Abstract
Background:
Liver regeneration following partial hepatectomy (PHx) is a complicated process involving multiple organs and several types of signaling networks. The bile acid-activated metabolic pathways occupy an auxiliary yet important chapter in the entire biochemical story. PHx is characterized by rapid but transient bile acid overload in the liver, which constitutes the first wave of proliferative signaling in the remnant hepatocytes. Bile acids trigger hepatocyte proliferation through activation of several nuclear receptors. Following biliary passage into the intestines, enterocytes reabsorb the bile acids, which results in the activation of farnesoid X receptor (FXR), the consequent excretion of fibroblast growth factor (FGF)19/FGF15, and its release into the enterohepatic circulation. FGF19/FGF15 subsequently binds to its cognate receptor, fibroblast growth factor receptor 4 (FGFR4) complexed with β-klotho, on the hepatocyte membrane, which initiates the second wave of proliferative signaling. Because some bile acids are toxic, the remnant hepatocytes must resolve the potentially detrimental state of bile acid excess. Therefore, the hepatocytes orchestrate a bile acid detoxification and elimination response as a protective mechanism in concurrence with the proliferative signaling. The response in part results in the excretion of (biotransformed) bile acids into the canalicular system, causing the bile acids to end up in the intestines.
Relevance for patients:
Recently, FXR agonists have been shown to promote regeneration via the gut-liver axis. This type of pharmacological intervention may prove beneficial for patients with hepatobiliary tumors undergoing PHx. In light of these developments, the review provides an in-depth account of the pathways that underlie post-PHx liver regeneration in the context of bile acid homeostasis in the liver and the gut-liver signaling axis.
Keywords: surgery, mitotic signaling, hepatocyte proliferation, arnesoid X receptor enteral, fibroblast growth factor, detoxification, transport and canalicular excretion
1. Introduction
The liver strictly maintains its size at a predefined setpoint in order to optimally fulfill its detoxification-, synthesis-, immunological-, and endocrinological functions [1]. Under pathological conditions, the host is not only able to sense a loss of viable liver tissue, but also to mount a regenerative response so as to rapidly restore original liver size and function. Factors that control the liver-to-body weight ratio, or the ‘hepatostat,’ are only partially understood [2].
The prototypical stimulus for liver regeneration is the surgical removal of part of the organ (partial hepatectomy, PHx), as is routinely performed by surgeons most often in case of hepatobiliary malignancies [3]. Rodent PHx models have been extensively employed to study the mechanisms that underlie post-PHx liver regeneration. Owing to these models we now know that, immediately after PHx, the activation of early response transcription factors [4,5] and mitogen-activated protein kinases (MAPKs) [6] leads to hepatocyte proliferation and compensatory liver regrowth. These growth signals are activated directly after PHx by hemodynamic changes [7], inflammation [8–10], and cell damage [11–14]. Proliferation is perpetuated until the liver size reaches a mass that complies with the hepatostat, at which point liver growth is terminated [15,16].
The regenerative capacity of the liver after PHx is not inexhaustible. As a reduced number of hepatocytes have to uphold all metabolic functions whilst the liver reclaims its original size, there is a risk of developing liver failure if the liver remnant is too small or too frail [17], a condition which is often referred to as small-for-size syndrome [ 18 ]. Several sensors involved in hemodynamic changes [7], inflammation [8–10], cell damage [11–14], and bile acid metabolism [19,20] are embedded to foster successful liver regeneration and prevent liver failure [21]. Sensors that are involved in bile acid metabolism are especially important in proregenerative signaling through the gut-liver axis [22,25]. Bile acid receptors such as farnesoid X receptor (FXR) promote liver regeneration and prevent liver failure by (i) modulating the bile acid pool size, composition, and compartmentalization [21,26], (ii) governing the production of mitogens such as fibroblast growth factor 15/19 (FGF15/19, signifying rodent/human orthologues) [22,23], and (iii) rewiring mitochondrial metabolism to fuel liver growth [27]. In addition to coordinating liver regeneration after PHx, bile acids are also able to override the hepatostat and consequently expand liver size to larger than normal without concurrent mitogenic triggers [28,29], underscoring their prolific role in compensatory liver regrowth.
As (semi-)synthetic selective agonists of bile acid- and other nuclear receptors have become available [30–33], the metabolic components of liver regeneration could be exploited to pharmacologically enhance liver growth, which in turn could benefit numerous medical scenarios. This review therefore summarizes the molecular pathways that lie at the basis of post-hepatectomy liver regeneration in the context of bile acid homeostasis and the gut-liver signaling axis.
2. General mechanisms of liver regeneration following partial hepatectomy
PHx-induced liver regeneration involves all cell types in the liver, including hepatocytes, Kupffer cells, stellate cells, biliary epithelial cells, and endothelial cells [34]. The time lapse of proliferation is distinct for every cell type. In the rat liver, hepatocytes begin DNA synthesis at 12 hours after PHx, which peaks at 24 hours. The onset of hepatocellular DNA synthesis is initiated in the vicinity of the portal veins and subsequently spreads towards the central veins [35]. The peak of DNA synthesis in non-parenchymal cells is later. Kupffer cells start to proliferate at 48 hours, and biliary epithelial cells and endothelial cells at 96 hours after PHx. The most profound increase in liver mass in rats occurs during the first 3 days after PHx, and restoration of the remnant liver to its original liver mass is completed within 7–14 days [24,36,37]. In humans, recovery of pre-operative liver function takes place within the first 10 days after PHx [38], but complete regrowth of the remnant human liver occurs 3–6 months after PHx [39]. Although all cell types contribute to the increase in liver mass after PHx, this review will primarily focus on the cell cycle progression and proliferation of hepatocytes.
Liver regeneration entails the activation of multiple regulatory pathways that include cytokine-, growth factor-, and metabolic networks [40]. More specifically, PHx induces differential regulation of genes that coordinate cell cycle regulation, chromatin reorganization, transcriptional regulation, signal transduction, protein targeting, metabolism, transport, xenobiotic metabolism, surface receptors, inflammation, and acute phase responses [41]. These pathways are well- coordinated to allow proper restoration of the tissue while maintaining vital liver functions. A global overview is provided in the next subsections. For more detailed information, interested readers are referred to specialized publications [36,37,42,43].
2.1. Initial triggers of liver regeneration: changes in hepatic hemodynamics, sterile and non-sterile inflammation, and a shift in intracellular redox state
The first physiological change during PHx is the redirection of portal and arterial blood supply to the remnant liver instead of the entire liver [44]. As a result, hepatocytes become exposed to a 3-fold greater amount of proregenerative factors [44], mainly supplied by the portal vein (Figure 1). The post-PHx hemodynamic heterogeneity [45–47] and portal hypertension [25] facilitate platelet-endothelial cell interactions during both stasis and flow [48,49]. Moreover, the fenestrations between the sinusoidal endothelial cells widen after PHx [50], allowing more facile passage of blood-borne signaling molecules but also platelets into the space of Disse [51,52].
Figure 1.
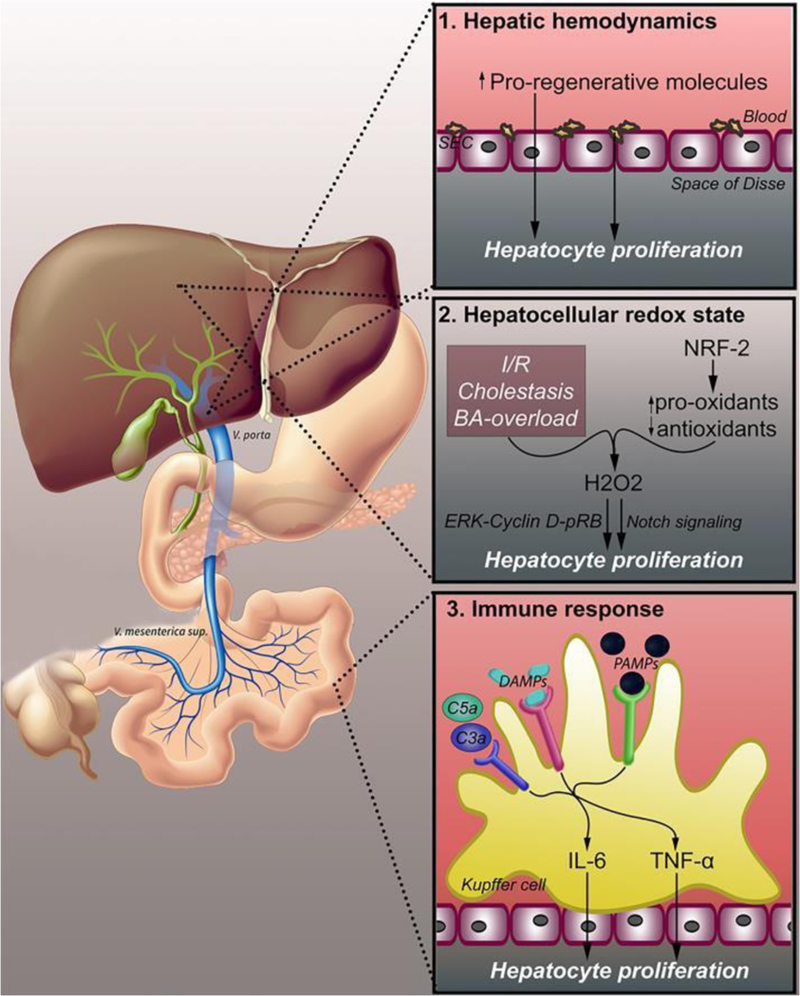
Changes in hepatic hemodynamics that lead to liver regeneration. Three physiological changes following PHx trigger liver regeneration. Altered hepatic hemodynamics (1) lead to increased hepatic exposure to pro-regenerative factors originating from the portal circulation. Additionally, platelets accumulate in the space of Disse and release pro- regenerative molecules. The hepatocellular redox state (2) shifts to a pro- oxidative state due to ischemia/reperfusion, cholestasis, and bile acid overload. NRF-2 and other redox-active enzymes upregulate pro-oxidant enzymes and downregulates antioxidant enzymes, leading to increased levels of H2O2, which promotes cell proliferation through both ERK-cyclin D1-p-RB and Notch signaling. PHx induces hepatocyte proliferation through an immune response (3), resulting from endotoxemia, intestine- derived PAMPs, and damaged cells leaking DAMPs. PAMPs and DAMPs bind PRRs on Kupffer cells, triggering the release of cytokines such as TNF-α and IL-6. Complement factors C3a and C5a are also triggered by the immune response and activate TNF-α and IL-6 release through complement receptors. Abbreviations: SEC, sinusoidal endothelial cell; BA, bile acid; NRF-2, nuclear factor (erythroid-derived 2)-like 2; H2O2, hydrogen peroxide; ERK, extracellular signal-regulated kinase; pRB, phosphorylated retinoblastoma protein; DAMPs, damage-associated molecular patterns; PRR, pattern recognition receptor; PAMPs, pathogen- associated molecular patterns; IL-6, interleukin 6; TNF-α, tumor necrosis factor alpha.
Secondly, the surgical trauma after PHx causes damaged and dying cells to leak their intracellular content [11,12], which contains damage-associated molecular patterns (DAMPs), into the extracellular compartment. The DAMPs bind pattern recognition receptors (PRR) such as Toll-like receptors (TLRs) on Kupffer cells and trigger a sterile immune response [13,14], characterized by the release of tumor necrosis factor-α (TNF-α) and interleukin-6 (IL-6) from Kupffer cells [8], as illustrated in Figure 1 and Figure 2. These cytokines trigger proliferative signaling in hepatocytes and subsequent liver regeneration [36,53–55 ]. PHx-induced injury also triggers the complement peptides C3a (mice and humans) and C5a (mice) complement activation [8–10], which bind to complement receptors on Kupffer cells and neutrophils and amplify the sterile immune response [8–10]. The consequent immune cell activation, cytokine production, and release of proregenerative factors contribute to liver regeneration through various cascades [2,36,56–59], as highlighted in sections 2.2 and 2.3.
Figure 2.
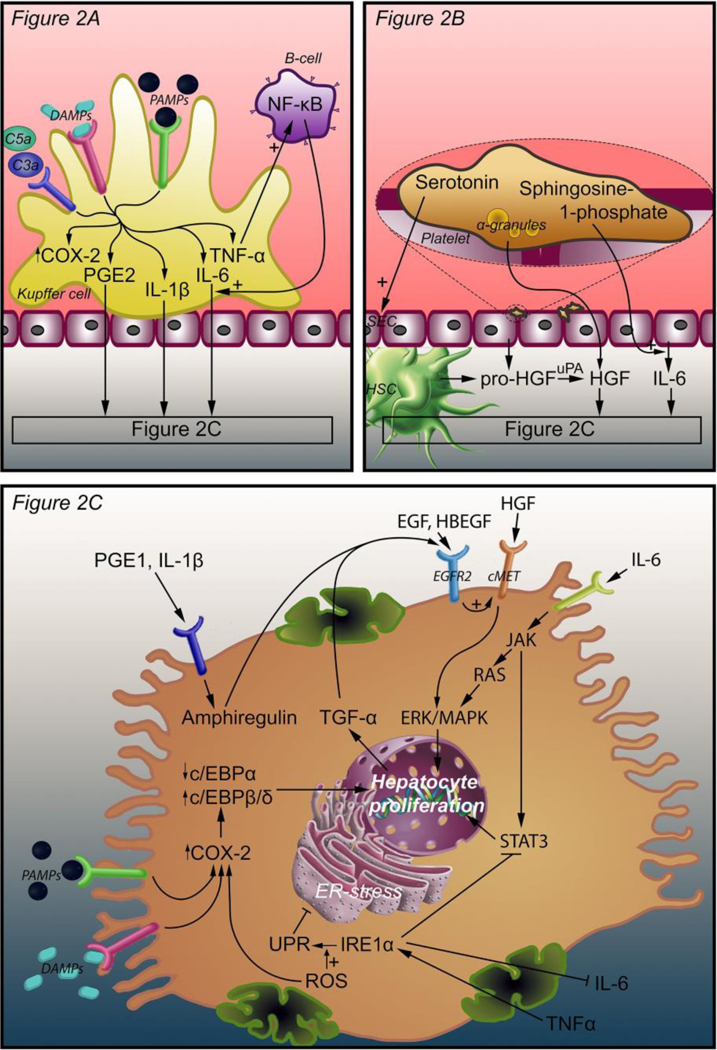
Intercellular and intracellular signals that initiate liver regeneration. Three mechanisms lead to hepatocyte proliferation. In response to complement factors C3a, C5a, PAMPs, and DAMPs, Kupffer cells release TNF-α, IL-6, PGE2, and IL-1β. TNF-α induces an autocrine loop through NF-κB production by B-cells. IL-6, PGE2, and IL-1β bind their cognate receptors on hepatocytes (A). Platelets accumulate in the space of Disse and release an armament of growth factors, including HGF, from their α-granules. Pro-HGF is released from SECs and HSCs and is converted to HGF by uPA, which is activated as a result of ECM damage after PHx. Platelets also release serotonin, which stimulates SECs, and sphringosine-1-phosphate to stimulate IL-6 release (B). PGE2 and IL-1β coming from Kupffer cells stimulate amphiregulin production in hepatocytes. HGF binds its receptor c-Met and activates hepatocyte proliferation through ERK-1/2 MAPK. IL-6 binds the IL-6 receptor and activates JAK, which induces hepatocyte proliferation through STAT3 and through the RAS-ERK-1/2 MAPK pathway. EGF, TGF-α, HBEGF, and amphiregulin enhance the effect of HGF through EGFR2. EGF is produced in the duodenum and reaches the liver through the portal circulation. TGF-α is produced by proliferating hepatocytes. HBEGF is released from monocytes and macrophages and converted to its active form by metalloproteinases. TNF-α (from Kupffer cells) and ROS enhance the UPR that is mediated by IRE1α. The UPR inhibits ER stress that in turn activates COX-2 expression. COX-2 is also stimulated by ROS, PAMPs, and DAMPs. Enhanced COX-2 expression increases C/EBPβ and C/EBPδ expression and decreases C/EBPα expression, thereby stimulating hepatocyte proliferation (C). Abbreviations: DAMPs, damage-associated molecular patterns; PAMPs, pathogen-associated molecular patterns; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B-cells; COX-2, cyclooxygenase 2; PGE2, prostaglandin E2; IL-1β, interleukin 1β; IL-6, interleukin 6; TNF-α, tumor necrosis factor alpha; SEC, sinusoidal endothelial cell; HSC, hepatic stellate cell; HGF, hepatocyte growth factor; uPA, urokinase plasminogen activator; C/EBPα, β, and δ, CCAAT enhancer binding protein alpha, beta, and delta; TGF- α, tumor growth factor alpha; EGF, epidermal growth factor; EGFR2, epidermal growth factor receptor 2; JAK, Janus kinase; ERK, extracellular signal- regulated kinase; MAPK, mitogen-activated protein kinase; STAT3, signal transducer and activator of transcription 3; IRE1α, inositol-requiring enzyme-1α; UPR, unfolded protein response; ROS, reactive oxygen species.
Simultaneously, the immune response is also fueled by pathogen-mediated inflammation emanating from the gut-liver axis [60,61]. PHx induces endotoxemia as a result of surgical ligation of part of the portal vein output, portal hypertension [25], and consequent perturbation of the intestinal mucosal barrier [ 62, 63 ]. This results in microbe-derived blood-borne lipopolysaccharide (LPS) – a pathogen-associated molecular pattern (PAMP) [ 64 ]- triggering liver regeneration by binding PRRs such as TLR4 on Kupffer cells [65]. LPS-PRR binding leads to Kupffer cell activation and the release of TNF-α and IL-6 [36,54,66]. The endotoxemia further facilitates the accumulation of platelets in the remnant liver [51,52], where the platelets locally orchestrate a pro-regenerative stimulus via degranulation and possibly sequestration by sinusoidal endothelial cells and hepatocytes [42] (section 2.3).
Thirdly, a shift in hepatocellular redox state is responsible for the onset of liver regeneration (Figure 1). The temporary pro-oxidative state post-PHx, which is characterized by an overproduction of reactive transients such as hydrogen peroxide (H2O2), can be caused by liver surgery encompassing ischemia/reperfusion [67,68], cholestasis [69] and other liver diseases [70], and a temporary post-PHx bile acid overload (section 3). Bai et al. showed that H2O2 at a specific intracellular concentration range promotes liver regeneration in rats during the first 24 hours after PHx [71], which has also been reported by others in mice [72,73], albeit with contradictory results [74].
Intracellular H2O2 acts as a cell cycle regulator that, depending on its intracellular concentration, signals quiescence or proliferation in hepatocytes. As illustrated in Figure 1, the process is regulated by the ERK-cyclin D1-pRB pathway [71] as well as Notch signaling [75]. ERK stands for extracellular regulated kinase (a MAPK), pRB signifies phosphorylated (i.e., activated) retinoblastoma protein (RB), and Notch is a cell surface protein that acts as an auxiliary mitogen in liver regeneration [76] (section 2.4). ERK signaling promotes cell cycle activity and ultimately proliferation, during which cyclin D1 and pRB, a cell cycle inhibitor in non-phosphorylated form [77,78], enable cell cycle progression and ultimately mitosis [79–81]. The transient increase in H2O2 levels is facilitated by temporary suppression of antioxidant enzymes and upregulation of pro-oxidant enzyme activity through nuclear factor erythroid 2-related factor 2 (NRF2) and other redox-active enzymes, particularly during the early regeneration phase [71]. In support of this, NRF2-null mice exhibit stalled liver regeneration [82]. To protect the hepatocytes from oxidative stress while undergoing reactive oxygen species (ROS)-mediated mitosis, heat shock proteins may be upregulated [83].
2.2. Cytokines in liver regeneration
Acting on the numerous environmental cues (section 2.1 and Figure 1), Kupffer cells become activated and release two important cytokines for liver regeneration, namely TNF-α and IL-6 [36,84,85]. While IL-6 is chiefly responsible for the mitogenic effects in hepatocytes, TNF-α predominantly serves an autocrine function in that it stimulates the production of IL-6 by Kupffer cells via nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) [36,86] (Figure 2). However, it was shown that quiescent rat liver epithelial (LE6) cells also exhibit mitogenic activity following TNF-α stimulation, which proceeds through NF-κB and concurs with the upregulation of IL-6, signal transducer and activator of transcription 3 (STAT3, see below), and c-myc (an immediate-early gene that regulates cell cycle progression) [84,87,88], suggesting direct mitogenic signaling by TNF-α. Furthermore, the release of sphingosine 1- phosphate from activated platelets [89] prompts human sinusoidal endothelial cells to secrete IL-6 and thereby amplify the proliferative response in hepatocytes [90,91].
Hepatocytes normally reside in the quiescent (G0) phase, but after PHx enter the G1 phase following a multitude of stimuli, which includes IL-6 binding to its cognate receptor on the hepatocyte membrane [92,93]. As is illustrated in Figure 2, this triggers Janus kinase (JAK) signaling and the consequent transcription and translation of immediate-early target genes involved in DNA synthesis, cell proliferation, cellular hypertrophy, metabolic homeostasis, and cell survival by two major pathways [36,37,94,95]. Firstly, JAK activates the ERK-1/2 MAPK cascade via RAS and its complexation partners, culminating in cell proliferation [6]. Secondly, JAK activates STAT3 and the transcription of a plethora of immediate-early target genes [84,95,96], which mediate numerous liver regeneration-related processes [97].
2.3. Growth factors (complete mitogens) in liver regeneration
Liver regeneration is propagated by growth factors, whereby hepatocyte growth factor (HGF), vascular endothelial growth factor (VEGF), and ligands that bind epidermal growth factor (EGF) receptor (EGFR) occupy key roles [36,37, 98–102]. Growth factor signaling encompasses several cell types and organs [36].
In the early phase of post-PHx liver regeneration, platelets accumulate in the remnant liver [103,104] via interactions with sinusoidal endothelial cells [42]. The platelets locally release an armament of mitogenic (HGF, EGF) and co-mitogenic growth factors (insulin-like growth factor-1 (IGF-1), VEGF, and platelet- derived growth factor (PDGF)) from their α-granules [47,105,106] as well as the proliferation-curtailing transforming growth factor β (TGF-β) [105,107]. Platelet degranulation also causes the release of non-growth factor mediators of liver regeneration [42], including serotonin [108] and nucleotides [105,106]. Moreover, extravasated platelets in the liver parenchyma induce proliferative AKT and ERK-1/2 signaling in hepatocytes through HGF, VEGF, and IGF [109,110].
An intricate signaling relationship exists between HGF and VEGF in the regenerating liver. HGF is released in its precursor form pro-HGF by stellate cells [111] and serotonin-activated sinusoidal endothelial cells [112]. Activated sinusoidal endothelial cells secrete VEGFA (hepatic VEGF) [112,113] that, upon autocrine binding to VEGF receptors (VEGFR1 [113] and VEGFR2 [114]), triggers the release of pro-HGF from sinusoidal endothelial cells [113]. At the same time, VEGFA drives the chemotaxis of bone marrow-derived sinusoidal progenitor cells to the liver, which are replete with HGF [ 115 ]. These progenitor cells not only differentiate into fenestrated sinusoidal endothelial cells as part of the regeneration process [99] but also locally release HGF [115]. Accordingly, rat plasma levels of (pro-)HGF increase rapidly by 10- to 20-fold following PHx [ 116 ]. Pro-HGF is converted to its active form by urokinase plasminogen activator (uPA) [117–119] that is hyperactivated after extracellular matrix (ECM) damage from the resection [2,34]. HGF acts in a paracrine and endocrine fashion with respect to hepatocytes, on which HGF binds its cognate receptor c-Met [120,122], inducing proliferative signaling and DNA synthesis through the ERK-1/2 MAPK pathway [123,124] and hepatoprotective signaling through AKT [125,126]. HGF-mediated proliferative signaling in rats was further shown to be amplified by the LPS [127] that is abundantly present in the enterohepatic circulation after PHx [60–64].
The HGF/c-Met pathway is amplified by ligands that bind epidermal growth factor receptor (EGFR), which include EGF, TGF-α, heparin-binding EGF-like growth factor (HBEGF), and amphiregulin [37,128,129], culminating in hepatocyte proliferation (Figure 2). Plasma levels of EGF, which is constitutively produced in the duodenum by the glands of Brunner [130], increase in response to elevated shear stress in the portal circulation [34] as well as norepinephrine signaling in the gut [131,132]. Plasma levels of norepinephrine increase within 20 min after PHx in rats [131,133] and may therefore fuel EGF signaling within the gut-liver signaling axis. TGF-α is produced by proliferating hepatocytes and relays proliferative signals to hepatocytes through an autocrine mechanism via EGFR [134–136]. HBEGF is produced by monocytes and macrophages [137,138] and converted to its active form by specific metalloproteinases [139]. In rats, plasma levels of this hepatocellular mitogen [140,141] are rapidly elevated after PHx [141,142] and expressed on or associated with sinusoidal endothelial cells and Kupffer cells as early as 90 minutes after PHx [141,143], reflecting early-onset proliferative cross-talk between the sinusoidal cells and EGFR-bearing hepatocytes via HBEGF. Amphiregulin is an autocrine growth factor and a mitogen for several cell types, including hepatocytes [144]. Accordingly, amphiregulin-null mice exhibit impaired hepatocellular proliferation [144]. The protein is induced in the early regeneration phase, triggered by prostaglandin E2 (PGE2) and IL-1β [144]. The latter was shown to be rapidly released after PHx in both mice and rats [145–147]. Similarly, PGE2 levels rise in the rat liver in the early phase of liver regeneration [148]. The release of IL-1β and PGE2 by Kupffer cells is stimulated by post- PHx endotoxemia [149]. Although both mediators induce amphiregulin and thus hepatocyte proliferation, IL-1β [145–147] and PGE2 concurrently inhibit liver regeneration. Whereas PGE2 inhibits liver regeneration through downmodulation of IL-6 by Kupffer cells [150], IL-1β inhibits expression of β-klotho and fibroblast growth factor receptor 4 (FGFR4), that together form the FGF15/19 receptor [151] (see section 3.3.1.3). Their inhibitory effects are evidently offset by the other proliferation - promoting processes after PHx [152].
Of the factors addressed above, HGF, EGF, TGF-α, HBEGF, and amphiregulin are classified as so-called ‘complete hepatic mitogens’ because these proteins trigger mitosis in cultured hepatocytes and induce liver hypertrophy and hepatocyte DNA synthesis in vivo [44]. By definition, ‘incomplete mitogens’ or ‘auxiliary mitogens’ are not mitogenic in cultured hepatocytes; they do not induce hepatocellular DNA synthesis and liver growth in vivo. Nonetheless, their inhibition (or inhibition of downstream targets) delays liver regeneration but does not abrogate it [44]. Some (potential) auxiliary mitogens are briefly addressed next.
2.4. Auxiliary mitogens in liver regeneration
The auxiliary mitogens TNF-α and IL-6 [36,84,85] (section 2.2), complement proteins [9] (section 2.1), serotonin [108] (section 2.3), and norepinephrine [153] (section 2.3) have been discussed above and elsewhere [2,36,37,44,56,98] in the framework of liver regeneration. Other putative auxiliary mitogens are the receptors of some of these compounds, which include VEGFR [113] (section 2.3), TNF receptor (TNFR) [93,154 ], serotonin receptors [108], and the norepinephrine receptor α1 adrenergic receptor [131]. Additional auxiliary mitogens that are thought to play a role in liver regeneration comprise FGF1 and FGF2 [155, 156], PDGF [157], macrophage inflammatory protein (MIP)-2 alpha (CXCL2) and its receptor IL-8 receptor beta (CXCR2) [158], the cell surface proteins NOTCH1 and JAG1 [76], leptin [159], insulin [160], hyaluronic acid [161–164], Wnt2 [114], and insulin-like growth- factor binding-protein 1 (IGFBP1) [165–167]. Their role has been reviewed in a broader context in [2,36,37,44,56,98]. The multiplicity of this non-exhaustive list of auxiliary mitogens clearly illustrates the complexity of signals that modulate post- PHx liver regeneration.
In addition to these putative auxiliary mitogens, some underexposed or more recently discovered auxiliary mitogens deserve to be highlighted. First, it was shown that PHx in mice leads to endoplasmic reticulum (ER) stress and the unfolded protein response (UPR) [168], which is triggered to resolve ER stress [169]. The UPR is mediated by inositol-requiring enzyme- 1α (IRE1α) [170] and exacerbated by TNF-α and ROS [171]. This is also shown in Figure 2. As addressed in sections 2.1 and 2.2, ROS and TNF-α are hallmarks of post-PHx liver regeneration and may therefore lie at the basis of the ER stress observed in hepatectomized mice [168]. More importantly, mice with Ire1α-null livers exhibit impaired hepatocyte proliferation and liver regeneration as a result of dampened IL-6-mediated STAT3 signaling [168]. It was further found that IRE1α interacts directly with STAT3 in the early phase of liver regeneration (6–12 hours), independently of IL-6 stimulation [168]. These results underscore the cytokine-driven as well as constitutive regulation of early, UPR-based mitogenic responses following PHx, and identify IRE1α as an auxiliary mitogen.
A related but relatively underexposed auxiliary mitogen is cyclooxygenase 2 (COX-2) [172], an enzyme that regulates prostaglandin synthesis. COX-2 is controlled by multiple processes [173] that occur during liver regeneration. These processes are also summarized in Figure 2. For instance, the post-PHx endotoxemia [60–63] drives COX-2 activation in rat Kupffer cells [174] that subsequently orchestrates cytokine signaling (section 2.2). Endotoxemia also positively affects hepatocyte COX-2 expression in mice [175]. Furthermore, ROS are a trigger for COX-2 upregulation [176,177]. COX-2-modulating pathways entail NF-κB [87] (section 2.2), ERK1/2 [36,178,179] (via e.g., transcriptional regulation of PTGS2 [173], the gene encoding COX-2), and MEK in the MAPK pathway [6,178,179] (through e.g., the phospholipase A2 → arachidonic acid connection [173]), which are all established in post-PHx liver regeneration. As a result, COX-2 promotes hepatocyte proliferation during early regeneration, with maximum hepatocellular expression at 16 hours after PHx in rats [172]. The upregulation of COX-2 in regenerating livers is associated with decreased CCAAT-enhancer binding protein (C/EBP)α levels and increased expression of C/ EBPβ and C/EBPδ [172,180]. C/EBPs are transcription factors involved in cell proliferation, growth, differentiation, and metabolism [181,182]. C/EBPα inhibits proliferation [183,184] while the β and δ isoforms promote proliferation [179,185,186]. There is also a potential link between ER stress and COX-2. ER stress has been shown to stimulate murine COX-2 expression via activation of NF-κB and p38 MAPK [187]. In turn, p38 MAPK is induced by LPS and pro-inflammatory cytokines (e.g., TNF-α, IL-1β) [188–191] as well as by reactive transients such as hydrogen peroxide [191,192] and nitric oxide [193]. Similarly, NF-κB is under positive cytokine [194] and redox control [195]. These mechanisms connect post-PHx ER stress to COX-2 signaling in terms of hepatocyte proliferation, which are stimulated by liver regeneration-specific inflammation and redox- modulated processes.
An auxiliary role in liver regeneration has further been ascribed to microRNAs (miRNAs) [196,197], whereby some miRNA types are overexpressed (usually mildly; e.g., miR-21, miR-33, miR-153, and miR-743b [198,199]) while others are underexpressed (usually intensely; e.g., let-7b, let-7f, let-7g, miR-22a, miR-23b, miR-26a, miR-30b, and miR-122a [200,201]) after PHx [197]. Experiments in mice lacking the enzyme dicer 1 in the liver, which is responsible for generating miRNA [202], demonstrated that these animals exhibit a proliferative liver phenotype [203], indicating that miRNAs are essentially inhibitors of liver regeneration [200,204,205]. This inverse correlation is biochemically logical given that some miRNAs inhibit the translation of messenger RNA (mRNA) to a functional protein; in many cases cell cycle regulators [206,207] and mediators of proliferation [200,208,209]. However, there are several examples of miRNAs that are upregulated during liver regeneration yet amplify the regenerative response, suggesting that these transcriptomic regulators inhibit repressors of hepatocellular proliferation. Specific examples include miR-21 [207,210–213], miR-221 [214], miR-378 [210], and miR-382 [215].
The expression patterns of miRNA are species-dependent [216], dynamic over time, and the peak expression levels do not temporally overlap [198,217,218], underscoring the phasic nature and pleiotropic signaling of miRNAs as has been reported for cytokines (section 2.2) and growth factors (section 2.3). In that respect, the temporal heterogeneity of intrahepatic and plasma cytokine levels [36,37,56,84] are partly responsible for the differential miRNA expression profiles inasmuch as cytokines modulate miRNA expression [219]. In addition to changes in their quantitative expression, the miRNAs, which are associated with polysomes (mRNA-ribosome complexes formed during active translation), also exhibit spatial translocation during liver regeneration [197]. Corroboratively, the expression levels of the miRNAs let-7a, miR-21, miR-195, and miR-215 increased in the membrane-bound polysomes relative to the free polysomes after PHx [197].
3. Bile acids co-regulate post-hepatectomy liver regeneration in the early phase
Studies in the recent years have shown that bile acid metabolism and signaling are directly involved in the liver regeneration process [21,28,220,222]. After PHx, the remaining liver is subject to acute overload of bile acids returning via the portal circulation [22,24–26]. Bile acids are complete mitogens by virtue of their binding to nuclear receptors or activating intracellular signaling pathways [19,20]. During the regeneration phase, the liver also activates multiple adaptive mechanisms to prevent bile acid toxicity and restore bile acid homeostasis since prolonged exposure to certain bile acids at higher concentrations may promote liver injury or tumorigenesis in chronic liver injury- repair processes [223]. The effect of bile acid metabolism on liver regeneration, regulation of liver proliferation by bile acid signaling, and the mechanisms regulating bile acid homeostasis during regeneration are further discussed in the following sections.
3.1. Bile acid synthesis and cycling through the enterohepatic circulation
3.1.1. Bile acid synthesis
Daily, 0.2 to 0.6 g of bile acids (Figure 3, placed at the end of the manuscript) is synthesized from cholesterol in the human liver via two pathways: the neutral pathway and the acidic pathway [224]. Bile acid synthesis pathways involve multi-step reactions catalyzed by enzymes in the endoplasmatic reticulum, mitochondria, cytoplasm, and peroxisomes. The enzyme cholesterol 7α-hydroxylase (CYP7A1) catalyzes the first rate- limiting step in the neutral pathway and converts cholesterol into 7α-hydroxycholesterol, which eventually leads to the synthesis of the primary bile acids cholic acid (CA) or chenodeoxycholic acid (CDCA) [225,226]. In the acidic pathway, the enzyme 27α-hydroxylase (CYP27A1) converts cholesterol to 27α-hydroxycholesterol, which leads to the synthesis of CDCA [224]. After excretion into the biliary system, bile acids are first deconjugated and then biotransformed by enteral bacteria through phase I reactions (oxidation, hydroxylation), forming secondary bile acids, including deoxycholic acid (DCA), lithocholic acid (LCA), and ursodeoxycholic acid (UDCA) [227–229]. Ninety five percent of all biliary excreted bile acids are resorbed and transported back to the liver via the portal vein, also referred to as the enterohepatic circulation [230]. However, most LCA is not recycled and the small amount of circulating LCA is rapidly conjugated by sulfation, a phase II reaction, and secreted into the biliary system [228]. Sulfation increases bile acid solubility, as a result of which sulfated LCA is less likely to be intestinally reabsorbed [231]. For this reason, the human bile acid pool mainly consists of the primary bile acids CA and CDCA and the secondary bile acid DCA. The liver efficiently conjugates both primary and secondary bile acids to taurine or glycine (ratio 3:1 in humans) by amidation, then commonly referred to as bile salts, which promotes solubility and prevents passive diffusion across cell membranes [232]. Throughout this review, the term ‘bile acids’ is used for both bile acids and bile salts.
Figure 3. Non-exhaustive list of bile acid species and bile acid analogues, chemical properties, and toxicity.
