period, medium was aspirated and cells were washed with phosphate buffered saline. Then resazurin solution (10 μg/ml in serum-free growth medium, 100 μl) was added, and after an incubation of 2–3 h, the conversion of resazurin to resorufin was determined in a fluorimeter (TECAN, exc. 530/em. 590 nm). After that, the supernatant was discarded, and the biomass was stained with crystal violet (0.5% crystal violet, 20% methanol) for 5 min. Excessive dye was rinsed off with tab water, and after drying of the plates, the dye was solubilized (with 0.05 M citrate in 50% ethanol) and quantified by spectrophotometry at 595 nm. IC50 values were usually determined by curve fitting of at least six different concentrations of test compound using GraphPad Prism software. In the case of combinatorial treatment of two active compounds, the combinatorial index (CI) was calculated as with a, b being the concentrations in the combination and A, B the single doses of the compounds to mediate a given effect (here: 50% inhibition). CI values >1 were indicative for antagonistic and CI values <1 for synergistic action (19–22).
Assessment of CPD and SA-β gal
Cells were counted using a ViCell cell viability analyzer (Beckman Coulter). Population doublings (PD) were calculated using the formula where N(t) is the number of cells per well at the time of harvest and N(t0) is the number of cells initially seeded. The sum of PD is cumulative population doublings (CPD) and can be plotted versus the duration of cultivation. Senescence-associated beta galactosidase (SA-β-gal) was visualized using X-gal (5-bromo-4-chloro-3-indolyl-B-d-galactopyranoside) (23). Stained cells were examined using a light microscope (and expressed as percentage of blue cells in at least five pictures of each sample).
Flow cytometric cell-cycle analysis
Cells were treated with test compounds as indicated. After detachment by accutase, cells were washed with phosphate buffered saline and then resuspended in a hypotonic propidium iodide (PI) solution containing 0.1% (v/v) Triton X-100, 0.1% (w/v) sodium citrate and 50 μg/ml PI. After incubation at 4°C overnight, PI-stained nuclei were analyzed on a FACSCalibur™ (BD Biosciences, Vienna, Austria) flow cytometer at an excitation wavelength of 488 nm and an emission wavelength of 585 nm.
Western blot analysis
After the desired cell treatment extraction of total cell lysates, SDS-polyacrylamide electrophoresis and immunoblot analysis were performed essentially as described previously (24). Enhanced chemoluminescence-based image acquisition was done on a Fuji LAS300 system coupled with densitometric analysis in the Multi Gauge software.
Determination of the cell energy phenotype by extracellular flux analysis
After treatment, as indicated, HCT1116 cells were spun on CellTak(Greiner)-coated 24-well cell culture plates (Agilent) at a density of 6–10 × 104 cells/well. Cells were kept in serum-free assay medium (DMEM; pH 7.3–7.4, 10 mM glucose, 1 mM pyruvate and 2 mM glutamine) at 37°C and ambient CO2 for 1 h prior to determination of the cell energy phenotype in a Seahorse 24XFe extracellular flux analyzer and evaluation with Wave software and proprietary XF Energy Phenotype Test Report Generator (details can be found on www.seahorsebio.com, Agilent). Briefly, basal extracellular acidification rates (ECAR in mpH/min) and oxygen consumption rates (OCR in pmol O2/min) were recorded over 25 min. Then addition of 1 μM oligomycin (an inhibitor of mitochondrial ATP synthase) and 0.5 μM FCCP (carbonylcyanid-p-trifluoromethoxy-phenylhydrazone, an uncoupler of respiration) elicited a shift to stressed ECAR and OCR, again recorded for another 35 min. At the end of the measurement, the plate was subjected to a crystal violet staining to ensure that cells had remained evenly attached to the plate during the experiment. The metabolic potential was then calculated as
Values for DMSO-treated cells were usually set as 1 and compared with urolithin A-treated cells in order to clearly dissect a differential influence of urolithin A on the metabolic potential of WT and p53−/− HCT116 cells, respectively.
siRNA-mediated knockdown
Cells were seeded in 6 cm dishes and grown to 75% confluency. Then cells were washed with Opti-MEM® (Gibco™,ThermoFisher Scientific, Vienna, Austria) and transfected with 200 nM siRNA-targeting human TIGAR or scrambled siRNA using oligofectamine according to the manufacturer’s instructions (Invitrogen, ThermoFisher Scientific). After recovery, cells were seeded for the planned experiments, i.e. into 96-well plates (for biomass staining) or 12-well plates (for protein extraction and immunoblot analysis to assess knockdown efficiency).
Statistics
Experiments were performed at least in three independent biological replicates. Error bars in the pictures represent the standard deviation (SD). Statistical significance was determined by using Student’s t-test (for two groups) or ANOVA (>2 groups) followed by Tukey’s post-test in the GraphPad Prism software. P-values <0.05 were considered as significant and are designated with * in the figures.
Results
Influence of microbial polyphenol metabolites on HCT116 colon cancer cell growth
In a first step, we tested enterolacton, S-equol and urolithin A for their impact on HCT116 colon cancer cell growth. In a concentration range of 0–60 μM, only urolithin A was able to inhibit cell growth by >50% with an IC50 value of ~39.2 μM after 48 h and 19.6 μM after 72 h, based on biomass staining. Enterolacton and S-equol showed ~30% inhibition at 60 μM, whereas lower concentrations remained ineffective (data not shown). The chemotherapeutic oxaliplatin was used as positive control and showed IC50 values of 2.8 and 1.6 μM after 48 and 72 h, respectively (Table 1A). Similar values were obtained when assessing residual metabolic activity of the treated cells via resazurin conversion (Supplementary Table 1A, available at Carcinogenesis Online). Also in comparison with its precursors, the exemplary ellagitannin geraniin (IC50 = 83 μM after 72-h incubation) and ellagic acid (IC50 = 44.2 μM), urolithin A excelled with regard to inhibition of cell growth (Supplementary Table 1B, available at Carcinogenesis Online). Next, we examined whether the metabolites showed any positive or negative cooperativity with oxaliplatin. Enterolacton and S-equol did not positively boost but rather dampen the activity of oxaliplatin, as suggested by the reproducibly elevated IC50 values of oxaliplatin in the presence of different (per se inactive) concentrations of either compound. In contrast, urolithin A showed positive cooperativity with oxaliplatin as apparent in combinatorial indices <1 (CI: 0.66–0.82) at all tested combinations (Table 1B).
