Abstract
The immune system has evolved to protect hosts from pathogens. T-cells represent a critical component of the immune system by their engagement in host defense mechanisms against microbial infections. Our knowledge of the molecular recognition by T-cells of pathogen derived-peptidic antigens that are presented by the Major Histocompatibility Complex (MHC) glycoproteins is now well established. However, lipids represent an additional, distinct chemical class of molecules that when presented by the family of CD1 antigen-presenting molecules can serve as antigens, and be recognized by specialized subsets of T-cells leading to antigen-specific activation. Over the past decades, numerous CD1-presented self- and bacterial lipid-based antigens have been isolated and characterized. However, our understanding at the molecular level of T-cell immunity to CD1 molecules presenting microbial lipid-based antigens is still largely unexplored. Here, we review the insights and the molecular basis underpinning the recognition of microbial lipid-based antigens by T-cells.
Keywords: T-cells, CD1-mediated immunity, microbial lipids, antigens, T-cell receptors, molecular recognition
Introduction
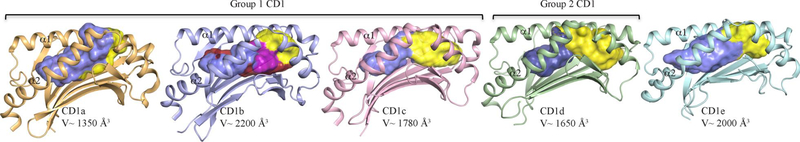
During the course of evolution, the immune system of vertebrates has consistently and remarkably developed to provide protection from pathogens such as bacteria, yeast, viruses, and parasites. To achieve this, the immune system of vertebrates comprises a complex network of cells, organs, tissues, proteins and other molecules that collectively defend the organism against various pathogens. The immune system consists of two major interconnected types of defense mechanisms to counter these microbial threats, namely, the innate and adaptive immune responses. The innate immune response is triggered by pathogen-associated molecular patterns and typically represents a rapid first line of defense that does not retain any long-lasting immunological memory. The weaponry of the innate arm includes physical epithelial barriers, antimicrobial peptides and enzymes, mast cells, innate lymphocytes, neutrophils, and macrophages. The molecular targets of the adaptive immune response are called antigens (Ags) that can consist of proteins, carbohydrates, metabolites and a wide range of chemically distinct lipids. Upon exposure to Ags, the adaptive immune response develops more slowly than the innate immune response, but will last longer. Two key immune cell populations within the adaptive arm of the immune system recognize Ags: T-cells and B-cells. T-cells express T-cell receptors (TCRs) on their cell surface that recognise small Ags or antigenic fragments from larger Ags. In T-cell immunity, Ags are presented on the cell surface of professional antigen presenting cells by specific antigen-presenting molecules. The subsequent recognition by T-cells leads to a cascade of immune responses that ultimately results in the clearance of the harmful pathogens. Presently, studies in the field of T-cell mediated immunity have largely focused on understanding the molecular presentation of antigenic peptides by the Major Histocompatibility Complex (MHC) proteins in humans and mice, and their subsequent molecular recognition by the TCR. MHC molecules are highly polymorphic glycoproteins that are able to bind peptides within their large and charged binding groove. The two classes of MHC molecules, class I and II MHC, bind small (8–13 residues long) or long (>15 residues) peptides, respectively. To cope with the wide array of pathogens that are encountered by the host, the immune system has enormous diversity in the TCRs expressed on T-cells and comprises two main classes of T-cells, the αβ and γδ T-cells. It was conventionally considered that αβ TCRs only recognised antigenic peptides complexed to the MHC (pMHC). The information gleaned from structural investigations on αβ TCR-pMHC complexes has been extremely informative in understanding how the TCR simultaneously, and specifically, focuses with host MHC and fragments of foreign peptide Ags [1,2]. However, peptides are not the only class of antigens that TCRs are able to recognize; indeed, lipids [3] and the recently discovered small vitamin B metabolites [4,5] can also activate T-cells. In addition to MHC molecules, it is now clear that there are other Ag-presenting molecules (termed MHC class I-like) [6] of the immune system that play a vital role in protective immunity, and that include the MR1 molecule and the Cluster of Differentiation 1 (CD1) family of glycoproteins that present the vitamin B metabolites [4] and lipid-based Ags to specialized subsets of T-cells, respectively [7–9]. The CD1 family represents an important cluster of largely monomorphic genes that have been classified, based on sequence identity and expression pattern, into 2 main groups of Ags presenting molecule, namely, the group 1 (CD1a, CD1b and CD1c) and group 2 (CD1d) (Figure 1). CD1e forms a third group and its function is still unclear but is believed to be involved in lipids transfer [10]. All CD1 molecules share structural similarities with the classical MHC-I molecules, but they have evolved to accommodate a chemically distinct class of Ags, namely, lipid-based Ags [11,12]. Typically, lipid-based Ags are amphipathic molecules with polar headgroups (e.g. carbohydrates, sulphates, and phosphates) and hydrophobic tails. The CD1 molecules present these lipid Ags by sequestering the tails within a hydrophobic groove while the polar headgroup is exposed at the CD1 surface for TCR recognition. Each CD1 isoform varies in terms of tissue distribution, intracellular trafficking and factors that modulate expression levels, signifying a specific function for each type of CD1 Ag-presenting molecule. For instance, while CD1c can be found in the splenic marginal zone B-cells, in the mantle zone B-cells of the lymph nodes and the tonsil [13]; human skin is the site of high density CD1a protein expression on Langerhans cells [14,15], the target of CD1a autoreactive T-cell responses [16,17]. Further, each CD1 isoform possesses distinct binding groove architecture and solvent accessibility that determines the repertoire of foreign and self-lipids that can be presented (Figure 1). For instance, the large CD1b antigen-binding cleft (Volume ~2200Å3) (Figure 1) can accommodate lipid-based antigens possessing long alkyl chains up to 80 carbons in length whereas CD1a possesses a more constricted binding groove (Volume ~1350Å3) that limits the size and diversity of antigens that it can bind (reviewed by [18]). These differences between the CD1 isoforms manifest in their ability to bind differing arrays of lipids, and their subsequent recognition by the TCRs. Here, we review the fundamental principles underscoring the molecular presentation of microbial lipid-based Ags by the family of CD1 molecules and their subsequent molecular recognition by specialized T-cell subsets.
Figure 1. The antigen binding cleft architecture of CD1 glycoproteins.
Cartoon representations of CD1a, light orange; CD1b, light blue; CD1c, pink; CD1d, light green; CD1e, cyan. For clarity, only the α1- and α2- domains for each CD1 are shown. The CD1 antigen binding clefts are shown as surface representations and have been generated using the CASTp program [124]. The A´-, F´-, C´- and T´-pockets are coloured in blue, yellow, magenta, and red respectively. The published calculated volumes (V, Å3) of the individual CD1 antigen-binding pocket are indicated [18]. This figure and all molecular graphics in subsequent figures were created with the PyMOL molecular visualization system [125].
Recognition of microbial lipid-based Ags by group 2 CD1-restricted T-cells
Natural Killer T-cells (NKT)
NKT cells are CD1d-specific innate-like T-cells that, when specifically activated via their TCR, produce an array of cytokines, including Th1-, Th2- and Th17-type cytokines, which enables them to influence immune outcomes in a broad range of diseases including a number of microbial infections [19,20]. Two main classes of NKT cells exist, namely, type I and II, which are distinguished by their TCR gene usage and Ag specificity [21]. Typically, the human type I NKT cells express an invariant TRAV10+ (T-cell Receptor Alpha Variable) TRAJ18+ (T-cell Receptor Alpha Joining) rearranged TCR α-chain and most express a TRBV25–1+ (T-cell Receptor Beta Variable) TCR β-chain. Type I NKT cells are also present in other mammalian species, including mice, whereby the NKT cells express an invariant TCR α-chain rearrangement TRAV11+TRAJ18+, and generally use 1 of 3 different TCR β-chain variable genes (TRBV13, TRBV29, or TRBV1). The type I NKT TCRs are also defined by their ability to recognize the prototypical glycosphingolipid α-Galactosylceramide (α-GalCer) as originally isolated from the marine sponge Agelas mauritianus [22] (Figure 2). As opposed to the type I NKT TCRs, the type II NKT TCRs are defined by their inability to respond to α-GalCer and are characterized by the expression of a more diverse TCR gene repertoire, but they share their specificity for CD1d with type I NKT cells [23–25].
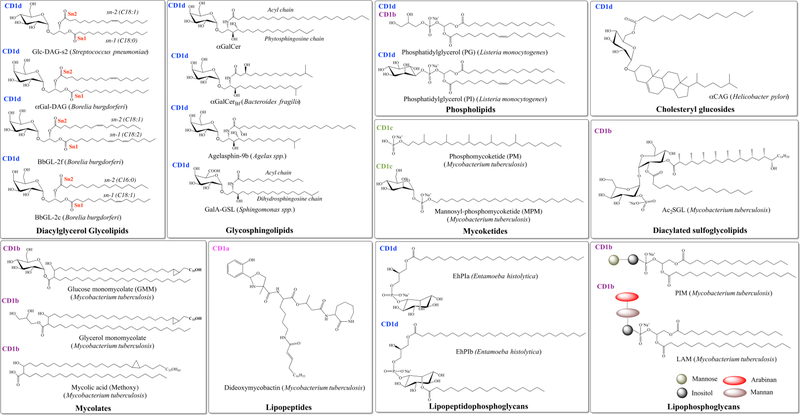
Figure 2. Chemical structures of the CD1-restricted microbial lipid-based antigens.
The lipids have been grouped by chemical classes and their binding to a specific CD1 molecule is indicated: CD1a (pink), CD1b (purple), CD1c (green), and CD1d (blue). For the lipophosphoglycan family, the carbohydrate headgroups are represented as coloured spheres (mannose, green; inositol, grey) and ovals (mannan, brown; arabinan, red). The chemical structures were prepared using ChemDraw Professional.
