Summary
CD4 T follicular helper (Tfh) cells are specialized in helping B cells during the germinal center (GC) reaction and ultimately promote long-term humoral immunity. Here we report that loss of the nuclear orphan receptor NR2F6 causes enhanced survival and accumulation of Tfh cells, GC B cells, and plasma cells (PCs) following T cell-dependent immunization. Nr2f6-deficient CD4 T cell dysfunction is the primary cause of cell accumulation. Cytokine expression in Nr2f6-deficient Tfh cells is dysregulated, and Il21 expression is enhanced. Mechanistically, NR2F6 binds directly to the interleukin 21 (IL-21) promoter and a conserved noncoding sequence (CNS) near the Il21 gene in resting CD4+ T cells. During Tfh cell differentiation, this direct NR2F6 DNA interaction is abolished. Enhanced Tfh cell accumulation in Nr2f6-deficient mice can be reverted by blocking IL-21R signaling. Thus, NR2F6 is a critical negative regulator of IL-21 cytokine production in Tfh cells and prevents excessive Tfh cell accumulation.
Introduction
The germinal center reaction is critical for the production of high-affinity, class-switched antibodies and effective responses to pathogens and vaccination. Germinal center (GC) formation depends on the differentiation of a specialized subset of CD4 T cells, T follicular helper cells, alongside antigen-specific B cell differentiation into GC B cells. Differentiation of both cell types depends on T cell-B cell interaction, co-receptor engagement, and cytokine expression, notably interleukin-4 (IL-4), IL-6, and IL-21 (De Silva and Klein, 2015; Victora and Nussenzweig, 2012). Within the GC, B cells undergo a process of random mutation of the B cell receptor genes known as somatic hypermutation (SHM) (Peled et al., 2008). B cells that acquire increased antigen affinity through SHM are selected for plasma cell differentiation, memory cell differentiation, or continued proliferation in the GC by T follicular helper (Tfh) cells and follicular dendritic cells (FDCs) (Mesin et al., 2016). However, because of the random nature of SHM, mutations causing self-reactivity are a potential hazard. It is thus critical that the differentiation and the number of Tfh cells are tightly regulated to prevent the expansion of dangerous B cell clones (Crotty, 2014; Pratama and Vinuesa, 2014). An additional T cell subset, T follicular regulatory (Tfr) cells, limits self-reactive T cells and GC B cells (Chung et al., 2011; Linterman et al., 2011).
Systemic lupus erythematosus (SLE) is a disease characterized by spontaneous GC formation, self-reactive antibodies, and increased Tfh cell activity and cell numbers (Tsokos, 2011; Yu and Vinuesa, 2010). IL-21 was shown to enhance pathogenesis in lupus mouse models, including BSXB-Yaa mice and MRL/MpJ-Faslpr mice (Herber et al., 2007; Ozaki et al., 2004). IL-21 overexpression has also been reported in human SLE patients (Wong et al., 2010). In the sanroque lupus mouse model, excessive expression of the co-receptor ICOS and production of the cytokine interferon gamma (IFNγ) can lead to accumulation of Tfh cells and contribute to disease pathology However, increased IL-21 levels in these mice does not contribute to pathogenesis (Linterman et al., 2009; Vinuesa et al., 2005; Yu et al., 2007).
The transcriptional activity of members of the nuclear receptor (NR) family has been shown to regulate both pro-and anti-inflammatory processes (Glass and Saijo, 2010; Huang and Glass, 2010). The orphan NRs of the chicken ovalbumin upstream promoter transcription factor (COUP-TF)/NR2F family (NR2F1, NR2F2, and NR2F6) have essential roles in cell differentiation and cell fate decisions (Cooney et al., 1992). As TFs, NR2F family members homo- or heterodimerize with retinoid X receptor (RXR/NR2B1), as well as other NRs, and bind to various response elements that contain imperfect TGACCT direct or inverted repeats (Cooney et al., 1992; Hermann-Kleiter et al., 2008). We have established NR2F6 as an intracellular immune checkpoint during cancer immune surveillance and experimental autoimmune responses (Hermann-Kleiter et al., 2012, 2015; Klepsch et al., 2018). Mechanistically, only sustained high-affinity antigen receptor-induced protein kinase C (PKC)-mediated phosphorylation inactivates the DNA binding capability of NR2F6, thereby displacing pre-bound NR2F6 from the DNA (Hermann-Kleiter et al., 2008, 2012).
We have previously shown that deletion of Nr2f6 in mice leads to a SLE-like immunopathology with enhanced titers of anti-double-stranded DNA (dsDNA) and anti-nuclear antibodies in aged animals (Hermann-Kleiter et al., 2008). Hutcheson et al. (2008) reported reduced Nr2f6 expression in SLE patients. Here we investigate whether and how the loss of Nr2f6 leads to enhanced GC responses and consequently antibody production in mice following T cell-dependent immunization. We provide evidence linking Nr2f6 deficiency to Tfh cell accumulation following ovalbumin (OVA)-aluminum potassium sulfate dodecahydrate (alum) immunization. This accumulation depends on increased IL-21 production by the CD4 T cell compartment, but Nr2f6 deficiency has no direct effect within the B cell compartment. NR2F6 directly binds to several Il21 regulatory regions in resting cells, but not Tfh-activated cells, and interruption of IL-21R signaling through blocking antibodies reduces Tfh cell accumulation.
Results
NR2F6 Loss Leads to Tfh Cell Accumulation and Increased GC B Cell and Plasma Cell Numbers
To test the role of NR2F6 in the GC, we first immunized Nr2f6+/+ or Nr2f6−/− mice intraperitoneally (i.p.) with OVA precipitated in 10% alum and harvested spleens on days 4 and 10 for analysis by flow cytometry (Figure 1A).
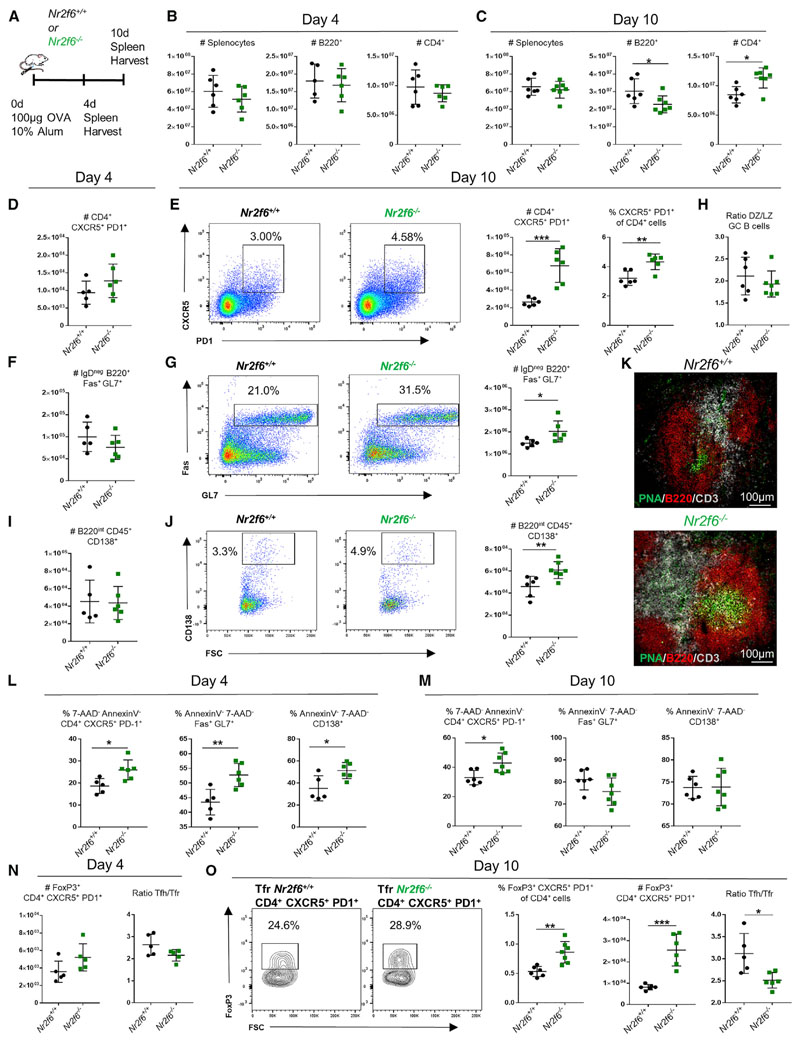
Figure 1. Nr2f6 Loss Leads to Tfh Cell Accumulation and Increased GC Responses following OVA-Alum Immunization.
(A) Experimental setup used for OVA-alum immunization of Nr2f6+/+ or Nr2f6−/− mice.
(B and C) Total counts of Nr2f6+/+ or Nr2f6−/− splenocytes, B220+ cells, and CD4+ cells on (B) day 4 or (C) day 10 after immunization.
(D and E) Splenic Tfh cell numbers on (D) day 4 or (E) day 10 after immunization; representative flow cytometry plots for Nr2f6+/+ and Nr2f6−/− Tfh cells, total Tfh cell counts, and Tfh cell frequency within CD4 cells. Total Tfh cell counts are shown as total live non-Tfr cells (annexin V−, FoxP3−, and 7-AAD−).
(F) Total germinal center B cell counts on day 4.
(G) Day 10 representative flow cytometry plots, with total cell counts.
(H) GC LZ and DZ distribution in Nr2f6+/+ or Nr2f6−/− GCs 10 days after immunization.
(I and J) PC counts on day 4 (I), and representative flow cytometry plots and cell counts on day 10 (J).
(K) Immunofluorescence staining for CD3 (white), B220 (red), and peanut agglutinin (PNA) (green) on 5 μM sections of spleens harvested day 10 after OVA immunization.
(L and M) Frequency of live Tfh cells, GC B cells, and PCs (annexin V− 7-AAD−) on (L) day 4 or (M) day 10 after OVA-alum immunization.
(N) Tfr (CD4+, CXCR5+, PD-1+, and FoxP3+) total cell counts on day 4 (left panel), and the Tfh cell to Tfr cell ratio (right panel).
(O) Representative FoxP3 staining of Nr2f6+/+ or Nr2f6−/− CD4+ CXCR5+ PD-1+ T cells, total Tfr cell counts, total Tfr frequency of all CD4 cells, and the Tfh cell to Tfr cell ratio on day 10 post-immunization.
Data shown are from at least two separate experiments with n ≥ 5. The middle bar represents the average for the dataset. Error bars represent SD, and an asterisk indicates statistically significant differences calculated using two-tailed Student’s t test or Mann-Whitney U test. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
Total splenocyte, B220+, and CD4+ cell numbers were comparable between genotypes on day 4. Similarly, total splenocyte counts were not different between genotypes on day 10. However, B220+ cell numbers were reduced and the CD4+ population significantly increased in Nr2f6−/− spleens (Figures 1B and 1C). We also investigated the Tfh cell population on day 4 and observed no differences in the number of Tfh cells (Figure 1D). However, we observed a significant increase in the total numbers of Tfh cells in Nr2f6−/− mice on day 10 after immunization (Figure 1E). This increase was not purely the consequence of the enlarged CD4+ population, because the frequency of Tfh cells was increased within the total CD4+ cell population (Figure 1E, right panel). GC B cell numbers at the early time point (day 4) were unchanged (Figure 1F), but the numbers and frequency of GC B cells increased significantly by day 10 in Nr2f6-deficient mice (Figure 1G). We stained GC B cells for the dark zone (DZ) and light zone (LZ) markers CXCR4 and CD86 (Victora et al., 2010) on day 10 post-immunization; no differences were observed in the ratio of DZ cells to LZ cells (Figure 1H). The numbers of plasma cells (PCs) were unchanged on day 4 (Figure 1I), but cell numbers were significantly increased in Nr2f6−/− mice on day 10 (Figure 1J). To corroborate the observed increase in Tfh cells and GC B cells of Nr2f6−/− mice, spleens were harvested on day 10 post-immunization, and sections were stained for CD3, B220, and peanut agglutinin (PNA). Larger GCs, as determined by PNA staining, were observed in Nr2f6−/− spleens (Figure 1K). These findings indicate a role for Nr2f6 in the regulation of Tfh cell, GC B cell, and PC numbers by day 10 after immunization and suggest that the early differentiation of these cells is unchanged. To exclude the possibility that enhanced GC reactions were confined to responses only against the OVA protein, we immunized Nr2f6+/+ or Nr2f6−/− mice i.p. with 5 × 108 sheep red blood cells (SRBCs) and harvested spleens 7 days later. Increases in the frequency of Tfh cells, GC B cells, and PC were observed in Nr2f6-deficient mice compared with wild-type controls (Figure S1).
