Abstract
Cancer is driven by genetic change, and the advent of massively parallel sequencing has enabled systematic documentation of this variation at whole genome scale1–3. We report the integrative analysis of >2,600 whole cancer genomes and their matching normal tissues across 38 tumour types, by the ICGC/TCGA Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium. We describe the generation of the PCAWG resource, facilitated by international data sharing using compute clouds. Cancer genomes contained 4-5 driver mutations on average when combining coding and non-coding genomic elements, but ~5% of cases had no drivers identified, suggesting that cancer driver discovery is not yet complete. Chromothripsis is frequently an early event in tumour evolution: in acral melanoma, for example, these clustered events precede most somatic point mutations and affect several cancer genes simultaneously. Cancers with abnormal telomere maintenance often originate in tissues with low replicative activity, with several different mechanisms of escaping critical telomere attrition. Common and rare germline variants affect patterns of somatic mutation, including point mutations, structural variants and somatic retrotransposition. PCAWG found few non-coding mutations that drive cancer beyond those in the TERT promoter4; identified new signatures of mutational processes causing base substitutions, indels and structural variation5,6; analysed timings and patterns of tumour evolution7; described the diverse transcriptional consequences of somatic mutation on splicing, expression levels, fusion genes and promoter activity8,9; and evaluated a range of more specialised features of cancer genomes8,10–18.
Cancer is the second most frequent cause of death worldwide, killing more than 8 million people every year and expected to increase by >50% over the coming decades19,20. ‘Cancer’ is a catch-all term used to denote a set of diseases characterised by autonomous expansion and spread of a somatic clone. To achieve this behaviour, the cancer clone must co-opt multiple cellular pathways that enable it to disregard the normal constraints on cell growth, to modify the local microenvironment favouring its own proliferation, to invade through tissue barriers, to spread to other organs, and to evade immune surveillance21. No single cellular programme directs these behaviours. Rather, there is a large pool of potential pathogenic abnormalities from which individual cancers draw their own combinations: the commonalities of macroscopic features across tumours belie a vastly heterogeneous landscape of cellular abnormalities.
This heterogeneity arises from the stochastic nature of Darwinian evolution. The preconditions for Darwinian evolution are three: characteristics must vary within a population; this variation must be heritable from parent to offspring; and there must be competition for survival within the population. In the context of somatic cells, heritable variation arises from mutations acquired stochastically throughout life, notwithstanding additional contributions from germline and epigenetic variation. A subset of these mutations alter cellular phenotype, and a small subset of those variants confer an advantage on clones in their competition to escape the tight physiological controls wired into somatic cells. Mutations providing selective advantage to the clone are termed ‘driver' mutations, as opposed to selectively neutral ‘passenger' mutations.
Initial studies using massively parallel sequencing demonstrated the feasibility of identifying every somatic point mutation, copy number change, and structural variant in a given cancer1–3. In 2008, recognising the opportunity this advance in technology provided, the global cancer genomics community established the International Cancer Genome Consortium (ICGC) with the goal of systematically documenting the somatic mutations driving common tumour types22.
Pan-Cancer Analysis of Whole Genomes
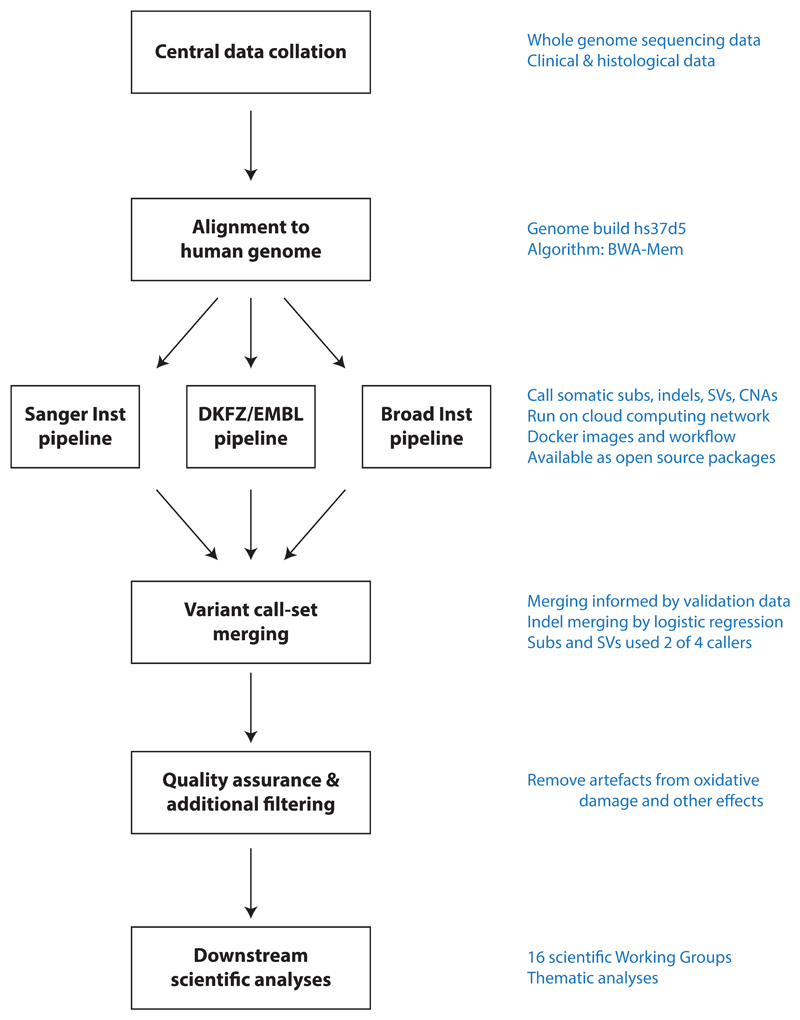
The maturing of whole genome sequencing studies from individual ICGC and TCGA working groups presented the opportunity to undertake a meta-analysis of genomic features across tumour types. To achieve this, the Pan-Cancer Analysis of Whole Genomes (PCAWG) consortium was established. A Technical Working Group implemented informatics analyses, aggregating the raw sequencing data from working groups studying individual tumour types, aligning to the human genome, and delivering a set of high-quality somatic mutation calls for downstream analysis (Extended Figure 1). Given the recent meta-analysis of exome data from the TCGA Pan-Cancer Atlas23–25, scientific working groups concentrated their efforts on analyses best informed by whole genome sequencing data.
We collected genome data from 2,834 donors (Extended Table 1), of which 176 were excluded after quality assurance. A further 75 had minor issues that could impact some analyses (grey-listed donors), and 2,583 had data of optimal quality (white-listed donors; Supplementary Table 1). Across the 2,658 white- and grey-listed donors, whole genome sequencing data were available from 2,605 primary tumours and 173 metastases or local recurrences. Mean read coverage was 39x for normal samples, while tumours had a bimodal coverage distribution with modes at 38x and 60x (Supplementary Figure 1). RNA-sequencing data was available for 1,222 donors. The final cohort comprised 1,469 males (55%) and 1,189 females (45%), with a mean age of 56 years (range, 1-90 years) across 38 tumour types (Extended Table 1; Supplementary Table 1).
In order to identify somatic mutations, we analysed all 6,835 samples using a uniform set of algorithms for alignment, variant calling and quality control (Extended Figure 1; Supplementary Figure 2; Supplementary Methods S2). We deployed three established pipelines to call somatic single nucleotide variations (SNVs), small insertions and deletions (indels), copy number alterations (CNAs), and structural variants (SVs). Somatic retrotransposition events, mitochondrial DNA mutations and telomere lengths were also called by bespoke algorithms. RNA-Sequencing data were uniformly processed to call transcriptomic alterations. Germline variants identified via three separate pipelines included single nucleotide polymorphisms (SNPs), indels, structural variants and mobile element insertions (Supplementary Table 2).
The requirement to uniformly realign and call variants on ~5,800 whole genomes presented significant computational challenges, and raised ethical issues due to the use of data from different jurisdictions (Box 1). We used cloud computing26,27 to distribute alignment and variant calling across 13 data centres in three continents (Supplementary Table 3). Core pipelines were packaged into Docker containers28 as reproducible, stand-alone packages, which we have made available for download. Data repositories for raw and derived datasets, together with portals for data visualisation and exploration have also been created (Box 1; Supplementary Table 4).
BOX 1. Online resources for data access, visualisation and analysis.
The PCAWG Landing Page at http://docs.icgc.org/pcawg provides links to several data resources for interactive online browsing, analysis and download of PCAWG data and results (Supplementary Table 4).
Direct download of PCAWG data
Aligned PCAWG read data in BAM format are also available at the European Genome Phenome Archive (EGA; https://www.ebi.ac.uk/ega/search/site/pcawg under accession EGAS00001001692). In addition, all open tier PCAWG genomics data, as well as reference data sets used for analysis, can be downloaded from the ICGC Data Portal at http://docs.icgc.org/pcawg/data/. Controlled tier genomic data, including SNVs and indels that originated from TCGA projects (in VCF format), and aligned reads (in BAM format) can be downloaded using the Score (https://www.overture.bio/) software package, which implements accelerated and secure file transfer, as well as BAM slicing facilities to selectively download defined regions of genomic alignments.
PCAWG computational pipelines
The core alignment, somatic variant-calling, quality control and variant consensus generation pipelines used by PCAWG have each been packaged into portable cross-platform images using the Dockstore system88 and released under an Open Source license that allows for unrestricted usage and redistribution. All PCAWG Dockstore images are available to the public at https://dockstore.org.
ICGC Data Portal (https://dcc.icgc.org).
The ICGC Data Portal89 serves as the main entry point for accessing PCAWG datasets with a single uniform web interface and a high-performance data download client. This uniform interface gives users easy access to the myriad of PCAWG sequencing data and variant calls that reside in many repositories and compute clouds worldwide. Streaming technology90 gives users high-level visualisations in real time of BAM and VCF files stored remotely on the Cancer Genome Collaboratory.
UCSC Xena (https://pcawg.xenahubs.net)
UCSC Xena91 visualises all PCAWG primary results, including copy number, gene expression, gene fusion, promoter usage, simple somatic mutations, large somatic structural variation, mutational signatures and phenotypic data. These open-access data are available through a public Xena hub, while consensus simple somatic mutations can be loaded into a user's local computer private Xena hub. Kaplan-Meier plots, histograms, boxplots, scatterplots and transcript-specific views offer additional visualisation options and statistical analyses.
Expression Atlas (https://www.ebi.ac.uk/gxa/home)
The Expression Atlas contains RNAseq and expression microarray data for querying gene expression across tissues, cell types, developmental stages and/or experimental conditions92. Two different views of the data are provided: summarised expression levels for each tumour type and gene expression at the level of individual samples, including reference gene expression datasets for matching normal tissues.
PCAWG-Scout (http://pcawgscout.bsc.es/)
PCAWG-Scout provides a framework for ‘omics workflow and website templating to make on-demand, in-depth analyses over the PCAWG data openly available to the whole research community. Views of protected data are available that still safeguard sensitive data. Through the PCAWG-Scout web interface, users can access an array of reports and visualisations that leverage on-demand bioinformatic computing infrastructure to produce results in real-time, allowing users to discover trends as well as form and test hypotheses.
Chromothripsis Explorer (http://compbio.med.harvard.edu/chromothripsis/)
Chromothripsis Explorer is a portal that allows structural variation in the PCAWG dataset to be explored on an individual patient basis through the use of circos plots. Patterns of chromothripsis can also be explored in aggregated formats.
Benchmarking of genetic variant calls
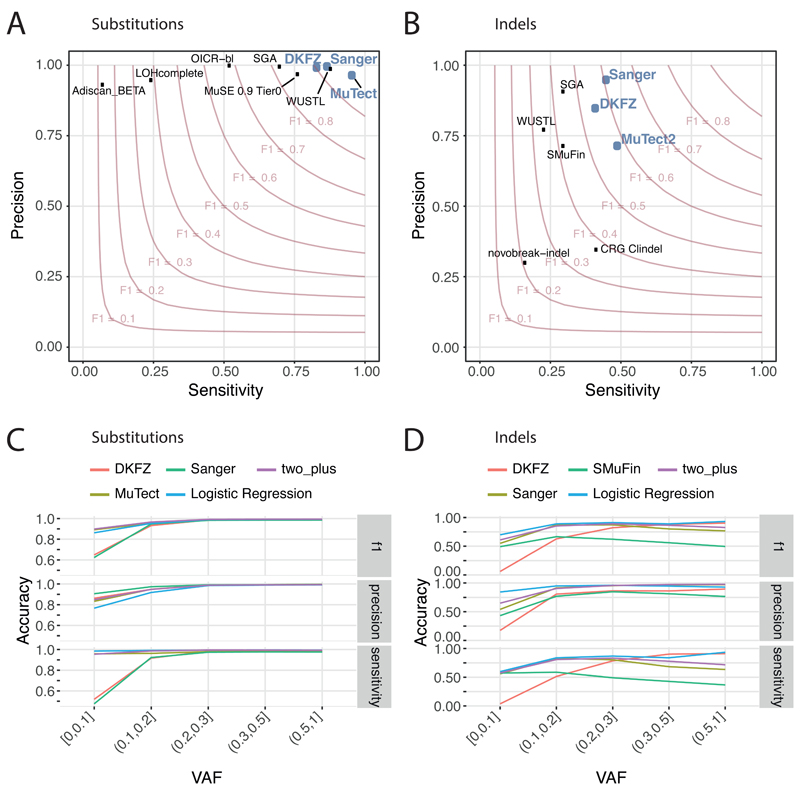
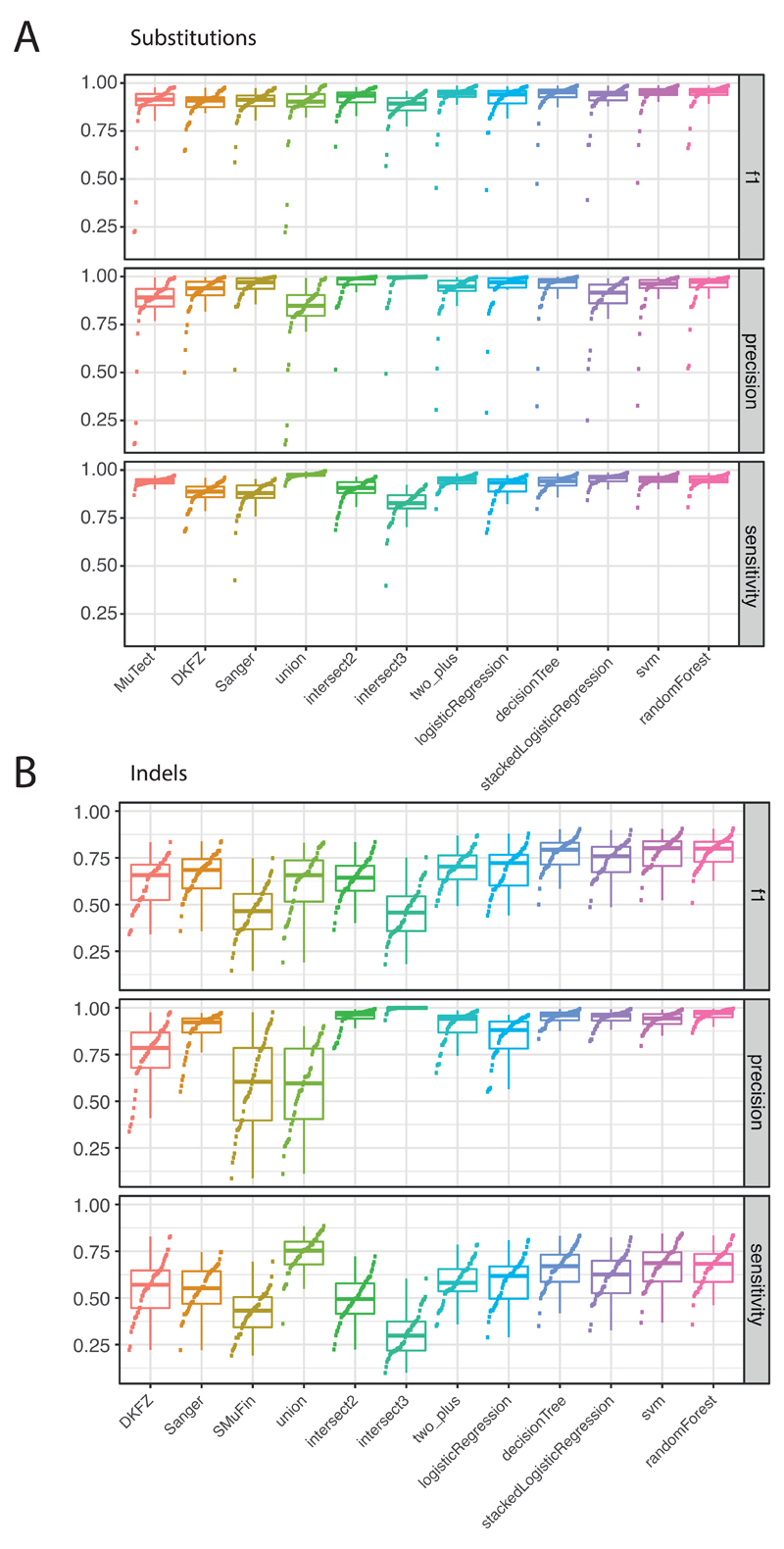
To benchmark mutation calling, we ran the three core pipelines, together with 10 additional pipelines, on 63 representative tumour/normal genome pairs (Supplementary Note 1). For 50 of these cases, we performed validation by hybridisation of tumour and matched normal DNA to a custom bait-set with deep sequencing29. The three core somatic variant-calling pipelines had individual estimates of sensitivity of 80-90% to detect a true somatic SNV called by any of the 13 pipelines; with >95% of SNV calls made by each of the core pipelines being genuine somatic variants (Figure 1A). For indels, a more challenging class of variants to identify with short-read sequencing, the three core algorithms had individual sensitivity estimates in the range 40-50%, with precision 70-95% (Figure 1B). For individual SV callers, we estimated precision to be in the range 80-95% for samples in the pilot-63 dataset.
Figure 1. Validation of variant-calling pipelines in PCAWG.
(A) Scatter plot of estimated sensitivity and precision for somatic SNVs across individual algorithms assessed in the validation exercise across n=63 PCAWG samples. Core algorithms included in the final PCAWG call-set are shown in blue. (B) Sensitivity and precision estimates across individual algorithms for somatic indels. (C) Accuracy (F1 score, precision and sensitivity) of somatic SNV calls across variant allele fractions (VAF) for the core algorithms. Also shown is the accuracy for two methods of combining variant calls (two-plus, which was used in the final data-set, and logistic regression). (D) Accuracy of indel calls across VAFs.
Next, we defined a strategy to merge results from the three pipelines into one final call-set to be used for downstream scientific analyses (Methods, Supplementary Note 2). Sensitivity and precision of consensus somatic variant calls were 95% (CI90%=88-98%) and 95% (CI90%=71-99%) respectively for SNVs (Extended Figure 2). For somatic indels, sensitivity and precision were 60% (34-72%) and 91% (73-96%) respectively (Extended Figure 2). Regarding somatic SVs, we estimate the sensitivity of merged calls to be 90% for true calls generated by any one caller; precision was estimated as 97.5%. The improvement in calling accuracy from combining different callers was most noticeable in variants having low variant allele fractions, which likely originate in tumour subclones (Figure 1C-D). Germline variant calls, phased using a haplotype-reference panel, displayed a precision >99% and sensitivity of 92%-98% (Supplementary Note 2).
Analysis of PCAWG data
The uniformly generated, high quality set of variant calls across >2,500 donors provided the springboard for a series of scientific working groups to explore the biology of cancer. A comprehensive suite of companion papers detailing the analyses and discoveries across these thematic areas is co-published with this paper (Extended Table 3).
Pan-cancer burden of somatic mutations
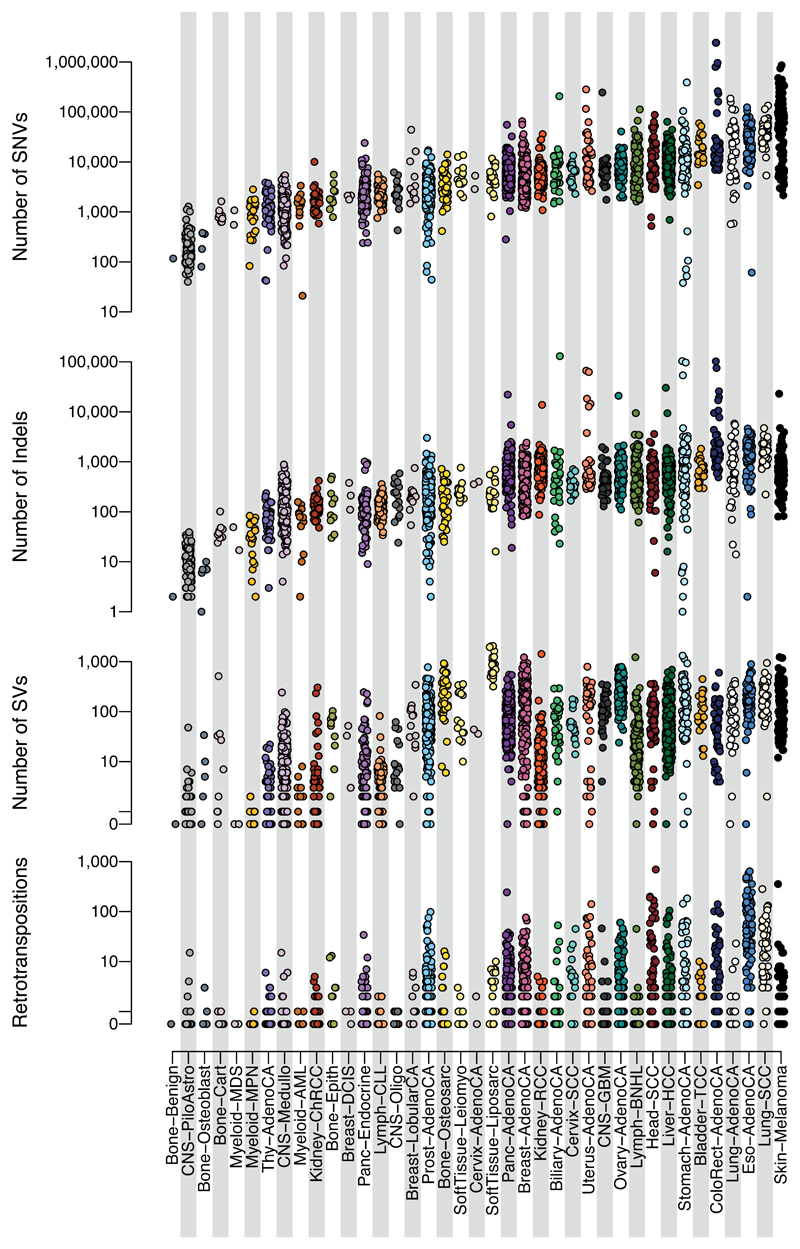
Across the 2,583 white-listed PCAWG donors, we called 43,778,859 somatic SNVs; 410,123 somatic multi-nucleotide variants; 2,418,247 somatic indels; 288,416 somatic structural variants; 19,166 somatic retrotransposition events; and 8,185 de novo mitochondrial DNA mutations (Supplementary Table 1). There was considerable heterogeneity in the burden of somatic mutations across patients and tumour types, with a broad correlation in mutation burden among different classes of somatic variation (Extended Figure 3). Analysed at a per-patient level, this correlation held, even when considering tumours with similar purity and ploidy (Supplementary Figure 3). Why such correlation should apply on a pan-cancer basis is unclear. It is likely that age plays some role, as we observe a correlation of most classes of somatic mutation with age at diagnosis (~190 SNVs/year, p=0.02; ~22 indels/year, p=5x10-5; 1.5 SVs/year, p<2x10-16; linear regression with likelihood ratio tests; Supplementary Figure 4). Other factors are also likely to contribute to the correlations among classes of somatic mutation, since there is evidence that some DNA repair defects can cause multiple types of somatic mutation30, and a single carcinogen can cause a range of DNA lesions31.
Panorama of driver mutations in cancer
We extracted the subset of somatic mutations in PCAWG tumours that have high confidence to be driver events, based on current knowledge. One challenge to pinpointing the specific driver mutations in an individual tumour is that not all point mutations in recurrently mutated cancer genes are drivers32. For genomic elements significantly mutated in the PCAWG, we developed a ‘rank-and-cut’ approach to identify the likely drivers (Supplementary Methods 8.1). This works by ranking the observed mutations in a given genomic element based on recurrence, estimated functional consequence, and expected pattern of drivers in that element. We then estimate the excess burden of somatic mutations in that genomic element above that expected for the background mutation rate, and cut the ranked mutations at this level. Mutations in that element with the highest driver ranking will then be assigned as likely drivers; those below the threshold will probably have arisen through chance, and be assigned as likely passengers. Improvements to features employed to rank the mutations and the methods used to measure them will contribute to further maturation of the rank-and-cut approach.
We also needed to account for the fact that some bona fide cancer genomic elements were not rediscovered in PCAWG data because of low statistical power. We therefore added previously known cancer genes to the discovery set, creating a ‘Compendium of Mutational Driver Elements’ (Supplementary Methods 8.2). Then, using stringent rules to nominate driver point mutations affecting these genomic elements on the basis of prior knowledge33, we separated likely driver from passenger point mutations. To cover all classes of variant, we also created a compendium of known driver SVs, using analogous rules to identify which somatic CNAs and SVs most likely act as drivers in each tumour. For likely pathogenic germline variants, we identified all truncating germline point mutations and SVs affecting high-penetrance germline cancer genes.
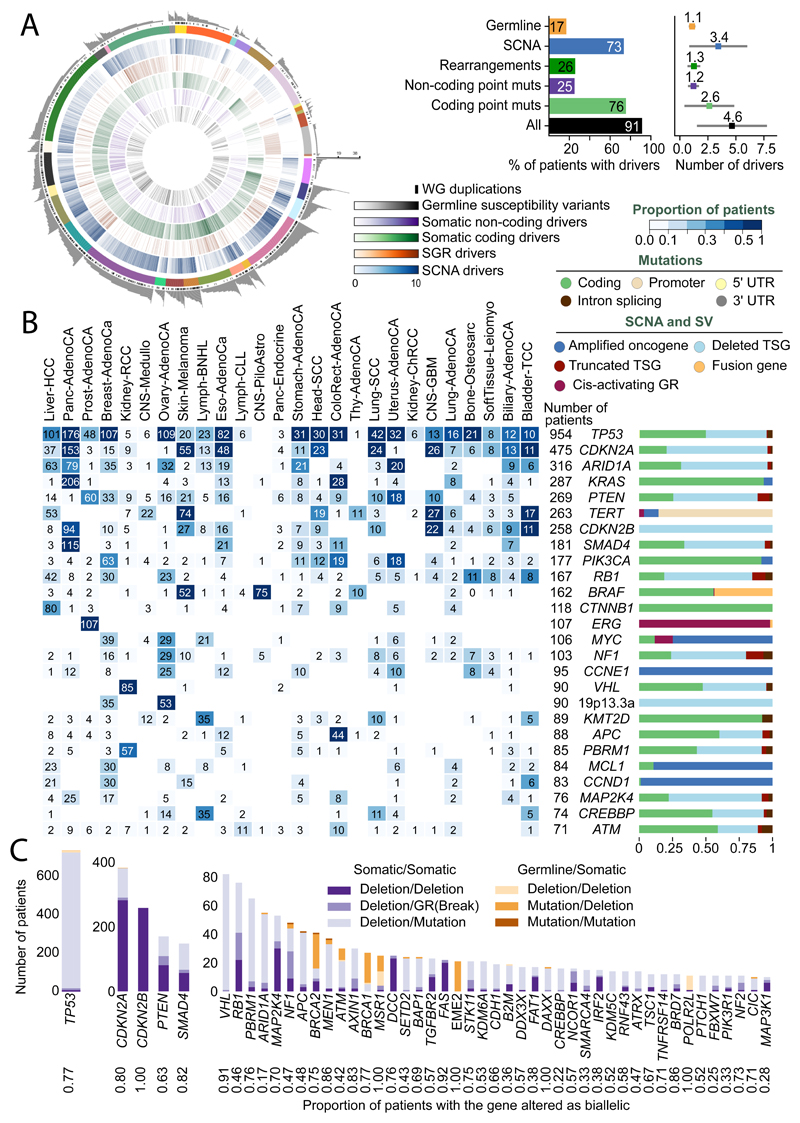
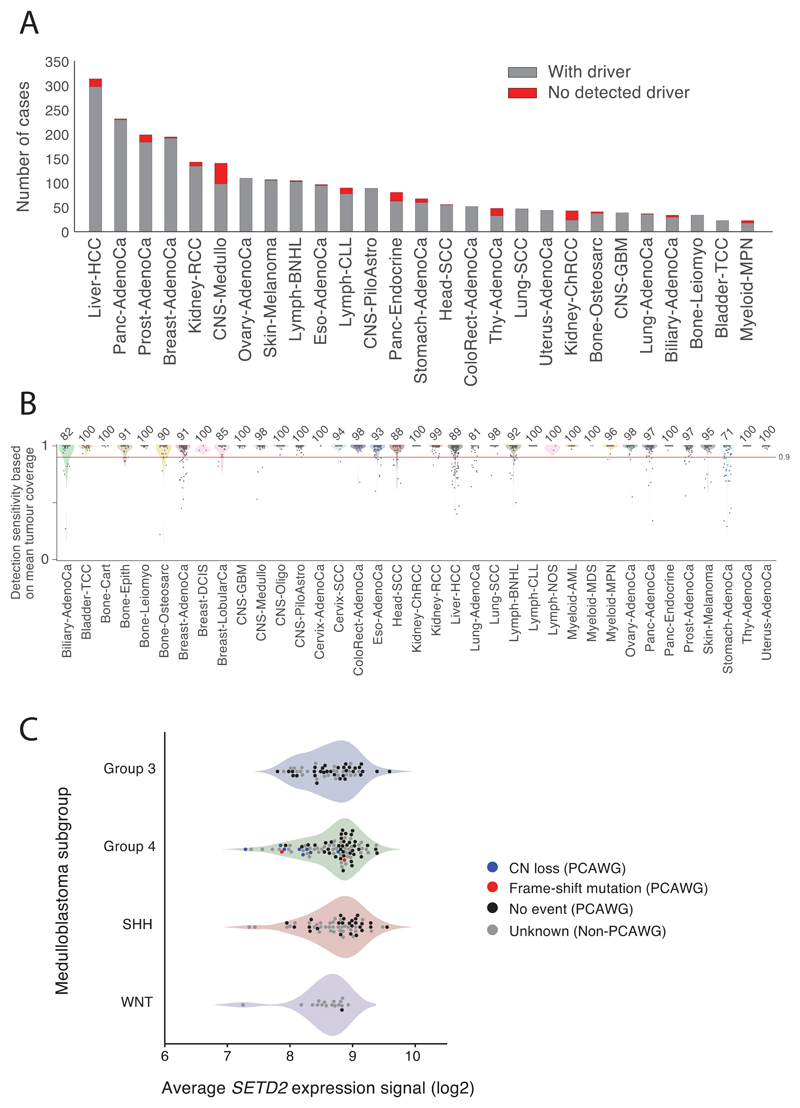
This analysis defined a set of mutations that we could confidently assert, based on current knowledge, drove tumorigenesis in the >2,500 tumours of PCAWG. We found that 91% of tumours had at least one identified driver mutation, with an average of 4.6 drivers per tumour identified, showing extensive variation across cancer types (Figure 2A). For coding point mutations, the average was 2.6 drivers per tumour, similar to numbers estimated in known cancer genes in TCGA tumours using similar approaches32.
Figure 2. Panorama of driver mutations in PCAWG.
(A) Left panel: putative driver mutations in PCAWG, represented as a circos plot. Each sector represents a tumour in the cohort. From the periphery to the centre of the plot the concentric rings represent: i) the total number of driver alterations; ii) presence of whole genome duplication; iii) the tumour type; iv) the number of driver copy number alterations; v) the number of driver genomic rearrangements; vi) driver coding point mutations; vii) driver non-coding point mutations; and viii) pathogenic germline variants. Right panel: Snapshots of the panorama of driver mutations. The horizontal bar plot at the left represents the proportion of patients with different types of drivers. The dot plot at the right represents the mean number of each type of driver mutation across tumours with at least one event (the square dot), and its standard deviation (gray whiskers), based on n=2,583 patients. (B) Genomic elements targeted by different types of mutations in the cohort in more than 65 tumours. Both germline and somatic variants are included. The heatmap shows the recurrence of alterations experienced across cancer types (with the colour indicating the proportion, and the number indicating the absolute count of mutated tumours); the barplot at the right reflects the proportion of each type of alteration affecting each genomic element. (C) Tumour suppressor genes with biallelic inactivation in 10 or more patients. The values quoted under the gene labels represent the proportions of patients who have biallelic mutations of the gene out of all patients with a somatic mutation in that gene.
To address the frequency of non-coding driver point mutations, we combined promoters and enhancers that are known targets of non-coding drivers34–37 with those newly discovered on PCAWG data, reported in a companion paper4. Using this approach, only 13% (785/5913) of driver point mutations were non-coding in PCAWG. Nonetheless, 25% of PCAWG tumours bear at least one putative non-coding driver point mutation, with one third (237/785) affecting the TERT promoter (9% of PCAWG tumours). Overall, then, non-coding driver point mutations are less frequent than coding drivers. With the exception of the TERT promoter, individual enhancers and promoters are only infrequent targets of driver mutations4.
Across tumour types, SVs and point mutations make different relative contribution to tumorigenesis. Driver SVs are more prevalent in breast adenocarcinomas (6.4±3.7 SVs vs. 2.2±1.3 point mutations on average±SD; p<10-16, Mann-Whitney U test) and ovary adenocarcinomas (5.8±2.6 SVs vs. 1.9±1.0 point mutations; p<10-16), while driver point mutations make a larger contribution in colorectal adenocarcinomas (2.4±1.4 SVs vs. 7.4±7.0 point mutations; p=4x10-10) and mature B-cell lymphomas (2.2±1.3 SVs vs. 6±3.8 point mutations; p<10-16), as shown previously38. Across tumour types, there are differences in which classes of mutation affect a given genomic element (Figure 2B).
We confirmed that many driver mutations affecting tumour suppressor genes are two-hit inactivation events (Figure 2C). For example, of the 954 tumours in the cohort with driver mutations in TP53, 736 (77%) had both alleles mutated, 96% of which (707/736) combined a somatic point mutation affecting one allele with somatic deletion of the other allele. Overall, 17% of patients harboured rare germline protein-truncating variants (PTVs) in cancer predisposition genes39, DNA damage response genes40 and somatic driver genes. Biallelic inactivation due to somatic alteration on top of a germline PTV was observed in 4.5% of patients overall, with 81% of these affecting known cancer predisposition genes (such as BRCA1, BRCA2 and ATM).
PCAWG tumours with no apparent drivers
Although >90% PCAWG cases had identified drivers, we found none in 181 tumours (Extended Figure 4A). Reasons for missing drivers have not yet been systematically evaluated in a pan-cancer cohort, and could arise from either technical or biological causes.
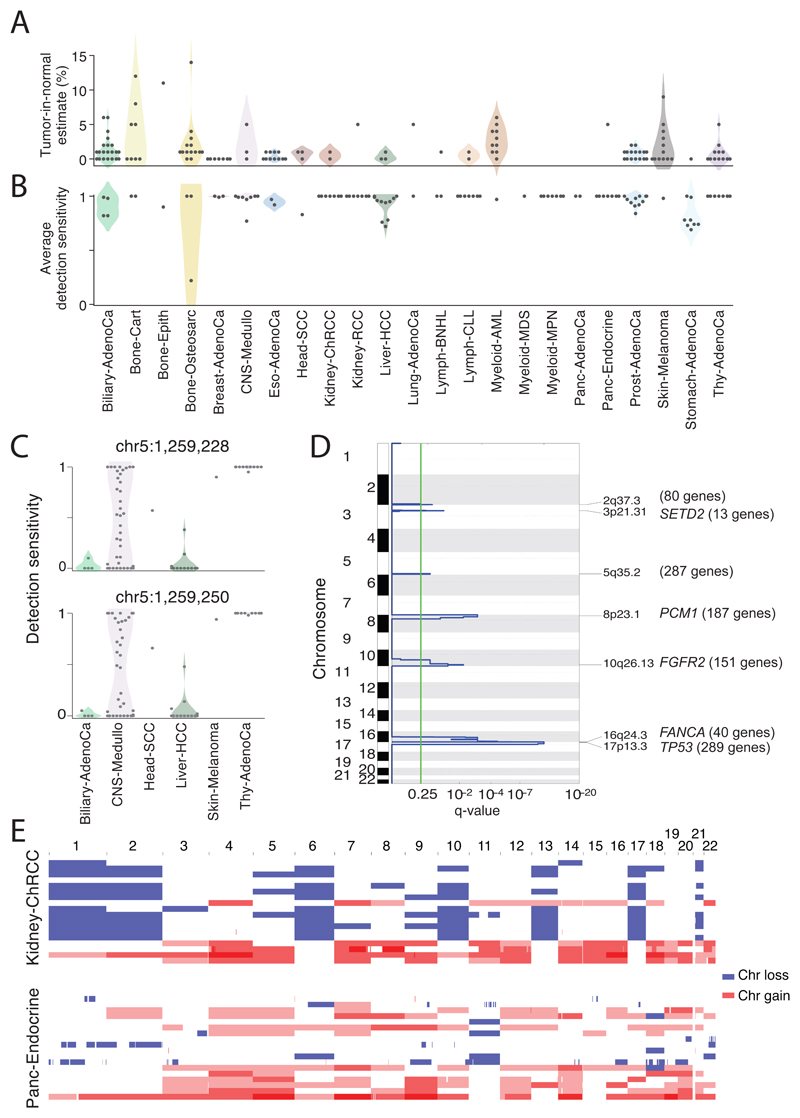
Technical explanations could include poor quality samples; inadequate sequencing; or failures in the bioinformatic algorithms deployed. Assessing the quality of samples, four of the 181 ‘missed-driver’ cases had >5% tumour DNA contamination in their matched normal (Figure 3A). Using an algorithm designed to correct for this contamination41, we identified previously missed mutations in genes relevant to the respective cancer types. Similarly, if the fraction of tumour cells in the cancer sample is low through stromal contamination, detection of driver mutations can be impaired. Most missed-driver tumours had an average power to detect mutations close to 100%, but a few had power in the 70-90% range (Figure 3B; Extended Figure 4B). Even in adequately sequenced genomes, lack of read depth at specific driver loci can impair mutation detection. For example, only ~50% of PCAWG tumours had sufficient coverage to call a mutation (≥90% power) at the two TERT promoter hotspots, likely because of the region’s high GC-content causing biased coverage (Figure 3C). In fact, six Liver-HCC and two Biliary-AdenoCa tumours among the 181 missed-driver cases actually did carry TERT mutations upon deep targeted sequencing42.
Figure 3. Analysis of patients with no detected driver mutations.
(A) Individual estimates of percentage of tumour-in-normal contamination across no-driver patients in PCAWG (n=181). No data were available for Myeloid-MDS and Myeloid-AML. Points represent estimates for individual patients, and the coloured areas beneath the points are estimated density distributions (violin plots). (B) Average detection sensitivity by tumour type for tumours without known drivers (n=181). Each dot represents a given sample and represents the average sensitivity for detecting clonal substitutions across the genome. Coloured areas beneath the points are estimated density distributions, shown for cohorts with ≥5 cases. (C) Detection sensitivity for TERT promoter hotspots in tumour types where TERT is frequently mutated. Coloured areas beneath the points are estimated density distributions. (D) Significant copy number losses identified by two-sided hypothesis testing using GISTIC2.0, corrected for multiple hypothesis testing. Numbers in parentheses are the number of genes in significant regions when analysing missing-driver tumours (n=181). Significant regions with known cancer genes are labelled with a representative cancer gene. (E) Aneuploidy in Kidney-ChRCC and Panc-Endocrine cancers without known drivers. Patients are ordered in the y axis by tumour type and then by presence of whole genome duplication (bottom) or not (top).
Finally, technical reasons for missing driver mutations include failures in the bioinformatic algorithms. This affected 35 myeloproliferative neoplasms in PCAWG, where the JAK2V617F driver mutation should have been called. Our somatic variant-calling algorithms rely on ‘panels of normals’, typically from blood samples, to remove recurrent sequencing artefacts. Since 2-5% healthy individuals carry occult haematopoietic clones43, recurrent driver mutations in these clones can enter panels of normals.
Turning to biological causes, tumours may be driven by mutations in cancer genes not yet discovered in that tumour type. Using driver discovery algorithms on missed-driver tumours, no individual genes reached significance for point mutations. However, we identified a recurrent CNA spanning SETD2 in medulloblastomas lacking known drivers (Figure 3D), indicating that restricting hypothesis-testing to missed-driver cases can improve power if undiscovered genes are enriched in such tumours. Inactivation of SETD2 in medulloblastoma significantly decreased gene expression (p=0.002; Extended Figure 4C). Interestingly, SETD2 mutations occurred exclusively in medulloblastoma group 4 tumours (p<1x10-4). Group 4 medulloblastomas are known for frequent mutations in other chromatin-modifying genes44, and our results suggest that SETD2 loss-of-function is an additional driver affecting chromatin regulators in this subgroup.