Molecular presentation of microbial CD1d-restricted lipid-based Ags
Together, the CD1d molecule from the different mammalian species form the group 2 CD1 family (Figure 1) and presents lipid-based antigens to the aforementioned invariant type I NKT cells that can express αβ TCRs [26] and clonally diverse T-cell subsets expressing αβ, γδ and δ/αβ TCRs [27–31,24,25]. Structurally, CD1d exhibits a medium sized antigen-binding groove that comprises two main antigen-binding pockets, namely, the A′- and F′-pockets (Figure 1) [32,33]. Here, whilst the A′-pocket is large, deeply buried and can accommodate acyl chain of lipids up to 29 carbons in length; the F′-pocket is smaller and thus is restricting its capacity to bind sphingosine chains to only ~18 carbons in length. However, its specialized binding-groove architecture and size has enabled CD1d to present a diverse range of exogenous lipid-based antigens to NKT cells that comprise chemically distinct classes of microbial lipids such as glycosphingolipids, glycerol-based lipids (DAG), phospholipids, and lysolipids (Figure 2) [34–36]. As such, the phosphatidylinositol mannoside (PIM) (Figure 2) and a lipophosphoglycan (LPG) isolated from the Mycobacterium bovis cell wall and Leishmania donovani, respectively, represented the first reported microbial lipid antigens recognized by NKT cells [37,38]. However, it was later shown that a chemically synthesized PIM4 failed to stimulate type I NKT cells [39]. Microbial glycosphingolipids (Figure 2) also represent a well-characterized class of lipid Ags for NKT cells. In particular, analogues of the prototypical iNKT antigen, α-GalCer, that comprise α-GalCerBf and Agelasphin-9b isolated from the gut bacterium Bacteroides fragilis and the marine sponge Agelas spp., respectively, were shown to be antigenic ligands for NKT cells [40,41]. Furthermore, the gram-negative bacteria Sphingomonas spp. [42] α-glucuronosylceramides (α-GlcACer) and α-galacturonosylceramides (α-GalACer) were also shown to be stimulating ligands for NKT and thereby inducing an increased production of IFN-γ and IL-4 [43,44,42,45,34]. Further studies identified the glycosphingolipid GalA-GSL produced by Sphingomonas spp. to activate NKT cells albeit to a lesser extent compared to α-GalCer. The numerous available crystal structures of the bound glycosphingolipid α-GalCer into CD1d in human and mouse [46,47] demonstrated a conserved mode of binding whereby its galactose headgroup is protruding out of the CD1d binding cleft to be exposed for interactions with the NKT TCRs while the phytosphingosine and the fatty acid chains (Figure 2) are typically buried within the F′- and A′-pockets of CD1d, respectively (Figure 3). The binary crystal structure of the Sphingomonas spp. GalA-GSL lipid bound to mCD1d [48] provided further molecular insights into the mode of binding of microbial glycosphingolipid into CD1d. Here, while α-GalCer and GalA-GSL differ by the chemical nature of their headgroup (Galactose vs. galacturonic acid, respectively) and their sphingosine chains (Figure 2), their overall positioning within the CD1d binding groove was highly conserved (Figure 3A).
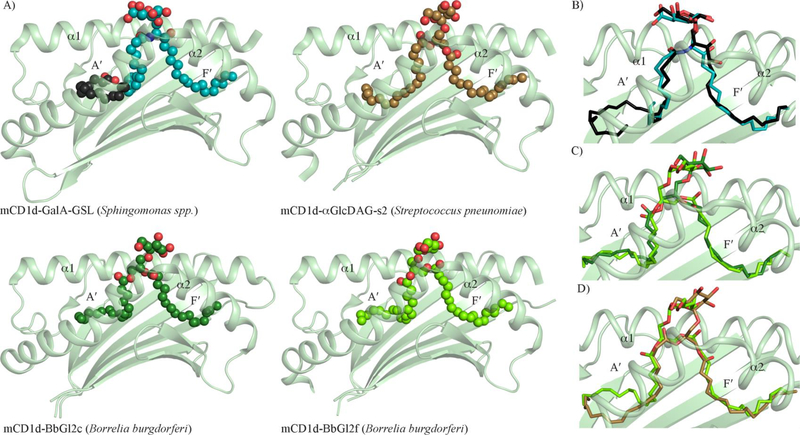
Figure 3. Molecular presentation of microbial lipid-based Ags by CD1d.
A) Cartoon representation of the crystal structure of mouse CD1d-microbial lipids binary complexes. For clarity, only the α1- and α2- domains of mouse CD1d (mCD1d) (light green) are shown. The microbial glycolipids GalA-GSL (cyan) from Sphingomonas spp., Glc-DAG-s2 (brown) from S. pneumoniae, BbGL2c (dark green) and BbGL2f (bright green) from B. burgdorferi are shown as spheres. For mCD1d-GalA-GSL, a spacer lipid is present in the A´- pocket and is shown as black spheres. B) Superposition of the glycosphingolipids GalA-GSL (cyan) and α-Galactosylceramide (α-GalCer) (black). C) Superposition of mCD1d presenting the diacylglycerol glycolipids BbGL2c (dark green) and BbGL2f (bright green). D) Superposition of mCD1d presenting the diacylglycerol glycolipids αGlcDAG-s2 (brown) and BbGL2f (bright green). For clarity, only the α1- and α2- domains of mouse CD1d (mCD1d) (light green) are shown and the lipids are shown as spheres. The oxygen and nitrogen atom are coloured in red and blue, respectively.
The important protective role played by NKT cells in microbial-mediated immunity was also highlighted by Olson et al.. who demonstrated clearance of the gram-negative bacteria Borrelia burgdorferi (the causative agent of Lyme disease) from mice through an NKT dependent activity, including the secretion of IFN-γ [49]. Further studies provided molecular insights into the nature of the B. burgdorferi CD1d-presented Ags through the characterization of a new chemical class of activating microbial lipid-based Ags (Diacylglycerol or DAG) for NKT cells (Figure 2) [39]. Indeed, two isolated glycoglycerol lipids (BbGL-2c and BbGL-2f) that structurally shared a galactose headgroup but differed in the chemical nature of their two fatty acid chains (Figure 2), were both able to stimulate mouse and human NKT cells albeit with different levels of potency [39]. In particular, BbGL-2c was significantly more potent than BbGL-2f for mouse invariant NKT cells. This latter intriguing observation led to suggest a possible role played by the different fatty acid chains into directing a different overall positioning of the DAG lipids into CD1d, and thereby affecting their subsequent molecular recognition by the NKT TCRs. The binary crystal structures of mouse CD1d presenting BbGL-2c and BbGL-2f (Figure 3A and Table 1) [50] subsequently supported this initial interpretation whereby the DAG adopted two distinct binding configurations within CD1d (Figure 3B). Here, the A′- and F′-pockets of CD1d accommodated the sn1-linked oleic acid (C18:1) and sn2-linked palmitic acid (C16:0) chains (Figure 2) of BbGL-2c, respectively, while BbGL-2f was sequestered within the CD1d cleft in a complete reverse orientation whereby the sn2-linked oleic acid (C18:1) and sn1-linked linoleic acid (C18:2) (Figure 2) were bound within the A′- and F′-pockets of CD1d, respectively (Figure 3B). Thus, this remarkable rearrangement of the fatty acid chains within CD1d also impacted on the overall positioning of the exposed galactose headgroup shared by both DAG lipid structures (Figure 3C) and which represents the key structural motif enabling NKT recognition. Collectively, these findings remarkably highlight how fine chemical modifications of the fatty acid chains of lipids such as the number of carbon unsaturations can drastically impact on their mode of presentation by CD1d and consequently influence their level of immunogenicity towards NKT cells. More recently, the repertoire of antigenic microbial DAG-derived lipids for type I NKT cells was extended to other bacterial species through the isolation and characterization of glucose-based DAG Ags (α-glucosyl-diacylglycerol) from Streptococcus pneumoniae (Figure 2) [51]. As observed for the B. burgdorferi galactose-based DAG Ags, the antigenic potency for NKT cells of the glucose-based DAG Ags was also greatly affected by the chemical nature and length of the aliphatic tails of the lipid Ags [51]. The binary crystal structure of the mouse CD1d in complex with α-GlcDAG-s2 (Figure 3A) revealed a very unusual orientation of the sn2-linked oleic acid (C18:1) bound within the A′-pocket whereby the aliphatic tail swirled around in opposite direction to what has been previously observed in other CD1d-Ags binary crystal structures [31,52–54] (Figure 3D).
Table 1:
Three-dimensional crystal structures of CD1 molecules in complex with microbial lipid-based Ags
| Lipid-based Ags | Microbial origin | CD1 isoform | PDB codes | References |
|---|---|---|---|---|
| Mycobactin lipopeptide | Mycobacterium spp. | CD1a | 1XZO | [106] |
| Diacylsulfoglycolipid (Ac2SGL) | Mycobacterium tuberculosis | CD1b | 3T8X | [103] |
| Glucose monomycolate-C54 (GMM-C54) | Nocardia spp. | CD1b | 1UQS | [102] |
| Glucose monomycolate-C32 (GMM-C32) | Rhodococcus spp. | CD1b | 5L2J | [101] |
| Mannosyl-β1-phosphomycoketide (MPM) | Mycobacterium tuberculosis | CD1c | 3OV6 | [109] |
| Phosphomycoketide (PM) | Mycobacterium tuberculosis | CD1c | 4ONO | [110] |
| α-galacturonosylceramide (GalA-GSL) | Sphingomonas spp. | CD1d | 2FIK | [43] |
| α-galactosyldiacylglycerols (BbGL-2c and BbGL-2f) | Borrelia burgdorferi | CD1d | 3ILQ 3ILP | [45] |
| α-glucosyldiacylcerol (αGlc-DAG-s2) | Streptococcus pneumoniae | CD1d | 3T1F | [46] |
Pathogenic lipopeptidophosphoglycans from Entamoeba histolytica (EhLPPG), the causative agent of amebiasis, were also reported to exhibit stimulatory effects for NKT cells through both TCR and Toll-like receptor (TLR) mediated pathways. Two phosphatidylinositol based-lipids (EhPIa and EhPIb) (Figure 2) were identified from the EhLPPG active fraction and interestingly, only EhPIb had the ability to produce IFN-γ [55]. Finally, cholesteryl α-glucoside (α-CAG) from the gastric pathogen Helicobacter pylori [56] (Figure 2) was also shown to be presented by CD1d and to activate NKT cells in both mice and humans [57,58].
