Abstract
Fever is one of the most common reasons for healthcare seeking globally and the majority of human pathogens are zoonotic. We conducted a systematic review to describe the occurrence and distribution of zoonotic causes of human febrile illness reported in malaria endemic countries. Articles included in the review yielded data from 53 (48·2%) of 110 malaria endemic countries. The 244 articles included described diagnosis of 30 zoonoses in febrile people. The majority of zoonoses were bacterial (n=17), with viruses (n=9), protozoa (n=3) and helminths (n=1) also identified. Leptospira spp. and nontyphoidal Salmonella serovars were the most frequently reported pathogens. Despite evidence of profound data gaps, this review reveals widespread distribution of a diverse range of zoonotic causes of febrile illness. Greater understanding of the epidemiology of zoonoses in different settings is needed to improve awareness and management of the multiple zoonotic causes of febrile illness.
Introduction
Fever is one of the most common symptoms prompting healthcare seeking globally.1–3 Fever has myriad causes and their non-specific clinical presentation means that clinical history and physical examination are often insufficient to accurately identify causal pathogens.1 Limitations in laboratory services and available diagnostic tools further contribute to diagnostic challenges.4 In malaria-endemic countries, fever is often assumed to be due to malaria.5 The mortality and morbidity attributable to malaria remains considerable, but there is also evidence of widespread over-diagnosis within malaria-endemic areas.6–8 The recognized over-diagnosis of malaria together with declines in malaria incidence since the peak in global malaria deaths in 20049,10 have prompted attention to non-malaria causes of fever in malaria-endemic areas.11,12 Zoonotic pathogens are likely to play a substantial role as causes of fever globally. Almost two-thirds of all human pathogens are zoonotic,13 and there is growing evidence that many zoonoses cause more cases of human febrile illness than previously appreciated.12,14–20 Improved understanding of the impacts and burdens of zoonotic causes of fever in malaria-endemic countries would provide the epidemiological evidence base for disease control program development and also influence diagnostic and treatment algorithms for fever, with the potential to improve clinical outcomes. The aim of this study was to systematically review the published literature to describe the occurrence and distribution of reported zoonotic causes of human febrile illness in countries where malaria is endemic.
Methods
Search strategy and selection criteria
The target literature for this systematic review was peer-reviewed published articles that described the testing of one or more febrile person from malaria-endemic countries for one or more zoonotic pathogen using robust diagnostic testing criteria to demonstrate acute infection. Literature searches of the Medline and Embase databases were run using the OvidSP gateway. Searches were limited to English language articles published in the period 2004 to 2019 inclusive, to span the period from the described peak of global malaria mortality in 2004 to present.9 The searches were last executed on 03 January 2019. Outputs of database searches were combined and de-duplicated using R.21 Additional details of searches, screening, review, and data extraction processes are given in the appendix.
Three search concepts for ‘fever,’ ‘zoonoses,’ and ‘malaria endemic countries’ were constructed. To construct the ‘fever’ concept the exploded subject heading and keywords were combined using database appropriate syntax (e.g., exp Fever/ OR fever$1.mp. OR febrile.mp.). For the ‘zoonoses’ concept, a reference list of eligible zoonotic pathogens was compiled using lists of zoonotic diseases from the World Health Organization (WHO)22 and World Organisation of Animal Health (OIE)23 as well as literature-based searches to identify frequently reported zoonotic causes of human fever. We conducted preliminary searches of Medline and Embase using the search syntax ‘(exp Fever/ OR fever.mp.) AND (exp Zoonoses/ OR zoonoses.mp OR zoonosis.mp)’ limited to humans. Additional details of search concept construction are given in the appendix. All pathogens identified through these approaches were mapped to existing subject headings and keywords at the lowest taxonomic level possible, typically genus or species. In instances where pathogen species or serovars within the same genus varied in their zoonotic status, search concepts were constructed to include all zoonotic and non-zoonotic species or serovars and articles relating to non-zoonotic species were excluded at the full text stage. The candidate pathogens were classified to differentiate pathogens normatively acquired by people through direct or indirect transmission from vertebrate animals to humans, as compared to pathogens where zoonotic transmission has been recorded but where the majority of human infections are not acquired through zoonotic transmission. We classified pathogens using the stages in the process towards human endemicity defined in Wolfe et al.24 Pathogens classified at stages one to three (normatively acquired through zoonotic transmission) were retained (appendix). The search concept for each pathogen or disease included exploded subject headings for both the pathogen and the diseases caused in humans and terms for both pathogen and disease were also included as keywords (e.g., exp anthrax/ OR anthrax.mp. OR exp Bacillus anthracis/ OR bacillus anthracis.mp.). The list of pathogen or disease specific searches was combined using OR syntax to generate the full ‘zoonoses’ search concept (appendix). The ‘malaria endemic countries’ concept was constructed by mapping country names for countries defined as malaria endemic in the WHO global malaria reports for the years 2005 and 2016 to Medline and Embase subject headings.10,25 Each country was searched for using both the exploded subject heading where possible and keywords in all cases (e.g., exp Kenya/OR Kenya.mp.). The three concepts, fever,’ ‘zoonoses,’ and ‘malaria endemic countries’ were combined using AND operators and database specific syntax (appendix).
Study selection and validity assessment
Articles that reported the diagnosis of a zoonotic pathogen in a population from a malaria endemic country defined on the basis of febrile illness were selected for full-text review. Conference proceedings and records that did not include any abstract text or an abstract in English were excluded. Abstracts and titles were screened by two independent reviewers (two of MC, MES, KJA, GAFL, DVH, JAC, SC and MPR) using pre-defined criteria (appendix table S1). Articles were selected for inclusion if the abstract or title described clinical and/or laboratory evaluation of a group of ≥ 2 people all of whom had fever and some of whom were diagnosed of one or more pathogens from the reference list of zoonotic pathogens (table 1). Abstracts referring to the use of blood culture were also retained at this stage even if a zoonosis was not explicitly mentioned in the abstract (appendix table S1). When two reviewers disagreed on article classification, a third independent reviewer (one of JEBH, MC, MES, GAFL, DVH or MPR) resolved the tiebreak. Full text articles were sought for all articles not excluded during abstract review steps. All articles were searched for using PubMed, Google and the libraries of the University of Glasgow, Duke University, Washington University in St. Louis, and US Centers for Disease Control and Prevention (US CDC). Articles were excluded if a full text for the citation could not be obtained. Two independent reviewers (two of, JEBH, MC, MES, JB and MPR) evaluated full text articles using pre-defined inclusion and exclusion criteria (table 2, appendix table S2). Strict diagnostic case definitions based on WHO and US CDC guidelines ensured that only studies reporting robust and specific diagnostic methods were retained (table 2). Articles were excluded if they did not meet one or more of the study inclusion criteria or if they did meet at least one of the study exclusion criteria (table 2). In cases where reviewers disagreed on article classification, discrepancies were checked and resolved by JEBH in discussion with other reviewers.