Two tumour types had a surprisingly high fraction of patients without identified driver mutations: chromophobe renal cell carcinoma (44%; 19/43) and pancreatic neuroendocrine cancers (22%; 18/81) (Extended Data Figure 4A). A striking feature of the missed-driver cases in both tumour types was a remarkably consistent profile of chromosomal aneuploidy, patterns that have been reported previously45,46 (Figure 3E). The absence of other identified driver mutations in these patients raises the intriguing hypothesis that certain combinations of whole chromosome gains and losses may be sufficient to initiate a cancer in the absence of more targeted driver events such as point mutations, fusion genes of focal CNAs.
Even after accounting for technical issues and novel drivers, 5.3% of PCAWG tumours still had no identifiable driver events. In a research setting, where we are interested in drawing conclusions about populations of patients, the consequences of technical issues affecting occasional samples will be mitigated by sample size. In a clinical setting, where we are interested in the driver mutations in a specific patient, these issues become substantially more important. Careful and critical appraisal of the whole pipeline, including sample acquisition, genome sequencing, mapping, variant calling, and driver annotation, as done here, should be required for laboratories offering clinical sequencing of cancer genomes.
Patterns of clustered mutations and SVs
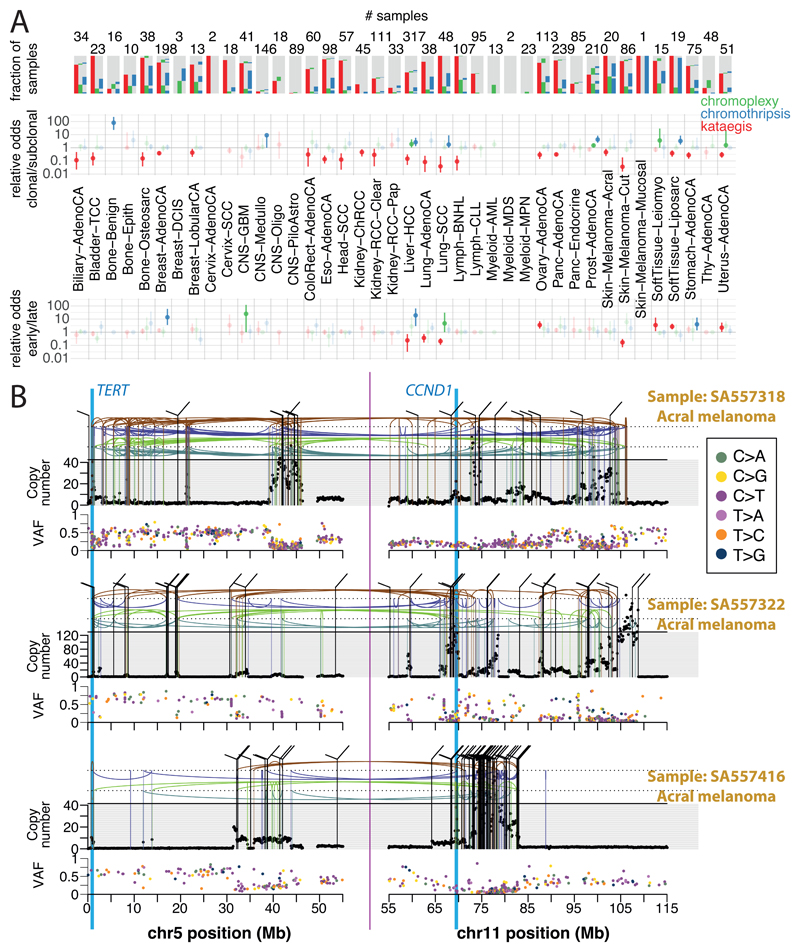
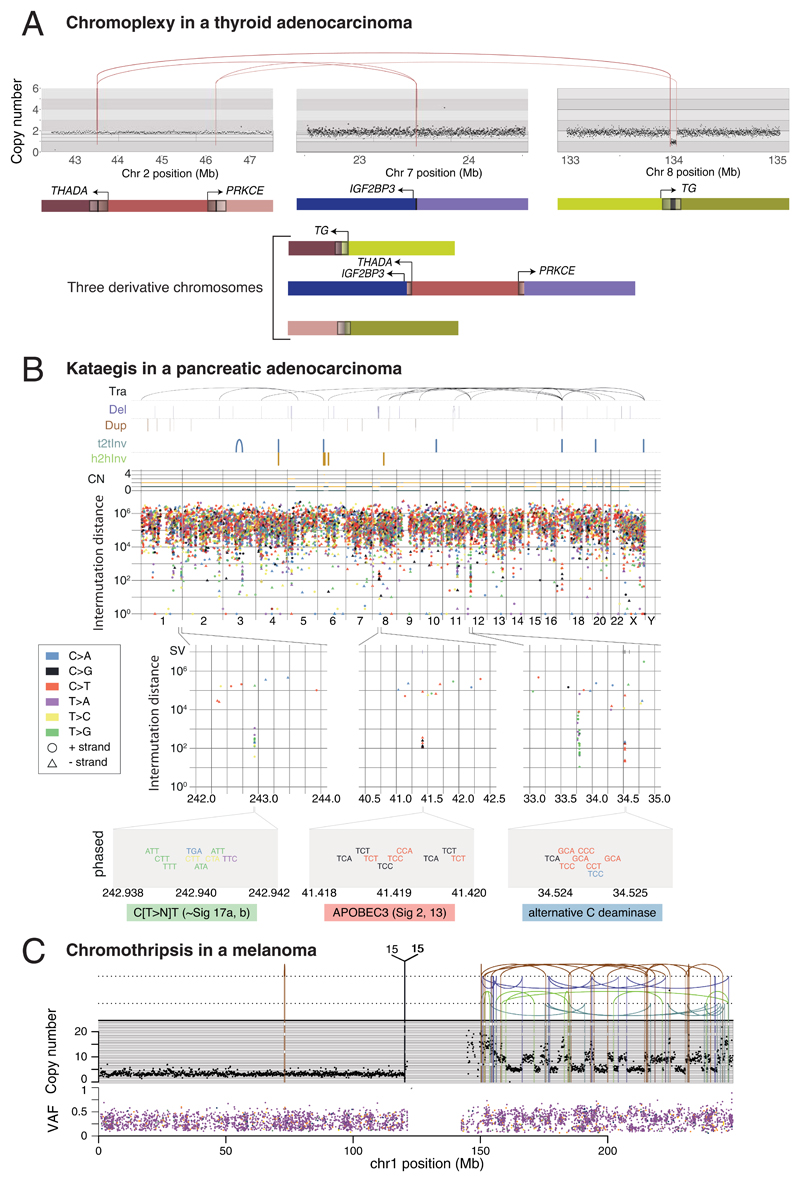
Some mutational processes generate multiple mutations in a single catastrophic event, typically clustered in genomic space, leading to substantial reconfiguration of the genome. Three such processes have been described: (i) chromoplexy, in which repair of co-occurring dsDNA breaks, typically on different chromosomes, results in shuffled chains of rearrangements47,48 (example in Extended Figure 5A); (ii) kataegis, a focal hypermutation process leading to locally clustered nucleotide substitutions, biased towards a single DNA strand49–51 (Extended Figure 5B); and (iii) chromothripsis, in which tens to hundreds of DNA breakages occur simultaneously, clustered on one or a few chromosomes, with near-random stitching together of the resulting fragments52–55 (Extended Figure 5C). We characterised the PCAWG genomes for these three processes (Figure 4).
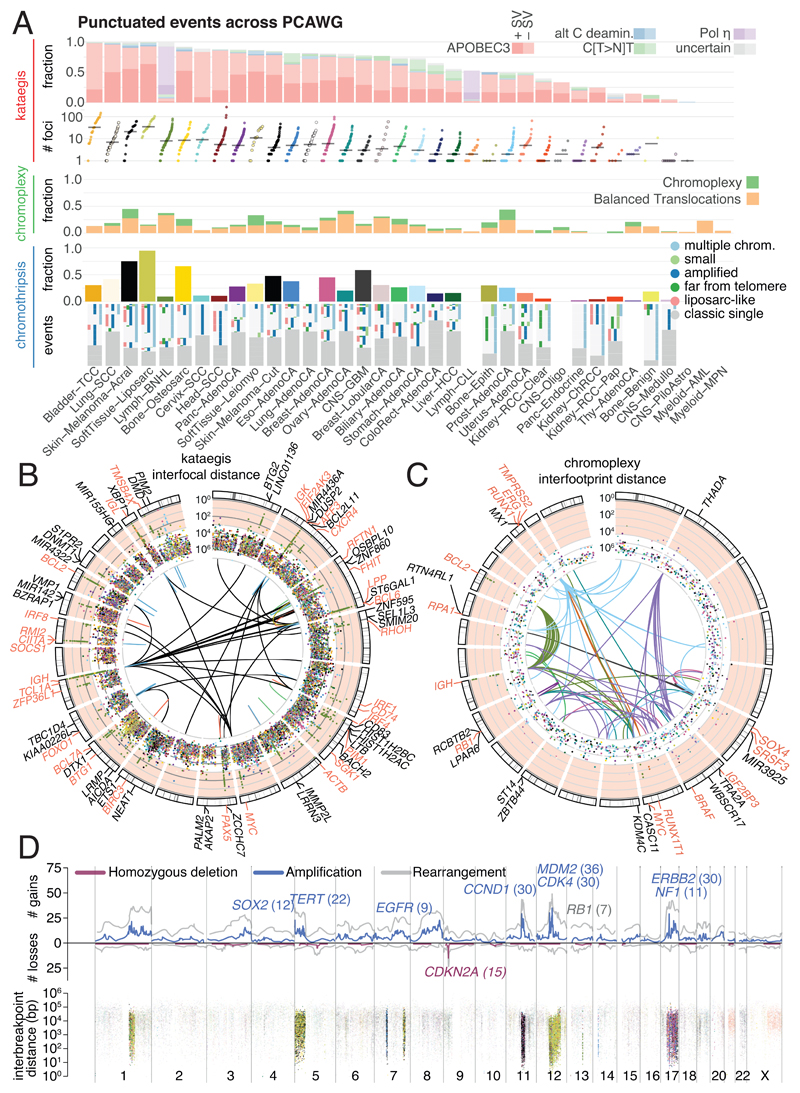
Figure 4. Patterns of clustered mutational processes in PCAWG.
(A) Kataegis. Prevalence of different types of kataegis and their association with SVs (≤1kb from the focus). The distribution of number of foci of kataegis per sample is given below. Chromoplexy. Prevalence of chromoplexy across cancer types, subdivided into balanced translocations and more complex events. Chromothripsis. Frequency of chromothripsis across cancer types. For each cancer type, a column is shown below, in which each row is a chromothripsis region represented by 5 coloured rectangles relating to its categorisation. (B) Circos rainfall plot showing the distances between consecutive kataegis events across PCAWG vs their genomic position. Lymphoid tumours (khaki for Lymph-BNHL and orange for Lymph-CLL) harbour hypermutation hot spots (≥3 foci with distance ≤1kb; pale red zone), many near known cancer genes (red annotations) and with associated SVs (≤10kb from the focus; shown as internal arcs). (C) Circos rainfall plot as in B showing distance vs position for consecutive chromoplexy and reciprocal translocation footprints across PCAWG. Lymphoid, prostate and thyroid cancers exhibit recurrent events (≥2 footprints with distance ≤10kb; pale red zone) likely to be driver structural variants and are annotated with nearby genes and associated SVs shown as bold and thin arcs for chromoplexy and reciprocal translocations, respectively (colours as in A). (D) Impact of chromothripsis along the genome and involvement of PCAWG driver genes. Top. Number of chromothripsis-induced gains/losses (grey) and amplifications/deletions (blue/red). Within the identified chromothripsis regions, selected recurrently rearranged (light grey), amplified (blue) and homozygously deleted (red) driver genes are indicated. Bottom. Inter-breakpoints distance between all subsequent breakpoints within chromothripsis regions across cancer types, coloured by cancer type. Regions with an average inter-breakpoint distance <10kb are highlighted.
Chromoplexy events and reciprocal translocations were identified in 467 (17.8%) samples (Figure 4A,C). Chromoplexy was prominent in prostate adenocarcinoma and lymphoid malignancies, as described previously47,48, and, unexpectedly, thyroid adenocarcinoma. Different genomic loci were recurrently rearranged by chromoplexy across the three tumour types, mediated by positive selection for particular fusion genes or enhancer-hijacking events. Of 13 fusion genes or enhancer hijacking events in 48 thyroid adenocarcinomas, at least 4 (31%) were caused by chromoplexy, with a further 4 (31%) part of complexes containing chromoplexy footprints (Extended Figure 5A). These generated fusion genes involving RET (2 cases) and NTRK3 (1 case)56, and juxtaposition of the oncogene IGF2BP3 with regulatory elements from highly expressed genes (5 cases).
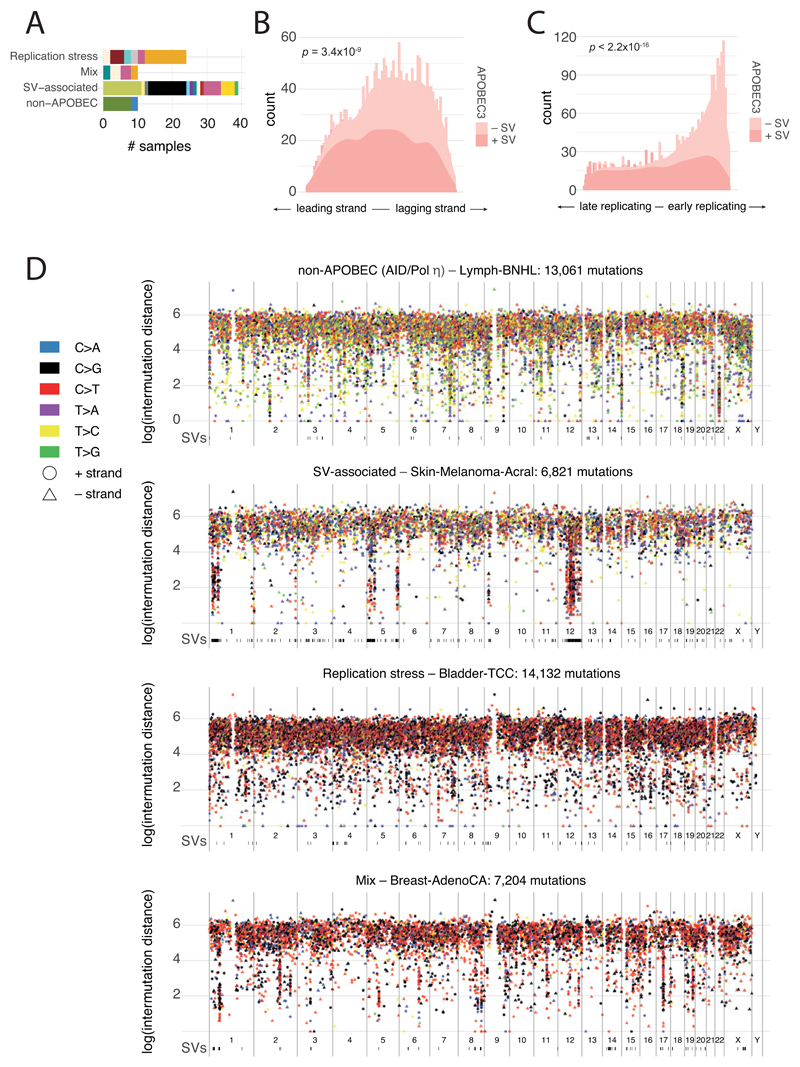
Kataegis events were seen in 60.5% of all cancers, with particularly high abundance in lung squamous cell carcinoma, bladder cancer, acral melanoma and sarcomas (Figure 4A,B). Typically, kataegis comprises C>N mutations in TpC context, likely due to APOBEC activity49–51, although a T>N at TpT or CpT process attributed to error-prone polymerases has recently been described57. The APOBEC signature accounted for 81.7% of kataegis events and correlated positively with APOBEC3B expression levels, somatic SV burden and age at diagnosis (Supplementary Figure 5). 5.7% of kataegis events involved the T>N error-prone polymerase signature and 2.3% of events, most notably in sarcomas, showed cytidine deamination in an alternative GpC or CpC context.
Kataegis events were frequently associated with somatic SV breakpoints (Figure 4A, Supplementary Figure 6A), as previously described50,51. Deletions and complex rearrangements were most strongly associated with kataegis, while tandem duplications and other simple SV classes were only infrequently associated (Supplementary Figure 6B). The C[T>N]T-type kataegis was enriched near deletions, specifically those in the 10-25kbp range (Supplementary Figure 6C).
Samples with extreme kataegis burden (>30 foci) comprise four types of focal hypermutation (Extended Figure 6): (i) off-target somatic hypermutation and C[T>N]T foci in B-cell non-Hodgkin lymphoma and oesophageal adenocarcinomas, respectively; (ii) APOBEC kataegis associated with complex rearrangements, notably in sarcoma and melanoma; (iii) rearrangement-independent APOBEC kataegis on the lagging strand and in early-replicating regions, mainly in bladder and head and neck cancer; (iv) a mix of the previous two types. Kataegis only occasionally led to driver mutations (Supplementary Table 5).
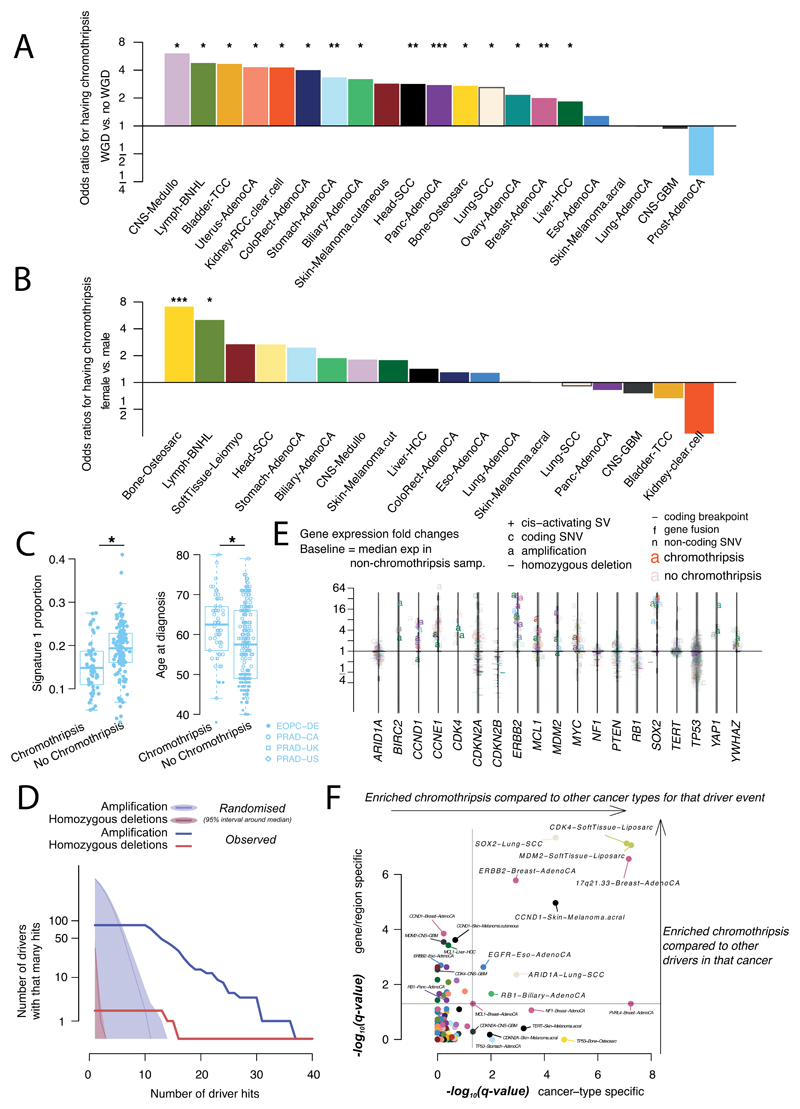
We identified chromothripsis in 587 samples (22.3%), most frequently amongst sarcoma, glioblastoma, lung squamous cell carcinoma, melanoma, and breast adenocarcinoma58. Chromothripsis increased with whole genome duplications in most cancer types (Extended Figure 7A), as previously shown in medulloblastoma59. The most recurrently associated driver was TP5352 (pan-cancer odds ratio=3.22; pan-cancer p=8.3x10-35; q<0.05 in breast lobular (OR=13), colorectal (OR=25), prostate (OR=2.6) and hepatocellular cancers (OR=3.9); Fisher-Boschloo tests). In two cancer types (osteosarcoma and B-cell lymphoma), females showed higher incidence of chromothripsis than males (Extended Figure 7B). In prostate cancer, we observed a higher incidence of chromothripsis in patients with late-onset than early-onset disease60 (Extended Figure 7C).
Chromothripsis regions coincided with 3.6% of all identified drivers in PCAWG and ~7% of copy number drivers (Figure 4D). These proportions are considerably enriched compared to expectation if selection were not acting on these events (Extended Figure 7D). The majority of coinciding driver events were amplifications (58%), followed by homozygous deletions (34%), and SVs within genes or promoter regions (8%). We frequently observed ≥2-fold increased or decreased expression of amplified or deleted drivers, respectively, when these loci were part of a chromothripsis event, compared to samples without chromothripsis (Extended Figure 7E).
Chromothripsis manifested in diverse patterns and frequencies across tumour types, which we categorised based on five characteristics (Figure 4A). In liposarcoma for example, chromothripsis events often involved multiple chromosomes, with universal MDM2 amplification61 and co-amplification of TERT in 4 of 19 cases (Figure 4D). In contrast, in glioblastoma, the events tended to affect a smaller region on a single chromosome, distant from the telomere, resulting in focal EGFR and MDM2 amplification, and CDKN2A loss. Acral melanomas frequently exhibited CCND1 amplification, and lung squamous cell carcinomas SOX2 amplifications. In both cases, these drivers were more frequently altered by chromothripsis compared to other drivers in the same cancer type, and to other cancer types for the same driver (Figure 4D, Extended Figure 7F). Finally, in chromophobe renal cell carcinoma, chromothripsis nearly always affected chromosome 5 (Supplementary Figure 7): these samples had breakpoints immediately adjacent to TERT, increasing TERT expression 80-fold on average over samples without rearrangements (p=0.0004; Mann-Whitney U test).
Timing clustered mutations in evolution
An unanswered question for clustered mutational processes is whether they occur early or late in cancer evolution. To address this, we used molecular clocks to define broad epochs in each tumour’s life history49,62. One transition point is between clonal and subclonal mutations: clonal mutations occurred before, and subclonal mutations after, emergence of the most recent common ancestor. In regions with copy number gains, molecular time can be further divided according to whether mutations preceded the copy number gain (and were themselves duplicated) or occurred after the gain (and therefore present on only one chromosomal copy)63.
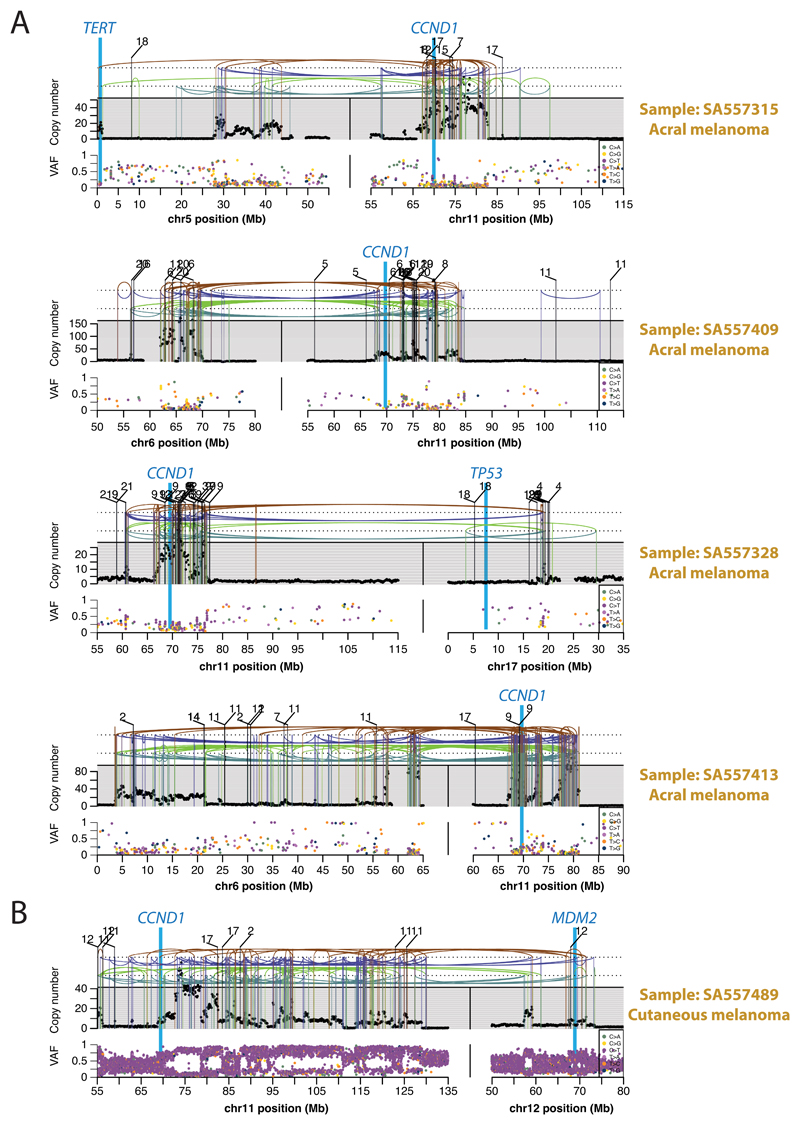
Chromothripsis tended to have greater relative odds of being clonal than subclonal, suggesting it occurs early in cancer evolution, especially in liposarcomas, prostate adenocarcinoma and squamous cell lung cancer, among others (Figure 5A). As previously reported, chromothripsis was especially common in melanomas64. We identified 89 separate chromothripsis events affecting 66 melanomas (61%), with 47/89 events affecting genes known to be recurrently altered in melanoma65 (Supplementary Table 6). Involvement of a region on chromosome 11 that includes the cell-cycle regulator CCND1 occurred in 21 cases (10/86 cutaneous, 11/21 acral or mucosal melanomas), typically combining chromothripsis with amplification (19/21 cases; Extended Figure 8). Co-involvement of other cancer genes in the same chromothripsis event was also frequent, including TERT (5 cases), CDKN2A (3 cases), TP53 (2 cases) and MYC (2 cases) (Figure 5B). In these co-amplifications, a chromothripsis event involving multiple chromosomes initiated the process, creating a derivative chromosome in which hundreds of fragments were stitched together in near-random order (Figure 5B). This derivative then rearranged further, leading to massive co-amplification of the multiple target oncogenes together with regions located nearby on the derivative chromosome.
Figure 5. Timing of clustered events in PCAWG.
(A) Extent and timing of chromothripsis, kataegis and chromoplexy across PCAWG. (top) Stacked bar-charts illustrate co-occurrence in samples. (middle) Relative odds of clustered events being clonal vs subclonal are plotted with bootstrapped 95% confidence intervals. Point estimates are highlighted when they do not overlap 1:1 odds. (bottom) Relative odds of the events being early vs late clonal are plotted as above. Sample sizes (number of patients) are shown across the top panel. (B) Three representative patients with melanoma and chromothripsis-induced amplification simultaneously affecting TERT and CCND1. The black points in the upper panel represent sequence coverage from individual genomic bins, with structural variants shown as coloured arcs (translocation in black, deletion in purple, duplication in brown, tail-to-tail inversion in cyan, head-to-head inversion in green). The lower panel shows the variant allele fraction (VAF) of somatic point mutations.
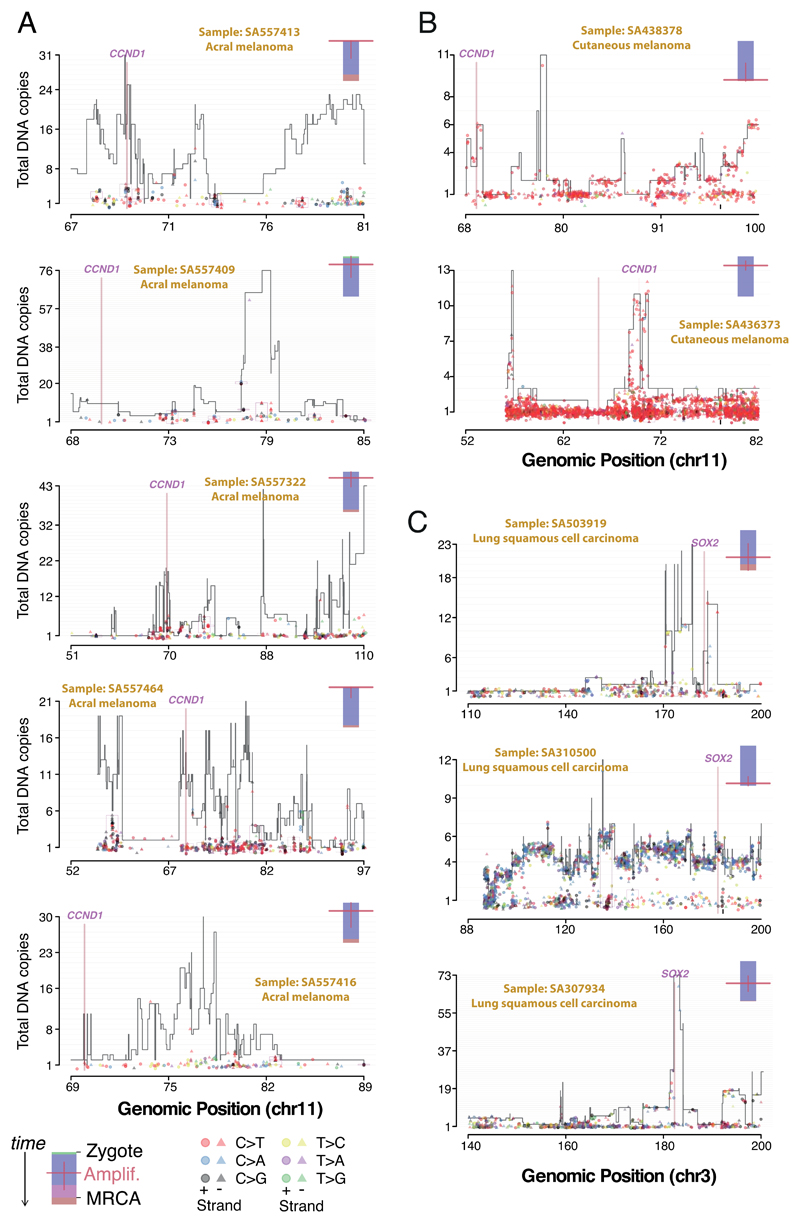
In these cases of amplified chromothripsis, we can use the inferred number of copies bearing each SNV to time the amplification process. SNVs present on the chromosome before amplification will themselves be amplified, and therefore reported in a high fraction of sequence reads (Figure 5B; Extended Figure 8). In contrast, late SNVs that occur after the amplification has concluded, will be present on only one chromosome copy out of many, and thus have low variant allele fraction. Regions of CCND1 amplification had few, sometimes zero, mutations at high variant allele fraction in acral melanomas, contrasting with later CCND1 amplifications in cutaneous melanomas (Figure 5B; Extended Figure 9A,B). Thus, both chromothripsis and the subsequent amplification generally occurred very early during the evolution of acral melanoma. By comparison, in lung squamous cell carcinomas, similar patterns of chromothripsis followed by SOX2 amplification are characterised by many amplified SNVs, suggesting a later event in the evolution of these cancers (Extended Figure 9C).
Interestingly, in cancer types where mutational load was sufficiently high, we could detect a larger than expected number of SNVs on an intermediate number of DNA copies, suggesting that they appeared during the amplification process (Supplementary Figure 8).
Germline effects on somatic mutations
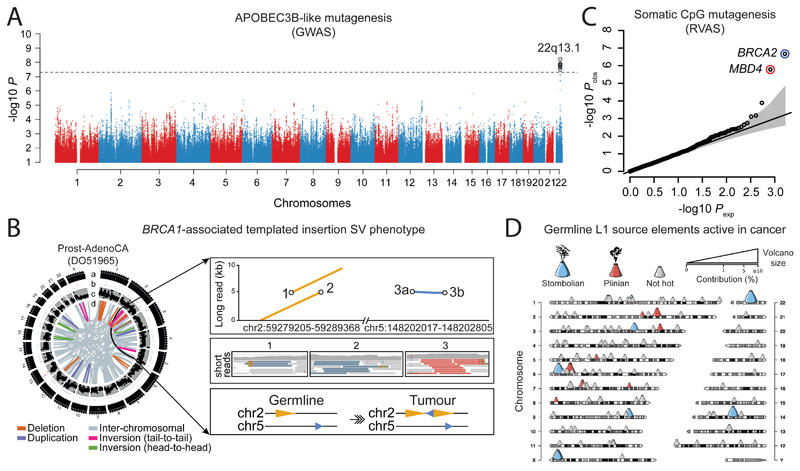
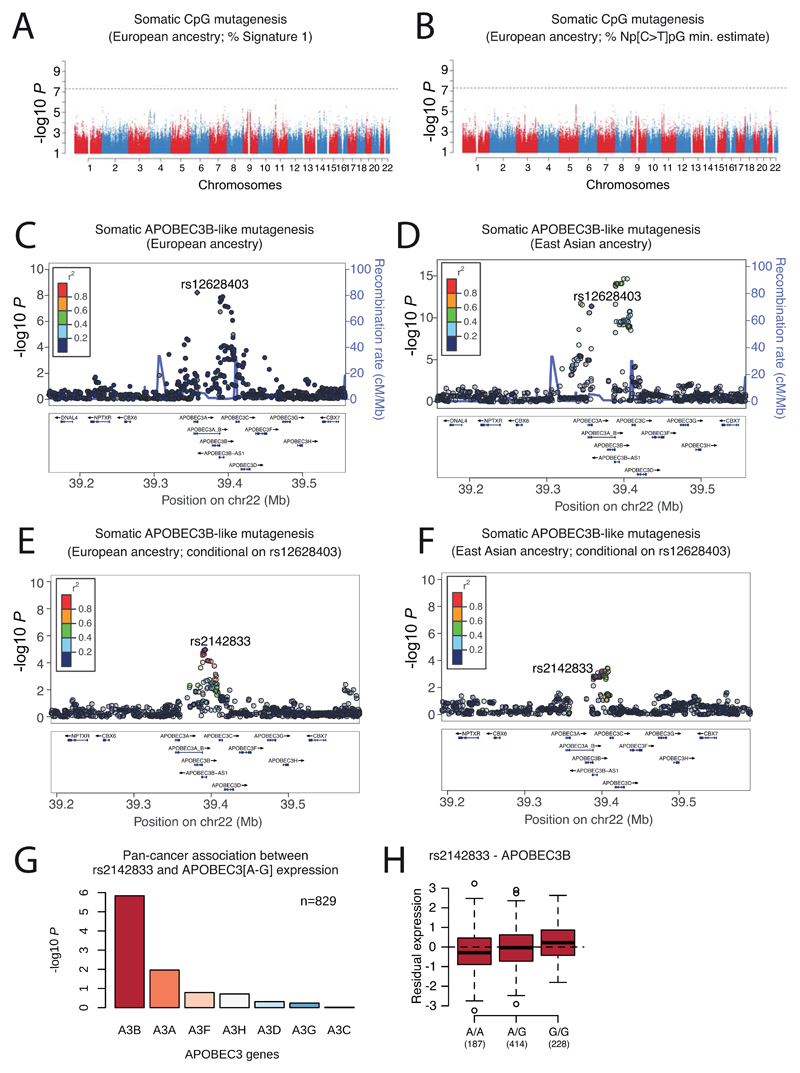
We integrated the set of 88 million germline genetic variant calls with somatic mutations in PCAWG, to study germline determinants of somatic mutation rates and patterns. First, we performed a genome-wide association study (GWAS) of somatic mutational processes with common germline variants (minor allele frequency (MAF) >5%) in individuals with inferred European ancestry. An independent GWAS was performed in East Asian individuals from Asian cancer genome projects. We focused on two prevalent endogenous mutational processes: spontaneous deamination of 5methyl-C at CpG dinucleotides66 (signature 1) and activity of the APOBEC3 family of cytidine deaminases67 (signatures 2 and 13). No locus reached genome-wide significance (p<5x10-8) for signature 1 (Extended Figure 10A,B). However, a locus at 22q13.1 predicted APOBEC3B-like mutagenesis at the pan-cancer level68 (Figure 6A). The strongest signal at 22q13.1 was driven by rs12628403, and the minor (non-reference) allele was protective against APOBEC3B-like mutagenesis (β=-0.43, p=5.6x10-9, MAF=8.2%, n=1,201 donors; Extended Figure 10C). This variant tags a common ~30kb germline SV that deletes the APOBEC3B coding sequence and fuses the APOBEC3B 3’-UTR with the coding sequence of APOBEC3A. The deletion is known to increase breast cancer risk and APOBEC mutagenesis in breast cancer genomes69,70. Here, we found that rs12628403 reduces APOBEC3B-like mutagenesis specifically in cancer types with low levels of APOBEC mutagenesis (βlow=-0.50, plow=1x10-8; βhigh=+0.17, phigh=0.2), and increases APOBEC3A-like mutagenesis in cancer types with high levels of APOBEC mutagenesis (βhigh=+0.44, phigh=8x10-4; βlow=-0.21, plow=0.02). Moreover, we identified a second, novel locus at 22q13.1 that associated with APOBEC3B-like mutagenesis across cancer types (rs2142833, β=+0.23, p=1.3x10-8). We independently validated the association between both loci and APOBEC3B-like mutagenesis using East Asian individuals from Asian cancer genome projects (βrs12628403=+0.57, prs12628403=4.2x10-12; βrs2142833=+0.58, prs2142833=8x10-15; Extended Figure 10D). Of note, in a conditional analysis that accounted for rs12628403, rs2142833 and rs12628403 are inherited independently in Europeans (r2<0.1), while rs2142833 remained significantly associated with APOBEC3B-like mutagenesis in Europeans (βEUR=+0.17, pEUR=3x10-5) and East Asians (βASN=+0.25, pASN=2x10-3) (Extended Figure 10E,F). Analysis of donor-matched expression data further suggests that rs2142833 is a cis-eQTL for APOBEC3B at the pan-cancer level (β=+0.19, p=2x10-6; Extended Figure 10G-H), consistent with cis-eQTL studies in normal cells71,72.
Figure 6. Germline determinants of the somatic mutation landscape.
(A) Association between common (MAF>5%) germline variants and somatic APOBEC3B-like mutagenesis in individuals of European ancestry (n=1201). Two-sided hypothesis testing was performed with PLINK v1.9. To mitigate multiple hypothesis-testing, the significance threshold was set at genome-wide significance (p<5x10-8). (B) Templated insertion SVs in a BRCA1-associated prostate cancer. Left panel: chromosome bands (a); SVs ≤10Mbp (b); 1kb read-depth from CN 0-6 (c); inter- and intra-chromosomal SVs (>10Mbp) (d). Right panel: complex somatic SV composed of a 2.2 kb tandem duplication on chr2 together with a 232 bp inverted templated insertion SV that is derived from chr5 and inserted in-between the tandem duplication (bottom panel). Consensus sequence alignment of locally assembled ONT long-reads to chrs 2 and 5 of the human reference genome (top panel). Breakpoints are circled and marked as 1 (beginning of tandem duplication), 2 (end of tandem duplication), and 3 (inverted templated insertion). For each breakpoint, the middle panel shows Illumina short reads at SV breakpoints. (C) Association between rare germline PTVs (MAF<0.5%) and somatic CpG mutagenesis (approx. with Signature 1) in individuals of European ancestry (n=1201). Genes highlighted in blue/red were associated with lower/higher somatic mutation rates. Two-sided hypothesis testing was performed using linear regression models with sex, age at diagnosis, and ICGC project as variables. To mitigate multiple hypothesis-testing, the significance threshold was set at exome-wide significance (p<2.5x10-6). The black line represents the identity line which would be followed if the observed p values followed the null expectation, with shaded area showing 95% confidence intervals. (D) Catalogue of polymorphic germline L1 source elements active in cancer. Chromosomal map shows germline source L1 elements as volcano symbols. Each volcano is colour-coded according to the type of source L1 activity. The contribution of each source locus (expressed as percentage) to the total number of transductions identified in PCAWG tumours is represented in a size gradient, with top contributing elements exhibiting larger sizes.
