To the Editor:
Atopic dermatitis (AD) is an itchy, inflammatory skin condition associated with multiple co-morbidities. Observational epidemiology suggests an increased prevalence of obesity in patients with AD, but it remains unclear (a) if there is a causal effect and (b) whether obesity leads to AD, or vice versa. Genetic predisposition to obesity has been shown to promote psoriasis,1 but dermatological disorders can also lead to reduced participation in physical activity, resulting in weight gain. We aimed to investigate evidence of causality in the association of AD with elevated body mass index (BMI).
We meta-analysed 33 published studies examining the association between obesity or elevated BMI and AD to summarise available observational data (Figure E1 in the online repository). The odds ratio (OR) for AD in overweight individuals was 1.05 (95% CI 0.94 to 1.19) in adults (n=51,008) and 1.08 (95% CI 1.00 to 1.16) in children (n=506,202) (Figure E2). For obese individuals the OR for having AD was 1.19 (95% CI 0.95 to 1.49) in adults (n=1,400,679) and 1.20 (95% CI 1.11 to 1.30) in children (n=796,514) (Figure E3); methods and results are in the Online Repository. We extended the observational analysis using two large population-based studies from the UK and Norway2, 3 (details in Online Repository). Among overweight individuals (BMI 25-30 kg/m2) the OR of AD was 1.02 per 1 kg/m2 higher BMI (95% CI 1.00 to 1.04; P=0.07; 4,820 cases; 130,776 controls) and a similar estimate was found among obese individuals (BMI >30 kg/m2), OR=1.02 (95% CI 1.01 to 1.03; P=3.3x10-4; 2,741 cases and 73,907 controls) (Figure E4).
Observational epidemiology has several limitations, including bias from confounding and reverse causation; this restricts its utility for causal inference. However, causality and the direction of effect can be investigated by Mendelian Randomization (MR). MR uses genetic variants as a proxy for the exposure (e.g. BMI) to estimate the effect upon an outcome (e.g. AD). Genetic variants are randomly allocated at fertilization, therefore avoiding confounding; they are not affected by outcomes later in life, therefore avoiding reverse causation. Genome-wide association studies (GWAS) have identified single-nucleotide polymorphisms (SNPs) associated with BMI (up to 941 loci4, 5) and AD (24 loci in European populations6). These SNPs can be combined into a genetic risk score (GRS) or ‘genetic instrument’ which acts as a proxy for the specified trait during MR.
We conducted MR analysis using data from the largest population-based studies in the UK (UK Biobank2) and Norway (Nord-Trøndelag Health Study, Norway3 (HUNT, 2006-08)) along with the largest published GWAS studies for BMI4, 5 and AD6 to date, representing a total of 742,611 individuals. One-sample MR was performed in the UK Biobank and HUNT datasets with the individuals’ BMI SNPs, measured BMI and AD status. Two-sample MR using published GWAS data4–6 was performed and meta-analysed with the one-sample estimate to obtain an overall causal estimate. Similarly, reverse MR was conducted to investigate the effect of AD genetic risk on BMI. Methodological details and sensitivity analysis are described in the Online Repository.
The BMI GRS was strongly associated with BMI in both UK Biobank and HUNT (Figures E6-E7 in this article’s Online Repository) supporting its use as a genetic instrument. Potential confounders of the GRS-BMI association were detected (Figures E6-E7) but the magnitudes were minimal in comparison to the strength of association with BMI. Similarly, the AD GRS was a good predictor of AD in both UK Biobank (OR=1.26; 95% CI 1.23 to 1.28, F-statistic=2036, R2=0.7%) and HUNT (OR=1.15; 95% CI 1.11 to 1.21, F-statistic=97, R2=0.4%) datasets, despite lacking a FLG null genotype (R501X/rs61816761), known to show strong association with AD.