Table 1. Overview over IC50 values and combinatorial indices of test compounds in HCT116 cells.
| (A) | IC50 (48 h) | IC50 (72 h) |
| Enterolacton | >60 μMw | >60 μM |
| S-Equol | >60 μM | >60 μM |
| Urolithin A | 39.2 ± 3.1 μM | 19.6 ± 2.8 μM |
| Oxaliplatin | 2.8 ± 0.4 μM | 1.6 ± 0.5 μM |
| (B) Compounds | Conc. of oxaliplatin to reach 50% inhibition | CI |
| Oxaliplatin | 1.6 ± 0.5 μM | |
| Oxaliplatin + Enterolacton (5 μM) | 2.61 ± 0.3 μM | |
| Oxaliplatin + Enterolacton (10 μM) | 2.75 ± 0.2 μM | |
| Oxaliplatin + Enterolacton (20 μM) | 2.99 ± 0.6 μM | |
| Oxaliplatin + Enterolacton (40 μM) | 2.27 ± 0.4 μM | |
| Oxaliplatin + S-Equol (5 μM) | 3.13 ± 0.1 μM | |
| Oxaliplatin + S-Equol (10 μM) | 3.52 ± 0.4 μM | |
| Oxaliplatin + S-Equol (20 μM) | 2.82 ± 0.2 μM | |
| Oxaliplatin + S-Equol (40 μM) | 2.34 ± 0.8 μM | |
| Oxaliplatin + Urolithin A (1.25 μM) | 1.22 ± 0.2 μM | 0.82 |
| Oxaliplatin + Urolithin A (2.5 μM) | 0.85 ± 0.1 μM | 0.66 |
| Oxaliplatin + Urolithin A (5 μM) | 0.67 ± 0.1 μM | 0.67 |
| Oxaliplatin + Urolithin A (10 μM) | 0.35 ± 0.05 μM | 0.73 |
| Oxaliplatin + Urolithin A (15 μM) | 0.015 ± 0.002 μM | 0.77 |
| (C) Compounds | IC50 (72 h) p53−/− | Combinatoral index |
| Urolithin A | 38.4 ± 1.63 μM | |
| Oxaliplatin | 6.8 ± 1.21 μM | |
| Oxaliplatin + Urolithin A (2.5 μM) | 5.8 ± 0.15 μM | 0.92 |
| Oxaliplatin + Urolithin A (5 μM) | 5.5 ± 0.54 μM | 0.94 |
| Oxaliplatin + Urolithin A (10 μM) | 5.7 ± 0.63 μM | 1.09 |
| Oxaliplatin + Urolithin A (15 μM) | 5.1 ± 0.32 μM | 1.14 |
| Oxaliplatin + Urolithin A (20 μM) | 5.1 ± 0.24 μM | 1.27 |
| Oxaliplatin + Urolithin A (30 μM) | 3.8±0.32 μM | 1.21 |
WT or p53−/− HCT116 cells were seeded in 96-well plates and incubated with different concentrations of test compounds for 48 or 72 h prior to biomass determination via crystal violet staining. IC50 and CI values were determined as described in the method section and are means ± SD of at least three independent experiments.
p53-dependence of the antiproliferative activity of urolithin A
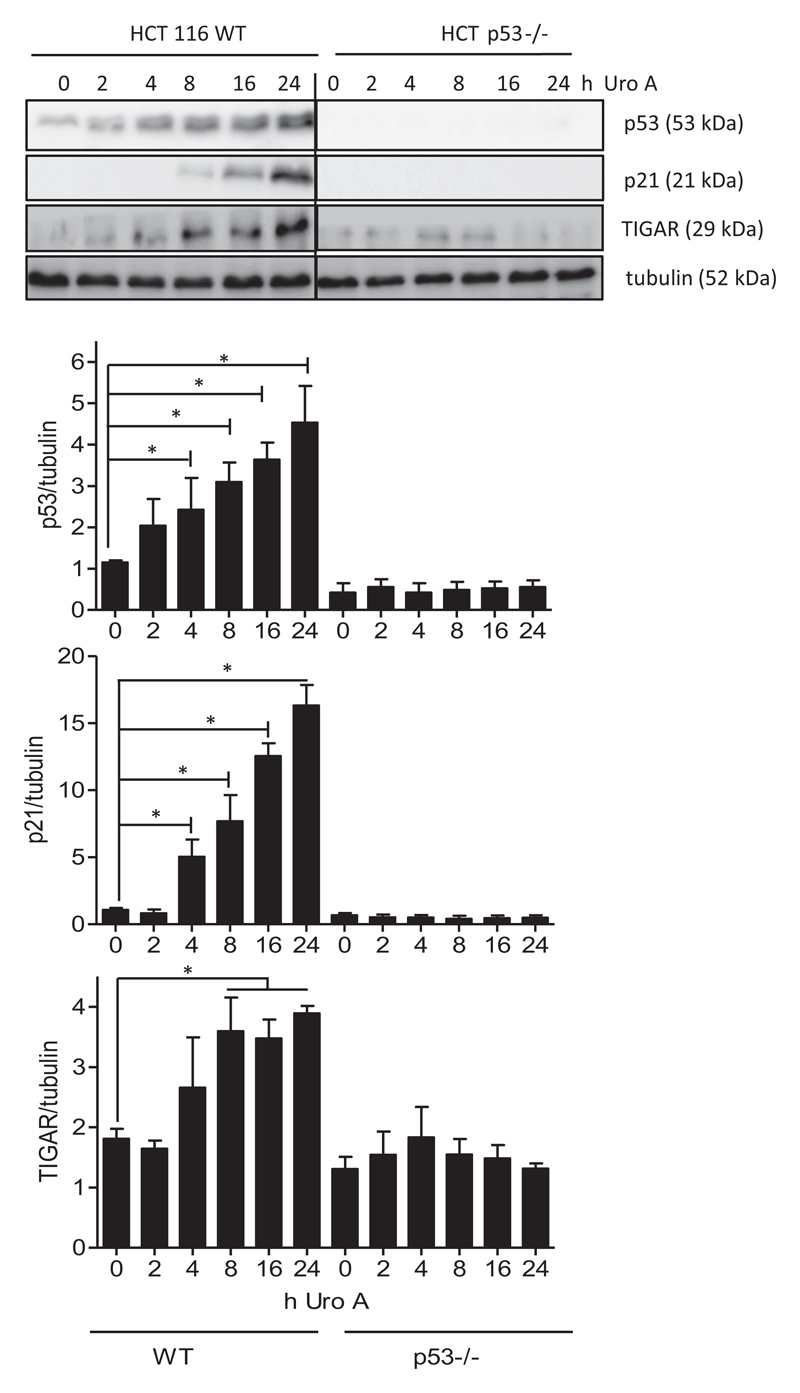
As urolithin A turned out to be the most promising of the tested compounds regarding antiproliferative activity, we focussed on this metabolite for a more detailed characterization of its mode of action. P53 is a tumor suppressor gene and mutated/dysfunctional in the majority of cancer cells, including colon carcinomas (25,26). Loss of p53 function allows cells to evade cell-cycle arrest or apoptosis, and the p53 status thus determines tumor progression and the efficacy of cytotoxic or cytostatic drugs (27,28). Urolithins, including urolithin A, have already been reported to stabilize p53 (29). However, to what extent p53 is actually needed for growth inhibition by urolithin A is not completely resolved. We therefore compared WT and p53−/− HCT cells in their susceptibility to urolithin A (Table 1C). Whereas WT cells are inhibited in their growth by urolithin A with an IC50 value of 19.6 μM (72h), their p53 knockout counterparts are still inhibited by urolithin A but at markedly higher IC50 values of around 38 μM (72 h). P53 dependency also applies for the antiproliferative activity of oxaliplatin that shows an IC50 value of 1.6 μM in WT and 6.6 μM in p53−/− cells, in line with previous observations (30). These findings suggest that urolithin A elicits p53-dependent and -independent signals that contribute to growth inhibition. Of note, the synergistic action between urolithin A and oxaliplatin seems to be diminished in the absence of p53 as the CI values only range