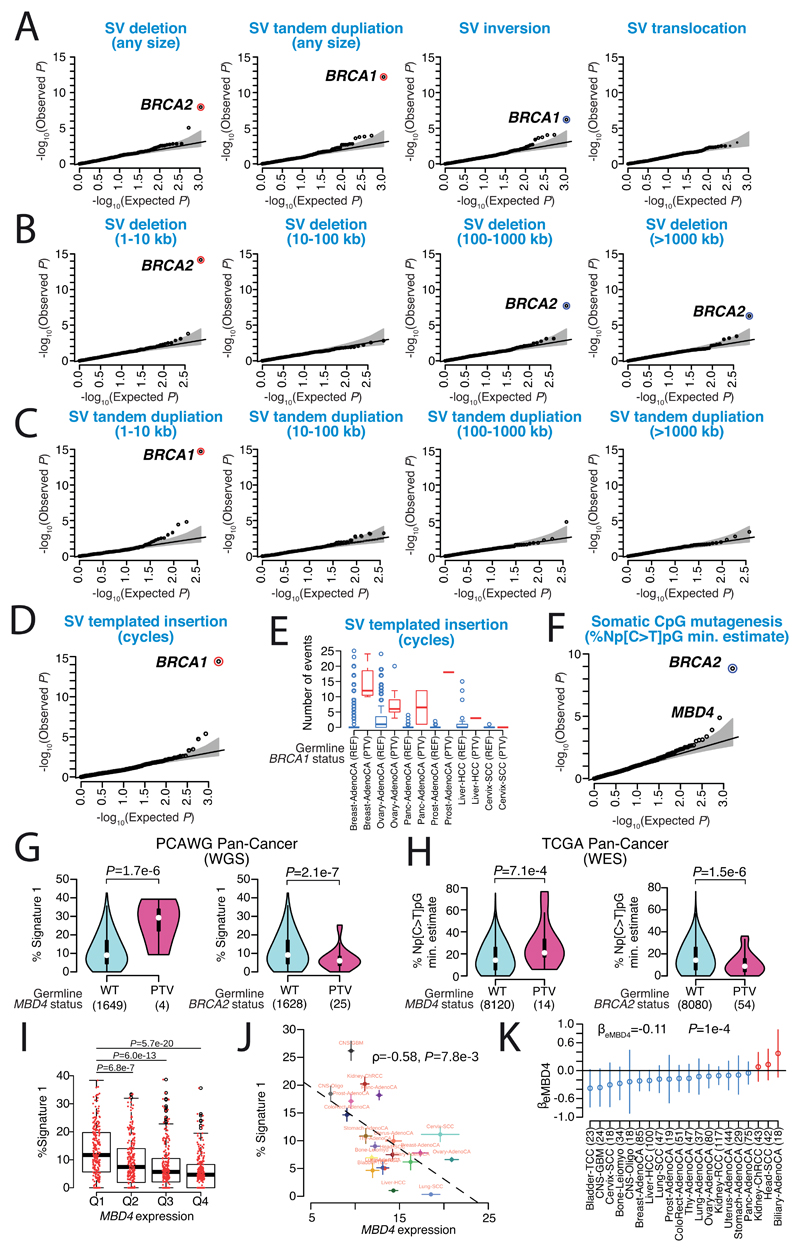
Second, we performed a rare variant association study (RVAS) (MAF<0.5%) to investigate the relationship between germline protein-truncating variants (PTVs) and somatic DNA rearrangements in individuals with European ancestry (Extended Figure 11A-C). Germline BRCA2 and BRCA1 PTVs associated with an increased burden of small (<10kb) somatic SV deletions (p=1x10-8) and tandem duplications (p=6x10-13), respectively, corroborating recent studies in breast and ovarian cancer30,73. In PCAWG data, this pattern extends to other tumour types as well, including adenocarcinomas of the prostate and pancreas6, typically in the setting of biallelic inactivation. In addition, tumours with high levels of small SV tandem duplications frequently exhibited a novel and distinct class of SVs termed ‘cycles of templated insertions’6. These complex SV events consist of DNA templates that are copied from across the genome, joined into one contiguous sequence, and inserted into a single derivative chromosome. We found a significant association between germline BRCA1 PTVs and templated insertions at the pan-cancer level (p=4x10-15; Extended Figure 11D,E). Whole genome long-read sequencing data generated for a BRCA1-deficient PCAWG prostate tumour verified the small tandem duplication and templated insertion SV phenotypes (Figure 6B). Virtually all (20/21) of BRCA1-associated tumours with a templated insertion SV phenotype displayed combined germline and somatic hits in the gene. Together, these data suggest that biallelic inactivation of BRCA1 is a driver of the templated insertion SV phenotype.
Third, rare variant association analysis revealed that patients with germline MBD4 PTVs exhibited increased rates of somatic C>T mutation rates at CpG dinucleotides (P<2.5x10-6; Figure 6C; Extended Figure 11F,G). Analysis of previously published TCGA WES samples (n=8,134) replicated the association between germline MBD4 PTVs and increased somatic CpG mutagenesis at the pan-cancer level (P=7.1x10-4; Extended Figure 11H). Moreover, gene expression profiling revealed a significant but modest correlation between MBD4 expression and somatic CpG mutation rates between and within PCAWG tumour types (Extended Figure 11I-K). MBD4 encodes a DNA repair gene that removes thymidines from T:G mismatches within methylated CpG sites74, a suggestive functionality for CpG mutational signatures in cancer.
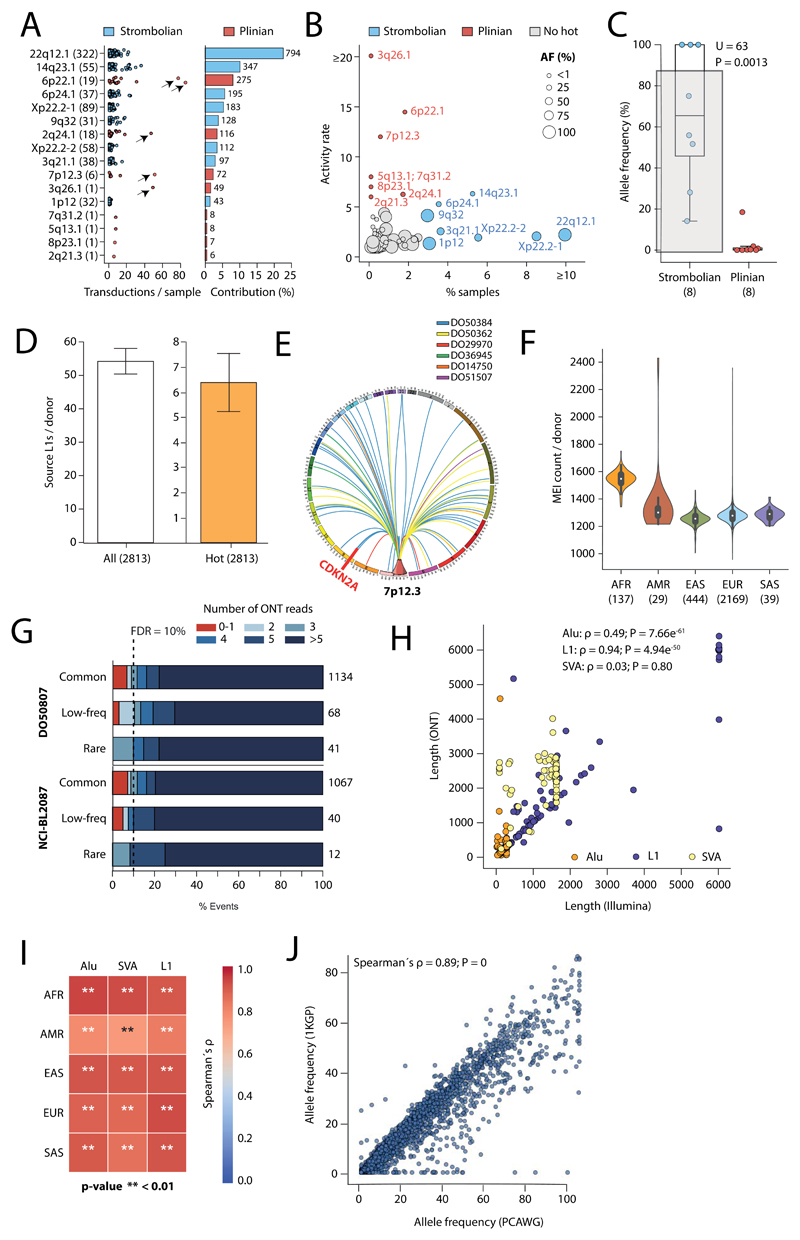
Fourth, we assessed LINE-1 (L1) elements that mediate somatic retrotransposition events75–77. We identified 114 germline source L1 elements capable of active somatic retrotransposition, including 70 that represent insertions with respect to the human reference genome (Figure 6D, Supplementary Table 7), and 53 that were tagged by SNPs in strong linkage disequilibrium (Supplementary Table 7). Only 16 germline L1 elements accounted for 67% (2,440/3,669) of all L1-mediated transductions10 detected in the PCAWG dataset (Extended Figure 12A). These 16 hot-L1 elements followed two broad patterns of somatic activity (8 of each), which we term Strombolian and Plinian in analogy to patterns of volcanic activity. Strombolian L1s are frequently active in cancer, but mediate only small to modest eruptions of somatic L1 activity in cancer samples (Extended Figure 12B). In contrast, Plinian L1s are more rarely seen, but display aggressive somatic activity. Whereas Strombolian elements are typically relatively common (MAF>2%) and sometimes even fixed in the human population, all Plinian elements were infrequent (MAF≤2%) in PCAWG donors (Extended Figure 12C; p=0.001; Mann-Whitney U test). This dichotomous pattern of activity and allele frequency may reflect differences in age and selective pressures, with Plinian elements potentially inserted into the human germline more recently. PCAWG donors bear on average between 50-60 L1 source elements and 5-7 elements with hot activity (Extended Figure 12D), but only 38% (1075/2814) of PCAWG donors carry ≥1 Plinian element. Some L1 germline source loci caused somatic loss of tumour suppressor genes (Extended Figure 12E). Many are restricted to individual continental population ancestries (Extended Figure 12F-J).
Replicative immortality
One of the hallmarks of cancer is its ability to evade cellular senescence21. Normal somatic cells typically have finite cell division potential, with telomere attrition one mechanism to limit numbers of mitoses78. Cancers enlist multiple strategies to achieve replicative immortality. Over-expression of the telomerase gene, TERT, which maintains telomere lengths, is especially prevalent. This can be achieved via point mutations in the promoter that lead to de novo transcription factor binding34,37; hitching TERT to highly active regulatory elements elsewhere in the genome46,79; insertions of viral enhancers upstream of the gene80,81; and increased dosage through chromosomal amplification, as we have seen in melanoma (Figure 5B). In addition, there is an ‘alternative lengthening of telomeres’ (ALT) pathway, in which telomeres are lengthened through homologous recombination, mediated by loss-of-function mutations in the ATRX and DAXX genes82.
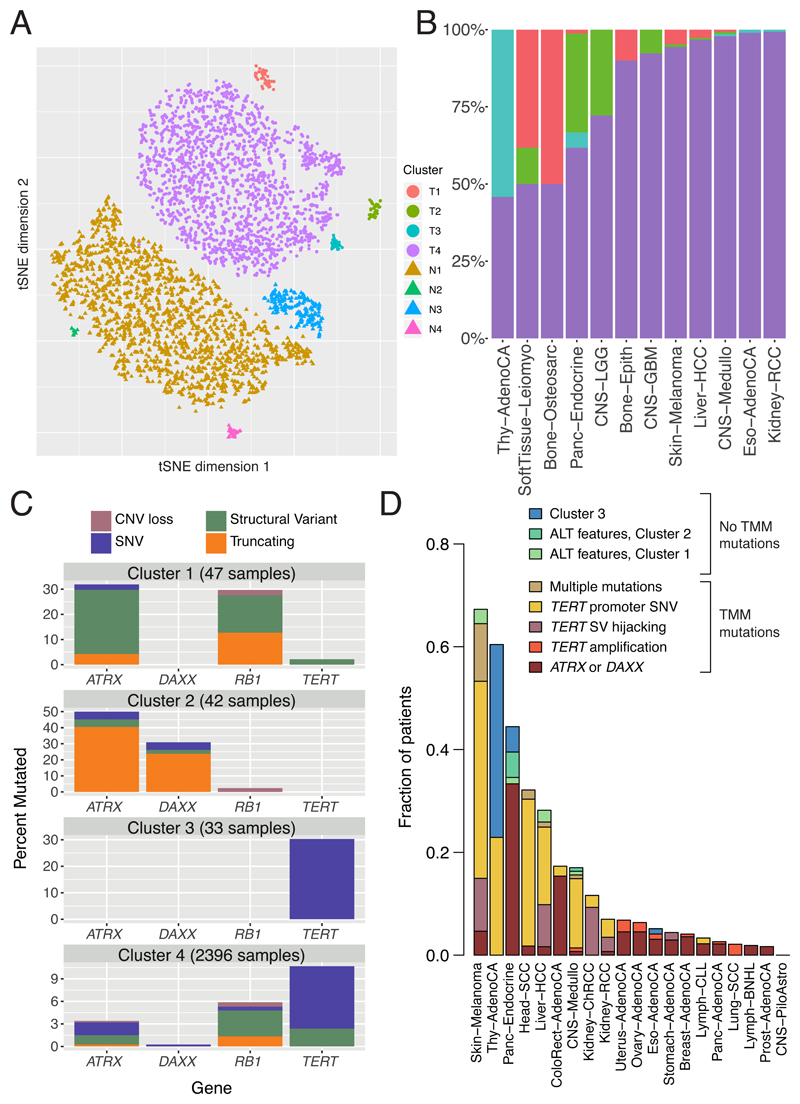
As reported in a companion paper, 16% of tumours in the PCAWG dataset exhibited somatic mutations in at least one of ATRX, DAXX and TERT83. TERT alterations were detected in 270 samples, whereas 128 tumours had alterations in ATRX or DAXX, of which 71 were protein-truncating. In the companion paper, which focused on describing patterns of ALT and TERT-mediated telomere maintenance83, twelve features of telomeric sequence were measured on the PCAWG cohort. These included counts of nine variants of the core hexameric sequence, the number of ectopic telomere-like insertions within the genome, the number of genomic breakpoints, and telomere length as a ratio between tumour and normal. Here we used the twelve features to overview telomere integrity across all tumours in the PCAWG dataset.
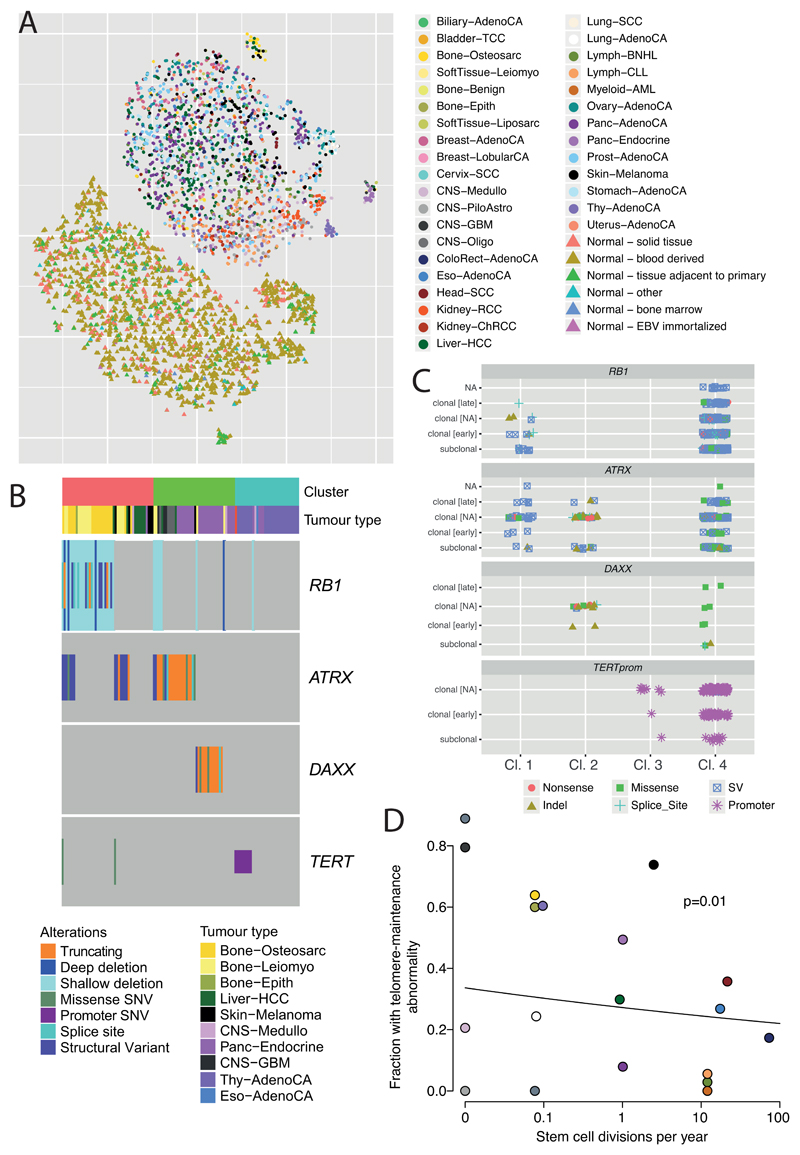
Based on these twelve features, tumour samples formed four distinct sub-clusters (Figure 7A, Extended Figure 13A), suggesting that telomere maintenance mechanisms are more diverse than the well-established TERT/ALT dichotomy. Clusters C1 (47 tumours) and C2 (42 tumours) were enriched for traits of the ALT pathway, having longer telomeres, more genomic breakpoints, more ectopic telomere insertions, and variant telomere sequence motifs (Supplementary Figure 9). C1 and C2 were distinguished from one another by the latter having striking elevation in the number of TTCGGG and TGAGGG variant motifs among the telomeric hexamers. Thyroid adenocarcinomas were strikingly enriched among C3 samples (26/33 C3 samples; p<10-16); the C1 cluster (ALT subtype 1) was common among sarcomas; and both pancreatic endocrine neoplasms and low-grade gliomas had a high proportion of samples in the C2 cluster (ALT subtype 2) (Figure 7B). Interestingly, some of the thyroid adenocarcinomas and pancreatic neuroendocrine tumours that cluster together (Cluster C3) had matched normals that also cluster together (Normal cluster N3, Extended Figure 13A), and which share common properties. For example, the GTAGGG repeat was overrepresented among samples in this group (Supplementary Figure 10).
Figure 7. Telomere sequence patterns across PCAWG.
(A) Scatter-plot showing clusters of telomere patterns identified across PCAWG by t-Distributed Stochastic Neighbour Embedding (tSNE), based on n=2,518 tumour samples and their matched normal samples. Axes have arbitrary dimension such that samples with similar telomere profiles are clustered together and samples with dissimilar telomere profiles are far apart with high probability. (B) Distribution of the four tumour-specific clusters of telomere patterns in selected tumour types from PCAWG. (C) Distribution of relevant driver mutations associated with alternative lengthening of telomere and normal telomere maintenance across the four clusters. (D) Distribution of telomere maintenance abnormalities across tumour types with more than 40 patients in PCAWG. Samples classified as tumour cluster 1-3 if they fall into a relevant cluster without mutations in TERT, ATRX or DAXX and have no ALT phenotype.
Somatic driver mutations were also unevenly distributed across the four clusters (Figure 7C). C1 tumours were enriched for RB1 mutations or structural variants (p=3x10-5), as well as frequent structural variants affecting ATRX (p=6x10-14), but not DAXX. RB1 and ATRX mutations were largely mutually exclusive (Extended Figure 13B). In contrast, C2 tumours were enriched for somatic point mutations in ATRX and DAXX (p=6x10-5), but not RB1. The enrichment of RB1 mutations in C1 remained significant when only leiomyosarcomas and osteosarcomas were considered, confirming that this enrichment is not merely a consequence of the different distribution of tumour types across clusters. C3 samples had frequent TERT promoter mutations (30%; p=2x10-6).
The predominance of RB1 mutations in C1 was striking. Nearly a third of the samples in C1 contained an RB1 alteration, evenly distributed across truncating SNVs, SVs and shallow deletions (Extended Figure 13C). Previous work has shown that RB1 mutations are associated with long telomeres in the absence of TERT mutations and ATRX inactivation84, and mouse models have revealed that knock-out of Rb-family proteins causes elongated telomeres85. The association with the C1 cluster here suggests that RB1 mutations can represent another route to activating the ALT pathway, with subtly different properties of telomeric sequence compared to inactivating DAXX, which fall almost exclusively in cluster C2.
Tumour types with the highest rates of abnormal telomere maintenance mechanisms often originate in tissues that have low endogenous replicative activity (Figure 7D). In support of this, we found an inverse correlation between previously estimated rates of stem cell division across tissues86 and the frequency of telomere maintenance abnormalities (p=0.01, Poisson regression; Extended Figure 13D). This suggests that restriction of telomere maintenance is a critical tumour suppression mechanism, particularly in tissues with low steady-state cellular proliferation, in which a clone must overcome this constraint to achieve replicative immortality.
Conclusions and future perspectives
The resource reported in this paper and its companion papers has yielded insights into the nature and timing of the many mutational processes that shape large and small-scale somatic variation in the cancer genome; the patterns of selection acting on these variations; the widespread impact of somatic variants on transcription; the complementary roles of coding and non-coding genome, for both germline and somatic mutations; the ubiquity of intratumoral heterogeneity; and the distinctive evolutionary trajectory of each cancer type. Many of these insights can only be obtained from an integrated analysis of all classes of somatic mutation on a whole genome scale, and would not be accessible with, for example, targeted exome sequencing.
The promise of precision medicine is to match patients to targeted therapies using genomics. A major barrier to its evidence-based implementation is the daunting heterogeneity of cancer chronicled in these pages, from tumour type to tumour type, from patient to patient, from clone to clone and from cell to cell. Building meaningful clinical predictors from genomic data can be achieved, but will require knowledge banks comprising tens of thousands of patients with comprehensive clinical characterisation87. Since these sample sizes will be too large for any single funding agency, pharmaceutical company or health system, international collaboration and data sharing will be required. The next phase of ICGC, ICGC-ARGO (https://icgc-argo.org/), will bring the cancer genomics community together with healthcare providers, pharma, data science and clinical trials groups to build comprehensive knowledge banks of clinical outcome and treatment data from patients with a wide variety of cancers, matched with detailed molecular profiling.
Extending the story begun by TCGA, ICGC and other cancer genomics projects, PCAWG has brought us closer to a comprehensive narrative of the causal biological changes that drive cancer phenotypes. We must now translate this knowledge into sustainable, meaningful clinical impacts.
Methods
Samples
We compiled an inventory of matched tumour/normal whole cancer genomes in the ICGC Data Coordinating Centre. Most samples came from treatment-naïve, primary cancers, but there were a small number of donors with multiple samples of primary, metastatic and/or recurrent tumours. Our inclusion criteria were: (i) matched tumour and normal specimen pair; (ii) a minimal set of clinical fields; and (iii) characterisation of tumour and normal whole genomes using Illumina HiSeq paired-end sequencing reads.
We collected genome data from 2,834 donors, representing all ICGC and TCGA donors that met these criteria at the time of the final data freeze in autumn 2014 (Extended Table 1). After quality assurance (Supplementary Methods S2.5), data from 176 donors were excluded as unusable, 75 had minor issues that could impact some analyses (grey-listed donors), and 2,583 had data of optimal quality (white-listed donors; Supplementary Table 1). Across the 2,658 white- and grey-listed donors, there were whole genome sequences from 2,605 primary tumours and 173 metastases or local recurrences. Matching normal samples were obtained from blood (2,064 donors), tissue adjacent to the primary (87 donors), or distant sites (507 donors). Whole genome sequencing data were available on tumour and normal DNA for the entire cohort. The mean read coverage was 39x for normal samples, while tumours had a bimodal coverage distribution with modes at 38x and 60x (Supplementary Figure 1). The majority of specimens (65.3%) were sequenced using 101 bp paired-end reads. An additional 28% were sequenced with 100 bp paired-end reads. Of the remaining specimens, 4.7% were sequenced with read lengths longer than 101 bp, and 1.9% with read lengths shorter than 100 bp. The distribution of read lengths by tumour cohort is shown in Supplementary Figure 11. Median read length for WGS paired end reads was 101 bp (mean=106.2, SD=16.7; min-max=50-151). RNA-sequencing data was collected and re-analysed centrally for 1,222 donors, including 1,178 primary tumours, 67 metastases or local recurrences, and 153 matched normal tissue samples adjacent to the primary tumour.
Demographically, the cohort included 1,469 males (55%) and 1,189 females (45%), with a mean age of 56 years (range, 1-90 years) (Supplementary Table 1). Using population ancestry-differentiated single nucleotide polymorphisms (SNPs), the ancestry distribution was heavily weighted towards donors of European descent (77% of total) followed by East Asians (16%), as expected for large contributions from European, North American and Australian projects (Supplementary Table 1).
We consolidated histopathology descriptions of the tumour samples, using the ICD-0-3 tumour site controlled vocabulary93. Overall, the PCAWG data set comprises 38 distinct tumour types (Extended Table 1; Supplementary Table 1). While the most common tumour types are included in the dataset, their distribution does not match the relative population incidences, largely due to differences among contributing ICGC/TCGA groups in numbers sequenced.
Uniform processing and somatic variant calling
In order to generate a consistent set of somatic mutation calls that could be used for cross-tumour analyses, we analysed all 6,835 samples using a uniform set of algorithms for alignment, variant calling, and quality control (Extended Figure 1; Supplementary Figure 2; Supplementary Table 3; Supplementary Methods S2). We used the BWA-MEM algorithm94 to align each tumour and normal sample to human reference build hs37d5 (as used in the 1000 Genomes Project95). Somatic mutations were identified in the aligned data using three established pipelines, run independently on each tumour/normal pair. Each of the three pipelines, labelled “Sanger”96–99, “EMBL/DKFZ”100,101 and “Broad”102–105 after the computational biology groups that created or assembled them, consisted of multiple software packages for calling somatic single nucleotide variations (SNVs), small insertions and deletions (indels), copy number alterations (CNAs), and somatic structural variants (SVs; with intrachromosomal SVs defined as those >100bp). Two additional variant callers106,107 were included to further improve accuracy across a broad range of clonal and subclonal mutations. We tested different merging strategies using validation data, choosing the optimal method for each variant type to generate a final consensus set of mutation calls (Supplementary Methods S2.4).
Somatic retrotransposition events, including Alu and LINE/L1 insertions75, L1-mediated transductions76 and pseudogene formation108, were called using a dedicated pipeline76. We removed these retrotransposition events from the somatic SV call-set. Mitochondrial DNA mutations were called using a published algorithm109. RNA-Sequencing data were uniformly processed to quantify normalised gene-level expression, splicing variation and allele-specific expression, and to identify fusion transcripts, alternative promoter usage and sites of RNA editing110.
Integration, phasing, and validation of germline variant call-sets
Calls of common (≥1% frequency in PCAWG) and rare (<1%) germline variants including single nucleotide polymorphisms (SNPs), indels, structural variants and mobile element insertions were generated using a population-scale genetic polymorphism detection approach95,111. The uniform germline data processing workflow comprised variant identification using six different variant callers100,112,113, and orchestrated via the Butler workflow system114.
We performed call-set benchmarking, merging, variant genotyping and statistical haplotype-block phasing95 (Supplementary Methods S3.4). Using this strategy, we identified 80.1 million germline SNPs, 5.9 million germline indels, 1.8 million multi-allelic short (<50bp-sized) germline variants, as well as germline SVs ≥50bp in size including 29,492 biallelic deletions and 27,254 mobile element insertions (MEIs) (Supplementary Table 2). We statistically phased this germline variant set utilising 1000 Genomes Project95 haplotypes as a reference panel, yielding an N50 phased block length of 265 kb based on haploid chromosomes from donor-matched tumour genomes. Precision estimates for germline SNVs and indels were >99% for the phased merged call-set, and sensitivity estimates ranged from 92% to 98%.
Core alignment and variant calling by cloud computing
The requirement to uniformly realign and call variants on nearly 5,800 whole genomes (tumour plus normal) presented significant computational challenges, and raised ethical issues due to the use of data from different jurisdictions (Box 1). To process the data, we adopted a cloud-computing architecture26 in which the alignment and variant calling was spread across 13 data centres in three continents, representing a mixture of commercial, infrastructure-as-a-service, academic cloud compute, and traditional academic high-performance computer clusters (Supplementary Table 3). Altogether, the effort used 10 million CPU core-hours.
To generate reproducible variant-calling across the 13 data centres, we built the core pipelines into Docker containers28, in which the workflow description, required code and all associated dependencies were packaged together in stand-alone packages. These heavily tested, extensively validated workflows are available for download (Box 1).
Validation, benchmarking and merging of somatic variant calls
In order to evaluate the performance of each of the mutation-calling pipelines and determine an integration strategy, we performed a large-scale deep sequencing validation experiment (Supplementary Notes 1). We selected a pilot set of 63 representative tumour/normal pairs, on which we ran the three core pipelines, together with a set of 10 additional somatic variant-calling pipelines contributed by members of the SNV Calling Working Group. Sufficient DNA remained for 50 of the 63 cases for validation, which was performed by hybridisation of tumour and matched normal DNA to a custom RNA bait-set, followed by deep sequencing, as described previously29. Although performed using the same sequencing chemistry as the original whole genome sequencing, the considerably greater depth achieved in the validation experiment enabled accurate assessment of sensitivity and precision of variant calls. Variant calls in repeat-masked regions were not tested due to the challenge of designing reliable validation probes in these areas.
The three core pipelines had individual estimates of sensitivity of 80-90% to detect a true somatic SNV called by any of the 13 pipelines; with >95% of SNV calls made by each of the core pipelines being genuine somatic variants (Figure 1A). For indels, a more challenging class of variants to identify in short read sequencing data, the three core algorithms had individual sensitivity estimates in the range 40-50%, with precision 70-95% (Figure 1B). Validation of SV calls is inherently more difficult because methods based on PCR or hybridisation to RNA baits often fails to isolate DNA spanning the breakpoint. To assess accuracy of SV calls, we therefore used the property that an SV must either generate a copy number change or be balanced, whereas artefactual calls will not respect this property. For individual SV callers, we estimated precision to be in the range 80-95% for samples in the pilot-63 dataset.
Next, we examined multiple methods for merging calls made by several algorithms into a single definitive call-set to be used for downstream analysis. The final consensus calls for SNVs were based on a simple approach that required two or more methods to agree on a call. For indels, because methods were less concordant, we used stacked logistic regression115,116 to integrate the calls. The merged SV set includes all calls made by two or more of the four primary SV callers100,104,117,118. Consensus CNA calls were obtained by joining the outputs of six individual CNA callers with SV consensus breakpoints to obtain base-pair resolution CNAs (Supplementary Methods 2.4.3). Consensus purity and ploidy were derived, and a multi-tier system was developed for consensus copy number calls (Supplementary Methods 2.4.3, and described in detail elsewhere63).
Overall, the sensitivity and precision of the consensus somatic variant calls were 95% (CI90%: 88-98%) and 95% (CI90%: 71-99%) respectively for SNVs (Extended Figure 2). For somatic indels, sensitivity and precision were 60% (34-72%) and 91% (73-96%) respectively. Regarding SVs, we estimate the sensitivity of the merging algorithm to be 90% for true calls generated by any one caller; precision was estimated as 97.5%. That is, 97.5% of SVs in the merged SV call-set have an associated copy number change or balanced partner rearrangement. The improvement in calling accuracy from combining different callers was most noticeable in variants having low variant allele fractions, which are likely to originate in subclonal populations of the tumour (Figure 1C-D). There remains much work to be done in improving indel callers; we still lack sensitivity for calling even fully clonal complex indels from short-read sequencing data.
Extended Data
Extended Figure 1. Flow-chart showing key steps in the analysis of PCAWG genomes.
Extended Figure 2. Distribution of accuracy estimates across algorithms and samples from validation data.
(A) F1 accuracy, precision and sensitivity estimates for somatic SNVs across the core algorithms and different approaches to merging the call sets. The box plots demarcate the interquartile range and median of estimates across the n=50 samples in the validation dataset. (B) F1 accuracy, precision and sensitivity estimates for somatic indels (n=50 samples). SVM, support vector machine; union, calls made by all variant callers; intersect2, calls made by any combination of two variant callers; intersect3, calls made by any three variant callers.
Extended Figure 3. Distribution of numbers of somatic mutations of different classes across tumour types.
The y axis is on a log scale. Plotted are the 2,583 donors with the highest quality metrics (white-listed donors). SNVs, single nucleotide variants (substitutions); Indels, insertions or deletions <100 base pairs in size; SVs, structural variants; Retrotranspositions, counts of somatic retrotransposon insertions, transductions and somatic pseudogene insertions combined.
Extended Figure 4. Patients with no detected driver mutations in PCAWG.
(A) Number (red) of patients without detected driver mutations distributed across the different tumour types studied. (B) Estimated sensitivity for detecting somatic point mutations genome-wide across tumour types (total sample size: n=2,583 patients). Each point represents the estimate for a single patient, layered on violin plots representing the estimated density distribution of sensitivity values for that tumour type (width proportional to density). (C) SETD2 expression levels across different medulloblastoma subtypes. Points represent individual patients, coloured by whether the gene exhibited focal copy number loss, truncating point mutation, or was wild-type. The coloured areas are violin plots representing the estimated density distribution of expression values for that medulloblastoma subtype (width proportional to density).
Extended Figure 5. Examples of clustered mutational processes.
(A) Chromoplexy example in a thyroid adenocarcinoma. Genes at the breakpoints are schematically depicted in their normal genomic context and again in the reconstructed derivative chromosomes below. (B) Distinct kataegis signatures in the genome of a pancreatic adenocarcinoma sample. SVs and their classification are shown above the main rainfall plot (Tra, translocation; Del, deletion; Dup, duplication; t2tInv, tail-to-tail inversion; h2hInv, head-to-head inversion), as well as the total and minor allele copy number. Zooming into three foci on chr1, chr8 and chr12, respectively, exemplifies distinct manifestations of kataegis: (left) a novel process similar to Signature 17 with T>N mutations at CT or TT dinucleotides; (middle) the prototypical APOBEC3A/B type with C>T (Signature 2) and/or C>G/A (Signature 13) substitutions at TpC; (right) an alternative cytidine deaminase(s) with a preference for substitutions at C/GpC. Most of the SNVs in each of these foci can be phased to the same allele and no evidence of anti-phasing is observed. (C) Example of a chromothripsis event in a melanoma. The black points in the upper panel represent copy number estimates from individual genomic bins, with structural variants shown as coloured arcs (translocation in black, deletion in purple, duplication in brown, tail-to-tail inversion in cyan, head-to-head inversion in green), mostly demarcating copy number changes. The mate chromosomes are displayed above translocations. The lower panel shows the variant allele fraction (VAF) of somatic mutations distributed along the relevant chromosomal region.
Extended Figure 6. Patterns of intense kataegis.
(A) Bar plot showing the tumour type distribution (colour-coded as in Extended Figure 3) of samples in the top 5% of kataegis intensity in each of the four genome-wide patterns identified: non-APOBEC, replication stress, rearrangement-associated and combination of the latter two. (B,C) Distribution of leading/lagging strand (B) and replication timing bias (C) for rearrangement-(in)dependent APOBEC kataegis, based on n=2,583 tumours. P-values were derived using a two-sided Mann–Whittney U test. (D) Example rainfall plots for each of the four kataegis patterns identified.
Extended Figure 7. Association of chromothripsis with covariates and driver events.
(A) Odds ratios per cancer type of harbouring chromothripsis in whole genome duplicated vs. diploid samples (n=2,583 patients). Asterisks represent significance level (***: q<0.001; **: q<0.01; *:q<0.05). Two-sided hypothesis testing was performed using Fisher-Boschloo tests, corrected for multiple-hypothesis testing. (B) Same as A for female vs. male. (C) Proportion of mutations explained by single base substitution signature 1 and age at diagnosis in prostate cancer samples (n=210 patients) with or without chromothripsis (q<0.05). The early-onset prostate cancer project drives the signal and was sequenced at lower depth. For the box-and-whisker plots, the box denotes the interquartile range, with the median marked as a horizontal line. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Two-sided hypothesis testing was performed using Mann-Whitney U tests. (D) Counts of co-occurrence of chromothripsis with amplification (blue) and homozygous deletions (red) in driver regions: observed (thick line) vs. randomised (shaded area and thin line). The cumulative number of drivers hit is plotted as a function of the number of times those drivers are hit. (E) For each sample where chromothripsis coincided with a driver event in those genes, we show the gene expression fold change compared to the median expression of the gene in non-chromothripsis samples of the same cancer type, coloured by cancer type and shaped by the type of driver event. We show with added transparency the fold changes calculated the same way for samples with driver mutations hitting the same driver genes but no evidence for chromothripsis. Analysis is based on n=1,222 patients with RNA-sequencing data. (F) Enrichment of co-occurrence of chromothripsis with driver events. The x-axis indicates the association of chromothripsis with a driver in a given cancer type compared to its rate of association with that driver in all other cancer types. The y-axis show the association of chromothripsis with a driver in a given cancer type compared to its rate of association with all other drivers in that type. Exact binomial tests are used and p-values are corrected for multiple testing according to Benjamini and Hochberg.
Extended Figure 8. Further examples of chromothripsis-induced amplification targeting multiple cancer genes simultaneously in melanoma.
(A) Examples of amplifications that occurred early in melanoma development. The black points in the upper panel represent copy number estimates from individual genomic bins, with structural variants shown as coloured arcs (translocation in black, deletion in purple, duplication in brown, tail-to-tail inversion in cyan, head-to-head inversion in green), mostly demarcating copy number changes. The mate chromosomes are displayed above translocations. The lower panel shows the variant allele fraction (VAF) of SNVs distributed along the relevant chromosomal region. The paucity of somatic mutations at high variant allele fraction in the most heavily amplified regions indicates that these amplifications began very early in tumour evolution, before the lineage had had opportunity to acquire many SNVs. (B) Example of an amplification that occurred late in melanoma development. The large numbers of somatic mutations at high variant allele fraction in the most heavily amplified regions indicates that these amplifications began late in tumour evolution, after the lineage had already acquired many SNVs.
Extended Figure 9. Timing the amplifications following chromothripsis in molecular time – 10 selected cases.
(A) Copy number plot of chromothriptic regions categorised as “liposarc-like” in 5 acral melanomas showing CCND1 amplification. Segments indicate the copy number of the major allele. Points represent SNV multiplicities, i.e. the estimated number of copies carrying them, coloured by base change and shaped by strand. Small vertical arrows link SNVs to their corresponding copy number segment. Kataegis foci are shown within black boxes, and show typical strand-specificity (all triangles or all circles), similar multiplicities and base changes of signatures 2 and 13 (red and black). A coloured bar on the top right represents the molecular timing of the amplification (red bar; high is early, low is late) and is coloured by the fraction of total SNVs assigned to timing categories clonal[early], clonal[NA], clonal[late] and subclonal. (B) Same as A in 2 cutaneous melanomas, one shows an early amplification, the other a late one. (C) Same as A-B for 3 lung squamous cell carcinomas and late amplification of SOX2.
Extended Figure 10. Association between common germline variants and endogenous mutational processes.
Genome-wide association of somatic CpG mutagenesis in individuals of European ancestry (n=1,201 patients) based on mutational signature analysis (A) and NpCpG motif analysis (B). Two-sided hypothesis-testing was performed using PLINK v1.9. To mitigate multiple hypothesis-testing, the significance threshold was set at genome-wide significance (p<5x10-8). (C,D) Locuszoom plot for somatic APOBEC3B-like mutagenesis association results, LD, and recombination rates around the genome-wide significant 22q13.1 locus in individuals with European (C) and East Asian (D) ancestry (n=1,201 and 318 patients respectively). Locuszoom plot for somatic APOBEC3B-like mutagenesis association results around the 22q13.1 locus in individuals of European (E) and East Asian (F) ancestry after conditioning on rs12628403. (G,H) Association between rs2142833 and expression of APOBEC3 genes in PCAWG tumour samples (adjusted for sex, age at diagnosis, histology, and population structure in linear regression models with two-sided hypothesis testing not corrected for multiple tests). For the box-and-whisker plot, the box denotes the interquartile range, with the median marked as a horizontal line. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Outlier patients are shown as points.
Extended Figure 11. Association between rare germline PTVs in protein-coding genes and somatic mutational phenotypes.
For panels A-D and F, the data are based on two-sided rare-variant association testing across n=2,583 patients, with a stringent p-value threshold of P<2.5x10-6 used to mitigate multiple hypothesis testing (significant genes marked with coloured circles). Blue/red circles mark genes that decrease/increase somatic mutation rates. The black line represents the identity line which would be followed if the observed p values followed the null expectation, with the shaded area showing 95% confidence intervals. (A) QQ plots for proportion of somatic SV deletions, tandem duplications, inversions, and translocation in cancer genomes. (B) QQ plots for proportion of somatic SV deletions in cancer genomes stratified by four size groups (1-10 kb; 10-100 kb; 100-1000 kb; >1000 kb). (C) QQ plots for proportion of somatic SV tandem duplications in cancer genomes stratified by four size groups (1-10 kb; 10-100 kb; 100-1000 kb; >1000 kb). (D) QQ plot for presence/absence of somatic SV templated insertion (cycles) in cancer genomes. (E) Number of SV templated insertion cycles in PCAWG tumours with germline BRCA1 PTVs. Only histologies with at least one germline BRCA1 PTV carrier are shown (n=1,095 patients combined). The box denotes the interquartile range, with the median marked as a horizontal line. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Outlier patients are shown as points. (F) QQ plot for somatic CpG mutagenesis in cancer genomes based on NpCpG motif analysis. (G) Violin plots show estimated density of the proportion of somatic CpG mutations in PCAWG donors with germline MBD4 and BRCA2 PTVs. The box denotes the interquartile range, with the median marked as a white point. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Two-sided hypothesis testing, not corrected for multiple testing, was performed using linear regression models. (H) Replication of germline MBD4 and BRCA2 PTV associations with somatic CpG mutagenesis in TCGA WES donors. Violin plots show estimated density of the proportion of somatic CpG mutations in TCGA exomes with germline MBD4 and BRCA2 PTVs. The box denotes the interquartile range, with the median marked as a white point. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Two-sided hypothesis testing, not corrected for multiple testing, was performed using linear regression models. (I) Correlation between MBD4 expression and somatic CpG mutagenesis in primary solid PCAWG tumours. Hypothesis testing was two-sided and not corrected for multiple testing, using linear regression models. The box denotes the interquartile range, with the median marked as a horizontal line. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. (J) Points represent means across n=20 tumour types and error bars represent standard error of the mean. The dashed black line shows the fitted line to the data, estimated with linear regression models. Hypothesis testing was two-sided and not corrected for multiple testing, performed with Spearman’s rank correlation method. (K) MBD4 effect sizes (open circles) with 95% confidence intervals (error bars) for individual cancer types were estimated with linear regression analysis after (if available) accounting for sex, age at diagnosis (low/high), and ICGC project. Hypothesis testing was two-sided and not corrected for multiple testing.