Whilst it is now clear that type I NKT cells play a central role in microbial mediated immunity, experimental evidence for type II NKT cells to fulfill a similar function are much more limited. However, Tatituri et al. recently reported that lipids isolated from the cell wall of Mycobacterial spp. that comprised phospholipids such phosphatidylglycerol (PG) and phosphatidylinositol (PI) were able to activate a range of type II NKT cell hybridomas (Figure 2) [59]. Similarly, PG isolated from the cell wall of Listeria monocytogenes exhibited reactivity towards type II NKT cells [60]. In both cases, IL-2 cytokine were produced upon the activation of type II NKT cell hybridomas by the bacterial PGs.
While it is becoming evident that the number of characterized microbial CD1dpresented lipid Ags is constantly growing (Figure 2), our current crystallography-based molecular insights into their presentation by CD1d have been essentially limited to two classes of lipids (Glycosphingolipids and diacylglycerol glycolipids) (Figure 2 and table 1) [48,50,51]. There is therefore significant scope to explore the molecular presentation of other relevant classes of microbial lipid-based Ags.
Molecular basis for the recognition of microbial lipid-based Ags by type I NKT TCRs
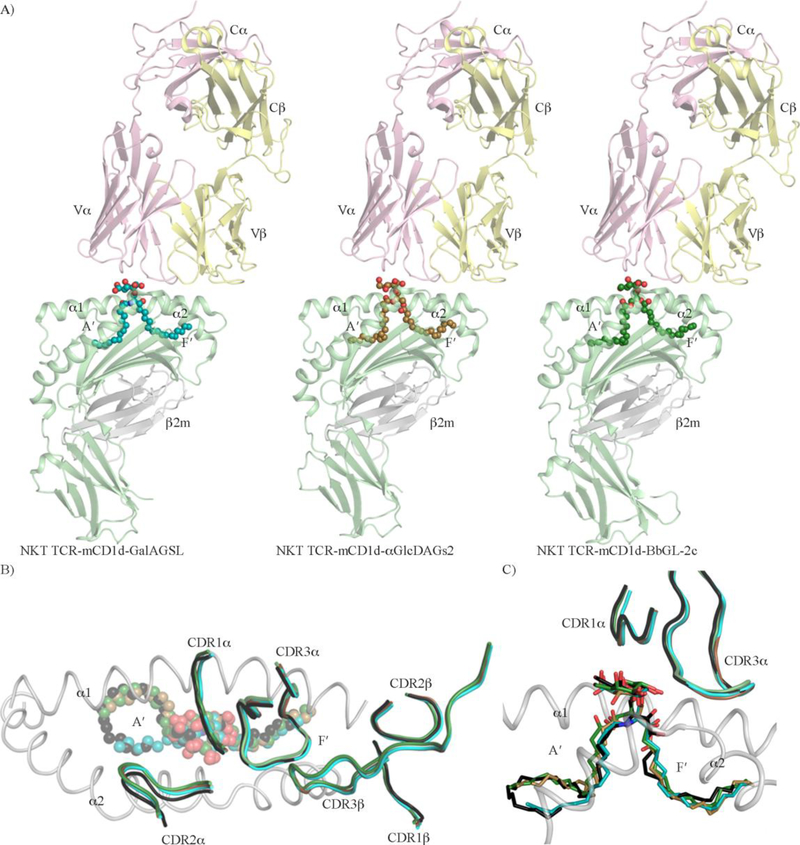
The molecular mechanism that underpins the recognition of microbial lipids by NKT TCRs has been rather surprisingly unexplored so far. Indeed, the crystal structures of the mouse type I NKT TCR in complex with mouse CD1d (mCD1d) presenting the lipids α-GalA-GSL, α-GalDAG-s2 [61] and BbGL-2c [62] (Figure 4 and table 2) only recently provided the first detailed insights into the molecular recognition of microbial lipid-based Ags by NKT TCRs. As observed previously in the crystal structures of type I NKT TCR-CD1d-lipids ternary complexes [63,47,23,64], the NKT TCR adopted a docking strategy whereby the TCR positioned in a parallel fashion over the F′-pocket of the CD1d binding cleft (Figure 4A). Here, at the NKT TCR/CD1d-microbial Ags interface, the CDR loops (Complementary Determining Region) of the TCR α-chain (CDR1α and CDR3α) were the main contributors to the molecular interactions [46].The carbohydrate headgroups of the Ags protruded from the CD1d binding cleft, and were exclusively contacted by residues belonging to the CDR1α and CDR3α loops (Asn30α, Arg95α and Gly96α). Interestingly, upon NKT TCR engagement, the overall orientation and positioning of the carbohydrate headgroups of the three microbial Ags was largely conserved and was similar to α-GalCer (Figure 4B). Furthermore, the position of all the NKT TCR CDR loops and particularly the CDR1α and CDR3α that are key contributors to the recognition of α-GalCer were also highly preserved between the 4 NKT TCR-CD1d-Ags ternary complexes (Figures 4B and 4C). However, as opposed to the conserved position of α-GalCer, upon TCR ligation, the orientation and positioning of the DAG-based microbial lipids were markedly affected. For instance, the acyl chain of α-GalDAG that encircles the A′-pocket of CD1d binary structure in clockwise direction preferred a counterclockwise direction in the ternary structure [48,61]. Furthermore, the carbohydrate headgroup of the microbial DAG lipids were all repositioned towards the center of the antigen binding cleft in the ternary structures [61] adopting an overall position similar to α-GalCer (Figure 4C) and thereby suggesting an “induced fit” molecular mechanism as an attributing feature for the type I NKT TCR recognition of microbial lipid-based antigens.
Figure 4. Molecular recognition of CD1d presenting microbial lipid-based Ags by NKT TCR.
A) Crystal structures of NKT TCR-mCD1d-GalAGSL (left panel), NKT TCR-mCD1d-αGlcDAGs2 (middle panel), and NKT TCR-mCD1d-BbGL-2c (right panel) ternary complexes. The mCD1d and β2-microglobulin molecules are coloured in light green and light grey, respectively. The NKT TCRα and TCRβ are coloured in pink and yellow, respectively. The microbial glycolipids GalAGSL (cyan), αGlcDAGs2 (brown) and BbGL-2c (dark green) are shown as spheres. B) View from the top of an overlay of the three NKT TCR-mCD1dmicrobial lipids crystal structures and NKT TCR-mCD1d-α-GalCer. For clarity, only the CDR loops are shown and coloured as for the respective lipids in each structure, GalAGSL (cyan), αGlcDAGs2 (brown), BbGL-2c (dark green) and α-Galactosylceramide (α-GalCer) (black). The lipids are shown as spheres. C) Overlay of three NKT TCR-mCD1d-microbial lipid crystal structures and NKT TCR-mCD1d-α-GalCer. For clarity, only the CDR1α and CDR3α loops are shown and coloured as for the respective lipids in each structure, GalAGSL (cyan), αGlcDAGs2 (brown), BbGL-2c (dark green) and α-Galactosylceramide (α-GalCer) (black). The lipids are shown as sticks.
Table 2:
Three-dimensional crystal structures of TCR-CD1-microbial lipid-based Ags ternary complexes
| Lipid-based Ags | Microbial origin | CD1 isoform | PDB codes | References |
|---|---|---|---|---|
| Glucose monomycolate-C32 (GMM-C32) | Mycobacterium tuberculosis | CD1b | 5L2K | [101] |
| α-glucosyldiacylcerol (αGlc-DAG-s2) | Streptococcus pneumoniae | CD1d | 3TA3 | [56] |
| α-galactosyldiacylglycerol (BbGL-2c) | Borrelia burgdorferi | CD1d | 3O9W | [55] |
| α-galacturonosylceramide (GalA-GSL) | Sphingomonas spp. | CD1d | 3O8X | [55] |
Recognition of lipid-based Ags by group 1 CD1-restricted T-cells
Intracellular trafficking and loading of microbial lipids
The intracellular trafficking pattern of CD1 molecules differs markedly from the classical MHC molecules as such that the MHC-I glycoproteins are typically loaded with peptidic Ags during their synthesis in the endoplasmic reticulum (ER). The MHC-II is blocked with the CLIP peptide (Class II-associated invariant chain peptide) directly after synthesis, which is then replaced in the late lysosome by exogenously captured peptidic antigens. Thus, MHC molecules generally do not traffic to the cell surface before they have gone through a cellular compartment where foreign peptides are loaded. By contrast, all the CD1 molecules are initially loaded with self- lipids that are present in the ER during synthesis, and before they are exposed to lipids from other cellular compartments, the newly synthesized CD1 molecules travel to the cell surface where they will encounter lipids from the cellular environment [65,66]. Thus, the antigen loading into CD1 molecules seems to take place by replacement of self- lipids. The lipids that are present in the ER and that are known to bind CD1 are phosphatidylinositol, phosphatidylethanolamine, phosphatidylcholine, phosphatidylserine and ceramides [67]. Lipid loading of CD1 at the cell surface is possible [68,69], but it is unclear what the physiologic role of this process is [70]. After this initial surfacing, each CD1 isoform travels to a specific cellular compartment, guided by signals in its cytoplasmic tail that interact with adaptor proteins [71–74]. While CD1a travels to the early endosome, CD1b and CD1c traffic to the late endosome and the intermediate endosome, respectively. Each of these compartments differs by their pH, and the nature of the enzymes and lipid transfer molecules that are present. Whereas the MHC molecules are completely dependent on the digestion of proteins in order to liberate antigenic peptides, most known lipid antigens do not need to be chemically cleaved or modified by the APC before they can be presented by CD1 molecules to T-cells, and this includes glucose-6-O-monomycolate (GMM) [75]. There are three examples of chemical cleavage of lipids Ags that are essential for antigenicity. The first one is the removal of a mannose moiety from mannosylphosphomycoketide (MPM) to form phosphomycoketide (PM), which is absolutely required to enable the recognition by the T-cell clone DN1 [76]. However, the mannose moiety is required for the recognition of mannosylphosphomycoketide (MPM) by another T-cell clone, CD8–1 [77,78,76]. The second example of cleavage of larger lipids to release antigenic lipid is the removal of two acyl chains from mycobacterial tetra-acylated phosphatidylinositol mannoside, as well as up till four mannosides to generate antigenic dimannosyl phosphatidylinositol mannoside with two acyl chains [79,80]. The third example is a chemically designed antigen: dihexosylceramide is dependent on the removal of a carbohydrate moiety to turn into antigenic α-GalCer [81]. Aside from these chemical alterations of lipids, the key factors that contribute to CD1 lipid loading and presentation are pH and lipid transfer molecules. Low pH of the late endosomal compartment enables CD1b to undergo conformational changes that facilitate the insertion of long lipids [82]. Lipid transfer molecules are thought to enable the extraction of lipids from lipid aggregates (membranes and cell walls) and to facilitate their transport through the aqueous environment of the cell. In the ER, the microsomal triglyceride transfer protein performs these functions [83], while in the endocytic pathway saposins are active [84–86]. The lysosomal lipid transfer molecule CD1e has been demonstrated to be required for the presentation of phosphatidylinositol mannoside as well as its chemical modification [79,87].