We next determined whether the increases in Tfh cells, GC B cells, and PCs from day 4 to day 10 after OVA-alum immunization depended on increased survival or proliferation. Splenocytes harvested from Nr2f6+/+ or Nr2f6−/− mice on day 4 or day 10 after immunization were costained with lineage-defining markers (as shown in Figures 1E, 1G, and 1J) and with annexin V and 7-amino-actinomycin D (7-AAD). The fraction of live Tfh cells, GC B cells, and PCs was significantly higher in Nr2f6−/− mice on day 4 (Figure 1L). In contrast, by day 10 post-immunization, only Nr2f6−/− Tfh cells, not GC B cells or PCs, showed enhanced survival, paralleling the relatively large numbers of Tfh cells seen by day 10 (Figure 1M). We investigated the proliferation of Tfh cells on day 4 by staining of Ki67. The frequency of CXCR5+ PD-1+ CD4+ Tfh cells costaining for Ki67 was unaltered at this time point (Figure S2A). The proliferation of Tfh cells, GC B cells, and PCs on day 10 was investigated by bromodeoxyuridine (BrdU) incorporation 7–12 h after pulse labeling of mice. A small decrease in BrdU incorporation was observed in Nr2f6-deficient Tfh cells and GC B cells, but not in PCs; these changes did not reach statistical significance (Figure S2B). Altogether, the data suggest that the increases in Tfh cells, GC B cells, and PCs from day 4 to day 10 post-immunization depend on increased survival and not an increase in proliferation.
GC reactions are controlled by Tfr cells, a unique regulatory cell type coexpressing Foxp3 and BCL6 (Chung et al., 2011; Linterman et al., 2011). On day 4, there was a trend toward higher Tfr total numbers in Nr2f6−/− spleens, and the ratio of Tfh cells to Tfr cells was slightly reduced in Nr2f6−/− compared with Nr2f6+/+ mice. However, these differences were not statistically significant (Figure 1N). Tfr numbers on day 10 were significantly increased in Nr2f6−/− mice, and the ratio of Tfh cells to Tfr cells was significantly reduced (Figure 1O). Frequency of Tfr from the total CD4 population was significantly higher in Nr2f6−/− mice, suggesting that increased Tfr is not the result of higher total CD4 numbers (Figure 1O). We also investigated CD4+ CXCR−PD-1− FoxP3+ regulatory T (Treg) cells. There was no significantly altered frequency or total cell count in this population in Nr2f6-deficient mice compared with wild-type controls (Figure S2C).
These data suggest that following immunization, along with Tfh cell, GC, and PC expansion, Tfr cells are significantly increased relative to the Tfh cell population in Nr2f6−/− mice.
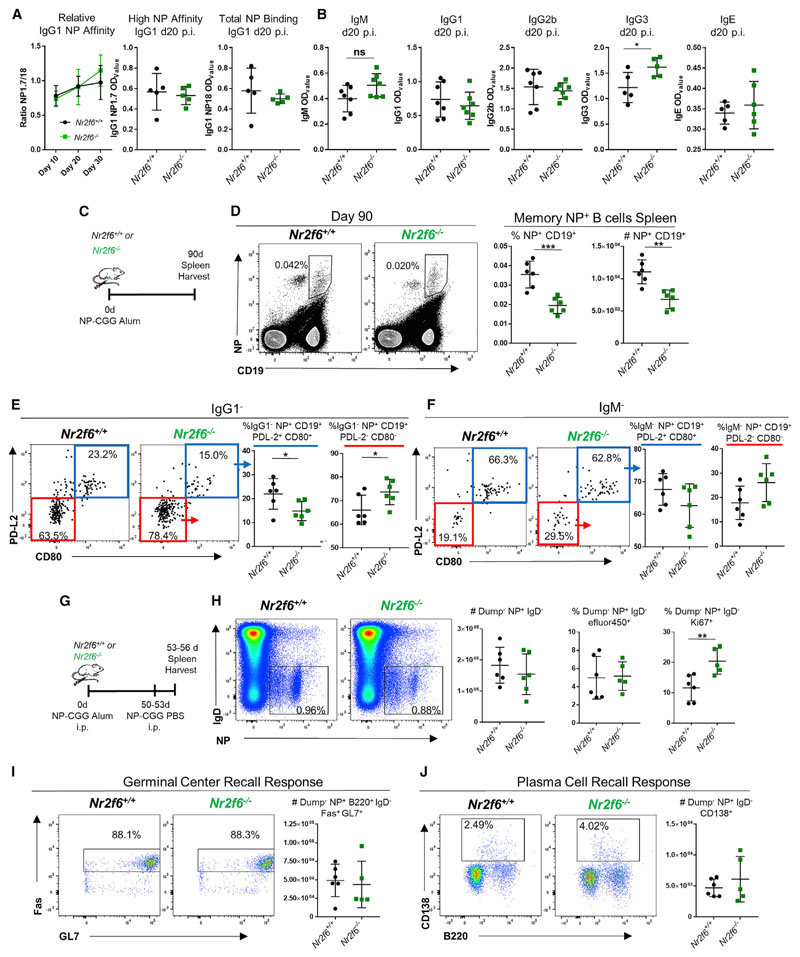
Nr2f6 Deficiency Does Not Alter Affinity Maturation but Affects Antigen-Specific Memory B Cells
To determine how Nr2f6 deficiency affects the GC, we investigated the antigen affinity of serum immunoglobulin (Ig)G1 collected from Nr2f6+/+ or Nr2f6−/− mice 10, 20, or 30 days after 4-hydroxy-3-nitrophenylacetyl (NP)-chicken gamma globulin (NP-CGG) alum immunization. IgG1 affinity for either high-affinity NP-specific binding (BSA:NP ratio 1.7 [NP1.7]) or total NP binding (BSA:NP ratio 18 [NP18]) was measured by ELISA. Relative binding of IgG1 was not significantly altered at any time point, nor were the titers of high-affinity IgG1 (NP1.7) or total IgG1 (NP18) (Figure 2A). We next investigated class switching on day 20 post-NP-CGG immunization; no significant changes were observed in total IgM, IgG1, IgG2b, or IgE. However, there was an increase in IgG3 titer (Figure 2B). GCs are also an important location for memory B cell formation (Weisel et al., 2016).We compared the population of memory B cells in Nr2f6−/− mice to those in wild-type controls. Spleens were harvested 90 days after immunization with NP-CGG, and single-cell suspensions were stained with NP-phycoerythrin (NP-PE) and CD19 to detect antigen-specific memory B cells (Figure 2C). Significant reduction in both frequency and total memory B cells was observed in Nr2f6−/− mice (Figure 2D). The substantial population of NP+CD19int cells were not PCs or memory B cells but may be the antigen-capturing cells described by Bell and Gray (2003), because these cells were negative for CD138, IgM, CD80, and PD-L2 but stained moderately for IgG1 (Figure S3A) (Bell and Gray, 2003). CD80+PD-L2+ memory B cells primarily differentiate into PCs during recall responses, while CD80−PD-L2− cells are more prone to populating GCs (Zuccarino-Catania et al., 2014). On day 90 post-immunization, an increase in the frequency of the IgG1−CD80−PD-L2− population in Nr2f6−/− mice compared with wild-type mice was observed (Figure 2E). Although the IgG1−CD80+PD-L2+ frequency was reduced in Nr2f6-deficient mice, no significant differences were observed in the IgM− memory B cell population despite a similar trend found in the IgG1− population (Figure 2F).
Figure 2. Nr2f6 Deficiency Does Not Alter Affinity Maturation but Affects Antigen-Specific Memory B Cells.
(A) Nr2f6+/+ or Nr2f6−/− mice were immunized with NP-CGG in alum; relative binding of high (18) or low (1.7) ratio haptenated NP-BSA-coated plates was measured by ELISA. IgG1 relative affinity is shown from serum collected 10, 20, or 30 days after immunization. High-affinity (NP1.7) or total IgG1 binding (NP18) titers from one representative ELISA experiment are shown.
(B) Sera was collected, and ELISA was performed for indicated antibodies 20 days after immunization, optical density 50 (OD50) is shown.
(C) Experimental scheme.
(D) Nr2f6+/+ or Nr2f6−/− mice were immunized with NP-CGG, and spleens were harvested on day 90; antigen-specific memory B cells were stained with NP-PE and CD19. Representative flow cytometry plots are shown in the left two panels, frequency and total cell count of this population are shown in the right two panels.
(E–G) CD80 and PD-L2 were investigated by flow cytometry on IgG1− (E) or IgM− (F) antigen-specific memory B cells from both Nr2f6+/+ or Nr2f6−/− genotypes. Recall responses were tested as described in the scheme (G).
(H) Total NP-specific, IgD−, and dump gate negative (Gr-1, F4/80, CD4, and CD8) cells, proliferation, and dead cell frequency were investigated by Ki67 and efluor450 fixable dye staining.
(I and J) GC (I) and PC (J) responses from this population were investigated by flow cytometry. Representative flow cytometry plots and total counts are shown. Memory results are representative of two independent experiments with n ≥ 5. The middle bar represents the dataset average. Error bars represent SD, and an asterisk indicates statistically significant differences calculated using two-tailed Student’s t test. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
B cell recall responses were investigated 3 days after antigen boost and >50 days after primary immunization (Figure 2G). Total NP+ IgD− cell numbers (Gr-1, F4/80, CD4, and CD8 negative [dump]) were unchanged between the two genotypes. However, the frequency of Ki67 expression in this population was significantly increased in Nr2f6−/− mice, while dead cell staining by fixable viability dye efluor450 was similar in both groups (Figure 2H). This suggests that despite decreased frequency and number, the Nr2f6-deficient NP-specific memory population may compensate by increasing proliferation upon recall. Total GC and PC counts were not significantly altered, although the frequency of PCs was consistently higher in Nr2f6−/− mice (Figures 2I and 2J).
Long-lived PCs (LLPCs) are essential antibody-secreting immune cells that develop from the GC. On day 90 after immunization, bone marrow and spleens were harvested and stained for CD138 and TACI, markers shown to label LLPCs (Pracht et al., 2017). No significant alterations in the frequency and total cell counts of LLPCs or their subpopulations, double-positive P1 (B220+CD19+), single-positive P2 (CD19+), or double-negative P3 (B220−CD19−) in either bone marrow or spleen were observed in Nr2f6−/− mice (Figures S3B–S3D).
Altogether, the loss of NR2F6 does not alter IgG1 affinity or class switch recombination with the exception of increased IgG3. Whereas LLPC numbers and subsets were unaltered, NP-specific memory responses in the spleen are significantly reduced in the absence of NR2F6. However, memory B cell recall responses were unchanged, possibly because of increased proliferation of these cells.
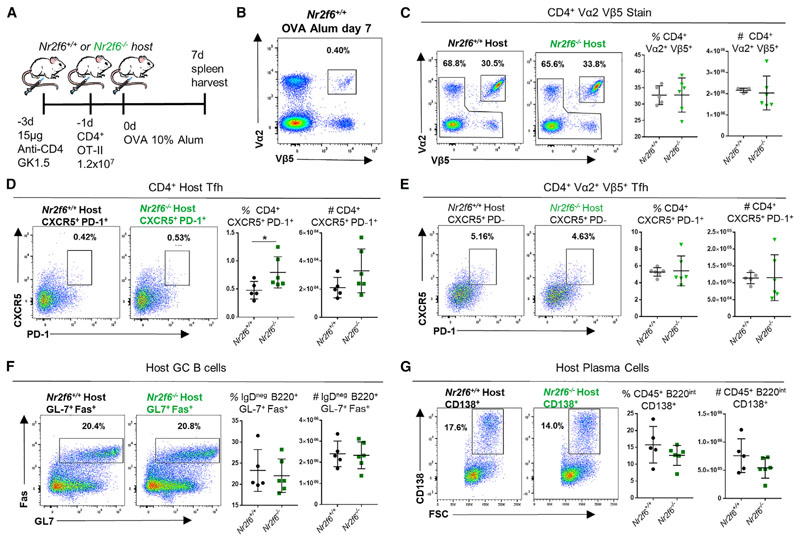
Increased Nr2f6-Deficient GC Reaction Is Not a B Cell-Autonomous Phenotype
To determine whether the loss of Nr2f6 phenotypically altered B cells, we employed an in vitro culture system that allows the generation of induced GC (iGC) B cells from primary splenic B cells (Nojima et al., 2011). However, loss of Nr2f6 did not have significant impact on B cell fold expansion or class switching to IgG1 and IgE in vitro (Figure S4).
