Abstract
Escherichia coli populations undergo repeated replacement of parental genotypes with fitter variants deep in stationary phase. We isolated one such variant, which emerged after 3 weeks of maintaining an E. coli K-12 population in stationary phase. This variant displayed a small colony phenotype and slow growth and was able to outcompete its ancestor over a narrow time window in stationary phase. The variant also shows tolerance to beta-lactam antibiotics, though not previously exposed to the antibiotic. We show that an RpoC(A494V) mutation confers the slow growth and small colony phenotype on this variant. The ability of this mutation to confer a growth advantage in stationary phase depends on the availability of the stationary-phase sigma factor σS. The RpoC(A494V) mutation upregulates the σS regulon. As shown over 20 years ago, early in prolonged stationary phase, σS attenuation, but not complete loss of activity, confers a fitness advantage. Our study shows that later mutations enhance σS activity, either by mutating the gene for σS directly or via mutations such as RpoC(A494V). The balance between the activities of the housekeeping major sigma factor and σS sets up a trade-off between growth and stress tolerance, which is tuned repeatedly during prolonged stationary phase.
Importance
An important general mechanism of a bacterium’s adaptation to its environment involves adjusting the balance between growing fast and tolerating stresses. One paradigm where this plays out is in prolonged stationary phase: early studies showed that attenuation, but not complete elimination, of the general stress response enables early adaptation of the bacterium E. coli to the conditions established about 10 days into stationary phase. We show here that this balance is not static and that it is tilted back in favor of the general stress response about 2 weeks later. This can be established by direct mutations in the master regulator of the general stress response or by mutations in the core RNA polymerase enzyme itself. These conditions can support the development of antibiotic tolerance although the bacterium is not exposed to the antibiotic. Further exploration of the growth-stress balance over the course of stationary phase will necessarily require a deeper understanding of the events in the extracellular milieu.
Keywords: E. coli, GASP, RNA polymerase, stationary phase, stress response
Under standard laboratory conditions, well-aerated cultures of Escherichia coli exhibit three distinct phases of growth within 24 h of inoculation: the initial slow-growing lag phase, followed by a period of exponential growth and finally the stationary phase during which no appreciable change in cell number is observed. Most of the cells in culture lose viability within 2 to 5 days after reaching stationary phase (1, 2). The small surviving subpopulation can remain viable for at least 60 months in a phase known as “prolonged stationary phase” or “long-term stationary phase” (LTSP) (1, 3, 4). Prolonged stationary phase might represent a laboratory approximation of different natural habitats populated by bacteria, many of which are nutrient limited (2).
Although the total number of viable cells does not appreciably change in this phase, prolonged stationary phase is dynamic and is characterized by repeated replacement of parental genotypes (and phenotypes) by a different set of genetic variants better adapted to the ambient environment (2). Populations from aged cultures outcompete younger cultures in prolonged stationary phase, a phenotype known as growth advantage in stationary phase (GASP) (4). In this phase, both genetic and phenotypic diversity increase over time (2, 5).
Mutations in different global regulators of gene expression confer GASP on their host cells. The most prominent among these are in the sigma factor σS, the master activator of the general stress response. An elongated and attenuated variant of σS, named rpoS819, was the first gene to be implicated in GASP (4). Other alleles conferring GASP include mutations in Lrp, a global regulator controlling amino acid metabolism (6); activation of the bgl operon (7) by mechanisms that attenuate H-NS, the global regulator responsible for transcriptional repression of horizontally acquired genes (8); and mutations in CpdA, the cyclic AMP phosphodiesterase (9), which might influence the hierarchy of carbon source utilization. Several mutations in the core RNA polymerase subunits have been observed in deep-sequencing-based studies of the genotype space explored in prolonged stationary phase (5, 10, 11). Previous research has described the emergence of diverse colony morphologies in prolonged stationary phase (3). Consistent with these observations, we isolated a variant characterized by its small colony size growing alongside cells forming “normal”-size colonies, after maintaining E. coli in stationary phase for over 3 weeks.
Slow-growing mutants of various bacteria can withstand a variety of stresses targeting growth-associated processes. For example, slow-growing E. coli variants deficient in the electron transport chain can tolerate aminoglycosides whose transport into the cells is an active process (12). A certain class of slow-growing bacteria, called “small colony variants” (SCV) emerge in response to stress in the environment (13) and have been discovered in pathogenic contexts. These variants arise from metabolic deficiencies, particularly in thymidine biosynthesis or in the electron transport chain. Some of these variants are known to evade host defense systems and the action of various antibiotics, while forming persister populations responsible for recalcitrant diseases (14).
In this study, we describe the phenotypic characteristics and genetic underpinnings of a slow-growing, small-colony-forming variant of Escherichia coli isolated 3 weeks into stationary phase in nutrient-rich medium (LB) and show that a mutation in the core RNA polymerase can confer GASP on its bearer by activating the σS regulon.
Results
Small colony phenotype evolved from ZK819 in prolonged stationary phase
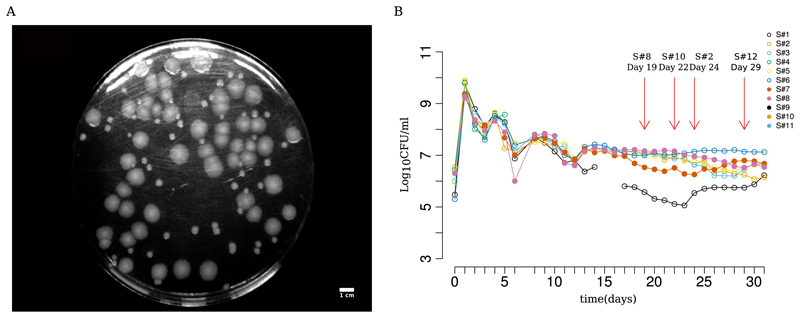
In a previous study (5), we had propagated 4 replicate populations of Escherichia coli ZK819 in rich medium (LB) for 30 days without the addition of external nutrients after inoculation. ZK819 is the pioneer GASP strain of E. coli, carrying rpoS819, an elongated and attenuated variant of the sigma factor for the general stress response σS. We repeated the long-term evolution experiment in LTSP with 11 replicates and followed the cultures daily for 30 days (Fig. 1B). In this experiment, colonies with different morphologies emerged over time: mucoid colonies, followed by fragmented colonies and small colonies. Variants with small colony (SC) phenotype first appeared on day 19 and remained for as long as the cultures were examined (Fig. 1) (5). We isolated an SC from these long-term stationary-phase experiments for further study and describe its characteristics here.
Fig 1.
Evolving a GASP-enabled strain (ZK819) in prolonged stationary phase and the emergence of small colony forms. (A) Representative picture of small colonies as seen on an LB agar plate spread with a 29-day-old culture of ZK819. (B) Daily colony count of 11 independent replicates of E. coli ZK819 for 30 days in prolonged stationary phase. First recorded emergences of small colonies in different replicate lines are marked by red arrows.
We briefly note here that the appearance of the SC phenotype is repeatable, and we observed its appearance, after 19 to 22 days postinoculation, in 4 out of 11 additional prolonged stationary-phase runs (see Table S2 in the supplemental material). These were not studied in greater detail here.