between 0.91 and 1.27 for the p53−/− cells (Table 1C). In line with the supportive effect of p53 for urolithin A action, we observed time-dependent stabilization of p53 and upregulation of the p53 target genes p21 and TIGAR (TP53-induced glycolytic regulatory phosphatase) in WT but not p53−/− cells upon treatment with urolithin A (Figure 1). Comparing the influence in WT- and mutant p53 breast (MCF7 versus MDA-MB231) and liver (HepG versus HCC1.2), cancer cell lines also suggested a p53-dependent and -independent activity of urolithin A with enhanced growth inhibition in the presence of functional p53 signaling (Supplementary Table 2, available at Carcinogenesis Online). However, the latter statement needs to be regarded with caution as the distinct susceptibility to urolithin A of the respective non-isogenic p53 WT and mutant cell lines may be also due to differences other than p53 status, such as e.g estrogen-receptor expression or rate of phase II metabolism, and also vary with growth conditions (see (31) where the authors observe better inhibition of MDA-MB231 than MCF7 cells by urolithin A).
Figure 1.
Urolithin A leads to stabilization of p53 and expression of the p53 target genes p21 and TIGAR. WT and p53−/− HCT 116 cells were treated with 30 μM urolithin A for the indicated periods of time. Total cell lysates were then subjected to immunoblot analysis for p53, p21, TIGAR and tubulin as loading control. Representative pictures and compiled densitometric analyses from three independent experiments are depicted (mean ± SD, *P < 0.05, ANOVA, Tukey’s post-test). The bands of WT and p53 knockout lysates originate from one membrane; the line indicates that interjacent bands of no interest (i.e. standard, empty space) were cut out.
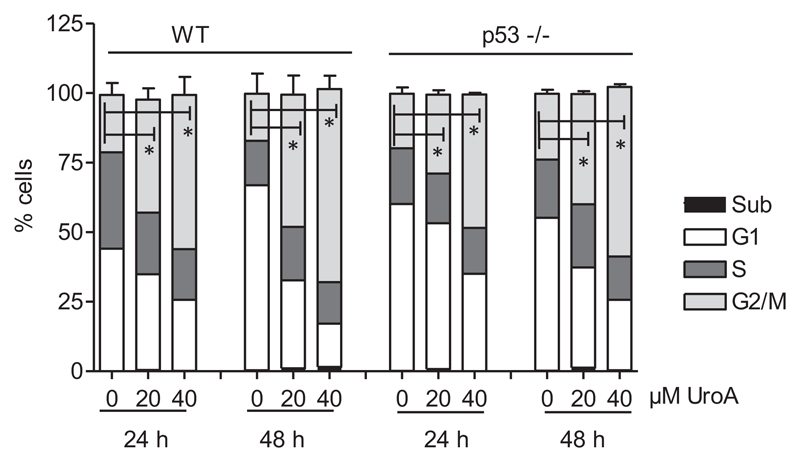
The role of p53 for urolithin A-mediated changes in cell-cycle progression
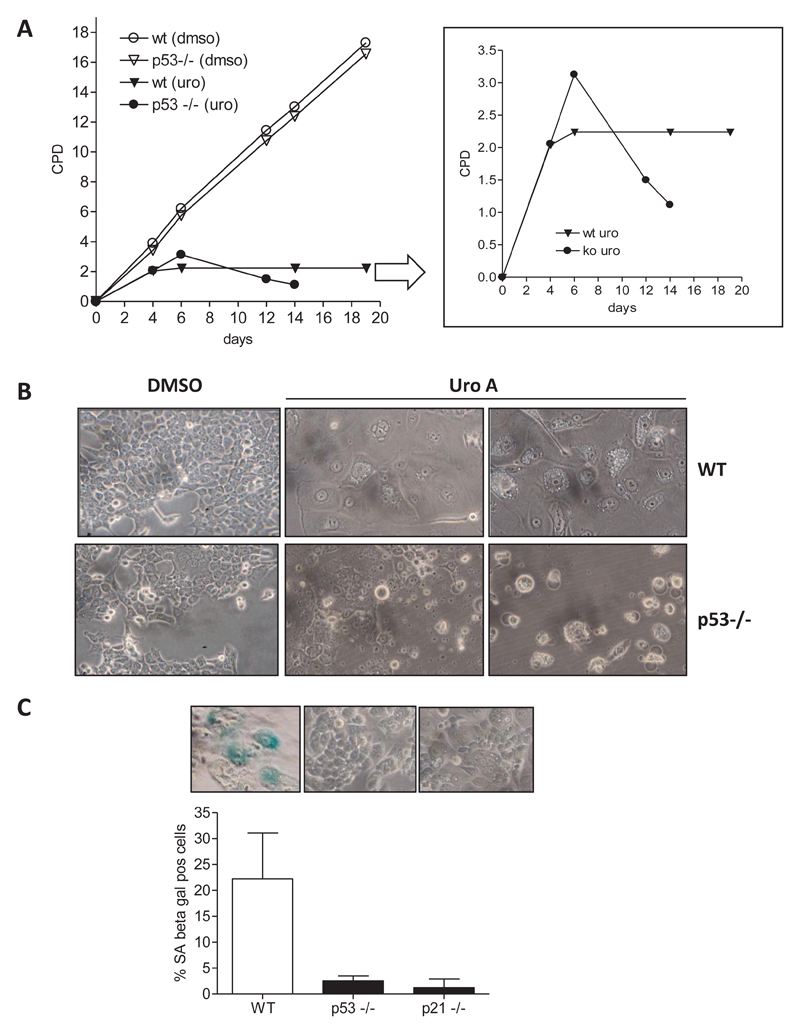
To get a clearer picture of the role of p53 in HCT116 cells, we compared cell-cycle progression, a process regulated by p53 and knowingly modulated by urolithin A (e.g (32,33)), in WT and p53−/− cells. Flow cytometric cell-cycle analysis after 24 and 48 h of treatment with urolithin A showed that both WT and p53−/− HCT116 cells accumulate in G2/M phase of the cell cycle without obviously inducing apoptosis (hardly any cell in the subG1 peak or detached/morphologically altered cells) (Figure 2). Likewise, after 48 h with 40 μM urolithin A, an average of 55% of WT and 47% of p53−/− HCT116 cells could be assigned to G2/M phase of the cell cycle compared with 20% and 24% in respective DMSO-treated control cells. Monitoring cell growth upon continuous treatment with urolithin A (renewal every other day) over several days revealed enhanced cell death in p53−/− cells, as evident in the declining number of CPD (Figure 3A). WT HCT cells reached a complete growth stop with urolithin A and appeared flat and bigger than solvent-treated cells. In line with these observed characteristics, the test for SA-β-gal was positive for those cells, but not for p53−/− counterparts (Figure 3B). Since the p53 target p21 is involved in senescence induction (34), we also tested p21−/− HCT116 cells for SA-β-gal activity upon long-term treatment with urolithin A. Like p53−/− cells also p21−/− cells did not show any signs of senescence (Figure 3B), indicating that the p53/p21 axis contributes to the growth arrest induced by urolithin A in WT cells.
Figure 2.
