Abstract
Memory formation is crucial for the survival of animals. Here, we study the effect of different crh-1 [Caenorhabditis elegans homolog of mammalian cAMP response element binding protein 1 (CREB1)] isoforms on the ability of C. elegans to form long-term memory (LTM). Null mutants in creb1/crh-1 are defective in LTM formation across phyla. We show that a specific isoform of CREB1/CRH-1, CRH-1e, is primarily responsible for memory related functions of the transcription factor in C. elegans. Silencing of CRH-1e-expressing neurons during training for LTM formation abolishes the LTM of the animal. Further, CRH-1e expression in RIM neurons is sufficient to rescue LTM defects of creb1/crh-1-null mutants. We go on to show that apart from being LTM defective, creb1/crh-1-null animals show defects in innate chemotaxis behavior. We further characterize the amino acids K247 and K266 as responsible for the LTM related functions of CREB1/CRH-1 while being dispensable for its innate chemotaxis behavior. These findings provide insight into the spatial and temporal workings of a crucial transcription factor that can be further exploited to find CREB1 targets involved in the process of memory formation.
Keywords: C. elegans, chemotaxis, CREB1/CRH-1, learning, reversals
Introduction
Innate behaviors are important to animals and many of these behaviors can be modified through experience. Innate behaviors, in part, represent the hardwired neural circuitry put in place during the development of an animal (Tierney, 1986; Bateson and Mameli, 2007). Mutations that affect the innate behavior could also alter the ability of animals to modify that particular behavior in response to experience. Hence, learning deficits that are attributed to certain alleles could sometimes arise because of underlying innate behavioral defects. Here, we show the effect of different crh-1 (homologs of mammalian CREB1) mutations on innate and learned chemotaxis behaviors in Caenorhabditis elegans.
C. elegans are compost worms living on microbes as their food source. Their nervous system consists of 302 neurons. They show a variety of robust and adaptable behaviors like chemotaxis and thermotaxis (for review, see Hart, 2006; Ardiel and Rankin, 2010). C. elegans move in a sinusoidal wave pattern. Under laboratory conditions, their continuous forward movement is punctuated by frequent stops and events of backward movement called reversals. After each reversal, the probability of change in direction of movement increases significantly. One of the strategies deployed by C. elegans to move in response to a chemical gradient is to change the frequency of reversals in response to a gradient (Pierce-Shimomura et al., 1999).
CREB1 (cAMP response element binding protein 1) was initially characterized for its role in long-term facilitation of the gill withdrawal reflex in Aplysia (Dash et al., 1990). Neural activitydependent CREB1 activation in a defined set of neurons is one of the requirements for the process of long-term memory (LTM) formation (for review, see Flavell and Greenberg, 2008). CREB1-null mutants are shown to be defective in LTM formation across phyla (for review, see Silva et al., 1998).
C. elegans have innate preferences for certain odors. They detect metabolic byproducts of microbes as cues for finding their food. For example, under well fed naive condition worms are attracted to AWC sensed odorants like isoamyl alcohol (for review, see Bargmann, 2006). However, this odor to behavior relationship can be modified if worms are exposed to periods of starvation along-with chemo-attractants (Pereira and van der Kooy, 2012). Likewise, neutral concentrations of butanone can be turned attractive when repetitively paired with food. These modified behaviors persist for 40 h after training (Kauffman et al., 2011). The role of crh-1 in LTM formation has been described using a variety of training paradigms using the crh-1-null mutant strain crh-1(tz2) (Kauffman et al., 2010; Nishida et al., 2011; Timbers and Rankin, 2011; Lau et al., 2013; Jin et al., 2016). Chemotaxis behavior is generally used to assess the performance of memory formation and retrieval assays. Chemotaxis in C. elegans is dependent on a large number of subtle behaviors like reversals and head bends (Pierce-Shimomura et al., 1999; Iino and Yoshida, 2009; Larsch et al., 2015). We have observed that the innate ability of worms to modulate reversals in response to external stimuli is a critical factor in determining the performance of C. elegans in chemotaxis assays, thereby affecting the readout of learning assays.
The role of crh-1 in defining LTM in C. elegans has only been partially understood. First, given its pleiotropy of function, it is unclear which crh-1 isoforms are involved in memory related functions. Second, the motifs on CRH-1 protein specifically involved in memory related processes are unknown. Third, the identity of neurons where crh-1 is required for memory formation is largely unknown. Here, we used CRISPR-based mutagenesis to dissect out memory related functions of CREB1/CRH-1. Our experiments show that two of the six potential isoforms of CRH1, CRH-1c, and CRH-1e, are responsible for these functions. Further, amino acid residues Lys247 and Lys266 are critical for memory related functions of CRH-1 while being dispensable for innate chemotaxis behavior. We have also identified RIM and/or AVE as the minimal set of neurons required for memory related functions of CRH-1.
Materials and Methods
Strains
C. elegans were maintained using standard methods (Brenner, 1974). Wild-type N2 and crh-1(tz2) mutant lines were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN). The primers, plasmids and strains used in this study are listed in Tables 1, 2, and 3, respectively.
Table 1. List of primers used in this study.
| Primer ID | Sequence | Primer type | Gene |
|---|---|---|---|
| qYD21 | GCCACTTCGCCGTTGATGATG | qPCR Forward | crh-1a |
| qYD20 | TCCTCCGGCTCCTTCTTCATC | qPCR Reverse | crh-1a |
| qYD179 | GTAGATGCTTCACCATTACAGTTT | qPCR Forward | crh-1b |
| qYD23 | GGCAAGACGAGTGGCTGATT | qPCR Reverse | crh-1b |
| qYD22 | ATGTCAGCGAAAGGTAACGGATC | qPCR Forward | crh-1c |
| qYD20 | TCCTCCGGCTCCTTCTTCATC | qPCR Reverse | crh-1c |
| qYD24 | GTACCCAACAGCAACACGG | qPCR Forward | crh-1d |
| qYD25 | CCGCCGTTTCTAGATCTTTGAG | qPCR Reverse | crh-1d |
| qYD19 | ATGATGTTCCTCAGGGCATTACAA | qPCR Forward | crh-1e |
| qYD20 | TCCTCCGGCTCCTTCTTCATC | qPCR Reverse | crh-1e |
| qYD26 | ATGGAGTCACTGGTTTTCAATGG | qPCR Forward | crh-1f |
| qYD27 | GTACGGATTGTTGTTGGGATGG | qPCR Reverse | crh-1f |
| qYD09 | TACTCTTTCACCACCACCGC | qPCR Forward | act-1 |
| qYD10 | ACGGTGATGACTTGTCCGTC | qPCR Reverse | act-1 |
| YD157 | TGGAAGGAGGAGGAGATGGAAA | Genotyping Forward (external) | crh-1(tz2) |
| YD158 | GCAGTACAGCTCTTTCAGCGTT | Genotyping Forward (internal) | crh-1(tz2) |
| YD159 | AATTCGGCACAACGGACTGG | Genotyping Reverse (external) | crh-1(tz2) |
| YD172 | ATTACCTGCAGGCTGCAGGTCGACTCTAGAG | Cloning Forward | phsp16.41 |
| YD173 | GGCAGCTAGCATTTTTTCTACCGGTACCAATAC | Cloning Reverse | phsp16.41 |
| YD165 | ATTAGCTAGCATGGCCACAATGGCGAGCAC | Cloning Forward | crh-1a |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1a |
| YD165 | ATTAGCTAGCATGGCCACAATGGCGAGCAC | Cloning Forward | crh-1b |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1b |
| YD166 | ATTAGCTAGCATGTCAGCGAAAGGTAACGGAT | Cloning Forward | crh-1c |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1c |
| YD167 | ATTAGCTAGCATGGCCACAATGGCGAGCAC | Cloning Forward | crh-1d |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1d |