Group 1 CD1-restricted T-cell repertoire
Hallmarks of the MHC system are the high level of the polymorphism among the MHC molecules associated with the high diversity of the TCR repertoire due to random genetic recombination. Somatic recombination generates TCRs that can interact with all possible allelic variants of MHC and an enormous diversity of pathogen-derived peptides. Invariant NKT cells also make use of the TCR recombination machinery, but in a different way: their TCR α-chains are formed by recombinations without or with very few insertion/deletions of nucleotides (N) that occur at such high frequency that all human beings form these NKT TCRs. Since they recognize a non-polymorphic molecule that is expressed in all humans, NKT cells are positively selected and activated in all humans. However, the monomorphic nature of the antigen-presenting molecule does not inherently limit the TCR repertoires to TCRs lacking N nucleotides, and in fact, many examples have emerged of CD1-specific TCRs with extensive N regions [88,89]. The question that is still unanswered is: how common is TCR conservation among non-polymorphic antigen presenting systems? This question is important to address because knowledge of invariant, microbial lipid-specific TCRs could potentially lead to diagnostics of microbial infections based on detection of expanded invariant TCRs in antigen-exposed humans. The discovery of invariant NKT TCRs took place long before the tetramer technology was available, and was facilitated by the relative abundance of NKT cells. Currently, lipid-loaded CD1 tetramers are available and allow for direct isolation and TCR sequencing of lipid-specific T-cells from blood. This technique was first applied to study TCRs that interact with CD1b-GMM tetramers [90,91] and led to the discovery of GEM (Germline-Encoded Mycolyl lipid-reactive) TCRs and LDN5-like TCRs as groups of TCRs that share structural features and that can be found in many blood donors.
GEM T-cells express an invariant α-chain defined as identical or nearly identical sequences derived from different clones and different blood donors, and a β-chain that typically uses TRBV6–2 or TRBV30, but without any apparent CDR3β length and sequence conservation. Expression of an invariant TCR chain is called type 3 TCR bias [92], and shared V gene usage without CDR3 conservation is called type 1 bias. Thus, type I NKT cells and GEM T-cells each express a defining, invariant (type 3-biased) α-chain, and a β-chain repertoire that consists of one or two Vβ genes without CDR3 conservation (type 1-biased).
LDN5-like cells were discovered alongside GEM T-cells as a result of analysis of TCRs that recognize CD1b-GMM tetramers. The first CD1b-GMM-specific TCR that was ever sequenced, LDN5, was initially considered to represent a single example of a diverse TCR repertoire for this antigen, mainly because it utilized many N nucleotides in α- and β-chain [88], but when many more TCRs with this specificity were sequenced using tetramers, the LDN5-like TCR pattern became apparent. LDN5-like expressed TRAV17 and/or TRBV4–1, which are the Vα and Vβ genes that are also used by LDN5, but did not share CDR3 sequences. Therefore, LDN5-like cells have type 1-biased α- and β-chains.
As described above, many well characterized group 1 CD1-microbial lipid specific T-cells belong to the αβ T-cell lineage. However, it is also established that group 1 CD1 molecules presenting self-lipid Ags can be bona fide ligands for specific γδ T-cell populations [93,94], and thus raising an important question: does this elusive subset of T-cells also play a key role in the group 1 CD1-mediated microbial surveillance? A recent report by Roy et al. [95] identified and biophysically characterized CD1c-Mycobacterium tuberculosis (Mtb) lipid specific γδ TCRs, and thus provided the first emerging insights into the microbial lipid reactivity of group 1 CD1-restricted γδ T-cells.
Because additional working CD1 tetramers loaded with microbial lipids have recently been developed, new invariant or biased αβ and γδ TCRs can now be discovered. Microbial lipid-CD1 combinations that have not yet been used to systematically study the human TCR repertoire are: CD1a-dideoxymycobactin [96], CD1b-sulfoglycolipid, CD1b-mycolic acid [97], CD1c-phosphomycoketide [76]. Though more sensitive to the bystander effect and to immunostimulatory properties that are independent from TCR activation, an alternative to tetramer-based identification of antigen-specific T-cells is T-cell isolation based on cytokine expression after stimulation with lipid antigen and CD1-expressing antigen presenting cells [98]. With two available, independent methods of detection of group 1 CD1-microbial lipid specific T-cells, it is now possible to follow these T-cells in humans and answer basic questions about their expansion and activation during disease. These questions could previously only be addressed using guinea pigs or mice transgenic for human group 1 CD1 molecules [99,100].
Molecular presentation of microbial lipid based Ags by group 1 CD1
The most well characterised microbial lipid antigens presented by the group 1 CD1 molecules are found within Mycobacterium tuberculosis (Mtb) [101], and those include free mycolic acids [3] and its derivatives, mannophosphoisoprenoids [102], mannosylated lipoarabinomannan [103], lipomannan [103] and phosphatidyl-myo-inositol mannosides [104]. The extreme complexity of the lipids rich cell wall of mycobacterial species is rather unique, and represents an ideal source for a wide range of chemically distinct class of lipids to be presented by group 1 CD1 molecules. Mycolic acid, a key component of the Mtb cell wall, was the first characterised lipid antigen presented by CD1b [3]. Mycolic acids are high molecular weight lipids that are comprised of two main components: A β-hydroxy fatty acid, and a long branched α-alkyl lipid tail [105] (Figure 2). Though not yet analysed at the molecular level, two groups have reported the influence of the lipid tail length or composition of mycolic acids on T-cell responses against CD1b-mycolic acid [106,97].
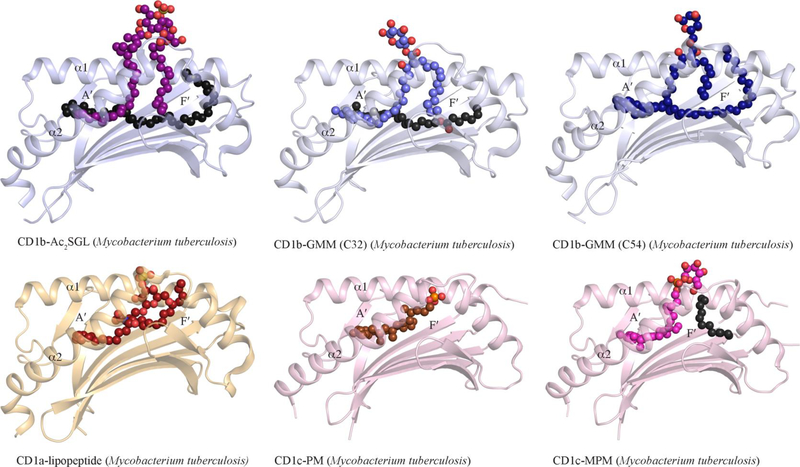
The list of CD1b-presented lipid Ags was further extended to derivatives of mycolic acids whereby additional headgroup moieties were incorporated, such as in GMM (Figure 2), and glycerol monomycolate [107–110]. Mycobacterial GMM is characterised by the presence of a glucose linked to the 6-position with a mycolyl β-hydroxy chain. Three forms of GMM, with varying mycolyl unit lengths of C32 (GMM-C32), C54 (GMM-C54) and C80 (GMM-C80), have been shown to activate GMM-reactive CD1b-restricted αβ T-cell lines and clones isolated from blood of patients infected with Mtb [111]. To date, the crystal structures of CD1b presenting the GMM-C32 [112] and GMM-C54 [113] isoforms have been determined (Figure 5). In both crystal structures, the β-hydroxy fatty acid and the α-alkyl lipid tail are bound within the C′-channel and A′-pockets, respectively. The β-hydroxy fatty acids are 10-carbons and 16-carbons in length for the GMM-C54 and GMM-C32 derivatives, respectively, and thereby enabling the GMM-C32 to be sequestered deeper into the C′-pocket of CD1b. By contrast, the α-alkyl lipid tails for GMM-C54 and GMM-C32 are 50 and 20-carbons in length, respectively, and CD1b is able to accommodate the entirety of the 50-carbons tail of GMM within its T′- tunnel and F′-pockets. Interestingly, whilst GMM-C32 does not extend throughout the entirety of the CD1b antigen binding pockets, a hydrophobic scaffold lipid is bound within the T′-tunnel to maintain the structural integrity of the glycoprotein. This scaffold lipid originated from the expression system that was utilised to produce the recombinantly expressed CD1b molecule; the presence of spacer lipids in CD1b has been previously reported [114,67] but the precise chemical natures of these scaffold lipids and their possible immunogenic roles remain unclear. In both crystal structures, the GMM glucose headgroup is protruding out of the CD1b binding groove to be exposed for GMM-restricted TCR recognition [112]. The exquisite specificity of the TCR for the glucose moiety explains the absence of an effect of mycolic acid chain length on the activation of GMM-specific T-cells [107].
Figure 5. Molecular presentation of microbial lipid-based Ags by group 1 CD1.