To test Nr2f6 deficiency in B cells independent of Nr2f6−/− CD4+ T cells in vivo, wild-type or Nr2f6-deficient mice were depleted of CD4 T cells using a low dose of GK1.5 anti-CD4 antibody. Two days later, 12 × 106 Nr2f6+/+ OT-II cells per mouse were transferred into depleted mice and immunized with OVAalum the following day (Figure 3A). Transferred cells were detected as double positive for both Vα2 and Vβ5. A Vα2+ Vβ5+ stain of immunized wild-type splenocytes (not depleted of CD4 cells or reconstituted with OT-II T cells) is shown in Figure 3B as a control. The Vα2+ Vβ5+ population was comparable in Nr2f6−/− mice, <0.5% of the total CD4 population (data not shown). Seven days after immunization, Nr2f6+/+ OT-II T cells in both wild-type and Nr2f6-deficient hosts made up approximately the same fraction of all CD4 T cells. Total OT-II cell numbers were not changed in either host (Figure 3C). In Nr2f6-deficient mice, host Tfh cell frequency was significantly higher than in the wild-type host; total host Tfh cell counts were similar in both genotypes. Host Tfh cell numbers from both genotypes were around five-fold lower (2 × 104 to 3 × 104) than those from the OT-II cells (1 × 105 to 2 × 105) (Figures 3D and 3E). Total counts and frequency of transferred Tfh OT-II cells were no different regardless of host mice (Figure 3E). No differences in either frequency or total cell counts of the GC B cell or PC compartment were observed in Nr2f6+/+ or Nr2f6−/− host mice when reconstituted with Nr2f6+/+ OT-II cells (Figures 3F and 3G). Altogether, our data suggest that the enhanced Tfh cell, GC, and PC numbers following immunization of Nr2f6-deficient mice do not result from B cell-intrinsic differences.
Figure 3. Nr2f6-Deficient B Cells Do Not Contribute to the GC Phenotype of Nr2f6−/− Mice.
(A) Experimental scheme showing CD4 depletion of Nr2f6+/+ or Nr2f6−/− host mice followed by Nr2f6+/+ OT-II transfer.
(B) Splenocyte stain from wild-type control mouse, with no depletion or OT-II transfer, harvested 7 days after OVA-alum immunization and stained for CD4, Vα2, and Vβ5.
(C) Representative plots of Vα2+ Vβ5+ CD4+ T cells 7 days after immunization and 8 days after OT-II transfer. Combined frequency and total cell counts are displayed in the right two panels.
(D) Vα2− Vβ5− CD4+ CXCR5+ PD-1+ T cell (host) frequency and total cell counts are displayed with representative flow cytometry plots.
(E) Frequency of the Tfh cell population from transferred OT-II cells (CD4+ Vα2+ Vβ5+) and total cell counts are displayed with representative flow cytometry plots.
(F and G) Host germinal center B cell (F) and plasma cell frequency (G), as well as total cell counts, are displayed.
Data shown are from two independent experiments with n ≥ 6. The middle bar represents the dataset average. Error bars represent SD, and an asterisk indicates statistically significant differences calculated using Student’s t test. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
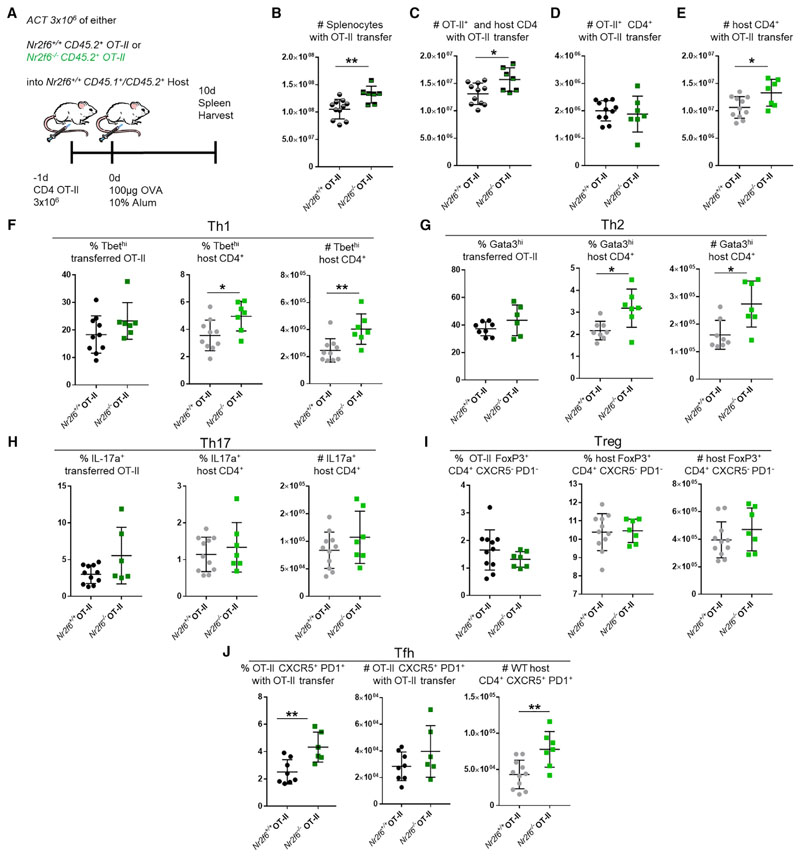
CD4 Nr2f6−/− OT-II Adoptive Cell Transfer into Wild-Type Hosts Enhances Wild-Type Helper Cell Responses
Our data from the iGC culture and CD4 depletion experiments suggested that CD4 T cells are likely the main contributor to the increased GCs in Nr2f6−/− mice. To test this hypothesis, we transferred either Nr2f6+/+ or Nr2f6−/− CD45.2+ OT-II T cells into wild-type CD45.1/45.2 congenic mice and 1 day later immunized these mice with OVA-alum. Spleens were harvested after 10 days (Figure 4A). Total spleen counts were significantly increased after host mice received Nr2f6−/− OT-II compared with Nr2f6+/+ OT-II cells (Figure 4B). Total CD4 cell counts were also significantly increased upon transfer of Nr2f6-deficient OT-II cells, although Nr2f6−/− OT-II total cell counts per se were not altered compared with wild-type OT-II controls (Figures 4C and 4D). Significant increases in wild-type host CD4 T cells were observed when Nr2f6−/− OT-II cells were present (Figure 4E).
Figure 4. Adoptive Cell Transfer of Nr2f6-Deficient OT-II T Cells into Wild-Type Hosts Results in Increased Host CD4 Tfh, Th1, and Th2 Subsets.
(A) Experimental scheme showing adoptive transfer of 3 × 106 OT-II CD4+CD45.2+ Nr2f6+/+ (black) or Nr2f6−/− (green) into Nr2f6+/+ CD45.1/CD45.2 congenic mice with OVA-alum immunization (n ≥ 7 per group).
(B) Total splenocytes, with Nr2f6+/+ or Nr2f6−/− OT-II adoptive cell transfer.
(C) Total CD4 T cell counts, including both host and transferred OT-II cells.
(D and E) Transferred OT-II T cell counts (D) and total host CD4 T cell counts (E).
(F) Frequency of Th1 cells from the indicated OT-II populations or host cells (left and middle panels). Total host Th1 cells in the spleen with indicated OT-II T cell transfer (right panel).
(G) Th2 as a frequency of OT-II or host CD4 T cell populations and total Th2 cells from the host CD4 T cells.
(H) Frequency and total cell count of Th17 cells.
(I) Treg subset defined as CXCR5− PD-1− CD4+ FoxP3+ from both OT-II transferred cells and host cells.
(J) Tfh cells defined as CXCR5+ PD-1+ Foxp3− CD4+ from OT-II and host cells.
Results shown are derived from three independent experiments (n ≥ 7). The middle bar shows an average of each dataset. Error bars represent SD, and an asterisk indicates statistically significant differences between genotypes calculated using Student’s t test or Mann-Whitney U test. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01.
Next, we examined CD4 T helper subset populations in detail. The frequency and counts of splenic Th1, Th2, Th17, and Treg cells were unaltered in either of the transferred OT-II genotypes (Figures 4F–4I). Host Th1 (T-bethi) frequency and cell numbers were significantly increased upon transfer of Nr2f6−/− OT-II cells compared with wild-type OT-II (Figure 4F). A similar change was seen in transferred Nr2f6−/− GATA3hi Th2 cells (Figure 4G). Th17 cells were only weakly induced. However, no changes in IL-17+ frequency or the number of total cells were seen in host CD4+ cells (Figure 4H). Similarly, no significant changes in Treg numbers or frequency were observed in host CD4 cells (Figure 4I).
In contrast to other T helper subsets, we observed an increased frequency, but not increased cell number, of CD4+ OT-II Tfh cells upon transfer of Nr2f6−/− OT-II compared with that of Nr2f6-sufficient OT-II (Figure 4J). As with Th1 and Th2 subsets, host Tfh cell numbers were significantly increased when Nr2f6−/− OT-II cells were present (Figure 4J). GC B cell and PC numbers were not altered despite increased numbers of Tfh cells upon OT-II Nr2f6−/− transfer. In addition, the memory NP+ CD19+ population was not affected by the transfer of these cells (Figure S5).
Our results indicate a role for Nr2f6 loss in the T cell compartment, in which paracrine factors derived from Nr2f6-deficient T cells may expand host-derived wild-type Tfh cells but also Th1 and Th2 cells.
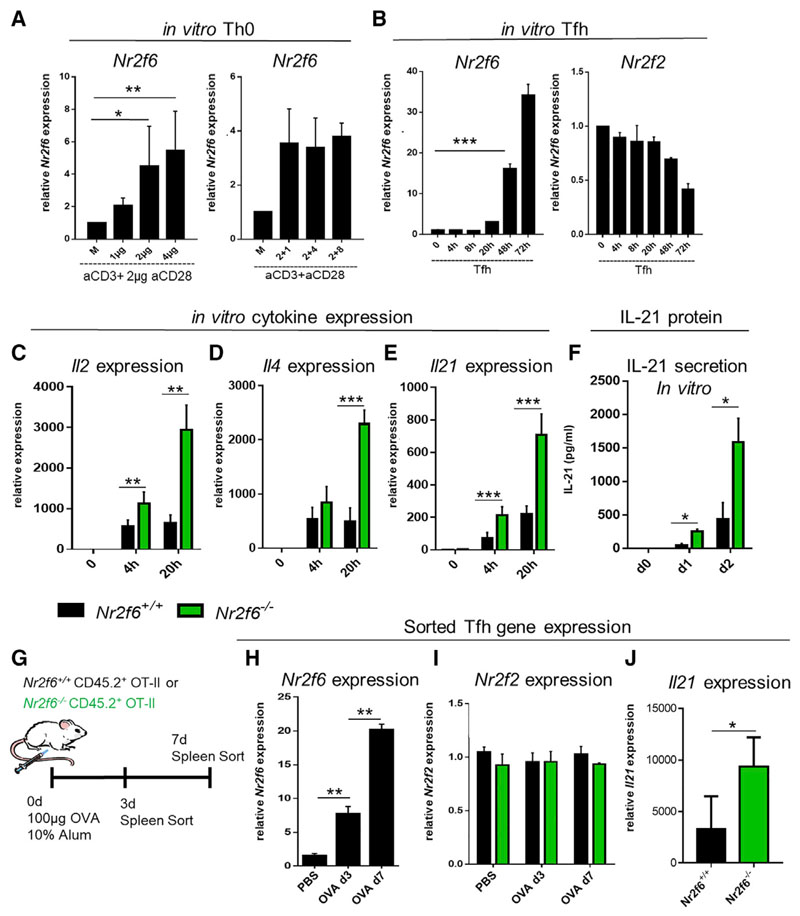
NR2F6 Suppresses Il21 Expression in Tfh Cells, Both In Vitro and In Vivo
To investigate the CD4 T cell-intrinsic role of NR2F6 in more detail, we first determined Nr2f6 expression in wild-type CD4 Th0 and Tfh cells and whether its expression is regulated upon T cell receptor (TCR) signaling or costimulation via CD28 in vitro. To test whether signals derived from TCR control of Nr2f6 expression, cells were stimulated with increasing concentrations of anti-CD3 (1–4 μg/mL) and a fixed concentration of anti-CD28 (2 μg/mL). Cells were harvested 24 h later, and Nr2f6 expression was measured by qRT-PCR and compared with unstimulated CD4 cells. Increasing concentrations of anti-CD3 enhanced Nr2f6 expression (Figure 5A). However, when the CD3 antibody amount was kept constant and anti-CD28 varied (1, 4, or 8 μg/mL), Nr2f6 expression remained unchanged (Figure 5A, right panel). Under Tfh cellpolarizing conditions, a strong time-dependent increase in Nr2f6 expression was observed (Figure 5B). The expression of the family member Nr2f2 was tested under the same conditions. It was neither differentially regulated under Tfh cellpolarizing conditions nor upregulated to compensate for the loss of Nr2f6 (Figure 5B).