| YD168 | ATTAGCTAGCATGATGTTCCTCAGGGCATTACA | Cloning Forward | crh-1e |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1e |
| YD169 | ATTAGCTAGCATGGAGTCACTGGTTTTCAATGG | Cloning Forward | crh-1f |
| YD164 | ATTACCATGGTCACATTCCGTCCTTTTCCTTTC | Cloning Reverse | crh-1f |
| YD180 | CATAATAACCGGTAAAATGTACCCATACGACGTTCCAG | Cloning Forward | 3xHA |
| YD181 | CATAGCTAGCAGCGTAATCTGGGACGTCA | Cloning Reverse | 3xHA |
| YD320 | ATCAGCATGCATCTTCAGATGGGAGCAGTGG | Cloning Forward | prab-3 |
| YD321 | GCATGGATCCAAACTTGTCATCTGAAAATAGGGCT | Cloning Reverse | prab-3 |
| YD260 | ATAGCCTGCAGGAAGTGGACACTGAGAGAGAGAG | Cloning Forward | pnmr-1 |
| YD261 | ATAGCCCGGGCTGTAACAAAACTAAAGTTTGTCGTGTTC | Cloning Reverse | pnmr-1 |
| AB139 | GCGTCGACAAGTGACACCACGCTCACA | Cloning Forward | prig-3 |
| AB140 | CCCCCCGGGAGCTGTGAAATTTTTAGGCAGT | Cloning Reverse | prig-3 |
| YD313 | ACGTGCATGCGTTAAATATGTGGCCACAATGACAATTAT | Cloning Forward | pgcy-13 |
| YD314 | ATCAGGATCCGTCCTGAAAAATTATTGAAAGTTTGTAATAAG | Cloning Reverse | pgcy-13 |
| YD253 | ATACCTGCAGGGAATTTAATAACTAATACATATTATTTGGCACACTC | Cloning Forward | pcrh-1e |
| YD254 | ATAGGATCCATTATTCAGAAATTGAATAGAGAATTAGAATTAGAAAC | Cloning Reverse | pcrh-1e |
| YD318 | CTGCAAGCTTCACTTTTCTACGTTTCCATAATAATTACATAG | Cloning Forward | pcrh-1a |
| YD319 | ATACGGATCCAGCCGTGAGATGTCCGC | Cloning Reverse | pcrh-1a |
| YD205 | AGTAAGCTTGTTTACGCTCGCGAGGAGAC | Cloning Forward | pstr-2 |
| YD206 | AGTCTGCAGTTTTATGGATCACGAGTATTCG | Cloning Reverse | pstr-2 |
| crYD35 | ATCAGGTCTCCTCTTCTTTTCTAATACTTACCCTGGTTTTAGAGCTAGAAATAGCAAG | Guide RNA | crh-1e deletion |
| crYD65 | ATCAGGTCTCCTCTTCAAAATGTCAGCGAAAGGTAAGTTTTAGAGCTAGAAATAGCAAG | Guide RNA | crh-1c deletion |
| crYD48 | ATCAGGTCTCCTCTTGAGCATCATCAACGGCGAAGGTTTTAGAGCTAGAAATAGCAAG | Guide RNA | crh-1a,b,d deletion |
| crYD106 | ATCAGGTCTCCTCTTTGGGCTCGAGCTGTAGCCGCGTTTTAGAGCTAGAAATAGCAAG | Guide RNA | crh1(K247R/K266R) |
| crYD01 | GTTTCTCGAGCCATGGTTACCGGTCTC | Guide RNA | Universal |
| crYD44 | ATACGCATGCACACCTAAAATTTTCAGATTGAGTCTGG | Forward | 5′ Homology arm crh-1e deletion |
| crYD45 | ATCAACTAGTTGCGCCCTGTTGAACTGGGA | Reverse | 5′ Homology arm crh-1e deletion |
| crYD46 | ATCAGGGCCCAGAAAAGATTAGTGTGTGTGTGTTTG | Forward | 3′ Homology arm crh-1e deletion |
| crYD47 | ATCACTTAAGCGAGTTGGTTTTATTACACAGCAGAG | Reverse | 3′ Homology arm crh-1e deletion |
| crYD59 | ATCAACATGTGCGAATCGCTGATTGGTTGCAATTC | Forward | 5′ Homology arm crh-1c deletion |
| crYD60 | ATCAACTAGTTTTATACTCGTTTTGGTGTGCAAAAAGGC | Reverse | 5′ Homology arm crh-1c deletion |
| crYD61 | ATCAGGGCCCAATGACACAAGAATGCGAAAACATCT | Forward | 3′ Homology arm crh-1c deletion |
| crYD62 | ATCACTTAAGTTCGGCCAATTTTGCGAATTTTAAG | Reverse | 3′ Homology arm crh-1c deletion |
| crYD50 | ATCAGCATGCTCTCTTTCACCGCCAATTTTTGTG | Forward | 5′ Homology arm crh- 1a,b,d deletion |
| crYD51 | ATCAACTAGTGAGATGTCCGCCACTTATTTTTGTTTA | Reverse | 5′ Homology arm crh- 1a,b,d deletion |
| crYD52 | ATCAGGGCCCAGTAGAGTTTTAGTGGAAAAATTTCGAG | Forward | 3′ Homology arm crh- 1a,b,d deletion |
| crYD53 | ATCACTTAAGCGATATGAGGGCCTCCTATTAAGTT | Reverse | 3′ Homology arm crh- 1a,b,d deletion |
| crYD74 | ACTCGCATGCGAAGTGCAATGAAGCCAATGTTGG | Forward | 5′ Homology arm crh- 1(K247R/K2 66R) |
| crYD90 | ACTAACTAGTCAAATTTCCGCTCCAAA | Reverse | 5′ Homology |
| AAATTACCTTACGTTTTCTGCGGCACTCTTTCGCTGCCTCTCGATT | arm crh- 1(K247R/K2 66R) | ||
| crYD91 | ACTCGGGCCCGAGGGAAAAAAATTATTTTTTGGCTGAAAAATTGA | Forward | 3′ Homology arm crh- 1(K247R/K2 66R) |
| crYD92 | ACGACTTAAGTGAGTGCTTTGTTCTGATTTTCCAGC | Reverse | 3′ Homology arm crh- 1(K247R/K2 66R) |
Table 2. List of plasmids used in this study.
| S. no. | Plasmid ID | Plasmid |
|---|---|---|
| 1 | pBAB713 | prab-3::CRH-1a |
| 2 | pBAB714 | prab-3::CRH-1b |
| 3 | pBAB715 | prab-3::CRH-1c |
| 4 | pBAB716 | prab-3::CRH-1d |
| 5 | pBAB717 | prab-3::CRH-1e |
| 6 | pBAB719 | prab-3::CRH-1f |
| 7 | pBAB712 | phsp16.41::CRH-1e |
| 8 | pBAB726 | pnmr-1::CRH-1e |
| 9 | pBAB746 | pcrh-1e::CRH-1e |
| 10 | pBAB748 | prig-3::mCherry |
| 11 | pBAB749 | popt-3::mCherry |
| 12 | pBAB750 | pgcy-13::mCherry |
| 13 | pBAB747 | prig-3::CRH-1e |
| 14 | pBAB745 | popt-3::CRH-1e |
| 15 | pBAB744 | pgcy-13::CRH-1e |
| 16 | pBAB761 | pstr-2::ChR2::YFP |
| 17 | pBAB751 | pcrh-1e::GFP |
| 18 | pBAB736 | pnmr-1::mCherry |
| 19 | pBAB752 | pcrh-1a::GFP |
| 20 | pBAB741 | pcrh-1e::HisCl1::sl2::GFP |
| 21 | pBAB709 | Repair template crh-1a,b,d deletion |
| 22 | pBAB711 | Repair template crh-1c deletion |
| 23 | pBAB708 | Repair template crh-1e deletion |
| 24 | pBAB760 | Repair template crh-1K247R/K266R point mutation |
| 25 | pBAB757 | Repair template 3xHA::CRH-1e |
| 26 | pBAB758 | Repair template 3xHA::CRH-1c |
| 27 | pBAB759 | gRNA crh-1a,b,d deletion |
| 28 | pBAB762 | gRNA crh-1c deletion |
| 29 | pBAB763 | gRNA crh-1e deletion |
| 30 | pBAB756 | gRNA crh-1K247R/266R point mutation |
| 31 | pBAB764 | gRNA 3xHA::CRH-1c |
| 32 | pBAB765 | gRNA 3xHA::CRH-1e |
Table 3. List of strains used in this study.
| Strain | Genotype | Description |
|---|---|---|
| BAB701 | crh-1(tz2) (CGC strain YT17) | From CGC (outcrossed 2X) |
| BAB713 | crh-1; prab-3::CRH-1a | Array no. IndEx713 |
| BAB714 | crh-1; prab-3::CRH-1b | Array no. IndEx714 |
| BAB715 | crh-1; prab-3::CRH-1c | Array no. IndEx715 |
| BAB716 | crh-1; prab-3::CRH-1d | Array no. IndEx716 |
| BAB717 | crh-1; prab-3::CRH-1e | Array no. IndEx717 |
| BAB708 | CRISPR based deletion corresponding to the first exon of crh-1e mRNA | crh-1(Ind708+loxP) |
| BAB709 | CRISPR deletion corresponding to the first exon of crh-1a,b,d mRNAs | crh-1(Ind709+loxP) |
| BAB710 | CRISPR based deletion corresponding to the first exon of crh-1c and crh-1e mRNAs | crh-1(Ind710+loxP) |
| BAB711 | CRISPR based deletion corresponding to the first exon of crh-1c mRNA | crh-1(Ind711+loxP) |
| BAB719 | prab-3::CRH-1f | Integrated line IndIs719 |
| BAB726 | crh-1; pnmr-1::CRH-1e | Array no. IndEx726 |
| BAB736 | pnmr-1::mCherry; pcrh-1e::GFP | Array no. IndEx736 |
| BAB703 | prig-3::mCherry; pcrh-1e::GFP | Array no. IndEx703 |
| BAB704 | popt-3::mCherry; pcrh-1e::GFP | Array no. IndEx704 |
| BAB705 | pgcy-13::mCherry; pcrh-1e::GFP | Array no. IndEx705 |
| BAB747 | crh-1; prig-3::CRH-1e | Array no. IndEx747 |
| BAB745 | crh-1; popt-3::CRH-1e | Array no. IndEx745 |
| BAB744 | crh-1; pgcy-13::CRH-1e | Array no. IndEx744 |
| BAB746 | crh-1; pcrh-1e::CRH-1e | Array no. IndEx746 |
| BAB741 | pcrh-1e::HISCl1::SL2::GFP | Array no. IndEx741 |
| BAB712 | crh-1; phsp16.41::CRH-1e | Array no. IndEx712 |
| BAB754 | CRISPR based addition of 3xHA::CRH-1c and e | crh-1(Ind754 [3xHA::crh-1c+loxP; 3xHA::crh-1e+loxP]) |
| BAB760 | CRISPR based substitution of K247R/266R | crh-1(Ind760 [K247R/K266R]+loxP) |
| BAB735 | pnmr-1::mCherry; pcrh-1a::GFP | Array no. IndEx735 |
| BAB761 | pstr-2::ChR2::YFP; prig-3::GCaMP5G | Array no. IndEx761 |
| BAB762 | BAB760; pstr-2::ChR2::YFP; prig-3::GCaMP5G | crh-1(Ind760 [K247R/K266R]+loxP) with array |
Constructs and transgenes
The crh-1 isoforms (crh-1a-f) were cloned into the pPD49.26 vector backbone. crh-1b and crh-1c cDNAs were synthesized from Sigma-Aldrich and the rest were obtained by reverse transcription and PCR using wild-type (WT) C. elegans RNA. N2 (WT) or crh-1(tz2) mutant lines were used for transforming the CRH-1 isoforms. Transformations were done as described previously (Mello et al., 1991). The rescue constructs and the promoter fusion constructs were injected in concentrations of 10–20 ng/μl. pmyo-2::mCherry (2 ng/μl) or pvha-6::mCherry (10 ng/μl) or punc-122::GFP (25 ng/μl) were used as coinjection markers. mCherry and GFP cDNA were cloned from pCFJ90 and pPD95.75, respectively. The primers used for the different cloning experiments are tabulated in Table 1.