SC exhibits slow growth and increased tolerance to beta-lactam antibiotics compared to ancestor and coexisting strains
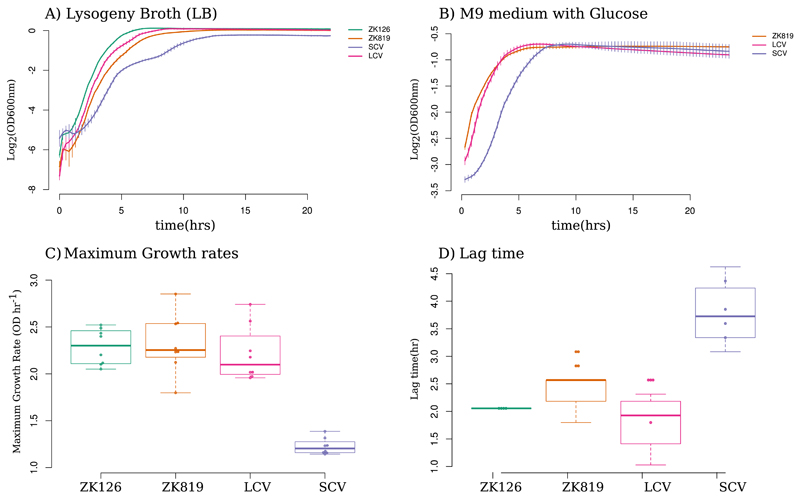
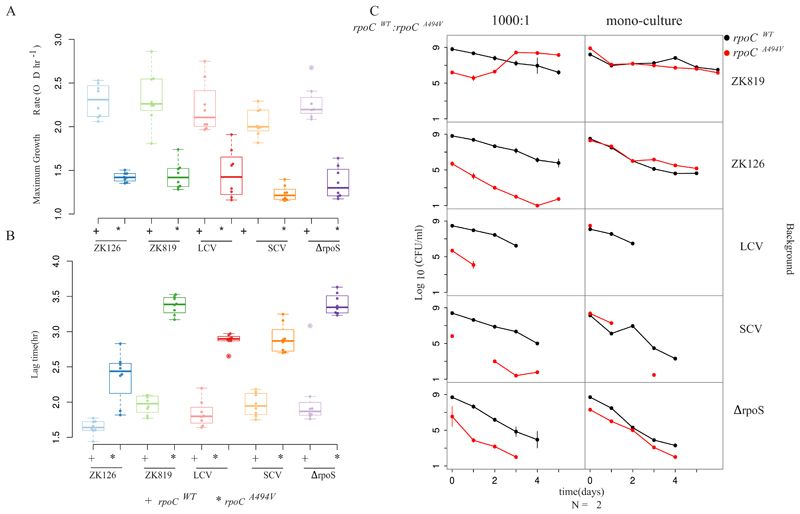
We measured the growth of SC, a coexisting variant with ancestral colony size (LC), ZK819 (the strain which was used to initiate our GASP experiment), and ZK126 (the parent of ZK819, used by the Kolter lab in reference 3) in LB (Fig. 2A). SC exhibited slow growth, in terms of both maximum growth rate (Fig. 2C) and lag times compared to ZK819 and LC (Fig. 2D). All three strains showed similar stationary-phase yields in rich media. In minimal medium supplemented with glucose as sole carbon source, SC retained its slow growth (Fig. 2B). Although spectroscopic measurements can overestimate the length of lag phase, significant differences in both maximum growth rate and length of lag periods between rpoCWT and rpoCA494V strains conclusively prove the slow-growth phenotype of rpoCA494V mutants.
Fig 2.
Growth characteristics of SC. (A and B) Growth curves of SC compared to coexisting strain (LCV), evolutionary ancestor (ZK819), and parent of ancestor (ZK126) in rich (LB) (A) and minimal (M9 plus glucose) (B) media. (C) Distribution of maximum growth rates of SCV, LCV, and ancestor strains. Error bars indicate standard deviations (n = 8). (D) Distribution of lag time for SCV, LCV, and ancestor strains. Error bars indicate standard deviations (n = 8).
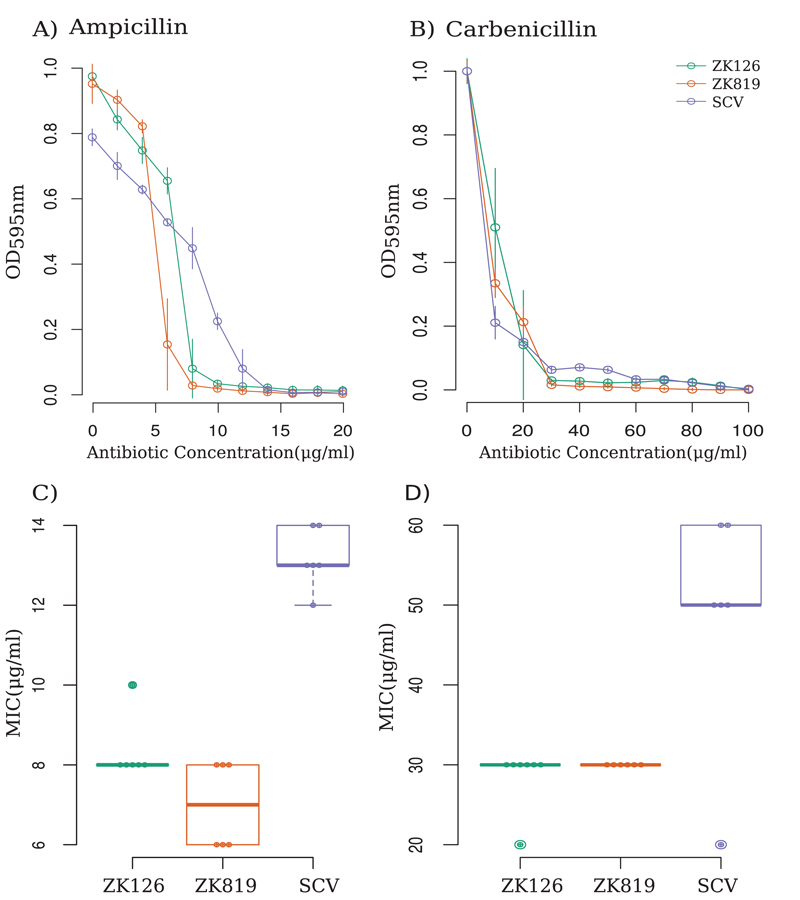
Small colony variant populations isolated in other bacteria—often in pathogenic contexts—show increased tolerance to antibiotics (15, 16). We tested if this would apply to the SC isolated here by comparing the MICs of three different antibiotics—ampicillin, kanamycin, and ciprofloxacin—against SC, ZK126, and ZK819. SC showed an increased resistance to ampicillin and the analogous carboxypenicillin antibiotic carbenicillin but not to the other two antibiotics (Fig. 3 and Fig. S1).
Fig 3.
SC exhibits tolerance against beta-lactam antibiotics. (A and B) Kill dynamics of ampicillin (A) and carbenicillin (B) against SC and ancestor strains. (C and D) Distribution of MICs of evolved mutant (SCV), evolutionary ancestor (ZK819), and parent of evolutionary ancestor (ZK126) strains against ampicillin (C) and carbenicillin (D) (n = 6).
We observed that ampicillin clears more SC than wild type when present in lower concentrations. At higher concentrations of the antibiotic, this trend is reversed. This type of concentration-dependent variable response requires different subpopulations to have variable affinity to the antibiotic and is known as “heteroresistance” (17).
SC shows GASP phenotype in a limited time window in prolonged stationary phase
Growth advantage in stationary phase (GASP) is defined as the ability of a strain to outcompete its ancestor in prolonged stationary phase (4). To examine the ability of SC to show GASP, we competed SC against its ancestor ZK819 using mixed culture competition experiments (4) in fresh LB and in media spent for different periods with the ancestral strain ZK819. In these experiments, the competing strains were inoculated in three different ratios: 1:1,000, 1:1, and 1,000:1.