Figure 5. NR2F6 Suppresses Il21 Expression in Tfh Cells In Vitro and In Vivo.
(A) Naive Nr2f6+/+ CD4+ T cells were stimulated for 24 h with indicated amounts of anti-CD3 and a fixed amount (2 μg/mL) of anti-CD28 (left panel). Alternatively, anti-CD3 was kept constant, and anti-CD28 was added at the indicated dose (right panel). qRT-PCR performed for Nr2f6 expression is displayed; values are shown relative to gapdh expression.
(B) Wild-type CD4 T cells were cultured under Tfh cell-polarizing conditions, and qRT-PCR was used to determine Nr2f6 expression at the indicated time points. Expression of the closely related nuclear receptor Nr2f2 was also determined under these conditions (right panel).
(C–E) Cytokine expression of (C) Il2, (D) Il4, and (E) Il21 was determined in in vitro Tfh cell culture by qRT-PCR at the indicated time points.
(F) IL-21 secretion into culture media was measured using Bioplex technology.
(G) Scheme for Nr2f6+/+ OT-II and Nr2f6−/− OT-II mouse immunization and Tfh cell sorting.
(H) Day 3 and day 7 Nr2f6 expression from sorted Nr2f6+/+ OT-II Tfh cells.
(I) Nr2f2 expression from Nr2f6+/+ OT-II and Nr2f6−/− OT-II Tfh cells sorted as in (H).
(J) Il21 expression from day 7 sorted Nr2f6+/+ OT-II or Nr2f6−/− OT-II Tfh cells.
Data shown are from two independent experiments with n ≥ 4. The middle bar represents the dataset average. The data are presented as the percentage of input samples before immunoprecipitation. Error bars represent SD, and an asterisk indicates statistically significant differences between genotypes calculated using Student’s t test. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
To analyze for transcriptional consequences of Nr2f6 deficiency in Tfh cells, we compared the gene expression profiles of Tfh cell-relevant TFs (Bcl6, Tbx21, Irf4, and Prdm1), cell surface receptors (Cxcr5, Icos, and Pd-1), and cytokines (Il21, Il4, Il2, Ifnγ, and Il17) in in vitro differentiated wild-type and Nr2f6−/− CD4+ T cells (Figures 5C–5E; data not shown). Because full Tfh cell differentiation is not possible in vitro (Choi et al., 2015; Nojima et al., 2011; Liu et al., 2014; McGuire et al., 2015), we focused on the initial phase of Tfh cell polarization in vitro, i.e., day 1 for qRT-PCR and days 1 and 2 for the analysis of cytokines secreted into the culture medium. The most substantial differences were observed in cytokine expression. Nr2f6 deficiency caused a significant increase in the expression of Il2, Il4, and Il21 after 20 h of culture, while Ifng and Il17 expression remained unaltered (Figures 5C–5E; data not shown). Increased secretion of IL-21 protein by Nr2f6-deficient cells in Tfh cell culture supernatant confirmed the qRT-PCR results for IL-21 (Figure 5F).
To confirm the in vitro data in vivo, Nr2f6+/+ OT-II or Nr2f6−/− OT-II mice were immunized with OVA-alum, and Tfh cells underwent fluorescence-activated cell sorting (FACS) 3 or 7 days later (Figure 5G). Wild-type Tfh cells were tested for Nr2f6 expression by qRT-PCR, and similar to the in vitro results, Nr2f6 expression was significantly upregulated in Tfh cells harvested on day 3 and enhanced on day 7 compared with sorted naive CD4 cells (Figure 5H). Nr2f2 expression in vivo was not altered in Nr2f6+/+ OT-II Tfh cells or Nr2f6−/− OT-II Tfh cells compared with naive cells (Figure 5I), suggesting that Nr2f2 is not differentially regulated in Nr2f6−/− Tfh cells to compensate for the loss of Nr2f6. We next investigated cytokine expression in sorted Tfh cells and could confirm enhanced expression of Il21 on day 7 but no increase of Il4 or Il2 expression (Figure 5J; data not shown). Altogether, Nr2f6 expression is regulated in a TCR-dependent manner in vitro and in vivo. Within Tfh cells, NR2F6 suppresses the expression of the cytokine IL-21.
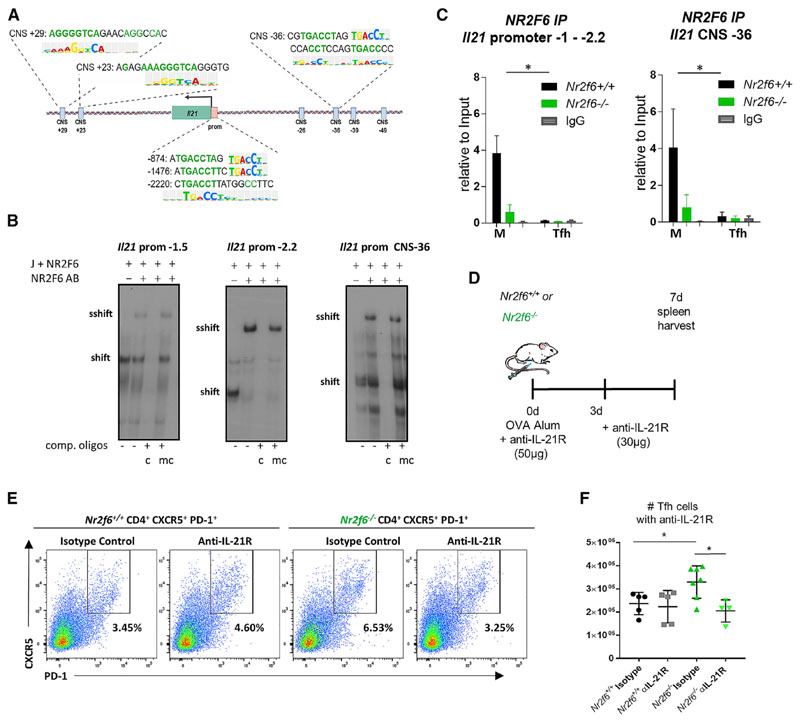
NR2F6 Directly Binds to the Il21 Promoter, and IL-21 Contributes to Tfh Cell Accumulation
We next examined whether increased expression of Il21 could be a result of the direct control of this gene by NR2F6. TF-binding analysis via TRANSFAC revealed several putative NR2F (COUP) (TGACCT) DNA binding sites within the murine promoter at −874, −1,476, and −2,220 bp from the transcriptional start site (Figure 6A; Figure S6A) (Matys et al., 2006). Park et al. (2016b) identified conserved noncoding sequences (CNSs) within the Il2-Il21 intergenic region, and putative NR2F (COUP) sites were detected in CNS regions +29, +23, and −36 (Figure 6A; Figure S6A) (Matys et al., 2006). We tested functional NR2F6 binding of these NR sites initially in nuclear extracts from Jurkat cells overexpressing NR2F6 via electromobility shift assays (EMSAs) (Table S1). We observed a specific NR2F6 super-shift for all three Il21 promoter sites; the strongest signal was observed with the oligonucleotide sequence of the −2,220-bp (−2.2) Il21 promoter region and the CNS −36 region (Figure 6B; Figure S6B; data not shown). In contrast, no NR2F6 binding to the CNS +29 or +23 sequences was observed (Figure S6B).
Figure 6. NR2F6 Directly Binds to the Il21 Promoter and CNS –36 in Resting CD4 T Cells, and Inhibition of IL-21 Signaling Reduces Tfh Cell Accumulation.
(A) Putative NR2F6 binding sites of the mouse Il21 promoter and CNS regions are shown.
(B) NR2F6 binding to the Il21 promoter at −1.5 kb, −2.2 kb, or CNS −36 by EMSA nuclear extracts from Jurkat cells (J+ NR2F6) transfected with pEFneo Nr2f6 plasmid. To validate binding specificity, consensus (c) or mutated (mc) oligonucleotides were added in excess as unlabeled competition oligonucleotides. Where indicated, an anti-NR2F6 antibody was added. One representative experiment out of three is shown.
(C) NR2F6 binding to the Il21 promoter at −1 to −2.2 kb or CNS −36 was investigated by ChIP. Nr2f6+/+ or Nr2f6–/– CD4 cells either resting or activated under Tfh-differentiating conditions were used with anti-NR2F6 or IgG control precipitation, Il21 promoter or CNS sequence was quantified by qPCR. Data are presented as the percentage of input. Differences between genotypes were calculated using log-transformed data following a linear mixed-effects model fit by restricted maximum likelihood (REML) and was analyzed via ANOVA; interaction plots are shown in Figures S6F–S6H.
(D) Scheme for anti-IL-21R block within OVA-alum-immunized Nr2f6+/+ or Nr2f6−/− mice. Wild-type or Nr2f6−/− mice were treated with anti-IL-21R antibody or isotype control at the same time as OVA immunization (50 μg) and 3 days later (30 μg).
(E and F) Representative Tfh cell flow cytometry data (E) and total Tfh cell numbers (F) are shown.
Data shown are from two independent experiments with n ≥ 4. Error bars represent SD, and an asterisk indicates statistically significant differences between genotypes calculated using Student’s t test, Mann-Whitney U test, or ANOVA. A p value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
To validate the EMSA results in a more physiological setting, we performed chromatin immunoprecipitation (ChIP) experiments on primary mouse CD4 cells. Nr2f6+/+ and Nr2f6−/− CD4 T cells were allowed to rest in media or were stimulated under Tfh cell-polarizing conditions. Stimulated cells were harvested at 48 or 72 h, and both sets of cells were subjected to ChIP using either anti-NR2F6 or isotype IgG control antibody. In agreement with our EMSA data, we detected NR2F6 binding in resting wildtype cells, but not Nr2f6-deficient cells, at the regions that capture the −874, −1,476, and −2,220 bp (−1 to −2.2) and the CNS −36 sites (Figure 6C; Figure S6C). Interaction-plot analysis of CD4 T cells under resting versus Tfh cell stimulation conditions revealed a highly significant reduction in NR2F6 binding to the Il21 promoter and the CNS −36 regions following stimulation, and within each region, regression curves of log-transformed data are shown (Figures S6F–S6H). We were again unable to detect NR2F6 binding at the CNS +23 region (Figure S6D). The CNS2 region of the Il17a promoter was included as a positive control (Figure S6E). These results support a role for a direct regulatory function of NR2F6 suppression of Il21 transcriptional activation.
To directly test the role of IL-21 in Tfh cell accumulation in Nr2f6-deficient mice, wild-type or Nr2f6-deficient mice were OVA immunized. IL-21R signaling was blocked via administration of 50 μg (day 0) and 30 μg (day 3) of an anti-IL-21R antibody (4A9) or an isotype control antibody (Figure 6D). Spleens were harvested on day 7, and Tfh cell numbers were calculated based on flow cytometry data. In line with previous immunization experiments, the frequency of Tfh cells was increased with isotype treatment of Nr2f6-deficient mice. Interruption of IL-21R signaling reduced Tfh cell frequency in these mice but had no effect on wild-type animals (Figure 6E). Similarly, Nr2f6 loss enhanced Tfh cell numbers in isotype control-treated animals. Blockade of IL-21R did not alter Tfh cell numbers in wild-type mice. In contrast, a significant reduction in Nr2f6-deficient Tfh cells was observed when IL-21 signaling was disrupted compared with isotype-treated Nr2f6−/− mice (Figure 6F).