Extended Figure 12. Germline mobile element insertion (MEI) callset.
(A) On the left, dots show the number of transductions promoted by each hot element in individual samples. Arrows highlight retrotransposition burst. On the right, the contribution of each hot locus is represented. The total number of transductions mediated by each source element is shown on the right side. (B) Source L1 activity rate (i.e., measured as the average number of transductions mediated by an element) versus the percentage of samples with retrotransposition activity in which the germline element is active. For visualization purposes, extreme points observed for a source L1 with an activity rate of 49 and for a L1 active in 31% of the samples are shown at “≥20” and “≥10”, respectively. (C) Contrasting allele frequencies for Strombolian and Plinian source loci (sample sizes shown under each axis label). The box denotes the interquartile range, with the median marked as a white point. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Hypothesis-testing was performed using two-sided Mann-Whitney tests without correction for multiple tests. (D) Number of active and hot source L1 elements per donor. The bar height represents the mean number of elements per donor and error bars the standard deviation. (E) Novel Plinian source element on 7p12.3 mediates 72 transductions amongst only six cancer samples. This includes a transduction that induces the deletion of the tumour suppressor gene CDKN2A. (F) Violin plots show estimated number of distinct germline MEI alleles per PCAWG donor. The box denotes the interquartile range, with the median marked as a white point. The whiskers extend as far as the range or 1.5x the interquartile range, whichever is less. Donors are grouped according to their genetic ancestry: AFR, African; AMR, Ad Mixed American; EAS, East Asian; EUR, European; SAS, South Asian. Sample sizes are shown under each axis label. (G) For each type of MEI (L1, Alu and SVA) identified both in PCAWG and in the 1000 Genomes Project (1KGP), the correlations between allele frequency estimates per ancestry derived from both projects are displayed in a blue (0) to a red (1) coloured gradient. Sample size was n=2,583 PCAWG patients. Two-sided hypothesis-testing was performed using Spearman’s rank correlation without correction for multiple tests. (H) Example displaying the correlation between PCAWG and 1KGP derived MEI allele frequencies in individuals with European ancestry (n=1201 patients in PCAWG). Two-sided hypothesis-testing was performed using Spearman’s rank correlation without correction for multiple tests. (I) Evaluation of TraFiC-mem false discovery rate on a liver hepatocellular carcinoma donor (DO50807) and a cell-line (NCI-BL2087) sequenced through single-molecule sequencing with MinION (Oxford Nanopore). For each allele frequency bin (common, >5%; low-freq, 1-5%; rare, <1%), the percentage of events supported by N long-reads is represented (N from 0-1 to more than 5). MEIs supported by at least two Nanopore reads were considered true positives (blue palette) and false positives (red), otherwise. The total number of germline MEIs per allele frequency bin is shown on the right side of the panels. (J) Correlation between predicted MEI lengths from Illumina and Nanopore data. Two-sided hypothesis testing was performed using Spearman’s rank correlation without correction for multiple testing.
Extended Figure 13. Different mechanisms of telomere lengthening in cancer.
(A) Scatter plot showing the four clusters of tumour-specific telomere patterns identified across PCAWG samples, together with the clusters of matched normal samples, generated by t-Distributed Stochastic Neighbour Embedding. Circles represent tumour samples and triangles represent matched normal samples. Points are coloured by tissue of origin. Data are based on n=2,518 tumour samples and their matched normal samples. (B) Patterns of co-mutation of the relevant driver mutations across individual patients. Columns in plot represent individual patients, coloured by type of abnormality observed. (C) Clonal [early] denotes clonal mutations occurring before duplications involving the relevant chromosome (including whole genome duplications); clonal [late] to clonal mutations occurring after such duplications; and clonal [NA] to mutations occurring when no duplication was observed. (D) Relationship between estimated number of stem cell divisions per year and rate of telomere maintenance abnormalities across tumour types. The analysis uses data on estimated rates of stem cell division per year across n=19 tissue types previously collated from the literature86. Tumour types are coloured according to the scheme shown in Extended Figure 3. Two-sided hypothesis testing was performed using likelihood ratio tests on Poisson regression models with no correction for multiple tests.
Extended Table 1. Overview of tumour types included in PCAWG project.
Med, Median; F, Female; M, Male; 10-90th, 10-90th centile; Adeno., Adenocarcinoma; Comb., Combined; SCC, squamous cell carcinoma; HCC, hepatocellular carcinoma; Ca. Carcinoma.
| Organ | Abbrevation | Included subtypes | Cases | Sex | Age | ||
|---|---|---|---|---|---|---|---|
| Neural Crest | Num. | F | M | Med. | 10th-90th | ||
| CNS | CNS-GBM | Glioblastoma | 41 | 13 | 28 | 60 | 43-72 |
| CNS | CNS-Medullo | Medulloblastoma and variants | 146 | 67 | 79 | 9 | 3-28 |
| CNS | CNS-Oligo | Oligodendroglioma | 18 | 9 | 9 | 41 | 21-62 |
| CNS | CNS-PiloAstro | Pilocytic astrocytoma | 89 | 47 | 42 | 8 | 2-17 |
| Skin | Skin-Melanoma | Malignant melanoma | 107 | 38 | 69 | 57 | 37-78 |
| Endoderm | |||||||
| Biliary | Biliary-AdenoCA | Papillary cholangiocarcinoma | 34 | 15 | 19 | 64 | 53-76 |
| Bladder | Bladder-TCC | Transitional cell carcinoma | 23 | 8 | 15 | 65 | 52-80 |
| Colon/Rectum | ColoRect-AdenoCA | Adenocarcinoma; Mucinous adeno. | 60 | 30 | 30 | 67 | 46-81 |
| Oesophagus | Eso-AdenoCA | Adenocarcinoma | 98 | 14 | 84 | 70 | 56-79 |
| Liver | Liver-HCC | Hepatocellular carcinoma; Comb. HCC/cholangio | 317 | 89 | 228 | 67 | 50-78 |
| Lung | Lung-AdenoCA | Adenocarcinoma; Adenocarcinoma in situ | 38 | 20 | 18 | 66 | 47-77 |
| Lung | Lung-SCC | Squamous cell carcinoma; Basaloid SCC | 48 | 10 | 38 | 68 | 54-77 |
| Pancreas | Panc-AdenoCA | Adeno.; Acinar cell Ca.; Mucinous adeno. | 239 | 119 | 120 | 67 | 50-79 |
| Pancreas | Panc-Endocrine | Neuroendocrine carcinoma | 85 | 30 | 55 | 59 | 38-75 |
| Prostate | Prost-AdenoCA | Adenocarcinoma | 210 | 0 | 210 | 59 | 47-71 |
| Stomach | Stomach-AdenoCA | Adenocarcinoma; Mucinous; Papillary; Tubular | 75 | 18 | 57 | 65 | 47-79 |
| Thyroid | Thy-AdenoCA | Adenocarcinoma; Columnar cell; Follicular type | 48 | 37 | 11 | 51 | 26-75 |
| Mesoderm | |||||||
| Bone/Soft Tissue | Bone-Benign | Osteoblastoma; Osteofibrous dysplasia | 7 | 4 | 3 | 18 | 12-30 |
| Bone/Soft Tissue | Bone-Benign | Chondroblastoma; Chrondromyxoid fibroma | 9 | 2 | 7 | 16 | 14-38 |
| Bone/Soft Tissue | Bone-Epith | Adamantinoma; Chordoma | 10 | 4 | 6 | 60 | 37-67 |
| Bone/Soft Tissue | Bone-Osteosarc | Osteosarcoma | 38 | 20 | 18 | 20 | 9-58 |
| Bone/Soft Tissue | SoftTissue-Leiomyo | Leiomyosarcoma | 15 | 10 | 5 | 61 | 51-78 |
| Bone/Soft Tissue | SoftTissue-Liposarc | Liposarcoma | 19 | 5 | 14 | n/a | n/a |
| Cervix | Cervix-AdenoCA | Adenocarcinoma | 2 | 2 | 0 | 39 | 33-46 |
| Cervix | Cervix-SCC | Squamous cell carcinoma | 18 | 18 | 0 | 39 | 25-58 |
| Head/Neck | Head-SCC | Squamous cell carcinoma | 57 | 10 | 47 | 53 | 34-71 |
| Kidney | Kidney-ChRCC | Adenocarcinoma, chromophobe type | 45 | 19 | 26 | 47 | 34-69 |
| Kidney | Kidney-RCC | Clear cell adenocarcinoma; papillary type | 144 | 54 | 90 | 60 | 48-75 |
| Lymphoid | Lymph-BNHL | Burkitt; Diffuse large B-cell; Follicular; Marginal | 107 | 51 | 56 | 57 | 10-74 |
| Lymphoid | Lymph-CLL | Chronic lymphocytic leukaemia | 95 | 31 | 64 | 62 | 46-78 |
| Myeloid | Myeloid-AML | Acute myeloid leukaemia | 10 | 3 | 7 | 50 | 35-56 |
| Myeloid | Myeloid-MDS | Myelodysplastic syndrome | 2 | 1 | 1 | 76 | 74-77 |
| Myeloid | Myeloid-MPN | Myeloproliferative neoplasm | 26 | 14 | 12 | 56 | 38-75 |
| Ovary | Ovary-AdenoCA | Adenocarcinoma; Serous cystadenocarcinoma | 113 | 113 | 0 | 60 | 48-74 |
| Uterus | Uterus-AdenoCA | Adeno., endometrioid; Serous cystadeno. | 51 | 51 | 0 | 69 | 57-81 |
| Ectoderm | |||||||
| Breast | Breast-AdenoCA | Infiltrating duct carcinoma; Medullary; Mucinous | 198 | 197 | 1 | 56 | 39-76 |
| Breast | Breast-DCIS | Duct micropapillary carcinoma | 3 | 3 | 0 | 55 | 43-60 |
| Breast | Breast-LobularCA | Lobular carcinoma | 13 | 13 | 0 | 53 | 42-69 |
| Total | 2658 | 1189 | 1469 | 59 | 21-76 | ||
Extended Table 2. Ethical considerations of genomic cloud computing.
| Ethical Considerations of Genomic Cloud Computing |
| The PCAWG project represents the first large-scale use of distributed cloud computing in genomics. The project involved the movement of large quantities of personal health information across multiple legal jurisdictions and responsible use of this data by several hundred international researchers. Donor consents were written to explicitly allow for broad research use of the data and for international data sharing. PCAWG was granted permission by the leads of each of the tumour data providers to store, analyse and distribute the data on academic and/or commercial compute clouds. |
| To ensure that the PCAWG personal data were handled in a manner consistent with the donor consents, authorised representatives of each of the academic clouds and high-performance computing facilities signed a commitment not to access controlled tier data beyond the minimum needed to administer it. We negotiated similar contractual terms with commercial cloud partners. Prior to accessing the data, each PCAWG researcher was required to obtain local Institutional Review Board approval for their proposed analytic projects, and obtained controlled tier authorisation from dbGaP (National Center for Biotechnology Information) and the ICGC DACO (Centre of Genomics and Policy at McGill University). To handle the data securely, we encrypted it while in motion and at rest. We used a central authentication and digital token generating system to enforce a strong data access protocol that required researchers to provide their TCGA and/or ICGC credentials prior to accessing controlled tier data. No data breach or other compromise of donor confidentiality is known to have occurred over the course of the PCAWG project, despite its extensive use of cloud computing. |
Extended Table 3. Scientific output using PCAWG data, in bite-size chunks.
| Scientific | Key findings | Citation |
|---|---|---|
| Driver mutations | ||
| Discovery of non-coding drivers |
|
4 |
| Drivers by pathways and networks |
|
16 |
| Evolution and heterogeneity | ||
| Timing of cancer evolution |
|
7 |
| Structural variants | ||
| Patterns of structural variation |
|
6 |
| Functional consequence of structural variation |
|
4 |
| Patterns of retrotransposition |
|
10 |
| Chromothripsis |
|
18 |
| Mutational signatures | ||
| Signatures of point mutations |
|
5 |
| Mutation distribution across genome |
|
11,12,15 |
| Transcriptional consequences of somatic mutation | ||
| RNA effects of somatic mutation |
|
8,9 |
| Others | ||
| Tumour subtypes from genome sequencing |
|
12 |
| Mitochondrial DNA mutations |
|
14 |
| Telomere biology and sequences |
|
4,13 |
Supplementary Material
Acknowledgements
We thank research participants who generously donated samples and data, the physicians and clinical staff who contributed to sample annotation and collection, and the numerous funding agencies that contributed to the collection and analysis of this data set.
Footnotes
Author contributions
A structured author contribution list is available in the online version of the paper.
Writing committee leads: Peter J Campbell, Gad Getz, Jan O Korbel, Joshua M Stuart, Jennifer L Jennings, Lincoln D Stein. Head of project management: Jennifer L Jennings. Sample Collection: Major contributions from Marc D Perry, Hardeep K Nahal-Bose; Led by BF Francis Ouellette. Histopathology harmonisation: Major contribution from Constance H Li; Further contributions from Esther Rheinbay, G Petur Nielsen, Dennis C Sgroi, Chin-Lee Wu, William C Faquin, Vikram Deshpande, Paul C Boutros, Alexander J Lazar, Katherine A Hoadley; Led by Lincoln D Stein, David N Louis. Uniform processing, somatic, germline variant calling: Major contribution from L Jonathan Dursi; Further contributions from Christina K Yung, Matthew H Bailey, Gordon Saksena, Keiran M Raine, Ivo Buchhalter, Kortine Kleinheinz, Matthias Schlesner, Junjun Zhang, Wenyi Wang, David A Wheeler; Led by Li Ding, Jared T Simpson. Core alignment, variant calling by cloud computing: Major contributions from Christina K Yung, Brian D O'Connor, Sergei Yakneen, Junjun Zhang; Further contributions from Kyle Ellrott, Kortine Kleinheinz, Naoki Miyoshi, Keiran M Raine, Adam P Butler, Romina Royo, Gordon Saksena, Matthias Schlesner, Solomon I Shorser, Miguel Vazquez. Integration, phasing,, validation of germline variant callsets: Major contributions from Tobias Rausch, Grace Tiao, Sebastian M Waszak, Bernardo Rodriguez-Martin, Suyash Shringarpure, Dai-Ying Wu; Further contributions from Sergei Yakneen, German M Demidov, Olivier Delaneau, Shuto Hayashi, Seiya Imoto, Nina Habermann, Ayellet V Segre, Erik Garrison, Andy Cafferkey, Eva G Alvarez, Jose Maria Heredia-Genestar, Francesc Muyas, Oliver Drechsel, Alicia L Bruzos, Javier Temes, Jorge Zamora, L Jonathan Dursi, Adrian Baez-Ortega, Hyung-Lae Kim, Matthew H Bailey, R Jay Mashl, Kai Ye, Ivo Buchhalter, Anthony DiBiase, Kuan-lin Huang, Ivica Letunic, Michael D McLellan, Steven J Newhouse, Matthias Schlesner, Tal Shmaya, Sushant Kumar, David C Wedge, Mark H Wright, Venkata D Yellapantula, Mark Gerstein, Ekta Khurana, Tomas Marques-Bonet, Arcadi Navarro, Carlos D Bustamante, Jared T Simpson, Li Ding, Reiner Siebert, Hidewaki Nakagawa, Douglas F Easton; Led by Stephan Ossowski, Jose MC Tubio, Gad Getz, Francisco M De La Vega, Xavier Estivill, Jan O Korbel. Validation, benchmarking, merging of somatic variant calls: Major contribution from L Jonathan Dursi; Further contributions from David A Wheeler, Christina K Yung; Led by Li Ding, Jared T Simpson. Data, code availability: Major contribution from Junjun Zhang; Further contributions from Christina K Yung, Sergei Yakneen, Denis Yuen, George L Mihaiescu, Larsson Omberg; Led by Vincent Ferretti. Pan-cancer burden of somatic mutations: Major contribution from Junjun Zhang; Led by Peter J Campbell. Panorama of driver mutations in human cancer: Led by Radhakrishnan Sabarinathan, Oriol Pich, Abel Gonzalez-Perez. PCAWG tumours with no apparent driver mutations: Major contribution from Esther Rheinbay; Further contributions from Amaro Taylor-Weiner, Radhakrishnan Sabarinathan; Led by Peter J Campbell, Gad Getz. Patterns, oncogenicity of kataegis, chromoplexy: Major contributions from Matthew W Fittall, Jonas Demeulemeester, Maxime Tarabichi; Further contributions from Nicola D Roberts, Peter J Campbell, Jan O Korbel; Led by Peter Van Loo. Patterns, oncogenicity of chromothripsis: Major-contributions from Maxime Tarabichi, Jonas Demeulemeester, Matthew W Fittall; Further contributions from Isidro Cortes-Ciriano, Lara Urban, Peter Park, Peter J Campbell, Jan O Korbel; Led by Peter Van Loo. Timing clustered mutational processes during tumour evolution: Major contributions from Jonas Demeulemeester, Maxime Tarabichi, Matthew W Fittall; Further contributions from Jan O Korbel, Peter J Campbell; Led by Peter Van Loo. Germline effects on somatic mutation: Major contributions from Sebastian M Waszak, Bin Zhu, Bernardo Rodriguez-Martin, Esa Pitkanen, Tobias Rausch; Further contributions from Yilong Li, Natalie Saini, Leszek J Klimczak, Joachim Weischenfeldt, Nikos Sidiropoulos, Ludmil B Alexandrov, Francesc Muyas, Raquel Rabionet, Georgia Escaramis, Adrian Baez-Ortega, Mattia Bosio, Aliaksei Z Holik, Hana Susak, Eva G Alvarez, Alicia L Bruzos, Javier Temes, Aparna Prasad, Nina Habermann, Serap Erkek, Lara Urban, Claudia Calabrese, Benjamin Raeder, Eoghan Harrington, Simon Mayes, Daniel Turner, Sissel Juul, Steven A Roberts, Lei Song, Roelof Koster, Lisa Mirabello, Xing Hua, Tomas J Tanskanen, Marta Tojo, David C Wedge, Jorge Zamora, Jieming Chen, Lauri A Aaltonen, Gunnar Ratsch, Roland F Schwarz, Atul J Butte, Alvis Brazma, Peter J Campbell, Stephen J Chanock, Nilanjan Chatterjee, Oliver Stegle, Olivier Harismendy; Led by G Steven Bova, Dmitry A Gordenin, Jose MC Tubio, Douglas F Easton, Xavier Estivill, Jan O Korbel. Replicative Immortality: Major contribution from David Haan; Further contributions from Lina Sieverling, Lars Feuerbach; Led by Lincoln D Stein, Joshua M Stuart. Ethical considerations of genomic cloud computing: Led by Don Chalmers, Yann Joly, Bartha Knoppers, Fruzsina Molnar-Gabor, Jan O Korbel, Mark Phillips, Adrian Thorogood, David Townend. Online resources for data access, visualisation, exploration, analysis: Major contributions from Mary Goldman, Junjun Zhang, Nuno A Fonseca; Further contributions from Qian Xiang, Brian Craft, Elena Pineiro-Yanez, Alfonso Munoz, Robert Petryszak, Anja Fullgrabe, Fatima Al-Shahrour, Maria Keays, David Haussler, John Weinstein, Wolfgang Huber, Alfonso Valencia, Irene Papatheodorou, Jingchun Zhu; Led by Brian O'Connor, Lincoln D Stein, Alvis Brazma, Vincent Ferretti, Miguel Vazquez. Pilot-63 Analysis Validation Process: Major contribution from L Jonathan Dursi; Further contributions from Christina K Yung, Matthew H Bailey, Gordon Saksena, Keiran M Raine, Ivo Buchhalter, Kortine Kleinheinz, Matthias Schlesner, Yu Fan, David Torrents, Matthias Bieg, Paul C Boutros, Ken Chen, Zechen Chong, Kristian Cibulskis, Oliver Drechsel, Roland Eils, Robert S Fulton, Josep Gelpi, Mark Gerstein, Santiago Gonzalez, Gad Getz, Ivo G Gut, Faraz Hach, Michael Heinold, Taobo Hu, Vincent Huang, Barbara Hutter, Hyung-Lae Kim, Natalie Jager, Jongsun Jung, Sushant Kumar, Yogesh Kumar, Christopher Lalansingh, Ignaty Leshchiner, Ivica Letunic, Dimitri Livitz, Eric Z Ma, Yosef Maruvka, R Jay Mashl, Michael D McLellan, Ana Milovanovic, Morten Muhlig Nielsen, Brian O’Connor, Stephan Ossowski, Nagarajan Paramasivam, Jakob Skou Pedersen, Marc D Perry, Montserrat Puiggros, Romina Royo, Esther Rheinbay, S Cenk Sahinalp, Iman Sarrafi, Chip Stewart, Miranda D Stobbe, Grace Tiao, Jeremiah A Wala, Jiayin Wang, Wenyi Wang, Sebastian M Waszak, Joachim Weischenfeldt, Michael Wendl, Johannes Werner, Zhenggang Wu, Hong Xue, Sergei Yakneen, Takafumi N Yamaguchi, Kai Ye, Venkata Yellapantula, Junjun Zhang, David A Wheeler; Led by Li Ding, Jared T Simpson. Processing of Validation Data: Major contributions from Christina K Yung, Brian D O'Connor, Sergei Yakneen, Junjun Zhang; Further contributions from Kyle Ellrott, Kortine Kleinheinz, Naoki Miyoshi, Keiran M Raine, Romina Royo, Gordon Saksena, Matthias Schlesner, Solomon I Shorser, Miguel Vazquez, Joachim Weischenfeldt, Denis Yuen, Adam P Butler, Brandi N Davis-Dusenbery, Roland Eils, Vincent Ferretti, Robert L Grossman, Olivier Harismendy, Youngwook Kim, Hidewaki Nakagawa, Steven J Newhouse, David Torrents; Led by Lincoln D Stein. Whole Genome Sequencing Somatic Variant Calling: Major contribution from Junjun Zhang; Further contributions from Christina K Yung, Solomon I Shorser. Whole Genome Alignment: Keiran M Raine, Junjun Zhang, Brian O’Connor. DKFZ Pipeline: Kortine Kleinheinz, Tobias Rausch, Jan O Korbel, Ivo Buchhalter, Michael C Heinold, Barbara Hutter, Natalie Jager, Nagarajan Paramasivam, Matthias Schlesner. EMBL Pipeline: Joachim Weischenfeldt. Sanger Pipeline: Keiran M Raine, Jonathan Hinton, David R Jones, Andrew Menzies, Lucy Stebbings, Adam P Butler. Broad Pipeline: Gordon Saksena, Dimitri Livitz, Esther Rheinbay, Julian M Hess, Ignaty Leshchiner, Chip Stewart, Grace Tiao, Jeremiah A Wala, Amaro Taylor-Weiner, Mara Rosenberg, Andrew J Dunford, Manaswi Gupta, Marcin Imielinski, Matthew Meyerson, Rameen Beroukhim, Gad Getz. MuSE Pipeline: Yu Fan, Wenyi Wang. Consensus Somatic SNV/Indel Annotation: Andrew Menzies, Matthias Schlesner, Juri Reimand, Priyanka Dhingra, Ekta Khurana. Somatic SNV, indel Merging: Major contribution from L Jonathan Dursi; Further contributions from Christina K Yung, Matthew H Bailey, Gordon Saksena, Keiran M Raine, Ivo Buchhalter, Kortine Kleinheinz, Matthias Schlesner, Yu Fan, David Torrents, Matthias Bieg, Paul C Boutros, Ken Chen, Zechen Chong, Kristian Cibulskis, Oliver Drechsel, Roland Eils, Robert S Fulton, Josep Gelpi, Mark Gerstein, Santiago Gonzalez, Gad Getz, Ivo G Gut, Faraz Hach, Michael Heinold, Taobo Hu, Vincent Huang, Barbara Hutter, Hyung-Lae Kim, Natalie Jager, Jongsun Jung, Sushant Kumar, Yogesh Kumar, Christopher Lalansingh, Ignaty Leshchiner, Ivica Letunic, Dimitri Livitz, Eric Z Ma, Yosef Maruvka, R Jay Mashl, Michael D McLellan, Ana Milovanovic, Morten Muhlig Nielsen, Brian O’Connor, Stephan Ossowski, Nagarajan Paramasivam, Jakob Skou Pedersen, Marc D Perry, Montserrat Puiggros, Romina Royo, Esther Rheinbay, S Cenk Sahinalp, Iman Sarrafi, Chip Stewart, Miranda D Stobbe, Grace Tiao, Jeremiah A Wala, Jiayin Wang, Wenyi Wang, Sebastian M Waszak, Joachim Weischenfeldt, Michael Wendl, Johannes Werner, Zhenggang Wu, Hong Xue, Sergei Yakneen, Takafumi N Yamaguchi, Kai Ye, Venkata Yellapantula, Junjun Zhang, David A Wheeler; Major contributions from Li Ding, Jared T Simpson. Somatic SV Merging: Joachim Weischenfeldt, Francesco Favero, Yilong Li. Somatic Copy Number Alteration Merging: Stefan Dentro, Jeff Wintersinger, Ignaty Leshchiner. Oxidative Artefact Filtration: Dimitri Livitz, Ignaty Leshchiner, Chip Stewart, Esther Rheinbay, Gordon Saksena, Gad Getz. Strand Bias Filtration: Matthias Bieg, Ivo Buchhalter, Johannes Werner, Matthias Schlesner. miniBAM generation: Jeremiah Wala, Gordon Saksena, Rameen Beroukhim, Gad Getz. Germline Variant Identification from WGS: Major contributions from Tobias Rausch, Grace Tiao, Sebastian M Waszak, Bernardo Rodriguez-Martin, Suyash Shringarpure, Dai-Ying Wu; Further contributions from Sergei Yakneen, German M Demidov, Olivier Delaneau, Shuto Hayashi, Seiya Imoto, Nina Habermann, Ayellet V Segre, Erik Garrison, Andy Cafferkey, Eva G Alvarez, Alicia L Bruzos, Jorge Zamora, Jose Maria Heredia-Genestar, Francesc Muyas, Oliver Drechsel, L Jonathan Dursi, Adrian Baez-Ortega, Hyung-Lae Kim, Matthew H Bailey, R Jay Mashl, Kai Ye, Ivo Buchhalter, Vasilisa Rudneva, Ji Wan Park, Eun Pyo Hong, Seong Gu Heo, Anthony DiBiase, Kuan-lin Huang, Ivica Letunic, Michael D McLellan, Steven J Newhouse, Matthias Schlesner, Tal Shmaya, Sushant Kumar, David C Wedge, Mark H Wright, Venkata D Yellapantula, Mark Gerstein, Ekta Khurana, Tomas Marques-Bonet, Arcadi Navarro, Carlos D Bustamante, Jared T Simpson, Li Ding, Reiner Siebert, Hidewaki Nakagawa, Douglas F Easton; Led by Stephan Ossowski, Jose MC Tubio, Gad Getz, Francisco M De La Vega, Xavier Estivill, Jan O Korbel. RNA-Seq analysis: Major contributions from Nuno A Fonseca, Andre Kahles, Kjong-Van Lehmann, Lara Urban, Cameron M Soulette, Yuichi Shiraishi, Fenglin Liu, Yao He, Deniz Demircioglu, Natalie R Davidson, Claudia Calabrese, Junjun Zhang, Marc D Perry, Qian Xiang; Further contributions from Liliana Greger, Siliang Li, Dongbing Liu, Stefan G Stark, Fan Zhang, Samirkumar B Amin, Peter Bailey, Aurelien Chateigner, Isidro Cortes-Ciriano, Brian Craft, Serap Erkek, Milana Frenkel-Morgenstern, Mary Goldman, Katherine A Hoadley, Yong Hou, Matthew R Huska, Ekta Khurana, Helena Kilpinen, Jan O Korbel, Fabien C Lamaze, Chang Li, Xiaobo Li, Xinyue Li, Xingmin Liu, Maximillian G Marin, Julia Markowski, Tannistha Nandi, Morten Muhlig Nielsen, Akinyemi I Ojesina, Qiang Pan-Hammarstrom, Peter J Park, Chandra Sekhar Pedamallu, Jakob Pedersen, Reiner Siebert, Hong Su, Patrick Tan, Bin Tean Teh, Jian Wang, Sebastian M Waszak, Heng Xiong, Sergei Yakneen, Chen Ye, Christina Yung, Xiuqing Zhang, Liangtao Zheng, Jingchun Zhu, Shida Zhu, Philip Awadalla, Chad J Creighton, Matthew Meyerson, BF Francis Ouellette, Kui Wu, Huanming Yang; Led by Jonathan Goke, Roland F Schwarz, Oliver Stegle, Zemin Zhang, Alvis Brazma, Gunnar Ratsch, Angela N Brooks. Clustering of tumour genomes based on telomere maintenance-related features: Major contribution from David Haan; Led by Lincoln D Stein, Joshua M Stuart. Clustered mutational processes in PCAWG: Major contributions from Jonas Demeulemeester, Maxime Tarabichi, Matthew W Fittall; Led by Peter J Campbell, Jan O Korbel, Peter Van Loo. Tumours without detected driver mutations: Esther Rheinbay, Amaro Taylor-Weiner, Radhakrishnan Sabarinathan, Peter J Campbell, Gad Getz. Panorama of driver mutations in human cancer: Major contributions from Radhakrishnan Sabarinathan, Oriol Pich; Further contributions from Inigo Martincorena, Carlota Rubio-Perez, Malene Juul, Jeremiah Wala, Steven Schumacher, Ofer Shapira, Nikos Sidiropoulos, Sebastian M Waszak, David Tamborero, Loris Mularoni, Esther Rheinbay, Henrik Hornshoj, Jordi Deu-Pons, Ferran Muinos, Johanna Bertl, Qianyun Guo, Chad J Creighton, Joachim Weischenfeldt, Jan O Korbel, Gad Getz, Peter J Campbell, Jakob Pedersen, Rameen Beroukhim; Led by Abel Gonzalez-Perez. Pilot- benchmarking, variant consensus development, validation: Major contribution from L Jonathan Dursi; Further contributions from Christina K Yung, Matthew H Bailey, Gordon Saksena, Keiran M Raine, Ivo Buchhalter, Kortine Kleinheinz, Matthias Schlesner, Yu Fan, David Torrents, Matthias Bieg, Paul C Boutros, Ken Chen, Zechen Chong, Kristian Cibulskis, Oliver Drechsel, Roland Eils, Robert S Fulton, Josep Gelpi, Mark Gerstein, Santiago Gonzalez, Gad Getz, Ivo G Gut, Faraz Hach, Michael Heinold, Taobo Hu, Vincent Huang, Barbara Hutter, Hyung-Lae Kim, Natalie Jager, Jongsun Jung, Sushant Kumar, Yogesh Kumar, Christopher Lalansingh, Ignaty Leshchiner, Ivica Letunic, Dimitri Livitz, Eric Z Ma, Yosef Maruvka, R Jay Mashl, Michael D McLellan, Ana Milovanovic, Morten Muhlig Nielsen, Brian O’Connor, Stephan Ossowski, Nagarajan Paramasivam, Jakob Skou Pedersen, Marc D Perry, Montserrat Puiggros, Romina Royo, Esther Rheinbay, S Cenk Sahinalp, Iman Sarrafi, Chip Stewart, Miranda D Stobbe, Grace Tiao, Jeremiah A Wala, Jiayin Wang, Wenyi Wang, Sebastian M Waszak, Joachim Weischenfeldt, Michael Wendl, Johannes Werner, Zhenggang Wu, Hong Xue, Sergei Yakneen, Takafumi N Yamaguchi, Kai Ye, Venkata Yellapantula, Junjun Zhang, David A Wheeler; Led by Li Ding, Jared T Simpson. Production somatic variant calling on the PCAWG Compute Cloud: Major contributions from Christina K Yung, Brian D O'Connor, Sergei Yakneen, Junjun Zhang; Further contributions from Kyle Ellrott, Kortine Kleinheinz, Naoki Miyoshi, Keiran M Raine, Romina Royo, Gordon Saksena, Matthias Schlesner, Solomon I Shorser, Miguel Vazquez, Joachim Weischenfeldt, Denis Yuen, Adam P Butler, Brandi N Davis-Dusenbery, Roland Eils, Vincent Ferretti, Robert L Grossman, Olivier Harismendy, Youngwook Kim, Hidewaki Nakagawa, Steven J Newhouse, David Torrents; Led by Lincoln D Stein. PCAWG data portals: Major contributions from Mary Goldman, Junjun Zhang, Nuno A Fonseca, Isidro Cortes-Ciriano; Further contributions from Qian Xiang, Brian Craft, Elena Pineiro-Yanez, Brian D O'Connor, Wojciech Bazant, Elisabet Barrera, Alfonso Munoz, Robert Petryszak, Anja Fullgrabe, Fatima Al-Shahrour, Maria Keays, David Haussler, John Weinstein, Wolfgang Huber, Alfonso Valencia, Irene Papatheodorou, Jingchun Zhu; Led by Vincent Ferretti, Miguel Vazquez.
The corresponding authors declare the following competing financial interests: G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect and POLYSOLVER. The other corresponding authors have no competing financial interests to declare. Other authors declare competing financial interests: details are available in the online version of the paper.
Data availability
The PCAWG-generated alignments, somatic variant calls, annotations and derived data sets are available for general research use for browsing and download at http://dcc.icgc.org/pcawg/ (Box 1; Supplementary Table 4). In accordance with the data access policies of the ICGC and TCGA projects, most molecular, clinical and specimen data are in an open tier which does not require access approval. To access potentially identifying information, such as germline alleles and underlying read data, researchers will need to apply to the TCGA Data Access Committee (DAC) via dbGaP (https://dbgap.ncbi.nlm.nih.gov/aa/wga.cgi?page=login) for access to the TCGA portion of the data set, and to the ICGC Data Access Compliance Office (DACO; http://icgc.org/daco) for the ICGC portion. In addition, to access somatic single nucleotide variants derived from TCGA donors, researchers will also need to obtain dbGaP authorisation.
Beyond the core sequence data and variant call-sets, the analyses in this paper used a number of datasets that were derived from the variant calls (Supplementary Table 4). The individual data sets are available at Synapse (https://www.synapse.org/), and are denoted with synXXXXX accession numbers; all these datasets are also mirrored at https://dcc.icgc.org, with full links, filenames, accession numbers and descriptions detailed in Supplementary Table 4. The datasets encompass: clinical data from each patient including demographics, tumour stage and vital status (syn10389158); harmonised tumour histopathology annotations using a standardised hierarchical ontology (syn1038916); inferred purity and ploidy values for each tumour sample (syn8272483); driver mutations for each patient from their cancer genome spanning all classes of variant, and coding versus non-coding drivers (syn11639581); mutational signatures inferred from PCAWG donors (syn11804065), including APOBEC mutagenesis (syn7437313); and transcriptional data from RNA-sequencing, including gene expression levels (syn5553985, syn5553991, syn8105922) and gene fusions (syn10003873, syn7221157).
Code availability
Computational pipelines for calling somatic mutations are available to the public at https://dockstore.org. A range of data visualisation and exploration tools are also available for PCAWG data (Box 1).
Author list: Participants in PCAWG Consortium
These authors jointly supervised this work:
Peter J Campbell, Gad Getz, Jan O Korbel, Joshua M Stuart and Lincoln D Stein.