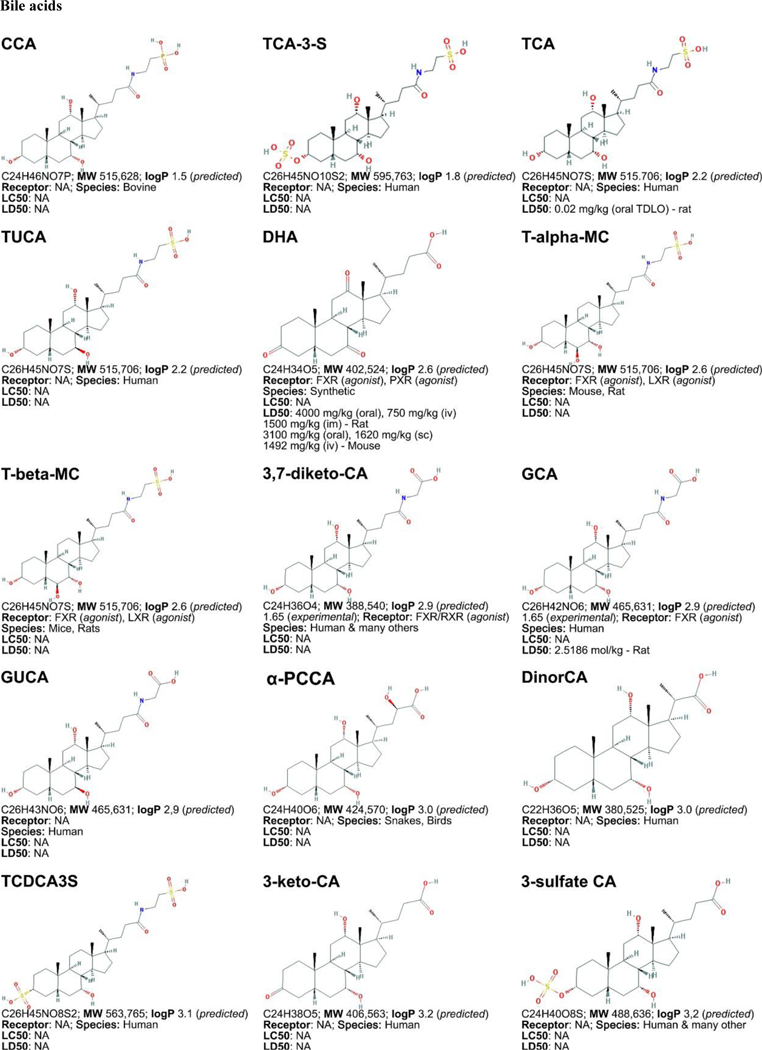
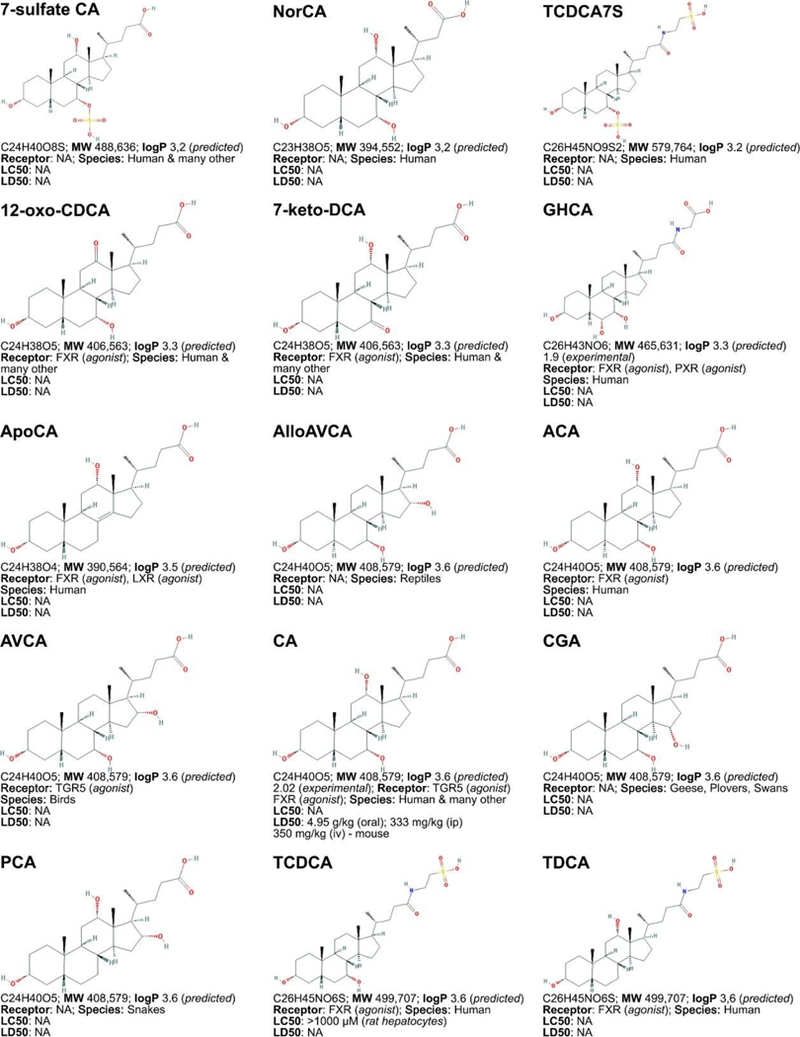
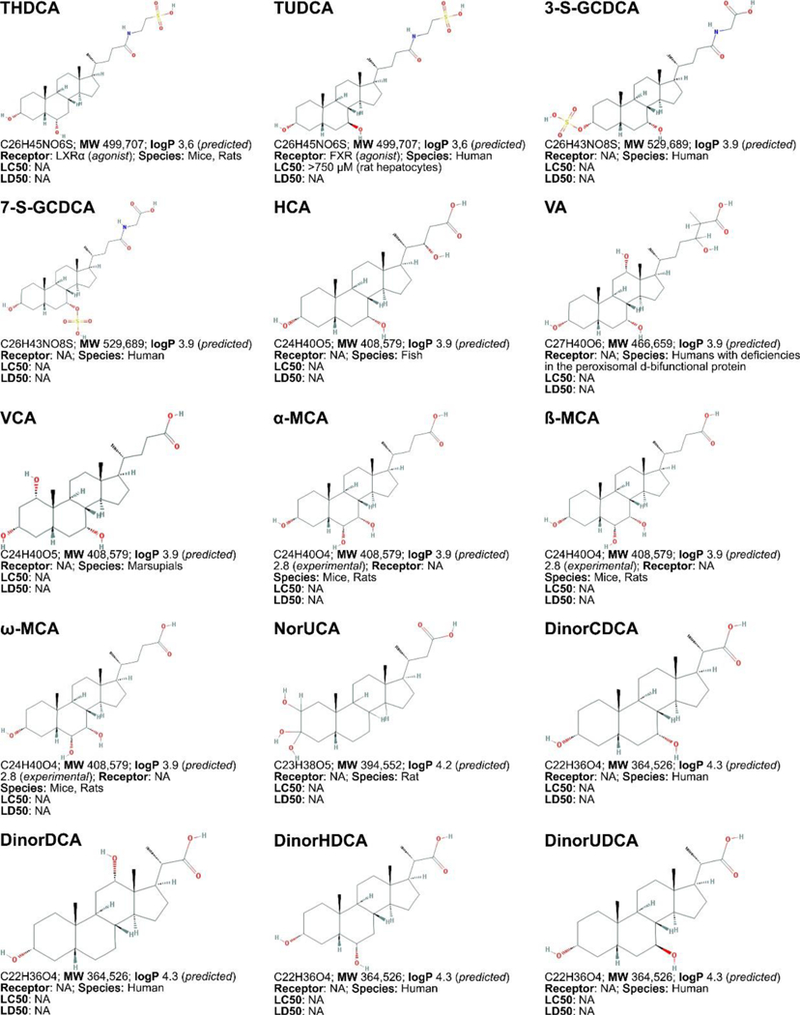
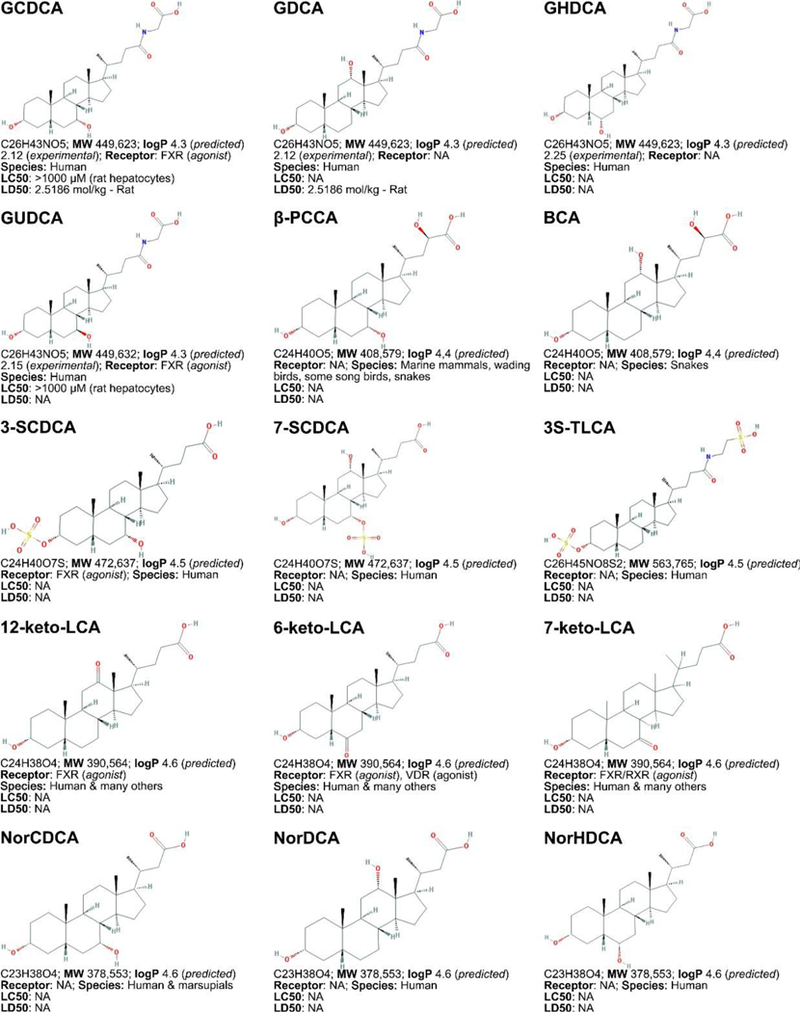
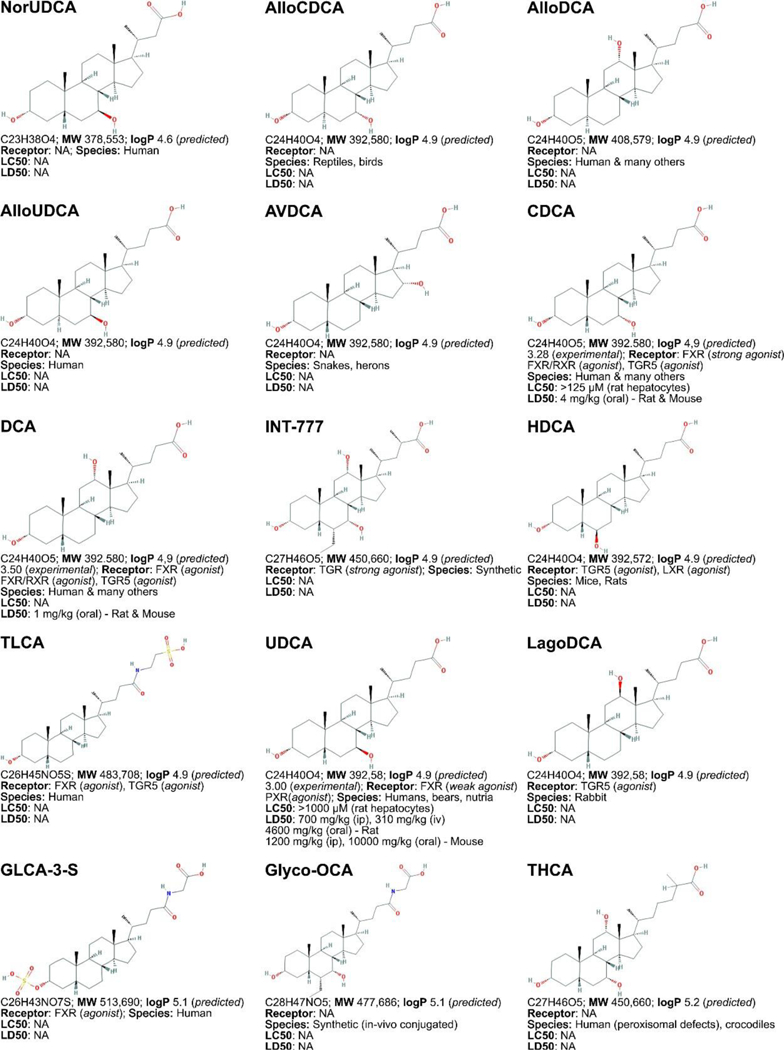
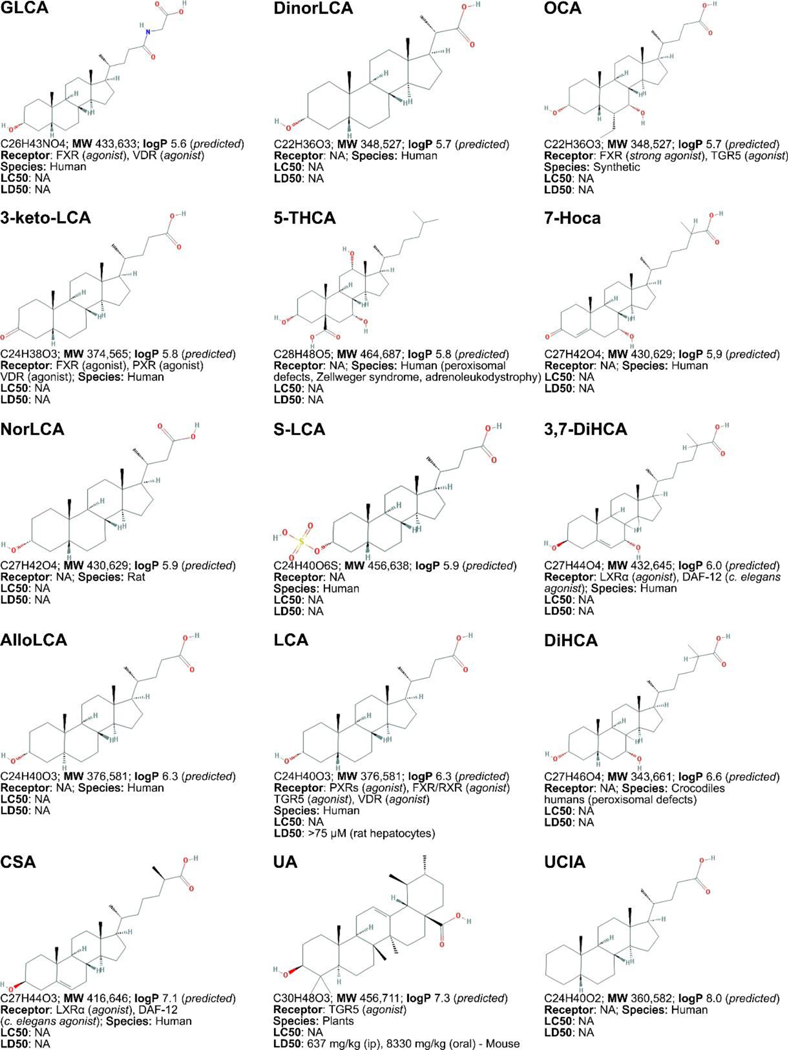
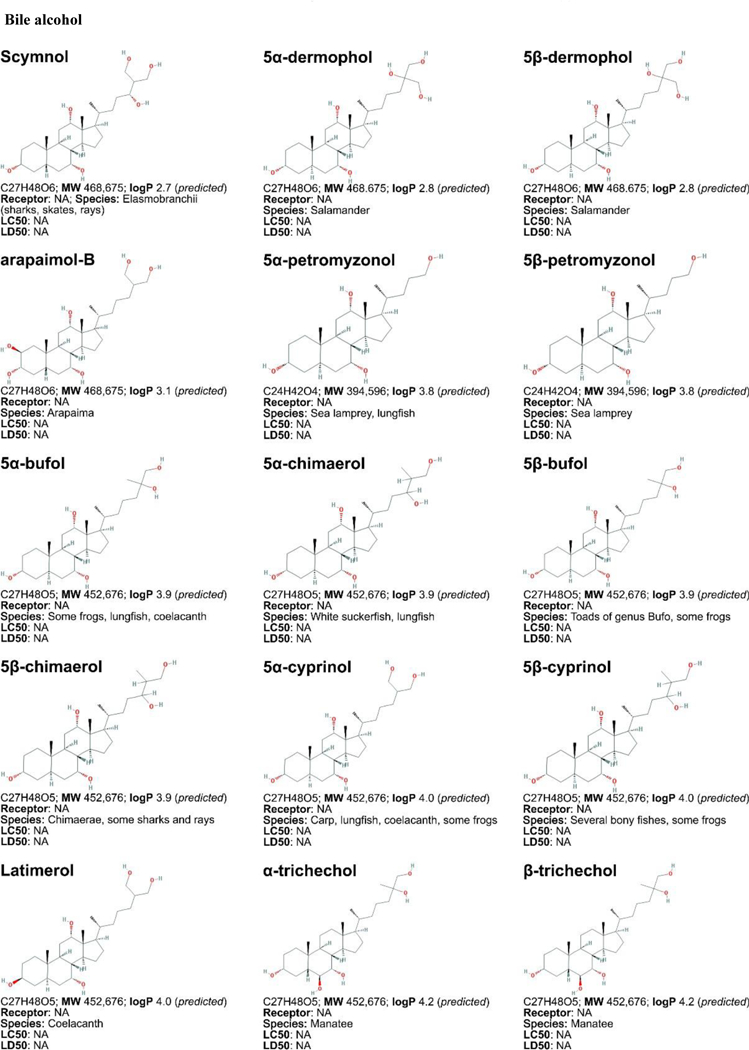
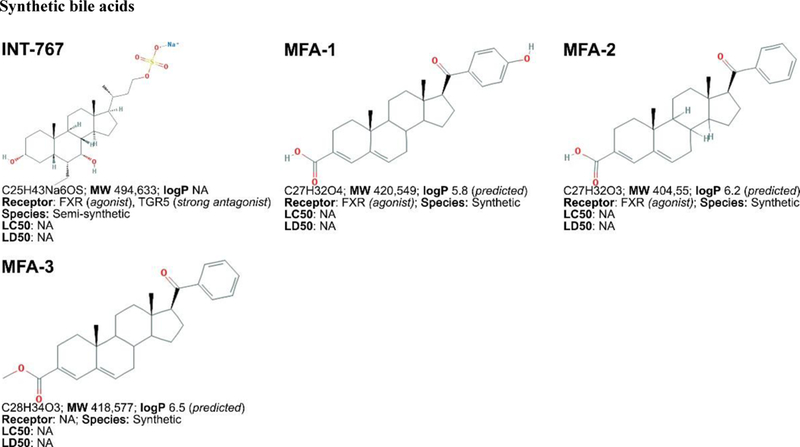
LogP (octanol:water partition coefficient) values were retrieved from PubChem and were predicted with XLogP2 or XLogP3 software. The 50% lethal concentration (LC50, used for in vitro data) and 50% lethal dose (LD50, used for in vivo data) were obtained from the material safety data sheets (retrieved from the Cayman Chemicals and Spectrum Chemical website) and the Toxicological Data Network (TOXNET, https://toxnet.nlm.nih.gov/) as well as available literature [544] [544].Abbreviations (bile acids excluded): NA: information not available; iv: intravenous; ip: intraperitoneal; MW: molecular weight; sc: subcutaneous; TDLO; the lowest dose causing a toxic effect.Abbreviations (bile acids): 12-keto-LCA: 12-ketolithocholic acid / 12-oxolithocholic acid; 12-oxo-CDCA: 12- oxochenodeoxycholate / 12-oxochenodeoxycholic acid; 3,7-DiHCA : 3,7-dihydroxy-5-cholestenoic acid; 3,7-diketo-CA :3,7-diketocholanic acid / 3,7-dioxhocholanoic acid; 3-keto-CA: 3-ketocholic acid / 3-oxocholic acid; 3-keto-LCA: 3-ketolithocholic acid / dehydrolithocholic acid; 3-SCDCA: chenodeoxycholic acid 3-sulfate; 3-S-GCDCA: glycochenodeoxycholic acid 3-sulfate; 3S-TLCA: taurolitocholate sulfate / taurolithocholic acid 3-sulfate; 3-sulfate CA: cholic acid 3-sulfate; 5-THCA: trihydrocoprostanic acid / (3alpha,5beta,7alpha,12alpha)-3,7,12-trihydroxycholestane-5-carboxylic acid; 6-keto-LCA: 6- ketolithocholic acid; 7-Hoca: 7α-hydroxy-3-oxo-4-cholestenoic acid; 7-keto-DCA: 7-ketodeoxycholic acid; 7-keto-LCA: 7- ketolithocholic acid / nutriacholic acid; 7-SCDCA: chenodeoxycholic acid 7-sulfate; 7-S-GCDCA: glycochenodeoxycholic acid 7- sulfate; 7-sulfate CA: cholic acid 7-sulfate; α-MCA: α-muricholate / α-muricholic acid / α-hyocholic acid; α-PCCA: α-phocaecholate/ alpha-phocaecholic acid; β-MCA: β-muricholate / β-muricholic acid / β-hyocholic acid; β-PCCA: β-phocaecholate / phocaecholicacid; ω-MCA: ω-muricholate / ω-muricholic acid / ω-hyocholic acid; ACA: allocholate / allocholic acid; AlloAVCA: alloavicholate/ alloavicholic acid; AlloCDCA: allochenodeoxycholate / allochenodeoxycholic acid; AlloDCA: allodeoxycholate / allodeoxycholic acid; AlloLCA: allolithocholic acid; AlloUDCA: alloursodeoxycholic acid; ApoCA: apocholate / apocholic acid; AVCA: avicholate/ avicholic acid; AVDCA: avideoxycholate / avideoxycholic acid; BCA: bitocholate / bitocholic acid; CA: cholate / cholic acid; CCA: ciliatocholate / ciliatocholic acid; CDCA: chenodeoxycholate / chenodeoxycholic acid; CGA: cygnocholate / cygnocholic acid; CSA: cholestenoic acid; DCA: deoxycholate / deoxycholic acid; DHA: dehydrocholate / dehydrocholic acid; DiHCA: dihydroxycoprostanoic acid; DinorCA: dinorcholic acid; DinorCDCA: dinorchenodeoxycholic acid; DinorDCA: dinordeoxycholic acid; DinorHDCA: dinorhyodeoxycholic acid; DinorLCA: dinorlithocholic acid; DinorUDCA: dinorursodeoxycholic acid; GCA: glycocholate / glycocholic acid; GCDCA: glycochenodeoxycholate / glycochenodeoxycholic acid; GDCA: glycodeoxycholate / glycodeoxycholic acid; GHCA: glycohyocholate / glycohyocholic acid; GHDCA: glycohyodeoxycholate / glycohyodeoxycholic acid; GLCA: glycolithocholate / glycolithocholic acid; GLCA-3-S: glycolithocholate 3-sulfate / glycolithocholic acid 3-sulfate; Glyco-OCA: glyco-obeticholic acid; GUCA:glycoursocholate / glycoursocholic acid; GUDCA: glycoursodeoxycholate / glycoursodeoxycholic acid; HCA: hemulcholate / hemulcholic acid; HDCA: hyodeoxycholic acid / murideoxycholic acid; LagoDCA: lagodeoxycholic acid; LCA: lithocholate / lithocholic acid; NorCA: norcholate / norcholic acid; NorCDCA: norchenodeoxycholic acid; NorDCA: nordeoxycholic acid; NorHDCA: norhyodeoxycholic acid; NorLCA: norlithocholic acid; NorUCA: noruroscholic acid; NorUDCA: norursodeoxycholic acid; OCA: obeticholic acid / ocaliva / 6-ethylchenodeoxycholic acid/ INT-747; PCA: pythocholic acid; S-LCA: lithocholic acid 3-sulfate; T-alpha-MC: tauro-α-muricholic acid; T-beta-MC: tauro-β- muricholic acid; TCA: taurocholate / taurocholic acid; TCA-3-S: taurocholate 3-sulfate / taurocholic acid 3-sulfate; TCDCA: taurochenodeoxycholate / taurochenodeoxycholic acid; TCDCA3S: taurochenodeoxycholic acid 3-sulfate; TCDCA7S: taurochenodeoxycholic acid 7-sulfate; TDC(A): taurodeoxycholate / taurodeoxycholic acid; THCA: trihydrocoprostanic acid / coprocholic acid / 3,7,12-trihydroxycholestan-26-oic acid; THDCA: taurohyodeoxycholate / taurohyodeoxycholic acid; TLCA: taurolithocholate / taurolithocholic acid; TUCA: tauroursocholate / tauroursocholic acid; TUDCA: tauroursodeoxycholate / tauroursodeoxycholic acid; UA: ursolic acid; UCA: ursocholic acid; UClA: ursocholanic acid; UDCA: ursodeoxycholate / ursodeoxycholic acid; VA: varanic acid; VCA: vulpecholate / vulpecholic acid.
In the mouse and rat liver, the majority of CDCA is further converted to the more hydrophilic muricholic acids (MCAs) α- muricholic acid and β-muricholic acid. Consequently, the mouse and rat bile acid pool consists of approximately equal amounts of CA and MCAs, with relatively low levels of CDCA. While in humans glycine-conjugated bile acids are most common, most bile acids in mice and rats are conjugated with taurine [233]. Thus, the mouse bile acid pool is much more hydrophilic compared to the human bile acid pool. This is the main reason for the direct cytotoxicity of bile acid during cholestasis in humans but not mice [234,235]. Nevertheless, similar mechanisms of bile acid metabolism likely apply to most species.
3.1.2. Bile acid transport in the enterohepatic circulation
Bile acids produced in hepatocytes are efficiently secreted into the bile and stored in the gallbladder. Upon food intake, the gallbladder contracts in response to cholecystokinin secreted by the epithelial cells in the duodenum, causing bile acids to be released into the small intestine [236]. In the small intestine, bile acids emulsify dietary lipids to form micelles, allowing pancreatic lipases to hydrolyze lipids for absorption.
After reabsorption by enterocytes, bile acids in the portal circulation are imported into hepatocytes across the basolateral membrane, after which they are secreted into the bile canaliculi, a process referred to as first pass metabolism (summarized in Figure 4). The liver first pass extraction rate for conjugated bile acids is about 90%, with little bile acids spilled into the systemic circulation. The Na+-dependent taurocholate transporter (NTCP) is a major bile acid uptake transporter in the basolateral membrane of hepatocytes [237–241]. In addition, organic anion transporter (OATP) isoforms mediate Na+-independent bile acid uptake at the basolateral membrane of the hepatocytes. At the canalicular side of the hepatocytes, the bile salt export pump (BSEP, ABCB11 / Abcb11; human / rodent gene) mediates bile acid secretion against a concentration gradient. Consequently, canalicular bile acid export is a rate-limiting step in bile formation [242]. The multidrug resistance-associated protein-2 (MRP2, ABCC2 / Abcc2) can also mediate the canalicular secretion of certain sulfated and conjugated bile acids, besides bilirubin conjugates, glutathione, and drugs [243]. Hepatobiliary free cholesterol secretion into the bile is mediated by the ATP-binding cassette transporters ABCG5 and ABCG8 [244]. Phosphatidylcholine, the major phospholipid in the bile, is secreted via the multi-drug resistance protein (MDR3, ABCB4 / Abcb4) [245]. Cholesterol, bile acids, and phospholipids are the major constituents of bile. They form micelles in the canaliculi to increase cholesterol solubility and decrease bile acid damage to the bile duct.
Figure 4. Hepatocellular bile acid transporters.
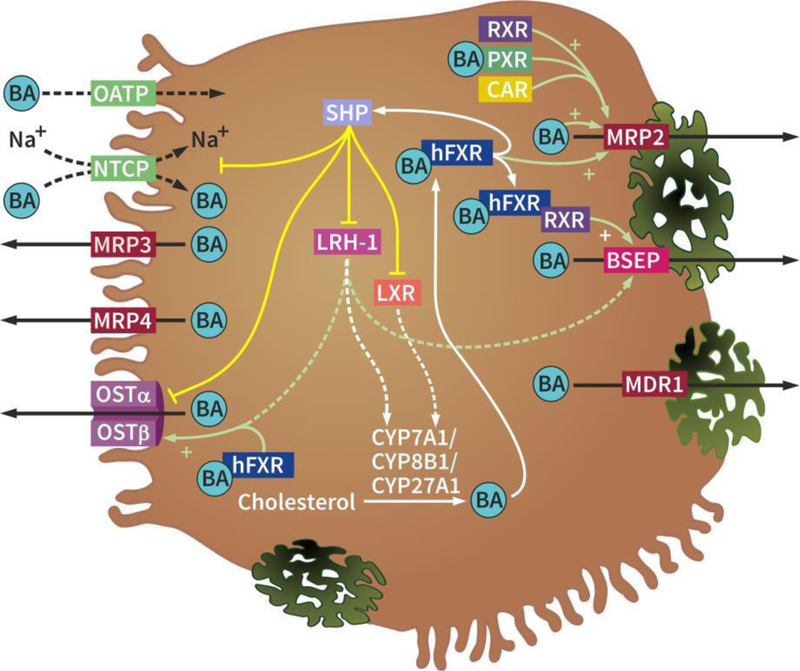
Basolateral import of bile acids is mediated by NTCP (Na+-dependent) and OATP isoforms (Na+- independent). Bile acids are exported through the basolateral exporters MRP3, MRP4, and the OSTα and OSTβ heterodimer and through the canalicular exporters BSEP, MRP2, and possibly MDR1. Bile acids regulate their own efflux through hepatic farnesoid X receptor (hFXR). Bile acid- activated hFXR induces the OSTα and OSTβ heterodimer and MRP2 and, as heterodimer with RXR, BSEP. hFXR also activates SHP that inhibits the importers NTCP, the OSTα and OSTβ heterodimer, LXR, and LRH-1. LXR and LRH-1 normally inhibit CYP7A1, CYP8B1, and CYP27A1, but because of SHP activation by hFXR and consequent inhibition of LRH-1 and LXR, bile acids are synthesized from cholesterol. Additionally, LRH-1 normally stimulates the expression of BSEP and the OSTα and OSTβ heterodimer, while induction of SHP by hFXR results in inhibition of those exporters. Abbreviations: BA, bile acid; BSEP, bile salt export pump; CAR, constitutively active/androstane receptor; CYP, cytochrome p450; hFXR, hepatic farnesoid X receptor; LRH-1, liver receptor homolog 1; LXR, liver X receptor; MDR1, multidrug resistance associated protein 1; MRP3/4, multidrug resistance protein 3 and 4; NTCP, Na+-taurocholate co-transporting polypeptide; OATP, organic anion transporting polypeptide; OSTα/β, organic solute transporter alpha/beta; PXR, pregnane X receptor; RXR, retinoid X receptor; SHP, small heterodimer partner.
In the intestine, bile acids are imported into enterocytes via the apical sodium-dependent bile acid transporter (ASBT, SCLC10A2 / Sclc10a2) [246] and subsequently excreted into the portal circulation via the organic solute and steroid transporter (OST)α and OSTβ heterodimer [247,248]. Whereas most of the conjugated bile acids are efficiently reabsorbed in the small intestine via active transport systems, some unconjugated primary bile acids and secondary bile acids, mainly DCA and to a much less extent LCA, can also be reabsorbed in the colon via passive diffusion and returned to the liver via the portal circulation.
3.1.3. Bile acid-activated signaling
Besides the digestive function, bile acids are also signaling molecules that regulate various physiological and pathophysiological processes, including metabolic homeostasis, tumorigenesis, and immunity. In the enterohepatic system, bile acids exert regulatory functions by activating either intracellular ligand-activated nuclear receptors or cell surface receptors that activate intracellular signaling [249–251]. One of the major functions of the nuclear receptors is to maintain bile acid homeostasis through coordinated regulation of bile acid synthesis, transport, and detoxification.
The best studied nuclear receptor, farnesoid X receptor (FXR), is primarily expressed in the liver and intestines and, as shown in Figure 5 and Figure 6, distinct pathways are initiated by hepatocellular FXR (hFXR) [252] and enterocytic FXR (eFXR) [253] that are involved in post-PHx liver regeneration and bile acid homeostasis [28,254]. In the liver, hFXR regulates bile acid metabolism through a feedback and feedfoward mechanism. Once activated by bile acids, hFXR mediates a negative feedback loop through inhibition of the bile acid synthesis genes CYP7A1, CYP8B1, CYP27A1, and the bile acid uptake transporter NTCP [224,255] (Table 1). Additionally, bile acid-activated hFXR regulates bile acid homeostasis through a feedfoward mechanism by stimulating the expression of the bile acid efflux transporter [256]. In the intestine, eFXR inhibits ASBT and induces OSTα and OSTβ to reduce bile acid accumulation in enterocytes. Bile acid activation of eFXR also induces the endocrine hormone fibroblast growth factor 15 (FGF15, FGF19 in humans) (Table 1). FGF15 can bind to its cognate receptor FGFR4 on the surface of hepatocytes and inhibits CYP7A1 and bile acid synthesis through various mechanisms activated by FGFR4, such as the MAPK-ERK pathway.
Figure 5.
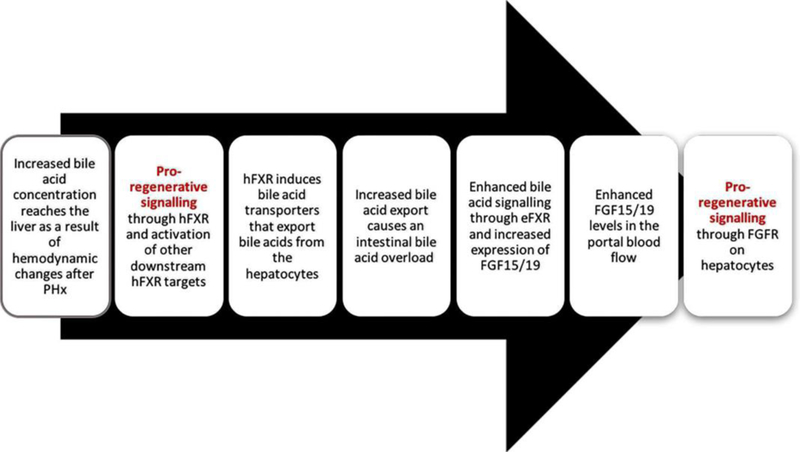
Chronological flowchart of mitogenic signaling by bile acids.
Figure 6.