Altogether, NR2F6 may dampen Il21 expression through direct suppression of the Il21 promoter. Increased IL-21 production of Nr2f6-deficient Tfh cells promotes the accumulation of Tfh cells, and IL-21R blockade reduces Tfh cell numbers in Nr2f6-deficient immunized mice.
Discussion
Spontaneous accumulation of Tfh cells is a hallmark of many SLE mouse models (Crotty, 2019; Linterman et al., 2009; Pratama and Vinuesa, 2014; Yu et al., 2007; Yu and Vinuesa, 2010). Increases in circulating Tfh-like cells have also been observed in humans with SLE (Simpson et al., 2010; Yu and Vinuesa, 2010). We document a role for Nr2f6 loss in Tfh cell accumulation during acute GC responses and present evidence indicating a paracrine component in the accumulation. Of interest is the late-onset immunopathology observed in Nr2f6-deficient mice, because the autoreactive antibodies and alterations in class switching are not observed in most mice until 10 months of age (Hermann-Kleiter et al., 2008). This suggests that loss of Nr2f6 alone is not sufficient for inducing spontaneous GC formation in young mice but that normal GC responses may become self-reactive over time and eventually escape control by the increased Tfr cells in Nr2f6−/− mice. It has been reported that excessive Tfh cell numbers can reduce overall antibody-antigen affinity. However, although significant increases in Tfh cells were observed in Nr2f6-deficient mice, no major alterations in antibody affinity were observed. This could be because of increased Tfr cell numbers and or activity (Baumjohann et al., 2013; Preite et al., 2015). It is also possible that increases in Tfh cell numbers are not sufficient to affect the overall affinity for antigen during acute responses. Total Tfr cell numbers depend on IL-2 and IL-21 signals, as well as a self-antigen presentation by B cells (Ritvo et al., 2017; Wu et al., 2006). It is plausible that several of these parameters are altered in Nr2f6-deficient mice; the exact role of NR2F6 in Tfr cells therefore requires further investigation.
Several groups have shown the importance of IL-21 signaling in B cells for proper GC formation and GC B cell differentiation (Linterman et al., 2010; Zotos et al., 2010), while others have reported on the critical effect of IL-21 signaling for the Tfh cell population (Vogelzang et al., 2008). Some groups have proposed redundant roles for IL-6 and IL-21 in Tfh cell differentiation (Eto et al., 2011; Nurieva et al., 2009). IL-6 alone is known to be a potent inducer of Tfh cell differentiation in the mouse and plays an important role in early Tfh cell differentiation (Choi et al., 2013; Crotty, 2014). This may be why both Zotos et al. (2010) and Linterman et al. (2010) did not observe a significant effect of IL-21 loss on the Tfh cell population. Like Eto et al. (2011), we observed no significant effect on wild-type Tfh cell populations when IL-21R signaling was disrupted; only Nr2f6-deficient Tfh cells affected our blockade of the IL-21R. The differential effect of IL-21R blockade on wild-type and Nr2f6−/− mice likely reflects the overlapping roles of IL-6 and IL-21 in the wild-type mouse and demonstrates that overexpression of IL-21 is largely responsible for the increased Tfh cell population in Nr2f6−/− mice.
In this study, we demonstrate that NR2F6 directly binds to several in silico-predicted NR2F (COUP) sites within the Il21 promoter and CNS regions. Independent data from the Encode Project, as well as the Immgen consortium, have identified putative NR2F binding sites within the Il21 genomic locus (Davis et al., 2018; Shay and Kang, 2013). These findings are in agreement with our previous work showing that NR2F6 can—in a TCR-dependent manner—negatively regulate the expression of cytokines such as Il2, Il17, or Ifng in various T helper subsets (Hermann-Kleiter et al., 2008, 2012; Hermann-Kleiter and Baier, 2014). Within these cytokine promoters, NR2F6 suppresses transcriptional activation via interference with NFAT DNA binding. In contrast, no such NFAT NR2F6 co-binding sites were predicted within the Il21 promoter or the CNS −36 region, suggesting an alternate mechanism.
Although further analysis, including corepressor recruitment, determination of histone marks, or even HiC approaches, will be necessary to fully unravel the detailed mode of interaction of NR2F6 within the Il21 promoter, as well as intergenic Il21-Il2 region, our data provide strong evidence for a critical functional role of NR2F6 in the Tfh cell effector compartment. DNA methylation and histone modification have not yet been investigated within this project, but their importance for the regulation of the Il2-Il21 region have been shown (Sin et al., 2019).
We demonstrate that enhanced IL-21 production by Nr2f6-deficient CD4 T cells promotes the accumulation of Nr2f6−/− Tfh cells. Choi et al. (2017) demonstrate a pathogenic role for IL-21 in the accumulation of Tfh cells using the B6.Sle1.Yaa mouse model of SLE. Confirming this role for IL-21, this group observed both reduced disease severity and Tfh cell accumulation when IL-21 signaling was interrupted (Choi et al., 2017). In parallel, Bubier et al. (2009) established a role for general IL-21 dysregulation with a similar mouse model of spontaneous early-onset SLE. We propose that enhanced Il21 production by Nr2f6-deficient Tfh cells during GC responses may contribute to the late-onset SLE like disease observed in these mice.
We observed reduced numbers of memory B cells in Nr2f6-deficient mice on day 90 post-immunization. Work by Zotos et al. (2010) demonstrated that IL-21 or IL-21R loss leads to enhanced memory formation after immunization through reduced BCL-6 expression in GC B cells. Increased IL-21 expression in Nr2f6−/− mice could thus be causative for the observed reduction in memory B cells. However, this scenario would require increased GC B cell numbers even at early time points, a phenotype that we did not observe on day 4. We suggest that maintenance of the memory B cell population is defective in Nr2f6-deficient mice or that loss of Nr2f6 has an intrinsic B cell effect on memory formation. This hypothesis is supported by our OT-II adoptive cell transfer experiments, which resulted in comparable NP+ CD19+ B cell populations on day 90 after immunization of wild-type host mice receiving either Nr2f6+/+ or Nr2f6−/− OT-II cells.
Rankin et al. (2011) described a role for IL-21 signaling in the proliferation of memory B cells and in the differentiation of PCs during recall responses. In line with these data, we observed increased proliferation of Nr2f6−/− memory B cells upon recall, an observation likely linked to increased Il21 expression in these mice. Therefore, Nr2f6-deficient mice mount sufficient memory B cell responses, in terms of cell numbers, but the antibody response and mechanisms involved require further investigation, particularly because Tfh memory cells play a key role in memory B cell recall responses (Ise et al., 2014).
Relatively little is known about the role of NRs during normal GC responses or in the autoimmune reactions leading to SLE. A suppressive role during GC formation has been validated for PPARγ/NR1C3 (Park et al., 2014, 2016a). PPARγ has been shown to play a minor role in mouse models of SLE and has been linked to human CD40-CD40L signaling in SLE patients (Aprahamian et al., 2014). Loss of the RORγt isoform results in aberrant Tfh cells and Tfr cells that, via enhanced production of IL-17 and IL-21, promote spontaneous GC formation and dysregulate B cell responses (Bubier et al., 2009).
The results presented here identify NR2F6 as a negative regulator of Tfh cells by restricting their excessive accumulation in the GC, partly by direct transcriptional regulation of the Il21 promoter. The NR superfamily is a primary class of therapeutic drug targets for human disease (Burris et al., 2013; Schulman, 2010). Understanding the complex interaction between Tfh and Tfr cells and GC B cell responses, the intensity and quality of an immune response, and the specific role of NRs might improve therapeutic options in autoimmune diseases, as well as future vaccination strategies.
Star⋆Methods
Key Resources Table.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4 (RM4-5) | BD Biosciences | Cat#560782; RRID:AB_1937315 |
| PD-1 (J43) | Thermo Fisher | Cat#11-9985-82; RRID:AB_465472 |
| Ki67 (SolA15) | Thermo Fisher | Cat#12-5698-80; RRID:AB_11149672 |
| CD45 (30-F11) | BD Biosciences | Cat#561487; RRID:AB_561487 |
| CD45.1 (A20) | Biolegend | Cat#110716; RRID:AB_10660302 |
| CD45.1 (A20) | Thermo Fisher | Cat#25-0453-81; RRID:AB_469628 |
| CD45.2 (clone 104) | BD Biosciences | Cat#562130; RRID:AB_10896649 |
| FoxP3 (FJK-15 s) | Thermo Fisher | Cat#11-5773-82; RRID:AB_465243 |
| CXCR5 (2G8) | BD Biosciences | Cat#551960; RRID:AB_394301 |
| CD19 (ID3) | Thermo Fisher | Cat#25-0193-82; RRID:AB_657663 |
| CD19 (ID3) | BD Biosciences | Cat#561113; RRID:AB_10563071 |
| B220 (RA3-6B2) | Thermo Fisher | Cat#12-0452-82; RRID:AB_465671 |
| B220 (RA3-6B2) | Biolegend | Cat#103236; RRID:AB_893354 |
| Fas (Jo2) | BD Biosciences | Cat#563647; RRID:AB_2738346 |
| GL7 (GL7) | BD Biosciences | Cat#553666; RRID:AB_394981 |
| CD86 (GL1) | Biolegend | Cat#105014; RRID:AB_439783 |
| CXCR4 (L276F12) | Biolegend | Cat#146509; RRID:AB_2562786 |
| CD138 (281-2) | Biolegend | Cat#142503; RRID:AB_10915989 |
| CD138 (281-2) | BD Biosciences | Cat#562610; RRID:AB_11153126 |
| IgD (11-26c.2a) | Biolegend | Cat#405711; RRID:AB_1937245 |
| IgG1 (A85-1) | BD Biosciences | Cat#550331; RRID:AB_2296342 |
| IgM (II/41) | Thermo Fisher | Cat#25-5790; RRID:AB_469655 |
| PD-L2/CD273 (122) | Thermo Fisher | Cat#11-9972-82; RRID:AB_465462 |
| CD80 (16-10A1) | Biolegend | Cat#104713; RRID:AB_1240733 |
| Tbet (4B10) | Thermo Fisher | Cat#50-5825-80; RRID:AB_10597459 |
| GATA3 (16E10A23) | Biolegend | Cat#653809; RRID:AB_2563216 |
| BCL6 (K112-91) | BD Biosciences | Cat#562198; RRID:AB_11153136 |
| IL-17a (TC11-18H10) | BD Biosciences | Cat#560220; RRID:AB_1645194 |
| CD38 (90) | Biolegend | Cat#102727 RRID:AB_2616967 |
| Gr-1 (RB6-8C5) | Biolegend | Cat#108437 RRID:AB_2562214 |
| F4/80 (BM8) | Biolegend | Cat#123135 RRID:AB_2562622 |
| IL-21R (2A9) | Bioxcell | Cat#BE0258; RRID:AB_2687737 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ovalbumin (OVA) | Hyglos | Cat#321001 |
| NP-chicken gamma globulin (NP-CGG) (Ratio 20) | Biosearch Technologies | Cat#N-5055C-1 |
| NP-Ovalbumin (NP-OVA) (Ratio 20 or 17) | Biosearch Technologies | Cat# N-5051-10-BS |
| Aluminum potassium sulfate dodecahydrate (Alum) | Sigma | Cat#31242-500G-D |
| Strepavidin APC | BD Biosciences | Cat#554067 |
| Strepavidin APC-Cy7 | Biolegend | Cat#405208 |
| NP-PE (Ratio 28) | Biosearch Technologies | Cat#N-5070-1 |
| 7-amino-actinomycin D (7-AAD) | Biolegend | Cat#420403 |
| AnnexinV-PE | Biolegend | Cat#640908 |
| Sodium Hydroxide | Fisher | Cat# S318-1 |
| Sheep Red Blood Cells (SRBCs) | Labor Dr. Merck, Ochsenhausen | Cat#E-400 |
| Critical Commercial Assays | ||
| Thermo Fisher FoxP3 Transcription Factor Staining Buffer Set | Fisher | Cat#00-5523-00 |
| RNA Isolation Kit | QIAGEN | Cat#GR8RNA |
| Omniscript RT Kit 200 | QIAGEN | Cat#205113 |
| qRT-PCR Kit (Rox) | Bio & SELL | Cat#BS76.520.5000 |
| Chromatrap® pro G ChIP spin column kit | Direct 7Bioscience | Cat#500190 |
| Bio-Plex Pro Mouse Cytokine IL-21 | Biorad | Cat#171GA003M |
| Experimental Models: Cell Lines | ||
| Jurkat T cells | ATCC | ATCC-TIB152 |
| Experimental Models: Organisms/Strains | ||
| Nr2f6-deficient mice (C57BL/6) | Hermann-Kleiter et al., 2008 | N/A |
| Wild-type, C57BL/6N | Charles River | Strain Code#027 |
| Nr2f6+/+ OT-II, C57BL/6-Tg(TcraTcrb) 425Cbn/Crl | Charles River | Strain Code#643 |
| Nr2f6−/− OT-II | In House | N/A |
| B6.SJL-Ptprca Pepcb/BoyJ (CD45.1) | Jackson | Stock# 002014; RRID:IMSR_JAX:002014 |
| B6.SJL x C57BL/6 (CD45.1/CD45.2) | In House | N/A |
| Oligonucleotides | ||
| See Table S1 for EMSA Oligonucleotide sequences | This paper | N/A |
| CNS +23 F-ACTTCCACTGGTCAGGGTCT | Synthesized by Eurofins | N/A |
| CNS +23 R-AGCAATGGTCAATTGTTGTGGG | Synthesized by Eurofins | N/A |
| CNS −36 F-CTCCAGTGACCCCATTCTCC | Synthesized by Eurofins | N/A |
| CNS −36 R-TTGCTCTCGCCAAGTGGAAA | Synthesized by Eurofins | N/A |
| Prom −1- −2.2kb F-CAGCAGCGTCCCTTACAGAA | Synthesized by Eurofins | N/A |
| Prom −1- −2.2kb R-CTTCTGCAGGAGATGAGCATT | Synthesized by Eurofins | N/A |
| qRT-PCR primers Il21 | Applied Biosystem | Cat#Mm00517640_m1 |
| qRT-PCR primers Il4 | Applied Biosystem | Cat#Mm00445259_m1 |
| qRT-PCR primers Il2 | Applied Biosystem | Cat#Mm00434256_m1 |
| Recombinant DNA | ||
| Nr2f6-pEFNeos construct | Hermann-Kleiter et al., 2008 | N/A |
| Software and Algorithms | ||
| FlowJo 10 | TreeStar | RRID:SCR_008520 |
| Fiji (ImageJ) | N/A | http://fiji.sc; RRID:SCR_002285 |
| Prism 7 | Graphpad | https://www.graphpad.com/; RRID:SCR_002798 |
| R | N/A | https://www.r-project.org/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Natascha Hermann-Kleiter (natascha.kleiter@i-med.ac.at).