Table 1. Zoonoses included in the review, with details of species and serovars excluded where appropriate.
| Pathogen | Species, subspecies, and serovars excluded | Pathogen type13 |
|---|---|---|
| Alphaviruses | All species excluded with the exception of Eastern equine encephalitis virus (EEEV) complex, Venezuelan equine encephalitis (VEEV) complex, and Western equine encephalitis (WEEV) complex | Virus |
| Anaplasma spp. | - | Bacteria |
| Aphthoviruses | All species excluded with the exception of Foot-and-mouth disease virus | Virus |
| Avulaviruses | All species excluded with the exception of Newcastle disease virus | Virus |
| Babesia spp. | - | Protozoa |
| Bacillus antrhracis | - | Bacteria |
| Bartonella spp. | B. bacilliformis and B. quintana excluded | Bacteria |
| Borrelia spp. | B. recurrentis excluded | Bacteria |
| Bovine spongiform encephalopathy | - | Prion |
| Brucella spp. | - | Bacteria |
| Burkholderia spp. | B. cepacia complex and B. pseudomallei excluded | Bacteria |
| Campylobacter spp. | - | Bacteria |
| Chlamydia spp. | All species excluded with the exception of C. psittaci | Bacteria |
| Coxiella burnetii | - | Bacteria |
| Cryptosporidium spp. | C. hominis excluded | Protozoa |
| Ebolavirus | - | Virus |
| Echinococcus spp. | - | Helminth |
| Ehrlichia spp. | - | Bacteria |
| Enteroviruses | All species excluded with the exception of Swine vesicular disease virus | Virus |
| Escherichia spp. | All species excluded with the exception of Shiga-toxin producing E. coli | Bacteria |
| Flaviviruses | All species excluded with the exception of Japanese encephalitis virus (JEV), West Nile virus (WNV), and Tick-borne-encephalitis virus. | Virus |
| Francisella spp. | All species excluded with the exception of F. tularensis | Bacteria |
| Hantavirus | - | Virus |
| Henipaviruses | - | Virus |
| Lassa virus | - | Virus |
| Leishmania spp. | L. donovani excluded if detected in India | Protozoa |
| Leptospira spp. | - | Bacteria |
| Listeria spp. | - | Bacteria |
| Lyssavirus | All species excluded with the exception of Rabies virus | Virus |
| Marburg virus | - | Virus |
| Mycobacterium | All species excluded with the exception of M. bovis and M. avis | Bacteria |
| Nairovirus | All species excluded with the exception of Crimean-Congo haemorrhagic fever virus | Virus |
| Orientia1 | - | Bacteria |
| Orthopox viruses | All species excluded with the exception of Cowpox virus, Monkeypox virus, and Vaccinia virus | Virus |
| Pasteurella spp. | - | Bacteria |
| Phleboviruses | All species excluded with the exception of Rift Valley fever (RVF) virus | Virus |
| Rickettsia spp.2 | R. prowazekii excluded | Bacteria |
| Salmonella spp. | All species, subspecies, and serovars excluded with the exception of nontyphoidal Salmonella serovars | Bacteria |
| Schistosoma spp. | S. haematobium, S. intercalatum, and S. mekongi.excluded | Helminth |
| Streptobacillus spp. | - | Bacteria |
| Streptococcus spp. | All species excluded with the exception of S. canis, S. suis, S. equi, and S. iniae | Bacteria |
| Taenia spp. | Helminth | |
| Toxocara | Helminth | |
| Toxoplasma gondii | - | Protozoa |
| Trichinella spp. | - | Helminth |
| Trypanosoma spp. | All species excluded with the exception of T. brucei rhodesiense and T. cruzi | Protozoa |
| Varicelloviruses | All species excluded with the exception of Pseudorabies virus | Virus |
| Vesiculoviruses | All species excluded with the exception of Vesicular Stomatitis virus | Virus |
| Yersinia spp. | All species excluded with the exception of Y. pestis, Y. enterocolitica and Y. pseudotuberculosis | Bacteria |
Orientia was covered by search syntax for Rickettsia.
For data extraction, data on Rickettsia were classified as Rickettsia (SFGR) or Rickettsia (TGR) where the data resolution allowed. When details on the species of Rickettsia were not given, these data were classified as Rickettsia spp.
Table 2. Inclusion and exclusion criteria for full text review.
| Outcome | Criterion |
|---|---|
| Inclusion: |
|
| Exclusion: |
|
The following met study criteria for valid diagnostics for pathogen detection based on single sera only: Leptospira spp. agglutination titer of ≥ 800 by microscopic agglutination test in one serum specimen 26; detection of Hantavirus-specific IgM in a serum sample 27; detection of virus-specific IgM antibodies in serum with confirmatory virus-specific neutralizing antibodies for Eastern equine encephalitis virus (EEEV), West Nile virus (WNV), Western equine encephalitis virus (WEEV), and Venezuelan equine encephalitis virus (VEEV) 28; identification of lyssavirus specific antibody by indirect fluorescent antibody test or complete rabies virus neutralization at 1:5 dilution in the serum of an unvaccinated person 29; detection of viral antigens in blood by enzyme-linked immunosorbent assay for Ebola 30,31, Marburg 31,32, Lassa 31,33, and Crimean-Congo haemorrhagic fever viruses 31; detection of Rift Valley fever antigens or IgM in blood by enzyme-linked Immunosorbent assay 34
Data extraction and bias assessment
Data extraction was conducted independently by one of two reviewers (JEBH and MC). Article-level data were extracted on the location (country and WHO regional classification), 36 study period (start and end year of data collection), and eligibility criteria used in the study. Each population was classified according to the clinical presentation as undifferentiated or differentiated. Differentiated febrile populations were further classified as: i) febrile neurologic; ii) febrile haemorrhagic; iii) febrile gastrointestinal; iv) febrile respiratory; v) specific febrile aetiology suspected; vi) febrile co-morbid group (i.e., malignancy, immunocompromise).37–39 Data extracted on each population included any demographic restriction of the study population, the age range of the study participants, whether the population was described as inpatient or outpatient, urban or rural, and whether data were collected during a reported disease outbreak or not. To extract data on zoonotic pathogens, every article was classified to record if the study reported looking for or diagnosing one or more febrile individuals with any of the zoonotic pathogens included in the study reference list (table 1), irrespective of the diagnostics used. Additional data were extracted when the article reported application of a diagnostic approach that met study validity criteria. For each combination of article and pathogen, details of the valid diagnostic methods used, the type and number of samples tested, and the number of positive samples were recorded (appendix table S3, S4). In instances where more than one valid diagnostic method was used in the same study for a given pathogen (e.g., culture-based and serologic case definitions), data on the total number of individuals tested and positive for each pathogen using valid methods were aggregated. Some articles contributed data on more than one pathogen but no data on participant numbers were extracted for pathogens not identified using diagnostic approaches that met study inclusion criteria.