Writing Committee
Writing Committee Leads
Peter J Campbell1,2, Gad Getz3,4,5,6, Jan O Korbel7,8, Joshua M Stuart9, Jennifer L Jennings10,11 and Lincoln D Stein12,13
Sample Collection
Marc D Perry14,15, Hardeep K Nahal-Bose15 and BF Francis Ouellette16,17
Histopathology harmonisation
Constance H Li12,18, Esther Rheinbay3,6,19, G Petur Nielsen19, Dennis C Sgroi19, Chin-Lee Wu19, William C Faquin19, Vikram Deshpande19, Paul C Boutros12,18,20,21, Alexander J Lazar22, Katherine A Hoadley23,24, Lincoln D Stein12,13 and David N Louis19
Uniform processing, somatic and germline variant calling
L Jonathan Dursi12,25, Christina K Yung15, Matthew H Bailey26,27, Gordon Saksena3, Keiran M Raine1, Ivo Buchhalter28,29,30, Kortine Kleinheinz28,30, Matthias Schlesner28,31, Junjun Zhang15, Wenyi Wang32, David A Wheeler33,34, Li Ding26,27,35 and Jared T Simpson12,36
Core alignment and variant calling by cloud computing
Christina K Yung15, Brian D O'Connor15,37, Sergei Yakneen8, Junjun Zhang15, Kyle Ellrott38, Kortine Kleinheinz28,30, Naoki Miyoshi39, Keiran M Raine1, Adam P Butler1, Romina Royo40, Gordon Saksena3, Matthias Schlesner28,31, Solomon I Shorser12 and Miguel Vazquez40,41
Integration, phasing, and validation of germline variant callsets
Tobias Rausch8, Grace Tiao3, Sebastian M Waszak8, Bernardo Rodriguez-Martin42,43,44, Suyash Shringarpure45, Dai-Ying Wu46, Sergei Yakneen8, German M Demidov47,48,49, Olivier Delaneau50,51,52, Shuto Hayashi39, Seiya Imoto39,39, Nina Habermann8, Ayellet V Segre3,53, Erik Garrison1, Andy Cafferkey7, Eva G Alvarez42,43,44, José María Heredia-Genestar54, Francesc Muyas47,48,49, Oliver Drechsel47,49, Alicia L Bruzos42,43,44, Javier Temes42,43, Jorge Zamora1,42,43,44, L Jonathan Dursi12,25, Adrian Baez-Ortega55, Hyung-Lae Kim56, Matthew H Bailey26,27, R Jay Mashl27,57, Kai Ye58,59, Ivo Buchhalter28,29,30, Anthony DiBiase60, Kuan-lin Huang27,61, Ivica Letunic62, Michael D McLellan26,27,35, Steven J Newhouse7, Matthias Schlesner28,31, Tal Shmaya46, Sushant Kumar63,64, David C Wedge1,65,66, Mark H Wright45, Venkata D Yellapantula67,68, Mark Gerstein63,64,69, Ekta Khurana70,71,72,73, Tomas Marques-Bonet74,75,76,77, Arcadi Navarro74,75,76, Carlos D Bustamante45,78, Jared T Simpson12,36, Li Ding26,27,35, Reiner Siebert79,80, Hidewaki Nakagawa81, Douglas F Easton82,83, Stephan Ossowski47,48,49, Jose MC Tubio42,43,44, Gad Getz3,4,5,6, Francisco M De La Vega45,46,78, Xavier Estivill47,84 and Jan O Korbel7,8
Validation, benchmarking and merging of somatic variant calls
L Jonathan Dursi12,25, David A Wheeler33,34, Christina K Yung15, Li Ding26,27,35 and Jared T Simpson12,36
Data and code availability
Junjun Zhang15, Christina K Yung15, Sergei Yakneen8, Denis Yuen12, George L Mihaiescu15, Larsson Omberg85 and Vincent Ferretti15,86
Pan-cancer burden of somatic mutations
Junjun Zhang15 and Peter J Campbell1,2
Panorama of driver mutations in human cancer
Radhakrishnan Sabarinathan87,88,89, Oriol Pich87,89 and Abel Gonzalez-Perez87,89
PCAWG tumours with no apparent driver mutations
Esther Rheinbay3,6,19, Amaro Taylor-Weiner90, Radhakrishnan Sabarinathan87,88,89, Peter J Campbell1,2 and Gad Getz3,4,5,6
Patterns and oncogenicity of kataegis and chromoplexy
Matthew W Fittall91, Jonas Demeulemeester91,92, Maxime Tarabichi1,91, Nicola D Roberts1, Peter J Campbell1,2, Jan O Korbel7,8 and Peter Van Loo91,92
Patterns and oncogenicity of chromothripsis
Maxime Tarabichi1,91, Jonas Demeulemeester91,92, Matthew W Fittall91, Isidro Cortés-Ciriano93,94,95, Lara Urban7,8, Peter Park94,95, Peter J Campbell1,2, Jan O Korbel7,8 and Peter Van Loo91,92
Timing clustered mutational processes during tumour evolution
Jonas Demeulemeester91,92, Maxime Tarabichi1,91, Matthew W Fittall91, Jan O Korbel7,8, Peter J Campbell1,2 and Peter Van Loo91,92
Germline genetic determinants of the somatic mutation landscape
Sebastian M Waszak8, Bin Zhu96, Bernardo Rodriguez-Martin42,43,44, Esa Pitkänen8, Tobias Rausch8, Yilong Li1, Natalie Saini97, Leszek J Klimczak98, Joachim Weischenfeldt8,99,100, Nikos Sidiropoulos100, Ludmil B Alexandrov1,101, Francesc Muyas47,48,49, Raquel Rabionet47,49,102, Georgia Escaramis47,103,104, Adrian Baez-Ortega55, Mattia Bosio40,47,49, Aliaksei Z Holik47, Hana Susak47,49, Eva G Alvarez42,43,44, Alicia L Bruzos42,43,44, Javier Temes42,43, Aparna Prasad49, Nina Habermann8, Serap Erkek8, Lara Urban7,8, Claudia Calabrese7,8, Benjamin Raeder8, Eoghan Harrington105, Simon Mayes106, Daniel Turner106, Sissel Juul105, Steven A Roberts107, Lei Song96, Roelof Koster108, Lisa Mirabello96, Xing Hua96, Tomas J Tanskanen109, Marta Tojo44, David C Wedge1,65,66, Jorge Zamora1,42,43,44, Jieming Chen64,110, Lauri A Aaltonen111, Gunnar Rätsch112,113,114,115,116,117, Roland F Schwarz7,118,119,120, Atul J Butte121, Alvis Brazma7, Peter J Campbell1,2, Stephen J Chanock96, Nilanjan Chatterjee122,123,123, Oliver Stegle7,8,124, Olivier Harismendy125, G Steven Bova126, Dmitry A Gordenin97, Jose MC Tubio42,43,44, Douglas F Easton82,83, Xavier Estivill47,84 and Jan O Korbel7,8
Replicative Immortality
David Haan9, Lina Sieverling127,128, Lars Feuerbach127, Lincoln D Stein12,13 and Joshua M Stuart9
Ethical considerations of genomic cloud computing
Don Chalmers129, Yann Joly130, Bartha Knoppers130, Fruzsina Molnár-Gábor131, Jan O Korbel7,8, Mark Phillips130 and Adrian Thorogood and David Townend130
Box 1: Online resources for data access, visualisation, exploration and analysis
Mary Goldman132, Junjun Zhang15, Nuno A Fonseca7,133, Qian Xiang134, Brian Craft132, Elena Piñeiro-Yáñez135, Alfonso Muñoz7, Robert Petryszak7, Anja Füllgrabe7, Fatima Al-Shahrour135, Maria Keays7, David Haussler132,136, John Weinstein137,138, Wolfgang Huber8, Alfonso Valencia40,76, Irene Papatheodorou7, Jingchun Zhu132, Brian O'Connor15,37, Lincoln D Stein12,13, Alvis Brazma7, Vincent Ferretti15,86 and Miguel Vazquez40,41
Methods 1.1 Validation Process
L Jonathan Dursi12,25, Christina K Yung15, Matthew H Bailey26,27, Gordon Saksena3, Keiran M Raine1, Ivo Buchhalter28,29,30, Kortine Kleinheinz28,30, Matthias Schlesner28,31, Yu Fan32, David Torrents40,76, Matthias Bieg139,140, Paul C Boutros12,18,20,21, Ken Chen141, Zechen Chong142, Kristian Cibulskis3, Oliver Drechsel47,49, Roland Eils28,30,143,144, Robert S Fulton26,27,35, Josep Gelpi40,145, Mark Gerstein63,64,69, Santiago Gonzalez7,8, Gad Getz3,4,5,6, Ivo G Gut49,74, Faraz Hach146,147, Michael Heinold28,30, Taobo Hu148, Vincent Huang12, Barbara Hutter140,149,150, Hyung-Lae Kim56, Natalie Jäger28, Jongsun Jung151, Sushant Kumar63,64, Yogesh Kumar148, Christopher Lalansingh12, Ignaty Leshchiner3, Ivica Letunic62, Dimitri Livitz3, Eric Z Ma148, Yosef Maruvka3,19,152, R Jay Mashl27,57, Michael D McLellan26,27,35, Ana Milovanovic40, Morten Muhlig Nielsen153, Brian O’Connor15,37, Stephan Ossowski47,48,49, Nagarajan Paramasivam28,140, Jakob Skou Pedersen153,154, Marc D Perry14,15, Montserrat Puiggròs40, Romina Royo40, Esther Rheinbay3,6,19, S Cenk Sahinalp147,155,156, Iman Sarrafi147,156, Chip Stewart3, Miranda D Stobbe49,74, Grace Tiao3, Jeremiah A Wala3,6,157, Jiayin Wang27,58,158, Wenyi Wang32, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Michael Wendl27,159,160, Johannes Werner28,161, Zhenggang Wu148, Hong Xue148, Sergei Yakneen8, Takafumi N Yamaguchi12, Kai Ye58,59, Venkata Yellapantula67,68, Junjun Zhang15, David A Wheeler33,34, Li Ding26,27,35 and Jared T Simpson12,36
Methods 1.2 Processing of Validation Data
Christina K Yung15, Brian D O'Connor15,37, Sergei Yakneen8, Junjun Zhang15, Kyle Ellrott38, Kortine Kleinheinz28,30, Naoki Miyoshi39, Keiran M Raine1, Romina Royo40, Gordon Saksena3, Matthias Schlesner28,31, Solomon I Shorser12, Miguel Vazquez40,41, Joachim Weischenfeldt8,99,100, Denis Yuen12, Adam P Butler1, Brandi N Davis-Dusenbery162, Roland Eils28,30,143,144, Vincent Ferretti15,86, Robert L Grossman163, Olivier Harismendy125, Youngwook Kim164,165, Hidewaki Nakagawa81, Steven J Newhouse7, David Torrents40,76 and Lincoln D Stein12,13
Methods 2. Whole Genome Sequencing Somatic Variant Calling
Junjun Zhang15, Christina K Yung15 and Solomon I Shorser12
Methods 2.1 Whole Genome Alignment
Keiran M Raine1, Junjun Zhang15 and Brian O’Connor15,37
Methods 2.2.1 DKFZ Pipeline
Kortine Kleinheinz28,30, Tobias Rausch8, Jan O Korbel7,8, Ivo Buchhalter28,29,30, Michael C Heinold28,30, Barbara Hutter140,149,150, Natalie Jäger28, Nagarajan Paramasivam28,140 and Matthias Schlesner28,31
Methods 2.2.2 EMBL Pipeline
Joachim Weischenfeldt8,99,100, Tobias Rausch8
Methods 2.2.3 Sanger Pipeline
Keiran M Raine1, Jonathan Hinton1, David R Jones1, Andrew Menzies1 and Lucy Stebbings1
Methods 2.2.4 Broad Pipeline
Gordon Saksena3, Dimitri Livitz3, Esther Rheinbay3,6,19, Julian M Hess3,152, Ignaty Leshchiner3, Chip Stewart3, Grace Tiao3, Jeremiah A Wala3,6,157, Amaro Taylor-Weiner90, Mara Rosenberg3,19, Andrew J Dunford3, Manaswi Gupta3, Marcin Imielinski166,167, Matthew Meyerson3,6,157, Rameen Beroukhim3,6,168 and Gad Getz3,4,5,6
Methods 2.2.5 MuSE Pipeline
Yu Fan32 and Wenyi Wang32
Methods 2.3 Consensus Somatic SNV/Indel Annotation
Andrew Menzies1, Matthias Schlesner28,31, Jüri Reimand12,18, Priyanka Dhingra71,73 and Ekta Khurana70,71,72,73
Methods 2.4.1 Somatic SNV and indel Merging
L Jonathan Dursi12,25, Christina K Yung15, Matthew H Bailey26,27, Gordon Saksena3, Keiran M Raine1, Ivo Buchhalter28,29,30, Kortine Kleinheinz28,30, Matthias Schlesner28,31, Yu Fan32, David Torrents40,76, Matthias Bieg139,140, Paul C Boutros12,18,20,21, Ken Chen141, Zechen Chong142, Kristian Cibulskis3, Oliver Drechsel47,49, Roland Eils28,30,143,144, Robert S Fulton26,27,35, Josep Gelpi40,145, Mark Gerstein63,64,69, Santiago Gonzalez7,8, Gad Getz3,4,5,6, Ivo G Gut49,74, Faraz Hach146,147, Michael Heinold28,30, Taobo Hu148, Vincent Huang12, Barbara Hutter140,149,150, Hyung-Lae Kim56, Natalie Jäger28, Jongsun Jung151, Sushant Kumar63,64, Yogesh Kumar148, Christopher Lalansingh12, Ignaty Leshchiner3, Ivica Letunic62, Dimitri Livitz3, Eric Z Ma148, Yosef Maruvka3,19,152, R Jay Mashl27,57, Michael D McLellan26,27,35, Ana Milovanovic40, Morten Muhlig Nielsen153, Brian O’Connor15,37, Stephan Ossowski47,48,49, Nagarajan Paramasivam28,140, Jakob Skou Pedersen153,154, Marc D Perry14,15, Montserrat Puiggròs40, Romina Royo40, Esther Rheinbay3,6,19, S Cenk Sahinalp147,155,156, Iman Sarrafi147,156, Chip Stewart3, Miranda D Stobbe49,74, Grace Tiao3, Jeremiah A Wala3,6,157, Jiayin Wang27,58,158, Wenyi Wang32, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Michael Wendl27,159,160, Johannes Werner28,161, Zhenggang Wu148, Hong Xue148, Sergei Yakneen8, Takafumi N Yamaguchi12, Kai Ye58,59, Venkata Yellapantula67,68, Junjun Zhang15, David A Wheeler33,34, Li Ding26,27,35 and Jared T Simpson12,36
Methods 2.4.2 Somatic SV Merging
Joachim Weischenfeldt8,99,100, Francesco Favero169 and Yilong Li1
Methods 2.4.3 Somatic Copy Number Alteration Merging
Stefan Dentro1,65,91, Jeff Wintersinger170,171,172 and Ignaty Leshchiner3
Methods 2.5.3 Oxidative Artefact Filtration
Dimitri Livitz3, Ignaty Leshchiner3, Chip Stewart3, Esther Rheinbay3,6,19, Gordon Saksena3 and Gad Getz3,4,5,6
Methods 2.5.4 Strand Bias Filtration
Matthias Bieg139,140, Ivo Buchhalter28,29,30, Johannes Werner28,161 and Matthias Schlesner28,31
Methods 2.6 miniBAM generation
Jeremiah Wala3,6,157, Gordon Saksena3, Rameen Beroukhim3,6,168 and Gad Getz3,4,5,6
Methods 3. Germline Variant Identification from WGS
Tobias Rausch8, Grace Tiao3, Sebastian M Waszak8, Bernardo Rodriguez-Martin42,43,44, Suyash Shringarpure45, Dai-Ying Wu46, Sergei Yakneen8, German M Demidov47,48,49, Olivier Delaneau50,51,52, Shuto Hayashi39, Seiya Imoto39,39, Nina Habermann8, Ayellet V Segre3,53, Erik Garrison1, Andy Cafferkey7, Eva G Alvarez42,43,44, Alicia L Bruzos42,43,44, Jorge Zamora1,42,43,44, José María Heredia-Genestar54, Francesc Muyas47,48,49, Oliver Drechsel47,49, L Jonathan Dursi12,25, Adrian Baez-Ortega55, Hyung-Lae Kim56, Matthew H Bailey26,27, R Jay Mashl27,57, Kai Ye58,59, Ivo Buchhalter28,29,30, Vasilisa Rudneva8, Ji Wan Park173, Eun Pyo Hong173, Seong Gu Heo173, Anthony DiBiase60, Kuan-lin Huang27,61, Ivica Letunic62, Michael D McLellan26,27,35, Steven J Newhouse7, Matthias Schlesner28,31, Tal Shmaya46, Sushant Kumar63,64, David C Wedge1,65,66, Mark H Wright45, Venkata D Yellapantula67,68, Mark Gerstein63,64,69, Ekta Khurana70,71,72,73, Tomas Marques-Bonet74,75,76,77, Arcadi Navarro74,75,76, Carlos D Bustamante45,78, Jared T Simpson12,36, Li Ding26,27,35, Reiner Siebert79,80, Hidewaki Nakagawa81, Douglas F Easton82,83, Stephan Ossowski47,48,49, Jose MC Tubio42,43,44, Gad Getz3,4,5,6, Francisco M De La Vega45,46,78 and Xavier Estivill and Jan O Korbel#47,84
Methods 4. RNA-Seq analysis
Nuno A Fonseca7,133, André Kahles, Kjong-Van Lehmann112,114,115,174,175, Lara Urban7,8, Cameron M Soulette37, Yuichi Shiraishi39, Fenglin Liu176,177, Yao He176, Deniz Demircioğlu178,179, Natalie R Davidson112,114,115,117,174, Claudia Calabrese7,8, Junjun Zhang15, Marc D Perry14,15, Qian Xiang134, Liliana Greger7, Siliang Li180,181, Dongbing Liu180,181, Stefan G Stark115,174,182,183, Fan Zhang176, Samirkumar B Amin184,185,186, Peter Bailey187, Aurélien Chateigner15, Isidro Cortés-Ciriano93,94,95, Brian Craft132, Serap Erkek8, Milana Frenkel-Morgenstern188, Mary Goldman132, Katherine A Hoadley23,24, Yong Hou180,181, Matthew R Huska118, Ekta Khurana70,71,72,73, Helena Kilpinen189, Jan O Korbel7,8, Fabien C Lamaze12, Chang Li180,181, Xiaobo Li180,181, Xinyue Li180, Xingmin Liu180,181, Maximillian G Marin37, Julia Markowski118, Tannistha Nandi190, Morten Muhlig Nielsen153, Akinyemi I Ojesina191,192,193, Qiang Pan-Hammarström180,194, Peter J Park94,95, Chandra Sekhar Pedamallu3,6,168, Jakob Pedersen153,154, Reiner Siebert79,80, Hong Su180,181, Patrick Tan190,195,196,197, Bin Tean Teh195,196,197,198,199, Jian Wang180, Sebastian M Waszak8, Heng Xiong180,181, Sergei Yakneen8, Chen Ye180,181, Christina Yung15, Xiuqing Zhang180, Liangtao Zheng176, Jingchun Zhu132, Shida Zhu180,181, Philip Awadalla12,13, Chad J Creighton200, Matthew Meyerson3,6,157, BF Francis Ouellette16,17, Kui Wu180,181, Huanming Yang180, Jonathan Göke178,201, Roland F Schwarz7,118,119,120, Oliver Stegle7,8,124, Zemin Zhang176,202, Alvis Brazma7, Gunnar Rätsch112,113,114,115,116,117 and Angela N Brooks3,37,157
Methods 5. Clustering of tumour genomes based on telomere maintenance-related features
David Haan9, Lincoln D Stein12,13 and Joshua M Stuart9
Methods 6. Clustered mutational processes in PCAWG
Jonas Demeulemeester91,92, Maxime Tarabichi1,91, Matthew W Fittall91, Peter J Campbell1,2, Jan O Korbel7,8 and Peter Van Loo91,92
Methods 7. Tumours without detected driver mutations
Esther Rheinbay3,6,19, Amaro Taylor-Weiner90, Radhakrishnan Sabarinathan87,88,89, Peter J Campbell1,2 and Gad Getz3,4,5,6
Methods 8. Panorama of driver mutations in human cancer
Radhakrishnan Sabarinathan87,88,89, Oriol Pich87,89, Iñigo Martincorena1, Carlota Rubio-Perez87,89,203, Malene Juul153, Jeremiah Wala3,6,157, Steven Schumacher3,204, Ofer Shapira3,157, Nikos Sidiropoulos100, Sebastian M Waszak8, David Tamborero87,89, Loris Mularoni87,89, Esther Rheinbay3,6,19, Henrik Hornshøj153, Jordi Deu-Pons89,205, Ferran Muiños87,89, Johanna Bertl153,206, Qianyun Guo154, Chad J Creighton200, Joachim Weischenfeldt8,99,100, Jan O Korbel7,8, Gad Getz3,4,5,6, Peter J Campbell1,2, Jakob Pedersen153,154, Rameen Beroukhim3,6,168 and Abel Gonzalez-Perez87,89,207
Notes 1. Pilot-63 benchmarking, variant consensus development and validation
L Jonathan Dursi12,25, Christina K Yung15, Matthew H Bailey26,27, Gordon Saksena3, Keiran M Raine1, Ivo Buchhalter28,29,30, Kortine Kleinheinz28,30, Matthias Schlesner28,31, Yu Fan32, David Torrents40,76, Matthias Bieg139,140, Paul C Boutros12,18,20,21, Ken Chen141, Zechen Chong142, Kristian Cibulskis3, Oliver Drechsel47,49, Roland Eils28,30,143,144, Robert S Fulton26,27,35, Josep Gelpi40,145, Mark Gerstein63,64,69, Santiago Gonzalez7,8, Gad Getz3,4,5,6, Ivo G Gut49,74, Faraz Hach146,147, Michael Heinold28,30, Taobo Hu148, Vincent Huang12, Barbara Hutter140,149,150, Hyung-Lae Kim56, Natalie Jäger28, Jongsun Jung151, Sushant Kumar63,64, Yogesh Kumar148, Christopher Lalansingh12, Ignaty Leshchiner3, Ivica Letunic62, Dimitri Livitz3, Eric Z Ma148, Yosef Maruvka3,19,152, R Jay Mashl27,57, Michael D McLellan26,27,35, Ana Milovanovic40, Morten Muhlig Nielsen153, Brian O’Connor15,37, Stephan Ossowski47,48,49, Nagarajan Paramasivam28,140, Jakob Skou Pedersen153,154, Marc D Perry14,15, Montserrat Puiggròs40, Romina Royo40, Esther Rheinbay3,6,19, S Cenk Sahinalp147,155,156, Iman Sarrafi147,156, Chip Stewart3, Miranda D Stobbe49,74, Grace Tiao3, Jeremiah A Wala3,6,157, Jiayin Wang27,58,158, Wenyi Wang32, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Michael Wendl27,159,160, Johannes Werner28,161, Zhenggang Wu148, Hong Xue148, Sergei Yakneen8, Takafumi N Yamaguchi12, Kai Ye58,59, Venkata Yellapantula67,68, Junjun Zhang15, David A Wheeler33,34, Li Ding26,27,35 and Jared T Simpson12,36
Notes 4. Production somatic variant calling on the PCAWG Compute Cloud
Christina K Yung15, Brian D O'Connor15,37, Sergei Yakneen8, Junjun Zhang15, Kyle Ellrott38, Kortine Kleinheinz28,30, Naoki Miyoshi39, Keiran M Raine1, Romina Royo40, Gordon Saksena3, Matthias Schlesner28,31, Solomon I Shorser12, Miguel Vazquez40,41, Joachim Weischenfeldt8,99,100, Denis Yuen12, Adam P Butler1, Brandi N Davis-Dusenbery162, Roland Eils28,30,143,144, Vincent Ferretti15,86, Robert L Grossman163, Olivier Harismendy125, Youngwook Kim164,165, Hidewaki Nakagawa81, Steven J Newhouse7, David Torrents40,76 and Lincoln D Stein12,13
Notes 5. PCAWG data portals
Mary Goldman132, Junjun Zhang15, Nuno A Fonseca7,133, Isidro Cortés-Ciriano93,94,95, Qian Xiang134, Brian Craft132, Elena Piñeiro-Yáñez135, Brian D O'Connor15,37, Wojciech Bazant7, Elisabet Barrera7, Alfonso Muñoz7, Robert Petryszak7, Anja Füllgrabe7, Fatima Al-Shahrour135, Maria Keays7, David Haussler132,136, John Weinstein137,138, Wolfgang Huber8, Alfonso Valencia40,76, Irene Papatheodorou7, Jingchun Zhu132, Vincent Ferretti15,86 and Miguel Vazquez40,41
ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Working Groups
Steering committee
Peter J Campbell1,2, Gad Getz3,4,5,6, Jan O Korbel7,8, Lincoln D Stein12,13 and Joshua M Stuart9
Executive committee
Sultan T Al-Sedairy208, Axel Aretz209, Cindy Bell210, Miguel Betancourt211, Christiane Buchholz212, Fabien Calvo213, Christine Chomienne214, Michael Dunn215, Stuart Edmonds216, Eric Green217, Shailja Gupta218, Carolyn M Hutter217, Karine Jegalian219, Jennifer L Jennings10,11, Nic Jones220, Hyung-Lae Kim56, Youyong Lu221,222,223, Hitoshi Nakagama224, Gerd Nettekoven225, Laura Planko225, David Scott220, Tatsuhiro Shibata226,227, Kiyo Shimizu228, Lincoln D Stein12,13, Michael Rudolf Stratton1, Takashi Yugawa228, Giampaolo Tortora229,230, K VijayRaghavan218, Huanming Yang180 and Jean C Zenklusen231
Ethics and legal working group
Yann Joly130, Fruzsina Molnár-Gábor131, Mark Phillips130, Adrian Thorogood130, David Townend232, Don Chalmers129 and Bartha M Knoppers130
Technical working group
Brice Aminou15, Javier Bartolome40, Keith A Boroevich81,233, Rich Boyce7, Alvis Brazma7, Angela N Brooks3,37,157, Alex Buchanan38, Ivo Buchhalter28,29,30, Adam P Butler1, Niall J Byrne15, Andy Cafferkey7, Peter J Campbell1,2, Zhaohong Chen234, Sunghoon Cho235, Wan Choi236, Peter Clapham1, Brandi N Davis-Dusenbery162, Francisco M De La Vega45,46,78, Jonas Demeulemeester91,92, Michelle T Dow234, Lewis Jonathan Dursi12,25, Juergen Eils143,144, Roland Eils28,30,143,144, Kyle Ellrott38, Claudiu Farcas234, Nodirjon Fayzullaev15, Vincent Ferretti15,86, Paul Flicek7, Nuno A Fonseca7,133, Josep Ll Gelpi40,145, Gad Getz3,4,5,6, Robert L Grossman163, Olivier Harismendy125, Allison P Heath237, Michael C Heinold28,30, Julian M Hess3,152, Oliver Hofmann238, Jongwhi H Hong239, Thomas J Hudson240,241, Barbara Hutter140,149,150, Carolyn M Hutter217, Daniel Hübschmann30,120,143,242,243, Seiya Imoto39,39, Sinisa Ivkovic244, Seung-Hyup Jeon236, Wei Jiao12, Jongsun Jung151, Rolf Kabbe28, Andre Kahles112,113,114,115,175, Jules NA Kerssemakers28, Hyung-Lae Kim56, Hyunghwan Kim236, Jihoon Kim245, Youngwook Kim164,165, Kortine Kleinheinz28,30, Jan O Korbel7,8, Michael Koscher246, Antonios Koures234, Milena Kovacevic244, Chris Lawerenz144, Ignaty Leshchiner3, Jia Liu247, Dimitri Livitz3, George L Mihaiescu15, Sanja Mijalkovic244, Ana Mijalkovic Mijalkovic-Lazic244, Satoru Miyano39, Naoki Miyoshi39, Hardeep K Nahal-Bose15, Hidewaki Nakagawa81, Mia Nastic244, Steven J Newhouse7, Jonathan Nicholson1, David Ocana7, Kazuhiro Ohi39, Lucila Ohno-Machado234, Larsson Omberg85, BF Francis Ouellette16,17, Nagarajan Paramasivam28,140, Marc D Perry14,15, Todd D Pihl248, Manuel Prinz28, Montserrat Puiggròs40, Petar Radovic244, Keiran M Raine1, Esther Rheinbay3,6,19, Mara Rosenberg3,19, Romina Royo40, Gunnar Rätsch112,113,114,115,116,117, Gordon Saksena3, Matthias Schlesner28,31, Solomon I Shorser12, Charles Short7, Heidi J Sofia217, Jonathan Spring163, Adam J Struck38, Grace Tiao3, Nebojsa Tijanic244, David Torrents40,76, Peter Van Loo91,92, Miguel Vazquez40,41, David Vicente40, Jeremiah A Wala3,6,157, Zhining Wang231, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Johannes Werner28,161, Ashley Williams234, Youngchoon Woo236, Adam J Wright12, Qian Xiang134, Liming Yang231, Denis Yuen12, Brian D O'Connor15,37, Lincoln D Stein12,13, Sergei Yakneen8, Christina K Yung15 and Junjun Zhang15
Annotations working group
Angela N Brooks3,37,157, Ivo Buchhalter28,29,30, Peter J Campbell1,2, Priyanka Dhingra71,73, Lars Feuerbach127, Mark Gerstein63,64,69, Gad Getz3,4,5,6, Mark P Hamilton249, Henrik Hornshøj153, Todd A Johnson233, Andre Kahles112,113,114,115,175, Abdullah Kahraman250,251,252, Manolis Kellis3,253, Jan O Korbel7,8, Morten Muhlig Nielsen153, Jakob Skou Pedersen153,154, Paz Polak3,4,6, Jüri Reimand12,18, Esther Rheinbay3,6,19, Nicola D Roberts1, Gunnar Rätsch112,113,114,115,116,117, Richard Sallari3, Nasa Sinnott-Armstrong3,45, Alfonso Valencia40,76, Miguel Vazquez40,41, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Christian von Mering252,254 and Ekta Khurana70,71,72,73
Quality control working group
Sergi Beltran49,74, Ivo Buchhalter28,29,30, Peter J Campbell1,2, Roland Eils28,30,143,144, Daniela S Gerhard255, Gad Getz3,4,5,6, Marta Gut49,74, Barbara Hutter140,149,150, Daniel Hübschmann30,120,143,242,243, Kortine Kleinheinz28,30, Jan O Korbel7,8, Dimitri Livitz3, Marc D Perry14,15, Keiran M Raine1, Esther Rheinbay3,6,19, Mara Rosenberg3,19, Gordon Saksena3, Matthias Schlesner28,31, Miranda D Stobbe49,74, Jean-Rémi Trotta74, Johannes Werner28,161, Justin P Whalley74 and Ivo G Gut49,74
Novel somatic mutation calling methods
Matthew H Bailey26,27, Beifang Niu256, Matthias Bieg139,140, Paul C Boutros12,18,20,21, Ivo Buchhalter28,29,30, Adam P Butler1, Ken Chen141, Zechen Chong142, Oliver Drechsel47,49, Lewis Jonathan Dursi12,25, Roland Eils28,30,143,144, Kyle Ellrott38, Shadrielle MG Espiritu12, Yu Fan32, Robert S Fulton26,27,35, Shengjie Gao180, Josep Ll Gelpi40,145, Mark Gerstein63,64,69, Gad Getz3,4,5,6, Santiago Gonzalez7,8, Ivo G Gut49,74, Faraz Hach146,147, Michael C Heinold28,30, Julian M Hess3,152, Jonathan Hinton1, Taobo Hu148, Vincent Huang12, Yi Huang158,257, Barbara Hutter140,149,150, David R Jones1, Jongsun Jung151, Natalie Jäger28, Hyung-Lae Kim56, Kortine Kleinheinz28,30, Sushant Kumar63,64, Yogesh Kumar148, Christopher M Lalansingh12, Ignaty Leshchiner3, Ivica Letunic62, Dimitri Livitz3, Eric Z Ma148, Yosef E Maruvka3,19,152, R Jay Mashl27,57, Michael D McLellan26,27,35, Andrew Menzies1, Ana Milovanovic40, Morten Muhlig Nielsen153, Stephan Ossowski47,48,49, Nagarajan Paramasivam28,140, Jakob Skou Pedersen153,154, Marc D Perry14,15, Montserrat Puiggròs40, Keiran M Raine1, Esther Rheinbay3,6,19, Romina Royo40, S Cenk Sahinalp147,155,156, Gordon Saksena3, Iman Sarrafi147,156, Matthias Schlesner28,31, Lucy Stebbings1, Chip Stewart3, Miranda D Stobbe49,74, Jon W Teague1, Grace Tiao3, David Torrents40,76, Jeremiah A Wala3,6,157, Jiayin Wang27,58,158, Wenyi Wang32, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Michael C Wendl27,159,160, Johannes Werner28,161, David A Wheeler33,34, Zhenggang Wu148, Hong Xue148, Sergei Yakneen8, Takafumi N Yamaguchi12, Kai Ye58,59, Venkata D Yellapantula67,68, Christina K Yung15, Junjun Zhang15, Li Ding26,27,35 and Jared T Simpson12,36
Drivers and functional interpretation
Federico Abascal1, Samirkumar B Amin184,185,186, Gary D Bader13, Pratiti Bandopadhayay3,258,259, Jonathan Barenboim12, Rameen Beroukhim3,6,168, Johanna Bertl153,206, Keith A Boroevich81,233, Søren Brunak260,261, Peter J Campbell1,2, Joana Carlevaro-Fita262,263,264, Dimple Chakravarty265,266, Calvin Wing Yiu Chan28,128, Ken Chen141, Jung Kyoon Choi267, Jordi Deu-Pons89,205, Priyanka Dhingra71,73, Klev Diamanti268, Lars Feuerbach127, J Lynn Fink40,269, Nuno A Fonseca7,133, Joan Frigola205, Carlo Gambacorti-Passerini270, Dale W Garsed271, Qianyun Guo154, Ivo G Gut49,74, David Haan9, Mark P Hamilton249, Nicholas J Haradhvala3,19, Arif O Harmanci64,272, Mohamed Helmy171, Carl Herrmann28,30,273, Julian M Hess3,152, Asger Hobolth154,206, Ermin Hodzic156, Chen Hong127,128, Henrik Hornshøj153, Keren Isaev12,18, Jose MG Izarzugaza260, Rory Johnson263,274, Todd A Johnson233, Malene Juul153, Randi Istrup Juul153, Andre Kahles112,113,114,115,175, Abdullah Kahraman250,251,252, Manolis Kellis3,253, Ekta Khurana70,71,72,73, Jaegil Kim3, Jong K Kim275, Youngwook Kim164,165, Jan Komorowski268,276, Jan O Korbel7,8, Sushant Kumar63,64, Andrés Lanzós263,264,274, Erik Larsson112, Donghoon Lee64, Kjong-Van Lehmann112,114,115,174,175, Shantao Li64, Xiaotong Li64, Ziao Lin3,277, Eric Minwei Liu71,73,278, Lucas Lochovsky63,64,186, Shaoke Lou63,64, Tobias Madsen153, Kathleen Marchal279,280, Iñigo Martincorena1, Alexander Martinez-Fundichely71,72,73, Yosef E Maruvka3,19,152, Patrick D McGillivray63, William Meyerson64,281, Ferran Muiños87,89, Loris Mularoni87,89, Hidewaki Nakagawa81, Morten Muhlig Nielsen153, Marta Paczkowska12, Keunchil Park282,283, Kiejung Park284, Tirso Pons285, Sergio Pulido-Tamayo279,280, Jüri Reimand12,18, Iker Reyes-Salazar87, Matthew A Reyna286, Esther Rheinbay3,6,19, Mark A Rubin274,287,288,289,290, Carlota Rubio-Perez87,89,203, S Cenk Sahinalp147,155,156, Gordon Saksena3, Leonidas Salichos63,64, Chris Sander112,157,291,292, Steven E Schumacher3,204, Mark Shackleton271, Ofer Shapira3,157, Ciyue Shen292,293, Raunak Shrestha147, Shimin Shuai12,13, Nikos Sidiropoulos100, Lina Sieverling127,128, Nasa Sinnott-Armstrong3,45, Lincoln D Stein12,13, David Tamborero87,89, Grace Tiao3, Tatsuhiko Tsunoda233,294,295,296, Husen M Umer268,297, Liis Uusküla-Reimand298,299, Alfonso Valencia40,76, Miguel Vazquez40,41, Lieven PC Verbeke280,300, Claes Wadelius301, Lina Wadi12, Jiayin Wang27,58,158, Jonathan Warrell63,64, Sebastian M Waszak8, Joachim Weischenfeldt8,99,100, Guanming Wu302, Jun Yu303, Jing Zhang64, Xuanping Zhang158,304, Yan Zhang64,305,306, Zhongming Zhao307, Lihua Zou308, Christian von Mering252,254, Mark Gerstein63,64,69, Gad Getz3,4,5,6, Michael S Lawrence3,19,233, Jakob Skou Pedersen153,154, Benjamin J Raphael286, Joshua M Stuart9 and David A Wheeler33,34
Integration of transcriptome and genome
Samirkumar B Amin184,185,186, Philip Awadalla12,13, Peter J Bailey187, Claudia Calabrese7,8, Aurélien Chateigner15, Isidro Cortés-Ciriano93,94,95, Brian Craft132, David Craft3,309, Chad J Creighton200, Natalie R Davidson112,114,115,117,174, Deniz Demircioğlu178,179, Serap Erkek8, Nuno A Fonseca7,133, Milana Frenkel-Morgenstern188, Mary J Goldman132, Liliana Greger7, Jonathan Göke178,201, Yao He176, Katherine A Hoadley23,24, Yong Hou180,181, Matthew R Huska118, Andre Kahles112,113,114,115,175, Ekta Khurana70,71,72,73, Helena Kilpinen189, Jan O Korbel7,8, Fabien C Lamaze12, Kjong-Van Lehmann112,114,115,174,175, Chang Li180,181, Siliang Li180,181, Xiaobo Li180,181, Xinyue Li180, Dongbing Liu180,181, Fenglin Liu176,177, Xingmin Liu180,181, Maximillian G Marin37, Julia Markowski118, Matthew Meyerson3,6,157, Tannistha Nandi190, Morten Muhlig Nielsen153, Akinyemi I Ojesina191,192,193, BF Francis Ouellette16,17, Qiang Pan-Hammarström180,194, Peter J Park94,95, Chandra Sekhar Pedamallu3,6,168, Jakob Skou Pedersen153,154, Marc D Perry14,15, Roland F Schwarz7,118,119,120, Yuichi Shiraishi39, Reiner Siebert79,80, Cameron M Soulette37, Stefan G Stark115,174,182,183, Oliver Stegle7,8,124, Hong Su180,181, Patrick Tan190,195,196,197, Bin Tean Teh195,196,197,198,199, Lara Urban7,8, Jian Wang180, Sebastian M Waszak8, Kui Wu180,181, Qian Xiang134, Heng Xiong180,181, Sergei Yakneen8, Huanming Yang180, Chen Ye180,181, Christina K Yung15, Fan Zhang176, Junjun Zhang15, Xiuqing Zhang180, Zemin Zhang176,202, Liangtao Zheng176, Jingchun Zhu132, Shida Zhu180,181, Alvis Brazma7, Angela N Brooks3,37,157 and Gunnar Rätsch112,113,114,115,116,117
Integration of epigenome and genome
Hiroyuki Aburatani310, Hans Binder311,312, Huy Q Dinh313, Lars Feuerbach127, Shengjie Gao180, Ivo G Gut49,74, Simon C Heath49,74, Steve Hoffmann311,312,314,315, Charles David Imbusch127, Ekta Khurana70,71,72,73, Helene Kretzmer312,315, Peter W Laird316, Jose I Martin-Subero76,317, Genta Nagae310,318, Paz Polak3,4,6, Hui Shen319, Reiner Siebert79,80, Nasa Sinnott-Armstrong3,45, Miranda D Stobbe49,74, Qi Wang246, Dieter Weichenhan320, Sergei Yakneen8, Wanding Zhou319, Benjamin P Berman313,321,322, Benedikt Brors127,150,323 and Christoph Plass320
Patterns of structural variations, signatures, genomic correlations, retrotransposons, mobile elements
Kadir C Akdemir141, Eva G Alvarez42,43,44, Adrian Baez-Ortega55, Paul C Boutros12,18,20,21, David D L Bowtell271, Benedikt Brors127,150,323, Kathleen H Burns324,325, John Busanovich3,326, Kin Chan327, Ken Chen141, Isidro Cortés-Ciriano93,94,95, Ana Dueso-Barroso40, Andrew J Dunford3, Paul A Edwards328,329, Xavier Estivill47,84, Dariush Etemadmoghadam271, Lars Feuerbach127, J Lynn Fink40,269, Milana Frenkel-Morgenstern188, Dale W Garsed271, Mark Gerstein63,64,69, Dmitry A Gordenin97, David Haan9, James E Haber330, Julian M Hess3,152, Barbara Hutter140,149,150, Marcin Imielinski166,167, David TW Jones331,332, Young Seok Ju1,267, Marat D Kazanov333,334,335, Leszek J Klimczak98, Youngil Koh336,337, Jan O Korbel7,8, Kiran Kumar3, Eunjung Alice Lee338, Jake June-Koo Lee94,95, Yilong Li1, Andy G Lynch328,329,339, Geoff Macintyre328, Florian Markowetz328,329, Iñigo Martincorena1, Alexander Martinez-Fundichely71,72,73, Matthew Meyerson3,6,157, Satoru Miyano39, Hidewaki Nakagawa81, Fabio CP Navarro63, Stephan Ossowski47,48,49, Peter J Park94,95, John V Pearson340,341, Montserrat Puiggròs40, Karsten Rippe120, Nicola D Roberts1, Steven A Roberts107, Bernardo Rodriguez-Martin42,43,44, Steven E Schumacher3,204, Ralph Scully342, Mark Shackleton271, Nikos Sidiropoulos100, Lina Sieverling127,128, Chip Stewart3, David Torrents40,76, Jose MC Tubio42,43,44, Izar Villasante40, Nicola Waddell340,341, Jeremiah A Wala3,6,157, Joachim Weischenfeldt8,99,100, Lixing Yang343, Xiaotong Yao166,344, Sung-Soo Yoon337, Jorge Zamora1,42,43,44, Cheng-Zhong Zhang3,6,157, Rameen Beroukhim3,6,168 and Peter J Campbell1,2
Mutation signatures and processes
Ludmil B Alexandrov1,101, Erik N Bergstrom345, Arnoud Boot196,346, Paul C Boutros12,18,20,21, Kin Chan327, Kyle Covington34, Akihiro Fujimoto81, Gad Getz3,4,5,6, Dmitry A Gordenin97, Nicholas J Haradhvala3,19, Mi Ni Huang196,346, S. M. Ashiqul Islam101, Marat D Kazanov333,334,335, Jaegil Kim3, Leszek J Klimczak98, Michael S Lawrence3,19,233, Iñigo Martincorena1, John R McPherson196,346, Sandro Morganella1, Ville Mustonen347,348,349, Hidewaki Nakagawa81, Alvin Wei Tian Ng350, Paz Polak3,4,6, Stephenie D Prokopec12, Steven A Roberts107, Radhakrishnan Sabarinathan87,88,89, Natalie Saini97, Tatsuhiro Shibata226,227, Yuichi Shiraishi39, Ignacio Vázquez-García1,67,354,355, Yang Wu196,346, Fouad Yousif12, Willie Yu356, Steven G Rozen196,197,346, Michael Rudolf Stratton1 and Bin Tean Teh195,196,197,198,199
Germline cancer genome
Ludmil B Alexandrov1,101, Eva G Alvarez42,43,44, Adrian Baez-Ortega55, Matthew H Bailey26,27, Mattia Bosio40,47,49, G Steven Bova126, Alvis Brazma7, Alicia L Bruzos42,43,44, Ivo Buchhalter28,29,30, Carlos D Bustamante45,78, Atul J Butte121, Andy Cafferkey7, Claudia Calabrese7,8, Peter J Campbell1,2, Stephen J Chanock96, Nilanjan Chatterjee122,123,123, Jieming Chen64,110, Francisco M De La Vega45,46,78, Olivier Delaneau50,51,52, German M Demidov47,48,49, Anthony DiBiase60, Li Ding26,27,35, Oliver Drechsel47,49, Lewis Jonathan Dursi12,25, Douglas F Easton82,83, Serap Erkek8, Georgia Escaramis47,103,104, Erik Garrison1, Mark Gerstein63,64,69, Gad Getz3,4,5,6, Dmitry A Gordenin97, Nina Habermann8, Olivier Harismendy125, Eoghan Harrington105, Shuto Hayashi39, Seong Gu Heo173, José María Heredia-Genestar54, Aliaksei Z Holik47, Eun Pyo Hong173, Xing Hua96, Kuan-lin Huang27,61, Seiya Imoto39,39, Sissel Juul105, Ekta Khurana70,71,72,73, Hyung-Lae Kim56, Youngwook Kim164,165, Leszek J Klimczak98, Roelof Koster108, Sushant Kumar63,64, Ivica Letunic62, Yilong Li1, Tomas Marques-Bonet74,75,76,77, R Jay Mashl27,57, Simon Mayes106, Michael D McLellan26,27,35, Lisa Mirabello96, Francesc Muyas47,48,49, Hidewaki Nakagawa81, Arcadi Navarro74,75,76, Steven J Newhouse7, Stephan Ossowski47,48,49, Ji Wan Park173, Esa Pitkänen8, Aparna Prasad49, Raquel Rabionet47,49,102, Benjamin Raeder8, Tobias Rausch8, Steven A Roberts107, Bernardo Rodriguez-Martin42,43,44, Vasilisa A Rudneva8, Gunnar Rätsch112,113,114,115,116,117, Natalie Saini97, Matthias Schlesner28,31, Roland F Schwarz7,118,119,120, Ayellet V Segre3,53, Tal Shmaya46, Suyash S Shringarpure45, Nikos Sidiropoulos100, Reiner Siebert79,80, Jared T Simpson12,36, Lei Song96, Oliver Stegle7,8,124, Hana Susak47,49, Tomas J Tanskanen109, Grace Tiao3, Marta Tojo44, Jose MC Tubio42,43,44, Daniel J Turner106, Lara Urban7,8, Sebastian M Waszak8, David C Wedge1,65,66, Joachim Weischenfeldt8,99,100, David A Wheeler33,34, Mark H Wright45, Dai-Ying Wu46, Tian Xia357, Sergei Yakneen8, Kai Ye58,59, Venkata D Yellapantula67,68, Jorge Zamora1,42,43,44, Bin Zhu96, Xavier Estivill47,84 and Jan O Korbel7,8