Behavioral assays
Conditioning with IAA/diacetyl and high temperature
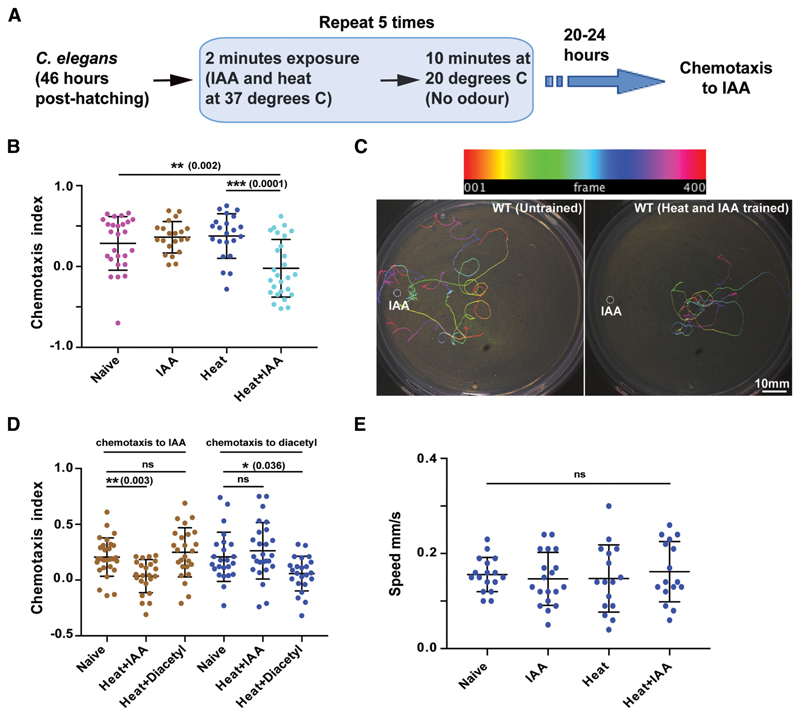
A dry heating block was maintained at 37°C. Twenty microliters of 10% isoamyl alcohol (IAA) or 1% diacetyl (both diluted in ethanol) was kept on a piece of cover glass on the heating block. A Petri-plate containing C. elegans on OP50 lawn was inverted onto the setup for 2 min. The training cycle was repeated five times with an inter-training interval of 10 min (illustrated in Fig. 1A).
Figure 1. Associative learning in C. elegans.
A, Schematic flowchart of the C. elegans training routine followed in this study. B, Scatter plot showing the effect of IAA and heat on WT C. elegans. C, Representative images of typical chemotaxis tracks. Top, Color-coding of tracks as a function of frame number. D, Scatter plot indicating the specificity of learning. Animals were trained with Heat + IAA or Heat + Diacetyl and their chemotaxis indices were plotted. E, Plot showing speed of worm movement when subjected to different training conditions. Error bars are SD. Multiple comparisons were done using one-way ANOVA and p values were adjusted using Dunnett’s correction method. In graphs of all figures significant p values are added in brackets next to the significance asterisks (*) and “ns” indicates not significant. See Figure 1-1 available at https://doi.org/10.1523/JNEUROSCI.0006-19.2019.f1-1.
Chemotaxis assay and behavioral analysis
Trained animals were kept at 20°C for 20–24 h unless mentioned otherwise. C. elegans were picked using an eyelash pick and allowed to crawl on an unseeded nematode growth medium (NGM) plates for 30 s. Four to six animals were gently transferred to the center of 90mm NGM agar plates (without food). One microliter of odorant [IAA (1%)/diacetyl (0.1%)/benzaldehyde (1%)] was kept at one end of the plate. Chemotaxis behavior was recorded for 10 min using a 5 megapixel CMOS USB camera (Mightex) at 2 frames/s using the Mightex Camera Demo v1.1.0 software. Recordings were done in a Peltier cooled incubator at 20° Celsius. Videos were analyzed using FIJI software (Schindelin et al., 2012). To quantify their chemotaxis behavior, a non-dimensional index based on individual C. elegans trajectory was used (Eq. 1). For the attractant, displacement was positive if the animal traveled up the gradient while it was negative if the animal traveled down the gradient (Luo et al., 2014). To quantify the reversal behavior of C. elegans, the worms were allowed to crawl in presence or absence of an IAA gradient for 10 min. Videos were recorded at 2 frames/s. Reversals were calculated manually by analyzing the videos.
| (1) |
Quantitative PCR
Total RNA was isolated using Trizol from WT animals grown to the young adult stage. Fifty nanograms of total RNA was used for cDNA synthesis. qPCR was performed using a Qiagen SYBR real time PCR kit. Isoform-specific primers were used for amplification of the different crh-1 isoforms. The primers used are listed in in Table 1. qPCR was done using the Roche Light Cycler 480. Ct values were calculated using Equation 2 with the Roche software. The data were analyzed using ΔC method (Eq. 3).
| (2) |
| (3) |
GOI indicates gene of interest, and HG indicates the housekeeping gene, act-1.
Fluorescence microscopy
Young adult hermaphrodites were used for imaging. Animals were mounted on 2% agarose pads with 10 mm sodium azide solution in M9 medium. Images were acquired on a Leica SP6 upright laser scanning confocal microscope using the 40× oil-immersion objective lens. All images were processed and analyzed by Fiji.
Histamine supplementation
Histamine-dihydrochloride (1 m; Sigma-Aldrich, catalog #53300) filter sterilized stock solution was prepared in distilled water. Final working concentrations (10 mm) were used for all experiments. Histaminesupplemented NGM plates were prepared as described previously (Pokala et al., 2014).
Optogenetics
The str-2 promoter was used to drive AWC neuron-specific expression of channelrhodopsin-2 (ChR2). Animals expressing ChR2 were grown on NGM agar plates seeded with OP50-containing 100 μm all-trans retinal. The C. elegans were grown in the dark until late L4/early adult stages. The assay was performed on freshly seeded NGM plates. During the assay, ChR2 was excited by blue light (460–490 nm) using an epifluorescence unit (U-HGLGPS, Olympus) attached to the Nikon SMZ2000 microscope. Blue light was illuminated for 3 s and reversal events were quantified. If any worm executed reverse movement for one body length, it was counted as a reversal event. Each worm was illuminated with blue light 8 times with 20 s intervals between subsequent stimulation. Reversal probability was calculated by dividing the number of reversal events with number of optogenetic stimulations. Each dot in the scatter plot indicates the reversal probability of a single C. elegans under observation. The results were plotted as mean SD and evaluated using the standard Student’s t test. The experimenter was blind to the genotypes of the strains while performing these experiments.
Calcium imaging
The genetically encoded Ca+2 indicator GCaMP-expressing strain prig-3::GCaMP5 was used to visualize Ca+2 transients in the AVA command interneuron (Larsch et al., 2013). A 0.2μm drop of polystyrene beads (0.1 μm) was added on the top of 10% agarose pad (prepared in M9).Asingle worm was kept on the drop containing beads. The animal was immobilized by putting a circular cover glass over it. ChR2 was excited by blue light (460–490 nm) using an epifluorescence unit (U-HGLGPS, Olympus) and simultaneously calcium transients were recorded for 3.0 s using Micro-Manager software at a speed of 10 frames/s (Edelstein et al., 2014). Image analysis was done using Fiji. A ROI was drawn over the AVA cell body. Fluorescence (F) was calculated by subtracting the background fluorescence (Fbkgd) value from the mean fluorescence (Fmean) of ROI. The fluorescence value was estimated for each frame by manual repositioning of the ROI. Calcium transients were plotted as F/Fo, where F is the change in the fluorescence value (F) from its baseline fluorescence (Fo). Fluorescence intensity of second frame (t=200 ms) was taken as baseline fluorescence (Fo).
CRISPR-based genome editing
CRISPR was used to create crh-1 isoform mutations as described previously (Dickinson and Goldstein, 2016). Briefly, the selection excision cassette (SEC) from the plasmid pDD287 was cloned along-with flanking loxP sites into pPD95.75. The resulting plasmid was used to clone homology arms (500–1500 bp) and the desired genetic modification using restriction enzyme-based cloning methods. A 20 bp guide RNA was cloned into pRB1017. The plasmid mixture containing repair template (50 ng/μl), sgRNA (20 ng/μl), pJW1259 (50 ng/μl), pCFJ90 (2.5 ng/μl), and pvha-6::mCherry (10 ng/μl) was injected into 20–30 adult hermaphrodite animals (containing 4–5 eggs). The C. elegans were kept at 20°C. After 60 h of injection, the antibiotic hygromycin (250 μg/ml) was added directly to the plates containing the injected worms. The C. elegans were left at 20°C for 10 d. After 10 d, 10–15 non-fluorescent roller animals were singled out onto regular NGM plates. To remove SEC 30–40 L1–L2 animals from plates with 100% roller progeny were kept at 34°C for 3–4h. Normal moving worms were isolated and target DNA was sequenced to analyze and confirm desired modifications.
Statistical analysis
All statistical analyses were done using GraphPad Prism 7. Outliers were identified using Grubbs method (α = 0.05). Mean values were compared using one-way ANOVA. Bonferroni or Dunnett correction was used to adjust p values for multiple comparisons. Error bars represent SD except for Figure 5E where they represent SEM. The level of significance was set as p ≤ 0.05.