Cartoon representation of the crystal structure of group 1 CD1-microbial lipids binary complexes. For clarity, only the α1- and α2- domains of CD1a (light orange), CD1b (light blue), and CD1c (pink) are shown. The microbial lipid antigens Ac2SGL (purple), glucose monomycolate (GMM C32) (light blue), GMM C54 (dark blue), synthetic dideoxymycobactin (JH-02215) (red), phosphomycoketide (PM) (brown), and mannose-phosphomycoketide (MPM) (pink) are represented as spheres. The spacer lipids are shown as black spheres The oxygen, nitrogen, sulphur and phosphate atoms are coloured in red, blue, yellow and orange, respectively.
Two other classes of unique mycobacterial lipids presented by group 1 CD1s are the lipopeptides and the sulfoglycolipids that are presented by CD1a [115–117] and CD1b [115,114], respectively. The identified lipopeptides belong to an iron-chelating subfamily of siderophores named mycobactins. CD1a can present to T-cells a naturally occurring modified form of mycobactins that lacks two hydroxyl groups, named didehydroxymycobactin (DDM) (Figure 2) [118]. Structural characterisation of CD1a presenting a synthetic mycobactin lipid analogue (JH-02215) [117] reveals that, in clear contrast to the CD1b-GMM structure, the lipopeptide is fully buried within the antigen-binding groove (Figure 5). Here, whilst the single acyl chain is bound within the A′-pocket, the peptidyl moiety and the lysine branch are bound along and into the F′-pocket, respectively [117]. The hydrophilic N-aryl group of the lipopeptide is solvent exposed, and is proximally positioned to Arg73 at the A′-roof [117]. Interestingly, DDM represents the sole identified CD1a-presented lipid-based Ag from Mtb.
Sulfoglycolipids form a group of compounds that is found only within Mtb, and not in other mycobacterial species [119]. The sulfoglycolipids (Figure 2) contain two to four acyl tails that are linked by a polar trehalose group harbouring a sulphate group that has been found to be essential for the immunogenic property of the Ag [115]. CD1b presents the diacylated form of sulfoglycolipid (Ac2SGL), whereas forms with one, three, or four acyl chains are not [115]. The three-dimensional structure of CD1b in complex with Ac2SGL (Figure 5) revealed that the C16-palmitoyl and hydroxyphthioceranic acid tails were bound within the C′- and A′-pockets, respectively [114]. CD1c has also the ability to present microbial lipid antigens to generate a T-cell mediated immune response. A major defining characteristic of microbial lipids presented by CD1c is their significantly methylated alkyl tails [78,108], and includes the mycobacterial lipids phosphomycoketide (PM) [76] and mannosyl-β1-phosphomycoketide (MPM) (Figure 2) [102,78]. Both lipid classes are comprised of a C32 methylated carbon tail, and a phosphate polar headgroup, with the most significant difference between the two lipids being the addition of a mannose sugar at the β1 position of the phosphate ion in MPM [120,76,121]. Structural characterisation of CD1c presenting both PM and MPM (Figure 5) shows that the lipid tail is bound within the A′-pocket, and penetrates in the D′/E′ back portal of CD1c [120,121]. Presentation of the polar headgroup differs between the two lipids, with the MPM headgroup being solvent exposed due to the presence of a C12 spacer lipid in the F′-pocket [120]. Studies performed on CD1crestricted T-cells that are specific for PM, MPM, or both lipids show that a mixture of CD4+, CD8+ and double negative T-cells are able to recognise those Ags presented by CD1c. Although there is no available crystal structure of a TCR-CD1c-PM or MPM ternary complex yet, the CD1c mutagenesis study revealed that despite a shared TRBV7–9 gene among the different TCRs, the latter may adopt different docking strategy to recognize the CD1c molecule presenting the PM or MPM lipid-based Ags [121]. Aside from the mycobacterial lipid-based antigens, lipids presented by group 1 CD1 molecules have been identified in several other bacteria, including Salmonella typhimurium, Staphylococcus aureus, and Brucella melitensis [122]. CD1b conjugated to dextramers presenting whole cell lipid extract from each of the bacterial species demonstrated significant binding to polyclonal T-cell populations, which were also found to stimulate cytokine production [122]. In each case, the identified immunodominant lipid-based Ag was phosphatidylglycerol (PG) (Figure 2), which while highly abundant in these bacterial species, is also found within mammalian cells in very low amounts. The PG species identified in both bacterial and mammalian cells that induce an immune response all retain the same phosphoglycerol headgroup, yet varied in acyl tail saturation levels, with carbon lengths limited to lengths of C15-C18 [59]. In the case of PG presentation by CD1b, variations in lipid tail were not distinguished by the T-cells [122], while in the case of PG presentation by CD1d, the different forms were distinguished by NKT cells [59]. Whilst binary crystal structures of CD1b presenting self- phospholipids have been determined [123,66], the structural characterisation of CD1b presenting bacterial PG is yet to be conducted.
Molecular basis for the recognition of CD1b-GMM by GEM TCRs
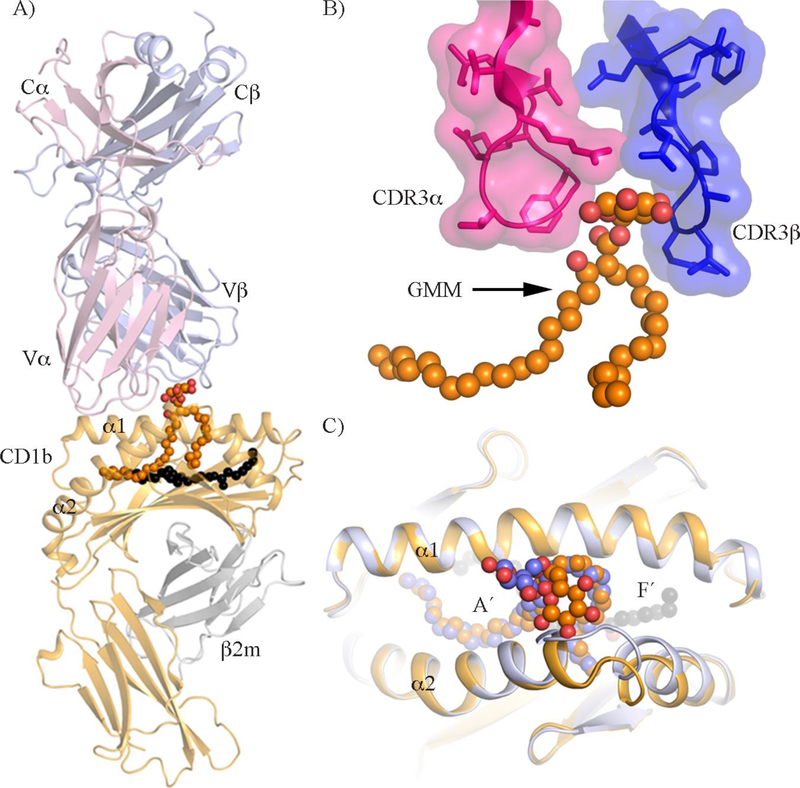
A structural characterization at atomic level of TCR recognition of a group 1 CD1 molecule presenting microbial lipid antigen is currently limited to a single example [112]. For microbial and non-microbial lipid Ags, the molecular basis of group 1 CD1 molecule antigen presentation and their subsequent TCR recognition has been understudied and therefore limited information is available. Recently, the crystal structure of the GEM42 TCR-CD1b-GMM ternary complex (Figure 6A and table 2) provided the first fundamental insights into the molecular mechanism that underpins the recognition of the mycobacterial lipid GMM by the GEM TCRs [112] (Figure 6A). Here, the structure of a GEM TCR, called GEM42, solved in complex with CD1b-GMM-C32 show how the high affinity TCR “caged” the sugar moiety of the microbial lipid Ag. The solvent exposed glucose moiety of the Ag is stabilized by the GEM42 CDR3 loops that form a “tweezers-like” structure around the carbohydrate (Figure 6B). Interestingly, the CDR3 loops are positioned underneath the lipid headgroup, thus enabling the entire glucose moiety and a section of the acyl tail to be contacted by the TCR. This high shape complementarity between the lipid and the GEM42 TCR might be responsible for the high affinity exhibited by those TCRs. In stark contrast to the previously determined crystal structures of TCR-CD1d-microbial lipids, the GEM42-CD1b-GMM ternary crystal structure remarkably revealed that the lipid Ag contribution (27%) to the overall interface was more than double. In addition to the stabilization of the conformation of the lipid headgroup, upon GEM42 TCR binding the cleft of CD1b was like “bulldozed” by the TCR allowing it to have a better grip on the Ag (Figure 6C). Now, it remains to be determined whether the LDN5-like TCRs will adopt distinct strategies to recognize the CD1b-GMM complex, resulting in a weaker affinity, and whether the observed “tweezers-like” mechanism is unique to the GEM TCRs.
Figure 6. Molecular recognition of CD1b-GMM by GEM TCR.
A) Overview of the GEM42 TCR-CD1b-GMM crystal structure shown as a cartoon representation. The α- and β-chains of GEM42 TCR are coloured in pink and blue, respectively. The CD1b and β2m are coloured in pale orange and grey, respectively. GMM is represented as orange spheres and the spacer lipid as black spheres. B) Close up view of the “tweezers-like” motif. The GEM42 TCR CDR3 loops, with CDR3α in pink and CDR3β in blue, surrounding the GMM represented as orange spheres. C) Structural changes in the CD1b-GMM upon GEM42 TCR binding. The free CD1b-GMM is coloured in light blue, while the CD1b-GMM in complex with the GEM42 TCR is coloured in orange.