Tumour subtypes and clinical translation
Fatima Al-Shahrour135, Gurnit Atwal12,13,172, Peter J Bailey187, Paul C Boutros12,18,20,21, Peter J Campbell1,2, David K Chang187,358, Susanna L Cooke187, Vikram Deshpande19, Bishoy M Faltas117, William C Faquin19, Gad Getz3,4,5,6, Syed Haider12, Wei Jiao12, Vera B Kaiser359, Rosa Karlić360, Mamoru Kato361, Kirsten Kübler3,6,19, Alexander J Lazar22, Constance H Li12,18, David N Louis19, Adam Margolin38, Sancha Martin1,362, Hardeep K Nahal-Bose15, G Petur Nielsen19, Serena Nik-Zainal1,351,352,353, Larsson Omberg85, Christine P'ng12, Marc D Perry14,15, Paz Polak3,4,6, Esther Rheinbay3,6,19, Mark A Rubin274,287,288,289,290, Colin A Semple359, Dennis C Sgroi19, Tatsuhiro Shibata226,227, Reiner Siebert79,80, Jaclyn Smith38, Miranda D Stobbe49,74, Ren X Sun12, Kevin Thai15, Derek W Wright363,364, Chin-Lee Wu19, Ke Yuan328,362,365, Junjun Zhang15, Andrew V Biankin187,358,366,367, Levi Garraway157, Sean M Grimmond368, Katherine A Hoadley23,24 and Lincoln D Stein12,13
Evolution and heterogeneity
David J Adams1, Pavana Anur369, Rameen Beroukhim3,6,168, Paul C Boutros12,18,20,21, David D L Bowtell271, Peter J Campbell1,2, Shaolong Cao32, Elizabeth L Christie271, Marek Cmero370,371,372, Yupeng Cun373, Kevin J Dawson1, Jonas Demeulemeester91,92, Stefan C Dentro1,65,91, Amit G Deshwar374, Nilgun Donmez147,156, Ruben M Drews328, Roland Eils28,30,143,144, Yu Fan32, Matthew W Fittall91, Dale W Garsed271, Moritz Gerstung7,8, Gad Getz3,4,5,6, Santiago Gonzalez7,8, Gavin Ha3, Kerstin Haase91, Marcin Imielinski166,167, Lara Jerman8,375, Yuan Ji376,377, Clemency Jolly91, Kortine Kleinheinz28,30, Juhee Lee378, Henry Lee-Six1, Ignaty Leshchiner3, Dimitri Livitz3, Geoff Macintyre328, Salem Malikic147,156, Florian Markowetz328,329, Iñigo Martincorena1, Thomas J Mitchell1,329,379, Quaid D Morris172,380, Ville Mustonen347,348,349, Layla Oesper381, Martin Peifer373, Myron Peto382, Benjamin J Raphael286, Daniel Rosebrock3, Yulia Rubanova36,172, S Cenk Sahinalp147,155,156, Adriana Salcedo12, Matthias Schlesner28,31, Steven E Schumacher3,204, Subhajit Sengupta383, Ruian Shi380, Seung Jun Shin183, Oliver Spiro3, Lincoln D Stein12,13, Maxime Tarabichi1,91, Shankar Vembu380,384, Ignacio Vázquez-García1,67,354,355, Wenyi Wang32, David A Wheeler33,34, Jeffrey A Wintersinger170,171,172, Tsun-Po Yang373, Xiaotong Yao166,344, Kaixian Yu385, Ke Yuan328,362,365, Hongtu Zhu386,387, Paul T Spellman388, Peter Van Loo91,92 and David C Wedge1,65,66
Portals, visualisation and software infrastructure
Fatima Al-Shahrour135, Elisabet Barrera7, Wojciech Bazant7, Alvis Brazma7, Isidro Cortés-Ciriano93,94,95, Brian Craft132, David Craft3,309, Vincent Ferretti15,86, Nuno A Fonseca7,133, Anja Füllgrabe7, Mary J Goldman132, Wolfgang Huber8, Maria Keays7, Alfonso Muñoz7, Brian D O'Connor15,37, Irene Papatheodorou7, Robert Petryszak7, Elena Piñeiro-Yáñez135, Alfonso Valencia40,76, John N Weinstein137,138, Qian Xiang134, Junjun Zhang15, David Haussler132,136, Miguel Vazquez40,41 and Jingchun Zhu132
Mitochondrial variants and HLA/immunogenicity
Peter J Campbell1,2, Yiwen Chen32, Chad J Creighton200, Li Ding26,27,35, Akihiro Fujimoto81, Masashi Fujita81, Gad Getz3,4,5,6, Leng Han304, Takanori Hasegawa39, Shuto Hayashi39, Seiya Imoto39,39, Young Seok Ju1,267, Hyung-Lae Kim56, Youngwook Kim164,165, Youngil Koh336,337, Mitsuhiro Komura39, Jun Li32, Iñigo Martincorena1, Satoru Miyano39, Shinichi Mizuno389, Keunchil Park282,283, Eigo Shimizu39, Yumeng Wang32,390, John N Weinstein137,138, Yanxun Xu391, Rui Yamaguchi39, Fan Yang380, Yang Yang304, Christopher J Yoon267, Sung-Soo Yoon337, Yuan Yuan32, Fan Zhang176, Zemin Zhang176,202, Han Liang32 and Hidewaki Nakagawa81
Pathogens
Malik Alawi392,393, Ivan Borozan12, Daniel S Brewer394,395, Colin S Cooper395,396,397, Nikita Desai15, Roland Eils28,30,143,144, Vincent Ferretti15,86, Adam Grundhoff392,398, Murat Iskar399, Kortine Kleinheinz28,30, Hidewaki Nakagawa81, Akinyemi I Ojesina191,192,193, Chandra Sekhar Pedamallu3,6,168, Matthias Schlesner28,31, Xiaoping Su400, Marc Zapatka399 and Peter Lichter149,399
Providers of tumour sequencing data
Tumour Specific Providers – Australia (Ovarian cancer)
Kathryn Alsop271, Australian Ovarian Cancer Study Group340,401,402, Timothy JC Bruxner269, Angelika N Christ269, Elizabeth L Christie271, Stephen M Cordner403, Prue A Cowin401, Ronny Drapkin404, Dariush Etemadmoghadam271, Sian Fereday401, Dale W Garsed271, Joshy George186, Sean M Grimmond368, Anne Hamilton401, Oliver Holmes340,341, Jillian A Hung405,406, Karin S Kassahn269,407, Stephen H Kazakoff340,341, Catherine J Kennedy408,409, Conrad R Leonard340,341, Linda Mileshkin271, David K Miller269,358,410, Gisela Mir Arnau401, Chris Mitchell401, Felicity Newell340,341, Katia Nones340,341, Ann-Marie Patch340,341, John V Pearson340,341, Michael C Quinn340,341, Mark Shackleton271, Darrin F Taylor269, Heather Thorne401, Nadia Traficante401, Ravikiran Vedururu401, Nick M Waddell341, Nicola Waddell340,341, Paul M Waring411, Scott Wood340,341, Qinying Xu340,341, Anna deFazio412,413,414 and David D L Bowtell271
Tumour Specific Providers – Australia (Pancreatic cancer)
Matthew J Anderson269, Davide Antonello415, Andrew P Barbour416,417, Claudio Bassi415, Samantha Bersani418, Timothy JC Bruxner269, Ivana Cataldo418,419, David K Chang187,358, Lorraine A Chantrill358,420, Yoke-Eng Chiew412, Angela Chou358,421, Angelika N Christ269, Sara Cingarlini229, Nicole Cloonan422, Vincenzo Corbo419,423, Maria Vittoria Davi424, Fraser R Duthie187,425, J Lynn Fink40,269, Anthony J Gill358,421, Janet S Graham187,426, Ivon Harliwong269, Oliver Holmes340,341, Nigel B Jamieson187,367,427, Amber L Johns358,410, Karin S Kassahn269,407, Stephen H Kazakoff340,341, James G Kench358,421,428, Luca Landoni415, Rita T Lawlor419, Conrad R Leonard340,341, Andrea Mafficini419, Neil D Merrett415,429, David K Miller269,358,410, Marco Miotto415, Elizabeth A Musgrove187, Adnan M Nagrial358, Felicity Newell340,341, Katia Nones340,341, Karin A Oien411,430, Marina Pajic358, Ann-Marie Patch340,341, John V Pearson340,341, Mark Pinese431, Michael C Quinn340,341, Alan J Robertson269, Ilse Rooman358, Borislav C Rusev419, Jaswinder S Samra415,421, Maria Scardoni418, Christopher J Scarlett358,432, Aldo Scarpa419, Elisabetta Sereni415, Katarzyna O Sikora419, Michele Simbolo423, Morgan L Taschuk15, Christopher W Toon358, Giampaolo Tortora229,230, Caterina Vicentini419, Nick M Waddell341, Nicola Waddell340,341, Scott Wood340,341, Jianmin Wu358, Qinying Xu340,341, Nikolajs Zeps433,434, Andrew V Biankin187,358,366,367 and Sean M Grimmond368
Tumour Specific Providers – Australia (Skin cancer)
Lauri A Aaltonen111, Andreas Behren435, Hazel Burke436, Jonathan Cebon435, Rebecca A Dagg437, Ricardo De Paoli-Iseppi438, Ken Dutton-Regester340, Matthew A Field439, Anna Fitzgerald440, Sean M Grimmond368, Peter Hersey436, Oliver Holmes340,341, Valerie Jakrot436, Peter A Johansson340, Hojabr Kakavand438, Stephen H Kazakoff340,341, Richard F Kefford441, Loretta MS Lau442, Conrad R Leonard340,341, Georgina V Long443, Felicity Newell340,341, Katia Nones340,341, Ann-Marie Patch340,341, John V Pearson340,341, Hilda A Pickett442, Antonia L Pritchard340, Gulietta M Pupo444, Robyn PM Saw443, Sarah-Jane Schramm445, Mark Shackleton271, Catherine A Shang440, Ping Shang443, Andrew J Spillane443, Jonathan R Stretch443, Varsha Tembe445, John F Thompson443, Ricardo E Vilain446, Nick M Waddell341, Nicola Waddell340,341, James S Wilmott443, Scott Wood340,341, Qinying Xu340,341, Jean Y Yang447, Nicholas K Hayward340,436, Graham J Mann448,449 and Richard A Scolyer413,443,446,450
Tumour Specific Providers – Canada (Pancreatic cancer)
John Bartlett451,452, Prashant Bavi453, Ivan Borozan12, Dianne E Chadwick454, Michelle Chan-Seng-Yue453, Sean Cleary453,455, Ashton A Connor455,456, Karolina Czajka241, Robert E Denroche453, Neesha C Dhani457, Jenna Eagles241, Vincent Ferretti15,86, Steven Gallinger453,455,456, Robert C Grant453,456, David Hedley457, Michael A Hollingsworth458, Gun Ho Jang453, Jeremy Johns241, Sangeetha Kalimuthu453, Sheng-Ben Liang459, Ilinca Lungu453,460, Xuemei Luo12, Faridah Mbabaali241, Treasa A McPherson456, Jessica K Miller241, Malcolm J Moore457, Faiyaz Notta453,461, Danielle Pasternack241, Gloria M Petersen462, Michael H A Roehrl18,453,463,464,465, Michelle Sam241, Iris Selander456, Stefano Serra411, Sagedeh Shahabi459, Morgan L Taschuk15, Sarah P Thayer458, Lee E Timms241, Gavin W Wilson12,453, Julie M Wilson453, Bradly G Wouters466, Thomas J Hudson240,241, John D McPherson241,453,467 and Lincoln D Stein12,13
Tumour Specific Providers – Canada (Prostate cancer)
Timothy A Beck15,468, Vinayak Bhandari12, Colin C Collins147, Shadrielle MG Espiritu12, Neil E Fleshner469, Natalie S Fox12, Michael Fraser12, Syed Haider12, Lawrence E Heisler470, Vincent Huang12, Emilie Lalonde12, Julie Livingstone12, John D McPherson241,453,467, Alice Meng471, Veronica Y Sabelnykova12, Adriana Salcedo12, Yu-Jia Shiah12, Theodorus Van der Kwast472, Takafumi N Yamaguchi12, Paul C Boutros12,18,20,21 and Robert G Bristow18,473,474,475,476
Tumour Specific Providers – China (Gastric cancer)
Shuai Ding477, Daiming Fan478, Yong Hou180,181, Yi Huang158,257, Lin Li180, Siliang Li180,181, Dongbing Liu180,181, Xingmin Liu180,181, Yongzhan Nie478,479, Hong Su180,181, Jian Wang180, Kui Wu180,181, Xiao Xiao158, Rui Xing222,480, Shanlin Yang477, Yingyan Yu481, Xiuqing Zhang180, Yong Zhou180, Shida Zhu180,181, Youyong Lu221,222,223 and Huanming Yang180
Tumour Specific Providers – EU: France (Renal cancer)
Rosamonde E Banks482, Guillaume Bourque483,484, Alvis Brazma7, Paul Brennan485, Louis Letourneau486, Yasser Riazalhosseini484, Ghislaine Scelo485, Naveen Vasudev487, Juris Viksna488, Mark Lathrop484 and Jörg Tost489
Tumour Specific Providers – EU: United Kingdom (Breast cancer)
Sung-Min Ahn490, Ludmil B Alexandrov1,101, Samuel Aparicio491, Laurent Arnould492, MR Aure493, Shriram G Bhosle1, E Birney7, Ake Borg494, S Boyault495, AB Brinkman496, JE Brock497, A Broeks498, Adam P Butler1, AL Børresen-Dale493, C Caldas499,500, Peter J Campbell1,2, Suet-Feung Chin499,500, Helen Davies1,351,352, C Desmedt501,502, L Dirix503, S Dronov1, Anna Ehinger504, JE Eyfjord505, A Fatima204, JA Foekens506, PA Futreal507, Øystein Garred508,509, Moritz Gerstung7,8, Dilip D Giri510, D Glodzik1, Dorthe Grabau511, Holmfridur Hilmarsdottir505, GK Hooijer512, Jocelyne Jacquemier513, SJ Jang514, Jon G Jonasson505, Jos Jonkers515, HY Kim513, Tari A King516,517, Stian Knappskog1,518,518, G Kong513, S Krishnamurthy519, SR Lakhani520, Anita Langerød493, Denis Larsimont521, HJ Lee514, JY Lee522, Ming Ta Michael Lee507, Yilong Li1, Ole Christian Lingjærde523, Gaetan MacGrogan524, JWM Martens506, Sancha Martin1,362, Iñigo Martincorena1, Andrew Menzies1, Sandro Morganella1, Ville Mustonen347,348,349, Serena Nik-Zainal1,351,352,353, Sarah O'Meara1, I Pauporté214, Sarah Pinder525, X Pivot526, Elena Provenzano527, CA Purdie528, Keiran M Raine1, M Ramakrishna1, K Ramakrishnan1, Jorge Reis-Filho510, AL Richardson204, M Ringnér494, Javier Bartolomé Rodriguez40, FG Rodríguez-González261, G Romieu529, Roberto Salgado411, Torill Sauer523, R Shepherd1, AM Sieuwerts506, PT Simpson520, M Smid506, C Sotiriou234, PN Span530, Lucy Stebbings1, Ólafur Andri Stefánsson531, Alasdair Stenhouse532, HG Stunnenberg181,533, Fred Sweep534, BK Tan535, Jon W Teague1, Gilles Thomas536, AM Thompson532, S Tommasi537, I Treilleux538,539, Andrew Tutt204, NT Ueno387, S Van Laere503, Peter Van Loo91,92, GG Van den Eynden503, P Vermeulen503, Alain Viari419, A Vincent-Salomon533, David C Wedge1,65,66, Bernice Huimin Wong540, Lucy Yates1, X Zou1, CHM van Deurzen541, MJ van de Vijver411, L van't Veer542 and Michael Rudolf Stratton1
Tumour Specific Providers – Germany (Malignant lymphoma)
Ole Ammerpohl543,544, Sietse Aukema545,546, Anke K Bergmann547, Stephan H Bernhart311,312,315, Hans Binder311,312, Arndt Borkhardt548, Christoph Borst549, Benedikt Brors127,150,323, Birgit Burkhardt550, Alexander Claviez551, Roland Eils28,30,143,144, Maria Elisabeth Goebler552, Andrea Haake543, Siegfried Haas549, Martin Hansmann553, Jessica I Hoell548, Steve Hoffmann311,312,314,315, Michael Hummel554, Daniel Hübschmann30,120,143,242,243, Dennis Karsch555, Wolfram Klapper545, Kortine Kleinheinz28,30, Michael Kneba555, Jan O Korbel7,8, Helene Kretzmer312,315, Markus Kreuz556, Dieter Kube557, Ralf Küppers558, Chris Lawerenz144, Dido Lenze554, Peter Lichter149,399, Markus Loeffler556, Cristina López80,543, Luisa Mantovani-Löffler559, Peter Möller560, German Ott561, Bernhard Radlwimmer399, Julia Richter543,545, Marius Rohde562, Philip C Rosenstiel563, Andreas Rosenwald564, Markus B Schilhabel563, Matthias Schlesner28,31, Stefan Schreiber565, Peter F Stadler311,312,315, Peter Staib566, Stephan Stilgenbauer567, Stephanie Sungalee8, Monika Szczepanowski545, Umut H Toprak30,568, Lorenz HP Trümper557, Rabea Wagener80,543, Thorsten Zenz150 and Reiner Siebert79,80
Tumour Specific Providers – Germany (Paediatric Brain cancer)
Ivo Buchhalter28,29,30, Juergen Eils143,144, Roland Eils28,30,143,144, Volker Hovestadt399, Barbara Hutter140,149,150, David TW Jones331,332, Natalie Jäger28, Christof von Kalle120, Marcel Kool246,331, Jan O Korbel7,8, Andrey Korshunov246, Pablo Landgraf569,570, Chris Lawerenz144, Hans Lehrach571, Paul A Northcott572, Stefan M Pfister246,331,573, Bernhard Radlwimmer399, Guido Reifenberger570, Matthias Schlesner28,31, Hans-Jörg Warnatz571, Joachim Weischenfeldt8,99,100, Stephan Wolf574, Marie-Laure Yaspo571, Marc Zapatka399 and Peter Lichter149,399
Tumour Specific Providers – Germany (Prostate cancer)
Yassen Assenov575, Benedikt Brors127,150,323, Juergen Eils143,144, Roland Eils28,30,143,144, Lars Feuerbach127, Clarissa Gerhauser320, Jan O Korbel7,8, Chris Lawerenz144, Hans Lehrach571, Sarah Minner576, Christoph Plass320, Thorsten Schlomm99,577, Nikos Sidiropoulos100, Ronald Simon578, Hans-Jörg Warnatz571, Dieter Weichenhan320, Joachim Weischenfeldt8,99,100, Marie-Laure Yaspo571, Guido Sauter578 and Holger Sültmann150,579
Tumour Specific Providers – India (Oral cancer)
Nidhan K Biswas580, Luca Landoni415, Arindam Maitra580, Partha P Majumder580 and Rajiv Sarin581
Tumour Specific Providers – Italy (Pancreatic cancer)
Davide Antonello415, Stefano Barbi423, Claudio Bassi415, Samantha Bersani418, Giada Bonizzato419, Cinzia Cantù419, Ivana Cataldo418,419, Sara Cingarlini229, Vincenzo Corbo419,423, Maria Vittoria Davi424, Angelo P Dei Tos582, Matteo Fassan583, Sonia Grimaldi419, Luca Landoni415, Rita T Lawlor419, Claudio Luchini418, Andrea Mafficini419, Giuseppe Malleo415, Giovanni Marchegiani415, Michele Milella229, Marco Miotto415, Salvatore Paiella415, Antonio Pea415, Paolo Pederzoli415, Borislav C Rusev419, Andrea Ruzzenente415, Roberto Salvia415, Maria Scardoni418, Elisabetta Sereni415, Michele Simbolo423, Nicola Sperandio419, Giampaolo Tortora229,230, Caterina Vicentini419 and Aldo Scarpa419
Tumour Specific Providers – Japan (Biliary tract cancer)
Yasuhito Arai226, Natsuko Hama226, Nobuyoshi Hiraoka584, Fumie Hosoda226,226, Mamoru Kato361, Hiromi Nakamura226, Hidenori Ojima585, Takuji Okusaka586, Yasushi Totoki226, Tomoko Urushidate227 and Tatsuhiro Shibata226,227
Tumour Specific Providers – Japan (Gastric cancer)
Yasuhito Arai226, Masashi Fukayama587, Natsuko Hama226, Fumie Hosoda226,226, Shumpei Ishikawa588, Hitoshi Katai589, Mamoru Kato361, Hiroto Katoh588, Daisuke Komura588, Genta Nagae310,318, Hiromi Nakamura226, Hirofumi Rokutan361, Mihoko Saito-Adachi361, Akihiro Suzuki310,590, Hirokazu Taniguchi591, Kenji Tatsuno310, Yasushi Totoki226, Tetsuo Ushiku587, Shinichi Yachida226,592, Shogo Yamamoto310, Hiroyuki Aburatani310 and Tatsuhiro Shibata226,227
Tumour Specific Providers – Japan (Liver cancer)
Hiroyuki Aburatani310, Hiroshi Aikata593, Koji Arihiro593, Shun-ichi Ariizumi594, Keith A Boroevich81,233, Kazuaki Chayama593, Akihiro Fujimoto81, Masashi Fujita81, Mayuko Furuta81, Kunihito Gotoh595, Natsuko Hama226, Takanori Hasegawa39, Shinya Hayami596, Shuto Hayashi39, Satoshi Hirano597, Seiya Imoto39,39, Mamoru Kato361, Yoshiiku Kawakami593, Kazuhiro Maejima81, Satoru Miyano39, Genta Nagae310,318, Hiromi Nakamura226, Toru Nakamura597, Kaoru Nakano81, Hideki Ohdan593, Aya Sasaki-Oku81, Yuichi Shiraishi39, Hiroko Tanaka39, Yasushi Totoki226, Tatsuhiko Tsunoda233,294,295,296, Masaki Ueno596, Rui Yamaguchi39, Masakazu Yamamoto594, Hiroki Yamaue596, Hidewaki Nakagawa81 and Tatsuhiro Shibata226,227
Tumour Specific Providers – Singapore (Biliary tract cancer)
Su Pin Choo598, Ioana Cutcutache196,346, Narong Khuntikeo415,599, John R McPherson196,346, Choon Kiat Ong600, Chawalit Pairojkul411, Irinel Popescu601, Steven G Rozen196,197,346, Patrick Tan190,195,196,197 and Bin Tean Teh195,196,197,198,199
Tumour Specific Providers – South Korea (Blood cancer)
Keun Soo Ahn602, Hyung-Lae Kim56, Youngil Koh336,337 and Sung-Soo Yoon337
Tumour Specific Providers – Spain (Chronic Lymphocytic Leukaemia)
Marta Aymerich603, Josep Ll Gelpi40,145, Ivo G Gut49,74, Marta Gut49,74, Armando Lopez-Guillermo604, Carlos López-Otín605, Xose S Puente605, Romina Royo40, David Torrents40,76 and Elias Campo606,607
Tumour Specific Providers – United Kingdom (Bone cancer)
Fernanda Amary608, Daniel Baumhoer609, Sam Behjati1, Bodil Bjerkehagen609,610, PA Futreal507, Ola Myklebost518, Nischalan Pillay611, Patrick Tarpey612, Roberto Tirabosco613, Olga Zaikova614, Peter J Campbell1,2 and Adrienne M Flanagan615
Tumour Specific Providers – United Kingdom (Chronic myeloid disorders)
Jacqueline Boultwood616, David T Bowen1, Adam P Butler1, Mario Cazzola617, Carlo Gambacorti-Passerini270, Anthony R Green329, Eva Hellstrom-Lindberg618, Luca Malcovati617, Sancha Martin1,362, Jyoti Nangalia619, Elli Papaemmanuil1, Paresh Vyas340,620 and Peter J Campbell1,2
Tumour Specific Providers – United Kingdom (Oesophageal cancer)
Yeng Ang621, Hugh Barr622, Duncan Beardsmore623, Matthew Eldridge328, James Gossage624, Nicola Grehan352, George B Hanna625, Stephen J Hayes626,627, Ted R Hupp628, David Khoo629, Jesper Lagergren618,630, Laurence B Lovat189, Shona MacRae137, Maria O'Donovan352, J Robert O'Neill631, Simon L Parsons632, Shaun R Preston633, Sonia Puig634, Tom Roques635, Grant Sanders24, Sharmila Sothi636, Simon Tavaré328, Olga Tucker637, Richard Turkington638, Timothy J Underwood639, Ian Welch640 and Rebecca C Fitzgerald352
Tumour Specific Providers – United Kingdom (Prostate cancer)
Daniel M Berney641, Johann S De Bono396, G Steven Bova126, Daniel S Brewer394,395, Adam P Butler1, Declan Cahill642, Niedzica Camacho396, Nening M Dennis642, Tim Dudderidge642,643, Sandra E Edwards396, Cyril Fisher642, Christopher S Foster644,645, Mohammed Ghori1, Pelvender Gill620, Vincent J Gnanapragasam379,646, Gunes Gundem278, Freddie C Hamdy647, Steve Hawkins328, Steven Hazell642, William Howat379, William B Isaacs648, Katalin Karaszi620, Jonathan D Kay189, Vincent Khoo642, Zsofia Kote-Jarai396, Barbara Kremeyer1, Pardeep Kumar642, Adam Lambert620, Daniel A Leongamornlert1,396, Naomi Livni642, Yong-Jie Lu641,649, Hayley J Luxton189, Andy G Lynch328,329,339, Luke Marsden620, Charlie E Massie328, Lucy Matthews396, Erik Mayer642,650, Ultan McDermott1, Sue Merson396, Thomas J Mitchell1,329,379, David E Neal328,379, Anthony Ng651, David Nicol642, Christopher Ogden642, Edward W Rowe642, Nimish C Shah379, Jon W Teague1, Sarah Thomas642, Alan Thompson642, Peter Van Loo91,92, Clare Verrill620,652, Tapio Visakorpi126, Anne Y Warren379,653, David C Wedge1,65,66, Hayley C Whitaker189, Jorge Zamora1,42,43,44, Hongwei Zhang649, Nicholas van As642, Colin S Cooper395,396,397 and Rosalind A Eeles396,642
Tumour Specific Providers – United States (TCGA)
Adam Abeshouse278, Nishant Agrawal163, Rehan Akbani352,654, Hikmat Al-Ahmadie278, Monique Albert452, Kenneth Aldape400,655, Adrian Ally656, Yeng Ang621, Elizabeth L Appelbaum27,189, Joshua Armenia657, Sylvia Asa632,658, J Todd Auman659, Matthew H Bailey26,27, Miruna Balasundaram656, Saianand Balu24, Jill Barnholtz-Sloan660,661, Hugh Barr622, John Bartlett451,452, Oliver F Bathe662,663, Stephen B Baylin123,643, Duncan Beardsmore623, Christopher Benz664, Andrew Berchuck665, Benjamin P Berman313,321,322, Rameen Beroukhim3,6,168, Mario Berrios666, Darell Bigner667, Michael Birrer19, Tom Bodenheimer24, Lori Boice634, Moiz S Bootwalla666, Marcus Bosenberg668, Reanne Bowlby656, Jeffrey Boyd669, Russell R Broaddus400, Malcolm Brock670, Denise Brooks656, Susan Bullman3,168, Samantha J Caesar-Johnson231, Thomas E Carey671, Rebecca Carlsen656, Robert Cerfolio672, Vishal S Chandan673, Hsiao-Wei Chen621,657, Andrew D Cherniack3,3,157,168, Jeremy Chien674, Juok Cho3, Eric Chuah656, Carrie Cibulskis3, Kristian Cibulskis3, Leslie Cope675, Matthew G Cordes27,635, Kyle Covington34, Erin Curley676, Bogdan Czerniak400,629, Ludmila Danilova675, Ian J Davis677, Timothy Defreitas3, John A Demchok231, Noreen Dhalla656, Rajiv Dhir678, Li Ding26,27,35, HarshaVardhan Doddapaneni34, Adel El-Naggar400,629, Ina Felau231, Martin L Ferguson679, Gaetano Finocchiaro680, Kwun M Fong681, Scott Frazer3, William Friedman682, Catrina C Fronick27,635, Lucinda A Fulton27, Robert S Fulton26,27,35, Stacey B Gabriel3, Jianjiong Gao657, Nils Gehlenborg3,683, Jeffrey E Gershenwald684,685, Gad Getz3,4,5,6, Ronald Ghossein510, Nasra H Giama686, Richard A Gibbs34, Carmen Gomez687, James Gossage624, Ramaswamy Govindan26, Nicola Grehan352, George B Hanna625, D Neil Hayes24,688,689, Stephen J Hayes626,627, Apurva M Hegde137,138, David I Heiman3, Zachary Heins278, Austin J Hepperla24, Katherine A Hoadley23,24, Andrea Holbrook666, Robert A Holt656, Alan P Hoyle24, Ralph H Hruban675,675, Jianhong Hu34, Mei Huang634, David Huntsman690, Ted R Hupp628, Jason Huse278, Christine A Iacobuzio-Donahue510, Michael Ittmann691,692, Joy C Jayaseelan34, Stuart R Jefferys24, Corbin D Jones693, Steven JM Jones694, Hartmut Juhl695, Koo Jeong Kang696, Beth Karlan697, Katayoon Kasaian694, Electron Kebebew698,699, David Khoo629, Hark Kyun Kim700, Jaegil Kim3, Tari A King516,517, Viktoriya Korchina34, Ritika Kundra621,657, Jesper Lagergren618,630, Phillip H Lai666, Peter W Laird316, Eric Lander3, Michael S Lawrence3,19,233, Alexander J Lazar22, Xuan Le701, Darlene Lee656, Douglas A Levine278,702, Lora Lewis34, Tim Ley703, Haiyan Irene Li656, Pei Lin3, W M Linehan704, Eric Minwei Liu71,73,278, Fei Fei Liu380, Laurence B Lovat189, Yiling Lu138, Lisa Lype705, Yussanne Ma656, Shona MacRae137, Dennis T Maglinte666,706, Elaine R Mardis27,669,707, Jeffrey Marks415,708, Marco A Marra656, Thomas J Matthew37, Michael Mayo656, Karen McCune709, Michael D McLellan26,27,35, Samuel R Meier3, Shaowu Meng24, Matthew Meyerson3,6,157, Piotr A Mieczkowski23, Tom Mikkelsen710, Christopher A Miller27, Gordon B Mills711, Richard A Moore656, Carl Morrison411,712, Lisle E Mose24, Catherine D Moser686, Andrew J Mungall656, Karen Mungall656, David Mutch713, Donna M Muzny714, Jerome Myers715, Yulia Newton37, Michael S Noble3, Peter O'Donnell716, Brian Patrick O'Neill717, Angelica Ochoa278, Akinyemi I Ojesina191,192,193, Joong-Won Park718, Joel S Parker719, Simon L Parsons632, Harvey Pass720, Alessandro Pastore112, Chandra Sekhar Pedamallu3,6,168, Nathan A Pennell721, Charles M Perou722, Gloria M Petersen462, Nicholas Petrelli723, Olga Potapova724, Shaun R Preston633, Sonia Puig634, Janet S Rader725, Suresh Ramalingam726, W Kimryn Rathmell727, Victor Reuter510, Sheila M Reynolds705, Matthew Ringel728, Jeffrey Roach729, Lewis R Roberts686, A Gordon Robertson656, Tom Roques635, Mark A Rubin274,287,288,289,290, Sara Sadeghi656, Gordon Saksena3, Charles Saller730, Francisco Sanchez-Vega621,657, Chris Sander112,157,291,292, Grant Sanders24, Dirk Schadendorf149,731, Jacqueline E Schein656, Heather K Schmidt27, Nikolaus Schultz657, Steven E Schumacher3,204, Richard A Scolyer413,443,446,450, Raja Seethala732, Yasin Senbabaoglu112, Troy Shelton676, Yan Shi24, Juliann Shih3,168, Ilya Shmulevich705, Craig Shriver733, Sabina Signoretti168,263,734, Janae V Simons24, Samuel Singer415,735, Payal Sipahimalani656, Tara J Skelly23, Karen Smith-McCune709, Nicholas D Socci112, Heidi J Sofia217, Matthew G Soloway719, Anil K Sood736, Sharmila Sothi636, Angela Tam656, Donghui Tan23, Roy Tarnuzzer231, Nina Thiessen656, R Houston Thompson737, Leigh B Thorne634, Ming Tsao632,658, Olga Tucker637, Richard Turkington638, Christopher Umbricht324,623,738, Timothy J Underwood639, David J Van Den Berg666, Erwin G Van Meir739, Umadevi Veluvolu23, Douglas Voet3, Jiayin Wang27,58,158, Linghua Wang34, Zhining Wang231, Paul Weinberger740, John N Weinstein137,138, Daniel J Weisenberger666, Ian Welch640, David A Wheeler33,34, Dennis Wigle741, Matthew D Wilkerson23, Richard K Wilson27,742, Boris Winterhoff743, Maciej Wiznerowicz744,745, Tina Wong27,656, Winghing Wong746, Liu Xi34, Liming Yang231, Christina Yau664, Venkata D Yellapantula67,68, Hailei Zhang3, Hongxin Zhang657, Jiashan Zhang231, Carolyn M Hutter217 and Jean C Zenklusen231
Author Affiliations
1. Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SA, UK.
2. Department of Haematology, University of Cambridge, Cambridge CB2 2XY, UK.
3. Broad Institute of MIT and Harvard, Cambridge, MA 02142, USA.
4. Center for Cancer Research, Massachusetts General Hospital, Boston, MA 02129, USA.
5. Department of Pathology, Massachusetts General Hospital, Boston, MA 02115, USA.
6. Harvard Medical School, Boston, MA 02115, USA.
7. European Molecular Biology Laboratory, European Bioinformatics Institute (EMBL-EBI), Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK.
8. Genome Biology Unit, European Molecular Biology Laboratory (EMBL), Heidelberg 69117, Germany.
9. Biomolecular Engineering Department, University of California Santa Cruz, Santa Cruz, CA 95064, USA.
10. Adaptive Oncology Initiative, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
11. International Cancer Genome Consortium (ICGC)/ICGC Accelerating Research in Genomic Oncology (ARGO) Secretariat, Toronto, ON M5G 0A3, Canada.
12. Computational Biology Program, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
13. Department of Molecular Genetics, University of Toronto, Toronto, ON M5S 1A8, Canada.
14. Department of Radiation Oncology, University of California San Francisco, San Francisco, CA 94518, USA.
15. Genome Informatics Program, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
16. Department of Cell and Systems Biology, University of Toronto, Toronto, ON M5S 3G5, Canada.
17. Genome Informatics, Ontario Institute for Cancer Research, Toronto, ON M5G 2C4, Canada.
18. Department of Medical Biophysics, University of Toronto, Toronto, ON M5S 1A8, Canada.
19. Massachusetts General Hospital, Boston, MA 02114, USA.
20. Department of Pharmacology, University of Toronto, Toronto, ON M5S 1A8, Canada.
21. University of California Los Angeles, Los Angeles, CA 90095, USA.
22. Departments of Pathology, Genomic Medicine and Translational Molecular Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
23. Department of Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
24. Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
25. The Hospital for Sick Children, Toronto, ON M5G 0A4, Canada.
26. Alvin J. Siteman Cancer Center, Washington University School of Medicine, St Louis, MO 63110, USA.
27. The McDonnell Genome Institute at Washington University, St Louis, MO 63108, USA.
28. Division of Theoretical Bioinformatics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
29. Heidelberg Center for Personalized Oncology (DKFZ-HIPO), German Cancer Research Center, Heidelberg 69120, Germany.
30. Institute of Pharmacy and Molecular Biotechnology and BioQuant, Heidelberg University, Heidelberg 69120, Germany.
31. Bioinformatics and Omics Data Analytics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
32. Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
33. Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX 77030, USA.
34. Human Genome Sequencing Center, Baylor College of Medicine, Houston, TX 77030, USA.
35. Department of Genetics, Department of Medicine, Washington University in St Louis, St Louis, MO 63110, USA.
36. Department of Computer Science, University of Toronto, Toronto, ON M5S 1A8, Canada.
37. University of California Santa Cruz, Santa Cruz, CA 95064, USA.
38. Oregon Health & Science University, Portland, OR 97239, USA.
39. The Institute of Medical Science, The University of Tokyo, Tokyo 108-8639, Japan.
40. Barcelona Supercomputing Center (BSC), Barcelona 08034, Spain.
41. Department of Clinical and Molecular Medicine, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology, Trondheim 7030, Norway.
42. Centre for Research in Molecular Medicine and Chronic Diseases (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela 15706, Spain.
43. Department of Zoology, Genetics and Physical Anthropology, (CiMUS), Universidade de Santiago de Compostela, Santiago de Compostela 15706, Spain.
44. The Biomedical Research Centre (CINBIO), Universidade de Vigo, Vigo 36310, Spain.
45. Department of Genetics, Stanford University School of Medicine, Stanford, CA 94305, USA.
46. Annai Systems, Inc, Carlsbad, CA 92013, USA.
47. Centre for Genomic Regulation (CRG), The Barcelona Institute of Science and Technology, Barcelona 08003, Spain.
48. Institute of Medical Genetics and Applied Genomics, University of Tübingen, Tübingen 72076, Germany.
49. Universitat Pompeu Fabra (UPF), Barcelona 08003, Spain.
50. Department of Computational Biology, University of Lausanne, Lausanne 1015, Switzerland.
51. Department of Genetic Medicine and Development, University of Geneva Medical School, Geneva CH 1211, Switzerland.
52. Swiss Institute of Bioinformatics, University of Geneva, Geneva CH 1211, Switzerland.
53. Department of Ophthalmology and Ocular Genomics Institute, Massachusetts Eye and Ear, Harvard Medical School, Boston, MA 02114, USA.
54. Institute of Evolutionary Biology (UPF-CSIC), Department of Experimental and Health Sciences, Universitat Pompeu Fabra, Barcelona 08003, Spain.
55. Transmissible Cancer Group, Department of Veterinary Medicine, University of Cambridge, Cambridge CB3 0ES, UK.
56. Department of Biochemistry, College of Medicine, Ewha Womans University, Seoul 07895, South Korea.
57. Division of Oncology, Washington University School of Medicine, St Louis, MO 63110, USA.
58. School of Electronic and Information Engineering, Xi'an Jiaotong University, Xi'an 710048, China.
59. The First Affiliated Hospital, Xi'an Jiaotong University, Xi'an 710049, China.
60. Independent Consultant, Wellesley 02481, USA.
61. Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
62. Biobyte solutions GmbH, Heidelberg 69126, Germany.
63. Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520, USA.
64. Program in Computational Biology and Bioinformatics, Yale University, New Haven, CT 06520, USA.
65. Big Data Institute, Li Ka Shing Centre, University of Oxford, Oxford OX3 7LF, UK.
66. Oxford NIHR Biomedical Research Centre, University of Oxford, Oxford OX4 2PG, UK.
67. Department of Epidemiology and Biostatistics, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
68. The McDonnell Genome Institute at Washington University, Department of Genetics, Department of Medicine, Siteman Cancer Center, Washington University in St Louis, St Louis, MO 63108, USA.
69. Department of Computer Science, Yale University, New Haven, CT 06520, USA.
70. Controlled Department and Institution, New York, NY 10065, USA.
71. Department of Physiology and Biophysics, Weill Cornell Medicine, New York, NY 10065, USA.
72. Englander Institute for Precision Medicine, Weill Cornell Medicine, New York, NY 10065, USA.
73. Institute for Computational Biomedicine, Weill Cornell Medicine, New York, NY 10021, USA.
74. CNAG-CRG, Centre for Genomic Regulation (CRG), Barcelona Institute of Science and Technology (BIST), Barcelona 08028, Spain.
75. Department of Experimental and Health Sciences, Institute of Evolutionary Biology (UPF-CSIC), Universitat Pompeu Fabra, Barcelona 08003, Spain.
76. Institució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona 08010, Spain.
77. Institut Català de Paleontologia Miquel Crusafont, Universitat Autònoma de Barcelona, Barcelona 08193, Spain.
78. Department of Biomedical Data Science, Stanford University School of Medicine, Stanford, CA 94305, USA.
79. Human Genetics, University of Kiel, Kiel 24118, Germany.
80. Institute of Human Genetics, Ulm University and Ulm University Medical Center, Ulm 89081, Germany.
81. RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa 230-0045, Japan.
82. Centre for Cancer Genetic Epidemiology, Department of Oncology, University of Cambridge, Cambridge CB1 8RN, UK.
83. Centre for Cancer Genetic Epidemiology, Department of Public Health and Primary Care, University of Cambridge, Cambridge CB1 8RN, UK.
84. Quantitative Genomics Laboratories (qGenomics), Barcelona 08950, Spain.
85. Sage Bionetworks, Seattle, WA 98109, USA.
86. Department of Biochemistry and Molecular Medicine, University of Montreal, Montreal, QC H3C 3J7, Canada.
87. Institute for Research in Biomedicine (IRB Barcelona), Barcelona 08028, Spain.
88. National Centre for Biological Sciences, Tata Institute of Fundamental Research, Bangalore 560065, India.
89. Research Program on Biomedical Informatics, Universitat Pompeu Fabra, Barcelona 08002, Spain.
90. Broad Institute of MIT and Harvard, Cambridge, MA 02124, USA.
91. The Francis Crick Institute, London NW1 1AT, UK.
92. University of Leuven, Leuven B-3000, Belgium.
93. Centre for Molecular Science Informatics, Department of Chemistry, University of Cambridge, Cambridge CB2 1EW, UK.
94. Department of Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
95. Ludwig Center at Harvard Medical School, Boston, MA 02115, USA.
96. Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
97. Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences (NIEHS), Durham, NC 27709, USA.
98. Integrative Bioinformatics Support Group, National Institute of Environmental Health Sciences (NIEHS), Durham, NC 27709, USA.