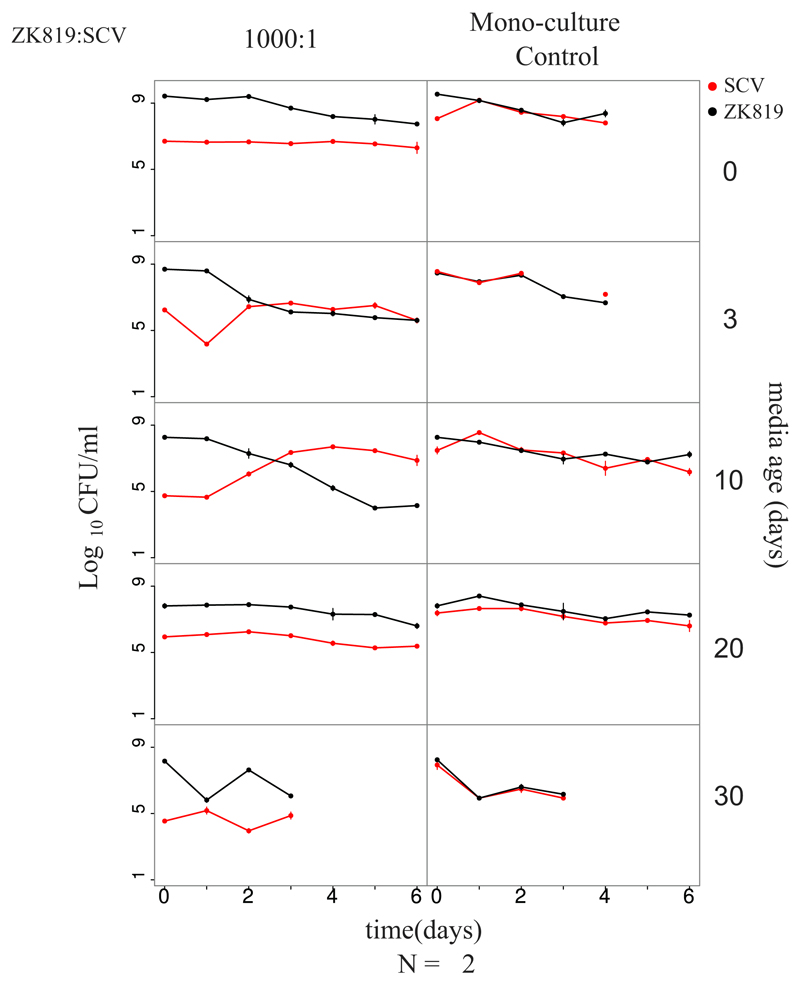
We found that SC does not show any competitive advantage against ZK819 in freshly autoclaved and 3-day-old spent medium. However, it outcompetes ZK819 in 10-day-old medium. Starting from a 1,000-fold minority, SC overtakes ZK819 after 3 days only in the 10-day-old spent medium (Fig. 4). Though it is not unreasonable to expect considerable batch-to-batch variation in spent media across trials, the above result was consistently found across three replicates. In 20-day- and 30-day-old media, however, the competitive advantage of SC was lost, indicating that SC can exhibit GASP only in a narrow time window in prolonged stationary phase (Fig. S2). This is consistent with the idea that the continuous wax and wane of different genetic variants is a reflection of the ever-changing biochemical properties of the prolonged stationary-phase medium (3).
Fig 4.
SC exhibits competitive advantage against its ancestor within a specific time window in prolonged stationary phase. Competition dynamics between ZK819 (ancestor) and SC in differently aged LB. Left panel shows competition experiments where ZK819 is inoculated in 1,000-times-higher frequency than SC. Right panel shows competing strains grown individually in the same-age medium as controls. Age of medium used for competition is shown on right. Competition dynamics with other starting frequencies are shown in Fig. S2.
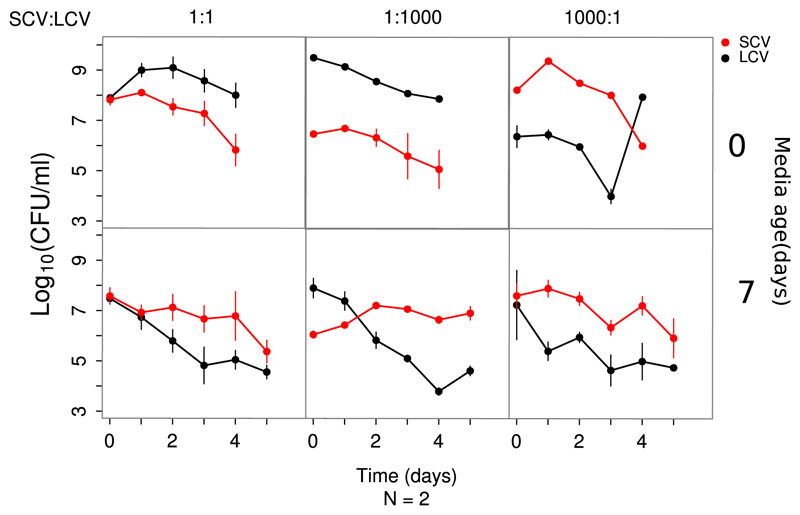
In light of the increasing genetic diversity of the population and the late emergence of SC, it is more reasonable to expect SC to compete with the coexisting LC than with the ancestral ZK819. We therefore investigated the competitive advantage of SC against LC in 10-day-old LB and found that SC, starting from a 1,000-fold minority, overtook LC after 2 days (Fig. 5). Therefore, SC shows the ability to outcompete both ZK819 and LC in prolonged stationary phase under conditions representing a narrow time window.
Fig 5.
Competitive dynamics between SCV and coexisting strain LCV in fresh (day 0) and 7-day-old spent (day 7) media. Population dynamics in all three possible inoculation ratios (indicated on top) of the competing species are shown in horizontal panels. Competitions performed in 0-day- and 7-day-old media are shown in different rows.
rpoCA494V is a unique mutation in SC that causes small colony size, irrespective of background
To identify and characterize the genetic basis of the small colony and the GASP phenotype of SC, we performed whole-genome sequencing of SC, LC, and ZK819. We found several mutations common to SC and LC and identified two loci at which the two diverged (for the complete mutation profile of SCV and LCV, please see Table S3). The first was at rpoS, encoding σS. Whereas ZK819 carried the attenuated rpoS819 allele, SC carried what we refer to as rpoS92. rpoS819 is defined by a 46-bp duplication at the 3′ end of the gene. This duplication results in a longer protein with reduced activity, which may in part be due to its reduced expression at the protein level. rpoS92 carries a reduplication of the 46-bp region. This introduces a stop codon prematurely for rpoS819, resulting in a σS protein variant of length intermediate between the wild type and rpoS819. rpoS92 partially restores the activity of σS that had been lost in rpoS819 (9). LC, in contrast to both SC and ZK819, carries the open reading frame (ORF) of wild-type rpoS.
The second locus at which these three strains diverged was rpoC, coding for the β′ subunit of the core RNA polymerase. Whereas ZK819 and LC carry the wild-type rpoC locus, SC has a G:A transition, resulting in the replacement of the alanine at residue 494 by a valine. Mutations in the core RNA polymerase have been found to confer adaptive advantages on E. coli under different conditions and have been found in at least two whole-genome sequencing studies of variants that emerge in prolonged stationary phase (5, 11). We also identified a unique synonymous mutation (H366H) in LC in the gene icd coding for isocitrate dehydrogenase, which we have not characterized in this study.
To test the phenotypic effect of rpoCA494V mutation, we introduced it in different relevant genetic backgrounds using phage-mediated transduction from a thiC39::Tn10 marker (CAG18500), as described in a previous study (18). The genetic backgrounds in which rpoCA494V was introduced were selected based on their σS status and their relevance to the strains used or isolated in this study; these were LC (evolved strain, rpoSWT), ZK819 (ancestor, rpoS819), ZK126 (rpoSWT, the parent of ZK819), and ΔrpoS (ZK819::ΔrpoS). In addition, the wild-type rpoC allele was transduced into SC, reverting the rpoCA494V mutation to the wild type. The presence of the rpoCA494V mutation was confirmed by Sanger sequencing with relevant primers (Table S1). Single-end whole-genome sequencing was performed on all transductant strains to check for the presence of any additional indels, copy number variations, and point mutations that may have been inadvertently introduced during transduction and found none apart from those introduced.
We observed that the rpoCA494V-bearing transductant in all backgrounds, including ΔrpoS, produced small colonies on LB agar plates and exhibited slow growth, characteristic of SC in liquid culture (Fig. 6A and B). Conversely, replacing the mutant rpoCA494V allele in the SC background with the wild-type rpoC allele ameliorated slow growth, thus reverting the small colony phenotype to ancestral colony size. These results indicate that the rpoCA494V mutation is sufficient to cause the small colony phenotype in its bearer, independent of the status of σS.
Fig 6.
rpoCA494V is necessary and sufficient to cause a slowdown in growth and confers GASP in a background with attenuated σS activity. (A) Comparison of maximum growth rates of strains carrying rpoCWT (marked +) and rpoCA494V (marked *) in different backgrounds. Maximum growth rates were calculated from growth data as described in Materials and Methods. (B) Comparison of lag times of strains carrying rpoCWT (+) and rpoCA494V (*) in different backgrounds. Lag times were calculated from growth curve data as in Materials and Methods. (C) Competition dynamics between strains carrying rpoCWT and rpoCA494V in different backgrounds. Left panel shows scenarios where the rpoCWT strain is inoculated in 1,000-times-higher frequency than the rpoCA494V strain. Right panel shows competing strains grown individually in the same-age medium as controls. Data shown only for competitions performed in 10-day-old spent medium. Other medium conditions and starting frequencies are provided at https://doi.org/10.6084/m9.figshare.12001212.v1.