The principal source of potential bias affecting the interpretation of the findings of this study is the lack of standardization of the febrile populations included in different studies. Criteria were defined to classify potential bias in study representativeness and prevalence estimate precision (appendix table S5).40–42 The representativeness bias criterion was designed to classify the representativeness of the study population, relative to the general population where the study was conducted. This was based on the description of the febrile population, the restriction (if any) of the study sample to specific clinical or demographic sub-populations and the reporting of disease outbreaks at the time of data collection. Each population was classified as follows: i) populations classified as undifferentiated febrile with no demographic restriction and no clinical aetiologies excluded were classified as low risk; ii) populations classified as undifferentiated febrile with demographic restriction and/or reporting exclusion of specific aetiologies or syndromes were classified as medium risk; iii) differentiated febrile populations and those from studies reporting disease outbreaks at the time of data collection were classified as high risk. The second, outcome-level, bias criterion was designed to classify risk of bias in the estimated precision of the proportion of fevers attributed to each pathogen. Thresholds used for this criterion are the sample sizes needed to estimate proportions of 50% and 10% with 95% confidence and 0·05 precision respectively, assuming an infinite population size. Each population was classified as follows: i) proportion estimates based on a sample size of greater than or equal to 385 were classified as low risk; ii) proportion estimates based on a sample size of greater than 385 but less than 139 were classified as medium risk; iii) proportion estimates based on a sample size of less than 139 were classified as high risk.
Additional potential sources of bias included variation in the pathogens tested for, and variation in the diagnostic approaches applied. For included studies, data on the pathogens tested for (with any diagnostic approach) were summarized alongside pathogens for which diagnostic test criteria were met to qualitatively evaluate the biases introduced by only extracting data on pathogens diagnosed using methods meeting study inclusion criteria.
Data analysis
Extracted data on the zoonotic pathogens diagnosed using valid methods, number of individuals tested for each pathogen, and number of individuals positive for each pathogen were used to estimate the proportion of fevers attributable to each pathogen for each unique pathogen and study combination. All analyses were conducted in R21 and plots were made using the package ggplot2.43
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Results
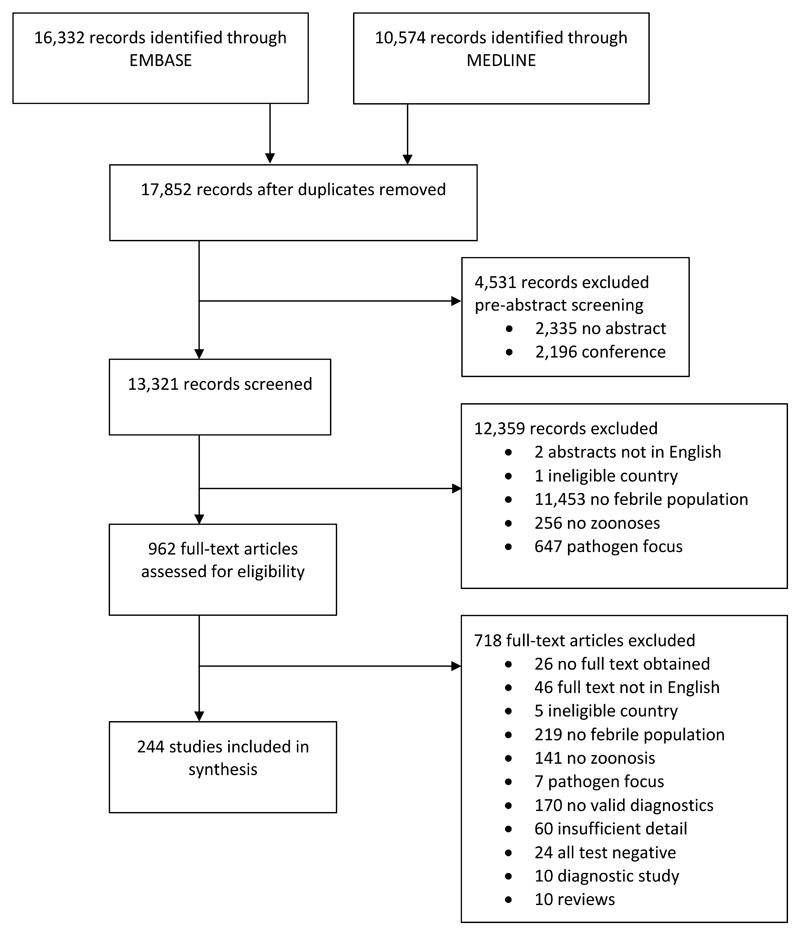
Database searches yielded a total of 16,332 and 10,574 records through Embase and Medline, respectively, resulting in a total of 17,852 unique records following de-duplication (figure 1). A total of 4,531 (25·4%) records were excluded during pre-screening, 13,321 (74·6%) records were screened and 962 (7·2%) of these were retained after title and abstract review. In total, 718 (74·6%) articles were excluded during full text review and 244 (25·4%) articles met all study inclusion criteria and were included (figure 1, appendix table S6).
Figure 1. Flow diagram of records and articles assessed for the review.
Among the 46 articles excluded because the full text was not accessible in English, the breakdown of languages was as follows: French (13 articles); Spanish (11 articles); Turkish (9 articles); Mandarin (6 articles); Portuguese (2 articles); Hebrew (2 articles); Arabic (1 article); Danish (1 article) and Russian (1 article).
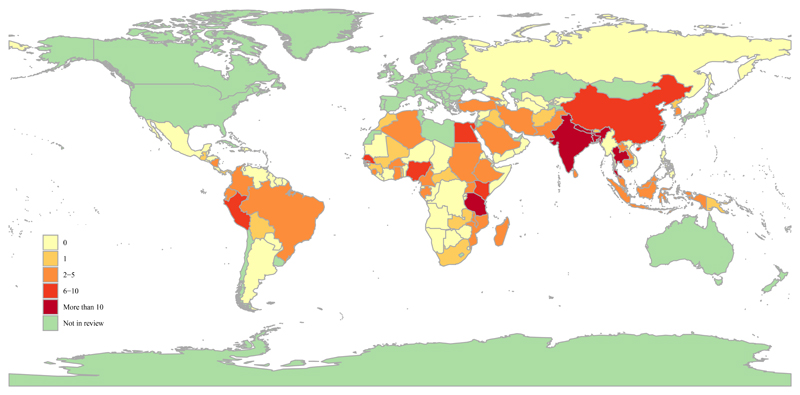
Articles included in the review yielded data from 53 (48·2%) of the 110 malaria endemic countries (figure 2). The majority of articles with a single country origin (n=235) reported data from Africa (83 of 235 articles, 35·3%) or South-East Asia (81 of 235 articles, 34·5%) (appendix table S7, figure S1). One hundred and six (45·1%) of the 235 articles with a single country origin were conducted in one of six dominant countries: India (n=31), United Republic of Tanzania (n=22), Thailand, (n=20), Nepal (n=12), Bangladesh (n=11), and Nigeria (n=10). The data reported in the review were gathered between 1994 and 2017 inclusive.
Figure 2. Map illustrating the malaria-endemic countries included in the study and number of articles contributing data for each country (indicated by colour shading).