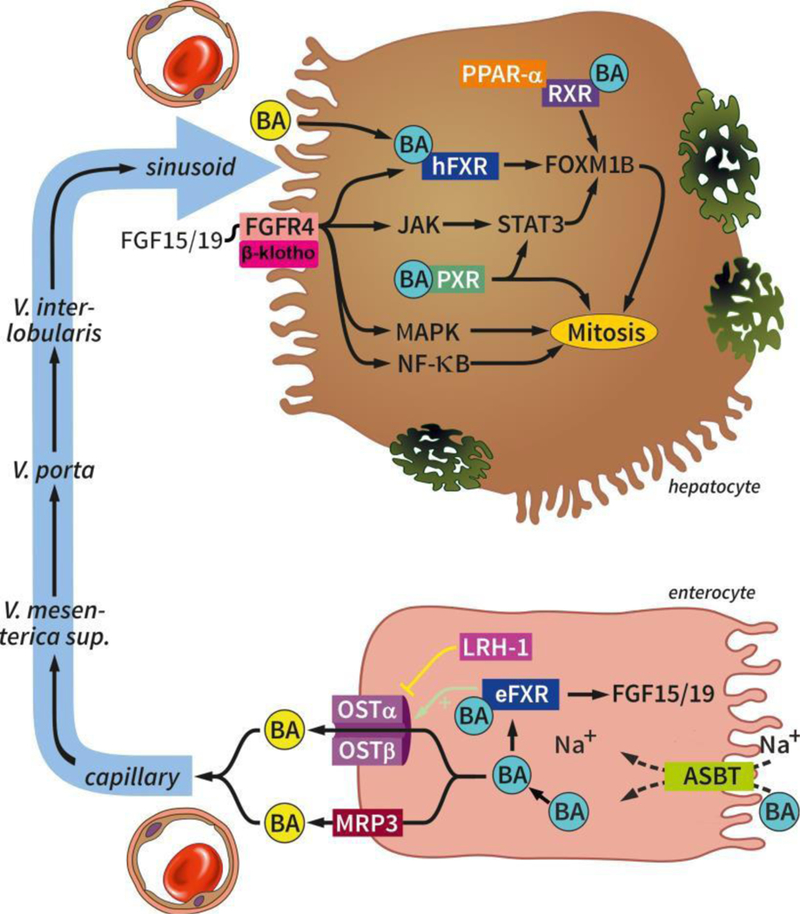
Hepatocyte-enterocyte interplay after PHx. Bile acids are taken up in the intestine by the enteral importer ASBT and exported into the portal circulation by the OSTα and OSTβ heterodimer and MRP3. In the enterocyte, bile acids activate eFXR that activates the OSTα and OSTβ heterodimer and induces the transcription of FGF15/19. FGF15/19 binds to the FGFR4/β-klotho receptor complex that in turn stimulates mitosis through pathways involving hFXR/FOXM1B, JAK/STAT3/FOXM1B, MAPK, and NF-κB. In the hepatocyte, bile acids can bind PXR and hFXR that stimulate mitosis through STAT3 and FOXM1B, respectively. Bile acids bound to RXR complexed with PPAR-α can also induce hepatocyte proliferation. Abbreviations: BA, bile acid; ASBT, apical sodium dependent bile acid transporter; FGF15/19, fibroblast growth factor 15/19; eFXR, enteral farnesoid X receptor; LRH-1, liver receptor homolog 1; OSTα/β, organic solute transporter alpha/beta; MRP3, multidrug resistance protein 3; FGFR4, fibroblast growth factor receptor 4; JAK, Janus kinase; ERK, extracellular signal-regulated kinase; MAPK, mitogen-activated protein kinase; STAT3, signal transducer and activator of transcription; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B-cells; FOXM1B, forkhead box M1B; hFXR, hepatic farnesoid X receptor; PXR, pregnane X receptor; RXR, retinoid X receptor; PPAR-α, peroxisome proliferator-activated receptor alpha.
Table 1.
Transcription factors and nuclear receptors involved in liver regeneration triggered by bile acids.
| Transcription factor | Regulator | Target | Function |
|---|---|---|---|
|
Liver receptor homologue 1 (LRH-1) fetoprotein AFP transcription factor (FTF), CYP7A promotor-binding factor (CPF) |
– CDCA [258], SHP [259,260] + TNF-α [261] |
– Ostα-Ostβ [262] – CYP7A1/ Cyp7a1 [260,263–266] – CYP8B1/Cyp8b1 [266–268] + ABCG5/Abcg5 and ABCG8/Abcg8 [ 269] + Abcc3 (MRP3) [258] + ABCB11 (BSEP) [269] |
Bile acid homeostasis [262,269] |
|
Enteral farnesoid X receptor (eFXR) |
+ bile acids [270–272] |
+ OSTα-OSTβ/Ostα-Ostβ [273–275] + Fgf15 [276] + FGF19 [277] |
Regulation of bile acid pool size, governing the production of mitogens, and regulation of lipid and glucose metabolism [17,22,23,26] |
|
Hepatic farnesoid X receptor (hFXR) |
+ bile acids [270,272] |
+ Ostα-Ostβ / OSTα-OSTβ [273,278] + ABCB11 (BSEP), as FXR/RXR heterodimer [256] + ABCC2/Abcc2 (MRP2), as FXR/RXR heterodimer [279] + UGT2B4 [280] – UGT2B7 [281] |
Regulation of bile acid pool size, governing the production of mitogens, and regulation of lipid and glucose metabolism [21–23,26,27] |
|
Forkhead box M1B (FOXM1B) |
+ hFXR [254,282–288] + STAT3 [289] |
+ Cdk1 and Cdk2 [288] + Cdc25b [22] + Ccnd1 (cyclin D1) [22] + Pcna [22] |
Regulation of DNA replication and mitosis [285,288] |
|
Retinoid X receptor (RXR) |
+ 9-cis retinoic acid |
+ Cdk1, Cdk2, Cdk 6, and cyclin D genes [290] + CYP3A4, as PXR/RXR heterodimer [291] + ABCB11 (BSEP), as FXR/RXR heterodimer [256] + ABCC2/Abcc2 (MRP2), as FXR/RXR, PXR/RXR, or CAR/RXR heterodimer [292] |
Activation of mitotic and proliferative activity [290,293] |
| Pregnane X receptor (PXR) steroid and xenobiotic sensing nuclear receptor |
+ bile acids [294,295] + pregnenolone 16α-carbonitrile [296] – IL-6 [297] |
+ CYP3A4, as PXR/RXR heterodimer [291] – Cyp7A1 [298] + ABCC2/Abcc2 (MRP2), as PXR/RXR heterodimer [279] + Abcc3 (MRP3) and Abcc3 (MRP4) [299] + Ppar-γ and Cd36 [300,301] |
Identification of toxic substances and upregulation of proteins involved in their detoxification and clearance [302] |
|
Constitutively active/androstane receptor (CAR) |
+ LCA [303] + TCPOBOP (selective) (phenobarbital) [304] |
+ Cyp2b, Cyp2c, Cyp3a sulfotransferases, glutathione transferases gene [299,305–311] + ABCC2/Abcc2 (MRP2), as CAR/RXR heterodimer [279] + Abcc4 (MRP4) [312] – UGT2B7 [313] |
Identification of toxic substances and upregulation of proteins involved in their detoxification and clearance [314] |
|
Growth arrest and DNA-damage- inducible beta (GADD45β) |
+ RXR, CAR, PPARs [315,316] |
Cell proliferation [317], anti- inflammatory signaling [318], and positive transcriptional regulation of nuclear receptors (RXR, CAR, PPARs) [315,316] |
|
|
Liver X receptor (LXR) |
the sterols 24(S),25-epoxycholesterol, 22(R)-hydroxycholesterol, 24(S)- hydroxycholesterol, several metabolized bile acid species, including hyodeoxycholic acid, taurohyodeoxycholic acid, and cholestenoic acid [ 319, 320], GW3965 (selective) [293] |
+ Abcg5, Abcg8 [321] |
Proliferation-repressive effects [293] |
|
Hepatocyte nuclear factor (HNF) - 4α |
– CAR [322] |
+ CYP7A1/Cyp7a1 [323,324] + CYP8B1/Cyp8b1 and CYP27A1/Cyp27a1 [325–327] + UGT2B7 [313] |
Lipid and glucose metabolism [328,329] |
|
Nuclear factor erythroid 2–related factor 2 (NRF2) |
+ Bsep [330,331] + Abcc3 (MRP3) and Abcc4 (MRP4) [331] + Ostα [331] – Cyp7a1 and Cyp8b1 [331] |
Regulation of bile acid pool size [331] |
|
| Peroxisome proliferator-activated receptor (PPAR)-α | + UGT2B4 [332] | Metabolic sensor [319] | |
| Signal transducer and activator of transcription 3 (STAT3) | + Foxm1b [289] | Activation of mitogenic pathways [23] | |
| Vitamin D receptor (VDR) | + LCA [333] | currently unknown | currently unknown |
Legend: – = downregulation, + = upregulation
Recent studies have shown that this bile acid activated gut-to-liver signaling axis plays an important part in the liver regeneration process [22,257] (Figure 5). In response to bile acid overload, pregnane X receptor (PXR, section 3.3.3) and constitutively active/andostane receptor (CAR, section 3.3.4) play important roles in activating numerous bile acid detoxification mechanisms. More recently, these two nuclear receptors have also been implicated in the regulation of liver regeneration.
3.2. Impact of bile acid pool alteration on liver regeneration
PHx causes acute but temporary bile acid overload in the liver [22,24–26] and the systemic circulation [21]. The bile acid overload in rat hepatocytes occurs within several hours after PHx [26] and peaks around 24 hours [22]. Bile acids are cytotoxic at high concentrations [334,335], and hepatic bile acid levels are quite quickly restored to pre-PHx status, namely within 24–48 hours after resection [22,24,26]. Initial evidence suggesting that bile acid signaling was involved in liver regeneration came from studies showing that experimentally altering the bile acid pool size could significantly modulate the liver regeneration rate in mice after PHx [28]. Rats that lack intestinal bile acid reabsorption due to external bile drainage showed lower proliferative activity in the liver and slower liver regrowth after PHx compared to rats that underwent bile drainage into the duodenum before PHx [21,221]. Huang et al. [28] further showed that mice that had been supplemented with 0.2% CA for 5 days exhibited more rapid liver regeneration, while mice fed the bile acid sequestrant cholestyramine for 5 days showed delayed liver regeneration after PH [28]. In rats that had undergone PHx, dietary supplementation with the bile acids UDCA and tauroursodeoxycholate induced hepatocyte proliferation [220]. A more recent clinical study showed that patients who underwent hemihepatectomy without external bile drainage had about ~3-fold more liver regrowth volume on day 7 than patients who underwent hemihepatectomy with external bile drainage [222,336].
The importance of bile acids in liver regeneration is also manifested in more circumstantial evidence. For example, liver regrowth elicited by CA feeding was reduced in mice lacking the basolateral bile acid exporter multidrug resistance- associated protein 3 (MRP3, Abcc3, Figure 6) [ 337, 338 ], and this was associated with decreased portal bile acid concentration and FXR activation [ 339 ]. Similarly, delayed regeneration was also reported in mice lacking CYP27A1 [340] and in mice lacking ASBT [21,339, 341 – 343 ]. Moreover, transcriptomic analysis revealed that activator protein 1 (AP-1), a heterodimeric early response transcription factor composed of c-Jun and c-Fos and in control of cell cycle activity and proliferation [344,345], is downregulated in the absence of bile acids in hepatectomized rats [21]. Finally, experiments with ‘hypertransgenic’ FRGN mice bearing humanized livers demonstrated a positive correlation between the size of the intrahepatic bile acid pool and the degree of liver growth [29]. These studies altogether support the prominent role of bile acids in post-PHx liver regeneration.
3.3. The role of nuclear receptors in post-hepatectomy liver regeneration and bile acid signaling
Nuclear receptors are transcription factors that serve as metabolic sensors in that they bind xenobiotics and metabolic intermediates (e.g., fatty acids, sterols, bile acids [319]) to regulate their levels and signaling effects as well as a plethora of metabolic pathways. A fraction of the mitogenic signals relayed by bile acids are moderated through nuclear receptors [19,20] - a process that is associated with translocation of specific bile acids from the cytoplasm to the nucleus [24]. The nuclear receptors involved in liver regeneration can exert bidirectional effects on hepatocellular proliferation, entailing both promotion and suppression of liver regeneration [19,20]. In terms of PHx, the promotors encompass FXR [28,254, 346 ], retinoid X receptor (RXR) [ 347, 348 ], PXR [ 349 ], and CAR [28, 350 ] and their common heterodimer partner, retinoid X receptor (RXR) [347,348]. Contrastingly, the suppressors encompass the peroxisome proliferator-activated receptors (PPARs) [ 351 ] PPAR-α [352] and PPAR-γ [353,354]. Genetic ablation of liver X receptor (LXR) in mice has no notable effect of post-PHx liver regeneration. However, the receptor is not necessarily mitogenically neutral since treatment with its agonist GW3965 in wild type mice hampers several effectors of proliferation and moderately retards liver regrowth [293]. LXR hence exhibits an inclination to repress liver regeneration. Liver receptor homologue 1 (LRH-1), alternatively referred to as fetoprotein AFP transcription factor (FTF) and CYP7A promoter-binding factor (CPF), is also a nuclear receptor in the liver [258]. The activities of LRH-1 revolve mainly around bile acid homeostasis [262,269] rather than induction of proliferation during post-PHx liver regeneration [355]. LRH-1 therefore fulfills a detoxification function during bile acid overload. A similar role is currently ascribed to the vitamin D receptor (VDR). This nuclear receptor is also expressed in hepatocytes [356] and regulates bile acid metabolism through CYP-based [333,357] and sulfotransferase-based bile acid detoxification [358–360] and bile acid transporter control [361].
FXR, LXR, PXR, PPARs, and LRH-1 are metabolic sensors in that they can be bound and activated by bile acids [319], and consequently regulate the levels and signaling intensity of the end-products through downstream effectors. Although CAR and VDR do not bind bile acids with the exception of LCA (in vivo) [303,333], the receptors are amenable to bile acid signaling since they are activated by ancillary bile acid-induced processes to mediate biological effects that are, or could be, pertinent in liver regeneration [319,333,357–363].
3.3.1. Farnesoid X receptor signaling
Of all nuclear receptors, FXR is the best studied in the context of post-PHx liver regeneration. The bile acids CDCA, DCA, and LCA are physiological ligands of FXR [270–272], with a relative FXR potency of CDCA > DCA = LCA > CA [271]. The prime function of FXR is to regulate bile acid metabolism and homeostasis [319,364–366]. On top of that, FXR governs lipid and glucose metabolism [319,364–366], and which are essential in liver regeneration [367,368]. With respect to bile acid homeostasis, the receptor regulates genes that control bile acid synthesis, secretion, uptake, transport, and endobiotic metabolism [369–372]. FXR is expressed at high levels in the liver [252] and intestines [253] and as such constitutes an integral part of enterohepatic communication, especially in case of liver regeneration. The individual effects of liver hFXR and intestinal eFXR will be described in detail in the following sections.
3.3.1.1. Mitogenic and metabolic signaling triggered by bile acids in the liver through hepatocellular farnesoid X receptor
Liver-specific FXR-null mice exhibit stalled liver regeneration compared to animals with functional hepatic FXR [28,254]. Similarly, FXR activity is significantly impaired and its downstream target genes Shp, Cyp7a1 (section 4.2), and Abcb11 (BSEP) (section 4.3.3) are afflicted in transgenic mice that overexpress the endogenous FXR inhibitor sirtuin 1 (SIRT1) [373]. PHx in these mice resulted in a debilitated regenerative response and bile acid-induced toxicity [373].
Mechanistically, activated hFXR signals proliferation through FOXM1B [254,282–288] (Table 1), a transcription factor that regulates DNA replication and mitosis by stimulating the expression of proteins that are responsible for cyclin-dependent kinase 2 (CDK2) and CDK1 cell cycle activity [285,288]. CDK2 is crucial for the G1/S transition [374], whereas CDK1 enables cell cycle progression from the S-phase to mitosis (M-phase) [375]. CDK2 complexes with cyclins and other CDKs to activate RB [376,377] (section 2.1) and is under positive control of CDC25a [376], which in turn is antagonized by the cyclin- dependent kinase inhibitor p21 3[78]. Hepatectomized Foxmlb mice showed increased hepatocellular nuclear p21 levels, reduced Cdc25a expression, and consequently decreased activation of CDK2 and RB [285]. Moreover, Foxm1b−/− mice exhibited increased levels of phosphorylated CDK1 [285], which is normally dephosphorylated by CDC5B to promote M-phase progression [379–381], as well as no expression of nuclear CDC25B protein [285]. These findings demonstrate that hepatocyte proliferation is controlled at the level of the cell cycle by hepatocellular FOXM1B, which is induced by hFXR that in turn is activated by bile acids (Table 1).
In addition to mitogenic signaling, hFXR-steered liver regeneration after PHx may be metabolically supported as a result of hFXR activation of the pyruvate dehydrogenase lipoamide kinase isozyme 4 (PDK4) [27,382] that resides in mitochondria and regulates pyruvate metabolism as part of gluconeogenesis [ 383 ]. PDK4 is induced by hyponutrition and facilitates the utilization of alternative carbon sources for gluconeogenesis [384]. PDK4 was upregulated following FXR activation and downregulated after FXR knockdown in HepG2 cells [27]. Induction of such a switch is analogous to the Warburg effect in highly proliferative cancer cells, typified by a metabolic shift towards converting glucose and certain amino acids into biomass [385,386]. Activation of FXR in human hepatocellular carcinoma (HepG2) cells by the agonists GW4064 and CDCA resulted in increased PDK4-mediated accumulation of lactic acid, pyruvic acid, and glucose-6- phostate (i.e., citric acid cycle metabolites), increased glucose uptake and glycolysis, and augmented production of glycine and serine [27], which are important in cell proliferation [387,388]. Conversely, siRNA knock-down of FXR abrogated or inverted these processes [27]. Cells in which FXR was knocked down proliferated poorly and did not exhibit the PDK4-mediated metabolic reprogramming [27]. It should be noted that, although FXR-induction of PDK4 is in line with anabolic demand during the liver regrowth phase, hitherto no study has shown that PDK4 enzyme activity is increased after PHx in wild type and FXR- null mice [27]. These studies are still needed to establish a role of the FXR-PDK4 axis in liver regeneration, especially given that PDK4 is transcriptionally regulated by complex signaling mechanisms, and that PDK4 enzyme activity is positively regulated by cellular ATP and NADH levels, which may be increased after PHx.
3.3.1.2. Mitogenic signaling triggered by bile acids in the intestines through enterocytic farnesoid X receptor
Hepatectomized mice in which eFxr was genetically deleted exhibit a considerable reduction in ileal Fgf15 transcript levels and increased hepatic transcript levels of downstream target Cyp7a1 [254] (Cyp7a1 is negatively regulated by hepatic FXR, sections 4.1 and 4.2 and Table 1). These effects coincided with elevated plasma bile acid concentrations and stalled hepatocyte proliferation [254], the latter as a result of impaired cell cycle progression [257]. The deficiency in hepatocyte proliferation could be restored in Fgf15−/− mice by adenoviral transduction of FGF15 [254], underpinning the involvement of intestinal FGF15 in liver regeneration. The results were reproduced in subsequent mouse PHx studies regarding ileal FGF15-mediated hepatocyte and cholangiocyte proliferation [22,257].
As mentioned earlier, activation of eFXR transcriptionally induces FGF15. Using primary hepatocytes, Holt et al. [277] showed that the promoter region of human FGF19 contains an FXR-responsive element in isolated primary hepatocytes. Inagaki et al. [276] followed up with an investigation in mice, demonstrating that, in conjunction with RXR, murine FXR binds to the Fgf15 IR1 motif, a conserved FXR binding site. FXR directly regulates Fgf15 transcription, leading upregulation in intestinal epithelium following oral administration of the FXR agonists GW4064 and CA. Upon production of the functional protein, FGF15 is secreted basolaterally into the portal circulation [276,389] and subsequently binds to FGFR4/β-klotho receptor complex on the outer hepatocyte membrane [390–393] (Figure 6).
Mechanistically, FGFR4-induced mitogenic pathways have been investigated in only a few studies in the context of post- PHx liver regeneration, although proliferative signaling by FGFR4 has been established outside of this context (e.g. [394]). Uriarte et al. [22] observed impaired hepatocyte proliferation in Fgf15−/− mice compared to wild type mice, which was associated with reduced hFxr (6–36 hours post-PHx) and Foxm1b transcript levels (44–48 hours post-PHx) as well as decreased levels of FOXM1B downstream gene targets, including Cdc25b (36–44 hours post-PHx), Ccnd1 (codes for cyclin D1, 24 hours post-PHx), and Pcna (proliferating cell nuclear antigen, involved in DNA replication and repair and chromatin remodeling [ 395 ], 36–44 hours post-PHx) compared to wild type mice (Table 1). Interestingly, transcript levels of the complete hepatic mitogens Hgf (44 and 72 hours post-PHx), Hbegf (6–44 hours post-PHx), and Areg (gene that encodes amphiregulin, 24–36 hours post-PHx) (section 2.3) were upregulated in Fgf15−/− mice [22]. The authors attributed this counter-regulation to the concomitant induction of alternative pro- proliferation pathways (e.g., EGFR signaling) as a reason for the observed, although latent, liver regeneration in Fgf15−/− mice.
Kong et al. [257] observed similar effects in Fgf15−/− mice and identified additional pathways. Fgf15−/− mice exhibited reduced protein expression of the MAPKs JNK and p38 and c- Myc (although p-ERK protein levels were elevated compared to wild type controls), confirming earlier reports on FGF15-induced MAPK signaling [396]. The knock-out mice also had decreased protein levels of JAK1, JAK2, and STAT3, indicating that FGF15 modulates proliferation through the JAK/STAT3 pathway [397,398]. As FXR, STAT3 is a positive regulator of Foxm1b [289] (Table 1), and is apparently activated by FGFR4 to induce cell cycle activity through FOXM1B in an IL-6- and TNF-α-independent manner [23]. Interestingly, STAT3 downregulation in Fgf15−/− mice occurred at augmented plasma IL-6 levels (a STAT3 inducer [84,95,96]) as well as increased hepatocellular SOCS3 levels (a STAT3 inhibitor [399]). Lastly, nuclear p65 protein (RELA, a functional unit of NF-κB [400]) was elevated in wild type animals but absent in Fgf15−/− mice, demonstrating that FGF15 promotes hepatocyte proliferation through NF-κB signaling [87,88] following PHx.
Tissue-specific knock-down and knock-out studies have also been performed for the FGF15 receptor FGFR4 to examine the underlying signaling pathways. Padrissa-Altés et al. [23] employed liposome-delivered siRNA to knock down hepatic FGFR4 in mice and observed comparable gross effects as in the Fgf15−/− mice [22,257] after PHx. Hepatocytes in the FGFR4 knock-down group exhibited reduced and latent proliferation and, besides decreased expression of Foxm1b (48 hours post-PHx) and Stat3 (24 hours post-PHx), decreased levels of Ccna1 and Ccnb1 (codes for cyclins A2 and B1, respectively, 48 hours post-PHx) were also reported. It should be noted that Padrissa- Altés et al. used liver-specific FGFR4 knock-down mice and that these results were not reproducible in Fgfr4−/− mice (which had genetically deleted FGFR4), which did not exhibit aberrant liver morphology or regeneration pattern [401].
Taken together, the studies suggest that at least 4 proliferative pathways are activated by eFXR-induced FGF15 in post-PHx liver regeneration (Figure 6 and Table 1):
1. FGF15/hFXR/FOXM1B → mitosis
2. FGF15/FGFR4/JAK/STAT3/FOXM1B → mitosis
3. FGF15/MAPK (JNK/p38/ERK?) → mitosis
4. FGF15/NF-κB → mitosis.
How FGFR4 is linked to FGF15 and STAT3-, MAPK-, and NF-κB signal transduction, and how FGF15 feeds (direct) signals into these pathways remains to be determined. Moreover, it is important to understand the influence of cytokines and growth factors on these pathways, as these play prominent roles in liver regeneration (sections 2.2 and 2.3).
Several pertinent inflammatory stimuli such as LPS and IL- 1β have been reported to inhibit hepatic β-klotho and FGFR4 expression in mice. In human cell lines (Huh-7 and HepG2), IL- 1β suppressed β-klotho transcription in a JNK- and NF-κB- dependent manner and blocked FGF19-induced ERK1/2 activation and cell proliferation [151]. Finally, a recent study[402] using human liver tissue slices demonstrated a 20-fold transcriptional upregulation of FGF19 following incubation with obeticholic acid, a potent FXR agonist [ 403 ]. To date, FGF19 expression in hepatocytes following FXR stimulation has been quite elusive. These new data, however, suggest that hepatocellular FGF19 may also facilitate autocrine proliferative signaling in the early phase of liver regeneration after PHx.
As a side note, FXR-activated downstream cascades may also amplify liver regeneration by peripheral mechanisms. For example, the human complement C3 gene contains FXR response elements in the proximal promoter region [404]. Complement C3a is elevated after PHx in mice and humans [8–10], possibly in part by the perturbed bile acid homeostasis that ensues after liver surgery [24–26]. C3a activates innate immune cells such as Kupffer cells and neutrophils to stimulate the sterile immune response [8–10] that triggers liver regeneration via IL-6 and TNF-α [36,84,85]. Complement proteins are therefore auxiliary hepatomitogens [9] (section 2.4) induced in part by bile acid-FXR signaling after PHx.
3.3.1.3. Mitogenic and metabolic signaling through hepatic farnesoid X receptor in humans
The promoter of the human FGF19 gene contains an FXR response element [277]. Accordingly, upon FXR stimulation by obeticholic acid, transcriptional upregulation of FGF19 was seen in human enterocytes [402]. Therefore, it is likely that, activated human eFXR induces FGF19 expression in enterocytes. It should be noted that, under cholestatic conditions, FGF19 expression in human hepatocytes has been reported [405]. FGF19 activates ERK1/2 through the FGFR4/ β-klotho receptor complex and subsequently induces hepatocyte proliferation, similar to FGF15-induced cell proliferation in rodents. Whereas β-klotho expression in mice is inhibited by LPS and IL-1β, human β-klotho expression is only affected by IL-1β but not by LPS [151].
Besides regulation of cell cycle progression, human hFXR has additional effects. Due to FXR response elements in the human complement C3 gene [404], complement C3a expression increases after PHx in hepatocytes, enhancing the effect of earlier induced cytokines. Also, FXR is essential for metabolic support of the liver after PHx. PDK4, an FXR target gene, regulates pyruvate metabolism and thus gluconeogenesis. Xie et al. [27] showed that FXR stimulation by both CDCA as well as GW4064 resulted in production of lactic acid, pyruvic acid, and glucose-6-phosphatase in human HepG2 cells. SiRNA knock-down inverted these processes. These results suggest that FXR is important for generation of glucose and amino acids and therefore essential for biomass generation.
3.3.2. Retinoid X receptor signaling in liver regeneration
RXR is a nuclear receptor comprising α, β, and γ isoforms and is activated by mainly 9-cis retinoic acid. Given that RXR is often an obligated heterodimer partner for other nuclear receptors, loss of RXR is expected to affect various nuclear receptor signals in the liver [ 406 ]. The proliferative and cell cycle regulatory properties of RXR have been corroborated in vitro [407], in rodents treated with RXR agonists [408,409], and in rodents with genetically deleted Rxr [348]. Principally, pharmacological induction of RXR leads to hepatomegaly, whereas genetic ablation of Rxr leads to hepatocyte hypoproliferation.
In terms of post-PHx liver regeneration in mice, genetic deletion of Rxra (codes for RXR-α) translated to a 20-hour delay in hepatocyte proliferation that coincided with reduced transcript and protein levels of hepatomitogens (HGF, FGF2 [410], and PDGF) and latent onset of pro-proliferative signaling [347]. Similar results were obtained in lecithin:retinol acyltransferase- deficient (Lrat−/−) mice [411], which lack hepatic retinoid stores and hence possess minimal RXR activation potential. In contrast to the Rxr−/− genotype [347], Lrat−/− mice had decreased levels of Tgfa (codes for TGF-α, a regeneration-promoting growth factor, section 2.3) [411]. Also, the replication forks were stalled in the G1 phase in Rxra−/− mice as a result of compromised PPAR-α/ BMAL1/REV-ERB/p21 cell cycle regulation. This stalled cell cycle was characterized by decreased levels of cyclins A2, -B1, - D1, and -E1 as well as CDK1, −2, and −4 at 48 hours after PHx compared to wild type animals [347]. In mice, PPAR-α is an obligate binding partner of RXR during hepatocyte proliferation [412], so the effects of RXR deletion on PPAR-α signaling and associated cell cycle regulators are warranted. Although supplementation with the endogenous ligand retinoic acid only increased cyclin E1 levels in Rxra−/− mice, wild type mice fed retinoic acid showed RXR-α/RXR-β binding to Cdk1, Cdk2, Cdk6, and cyclin D genes [290], suggesting genetic and epigenetic control of liver regeneration by RXR when all results are considered. Moreover, PHx-induced liver regeneration in wild type mice was associated with RXR-β activation [290] and increased mitotic and proliferative activity via upregulation of cyclins D and E, Cdc25b, c-Myc/c-Myc, and Foxm1b [290,293] (Table 1), despite the fact that retinoic acid levels in the liver drop somewhat after PHx [411]. At this point, no data are available on the role of bile acids in RXR-induced liver regeneration after PHx.
3.3.3. Pro-regenerative signaling by pregnane X receptor
PXR, also referred to as steroid and xenobiotic sensing nuclear receptor, has a plethora of endogenous and exogenous ligands, including bile acids [413]. As the name implies, the chief function of this receptor is to identify toxic substances and consequently upregulate the expression of proteins involved in their detoxification and clearance [302]. The detoxification machinery predominantly entails CYP3A4, which is upregulated by PXR in conjunction with RXR [291]. The potency with which bile acids activate PXR is DCA > LCA > CDCA [414] (Table 1). PXR activation leads to hepatocyte proliferation [295,298].
Dai et al. [349] reported a temporary delay of liver growth in hepatectomized Pxr−/− mice at 36 hours post-PHx and persistently slowed liver growth from post-PHx day 5 onward. This culminated in a 17% reduction in liver mass on post-PHx day 10, which was associated with significantly reduced hepatocyte proliferation and decreased STAT3 protein levels. Wild type livers exhibited transient steatosis post-PHx, which was absent in Pxr−/− mice. PXR is known to positively regulate hepatic lipogenesis via transcriptional induction of PPARγ and CD36 [300,301] (Table 1). Expression of genes that mediate lipid metabolism (Ppar-α, Ppar-γ, fatty acid translocase, acetyl-CoA- carboxylase 1, and long-chain free fatty acid elongase) was lower in Pxr−/− mice after PHx, indicating that impaired fatty acid uptake and lipogenesis may account for delayed liver regeneration in PXR-null mice. Furthermore, a recent liver regeneration study in mice suggested that PXR activation by pregnenolone 16α-carbonitrile accelerated cell cycle activity via inhibition of forkhead box O3 (FOXO3) [296], a transcription factor that negatively regulates cell growth, proliferation, and differentiation in the phosphoinositide 3-kinase (PI3K) pathway [415]. Presently, no data are available on the role of bile acids in PXR-induced liver regeneration after PHx. Nevertheless taken that PHx is followed by a transient bile acid overload [22,24–26] and PXR is activated by bile acids [414], enhanced PXR activity after PHx is not unlikely. However, PXR is under negative control of IL-6 [297] (Table 1), so the proliferative effects of activated PXR may be tuned down by certain cytokines. IL-6 itself is also a mitogen after PHx (section 2.4), and therefore the exact interplay remains to be investigated.
3.3.4. Pro-regenerative signaling by constitutively active/ androstane receptor
CAR is a xenobiotic and endobiotic sensor and therefore transcriptionally regulates genes including CYP2B, CYP2C, CYP3A, sulfotransferases, and glutathione-S-transferases, amongst others involved in detoxification and elimination of such compounds [305–308,314] (Table 1). A unique feature of CAR, as opposed to other nuclear receptors, is that the receptor shows basal activity in the absence of ligands in human hepatocytes, which can be enhanced by the binding of agonists, such as LCA. Following ligand binding, CAR migrates to the nucleus and binds to DNA as a monodimer or CAR/RXR heterodimer to activate transcription of target genes [416]. Mice treated with CAR agonists develop hepatomegaly [417,418], but Car−/− mice paradoxically exhibit only modestly impaired liver regeneration after PHx [28]. CAR also regulates the biogenesis of critical cell components in mice after PHx [419].
Several pathways lie at the basis of CAR-mediated proliferative signaling. One of the better studied pathways entails growth arrest and DNA-damage-inducible, beta (GADD45β), which is a transcription factor with pleiotropic functions that encompass cell proliferation [317], pro-inflammatory signaling [318], and positive transcriptional regulation of nuclear receptors (RXR, CAR, PPARs) [315,316] (Table 1), albeit strongly dependent on the cell type. In the murine liver, GADD45β is profoundly expressed during early compensatory regeneration [419]. Studies in wild type and Tnfr−/− mice demonstrated that CAR activation with the selective CAR agonist TCPOBOP induced GADD45β and cyclin D1 in a TNF(R)-independent manner, which was abolished in Car−/− mice [304]. Furthermore, TCPOBOP-primed mice with deleted Gadd45b (rodent gene for GADD45β) exhibited stalled liver regeneration and transcriptional repression of downstream CAR target genes after PHx, despite intact proliferative signaling [420]. This study further unveiled that both CAR and GADD45β bind the CAR regulatory element of the Cyp2b10 gene, underpinning the signal-amplifying role of GADD45β in terms of CAR signaling [420] (Table 1).
Another CAR signal amplifier is steroid receptor co-activator-3 (SRC-3) [421], a transcriptional co-activator that assists nuclear receptors in the upregulation of gene expression [422]. Src−/− mice present with reduced liver hyperplasia and decreased c-Myc (section 2.2) and FOXM1B (section 3.1.1) expression upon CAR activation with TCPOBOP [421] (Table 1). The same applies to β-catenin that, when co-activated with CAR, induces hepatocyte proliferation and hepatomegaly in mice [423]. An additional proliferative trigger for CAR-controlled mitogenesis may be oxidative stress [424], which is induced in the early regeneration phase (section 2.1). Finally, the inhibition of the nuclear receptor hepatocyte nuclear factor 4 (HNF-4)α by TCPOBOP-activated CAR was shown to downregulate miR-122 and upregulate corresponding promitogenic target genes (the transcription factor E2f1 and its downstream target c-Myc) in the murine liver [322]. This is in agreement with the data presented in section 2.4, where miR-122 downregulation was associated with a post-PHx pro-regenerative response [200,201]. It should be noted, however, that HNF-4α levels remain relatively stable during the early phase of post-PHx regeneration, as measured in rats [180].
Unfortunately, all abovementioned studies were conducted in the absence of PHx and relied solely on CAR activation, usually by TCPOBOP, as a result of which no definitive conclusions can be drawn on the role of CAR in post-PHx liver regeneration. The only study that used PHx in combination with Car−/− mice, aside from Huang et al. [28], examined the role of type 1 deiodinase and thyroid hormone activity in liver regeneration [350]. This investigation demonstrated that reverse tri-iodothyronine (rT3) levels rise after PHx in wild type and Car−/− mice, which could be reversed in wild type animals but not Car−/− mice with phenobarbital, an indirect CAR agonist. Levels of T3, a thyroid hormone that acts as a hepatic (auxiliary) mitogen in the post-PHx liver regeneration setting [ 425, 426 ], remained unaltered independently of CAR. The activity and expression of type 1 deiodinase, the enzyme that catalyzes the production of the endocrinologically potent T3 as well as the endocrinologically less potent rT3 from thyroxine (T4), was reduced by PHx and increased by phenobarbital in a CAR-dependent manner. The same applied to promitogenic T3 target genes, which were repressed after PHx as well as rT3 infusion but reactivated by phenobarbital, altogether implicating CAR in hormonal regulation of liver regeneration in mice. To add to the regulatory complexity, the proliferative activity of T3 is enabled by β-catenin [ 427 ], which in turn potentiates the mitogenic signaling of CAR in mice [423] as described in the previous paragraph. Evidently, the thyroid hormone/hepatic nuclear receptor signaling axes are still to be fully unraveled. How bile acids fit into these signaling networks is most elusive in terms of post-PHx liver regeneration.
3.3.5. Pro-regenerative signaling by suppression of liver X receptor
LXR exists as an α and β isoform [428] that forms obligate heterodimers with RXR upon ligand activation to regulate gene expression [ 429 ]. The cognate LXR ligands are the sterols 24(S),25-epoxycholesterol, 22(R)-hydroxycholesterol and 24(S)-hydroxycholesterol, but the receptor also binds several metabolized bile acid species, including hyodeoxycholic acid, taurohyodeoxycholic acid, and cholestenoic acid [319,320] (Table 1).