Urolithin A leads to accumulation of HCT116 cells in G2/M phase of the cell cycle. WT and p53−/− HCT 116 cells were treated with solvent (0.1% DMSO), 20 μM or 40 μM urolithin A for 24 and 48 h prior to PI staining of their nuclei and flow cytometric analysis of cell-cycle distribution. Bar graphs depict compiled data of three independent biological replicates. (mean ± SD, *P < 0.05, ANOVA, Tukey’s post-test).
Figure 3.
Urolithin A leads to p53/p21-dependent senescence-like growth arrest in HCT116 cells. (A) WT and p53−/− HCT 116 cells were subcultivated in the continuous presence of solvent (0.1% DMSO) or 20 μM urolithin A (medium exchange every other day), and CPD were plotted against time of cultivation. Representative curves of three independent experiments with consistent results are depicted. (B) Cells after 25 days of cultivation in DMSO or 20 μM Uro A containing growth medium were photographed. (C) WT, p53−/− and p21−/− were chronically treated with 20 μM urolithin A prior to fixation and staining for SA-β-gal. The percentage of blue (here: dark) stained and hence SA-Gal positive cells from at least five different samples is summarized and depicted in the bar graph. Only urolithin A-treated WT, p53−/− or p21−/− HCT116 cells are shown.
The role of p53 for urolithin A-mediated changes in cell energy phenotype
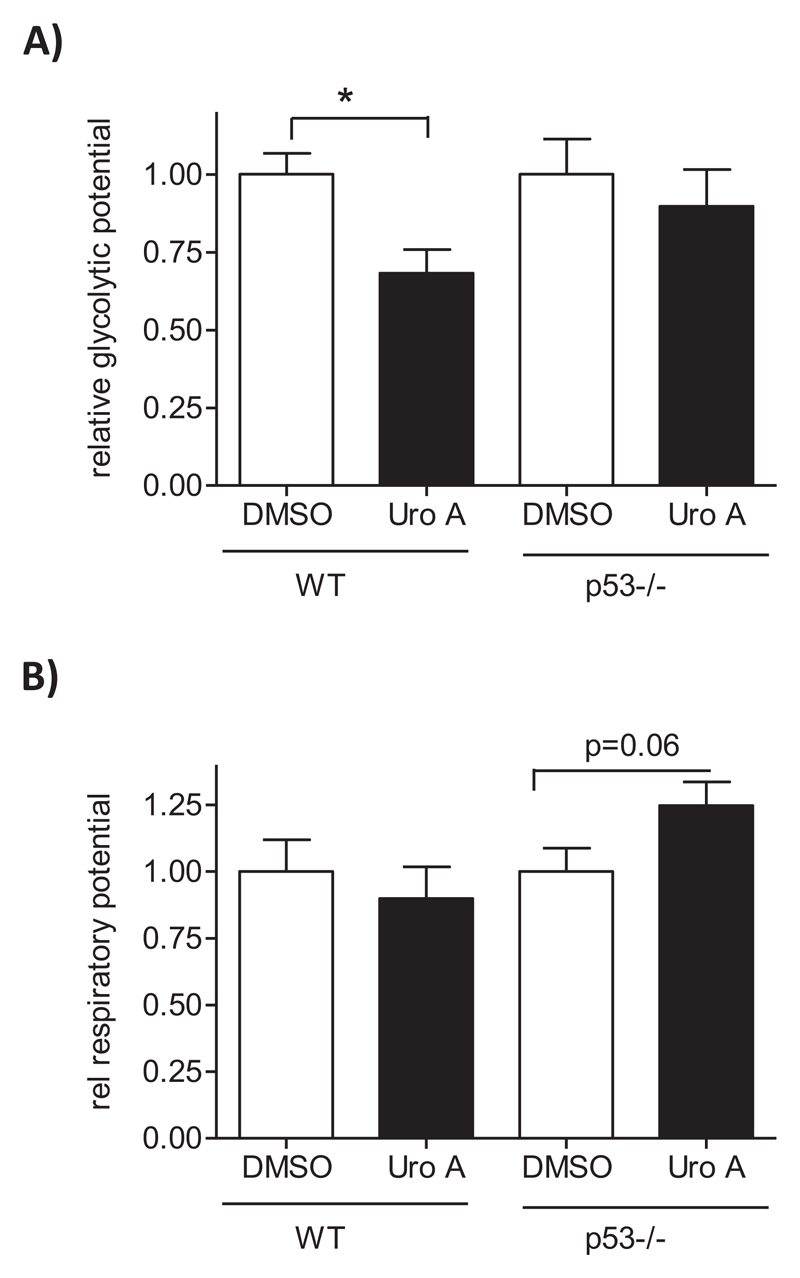
Besides cell-cycle control, p53 is involved in the regulation of cellular bioenergetics, among others shifting cellular energy metabolism from aerobic glycolysis to oxidative phosphorylation and hereby counteracting the cancer-typical Warburg effect (35). In order to see whether urolithin A also influences cellular bioenergetics via p53, we determined the cell energy phenotype via extracellular flux analysis. For this, cells were treated with solvent or urolithin A (24 h) and subjected to the assessment of basal extracellular acidification (ECAR; as surrogate readout for glycolysis) and oxygen consumption (OCR; as readout for respiration) rates. Then oligomycin (inhibitor of ATP synthase) and FCCP (uncoupler of mitochondrial respiration) were added as stressors in order to push cells to their glycolytic and respiratory maxima, respectively. The metabolic potential was defined as to what extent stressed cells are still capable of elevating glycolysis or respiration above baseline (see also Methods section). As depicted in Figure 4A, urolithin-treated WT cells displayed a reduced glycolytic potential compared with solvent controls. In contrast, p53-deficient counterparts showed a comparable glycolytic potential after DMSO and urolithin A treatment, indicating that the reduced glycolytic potential by urolithin is p53-dependent. Based on the employed metabolic phenotype assay and used experimental setting, urolithin A did not have a significant influence on the oxidative potential in WT or p53−/− cells (Figure 4B). The reduced glycolytic potential in WT cells appears in line with the p53-mediated induction of TIGAR (see Figure 1), a known negative regulator of glycolysis reducing fructose-2,6-bisphosphate levels (36).
Figure 4.
Urolithin A reduces the glycolytic potential in WT but not p53−/− HCT116 cells. WT and p53−/− HCT 116 cells were treated with DMSO (0.1%) or urolithin A (30 μM) for 24 h before they were subjected to extracellular flux analysis and assessment of the cell energy phenotype and metabolic potential. Data of four independent biological replicates were compiled and presented relative to the overall mean of the DMSO controls. (A) shows the glycolytic, (B) the respiratory potential. (mean ± SD,*P < 0.05, ANOVA, Tukey’s).