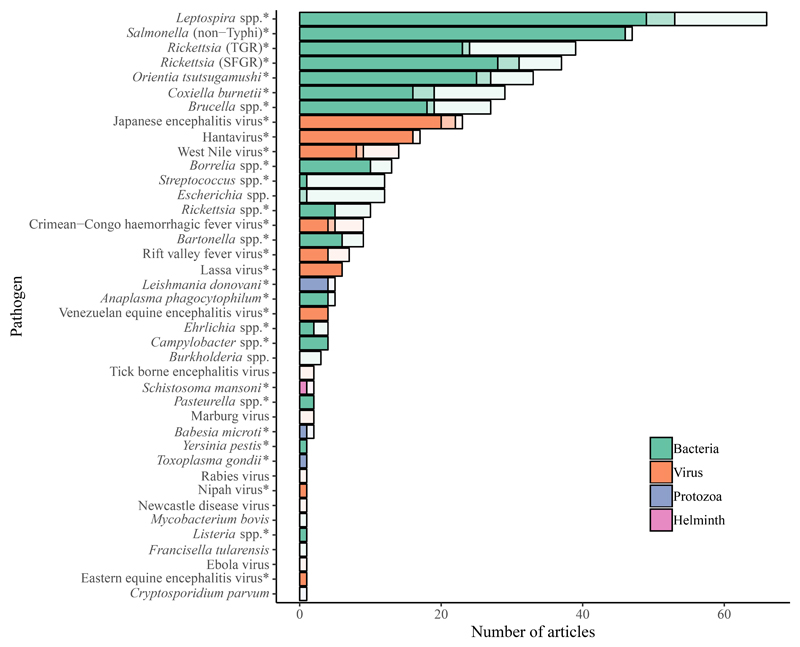
The 244 articles included for data extraction reported looking for and diagnosing 40 and 31 zoonoses, respectively, in these populations (figure 3). The number of included zoonoses was reduced to 30 after the criteria for diagnostic testing approach were applied. The 244 articles yielded data that met diagnostic test criteria for 30 zoonoses that included 17 bacterial pathogens (56·7%), nine viruses (30·0%), three protozoa (10·0%), and one helminth (3·3%). Leptospira spp., nontyphoidal Salmonella serovars (NTS) and rickettsioses were the most frequently reported bacteria, while Japanese encephalitis virus (JEV), Hantavirus, and West Nile virus (WNV) dominated among reported viruses (figures 3, 4).
Figure 3. Barchart showing the number of articles that looked for, reported diagnosis of and contributed data for each of 40, 31 and 30 zoonoses respectively.
These data were tabulated for all zoonoses (n=40) and articles included in the review (n=244). Bar colour indicates pathogen type and shading differentiates studies that i) contribute data meeting study diagnostic criteria (left hand bar sections with darkest shading, n=30 pathogens indicated by *), ii) report diagnosis with approaches that do not meet study diagnostic criteria (central bar sections with lighter shading, n=31 pathogens that comprised the 30 with extracted data and Escherichia coli), iii) report looking for but not diagnosing a zoonosis (right hand bar section with lightest shading, n=40 pathogens, also including Burkholderia spp. Tick borne encephalitis virus, Marburg virus, Rabies virus, Newcastle Disease virus, Mycobacterium bovis, Francisella tularensis, Ebola virus and Cryptosporidium parvum).
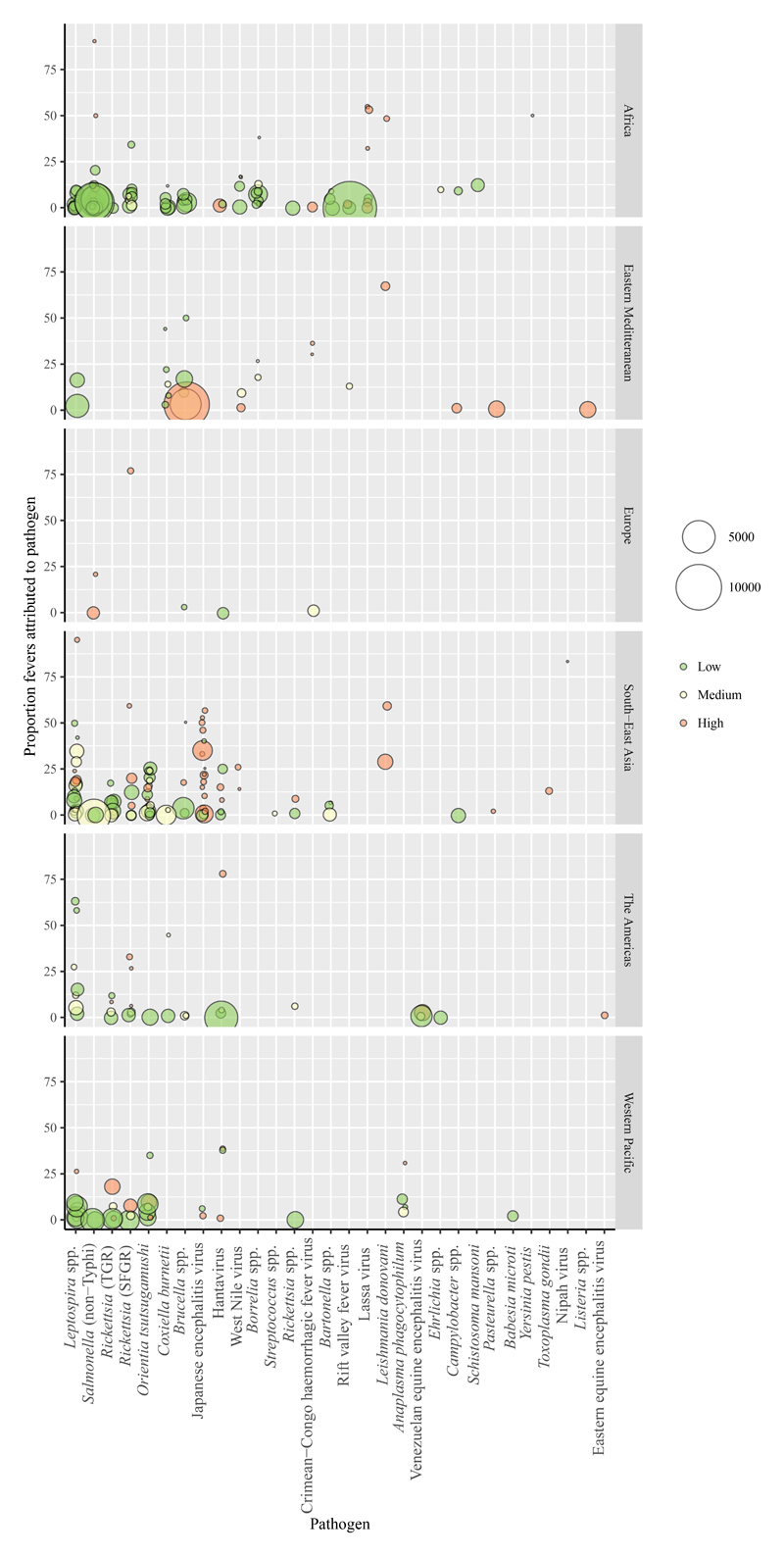
Figure 4. Proportion of fevers attributed to each zoonosis.
The plot includes one data point per study and pathogen combination. The different panels include data from different WHO regions. Point colour indicates the coding for the risk of bias for the representativeness of the febrile population and point size is proportional to the number of individuals tested. Points are jittered on the x axis and shaded to visualize overlapping points.