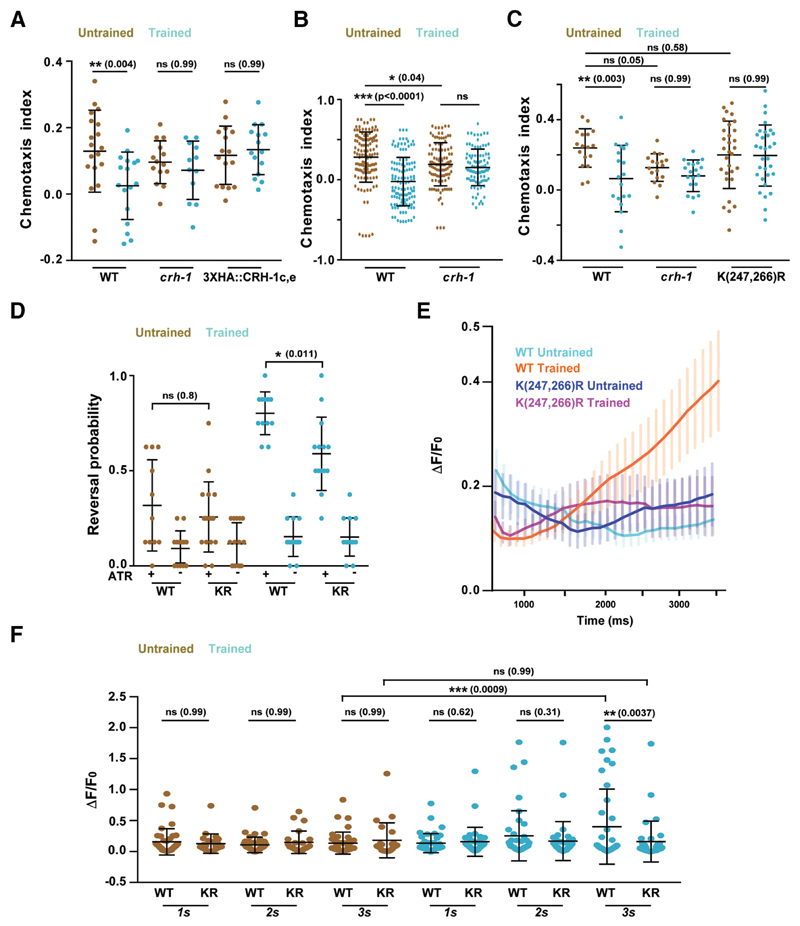
Figure 5. Lysine 247 and 266 residues are required for normal learning but not required for innate chemotaxis in C. elegans.
A, Chemotaxis indices of WT, crh-1-null mutants, and N-terminal 3xHA tagged. CRH-1c and CRH-1e. The tag was appended using CRISPR on N-terminal of CRH1c and CRH-1e translation start codon. B, Scatter plot showing collated chemotaxis index values of WT and crh-1-null C. elegans used in this study. Data were pooled only form the experiments that used both strains. C, Chemotaxis indices of WT, crh1-null mutants, and crh-1(K247R/K266R) mutants. D, Reversal probability of C. elegans when AWC is excited with blue light. Blue light was illuminated for 3 s and reversal events were quantified. If the worm executed reverse movement for one body length, it was counted as a reversal event. Each C. elegans was illuminated with blue light 8 times with 20 s (s) intervals between subsequent stimulations. Each dot in the scatter plot indicates the reversal probability of a single animal under observation. E, Plot showing Calcium traces of the AVA neuron in responseto ChR2-mediated AWC activation. Mean values are plotted against time. Error bars are SEM. n = 25–41 for each genotype. F, Scatter plot comparing AVA excitation in response to AWC activation at three time points. The figure represents a subset of data (1, 2, 3 s) from E. Error bars are SD except in E (SEM). Multiple comparisons were done using one-way ANOVA and p values were adjusted using Bonferroni (A, D, F) and Dunnett’s (B, C) method. *p < 0.05, **p < 0.01, ***p < 0.001, ns, p > 0.05.
Results
Associative learning in C. elegans using isoamyl alcohol and high temperature
Previous studies have largely examined associative LTM formation in C. elegans by pairing the presence/absence of food (unconditioned stimulus) with a variety of cues (Amano and Maruyama, 2011; Kauffman et al., 2011; Nishida et al., 2011). Under these previously reported training conditions C. elegans could retain memory for upto 40 h (Kauffman et al., 2011). Be-cause studies have shown that starvation alone in the absence of any external stimulus is sufficient to induce the expression of CREB1/CRH-1 in C. elegans (Suo et al., 2006, 2009), we were interested in developing a training paradigm that was independent of the feeding state of the animal. To this end we went on to develop a learning paradigm using IAA and high temperature.
C. elegans were exposed to the vapors of IAA at high temperatures (37°C) for 2 min followed by a rest period of 10 min at 20°C, repeated five times (illustrated in Fig. 1A). After 20–24 h, the animals were allowed to crawl on a 90 mm NGM agar plate in response to an IAA gradient. We observed significantly reduced chemotaxis index (CI) values for C. elegans exposed to IAA at 37°C (Figs. 1B, plate images shown in C, and Fig. 1-1A, B, available at https://doi.org/10.1523/JNEUROSCI.0006-19.2019.f1-1), whereas the CI values for C. elegans exposed only to 37°C temperature or IAA alone were comparable to naive animals (Fig. 1B). To understand whether learning was specific to the cues provided during training, we trained worms with heat and IAA followed by chemotaxis to diacetyl after 20–24 h. We also did experiments where we used the training paradigm with heat and diacetyl and then went on to test for chemotaxis to IAA after 20–24 h. IAA when paired with heat resulted in reduced chemotaxis to IAA, whereas chemotaxis to diacetyl was unaffected and diacetyl when paired with heat resulted in reduced chemotaxis to diacetyl while chemotaxis to IAA was unaffected (Fig. 1D). Animals trained with heat and IAA showed normal chemotaxis in response to 1% benzaldehyde, another odorant sensed by AWC neurons, negating a generic functional downregulation of AWC neurons in response to the training paradigm (Fig. 1-1C, available at https://doi.org/10.1523/JNEUROSCI.0006-19.2019.f1-1). To negate the potentially harmful effects of the chemicals/high temperature used in training we measured the average crawling speed of C. elegans that were subjected to various training conditions, these animals crawled at speeds comparable to naive animals (Fig. 1E).
LTM formation in C. elegans requires CRH-1c and CRH-1e
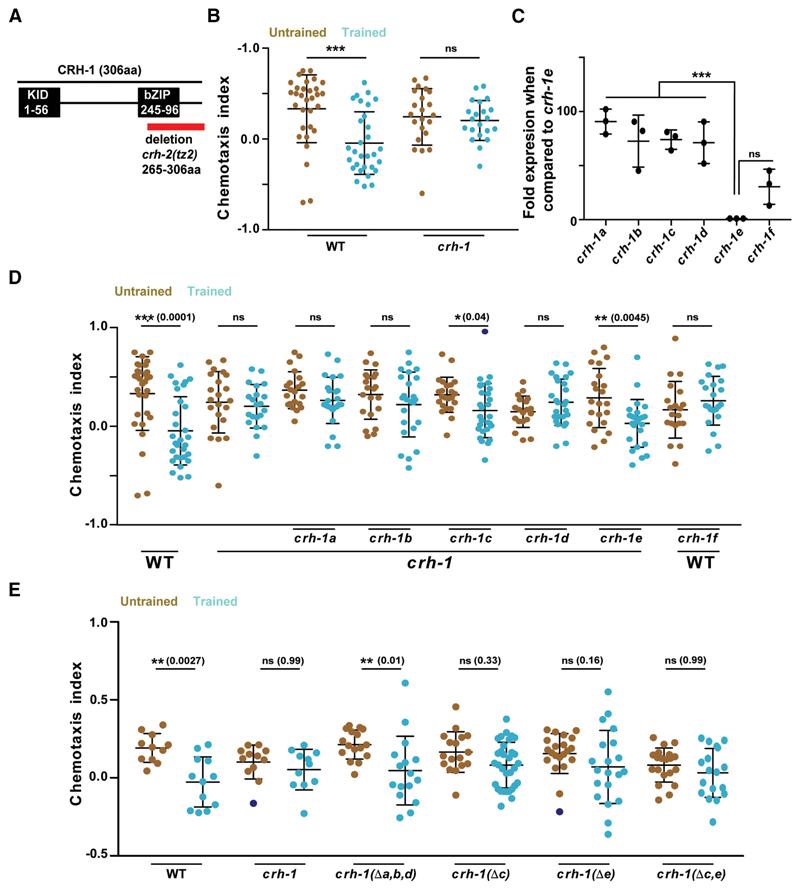
The creb1/crh-1 gene is pleotropic in function, known to be involved in a variety of signaling pathways like cellular energy metabolism, aging, memory formation, circadian rhythm etc. (for review, see Johannessen et al., 2004). Using multiple isoforms of a protein to achieve functional diversity is a widely used strategy during the course of molecular evolution (for review, see Graveley, 2001; Touriol et al., 2003; Perrin and Ervasti, 2010). To test whether the above learning paradigm would allow us to parse out the isoform/s of CRH-1 required for LTM, we first tested the creb1/crh-1-null mutant, crh-1(tz2) (depicted in Fig. 2A). We observed that trained WT worms showed significantly reduced CI compared with creb1/crh-1-null mutants (Fig. 2B).
Figure 2. CRH-1c and e are required for associative learning in C. elegans.