99. Department of Urology, Charité Universitätsmedizin Berlin, Berlin 10117, Germany.
100. Finsen Laboratory and Biotech Research & Innovation Centre (BRIC), University of Copenhagen, Copenhagen 2200, Denmark.
101. Department of Cellular and Molecular Medicine and Department of Bioengineering and Moores Cancer Center, University of California San Diego, La Jolla, CA 92093, USA.
102. Department of Genetics, Microbiology and Statistics, University of Barcelona, IRSJD, IBUB, Barcelona 08028, Spain.
103. CIBER Epidemiología y Salud Pública (CIBERESP), Madrid 28029, Spain.
104. Research Group on Statistics, Econometrics and Health (GRECS), UdG, Barcelona 8041, Spain.
105. Oxford Nanopore Technologies, New York, NY 10013, USA.
106. Applications Department, Oxford Nanopore Technologies, Oxford OX4 4DQ, UK.
107. School of Molecular Biosciences and Center for Reproductive Biology, Washington State University, Pullman, WA 99164, USA.
108. Laboratory of Translational Genomics, Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
109. Department of Medical and Clinical Genetics, Genome-Scale Biology Research Program, University of Helsinki, Helsinki 00100, Finland.
110. Integrated Graduate Program in Physical and Engineering Biology, Yale University, New Haven, CT 06520, USA.
111. Applied Tumor Genomics Research Program, Research Programs Unit, University of Helsinki, Helsinki 00290, Finland.
112. Computational Biology Center, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
113. Department of Biology, ETH Zurich, Zürich 8093, Switzerland.
114. Department of Computer Science, ETH Zurich, Zurich 8092, Switzerland.
115. SIB Swiss Institute of Bioinformatics, Lausanne 1015, Switzerland.
116. University Hospital Zurich, Zurich 8091, Switzerland.
117. Weill Cornell Medical College, New York, NY 10065, USA.
118. Berlin Institute for Medical Systems Biology, Max Delbrück Center for Molecular Medicine, Berlin 13125, Germany.
119. German Cancer Consortium (DKTK), Partner site Berlin.
120. German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
121. Bakar Computational Health Sciences Institute and Department of Pediatrics, University of California, San Francisco, CA 94158-2549, USA.
122. Department of Biostatistics, Bloomberg School of Public Health, Johns Hopkins University, Baltimore, MD 21205, USA.
123. Department of Oncology, The Johns Hopkins School of Medicine, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, MD 21230, USA.
124. Division of Computational Genomics and Systems Genetics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
125. Division of Biomedical Informatics, Department of Medicine, & Moores Cancer Center, UC San Diego School of Medicine, San Diego, CA 92093, USA.
126. Faculty of Medicine and Health Technology, Tampere University and Tays Cancer Center, Tampere University Hospital, Tampere Fl-33014, Finland.
127. Division of Applied Bioinformatics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
128. Faculty of Biosciences, Heidelberg University, Heidelberg 69120, Germany.
129. Centre for Law and Genetics, University of Tasmania, Sandy Bay Campus, Hobart, TAS 7001, Australia.
130. Centre of Genomics and Policy, McGill University and Génome Québec Innovation Centre, Montreal, QC H3A 1A4, Canada.
131. Heidelberg Academy of Sciences and Humanities, Heidelberg 69120, Germany.
132. UC Santa Cruz Genomics Institute, University of California Santa Cruz, Santa Cruz, CA 95064, USA.
133. CIBIO/InBIO - Research Center in Biodiversity and Genetic Resources, Universidade do Porto, Vairão 4485-601, Portugal.
134. Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
135. Bioinformatics Unit, Spanish National Cancer Research Centre (CNIO), Madrid 28029, Spain.
136. Howard Hughes Medical Institute, University of California Santa Cruz, Santa Cruz, CA 95065, USA.
137. Cancer Unit, MRC University of Cambridge, Cambridge CB2 0XZ, UK.
138. Department of Bioinformatics and Computational Biology and Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
139. Center for Digital Health, Berlin Institute of Health and Charitè - Universitätsmedizin Berlin, Berlin 10117, Germany.
140. Heidelberg Center for Personalized Oncology (DKFZ-HIPO), German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
141. University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
142. Department of Genetics and Informatics Institute, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
143. Heidelberg University, Heidelberg 69120, Germany.
144. New BIH Digital Health Center, Berlin Institute of Health (BIH) and Charité - Universitätsmedizin Berlin, Berlin 10117, Germany.
145. Department Biochemistry and Molecular Biomedicine, University of Barcelona, Barcelona 08028, Spain.
146. Department of Urologic Sciences, University of British Columbia, Vancouver, BC V5Z 1M9, Canada.
147. Vancouver Prostate Centre, Vancouver, BC V6H 3Z6, Canada.
148. Division of Life Science and Applied Genomics Center, Hong Kong University of Science and Technology, Clear Water Bay, Hong Kong, China.
149. German Cancer Consortium (DKTK), Heidelberg 69120, Germany.
150. National Center for Tumor Diseases (NCT) Heidelberg, Heidelberg 69120, Germany.
151. Genome Integration Data Center, Syntekabio, Inc, Daejon, 34025, South Korea.
152. Massachusetts General Hospital Center for Cancer Research, Charlestown, MA 02129, USA.
153. Department of Molecular Medicine (MOMA), Aarhus University Hospital, Aarhus N 8200, Denmark.
154. Bioinformatics Research Centre (BiRC), Aarhus University, Aarhus 8000, Denmark.
155. Indiana University, Bloomington, IN 47405, USA.
156. Simon Fraser University, Burnaby, BC V5A 1S6, Canada.
157. Dana-Farber Cancer Institute, Boston, MA 02215, USA.
158. School of Computer Science and Technology, Xi'an Jiaotong University, Xi'an 710048, China.
159. Department of Genetics, Washington University School of Medicine, St Louis, MO 63110, USA.
160. Department of Mathematics, Washington University in St Louis, St Louis, MO 63130, USA.
161. Department of Biological Oceanography, Leibniz Institute of Baltic Sea Research, Rostock 18119, Germany.
162. Seven Bridges Genomics, Charlestown, MA 02129, USA.
163. University of Chicago, Chicago, IL 60637, USA.
164. Department of Health Sciences and Technology, Sungkyunkwan University School of Medicine, Seoul 06351, South Korea.
165. Samsung Genome Institute, Seoul 06351, South Korea.
166. New York Genome Center, New York, NY 10013, USA.
167. Weill Cornell Medicine, New York, NY 10065, USA.
168. Department of Medical Oncology, Dana-Farber Cancer Institute, Boston, MA 02115, USA.
169. Rigshospitalet, Copenhagen 2200, Denmark.
170. Department of Computer Science, University of Toronto, Toronto, ON M5S 2E4, Canada.
171. The Donnelly Centre, University of Toronto, Toronto, ON M5S 3E1, Canada.
172. Vector Institute, Toronto, ON M5G 0A3, Canada.
173. Department of Medical Genetics, College of Medicine, Hallym University, Chuncheon 24252, South Korea.
174. Department of Biology, ETH Zurich, Wolfgang-Pauli-Strasse 27, 8093 Zürich, Switzerland.
175. University Hospital Zurich, Zurich, 8091, Switzerland.
176. Peking University, Beijing 100871, China.
177. School of Life Sciences, Peking University, Beijing 100180, China.
178. Computational and Systems Biology, Genome Institute of Singapore, Singapore 138672, Singapore.
179. School of Computing, National University of Singapore, Singapore 117417, Singapore.
180. BGI-Shenzhen, Shenzhen 518083, China.
181. China National GeneBank-Shenzhen, Shenzhen 518083, China.
182. Computational & Systems Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
183. Korea University, Seoul 02481, South Korea.
184. Department of Genomic Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
185. Quantitative & Computational Biosciences Graduate Program, Baylor College of Medicine, Houston, TX 77030, USA.
186. The Jackson Laboratory for Genomic Medicine, Farmington, CT 06032, USA.
187. Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Bearsden, Glasgow G61 1QH, UK.
188. The Azrieli Faculty of Medicine, Bar-Ilan University, Safed 13195, Israel.
189. University College London, London WC1E 6BT, UK.
190. Genome Institute of Singapore, Singapore 138672, Singapore.
191. Department of Epidemiology, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
192. HudsonAlpha Institute for Biotechnology, Huntsville, AL 35806, USA.
193. O'Neal Comprehensive Cancer Center, University of Alabama at Birmingham, Birmingham, AL 35294, USA.
194. Department of Biosciences and Nutrition, Karolinska Institutet, Stockholm 14183, Sweden.
195. Cancer Science Institute of Singapore, National University of Singapore, Singapore 169609, Singapore.
196. Programme in Cancer & Stem Cell Biology, Duke-NUS Medical School, Singapore 169857, Singapore.
197. SingHealth, Duke-NUS Institute of Precision Medicine, National Heart Centre Singapore, Singapore 169609, Singapore.
198. Institute of Molecular and Cell Biology, Singapore 169609, Singapore.
199. Laboratory of Cancer Epigenome, Division of Medical Science, National Cancer Centre Singapore, Singapore 169610, Singapore.
200. Department of Medicine, Baylor College of Medicine, Houston, TX 77030, USA.
201. National Cancer Centre Singapore, Singapore 169610, Singapore.
202. BIOPIC, ICG and College of Life Sciences, Peking University, Beijing 100871, China.
203. Vall d'Hebron Institute of Oncology: VHIO, Barcelona 08035, Spain.
204. Department of Cancer Biology, Dana-Farber Cancer Institute, Boston, MA 02215, USA.
205. Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona 8003, Spain.
206. Department of Mathematics, Aarhus University, Aarhus 8000, Denmark.
207. Institut Hospital del Mar d'Investigacions Mèdiques (IMIM), Barcelona 08003, Spain.
208. King Faisal Specialist Hospital and Research Centre, Al Maather, Riyadh 12713, Saudi Arabia.
209. DLR Project Management Agency, Bonn 53227, Germany.
210. Genome Canada, Ottawa, ON K2P 1P1, Canada.
211. Instituto Carlos Slim de la Salud, Mexico City, Mexico.
212. Federal Ministry of Education and Research, Berlin 10117, Germany.
213. Institut Gustave Roussy, Villejuif 94805, France.
214. Institut National du Cancer (INCA), Boulogne-Billancourt 92100, France.
215. The Wellcome Trust, London NW1 2BE, UK.
216. Prostate Cancer Canada, Toronto, ON M5C 1M1, Canada.
217. National Human Genome Research Institute, National Institutes of Health, Bethesda, MD 20892, USA.
218. Department of Biotechnology, Ministry of Science & Technology, Government of India, New Delhi, Delhi 110003, India.
219. Science Writer, Garrett Park, MD 20896, USA.
220. Cancer Research UK, London EC1V 4AD, UK.
221. Chinese Cancer Genome Consortium, Shenzhen 518083, China.
222. Laboratory of Molecular Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital & Institute, Beijing 100142, China.
223. Peking University Cancer Hospital & Institute, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital & Institute, Beijing 100142, China.
224. National Cancer Center, Tokyo 104-0045, Japan.
225. German Cancer Aid, Bonn 53113, Germany.
226. Division of Cancer Genomics, National Cancer Center Research Institute, National Cancer Center, Tokyo 104-0045, Japan.
227. Laboratory of Molecular Medicine, Human Genome Center, The Institute of Medical Science, The University of Tokyo, Minato-ku, Tokyo 108-8639, Japan.
228. Japan Agency for Medical Research and Development, Chiyoda-ku, Tokyo 100-0004, Japan.
229. Medical Oncology, University and Hospital Trust of Verona, Verona 37134, Italy.
230. University of Verona, Verona 37129, Italy.
231. National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
232. CAPHRI Research School, Maastricht University, Maastricht, ER 6200MD, The Netherlands.
233. Laboratory for Medical Science Mathematics, RIKEN Center for Integrative Medical Sciences, Yokohama, Kanagawa 230-0045, Japan.
234. University of California San Diego, San Diego, CA 92093, USA.
235. PDXen Biosystems Inc, Seoul 4900, South Korea.
236. Electronics and Telecommunications Research Institute, Daejeon 34129, South Korea.
237. Children's Hospital of Philadelphia, Philadelphia, PA 19146, USA.
238. University of Melbourne Centre for Cancer Research, Melbourne, VIC 3010, Australia.
239. Syntekabio Inc, Daejon 34025, South Korea.
240. AbbVie, North Chicago, IL 60064, USA.
241. Genomics Research Program, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
242. Department of Pediatric Immunology, Hematology and Oncology, University Hospital, Heidelberg 69120, Germany.
243. Heidelberg Institute for Stem Cell Technology and Experimental Medicine (HI-STEM), Heidelberg 69120, Germany.
244. Seven Bridges, Charlestown, MA 02129, USA.
245. Health Sciences Department of Biomedical Informatics, University of California San Diego, La Jolla, CA 92093, USA.
246. Functional and Structural Genomics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
247. Leidos Biomedical Research, Inc, McLean, VA 22102, USA.
248. CSRA Incorporated, Fairfax, VA 22042, USA.
249. Department of Internal Medicine, Stanford University, Stanford, CA 94305, USA.
250. Clinical Bioinformatics, Swiss Institute of Bioinformatics, Geneva 1202, Switzerland.
251. Institute for Pathology and Molecular Pathology, University Hospital Zurich, Zurich 8091, Switzerland.
252. Institute of Molecular Life Sciences, University of Zurich, Zurich 8057, Switzerland.
253. MIT Computer Science and Artificial Intelligence Laboratory, Massachusetts Institute of Technology, Cambridge, MA 02139, USA.
254. Institute of Molecular Life Sciences and Swiss Institute of Bioinformatics, University of Zurich, Zurich 8057, Switzerland.
255. Office of Cancer Genomics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
256. Computer Network Information Center, Chinese Academy of Sciences, Beijing 100190, China.
257. Geneplus-Shenzhen, Shenzhen 518122, China.
258. Dana-Farber/Boston Children's Cancer and Blood Disorders Center, Boston, MA 02215, USA.
259. Department of Pediatrics, Harvard Medical School, Boston, MA 02115, USA.
260. Technical University of Denmark, Lyngby 2800, Denmark.
261. University of Copenhagen, Copenhagen 2200, Denmark.
262. Department for BioMedical Research, University of Bern, Bern 3008, Switzerland.
263. Department of Medical Oncology, Inselspital, University Hospital and University of Bern, Bern 3010, Switzerland.
264. Graduate School for Cellular and Biomedical Sciences, University of Bern, Bern 3012, Switzerland.
265. Department of Genitourinary Medical Oncology - Research, Division of Cancer Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
266. Department of Urology, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA.
267. Korea Advanced Institute of Science and Technology, Daejeon 34141, South Korea.
268. Science for Life Laboratory, Department of Cell and Molecular Biology, Uppsala University, Uppsala SE-75124, Sweden.
269. Queensland Centre for Medical Genomics, Institute for Molecular Bioscience, The University of Queensland, St Lucia, Brisbane, QLD 4072, Australia.
270. University of Milano Bicocca, Monza 20052, Italy.
271. Sir Peter MacCallum Department of Oncology, Peter MacCallum Cancer Centre, University of Melbourne, Melbourne, VIC 3000, Australia.
272. Center for Precision Health, School of Biomedical Informatics, The University of Texas Health Science Center, Houston, TX 77030, USA.
273. Health Data Science Unit, University Clinics, Heidelberg 69120, Germany.
274. Department for Biomedical Research, University of Bern, Bern 3008, Switzerland.
275. Research Core Center, National Cancer Centre Korea, Goyang-si 410-769, South Korea.
276. Institute of Computer Science, Polish Academy of Sciences, Warsawa 01-248, Poland.
277. Harvard University, Cambridge, MA 02138, USA.
278. Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
279. Department of Information Technology, Ghent University, Ghent B-9000, Belgium.
280. Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent B-9000, Belgium.
281. Yale School of Medicine, Yale University, New Haven, CT 06520, USA.
282. Division of Hematology-Oncology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul 06351, South Korea.
283. Samsung Advanced Institute for Health Sciences and Technology, Sungkyunkwan University School of Medicine, Seoul 06351, South Korea.
284. Cheonan Industry-Academic Collaboration Foundation, Sangmyung University, Cheonan 31066, South Korea.
285. Spanish National Cancer Research Centre, Madrid 28029, Spain.
286. Department of Computer Science, Princeton University, Princeton, NJ 08540, USA.
287. Bern Center for Precision Medicine, University Hospital of Bern, University of Bern, Bern 3008, Switzerland.
288. Englander Institute for Precision Medicine, Weill Cornell Medicine and New York Presbyterian Hospital, New York, NY 10021, USA.
289. Meyer Cancer Center, Weill Cornell Medicine, New York, NY 10065, USA.
290. Pathology and Laboratory, Weill Cornell Medical College, New York, NY 10021, USA.
291. cBio Center, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA 02115, USA.
292. Department of Cell Biology, Harvard Medical School, Boston, MA 02115, USA.
293. cBio Center, Dana-Farber Cancer Institute, Boston, MA 02215, USA.
294. CREST, Japan Science and Technology Agency, Tokyo 113-0033, Japan.
295. Department of Medical Science Mathematics, Medical Research Institute, Tokyo Medical and Dental University, Bunkyo-ku, Tokyo 113-8510, Japan.
296. Laboratory for Medical Science Mathematics, Department of Biological Sciences, Graduate School of Science, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan.
297. Science for Life Laboratory, Department of Oncology-Pathology, Karolinska Institutet, Stockholm 17121, Sweden.
298. Department of Gene Technology, Tallinn University of Technology, Tallinn 12616, Estonia.
299. Genetics & Genome Biology Program, SickKids Research Institute, The Hospital for Sick Children, Toronto, ON M5G 1X8, Canada.
300. Department of Information Technology, Ghent University, Interuniversitair Micro-Electronica Centrum (IMEC), Ghent B-9000, Belgium.
301. Science for Life Laboratory, Department of Immunology, Genetics and Pathology, Uppsala University, Uppsala SE-75108, Sweden.
302. Oregon Health & Sciences University, Portland, OR 97239, USA.
303. Department of Medicine and Therapeutics, The Chinese University of Hong Kong, Shatin, NT, Hong Kong, China.
304. The University of Texas Health Science Center at Houston, Houston, TX 77030, USA.
305. Department of Biomedical Informatics, College of Medicine, The Ohio State University, Columbus, OH 43210, USA.
306. The Ohio State University Comprehensive Cancer Center (OSUCCC – James), Columbus, OH 43210, USA.
307. The University of Texas School of Biomedical Informatics (SBMI) at Houston, Houston, TX 77030, USA.
308. Department of Biochemistry and Molecular Genetics, Feinberg School of Medicine, Northwestern University, Chicago, IL 60637, USA.
309. Physics Division, Optimization and Systems Biology Lab, Massachusetts General Hospital, Boston, MA 02114, USA.
310. Genome Science Division, Research Center for Advanced Science and Technology, The University of Tokyo, Tokyo 153-8904, Japan.
311. Bioinformatics Group, Department of Computer Science, University of Leipzig, Leipzig 04109, Germany.
312. Interdisciplinary Center for Bioinformatics, University of Leipzig, Leipzig 04109, Germany.
313. Center for Bioinformatics and Functional Genomics, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
314. Computational Biology, Leibniz Institute on Aging - Fritz Lipmann Institute (FLI), Jena 07745, Germany.
315. Transcriptome Bioinformatics, LIFE Research Center for Civilization Diseases, University of Leipzig, Leipzig 04109, Germany.
316. Center for Epigenetics, Van Andel Research Institute, Grand Rapids, MI 49503, USA.
317. Institut d’Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), Barcelona 08036, Spain.
318. Research Center for Advanced Science and Technology, The University of Tokyo, Minato-ku, Tokyo 108-8639, Japan.
319. Van Andel Research Institute, Grand Rapids, MI 49503, USA.
320. Cancer Epigenomics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
321. Department of Biomedical Sciences, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
322. The Hebrew University Faculty of Medicine, Jerusalem 91120, Israel.
323. German Cancer Consortium (DKTK), German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
324. Department of Pathology, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
325. McKusick-Nathans Institute of Genetic Medicine, Sidney Kimmel Comprehensive Cancer Center, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
326. Foundation Medicine, Inc, Cambridge, MA 02141, USA.
327. University of Ottawa Faculty of Medicine, Department of Biochemistry, Microbiology and Immunology, Ottawa, ON K1H 8M5, Canada.
328. Cancer Research UK Cambridge Institute, University of Cambridge, Cambridge CB2 0RE, UK.
329. University of Cambridge, Cambridge CB2 1TN, UK.
330. Brandeis University, Waltham, MA 02254, USA.
331. Hopp Children's Cancer Center (KiTZ), Heidelberg 69120, Germany.
332. Pediatric Glioma Research Group, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
333. A.A. Kharkevich Institute of Information Transmission Problems, Moscow 127051, Russia.
334. Oncology and Immunology, Dmitry Rogachev National Research Center of Pediatric Hematology, Moscow 117997, Russia.
335. Skolkovo Institute of Science and Technology, Moscow 121205, Russia.
336. Center For Medical Innovation, Seoul National University Hospital, Seoul 03080, South Korea.
337. Department of Internal Medicine, Seoul National University Hospital, Seoul 03080, South Korea.
338. Division of Genetics and Genomics, Boston Children's Hospital, Harvard Medical School, Boston, MA 02115, USA.
339. School of Medicine/School of Mathematics and Statistics, University of St Andrews, St Andrews, Fife KY16 9SS, UK.
340. Department of Genetics and Computational Biology, QIMR Berghofer Medical Research Institute, Brisbane, QLD 4006, Australia.
341. Institute for Molecular Bioscience, University of Queensland, St Lucia, Brisbane, QLD 4072, Australia.
342. Cancer Research Institute, Beth Israel Deaconess Medical Center, Boston, MA 02215, USA.
343. Ben May Department for Cancer Research, Department of Human Genetics, The University of Chicago, Chicago, IL 60637, USA.
344. Tri-Institutional PhD Program in Computational Biology and Medicine, Weill Cornell Medicine, New York, NY 10065, USA.
345. Department of Cellular and Molecular Medicine and Department of Bioengineering and Moores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA.
346. Centre for Computational Biology, Duke-NUS Medical School, Singapore 169857, Singapore.
347. Department of Computer Science, University of Helsinki, Helsinki 00014, Finland.
348. Institute of Biotechnology, University of Helsinki, Helsinki 00014, Finland.
349. Organismal and Evolutionary Biology Research Programme, University of Helsinki, Helsinki 00014, Finland.
350. Programme in Cancer & Stem Cell Biology, Centre for Computational Biology, Duke-NUS Medical School, Singapore 169857, Singapore.
351. Academic Department of Medical Genetics, University of Cambridge, Addenbrooke’s Hospital, Cambridge CB2 0QQ, UK.
352. MRC Cancer Unit, University of Cambridge, Cambridge CB2 0XZ, UK.
353. The University of Cambridge School of Clinical Medicine, Cambridge CB2 0SP, UK.
354. Department of Applied Mathematics and Theoretical Physics, Centre for Mathematical Sciences, University of Cambridge, Cambridge CB3 0WA, UK.
355. Department of Statistics, Columbia University, New York, NY 10027, USA.
356. Duke-NUS Medical School, Singapore 169857, Singapore.
357. School of Electronic Information and Communications, Huazhong University of Science and Technology, Wuhan, Hubei 430074, China.
358. The Kinghorn Cancer Centre, Cancer Division, Garvan Institute of Medical Research, University of NSW, Sydney, NSW 2010, Australia.
359. MRC Human Genetics Unit, MRC IGMM, University of Edinburgh, Edinburgh EH4 2XU, UK.
360. Bioinformatics Group, Division of Molecular Biology, Department of Biology, Faculty of Science, University of Zagreb, Zagreb 10000, Croatia.
361. Department of Bioinformatics, Division of Cancer Genomics, National Cancer Center Research Institute, National Cancer Center, Tokyo 104-0045, Japan.
362. University of Glasgow, Glasgow G61 1BD, UK.
363. MRC-University of Glasgow Centre for Virus Research, Glasgow G61 1QH, UK.
364. Wolfson Wohl Cancer Research Centre, Institute of Cancer Sciences, University of Glasgow, Bearsden, Glasgow G61 1BD, UK.
365. School of Computing Science, University of Glasgow, Glasgow G12 8RZ, UK.
366. South Western Sydney Clinical School, Faculty of Medicine, University of NSW, Liverpool, NSW 2170, Australia.
367. West of Scotland Pancreatic Unit, Glasgow Royal Infirmary, Glasgow G31 2ER, UK.
368. University of Melbourne Centre for Cancer Research, Melbourne, VIC 3052, Australia.
369. Molecular and Medical Genetics, Oregon Health and Science University, Portland, OR 97201, USA.
370. Department of Surgery, University of Melbourne, Parkville, VIC 3010, Australia.
371. The Murdoch Children's Research Institute, Royal Children’s Hospital, Parkville, VIC 3052, Australia.
372. Walter + Eliza Hall Institute, Parkville, VIC 3052, Australia.
373. University of Cologne, Cologne 50931, Germany.
374. The Edward S. Rogers Sr. Department of Electrical and Computer Engineering, University of Toronto, Toronto, ON M5S 3G4, Canada.
375. University of Ljubljana, Ljubljana 1000, Slovenia.
376. Department of Public Health Sciences, The University of Chicago, Chicago, IL 60637, USA.
377. Research Institute, NorthShore University HealthSystem, Evanston, IL 60201, USA.
378. Department of Statistics, University of California Santa Cruz, Santa Cruz, CA 95064, USA.
379. Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK.
380. University of Toronto, Toronto, ON M5G 2M9, Canada.
381. Department of Computer Science, Carleton College, Northfield, MN 55057, USA.
382. Molecular and Medical Genetics, Oregon Health & Science University, Portland, OR 97239, USA.
383. Center for Psychiatric Genetics, NorthShore University HealthSystem, Evanston, IL 60201, USA.
384. Argmix Consulting, North Vancouver, BC V7M 2J5, Canada.
385. Department of Biostatistics, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
386. Department of Biostatistics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
387. The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
388. Molecular and Medical Genetics, Knight Cancer Institute, Oregon Health & Science University, Portland, OR 97219, USA.
389. Department of Health Sciences, Faculty of Medical Sciences, Kyushu University, Fukuoka 812-8582, Japan.
390. Baylor College of Medicine, Houston, TX 77030, USA.
391. Department of Applied Mathematics and Statistics, Johns Hopkins University, Baltimore, MD 21218, USA.
392. Heinrich Pette Institute, Leibniz Institute for Experimental Virology, Hamburg 20251, Germany.
393. University Medical Center Hamburg-Eppendorf, Bioinformatics Core, Hamburg 20246, Germany.
394. Earlham Institute, Norwich NR4 7UZ, UK.
395. Norwich Medical School, University of East Anglia, Norwich NR4 7TJ, UK.
396. The Institute of Cancer Research, London SW7 3RP, UK.
397. University of East Anglia, Norwich NR4 7TJ, UK.
398. German Center for Infection Research (DZIF), Partner Site Hamburg-Borstel-Lübeck-Riems, Hamburg, Germany.
399. Division of Molecular Genetics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
400. Department of Pathology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
401. Peter MacCallum Cancer Centre, University of Melbourne, Melbourne, VIC 3000, Australia.
402. QIMR Berghofer Medical Research Institute, Brisbane, QLD 4006, Australia.
403. Victorian Institute of Forensic Medicine, Southbank, VIC 3006, Australia.
404. University of Pennsylvania, Philadelphia, PA 19104, USA.
405. Centre for Cancer Research, The Westmead Institute for Medical Research, Sydney, NSW 2145, Australia.
406. Department of Gynaecological Oncology, Westmead Hospital, Sydney, NSW 2145, Australia.
407. Genetics and Molecular Pathology, SA Pathology, Adelaide, SA 5000, Australia.
408. Centre for Cancer Research, The Westmead Institute for Medical Research, The University of Sydney, Sydney, NSW 2145, Australia.
409. Department of Gynaecological Oncology, Westmead Hospital, Sydney, NSW 2006, Australia.
410. Garvan Institute of Medical Research, Darlinghurst, NSW 2010 Australia.
411. Department of Clinical Pathology, University of Melbourne, Melbourne, VIC 3052, Australia.
412. Centre for Cancer Research, The Westmead Institute for Medical Research, and Department of Gynaecological Oncology, Westmead Hospital, Sydney, NSW 2145, Australia.
413. The University of Sydney, Sydney, NSW 2006, Australia.
414. The Westmead Institute for Medical Research. The University of Sydney. The Department of Gynaecological Oncology, Westmead Hospital, Westmead, NSW 2145, Australia.
415. Department of Surgery, Pancreas Institute, University and Hospital Trust of Verona, Verona 37134, Italy.
416. Department of Surgery, Princess Alexandra Hospital, Woolloongabba QLD 4102, Australia.
417. Surgical Oncology Group, Diamantina Institute, The University of Queensland, Woolloongabba, Brisbane, QLD 4102, Australia.
418. Department of Diagnostics and Public Health, University and Hospital Trust of Verona, Verona 37134, Italy.
419. ARC-Net Centre for Applied Research on Cancer, University and Hospital Trust of Verona, Verona 37134, Italy.
420. Illawarra Shoalhaven Local Health District L3 Illawarra Cancer Care Centre, Wollongong Hospital, Wollongong NSW 2500, Australia.
421. University of Sydney, Sydney, NSW 2006, Australia.
422. School of Biological Sciences, The University of Auckland, Auckland 1010, New Zealand.
423. Department of Pathology and Diagnostics, University and Hospital Trust of Verona, Verona 37134, Italy.
424. Department of Medicine, Section of Endocrinology, University and Hospital Trust of Verona, Verona 37134, Italy.
425. Department of Pathology, Queen Elizabeth University Hospital, Glasgow G51 4TF, UK.
426. Department of Medical Oncology, Beatson West of Scotland Cancer Centre, Glasgow G12 0YN, UK.
427. Academic Unit of Surgery, School of Medicine, College of Medical, Veterinary and Life Sciences, University of Glasgow, Glasgow Royal Infirmary, Glasgow G4 OSF, UK.
428. Tissue Pathology and Diagnostic Oncology, Royal Prince Alfred Hospital, Camperdown, NSW 2050, Australia.
429. Discipline of Surgery, Western Sydney University, Penrith, NSW 2751, Australia.
430. Institute of Cancer Sciences, College of Medical Veterinary and Life Sciences, University of Glasgow, Glasgow G12 8QQ, UK.
431. The Kinghorn Cancer Centre, Cancer Division, Garvan Institute of Medical Research, University of NSW, Sydney, NSW 2109, Australia.
432. School of Environmental and Life Sciences, Faculty of Science, The University of Newcastle, Ourimbah, NSW 2258, Australia.
433. Eastern Clinical School, Monash University, Melbourne, VIC 3128, Australia.
434. Epworth HealthCare, Richmond, VIC 3121, Australia.
435. Olivia Newton-John Cancer Research Institute, La Trobe University, Heidelberg, VIC 3084, Australia.
436. Melanoma Institute Australia, The University of Sydney, Wollstonecraft NSW 2065, Australia.
437. Children’s Hospital at Westmead, The University of Sydney, Westmead, NSW 2145, Australia.
438. Melanoma Institute Australia, The University of Sydney, Sydney, NSW 2065, Australia.
439. Australian Institute of Tropical Health and Medicine, James Cook University, Douglas, QLD 4814, Australia.
440. Bioplatforms Australia, North Ryde, NSW 2109, Australia.
441. Melanoma Institute Australia, Macquarie University, Wollstonecraft NSW, 2109, Australia.
442. Children’s Medical Research Institute, Westmead, NSW 2145 Australia.
443. Melanoma Institute Australia, The University of Sydney, Wollstonecraft 2065, NSW, Australia.
444. Centre for Cancer Research, The Westmead Millennium Institute for Medical Research, University of Sydney, Westmead Hospital, Westmead, NSW 2145, Australia.
445. Centre for Cancer Research, Westmead Institute for Medical Research, Westmead, NSW 2145, Australia.
446. Discipline of Pathology, Sydney Medical School, The University of Sydney, Sydney, NSW 2065, Australia.
447. School of Mathematics and Statistics, The University of Sydney, Sydney, NSW 2006 Australia.
448. Melanoma Institute Australia, The University of Sydney, Wollstonecraft, NSW 2065, Australia.
449. Westmead Institute for Medical Research, University of Sydney, Westmead, NSW 2145 Australia.
450. Royal Prince Alfred Hospital, Sydney, NSW 2050, Australia.
451. Diagnostic Development, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
452. Ontario Tumour Bank, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
453. PanCuRx Translational Research Initiative, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
454. BioSpecimen Sciences Program, University Health Network, Toronto, ON M5G 2C4, Canada, Toronto, ON M5G 2C4, Canada.
455. Hepatobiliary/Pancreatic Surgical Oncology Program, University Health Network, Toronto, ON M5G 2C4, Canada.
456. Lunenfeld-Tanenbaum Research Institute, Mount Sinai Hospital, Toronto, ON M5G 1X5, Canada.
457. Division of Medical Oncology, Princess Margaret Cancer Centre, Toronto, ON M5G 2M9, Canada.
458. University of Nebraska Medical Center, Omaha, NE 68198-6880, USA.
459. BioSpecimen Sciences Program, University Health Network, Toronto, ON M5G 2C4, Canada.
460. Transformative Pathology, Ontario Institute for Cancer Research, Toronto, ON M5G 0A3, Canada.
461. University Health Network, Princess Margaret Cancer Centre, Toronto, ON M5G 1L7, Canada.
462. Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA.
463. BioSpecimen Sciences, Laboratory Medicine (Toronto), Medical Biophysics, PanCuRX, Toronto, ON M5S 1A8, Canada.
464. Department of Laboratory Medicine and Pathobiology, University of Toronto, Toronto, ON M5S 1A8, Canada.
465. Department of Pathology, Human Oncology and Pathogenesis Program, Memorial Sloan Kettering Cancer Center, New York, NY 10053, USA.
466. Department of Medical Biophysics, University of Toronto, Toronto, ON M5G 1L7, Canada.
467. Department of Biochemistry and Molecular Medicine, University California at Davis, Sacramento, CA 95817, USA.
468. Human Longevity Inc, San Diego, CA 92121, USA.
469. Department of Surgical Oncology, Princess Margaret Cancer Centre, Toronto, ON M5G 2M9, Canada.
470. Genome Informatics Program, Ontario Institute for Cancer Research, Toronto, ON M5G 2C4, Canada.
471. STTARR Innovation Facility, Princess Margaret Cancer Centre, Toronto, ON M5G 1L7, Canada.
472. Department of Pathology, Toronto General Hospital, Toronto, ON M5G 2C4, Canada.
473. CRUK Manchester Institute and Centre, Manchester M20 4GJ, UK.
474. Department of Radiation Oncology, University of Toronto, Toronto, ON M5S 1A8, Canada.
475. Manchester Cancer Research Centre, Cancer Division, FBMH, University of Manchester, Manchester M20 4GJ, UK.
476. Radiation Medicine Program, Princess Margaret Cancer Centre, Toronto, ON M5G 2M9, Canada.
477. Hefei University of Technology, Anhui 230009, China.
478. State Key Laboratory of Cancer Biology, and Xijing Hospital of Digestive Diseases, Fourth Military Medical University, Shaanxi 710032, China.
479. Fourth Military Medical University, Shaanxi 710032, China.
480. Laboratory of Molecular Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital & Institute, Beijing,100142, China.
481. Department of Surgery, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200025, China.
482. Leeds Institute of Medical Research, University of Leeds, St James's University Hospital, Leeds LS9 7TF, UK.
483. Canadian Center for Computational Genomics, McGill University, Montreal, QC H3A 0G1, Canada.
484. Department of Human Genetics, McGill University, Montreal, QC H3A 1B1, Canada.
485. International Agency for Research on Cancer, Lyon 69008, France.
486. McGill University and Genome Quebec Innovation Centre, Montreal, QC H3A 0G1, Canada.
487. Leeds Institute of Medical Research @ St James's, University of Leeds, St James’s University Hospital, Leeds LS9 7TF, UK.
488. Institute of Mathematics and Computer Science, University of Latvia, Riga LV 1459, Latvia.
489. Centre National de Génotypage, CEA - Institute de Génomique, Evry 91000, France.
490. Department of Oncology, Gil Medical Center, Gachon University, Incheon 405-760, South Korea.
491. Department of Molecular Oncology, BC Cancer Agency, Vancouver, BC V5Z 1L3, Canada.
492. Los Alamos National Laboratory, Los Alamos, NM 87545, USA.
493. Department of Genetics, Institute for Cancer Research, Oslo University Hospital, The Norwegian Radium Hospital, Oslo O310, Norway.
494. Lund University, Lund 223 62, Sweden.
495. Translational Research Lab, Centre Léon Bérard, Lyon 69373, France.
496. Department of Molecular Biology, Faculty of Science, Radboud Institute for Molecular Life Sciences, Radboud University, Nijmegen 6500 HB, The Netherlands.
497. Department of Pathology, Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
498. Department of Molecular Pathology, The Netherlands Cancer Institute, Amsterdam 1066 CX, The Netherlands.
499. Cancer Research UK Cambridge Institute, University of Cambridge, Li Ka Shing Centre, Cambridge CB2 0RE, UK.
500. Department of Oncology, University of Cambridge, Cambridge CB2 1TN, UK.
501. Breast Cancer Translational Research Laboratory JC Heuson, Institut Jules Bordet, Brussels 1000, Belgium.
502. Laboratory for Translational Breast Cancer Research, Department of Oncology, KU Leuven, Leuven 3000, Belgium.
503. Translational Cancer Research Unit, GZA Hospitals St.-Augustinus, Center for Oncological Research, Faculty of Medicine and Health Sciences, University of Antwerp, Antwerp 2000, Belgium.
504. Department of Gynecology & Obstetrics, Department of Clinical Sciences, Skåne University Hospital, Lund University, Lund SE-221 85, Sweden.
505. Icelandic Cancer Registry, Icelandic Cancer Society, Reykjavik 125, Iceland.
506. Department of Medical Oncology, Josephine Nefkens Institute and Cancer Genomics Centre, Erasmus Medical Center, Rotterdam 3015 CN, The Netherlands.
507. National Genotyping Center, Institute of Biomedical Sciences, Academia Sinica, Taipei 115, Taiwan.
508. Department of Pathology, Oslo University Hospital Ulleval, Oslo 0450, Norway.
509. Faculty of Medicine and Institute of Clinical Medicine, University of Oslo, Oslo NO-0316, Norway.
510. Department of Pathology, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
511. Department of Pathology, Skåne University Hospital, Lund University, Lund SE-221 85, Sweden.
512. Department of Pathology, Academic Medical Center, Amsterdam 1105 AZ, The Netherlands.
513. Department of Pathology, College of Medicine, Hanyang University, Seoul 133-791, South Korea.
514. Department of Pathology, Asan Medical Center, College of Medicine, Ulsan University, Songpa-gu, Seoul 05505, South Korea.
515. The Netherlands Cancer Institute, Amsterdam 1066 CX, The Netherlands.
516. Department of Surgery, Brigham and Women’s Hospital, Dana-Farber Cancer Institute, Boston, MA 02115, USA.
517. Department of Surgery, Memorial Sloan-Kettering Cancer Center, New York, NY 10065, USA.
518. Department of Clinical Science, University of Bergen, Bergen 5020, Norway.
519. Morgan Welch Inflammatory Breast Cancer Research Program and Clinic, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
520. The University of Queensland Centre for Clinical Research, The Royal Brisbane & Women's Hospital, Herston, QLD 4029, Australia.
521. Department of Pathology, Institut Jules Bordet, Brussels 1000, Belgium.
522. Institute for Bioengineering and Biopharmaceutical Research (IBBR), Hanyang University, Seoul 133-791, South Korea.
523. University of Oslo, Oslo 0316, Norway.
524. Institut Bergonié, Bordeaux 33076, France.
525. Department of Research Oncology, Guy’s Hospital, King’s Health Partners AHSC, King’s College London School of Medicine, London SE1 9RT, UK.
526. University Hospital of Minjoz, INSERM UMR 1098, Besançon 25000, France.
527. Cambridge Breast Unit, Addenbrooke’s Hospital, Cambridge University Hospital NHS Foundation Trust and NIHR Cambridge Biomedical Research Centre, Cambridge CB2 2QQ, UK.
528. East of Scotland Breast Service, Ninewells Hospital, Aberdeen AB25 2XF, UK.
529. Oncologie Sénologie, ICM Institut Régional du Cancer, Montpellier 34298, France.
530. Department of Radiation Oncology, Radboud University Nijmegen Medical Centre, Nijmegen 6525 GA, The Netherlands.
531. University of Iceland, Reykjavik 101, Iceland.
532. Dundee Cancer Centre, Ninewells Hospital, Dundee DD2 1SY, UK.
533. Institut Curie, INSERM Unit 830, Paris 75248, France.
534. Department of Laboratory Medicine, Radboud University Nijmegen Medical Centre, Nijmegen 6525 GA, The Netherlands.
535. Department of General Surgery, Singapore General Hospital, Singapore 169608, Singapore.
536. Universite Lyon, INCa-Synergie, Centre Léon Bérard, Lyon 69008, France.
537. Giovanni Paolo II / I.R.C.C.S. Cancer Institute, Bari BA 70124, Italy.
538. Department of Biopathology, Centre Léon Bérard, Lyon 69008, France.
539. Université Claude Bernard Lyon 1, Villeurbanne 69100, France.
540. NCCS-VARI Translational Research Laboratory, National Cancer Centre Singapore, Singapore 169610, Singapore.
541. Department of Pathology, Erasmus Medical Center Rotterdam, Rotterdam 3015 GD, The Netherlands.
542. Division of Molecular Carcinogenesis, The Netherlands Cancer Institute, Amsterdam 1066 CX, The Netherlands.
543. Institute of Human Genetics, Christian-Albrechts-University, Kiel 24118, Germany.
544. Institute of Human Genetics, Ulm University and Ulm University Medical Center of Ulm, Ulm 89081, Germany.
545. Hematopathology Section, Institute of Pathology, Christian-Albrechts-University, Kiel 24118, Germany.
546. Institute of Human Genetics, University of Ulm and University Hospital of Ulm, Ulm 89081, Germany.
547. Department of Human Genetics, Hannover Medical School, Hannover 30625, Germany.
548. Department of Pediatric Oncology, Hematology and Clinical Immunology, Heinrich-Heine-University, Düsseldorf 40225, Germany.