This study did not generate new unique reagents.
Experimental Model and Subject Details
Mouse Strains
Nr2f6-deficient mice on the C57BL/6 background (Hermann-Kleiter et al., 2008, 2012, 2015), Nr2f6+/+-OT-II (C57BL/6-Tg(TcraTcrb) 425Cbn/Crl; Nr2f6+/+-OT-II mice) purchased from Charles River and crossed into the Nr2f6-deficient background (Nr2f6−/− -OT-II mice). B6.SJL (CD45.1) on a C57BL/6 background, or B6.SJL crossed to C57BL/6 (CD45.1/CD45.2) were used for adoptive transfer experiments. Mice were maintained under specific pathogen-free conditions (SPF following FELASA guidelines) and used at the age of 6-14 weeks of age (with the exception of the memory experiments). All animal experiments were performed in accordance with national and European guidelines and reviewed and authorized by the committee on animal experiments (BMWFW-66.011/0064-WF/V/3b/2016; BMWFW-66.011/0112-WF/V/3b/2017).
Cell Lines
Jurkat-Tag (Nr2f6+/+) cells were obtained from the American Type Culture Collection (ATCC-TIB152). These cells were maintained in RPMI supplemented with 10% fetal calf serum, 100U/ml penicillin, 100μg/ml streptomycin and 2mML-glutamine in a 37°C humidified incubator with 5% CO2. Cells were split to new media as needed.
Method Details
Immunizations
For T cell-dependent immunization, 6-14-week-old mice were age and sex matched, injected i.p. with 5 × 108 sheep red blood cells (SRBCs) in PBS or 100 μg of endotoxin-free chicken ovalbumin (OVA) (Hyglos), NP-CGG (Biosearch Technologies) or NP-OVA (Biosearch Technologies), precipitated in 10% aluminum potassium sulfate dodecahydrate (Alum). Briefly, required volume of antigen suspended in PBS was mixed with an equal volume of 10% Alum and pH adjusted to seven with 10N NaOH (Fisher). Precipitated antigen-alum mix was centrifuged for 15sec at 5000 g, washed with PBS three times and re-suspended in the required volume (200μl/mouse) of sterile PBS for i.p. injection. For recall experiments NP-CGG immunized mice (see above) were boosted with 60μg of NP-CGG suspended in sterile PBS administered i.p. > 50 days after priming immunization. Recall PC gating strategy was performed as previously described (Rankin et al., 2011). Animals were analyzed at the indicated time points post immunization.
Adoptive transfer experiments
For experiments where CD4 cells were not depleted, 3x106 OT-II CD45.2+ CD4 T cells of either Nr2f6+/+-OT-II or Nr2f6−/− -OT-II mice were isolated via negative selection using CD4 T Cell Isolation Kit (Miltenyi Biotec) and transferred intraperitoneally into CD45.1/CD45.2 heterozygous congenic mice or CD45.1 only congenic mice, 24 hours before OVA-alum or NP-OVA (memory) immunizations. Harvests were performed at indicated time points post immunization. For CD4 depletion experiments followed by adoptive cell transfer of Nr2f6+/+-OT-II isolation was performed as above, with a larger total number of OT-II cells (12x106).
Cell sorting
Splenic single-cell suspensions were prepared and incubated with fluorochrome-labeled antibodies as described above. The sorted cell subsets were defined as follows: T cell populations: Tfh cells: (CD45+CD4+PD-1+CXCR5+). Cells were sorted using a FACSAria (BD Biosciences). Non-singlet events were typically excluded from analyses based on characteristics of forward and side scatter.
Flow Cytometry
Spleens were crushed through a 100-μm filter, and the resulting single-cell suspensions were depleted of erythrocytes using the mouse erythrocyte lysing kit (R&D, WL2000). For flow cytometry analysis, cells were incubated with FcR Block (BD Biosciences, 553142) before staining with cell surface antibody cocktails for 30 min at 4°C, washed with PBS+2% FCS. Cells requiringly only surface stains were than run on a flow cytometer. For transcription factor staining, cells were then fixed with FoxP3 staining buffer set (eBioscience, 00-5523), > 30 minutes, 4°C, and permeabilized according to manufacturer’s instructions. Subsequently, cells were stained with intracellular antibodies. The following staining panels were used: for Tfh cells (CD45+CD4+CXCR5+PD-1+Foxp3-), GC B cells (CD45+CD19+B220+FashiGL7hi) - DZ- and LZ- GC B cells were distinguished by CD86 and CXCR4, plasmablasts/cells (CD45+B220-/lo CD138hi). Biotinylated antibodies were fluorescently labeled using either Streptavidin conjugated APC or APC-Cy7 (BD). Data was acquired on a FACSVerse (BD), a FACSFortessa (BD) or a FACSCantoII (BD) and analyzed with the FlowJo software (Treestar) as published previously (Hermann-Kleiter et al., 2015).
Live Dead Staining
Single cell suspensions were washed once with Hank’s Salt solution, containing Calcium and Magnesium (HSS) (Millipore, L 2035) and supplemented with 2% FCS. Cells were resuspended in 100μl of wash buffer containing the manufacturers recommended concentration of AnnexinV-PE (Biolegend, 640908). Samples were incubated for 20 minutes at 4°C along with surface marker antibody stains. Cells were then washed with 800μl of HSS plus 2% FCS and resuspended in 200μl. 7-AAD (Biolegend, 420404) was added to the 200μl cell suspension shortly before analysis on the flow cytometer at the manufacturers recommended concentration. Live cells were considered 7-AAD- and AnnexinV-.
In vitro iGC B cell culture
Splenic naive B cells were purified with the MagniSort Streptavidin Negative Selection Beads (eBioscience) following the manufacturer’s instructions, using TCRb-Bio (BioLegend, 109204), CD11b-Bio (Biolegend, 101204) and Ter119-Bio (Biolegend, 116204). iGC B cell culture was performed following (Nojima et al., 2011). Briefly, B cells were plated (5 × 106 cells per 10 cm dish) on mitomycin C (Sigma, M0305)-treated 40LB feeder cells (3 × 106 cells per 10 cm dish) in 40 mL B cell medium: DMEM (Sigma, WHMISDZB)) supplemented with 10% (v/v) FBS (GIBCO, 10270-106), 2 mM L-glutamine (Sigma, G7513), 10 mM HEPES (LONZA, BE17-737E), 1 mM sodium pyruvate (GIBCO, 13360-039), 1 x non-essential amino acids (GIBCO, 11140-035), 100 U/ml penicillin, 100 μg/ml streptomycin (Sigma, 0781), 50 μM β-mercaptoethanol (Sigma, M3148) and 10 ng/ml rIL-4 (Peprotech, 214-14); 30 mL of medium were changed on day 3. On day 4, iGC B cells were harvested, analyzed and 1,5 × 106 cells replated per 10 cm dish onto a new Mitomycin C-treated feeder layer in B cell medium supplemented with either 10 ng/ml rIL-4 or 10 ng/ml rIL-21 (Peprotech, 210-21); 30 mL of medium were changed on day 7 and the final analysis was performed on day 8. The analysis included cell count with the trypan blue exclusion method using a hemocytometer (Neubauer) and flow cytometric analysis, as described above.
In vitro T cell culture
Naive CD4+ T cells were isolated from RBC-lysed single-cell suspensions via negative selection using CD4 T Cell Isolation Kit (Miltenyi Biotec). The stimulation of naive CD4+ T cells under Th0 or Tfh conditions was performed in complete IMDM medium in the presence of plate-bound 2C11 (antiCD3) and soluble antiCD28 (Th0 5 μg/ml + 1 μg/ml; Tfh 8 μg/ml + 8 μg/ml) cells supplemented with a follicular T helper cell-promoting cytokine milieu containing IL-21 (50 ng/ml), and/or IL-6 (50 ng/ml), anti-TGFβ (20 μg/ml); anti-IL-4; (20 μg/ml) anti-IFNγ (20 μg/ml). Expression (qRT-PCR) and cytokine protein secretion (via Bioplex technology) was determined at the indicated time points.
Gene expression analysis
Total RNA was isolated using the RNeasy® Mini Kit (QIAGEN). First-strand cDNA synthesis was performed using oligo (dT) primers (Promega) with the QIAGEN Omniscript RT kit, according to the instructions of the supplemented and as described previously (Hermann-Kleiter et al., 2015). Expression analysis was performed using real-time PCR with an ABI PRIM 7000 or ABI PRIM 7500fast Sequence Detection System with TaqMan gene expression assays (Applied Biosystems); all mouse expression patterns were normalized to Gapdh.