A, Schematic showing C. elegans CRH-1 and the deletion in the mutant. Two major functional domains KID and bZIP (DNA-binding domain) domains are depicted. The red bar shows the deletion found in the crh-1-null mutant tz2. This deletion affects all CRH-1 isoforms. B, Scatter plot showing chemotaxis indices of WTand crh-1-null mutants. C, Graph showing quantitative PCR results comparing mRNA expression levels of different CRH-1 isoforms to the CRH-1eisoform in young adult WT C. elegans. D, Scatter plot showing chemotaxis indices of WT, crh-1-null mutants, and pan-neuronal CRH-1 rescue lines using different crh-1 isoforms. E, Scatter plot showing chemotaxis indices of WT, crh-1-null mutants and crh-1 isoform deletion lines. Error bars are SD. Multiple comparisons were done using one-way ANOVA and p values were adjusted using Bonferroni (B, D, E) and Dunnett’s (C) methods. Data points represented with dark blue dots in the scatter plots represents outliers identified using Grubb’s method (α. = 0.05). **p < 0.01, ***p < 0.001, ns, p > 0.05.
The crh-1 gene encodes seven different isoforms (crh-1 a-g) (Wormbase gene: WBGene00000793). We were able to clone cDNA for six of the seven isoforms (crh1 a-f). We performed a quantitative PCR experiment to examine the expression levels of the different isoforms of CRH-1 during the early adult stage of C. elegans. We observed strikingly variable expression levels for the various isoforms indicating a possibility of functional diversity among the different isoforms (Fig. 2C). To further investigate the function of CREB1/CRH-1 in memory, different CRH-1 isoforms, i.e., crh-1 (a-f) were cloned under the pan-neuronal rab-3 promoter and their ability to rescue the LTM defect in creb1/crh-1-null mutants was tested (Fig. 2D). Five of the six CRH-1 isoforms (CRH-1a-e) are full-length proteins having both the KID (kinase inducible domain) and the bZIP (DNA binding) domain, these five isoforms differ only in the N-terminal 30 aa, whereas CRH-1f is a truncated protein lacking the N-terminal KID motif (Fig. 1-1D, available at https://doi.org/10.1523/JNEUROSCI.0006-19.2019.f1-1). We observed rescue of LTM formation in case of CRH-1c and CRH-1e-expressing animals. Moreover, pan-neuronal expression of the repressor isoform CRH-1f in WT animals inhibited memory formation (Fig. 2D).
To negate the potential artifacts of spatiotemporal misexpression and/or overexpression because of extrachromosomal arrays, we generated CRISPR mutants for the different crh-1 isoforms and tested them for LTM defects. Deletion of a, b, and d isoforms by removing their first exon had no effect on LTM formation, whereas C. elegans without c and e isoforms were defective in LTM formation (Fig. 2E).
Expression of CRH-1e in the RIM neurons rescues the learning defects of creb1/crh-1 mutants
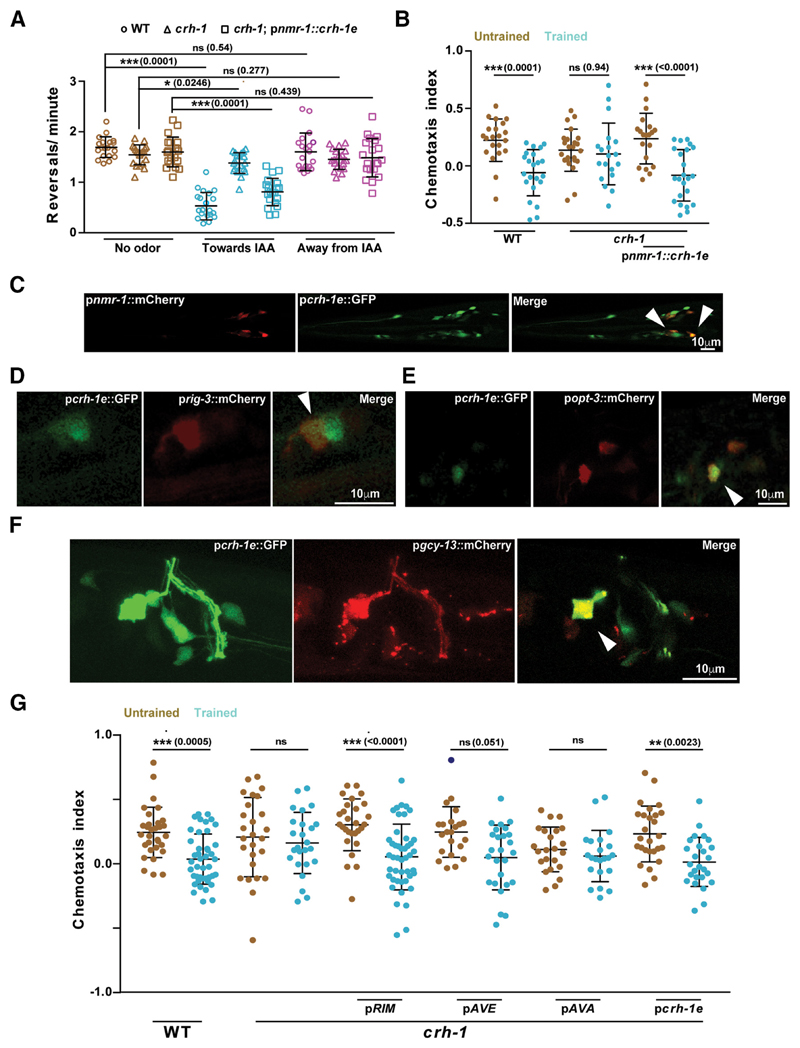
Under stimulus-deficient environments, the movement of C. elegans can be described as a random walk. The presence of an attractant/repellent introduces a bias in this random walk strategy whereby the animal can suppress or enhance its reversal and turn frequency depending on whether it is moving toward or away from the attractant/repellent (Pierce-Shimomura et al., 1999). To test whether naive creb1/crh-1-null mutants showed any differences in their random walk pattern, we compared the reversal frequency of WT and creb1/crh-1-null mutants while they were moving on an NGM agar plate in response to an IAA gradient. Our results indicated that creb1/crh-1-null mutants were defective in modifying their reversal behavior in response to the varying attractant concentrations that they were experiencing while moving (Fig. 3A). Reversal behavior in C. elegans is controlled by the command interneurons (AVA, AVD, AVE) and the RIM interneurons (Chalfie et al., 1985; Gray et al., 2005; Piggott et al., 2011). To test whether CRH-1e is required in these neurons we tested a transgenic line expressing CRH-1e under the control of nmr-1 promoter, which is expressed in five sets of neurons including AVA, AVD, AVE, and RIM (Brockie et al., 2001). This line could largely rescue the reversal defect as well as the memory deficits observed in the creb1/crh-1-null mutants (Fig. 3A,B).
Figure 3. CRH-1e expression in the RIM or AVE interneurons is sufficient to rescue the associative learning phenotype seen in crh-1 mutants.
A, Scatter plot showing reversal frequency of worms while they were moving either in odor free conditions, toward or away from the IAA point source. Each data point represents average reversal events per minute for individual animals counted over 10 min of chemotactic movement. B, Chemotaxis indices for WT, crh-1, and pnmr-1::crh-1e rescue line. C, Confocal microscope images showing overlapping expression of crh-1e promoter with neurons showing expression with the nmr-1 promoter. Arrowheads indicate points of coexpression. D–F, Confocal microscope images showing overlapping expression of crh-1e promoter with neuron-specific promoters; prig-3 (AVA), popt-3 (AVE), and pgcy-13 (RIM). G, Scatter plot showing chemotaxis indices of WT, crh-1, and rescue lines expressing CRH-1e under the promoters; gcy-13 (RIM), opt-3 (AVE), rig-3 (AVA), and crh-1e. Error bars are SD. Multiple comparisons were done using one-way ANOVA and p values were adjusted using Bonferroni method. Data points represented with dark blue dots in the scatter plots represents outliers identified using Grubb’s method (α. = 0.05). *p < 0.05, **p < 0.01, ***p < 0.001, ns, p > 0.05.
The presence of a unique 5′ UTR on CRH-1e mRNA along with its low mRNA expression levels (Wormbase gene: WBGene00000793; Fig. 2C) suggests that this promoter sequence may be different from the other crh-1 isoforms. We expressed GFP under the control of a 2.7 kb DNA sequence upstream of the CRH-1e translation start site (henceforth termed pcrh-1e). Consistent with remarkably low levels of expression in qPCR measurements (Fig. 2C), we could observe the GFP expression in the pcrh-1e::GFP line in only a few head neurons, expression in the head neurons was also seen previously by Kimura et al. (2002) who had done in situ hybridization of the crh-1 gene. Colocalization experiments showed that the overlap in expression pattern of pnmr-1 was largely restricted to three pairs of neurons (Fig. 3C). Based on the position of the overlapping neuron, we assessed that localization was seen in the AVA, AVE, and RIM interneurons. The same was confirmed by coexpression experiments using neuron-specific promoter marker lines for AVA (prig3::mCherry), AVE (popt-3::mCherry), and RIM (pgcy-13::mCherry; Fei et al., 2000; Ortiz et al., 2006; Feinberg et al., 2008). We found that pcrh-1e::GFP localized with all these neurons (Fig. 3D–F). To test where CRH-1e is required for memory formation we expressed CRH-1e under the control of crh-1e, rig-3, opt-3, and gcy-13 promoters. When tested for rescue of the associated memory phenotype with IAA and heat, we found that the crh-1e, the gcy-13 and to a lesser extent the opt-3 promoters could rescue the learning defects seen in creb1/crh-1 mutants (Fig. 3G). Expressing CRH-1e in AVA using the rig-3 promoter did not rescue the associative memory defects of creb1/crh-1 mutants (Fig. 3G).