Although hepatectomized Lxr−/− mice do not exhibit impaired hepatocyte proliferation and liver regeneration compared to wild type controls, LXR activation in wild type mice with the selective LXR agonist GW3965 (Table 1) reduced hepatic levels of pro-proliferative STAT3, the cell cycle regulators FOXM1B and CDC25B (section 3.3.1.1), and the tissue remodelling protein matrix metalloproteinase 9 (MMP-9) [239], indicating that LXR exerts proliferation-repressive effects. The cause for LXR inactivation after PHx is rooted in the considerable decrease in sterol-based LXR ligands, namely 24(S),25- epoxycholesterol, 24(S)-hydroxycholesterol, and 27- hydroxycholesterol (Table 1), which occurs in the early phase of post-PHx liver regeneration in mice [239].
How bile acids mediate post-PHx liver regeneration in terms of LXR signaling is unclear, although one study suggests that LXR downregulation in mice may be mediated by bile acid overload-induced overexpression of nuclear receptor-interacting protein 1 (NRIP1) [401]. NRIP1 is a nuclear receptor that represses the activation of transcription by LXRα/RXRα as well as PPAR-α/RXRα heterodimers through its interaction with LXRα and PPAR-α [430]. In a mouse model, NRIP1 is considerably induced by dietary CA [401], a bile acid that is substantially elevated after PHx during the early phase of liver regeneration [24]. Consequently, bile acid-induced overexpression of NRIP1 after PHx may contribute to suppression of LXR-mediated gene transcription and consequent promotion of hepatocyte proliferation and liver regeneration.
3.3.6. Pro-regenerative signaling by non-farnesoid X receptor nuclear receptors in humans
Pascussi et al. [297] showed that IL-6 inhibits expression of PXR and CAR in primary human hepatocytes. However, in contrast to FXR signaling, there is hardly any information on the links between non-FXR nuclear receptors in humans and PHx. In few studies, human cell lines were used to investigate nuclear receptor activity. For example, Frank et al. [416] showed that, in contrast to other nuclear receptors, CAR is continuously expressed in the cytoplasm of human breast cancer cells. Upon ligand binding, CAR translocates to the nucleus and activates transcription of target genes. Osabe and Negishi [ 431 ] used hepatocellular carcinoma cells to study ERK1/2-CAR interaction. Their results show that the ERK1/2 protein prevents CAR phosphorylation and subsequently avoids nuclear translocation of CAR. Yamamoto and Negishi [316] also studied CAR in the context of cell cycle progression. When CAR is stimulated, GADD45B gene expression in HepG2 cells is decreased. As described in section 3.3.4, GADD45β is a transcription factor involved in cell cycle progression [317] and expression of several nuclear receptors [315,316] (Table 1).
3.4. Role of bile acid-activated TGR5 in post-PHx liver regeneration
In addition to intracellular nuclear receptors, bile acids also activate a cell surface G protein coupled receptor TGR5 [432]. Activation of TGR5 in most target cells results in increased intracellular cAMP and activation of protein kinase A (PKA). TGR5 is expressed in the brown adipose tissue, muscle and the intestine where activation of TGR5 by agonists promotes energy expenditure and glucose homeostasis [433,434]. TGR5 is highly expressed in macrophages and activation of TGR5 inhibits cytokine production and inflammation [435–437]. Pharmacological activation of TGR5 has been shown to decrease inflammation and prevent atherosclerosis in mice [435]. In the liver and biliary system, TGR5 is highly expressed in cholangiocytes, Kupffer cells, and gallbladder epithelial cells, but not hepatocytes [437–440 ]. Activation of TGR5 is known to decrease cytokine production and Kupffer cell activation to decrease hepatic inflammation in mice [436]. Compared to adipocytes, myocytes, and macrophages that are usually exposed to low levels of bile acids in the systemic circulation, cholangiocytes are routinely exposed to high levels of bile acids, and bile acid signaling through TGR5 in cholangiocytes may be more pathophysiologically relevant. So far, studies have shown that TGR5 mediates the proliferative and anti-apoptotic role of bile acids in cholestatic liver diseases [26,441], which may on one hand protect cholangiocytes against bile acid toxicity but on the other hand promote cholangiocarcimona development and progression. After PHx, TGR5 KO mice showed significantly impaired regeneration, with increased liver injury and inflammation. Such detrimental effects of TGR5 knockout could be attributed to cholestatic liver injury upon bile acid overload after PHx, suggesting an important role of TGR5 in regulating bile flow and cholangiocyte cell death [26]. In addition, TGR5 KO mice have been shown to have a more hydrophobic bile acid pool, which may further predispose TGR5 KO mice to bile acid- induced toxicity after PHx. The mechanisms by which alterations in bile acid composition in TGR5 KO mice are caused still have to be elucidated.
4. Containment of post-hepatectomy bile acid toxicity in hepatocytes
To protect the liver from bile acid-induced hepatotoxicity [223], bile acid-laden hepatocytes have several mechanisms in place to curtail prolonged exposure to supraphysiological intrahepatic levels of bile acids. These mechanisms, which are an integral part of hepatocyte function [1,442–444] in terms of phase I-III endobiotic metabolism, entail:
-
–
chemically altering the composition of the bile acid pool towards less toxic (more hydrophilic) species (phases I and II, section 4.1);
-
–
regulation of substrate (cholesterol) availability for de novo bile acid synthesis (section 4.2);
-
–
hepatobiliary transport (phase III, section 4.3).
These processes work in concert to gradually remove excessive bile acids from hepatocytes after PHx once the mitogenic signals have been relayed.
4.1. Modulation of hepatocellular bile acid composition during liver regeneration
Due to the transient bile acid overload after PHx [25,26], several nuclear receptors are activated and engage phase I and II metabolic processes to detoxify bile acids by chemical modification. Phase I bile acid detoxification is mediated mainly by CYP3A4, which hydroxylates the bile acids to more hydrophilic entities that are subsequently readily eliminated by the liver [445–448]. CYP3A4 expression is regulated by FXR [449] (section 3.3.1), PXR [291,295,414, 450 ] (section 3.3.3), CAR [299,309–311] (section 3.3.4), and VDR [333,451]. The nuclear receptors are activated after PHx as described in the referenced sections. CYP3A11 is also induced by bile acids such as LCA [295,414, 452 ], while CYP3A4 is induced by CDCA [449] - effects that have been mirrored in obstructive cholestasis in mice and human livers [453,454] - as a feed-forward protective mechanism against bile acid-induced hepatotoxicity.
Phase II metabolic neutralization of bile acid toxicity occurs through primarily sulfation and glucuronidation at the sterol’s hydroxyl groups [455–457]. Sulfation is catalyzed by sulfotransferase (SULT)2A1 [458,459] that is under positive transcriptional control of PXR and CAR [312,358, 460 – 463 ], although negative regulation of SULT2A1 by FXR has been reported in mice [459] (Table 1). Glucuronidation is restricted to LCA- and CDCA-derived 6α-hydroxylated bile acids [295,414] and is catalyzed by UDP-glucuronosyltransferase (UGT)2B4 and UGT2B7 [464,465]. Several UGT isoforms in the human liver are under control of PXR and CAR [ 466 ], whereby UGT2B4 is positively regulated by FXR [280] and PPARα [332]. UGT2B7 seems to be repressed by bile acid- activated FXR [281]. In transgenic mice, the human UGT2B7 gene was suppressed by CAR, which was subsequently shown to proceed via CAR-mediated inhibition of HNF-4α binding to the UGT2B7 promoter in HepG2 cells [313] (Table 1). Accordingly, CYP3A4 and UGT2B4 seem to contribute to hepatoprotection during post-PHx bile acid overload.
Some of the nuclear receptors that govern the detoxification processes also regulate the excretion of the biotransformed bile acids (transporters are discussed in detail in section 4.3). For example, CAR activation leads to the overexpression of MRP4 (basolateral export of sulfated sterols) [312] and MRP2 (canalicular export of glucuronidated bile acids [467]) in mice [299], while activated PXR upregulates the murine basolateral exporters MRP3 and MRP4 [299], which transport sulfate- and/or glucuronite- conjugated bile acids out of hepatocytes [312,338, 468 ]. The regulation of bile acid transporters by nuclear receptors is also illustrated in Figure 4 and Table 1.
Finally, bile acid composition in mice is regulated by LRH-1 [263,267], but this regulation is expected to offset the hepatoprotection conferred by the nuclear receptors and TGR5. LRH-1 is a nuclear receptor in the liver [258] and intestines [469] that regulates bile acid homeostasis [258,260,263–267,470,471] through CYP7A1 [260,263–266] and CYP8B1 [267] (Table 1). CYP7A1 is the rate-limiting enzyme that catalyzes hepatic bile acid synthesis using cholesterol as substrate [225, 472 – 474 ]. CYP8B1, abundantly expressed in hepatocytes [475], is also responsible for bile acid synthesis [225,472]. Murine LRH-1 is repressed by CDCA [258] but upregulated by TNF-α under cholestatic conditions [261] (Table 1). Accordingly, LRH-1 is expected to affect bile acid composition after PHx. Mice in which Lrh1 has been deleted exhibit increased relative amounts of hydrophobic bile acid species at the expense of reduced CA and taurocholic acid (TCA) in the bile acid pool [267]. In light of the transient bile acid overload after PHx, LRH-1 levels are expected to drop because of augmented FXR and small heterodimer partner (SHP) activation by bile acids (next section). The subsequent SHP-induced repression of LRH-1 [259,260] may therefore lead to a shift in bile acids to more hydrophobic species and skew the beneficial effects imparted by nuclear receptors and TGR5. However, numerous bile acid exporters are also hyperactivated during bile acid overload, as explained in section 4.3, so saturated hepatocytes may not necessarily have to be afflicted by the LRH-1 downregulation and the more toxic bile acid pool. At this point the exact function of LRH-1 during post-PHx liver regeneration warrants further investigation, particularly in the context of bile acid composition and hepatotoxicity.
4.2. Regulation of bile acid synthesis during liver regeneration
PHx results in transient bile acid overload in hepatocytes [25,26] as well as extensive hepatocellular cholesterol accumulation [293, 476 ]. While bile acids are instrumental in proliferative signaling and liver regeneration (section 3), they are also toxic to hepatocytes. This biochemical ambiguity also applies to cholesterol, the metabolic substrate for bile acid production [477]. On the one hand, cholesterol is exacted by mitotic hepatocytes for biomass generation and sustenance of various metabolic functions [412, 478 ]. That mitotic hepatocytes increase their chromatin cholesterol levels in order to facilitate proliferation (8–16 hours after PHx) [ 479 ], which is why cholesterol metabolic pathways are hyperactivated after PHx [209,293,476, 480, 481 ]. On the other hand, high cholesterol levels feed into the production of (potentially toxic) bile acids in hepatocytes, especially since CYP7A1 (the protein product of the CYP7A1/ Cyp7a1 gene) is strongly upregulated 24 hours after PHx [482]. CYP7A1 is the rate-limiting enzyme in the bile acid synthesis pathway, using cholesterol as substrate [225]. Its upregulation in combination with elevated cholesterol levels [293, 476] may therefore account for the temporarily increased bile acid pool early after PHx.
4.2.1. Increased hepatocellular cholesterol levels after partial hepatectomy
Hepatocytes control intracellular levels of cholesterol via synthesis and non-synthesis routes. The non-synthesis routes pertain to cholesterol import into and export out of hepatocytes. A study in rats found that the binding and uptake of cholesterol- enriched lipoproteins by hepatocytes isolated from the remnant liver was decreased early (16 hours) after PHx and restored to baseline levels in later phases of regeneration [483], suggesting that cholesterol uptake from the systemic circulation does not lie at the basis of post-PHx cholesterol loading in the early stages. However, there are at least 3 metabolic switches manifested at the level of synthesis and export during post-PHx liver regeneration.
In terms of the first metabolic switch, the increase in hepatocellular cholesterol is mediated by suppression of LXR signaling after PHx in mice [293]. As addressed in section 3.3.5, LXR is inactivated after PHx in mice, associated with the promotion of liver regeneration. One potentially contributing factor to the inactivation of LXR is upregulation of NRIP1 [401] (section 3.3.5), while another contributing factor is the substantial post-PHx reduction in LXR-activating ligands in hepatocytes [293]. The inactivation of LXR leads to increased cholesterol levels because LXR is a master regulator of cholesterol catabolism [484] and an inhibitor of cholesterol synthesis [485]. Second, intracellular cholesterol levels are coordinated by LRH-1, which positively modulates sterols through ABCG5 and ABCG8 [269]. Importantly, LXR also positively regulates the expression of Abcg5 and Abcg8 in mice [321] (Table 1). These ATP binding cassette half-transporters are replete in the liver and small intestine [244,486,487] and jointly transport cholesterol across the hepatocellular cell membrane into the canalicular system [319,321,488,489]. Indeed, ABCG5/ABCG8 deficiency in mice leads to augmented plasma sitosterol (a plant-derived cholesterol analogue) concentration and decreased cholesterol levels in bile [490]. Analogously, ABCG5/ABCG8 overexpression increases biliary cholesterol secretion and decreases cholesterol absorption [491]. Given that LRH-1 is negatively regulated by bile acid-activated FXR/SHP signaling [260,266] (Table 1), bile acids (e.g., DCA) may downregulate Abcg5 and Abcg8 [269] through FXR/SHP/LRH-1 during bile acid overload, a process that is aided by the concomitant downregulation of LXR in mice [293]. Although no information is available on LHR-1 in connection with ABCG5 and ABCG8 during post-PHx liver regeneration, it is anticipated that ABCG5 and ABCG8 are downregulated via bile acid-mediated suppression of LRH-1 signaling in early phases. Accordingly, Lo Sasso et al. demonstrated that Abcg5 and Abcg8 levels were dramatically decreased in hepatectomized mice [293], although in that study the repression was ascribed to LXR.
The third, albeit converse metabolic switch, is that hepatocytes reduce their cholesterol synthesis rate in the early phase of post- PHx liver regeneration (first 24 hours) [481,492]. Evidently, the net effect of the rate reduction is in itself not sizeable enough to decrease the hepatocellular cholesterol concentration in the very short term inasmuch as cholesterol levels peak at 1–3 days after PHx (in mice) [293]. However, in the longer term (72 hours after PHx and beyond) the cholesterol concentrations decline [293], as do intrahepatic bile salt levels [24].
In summary, the three metabolic switches after PHx are:
-
–
ligand ↓ / NRIP1 ↑ → LXR ↓ → cholesterol synthesis ↑ / cholesterol catabolism ↓;
-
–
bile acids ↑ → FXR ↑ → SHP ↑ → LXR ↓ / LRH-1 ↓ → ABCG5 ↓ / ABCG8 ↓ → cholesterol export;
-
–
cholesterol synthesis ↓.
In light of these pathways, it is clear that hepatocytes regulate bile acid synthesis by mainly enzymological means (i.e., regulation of CYP7A1, next section) and transport (section 4.3) rather than through substrate control.
4.2.2. Inhibition of bile acid synthesis during liver regeneration.
Bile acid production is susceptible to a negative feedback loop that that reduces bile acid accumulation and decreases the bile acid pool size [260,265,266,493]. This is mainly achieved by transcriptional inhibition of Cyp7a1/CYP7A1, which encodes the rate-limiting enzyme in the neutral bile acid synthesis pathway[494,495]. As mentioned earlier, bile acid activation of hFXR induces SHP, which inhibits the transactivational activity of LRH-1 and HNF4-α that are key stimulators of Cyp7a1/CYP7A1 transcription [259,260,266] (Table 1). In addition, elevated intestinal bile acid levels inhibit hepatic CYP7A1 via the FXR- FGF15/19 gut-to-liver signaling axis [266,276,277,496]. Accordingly, the post-PHx bile acid overload in the liver [25,26] coincides with pleiotropic transcriptional repression of Cyp7a1 [28]. In vivo, intestinal FXR/FGF15/19 appears to be the dominant route by which bile acids repress CYP7A1 in comparison to the hepatic FXR/SHP/LRH-1 route. Using tissue- specific knockout mice, Kim et al. demonstrated that FXR activation in the intestines but not the liver is necessary for short- term downregulation of hepatocellular Cyp7a1 in mice [496].
In addition, murine Cyp7a1 downregulation is also controlled by PXR, another nuclear receptor that is activated by LCA [295]. Correspondingly, bile acid species that have been reported to induce Cyp7a1 repression include CDCA [324,497], DCA [498,499], and LCA [295], which constitute physiological ligands of FXR and/or PXR (section 3.3).
The bile acid response element of CYP7A1/Cyp7a1 [323,324] is also targeted by HNF-4α, a nuclear receptor involved in lipid and glucose metabolism [328,329] (Table 1). CDCA can decrease HNF-4α-regulated downstream genes [325,326,500 ], indicating that this nuclear factor is involved in bile acid-mediated repression of CYP7A1. Despite a single study reporting unperturbed HNF-4α levels in the early post-PHx regeneration phase [180], mice in which Hnf4a (encodes HNF-4α) was genetically deleted exhibit derailed bile acid homeostasis [328,329]. HNF-4α further regulates CYP8B1/Cyp8b1 and CYP27A1/Cyp27a1, that encode the acidic pathway regulating enzyme CYP27A1, via a HNF-4α binding site in the promotor region [325–327]. The promotor region of CYP8B1 also contains a binding site for LRH-1 [268], which is increased on day 2–3 after PHx in mice [482]. Elevated levels of LRH-1 are therefore expected to augment CYP8B1 expression and promote bile acid synthesis. However, this signaling axis may be offset by negative CYP8B1 regulation by FXR/SHP-dependent and independent pathways [501]. In vivo, HNF-4α and LRH-1 facilitate SHP binding to the Cyp7a1 promoter and drive FGF15-mediated repression of bile acid synthesis [471]. Taken together, bile acid synthesis is suppressed during the early phase of post-PHx liver regeneration (Table 1).
Other negative mediators of Cyp7a1 include NRF2 [331] (Table 1) that is overexpressed during the early phase of liver regeneration following PHx in mice [71–73,82] (section 2.1) and in part activated by certain bile acids such as UDCA [502]. Cytokines such as TNF-α [498] and IL-1β [497] (section 2.2) downmodulate Cyp7a1 in rats through pathways involving FAS receptor/JNK/c-Jun [498,499] and HNF-4α [324,497]. Additionally, the growth factors TGF-β1 [ 503 ] and HGF [ 504 ] (section 2.3) downregulate CYP7A1/Cyp7a1 through signaling cascades involving HNF-4α (induced by TGF-β1) [503] and protein kinase C (PKC)/ERK1 and 2/JNK (induced by HGF) [504], at least in human hepatocytes [505].
4.3. Regulation of bile acid transport during liver regeneration
To further deter the toxicity of bile acids [223], overloaded hepatocytes instate protective routes to limit bile acid influx at the basolateral end and eliminate excessive bile acids from the intracellular milieu via basolateral and canalicular exporters [355,506]. This process is also referred to as phase III metabolism.
4.3.1. Regulation of basolateral bile acid import
As described before, the extraction of bile acids from the portal circulation by hepatocytes is facilitated by NTCP (encoded by SLC10A1/Slc10a1), and OATP isoforms OATP1B1 (SLCO1B1), OATP1B3 (SLCO1B3), and OATP2B1 (SLCO2B1)[507–510], all located at the basolateral membrane of hepatocytes [238,509,510]. Studies in rats have shown that the expression of NTCP [506,511–513], OATP1, and OATP2 [506,512]considerably decline during post-PHx, early-phase liver regeneration (0.5–2 days). Additionally, expression of the specific isoforms OATP1B1 and OATP1B3 is decreased under cholestatic conditions [437,514]. Therefore, NTCP and OATP isoforms may prevent bile acid loading from the enterohepatic circulation. Accordingly, a significant rise in plasma bile acids has been observed in mice and rats within several hours after PHx [25,26]. Similar to regulation of CYP7A1, NTCP is transcriptionally downregulated by bile acids (Figure 4). The route by which bile acids suppress NTCP is multilayered and complex, involving FXR/SHP/LRH-1, HNF-1α, and HNF-4α [355,515]. In addition, NTCP is negatively regulated by TNF-α and IL-1β [ 516 ] but not NRF2 [331]. The same applies to OAT1B isoforms in terms of IL-1β [517] (Table 1.). It therefore seems that, during regeneration, the net expressional effect on these transporters is chiefly dictated by cytokine networks rather than metabolic networks (i.e., bile acid signaling).
4.3.2. Regulation of basolateral bile acid export
Under normophysiological conditions, basolateral bile acid efflux is maintained at a very low rate, as most of the bile acids undergo canalicular secretion. Upregulation of basolateral bile acid efflux transporters is considered as an adaptive response to secrete bile acids into the systemic circulation for renal excretion. As is shown in Figure 4, the basolateral efflux of bile acids from hepatocytes is mediated by the transporters MRP3 (ABCC3/ Abcc3) [518–520] and MRP4 (ABCC4/Abcc4) [468], both located at the basolateral membrane of hepatocytes [521,522] and both under positive control of e.g., NRF2 in mice [331] (section 2.1 and Table 1). Murine Abcc3 is under negative control of IL-1β [516] and positive control of CAR [523] (section 3.3.4 and Table 1) and TNF-α via LRH-1 [261] (section 3.3 and Table 1), both implicated in post-PHx liver regeneration, albeit to different degrees. Following PHx in mice, MRP3 and MRP4 levels were shown to have increased up to 3.1-fold [524,525], which was associated with increased plasma bile acid levels [524]. Similar results were found for MRP4 in both mice and rats [273,513]. Since OATPs are bilateral exchangers, the previously mentioned A and B isoforms may also mediate basolateral efflux of bile acids [355], albeit to a limited extent because of their post-PHx downregulation [506].
The OSTα-OSTβ heterodimer is also located at the basolateral membrane of hepatocytes [278, 526 ] and has been shown to transport bile acids [247] out of bile acid-overloaded hepatocytes [278]. As in the intestines (section 3.1.1), OSTα and OSTβ are under positive transcriptional control by several bile acid-activated nuclear receptors, including FXR and LRH-1 [262,273,278] (section 3.3 and Figure 4). The transporter complex is further induced by nuclear receptor ligands that are not activated by bile acids but do play a role in post-PHx liver regeneration, including glucocorticoid receptor (GR) [526]. Hepatic GR levels exhibit a considerable increase during the first 24 hours after PHx in rats [527], suggesting concomitant upregulation of OSTα-OSTβ and facilitated export of (toxic) bile acids. Contrastingly, OSTα-OSTβ are repressed by SHP [262] and IL-1β in mice [516] (section 2.2). The positive and negative regulation notwithstanding, the involvement of OSTα-OSTβ in bile acid flux modulation under conditions of PHx remains to be proven experimentally. Based on our knowledge and expertise, OSTα- OSTβ are induced by FXR and bile acids, where LCA plays a marginal-to-no role in terms of liver regeneration. LRH likely maintains basal transcriptional activity of the OST isoforms, but does not play an important role in response to bile acids.
In sum, these basolateral transporters, or at least MRP3 and MRP4, ensure that bile acids are pumped out of the cell back into the systemic circulation. All the abovementioned basolateral transporters are capable of also removing sulfated and glucuronidated bile acids from hepatocytes [528], indicating that the transporters work in concert with the hepatocyte detoxification machinery (section 4.1) to remove (conjugated) bile acids. The conjugated bile acids can subsequently be removed from the circulation by renal clearance [529–531].
4.3.3. Regulation of canalicular bile acid export
Expression of BSEP, an export pump that traffics bile acids from hepatocytes into the biliary tree, is positively regulated by FXR [256,532,533], which heterodimerizes with RXR to bind the promotor region of ABCB11 [256], as well as by NRF2 [330,331] and LRH-1 [259] (Table 1). While one study reported only slightly increased levels of BSEP after PHx in rats [506], FXR and NRF2 are activated during post-PHx liver regeneration [71–73,82,254,534] and thus may promote canalicular export of bile acids via BSEP in bile acid-congested hepatocytes post-PHx [370,506,534,535]. In support of the BSEP-mediated canalicular export of bile acids, Monte et al. demonstrated that bile acid levels in bile rise during the first 1–3 days after PHx in rats, and subsequently decline to near-baseline levels during the subsequent 11–13 days [24,482]. Correspondingly, mice fed CA and UDCA exhibit increased BSEP expression and elevated bile acid levels in bile [535,536].
In rats, increased canalicular export of bile acids from bile acid-overloaded hepatocytes also applies to MRP2 [506]. The same effect of CA and UDCA feeding to mice has been observed for hepatocellular MRP2 expression as was reported for BSEP expression [535,536]. Although this transporter is under negative control of IL-1β [516] and only one study reported unaltered or slightly increased BSEP mRNA levels after PHx [506], Abcc2 (MRP2) is under positive control of several nuclear receptors that are activated during post-PHx liver regeneration (section 3.1), including RXR [292], FXR [279], PXR [279], and CAR [279,537], as well as the bile acids that activate these receptors and/or Abcc2 directly [279,535,538,539] (Table 1). It therefore stands to argue that MRP2 is upregulated during the bile acid overload following PHx and assists in canalicular export of (toxic) bile acids. Finally, PHx in rats results in a considerable (up to 20-fold) increase in Mdr1 mRNA levels in the remnant liver [540,541], but the significance of this upregulation in the context of bile acid trafficking during post-PHx liver regeneration is presently elusive.
4.4. Containment of post-hepatectomy bile acid toxicity in human hepatocytes
The modulation of hepatocellular bile acid composition during liver regeneration occurs through various enzymes that mediate phase I and II metabolic processes [445–448]. Phase I, hydroxylation, is mediated CYP3A4 that is under positive control of both bile acids and FXR in human hepatocytes [449]. Phase II metabolic reactions include sulfation, mediated by SULT2A1 [458,459] isoforms, and glucuronidiation, mediated by the UGT isoforms UGT2B4 and UGT2B7 [464,465]. While FXR [280] and PPAR-α[332] positively regulate UGT2B4 expression, FXR negatively regulates UGT2B7 expression [281]. Inhibition of UGT2B7 was also seen as a result of CAR-mediated HNF-4α inhibition in HepG2 cells [313] (Table 1). These results suggest that bile acid metabolism is tightly regulated after PHx in humans in order to protect the liver from the post-PHx relative bile-acid overload. Besides, CYP3A4 and CYP3A11, CYP7A14 (the gene that encodes the enzyme that regulates bile acid synthesis from cholesterol) also contains a bile acid response element in its promotor region, suggesting that human hepatocytes also regulate bile acid synthesis after PHx though a negative feedback loop.
Additionally, basolateral bile acid import and export are well-controlled after PHx in humans. Human bile acid transporters have been identified and classified [239,508,510]. However, the expression of these transporters in humans has only been investigated in the context of cholestasis [278,542], and expression levels have not yet been measured after PHx. Chen et al. [542] investigated the expression of hepatocyte transporters and nuclear receptors in children with biliary atresia, a disease that leads to cholestasis. In early-stage cholestasis, expression levels of ABCB11 (BSEP), ABCC2 (MRP2), and SCLO-isoforms (OATP) where increased, suggesting that in situations of increased relative exposure to bile acids, human hepatocytes downregulate bile acid resorption. In HepG2 cells, FXR/RXR heterodimers positively regulate ABCB11 (BSEP) [256,533]. CDCA induces ABCB11 expression through FXR, whilst LCA works as a CDCA antagonist and decreases BSEP expression [533]. As a result of FXR activation, expression levels of OSTα-OSTβ also increase due to cholestasis in humans [278]. Human OSTα-OSTβ are also under positive control of GR, which is important after PHx [526].
5. Concluding remarks
Liver regeneration following PHx is a highly complicated process involving multiple organs and several types of signaling networks. The metabolic pathways that revolve around bile acid signaling occupy an ancillary yet important part in post-PHx liver regeneration.
The metabolic cues for liver regeneration are given off by bile acids directly after PHx and are hence facilitated by the temporary bile acid overload that lasts for up to 48–72 hours. The bile acids induce hepatocyte proliferation and liver regeneration through several nuclear receptors, either by direct binding or by indirect activation. Inasmuch as bile acids are inherently toxic, the state of bile acid hypersaturation cannot be sustained by the remnant hepatocytes that, while proliferating, also have to properly execute all aspects of liver function. Consequently, once the proliferative signals have been relayed by the bile acids, hepatocytes activate a bile acid detoxification and elimination machinery in order to protect themselves. These processes, induced in part by bile acids and nuclear receptors, encompass phase I-III endobiotic metabolism that predominantly results in the excretion of (biotransformed) bile acids into the canalicular system.
The excreted bile acids subsequently translocate to the intestines, where they mount a secondary wave of proliferative signaling in the liver. This system is based on the enterocytic reabsorption of bile acids, activation of FXR and consequent production of FGF19/FGF15, its release into the enterohepatic circulation, and the basolateral binding of FGF19/FGF15 to its cognate receptor, a complex of FGFR4/FGFR4 and β-klotho, on hepatocytes.
6. Future implications
Further research on the abovementioned pathways may have beneficial implications for various liver diseases and surgical procedures such as PHx. This last chapter will highlight points of interest for further research.
Firstly, no data are available on the role of several nuclear receptors in liver regeneration after PHx. Additional research on the effects of RXR, PXR, CAR, and LRH-1 in PHx-induced liver regeneration could help with understanding the role of bile acids in liver regeneration. It was shown that CAR influences thyroid hormone signaling [28,350], but further research has to be done to find out how the thyroid hormone/hepatic nuclear receptor signaling axis works. The kinetics of LRH-1 still have to be investigated, especially in the context of bile acid composition and hepatotoxicity. How bile acids influence liver regeneration remains unclear for all nuclear receptors except for FXR.
Secondly, several implications have been made on how bile acid flux is modulated under conditions of PHx. The significance of bile acid transporters in liver regeneration such as NTCP, OATP isoforms, MRP-2, −3 and −4, OSTα-OSTβ, BSEP, and MDR-1 remains to be proven experimentally.
Thirdly, it remains unclear how FGFR4, the FGF15/ FGF19 receptor, feeds signals into STAT3-, MAPK-, and NF-κB pathways and how cytokines and growth factors fit into these pathways. Ligand binding to this receptor initiates proliferative signaling that has only been partially elucidated in terms of mechanisms. This aspect of liver regeneration therefore warrants further research, especially since several FXR agonists have recently become available that promote liver regeneration via the gut-liver axis. Since cytokines and growth factors play prominent roles in liver regeneration (section 2.2), it is important to elucidate these pathways.
Finally, it was shown that downregulation of PDK4, a protein that regulates pyruvate metabolism, promotes liver regeneration [543]. Another study reported upregulation of PDK4 as a result of bile acid induced FXR activation [27], a mechanism that is in line with the anabolic demand during the liver regrowth face. It would be useful to further investigate the effects of PDK4 on liver regeneration in a PHx model since no study has shown that PDK4 enzyme activity is increased after PHx. Other signals that need further investigation entail the mechanism through which TGR5 confers biochemical protection during liver regeneration.
Both in vitro and in vivo studies are required for a better understanding of the importance of bile acids and their effects and effectors in the context of liver regeneration after PHx. Further elucidation of these pathways could be beneficial for patients with various liver diseases, including patients that have undergone a PHx.
Acknowledgements
The authors are grateful to Ms. Inge Kos for the illustrations and the reviewers for providing instrumental comments. MH is sponsored by a grant from the Dutch Anti-Cancer Society (grant #10666).
List of abbreviations:
- AKT
protein kinase B
- AP-1
activator protein 1
- ASBT
apical sodium-dependent bile acid transporter
- ATF2
activating transcription factor 2
- BSEP
bile salt export pump
- CA
cholic acid / cholate
- CAR
constitutive androstane receptor
- CDCA
chenodeoxycholic acid / chenodeoxycholate
- CDK
cyclin-dependent kinase
- C/EBP
CCAAT-enhancer-binding protein
- c- Met
hepatocyte growth factor receptor
- COX
cyclooxygenase
- CPF
CYP7A promoter-binding factor
- CREB
cAMP response element-binding protein
- CYP7A1
cholesterol 7 alpha-hydroxylase
- DAMPs
damage-associated molecular patterns
- DCA
deoxycholic acid / deoxycholate
- E2f
E2f transcription factor
- ECM
extracellular matrix
- EGF
epidermal growth factor
- ER
endoplasmic reticulum
- ERK
extracellular signal-regulated kinases
- FAS receptor
apoptosis antigen I
- FGF
fibroblast growth factor
- FGFR
fibroblast growth factor receptor
- FOXM1B
forkhead box M1B
- FOXO3
forkhead box O3
- FTF
fetoprotein AFP transcription factor
- FXR
farnesoid X receptor
- GADD45β
growth arrest and DNA damage-inducible, beta
- GR
glucocorticoid receptor
- H2O2
hydrogen peroxide
- HBEGF
heparin-binding EGF-like growth factor
- HGF
hepatocyte growth factor
- HNF
hepatocyte nuclear factor
- HSP
heat shock protein
- IGF
insulin-like growth factor
- IGFBP1
insulin-like growth factor binding- protein 1
- IL
interleukin
- IRE1α
inositol-requiring enzyme-1α
- JNK
c-Jun amino-terminal kinase
- JUN
JUN proto oncogene
- JAK
Janus kinase
- LCA
lithocholic acid / lithocholate
- LPS
lipopolysaccharide
- LRH-1
liver receptor homolog 1
- LXR
liver X receptor
- MAPK
mitogen-activated protein kinase
- MDR-1
multidrug resistance protein 1
- miRNA
microRNA
- MMP-9
matrix metalloproteinase 9
- mRNA
messenger ribonucleic acid
- MRP
multidrug resistance protein
- mTOR
mammalian target of rapamycin
- NF-κB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NRF2
nuclear factor (erythroid-derived 2)-like 2
- NRIP1
nuclear receptor-interacting protein 140
- NTCP
Na+-taurocholate co-transporting polypeptide
- OATP
organic anion transporting polypeptide
- OST
organic solute transporter
- p21
cyclin-dependent kinase inhibitor 1
- PAMPs
pathogen-associated molecular patterns
- PCNA
proliferating cell nuclear antigen
- PDGF
platelet-derived growth factor
- PDK4
pyruvate dehydrogenase lipoamide kinase isozyme 4
- PG
prostaglandin
- PHx
partial hepatectomy
- PI3K
phosphoinositide 3-kinase
- PKA/C
protein kinase A/C
- PPAR
peroxisome proliferator-activated receptor
- pRB
phosphorylated retinoblastoma protein
- PRR
pattern recognition receptor
- PXR
pregnane X receptor
- ROS
reactive oxygen species
- RXR
retinoid X receptor
- SHP
small heterodimer partner
- SIRT1
sirtuin1
- SOCS
suppressor of cytokine signaling
- SRC3
steroid receptor co-activator-3
- STAT3
signal transducer and activator of transcription 3
- SULT
sulfotransferase
- TCA
taurocholic acid / taurocholate
- TGF
transforming growth factor
- TGR5
G protein coupled bile acid receptor
- TLR4
Toll-like receptor 4; TNF-α, tumor necrosis factor alpha
- TNF-α
tumor necrosis factor alpha
- VDR
vitamin D receptor
- VEGF
vascular endothelial growth factor
- VEGFR
vascular endothelial growth factor receptor
- UDCA
ursodeoxycholic acid / ursodeoxycholate
- UGT
UDP-glucuronosyltransferase
- uPA
urokinase plasminogen activator
- UPR
unfolded protein response
Footnotes
Disclosures
The authors declare that they do not have any conflict of interest. MH, RFvG, and PBO are conducting 2 liver regeneration-related projects with obeticholic acid that was provided by Intercept Pharmaceuticals under a non-commercial license agreement.