The number of febrile individuals included in each study population ranged from 4 to 13,845, with a median of 300 (IQR: 120 – 812). In total, 309 records of zoonotic pathogens causing fever were extracted from the 244 articles. The proportion of fevers attributed to each pathogen reported ranged from <1·0% to 95·0% (figure 4). The risk of bias classification in the precision of the proportion of fevers attributed to each zoonosis was 136 (44·0%) of 309 low risk, 79 (25·6%) of 309 medium risk, and 94 (30·4%) of 309 high risk.
Of the 244 studies, 87 (35·7%) described the clinical setting as inpatient, 36 (14·8%) as outpatient, 39 (16·0%) as mixed, and 82 (33·6%) gave no clear classification of the clinical setting. Thirty (12·3%) studies described the study area as urban, 59 (24·2%) as rural, 45 (18·4%) mixed or both, and 110 (45·1%) gave no clear classification of the study area. Eighteen (7·4%) studies included adult participants, 43 (17·6%) included children, 153 (62·7%) included both adults and children and 30 (12·3%) gave no clear classification of the ages included. Of the 244 studies, twelve (4·9%) described a demographically restricted population, 55 (22·5%) reported some exclusions from the population, and 32 (13·1%) mentioned exclusion of malaria-infected individuals specifically (appendix table S6). Of the 244 studies, 73 (29·9%) reported looking for more than one zoonosis, 43 (17·6%) diagnosing more than one zoonosis and 37 (15·2%) contributing data on more than one zoonosis. Of the 244 studies, 10 (4·1%) were described as outbreak investigations and 169 (69·3%) populations were classified as undifferentiated febrile populations. Among the 75 differentiated populations, 36 (48·0%) had specific febrile aetiologies suspected, 17 (22·7%) were classified as febrile neurological, eight (10·7%) as comorbid populations, eight (10·7%) as febrile haemorrhagic, five (6·7%) as febrile gastrointestinal and one (1·3%) as febrile respiratory. The associations between clinical presentation of febrile populations and the subset of 25 pathogens identified in the differentiated populations are shown in figure 5. The risk of bias classification in the representativeness of febrile populations was 121 (49·6%,) of 244 low risk, 45 (18·4%,) of 244 medium risk, and 78 (32·0%,) of 244 high risk.
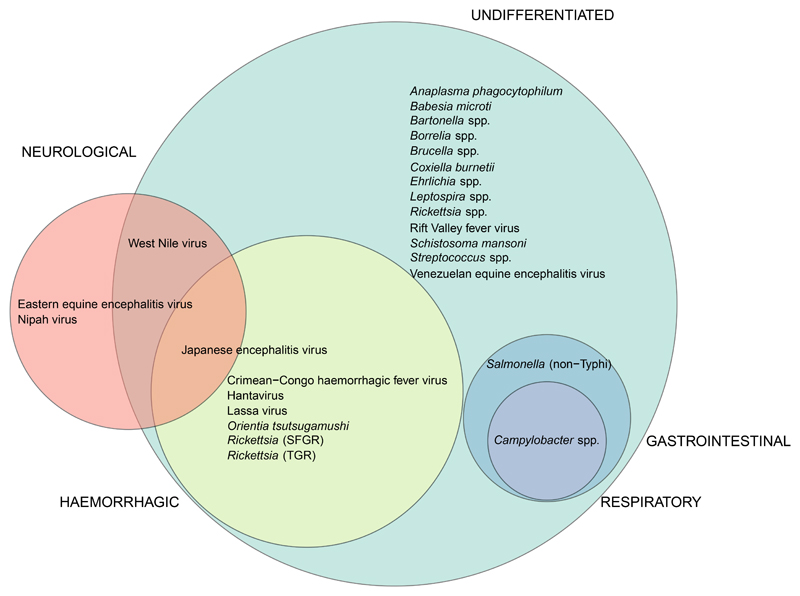
Figure 5. Venn diagram illustrating the associations between febrile population clinical presentation and pathogens identified.
Circles are scaled to the number of pathogens detected in each type of febrile population. Undifferentiated, shown in green, 23 pathogens (including pathogens also seen in other populations); febrile neurological, shown in red, four pathogens; febrile gastrointestinal, shown in blue, two pathogens; febrile respiratory, shown in purple, one pathogen, febrile haemorrhagic, shown in yellow, seven pathogens. Five pathogens are not represented in the figure as they were only detected in febrile populations classified as co-morbid (Listeria spp., Pasteurella spp. and Toxoplasma gondii) or in febrile populations with a specific febrile aetiology suspected (Leishmania donavani, and Yersinia pestis).
Discussion
This systematic review reveals diverse zoonoses causing febrile illness within multiple malaria-endemic countries, often at high prevalence. However, sparse and patchy reporting suggests that the prevalence of zoonoses is widely under-estimated. Knowledge of probable infecting pathogen is crucial to inform clinical management of febrile illness and there is a clear need for further investigation of the zoonotic causes of febrile illness to generate data relevant to clinicians, epidemiologists, and health policy makers globally. This study should generate greater awareness of the clinical importance of zoonoses and provide a pragmatic starting point for actions to better manage these diseases, for example through improved diagnostic and clinical treatment algorithms. These findings demonstrate the need for enhanced epidemiological understanding of multiple zoonoses to inform disease prevention.
This review reveals substantial gaps in the evidence base, including a complete absence of eligible studies from more than half of the 110 countries included in the review (figure 2). There are multiple steps and biases in the processes from a patient seeking care with febrile illness to the publication of an English language scientific paper on the occurrence and prevalence of a specific zoonosis that could be included in this review. The underlying distribution and relative clinical importance of individual pathogens varies, as do patient healthcare seeking behaviour, clinical, and patient awareness of different pathogens, diagnostic capacities, and probability of publication. It is therefore not plausible to expect this review to yield data on all zoonoses in all countries. However, considering the inclusion of 110 countries and construction of searches for 50 pathogens or pathogen groups, the identification of just 244 eligible studies underscores the profound overall shortage of robust quantitative data describing the role of any zoonoses as causes of fever in most malaria-endemic countries.
The geographic variation in the distribution of studies by country (figure 2) and region (appendix table S7, figure S2) is likely to be strongly influenced by variation in research and publication effort. There is noticeable geographic segregation for some zoonoses, with NTS and SFGR reported more frequently in Africa, and Leptospira spp., Orientia tsutsugamushi, and typhus-group rickettsioses (TGR) reported more frequently in South-East Asia and Western Pacific regions (appendix figure S2). For viruses, Lassa virus was reported only in Africa and JEV predominantly in South-East Asia. The distribution of studies cannot be interpreted as an accurate reflection of the underlying distribution of zoonotic pathogens, their prevalence or clinical importance. The pathogens that are looked for depend on factors such as the diagnostic capacity available, existing data, and local assessment of the likely causes of febrile illness in a specific location. Once pathogens are identified in any location there will likely be increased clinical, patient, and community awareness of those pathogens, as well as improved diagnostic capacity to detect them. In this way, dogma about the ‘known’ important causes of febrile illness in specific locations can arise and contribute to the neglect of other pathogens. The findings of this review may help indicate potential gaps in what is looked for and can highlight pathogens and locations where these dogmas should be questioned.