A recent study has shown that the CRH-1a isoform is broadly expressed along the C. elegans body and rescued the aging defects seen in creb1/crh-1 mutants (Chen et al., 2016). We were interested in seeing whether CRH-1a was expressed in the command interneurons and in the RIM interneuron. To look for the localization of the crh-1a promoter in these neurons we made a pcrh-1a::GFP construct and assessed for the colocalization between pcrh-1a::GFP and pnmr-1::mCherry. Our results indicated that there was no overlap between pcrh-1a expression and the command interneurons (data not shown) corroborating our hypothesis of functional diversity among CRH-1 isoforms.
CRH-1 is required at the time of training
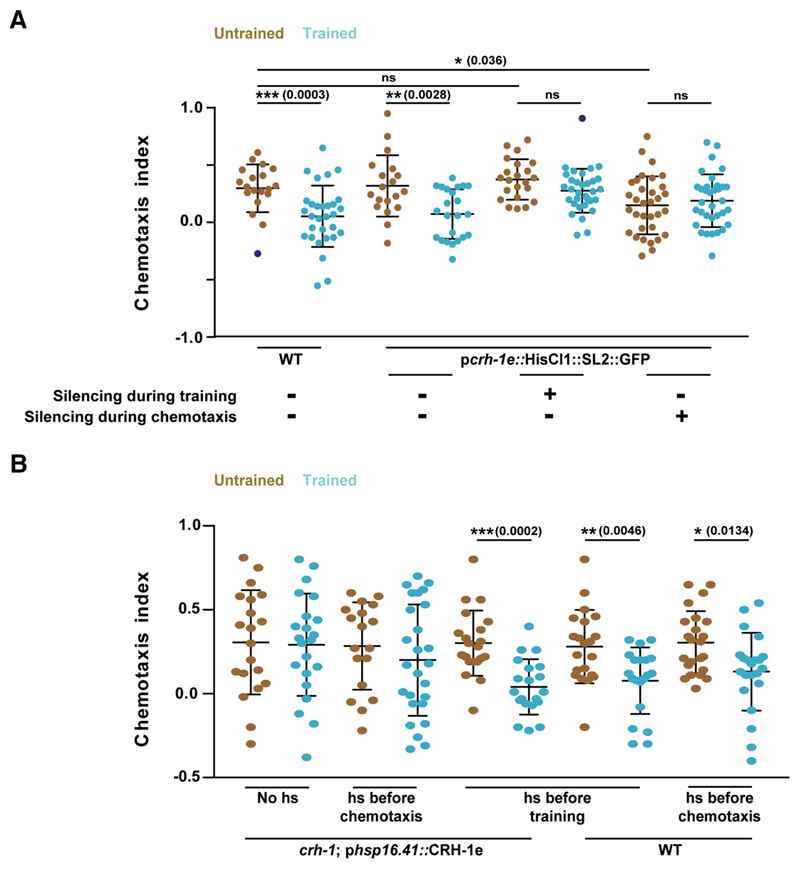
To study the temporal requirement of CRH-1e, we asked whether the activity of the CRH-1e-expressing neurons is required at the time of training. We expressed HisCl1 under the control of pcrh-1e in WT animals and silenced the HisCl1-expressing neurons by growing the C. elegans on histamine-containing plates while executing the training protocol (from 10 min before training to 2 h after training) or during chemotaxis (20 h after training till the end of the assay). Memory formation was completely abolished in the animals with silenced neurons during training while silencing during chemotaxis resulted in defective chemotaxis even in naive animals (Fig. 4A).
Figure 4. CRH-1e-expressing neurons are required for normal associative learning.
A, Scatter plot showing chemotaxis indices of C. elegans under the condition of histamine mediated neuronal silencing. B, Chemotaxis indices of WT and crh-1-null mutants expressing CRH-1e under the control of the heat shock promoter hsp-16.41. Error bars are SD. Multiple comparisons were done using one-way ANOVA and p values were adjusted using Bonferroni method. Data points represented with dark blue dots in the scatter plots represents outliers identified using Grubb’s method (α. = 0.05). hs, Heat Shock. *p < 0.05, **p < 0.01, ***p < 0.001, ns, p > 0.05.
To test the requirement of CRH-1e during the acquisition/consolidation phase of memory formation, we expressed CRH-1e under the control of the heat shock promoter (phsp16.41) and used these animals to rescue the creb1/crh-1 mutant phenotype. CRH-1e induction was done either 3 h before training or 3 h before chemotaxis. We observed that CRH-1e induction before training could largely rescue the learning defects, whereas induction before chemotaxis could not rescue the learning defect (Fig. 4B). These results suggest that CRH-1e is required at the time of acquisition and/or consolidation phase of memory formation in creb1/crh-1 mutant animals.
Lysine247 and lysine266 are key amino acid residues required for memory related functions of CRH-1
Most of the critical amino acid residues implicated in CREB1 activation are present in CRH-1a, b, and d isoforms. CRH-1c and e differ from CRH-1a, b, and d isoforms in having a much smaller and featureless N-terminal region. We hypothesized that the absence of these additional N-terminal residues is crucial for the functioning of CRH1c and CRH-1e in LTM related processes. We added an inert 3xHA tag to the N-terminal of CRH-1c and e (3xHA::CRH-1c,e) using CRISPR and tested these worms for the formation of LTM. Our experiments suggest defective memory formation in C. elegans with a modified N-terminal in CRH-1c and e (Fig. 5A).
Null mutants of creb1/crh-1 are defective in LTM formation. Naive creb1/crh-1 C. elegans also show reduced chemotaxis indices compared with WT animals (Fig. 5B). This defect is partially because of the inability of creb1/crh-1 mutant animals to modulate reversal frequency in response to chemical gradients (Fig. 3A). Therefore, observed learning defects in the creb1/crh-1 mutant C. elegans appears to be the compounded effect of learning deficit as well as defective innate chemotactic behavior.
There are reports of dendritic localization of CREB1 (Crino et al., 1998). This increases the possibility of nuclear localization of CREB1 in response to dendritic stimulation. Moreover, SUMOylation-dependent nuclear localization of CREB1 has been found to be critical for LTM formation in mouse (Chen et al., 2014). K285 and K304 have been identified as CREB1 SUMO acceptor sites in mouse (Comerford et al., 2003). To test for the possible involvement of these residues we generated K247R/K266R (corresponding to K285 and K304 in mouse CREB1) C. elegans using CRISPR. These mutant animals were trained and tested for the LTM phenotype. We observed that K247R/K266R mutant worms were defective in LTM formation, whereas their naive behavior was comparable to that seen in WT animals (Fig. 5C). Because CRH-1c and e are the only isoforms that appear to be involved in the learning process in our experiments, it is conceivable that K247R/K266R mutations affect the function of these isoforms in the learning process.
The ability to modulate reversal frequency in response to chemical gradients is a primary factor in mediating chemotactic behaviors in worms. We tested the effect of K247R/K266R mutations on the ability of C. elegans to initiate reversals in response to AWC activity. We optogenetically activated the AWC neuron and recorded the reversal probability of WT and K247R/K266R animals under naive and trained conditions. Naive WT and K247R/K266R animals showed similar reversal frequencies while trained worms showed significantly increased reversal frequencies with the K247R/K266R animals showing lower reversal probability compared with WT control animals (Fig. 5D). To understand whether K247R/K266R mutations have any effect on AVA activity in response to AWC activity we optogenetically activated AWC for 3.0 s while simultaneously imaging AVA using GCaMP5G. While trained WT C. elegans showed significant increase in the probability of AVA firing upon AWC activation there was no such increase seen in case of K247R/K266R (Fig. 5 E,F, KR) animals. These data suggest the involvement of K247 and K266 residues in regulating the AVA firing and reversal probability of worms under specific training conditions.
Discussion
The role of CREB1 is well documented in the studies on learning and memory across phyla. It has been described as one of the main inducers of immediate early genes (IEGs) expressed in response to experience-dependent neural activation (for review, see Alberini, 1999). However, functional pleiotropy of CREB1 remains a major bottleneck in studying CREB1-dependent IEGs that are activated specifically in response to experience-dependent neural activation. Here, we describe the functional specialization of CRH-1 isoforms in C. elegans. Our experiments show that C. elegans lacking CRH-1c and CRH-1e are defective in LTM formation while having normal innate chemotaxis function. Null mutants of creb1/crh-1 are defective in showing innate as well as learned behavior. Restricted expression of CRH-1e in a small subset of neurons indicates functional specialization of the isoform in C. elegans. In Aplysia and Drosophila, it has been shown that different isoforms of CREB1 can repress and facilitate the process of memory formation (Yin et al., 1995; Yin and Tully, 1996; Bartsch et al., 1998). However, what separates CRH-1 isoforms from their CREB1 orthologs is their remarkable sequence similarity yet striking functional diversity. Spatial segregation is an efficient way of exploiting different functional properties of proteins. CRH-1e and CRH-1a have different expression patterns (this work; Chen et al., 2016) and this spatial segregation can explain their functional segregation. However, this might not be the whole story here because pan-neuronal expression of CRH-1e cDNA could rescue the memory defects of creb1/crh-1-null mutants; while expressing CRH-1a, the isoform that could rescue aging defect of null animals (Chen et al., 2016), could not rescue memory defects. Further, a small featureless sequence like a 3xHA epitope tag that is routinely used in various biochemical and immunohistochemical assays without affecting the function of most proteins could disrupt the memory related functions of CRH-1c and CRH-1e. It is possible that these small differences result in significantly altered tertiary structures as seen in other proteins (Goda et al., 2000; Korepanova et al., 2001), hence affecting activation dynamics or subcellular localization of the different protein isoforms (Crino et al., 1998). It appears that the absence of these additional N-terminal amino acids from CRH-1a, b, and d enables the CRH-1c and e isoforms to interact with new binding partners either by allowing CRH-1c and e a different tertiary structure or by removing steric hindrance. CRISPR mutants with K247R/K266R are defective in learned responses while having normal innate response to IAA gradient. SUMOylation of homologous residues has been shown to be associated with LTM defects in rats undergoing water maze test. It is yet to be seen whether K247 and K266 undergo SUMOylation in C. elegans.