References
- [1].Hoekstra LT, de Graaf W, Nibourg GA, Heger M, Bennink RJ, Stieger B, van Gulik TM. Physiological and biochemical basis of clinical liver function tests: A review. Ann Surg 2013;257:27–36. [DOI] [PubMed] [Google Scholar]
- [2].Michalopoulos GK. Advances in liver regeneration. Exp Rev Gastroenterol Hepatol 2014;8:897–907. [DOI] [PubMed] [Google Scholar]
- [3].Bismuth H, Majno PE. Hepatobiliary surgery. J Hepatol 2000;32:208–224. [DOI] [PubMed] [Google Scholar]
- [4].Togo S, Makino H, Kobayashi T, Morita T, Shimizu T, Kubota T, Ichikawa Y, Ishikawa T, Okazaki Y, Hayashizaki Y, Shimada H. Mechanism of liver regeneration after partial hepatectomy using mouse cdna microarray. J Hepatol 2004;40:464–471. [DOI] [PubMed] [Google Scholar]
- [5].Liao Y, Shikapwashya ON, Shteyer E, Dieckgraefe BK, Hruz PW, Rudnick DA. Delayed hepatocellular mitotic progression and impaired liver regeneration in early growth response-1- deficient mice. J Biol Chem 2004;279:43107–43116. [DOI] [PubMed] [Google Scholar]
- [6].Talarmin H, Rescan C, Cariou S, Glaise D, Zanninelli G, Bilodeau M, Loyer P, Guguen-Guillouzo C, Baffet G. The mitogen-activated protein kinase kinase/extracellular signal- regulated kinase cascade activation is a key signalling pathway involved in the regulation of g(1) phase progression in proliferating hepatocytes. Mol Cell Biol 1999;19:6003–6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Markiewski MM, DeAngelis RA, Strey CW, Foukas PG, Gerard C, Gerard N, Wetsel RA, Lambris JD. The regulation of liver cell survival by complement. J Immunol 2009;182:5412–5418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Strey CW, Markiewski M, Mastellos D, Tudoran R, Spruce LA, Greenbaum LE, Lambris JD. The proinflammatory mediators c3a and c5a are essential for liver regeneration. J Exp Med 2003;198:913–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Strey CW, Siegmund B, Rosenblum S, Marquez-Pinilla RM, Oppermann E, Huber-Lang M, Lambris JD, Bechstein WO. Complement and neutrophil function changes after liver resection in humans. World J Surg 2009;33:2635–2643. [DOI] [PubMed] [Google Scholar]
- [10].Strey CW, Siegmund B, Rosenblum S, Marquez-Pinilla RM, Oppermann E, Huber-Lang M, Lambris JD, Bechstein WO. Complement and neutrophil function changes after liver resection in humans. World J Surg 2009;33:2635–2643. [DOI] [PubMed] [Google Scholar]
- [11].van de Poll MC, Derikx JP, Buurman WA, Peters WH, Roelofs HM, Wigmore SJ, Dejong CH. Liver manipulation causes hepatocyte injury and precedes systemic inflammation in patients undergoing liver resection. World J Surg 2007;31:2033–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].van den Broek MA, Shiri-Sverdlov R, Schreurs JJ, Bloemen JG, Bieghs V, Rensen SS, Dejong CH, Olde Damink SW. Liver manipulation during liver surgery in humans is associated with hepatocellular damage and hepatic inflammation. Liver Int 2013;33:633–641. [DOI] [PubMed] [Google Scholar]
- [13].van Golen RF, van Gulik TM, Heger M. The sterile immune response during hepatic ischemia/reperfusion. Cytokine Growth Factor Rev 2012;23:69–84. [DOI] [PubMed] [Google Scholar]
- [14].Chen GY, Nunez G. Sterile inflammation: Sensing and reacting to damage. Nat Rev Immunol 2010;10:826–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Oe S, Lemmer ER, Conner EA, Factor VM, Leveen P, Larsson J, Karlsson S, Thorgeirsson SS. Intact signaling by transforming growth factor beta is not required for termination of liver regeneration in mice. Hepatology 2004;40:1098–1105. [DOI] [PubMed] [Google Scholar]
- [16].Romero-Gallo J, Sozmen EG, Chytil A, Russell WE, Whitehead R, Parks WT, Holdren MS, Her MF, Gautam S, Magnuson M, Moses HL, Grady WM. Inactivation of tgf-beta signaling in hepatocytes results in an increased proliferative response after partial hepatectomy. Oncogene 2005;24:3028–3041. [DOI] [PubMed] [Google Scholar]
- [17].Hammond JS, Guha IN, Beckingham IJ, Lobo DN. Prediction, prevention and management of postresection liver failure. Br J Surg 2011;98:1188–1200. [DOI] [PubMed] [Google Scholar]
- [18].Dahm F, Georgiev P, Clavien PA. Small-for-size syndrome after partial liver transplantation: Definition, mechanisms of disease and clinical implications. Am J Transplant 2005;5:2605–2610. [DOI] [PubMed] [Google Scholar]
- [19].Columbano A, Ledda-Columbano GM. Mitogenesis by ligands of nuclear receptors: An attractive model for the study of the molecular mechanisms implicated in liver growth. Cell Death Differ 2003;10 Suppl 1:S19–21. [DOI] [PubMed] [Google Scholar]
- [20].Vacca M, Degirolamo C, Massafra V, Polimeno L, Mariani- Costantini R, Palasciano G, Moschetta A. Nuclear receptors in regenerating liver and hepatocellular carcinoma. Mol Cell Endocrinol 2013;368:108–119. [DOI] [PubMed] [Google Scholar]
- [21].Naugler WE. Bile acid flux is necessary for normal liver regeneration. PLoS One 2014;9:e97426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Uriarte I, Fernandez-Barrena MG, Monte MJ, Latasa MU, Chang HC, Carotti S, Vespasiani-Gentilucci U, Morini S, Vicente E, Concepcion AR, Medina JF, Marin JJ, Berasain C, Prieto J, Avila MA. Identification of fibroblast growth factor 15 as a novel mediator of liver regeneration and its application in the prevention of post-resection liver failure in mice. Gut 2013;62:899–910. [DOI] [PubMed] [Google Scholar]
- [23].Padrissa-Altes S, Bachofner M, Bogorad RL, Pohlmeier L, Rossolini T, Bohm F, Liebisch G, Hellerbrand C, Koteliansky V, Speicher T, Werner S. Control of hepatocyte proliferation and survival by fgf receptors is essential for liver regeneration in mice. Gut 2015;64:1444–1453. [DOI] [PubMed] [Google Scholar]
- [24].Monte MJ, Martinez-Diez MC, El-Mir MY, Mendoza ME, Bravo P, Bachs O, Marin JJ. Changes in the pool of bile acids in hepatocyte nuclei during rat liver regeneration. J Hepatol 2002;36:534–542. [DOI] [PubMed] [Google Scholar]
- [25].Doignon I, Julien B, Serriere-Lanneau V, Garcin I, Alonso G, Nicou A, Monnet F, Gigou M, Humbert L, Rainteau D, Azoulay D, Castaing D, Gillon MC, Samuel D, Duclos-Vallee JC, Tordjmann T. Immediate neuroendocrine signaling after partial hepatectomy through acute portal hyperpressure and cholestasis. J Hepatol 2011;54:481–488. [DOI] [PubMed] [Google Scholar]
- [26].Pean N, Doignon I, Garcin I, Besnard A, Julien B, Liu B, Branchereau S, Spraul A, Guettier C, Humbert L, Schoonjans K, Rainteau D, Tordjmann T. The receptor tgr5 protects the liver from bile acid overload during liver regeneration in mice. Hepatology 2013;58:1451–1460. [DOI] [PubMed] [Google Scholar]
- [27].Xie Y, Wang H, Cheng X, Wu Y, Cao L, Wu M, Xie W, Wang G, Hao H. Farnesoid x receptor activation promotes cell proliferation via pdk4-controlled metabolic reprogramming. Sci Rep 2016;6:18751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Huang W, Ma K, Zhang J, Qatanani M, Cuvillier J, Liu J, Dong B, Huang X, Moore DD. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science 2006;312:233–236. [DOI] [PubMed] [Google Scholar]
- [29].Naugler WE, Tarlow BD, Fedorov LM, Taylor M, Pelz C, Li B, Darnell J, Grompe M. Fibroblast growth factor signaling controls liver size in mice with humanized livers. Gastroenterology 2015;149:728–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Jansen PL, Schaap FG. Pandora’s box opens for cholestatic liver disease. Hepatology 2016;63:694–696. [DOI] [PubMed] [Google Scholar]
- [31].Schaap FG, Trauner M, Jansen PL. Bile acid receptors as targets for drug development. Nat Rev Gastroenterol Hepatol 2014;11:55–67. [DOI] [PubMed] [Google Scholar]
- [32].Olthof PB, Huisman F, Schaap FG, van Lienden KP, Bennink RJ, van Golen RF, Heger M, Verheij J, Jansen PL, Olde Damink SW, van Gulik TM. Effect of obeticholic acid on liver regeneration following portal vein embolization in an experimental model. Br J Surg 2017;104:590–599. [DOI] [PubMed] [Google Scholar]
- [33].Beuers U, Trauner M, Jansen P, Poupon R. New paradigms in the treatment of hepatic cholestasis: From udca to fxr, pxr and beyond. J Hepatol 2015;62:S25–37. [DOI] [PubMed] [Google Scholar]
- [34].Michalopoulos GK, DeFrances MC. Liver regeneration. Science 1997;276:60–66. [DOI] [PubMed] [Google Scholar]
- [35].Grisham JW. A morphologic study of deoxyribonucleic acid synthesis and cell proliferation in regenerating rat liver; autoradiography with thymidine-h3. Cancer Res 1962;22:842–849. [PubMed] [Google Scholar]
- [36].Taub R Liver regeneration: From myth to mechanism. Nat Rev Mol Cell Biol 2004;5:836–847. [DOI] [PubMed] [Google Scholar]
- [37].Mao SA, Glorioso JM, Nyberg SL. Liver regeneration. Transl Res 2014;163:352–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lock JF, Malinowski M, Seehofer D, Hoppe S, Rohl RI, Niehues SM, Neuhaus P, Stockmann M. Function and volume recovery after partial hepatectomy: Influence of preoperative liver function, residual liver volume, and obesity. Langenbecks Arch Surg 2012;397:1297–1304. [DOI] [PubMed] [Google Scholar]
- [39].Nagasue N, Yukaya H, Ogawa Y, Kohno H, Nakamura T. Human liver regeneration after major hepatic resection. A study of normal liver and livers with chronic hepatitis and cirrhosis. Ann Surg 1987;206:30–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Riehle KJ, Dan YY, Campbell JS, Fausto N. New concepts in liver regeneration. J Gastroenterol Hepatol 2011;26:203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Satyanarayana A, Geffers R, Manns MP, Buer J, Rudolph KL. Gene expression profile at the g1/s transition of liver regeneration after partial hepatectomy in mice. Cell Cycle 2004;3:1405–1417. [DOI] [PubMed] [Google Scholar]
- [42].Meyer J, Lejmi E, Fontana P, Morel P, Gonelle-Gispert C, Buhler L. A focus on the role of platelets in liver regeneration: Do platelet-endothelial cell interactions initiate the regenerative process? J Hepatol 2015;63:1263–1271. [DOI] [PubMed] [Google Scholar]
- [43].Kang LI, Mars WM, Michalopoulos GK. Signals and cells involved in regulating liver regeneration. Cells 2012;1:1261–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Michalopoulos GK. Liver regeneration after partial hepatectomy: Critical analysis of mechanistic dilemmas. Am J Pathol 2010;176:2–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Marubashi S, Sakon M, Nagano H, Gotoh K, Hashimoto K, Kubota M, Kobayashi S, Yamamoto S, Miyamoto A, Dono K, Nakamori S, Umeshita K, Monden M. Effect of portal hemodynamics on liver regeneration studied in a novel portohepatic shunt rat model. Surgery 2004;136:1028–1037. [DOI] [PubMed] [Google Scholar]
- [46].Lee S, Broelsch CE, Flamant YM, Chandler JG, Charters AC 3rd, Orloff MJ. Liver regeneration after portacaval transportation in rats. Surgery 1975;77:144–149. [PubMed] [Google Scholar]
- [47].Starlinger P, Haegele S, Offensperger F, Oehlberger L, Pereyra D, Kral JB, Schrottmaier WC, Badrnya S, Reiberger T, Ferlitsch A, Stift J, Luf F, Brostjan C, Gruenberger T, Assinger A,.The profile of platelet alpha-granule released molecules affects postoperative liver regeneration. Hepatology 2016;63:1675–1688. [DOI] [PubMed] [Google Scholar]
- [48].Zhang JN, Bergeron AL, Yu Q, Sun C, McIntire LV, Lopez JA, Dong JF. Platelet aggregation and activation under complex patterns of shear stress. Thromb Haemost 2002;88:817–821. [PubMed] [Google Scholar]
- [49].van Golen RF, Stevens KM, Colarusso P, Jaeschke H, Heger M.Platelet aggregation but not activation and degranulation during the acute post-ischemic reperfusion phase in livers with no underlying disease. J Clin Transl Res 2015;1:107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wack KE, Ross MA, Zegarra V, Sysko LR, Watkins SC, Stolz DB. Sinusoidal ultrastructure evaluated during the revascularization of regenerating rat liver. Hepatology 2001;33:363–378. [DOI] [PubMed] [Google Scholar]
- [51].Nakamura M, Shibazaki M, Nitta Y, Endo Y. Translocation of platelets into disse spaces and their entry into hepatocytes in response to lipopolysaccharides, interleukin-1 and tumour necrosis factor: The role of kupffer cells. J Hepatol 1998;28:991–999. [DOI] [PubMed] [Google Scholar]
- [52].Endo Y, Nakamura M. The effect of lipopolysaccharide, interleukin-1 and tumour necrosis factor on the hepatic accumulation of 5-hydroxytryptamine and platelets in the mouse. Br J Pharmacol 1992;105:613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Clavien PA, Petrowsky H, DeOliveira ML, Graf R. Strategies for safer liver surgery and partial liver transplantation. N Engl J Med 2007;356:1545–1559. [DOI] [PubMed] [Google Scholar]
- [54].Wuestefeld T, Klein C, Streetz KL, Betz U, Lauber J, Buer J, Manns MP, Muller W, Trautwein C. Interleukin-6/glycoprotein 130-dependent pathways are protective during liver regeneration. J Biol Chem 2003;278:11281–11288. [DOI] [PubMed] [Google Scholar]
- [55].Iimuro Y, Fujimoto J. Tlrs, nf-kappab, jnk, and liver regeneration. Gastroenterol Res Pract 2010;598109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Michalopoulos GK. Principles of liver regeneration and growth homeostasis. Compr Physiol 2013;3:485–513. [DOI] [PubMed] [Google Scholar]
- [57].Marshall KM, He S, Zhong Z, Atkinson C, Tomlinson S. Dissecting the complement pathway in hepatic injury and regeneration with a novel protective strategy. J Exp Med 2014;211:1793–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].DeAngelis RA, Markiewski MM, Kourtzelis I, Rafail S, Syriga M, Sandor A, Maurya MR, Gupta S, Subramaniam S, Lambris JD. A complement-il-4 regulatory circuit controls liver regeneration. J Immunol 2012;188:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].DeAngelis RA, Markiewski MM, Lambris JD. Liver regeneration: A link to inflammation through complement. Adv Exp Med Biol 2006;586:17–34. [DOI] [PubMed] [Google Scholar]
- [60].Liu HX, Keane R, Sheng L, Wan YJ. Implications of microbiota and bile acid in liver injury and regeneration. J Hepatol 2015;63:1502–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Liu HX, Rocha CS, Dandekar S, Wan YY. Functional analysis of the relationship between intestinal microbiota and the expression of hepatic genes and pathways during the course of liver regeneration. J Hepatol 2016;64:641–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Sakaguchi T, Nakamura S, Suzuki S, Baba S, Nakashima M. Endogenous endotoxemia after massive hepatectomy and portal vein stenosis: Beneficial effect of a prostaglandin i2 analogue on intestinal permeability. Eur Surg Res 1996;28:341–350. [DOI] [PubMed] [Google Scholar]
- [63].Cornell RP. Gut-derived endotoxin elicits hepatotrophic factor secretion for liver regeneration. Am J Physiol 1985;249:R551–562. [DOI] [PubMed] [Google Scholar]
- [64].Gao B, Jeong WI, Tian Z. Liver: An organ with predominant innate immunity. Hepatology 2008;47:729–736. [DOI] [PubMed] [Google Scholar]
- [65].Cornell RP, Liljequist BL, Bartizal KF. Depressed liver regeneration after partial hepatectomy of germ-free, athymic and lipopolysaccharide-resistant mice. Hepatology 1990;11:916–922. [DOI] [PubMed] [Google Scholar]
- [66].Cienfuegos JA, Rotellar F, Baixauli J, Martínez-Regueira F, Pardo F, Hernández-Lizoáin JL. Liver regeneration--the best kept secret. A model of tissue injury response. Rev Esp Enferm Dig 2014;106:171–194. [PubMed] [Google Scholar]
- [67].van Golen RF, van Gulik TM, Heger M. Mechanistic overview of reactive species-induced degradation of the endothelial glycocalyx during hepatic ischemia/reperfusion injury. Free Radic Biol Med 2012;52:1382–1402. [DOI] [PubMed] [Google Scholar]
- [68].van Golen RF, Reiniers MJ, Vrisekoop N, Zuurbier CJ, Olthof PB, van Rheenen J, van Gulik TM, Parsons BJ, Heger M. The mechanisms and physiological relevance of glycocalyx degradation in hepatic ischemia/reperfusion injury. Antioxid Redox Signal 2014;21:1098–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Kloek JJ, Marechal X, Roelofsen J, Houtkooper RH, van Kuilenburg AB, Kulik W, Bezemer R, Neviere R, van Gulik TM, Heger M. Cholestasis is associated with hepatic microvascular dysfunction and aberrant energy metabolism before and during ischemia-reperfusion. Antioxid Redox Signal 2012;17:1109–1123. [DOI] [PubMed] [Google Scholar]
- [70].Reiniers MJ, van Golen RF, van Gulik TM, Heger M. Reactive oxygen and nitrogen species in steatotic hepatocytes: A molecular perspective on the pathophysiology of ischemia- reperfusion injury in the fatty liver. Antioxid Redox Signal 2014;21:1119–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bai H, Zhang W, Qin XJ, Zhang T, Wu H, Liu JZ, Hai CX. Hydrogen peroxide modulates the proliferation/quiescence switch in the liver during embryonic development and posthepatectomy regeneration. Antioxid Redox Signal 2015;22:921–937. [DOI] [PubMed] [Google Scholar]
- [72].Dayoub R, Vogel A, Schuett J, Lupke M, Spieker SM, Kettern N, Hildt E, Melter M, Weiss TS. Nrf2 activates augmenter of liver regeneration (alr) via antioxidant response element and links oxidative stress to liver regeneration. Mol Med 2013;19:237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Hu M, Zou Y, Nambiar SM, Lee J, Yang Y, Dai G. Keap1 modulates the redox cycle and hepatocyte cell cycle in regenerating liver. Cell Cycle 2014;13:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kohler UA, Kurinna S, Schwitter D, Marti A, Schafer M, Hellerbrand C, Speicher T, Werner S. Activated nrf2 impairs liver regeneration in mice by activation of genes involved in cell-cycle control and apoptosis. Hepatology 2014;60:670–678. [DOI] [PubMed] [Google Scholar]
- [75].Wakabayashi N, Shin S, Slocum SL, Agoston ES, Wakabayashi J, Kwak MK, Misra V, Biswal S, Yamamoto M, Kensler TW. Regulation of notch1 signaling by nrf2: Implications for tissue regeneration. Sci Signal 2010;3:ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kohler C, Bell AW, Bowen WC, Monga SP, Fleig W, Michalopoulos GK. Expression of notch-1 and its ligand jagged-1 in rat liver during liver regeneration. Hepatology 2004;39:1056–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Goodrich DW, Wang NP, Qian YW, Lee EY, Lee WH. The retinoblastoma gene product regulates progression through the g1 phase of the cell cycle. Cell 1991;67:293–302. [DOI] [PubMed] [Google Scholar]
- [78].Weintraub SJ, Prater CA, Dean DC. Retinoblastoma protein switches the e2f site from positive to negative element. Nature 1992;358:259–261. [DOI] [PubMed] [Google Scholar]
- [79].Nelsen CJ, Rickheim DG, Timchenko NA, Stanley MW, Albrecht JH. Transient expression of cyclin d1 is sufficient to promote hepatocyte replication and liver growth in vivo. Cancer Res 2001;61:8564–8568. [PubMed] [Google Scholar]
- [80].Lavoie JN, Rivard N, L’Allemain G, Pouyssegur J. A temporal and biochemical link between growth factor-activated map kinases, cyclin d1 induction and cell cycle entry. Prog Cell Cycle Res 1996;2:49–58. [DOI] [PubMed] [Google Scholar]
- [81].Lavoie JN, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin d1 expression is regulated positively by the p42/p44mapk and negatively by the p38/hogmapk pathway. J Biol Chem 1996;271:20608–20616. [DOI] [PubMed] [Google Scholar]
- [82].Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, Thiery J, Kan YW, Werner S. Impaired liver regeneration in nrf2 knockout mice: Role of ros-mediated insulin/igf-1 resistance. EMBO J 2008;27:212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Yan LJ, Rajasekaran NS, Sathyanarayanan S, Benjamin IJ. Mouse hsf1 disruption perturbs redox state and increases mitochondrial oxidative stress in kidney. Antioxid Redox Signal 2005;7:465–471. [DOI] [PubMed] [Google Scholar]
- [84].Taub R Liver regeneration 4: Transcriptional control of liver regeneration. FASEB J 1996;10:413–427. [PubMed] [Google Scholar]
- [85].Aldeguer X, Debonera F, Shaked A, Krasinkas AM, Gelman AE, Que X, Zamir GA, Hiroyasu S, Kovalovich KK, Taub R, Olthoff KM. Interleukin-6 from intrahepatic cells of bone marrow origin is required for normal murine liver regeneration. Hepatology 2002;35:40–48. [DOI] [PubMed] [Google Scholar]
- [86].Akerman P, Cote P, Yang SQ, McClain C, Nelson S, Bagby GJ, Diehl AM. Antibodies to tumor necrosis factor-alpha inhibit liver regeneration after partial hepatectomy. Am J Physiol 1992;263:G579–585. [DOI] [PubMed] [Google Scholar]
- [87].Kirillova I, Chaisson M, Fausto N. Tumor necrosis factor induces DNA replication in hepatic cells through nuclear factor kappab activation. Cell Growth Differ 1999;10:819–828. [PubMed] [Google Scholar]
- [88].FitzGerald MJ, Webber EM, Donovan JR, Fausto N. Rapid DNA binding by nuclear factor kappa b in hepatocytes at the start of liver regeneration. Cell Growth Differ 1995;6:417–427. [PubMed] [Google Scholar]
- [89].Yatomi Y, Ohmori T, Rile G, Kazama F, Okamoto H, Sano T, Satoh K, Kume S, Tigyi G, Igarashi Y, Ozaki Y. Sphingosine 1-phosphate as a major bioactive lysophospholipid that is released from platelets and interacts with endothelial cells. Blood 2000;96:3431–3438. [PubMed] [Google Scholar]
- [90].Kawasaki T, Murata S, Takahashi K, Nozaki R, Ohshiro Y, Ikeda N, Pak S, Myronovych A, Hisakura K, Fukunaga K, Oda T, Sasaki R, Ohkohchi N. Activation of human liver sinusoidal endothelial cell by human platelets induces hepatocyte proliferation. J Hepatol 2010;53:648–654. [DOI] [PubMed] [Google Scholar]
- [91].Nowatari T, Murata S, Nakayama K, Sano N, Maruyama T, Nozaki R, Ikeda N, Fukunaga K, Ohkohchi N. Sphingosine 1- phosphate has anti-apoptotic effect on liver sinusoidal endothelial cells and proliferative effect on hepatocytes in a paracrine manner in human. Hepatol Res 2015;45:1136–1145. [DOI] [PubMed] [Google Scholar]
- [92].Cressman DE, Greenbaum LE, DeAngelis RA, Ciliberto G, Furth EE, Poli V, Taub R. Liver failure and defective hepatocyte regeneration in interleukin-6-deficient mice. Science 1996;274:1379–1383. [DOI] [PubMed] [Google Scholar]
- [93].Yamada Y, Kirillova I, Peschon JJ, Fausto N. Initiation of liver growth by tumor necrosis factor: Deficient liver regeneration in mice lacking type i tumor necrosis factor receptor. Proc Natl Acad Sci U S A 1997;94:1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Li W, Liang X, Leu JI, Kovalovich K, Ciliberto G, Taub R. Global changes in interleukin-6-dependent gene expression patterns in mouse livers after partial hepatectomy. Hepatology 2001;33:1377–1386. [DOI] [PubMed] [Google Scholar]
- [95].Haber BA, Mohn KL, Diamond RH, Taub R. Induction patterns of 70 genes during nine days after hepatectomy define the temporal course of liver regeneration. J Clin Invest 1993;91:1319–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Levy DE, Lee CK. What does stat3 do? J Clin Invest 2002;109:1143–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Columbano A, Shinozuka H. Liver regeneration versus direct hyperplasia. FASEB J 1996;10:1118–1128. [DOI] [PubMed] [Google Scholar]
- [98].Michalopoulos GK, Khan Z. Liver regeneration, growth factors, and amphiregulin. Gastroenterology 2005;128:503–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wang L, Wang X, Wang L, Chiu JD, van de Ven G, Gaarde WA, Deleve LD. Hepatic vascular endothelial growth factor regulates recruitment of rat liver sinusoidal endothelial cell progenitor cells. Gastroenterology 2012;143:1555–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Shimizu H, Miyazaki M, Wakabayashi Y, Mitsuhashi N, Kato A, Ito H, Nakagawa K, Yoshidome H, Kataoka M, Nakajima N. Vascular endothelial growth factor secreted by replicating hepatocytes induces sinusoidal endothelial cell proliferation during regeneration after partial hepatectomy in rats. J Hepatol 2001;34:683–689. [DOI] [PubMed] [Google Scholar]
- [101].Taniguchi E, Sakisaka S, Matsuo K, Tanikawa K, Sata M. Expression and role of vascular endothelial growth factor in liver regeneration after partial hepatectomy in rats. J Histochem Cytochem 2001;49:121–130. [DOI] [PubMed] [Google Scholar]
- [102].Mochida S, Ishikawa K, Inao M, Shibuya M, Fujiwara K. Increased expressions of vascular endothelial growth factor and its receptors, flt-1 and kdr/flk-1, in regenerating rat liver. Biochem Biophys Res Commun 1996;226:176–179. [DOI] [PubMed] [Google Scholar]
- [103].Matsuo R, Nakano Y, Ohkohchi N. Platelet administration via the portal vein promotes liver regeneration in rats after 70% hepatectomy. Ann Surg 2011;253:759–763. [DOI] [PubMed] [Google Scholar]
- [104].Murata S, Matsuo R, Ikeda O, Myronovych A, Watanabe M, Hisakura K, Nakano Y, Hashimoto I, Ohkohchi N. Platelets promote liver regeneration under conditions of kupffer cell depletion after hepatectomy in mice. World J Surg 2008;32:1088–1096. [DOI] [PubMed] [Google Scholar]
- [105].Rendu F, Brohard-Bohn B. The platelet release reaction: Granules’ constituents, secretion and functions. Platelets 2001;12:261–273. [DOI] [PubMed] [Google Scholar]
- [106].Nurden AT, Nurden P, Sanchez M, Andia I, Anitua E. Platelets and wound healing. Front Biosci 2008;13:3532–3548. [DOI] [PubMed] [Google Scholar]
- [107].Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. Transforming growth factor-beta in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem 1983;258:7155–7160. [PubMed] [Google Scholar]
- [108].Lesurtel M, Graf R, Aleil B, Walther DJ, Tian Y, Jochum W, Gachet C, Bader M, Clavien PA. Platelet-derived serotonin mediates liver regeneration. Science 2006;312:104–107. [DOI] [PubMed] [Google Scholar]
- [109].Hong F, Nguyen VA, Shen X, Kunos G, Gao B. Rapid activation of protein kinase b/akt has a key role in antiapoptotic signaling during liver regeneration. Biochem Biophys Res Commun 2000;279:974–979. [DOI] [PubMed] [Google Scholar]
- [110].Matsuo R, Ohkohchi N, Murata S, Ikeda O, Nakano Y, Watanabe M, Hisakura K, Myronovych A, Kubota T, Narimatsu H, Ozaki M. Platelets strongly induce hepatocyte proliferation with igf-1 and hgf in vitro. J Surg Res 2008;145:279–286. [DOI] [PubMed] [Google Scholar]
- [111].Nejak-Bowen K, Orr A, Bowen WC Jr., Michalopoulos GK Conditional genetic elimination of hepatocyte growth factor in mice compromises liver regeneration after partial hepatectomy. PLoS One 2013;8:e59836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Furrer K, Rickenbacher A, Tian Y, Jochum W, Bittermann AG, Kach A, Humar B, Graf R, Moritz W, Clavien PA. Serotonin reverts age-related capillarization and failure of regeneration in the liver through a vegf-dependent pathway. Proc Natl Acad Sci U S A 2011;108:2945–2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].LeCouter J, Moritz DR, Li B, Phillips GL, Liang XH, Gerber HP, Hillan KJ, Ferrara N. Angiogenesis-independent endothelial protection of liver: Role of vegfr-1. Science 2003;299:890–893. [DOI] [PubMed] [Google Scholar]
- [114].Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, Mittal V, Kobayashi H, Shido K, Lyden D, Sato TN, Rabbany SY, Rafii S. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature 2010;468:310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].DeLeve LD. Liver sinusoidal endothelial cells and liver regeneration. J Clin Invest 2013;123:1861–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Lindroos PM, Zarnegar R, Michalopoulos GK. Hepatocyte growth factor (hepatopoietin a) rapidly increases in plasma before DNA synthesis and liver regeneration stimulated by partial hepatectomy and carbon tetrachloride administration. Hepatology 1991;13:743–750. [PubMed] [Google Scholar]
- [117].Currier AR, Sabla G, Locaputo S, Melin-Aldana H, Degen JL, Bezerra JA. Plasminogen directs the pleiotropic effects of upa in liver injury and repair. Am J Physiol Gastrointest Liver Physiol 2003;284:G508–515. [DOI] [PubMed] [Google Scholar]
- [118].Shimizu M, Hara A, Okuno M, Matsuno H, Okada K, Ueshima S, Matsuo O, Niwa M, Akita K, Yamada Y, Yoshimi N, Uematsu T, Kojima S, Friedman SL, Moriwaki H, Mori H. Mechanism of retarded liver regeneration in plasminogen activator-deficient mice: Impaired activation of hepatocyte growth factor after fas-mediated massive hepatic apoptosis. Hepatology 2001;33:569–576. [DOI] [PubMed] [Google Scholar]
- [119].Pediaditakis P, Lopez-Talavera JC, Petersen B, Monga SP, Michalopoulos GK. The processing and utilization of hepatocyte growth factor/scatter factor following partial hepatectomy in the rat. Hepatology 2001;34:688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Mars WM, Zarnegar R, Michalopoulos GK. Activation of hepatocyte growth factor by the plasminogen activators upa and tpa. Am J Pathol 1993;143:949–958. [PMC free article] [PubMed] [Google Scholar]
- [121].Maroun CR, Rowlands T. The met receptor tyrosine kinase: A key player in oncogenesis and drug resistance. Pharmacol Ther 2014;142:316–338. [DOI] [PubMed] [Google Scholar]
- [122].Nakamura T, Mizuno S. The discovery of hepatocyte growth factor (hgf) and its significance for cell biology, life sciences and clinical medicine. Proc Jpn Acad Ser B Phys Biol Sci 2010;86:588–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Borowiak M, Garratt AN, Wustefeld T, Strehle M, Trautwein C, Birchmeier C. Met provides essential signals for liver regeneration. Proc Natl Acad Sci U S A 2004;101:10608–10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Okano J, Shiota G, Matsumoto K, Yasui S, Kurimasa A, Hisatome I, Steinberg P, Murawaki Y. Hepatocyte growth factor exerts a proliferative effect on oval cells through the pi3k/akt signaling pathway. Biochem Biophys Res Commun 2003;309:298–304. [DOI] [PubMed] [Google Scholar]
- [125].Ozaki M, Haga S, Zhang HQ, Irani K, Suzuki S. Inhibition of hypoxia/reoxygenation-induced oxidative stress in hgf- stimulated antiapoptotic signaling: Role of pi3-k and akt kinase upon rac1. Cell Death Differ 2003;10:508–515. [DOI] [PubMed] [Google Scholar]
- [126].Xiao GH, Jeffers M, Bellacosa A, Mitsuuchi Y, Vande Woude GF, Testa JR. Anti-apoptotic signaling by hepatocyte growth factor/met via the phosphatidylinositol 3-kinase/akt and mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 2001;98:247–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Gao C, Jokerst R, Gondipalli P, Cai SR, Kennedy S, Flye MW, Ponder KP. Lipopolysaccharide potentiates the effect of hepatocyte growth factor on hepatocyte replication in rats by augmenting ap-1 activity. Hepatology 1999;30:1405–1416. [DOI] [PubMed] [Google Scholar]
- [128].Puri N, Salgia R. Synergism of egfr and c-met pathways, cross- talk and inhibition, in non-small cell lung cancer. J Carcinog 2008;7:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Jo M, Stolz DB, Esplen JE, Dorko K, Michalopoulos GK, Strom SC. Cross-talk between epidermal growth factor receptor and c-met signal pathways in transformed cells. J Biol Chem 2000;275:8806–8811. [DOI] [PubMed] [Google Scholar]
- [130].Skov Olsen P, Boesby S, Kirkegaard P, Therkelsen K, Almdal T, Poulsen SS, Nexo E. Influence of epidermal growth factor on liver regeneration after partial hepatectomy in rats. Hepatology 1988;8:992–996. [DOI] [PubMed] [Google Scholar]
- [131].Cruise JL, Knechtle SJ, Bollinger RR, Kuhn C, Michalopoulos G. Alpha 1-adrenergic effects and liver regeneration. Hepatology 1987;7:1189–1194. [DOI] [PubMed] [Google Scholar]
- [132].Olsen PS, Poulsen SS, Kirkegaard P. Adrenergic effects on secretion of epidermal growth factor from brunner’s glands. Gut 1985;26:920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Knopp J, Jezova D, Rusnak M, Jaroscakova I, Farkas R, Kvetnansky R. Changes in plasma catecholamine and corticosterone levels and gene expression of key enzymes of catecholamine biosynthesis in partially hepatectomized rats. Endocr Regul 1999;33:145–153. [PubMed] [Google Scholar]
- [134].Mead JE, Fausto N. Transforming growth factor alpha may be a physiological regulator of liver regeneration by means of an autocrine mechanism. Proc Natl Acad Sci U S A 1989;86:1558–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Webber EM, Wu JC, Wang L, Merlino G, Fausto N. Overexpression of transforming growth factor-alpha causes liver enlargement and increased hepatocyte proliferation in transgenic mice. Am J Pathol 1994;145:398–408. [PMC free article] [PubMed] [Google Scholar]
- [136].Russell WE, Kaufmann WK, Sitaric S, Luetteke NC, Lee DC. Liver regeneration and hepatocarcinogenesis in transforming growth factor-alpha-targeted mice. Mol Carcinog 1996;15:183–189. [DOI] [PubMed] [Google Scholar]