Causal contribution of the p53/TIGAR axis to the urolithin A-mediated growth inhibition
To elucidate whether induced TIGAR expression causally accounts for the stronger antiproliferative effect of urolithin A in WT than p53−/− cells, we employed a siRNA-mediated knockdown approach. Transfection with different siRNA sequences designed to target TIGAR prevented the urolithin A-triggered induction of TIGAR in WT HCT116 cells. The combination of different siRNA sequences exerted almost complete inhibition of TIGAR and had no (unspecific) impact on p21 induction compared with cells transfected with scrambled control siRNA (Figure 5A). Subjecting siRNA (scrambled or combination of #1–3, as indicated)-transfected WT cells to a biomass staining revealed that reduced TIGAR upregulation comes along with significantly (P = 0.04) reduced antiproliferative activity of urolithin A (Figure 5B). The IC50 (72 h) value in control WT (transfected with scrambled siRNA) cells is 25.9 ± 4.1 μM, whereas in cells with downregulated TIGAR, it is around 45.3 ± 1.7 μM. Analogous to HCT116 cells, also HepG2 cells transfected with scrambled siRNA show a lower IC50 value (21 μM) than HepG2 cells transfected with siRNA targeting TIGAR (38 μM) (Supplementary Table 2, available at Carcinogenesis Online; immunoblots controlling for successful knockdown are displayed in Supplementary Figure 1, available at Carcinogenesis Online). In contrast, p53 knockout cells that were transfected with scrambled or anti-TIGAR siRNA show comparable IC50 values of around 47 and 50 μM, respectively (Supplementary Table 2, available at Carcinogenesis Online). This data confirm a causal contribution of glycolysis impediment by the p53/TIGAR axis to the observed growth inhibition by urolithin A. Also, the synergistic action between urolithin A and oxaliplatin is dampened by the knockdown of TIGAR induction (Figure 5C): WT HCT116 cells transfected with scrambled siRNA show CI values of urolithin A and oxaliplatin between 0.56 and 0.69, whereas cells experiencing a siRNA-mediated blockade of TIGAR upregulation display CI values ranging between 0.91 and 0.98.
Figure 5.
Knockdown of TIGAR by siRNA reduces the antiproliferative properties of urolithin A. (A) HCT116 WT cells were left untreated (ctrl) or transfected with siRNA (200 nM final concentration, scrambled (scr) or TIGAR-specific sequences #1–3, as indicated). After recovery and 24-h treatment with solvent (0.1% DMSO) or 30 μM urolithin A cells were lysed and subjected to western blot analysis for TIGAR, p21 and actin as loading control. Representative blots and densitometric analyses of three biological replicates are depicted. (B) HCT116 WT cells were transfected with the siRNAs (scr/mix#1–3) as indicated. Eight hours after transfection, cells were seeded into 96-well plates and treated with different concentrations of urolithin A. After 72-h incubation, a biomass staining via crystal violet was performed. Residual biomass was plotted against the concentration of urolithin A, and IC50 values were derived from the fitted curve (n = 3, mean ± SD, *P < 0.05; scr versus si). (C) HCT WT cells were transfected with scrambled siRNA or siRNAs (mix #1–3) targeting TIGAR, seeded in 96-well plates and treated with urolithin A and oxaliplatin at the indicated concentrations for 72 h. After biomass staining isobolograms for 50% reduction in biomass were drawn and combinatorial indices determined. Presented data are representative for three independent experiments with consistent results.