Conclusions
While lipid reactive T-cells were first identified 25 years ago, understanding the general molecular basis of lipid antigen recognition and their role in human T-cell immunity has lagged behind that of the MHC-restricted T-cells. Indeed, CD1-restricted T-cells have only recently emerged as central players in host protection. In the context of group 2 CD1, we have gained fundamental insights in recent years into the molecular basis that underpins the recognition of two main classes of microbial lipid-based Ags by NKT cells. However, the growing list of newly identified CD1d-presented microbial lipids offers opportunities to further investigate the mode of recognition of microbial lipids by NKT TCRs. In the context of the CD1 group 1 system, the lack of mouse-based models coupled with the difficulty in working with, and identifying, lipid-based Ags, has hampered progress in this exciting field. However, the development of the tetramers and dextramers technology has been key to the identification of group 1 CD1-restricted T-cell subsets and thus enabled to further advance our understanding of the role played by group 1 CD1 molecules and restricted T-cells in human antimicrobial immunity. Our understanding of the molecular basis that underpins the recognition of microbial lipid Ags by group 1 CD1-restricted TCRs is emerging with the first crystal structure of a TCR-CD1b-microbial lipid Ag recently determined. Finally, unlike the genetically diverse MHC molecules, CD1a, CD1b, CD1c and CD1d proteins exhibit limited polyphormism, allowing CD1 proteins to be targeted pharmacologically with lipid ligands or small molecules. Therefore, the pursuit of our efforts to gain general molecular insights into their mode of binding to CD1 and TCRs offers exciting perspectives to design novel therapeutics to augment the protective immune response against microbial infections.
Acknowledgments
Our research is supported by the US National Institutes of Health (NIH) (AI 111224), Monash University (Australia), the Australian National Health and Medical Research (NHMRC) and the Australian Research Council (ARC). SG is a Monash Senior Research Fellow and J.L.N. is an ARC Future Fellow (FT160100074).
Footnotes
Conflict of interest statement
The authors declare no competing financial interests.
References
- 1.Miles JJ, McCluskey J, Rossjohn J, Gras S (2015) Understanding the complexity and malleability of T-cell recognition. Immunology and cell biology 93 (5):433–441. doi: 10.1038/icb.2014.112 [DOI] [PubMed] [Google Scholar]
- 2.Rossjohn J, Gras S, Miles JJ, Turner SJ, Godfrey DI, McCluskey J (2015) T cell antigen receptor recognition of antigen-presenting molecules. Annual review of immunology 33:169–200. doi: 10.1146/annurev-immunol-032414-112334 [DOI] [PubMed] [Google Scholar]
- 3.Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB (1994) Recognition of a lipid antigen by CD1-restricted alpha beta+ T cells. Nature 372 (6507):691–694. doi: 10.1038/372691a0 [DOI] [PubMed] [Google Scholar]
- 4.Kjer-Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L, Bhati M, Chen Z, Kostenko L, Reantragoon R, Williamson NA, Purcell AW, Dudek NL, McConville MJ, O’Hair RA, Khairallah GN, Godfrey DI, Fairlie DP, Rossjohn J, McCluskey J (2012) MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 491 (7426):717–723. doi: 10.1038/nature11605 [DOI] [PubMed] [Google Scholar]
- 5.Patel O, Kjer-Nielsen L, Le Nours J, Eckle SB, Birkinshaw R, Beddoe T, Corbett AJ, Liu L, Miles JJ, Meehan B, Reantragoon R, Sandoval-Romero ML, Sullivan LC, Brooks AG, Chen Z, Fairlie DP, McCluskey J, Rossjohn J (2013) Recognition of vitamin B metabolites by mucosal-associated invariant T cells. Nature communications 4:2142. doi: 10.1038/ncomms3142 [DOI] [PubMed] [Google Scholar]
- 6.Adams EJ, Luoma AM (2013) The adaptable major histocompatibility complex (MHC) fold: structure and function of nonclassical and MHC class I-like molecules. Annual review of immunology 31:529–561. doi: 10.1146/annurev-immunol-032712-095912 [DOI] [PubMed] [Google Scholar]
- 7.Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB (2015) Lipid and small-molecule display by CD1 and MR1. Nature reviews Immunology 15 (10):643–654. doi: 10.1038/nri3889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zajonc DM, Flajnik MF (2016) CD1, MR1, NKT, and MAIT: evolution and origins of non-peptidic antigen recognition by T lymphocytes. Immunogenetics 68 (8):489–490. doi: 10.1007/s00251-016-0941-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Godfrey DI, Uldrich AP, McCluskey J, Rossjohn J, Moody DB (2015) The burgeoning family of unconventional T cells. Nature immunology 16 (11):1114–1123. doi: 10.1038/ni.3298 [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Alles LF, Giacometti G, Versluis C, Maveyraud L, de Paepe D, Guiard J, Tranier S, Gilleron M, Prandi J, Hanau D, Heck AJ, Mori L, De Libero G, Puzo G, Mourey L, de la Salle H (2011) Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proceedings of the National Academy of Sciences of the United States of America 108 (32):13230–13235. doi: 10.1073/pnas.1105627108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zajonc DM (2016) The CD1 family: serving lipid antigens to T cells since the Mesozoic era. Immunogenetics 68 (8):561–576. doi: 10.1007/s00251-016-0931-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moody DB, Zajonc DM, Wilson IA (2005) Anatomy of CD1-lipid antigen complexes. Nature reviews Immunology 5 (5):387–399. doi: 10.1038/nri1605 [DOI] [PubMed] [Google Scholar]
- 13.Smith ME, Thomas JA, Bodmer WF (1988) CD1c antigens are present in normal and neoplastic B-cells. The Journal of pathology 156 (2):169–177. doi: 10.1002/path.1711560212 [DOI] [PubMed] [Google Scholar]
- 14.Pena-Cruz V, Ito S, Oukka M, Yoneda K, Dascher CC, Von Lichtenberg F, Sugita M (2001) Extraction of human Langerhans cells: a method for isolation of epidermis-resident dendritic cells. Journal of immunological methods 255 (1–2):83–91 [DOI] [PubMed] [Google Scholar]
- 15.Caux C, Dezutter-Dambuyant C, Schmitt D, Banchereau J (1992) GM-CSF and TNF-alpha cooperate in the generation of dendritic Langerhans cells. Nature 360 (6401):258–261. doi: 10.1038/360258a0 [DOI] [PubMed] [Google Scholar]
- 16.de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG, Van Rhijn I, Pena-Cruz V, Ruan DT, Altman JD, Rossjohn J, Moody DB (2014) CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nature immunology 15 (2):177–185. doi: 10.1038/ni.2790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong A, Pena-Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB (2010) CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nature immunology 11 (12):1102–1109. doi: 10.1038/ni.1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ly D, Moody DB (2014) The CD1 size problem: lipid antigens, ligands, and scaffolds. Cellular and molecular life sciences : CMLS 71 (16):3069–3079. doi: 10.1007/s00018-014-1603-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skold M, Behar SM (2003) Role of CD1d-restricted NKT cells in microbial immunity. Infection and immunity 71 (10):5447–5455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Godfrey DI, McCluskey J, Rossjohn J (2005) CD1d antigen presentation: treats for NKT cells. Nature immunology 6 (8):754–756. doi: 10.1038/ni0805-754 [DOI] [PubMed] [Google Scholar]
- 21.Godfrey DI, MacDonald HR, Kronenberg M, Smyth MJ, Van Kaer L (2004) NKT cells: what’s in a name? Nature reviews Immunology 4 (3):231–237. doi: 10.1038/nri1309 [DOI] [PubMed] [Google Scholar]
- 22.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, Koseki H, Taniguchi M (1997) CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science 278 (5343):1626–1629 [DOI] [PubMed] [Google Scholar]
- 23.Rossjohn J, Pellicci DG, Patel O, Gapin L, Godfrey DI (2012) Recognition of CD1d-restricted antigens by natural killer T cells. Nature reviews Immunology 12 (12):845–857. doi: 10.1038/nri3328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel O, Pellicci DG, Gras S, Sandoval-Romero ML, Uldrich AP, Mallevaey T, Clarke AJ, Le Nours J, Theodossis A, Cardell SL, Gapin L, Godfrey DI, Rossjohn J (2012) Recognition of CD1d-sulfatide mediated by a type II natural killer T cell antigen receptor. Nature immunology 13 (9):857–863. doi: 10.1038/ni.2372 [DOI] [PubMed] [Google Scholar]
- 25.Girardi E, Maricic I, Wang J, Mac TT, Iyer P, Kumar V, Zajonc DM (2012) Type II natural killer T cells use features of both innate-like and conventional T cells to recognize sulfatide self antigens. Nature immunology 13 (9):851–856. doi: 10.1038/ni.2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brigl M, Brenner MB (2004) CD1: antigen presentation and T cell function. Annual review of immunology 22:817–890. doi: 10.1146/annurev.immunol.22.012703.104608 [DOI] [PubMed] [Google Scholar]
- 27.Uldrich AP, Patel O, Cameron G, Pellicci DG, Day EB, Sullivan LC, Kyparissoudis K, Kjer-Nielsen L, Vivian JP, Cao B, Brooks AG, Williams SJ, Illarionov P, Besra GS, Turner SJ, Porcelli SA, McCluskey J, Smyth MJ, Rossjohn J, Godfrey DI (2011) A semi-invariant Valpha10+ T cell antigen receptor defines a population of natural killer T cells with distinct glycolipid antigen-recognition properties. Nature immunology 12 (7):616–623. doi: 10.1038/ni.2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pellicci DG, Uldrich AP, Le Nours J, Ross F, Chabrol E, Eckle SB, de Boer R, Lim RT, McPherson K, Besra G, Howell AR, Moretta L, McCluskey J, Heemskerk MH, Gras S, Rossjohn J, Godfrey DI (2014) The molecular bases of delta/alphabeta T cell-mediated antigen recognition. The Journal of experimental medicine 211 (13):2599–2615. doi: 10.1084/jem.20141764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zajonc DM, Girardi E (2014) A gammadelta T-cell glimpse of glycolipids. Immunology and cell biology 92 (2):99–100. doi: 10.1038/icb.2013.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uldrich AP, Le Nours J, Pellicci DG, Gherardin NA, McPherson KG, Lim RT, Patel O, Beddoe T, Gras S, Rossjohn J, Godfrey DI (2013) CD1d-lipid antigen recognition by the gammadelta TCR. Nature immunology 14 (11):1137–1145. doi: 10.1038/ni.2713 [DOI] [PubMed] [Google Scholar]