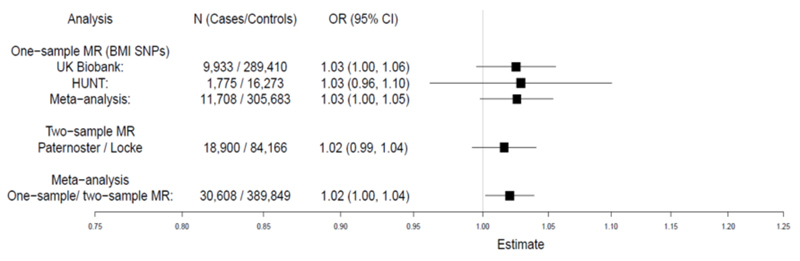
Meta-analysed one- and two-sample MR show evidence of a small causal effect of higher BMI increasing the risk of AD, OR=1.02 (95% CI 1.00 to 1.04; P=0.03) (Figure 1). This represents an increase in AD risk by ~ 2% for each 1kg/m2 increase in BMI, which is remarkably similar to the observational estimate (Figures E2 and E3 in the Online Repository). Importantly, sensitivity analyses showed little evidence of pleiotropy (detailed in the Online Repository, Figure E10 and Table E6) or heterogeneity among the individual SNP effect estimates (UK Biobank Q=101.07, P=0.32; HUNT Q=100.04, P=0.37). Two-sample MR using the larger number of recently published BMI SNP estimates (941 SNPs)4 gave similar evidence of a causal effect upon AD risk (OR=1.08; 95% CI 1.01 to 1.16; P=0.02).
Figure 1. MR analysis of the causal effect of BMI upon AD.
Meta-analysis of one-sample and two-sample MR estimates using individual BMI SNPs as instruments. Estimates are given per 1 kg/m2 increase in BMI. CI, confidence interval; MR, Mendelian Randomization.
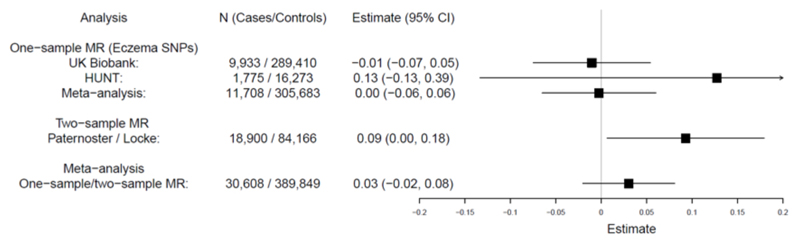
In the reverse direction, meta-analysis gave weak evidence of a very small causal effect (Figure 2): 0.03kg/m2 change in BMI per doubling odds of AD (95% CI -0.02 to 0.08, P=0.24). There was little evidence of pleiotropy but modest heterogeneity among the individual SNP effects (Figure E12 and Table E7 in the Online Repository). The difference in BMI between AD cases and controls estimated in one-sample MR (0.15 kg/m2, 95% CI -1.97 to 2.27) was small compared to observational estimates (Tables E1-E2) indicating that the association is mainly explained by the causal effect of BMI upon AD.
Figure 2. Reverse direction MR analysis: effect of AD genetic risk upon BMI.
Meta-analysis of one-sample and two-sample MR estimates using individual AD SNPs as instrumental variables. Estimates represent change in BMI (kg/m2) per doubling odds of AD.
The association of obesity with cardiometabolic disease and systemic inflammation is now well recognised and clinical guidelines recommend screening patients with psoriasis for obesity. The presence of a causal effect and the direction of effect are both clinically relevant, to define a primary target (i.e. obesity) for intervention. Our MR analysis shows evidence that higher BMI increases the risk of AD: a 2% increase in disease risk for each 1kg/m2 increase in BMI. Conversely, there was no strong evidence of a causal effect of AD genetic risk upon BMI; the estimate of 0.03kg/m2 suggests that genetic risk for AD has little meaningful influence on an individual’s BMI. These findings may be compared to the causal effect of BMI on psoriasis and lack of effect of psoriasis genetic risk on BMI.1 The effect of BMI on AD is more modest than the effect size observed in psoriasis but the high prevalence of obesity (over one-third of US adults) and AD (up to 10% of adults) demonstrate the potential importance of this causal effect on a population scale.