Discussion
Main finding of this study is the decisive role of the p53 status and an altered glycolytic capacity for the antiproliferative efficacy of urolithin A in colon cancer cells.
Out of the tested microbial metabolites of ellagitannins, lignans or isoflavones, only urolithin A exhibited antiproliferative activity in HCT116 cells with IC50 values in the two-digit micromolar range which is in the range of previously reported values for cancer cell growth inhibition (e.g (37)). It furthermore displayed synergism with oxaliplatin, extending its potentiation of chemotherapy from 5-fluoruracil to another commonly used drug (38). In contrast, enterolacton or S-equol did not markedly impair proliferation of colon cancer cells at the employed concentrations and even seemed to antagonize the effect of oxaliplatin. The previously reported chemopreventive potential of enterolacton and S-equol (e.g (39,40)) may be based on targeted steps in cancer progression other than cell proliferation (e.g. increased phase II detoxification) or achieved at concentrations beyond the range used in this study. Of note, a case-control study nested within the European Prospective Investigation into Cancer and Nutrition (EPIC) assessed the relationship between prediagnostic polyphenol plasma levels and colon cancer risk. Of the examined 35 polyphenols (including equol and enterolacton but not urolithins), only equol concentrations turned out to be inversely associated with colon cancer risk (41). Future studies will need to investigate whether equol directly reduces the colon cancer risk or is rather a biomarker of a particular intestinal microflora that may be protective against colon cancer. Also, the here suggested potential adverse effect of equol and enterolacton during oxaliplatin therapy may be worthwhile to follow-up with a particular eye on effective and achievable plasma and tissue levels. The indication that a diet rich in ellagitannins aids the prevention or oxaliplatin therapy of colon cancer by fueling the microbial production of urolithin A also awaits thorough corroboration in vivo. The translation to the in vivo setting can then fully appreciate the impact and consequences of (i) interindividually different achievable levels of urolithin A which quite probably may go below the 20–30 μM determined as IC50 in vitro, of (ii) metabolization of urolithin A to (less active) phase II metabolites, such as glucuronides or sulfates (31,37), but also (iii) the possible cooperativity with other ellagitannin metabolites, such as urolithin B or isourolithins, or with the parent compounds that may resist microbial metabolization to some extent.
P53 turned out to be an important determinant for the antiproliferative influence of urolithin A in the used cell system. Initially presumed to be an oncogene, p53 has been recognized as an important tumor suppressor in 1989 which is mutated in almost 50% of all tumors. Also known as the guardian of the genome, p53 is a labile transcription factor that, when stabilized upon endogenous and exogenous signals (DNA damage, viruses, radiation or chemicals), regulates the expression of various proteins that finally contribute to tumor suppression (42). P53 target genes include those involved in DNA repair, cell-cycle arrest, apoptosis or primary metabolism (43,44). A comparison between WT and p53−/− HCT116 cells revealed that urolithin A is able to stabilize p53, in line with findings in HepG2 hepatocarcinoma cells (29), and to trigger expression of the p53 target genes p21 and TIGAR. P21 upregulation has already been reported to be commonly involved in the antiproliferative activities of urolithins (14,33), partly due to altered levels of specific microRNAs. It needs further examination how urolithin A exactly leads to p53 stabilization and to what extent microRNAs and p53 take their share in p21 upregulation. Our study revealed that an intact p53 markedly supports the growth inhibition by urolithin A as evident by a significantly higher IC50 value and a lacking positive cooperativity with oxaliplatin in p53−/− cells. However, the accumulation/retardation of cells in the G2/M phase of the cell cycle occurred independently of the p53 status and may be responsible for the reduced growth also in p53−/− cells upon urolithin A exposure. Nonetheless, after long-term incubation with urolithin A, a functional p53 seemed to shift the cell fate: whereas WT cells were driven to a senescence-like growth arrest, cells lacking p53 encountered cell death to a major extent. Whether the induction of senescence or cell death by urolithin A would contribute to distinct p53-dependent treatment outcomes still needs to be investigated. P53 also appeared crucial for reduction in the glycolytic potential of the HCT 116 cells by urolithin A, probably due to the observed induction of the p53-dependent glycolytic inhibitor TIGAR. It has been shown that several cancer cells drive an elevated glycolytic program (despite a functional OXPHOS) to best cover for their needs in ATP, building blocks (such as nucleotides) and reducing equivalents during excessive proliferation (45), a metabolic phenotype known as Warburg effect (46). By blocking the glycolytic potential via p53 stabilization and TIGAR induction, urolithin A may thus interfere with this optimal supply and limit cell growth. In line with this finding, urolithin A lost part of its inhibitory potential and synergism with oxaliplatin when TIGAR induction was blocked by siRNA. Interestingly, Liu et al. (47) recently showed protection of cancer cells from the proapoptotic action of cisplatin by increased levels of fructose-2,6-bisphosphate and elevated glycolytic activity, bringing forward the concept of a potential metabolic (de)sensitization of cancer cells to chemotherapy.
Urolithin A is reported to affect a number of cellular signaling pathways, including Wnt-, PI3K- and autophagic signaling, that could also contribute to the observed growth inhibition of colon cancer cells (17,32,48). Pilot experiments, however, did not reveal any major changes in these signaling pathways in our HCT116 cells (data not shown). Urolithin A also mediates inhibition of pro-inflammatory nuclear factor κB (NF-κB) signaling (16,17,29). As intestinal cancers possess an inflammatory aspect in their etiology (49), the chemopreventive features of dietary ellagitannins may thus indeed result from polypharmacological targeting of several “tumor hubs,” including both NF-κB and p53. P53 and NF-κB engage in a vivid crosstalk with often mutually exclusive activities and antagonistic cellular responses. Simply put, active NF-kB is accompanied by p53 inactivity and favors glycolysis and growth by boosting inflammation and protection from apoptosis. Active p53 with concomitant inactive NF-κB impedes glycolysis and proliferation (50,51). Future studies will need to show whether inhibition of NF-κB signaling and activation of p53 signaling by urolithin A can be causally and hierarchically connected, and whether the antiproliferative, anti-inflammatory and bioenergetic features of this compound may be finally assigned one common signal or hub. Interestingly, the p53 target TIGAR was lately unraveled to be an inhibitor of NF-κB signaling (52).