The rpoCA494V mutant might cause inefficient transcription
The ability of rpoCA494V to slow growth is independent of the status of σS. This being a mutation in the core RNA polymerase, we suspected that it would have a broad impact on global gene expression levels.
To test whether the mutation causes any alteration to transcription, we tested induction kinetics of the chromosomal ara operon in rpoC mutant and wild-type strains in the ZK819 background. Note that the ara operon comes under the control of the housekeeping sigma factor σD and not σS. We measured the induction of the araB gene over time following the addition of arabinose to an exponentially growing culture in fresh LB by performing RT-PCR using primers amplifying a 134-bp region at the 5′ end of the gene (Table S1). To limit any population-level heterogeneity in ara induction (19), we used a nearly 100-fold excess of arabinose for induction.
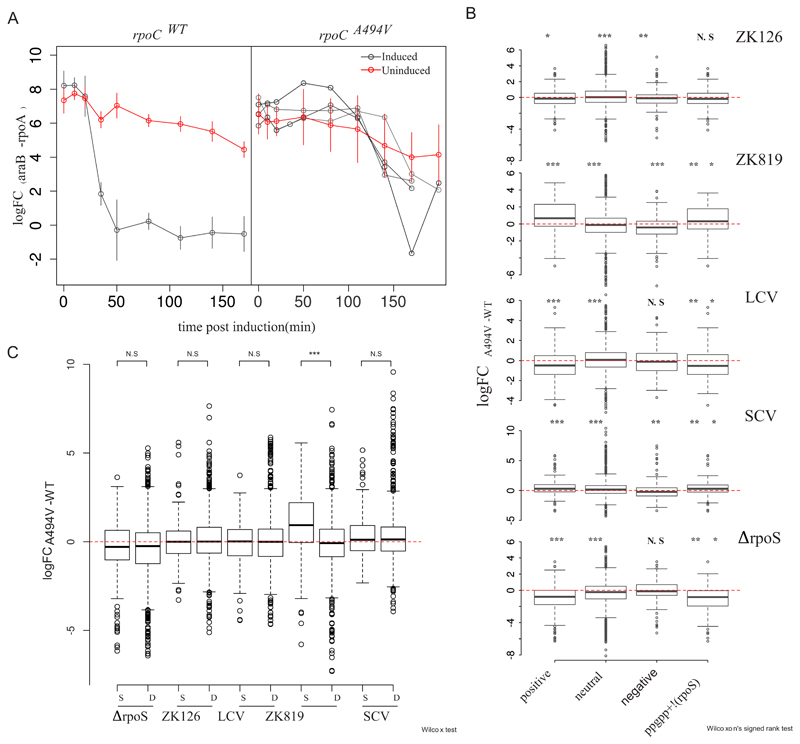
In the rpoCWT mutant, araB levels increase steeply (difference in CT values between mutant and wild-type strains in induced and uninduced conditions [ΔΔFCinduced-uninduced] = -6.05) within 50 min of adding arabinose. In the rpoCA494V mutant, the increase in mRNA is muted (ΔΔFCinduced-uninduced = −0.74 to −4.54), more variable across biological replicates, and delayed, occurring after 110 min postinduction (Fig. 7A). This delayed and repressed response to arabinose induction in the rpoCA494V mutant suggests that the mutation reduces the activity of RNA polymerase (RNAP). This is unlikely to be an effect of a possible reduced arabinose transporter expression in the mutant: transcriptome sequencing (RNA-seq) data (see below) showed no significant difference in the basal expression level of araE between the wild-type and mutant rpoC strains. Thus, slow transcription might underpin the ability of rpoCA494V to cause slow growth.
Fig 7.
rpoCA494V mutation affects global gene expression states. (A) Semiquantitative estimation of araB expression by RT-PCR in rpoCWT (left) and rpoCA494V (right) in the ZK819 background, after induction with 100 mM L-arabinose. Error bars indicate standard deviations (n = 4). Individual replicates have been shown separately for the rpoCA494V strain to emphasize the biological variation observed in the induction pattern in mutant strain compared to wild type. Threshold cycle values from araB mRNA were normalized with that of constitutively expressing rpoA gene. araB measurements from uninduced cultures as controls are shown in red. (B) Differential expression of positive, negative, and neutrally regulated targets of ppGpp between rpoCA494V and rpoCWT in different backgrounds (noted in top right of each figure). To control for the behavior of a large number of genes controlled by both σS and ppGpp, differential expression of genes which are upregulated by ppGpp but are not σS targets was taken into account. (C) Differential expression of σS and σD target genes between rpoCA494V and rpoCWT in different backgrounds. Wilcoxon’s test was used to test significance of the difference in median expression values. P value scale: ***, <0.001; **, <0.01; *, <0.05.
The competitive advantage conferred by the rpoCA494V mutation depends on the status of σS
Having established that the rpoCA494V mutation can cause the small colony phenotype independently of the status of σS, we investigated the ability of this mutation to confer GASP on its bearer. To do this, we performed mixed culture competition experiments of rpoCA494V strains against rpoCWT strains in all relevant genetic backgrounds in both fresh and spent media.
We found that rpoCA494V confers GASP on its bearer only in the ZK819 background, carrying the attenuated rpoS819 allele, and not others (Fig. 6C) (https://doi.org/10.6084/m9.figshare.12001212.v1). In 10-day-old spent medium, ZK819+rpoCA494V overtook ZK819+rpoCWT from a 1,000-fold-lower starting frequency after 2 days. However, SC with rpoS92 and rpoCA494V did not outcompete SC with rpoS92 and rpoCWT. This suggests, under the assumption that these mutations emerged under a regime of natural selection, that the rpoCA494V mutant might have arisen in the rpoS819 background, with rpoS92 emerging later. We also found that in the ZK819ΔrpoS background, both rpoCA494V and rpoCWT exhibited precipitous drops in cell count with no viable colonies remaining after 3 days of growth, highlighting the importance of rpoS in survival and growth under nutrient-depleted/altered conditions (https://doi.org/10.6084/m9.figshare.12001212.v1 [ΔrpoS background]).
These data suggest that rpoCA494V confers GASP only when σS is severely attenuated, as seen in rpoS819. However, its ability to confer GASP does require some level of functional σS, bringing about the suggestion that it acts through σS.
rpoCA494V induces expression of the σS regulon depending on the rpoS genetic background
Toward understanding the mechanisms by which rpoCA494V causes the small colony and the GASP phenotypes, we performed RNA-seq-based transcriptome experiments comparing rpoCA494V and rpoCWT in different genetic backgrounds differing in their σS activities (ZK126, ZK819, SC, LC, and ΔrpoS). We identified differentially expressed genes that showed a log2 fold change of over 1.5 at a P value of less than 0.05.
The number of differentially expressed genes across these five backgrounds ranges from 407 to 654. However, the number of genes differentially expressed irrespective of the genetic background was only 18. This shows that the effect of rpoCA494V on the transcriptome is dependent on the background expression state established at least in part by σS activity.
Because the ability of rpoCA494V to confer GASP—as well as alter gene expression states—seemed to depend on the status of σS, we tested the effect of the mutation on the expression levels of σS targets. Toward this, we compared the fold changes—between rpoCA494V and rpoCWT—of targets of σS with those of a set of targets of the housekeeping sigma factor σD. In a second analysis, we tested for the overlap between σS and σD target genes that are differentially expressed in rpoCA494V over rpoCWT. When we looked at all genes, σS targets showed higher positive fold changes in rpoCA494V than rpoCWT only in the ZK819 background carrying the attenuated and elongated rpoS819 allele (Fig. 7B). Consistent with this, sets of differentially expressed gene sets between rpoCA494V and rpoCWT were significantly enriched for σS targets in the ZK819 (rpoS819) background, but not the other backgrounds (data available at https://doi.org/10.6084/m9.figshare.12005166.v1).