The majority of the 30 zoonotic causes of fever contributing data for this review were bacteria (56·7%). This proportion is greater than expected from the taxonomic distribution of all zoonotic pathogens, which comprise 30·1% bacteria44 and also contrasts with the taxonomic distribution of emerging zoonoses, which are dominated by viruses.13 This finding reinforces the clinical importance of endemic bacterial zoonoses. The comparisons between the number of articles that looked for, diagnosed, and contributed data for each of 40 zoonoses reveals the range of zoonotic pathogens investigated and indicates the relative investigative effort used for each pathogen (figure 3). However, the figures for number of articles where a pathogen was looked for but not identified must be interpreted with caution given the high probability of reporting bias and how rarely negative results are reported. For several pathogens, the number and proportion of articles that reported a zoonotic diagnosis but did not contribute further data for analysis (because the diagnostic approaches described did not meet study quality criteria) are substantial (figure 3). This demonstrates that for many, predominantly bacterial pathogens, suboptimal diagnostic tests or imprecise case definitions are in widespread use, highlighting the challenges of accurately quantifying disease prevalence and comparing studies.
Persistent challenges in the diagnosis of febrile patients include limited laboratory capacity, reliance on demonstration of seroconversion for confirmed diagnosis of many pathogens, unsustainable costs associated with more advanced diagnostic technologies, and lack of simple and affordable tests for the accurate and timely diagnosis of several zoonotic pathogens. In addition, the delays in patient presentation that are typical in many resource limited settings, low magnitude bacteraemia at presentation and, presentation of patients during the immune phase of illness, all limit the sensitivity of culture or PCR-based diagnostic approaches when available. These challenges necessitate syndromic approaches to patient management and broad-spectrum treatment. One specific issue relates to tetracycline use. This study identified rickettsioses and O. tsutsugamushi as common causes of fever. These would benefit from treatment with tetracyclines, which are not currently included in the Integrated Management of Adolescent and Adult Illness (IMAI) algorithms for septic shock and severe respiratory distress without shock.45 In light of the extensive contribution of tetracycline-responsive infections to fever in malaria-endemic countries, revisions to clinical guidelines may be warranted to suggest the empirical use of tetracyclines in addition to beta-lactams in scenarios where the infection with tetracycline-responsive pathogens cannot be excluded.
The findings of this review show that one or more zoonotic causes of fever are likely to present a threat to health in all of the countries included in this review. Only a small proportion of the febrile populations included in the study were defined as demographically restricted and most were not clinically differentiated. Even zoonoses commonly linked with specific syndromes (e.g., Crimean-Congo haemorrhagic fever virus and JEV) were diagnosed in undifferentiated populations and should thus be considered in the differential diagnosis of undifferentiated febrile illness. Within populations at risk, it is important that aetiologic studies are followed by epidemiologic risk factor studies to determine whether certain sub-groups are at higher risk for specific zoonotic diseases. Robust febrile illness surveillance systems help inform local epidemiology and febrile illness management, and are also essential for detection of disease outbreaks.46
There are several important limitations to this study. We examined the contribution of zoonotic pathogens to febrile illness only in malaria-endemic countries and excluded articles not available in English from our analysis. The restriction of this review to English language texts will have reduced the probability that studies from French and Spanish speaking countries were included and may partially account for some gaps, such as the 23 countries in Africa and 15 in the Americas for which no eligible studies were identified. Studies reporting all negative test results were excluded. This strategy was motivated by the inevitable influence of publication bias and challenges of systematically quantifying the non-reporting of either diagnostic test performance or the non-detection of specific pathogens. Biases in testing practices for different pathogens in different locations and with different clinical febrile presentations will influence the pathogens looked for, detected and reported. The application of diagnostic criteria that are strictly comparable across pathogens is not feasible. In this study, strict diagnostic criteria were applied, preferentially including diagnostic approaches with a high specificity, to minimize the influence of false positives within the analyses. The bias assessments for study representativeness and precision in the estimates of proportion of fevers attributable to a given pathogen both reveal that the majority of data points had medium or high risk of one or both types of bias. This emphasizes the need for cautious and essentially non-quantitative interpretation of the data extracted from these studies. Many studies with risk of precision bias due to smaller sample size tended to report the highest prevalences of disease attribution to a given pathogen (figure 5); and, interestingly, these studies were often also classified as high risk for representativeness bias. Figure 5 shows clear variation in risk of representativeness bias across pathogens, potentially linked to variation in clinical presentation. For example, the majority of data points for Japanese encephalitis virus and indeed all data points for Leishmania donovanii are classified as high risk of representativeness bias. This review focused on studies reporting diagnostic investigation of patient populations that were principally defined by fever and populations principally defined by a common aetiological diagnosis were excluded (e.g,. populations defined by presence or suspicion of one or more zoonosis, some of whom were febrile). This review therefore had an inherently low sensitivity for studies describing disease outbreaks. This focus explains, for example, the absence of studies describing the 2014-2016 Ebola West Africa outbreak. The design of this review did not allow explicit investigation of co-infections, either of zoonoses with malaria or of multiple zoonoses. Co-infections are likely to be an important factor underlying both the distribution and prevalence of some zoonotic pathogens, including for example nontyphoidal Salmonella serovars.47 Serological diagnosis of acute infection based on testing of both acute and convalescent phase sera is central to the confirmed diagnosis of multiple pathogens included in the study. As a consequence, individuals who die prior to the collection of convalescent samples are unlikely to contribute data (in the absence of other valid test options) and the proportions of fevers attributable to pathogens with high probability of acute fatality will be under-estimated. Furthermore, no validity criteria regarding the timing of sample collection for acute and convalescent samples were imposed, leading potentially to false negative results (e.g., seroconversion not detected because of premature convalescent sampling). For these reasons, our findings are unlikely to capture the full extent of morbidity and mortality attributable to zoonoses.
The data compiled in this review demonstrate the need to consider multiple zoonoses among the potential causes of febrile illnesses in malaria-endemic countries. Different zoonoses are likely to be important in different settings. Our study provides a starting point for improving awareness of first the zoonoses that are known to contribute to febrile illness in different malaria-endemic regions and second the fever-causing zoonoses with widespread distribution that should be considered in patient evaluation. The demonstration of major data gaps should encourage a more open-minded approach when considering zoonoses as a potential cause of febrile illness. Continued efforts are needed to develop multi-pathogen diagnostics, ideally with formats appropriate for point of care use. To avoid perpetuation of self-fulfilling prophesies that can arise when only pathogens tested for (and detected) are assumed to be present, the development and evaluation of such diagnostics should be informed by data describing the pathogens present in specific settings and also the wider context. Untapped sources of information on the distribution and occurrence of fever-causing zoonoses almost certainly exist, particularly in the animal health sector. One Health efforts to share data and knowledge between animal and human health sectors could help raise clinician awareness of locally relevant zoonoses, inform history taking, and guide diagnostic and management decision making. Control of disease in animal populations and prevention of transmission from animals to humans are likely to be the most effective ways to reduce human disease risk with many zoonoses, necessitating active engagement with populations at risk to develop sustainable disease control interventions. There are substantial challenges to clinicians and epidemiologists in revealing the true impacts of many zoonoses. The enormous global burden of febrile illness and scope for improvements in the diagnosis and treatment of zoonotic pathogens necessitate efforts to overcome these challenges and translate findings into important public health gains.