- [137].Nakano T, Raines EW, Abraham JA, Klagsbrun M, Ross R. Lysophosphatidylcholine upregulates the level of heparin- binding epidermal growth factor-like growth factor mrna in human monocytes. Proc Natl Acad Sci U S A 1994;91:1069–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Edwards JP, Zhang X, Mosser DM. The expression of heparin- binding epidermal growth factor-like growth factor by regulatory macrophages. J Immunol 2009;182:1929–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Higashiyama S Metalloproteinase-mediated shedding of heparin-binding egf-like growth factor and its pathophysiological roles. Protein Pept Lett 2004;11:443–450. [DOI] [PubMed] [Google Scholar]
- [140].Ito N, Kawata S, Tamura S, Kiso S, Tsushima H, Damm D, Abraham JA, Higashiyama S, Taniguchi N, Matsuzawa Y. Heparin-binding egf-like growth factor is a potent mitogen for rat hepatocytes. Biochem Biophys Res Commun 1994;198:25–31. [DOI] [PubMed] [Google Scholar]
- [141].Kiso S, Kawata S, Tamura S, Higashiyama S, Ito N, Tsushima H, Taniguchi N, Matsuzawa Y. Role of heparin-binding epidermal growth factor-like growth factor as a hepatotrophic factor in rat liver regeneration after partial hepatectomy. Hepatology 1995;22:1584–1590. [PubMed] [Google Scholar]
- [142].Yamada A, Kawata S, Tamura S, Kiso S, Higashiyama S, Umeshita K, Sakon M, Taniguchi N, Monden M, Matsuzawa Y. Plasma heparin-binding egf-like growth factor levels in patients after partial hepatectomy as determined with an enzyme-linked immunosorbent assay. Biochem Biophys Res Commun 1998;246:783–787. [DOI] [PubMed] [Google Scholar]
- [143].Sakuda S, Tamura S, Yamada A, Miyagawa J, Yamamoto K, Kiso S, Ito N, Higashiyama S, Taniguchi N, Kawata S, Matsuzawa Y. Nf-kappab activation in non-parenchymal livercells after partial hepatectomy in rats: Possible involvement in expression of heparin-binding epidermal growth factor-like growth factor. J Hepatol 2002;36:527–533. [DOI] [PubMed] [Google Scholar]
- [144].Berasain C, Garcia-Trevijano ER, Castillo J, Erroba E, Lee DC, Prieto J, Avila MA. Amphiregulin: An early trigger of liver regeneration in mice. Gastroenterology 2005;128:424–432. [DOI] [PubMed] [Google Scholar]
- [145].Hou Z, Yanaga K, Kamohara Y, Eguchi S, Tsutsumi R, Furui J, Kanematsu T. A new suppressive agent against interleukin- 1beta and tumor necrosis factor-alpha enhances liver regeneration after partial hepatectomy in rats. Hepatol Res 2003;26:40–46. [DOI] [PubMed] [Google Scholar]
- [146].Tsutsumi R, Kamohara Y, Eguchi S, Azuma T, Fujioka H, Okudaira S, Yanaga K, Kanematsu T. Selective suppression of initial cytokine response facilitates liver regeneration after extensive hepatectomy in rats. Hepatogastroenterology 2004;51:701–704. [PubMed] [Google Scholar]
- [147].Ogiso T, Nagaki M, Takai S, Tsukada Y, Mukai T, Kimura K, Moriwaki H. Granulocyte colony-stimulating factor impairs liver regeneration in mice through the up-regulation of interleukin-1beta. J Hepatol 2007;47:816–825. [DOI] [PubMed] [Google Scholar]
- [148].Tsujii H, Okamoto Y, Kikuchi E, Matsumoto M, Nakano H. Prostaglandin e2 and rat liver regeneration. Gastroenterology 1993;105:495–499. [DOI] [PubMed] [Google Scholar]
- [149].Goss JA, Mangino MJ, Flye MW. Kupffer cell autoregulation of il-1 production by pge2 during hepatic regeneration. J Surg Res 1992;52:422–428. [DOI] [PubMed] [Google Scholar]
- [150].Goss JA, Mangino MJ, Flye MW. Prostaglandin e2 production during hepatic regeneration downregulates kupffer cell il-6 production. Ann Surg 1992;215:553–559; discussion 559–560. [PMC free article] [PubMed] [Google Scholar]
- [151].Zhao Y, Meng C, Wang Y, Huang H, Liu W, Zhang JF, Zhao H, Feng B, Leung PS, Xia Y. Interleukin (il)-1beta inhibits beta-klotho expression and fgf19 signaling in hepatocytes. Am J Physiol Endocrinol Metab 2016;310:E289–300. [DOI] [PubMed] [Google Scholar]
- [152].Ledda-Columbano GM, Curto M, Piga R, Zedda AI, Menegazzi M, Sartori C, Shinozuka H, Bluethmann H, Poli V, Ciliberto G, Columbano A. In vivo hepatocyte proliferation is inducible through a tnf and il-6-independent pathway. Oncogene 1998;17:1039–1044. [DOI] [PubMed] [Google Scholar]
- [153].Cruise JL, Houck KA, Michalopoulos GK. Induction of DNA synthesis in cultured rat hepatocytes through stimulation of alpha 1 adrenoreceptor by norepinephrine. Science 1985;227:749–751. [DOI] [PubMed] [Google Scholar]
- [154].Yamada Y, Webber EM, Kirillova I, Peschon JJ, Fausto N. Analysis of liver regeneration in mice lacking type 1 or type 2 tumor necrosis factor receptor: Requirement for type 1 but not type 2 receptor. Hepatology 1998;28:959–970. [DOI] [PubMed] [Google Scholar]
- [155].Kan M, Huang JS, Mansson PE, Yasumitsu H, Carr B, McKeehan WL. Heparin-binding growth factor type 1 (acidic fibroblast growth factor): A potential biphasic autocrine and paracrine regulator of hepatocyte regeneration. Proc Natl Acad Sci U S A 1989;86:7432–7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Steiling H, Wustefeld T, Bugnon P, Brauchle M, Fassler R, Teupser D, Thiery J, Gordon JI, Trautwein C, Werner S. Fibroblast growth factor receptor signalling is crucial for liver homeostasis and regeneration. Oncogene 2003;22:4380–4388. [DOI] [PubMed] [Google Scholar]
- [157].Campbell JS, Hughes SD, Gilbertson DG, Palmer TE, Holdren MS, Haran AC, Odell MM, Bauer RL, Ren HP, Haugen HS, Yeh MM, Fausto N. Platelet-derived growth factor c induces liver fibrosis, steatosis, and hepatocellular carcinoma. Proc Natl Acad Sci U S A 2005;102:3389–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Ren X, Carpenter A, Hogaboam C, Colletti L. Mitogenic properties of endogenous and pharmacological doses of macrophage inflammatory protein-2 after 70% hepatectomy in the mouse. Am J Pathol 2003;163:563–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Yamauchi H, Uetsuka K, Okada T, Nakayama H, Doi K. Impaired liver regeneration after partial hepatectomy in db/db mice. Exp Toxicol Pathol 2003;54:281–286. [DOI] [PubMed] [Google Scholar]
- [160].Starzl TE, Porter KA, Putnam CW. Insulin, glucagon, and the control of hepatic structure, function, and capacity for regeneration. Metabolism 1976;25:1429–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Furukawa K, Terayama H. Pattern of glycosaminoglycans and glycoproteins associated with nuclei of regenerating liver of rat. Biochim Biophys Acta 1979;585:575–588. [DOI] [PubMed] [Google Scholar]
- [162].Vrochides D, Papanikolaou V, Pertoft H, Antoniades AA, Heldin P. Biosynthesis and degradation of hyaluronan by nonparenchymal liver cells during liver regeneration. Hepatology 1996;23:1650–1655. [DOI] [PubMed] [Google Scholar]
- [163].Della Fazia MA, Pettirossi V, Ayroldi E, Riccardi C, Magni MV, Servillo G. Differential expression of cd44 isoforms during liver regeneration in rats. J Hepatol 2001;34:555–561. [DOI] [PubMed] [Google Scholar]
- [164].Ogata T, Okuda K, Ueno T, Saito N, Aoyagi S. Serum hyaluronan as a predictor of hepatic regeneration after hepatectomy in humans. Eur J Clin Invest 1999;29:780–785. [DOI] [PubMed] [Google Scholar]
- [165].Ghahary A, Minuk GY, Luo J, Gauthier T, Murphy LJ. Effects of partial hepatectomy on hepatic insulinlike growth factor binding protein-1 expression. Hepatology 1992;15:1125–1131. [DOI] [PubMed] [Google Scholar]
- [166].Weir E, Chen Q, DeFrances MC, Bell A, Taub R, Zarnegar R. Rapid induction of mrnas for liver regeneration factor and insulin-like growth factor binding protein-1 in primary cultures of rat hepatocytes by hepatocyte growth factor and epidermal growth factor. Hepatology 1994;20:955–960. [DOI] [PubMed] [Google Scholar]
- [167].Leu JI, Crissey MA, Leu JP, Ciliberto G, Taub R. Interleukin- 6-induced stat3 and ap-1 amplify hepatocyte nuclear factor 1- mediated transactivation of hepatic genes, an adaptive response to liver injury. Mol Cell Biol 2001;21:414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Liu Y, Shao M, Wu Y, Yan C, Jiang S, Liu J, Dai J, Yang L, Li J, Jia W, Rui L, Liu Y. Role for the endoplasmic reticulum stress sensor ire1alpha in liver regenerative responses. J Hepatol 2015;62:590–598. [DOI] [PubMed] [Google Scholar]
- [169].Lee AH, Iwakoshi NN, Anderson KC, Glimcher LH. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc Natl Acad Sci U S A 2003;100:9946–9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Gu F, Nguyen DT, Stuible M, Dube N, Tremblay ML, ChevetE. Protein-tyrosine phosphatase 1b potentiates ire1 signaling during endoplasmic reticulum stress. J Biol Chem 2004;279:49689–49693. [DOI] [PubMed] [Google Scholar]
- [171].Xue X, Piao JH, Nakajima A, Sakon-Komazawa S, Kojima Y, Mori K, Yagita H, Okumura K, Harding H, Nakano H. Tumor necrosis factor alpha (tnfalpha) induces the unfolded protein response (upr) in a reactive oxygen species (ros)-dependent fashion, and the upr counteracts ros accumulation by tnfalpha. J Biol Chem 2005;280:33917–33925. [DOI] [PubMed] [Google Scholar]
- [172].Casado M, Callejas NA, Rodrigo J, Zhao X, Dey SK, Bosca L, Martin-Sanz P. Contribution of cyclooxygenase 2 to liver regeneration after partial hepatectomy. FASEB J 2001;15:2016–2018. [DOI] [PubMed] [Google Scholar]
- [173].Heger M, van Golen RF, Broekgaarden M, Michel MC. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol Rev 2014;66:222–307. [DOI] [PubMed] [Google Scholar]
- [174].Dieter P, Hempel U, Kamionka S, Kolada A, Malessa B, Fitzke E, Tran-Thi TA. Prostaglandin e2 affects differently the release of inflammatory mediators from resident macrophages by lps and muramyl tripeptides. Mediators Inflamm 1999;8:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Yang Y, Zhao Z, Liu Y, Kang X, Zhang H, Meng M. Suppression of oxidative stress and improvement of liver functions in mice by ursolic acid via lkb1-amp-activated protein kinase signaling. J Gastroenterol Hepatol 2015;30:609–618. [DOI] [PubMed] [Google Scholar]
- [176].Hernanz R, Briones AM, Salaices M, Alonso MJ. New roles for old pathways? A circuitous relationship between reactive oxygen species and cyclo-oxygenase in hypertension. Clin Sci (Lond) 2014;126:111–121. [DOI] [PubMed] [Google Scholar]
- [177].Korbecki J, Baranowska-Bosiacka I, Gutowska I, Chlubek D. The effect of reactive oxygen species on the synthesis of prostanoids from arachidonic acid. J Physiol Pharmacol 2013;64:409–421. [PubMed] [Google Scholar]
- [178].Rescan C, Coutant A, Talarmin H, Theret N, Glaise D, Guguen-Guillouzo C, Baffet G. Mechanism in the sequential control of cell morphology and s phase entry by epidermal growth factor involves distinct mek/erk activations. Mol Biol Cell 2001;12:725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Coutant A, Rescan C, Gilot D, Loyer P, Guguen-Guillouzo C, Baffet G. Pi3k-frap/mtor pathway is critical for hepatocyte proliferation whereas mek/erk supports both proliferation and survival. Hepatology 2002;36:1079–1088. [DOI] [PubMed] [Google Scholar]
- [180].Flodby P, Antonson P, Barlow C, Blanck A, Porsch-Hallstrom I, Xanthopoulos KG. Differential patterns of expression of three c/ebp isoforms, hnf-1, and hnf-4 after partial hepatectomy in rats. Exp Cell Res 1993;208:248–256. [DOI] [PubMed] [Google Scholar]
- [181].Ramji DP, Foka P. Ccaat/enhancer-binding proteins: Structure, function and regulation. Biochem J 2002;365:561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Diehl AM. Roles of ccaat/enhancer-binding proteins in regulation of liver regenerative growth. J Biol Chem 1998;273:30843–30846. [DOI] [PubMed] [Google Scholar]
- [183].Hendricks-Taylor LR, Darlington GJ. Inhibition of cell proliferation by c/ebp alpha occurs in many cell types, does not require the presence of p53 or rb, and is not affected by large t- antigen. Nucleic Acids Res 1995;23:4726–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Diehl AM, Johns DC, Yang S, Lin H, Yin M, Matelis LA, Lawrence JH. Adenovirus-mediated transfer of ccaat/enhancer- binding protein-alpha identifies a dominant antiproliferative role for this isoform in hepatocytes. J Biol Chem 1996;271:7343–7350. [DOI] [PubMed] [Google Scholar]
- [185].Greenbaum LE, Li W, Cressman DE, Peng Y, Ciliberto G, Poli V, Taub R. Ccaat enhancer- binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J Clin Invest 1998;102:996–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Yu X, Si J, Zhang Y, Dewille JW. Ccaat/enhancer binding protein-delta (c/ebp-delta) regulates cell growth, migration and differentiation. Cancer Cell Int 2010;10:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Hung JH, Su IJ, Lei HY, Wang HC, Lin WC, Chang WT, Huang W, Chang WC, Chang YS, Chen CC, Lai MD. Endoplasmic reticulum stress stimulates the expression of cyclooxygenase-2 through activation of nf-kappab and pp38 mitogen-activated protein kinase. J Biol Chem 2004;279:46384–46392. [DOI] [PubMed] [Google Scholar]
- [188].Han J, Lee JD, Bibbs L, Ulevitch RJ. A map kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science 1994;265:808–811. [DOI] [PubMed] [Google Scholar]
- [189].Lee JC, Young PR. Role of csb/p38/rk stress response kinase in lps and cytokine signaling mechanisms. J Leukoc Biol 1996;59:152–157. [DOI] [PubMed] [Google Scholar]
- [190].Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem 1995;270:7420–7426. [DOI] [PubMed] [Google Scholar]
- [191].Moriguchi T, Toyoshima F, Gotoh Y, Iwamatsu A, Irie K, Mori E, Kuroyanagi N, Hagiwara M, Matsumoto K, Nishida E. Purification and identification of a major activator for p38 from osmotically shocked cells. Activation of mitogen-activated protein kinase kinase 6 by osmotic shock, tumor necrosis factor-alpha, and h2o2. J Biol Chem 1996;271:26981–26988. [DOI] [PubMed] [Google Scholar]
- [192].Guyton KZ, Liu Y, Gorospe M, Xu Q, Holbrook NJ. Activation of mitogen-activated protein kinase by h2o2. Role in cell survival following oxidant injury. J Biol Chem 1996;271:4138–4142. [DOI] [PubMed] [Google Scholar]
- [193].Lander HM, Jacovina AT, Davis RJ, Tauras JM. Differential activation of mitogen-activated protein kinases by nitric oxide- related species. J Biol Chem 1996;271:19705–19709. [DOI] [PubMed] [Google Scholar]
- [194].Li H, Lin X. Positive and negative signaling components involved in tnfalpha-induced nf-kappab activation. Cytokine 2008;41:1–8. [DOI] [PubMed] [Google Scholar]
- [195].Janssen-Heininger YM, Poynter ME, Baeuerle PA. Recent advances towards understanding redox mechanisms in the activation of nuclear factor kappab. Free Radic Biol Med 2000;28:1317–1327. [DOI] [PubMed] [Google Scholar]
- [196].Chen X, Zhao Y, Wang F, Bei Y, Xiao J, Yang C. Micrornas in liver regeneration. Cell Physiol Biochem 2015;37:615–628. [DOI] [PubMed] [Google Scholar]
- [197].Kren BT, Wong PY, Shiota A, Zhang X, Zeng Y, Steer CJ. Polysome trafficking of transcripts and micrornas in regenerating liver after partial hepatectomy. Am J Physiol Gastrointest Liver Physiol 2009;297:G1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Raschzok N, Werner W, Sallmon H, Billecke N, Dame C, Neuhaus P, Sauer IM. Temporal expression profiles indicate a primary function for microrna during the peak of DNA replication after rat partial hepatectomy. Am J Physiol Regul Integr Comp Physiol 2011;300:R1363–1372. [DOI] [PubMed] [Google Scholar]
- [199].Chaveles I, Zaravinos A, Habeos IG, Karavias DD, Maroulis I, Spandidos DA, Karavias D. Microrna profiling in murine liver after partial hepatectomy. Int J Mol Med 2012;29:747–755. [DOI] [PubMed] [Google Scholar]
- [200].Chen X, Murad M, Cui YY, Yao LJ, Venugopal SK, Dawson K, Wu J. Mirna regulation of liver growth after 50% partial hepatectomy and small size grafts in rats. Transplantation 2011;91:293–299. [DOI] [PubMed] [Google Scholar]
- [201].Yuan B, Dong R, Shi D, Zhou Y, Zhao Y, Miao M, Jiao B. Down-regulation of mir-23b may contribute to activation of the tgf-beta1/smad3 signalling pathway during the termination stage of liver regeneration. FEBS Lett 2011;585:927–934. [DOI] [PubMed] [Google Scholar]
- [202].Lee YS, Nakahara K, Pham JW, Kim K, He Z, Sontheimer EJ, Carthew RW. Distinct roles for drosophila dicer-1 and dicer-2 in the sirna/mirna silencing pathways. Cell 2004;117:69–81. [DOI] [PubMed] [Google Scholar]
- [203].Hand NJ, Master ZR, Le Lay J, Friedman JR. Hepatic function is preserved in the absence of mature micrornas. Hepatology 2009;49:618–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Zhou J, Ju W, Wang D, Wu L, Zhu X, Guo Z, He X. Down- regulation of microrna-26a promotes mouse hepatocyte proliferation during liver regeneration. PLoS One 2012;7:e33577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Pan C, Chen H, Wang L, Yang S, Fu H, Zheng Y, Miao M, Jiao B. Down-regulation of mir-127 facilitates hepatocyte proliferation during rat liver regeneration. PLoS One 2012;7:e39151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Zhang B, Pan X, Cobb GP, Anderson TA. Micrornas as oncogenes and tumor suppressors. Dev Biol 2007;302:1–12. [DOI] [PubMed] [Google Scholar]
- [207].Ng R, Song G, Roll GR, Frandsen NM, Willenbring H. A microrna-21 surge facilitates rapid cyclin d1 translation and cell cycle progression in mouse liver regeneration. J Clin Invest 2012;122:1097–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Chen H, Sun Y, Dong R, Yang S, Pan C, Xiang D, Miao M, Jiao B. Mir-34a is upregulated during liver regeneration in rats and is associated with the suppression of hepatocyte proliferation. PLoS One 2011;6:e20238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, Baldan A, Esplugues E, Fernandez-Hernando C. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 2012;11:922–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Song G, Sharma AD, Roll GR, Ng R, Lee AY, Blelloch RH, Frandsen NM, Willenbring H. Micrornas control hepatocyte proliferation during liver regeneration. Hepatology 2010;51:1735–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Marquez RT, Wendlandt E, Galle CS, Keck K, McCaffrey AP. Microrna-21 is upregulated during the proliferative phase of liver regeneration, targets pellino-1, and inhibits nf-kappab signaling. Am J Physiol Gastrointest Liver Physiol 2010;298:G535–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Castro RE, Ferreira DM, Zhang X, Borralho PM, Sarver AL, Zeng Y, Steer CJ, Kren BT, Rodrigues CM. Identification of micrornas during rat liver regeneration after partial hepatectomy and modulation by ursodeoxycholic acid. Am J Physiol Gastrointest Liver Physiol 2010;299:G887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Yan-nan B, Zhao-yan Y, Li-xi L, jiang Y, Qing-jie X, Yong Z. Microrna-21 accelerates hepatocyte proliferation in vitro via pi3k/akt signaling by targeting pten. Biochem Biophys Res Commun 2014;443:802–807. [DOI] [PubMed] [Google Scholar]
- [214].Yuan Q, Loya K, Rani B, Mobus S, Balakrishnan A, Lamle J, Cathomen T, Vogel A, Manns MP, Ott M, Cantz T, Sharma AD. Microrna-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 2013;57:299–310. [DOI] [PubMed] [Google Scholar]
- [215].Bei Y, Song Y, Wang F, Dimitrova-Shumkovska J, Xiang Y, Zhao Y, Liu J, Xiao J, Yang C. Mir-382 targeting pten-akt axis promotes liver regeneration. Oncotarget 2016;7:1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Raschzok N, Sallmon H, Dame C, Sauer IM. Liver regeneration after partial hepatectomy: Inconsistent results of expression screenings for human, mouse, and rat micrornas. Am J Physiol Gastrointest Liver Physiol 2012;302:G470–471. [DOI] [PubMed] [Google Scholar]
- [217].Shu J, Kren BT, Xia Z, Wong PY, Li L, Hanse EA, Min MX, Li B, Albrecht JH, Zeng Y, Subramanian S, Steer CJ. Genomewide microrna down-regulation as a negative feedback mechanism in the early phases of liver regeneration. Hepatology 2011;54:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Schug J, McKenna LB, Walton G, Hand N, Mukherjee S, Essuman K, Shi Z, Gao Y, Markley K, Nakagawa M, Kameswaran V, Vourekas A, Friedman JR, Kaestner KH, Greenbaum LE. Dynamic recruitment of micrornas to their mrna targets in the regenerating liver. BMC Genomics 2013;14:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Benes V, Collier P, Kordes C, Stolte J, Rausch T, Muckentaler MU, Haussinger D, Castoldi M. Identification of cytokine- induced modulation of microrna expression and secretion as measured by a novel microrna specific qpcr assay. Sci Rep 2015;5:11590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Barone M, Francavilla A, Polimeno L, Ierardi E, Romanelli D, Berloco P, Di Leo A, Panella C. Modulation of rat hepatocyte proliferation by bile salts: In vitro and in vivo studies. Hepatology 1996;23:1159–1166. [DOI] [PubMed] [Google Scholar]
- [221].Ueda J, Chijiiwa K, Nakano K, Zhao G, Tanaka M. Lack of intestinal bile results in delayed liver regeneration of normal rat liver after hepatectomy accompanied by impaired cyclin e- associated kinase activity. Surgery 2002;131:564–573. [DOI] [PubMed] [Google Scholar]
- [222].Otao R, Beppu T, Isiko T, Mima K, Okabe H, Hayashi H, Masuda T, Chikamoto A, Takamori H, Baba H. External biliary drainage and liver regeneration after major hepatectomy. Br J Surg 2012;99:1569–1574. [DOI] [PubMed] [Google Scholar]
- [223].Rodrigues AD, Lai Y, Cvijic ME, Elkin LL, Zvyaga T, Soars MG. Drug-induced perturbations of the bile acid pool, cholestasis, and hepatotoxicity: Mechanistic considerations beyond the direct inhibition of the bile salt export pump. Drug Metab Dispos 2014;42:566–574. [DOI] [PubMed] [Google Scholar]
- [224].Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev 2014;66:948–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Chiang JY. Bile acids: Regulation of synthesis. J Lipid Res 2009;50:1955–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Myant NB, Mitropoulos KA. Cholesterol 7 alpha-hydroxylase. J Lipid Res 1977;18:135–153. [PubMed] [Google Scholar]
- [227].Bjorkhem I, Einarsson K, Melone P, Hylemon P. Mechanism of intestinal formation of deoxycholic acid from cholic acid in humans: Evidence for a 3-oxo-delta 4-steroid intermediate. J Lipid Res 1989;30:1033–1039. [PubMed] [Google Scholar]
- [228].Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res 2006;47:241–259. [DOI] [PubMed] [Google Scholar]
- [229].Hylemon PB, Melone PD, Franklund CV, Lund E, Bjorkhem I. Mechanism of intestinal 7 alpha-dehydroxylation of cholic acid: Evidence that allo-deoxycholic acid is an inducible side- product. J Lipid Res 1991;32:89–96. [PubMed] [Google Scholar]
- [230].Visschers RG, Luyer MD, Schaap FG, Olde Damink SW, Soeters PB. The gut-liver axis. Curr Opin Clin Nutr Metab Care 2013;16:576–581. [DOI] [PubMed] [Google Scholar]
- [231].Alnouti Y Bile acid sulfation: A pathway of bile acid elimination and detoxification. Toxicol Sci 2009;108:225–246. [DOI] [PubMed] [Google Scholar]
- [232].Chiang JYL. Bile acid metabolism and signaling. Compr Physiol 2013;3:1191–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Hofmann AF. The enterohepatic circulation of bile acids in mammals: Form and functions. Front Biosci (Landmark Ed) 2009;14:2584–2598. [DOI] [PubMed] [Google Scholar]
- [234].Woolbright BL, Jaeschke H. Critical factors in the assessment of cholestatic liver injury in vitro. Methods Mol Biol 2015;1250:363–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Zhang Y, Hong JY, Rockwell CE, Copple BL, Jaeschke H, Klaassen CD. Effect of bile duct ligation on bile acid composition in mouse serum and liver. Liver Int 2012;32:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Otsuki M Pathophysiological role of cholecystokinin in humans. J Gastroenterol Hepatol 2000;15 Suppl:D71–83. [DOI] [PubMed] [Google Scholar]
- [237].Hagenbuch B, Stieger B, Foguet M, Lubbert H, Meier PJ. Functional expression cloning and characterization of the hepatocyte na+/bile acid cotransport system. Proc Natl Acad Sci U S A 1991;88:10629–10633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Stieger B, Hagenbuch B, Landmann L, Hochli M, Schroeder A, Meier PJ. In situ localization of the hepatocytic na+/taurocholate cotransporting polypeptide in rat liver. Gastroenterology 1994;107:1781–1787. [DOI] [PubMed] [Google Scholar]
- [239].Meier PJ, Stieger B. Bile salt transporters. Annu Rev Physiol 2002;64:635–661. [DOI] [PubMed] [Google Scholar]
- [240].Vaz FM, Paulusma CC, Huidekoper H, de Ru M, Lim C, Koster J, Ho-Mok K, Bootsma AH, Groen AK, Schaap FG, Oude Elferink RP, Waterham HR, Wanders RJ. Sodium taurocholate cotransporting polypeptide (slc10a1) deficiency: Conjugated hypercholanemia without a clear clinical phenotype. Hepatology 2015;61:260–267. [DOI] [PubMed] [Google Scholar]
- [241].Slijepcevic D, Kaufman C, Wichers CG, Gilglioni EH, Lempp FA, Duijst S, de Waart DR, Elferink RP, Mier W, Stieger B, Beuers U, Urban S, van de Graaf SF. Impaired uptake of conjugated bile acids and hepatitis b virus pres1-binding in na(+) -taurocholate cotransporting polypeptide knockout mice. Hepatology 2015;62:207–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Boyer JL. Bile formation and secretion. Compr Physiol 2013;3:1035–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Akita H, Suzuki H, Ito K, Kinoshita S, Sato N, Takikawa H, Sugiyama Y. Characterization of bile acid transport mediated by multidrug resistance associated protein 2 and bile salt export pump. Biochim Biophys Acta 2001;1511:7–16. [DOI] [PubMed] [Google Scholar]
- [244].Berge KE, Tian H, Graf GA, Yu L, Grishin NV, Schultz J, Kwiterovich P, Shan B, Barnes R, Hobbs HH. Accumulation of dietary cholesterol in sitosterolemia caused by mutations in adjacent abc transporters. Science 2000;290:1771–1775. [DOI] [PubMed] [Google Scholar]
- [245].Smit JJ, Schinkel AH, Oude Elferink RP, Groen AK, Wagenaar E, van Deemter L, Mol CA, Ottenhoff R, van der Lugt NM, van Roon MA, et al. Homozygous disruption of the murine mdr2 p-glycoprotein gene leads to a complete absence of phospholipid from bile and to liver disease. Cell 1993;75:451–462. [DOI] [PubMed] [Google Scholar]
- [246].Shneider BL. Expression cloning of the ileal sodium-dependent bile acid transporter. J Pediatr Gastroenterol Nutr 1995;20:233–235. [DOI] [PubMed] [Google Scholar]
- [247].Ballatori N, Christian WV, Lee JY, Dawson PA, Soroka CJ, Boyer JL, Madejczyk MS, Li N. Ostalpha-ostbeta: A major basolateral bile acid and steroid transporter in human intestinal, renal, and biliary epithelia. Hepatology 2005;42:1270–1279. [DOI] [PubMed] [Google Scholar]
- [248].Rao A, Haywood J, Craddock AL, Belinsky MG, Kruh GD, Dawson PA. The organic solute transporter alpha-beta, ostalpha-ostbeta, is essential for intestinal bile acid transport and homeostasis. Proc Natl Acad Sci U S A 2008;105:3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Mangelsdorf DJ, Evans RM. The rxr heterodimers and orphan receptors. Cell 1995;83:841–850. [DOI] [PubMed] [Google Scholar]
- [250].Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell 1995;83:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Chiang JY. Bile acid regulation of gene expression: Roles of nuclear hormone receptors. Endocr Rev 2002;23:443–463. [DOI] [PubMed] [Google Scholar]
- [252].Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T, Noonan DJ, Burka LT, McMorris T, Lamph WW, Evans RM, Weinberger C. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995;81:687–693. [DOI] [PubMed] [Google Scholar]
- [253].Higashiyama H, Kinoshita M, Asano S. Immunolocalization of farnesoid x receptor (fxr) in mouse tissues using tissue microarray. Acta Histochem 2008;110:86–93. [DOI] [PubMed] [Google Scholar]
- [254].Zhang L, Wang YD, Chen WD, Wang X, Lou G, Liu N, Lin M, Forman BM, Huang W. Promotion of liver regeneration/repair by farnesoid x receptor in both liver and intestine in mice. Hepatology 2012;56:2336–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Gartung C, Matern S. Molecular regulation of sinusoidal liver bile acid transporters during cholestasis. Yale J Biol Med 1997;70:355–363. [PMC free article] [PubMed] [Google Scholar]
- [256].Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid x receptor/bile acid receptor. J Biol Chem 2001;276:28857–28865. [DOI] [PubMed] [Google Scholar]
- [257].Kong B, Huang J, Zhu Y, Li G, Williams J, Shen S, Aleksunes LM, Richardson JR, Apte U, Rudnick DA, Guo GL. Fibroblast growth factor 15 deficiency impairs liver regeneration in mice. Am J Physiol Gastrointest Liver Physiol 2014;306:G893–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Xu Z, Ouyang L, Del Castillo-Olivares A, Pandak WM, Gil G. Alpha(1)-fetoprotein transcription factor (ftf)/liver receptor homolog-1 (lrh-1) is an essential lipogenic regulator. Biochim Biophys Acta 2010;1801:473–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Nitta M, Ku S, Brown C, Okamoto AY, Shan B. Cpf: An orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A 1999;96:6660–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Lu TT, Makishima M, Repa JJ, Schoonjans K, Kerr TA, Auwerx J, Mangelsdorf DJ. Molecular basis for feedback regulation of bile acid synthesis by nuclear receptors. Mol Cell 2000;6:507–515. [DOI] [PubMed] [Google Scholar]
- [261].Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of lrh-1 and mrp3(abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem 2003;278:36688–36698. [DOI] [PubMed] [Google Scholar]
- [262].Frankenberg T, Rao A, Chen F, Haywood J, Shneider BL, Dawson PA. Regulation of the mouse organic solute transporter alpha-beta, ostalpha-ostbeta, by bile acids. Am J Physiol Gastrointest Liver Physiol 2006;290:G912–922. [DOI] [PubMed] [Google Scholar]
- [263].Out C, Hageman J, Bloks VW, Gerrits H, Sollewijn Gelpke MD, Bos T, Havinga R, Smit MJ, Kuipers F, Groen AK. Liver receptor homolog-1 is critical for adequate up-regulation of cyp7a1 gene transcription and bile salt synthesis during bile salt sequestration. Hepatology 2011;53:2075–2085. [DOI] [PubMed] [Google Scholar]
- [264].Lee YK, Schmidt DR, Cummins CL, Choi M, Peng L, Zhang Y, Goodwin B, Hammer RE, Mangelsdorf DJ, Kliewer SA. Liver receptor homolog-1 regulates bile acid homeostasis but is not essential for feedback regulation of bile acid synthesis. Mol Endocrinol 2008;22:1345–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Chawla A, Saez E, Evans RM. “Don’t know much bile-ology”. Cell 2000;103:1–4. [DOI] [PubMed] [Google Scholar]
- [266].Goodwin B, Jones SA, Price RR, Watson MA, McKee DD, Moore LB, Galardi C, Wilson JG, Lewis MC, Roth ME, Maloney PR, Willson TM, Kliewer SA. A regulatory cascade of the nuclear receptors fxr, shp-1, and lrh-1 represses bile acid biosynthesis. Mol Cell 2000;6:517–526. [DOI] [PubMed] [Google Scholar]
- [267].Mataki C, Magnier BC, Houten SM, Annicotte JS, Argmann C, Thomas C, Overmars H, Kulik W, Metzger D, Auwerx J, Schoonjans K. Compromised intestinal lipid absorption in mice with a liver-specific deficiency of liver receptor homolog 1. Mol Cell Biol 2007;27:8330–8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].del Castillo-Olivares A, Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha -hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem 2000;275:17793–17799. [DOI] [PubMed] [Google Scholar]
- [269].Freeman LA, Kennedy A, Wu J, Bark S, Remaley AT, Santamarina-Fojo S, Brewer HB, Jr. The orphan nuclear receptor lrh-1 activates the abcg5/abcg8 intergenic promoter. J Lipid Res 2004;45:1197–1206. [DOI] [PubMed] [Google Scholar]
- [270].Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, Lustig KD, Mangelsdorf DJ, Shan B. Identification of a nuclear receptor for bile acids. Science 1999;284:1362–1365. [DOI] [PubMed] [Google Scholar]
- [271].Parks DJ, Blanchard SG, Bledsoe RK, Chandra G, Consler TG, Kliewer SA, Stimmel JB, Willson TM, Zavacki AM, Moore DD, Lehmann JM. Bile acids: Natural ligands for an orphan nuclear receptor. Science 1999;284:1365–1368. [DOI] [PubMed] [Google Scholar]
- [272].Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor fxr/bar. Mol Cell 1999;3:543–553. [DOI] [PubMed] [Google Scholar]
- [273].Zollner G, Wagner M, Moustafa T, Fickert P, Silbert D, Gumhold J, Fuchsbichler A, Halilbasic E, Denk H, Marschall HU, Trauner M. Coordinated induction of bile acid detoxification and alternative elimination in mice: Role of fxr- regulated organic solute transporter-alpha/beta in the adaptive response to bile acids. Am J Physiol Gastrointest Liver Physiol 2006;290:G923–932. [DOI] [PubMed] [Google Scholar]
- [274].Lee H, Zhang Y, Lee FY, Nelson SF, Gonzalez FJ, Edwards PA. Fxr regulates organic solute transporters alpha and beta in the adrenal gland, kidney, and intestine. J Lipid Res 2006;47:201–214. [DOI] [PubMed] [Google Scholar]