A previous study had identified rpoCA494V as a mutation that enables RNAP to bypass the requirement of the small molecule ppGpp in transcribing a ppGpp-dependent, σ54 promoter (18). Following from this work, we analyzed the expression patterns of genes known to respond to ppGpp (20) in rpoCA494V compared to rpoCWT. We observed that positively regulated targets of ppGpp are upregulated and negatively regulated targets are downregulated in rpoCA494V compared to rpoCWT in the ZK819 and SC backgrounds, which harbor elongated rpoS alleles with less than wild-type σS activity (Fig. 7C). This applies for genes that are known to respond to ppGpp but not known to be regulated by σS as well. While this might indicate that this effect may not be mediated by σS, we observed an opposite pattern in rpoCA494V compared to rpoCWT in the ΔrpoS background (Fig. 7C). We did not observe any significant effect on ppGpp targets in the rpoSWT background.
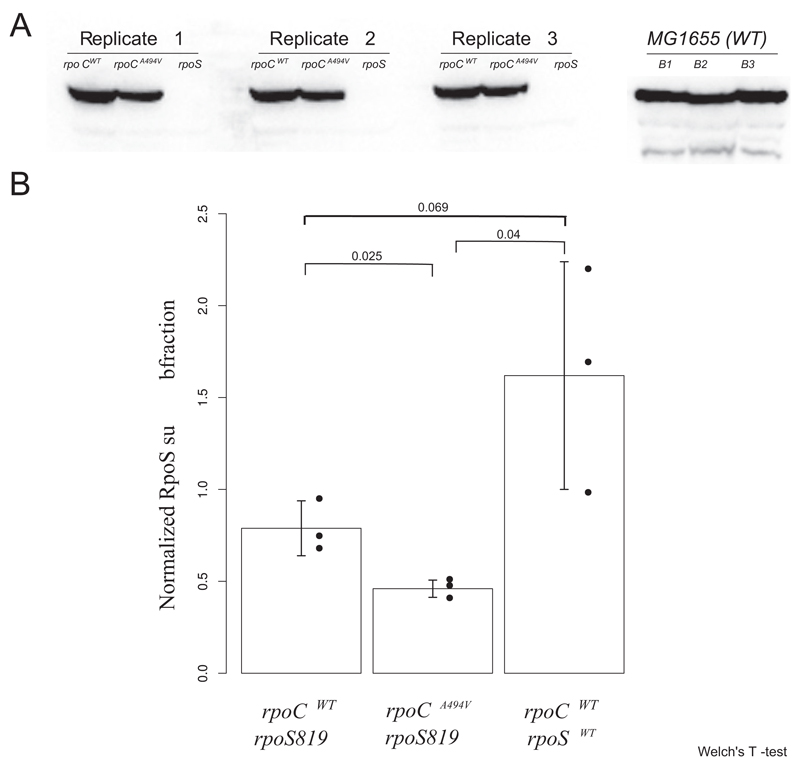
rpoCA494V does not seem to induce the σS regulon by increasing the expression levels of σS, on the basis of our observation that there is little differential expression of the rpoS gene in the RNA-seq data, nor is there any increase in σS protein levels as measured by Western blotting. In fact, Western blots indicate a decrease in the levels of σS in rpoCA494V (Fig. 8). This suggests that rpoCA494V may activate the σS regulon by affecting σ-factor competition for the core RNA polymerase. There is evidence (21) that ppGpp reduces the ability of σD to compete with at least two alternative sigma factors, including σS. Whether the induction of the σS regulon by rpoCA494V operates through this mechanism, and whether this might even require the extended C-terminal tail that rpoS819 and rpoS92 alleles add to the wild-type rpoS, remains to be tested.
Fig 8.
σS protein level in rpoCWT and rpoCA494V strains in ZK819 background. (A) Nitrocellulose blot containing σS protein in rpoCWT and rpoCA494V strains in the rpoS819 background. The ΔrpoS strain, with the deletion introduced into ZK819, is shown as a control. On the right, σS protein derived from the wild-type MG1655 strain containing the wild-type rpoS allele is shown as positive control. A part of the gel was stained with Coomassie blue, and total protein intensities from each lane were quantified, which were then used for normalizing the RpoS protein intensity (Fig. S3). (B) Normalized intensity of σS protein was quantified for each strain, and average values are shown. Error bars represent standard deviations over three replicates. Statistical significance of difference of intensity between each pair of strains was calculated by Welch’s t test, and P values are shown.
In summary, though rpoCA494V increases the expression of the σS regulon in backgrounds with low σS activity, this effect requires the presence and expression of σS. The manifestation of GASP in the regime in which rpoCA494V displays a selective advantage might require higher σS activity, in contrast to earlier time points in which attenuated variants of σS such as rpoS819 are favored. In backgrounds with elongated rpoS alleles (those with extended C-terminal ends, i.e., rpoS819 and rpoS92), the effect of the rpoCA494V allele on the transcriptome shows similarities to that of higher intracellular ppGpp. This effect also seems to be mediated by σS as in the ΔrpoS background; the effect of the rpoCA494V mutation flips and resembles that of lower intracellular ppGpp.
Discussion
This work highlights the global effect of a single nucleotide substitution in the β′ subunit of bacterial RNA polymerase emerging a few weeks into prolonged stationary phase. We show that in this phase, slow growth is advantageous within a certain time window. Our experiments demonstrate that the rpoCA494V mutation is necessary and sufficient to confer slow growth on the bacteria. However, to show the GASP phenotype, an attenuated rpoS819 allele is additionally required.
Genetic and phenotypic diversification of an initially isogenic inoculum is a characteristic of prolonged stationary phase. Although we cannot reject the assumption that some alleles gaining prominence up to detection levels later in long-term stationary phase (LTSP) could be present in the initial inoculum, we know novel mutations arise in prolonged stationary phase based on the ambient environmental conditions and largely under selection (5).
Constantly changing physicochemical conditions of the media result in different subpopulations arising over time. This is orchestrated by different allelic forms of σS that sequentially emerge with time. Within the first 10 days, the elongated and attenuated rpoS819 allele emerges, giving rise to the first GASP population (4). A constant competition for occupying the RNA polymerase holoenzyme exists between the housekeeping sigma factor σD driving growth and the stationary-phase sigma factor σS involved in dormancy and maintenance (22, 23). There is always a trade-off between growth and stress response, which is determined by this competition between the two sigma factors. The expression and activity of σS are tightly regulated at multiple levels, and adjusting this switch helps the bacterial cell modulate global gene expression states according to environmental cues. (24, 25). Additionally, the gene for σS is highly mutable, thus enabling genetic alterations of the growth-stress tolerance trade-off (26).
The attenuated rpoS819 allele alters this growth-stress response trade-off in favor of growth. In our experiments, we found that the rpoS819 allele, when evolved in LB medium, suffers another reduplication at the C-terminal end around 19 to 22 days postinoculation, giving rise to the rpoS92 allele, which has activity closer to that of the wild-type rpoS allele (9). The rpoCA494V mutation also enhances the activity of σS. This suggests that stress response takes some precedence in later stages of stationary phase. GASP outcomes depend on initial medium conditions (27), and this can interplay with the balance between stress tolerance and growth.