Supplementary Material
Acknowledgements
This work was supported by US National Institutes of Health-National (NIH) Science Foundation Ecology and Evolution of Infectious Disease program (R01 TW009237) and the UK Biotechnology and Biological Sciences Research Council (BBSRC) (BB/J010367). Additional support was provided by: BBSRC grants BB/L018845/1 (JAC and JEBH) and BB/L018926/1 (SC, and JAC); Medical Research Council (MRC) grant MR/K500847/1 (GAFL); the Leverhulme Royal Society Africa Award AA130131 (JEBH); Wellcome Trust 096400/Z/11/Z (KJA); National Institute of Allergy & Infectious Diseases K23AI116869 (MPR), R01AI121378 (JAC) and Fogarty International Center Global Health Fellowship R25TW009343 (MPR).
Footnotes
Contributors
The author contributions are as follows. Study design: JEBH, KJA, JAC, SC, and MPR. Searches, screening and article review: JEBH, MC, MES, KJA, JB, GAFL, DVH, PH, JAC, SC, and MPR. Data extraction: JEBH and MC. Data analysis: JEBH. Manuscript writing: JEBH, MC, MES, KJA, JAC, SC, and MPR.
Declaration of interests
JEBH reports grants from the Biotechnology and Biological Sciences Research Council, UK, and collaboration with Arbor biosciences outside the submitted work. JAC reports grants from United States National Institutes of Health and Biotechnology and Biological Sciences Research Council, UK. MPR reports grants from United States National Institute for Allergy and Infectious Diseases and contracted research with BioFire Defense, LLC, outside the submitted work. Other authors declare they have no conflicts of interest.
Contributor Information
Jo E B Halliday, Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, United Kingdom.
Manuela Carugati, Division of Infectious Diseases, Duke University Medical Center, Durham, North Carolina, United States of America; Kilimanjaro Christian Medical Centre, Moshi, Tanzania; Division of Infectious Diseases, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan, Italy.
Michael E Snavely, Duke Global Health Institute, Duke University, Durham, North Carolina, United States of America.
Kathryn J Allan, Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, United Kingdom.
Julia Beamesderfer, Perelman School of Medicine, University of Pennsylvania, Philadelphia, Pennsylvania, USA.
Georgia A F Ladbury, Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, United Kingdom.
Deborah V Hoyle, Roslin Institute and Royal (Dick) School of Veterinary Studies, Edinburgh, UK.
Paul Holland, Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, United Kingdom.
John A Crump, Centre for International Health, University of Otago, Dunedin, New Zealand; Division of Infectious Diseases, Duke University Medical Center, Durham, North Carolina, United States of America; Duke Global Health Institute, Duke University, Durham, North Carolina, United States of America; Kilimanjaro Christian Medical University College, Moshi, Tanzania.
Sarah Cleaveland, Boyd Orr Centre for Population and Ecosystem Health, Institute of Biodiversity, Animal Health and Comparative Medicine, University of Glasgow, Glasgow, United Kingdom.
Matthew P Rubach, Division of Infectious Diseases, Duke University Medical Center, Durham, North Carolina, United States of America; Kilimanjaro Christian Medical Centre, Moshi, Tanzania; Duke Global Health Institute, Duke University, Durham, North Carolina, United States of America. Programme in Emerging Infectious Diseases, Duke-National University of Singapore Medical School, Singapore.
References
- 1.Crump JA. Typhoid Fever and the challenge of nonmalaria febrile illness in sub-saharan Africa. Clin Infect Dis. 2012;54:1107–9. doi: 10.1093/cid/cis024. [DOI] [PubMed] [Google Scholar]
- 2.Feikin DR, Olack B, Bigogo GM, et al. The burden of common infectious disease syndromes at the clinic and household level from population-based surveillance in rural and urban Kenya. PLoS One. 2011;6:e16085. doi: 10.1371/journal.pone.0016085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Institute for Health Metrics and Evaluation. Global Health Data Exchange. GBD Results Tool. [Accessed 18 June 2018];2018 http://ghdx.healthdata.org/gbd-results-tool.
- 4.Prasad N, Murdoch DR, Reyburn H, Crump JA. Etiology of severe febrile illness in low- and middle-income countries: A systematic review. PLoS One. 2015;10:e0127962. doi: 10.1371/journal.pone.0127962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crump JA, Ramadhani HO, Morrissey AB, et al. Invasive bacterial and fungal infections among hospitalized HIV-infected and HIV-uninfected adults and adolescents in northern Tanzania. Clin Infect Dis. 2011;52:341–8. doi: 10.1093/cid/ciq103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reyburn H, Mbatia R, Drakeley C, et al. Overdiagnosis of malaria in patients with severe febrile illness in Tanzania: a prospective study. 2004;329:1212. doi: 10.1136/bmj.38251.658229.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chandler CI, Chonya S, Boniface G, Juma K, Reyburn H, Whitty CJ. The importance of context in malaria diagnosis and treatment decisions - a quantitative analysis of observed clinical encounters in Tanzania. Trop Med Int Health. 2008;13:1131–42. doi: 10.1111/j.1365-3156.2008.02118.x. [DOI] [PubMed] [Google Scholar]
- 8.Amexo M, Tolhurst R, Barnish G, Bates I. Malaria misdiagnosis: effects on the poor and vulnerable. Lancet. 2004;364:1896–8. doi: 10.1016/S0140-6736(04)17446-1. [DOI] [PubMed] [Google Scholar]
- 9.Murray CJ, Rosenfeld LC, Lim SS, et al. Global malaria mortality between 1980 and 2010: a systematic analysis. Lancet. 2012;379:413–31. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization. World Malaria Report 2016. [Accessed 1 June 2018];2016 http://www.who.int/malaria/publications/world-malaria-report-2016/report/en/
- 11.D'Acremont Vr, Lengeler C, Mshinda H, Mtasiwa D, Tanner M, Genton B. Time to move from presumptive malaria treatment to laboratory-confirmed diagnosis and treatment in African children with fever. PLoS Med. 2009;6:e252. doi: 10.1371/journal.pmed.0050252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crump JA, Morrissey AB, Nicholson WL, et al. Etiology of severe non-malaria febrile illness in Northern Tanzania: a prospective cohort study. PLoS Negl Trop Dis. 2013;7:e2324. doi: 10.1371/journal.pntd.0002324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor LH, Latham SM, Woolhouse ME. Risk factors for human disease emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:983–9. doi: 10.1098/rstb.2001.0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allan KJ, Biggs HM, Halliday JEB, et al. Epidemiology of leptospirosis in Africa: A systematic review of a neglected zoonosis and a paradigm for 'One Health' in Africa. 2015;9:e0003899. doi: 10.1371/journal.pntd.0003899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vanderburg S, Rubach MP, Halliday JEB, Cleaveland S, Reddy EA, Crump JA. Epidemiology of Coxiella burnetii infection in Africa: a OneHealth systematic review. PLoS Negl Trop Dis. 2014;8:e2787. doi: 10.1371/journal.pntd.0002787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parola P, Paddock CD, Socolovschi C, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2013;26:657–702. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costa F, Hagan JE, Calcagno J, et al. Global morbidity and mortality of leptospirosis: A systematic review. PLoS Negl Trop Dis. 2015;9:e0003898. doi: 10.1371/journal.pntd.0003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torgerson PR, Hagan JE, Costa F, et al. Global Burden of Leptospirosis: Estimated in Terms of Disability Adjusted Life Years. PLoS Negl Trop Dis. 2015;9:e0004122. doi: 10.1371/journal.pntd.0004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maina AN, Farris CM, Odhiambo A, et al. Q Fever, Scrub Typhus, and Rickettsial diseases in children, Kenya, 2011-2012. Emerg Infect Dis. 2016;22:883–6. doi: 10.3201/eid2205.150953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.ILRI. Mapping of poverty and likely zoonoses hotspots. Zoonoses Project 4. Report to Department for International Development, UK. [Accessed 1 June 2018];2012 http://www.dfid.gov.uk/r4d/pdf/outputs/livestock/ZooMapDFIDreport18June2012FINALsm.pdf.