- 31.Luoma AM, Castro CD, Mayassi T, Bembinster LA, Bai L, Picard D, Anderson B, Scharf L, Kung JE, Sibener LV, Savage PB, Jabri B, Bendelac A, Adams EJ (2013) Crystal structure of Vdelta1 T cell receptor in complex with CD1d-sulfatide shows MHC-like recognition of a self-lipid by human gammadelta T cells. Immunity 39 (6):1032–1042. doi: 10.1016/j.immuni.2013.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G, Fersht AR, Besra GS, Schmidt RR, Jones EY, Cerundolo V (2005) The crystal structure of human CD1d with and without alpha-galactosylceramide. Nature immunology 6 (8):819–826. doi: 10.1038/ni1225 [DOI] [PubMed] [Google Scholar]
- 33.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA (1997) Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science 277 (5324):339–345 [DOI] [PubMed] [Google Scholar]
- 34.Kinjo Y, Ueno K (2011) iNKT cells in microbial immunity: recognition of microbial glycolipids. Microbiology and immunology 55 (7):472–482. doi: 10.1111/j.1348-0421.2011.00338.x [DOI] [PubMed] [Google Scholar]
- 35.Zajonc DM, Girardi E (2015) Recognition of Microbial Glycolipids by Natural Killer T Cells. Frontiers in immunology 6:400. doi: 10.3389/fimmu.2015.00400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Van Kaer L, Parekh VV, Wu L (2015) The Response of CD1d-Restricted Invariant NKT Cells to Microbial Pathogens and Their Products. Frontiers in immunology 6:226. doi: 10.3389/fimmu.2015.00226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S, Hurwitz R, Kursar M, Bonneville M, Kaufmann SH, Schaible UE (2004) Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d-restricted T cells. Proceedings of the National Academy of Sciences of the United States of America 101 (29):10685–10690. doi: 10.1073/pnas.0403787101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Venkataswamy MM, Porcelli SA (2010) Lipid and glycolipid antigens of CD1d-restricted natural killer T cells. Seminars in immunology 22 (2):68–78. doi: 10.1016/j.smim.2009.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kinjo Y, Tupin E, Wu D, Fujio M, Garcia-Navarro R, Benhnia MR, Zajonc DM, Ben-Menachem G, Ainge GD, Painter GF, Khurana A, Hoebe K, Behar SM, Beutler B, Wilson IA, Tsuji M, Sellati TJ, Wong CH, Kronenberg M (2006) Natural killer T cells recognize diacylglycerol antigens from pathogenic bacteria. Nature immunology 7 (9):978–986. doi: 10.1038/ni1380 [DOI] [PubMed] [Google Scholar]
- 40.An D, Oh SF, Olszak T, Neves JF, Avci FY, Erturk-Hasdemir D, Lu X, Zeissig S, Blumberg RS, Kasper DL (2014) Sphingolipids from a symbiotic microbe regulate homeostasis of host intestinal natural killer T cells. Cell 156 (1–2):123–133. doi: 10.1016/j.cell.2013.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wieland Brown LC, Penaranda C, Kashyap PC, Williams BB, Clardy J, Kronenberg M, Sonnenburg JL, Comstock LE, Bluestone JA, Fischbach MA (2013) Production of alpha-galactosylceramide by a prominent member of the human gut microbiota. PLoS biology 11 (7):e1001610. doi: 10.1371/journal.pbio.1001610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C 3rd, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, Hoebe K, Schneewind O, Walker D, Beutler B, Teyton L, Savage PB, Bendelac A (2005) Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature 434 (7032):525–529. doi: 10.1038/nature03408 [DOI] [PubMed] [Google Scholar]
- 43.Kinjo Y, Wu D, Kim G, Xing GW, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong CH, Kronenberg M (2005) Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 434 (7032):520–525. doi: 10.1038/nature03407 [DOI] [PubMed] [Google Scholar]
- 44.Kawahara K, Moll H, Knirel YA, Seydel U, Zahringer U (2000) Structural analysis of two glycosphingolipids from the lipopolysaccharide-lacking bacterium Sphingomonas capsulata. European journal of biochemistry / FEBS 267 (6):1837–1846 [DOI] [PubMed] [Google Scholar]
- 45.Florence WC, Bhat RK, Joyce S (2008) CD1d-restricted glycolipid antigens: presentation principles, recognition logic and functional consequences. Expert reviews in molecular medicine 10:e20. doi: 10.1017/S1462399408000732 [DOI] [PubMed] [Google Scholar]
- 46.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J (2007) CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature 448 (7149):44–49. doi: 10.1038/nature05907 [DOI] [PubMed] [Google Scholar]
- 47.Pellicci DG, Patel O, Kjer-Nielsen L, Pang SS, Sullivan LC, Kyparissoudis K, Brooks AG, Reid HH, Gras S, Lucet IS, Koh R, Smyth MJ, Mallevaey T, Matsuda JL, Gapin L, McCluskey J, Godfrey DI, Rossjohn J (2009) Differential recognition of CD1d-alpha-galactosyl ceramide by the V beta 8.2 and V beta 7 semi-invariant NKT T cell receptors. Immunity 31 (1):47–59. doi: 10.1016/j.immuni.2009.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu D, Zajonc DM, Fujio M, Sullivan BA, Kinjo Y, Kronenberg M, Wilson IA, Wong CH (2006) Design of natural killer T cell activators: structure and function of a microbial glycosphingolipid bound to mouse CD1d. Proceedings of the National Academy of Sciences of the United States of America 103 (11):3972–3977. doi: 10.1073/pnas.0600285103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olson CM Jr., Bates TC, Izadi H, Radolf JD, Huber SA, Boyson JE, Anguita J (2009) Local production of IFN-gamma by invariant NKT cells modulates acute Lyme carditis. Journal of immunology 182 (6):3728–3734. doi: 10.4049/jimmunol.0804111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang J, Li Y, Kinjo Y, Mac TT, Gibson D, Painter GF, Kronenberg M, Zajonc DM (2010) Lipid binding orientation within CD1d affects recognition of Borrelia burgorferi antigens by NKT cells. Proceedings of the National Academy of Sciences of the United States of America 107 (4):1535–1540. doi: 10.1073/pnas.0909479107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kinjo Y, Illarionov P, Vela JL, Pei B, Girardi E, Li X, Li Y, Imamura M, Kaneko Y, Okawara A, Miyazaki Y, Gomez-Velasco A, Rogers P, Dahesh S, Uchiyama S, Khurana A, Kawahara K, Yesilkaya H, Andrew PW, Wong CH, Kawakami K, Nizet V, Besra GS, Tsuji M, Zajonc DM, Kronenberg M (2011) Invariant natural killer T cells recognize glycolipids from pathogenic Gram-positive bacteria. Nature immunology 12 (10):966–974. doi: 10.1038/ni.2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallevaey T, Clarke AJ, Scott-Browne JP, Young MH, Roisman LC, Pellicci DG, Patel O, Vivian JP, Matsuda JL, McCluskey J, Godfrey DI, Marrack P, Rossjohn J, Gapin L (2011) A molecular basis for NKT cell recognition of CD1d-self-antigen. Immunity 34 (3):315–326. doi: 10.1016/j.immuni.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zajonc DM, Ainge GD, Painter GF, Severn WB, Wilson IA (2006) Structural characterization of mycobacterial phosphatidylinositol mannoside binding to mouse CD1d. Journal of immunology 177 (7):4577–4583 [DOI] [PubMed] [Google Scholar]
- 54.Giabbai B, Sidobre S, Crispin MD, Sanchez-Ruiz Y, Bachi A, Kronenberg M, Wilson IA, Degano M (2005) Crystal structure of mouse CD1d bound to the self ligand phosphatidylcholine: a molecular basis for NKT cell activation. Journal of immunology 175 (2):977–984 [DOI] [PubMed] [Google Scholar]
- 55.Lotter H, Gonzalez-Roldan N, Lindner B, Winau F, Isibasi A, Moreno-Lafont M, Ulmer AJ, Holst O, Tannich E, Jacobs T (2009) Natural killer T cells activated by a lipopeptidophosphoglycan from Entamoeba histolytica are critically important to control amebic liver abscess. PLoS pathogens 5 (5):e1000434. doi: 10.1371/journal.ppat.1000434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hirai Y, Haque M, Yoshida T, Yokota K, Yasuda T, Oguma K (1995) Unique cholesteryl glucosides in Helicobacter pylori: composition and structural analysis. Journal of bacteriology 177 (18):5327–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ito Y, Vela JL, Matsumura F, Hoshino H, Tyznik A, Lee H, Girardi E, Zajonc DM, Liddington R, Kobayashi M, Bao X, Bugaytsova J, Boren T, Jin R, Zong Y, Seeberger PH, Nakayama J, Kronenberg M, Fukuda M (2013) Helicobacter pylori cholesteryl alpha-glucosides contribute to its pathogenicity and immune response by natural killer T cells. PloS one 8 (12):e78191. doi: 10.1371/journal.pone.0078191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang YJ, Kim HY, Albacker LA, Lee HH, Baumgarth N, Akira S, Savage PB, Endo S, Yamamura T, Maaskant J, Kitano N, Singh A, Bhatt A, Besra GS, van den Elzen P, Appelmelk B, Franck RW, Chen G, DeKruyff RH, Shimamura M, Illarionov P, Umetsu DT (2011) Influenza infection in suckling mice expands an NKT cell subset that protects against airway hyperreactivity. The Journal of clinical investigation 121 (1):57–69. doi: 10.1172/JCI44845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tatituri RV, Watts GF, Bhowruth V, Barton N, Rothchild A, Hsu FF, Almeida CF, Cox LR, Eggeling L, Cardell S, Rossjohn J, Godfrey DI, Behar SM, Besra GS, Brenner MB, Brigl M (2013) Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proceedings of the National Academy of Sciences of the United States of America 110 (5):1827–1832. doi: 10.1073/pnas.1220601110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolf BJ, Tatituri RV, Almeida CF, Le Nours J, Bhowruth V, Johnson D, Uldrich AP, Hsu FF, Brigl M, Besra GS, Rossjohn J, Godfrey DI, Brenner MB (2015) Identification of a Potent Microbial Lipid Antigen for Diverse NKT Cells. J Immunol 195 (6):2540–2551. doi: 10.4049/jimmunol.1501019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y, Girardi E, Wang J, Yu ED, Painter GF, Kronenberg M, Zajonc DM (2010) The Valpha14 invariant natural killer T cell TCR forces microbial glycolipids and CD1d into a conserved binding mode. The Journal of experimental medicine 207 (11):2383–2393. doi: 10.1084/jem.20101335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Girardi E, Yu ED, Li Y, Tarumoto N, Pei B, Wang J, Illarionov P, Kinjo Y, Kronenberg M, Zajonc DM (2011) Unique interplay between sugar and lipid in determining the antigenic potency of bacterial antigens for NKT cells. PLoS biology 9 (11):e1001189. doi: 10.1371/journal.pbio.1001189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pellicci DG, Clarke AJ, Patel O, Mallevaey T, Beddoe T, Le Nours J, Uldrich AP, McCluskey J, Besra GS, Porcelli SA, Gapin L, Godfrey DI, Rossjohn J (2011) Recognition of beta-linked self glycolipids mediated by natural killer T cell antigen receptors. Nat Immunol 12 (9):827–833. doi: 10.1038/ni.