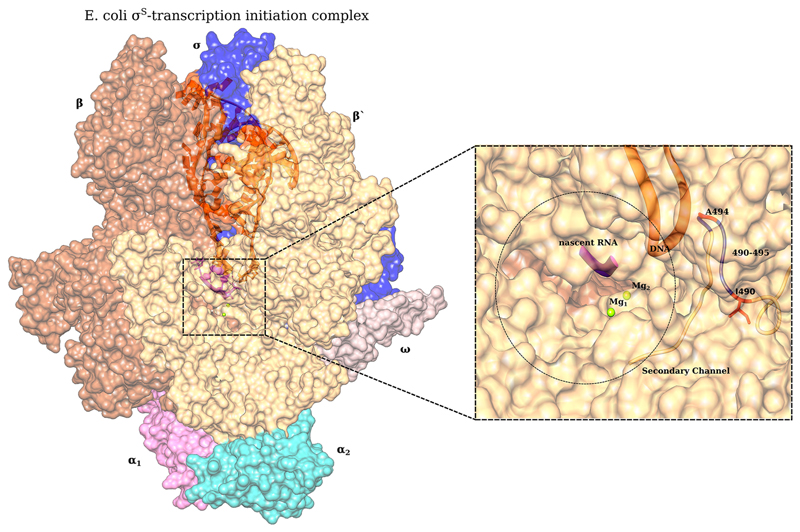
Mutations in the rpoABC operon result in a wide variety of pleiotropic phenotypes, by affecting different steps of the transcription initiation process (28). In a study by Mukhopadhyay et al., mutations mapping near the locus of our mutation of interest on the β′ subunit were found to confer resistance to antibacterial micropeptide microcin J25 (29). This region lies in the secondary channel of RNA polymerase in the shape of a hollow cylinder. Our mutation of interest rpoCA494V could alter the local architecture of the secondary channel, reducing the rate of influx of ribonucleotides to be added to a nascent RNA chain at the catalytic site of RNA polymerase. Other mutations like A490E (30) and Δ490–495 (31) in RpoC alter nucleoside triphosphate (NTP) sensing and uptake and finally to delay stable elongation complex (SEC) formation (Fig. 9). This would lead to slow transcription by the mutated polymerase and explain the slow growth exhibited by rpoCA494V in different rpoS backgrounds. Further, this slow transcription could internally trigger a stress response, initiating the upregulation of σS targets that we observe in ZK819 and SC backgrounds. Bacterial sigma factors are known to interact with the β and β′ subunits of RNA polymerase (32), and recently, it has been shown that σS directly interacts with the β′ subunit of RNA polymerase, diminishing the pore size of the secondary channel (32). This interaction could be affected by different allelic variants of rpoS (rpoS819 and rpoS92).
Fig 9.
Structural representation of E. coli RNA polymerase holoenzyme (PDB 5IPL) along with DNA (orange) and nascent RNA (purple). (Inset) Secondary channel and the locus of interest A494 along with other reported mutations known to cause slow transcription are indicated: A490E (25) and Δ490–495 (26). This figure was generated by Aravind Ravichandran.
In recent literature, there has been a substantial focus on SC of pathogenic origin. Different strains of Staphylococcus aureus (33), Pseudomonas aeruginosa (34), Burkholderia cepacia (35), and E. coli (36), have been described to form small colony variants, some of which are genetically stable. Among the genetically stable variants of SC, most are metabolic mutants, defective either in the electron transport chain or in the thymidine biosynthesis pathway (14). Early studies have found nonpathogenic E. coli to unstably form small colonies under prolonged exposure to chemicals like phenol or anthraquinone (37, 38). Under our experimental framework, we observed that E. coli diversifies into stable, regulatory mutants which phenotypically produce small colonies with enhanced stress tolerance after 3 weeks of incubation in rich spent medium.
In summary, we show that a mutation in the core RNA polymerase provides a competitive advantage to its bearer in deep stationary phase, by altering the balance between growth and stress tolerance.
Materials and Methods
Bacterial strains and culture conditions
Escherichia coli K-12 strain W3110 is the parent strain of Escherichia coli ZK126. The latter, when evolved in prolonged stationary phase for 10 days, gave rise to Escherichia coli ZK819, which has an elongated and attenuated rpoS allele (4) called rpoS819. ZK819 was evolved in prolonged stationary phase for 28 days (5). The small colony form isolated is referred to as SC, and its coexisting ancestral-colony-sized variant is referred to as LC. Strains are described in Table 1.
Table 1. Strains used in this study.
| Strain | Description | Source or reference |
|---|---|---|
| Escherichia coli W3110 | Wild-type strain | CGSC 4474 |
| ZK126 | W3110 Δlac U169 tna2, parent of ZK819 | 5 |
| ZK819 | ZK126 rpoS819; evolutionary ancestor of SCV and LCV | 5 |
| Large colony variant (LCV) | Evolved strain, colony size comparable to wild-type strain, rpoSWT icdH366H | This study |
| Small colony variant (SCV) | Evolved strain, small colony size, rpoS92 rpoCA494V | This study |
Additional long-term stationary-phase experiments were performed to test the repeatability of the appearance of small colony forms. Twelve replicate lines of E. coli were propagated in 500 ml of LB in 1.5-liter Erlenmeyer flasks. They were grown at 37°C with circular agitation at 200 rpm. Every day, 1-ml aliquots were taken from each flask. Five hundred microliters of this sample was used to prepare dilutions in an 0.9% NaCl solution which were then plated on LB plates in two technical replicates. The remaining 500 μl solution was mixed with an equal volume of 50% glycerol and saved at −80°C as stocks (Fig. 1). The plates were incubated at 37°C for 16 to 18 h, and the colonies were counted to track population changes over time and to check for contamination. Images were taken with the help of the Gel-Doc system. Putative small colonies, whenever seen, were restreaked to verify the stability of the phenotype, and a separate count of them was maintained daily.
For measuring growth characteristics of various strains, 5 ml of LB in a 50-ml centrifuge tube (Tarsons) was inoculated with single colonies of respective strains, grown for 12 to 16 h at 37°C, and agitated at 200 rpm to form the starter culture. This culture was diluted 1,000-fold in 30 ml LB contained in 100-ml Erlenmeyer flasks (Pyrex; NC-0400). Cultures were maintained at 37°C, with circular shaking at 200 rpm to maintain aeration. Growth dynamics were tested in rich medium (Luria broth [HiMedia], containing 5 g/liter NaCl), as well as minimal medium (M9) with 0.5% glucose as carbon source. Bacterial growth was measured by tracking change in optical density (OD) at 600 nm.
Calculation of maximum growth rate and lag time
To estimate the maximum growth rate, specific growth rates of each strain were calculated from turbidimetric change for every 15 min over 20 h. Based on the extent of initial noise, a sliding window of 5 to 8 time points in size was taken for each strain, and the median growth rate was ascertained for each window. The element of the window showing maximum median was defined as corresponding to the maximum growth rate for a specific strain.
To calculate time spent by each culture in lag phase, the last point of lag was determined according to the method of Bertrand (39). The intersection of nongrowing lag phase and exponentially growing log phase was ascertained by fitting linear models to growth dynamics. The time obtained from this intersecting point was considered the duration of the lag phase for a specific strain.
Construction of strains with rpoCA494V mutation
Transferring and reverting the point mutation rpoCA494V in different genetic backgrounds were performed using Escherichia coli strain CAG18500 (CGSC; thiC39) (18). In this strain, thiC, a gene 4.6 kb upstream of rpoC, is tagged with Tn10 carrying a tetracycline resistance marker. The cotransduction frequency of rpoC in this strain has been previously reported as 65% (18). P1 transduction, using this strain as donor, was used to first transfer the selection marker to the rpoCA494V-carrying strain. Among transductants having the selection marker, one set carried rpoCA494V and the rest rpoCWT. The former were used as donors for further P1 transduction of the mutation into different genetic backgrounds. The transductants were plated on LB agar having 12.5 mg/ml of tetracycline and were screened for the presence or reversion of the point mutation by Sanger sequencing (see Table S2 in the supplemental material). Marked strains carrying the wild-type rpoC gene were utilized as donor to revert the mutant allele where applicable.
Determination of MICs of different antibiotics against SC and ancestor strains
Stock solutions (100 μg/ml) of relevant antibiotics were prepared, and serial dilutions were made for each antibiotic within a range of 0 to 100 μg/ml in 300 μl of LB. Two hundred microliters of LB with the appropriate antibiotics was dispensed into wells of a 96-well plate, which was then inoculated with overnight-grown cultures of appropriate strains in 1:100 dilution. The plates were incubated for 24 h at 37°C, 200 rpm. The turbidity of the cultures was checked by a spectrophotometer at 600 nm. OD at 600 nm was plotted as a function of antibiotic concentration, and the lowest concentration of the antibiotic which produced no perceptible growth at a threshold of OD 0.05 was taken as the MIC for that antibiotic. The experiment was repeated over 6 independent replicates for each strain.