Lau et al. have shown that the NMDA-type glutamate receptor-dependent associative memory defects seen is nmr-1 mutants is rescued by expressing NMR-1 in the RIM interneuron among other neurons (Lau et al., 2013). Previous work has shown that the glutamate receptor, GLR-1 function is necessary for LTM formation in C. elegans (Rose et al., 2003). AVE and RIM both express NMR-1 and GLR-1 and are connected to each other through gap junctions (Brockie et al., 2001; Kano et al., 2008; Kawano et al., 2011; Piggott et al., 2011; Lau et al., 2013). This along with our data makes it conceivable that RIM and AVE could be functioning together in the process of memory formation. Further, Jin et al. (2016) have recently shown that the interneurons RIM and AIB are required for CREB/CRH-1-mediated imprinted memory formation, whereas the interneurons AIY and RIA are required for retrieval of the memory. Our results also show that presence of functional CRH-1e in RIM or AVE is sufficient for overcoming LTM formation defect in creb1/crh-1-null worms. Both long-term associative memory and longterm habituation implicate CREB1/CRH-1 in RIM neurons (Timbers and Rankin, 2011). Even though different neural circuits are activated by the training paradigms used in these studies, both paradigms operate in part by modulating the naive reversal behavior in response to a cue. Consistent with these observations our experiments implicate K247 and K266 in modulating reversal probability in trained worms upon AWC activation through channel rhodopsin. Moreover, CRH-1 (K247R/K266R) trained animals have diminished AWC-dependent AVA activation as measured by GCaMP5-mediated calcium imaging. This suggests that CRH-1-dependent genes might provide substrates for experience-dependent modulation of reversal behaviors in C. elegans.
Channelrhodopsin-mediated AWC activation highlights the importance of K247R/K266R residues in the modulation of reversal probability/AVA excitability exclusively in an experiencedependent manner. Under these experimental conditions, these mutations express themselves exclusively when worms are subjected to the training protocol. However, these results may not be true representative of natural phenomena operating during chemotactic movement of worms. Physiologically AWC are OFF type neurons, they are activated in response to step down of attractant concentration (Troemel et al., 1999). This makes the results of our optogenetics experiments counterintuitive. Our optogenetics experiment suffers from two technical problems; the promoter used for expression of ChR2, pstr-2, only expresses in one AWC neuron, AWCON (Troemel et al., 1999). Hence, we could only activate one AWC neuron, which is unnatural for worms sensing IAA. Second is our inability to localize ChR2 to the subcellular position of chemosensors. The receptor for IAA is unknown. However, we know that ODR-10, a GPCR for diacetyl, is primarily localized to the ciliated endings of AWA neurons (Sengupta et al., 1996). We can assume similar localization patterns for IAA receptors. Sensory neurons in worms also receive information from other neurons hence technically also act as interneurons and due to graded potential, they process and transmit this information in a complex way. Because of localization of channelrhodopsin throughout the surface of the neuron the output of optogenetically activated neuron lacks any resemblance to the output of ligand-activated neuron. However, we do observe reversal probability of optogenetically activated naive worms that are consistent with the previous studies making the results that we got from trained group of animals difficult to explain in terms of experimental artifacts alone (Cho et al., 2016). These problems could be overcome by using a more natural method using ligandbased neuron activation in microfluidics chambers.
AVA activity is highly correlated with reversal behavior in C. elegans (Gray et al., 2005; Ben Arous et al., 2010; Bhardwaj et al., 2018). The ability to modulate AVA activity in response to sensory neuron activation provides for an important control point of behavioral modification through reversal modulation. In a parallel set of experiments, we have found that learning in worms is specific to the cues that are presented at the time of training. Therefore, given the strong correlation of AVA activity and reversal event it was surprising to find that expression of CRH-1e RIM and AVE was sufficient to restore LTM defect in creb1/crh-1-null mutants. AVA is heavily connected with AVE (44 incoming chemical synapses) and RIM (6 electrical synapses, 3 incoming chemical synapses; White et al., 1986). Any change in RIM/AVE due to activity-dependent CRH-1 activation in response to different sensory cues is likely to produce similar effects on AVA neurons. There are two probable explanations. One, the rescue of LTM defect that we observed in our experiments is an artifact of the experimental methods used. Because specificity of promoters is largely determined by visible fluorescent protein reporter gene expression in a particular cell, it is likely that subvisible expression at other sites is responsible for the LTM defect rescue. Second, learning but not its specificity is a function of CRH-1. If this is true, then it is likely that by studying CRH-1-dependent IEGs in C. elegans we will only be able to study the somewhat mechanical aspect of memory manifesting at the terminal end of neural hierarchy just before behavior execution.
Supplementary Material
Significance Statement.
This study elucidates the role of a specific isoform of CREB1/CRH-1, CRH-1e, in Caenorhabditis elegans memory formation and chemosensation. Removal of this single isoform of creb1/crh-1 shows defects in long-term memory formation in the animal and expression of CREB1/CRH-1e in a single pair of neurons is sufficient to rescue the memory defects seen in the mutant animals. We further show that two specific amino acids of CRH-1 are required for the process of memory formation in the animal.
Acknowledgments
This work was supported by Wellcome Trust/DBT India Alliance Fellowships awarded to Y.D. (Grant no. IA/E/13/1/501257) and K.B. (Grant no. IA/I/12/1/500516); K.B. was also supported by DBT-IYBA (Grant BT/05/IYBA/2011), DST-SERB (Grant EMR/2016/005332), and IISER Mohali; S.B. was supported by a KVPY fellowship; M.P. and S.R. by DST-INSPIRE fellowships; and S.T. by a CSIR fellowship. The crh1 mutants were provided by CGC, which is funded by the National Institutes of Health Office of Research Infrastructure Program P40 OD010440. All vectors used in this study were obtained from Addgene. We thank members of K.B.’s laboratory for comments on the paper; Cori Bargmann and Hernan Jaramillo for the prig3::GCCaMP5.0 array and the CX15141 strain that we used to amplify HisCl1; Ankit; Negi for routine help; Ashwani Bhardwaj and Saurabh Thapliyal for discussion; Arjit Kant Gupta for help in making the chemotaxis video recording setup and the IISER Mohali confocal facility; and the people contributing to and maintaining open source software; Libre Office, Inkscape, and Zotero.
Footnotes
Yogesh Dahiya: 0000-0002-8604-739X
Kavita Babu: 0000-0002-7527-3666
The authors declare no competing financial interests.
References
- Alberini CM. Genes to remember. J Exp Biol. 1999;202:2887–2891. doi: 10.1242/jeb.202.21.2887. [DOI] [PubMed] [Google Scholar]
- Amano H, Maruyama IN. Aversive olfactory learning and associative long-term memory in Caenorhabditis elegans. Learn Mem. 2011;18:654–665. doi: 10.1101/lm.2224411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardiel EL, Rankin CH. An elegant mind: learning and memory in Caenorhabditis elegans. Learn Mem. 2010;17:191–201. doi: 10.1101/lm.960510. [DOI] [PubMed] [Google Scholar]
- Bargmann CI. The C. elegans Research Community, WormBook. 2006. Chemosensation in C. elegans. Available at http://wormbook.org/chapters/www_chemosensation/chemosensation.html. [Google Scholar]
- Bartsch D, Casadio A, Karl KA, Serodio P, Kandel ER. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell. 1998;95:211–223. doi: 10.1016/s0092-8674(00)81752-3. [DOI] [PubMed] [Google Scholar]
- Bateson P, Mameli M. The innate and the acquired: useful clusters or a residual distinction from folk biology? Dev Psychobiol. 2007;49:818–831. doi: 10.1002/dev.20277. [DOI] [PubMed] [Google Scholar]
- Ben Arous J, Tanizawa Y, Rabinowitch I, Chatenay D, Schafer WR. Automated imaging of neuronal activity in freely behaving Caenorhabditis elegans. J Neurosci Methods. 2010;187:229–234. doi: 10.1016/j.jneumeth.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Bhardwaj A, Thapliyal S, Dahiya Y, Babu K. FLP-18 functions through the G-protein-coupled receptors NPR-1 and NPR-4 to modulate reversal length in Caenorhabditis elegans. J Neurosci. 2018;38:4641–4654. doi: 10.1523/JNEUROSCI.1955-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockie PJ, Madsen DM, Zheng Y, Mellem J, Maricq AV. Differential expression of glutamate receptor subunits in the nervous system of Caenorhabditis elegans and their regulation by the homeodomain protein UNC-42. J Neurosci. 2001;21:1510–1522. doi: 10.1523/JNEUROSCI.21-05-01510.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalfie M, Sulston JE, White JG, Southgate E, Thomson JN, Brenner S. The neural circuit for touch sensitivity in Caenorhabditis elegans. J Neurosci. 1985;5:956–964. doi: 10.1523/JNEUROSCI.05-04-00956.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Hsu WL, Ma YL, Tai DJ, Lee EH. CREB SUMOylation by the E3 ligase PIAS1 enhances spatial memory. J Neurosci. 2014;34:9574–9589. doi: 10.1523/JNEUROSCI.4302-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Chen HJ, Tseng WC, Hsu JM, Huang TT, Chen CH, Pan CL. A C. elegans thermosensory circuit regulates longevity through crh1/CREB-dependent flp-6 neuropeptide signaling. Dev Cell. 2016;39:209–223. doi: 10.1016/j.devcel.2016.08.021. [DOI] [PubMed] [Google Scholar]