- 21.R Core Team. R: A Language and Environment for Statistical Computing. [Accessed 01 October 2019];2018 http://www.R-project.org.
- 22.WHO. Zoonoses. [Accessed 01 June 2016];2016 http://www.who.int/zoonoses/diseases/en/
- 23.OIE. OIE-Listed diseases, infections and infestations in force in 2016. [Accessed 01 Jun 2016];2016 http://www.oie.int/animal-health-in-the-world/oie-listed-diseases-2016/
- 24.Wolfe ND, Dunavan CP, Diamond J. Origins of major human infectious diseases. 2007;447:279–83. doi: 10.1038/nature05775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.World Health Organization. World Malaria Report 2005. [Accessed 01 June 2018];2005 http://www.who.int/malaria/publications/atoz/9241593199/en/
- 26.Centers for Disease Control and Prevention. Leptospirosis (Leptospira interrogans) 2013 Case Definition. [Accessed 12 June];2013 https://wwwn.cdc.gov/nndss/conditions/leptospirosis/case-definition/2013/
- 27.Centers for Disease Control and Prevention. Hantavirus Pulmonary Syndrome (HPS) Case Definition. [Accessed 12 Jun];1996 https://www.cdc.gov/hantavirus/health-care-workers/hps-case-definition.html.
- 28.Centers for Disease Control and Prevention. Arboviral Diseases, Neroinvasive and Non-neuroinvasive 2015 Case definition. [Accessed 12 June];2015 https://wwwn.cdc.gov/nndss/conditions/west-nile-virus-disease/case-definition/2015/
- 29.Centers for Disease Control and Prevention. Rabies, Human 2011 Case Definition. [Accessed 12 June];2011 https://wwwn.cdc.gov/nndss/conditions/rabies-human/case-definition/2011/
- 30.Broadhurst MJ, Kelly JD, Miller A, et al. ReEBOV Antigen Rapid Test kit for point-of-care and laboratory-based testing for Ebola virus disease: a field validation study. Lancet. 2015;386:867–74. doi: 10.1016/S0140-6736(15)61042-X. [DOI] [PubMed] [Google Scholar]
- 31.Centers for Disease Control and Prevention. Viral Hemorrhagic Fever (VHF) 2011 Case Definition. [Accessed 12 June];2011 https://wwwn.cdc.gov/nndss/conditions/viral-hemorrhagic-fever/case-definition/2011/
- 32.Saijo M, Niikura M, Ikegami T, Kurane I, Kurata T, Morikawa S. Laboratory diagnostic systems for Ebola and Marburg hemorrhagic fevers developed with recombinant proteins. Clin Vaccine Immunol. 2006;13:444–51. doi: 10.1128/CVI.13.4.444-451.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bausch DG, Rollin PE, Demby AH, et al. Diagnosis and clinical virology of Lassa fever as evaluated by enzyme-linked immunosorbent assay, indirect fluorescent-antibody test, and virus isolation. J Clin Microbiol. 2000;38:2670–7. doi: 10.1128/jcm.38.7.2670-2677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Centers for Disease Control and Prevention. Rift Valley Fever (RVF). Diagnosis. [Accessed 12 June];2013 https://www.cdc.gov/vhf/rvf/diagnosis/index.html.
- 35.Solomon T, Thao TT, Lewthwaite P, et al. A cohort study to assess the new WHO Japanese encephalitis surveillance standards. Bull World Health Organ. 2008;86:178–86. doi: 10.2471/BLT.07.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.World Health Organization. WHO regional offices. [Accessed 12 June 2018];2018 http://www.who.int/about/regions/en/
- 37.Southeast Asia Infectious Disease Clinical Research Network. Causes and outcomes of sepsis in southeast Asia: a multinational multicentre cross-sectional study. Lancet Glob Health. 2017;5:e157–e67. doi: 10.1016/S2214-109X(17)30007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.D'Acremont V, Kilowoko M, Kyungu E, et al. Beyond malaria--causes of fever in outpatient Tanzanian children. N Engl J Med. 2014;370:809–17. doi: 10.1056/NEJMoa1214482. [DOI] [PubMed] [Google Scholar]
- 39.Wang TH, Wei KC, Jiang DD, Chiu CH, Chang SC, Wang JD. Unexplained deaths and critical illnesses of suspected infectious cause, Taiwan, 2000-2005. Emerg Infect Dis. 2008;14:1653–5. doi: 10.3201/eid1410.061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chapter 8: Assessing risk of bias in included studies. In: Higgins JPT, Altman DG, JAC S, editors; Higgins JPT, editor. SGCochrane Handbook for Systematic Reviews of Interventions. Cochrane; 2017. [Accessed 1 October 2019]. Version 5.2.0 (updated June 2017), http://handbook-5-1.cochrane.org. [Google Scholar]
- 41.Whiting PF, Rutjes AW, Westwood ME, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 42.Wells GA, Shea B, O’Connell D, et al. The Newcastle-Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses. [Accessed 01 October 2019];2019 http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp.
- 43.Wickham H. ggplot2: Elegant Graphics for Data Analysis. [Accessed 01 October 2019];2016 https://ggplot2.tidyverse.org.
- 44.Cleaveland S, Laurenson MK, Taylor LH. Diseases of humans and their domestic mammals: pathogen characteristics, host range and the risk of emergence. Philos Trans R Soc Lond B Biol Sci. 2001;356:991–9. doi: 10.1098/rstb.2001.0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rubach MP, Maro VP, Bartlett JA, Crump JA. Etiologies of illness among patients meeting integrated management of adolescent and adult illness district clinician manual criteria for severe infections in northern Tanzania: implications for empiric antimicrobial therapy. Am J Trop Med Hyg. 2015;92:454–62. doi: 10.4269/ajtmh.14-0496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keusch GT, Pappaioanou M, Gonzalez MC, Scott KA, Tsai P, editors. Sustaining Global Surveillance and Response to Emerging Zoonotic Diseases. Washington (DC): 2009. [PubMed] [Google Scholar]
- 47.Park SE, Pak GD, Aaby P, et al. The Relationship Between Invasive Nontyphoidal Salmonella Disease, Other Bacterial Bloodstream Infections, and Malaria in Sub-Saharan Africa. Clin Infect Dis. 2016;62(Suppl 1):S23–31. doi: 10.1093/cid/civ893. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.