- [275].Landrier JF, Eloranta JJ, Vavricka SR, Kullak-Ublick GA. The nuclear receptor for bile acids, fxr, transactivates human organic solute transporter-alpha and -beta genes. Am J Physiol Gastrointest Liver Physiol 2006;290:G476–485. [DOI] [PubMed] [Google Scholar]
- [276].Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab 2005;2:217–225. [DOI] [PubMed] [Google Scholar]
- [277].Holt JA, Luo G, Billin AN, Bisi J, McNeill YY, Kozarsky KF, Donahee M, Wang DY, Mansfield TA, Kliewer SA, Goodwin B, Jones SA. Definition of a novel growth factor-dependent signal cascade for the suppression of bile acid biosynthesis. Genes Dev 2003;17:1581–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Boyer JL, Trauner M, Mennone A, Soroka CJ, Cai SY, Moustafa T, Zollner G, Lee JY, Ballatori N. Upregulation of a basolateral fxr-dependent bile acid efflux transporter ostalpha- ostbeta in cholestasis in humans and rodents. Am J Physiol Gastrointest Liver Physiol 2006;290:G1124–1130. [DOI] [PubMed] [Google Scholar]
- [279].Kast HR, Goodwin B, Tarr PT, Jones SA, Anisfeld AM, Stoltz CM, Tontonoz P, Kliewer S, Willson TM, Edwards PA. Regulation of multidrug resistance-associated protein 2 (abcc2) by the nuclear receptors pregnane x receptor, farnesoid x- activated receptor, and constitutive androstane receptor. J Biol Chem 2002;277:2908–2915. [DOI] [PubMed] [Google Scholar]
- [280].Barbier O, Torra IP, Sirvent A, Claudel T, Blanquart C, Duran- Sandoval D, Kuipers F, Kosykh V, Fruchart JC, Staels B. Fxr induces the ugt2b4 enzyme in hepatocytes: A potential mechanism of negative feedback control of fxr activity. Gastroenterology 2003;124:1926–1940. [DOI] [PubMed] [Google Scholar]
- [281].Lu Y, Heydel JM, Li X, Bratton S, Lindblom T, Radominska- Pandya A. Lithocholic acid decreases expression of ugt2b7 in caco-2 cells: A potential role for a negative farnesoid x receptor response element. Drug Metab Dispos 2005;33:937–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Chen WD, Wang YD, Zhang L, Shiah S, Wang M, Yang F, Yu D, Forman BM, Huang W. Farnesoid x receptor alleviates age- related proliferation defects in regenerating mouse livers by activating forkhead box m1b transcription. Hepatology 2010;51:953–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Wang X, Quail E, Hung NJ, Tan Y, Ye H, Costa RH. Increased levels of forkhead box m1b transcription factor in transgenic mouse hepatocytes prevent age-related proliferation defects in regenerating liver. Proc Natl Acad Sci U S A 2001;98:11468–11473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Wang X, Krupczak-Hollis K, Tan Y, Dennewitz MB, Adami GR, Costa RH. Increased hepatic forkhead box m1b (foxm1b) levels in old-aged mice stimulated liver regeneration through diminished p27kip1 protein levels and increased cdc25b expression. J Biol Chem 2002;277:44310–44316. [DOI] [PubMed] [Google Scholar]
- [285].Wang X, Kiyokawa H, Dennewitz MB, Costa RH. The forkhead box m1b transcription factor is essential for hepatocyte DNA replication and mitosis during mouse liver regeneration. Proc Natl Acad Sci U S A 2002;99:16881–16886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Krupczak-Hollis K, Wang X, Dennewitz MB, Costa RH. Growth hormone stimulates proliferation of old-aged regenerating liver through forkhead box m1b. Hepatology 2003;38:1552–1562. [DOI] [PubMed] [Google Scholar]
- [287].Wang X, Bhattacharyya D, Dennewitz MB, Kalinichenko VV, Zhou Y, Lepe R, Costa RH. Rapid hepatocyte nuclear translocation of the forkhead box m1b (foxm1b) transcription factor caused a transient increase in size of regenerating transgenic hepatocytes. Gene Expr 2003;11:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Major ML, Lepe R, Costa RH. Forkhead box m1b transcriptional activity requires binding of cdk-cyclin complexes for phosphorylation-dependent recruitment of p300/cbp coactivators. Mol Cell Biol 2004;24:2649–2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Mencalha AL, Binato R, Ferreira GM, Du Rocher B, Abdelhay E. Forkhead box m1 (foxm1) gene is a new stat3 transcriptional factor target and is essential for proliferation, survival and DNA repair of k562 cell line. PLoS One 2012;7:e48160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Liu HX, Ly I, Hu Y, Wan YJ. Retinoic acid regulates cell cycle genes and accelerates normal mouse liver regeneration. Biochem Pharmacol 2014;91:256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. The human orphan nuclear receptor pxr is activated by compounds that regulate cyp3a4 gene expression and cause drug interactions. J Clin Invest 1998;102:1016–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Denson LA, Auld KL, Schiek DS, McClure MH, Mangelsdorf DJ, Karpen SJ. Interleukin-1beta suppresses retinoid transactivation of two hepatic transporter genes involved in bile formation. J Biol Chem 2000;275:8835–8843. [DOI] [PubMed] [Google Scholar]
- [293].Lo Sasso G, Celli N, Caboni M, Murzilli S, Salvatore L, Morgano A, Vacca M, Pagliani T, Parini P, Moschetta A. Down-regulation of the lxr transcriptome provides the requisite cholesterol levels to proliferating hepatocytes. Hepatology 2010;51:1334–1344. [DOI] [PubMed] [Google Scholar]
- [294].Carnahan VE, Redinbo MR. Structure and function of the human nuclear xenobiotic receptor pxr. Curr Drug Metab 2005;6:357–367. [DOI] [PubMed] [Google Scholar]
- [295].Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, Liu Y, Klaassen CD, Brown KK, Reinhard J, Willson TM, Koller BH, Kliewer SA. The nuclear receptor pxr is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci U S A 2001;98:3369–3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Shizu R, Abe T, Benoki S, Takahashi M, Kodama S, Miayata M, Matsuzawa A, Yoshinari K. Pxr stimulates growth factor- mediated hepatocyte proliferation by cross-talk with the foxo transcription factor. Biochem J 2016;473:257–266. [DOI] [PubMed] [Google Scholar]
- [297].Pascussi JM, Gerbal-Chaloin S, Pichard-Garcia L, Daujat M, Fabre JM, Maurel P, Vilarem MJ. Interleukin-6 negatively regulates the expression of pregnane x receptor and constitutively activated receptor in primary human hepatocytes. Biochem Biophys Res Commun 2000;274:707–713. [DOI] [PubMed] [Google Scholar]
- [298].Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen CD. Coordinate regulation of xenobiotic and bile acid homeostasis by pregnane x receptor. Drug Metab Dispos 2001;29:1467–1472. [PubMed] [Google Scholar]
- [299].Li CY, Renaud HJ, Klaassen CD, Cui JY. Age-specific regulation of drug-processing genes in mouse liver by ligands of xenobiotic-sensing transcription factors. Drug Metab Dispos 2016;44:1038–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Zhou J, Zhai Y, Mu Y, Gong H, Uppal H, Toma D, Ren S, Evans RM, Xie W. A novel pregnane x receptor-mediated and sterol regulatory element-binding protein-independent lipogenic pathway. J Biol Chem 2006;281:15013–15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Zhou J, Febbraio M, Wada T, Zhai Y, Kuruba R, He J, Lee JH, Khadem S, Ren S, Li S, Silverstein RL, Xie W. Hepatic fatty acid transporter cd36 is a common target of lxr, pxr, and ppargamma in promoting steatosis. Gastroenterology 2008;134:556–567. [DOI] [PubMed] [Google Scholar]
- [302].Kliewer SA, Willson TM. Regulation of xenobiotic and bile acid metabolism by the nuclear pregnane x receptor. J Lipid Res 2002;43:359–364. [PubMed] [Google Scholar]
- [303].Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane x receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem 2004;279:49517–49522. [DOI] [PubMed] [Google Scholar]
- [304].Columbano A, Ledda-Columbano GM, Pibiri M, Cossu C, Menegazzi M, Moore DD, Huang W, Tian J, Locker J. Gadd45beta is induced through a car-dependent, tnf- independent pathway in murine liver hyperplasia. Hepatology 2005;42:1118–1126. [DOI] [PubMed] [Google Scholar]
- [305].Yang H, Wang H. Signaling control of the constitutive androstane receptor (car). Protein Cell 2014;5:113–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Gerbal-Chaloin S, Daujat M, Pascussi JM, Pichard-Garcia L, Vilarem MJ, Maurel P. Transcriptional regulation of cyp2c9 gene. Role of glucocorticoid receptor and constitutive androstane receptor. J Biol Chem 2002;277:209–217. [DOI] [PubMed] [Google Scholar]
- [307].Waxman DJ. P450 gene induction by structurally diverse xenochemicals: Central role of nuclear receptors car, pxr, and ppar. Arch Biochem Biophys 1999;369:11–23. [DOI] [PubMed] [Google Scholar]
- [308].Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor car in regulating hepatic genes in response to phenobarbital. Mol Pharmacol 2002;61:1–6. [DOI] [PubMed] [Google Scholar]
- [309].Sueyoshi T, Kawamoto T, Zelko I, Honkakoski P, Negishi M. The repressed nuclear receptor car responds to phenobarbital in activating the human cyp2b6 gene. J Biol Chem 1999;274:6043–6046. [DOI] [PubMed] [Google Scholar]
- [310].Goodwin B, Hodgson E, D’Costa DJ, Robertson GR, Liddle C. Transcriptional regulation of the human cyp3a4 gene by the constitutive androstane receptor. Mol Pharmacol 2002;62:359–365. [DOI] [PubMed] [Google Scholar]
- [311].Robertson GR, Field J, Goodwin B, Bierach S, Tran M, Lehnert A, Liddle C. Transgenic mouse models of human cyp3a4 gene regulation. Mol Pharmacol 2003;64:42–50. [DOI] [PubMed] [Google Scholar]
- [312].Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic mrp4 and sult2a as revealed by the constitutive androstane receptor and mrp4 knockout mice. J Biol Chem 2004;279:22250–22257. [DOI] [PubMed] [Google Scholar]
- [313].Yueh MF, Mellon PL, Tukey RH. Inhibition of human ugt2b7 gene expression in transgenic mice by the constitutive androstane receptor. Mol Pharmacol 2011;79:1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Wada T, Gao J, Xie W. Pxr and car in energy metabolism. Trends Endocrinol Metab 2009;20:273–279. [DOI] [PubMed] [Google Scholar]
- [315].Yi YW, Kim D, Jung N, Hong SS, Lee HS, Bae I. Gadd45 family proteins are coactivators of nuclear hormone receptors. Biochem Biophys Res Commun 2000;272:193–198. [DOI] [PubMed] [Google Scholar]
- [316].Yamamoto Y, Negishi M. The antiapoptotic factor growth arrest and DNA-damage-inducible 45 beta regulates the nuclear receptor constitutive active/androstane receptor-mediated transcription. Drug Metab Dispos 2008;36:1189–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Liebermann DA, Hoffman B. Gadd45 in the response of hematopoietic cells to genotoxic stress. Blood Cells Mol Dis 2007;39:329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Lu B, Ferrandino AF, Flavell RA. Gadd45beta is important for perpetuating cognate and inflammatory signals in t cells. Nat Immunol 2004;5:38–44. [DOI] [PubMed] [Google Scholar]
- [319].Wollam J, Antebi A. Sterol regulation of metabolism, homeostasis, and development. Annu Rev Biochem 2011;80:885–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [320].Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor lxr alpha. Nature 1996;383:728–731. [DOI] [PubMed] [Google Scholar]
- [321].Yu L, Gupta S, Xu F, Liverman AD, Moschetta A, Mangelsdorf DJ, Repa JJ, Hobbs HH, Cohen JC. Expression of abcg5 and abcg8 is required for regulation of biliary cholesterol secretion. J Biol Chem 2005;280:8742–8747. [DOI] [PubMed] [Google Scholar]
- [322].Kazantseva YA, Yarushkin AA, Mostovich LA, Pustylnyak YA, Pustylnyak VO. Xenosensor car mediates down-regulation of mir-122 and up-regulation of mir-122 targets in the liver. Toxicol Appl Pharmacol 2015;288:26–32. [DOI] [PubMed] [Google Scholar]
- [323].Crestani M, Sadeghpour A, Stroup D, Galli G, Chiang JY. Transcriptional activation of the cholesterol 7alpha- hydroxylase gene (cyp7a) by nuclear hormone receptors. J Lipid Res 1998;39:2192–2200. [PubMed] [Google Scholar]
- [324].De Fabiani E, Mitro N, Anzulovich AC, Pinelli A, Galli G, Crestani M. The negative effects of bile acids and tumor necrosis factor-alpha on the transcription of cholesterol 7alpha- hydroxylase gene (cyp7a1) converge to hepatic nuclear factor- 4: A novel mechanism of feedback regulation of bile acid synthesis mediated by nuclear receptors. J Biol Chem 2001;276:30708–30716. [DOI] [PubMed] [Google Scholar]
- [325].Yang Y, Zhang M, Eggertsen G, Chiang JY. On the mechanism of bile acid inhibition of rat sterol 12alpha-hydroxylase gene (cyp8b1) transcription: Roles of alpha-fetoprotein transcription factor and hepatocyte nuclear factor 4alpha. Biochim Biophys Acta 2002;1583:63–73. [DOI] [PubMed] [Google Scholar]
- [326].Zhang M, Chiang JY. Transcriptional regulation of the human sterol 12alpha-hydroxylase gene (cyp8b1): Roles of heaptocyte nuclear factor 4alpha in mediating bile acid repression. J Biol Chem 2001;276:41690–41699. [DOI] [PubMed] [Google Scholar]
- [327].Chen W, Chiang JY. Regulation of human sterol 27- hydroxylase gene (cyp27a1) by bile acids and hepatocyte nuclear factor 4alpha (hnf4alpha). Gene 2003;313:71–82. [DOI] [PubMed] [Google Scholar]
- [328].Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4alpha (nuclear receptor 2a1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol Cell Biol 2001;21:1393–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Nagaki M, Moriwaki H. Transcription factor hnf and hepatocyte differentiation. Hepatol Res 2008;38:961–969. [DOI] [PubMed] [Google Scholar]
- [330].Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Nuclear factor erythroid 2-related factor 2 is a positive regulator of human bile salt export pump expression. Hepatology 2009;50:1588–1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Weerachayaphorn J, Mennone A, Soroka CJ, Harry K, Hagey LR, Kensler TW, Boyer JL. Nuclear factor-e2-related factor 2 is a major determinant of bile acid homeostasis in the liver and ntestine. Am J Physiol Gastrointest Liver Physiol 2012;302:G925–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Barbier O, Duran-Sandoval D, Pineda-Torra I, Kosykh V, Fruchart JC, Staels B. Peroxisome proliferator-activated receptor alpha induces hepatic expression of the human bile acid glucuronidating udp-glucuronosyltransferase 2b4 enzyme. J Biol Chem 2003;278:32852–32860. [DOI] [PubMed] [Google Scholar]
- [333].Makishima M, Lu TT, Xie W, Whitfield GK, Domoto H, Evans RM, Haussler MR, Mangelsdorf DJ. Vitamin d receptor as an intestinal bile acid sensor. Science 2002;296:1313–1316. [DOI] [PubMed] [Google Scholar]
- [334].Rust C, Wild N, Bernt C, Vennegeerts T, Wimmer R, Beuers U. Bile acid-induced apoptosis in hepatocytes is caspase-6- dependent. J Biol Chem 2009;284:2908–2916. [DOI] [PubMed] [Google Scholar]
- [335].Zucchini-Pascal N, de Sousa G, Pizzol J, Rahmani R. Pregnane x receptor activation protects rat hepatocytes against deoxycholic acid-induced apoptosis. Liver Int 2010;30:284–297. [DOI] [PubMed] [Google Scholar]
- [336].Olthof PB, Coelen RJ, Wiggers JK, Besselink MG, Busch OR, van Gulik TM. External biliary drainage following major liver resection for perihilar cholangiocarcinoma: Impact on development of liver failure and biliary leakage. HPB (Oxford) 2016;18:348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Ortiz DF, Li S, Iyer R, Zhang X, Novikoff P, Arias IM. Mrp3, a new atp-binding cassette protein localized to the canalicular domain of the hepatocyte. Am J Physiol 1999;276:G1493–1500. [DOI] [PubMed] [Google Scholar]
- [338].Hirohashi T, Suzuki H, Takikawa H, Sugiyama Y. Atp- dependent transport of bile salts by rat multidrug resistance- associated protein 3 (mrp3). J Biol Chem 2000;275:2905–2910. [DOI] [PubMed] [Google Scholar]
- [339].Fernandez-Barrena MG, Monte MJ, Latasa MU, Uriarte I, Vicente E, Chang HC, Rodriguez-Ortigosa CM, Elferink RO, Berasain C, Marin JJ, Prieto J, Avila MA. Lack of abcc3 expression impairs bile-acid induced liver growth and delays hepatic regeneration after partial hepatectomy in mice. J Hepatol 2012;56:367–373. [DOI] [PubMed] [Google Scholar]
- [340].Holmberg-Betsholtz I, Lund E, Bjorkhem I, Wikvall K. Sterol 27-hydroxylase in bile acid biosynthesis. Mechanism of oxidation of 5 beta-cholestane-3 alpha,7 alpha,12 alpha,27- tetrol into 3 alpha,7 alpha,12 alpha-trihydroxy-5 beta- cholestanoic acid. J Biol Chem 1993;268:11079–11085. [PubMed] [Google Scholar]
- [341].Meng Z, Liu N, Fu X, Wang X, Wang YD, Chen WD, Zhang L, Forman BM, Huang W. Insufficient bile acid signaling impairs liver repair in cyp27(−/−) mice. J Hepatol 2011;55:885–895. [DOI] [PubMed] [Google Scholar]
- [342].Dawson PA, Haywood J, Craddock AL, Wilson M, Tietjen M, Kluckman K, Maeda N, Parks JS. Targeted deletion of the ileal bile acid transporter eliminates enterohepatic cycling of bile acids in mice. J Biol Chem 2003;278:33920–33927. [DOI] [PubMed] [Google Scholar]
- [343].Oelkers P, Kirby LC, Heubi JE, Dawson PA. Primary bile acid malabsorption caused by mutations in the ileal sodium- dependent bile acid transporter gene (slc10a2). J Clin Invest 1997;99:1880–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Shaulian E, Karin M. Ap-1 in cell proliferation and survival. Oncogene 2001;20:2390–2400. [DOI] [PubMed] [Google Scholar]
- [345].Shaulian E, Karin M. Ap-1 as a regulator of cell life and death. Nat Cell Biol 2002;4:E131–136. [DOI] [PubMed] [Google Scholar]
- [346].Chen WD, Wang YD, Meng Z, Zhang L, Huang W. Nuclear bile acid receptor fxr in the hepatic regeneration. Biochim Biophys Acta 2011;1812:888–892. [DOI] [PubMed] [Google Scholar]
- [347].Yang X, Guo M, Wan YJ. Deregulation of growth factor, circadian clock, and cell cycle signaling in regenerating hepatocyte rxralpha-deficient mouse livers. Am J Pathol 2010;176:733–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [348].Imai T, Jiang M, Kastner P, Chambon P, Metzger D. Selective ablation of retinoid x receptor alpha in hepatocytes impairs their lifespan and regenerative capacity. Proc Natl Acad Sci U S A 2001;98:4581–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Dai G, He L, Bu P, Wan YJ. Pregnane x receptor is essential for normal progression of liver regeneration. Hepatology 2008;47:1277–1287. [DOI] [PubMed] [Google Scholar]
- [350].Tien ES, Matsui K, Moore R, Negishi M. The nuclear receptor constitutively active/androstane receptor regulates type 1 deiodinase and thyroid hormone activity in the regenerating mouse liver. J Pharmacol Exp Ther 2007;320:307–313. [DOI] [PubMed] [Google Scholar]
- [351].Yuan X, Yan S, Zhao J, Shi D, Yuan B, Dai W, Jiao B, Zhang W, Miao M. Lipid metabolism and peroxisome proliferator- activated receptor signaling pathways participate in late-phase liver regeneration. J Proteome Res 2011;10:1179–1190. [DOI] [PubMed] [Google Scholar]
- [352].Anderson SP, Yoon L, Richard EB, Dunn CS, Cattley RC, Corton JC. Delayed liver regeneration in peroxisome proliferator-activated receptor-alpha-null mice. Hepatology 2002;36:544–554. [DOI] [PubMed] [Google Scholar]
- [353].Yamamoto Y, Ono T, Dhar DK, Yamanoi A, Tachibana M, Tanaka T, Nagasue N. Role of peroxisome proliferator- activated receptor-gamma (ppargamma) during liver regeneration in rats. J Gastroenterol Hepatol 2008;23:930–937. [DOI] [PubMed] [Google Scholar]
- [354].Gazit V, Huang J, Weymann A, Rudnick DA. Analysis of the role of hepatic ppargamma expression during mouse liver regeneration. Hepatology 2012;56:1489–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Geier A, Wagner M, Dietrich CG, Trauner M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochim Biophys Acta 2007;1773:283–308. [DOI] [PubMed] [Google Scholar]
- [356].Barchetta I, Carotti S, Labbadia G, Gentilucci UV, Muda AO, Angelico F, Silecchia G, Leonetti F, Fraioli A, Picardi A, Morini S, Cavallo MG. Liver vitamin d receptor, cyp2r1, and cyp27a1 expression: Relationship with liver histology and vitamin d3 levels in patients with nonalcoholic steatohepatitis or hepatitis c virus. Hepatology 2012;56:2180–2187. [DOI] [PubMed] [Google Scholar]
- [357].Schmidt DR, Holmstrom SR, Fon Tacer K, Bookout AL, Kliewer SA, Mangelsdorf DJ. Regulation of bile acid synthesis by fat-soluble vitamins a and d. J Biol Chem 2010;285:14486–14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Echchgadda I, Song CS, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase is a target for transcriptional induction by the vitamin d receptor. Mol Pharmacol 2004;65:720–729. [DOI] [PubMed] [Google Scholar]
- [359].Song CS, Echchgadda I, Seo YK, Oh T, Kim S, Kim SA, Cho S, Shi L, Chatterjee B. An essential role of the caat/enhancer binding protein-alpha in the vitamin d-induced expression of the human steroid/bile acid-sulfotransferase (sult2a1). Mol Endocrinol 2006;20:795–808. [DOI] [PubMed] [Google Scholar]
- [360].Chatterjee B, Echchgadda I, Song CS. Vitamin d receptor regulation of the steroid/bile acid sulfotransferase sult2a1. Methods Enzymol 2005;400:165–191. [DOI] [PubMed] [Google Scholar]
- [361].Chow EC, Maeng HJ, Liu S, Khan AA, Groothuis GM, Pang KS. 1alpha,25-dihydroxyvitamin d(3) triggered vitamin d receptor and farnesoid x receptor-like effects in rat intestine and liver in vivo. Biopharm Drug Dispos 2009;30:457–475. [DOI] [PubMed] [Google Scholar]
- [362].Columbano A, Ledda-Columbano GM, Pibiri M, Concas D, Reddy JK, Rao MS. Peroxisome proliferator-activated receptor-alpha mice show enhanced hepatocyte proliferation in response to the hepatomitogen 1,4-bis [2-(3,5- dichloropyridyloxy)] benzene, a ligand of constitutive androstane receptor. Hepatology 2001;34:262–266. [DOI] [PubMed] [Google Scholar]
- [363].Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor car mediates specific xenobiotic induction of drug metabolism. Nature 2000;407:920–923. [DOI] [PubMed] [Google Scholar]
- [364].Claudel T, Staels B, Kuipers F. The farnesoid x receptor: A molecular link between bile acid and lipid and glucose metabolism. Arterioscler Thromb Vasc Biol 2005;25:2020–2030. [DOI] [PubMed] [Google Scholar]
- [365].Houten SM, Watanabe M, Auwerx J. Endocrine functions of bile acids. EMBO J 2006;25:1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Lefebvre P, Cariou B, Lien F, Kuipers F, Staels B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol Rev 2009;89:147–191. [DOI] [PubMed] [Google Scholar]
- [367].Bechmann LP, Hannivoort RA, Gerken G, Hotamisligil GS, Trauner M, Canbay A. The interaction of hepatic lipid and glucose metabolism in liver diseases. J Hepatol 2012;56:952–964. [DOI] [PubMed] [Google Scholar]
- [368].Trauner M, Halilbasic E. Nuclear receptors as new perspective for the management of liver diseases. Gastroenterology 2011;140:1120–1125. [DOI] [PubMed] [Google Scholar]
- [369].Cariou B, Staels B. Fxr: A promising target for the metabolic syndrome? Trends Pharmacol Sci 2007;28:236–243. [DOI] [PubMed] [Google Scholar]
- [370].Sinal CJ, Tohkin M, Miyata M, Ward JM, Lambert G, Gonzalez FJ. Targeted disruption of the nuclear receptor fxr/bar impairs bile acid and lipid homeostasis. Cell 2000;102:731–744. [DOI] [PubMed] [Google Scholar]
- [371].Edwards PA, Kast HR, Anisfeld AM. Bareing it all: The adoption of lxr and fxr and their roles in lipid homeostasis. J Lipid Res 2002;43:2–12. [PubMed] [Google Scholar]
- [372].Zhang Y, Castellani LW, Sinal CJ, Gonzalez FJ, Edwards PA. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (pgc-1alpha) regulates triglyceride metabolism by activation of the nuclear receptor fxr. Genes Dev 2004;18:157–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Garcia-Rodriguez JL, Barbier-Torres L, Fernandez-Alvarez S, Gutierrez-de Juan V, Monte MJ, Halilbasic E, Herranz D, Alvarez L, Aspichueta P, Marin JJ, Trauner M, Mato JM, Serrano M, Beraza N, Martinez-Chantar ML. Sirt1 controls liver regeneration by regulating bile acid metabolism through farnesoid x receptor and mammalian target of rapamycin signaling. Hepatology 2014;59:1972–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].Lees E, Faha B, Dulic V, Reed SI, Harlow E. Cyclin e/cdk2 and cyclin a/cdk2 kinases associate with p107 and e2f in a temporally distinct manner. Genes Dev 1992;6:1874–1885. [DOI] [PubMed] [Google Scholar]
- [375].Rizzardi LF, Coleman KE, Varma D, Matson JP, Oh S, Cook JG. Cdk1-dependent inhibition of the e3 ubiquitin ligase crl4cdt2 ensures robust transition from s phase to mitosis. J Biol Chem 2015;290:556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].Harbour JW, Dean DC. The rb/e2f pathway: Expanding roles and emerging paradigms. Genes Dev 2000;14:2393–2409. [DOI] [PubMed] [Google Scholar]
- [377].Ishida S, Huang E, Zuzan H, Spang R, Leone G, West M, Nevins JR. Role for e2f in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol Cell Biol 2001;21:4684–4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [378].Jaime M, Pujol MJ, Serratosa J, Pantoja C, Canela N, Casanovas O, Serrano M, Agell N, Bachs O. The p21(cip1) protein, a cyclin inhibitor, regulates the levels and the intracellular localization of cdc25a in mice regenerating livers. Hepatology 2002;35:1063–1071. [DOI] [PubMed] [Google Scholar]
- [379].Nilsson I, Hoffmann I. Cell cycle regulation by the cdc25 phosphatase family. Prog Cell Cycle Res 2000;4:107–114. [DOI] [PubMed] [Google Scholar]
- [380].Sebastian B, Kakizuka A, Hunter T. Cdc25m2 activation of cyclin-dependent kinases by dephosphorylation of threonine-14 and tyrosine-15. Proc Natl Acad Sci U S A 1993;90:3521–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [381].Trembley JH, Ebbert JO, Kren BT, Steer CJ. Differential regulation of cyclin b1 rna and protein expression during hepatocyte growth in vivo. Cell Growth Differ 1996;7:903–916. [PubMed] [Google Scholar]
- [382].Savkur RS, Bramlett KS, Michael LF, Burris TP. Regulation of pyruvate dehydrogenase kinase expression by the farnesoid x receptor. Biochem Biophys Res Commun 2005;329:391–396. [DOI] [PubMed] [Google Scholar]
- [383].Jeoung NH, Wu P, Joshi MA, Jaskiewicz J, Bock CB, Depaoli- Roach AA, Harris RA. Role of pyruvate dehydrogenase kinase isoenzyme 4 (pdhk4) in glucose homoeostasis during starvation. Biochem J 2006;397:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [384].Roche TE, Hiromasa Y. Pyruvate dehydrogenase kinase regulatory mechanisms and inhibition in treating diabetes, heart ischemia, and cancer. Cell Mol Life Sci 2007;64:830–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Vander Heiden MG, Cantley LC, Thompson CB. Understanding the warburg effect: The metabolic requirements of cell proliferation. Science 2009;324:1029–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [386].Hamanaka RB, Chandel NS. Targeting glucose metabolism for cancer therapy. J Exp Med 2012;209:211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Jain M, Nilsson R, Sharma S, Madhusudhan N, Kitami T, Souza AL, Kafri R, Kirschner MW, Clish CB, Mootha VK. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science 2012;336:1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [388].Locasale JW. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat Rev Cancer 2013;13:572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [389].Potthoff MJ, Kliewer SA, Mangelsdorf DJ. Endocrine fibroblast growth factors 15/19 and 21: From feast to famine. Genes Dev 2012;26:312–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Lin BC, Wang M, Blackmore C, Desnoyers LR. Liver-specific activities of fgf19 require klotho beta. J Biol Chem 2007;282:27277–27284. [DOI] [PubMed] [Google Scholar]
- [391].Wu X, Ge H, Gupte J, Weiszmann J, Shimamoto G, Stevens J, Hawkins N, Lemon B, Shen W, Xu J, Veniant MM, Li YS, Lindberg R, Chen JL, Tian H, Li Y. Co-receptor requirements for fibroblast growth factor-19 signaling. J Biol Chem 2007;282:29069–29072. [DOI] [PubMed] [Google Scholar]
- [392].Kurosu H, Choi M, Ogawa Y, Dickson AS, Goetz R, Eliseenkova AV, Mohammadi M, Rosenblatt KP, Kliewer SA, Kuro-o M. Tissue-specific expression of betaklotho and fibroblast growth factor (fgf) receptor isoforms determines metabolic activity of fgf19 and fgf21. J Biol Chem 2007;282:26687–26695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [393].Wu X, Lemon B, Li X, Gupte J, Weiszmann J, Stevens J, Hawkins N, Shen W, Lindberg R, Chen JL, Tian H, Li Y. C- terminal tail of fgf19 determines its specificity toward klotho co-receptors. J Biol Chem 2008;283:33304–33309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [394].Wu X, Ge H, Lemon B, Vonderfecht S, Baribault H, Weiszmann J, Gupte J, Gardner J, Lindberg R, Wang Z, Li Y. Separating mitogenic and metabolic activities of fibroblast growth factor 19 (fgf19). Proc Natl Acad Sci U S A 2010;107:14158–14163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [395].Moldovan GL, Pfander B, Jentsch S. Pcna, the maestro of the replication fork. Cell 2007;129:665–679. [DOI] [PubMed] [Google Scholar]
- [396].Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid x receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 2012;56:1034–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Cressman DE, Diamond RH, Taub R. Rapid activation of the stat3 transcription complex in liver regeneration. Hepatology 1995;21:1443–1449. [PubMed] [Google Scholar]
- [398].Li W, Liang X, Kellendonk C, Poli V, Taub R. Stat3 contributes to the mitogenic response of hepatocytes during liver regeneration. J Biol Chem 2002;277:28411–28417. [DOI] [PubMed] [Google Scholar]
- [399].Kim JH, Kim JE, Liu HY, Cao W, Chen J. Regulation of interleukin-6-induced hepatic insulin resistance by mammalian target of rapamycin through the stat3-socs3 pathway. J Biol Chem 2008;283:708–715. [DOI] [PubMed] [Google Scholar]
- [400].Duckett CS, Perkins ND, Kowalik TF, Schmid RM, Huang ES, Baldwin AS Jr., Nabel GJ Dimerization of nf-kb2 with rela(p65) regulates DNA binding, transcriptional activation, and inhibition by an i kappa b-alpha (mad-3). Mol Cell Biol 1993;13:1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor fgfr4. J Biol Chem 2000;275:15482–15489. [DOI] [PubMed] [Google Scholar]
- [402].Ijssennagger N, Janssen AW, Milona A, Ramos Pittol JM, Hollman DA, Mokry M, Betzel B, Berends FJ, Janssen IM, van Mil SW, Kersten S. Gene expression profiling in human precision cut liver slices upon treatment with the fxr agonist obeticholic acid. J Hepatol 2016;64:1158–1166. [DOI] [PubMed] [Google Scholar]
- [403].Mudaliar S, Henry RR, Sanyal AJ, Morrow L, Marschall HU, Kipnes M, Adorini L, Sciacca CI, Clopton P, Castelloe E, Dillon P, Pruzanski M, Shapiro D. Efficacy and safety of the farnesoid x receptor agonist obeticholic acid in patients with type 2 diabetes and nonalcoholic fatty liver disease. Gastroenterology 2013;145:574–582. [DOI] [PubMed] [Google Scholar]
- [404].Li J, Pircher PC, Schulman IG, Westin SK. Regulation of complement c3 expression by the bile acid receptor fxr. J Biol Chem 2005;280:7427–7434. [DOI] [PubMed] [Google Scholar]
- [405].Schaap FG, van der Gaag NA, Gouma DJ, Jansen PL. High expression of the bile salt-homeostatic hormone fibroblast growth factor 19 in the liver of patients with extrahepatic cholestasis. Hepatology 2009;49:1228–1235. [DOI] [PubMed] [Google Scholar]
- [406].Dawson MI, Xia Z. The retinoid x receptors and their ligands. Biochim Biophys Acta 2012;1821:21–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Wu Y, Cai Y, Aguilo J, Dai T, Ao Y, Wan YJ. Rxralpha mrna expression is associated with cell proliferation and cell cycle regulation in hep3b cell. Exp Mol Pathol 2004;76:24–28. [DOI] [PubMed] [Google Scholar]
- [408].Standeven AM, Escobar M, Beard RL, Yuan YD, Chandraratna RA. Mitogenic effect of retinoid x receptor agonists in rat liver. Biochem Pharmacol 1997;54:517–524. [DOI] [PubMed] [Google Scholar]
- [409].Ledda-Columbano GM, Pibiri M, Molotzu F, Cossu C, Sanna L, Simbula G, Perra A, Columbano A. Induction of hepatocyte proliferation by retinoic acid. Carcinogenesis 2004;25:2061–2066. [DOI] [PubMed] [Google Scholar]
- [410].Sturm J, Keese M, Zhang H, Bonninghoff R, Magdeburg R, Vajkoczy P, Dono R, Zeller R, Gretz N. Liver regeneration in fgf-2-deficient mice: Vegf acts as potential functional substitute for fgf-2. Liver Int 2004;24:161–168. [DOI] [PubMed] [Google Scholar]
- [411].Shmarakov IO, Jiang H, Yang KJ, Goldberg IJ, Blaner WS. Hepatic retinoid stores are required for normal liver regeneration. J Lipid Res 2013;54:893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [412].Anderson SP, Dunn C, Laughter A, Yoon L, Swanson C, Stulnig TM, Steffensen KR, Chandraratna RA, Gustafsson JA, Corton JC. Overlapping transcriptional programs regulated by the nuclear receptors peroxisome proliferator-activated receptor alpha, retinoid x receptor, and liver x receptor in mouse liver. Mol Pharmacol 2004;66:1440–1452. [DOI] [PubMed] [Google Scholar]