Additionally, LB-agar plates containing a specific antibiotic were prepared at concentrations ranging from 0 to 100 μg/ml. Overnight-grown cultures were diluted 6- to 8 fold in saline solution and plated on LB plates with different antibiotic concentrations. After incubation at 37°C for 16 to 18 h, the resulting colonies were counted. From each plate, the number of CFU was determined per unit volume and the same was plotted as a function of antibiotic concentrations. The lowest concentration of antibiotic which did not exhibit any colony growth was taken as the MIC of that antibiotic for a specific strain.
Competition experiment in fresh and spent media
Mixed culture competition experiments (4) were carried out to test the fitness advantage of the mutant strain compared to wild-type strains in different backgrounds. Competing strains were tagged with resistance markers for kanamycin or tetracycline in a featureless genomic region between htrC and rpoC. They were then inoculated in 5 ml of LB without antibiotics in 50-ml centrifuge tubes. Competing strains were inoculated either in equal frequencies (500 μl of each in 5 ml LB) or with one of them in 1,000-fold excess (1 ml and 1 μl in 5 ml medium). As a control, they were inoculated in isolation to probe population dynamics under no competition. These centrifuge tubes were then maintained for 5 days under standard growth conditions. Every day, cultures were appropriately diluted in saline and plated on kanamycin (50 μg/ml)- and tetracycline (12.5 μg/ml)-supplemented LB agar plates. Colonies developed after overnight incubation at 37°C were counted.
In addition to freshly autoclaved medium, competition experiments were carried out in medium spent by the ancestor ZK819. Three-, 10-, 20-, and 30-day-old media were used for competitions for all backgrounds. To prepare spent media, 500 ml of LB in a 1-liter flask was inoculated with overnight-grown culture of ZK819 in 1:100 dilution. This flask was incubated at 37°C, with circular shaking at 200 rpm. After appropriate growth, 100 ml of the culture was taken from the flask. Bacterial cells were pelleted by spinning the culture at 4,000 rpm for 15 min. The supernatant was then passed through a 0.45-μm membrane filter (Sartorius) and stored in sterile 500-ml bottles.
Arabinose induction and estimation of araB mRNA by qPCR
Appropriate cultures were incubated overnight in rich medium and were diluted 100-fold in 100 ml of LB. They were allowed to grow until mid-exponential phase. Induction was performed at maximum growth rate stage with L-arabinose (HiMedia) added to a final concentration of 100 mM in the culture. Transcription “Stop” solution was prepared according to the method of Vogel et al. and prechilled in a −30°C bath (40). Postinduction, 500-μl samples were periodically collected and mixed with equal volumes of stop solution in a −30°C bath. After a 30-min incubation, a whole-RNA fraction was isolated from each sample by TRIzol (Ambion)-based extraction.
qPCR was performed to amplify specific regions of araB and rpoA transcripts with relevant primers (Table S1). The One Step TB green PrimeScript RT-PCR kit II (TaKaRa, RR086B) was used for qPCR on the QuantStudio 6 Flex system (Applied Biosystems). Fifty micrograms of total RNA was used for each reaction. araB transcripts were estimated and internally normalized by rpoA transcripts.
Whole-genome and whole-transcriptome sequencing
Whole genomes of SC, LC, and ZK819 were isolated from single colonies in replicates and grown overnight in LB. Whole-genome isolation was performed using the GenElute bacterial DNA isolation kit (Sigma-Aldrich). Paired-end sequencing was done by using the Illumina MiSeq platform; libraries for the same were prepared with the TruSeq DNA Nano library preparation kit (Illumina) as suggested by the manufacturers.
Sequence data were mapped to the reference sequence of Escherichia coli K-12 strain W3110 (GenBank accession no. NC007779.1). The Breseq pipeline (41; https://barricklab.org/twiki/bin/view/Lab/ToolsBacterialGenomeResequencing), which uses Bowtie for read alignment, was used to call single nucleotide polymorphisms (SNPs), indels, and structural variants (33). Mutations present in only SC at 100% frequency and not present in either LC or ZK819 were considered unique to SC.
To verify whether rpoCA494V transductant strains harbored only the said mutation compared to donors, whole genomes of transductants from five different rpoS backgrounds were sequenced. rpoCWT and rpoCA494V strains in ZK126, ZK819, LC, SC, and ΔrpoS backgrounds were sequenced in duplicates in two different stages of growth. Cells were isolated during maximum growth rate, which occurs just before the population enters exponential growth, and during the stationary phase of growth.
To look at the effects of the rpoCA494V mutation on gene expression, whole-RNA fractions were isolated from the cell lysate of wild-type and rpoCA494V transductants in different backgrounds. Whole-RNA isolation was performed after the bacterial cultures entered stationary phase, upon 16 h of growth. RNA isolation was performed using TRIzol reagent (Ambion). The mRNA fraction from whole RNA was enriched using the MICROBExpress kit (Ambion). The purity of whole RNA was checked by assessing the A260/A280 ratio in NanoDrop and further on Bioanalyzer. rRNA contamination was checked on a 1% agarose gel and on Bioanalyzer. Sequencing was carried out on an Illumina HiSeq2500 platform, and library preparation was done using the TruSeq stranded mRNA library preparation kit (Illumina).
RNA-seq data were aligned to the genome of Escherichia coli K-12 strain W3110 (GenBank accession no. NC007779.1) using BWA (42). Read coverage per gene was calculated using SAMtools (v.1.2) and BEDTools (v2.25.0). EdgeR pipeline (Bioconductor) was used to find differentially expressed genes in mutants (rpoCA494V) compared to wild type (rpoCWT) in different backgrounds. Normalization and calling of differentially expressed genes were done according to the method of Chen et al. (43). A P value cutoff of 0.05 was applied to identify differentially expressed genes.
Estimating CTS levels with Western blotting
Intracellular levels of rpoS protein were estimated between rpoCWT and rpoCA494V strains in the ZK819 background. Whole protein was isolated from respective cultures grown overnight by trichloroacetic acid (TCA) extraction. One hundred micrograms of whole-cell lysate from each strain was run on a NuPage 4 to 12% Bis-Tris protein gel (Thermo Scientific) at 120 V for 3 h. rpoS protein was probed by anti-mouse IgG anti-E. coli RNA σS clone-1RS1 antibody (BioLegend; batch B274469). The protein gel was cut at the 50-kDa ladder mark. One part was used to probe the protein of interest by transferring to a nitrocellulose membrane, and the rest was stained with Coomassie blue and imaged under UV fluorescence. Horseradish peroxidase (HRP)-conjugated IgG (Sigma) was used to develop the blot. HRP signal was detected using SuperSignal West Dura extended-duration substrate (Thermo Scientific). RpoS signal from each lane was normalized by the total protein for that lane.
Supplementary Material
Acknowledgments
We thank Awadhesh Pandit and Tejali Naik from Genomics Facility, NCBS, for helping with next-generation sequencing. We thank Vineeth Vengayil for help with Western blotting experiments and Farhan Ali for help with growth rate and lag-phase analysis codes. We acknowledge Aravind Ravichandran and Ranabir Das for their inputs in understanding the structure of RNA polymerase. We are grateful to Victoria Shingler for her valuable technical inputs for transferring the rpoCA494V mutation to different genetic backgrounds and pointing toward the relevant donor strain. We thank The Coli Genetic Stock Center (CGSC) for supplying us strain CAG18500.
This work was supported by a DBT grant (BT/PR3695/BRB/10/979/201) and a Wellcome Trust-DBT India Alliance Intermediate fellowship (IA/I/16/2/502711), and P.N. is supported by an NCBS graduate program scholarship. S.C. is supported by DST-SERB Young Scientist grant SB/YS/LS-148/2014. We acknowledge support from the Department of Atomic Energy, Government of India, under project no. 12-R&D-TFR-5.04-0800.