The molecular mechanisms by which obesity contributes to skin inflammation remain unclear. Excess adipose tissue secretes pro-inflammatory cytokines and hormones7 and atopic inflammation may be promoted by disruption of the epidermal barrier in obese individuals.8 Changes in the adipocytes and lymphatic vessels may also contribute to obesity-related skin inflammation.8 Research to define mechanisms underlying the causal relationship demonstrated by MR may identify novel therapeutic targets.
Our MR analyses have various strengths as well as weaknesses. The large sample size is powerful and the genetic instruments are strong. The two-sample analysis included an overlap of data sources (from the HUNT study), which has the potential to bias the causal estimate, but this bias would be in the direction of the null. There is the possibility of misclassification of AD and since AD often shows remission in childhood this phenotype may be particularly susceptible to recall bias in adult studies, however this would likely drive any estimate towards the null. It is also important to note that the MR methodology applied here does not define temporal relationship. Genetic risk has a lifetime effect and therefore pre-dates disease onset, but a causal effect determined by MR does not rely on obesity occurring before the onset of AD. However, we attempted to mitigate this issue by repeating the MR analyses using SNPs that are strongly associated with childhood BMI9 and a causal estimate with the same direction of effect was obtained (OR=1.04; 95% CI 1.01 to 1.07; P=0.01). Nevertheless, replication of these analyses within paediatric cohorts with large sample sizes would be valuable future work.
In conclusion, we have found evidence of a small but potentially important causal effect of BMI upon AD. Clinical trials have shown that interventions to promote weight loss can lead to improvement in psoriasis, but this approach has not been tested in AD. The results of our study provide support for the investigation of obesity management strategies and/or targeting of the adipocyte-keratinocyte cross-talk as therapeutic opportunities for AD. This may contribute to the prevention of AD, as well as a reduction in the population prevalence of this chronic disease.
Supplementary Material
Capsule summary.
Mendelian Randomization shows a small causal effect of higher BMI on AD risk, but no effect of AD on BMI. Further work is needed to determine whether obesity reduction could contribute to AD management.
Acknowledgements
This research has been conducted using data from the UK Biobank Resource (application number 10074) and the Nord-Trøndelag Health Study (the HUNT Study). Details of patient and public involvement in the UK Biobank are available online (http://www.ukbiobank.ac.uk/about-biobank-uk/ and https://www.ukbiobank.ac.uk/wp-content/uploads/2011/07/Summary-EGF-consultation.pdf?phpMyAdmin=trmKQlYdjjnQIgJ%2CfAzikMhEnx6). No patients were specifically involved in setting the research question or the outcome measures, nor were they involved in developing plans for recruitment, design, or implementation of this study. No patients were asked to advise on interpretation or writing up of results. There are no specific plans to disseminate the results of the research to study participants, but the UK Biobank and the HUNT Study disseminate key findings from projects on their websites. The HUNT Study is a collaboration between HUNT Research Centre (Faculty of Medicine and Health Sciences, NTNU, Norwegian University of Science and Technology), Nord-Trøndelag County Council, Central Norway Regional Health Authority, and the Norwegian Institute of Public Health. We acknowledge the permission of the EAGLE consortium (including 23andMe) to use results from their previous GWAS of atopic dermatitis (Paternoster et al., 2015).