549. Department of Internal Medicine/Hematology, Friedrich-Ebert-Hospital, Neumünster 24534, Germany.
550. Pediatric Hematology and Oncology, University Hospital Muenster, Muenster 24534, Germany.
551. Department of Pediatrics, University Hospital Schleswig-Holstein, Kiel 24105, Germany.
552. Department of Medicine II, University of Würzburg, Würzburg, Germany.
553. Senckenberg Institute of Pathology, University of Frankfurt Medical School, Frankfurt 60596, Germany.
554. Institute of Pathology, Charité – University Medicine Berlin, Berlin 10117, Germany.
555. Department for Internal Medicine II, University Hospital Schleswig-Holstein, Kiel 24105, Germany.
556. Institute for Medical Informatics Statistics and Epidemiology, University of Leipzig, Leipzig 04109, Germany.
557. Department of Hematology and Oncology, Georg-Augusts-University of Göttingen, Göttingen 37073, Germany.
558. Institute of Cell Biology (Cancer Research), University of Duisburg-Essen, Essen D-45147, Germany.
559. MVZ Department of Oncology, PraxisClinic am Johannisplatz, Leipzig 04109, Germany.
560. Institute of Pathology, Ulm University and University Hospital of Ulm, Ulm 89081, Germany.
561. Department of Pathology, Robert-Bosch-Hospital, Stuttgart, Germany, Stuttgart 70376, Germany.
562. University Hospital Giessen, Pediatric Hematology and Oncology, Giessen 35392, Germany.
563. Institute of Clinical Molecular Biology, Christian-Albrechts-University, Kiel 24118, Germany.
564. Institute of Pathology, University of Wuerzburg, Wuerzburg 97070, Germany.
565. Department of General Internal Medicine, University Kiel, Kiel 24118, Germany.
566. Clinic for Hematology and Oncology, St.-Antonius-Hospital, Eschweiler D-52249, Germany.
567. Department for Internal Medicine III, University of Ulm and University Hospital of Ulm, Ulm 89081, Germany.
568. Neuroblastoma Genomics, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
569. Department of Pediatric Oncology and Hematology, University of Cologne, Cologne 50937, Germany.
570. University of Düsseldorf, Düsseldorf 40225, Germany.
571. Department of Vertebrate Genomics/Otto Warburg Laboratory Gene Regulation and Systems Biology of Cancer, Max Planck Institute for Molecular Genetics, Berlin 14195, Germany.
572. St. Jude Children's Research Hospital, Memphis, TN 38105-3678, USA.
573. Heidelberg University Hospital, Heidelberg 69120, Germany.
574. Genomics and Proteomics Core Facility High Throughput Sequencing Unit, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
575. Epigenomics and Cancer Risk Factors, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
576. University Medical Center Hamburg-Eppendorf, Hamburg 20251, Germany.
577. Martini-Clinic, Prostate Cancer Center, University Medical Center Hamburg-Eppendorf, Hamburg 20095, Germany.
578. Institute of Pathology, University Medical Center Hamburg-Eppendorf, Hamburg 20251, Germany.
579. Division of Cancer Genome Research, German Cancer Research Center (DKFZ), Heidelberg 69120, Germany.
580. National Institute of Biomedical Genomics, Kalyani 741235, West Bengal, India.
581. Advanced Centre for Treatment Research & Education in Cancer, Tata Memorial Centre, Navi Mumbai, Maharashtra 410210, India.
582. Department of Pathology, General Hospital of Treviso, Department of Medicine, University of Padua, Treviso 31100, Italy.
583. Department of Medicine (DIMED), Surgical Pathology Unit, University of Padua, Padua 35121, Italy.
584. Department of Hepatobiliary and Pancreatic Oncology, Hepatobiliary and Pancreatic Surgery Division, Division of Pathology and Clinical Laboratories, National Cancer Center Hospital, Chuo-ku, Tokyo, 104-0045, Japan.
585. Department of Pathology, Keio University School of Medicine, Tokyo 160-8582, Japan.
586. Department of Hepatobiliary and Pancreatic Oncology, National Cancer Center Hospital, Tokyo, 104-0045 Japan.
587. Department of Pathology, Graduate School of Medicine, The University of Tokyo, Bunkyo-ku, Tokyo 113-0033, Japan.
588. Preventive Medicine, Graduate School of Medicine, The University of Tokyo, Tokyo 113-0033, Japan.
589. Gastric Surgery Division, Division of Pathology and Clinical Laboratories, National Cancer Center Hospital, Tokyo 104-0045, Japan.
590. Department of Gastroenterology and Hepatology, Yokohama City University Graduate School of Medicine, Kanagawa 236-0004, Japan.
591. Laboratory of Molecular Medicine, Human Genome Center, The Institute of Medical Science, University of Tokyo, Tokyo 108-8639, Japan.
592. Department of Cancer Genome Informatics, Graduate School of Medicine, Osaka University, Osaka 565-0871, Japan.
593. Hiroshima University, Hiroshima 734-8553, Japan.
594. Tokyo Women’s Medical University, Tokyo 162-8666, Japan.
595. Osaka International Cancer Center, Osaka 541-8567, Japan.
596. Wakayama Medical University, Wakayama 641-8509, Japan.
597. Hokkaido University, Sapporo 060-8648, Japan.
598. Division of Medical Oncology, National Cancer Centre, Singapore 169610, Singapore.
599. Cholangiocarcinoma Screening and Care Program and Liver Fluke and Cholangiocarcinoma Research Centre, Faculty of Medicine, Khon Kaen University, Khon Kaen 40002, Thailand.
600. Lymphoma Genomic Translational Research Laboratory, National Cancer Centre, Singapore 169610, Singapore.
601. Center of Digestive Diseases and Liver Transplantation, Fundeni Clinical Institute, Bucharest 022328, Romania.
602. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, School of Medicine, Keimyung University Dongsan Medical Center, Daegu 41931, South Korea.
603. Pathology, Hospital Clinic, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona 8034, Spain.
604. Hematology, Hospital Clinic, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona 8034, Spain.
605. Department of Biochemistry and Molecular Biology, Faculty of Medicine, University Institute of Oncology-IUOPA, Oviedo 33006, Spain.
606. Anatomia Patológica, Hospital Clinic, Institut d'Investigacions Biomèdiques August Pi i Sunyer (IDIBAPS), University of Barcelona, Barcelona 8036, Spain.
607. Spanish Ministry of Science and Innovation, Madrid 28046, Spain.
608. Royal National Orthopaedic Hospital - Bolsover, London W1W 5AQ, UK.
609. Department of Pathology, Oslo University Hospital, The Norwegian Radium Hospital, Oslo O310, Norway.
610. Institute of Clinical Medicine and Institute of Oral Biology, University of Oslo, Oslo O310, Norway.
611. Research Department of Pathology, University College London Cancer Institute, London, WC1E 6BT, UK.
612. East Anglian Medical Genetics Service, Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK.
613. Royal National Orthopaedic Hospital - Stanmore, Stanmore, Middlesex HA7 4LP, UK.
614. Division of Orthopaedic Surgery, Oslo University Hospital, Oslo 0379, Norway.
615. Department of Pathology (Research), University College London Cancer Institute, London WC1E 6BT, UK.
616. Radcliffe Department of Medicine, University of Oxford, Oxford OX3 9DU, UK.
617. University of Pavia, Pavia 27100, Italy.
618. Karolinska Institute, Stockholm SE-171 76, Sweden.
619. Wellcome Sanger Institute, Wellcome Genome Campus, Hinxton, Cambridge, CB10 1SD, UK.
620. University of Oxford, Oxford OX3 9DU, UK.
621. Salford Royal NHS Foundation Trust, Salford M6 8HD, UK.
622. Gloucester Royal Hospital, Gloucester GL1 3NL, UK.
623. Royal Stoke University Hospital, Stoke-on-Trent ST4 6QG, UK.
624. St Thomas's Hospital, London SE1 7EH, UK.
625. Imperial College NHS Trust, Imperial College, London W2 INY, UK.
626. Department of Histopathology, Salford Royal NHS Foundation Trust, Salford M6 8HD, UK.
627. Faculty of Biology, Medicine and Health, The University of Manchester, Manchester M13 9PL, UK.
628. Edinburgh Royal Infirmary, Edinburgh EH16 4SA, UK.
629. Barking Havering and Redbridge University Hospitals NHS Trust, Romford, RM7 0AG, UK.
630. King's College London and Guy's and St Thomas' NHS Foundation Trust, London SE1 7EH, UK.
631. Cambridge Oesophagogastric Centre, Cambridge University Hospitals NHS Foundation Trust, Cambridge, CB2 0QQ.
632. Nottingham University Hospitals NHS Trust, Nottingham NG7 2UH, UK.
633. St Luke's Cancer Centre, Royal Surrey County Hospital NHS Foundation Trust, Guildford GU2 7XX, UK.
634. University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
635. Norfolk and Norwich University Hospital NHS Trust, Norwich NR4 7UY, UK.
636. University Hospitals Coventry and Warwickshire NHS Trust, Coventry CV2 2DX, UK.
637. University Hospitals Birmingham NHS Foundation Trust, Birmingham B15 2GW, UK.
638. Centre for Cancer Research and Cell Biology, Queen's University, Belfast BT9 7AB, UK.
639. School of Cancer Sciences, Faculty of Medicine, University of Southampton, Southampton SO17 1BJ, UK.
640. Wythenshawe Hospital, Manchester M23 9LT, UK.
641. Barts Cancer Institute, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London EC1M 6BQ, UK.
642. Royal Marsden NHS Foundation Trust, London and Sutton SW3 6JJ, UK.
643. University Hospital Southampton NHS Foundation Trust, Southampton SO16 6YD, UK.
644. HCA Laboratories, London W1G 8AQ, UK.
645. University of Liverpool, Liverpool L69 3BX, UK.
646. Academic Urology Group, Department of Surgery, University of Cambridge, Cambridge CB2 0QQ, UK.
647. University of Oxford, Oxford, OX3 9DU, UK.
648. Department of Urology, James Buchanan Brady Urological Institute, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
649. Second Military Medical University, Shanghai 200433, China.
650. Department of Surgery and Cancer, Imperial College, London W2 INY, UK.
651. The Chinese University of Hong Kong, Shatin, NT, Hong Kong, China.
652. Nuffield Department of Surgical Sciences, John Radcliffe Hospital, University of Oxford, Headington, Oxford OX3 9DU, UK.
653. Department of Histopathology, Cambridge University Hospitals NHS Foundation Trust, Cambridge CB2 0QQ, UK.
654. Department of Bioinformatics and Computational Biology / Department of Systems Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
655. Laboratory of Pathology, Center for Cancer Research, National Cancer Institute, Bethesda, MD 20892, USA.
656. Canada's Michael Smith Genome Sciences Center, BC Cancer Agency, Vancouver, BC V5Z 4S6, Canada.
657. Center for Molecular Oncology, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
658. University Health Network, Toronto, ON M5G 2C4, Canada.
659. Department of Pathology and Laboratory Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
660. Department of Population and Quantitative Health Sciences, Case Western Reserve University School of Medicine, Cleveland, OH 44016, USA.
661. Research Health Analytics and Informatics, University Hospitals Cleveland Medical Center, Cleveland, OH 44106, USA.
662. Arnie Charbonneau Cancer Institute, University of Calgary, Calgary, AB T2N 4N2, Canada.
663. Departments of Surgery and Oncology, University of Calgary, Calgary, AB T2N 4N2, Canada.
664. Buck Institute for Research on Aging, Novato, CA 94945, USA.
665. Duke University Medical Center, Durham, NC 27710, USA.
666. USC Norris Comprehensive Cancer Center, University of Southern California, Los Angeles, CA 90033, USA.
667. The Preston Robert Tisch Brain Tumor Center, Duke University Medical Center, Durham, NC 27710, USA.
668. Departments of Dermatology and Pathology, Yale University, New Haven, CT 06510, USA.
669. Fox Chase Cancer Center, Philadelphia, PA 19111, USA.
670. Department of Surgery, Division of Thoracic Surgery, The Johns Hopkins University School of Medicine, Baltimore, MD 21287, USA.
671. University of Michigan Comprehensive Cancer Center, Ann Arbor, MI 48109, USA.
672. University of Alabama at Birmingham, Birmingham, AL 35294, USA.
673. Division of Anatomic Pathology, Mayo Clinic, Rochester, MN 55905, USA.
674. Division of Experimental Pathology, Mayo Clinic, Rochester, MN 55905, USA.
675. Department of Oncology, The Johns Hopkins School of Medicine, The Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, Baltimore, MD 21287, USA.
676. International Genomics Consortium, Phoenix, AZ 85004, USA.
677. Departments of Pediatrics and Genetics, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
678. Department of Pathology, UPMC Shadyside, Pittsburgh, PA 15232, USA.
679. Center for Cancer Genomics, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
680. Istituto Neurologico Besta, Department of Neuro-Oncology, Milano 20133, Italy.
681. University of Queensland Thoracic Research Centre, The Prince Charles Hospital, Brisbane, QLD 4032, Australia.
682. Department of Neurosurgery, University of Florida, Gainesville, FL 32610, USA.
683. Center for Biomedical Informatics, Harvard Medical School, Boston, MA 02115, USA.
684. Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
685. Department of Surgical Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
686. Division of Gastroenterology and Hepatology, Mayo Clinic, Rochester, MN 55905, USA.
687. University of Miami, Sylvester Comprehensive Cancer Center, Miami, FL 33136, USA.
688. Department of Internal Medicine, Division of Medical Oncology, Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
689. University of Tennessee Health Science Center for Cancer Research, Memphis, TN 38163, USA.
690. Centre for Translational and Applied Genomics, British Columbia Cancer Agency, Vancouver, BC V5Z 1L3, Canada.
691. Department of Pathology & Immunology, Baylor College of Medicine, Houston, TX 77030, USA.
692. Michael E. DeBakey Veterans Affairs Medical Center, Houston, TX 77030, USA.
693. Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
694. Canada’s Michael Smith Genome Sciences Centre, BC Cancer Agency, Vancouver, BC V5Z 4S6, Canada.
695. Indivumed GmbH, Hamburg 20251, Germany.
696. Division of Hepatobiliary and Pancreatic Surgery, Department of Surgery, School of Medicine, Keimyung University Dong-san Medical Center, Daegu 41931, South Korea.
697. Women's Cancer Program at the Samuel Oschin Comprehensive Cancer Institute, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
698. Department of Surgery, The George Washington University, School of Medicine and Health Science, Washington, DC 20052, USA.
699. Endocrine Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
700. National Cancer Center, Gyeonggi 10408, South Korea.
701. ILSbio, LLC Biobank, Chestertown, MD 21620, USA.
702. Gynecologic Oncology, NYU Laura and Isaac Perlmutter Cancer Center, New York University, New York, NY 10016, USA.
703. Division of Oncology, Stem Cell Biology Section, Washington University School of Medicine, St. Louis, MO 63110, USA.
704. Urologic Oncology Branch, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892, USA.
705. Institute for Systems Biology, Seattle, WA 98109, USA.
706. Center for Personalized Medicine, Department of Pathology and Laboratory Medicine, Children’s Hospital Los Angeles, Los Angeles, CA 90027, USA.
707. Institute for Genomic Medicine, Nationwide Children's Hospital, Columbus, OH 43215, USA.
708. Department of Surgery, Duke University, Durham, NC 27710, USA.
709. Department of Obstetrics, Gynecology and Reproductive Services, University of California San Francisco, San Francisco, CA 94143, USA.
710. Departments of Neurology and Neurosurgery, Henry Ford Hospital, Detroit, MI 48202, USA.
711. Oregon Health & Science University (OHSU) Knight Cancer Institute, Portland, OR 97210, USA.
712. Department of Pathology, Roswell Park Cancer Institute, Buffalo, NY 14263, USA.
713. Department of Obstetrics and Gynecology, Division of Gynecologic Oncology, Washington University School of Medicine, St. Louis, MO 63110, USA.
714. Department of Palliative, Rehabilitation and Integrative Medicine, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
715. Penrose St. Francis Health Services, Colorado Springs, CO 80907, USA.
716. The University of Chicago, Chicago, IL 60637, USA.
717. Department of Neurology, Mayo Clinic, Rochester, MN 55905, USA.
718. Center for Liver Cancer, Research Institute and Hospital, National Cancer Center, Gyeonggi 410-769, South Korea.
719. Department of Genetics and Lineberger Comprehensive Cancer Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
720. NYU Langone Medical Center, New York, NY 10016, USA.
721. Department of Hematology and Medical Oncology, Cleveland Clinic, Cleveland, OH 44195, USA.
722. Department of Genetics, Department of Pathology and Laboratory Medicine, School of Medicine, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
723. Helen F. Graham Cancer Center at Christiana Care Health Systems, Newark, DE 19713, USA.
724. Cureline, Inc, South San Francisco, CA 94080, USA.
725. Department of Obstetrics and Gynecology, Medical College of Wisconsin, Milwaukee, WI 53226, USA.
726. Hematology and Medical Oncology, Winship Cancer Institute of Emory University, Atlanta, GA 30322, USA.
727. Vanderbilt Ingram Cancer Center, Vanderbilt University, Nashville, TN 37232, USA.
728. Ohio State University College of Medicine and Arthur G. James Comprehensive Cancer Center, Columbus, OH 43210, USA.
729. Research Computing Center, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
730. Analytical Biological Services, Inc, Wilmington, DE 19801, USA.
731. Department of Dermatology, University Hospital Essen, Westdeutsches Tumorzentrum & German Cancer Consortium, Essen 45122, Germany.
732. University of Pittsburgh, Pittsburgh, PA 15213, USA.
733. Murtha Cancer Center, Walter Reed National Military Medical Center, Bethesda, MD 20889, USA.
734. Brigham and Women's Hospital, Harvard Medical School, Boston, MA 02115, USA.
735. Department of Surgery, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA.
736. Department of Gynecologic Oncology and Reproductive Medicine, and Center for RNA Interference and Non-Coding RNA, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
737. Department of Urology, Mayo Clinic, Rochester, MN 55905, USA.
738. Department of Surgery, Johns Hopkins University School of Medicine, Baltimore, MD 21205, USA.
739. Departments of Neurosurgery and Hematology and Medical Oncology, Winship Cancer Institute and School of Medicine, Emory University, Atlanta, GA 30322, USA.
740. Georgia Regents University Cancer Center, Augusta, GA 30912, USA.
741. Thoracic Oncology Laboratory, Mayo Clinic, Rochester, MN 55905, USA.
742. Institute for Genomic Medicine, Nationwide Children’s Hospital, Columbus, OH 43205, USA.
743. Department of Obstetrics & Gynecology, Division of Gynecologic Oncology, Mayo Clinic, Rochester, MN 55905, USA.
744. International Institute for Molecular Oncology, Poznań 60-203, Poland.
745. Poznan University of Medical Sciences, Poznań 61-701, Poland.
746. Edison Family Center for Genome Sciences and Systems Biology, Washington University, St. Louis, MO 63110, USA.
Competing interests
The following authors declare that they have competing interests: Hikmat Al-Ahmadie (H. A. is consultant to AstraZeneca and Bristol-Myers-Squibb); Samuel Aparicio (Founder and shareholder of Contextual Genomics Inc.); Pratiti Bandopadhayay (P.B. receives grant funding from Novartis from an unrelated project.); Rameen Beroukhim (R.B. owns equity in Ampressa Therapeutics); Andrew Biankin (Grant funding from Celgene, AstraZeneca; Consultancies/Advisory boards: AstraZeneca, Celgene, Elstar Therapeutics, Clovis Oncology, Roche.); E Birney (Consultant to Oxford Nanopore, Dovetail and GSK); Marcus Bosenberg (Eli Lilly and Company); Atul Butte (A.B. is a co-founder and consultant to Personalis, NuMedii; consultant to Samsung, Geisinger Health, Mango Tree Corporation, Regenstrief Institute, and in the recent past 10x Genomics and Helix; shareholder in Personalis; minor shareholder in Apple, Twitter, Facebook, Google, Microsoft, Sarepta, 10x Genomics, Amazon, Biogen, CVS, Illumina, Snap, and Sutro; and has received honoraria and travel reimbursement for invited talks from Genentech, Roche, Pfizer, Optum, AbbVie, and many academic institutions and health systems.); C Caldas (C.C. has served on the Scientific Advisory Board of Illumina.); Lorraine Chantrill (L.C. acted on an advisory board for AMGEN Australia in the last 2 years.); Andrew D Cherniack (A.D.C. receives research funding from Bayer AG.); Helen Davies (Helen Davies is an inventor on a number of patent filings encompassing the use of mutational signatures); Francisco De La Vega (Employment at Annai Systems Inc. during part of the project.); Ronny Drapkin (R.D. serves on the SAB of Repare Therapeutics and Siamab Therapeutics.); Rosalind Eeles (GU-ASCO meeting in San Francisco – Jan 2016 – Honorarium as speaker $500. 2. RMH FR meeting – Nov 2017 – support from Janssen, honorarium as speaker £1100 (Title: Genetics and Prostate Cancer). 3. University of Chicago invited talk May 2018 – Honorarium as speaker $1000. 4. EUR 200 educational honorarium paid by Bayer & Ipsen to attend GU Connect “Treatment sequencing for mCRPC patients within the changing landscape of mHSPC” at a venue at ESMO, Barcelona, 28 September 2019.); Paul Flicek (Member of the Scientific Advisory Boards of Fabric Genomics, Inc., and Eagle Genomics, Ltd.); Gad Getz (G.G. receives research funds from IBM and Pharmacyclics and is an inventor on patent applications related to MuTect, ABSOLUTE, MutSig, MSMuTect, MSMutSig and POLYSOLVER); Ronald Ghossein (veracyte, inc); D Glodzik (D.G. is an inventor on a number of patent filings encompassing the use of mutational signatures.); Eoghan Harrington (Eoghan Harrington is a full-time employee of Oxford Nanopore Technologies Inc. and is a stock option holder); Yann Joly (Responsible for the Data Access Compliance Office (DACO) of ICGC 2009-2018.); Sissel Juul (SJ is a full-time employee of Oxford Nanopore Technologies Inc. and is a stock option holder); Vincent Khoo (VK has received personal fees and non-financial support from Accuray, Astellas, Bayer, Boston Scientific, and Janssen.); Stian Knappskog (co-PI on clinical trial receiving research funding from AstraZeneca and Pfizer); Ignaty Leshchiner (Consulting-PACT Pharma); Yong-Jie Lu (CA16672 and R50CA221675); Carlos López-Otín (CLO has ownership interest (including stock, patents, etc.) from DREAMgenics); Matthew Meyerson (Scientific advisory board chair of, and consultant for, OrigiMed. Research funding from Bayer and Ono Pharma. Patent royalties from LabCorp.); Serena Nik-Zainal (S. N.-Z. is an inventor on a number of patent filings encompassing the use of mutational signatures.); Nathan Pennell (N.P. has done consulting work with Merck, Astrazeneca, Eli Lilly, and BMS.); Xose Puente (XSP has ownership interest (including stock, patents, etc.) from DREAMgenics); Benjamin Raphael (BJR is a consultant at and has ownership interest (including stock, patents, etc.) in Medley Genomics.); Jorge Reis-Filho (Consultant of Goldman Sachs and REPARE Therapeutics; member of the scientific advisory board of Volition RX and Paige.AI; ad hoc member of the scientific advisory board of Ventana Medical Systems, Roche Tissue Diagnostics, InVicro, Roche, Genentech and Novartis); Lewis Roberts (LRR has received grant support from ARIAD Pharmaceuticals, Bayer, BTG International, Exact Sciences, Gilead Sciences, Glycotest, Inc., RedHill Biopharma, Inc., Target PharmaSolutions, and Wako Diagnostics; he has provided advisory services to Bayer, Exact Sciences, Gilead Sciences, GRAIL, Inc., QED Therapeutics and TAVEC Pharmaceuticals Inc.); Richard Scolyer (Richard A. Scolyer reports receiving fees for professional services from Merck Sharp & Dohme, GlaxoSmithKline Australia, Bristol-Myers Squibb, Dermpedia, Novartis Pharmaceuticals Australia Pty Ltd, Myriad, NeraCare GmbH and Amgen); Tal Shmaya (Employment at Annai Systems, Inc.); Reiner Siebert (Speakers Honorary Roche, AstraZeneca); Sabina Signoretti (Consulting: Bristol-Myers Squibb, AstraZeneca, Merck, AACR, NCI. Funding: Bristol-Myers Squibb, AstraZeneca, Exelixis. Royalties: Biogenex.); Jared Simpson (Research funding and travel support from Oxford Nanopore Technologies); Anil K Sood (Consulting – Merck, Kiyatec); Research funding (M-Trap); Shareholder (BioPath)); Simon Tavaré (I am on the SAB of Ipsen, and a consultant for Kallyope Inc.); John Thompson (J.F.T. has received honoraria and travel support for attending advisory board meetings of GlaxoSmithKline and Provectus Inc. He has received honoraria for participation in advisory boards for MSD Australia and BMS Australia.); Daniel Turner (DT is a full-time employee of Oxford Nanopore Technologies Ltd. and is a stock option holder); Naveen Vasudev (Speaker honoraria and/or consultancy fees from Bristol Myers Squibb, Pfizer, EUSA pharma, MSD, Novartis); Jeremiah Wala (J.A.W. is a consultant for Nference Inc.); Daniel Weisenberger (D.J.W. is a consultant for Zymo Research Corporation.); Dai-Ying Wu (Employment at Annai Systems, Inc.); Cheng-Zhong Zhang (C.-Z.Z. is a co-founder and equity holder of Pillar Biosciences, a for-profit company specializing in the development of targeted sequencing assays.)
References
- 1.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pleasance ED, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008 doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rheinbay E, et al. Analyses of non-coding somatic drivers in 2,693 cancer whole genomes. Nature. 2019 doi: 10.1038/s41586-020-1965-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexandrov LB, et al. The Repertoire of Mutational Signatures in Human Cancer. Nature. 2019 doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, et al. Patterns of somatic structural variation in human cancer genomes. Nature. 2019 doi: 10.1038/s41586-019-1913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerstung M, et al. The evolutionary history of 2,658 cancers. Nature. 2019 doi: 10.1038/s41586-019-1907-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.PCAWG Transcriptome Core Group et al. Genomic basis of RNA alterations in cancer. Nature. 2019 doi: 10.1038/s41586-020-1970-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Y, et al. High-coverage whole-genome analysis of 1220 cancers reveals hundreds of genes deregulated by rearrangement-mediated cis-regulatory alterations. Nat Commun. 2019 doi: 10.1038/s41467-019-13885-w. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Martin B, et al. Pan-cancer analysis of whole genomes identifies driver rearrangements promoted by LINE-1 retrotransposition. Nat Genet. 2019 doi: 10.1038/s41588-019-0562-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kübler K, et al. Tumor mutational landscape is a record of the pre-malignant state. Nature. 2019 doi: 10.1101/517565. [DOI] [Google Scholar]
- 12.Jiao W, et al. A deep learning system can accurately classify primary and metastatic cancers based on patterns of passenger mutations. Nat Commun. 2019 [Google Scholar]
- 13.Sieverling L, et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Nat Commun. 2019 doi: 10.1038/s41467-019-13824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan Y, et al. Comprehensive Molecular Characterization of Mitochondrial Genomes in Human Cancers. Nat Genet. 2019 doi: 10.1038/s41588-019-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akdemir KC, et al. Chromatin Folding Domains Disruptions by Somatic Genomic Rearrangements in Human Cancers. Nat Genet. 2019 doi: 10.1038/s41588-019-0564-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyna MA, et al. Pathway and network analysis of more than 2,500 whole cancer genomes. Nat Commun. 2019 doi: 10.1038/s41467-020-14367-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bailey MH, et al. Retrospective evaluation of whole exome and genome mutation calls in 746 cancer samples. Nat Commun. 2019 doi: 10.1038/s41467-020-18151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cortes-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Nature Genetics. 2019 doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bray F, Ren J-S, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 20.Tarver T. Cancer Facts & Figures 2012. American Cancer Society (ACS) Journal of Consumer Health On the Internet. 2012;16:366–367. [Google Scholar]
- 21.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 22.International Cancer Genome Consortium et al. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bailey MH, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell. 2018;173:371–385.e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Vega F, et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoadley KA, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell. 2018;173:291–304.e6. doi: 10.1016/j.cell.2018.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein LD, Knoppers BM, Campbell P, Getz G, Korbel JO. Data analysis: Create a cloud commons. Nature. 2015;523:149–151. doi: 10.1038/523149a. [DOI] [PubMed] [Google Scholar]
- 27.Phillips M, et al. Of Clouds and Genomic Data Protection. Nature. 2019 [Google Scholar]
- 28.Krochmalski J. Developing with Docker. Packt Publishing Ltd; 2016. [Google Scholar]
- 29.Welch JS, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150:264–278. doi: 10.1016/j.cell.2012.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nik-Zainal S, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meier B, et al. C. elegans whole-genome sequencing reveals mutational signatures related to carcinogens and DNA repair deficiency. Genome Research. 2014;24:1624–1636. doi: 10.1101/gr.175547.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martincorena I, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2018;173:1823. doi: 10.1016/j.cell.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamborero D, et al. Cancer Genome Interpreter annotates the biological and clinical relevance of tumor alterations. Genome Med. 2018;10:25. doi: 10.1186/s13073-018-0531-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang FW, et al. Highly recurrent TERT promoter mutations in human melanoma. Science. 2013;339:957–959. doi: 10.1126/science.1229259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rheinbay E, et al. Recurrent and functional regulatory mutations in breast cancer. Nature. 2017;547:55–60. doi: 10.1038/nature22992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fredriksson NJ, Ny L, Nilsson JA, Larsson E. Systematic analysis of noncoding somatic mutations and gene expression alterations across 14 tumor types. Nat Genet. 2014;46:1258–1263. doi: 10.1038/ng.3141. [DOI] [PubMed] [Google Scholar]
- 37.Horn S, et al. TERT promoter mutations in familial and sporadic melanoma. Science. 2013;339:959–961. doi: 10.1126/science.1230062. [DOI] [PubMed] [Google Scholar]
- 38.Ciriello G, et al. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahman N. Realizing the promise of cancer predisposition genes. Nature. 2014;505:302–308. doi: 10.1038/nature12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pearl LH, Schierz AC, Ward SE, Al-Lazikani B, Pearl FMG. Therapeutic opportunities within the DNA damage response. Nat Rev Cancer. 2015;15:166–180. doi: 10.1038/nrc3891. [DOI] [PubMed] [Google Scholar]
- 41.Taylor-Weiner A, et al. DeTiN: overcoming tumor-in-normal contamination. Nat Methods. 2018;15:531–534. doi: 10.1038/s41592-018-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fujimoto A, et al. Whole-genome mutational landscape and characterization of noncoding and structural mutations in liver cancer. Nature Genetics. 2016;48:500–509. doi: 10.1038/ng.3547. [DOI] [PubMed] [Google Scholar]
- 43.Shlush LI. Age-related clonal hematopoiesis. Blood. 2018;131:496–504. doi: 10.1182/blood-2017-07-746453. [DOI] [PubMed] [Google Scholar]
- 44.Northcott PA, et al. The whole-genome landscape of medulloblastoma subtypes. Nature. 2017;547:311–317. doi: 10.1038/nature22973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scarpa A, et al. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543:65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 46.Davis CF, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26:319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baca SC, et al. Punctuated evolution of prostate cancer genomes. Cell. 2013;153:666–677. doi: 10.1016/j.cell.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nik-Zainal S, et al. The Life History of 21 Breast Cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nik-Zainal S, et al. Mutational processes molding the genomes of 21 breast cancers. Cell. 2012;149:979–993. doi: 10.1016/j.cell.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts SA, et al. Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions. Mol Cell. 2012;46:424–435. doi: 10.1016/j.molcel.2012.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rausch T, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stephens PJ, et al. Massive Genomic Rearrangement Acquired in a Single Catastrophic Event during Cancer Development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Korbel JO, Campbell PJ. Criteria for inference of chromothripsis in cancer genomes. Cell. 2013;152:1226–1236. doi: 10.1016/j.cell.2013.02.023. [DOI] [PubMed] [Google Scholar]
- 55.Zhang C-Z, et al. Chromothripsis from DNA damage in micronuclei. Nature. 2015;522:179–184. doi: 10.1038/nature14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cancer Genome Atlas Research Network. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Supek F, Lehner B. Clustered Mutation Signatures Reveal that Error-Prone DNA Repair Targets Mutations to Active Genes. Cell. 2017;170:534–547.e23. doi: 10.1016/j.cell.2017.07.003. [DOI] [PubMed] [Google Scholar]
- 58.Cortes-Ciriano I, et al. Comprehensive analysis of chromothripsis in 2,658 human cancers using whole-genome sequencing. Genomics. 2018 doi: 10.1038/s41588-019-0576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mardin BR, et al. A cell-based model system links chromothripsis with hyperploidy. Mol Syst Biol. 2015;11:828. doi: 10.15252/msb.20156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weischenfeldt J, et al. Integrative genomic analyses reveal an androgen-driven somatic alteration landscape in early-onset prostate cancer. Cancer Cell. 2013;23:159–170. doi: 10.1016/j.ccr.2013.01.002. [DOI] [PubMed] [Google Scholar]
- 61.Garsed DW, et al. The architecture and evolution of cancer neochromosomes. Cancer Cell. 2014;26:653–667. doi: 10.1016/j.ccell.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 62.Durinck S, et al. Temporal dissection of tumorigenesis in primary cancers. Cancer Discov. 2011;1:137–143. doi: 10.1158/2159-8290.CD-11-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dentro SC, et al. Portraits of genetic intra-tumour heterogeneity and subclonal selection across cancer types. doi: 10.1101/312041. [DOI] [Google Scholar]
- 64.Hayward NK, et al. Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545:175–180. doi: 10.1038/nature22071. [DOI] [PubMed] [Google Scholar]
- 65.Cancer Genome Atlas Network. Genomic Classification of Cutaneous Melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alexandrov LB, et al. The Repertoire of Mutational Signatures in Human Cancer. Nature. 2019 doi: 10.1038/s41586-020-1943-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexandrov LB, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chan K, et al. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nik-Zainal S, et al. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Middlebrooks CD, et al. Association of germline variants in the APOBEC3 region with cancer risk and enrichment with APOBEC-signature mutations in tumors. Nat Genet. 2016;48:1330–1338. doi: 10.1038/ng.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Menghi F, et al. The tandem duplicator phenotype as a distinct genomic configuration in cancer. Proc Natl Acad Sci U S A. 2016;113:E2373–82. doi: 10.1073/pnas.1520010113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hendrich B, Hardeland U, Ng HH, Jiricny J, Bird A. The thymine glycosylase MBD4 can bind to the product of deamination at methylated CpG sites. Nature. 1999;401:301–304. doi: 10.1038/45843. [DOI] [PubMed] [Google Scholar]
- 75.Lee E, et al. Landscape of Somatic Retrotransposition in Human Cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tubio JMC, et al. Extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345 doi: 10.1126/science.1251343. 1251343–1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Helman E, et al. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–1063. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shay JW, Wright WE. Hayflick, his limit, and cellular ageing. Nat Rev Mol Cell Biol. 2000;1:72–76. doi: 10.1038/35036093. [DOI] [PubMed] [Google Scholar]
- 79.Peifer M, et al. Telomerase activation by genomic rearrangements in high-risk neuroblastoma. Nature. 2015;526:700–704. doi: 10.1038/nature14980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Totoki Y, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet. 2014;46:1267–1273. doi: 10.1038/ng.3126. [DOI] [PubMed] [Google Scholar]
- 81.Paterlini-Bréchot P, et al. Hepatitis B virus-related insertional mutagenesis occurs frequently in human liver cancers and recurrently targets human telomerase gene. Oncogene. 2003;22:3911–3916. doi: 10.1038/sj.onc.1206492. [DOI] [PubMed] [Google Scholar]
- 82.Heaphy CM, et al. Prevalence of the alternative lengthening of telomeres telomere maintenance mechanism in human cancer subtypes. Am J Pathol. 2011;179:1608–1615. doi: 10.1016/j.ajpath.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sieverling L, et al. Genomic footprints of activated telomere maintenance mechanisms in cancer. Genomics. 2017 doi: 10.1038/s41467-019-13824-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Barthel FP, et al. Systematic analysis of telomere length and somatic alterations in 31 cancer types. Nat Genet. 2017;49:349–357. doi: 10.1038/ng.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.García-Cao M, Gonzalo S, Dean D, Blasco MA. A role for the Rb family of proteins in controlling telomere length. Nat Genet. 2002;32:415–419. doi: 10.1038/ng1011. [DOI] [PubMed] [Google Scholar]
- 86.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gerstung M, et al. Precision oncology for acute myeloid leukemia using a knowledge bank approach. Nat Genet. 2017;49:332–340. doi: 10.1038/ng.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.O’Connor BD, et al. The Dockstore: enabling modular, community-focused sharing of Docker-based genomics tools and workflows. F1000Research. 2017;6:52. doi: 10.12688/f1000research.10137.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang J, et al. The International Cancer Genome Consortium Data Portal. Nat Biotechnol. 2019;37:367–369. doi: 10.1038/s41587-019-0055-9. [DOI] [PubMed] [Google Scholar]
- 90.Miller CA, Qiao Y, DiSera T, D’Astous B, Marth GT. bam.iobio: a web-based, real-time, sequence alignment file inspector. Nature Methods. 2014;11:1189–1189. doi: 10.1038/nmeth.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Goldman M, et al. The UCSC Xena platform for public and private cancer genomics data visualization and interpretation. doi: 10.1101/326470. [DOI] [Google Scholar]
- 92.Papatheodorou I, et al. Expression Atlas: gene and protein expression across multiple studies and organisms. Nucleic Acids Research. 2018;46:D246–D251. doi: 10.1093/nar/gkx1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.SEER ICD-O-3 Coding Materials. Available at: https://seer.cancer.gov/icd-o-3/
- 94.Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raine KM, et al. ascatNgs: Identifying Somatically Acquired Copy-Number Alterations from Whole-Genome Sequencing Data. Curr Protoc Bioinformatics. 2016;56:15.9.1–15.9.17. doi: 10.1002/cpbi.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jones D, et al. cgpCaVEManWrapper: Simple Execution of CaVEMan in Order to Detect Somatic Single Nucleotide Variants in NGS Data. Curr Protoc Bioinformatics. 2016;56:15.10.1–15.10.18. doi: 10.1002/cpbi.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raine KM, et al. cgpPindel: Identifying Somatically Acquired Insertion and Deletion Events from Paired End Sequencing. Curr Protoc Bioinformatics. 2015;52:15.7.1–12. doi: 10.1002/0471250953.bi1507s52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rausch T, et al. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339. doi: 10.1093/bioinformatics/bts378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nature Genetics. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Drier Y, et al. Somatic rearrangements across cancer reveal classes of samples with distinct patterns of DNA breakage and rearrangement-induced hypermutability. Genome Research. 2013;23:228–235. doi: 10.1101/gr.141382.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ramos AH, et al. Oncotator: Cancer Variant Annotation Tool. Human Mutation. 2015;36:E2423–E2429. doi: 10.1002/humu.22771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Moncunill V, et al. Comprehensive characterization of complex structural variations in cancer by directly comparing genome sequence reads. Nat Biotechnol. 2014;32:1106–1112. doi: 10.1038/nbt.3027. [DOI] [PubMed] [Google Scholar]
- 107.Fan Y, et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol. 2016;17:178. doi: 10.1186/s13059-016-1029-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cooke SL, et al. Processed pseudogenes acquired somatically during cancer development. Nature Communications. 2014;5 doi: 10.1038/ncomms4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ju YS, et al. Origins and functional consequences of somatic mitochondrial DNA mutations in human cancer. eLife. 2014;3 doi: 10.7554/eLife.02935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Group PTC, et al. Genomic basis for RNA alterations revealed by whole-genome analyses of 27 cancer types. doi: 10.1101/183889. [DOI] [Google Scholar]
- 111.Sudmant PH, et al. An integrated map of structural variation in 2,504 human genomes. Nature. 2015;526:75–81. doi: 10.1038/nature15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. arXiv [q-bio.GN] 2012 [Google Scholar]
- 113.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yakneen S, Waszak SM, Gertz M, Korbel JO, PCAWG Consortium Enabling rapid cloud-based analysis of thousands of human genomes via Butler. Nat Biotechnol. 2019 [Google Scholar]
- 115.Kim SY, Jacob L, Speed TP. Combining calls from multiple somatic mutation-callers. BMC Bioinformatics. 2014;15:154. doi: 10.1186/1471-2105-15-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Breiman L. Stacked regressions. Mach Learn. 1996;24:49–64. [Google Scholar]
- 117.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wala JA, et al. SvABA: genome-wide detection of structural variants and indels by local assembly. Genome Research. 2018;28:581–591. doi: 10.1101/gr.221028.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.