Footnotes
References
- 1.Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- 2.Zinser ER, Kolter R. Escherichia coli evolution during stationary phase. Res Microbiol. 2004;155:328–336. doi: 10.1016/j.resmic.2004.01.014. [DOI] [PubMed] [Google Scholar]
- 3.Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci U S A. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zambrano M, Siegele D, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 5.Chib S, Ali F, Seshasayee A. Genomewide mutational diversity in Escherichia coli population evolving in prolonged stationary phase. mSphere. 2017;2:e00059–17. doi: 10.1128/mSphere.00059-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zinser ER, Kolter R. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J Bacteriol. 2000;182:4361–4365. doi: 10.1128/jb.182.15.4361-4365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madan R, Kolter R, Mahadevan S. Mutations that activate the silent bgl operon of Escherichia coli confer a growth advantage in stationary phase. J Bacteriol. 2005;187:7912–7917. doi: 10.1128/JB.187.23.7912-7917.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chib S, Mahadevan S. Involvement of the global regulator H-NS in the survival of Escherichia coli in stationary phase. J Bacteriol. 2012;194:5285–5293. doi: 10.1128/JB.00840-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chib S, Seshasayee AS. Modulation of rpoS fitness by loss of cpdA activity during stationary-phase in Escherichia coli. bioRxiv. 2018 doi: 10.1101/460451. [DOI] [Google Scholar]
- 10.LaCroix RA, Sandberg TE, O’Brien EJ, Utrilla J, Ebrahim A, Guzman GI, Szubin R, Palsson BO, Feist AM. Use of Adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl Environ Microbiol. 2015;81:17–30. doi: 10.1128/AEM.02246-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avrani S, Bolotin E, Katz S, Hershberg R. Rapid genetic adaptation during the first four months of survival under resource exhaustion. Mol Biol Evol. 2017;34:1758–1769. doi: 10.1093/molbev/msx118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lázár V, Pal Singh G, Spohn R, Nagy I, Horváth B, Hrtyan M, Busa-Fekete R, Bogos B, Méhi O, Csörgȍ B, Pósfai G, et al. Bacterial evolution of antibiotic hypersensitivity. Mol Syst Biol. 2013;9:700. doi: 10.1038/msb.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johns BE, Purdy KJ, Tucker NP, Maddocks SE. Phenotypic and genotypic characteristics of small colony variants and their role in chronic infection. Microbiol Insights. 2015;8:15–23. doi: 10.4137/MBI.S25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 15.Suwantarat N, Rubin M, Bryan L, Tekle T, Boyle MP, Carroll KC, Jennings MT. Frequency of small-colony variants and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus in cystic fibrosis patients. Diagn Microbiol Infect Dis. 2018;90:296–299. doi: 10.1016/j.diagmicrobio.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 16.Singh R, Ray P, Das A, Sharma M. Role of persisters and small-colony variants in antibiotic resistance of planktonic and biofilm-associated Staphylococcus aureus: an in vitro study. J Med Microbiol. 2009;58:1067–1073. doi: 10.1099/jmm.0.009720-0. [DOI] [PubMed] [Google Scholar]
- 17.Andersson DI, Nicoloff H, Hjort K. Mechanisms and clinical relevance of bacterial heteroresistance. Nat Rev Microbiol. 2019;17:479–496. doi: 10.1038/s41579-019-0218-1. [DOI] [PubMed] [Google Scholar]
- 18.Szalewska-Palasz A, Johansson LUM, Bernardo LMD, Skärfstad E, Stec E, Brännström K, Shingler V. Properties of RNA polymerase bypass mutants: implications for the role of ppGpp and its co-factor DksA in controlling transcription dependent on σ54. J Biol Chem. 2007;282:18046–18056. doi: 10.1074/jbc.M610181200. [DOI] [PubMed] [Google Scholar]
- 19.Siegele DA, Hu JC. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci U S A. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Traxler MF, Summers SM, Nguyen H-T, Zacharia VM, Hightower GA, Smith JT, Conway T. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jishage M, Kvint K, Shingler V, Nyström T. Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cho B-K, Kim D, Knight EM, Zengler K, Palsson BO. Genome-scale reconstruction of the sigma factor network in Escherichia coli: topology and functional states. BMC Biol. 2014;12:4. doi: 10.1186/1741-7007-12-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramnaresh Gupta K, Chatterji D. Sigma factor competition in Escherichia Coli: kinetic and thermodynamic perspectives. In: de Bruijn FJ, editor. Stress and environmental regulation of gene expression and adaptation in bacteria. John Wiley & Sons, Inc; Hoboken, NJ: 2016. pp. 196–202. [Google Scholar]
- 24.Sharma UK, Chatterji D. Transcriptional switching in Escherichia coli during stress and starvation by modulation of σ 70 activity. FEMS Microbiol Rev. 2010;34:646–657. doi: 10.1111/j.1574-6976.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- 25.Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- 26.Ferenci T. What is driving the acquisition of mutS and rpoS polymorphisms in Escherichia coli? Trends Microbiol. 2003;11:457–461. doi: 10.1016/j.tim.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 27.Farrell MJ, Finkel SE. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol. 2003;185:7044–7052. doi: 10.1128/jb.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinh V, Langelier M-F, Archambault J, Coulombe B. Structural perspective on mutations affecting the function of multisubunit RNA polymerases. Microbiol Mol Biol Rev. 2006;70:12–36. doi: 10.1128/MMBR.70.1.12-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mukhopadhyay J, Sineva E, Knight J, Levy RM, Ebright RH. Antibacterial peptide microcin J25 inhibits transcription by binding within and obstructing the RNA polymerase secondary channel. Mol Cell. 2004;14:739–751. doi: 10.1016/j.molcel.2004.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trautinger BW, Lloyd RG. Modulation of DNA repair by mutations flanking the DNA channel through RNA polymerase. EMBO J. 2002;21:6944–6953. doi: 10.1093/emboj/cdf654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bartlett MS, Gaal T, Ross W, Gourse RL. RNA polymerase mutants that destabilize RNA polymerase-promoter complexes alter NTP-sensing by rrn P1 promoters. J Mol Biol. 1998;279:331–345. doi: 10.1006/jmbi.1998.1779. [DOI] [PubMed] [Google Scholar]
- 32.Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase σ factor activity: a structural perspective. Curr Opin Microbiol. 2008;11:121–127. doi: 10.1016/j.mib.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulger RJ. A methicillin-resistant strain of Staphylococcus aureus: clinical and laboratory experience. Ann Intern Med. 1967;67:81–89. doi: 10.7326/0003-4819-67-1-81. [DOI] [PubMed] [Google Scholar]
- 34.Bryan LE, Kwan S. Aminoglycoside-resistant mutants of Pseudomonas aeruginosa deficient in cytochrome d, nitrite reductase, and aerobic transport. Antimicrob Agents Chemother. 1981;19:958–964. doi: 10.1128/aac.19.6.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Häussler S, Lehmann C, Breselge C, Rohde M, Classen M, Tümmler B, Vandamme P, Steinmetz I. Fatal outcome of lung transplantation in cystic fibrosis patients due to small-colony variants of the Burkholderia cepacia complex. Eur J Clin Microbiol Infect Dis. 2003;22:249–253. doi: 10.1007/s10096-003-0901-y. [DOI] [PubMed] [Google Scholar]
- 36.Roggenkamp A, Sing A, Hornef M, Brunner U, Autenrieth IB, Heesemann J. Chronic prosthetic hip infection caused by a small-colony variant of Escherichia coli. J Clin Microbiol. 1998;36:2530–2534. doi: 10.1128/JCM.36.9.2530-2534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Colwell CA. Small colony variants of Escherichia coli. J Bacteriol. 1946;52:417–422. doi: 10.1128/JB.52.4.417-422.1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Clowes RC, Rowley D. Genetic studies on small-colony variants of Escherichia coli K-12. J Gen Microbiol. 1955;13:461–473. doi: 10.1099/00221287-13-3-461. [DOI] [PubMed] [Google Scholar]
- 39.Bertrand RL. Lag phase is a dynamic, organized, adaptive, and evolvable period that prepares bacteria for cell division. J Bacteriol. 2019;201:e00697–18. doi: 10.1128/JB.00697-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel U, Sørensen M, Pedersen S, Jensen KF, Kilstrup M. Decreasing transcription elongation rate in Escherichia coli exposed to amino acid starvation. Mol Microbiol. 1992;6:2191–2200. doi: 10.1111/j.1365-2958.1992.tb01393.x. [DOI] [PubMed] [Google Scholar]
- 41.Deatherage DE, Barrick JE. Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol Biol. 2014;1151:165–188. doi: 10.1007/978-1-4939-0554-6_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Lun ATL, Smyth GK. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res. 2016;5:1438. doi: 10.12688/f1000research.8987.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.