Funding
AB-A is funded by a grant awarded to LP by the British Skin Foundation (8010 Innovative Project). AB-A, LP, SW & GDS work in a research unit funded by the UK Medical Research Council (MC_UU_00011/1). BB, ML, LGF & BOA work in a research unit funded by Stiftelsen Kristian Gerhard Jebsen; Faculty of Medicine and Health Sciences, NTNU; The Liaison Committee for education, research and innovation in Central Norway; and the Joint Research Committee between St. Olavs hospital and the Faculty of Medicine and Health Sciences, NTNU. JT is supported by an Academy of Medical Sciences (AMS) Springboard award, which is supported by the AMS, the Wellcome Trust, GCRF, the Government Department of Business, Energy and Industrial strategy, the British Heart Foundation and Diabetes UK. EHM was supported by a research grant from the Liaison Committee for education, research and innovation in Central Norway. GAV is supported by a research grant from the Norwegian Research Council, grant number 250335. ML is supported by research grants from the Liaison Committee for education, research and innovation in Central Norway. JBN was supported by grants from the Danish Heart Foundation and the Lundbeck Foundation. LP is supported by an Academy of Medical Sciences Springboard Award, which is supported by the Wellcome Trust, The Government Department for Business, Energy and Industrial Strategy, the Global Challenges Research Fund and the British Heart Foundation [SBF003\1094]. SJB holds a Wellcome Trust Senior Research Fellowship in Clinical Science (106865/Z/15/Z) and a British Skin Foundation large project grant. The genotyping in HUNT was financed by the National Institutes of Health (NIH); University of Michigan; The Research Council of Norway; The Liaison Committee for education, research and innovation in Central Norway; and the Joint Research Committee between St. Olavs hospital and the Faculty of Medicine and Health Sciences, NTNU. The funders had no influence on study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Contributors
LP, BOA and SJB conceived the study concept and managed the project. AB-A and JT performed statistical analysis with the UK Biobank Resource. BB performed statistical analysis with the HUNT dataset, whose genotypes were quality controlled and prepared primarily by LGF. SW conducted the literature review with support from AB-A, BB, SJB and LP. TP contributed towards the statistical analysis performed. AB-A, SW, SJB, LP, and BB drafted the manuscript. All authors were involved in the interpretation of the data and contributed to and approved the final version of the manuscript.
Competing interests
LP has received personal fees from Merck for Scientific Input Engagement related to MR methodology.
Data sharing
The UK Biobank dataset used to conduct the research in this paper is available via application directly to the UK Biobank. Applications are assessed for meeting the required criteria for access, including legal and ethics standards. More information regarding data access can be found here: http://www.ukbiobank.ac.uk/scientists-3/. Data from the HUNT Study used in research projects will when reasonably requested by others be made available upon request to the HUNT Data Access Committee (hunt@medisin.ntnu.no). The HUNT data access information (available here: http://www.ntnu.edu/hunt/data) describes in detail the policy regarding data availability.
References
- 1.Budu-Aggrey A, Brumpton B, Tyrrell J, Watkins S, Modalsli EH, Celis-Morales C, et al. Evidence of a causal relationship between body mass index and psoriasis: A mendelian randomization study. PLoS Med. 2019;16:e1002739. doi: 10.1371/journal.pmed.1002739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krokstad S, Langhammer A, Hveem K, Holmen TL, Midthjell K, Stene TR, et al. Cohort Profile: the HUNT Study, Norway. Int J Epidemiol. 2013;42:968–77. doi: 10.1093/ije/dys095. [DOI] [PubMed] [Google Scholar]
- 4.Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, et al. Meta-analysis of genome-wide association studies for height and body mass index in approximately 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–9. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Paternoster L, Standl M, Waage J, Baurecht H, Hotze M, Strachan DP, et al. Multi-ancestry genome-wide association study of 21,000 cases and 95,000 controls identifies new risk loci for atopic dermatitis. Nat Genet. 2015;47:1449–56. doi: 10.1038/ng.3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakamizo S, Honda T, Kabashima K. Obesity and inflammatory skin diseases. Trends in Immunotherapy. 2017;1:67–74. [Google Scholar]
- 8.Yosipovitch G, DeVore A, Dawn A. Obesity and the skin: skin physiology and skin manifestations of obesity. J Am Acad Dermatol. 2007;56:901–16. doi: 10.1016/j.jaad.2006.12.004. quiz 17-20. [DOI] [PubMed] [Google Scholar]
- 9.Felix JF, Bradfield JP, Monnereau C, van der Valk RJ, Stergiakouli E, Chesi A. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet. 2016;25:389–403. doi: 10.1093/hmg/ddv472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.