- [413].Iyer M, Reschly EJ, Krasowski MD. Functional evolution of the pregnane x receptor. Expert Opin Drug Metab Toxicol 2006;2:381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].Xie W, Radominska-Pandya A, Shi Y, Simon CM, Nelson MC, Ong ES, Waxman DJ, Evans RM. An essential role for nuclear receptors sxr/pxr in detoxification of cholestatic bile acids. Proc Natl Acad Sci U S A 2001;98:3375–3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [415].Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a forkhead transcription factor. Cell 1999;96:857–868. [DOI] [PubMed] [Google Scholar]
- [416].Frank C, Gonzalez MM, Oinonen C, Dunlop TW, Carlberg C. Characterization of DNA complexes formed by the nuclear receptor constitutive androstane receptor. J Biol Chem 2003;278:43299–43310. [DOI] [PubMed] [Google Scholar]
- [417].Blanco-Bose WE, Murphy MJ, Ehninger A, Offner S, Dubey C, Huang W, Moore DD, Trumpp A. C-myc and its target foxm1 are critical downstream effectors of constitutive androstane receptor (car) mediated direct liver hyperplasia. Hepatology 2008;48:1302–1311. [DOI] [PubMed] [Google Scholar]
- [418].Donthamsetty S, Bhave VS, Kliment CS, Bowen WC, Mars WM, Bell AW, Stewart RE, Orr A, Wu C, Michalopoulos GK. Excessive hepatomegaly of mice with hepatocyte-targeted elimination of integrin linked kinase following treatment with 1,4-bis [2-(3,5-dichaloropyridyloxy)] benzene. Hepatology 2011;53:587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [419].Locker J, Tian J, Carver R, Concas D, Cossu C, Ledda- Columbano GM, Columbano A. A common set of immediate- early response genes in liver regeneration and hyperplasia. Hepatology 2003;38:314–325. [DOI] [PubMed] [Google Scholar]
- [420].Tian J, Huang H, Hoffman B, Liebermann DA, Ledda- Columbano GM, Columbano A, Locker J. Gadd45beta is an inducible coactivator of transcription that facilitates rapid liver growth in mice. J Clin Invest 2011;121:4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Chen T, Chen Q, Xu Y, Zhou Q, Zhu J, Zhang H, Wu Q, Xu J, Yu C. Src-3 is required for car-regulated hepatocyte proliferation and drug metabolism. J Hepatol 2012;56:210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [422].Suen CS, Berrodin TJ, Mastroeni R, Cheskis BJ, Lyttle CR, Frail DE. A transcriptional coactivator, steroid receptor coactivator-3, selectively augments steroid receptor transcriptional activity. J Biol Chem 1998;273:27645–27653. [DOI] [PubMed] [Google Scholar]
- [423].Dong B, Lee JS, Park YY, Yang F, Xu G, Huang W, Finegold MJ, Moore DD. Activating car and beta-catenin induces uncontrolled liver growth and tumorigenesis. Nat Commun 2015;6:5944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].Kakehashi A, Hagiwara A, Imai N, Nagano K, Nishimaki F, Banton M, Wei M, Fukushima S, Wanibuchi H. Mode of action of ethyl tertiary-butyl ether hepatotumorigenicity in the rat: Evidence for a role of oxidative stress via activation of car, pxr and ppar signaling pathways. Toxicol Appl Pharmacol 2013;273:390–400. [DOI] [PubMed] [Google Scholar]
- [425].Bockhorn M, Frilling A, Benko T, Best J, Sheu SY, Trippler M, Schlaak JF, Broelsch CE. Tri-iodothyronine as a stimulator of liver regeneration after partial and subtotal hepatectomy. Eur Surg Res 2007;39:58–63. [DOI] [PubMed] [Google Scholar]
- [426].Francavilla A, Carr BI, Azzarone A, Polimeno L, Wang Z, Van Thiel DH, Subbotin V, Prelich JG, Starzl TE. Hepatocyte proliferation and gene expression induced by triiodothyronine in vivo and in vitro. Hepatology 1994;20:1237–1241. [PubMed] [Google Scholar]
- [427].Fanti M, Singh S, Ledda-Columbano GM, Columbano A, Monga SP. Tri-iodothyronine induces hepatocyte proliferation by protein kinase a-dependent beta-catenin activation in rodents. Hepatology 2014;59:2309–2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [428].Ulven SM, Dalen KT, Gustafsson JA, Nebb HI. Lxr is crucial in lipid metabolism. Prostaglandins Leukot Essent Fatty Acids 2005;73:59–63. [DOI] [PubMed] [Google Scholar]
- [429].Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. Lxr, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev 1995;9:1033–1045. [DOI] [PubMed] [Google Scholar]
- [430].Miyata KS, McCaw SE, Meertens LM, Patel HV, Rachubinski RA, Capone JP. Receptor-interacting protein 140 interacts with and inhibits transactivation by, peroxisome proliferator- activated receptor alpha and liver-x-receptor alpha. Mol Cell Endocrinol 1998;146:69–76. [DOI] [PubMed] [Google Scholar]
- [431].Osabe M, Negishi M. Active erk1/2 protein interacts with the phosphorylated nuclear constitutive active/androstane receptor (car; nr1i3), repressing dephosphorylation and sequestering car in the cytoplasm. J Biol Chem 2011;286:35763–35769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [432].Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A g protein-coupled receptor responsive to bile acids. J Biol Chem 2003;278:9435–9440. [DOI] [PubMed] [Google Scholar]
- [433].Watanabe M, Houten SM, Mataki C, Christoffolete MA, Kim BW, Sato H, Messaddeq N, Harney JW, Ezaki O, Kodama T, Schoonjans K, Bianco AC, Auwerx J. Bile acids induce energy expenditure by promoting intracellular thyroid hormone activation. Nature 2006;439:484–489. [DOI] [PubMed] [Google Scholar]
- [434].Thomas C, Gioiello A, Noriega L, Strehle A, Oury J, Rizzo G, Macchiarulo A, Yamamoto H, Mataki C, Pruzanski M, Pellicciari R, Auwerx J, Schoonjans K. Tgr5-mediated bile acid sensing controls glucose homeostasis. Cell Metab 2009;10:167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [435].Pols TW, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor tgr5: A valuable metabolic target. Dig Dis 2011;29:37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [436].Wang YD, Chen WD, Yu D, Forman BM, Huang W. The g- protein-coupled bile acid receptor, gpbar1 (tgr5), negatively regulates hepatic inflammatory response through antagonizing nuclear factor kappa light-chain enhancer of activated b cells (nf-kappab) in mice. Hepatology 2011;54:1421–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [437].Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor tgr5 in kupffer cells. Biochem Biophys Res Commun 2008;372:78–84. [DOI] [PubMed] [Google Scholar]
- [438].Keitel V, Ullmer C, Haussinger D. The membrane-bound bile acid receptor tgr5 (gpbar-1) is localized in the primary cilium of cholangiocytes. Biol Chem 2010;391:785–789. [DOI] [PubMed] [Google Scholar]
- [439].Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, Haussinger D, Kubitz R. The g-protein coupled bile salt receptor tgr5 is expressed in liver sinusoidal endothelial cells. Hepatology 2007;45:695–704. [DOI] [PubMed] [Google Scholar]
- [440].Reich M, Deutschmann K, Sommerfeld A, Klindt C, Kluge S, Kubitz R, Ullmer C, Knoefel WT, Herebian D, Mayatepek E, Haussinger D, Keitel V. Tgr5 is essential for bile acid- dependent cholangiocyte proliferation in vivo and in vitro. Gut 2016;65:487–501. [DOI] [PubMed] [Google Scholar]
- [441].Reich M, Deutschmann K, Sommerfeld A, Klindt C, Kluge S, Kubitz R, Ullmer C, Knoefel WT, Herebian D, Mayatepek E, Haussinger D, Keitel V. Tgr5 is essential for bile acid- dependent cholangiocyte proliferation in vivo and in vitro. Gut 2016;65:487–501. [DOI] [PubMed] [Google Scholar]
- [442].de Graaf W, Hausler S, Heger M, van Ginhoven TM, van Cappellen G, Bennink RJ, Kullak-Ublick GA, Hesselmann R, van Gulik TM, Stieger B. Transporters involved in the hepatic uptake of (99m)tc-mebrofenin and indocyanine green. J Hepatol 2011;54:738–745. [DOI] [PubMed] [Google Scholar]
- [443].de Graaf W, Bennink RJ, Heger M, Maas A, de Bruin K, van Gulik TM. Quantitative assessment of hepatic function during liver regeneration in a standardized rat model. J Nucl Med 2011;52:294–302. [DOI] [PubMed] [Google Scholar]
- [444].Stieger B, Heger M, de Graaf W, Paumgartner G, van Gulik T. The emerging role of transport systems in liver function tests. Eur J Pharmacol 2012;675:1–5. [DOI] [PubMed] [Google Scholar]
- [445].Handschin C, Meyer UA. Induction of drug metabolism: The role of nuclear receptors. Pharmacol Rev 2003;55:649–673. [DOI] [PubMed] [Google Scholar]
- [446].Bodin K, Lindbom U, Diczfalusy U. Novel pathways of bile acid metabolism involving cyp3a4. Biochim Biophys Acta 2005;1687:84–93. [DOI] [PubMed] [Google Scholar]
- [447].Araya Z, Wikvall K. 6alpha-hydroxylation of taurochenodeoxycholic acid and lithocholic acid by cyp3a4 in human liver microsomes. Biochim Biophys Acta 1999;1438:47–54. [DOI] [PubMed] [Google Scholar]
- [448].Bremmelgaard A, Sjovall J. Hydroxylation of cholic, chenodeoxycholic, and deoxycholic acids in patients with intrahepatic cholestasis. J Lipid Res 1980;21:1072–1081. [PubMed] [Google Scholar]
- [449].Gnerre C, Blattler S, Kaufmann MR, Looser R, Meyer UA. Regulation of cyp3a4 by the bile acid receptor fxr: Evidence for functional binding sites in the cyp3a4 gene. Pharmacogenetics 2004;14:635–645. [DOI] [PubMed] [Google Scholar]
- [450].Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Identification of a human nuclear receptor defines a new signaling pathway for cyp3a induction. Proc Natl Acad Sci U S A 1998;95:12208–12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [451].Drocourt L, Ourlin JC, Pascussi JM, Maurel P, Vilarem MJ. Expression of cyp3a4, cyp2b6, and cyp2c9 is regulated by the vitamin d receptor pathway in primary human hepatocytes. J Biol Chem 2002;277:25125–25132. [DOI] [PubMed] [Google Scholar]
- [452].Goodwin B, Redinbo MR, Kliewer SA. Regulation of cyp3a gene transcription by the pregnane x receptor. Annu Rev Pharmacol Toxicol 2002;42:1–23. [DOI] [PubMed] [Google Scholar]
- [453].Marschall HU, Wagner M, Bodin K, Zollner G, Fickert P, Gumhold J, Silbert D, Fuchsbichler A, Sjovall J, Trauner M. Fxr(−/−) mice adapt to biliary obstruction by enhanced phase i detoxification and renal elimination of bile acids. J Lipid Res 2006;47:582–592. [DOI] [PubMed] [Google Scholar]
- [454].Stedman C, Robertson G, Coulter S, Liddle C. Feed-forward regulation of bile acid detoxification by cyp3a4: Studies in humanized transgenic mice. J Biol Chem 2004;279:11336–11343. [DOI] [PubMed] [Google Scholar]
- [455].Negishi M, Pedersen LG, Petrotchenko E, Shevtsov S, Gorokhov A, Kakuta Y, Pedersen LC. Structure and function of sulfotransferases. Arch Biochem Biophys 2001;390:149–157. [DOI] [PubMed] [Google Scholar]
- [456].Tephly TR, Burchell B. Udp-glucuronosyltransferases: A family of detoxifying enzymes. Trends Pharmacol Sci 1990;11:276–279. [DOI] [PubMed] [Google Scholar]
- [457].Radominska-Pandya A, Czernik PJ, Little JM, Battaglia E, Mackenzie PI. Structural and functional studies of udp- glucuronosyltransferases. Drug Metab Rev 1999;31:817–899. [DOI] [PubMed] [Google Scholar]
- [458].Huang J, Bathena SP, Tong J, Roth M, Hagenbuch B, Alnouti Y. Kinetic analysis of bile acid sulfation by stably expressed human sulfotransferase 2a1 (sult2a1). Xenobiotica 2010;40:184–194. [DOI] [PubMed] [Google Scholar]
- [459].Kitada H, Miyata M, Nakamura T, Tozawa A, Honma W, Shimada M, Nagata K, Sinal CJ, Guo GL, Gonzalez FJ, Yamazoe Y. Protective role of hydroxysteroid sulfotransferase in lithocholic acid-induced liver toxicity. J Biol Chem 2003;278:17838–17844. [DOI] [PubMed] [Google Scholar]
- [460].Song CS, Echchgadda I, Baek BS, Ahn SC, Oh T, Roy AK, Chatterjee B. Dehydroepiandrosterone sulfotransferase gene induction by bile acid activated farnesoid x receptor. J Biol Chem 2001;276:42549–42556. [DOI] [PubMed] [Google Scholar]
- [461].Runge-Morris M, Wu W, Kocarek TA. Regulation of rat hepatic hydroxysteroid sulfotransferase (sult2–40/41) gene expression by glucocorticoids: Evidence for a dual mechanism of transcriptional control. Mol Pharmacol 1999;56:1198–1206. [DOI] [PubMed] [Google Scholar]
- [462].Sonoda J, Xie W, Rosenfeld JM, Barwick JL, Guzelian PS, Evans RM. Regulation of a xenobiotic sulfonation cascade by nuclear pregnane x receptor (pxr). Proc Natl Acad Sci U S A 2002;99:13801–13806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [463].Saini SP, Sonoda J, Xu L, Toma D, Uppal H, Mu Y, Ren S, Moore DD, Evans RM, Xie W. A novel constitutive androstane receptor-mediated and cyp3a-independent pathway of bile acid detoxification. Mol Pharmacol 2004;65:292–300. [DOI] [PubMed] [Google Scholar]
- [464].Gall WE, Zawada G, Mojarrabi B, Tephly TR, Green MD, Coffman BL, Mackenzie PI, Radominska-Pandya A. Differential glucuronidation of bile acids, androgens and estrogens by human ugt1a3 and 2b7. J Steroid Biochem Mol Biol 1999;70:101–108. [DOI] [PubMed] [Google Scholar]
- [465].King CD, Rios GR, Green MD, Tephly TR. Udp- glucuronosyltransferases. Curr Drug Metab 2000;1:143–161. [DOI] [PubMed] [Google Scholar]
- [466].Liu W, Ramirez J, Gamazon ER, Mirkov S, Chen P, Wu K, Sun C, Cox NJ, Cook E Jr., Das S, Ratain MJ. Genetic factors affecting gene transcription and catalytic activity of udp- glucuronosyltransferases in human liver. Hum Mol Genet 2014;23:5558–5569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [467].Kullak-Ublick GA, Stieger B, Hagenbuch B, Meier PJ. Hepatic transport of bile salts. Semin Liver Dis 2000;20:273–292. [DOI] [PubMed] [Google Scholar]
- [468].Zelcer N, Reid G, Wielinga P, Kuil A, van der Heijden I, Schuetz JD, Borst P. Steroid and bile acid conjugates are substrates of human multidrug-resistance protein (mrp) 4 (atp- binding cassette c4). Biochem J 2003;371:361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [469].Mueller M, Cima I, Noti M, Fuhrer A, Jakob S, Dubuquoy L, Schoonjans K, Brunner T. The nuclear receptor lrh-1 critically regulates extra-adrenal glucocorticoid synthesis in the intestine. J Exp Med 2006;203:2057–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [470].Fayard E, Auwerx J, Schoonjans K. Lrh-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol 2004;14:250–260. [DOI] [PubMed] [Google Scholar]
- [471].Kir S, Zhang Y, Gerard RD, Kliewer SA, Mangelsdorf DJ. Nuclear receptors hnf4alpha and lrh-1 cooperate in regulating cyp7a1 in vivo. J Biol ]Chem 2012;287:41334–41341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [472].Li T, Francl JM, Boehme S, Chiang JY. Regulation of cholesterol and bile acid homeostasis by the cholesterol 7alpha- hydroxylase/steroid response element-binding protein 2/microrna-33a axis in mice. Hepatology 2013;58:1111–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [473].Li H, Xu G, Shang Q, Pan L, Shefer S, Batta AK, Bollineni J, Tint GS, Keller BT, Salen G. Inhibition of ileal bile acid transport lowers plasma cholesterol levels by inactivating hepatic farnesoid x receptor and stimulating cholesterol 7 alpha-hydroxylase. Metabolism 2004;53:927–932. [DOI] [PubMed] [Google Scholar]
- [474].Chiang JY. Regulation of bile acid synthesis. Front Biosci 1998;3:d176–193. [DOI] [PubMed] [Google Scholar]
- [475].Wang J, Greene S, Eriksson LC, Rozell B, Reihner E, Einarsson C, Eggertsen G, Gafvels M. Human sterol 12a- hydroxylase (cyp8b1) is mainly expressed in hepatocytes in a homogenous pattern. Histochem Cell Biol 2005;123:441–446. [DOI] [PubMed] [Google Scholar]
- [476].Bollard ME, Contel NR, Ebbels TM, Smith L, Beckonert O, Cantor GH, Lehman-McKeeman L, Holmes EC, Lindon JC, Nicholson JK, Keun HC. Nmr-based metabolic profiling identifies biomarkers of liver regeneration following partial hepatectomy in the rat. J Proteome Res 2010;9:59–69. [DOI] [PubMed] [Google Scholar]
- [477].Schwartz CC, Berman M, Vlahcevic ZR, Halloran LG, Gregory DH, Swell L. Multicompartmental analysis of cholesterol metabolism in man. Characterization of the hepatic bile acid and biliary cholesterol precursor sites. J Clin Invest 1978;61:408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [478].Abel S, Smuts CM, de Villiers C, Gelderblom WC. Changes in essential fatty acid patterns associated with normal liver regeneration and the progression of hepatocyte nodules in rat hepatocarcinogenesis. Carcinogenesis 2001;22:795–804. [DOI] [PubMed] [Google Scholar]
- [479].Albi E, Magni MV. The presence and the role of chromatin cholesterol in rat liver regeneration. J Hepatol 2002;36:395–400. [DOI] [PubMed] [Google Scholar]
- [480].Xu C, Lin F, Qin S. Relevance between lipid metabolism- associated genes and rat liver regeneration. Hepatol Res 2008;38:825–837. [DOI] [PubMed] [Google Scholar]
- [481].Field FJ, Mathur SN, LaBrecque DR. Cholesterol metabolism in regenerating liver of the rat. Am J Physiol 1985;249:G679–684. [DOI] [PubMed] [Google Scholar]
- [482].Monte MJ, Fernandez-Tagarro M, Marin JJ. Transient changes in the expression pattern of key enzymes for bile acid synthesis during rat liver regeneration. Biochim Biophys Acta 2005;1734:127–135. [DOI] [PubMed] [Google Scholar]
- [483].Trentalance A, Bruscalupi G, Conti Devirgiliis L, Leoni S, Mangiantini MT, Rossini L, Spagnuolo S, Erickson SK. Changes in lipoprotein binding and uptake by hepatocytes during rat liver regeneration. Biosci Rep 1989;9:231–241. [DOI] [PubMed] [Google Scholar]
- [484].Zhao C, Dahlman-Wright K. Liver x receptor in cholesterol metabolism. J Endocrinol 2010;204:233–240. [DOI] [PubMed] [Google Scholar]
- [485].Wang Y, Rogers PM, Su C, Varga G, Stayrook KR, Burris TP. Regulation of cholesterologenesis by the oxysterol receptor, lxralpha. J Biol Chem 2008;283:26332–26339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [486].Lee MH, Lu K, Hazard S, Yu H, Shulenin S, Hidaka H, Kojima H, Allikmets R, Sakuma N, Pegoraro R, Srivastava AK, Salen G, Dean M, Patel SB. Identification of a gene, abcg5, important in the regulation of dietary cholesterol absorption. Nat Genet 2001;27:79–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [487].Lu K, Lee MH, Hazard S, Brooks-Wilson A, Hidaka H, Kojima H, Ose L, Stalenhoef AF, Mietinnen T, Bjorkhem I, Bruckert E, Pandya A, Brewer HB, Jr., Salen G, Dean M, Srivastava A, Patel SB. Two genes that map to the stsl locus cause sitosterolemia: Genomic structure and spectrum of mutations involving sterolin-1 and sterolin-2, encoded by abcg5 and abcg8, respectively. Am J Hum Genet 2001;69:278–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [488].Graf GA, Li WP, Gerard RD, Gelissen I, White A, Cohen JC, Hobbs HH. Coexpression of atp-binding cassette proteins abcg5 and abcg8 permits their transport to the apical surface. J Clin Invest 2002;110:659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [489].Graf GA, Yu L, Li WP, Gerard R, Tuma PL, Cohen JC, Hobbs HH. Abcg5 and abcg8 are obligate heterodimers for protein trafficking and biliary cholesterol excretion. J Biol Chem 2003;278:48275–48282. [DOI] [PubMed] [Google Scholar]
- [490].Yu L, Hammer RE, Li-Hawkins J, Von Bergmann K, Lutjohann D, Cohen JC, Hobbs HH. Disruption of abcg5 and abcg8 in mice reveals their crucial role in biliary cholesterol secretion. Proc Natl Acad Sci U S A 2002;99:16237–16242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [491].Yu L, Li-Hawkins J, Hammer RE, Berge KE, Horton JD, Cohen JC, Hobbs HH. Overexpression of abcg5 and abcg8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J Clin Invest 2002;110:671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [492].Cheng HC, Yang CM, Shiao MS. Zonation of cholesterol and glycerolipid synthesis in regenerating rat livers. Hepatology 1993;17:280–286. [PubMed] [Google Scholar]
- [493].Miyake JH, Wang SL, Davis RA. Bile acid induction of cytokine expression by macrophages correlates with repression of hepatic cholesterol 7alpha-hydroxylase. J Biol Chem 2000;275:21805–21808. [DOI] [PubMed] [Google Scholar]
- [494].Stroup D, Crestani M, Chiang JY. Identification of a bile acid response element in the cholesterol 7 alpha-hydroxylase gene cyp7a. Am J Physiol 1997;273:G508–517. [DOI] [PubMed] [Google Scholar]
- [495].Chiang JY, Kimmel R, Weinberger C, Stroup D. Farnesoid x receptor responds to bile acids and represses cholesterol 7alpha-hydroxylase gene (cyp7a1) transcription. J Biol Chem 2000;275:10918–10924. [DOI] [PubMed] [Google Scholar]
- [496].Li T, Jahan A, Chiang JY. Bile acids and cytokines inhibit the human cholesterol 7 alpha-hydroxylase gene via the jnk/c-jun pathway in human liver cells. Hepatology 2006;43:1202–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [497].Gupta S, Stravitz RT, Dent P, Hylemon PB. Down-regulation of cholesterol 7alpha-hydroxylase (cyp7a1) gene expression by bile acids in primary rat hepatocytes is mediated by the c-jun n- terminal kinase pathway. J Biol Chem 2001;276:15816–15822. [DOI] [PubMed] [Google Scholar]
- [498].Gupta S, Natarajan R, Payne SG, Studer EJ, Spiegel S, Dent P, Hylemon PB. Deoxycholic acid activates the c-jun n-terminal kinase pathway via fas receptor activation in primary hepatocytes. Role of acidic sphingomyelinase-mediated ceramide generation in fas receptor activation. J Biol Chem 2004;279:5821–5828. [DOI] [PubMed] [Google Scholar]
- [499].Jung D, Kullak-Ublick GA. Hepatocyte nuclear factor 1 alpha: A key mediator of the effect of bile acids on gene expression. Hepatology 2003;37:622–631. [DOI] [PubMed] [Google Scholar]
- [500].Chiang JY. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J Hepatol 2004;40:539–551. [DOI] [PubMed] [Google Scholar]
- [501].Peng S, Huo X, Rezaei D, Zhang Q, Zhang X, Yu C, Asanuma K, Cheng E, Pham TH, Wang DH, Chen M, Souza RF, Spechler SJ. In barrett’s esophagus patients and barrett’s cell lines, ursodeoxycholic acid increases antioxidant expression and prevents DNA damage by bile acids. Am J Physiol Gastrointest Liver Physiol 2014;307:G129–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [502].Li T, Chiang JY. A novel role of transforming growth factor beta1 in transcriptional repression of human cholesterol 7alpha- hydroxylase gene. Gastroenterology 2007;133:1660–1669. [DOI] [PubMed] [Google Scholar]
- [503].Song KH, Ellis E, Strom S, Chiang JY. Hepatocyte growth factor signaling pathway inhibits cholesterol 7alpha- hydroxylase and bile acid synthesis in human hepatocytes. Hepatology 2007;46:1993–2002. [DOI] [PubMed] [Google Scholar]
- [504].Li T, Ma H, Chiang JY. Tgfbeta1, tnfalpha, and insulin signaling crosstalk in regulation of the rat cholesterol 7alpha- hydroxylase gene expression. J Lipid Res 2008;49:1981–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [505].Gerloff T, Geier A, Stieger B, Hagenbuch B, Meier PJ, Matern S, Gartung C. Differential expression of basolateral and canalicular organic anion transporters during regeneration of rat liver. Gastroenterology 1999;117:1408–1415. [DOI] [PubMed] [Google Scholar]
- [506].Gerloff T, Geier A, Stieger B, Hagenbuch B, Meier PJ, Matern S, Gartung C. Differential expression of basolateral and canalicular organic anion transporters during regeneration of rat liver. Gastroenterology 1999;117:1408–1415. [DOI] [PubMed] [Google Scholar]
- [507].Meier PJ. Molecular mechanisms of hepatic bile salt transport from sinusoidal blood into bile. Am J Physiol 1995;269:G801–812. [DOI] [PubMed] [Google Scholar]
- [508].Hagenbuch B, Meier PJ. Organic anion transporting polypeptides of the oatp/ slc21 family: Phylogenetic classification as oatp/ slco superfamily, new nomenclature and molecular/functional properties. Pflugers Arch 2004;447:653–665. [DOI] [PubMed] [Google Scholar]
- [509].Konig J, Klatt S, Dilger K, Fromm MF. Characterization of ursodeoxycholic and norursodeoxycholic acid as substrates of the hepatic uptake transporters oatp1b1, oatp1b3, oatp2b1 and ntcp. Basic Clin Pharmacol Toxicol 2012;111:81–86. [DOI] [PubMed] [Google Scholar]
- [510].Roth M, Obaidat A, Hagenbuch B. Oatps, oats and octs: The organic anion and cation transporters of the slco and slc22a gene superfamilies. Br J Pharmacol 2012;165:1260–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [511].Green RM, Gollan JL, Hagenbuch B, Meier PJ, Beier DR. Regulation of hepatocyte bile salt transporters during hepatic regeneration. Am J Physiol 1997;273:G621–627. [DOI] [PubMed] [Google Scholar]
- [512].Vos TA, Ros JE, Havinga R, Moshage H, Kuipers F, Jansen PL, Muller M. Regulation of hepatic transport systems involved in bile secretion during liver regeneration in rats. Hepatology 1999;29:1833–1839. [DOI] [PubMed] [Google Scholar]
- [513].Miura T, Kimura N, Yamada T, Shimizu T, Nanashima N, Yamana D, Hakamada K, Tsuchida S. Sustained repression and translocation of ntcp and expression of mrp4 for cholestasis after rat 90% partial hepatectomy. J Hepatol 2011;55:407–414. [DOI] [PubMed] [Google Scholar]
- [514].Keitel V, Burdelski M, Warskulat U, Kuhlkamp T, Keppler D, Haussinger D, Kubitz R. Expression and localization of hepatobiliary transport proteins in progressive familial intrahepatic cholestasis. Hepatology 2005;41:1160–1172. [DOI] [PubMed] [Google Scholar]
- [515].Dawson PA, Lan T, Rao A. Bile acid transporters. J Lipid Res 2009;50:2340–2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [516].Geier A, Dietrich CG, Voigt S, Ananthanarayanan M, Lammert F, Schmitz A, Trauner M, Wasmuth HE, Boraschi D, Balasubramaniyan N, Suchy FJ, Matern S, Gartung C. Cytokine-dependent regulation of hepatic organic anion transporter gene transactivators in mouse liver. Am J Physiol Gastrointest Liver Physiol 2005;289:G831–841. [DOI] [PubMed] [Google Scholar]
- [517].Le Vee M, Gripon P, Stieger B, Fardel O. Down-regulation of organic anion transporter expression in human hepatocytes exposed to the proinflammatory cytokine interleukin 1beta. Drug Metab Dispos 2008;36:217–222. [DOI] [PubMed] [Google Scholar]
- [518].Zeng H, Liu G, Rea PA, Kruh GD. Transport of amphipathic anions by human multidrug resistance protein 3. Cancer Res 2000;60:4779–4784. [PubMed] [Google Scholar]
- [519].Inokuchi A, Hinoshita E, Iwamoto Y, Kohno K, Kuwano M, Uchiumi T. Enhanced expression of the human multidrug resistance protein 3 by bile salt in human enterocytes. A transcriptional control of a plausible bile acid transporter. J Biol Chem 2001;276:46822–46829. [DOI] [PubMed] [Google Scholar]
- [520].Zelcer N, Saeki T, Bot I, Kuil A, Borst P. Transport of bile acids in multidrug-resistance-protein 3-overexpressing cells co- transfected with the ileal na+-dependent bile-acid transporter. Biochem J 2003;369:23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [521].Konig J, Rost D, Cui Y, Keppler D. Characterization of the human multidrug resistance protein isoform mrp3 localized to the basolateral hepatocyte membrane. Hepatology 1999;29:1156–1163. [DOI] [PubMed] [Google Scholar]
- [522].Rius M, Nies AT, Hummel-Eisenbeiss J, Jedlitschky G, Keppler D. Cotransport of reduced glutathione with bile salts by mrp4 (abcc4) localized to the basolateral hepatocyte membrane. Hepatology 2003;38:374–384. [DOI] [PubMed] [Google Scholar]
- [523].Cherrington NJ, Hartley DP, Li N, Johnson DR, Klaassen CD. Organ distribution of multidrug resistance proteins 1, 2, and 3 (mrp1, 2, and 3) mrna and hepatic induction of mrp3 by constitutive androstane receptor activators in rats. J Pharmacol Exp Ther 2002;300:97–104. [DOI] [PubMed] [Google Scholar]
- [524].Csanaky IL, Aleksunes LM, Tanaka Y, Klaassen CD. Role of hepatic transporters in prevention of bile acid toxicity after partial hepatectomy in mice. Am J Physiol Gastrointest Liver Physiol 2009;297:G419–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [525].Chang TH, Hakamada K, Toyoki Y, Tsuchida S, Sasaki M. Expression of mrp2 and mrp3 during liver regeneration after 90% partial hepatectomy in rats. Transplantation 2004;77:22–27. [DOI] [PubMed] [Google Scholar]
- [526].Khan AA, Chow EC, Porte RJ, Pang KS, Groothuis GM. Expression and regulation of the bile acid transporter, ostalpha- ostbeta in rat and human intestine and liver. Biopharm Drug Dispos 2009;30:241–258. [DOI] [PubMed] [Google Scholar]
- [527].Karabelyos C, Dobozy O, Szalai C, Klenjanszki K, Varju K, Hadhazi A, Kiss A, Fulop AK, Madarasz B, Falus A. Elevated hepatic glucocorticoid receptor expression during liver regeneration in rats. Pathol Oncol Res 1999;5:107–109. [DOI] [PubMed] [Google Scholar]
- [528].Zollner G, Marschall HU, Wagner M, Trauner M. Role of nuclear receptors in the adaptive response to bile acids and cholestasis: Pathogenetic and therapeutic considerations. Mol Pharm 2006;3:231–251. [DOI] [PubMed] [Google Scholar]
- [529].Bremmelgaard A, Sjovall J. Bile acid profiles in urine of patients with liver diseases. Eur J Clin Invest 1979;9:341–348. [DOI] [PubMed] [Google Scholar]
- [530].Alme B, Sjovall J. Analysis of bile acid glucuronides in urine. Identification of 3 alpha, 6 alpha, 12 alpha-trihydroxy-5 beta- cholanoic acid. J Steroid Biochem 1980;13:907–916. [DOI] [PubMed] [Google Scholar]
- [531].Alme B, Bremmelgaard A, Sjovall J, Thomassen P. Analysis of metabolic profiles of bile acids in urine using a lipophilic anion exchanger and computerized gas-liquid chromatorgaphy-mass spectrometry. J Lipid Res 1977;18:339–362. [PubMed] [Google Scholar]
- [532].Gerloff T, Geier A, Roots I, Meier PJ, Gartung C. Functional analysis of the rat bile salt export pump gene promoter. Eur J Biochem 2002;269:3495–3503. [DOI] [PubMed] [Google Scholar]
- [533].Plass JR, Mol O, Heegsma J, Geuken M, Faber KN, Jansen PL, Muller M. Farnesoid x receptor and bile salts are involved in transcriptional [regulation of the gene encoding the human bile salt export pump. Hepatology 2002;35:589–596. [DOI] [PubMed] [Google Scholar]
- [534].Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, Venkataramanan R, Cai H, Sinal CJ, Gonzalez FJ, Schuetz JD. Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome p450. J Biol Chem 2001;276:39411–39418. [DOI] [PubMed] [Google Scholar]
- [535].Zollner G, Fickert P, Fuchsbichler A, Silbert D, Wagner M, Arbeiter S, Gonzalez FJ, Marschall HU, Zatloukal K, Denk H, Trauner M. Role of nuclear bile acid receptor, fxr, in adaptive abc transporter regulation by cholic and ursodeoxycholic acid in mouse liver, kidney and intestine. J Hepatol 2003;39:480–488. [DOI] [PubMed] [Google Scholar]
- [536].Fickert P, Zollner G, Fuchsbichler A, Stumptner C, Pojer C, Zenz R, Lammert F, Stieger B, Meier PJ, Zatloukal K, Denk H, Trauner M. Effects of ursodeoxycholic and cholic acid feeding on hepatocellular transporter expression in mouse liver. Gastroenterology 2001;121:170–183. [DOI] [PubMed] [Google Scholar]
- [537].Courtois A, Payen L, Le Ferrec E, Scheffer GL, Trinquart Y, Guillouzo A, Fardel O. Differential regulation of multidrug resistance-associated protein 2 (mrp2) and cytochromes p450 2b1/2 and 3a1/2 in phenobarbital-treated hepatocytes. Biochem Pharmacol 2002;63:333–341. [DOI] [PubMed] [Google Scholar]
- [538].Guo GL, Lambert G, Negishi M, Ward JM, Brewer HB Jr., Kliewer SA, Gonzalez FJ, Sinal CJ. Complementary roles of farnesoid x receptor, pregnane x receptor, and constitutive androstane receptor in protection against bile acid toxicity. J Biol Chem 2003;278:45062–45071. [DOI] [PubMed] [Google Scholar]
- [539].Wagner M, Halilbasic E, Marschall HU, Zollner G, Fickert P, Langner C, Zatloukal K, Denk H, Trauner M. Car and pxr agonists stimulate hepatic bile acid and bilirubin detoxification and elimination pathways in mice. Hepatology 2005;42:420–430. [DOI] [PubMed] [Google Scholar]
- [540].Marino PA, Gottesman MM, Pastan I. Regulation of the multidrug resistance gene in regenerating rat liver. Cell Growth Differ 1990;1:57–62. [PubMed] [Google Scholar]
- [541].Teeter LD, Estes M, Chan JY, Atassi H, Sell S, Becker FF, Kuo MT. Activation of distinct multidrug-resistance (p- glycoprotein) genes during rat liver regeneration and hepatocarcinogenesis. Mol Carcinog 1993;8:67–73. [DOI] [PubMed] [Google Scholar]
- [542].Chen HL, Liu YJ, Chen HL, Wu SH, Ni YH, Ho MC, Lai HS, Hsu WM, Hsu HY, Tseng HC, Jeng YM, Chang MH. Expression of hepatocyte transporters and nuclear receptors in children with early and late-stage biliary atresia. Pediatr Res 2008;63:667–673. [DOI] [PubMed] [Google Scholar]
- [543].Grassian AR, Metallo CM, Coloff JL, Stephanopoulos G, Brugge JS. Erk regulation of pyruvate dehydrogenase flux through pdk4 modulates cell proliferation. Genes Dev 2011;25:1716–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [544].Rolo AP, Palmeira CM, Wallace KB. Interactions of combined bile acids on hepatocyte viability: Cytoprotection or synergism. Toxicol Lett 2002;126:197–203. [DOI] [PubMed] [Google Scholar]