Chromatin immunoprecipitation (ChIP)
ChIP assay was performed with a ChIP assay kit according to the recommendations of the manufacturer Chromatrap® ChIP SEQ kitsPremium //Chromatrap® pro G ChIP spin column kit 24 (500190) and previously described methods (Hermann-Kleiter et al., 2015). Briefly, Nr2f6+/+ and Nr2f6-deficient CD4+ T cells were isolated, and either rested for 2 hr in X-Vivo medium or Tfh differentiated as described above. On day 2 or 3, cells were washed once in IMDM for subsequent fixation in 1% methanol free formaldehyde at 37°C for 6 min, and quenching of cross-linking was performed by the addition of 1.375 M glycine. Cells were lysed, and subsequently, sonication was performed with 15x 30 s pulses using a Bioruptor Next Generation (Diagenode). The sheared chromatin was used to set up immune precipitation reactions with 5-10 μg of the indicated antibodies (IgG2b, BioXell; NR2F6 (R&D PP-N2025-00) at 4°C for 1h in a 1:1 antibody:chromatin ratio. Immunoprecipitation was subsequently performed with Chromatrap spin columns followed by reverse cross-linking and clean-up of the DNA with QIAGEN MinElute columns. Real-time PCR was performed using an ABI PRIM 7000 Sequence Detection System (Applied Biosystems). Ct values obtained were normalized against the total input to obtain the percentage of amplified ChIP DNA per input. Primer sequences of the Il21 genomic region were either from literature of designed using Blast Primer and can be found in the Key Resources Table (Park et al., 2016b).
Electrophoretic Mobility Shift Assay (EMSA)
Single-stranded oligonucleotides were synthesized by Eurofins MWG Operon and annealed.
Jurkat-TAg cells were transiently transfected with NR2F6 in pEFneo by electroporation, 3 days later cells were lysed and nuclear extracts were prepared as described previously (Hermann-Kleiter et al., 2008).
Binding reactions and supershift assays were performed for 30 min at 4−8°C using the Binding Buffer B-1 (Active Motif 37480) together with Stabilizing Solution D (Active Motif 37488) containing 5 μg nuclear extracts (Active Motif 36042) and 1 μg of NR2F6 (R&D PP-N2025-00) antibody for the supershift (Hermann-Kleiter et al., 2012). Binding reactions with the 3 × 105 cpm labeled probe were performed for 20 min at 4-8°C using Binding Buffer C-1 (Active Motif 37484) together with Stabilizing Solution D (Active Motif 37488). Samples were run on a 4% native polyacrylamide gel in 0.5 × TBE for 3 h at 250 V. For competition assays, 10-fold unlabeled oligonucleotides either identical or with a mutant nuclear receptor site were added to the binding reaction. Oligonucleotide sequences can be found in Table S1.
Immunohistochemistry
Cryosections (4μm) of spleens from Nr2f6+/+ and Nr2f6−/− immunized mice were fixed with 1% paraformaldehyde for 10 min at room temperature (RT). Slides were then blocked with 2% BSA/PBS for 30 min at RT, followed by incubation with mouse monoclonal antibodies against CD3 (eBioscience 17-0031-82, APC), B220 (eBioscience 12-0425-82, PE) and a biotin-labeled PNA (Vector Labs B-1085) for 45 min at RT. A kit for endogenous biotin and avidin blocking was used according to the manufacturer’s instructions (Vector Labs SP-2002). Subsequently, slides were incubated with streptavidin A488 (Invitrogen S32354) for 45 min at RT. After washing, slides were mounted with Diamond Antifade medium (Invitrogen P36965), images were acquired on a Leica DMi8 inverted microscope, and processed using Fiji software (Schindelin et al., 2012).
Affinity and Class switching, Enzyme-linked Immunosorbent Assay (ELISA)
Serum was collected on day 10, 20 and 30 after NP-CGG immunization, for quantification of NP-specific IgG1. NP-BSA (Biosearch Technologies) in ratios of 18 and 1.7 to 1 were diluted in PBS to 50 μg/μl and coated a 96-well enzyme-linked immunosorbent assay plates (Sigma CLS3590) overnight at 4°C. Coated plates were washed three times with PBS containing 0.05% TWEEN 20, followed by blocking using 100 μl blocking buffer (1% BSA in PBS). 100 μl of sera (diluted 1:102,400) was added to the plate and incubated overnight at 4°C. Plates were washed three times with PBS containing 0.05% TWEEN 20, 100 μl of anti-mouse IgG1 conjugated to HRP (Southern Biotech 1070-05), diluted 1:5000 in 1% BSA/PBS was incubated per well for 4 hours at room temperature. Plates were washed three times with PBS containing 0.05% TWEEN 20. For HRP detection 100 μl of ABTS ((200 μL ABTS (Stock: 15 mg/ml in water), 10 mL citrate-phosphate buffer (574 mg citric acid monohydrate in 50 mL water), 10 μL H2O2) was incubated per well for 20 minutes. Absorbance was measured at 405nm (Tecan Sunrise). Data are displayed as the OD50 value.
BrdU Incorporation
BrdU incorporation was measured using a BrdU flow kit from BD (559619) according to the manufacturer’s instructions. Briefly, mice were pulse labeled by i.p. injection of 100 mg (100 μl) of BrdU (stock: 10mg/ml). Spleens were then removed 7-12 hours post-injection. Single cell suspension from spleen was surface stained and labeled with fixable viability dye efluor450 (eBioscience, 65-0863-14). Cells were fixed 100 μl of BD Cytofix/Cytoperm for 30 min on ice and washed with 1 mL of Perm/Wash buffer. Cells were resuspended in 100 μl of BD Cytoperm for 10 min on ice and washed with 1 mL of Perm/Wash buffer. Samples were re-fixed with 100 μl of Cytofix/Cytoperm for 5 min on ice and washed with 1 mL of Perm/Wash buffer. Samples were treated with DNase I (300 μg/ml) for 1 hour at 37°C and washed as in previous steps. 50 μl of BrdU antibody (diluted 1:50) was incubated with the cells for 20 min at room temperature and washed as before. Cells were resuspended into 1 mL of staining buffer (2% FCS/PBS). Data was acquired on a BDVerse at a low flow rate (500 events/second).
IL-21R blocking
Mice were injected i.p. with 50μg of anti-IL21R (Biolegend) antibody or isotype control (Bioxcell) at the same time as OVA alum immunization (50μg) and 3 days later with 30μg of the appropriate antibody according to Hermann-Kleiter et al. (2015).
Quantification and Statistical Analysis
Statistical analysis was performed using Prism 5.03 and R. Experiments were repeated at least two times. Normality was tested using a Shapiro-Wilk test. Differences between means were investigated by Student’s t test. The few variables that were not normally distributed were tested by Mann-Whitney U test.
For the statistical analyses of ChIP data we used the percent input method (Lacazette, 2017). The frequency distribution of percent input data was clearly right skewed. We therefore log-transformed the values, inducing a distribution much closer to a normal distribution. The data were analyzed (i) by linear regression models for mixed effects and (ii) by analyses of variance models. The lme and the aov (Weisberg and Fox, 2019) packages of R were employed, respectively.
In the linear regression model fixed effects factors were Tfh stimulation (M/Tfh), and the type (wild-type/Nr2f6−/−). The eight mice were a random factor. Measurements for region −1 - 2.2 and region CNS-36 were of interest. The main results are shown in the interaction-plot (Figure 6SG). While NR2F6 binding to the relevant Il21 promoter regions could not be detected in Nr2f6-deficient cells neither in the resting nor stimulated condition (estimated means −2.36 and −2.29, −2.34 and −2.26 respectively). NR2F6 binding to the relevant Il21 promoter regions in wild-type was much higher in the resting cells and dropped to nearly the same level as in the Nr2f6-deficient cells in the Tfh stimulation condition (estimated means 0.9 and −2.32).
The regression coefficients were 0.07 for the Nr2f6-deficient group (not significantly different from zero, t = 0.18) and −2.41 in the wild-type mice (clearly different from zero, t = −5.6***). In the analysis of variance the interaction term was highly significant with a high effect size (F = 14.25***; df = 2/52, η2 = 0.27, credible interval = [.201, 0.298]). Error bars represent SD and an asterisk (*) indicates statistically significant differences between genotypes p-value of <0.05 was considered statistically significant. *p < 0.05; **p < 0.01; ***p < 0.001.
Data and Code Availability
This study did not generate new datasets or code.
Supplementary Material
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.08.024.
Highlights.
Loss of NR2F6 results in accumulation of Tfh, GC B, and plasma cells after immunization
Increased GC populations depend on Nr2f6 loss within the CD4 compartment
NR2F6 directly binds to several sites within the Il21 promoter and CNS −36
NR2F6 restrains Il21 expression in CD4 cells; IL-21R blockade reduces Tfh accumulation
Acknowledgments
The FWF Austrian Science Fund supplemented this work with grants (W1101-B18 and P28694-B30 to N.-H.K., P30324-B21 and P31383-B30 to G.B.), the ERC ADG 786462-HOPE, and the Christian Doppler [CD] Society CD Laboratory I-CARE), as well as Daiichi Sankyo Co., Japan. K.S. is supported by a DOC PhD fellowship by the Austrian Academy of Sciences (ÖAW). We are grateful to Gernot Kleiter for help with statistical analysis and using R, Nadja Haas and Irene Gaggl for technical assistance, Martin Offterdinger for his help with the microscopes at the MUI Biooptics core facility, and Francesco Ferraguti for letting us use his cryocut.