- Cho CE, Brueggemann C, L’Etoile ND, Bargmann CI. Parallel encoding of sensory history and behavioral preference during Caenorhabditis elegans olfactory learning. eLife. 2016;5:e14000. doi: 10.7554/eLife.14000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comerford KM, Leonard MO, Karhausen J, Carey R, Colgan SP, Taylor CT. Small ubiquitin-related modifier-1 modification mediates resolution of CREB-dependent responses to hypoxia. Proc Natl Acad Sci U S A. 2003;100:986–991. doi: 10.1073/pnas.0337412100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crino P, Khodakhah K, Becker K, Ginsberg S, Hemby S, Eberwine J. Presence and phosphorylation of transcription factors in developing dendrites. Proc Natl Acad Sci U S A. 1998;95:2313–2318. doi: 10.1073/pnas.95.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Goldstein B. CRISPR-based methods for Caenorhabditis elegans genome engineering. Genetics. 2016;202:885–901. doi: 10.1534/genetics.115.182162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N. Advanced methods of microscope control using μManager software. J Biol Methods. 2014;1:e10. doi: 10.14440/jbm.2014.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei YJ, Romero MF, Krause M, Liu JC, Huang W, Ganapathy V, Leibach FH. A novel H+ -coupled oligopeptide transporter (OPT3) from Caenorhabditis elegans with a predominant function as a H+ channel and an exclusive expression in neurons. J Biol Chem. 2000;275:9563–9571. doi: 10.1074/jbc.275.13.9563. [DOI] [PubMed] [Google Scholar]
- Feinberg EH, Vanhoven MK, Bendesky A, Wang G, Fetter RD, Shen K, Bargmann CI. GFP reconstitution across synaptic partners (GRASP) defines cell contacts and synapses in living nervous systems. Neuron. 2008;57:353–363. doi: 10.1016/j.neuron.2007.11.030. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda S, Takano K, Yamagata Y, Katakura Y, Yutani K. Effect of extra N-terminal residues on the stability and folding of human lysozyme expressed in Pichia pastoris. Protein Eng. 2000;13:299–307. doi: 10.1093/protein/13.4.299. [DOI] [PubMed] [Google Scholar]
- Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Gray JM, Hill JJ, Bargmann CI. A circuit for navigation in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2005;102:3184–3191. doi: 10.1073/pnas.0409009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC. The C. elegans Research Community, WormBook. 2006. Behavior. Available at http://wormbook.org/chapters/www_behavior/behavior.html. [Google Scholar]
- Iino Y, Yoshida K. Parallel use of two behavioral mechanisms for chemotaxis in Caenorhabditis elegans. J Neurosci. 2009;29:5370–5380. doi: 10.1523/JNEUROSCI.3633-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X, Pokala N, Bargmann CI. Distinct circuits for the formation and retrieval of an imprinted olfactory memory. Cell. 2016;164:632–643. doi: 10.1016/j.cell.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Kano T, Brockie PJ, Sassa T, Fujimoto H, Kawahara Y, Iino Y, Mellem JE, Madsen DM, Hosono R, Maricq AV. Memory in Caenorhabditis elegans is mediated by NMDA-type ionotropic glutamate receptors. Curr Biol. 2008;18:1010–1015. doi: 10.1016/j.cub.2008.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman AL, Ashraf JM, Corces-Zimmerman MR, Landis JN, Murphy CT. Insulin signaling and dietary restriction differentially influence the decline of learning and memory with age. PLoS Biol. 2010;8:e1000372. doi: 10.1371/journal.pbio.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffman A, Parsons L, Stein G, Wills A, Kaletsky R, Murphy C. C. elegans positive butanone learning, short-term, and long-term associative memory assays. J Vis Exp. 2011;49:2490. doi: 10.3791/2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Po MD, Gao S, Leung G, Ryu WS, Zhen M. An imbalancing act: gap junctions reduce the backward motor circuit activity to bias C. elegans for forward locomotion. Neuron. 2011;72:572–586. doi: 10.1016/j.neuron.2011.09.005. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Corcoran EE, Eto K, Gengyo-Ando K, Muramatsu MA, Kobayashi R, Freedman JH, Mitani S, Hagiwara M, Means AR, Tokumitsu H. A CaMK cascade activates CRE-mediated transcription in neurons of Caenorhabditis elegans. EMBO Rep. 2002;3:962–966. doi: 10.1093/embo-reports/kvf191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korepanova A, Douglas C, Leyngold I, Logan TM. N-terminal extension changes the folding mechanism of the FK506-binding protein. Protein Sci. 2001;10:1905–1910. doi: 10.1110/ps.14801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsch J, Flavell SW, Liu Q, Gordus A, Albrecht DR, Bargmann CI. A circuit for gradient climbing in C. elegans chemotaxis. Cell Rep. 2015;12:1748–1760. doi: 10.1016/j.celrep.2015.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsch J, Ventimiglia D, Bargmann CI, Albrecht DR. High-throughput imaging of neuronal activity in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2013;110:E4266–4273. doi: 10.1073/pnas.1318325110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau HL, Timbers TA, Mahmoud R, Rankin CH. Genetic dissection of memory for associative and non-associative learning in Caenorhabditis elegans. Genes Brain Behav. 2013;12:210–223. doi: 10.1111/j.1601-183X.2012.00863.x. [DOI] [PubMed] [Google Scholar]
- Luo L, Wen Q, Ren J, Hendricks M, Gershow M, Qin Y, Greenwood J, Soucy ER, Klein M, Smith-Parker HK, Calvo AC, et al. Dynamic encoding of perception, memory, and movement in a C. elegans chemotaxis circuit. Neuron. 2014;82:1115–1128. doi: 10.1016/j.neuron.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y, Sugi T, Nonomura M, Mori I. Identification of the AFD neuron as the site of action of the CREB protein in Caenorhabditis elegans thermotaxis. EMBO Rep. 2011;12:855–862. doi: 10.1038/embor.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz CO, Etchberger JF, Posy SL, Frøkjaer-Jensen C, Lockery S, Honig B, Hobert O. Searching for neuronal left/right asymmetry: genomewide analysis of nematode receptor-type guanylyl cyclases. Genetics. 2006;173:131–149. doi: 10.1534/genetics.106.055749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira S, van der Kooy D. Two forms of learning following training to a single odorant in Caenorhabditis elegans AWC neurons. J Neurosci. 2012;32:9035–9044. doi: 10.1523/JNEUROSCI.4221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin BJ, Ervasti JM. The actin gene family: function follows isoform. Cytoskeleton. 2010;67:630–634. doi: 10.1002/cm.20475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce-Shimomura JT, Morse TM, Lockery SR. The fundamental role of pirouettes in Caenorhabditis elegans chemotaxis. J Neurosci. 1999;19:9557–9569. doi: 10.1523/JNEUROSCI.19-21-09557.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggott BJ, Liu J, Feng Z, Wescott SA, Xu XZ. The neural circuits and synaptic mechanisms underlying motor initiation in C. elegans. Cell. 2011;147:922–933. doi: 10.1016/j.cell.2011.08.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokala N, Liu Q, Gordus A, Bargmann CI. Inducible and titratable silencing of Caenorhabditis elegans neurons in vivo with histamine-gated chloride channels. Proc Natl Acad Sci U S A. 2014;111:2770–2775. doi: 10.1073/pnas.1400615111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JK, Kaun KR, Chen SH, Rankin CH. GLR-1, a non-NMDA glutamate receptor homolog, is critical for long-term memory in Caenorhabditis elegans. J Neurosci. 2003;23:9595–9599. doi: 10.1523/JNEUROSCI.23-29-09595.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, et al. Fiji: an open source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta P, Chou JH, Bargmann CI. odr-10 Encodes a seven transmembrane domain olfactory receptor required for responses to the odorant diacetyl. Cell. 1996;84:899–909. doi: 10.1016/s0092-8674(00)81068-5. [DOI] [PubMed] [Google Scholar]
- Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- Suo S, Kimura Y, Van Tol HH. Starvation induces cAMP response element-binding protein-dependent gene expression through octopamine-Gq signaling in Caenorhabditis elegans. J Neurosci. 2006;26:10082–10090. doi: 10.1523/JNEUROSCI.0819-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo S, Culotti JG, Van Tol HH. Dopamine counteracts octopamine signalling in a neural circuit mediating food response in C. elegans. EMBO J. 2009;28:2437–2448. doi: 10.1038/emboj.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tierney AJ. The evolution of learned and innate behavior: contributions from genetics and neurobiology to a theory of behavioral evolution. Anim Learn Behav. 1986;14:339–348. [Google Scholar]
- Timbers TA, Rankin CH. Tap withdrawal circuit interneurons require CREB for long-term habituation in Caenorhabditis elegans. Behav Neurosci. 2011;125:560–566. doi: 10.1037/a0024370. [DOI] [PubMed] [Google Scholar]
- Touriol C, Bornes S, Bonnal S, Audigier S, Prats H, Prats AC, Vagner S. Generation of protein isoform diversity by alternative initiation of translation at non-AUG codons. Biol Cell. 2003;95:169–178. doi: 10.1016/s0248-4900(03)00033-9. [DOI] [PubMed] [Google Scholar]
- Troemel ER, Sagasti A, Bargmann CI. Lateral signaling mediated by axon contact and calcium entry regulates asymmetric odorant receptor expression in C. elegans. Cell. 1999;99:387–398. doi: 10.1016/s0092-8674(00)81525-1. [DOI] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos Trans R Soc Lond B Biol Sci. 1986;314:1–340. doi: 10.1098/rstb.1986.0056. [DOI] [PubMed] [Google Scholar]
- Yin JC, Tully T. CREB and the formation of long-term memory. Curr Opin Neurobiol. 1996;6:264–268. doi: 10.1016/s0959-4388(96)80082-1. [DOI] [PubMed] [Google Scholar]
- Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances longterm memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.