Overall, we show that urolithin A, a microbial gut metabolite of ellagitannins, inhibits colon cancer cell growth in vitro and is hereby markedly supported by a functional p53 and TIGAR induction. This data can be basis and motivation for further investigations aiming at complementing our knowledge on urolithins. For instance, next to considering the individual’s microbiome and metabolome as done in a previous study (53), personalized in vivo studies should include a prior stratification of subjects according to their p53 status (and other defined factors crucial for the bioactivity of urolithins) in order to fully capture and predict the cancer chemopreventive or adjuvant potential of dietary ellagitannins.
Supplementary Material
Supplementary data can be found at Carcinogenesis online.
Acknowledgements
The authors are grateful to Dr Giang Thanh Thi Ho for performing pilot western blot experiments on p53 target genes and thank H. Beres, S. Hummelbrunner and D. Schachner for excellent technical assistance.
Funding
This study was supported by grants from the Austrian Science Fund (FWF P29392) and the Herzfelder´sche Familienstiftung (both to E.H.H.).
Abbreviations
- CI
combinatorial index
- CPD
cumulative population doublings
- NF-κB
nuclear factor κB
- PD
population doublings
- PI
propidium iodide
- SA-β-gal
senescence-associated beta galactosidase
Footnotes
Conflict of Interest Statement: None declared.
References
- 1.Pandey MK, et al. Regulation of cell signaling pathways by dietary agents for cancer prevention and treatment. Semin Cancer Biol. 2017;46:158–181. doi: 10.1016/j.semcancer.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Kajla P, et al. Flaxseed-a potential functional food source. J Food Sci Technol. 2015;52:1857–1871. doi: 10.1007/s13197-014-1293-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clavel T, et al. Bioavailability of lignans in human subjects. Nutr Res Rev. 2006;19:187–196. doi: 10.1017/S0954422407249704. [DOI] [PubMed] [Google Scholar]
- 4.Edel AL, et al. The effect of flaxseed dose on circulating concentrations of alpha-linolenic acid and secoisolariciresinol diglucoside derived enterolignans in young, healthy adults. Eur J Nutr. 2016;55:651–663. doi: 10.1007/s00394-015-0885-2. [DOI] [PubMed] [Google Scholar]
- 5.Adolphe JL, et al. Health effects with consumption of the flax lignan secoisolariciresinol diglucoside. Br J Nutr. 2010;103:929–938. doi: 10.1017/S0007114509992753. [DOI] [PubMed] [Google Scholar]
- 6.Rafii F. The role of colonic bacteria in the metabolism of the natural isoflavone daidzin to equol. Metabolites. 2015;5:56–73. doi: 10.3390/metabo5010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Setchell KD, et al. Equol: history, chemistry, and formation. J Nutr. 2010;140:1355S–1362S. doi: 10.3945/jn.109.119776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Landete JM, et al. Bioactivation of phytoestrogens: intestinal bacteria and health. Crit Rev Food Sci Nutr. 2016;56:1826–1843. doi: 10.1080/10408398.2013.789823. [DOI] [PubMed] [Google Scholar]
- 9.Vergne S, et al. Bioavailability and urinary excretion of isoflavones in humans: effects of soy-based supplements formulation and equol production. J Pharm Biomed Anal. 2007;43:1488–1494. doi: 10.1016/j.jpba.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 10.Shinkaruk S, et al. Bioavailability of glycitein relatively to other soy isoflavones in healthy young Caucasian men. Food Chem. 2012;135:1104–1111. doi: 10.1016/j.foodchem.2012.03.135. [DOI] [PubMed] [Google Scholar]
- 11.Hedlund TE, et al. Soy isoflavonoid equol modulates the growth of benign and malignant prostatic epithelial cells in vitro. Prostate. 2003;54:68–78. doi: 10.1002/pros.10137. [DOI] [PubMed] [Google Scholar]
- 12.Tomas-Barberan FA, et al. Urolithins, the rescue of “old” metabolites to understand a “new” concept: metabotypes as a nexus among phenolic metabolism, microbiota dysbiosis, and host health status. Mol Nutr Food Res. 2017;61 doi: 10.1002/mnfr.201500901. 1500901. [DOI] [PubMed] [Google Scholar]
- 13.Espín JC, et al. Biological significance of urolithins, the gut microbial ellagic Acid-derived metabolites: the evidence so far. Evid Based Complement Alternat Med. 2013;2013 doi: 10.1155/2013/270418. 270418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.González-Sarrías A, et al. Comprehensive characterization of the effects of ellagic acid and urolithins on colorectal cancer and key-associated molecular hallmarks: microRNA cell specific induction of CDKN1A (p21) as a common mechanism involved. Mol Nutr Food Res. 2016;60:701–716. doi: 10.1002/mnfr.201500780. [DOI] [PubMed] [Google Scholar]
- 15.Nunez-Sanchez MA, et al. In vivo relevant mixed urolithins and ellagic acid inhibit phenotypic and molecular colon cancer stem cell features: a new potentiality for ellagitannin metabolites against cancer. Food Chem Toxicol. 2016;92:8–16. doi: 10.1016/j.fct.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 16.Piwowarski JP, et al. Urolithins, gut microbiota-derived metabolites of ellagitannins, inhibit LPS-induced inflammation in RAW 264.7 murine macrophages. Mol Nutr Food Res. 2015;59:2168–2177. doi: 10.1002/mnfr.201500264. [DOI] [PubMed] [Google Scholar]
- 17.Boakye YD, et al. An increased autophagic flux contributes to the anti-inflammatory potential of urolithin A in macrophages. Biochim Biophys Acta Gen Subj. 2018;1862:61–70. doi: 10.1016/j.bbagen.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryu D, et al. Urolithin A induces mitophagy and prolongs lifespan in C. elegans and increases muscle function in rodents. Nat Med. 2016;22:879–888. doi: 10.1038/nm.4132. [DOI] [PubMed] [Google Scholar]
- 19.Foucquier J, et al. Analysis of drug combinations: current methodological landscape. Pharmacol Res Perspect. 2015;3:e00149. doi: 10.1002/prp2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berenbaum MC. Synergy, additivism and antagonism in immunosuppression. A critical review. Clin Exp Immunol. 1977;28:1–18. [PMC free article] [PubMed] [Google Scholar]