Footnotes
Author Contributions
This study was designed by N.-H.K. and W.J.O. N.-H.K. directed the work. W.J.O. wrote the manuscript, along with N.-H.K. W.J.O. performed most experiments, analyzed data, and interpreted results. N.H.-K., B.J., V.L., and K.S. performed experiments and analyzed data. V.K. and M.K. contributed with technical help and materials. N.H.-K., W.J.O., V.L., and G.B. discussed and interpreted results. All authors read, edited, and approved the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
References
- Aprahamian TR, Bonegio RG, Weitzner Z, Gharakhanian R, Rifkin IR. Peroxisome proliferator-activated receptor gamma agonists in the prevention and treatment of murine systemic lupus erythematosus. Immunology. 2014;142:363–373. doi: 10.1111/imm.12256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, Sallusto F. Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity. 2013;38:596–605. doi: 10.1016/j.immuni.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Bell J, Gray D. Antigen-capturing cells can masquerade as memory B cells. J Exp Med. 2003;197:1233–1244. doi: 10.1084/jem.20020270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bubier JA, Sproule TJ, Foreman O, Spolski R, Shaffer DJ, Morse HC, 3rd, Leonard WJ, Roopenian DC. A critical role for IL-21 receptor signaling in the pathogenesis of systemic lupus erythematosus in BXSB-Yaa mice. Proc Natl Acad Sci USA. 2009;106:1518–1523. doi: 10.1073/pnas.0807309106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, Lundasen T, Hughes T, Kojetin DJ. Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev. 2013;65:710–778. doi: 10.1124/pr.112.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Eto D, Yang JA, Lao C, Crotty S. Cutting edge: STAT1 is required for IL-6-mediated Bcl6 induction for early follicular helper cell differentiation. J Immunol. 2013;190:3049–3053. doi: 10.4049/jimmunol.1203032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Gullicksrud JA, Xing S, Zeng Z, Shan Q, Li F, Love PE, Peng W, Xue HH, Crotty S. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol. 2015;16:980–990. doi: 10.1038/ni.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Seth A, Kashgarian M, Terrillon S, Fung E, Huang L, Wang LC, Craft J. Disruption of Pathogenic Cellular Networks by IL-21 Blockade Leads to Disease Amelioration in Murine Lupus. J Immunol. 2017;198:2578–2588. doi: 10.4049/jimmunol.1601687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung Y, Tanaka S, Chu F, Nurieva RI, Martinez GJ, Rawal S, Wang YH, Lim H, Reynolds JM, Zhou XH, et al. Follicular regulatory T cells expressing Foxp3 and Bcl-6 suppress germinal center reactions. Nat Med. 2011;17:983–988. doi: 10.1038/nm.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney AJ, Tsai SY, O’Malley BW, Tsai MJ. Chicken oval-bumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol Cell Biol. 1992;12:4153–4163. doi: 10.1128/mcb.12.9.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–542. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. T Follicular Helper Cell Biology: A Decade of Discovery and Diseases. Immunity. 2019;50:1132–1148. doi: 10.1016/j.immuni.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Hitz BC, Sloan CA, Chan ET, Davidson JM, Gabdank I, Hilton JA, Jain K, Baymuradov UK, Narayanan AK, et al. The Encyclopedia of DNA elements (ENCODE): data portal update. Nucleic Acids Res. 2018;46(D1):D794–D801. doi: 10.1093/nar/gkx1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol. 2015;15:137–148. doi: 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D, Lao C, DiToro D, Barnett B, Escobar TC, Kageyama R, Yusuf I, Crotty S. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 2011;6:e17739. doi: 10.1371/journal.pone.0017739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K. Nuclear receptor transrepression pathways that regulate inflammation in macrophages and T cells. Nat Rev Immunol. 2010;10:365–376. doi: 10.1038/nri2748. [DOI] [PubMed] [Google Scholar]
- Herber D, Brown TP, Liang S, Young DA, Collins M, Dunussi-Joan-nopoulos K. IL-21 has a pathogenic role in a lupus-prone mouse model and its blockade with IL-21R.Fc reduces disease progression. J Immunol. 2007;178:3822–3830. doi: 10.4049/jimmunol.178.6.3822. [DOI] [PubMed] [Google Scholar]
- Hermann-Kleiter N, Baier G. Orphan nuclear receptor NR2F6 acts as an essential gatekeeper of Th17 CD4+ T cell effector functions. Cell Commun Signal. 2014;12:38. doi: 10.1186/1478-811X-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Kleiter N, Gruber T, Lutz-Nicoladoni C, Thuille N, Fresser F, Labi V, Schiefermeier N, Warnecke M, Huber L, Villunger A, et al. The nuclear orphan receptor NR2F6 suppresses lymphocyte activation and T helper 17-dependent autoimmunity. Immunity. 2008;29:205–216. doi: 10.1016/j.immuni.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Kleiter N, Meisel M, Fresser F, Thuille N, Müller M, Roth L, Katopodis A, Baier G. Nuclear orphan receptor NR2F6 directly antagonizes NFAT and RORγt binding to the Il17a promoter. J Autoimmun. 2012;39:428–440. doi: 10.1016/j.jaut.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann-Kleiter N, Klepsch V, Wallner S, Siegmund K, Klepsch S, Tuzlak S, Villunger A, Kaminski S, Pfeifhofer-Obermair C, Gruber T, et al. The Nuclear Orphan Receptor NR2F6 Is a Central Checkpoint for Cancer Immune Surveillance. Cell Rep. 2015;12:2072–2085. doi: 10.1016/j.celrep.2015.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Glass CK. Nuclear receptors and inflammation control: molecular mechanisms and pathophysiological relevance. Arterioscler Thromb Vasc Biol. 2010;30:1542–1549. doi: 10.1161/ATVBAHA.109.191189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutcheson J, Scatizzi JC, Siddiqui AM, Haines GK, 3rd, Wu T, Li QZ, Davis LS, Mohan C, Perlman H. Combined deficiency of proapoptotic regulators Bim and Fas results in the early onset of systemic autoimmunity. Immunity. 2008;28:206–217. doi: 10.1016/j.immuni.2007.12.015. [DOI] [PubMed] [Google Scholar]
- Ise W, Inoue T, McLachlan JB, Kometani K, Kubo M, Okada T, Kurosaki T. Memory B cells contribute to rapid Bcl6 expression by memory follicular helper T cells. Proc Natl Acad Sci USA. 2014;111:11792–11797. doi: 10.1073/pnas.1404671111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepsch V, Hermann-Kleiter N, Do-Dinh P, Jakic B, Offermann A, Efremova M, Sopper S, Rieder D, Krogsdam A, Gamerith G, et al. Nuclear receptor NR2F6 inhibition potentiates responses to PD-L1/PD-1 cancer immune checkpoint blockade. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04004-2. 1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacazette E. A laboratory practical illustrating the use of the ChIP-qPCR method in a robust model: Estrogen receptor alpha immunoprecipitation using Mcf-7 culture cells. Biochem Mol Biol Educ. 2017;45:152–160. doi: 10.1002/bmb.20999. [DOI] [PubMed] [Google Scholar]
- Linterman MA, Rigby RJ, Wong RK, Yu D, Brink R, Cannons JL, Schwartzberg PL, Cook MC, Walters GD, Vinuesa CG. Follicular helper T cells are required for systemic autoimmunity. J Exp Med. 2009;206:561–576. doi: 10.1084/jem.20081886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Beaton L, Yu D, Ramiscal RR, Srivastava M, Hogan JJ, Verma NK, Smyth MJ, Rigby RJ, Vinuesa CG. IL-21 acts directly on B cells to regulate Bcl-6 expression and germinal center responses. J Exp Med. 2010;207:353–363. doi: 10.1084/jem.20091738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman MA, Pierson W, Lee SK, Kallies A, Kawamoto S, Rayner TF, Srivastava M, Divekar DP, Beaton L, Hogan JJ, et al. Foxp3+ follicular regulatory T cells control the germinal center response. Nat Med. 2011;17:975–982. doi: 10.1038/nm.2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Chen X, Zhong B, Wang A, Wang X, Chu F, Nurieva RI, Yan X, Chen P, van der Flier LG, et al. Transcription factor achaete-scute homologue 2 initiates follicular T-helper-cell development. Nature. 2014;507:513–518. doi: 10.1038/nature12910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire HM, Vogelzang A, Warren J, Loetsch C, Natividad KD, Chan TD, Brink R, Batten M, King C. IL-21 and IL-4 Collaborate To Shape T-Dependent Antibody Responses. J Immunol. 2015;195:5123–5135. doi: 10.4049/jimmunol.1501463. [DOI] [PubMed] [Google Scholar]
- Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity. 2016;45:471–482. doi: 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, Azuma T, Kitamura D. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun. 2011;2 doi: 10.1038/ncomms1475. 465. [DOI] [PubMed] [Google Scholar]
- Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, Matskevitch TD, Wang Y-H, Dong C. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Spolski R, Ettinger R, Kim HP, Wang G, Qi CF, Hwu P, Shaffer DJ, Akilesh S, Roopenian DC, et al. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol. 2004;173:5361–5371. doi: 10.4049/jimmunol.173.9.5361. [DOI] [PubMed] [Google Scholar]
- Park HJ, Kim DH, Choi JY, Kim WJ, Kim JY, Senejani AG, Hwang SS, Kim LK, Tobiasova Z, Lee GR, et al. PPARγ Negatively Regulates T Cell Activation to Prevent Follicular Helper T Cells and Germinal Center Formation. PLoS ONE. 2014;9:e99127. doi: 10.1371/journal.pone.0099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Park HS, Lee JU, Bothwell AL, Choi JM. Gender-specific differences in PPARγ regulation of follicular helper T cell responses with estrogen. Sci Rep. 2016a;6 doi: 10.1038/srep28495. 28495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Choi Y, Song MJ, Park K, Lee JJ, Kim HP. Dynamic Long-Range Chromatin Interaction Controls Expression of IL-21 in CD4+ T Cells. J Immunol. 2016b;196:4378–4389. doi: 10.4049/jimmunol.1500636. [DOI] [PubMed] [Google Scholar]
- Peled JU, Kuang FL, Iglesias-Ussel MD, Roa S, Kalis SL, Goodman MF, Scharff MD. The biochemistry of somatic hypermutation. Annu Rev Immunol. 2008;26:481–511. doi: 10.1146/annurev.immunol.26.021607.090236. [DOI] [PubMed] [Google Scholar]
- Pracht K, Meinzinger J, Daum P, Schulz SR, Reimer D, Hauke M, Roth E, Mielenz D, Berek C, Côrte-Real J, et al. A new staining protocol for detection of murine antibody-secreting plasma cell subsets by flow cytometry. Eur J Immunol. 2017;47:1389–1392. doi: 10.1002/eji.201747019. [DOI] [PubMed] [Google Scholar]
- Pratama A, Vinuesa CG. Control of TFH cell numbers: why and how? Immunol Cell Biol. 2014;92:40–48. doi: 10.1038/icb.2013.69. [DOI] [PubMed] [Google Scholar]
- Preite S, Baumjohann D, Foglierini M, Basso C, Ronchi F, Fernandez Rodriguez BM, Corti D, Lanzavecchia A, Sallusto F. Somatic mutations and affinity maturation are impaired by excessive numbers of T follicular helper cells and restored by Treg cells or memory T cells. Eur J Immunol. 2015;45:3010–3021. doi: 10.1002/eji.201545920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin AL, MacLeod H, Keegan S, Andreyeva T, Lowe L, Bloom L, Collins M, Nickerson-Nutter C, Young D, Guay H. IL-21 receptor is critical for the development of memory B cell responses. J Immunol. 2011;186:667–674. doi: 10.4049/jimmunol.0903207. [DOI] [PubMed] [Google Scholar]
- Ritvo PG, Churlaud G, Quiniou V, Florez L, Brimaud F, Fourcade G, Mariotti-Ferrandiz E, Klatzmann D. Tfr cells lack IL-2Rα but express decoy IL-1R2 and IL-1Ra and suppress the IL-1-dependent activation of Tfh cells. Sci Immunol. 2017;2:eaan0368. doi: 10.1126/sciimmunol.aan0368. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG. Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev. 2010;62:1307–1315. doi: 10.1016/j.addr.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shay T, Kang J. Immunological Genome Project and systems immunology. Trends Immunol. 2013;34:602–609. doi: 10.1016/j.it.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson N, Gatenby PA, Wilson A, Malik S, Fulcher DA, Tangye SG, Manku H, Vyse TJ, Roncador G, Huttley GA, et al. Expansion of circulating T cells resembling follicular helper T cells is a fixed phenotype that identifies a subset of severe systemic lupus erythematosus. Arthritis Rheum. 2010;62:234–244. doi: 10.1002/art.25032. [DOI] [PubMed] [Google Scholar]
- Sin JH, Zuckerman C, Cortez JT, Eckalbar WL, Erle DJ, Anderson MS, Waterfield MR. The epigenetic regulator ATF7ip inhibits Il2 expression, regulating Th17 responses. J Exp Med. 2019 doi: 10.1084/jem.20182316. Published online June 19, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol. 2012;30:429–457. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- Victora GD, Schwickert TA, Fooksman DR, Kamphorst AO, Meyer-Hermann M, Dustin ML, Nussenzweig MC. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010;143:592–605. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuesa CG, Cook MC, Angelucci C, Athanasopoulos V, Rui L, Hill KM, Yu D, Domaschenz H, Whittle B, Lambe T, et al. A RING-type ubiquitin ligase family member required to repress follicular helper T cells and autoimmunity. Nature. 2005;435:452–458. doi: 10.1038/nature03555. [DOI] [PubMed] [Google Scholar]
- Vogelzang A, McGuire HM, Yu D, Sprent J, Mackay CR, King C. A fundamental role for interleukin-21 in the generation of T follicular helper cells. Immunity. 2008;29:127–137. doi: 10.1016/j.immuni.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Weisberg S, Fox J. An R Companion to Applied Regression. Third Edition. Sage Publishing; 2019. [Google Scholar]
- Weisel FJ, Zuccarino-Catania GV, Chikina M, Shlomchik MJ. A Temporal Switch in the Germinal Center Determines Differential Output of Memory B and Plasma Cells. Immunity. 2016;44:116–130. doi: 10.1016/j.immuni.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CK, Wong PTY, Tam LS, Li EK, Chen DP, Lam CWK. Elevated production of B cell chemokine CXCL13 is correlated with systemic lupus erythematosus disease activity. J Clin Immunol. 2010;30:45–52. doi: 10.1007/s10875-009-9325-5. [DOI] [PubMed] [Google Scholar]
- Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- Yu D, Vinuesa CG. Multiple checkpoints keep follicular helper T cells under control to prevent autoimmunity. Cell Mol Immunol. 2010;7:198–203. doi: 10.1038/cmi.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Tan AH, Hu X, Athanasopoulos V, Simpson N, Silva DG, Hutloff A, Giles KM, Leedman PJ, Lam KP, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450:299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- Zotos D, Coquet JM, Zhang Y, Light A, D’Costa K, Kallies A, Corcoran LM, Godfrey DI, Toellner KM, Smyth MJ, et al. IL-21 regulates germinal center B cell differentiation and proliferation through a B cell-intrinsic mechanism. J Exp Med. 2010;207:365–378. doi: 10.1084/jem.20091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarino-Catania GV, Sadanand S, Weisel FJ, Tomayko MM, Meng H, Kleinstein SH, Good-Jacobson KL, Shlomchik MJ. CD80 and PD-L2 define functionally distinct memory B cell subsets that are independent of antibody isotype. Nat Immunol. 2014;15:631–637. doi: 10.1038/ni.2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new datasets or code.