- 21.Chou TC, et al. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 1984;22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 22.Greco WR, et al. The search for synergy: a critical review from a response surface perspective. Pharmacol Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- 23.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heiss EH, et al. Glucose availability is a decisive factor for Nrf2-mediated gene expression. Redox Biol. 2013;1:359–365. doi: 10.1016/j.redox.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller PA, et al. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25:304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi K, et al. p53 expression as a diagnostic biomarker in ulcerative colitis-associated cancer. Int J Mol Sci. 2017;18:E1284. doi: 10.3390/ijms18061284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo A, et al. Prognostic and predictive factors in colorectal cancer: kirsten Ras in CRC (RASCAL) and TP53CRC collaborative studies. Ann Oncol. 2005;16(suppl 4):iv44–iv49. doi: 10.1093/annonc/mdi907. [DOI] [PubMed] [Google Scholar]
- 28.Munro AJ, et al. P53 abnormalities and outcomes in colorectal cancer: a systematic review. Br J Cancer. 2005;92:434–444. doi: 10.1038/sj.bjc.6602358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang Y, et al. In vitro antiproliferative and antioxidant effects of urolithin A, the colonic metabolite of ellagic acid, on hepatocellular carcinomas HepG2 cells. Toxicol In Vitro. 2015;29:1107–1115. doi: 10.1016/j.tiv.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 30.Toscano F, et al. p53 dependent and independent sensitivity to oxaliplatin of colon cancer cells. Biochem Pharmacol. 2007;74:392–406. doi: 10.1016/j.bcp.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 31.Ávila-Gálvez MÁ, et al. Physiological relevance of the antiproliferative and estrogenic effects of dietary polyphenol aglycones versus their phase-ii metabolites on breast cancer cells: a call of caution. J Agric Food Chem. 2018;66:8547–8555. doi: 10.1021/acs.jafc.8b03100. [DOI] [PubMed] [Google Scholar]
- 32.Liberal J, et al. Urolithins impair cell proliferation, arrest the cell cycle and induce apoptosis in UMUC3 bladder cancer cells. Invest New Drugs. 2017;35:671–681. doi: 10.1007/s10637-017-0483-7. [DOI] [PubMed] [Google Scholar]
- 33.Sánchez-González C, et al. Urolithin A causes p21 up-regulation in prostate cancer cells. Eur J Nutr. 2016;55:1099–1112. doi: 10.1007/s00394-015-0924-z. [DOI] [PubMed] [Google Scholar]
- 34.Romanov VS, et al. Cyclin-dependent kinase inhibitor p21(Waf1): contemporary view on its role in senescence and oncogenesis. Biochemistry (Mosc) 2012;77:575–584. doi: 10.1134/S000629791206003X. [DOI] [PubMed] [Google Scholar]
- 35.Berkers CR, et al. Metabolic regulation by p53 family members. Cell Metab. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensaad K, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 37.González-Sarrías A, et al. Antiproliferative activity of the ellagic acid-derived gut microbiota isourolithin A and comparison with its urolithin A isomer: the role of cell metabolism. Eur J Nutr. 2017;56:831–841. doi: 10.1007/s00394-015-1131-7. [DOI] [PubMed] [Google Scholar]
- 38.González-Sarrías A, et al. The ellagic acid-derived gut microbiota metabolite, urolithin A, potentiates the anticancer effects of 5-fluorouracil chemotherapy on human colon cancer cells. Food Funct. 2015;6:1460–1469. doi: 10.1039/c5fo00120j. [DOI] [PubMed] [Google Scholar]
- 39.Brown NM, et al. The chemopreventive action of equol enantiomers in a chemically induced animal model of breast cancer. Carcinogenesis. 2010;31:886–893. doi: 10.1093/carcin/bgq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.van Duursen MB, et al. Chemopreventive actions by enterolactone and 13 VIOXX-related lactone derivatives in H295R human adrenocortical carcinoma cells. Toxicol Lett. 2010;192:271–277. doi: 10.1016/j.toxlet.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Murphy N, et al. A prospective evaluation of plasma polyphenol levels and colon cancer risk. Int J Cancer. 2018 doi: 10.002/ijc.31563. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horn HF, et al. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 43.Ryan KM, et al. Regulation and function of the p53 tumor suppressor protein. Curr Opin Cell Biol. 2001;13:332–337. doi: 10.1016/s0955-0674(00)00216-7. [DOI] [PubMed] [Google Scholar]
- 44.Fischer M. Census and evaluation of p53 target genes. Oncogene. 2017;36:3943–3956. doi: 10.1038/onc.2016.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vander Heiden MG, et al. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 47.Li FL, et al. Acetylation accumulates PFKFB3 in cytoplasm to promote glycolysis and protects cells from cisplatin-induced apoptosis. Nat Commun. 2018;9 doi: 10.1038/s41467-018-02950-5. 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma M, et al. Effects of fruit ellagitannin extracts, ellagic acid, and their colonic metabolite, urolithin A, on Wnt signaling. J Agric Food Chem. 2010;58:3965–3969. doi: 10.1021/jf902857v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shawki S, et al. Colon cancer: inflammation-associated cancer. Surg Oncol Clin N Am. 2018;27:269–287. doi: 10.1016/j.soc.2017.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schneider G, et al. NFκB/p53 crosstalk-a promising new therapeutic target. Biochim Biophys Acta. 2011;1815:90–103. doi: 10.1016/j.bbcan.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Johnson RF, et al. Nuclear factor-κB, p53, and mitochondria: regulation of cellular metabolism and the Warburg effect. Trends Biochem Sci. 2012;37:317–324. doi: 10.1016/j.tibs.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 52.Tang Y, et al. The fructose-2,6-bisphosphatase TIGAR suppresses NF-κB signaling by directly inhibiting the linear ubiquitin assembly complex LUBAC. J Biol Chem. 2018;293:7578–7591. doi: 10.1074/jbc.RA118.002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nuñez-Sánchez MA, et al. Gene expression changes in colon tissues from colorectal cancer patients following the intake of an ellagitannin-containing pomegranate extract: a randomized clinical trial. J Nutr Biochem. 2017;42:126–133. doi: 10.1016/j.jnutbio.2017.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.