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Girardi E, Zajonc DM (2012) Molecular basis of lipid antigen presentation by CD1d and recognition by natural killer T cells. Immunological reviews 250 (1):167–179. doi: 10.1111/j.1600-065X.2012.01166.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Silva AD, Park JJ, Matsuki N, Stanic AK, Brutkiewicz RR, Medof ME, Joyce S (2002) Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol 168 (2):723–733 [DOI] [PubMed] [Google Scholar]
- 66.Garcia-Alles LF, Versluis K, Maveyraud L, Vallina AT, Sansano S, Bello NF, Gober HJ, Guillet V, de la Salle H, Puzo G, Mori L, Heck AJ, De Libero G, Mourey L (2006) Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. The EMBO journal 25 (15):3684–3692. doi: 10.1038/sj.emboj.7601244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J, Cerundolo V, Altman JD, Moody DB (2011) Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove-blocking lipids of the human CD1 system. Proceedings of the National Academy of Sciences of the United States of America 108 (48):19335–19340. doi: 10.1073/pnas.1112969108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shamshiev A, Donda A, Prigozy TI, Mori L, Chigorno V, Benedict CA, Kappos L, Sonnino S, Kronenberg M, De Libero G (2000) The alphabeta T cell response to self-glycolipids shows a novel mechanism of CD1b loading and a requirement for complex oligosaccharides. Immunity 13 (2):255–264 [DOI] [PubMed] [Google Scholar]
- 69.Moody DB, Briken V, Cheng TY, Roura-Mir C, Guy MR, Geho DH, Tykocinski ML, Besra GS, Porcelli SA (2002) Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nature immunology 3 (5):435–442. doi: 10.1038/ni780 [DOI] [PubMed] [Google Scholar]
- 70.Manolova V, Kistowska M, Paoletti S, Baltariu GM, Bausinger H, Hanau D, Mori L, De Libero G (2006) Functional CD1a is stabilized by exogenous lipids. European journal of immunology 36 (5):1083–1092. doi: 10.1002/eji.200535544 [DOI] [PubMed] [Google Scholar]
- 71.Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA (2000) Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. The Journal of experimental medicine 192 (2):281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugita M, van Der WN, Rogers RA, Peters PJ, Brenner MB (2000) CD1c molecules broadly survey the endocytic system. ProcNatlAcadSciUSA 97 (15):8445–8450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Briken V, Jackman RM, Dasgupta S, Hoening S, Porcelli SA (2002) Intracellular trafficking pathway of newly synthesized CD1b molecules. The EMBO journal 21 (4):825–834. doi: 10.1093/emboj/21.4.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moody DB, Porcelli SA (2003) Intracellular pathways of CD1 antigen presentation. NatRevImmunol 3 (1):11–22 [DOI] [PubMed] [Google Scholar]
- 75.Cheng TY, Relloso M, Van Rhijn I, Young DC, Besra GS, Briken V, Zajonc DM, Wilson IA, Porcelli S, Moody DB (2006) Role of lipid trimming and CD1 groove size in cellular antigen presentation. The EMBO journal 25 (13):2989–2999. doi: 10.1038/sj.emboj.7601185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S, Bhatt A, van Summeren RP, Altman JD, Jacobs WR Jr., Adams EJ, Minnaard AJ, Porcelli SA, Moody DB (2013) CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. The Journal of experimental medicine 210 (4):729–741. doi: 10.1084/jem.20120624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA (2000) CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 20;404 (6780):884–888 [DOI] [PubMed] [Google Scholar]
- 78.de Jong A, Arce EC, Cheng TY, van Summeren RP, Feringa BL, Dudkin V, Crich D, Matsunaga I, Minnaard AJ, Moody DB (2007) CD1c presentation of synthetic glycolipid antigens with foreign alkyl branching motifs. Chemistry & biology 14 (11):1232–1242. doi: 10.1016/j.chembiol.2007.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.de la Salle H, Mariotti S, Angenieux C, Gilleron M, Garcia-Alles LF, Malm D, Berg T, Paoletti S, Maitre B, Mourey L, Salamero J, Cazenave JP, Hanau D, Mori L, Puzo G, De Libero G (2005) Assistance of microbial glycolipid antigen processing by CD1e. Science 310 (5752):1321–1324. doi: 10.1126/science.1115301 [DOI] [PubMed] [Google Scholar]
- 80.Gilleron M, Lepore M, Layre E, Cala-De Paepe D, Mebarek N, Shayman JA, Canaan S, Mori L, Carriere F, Puzo G, De Libero G (2016) Lysosomal Lipases PLRP2 and LPLA2 Process Mycobacterial Multi-acylated Lipids and Generate T Cell Stimulatory Antigens. Cell chemical biology 23 (9):1147–1156. doi: 10.1016/j.chembiol.2016.07.021 [DOI] [PubMed] [Google Scholar]
- 81.Prigozy TI, Naidenko O, Qasba P, Elewaut D, Brossay L, Khurana A, Natori T, Koezuka Y, Kulkarni A, Kronenberg M (2001) Glycolipid antigen processing for presentation by CD1d molecules. Science 291 (5504):664–667. doi: 10.1126/science.291.5504.664 [DOI] [PubMed] [Google Scholar]
- 82.Relloso M, Cheng TY, Im JS, Parisini E, Roura-Mir C, DeBono C, Zajonc DM, Murga LF, Ondrechen MJ, Wilson IA, Porcelli SA, Moody DB (2008) pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity 28 (6):774–786. doi: 10.1016/j.immuni.2008.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Brozovic S, Nagaishi T, Yoshida M, Betz S, Salas A, Chen D, Kaser A, Glickman J, Kuo T, Little A, Morrison J, Corazza N, Kim JY, Colgan SP, Young SG, Exley M, Blumberg RS (2004) CD1d function is regulated by microsomal triglyceride transfer protein. Nature medicine 10 (5):535–539. doi: 10.1038/nm1043 [DOI] [PubMed] [Google Scholar]
- 84.Zhou D, Cantu C 3rd, Sagiv Y, Schrantz N, Kulkarni AB, Qi X, Mahuran DJ, Morales CR, Grabowski GA, Benlagha K, Savage P, Bendelac A, Teyton L (2004) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303 (5657):523–527. doi: 10.1126/science.1092009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yuan W, Qi X, Tsang P, Kang SJ, Illarionov PA, Besra GS, Gumperz J, Cresswell P (2007) Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proceedings of the National Academy of Sciences of the United States of America 104 (13):5551–5556. doi: 10.1073/pnas.0700617104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Winau F, Schwierzeck V, Hurwitz R, Remmel N, Sieling PA, Modlin RL, Porcelli SA, Brinkmann V, Sugita M, Sandhoff K, Kaufmann SH, Schaible UE (2004) Saposin C is required for lipid presentation by human CD1b. Nature immunology 5 (2):169–174. doi: 10.1038/ni1035 [DOI] [PubMed] [Google Scholar]
- 87.Cala-De Paepe D, Layre E, Giacometti G, Garcia-Alles LF, Mori L, Hanau D, de Libero G, de la Salle H, Puzo G, Gilleron M (2012) Deciphering the role of CD1e protein in mycobacterial phosphatidyl-myo-inositol mannosides (PIM) processing for presentation by CD1b to T lymphocytes. J Biol Chem 287 (37):31494–31502. doi: 10.1074/jbc.M112.386300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB (1999) Molecular recognition of lipid antigens by T cell receptors. The Journal of experimental medicine 189 (1):195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Van Rhijn I, Young DC, Im JS, Levery SB, Illarionov PA, Besra GS, Porcelli SA, Gumperz J, Cheng TY, Moody DB (2004) CD1d-restricted T cell activation by nonlipidic small molecules. Proceedings of the National Academy of Sciences of the United States of America 101 (37):13578–13583. doi: 10.1073/pnas.0402838101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, Rossjohn J, Moody DB (2013) A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol 14 (7):706–713. doi: 10.1038/ni.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Van Rhijn I, Gherardin NA, Kasmar A, de Jager W, Pellicci DG, Kostenko L, Tan LL, Bhati M, Gras S, Godfrey DI, Rossjohn J, Moody DB (2014) TCR Bias and Affinity Define Two Compartments of the CD1b-Glycolipid-Specific T Cell Repertoire. J Immunol 193 (10):5338–5344. doi: 10.4049/jimmunol.1400158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turner SJ, Doherty PC, McCluskey J, Rossjohn J (2006) Structural determinants of T-cell receptor bias in immunity. Nature reviews Immunology 6 (12):883–894. doi: 10.1038/nri1977 [DOI] [PubMed] [Google Scholar]
- 93.Spada FM, Grant EP, Peters PJ, Sugita M, Melian A, Leslie DS, Lee HK, van Donselaar E, Hanson DA, Krensky AM, Majdic O, Porcelli SA, Morita CT, Brenner MB (2000) Self-recognition of CD1 by gamma/delta T cells: implications for innate immunity. The Journal of experimental medicine 191 (6):937–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Russano AM, Bassotti G, Agea E, Bistoni O, Mazzocchi A, Morelli A, Porcelli SA, Spinozzi F (2007) CD1-restricted recognition of exogenous and self-lipid antigens by duodenal gammadelta+ T lymphocytes. Journal of immunology 178 (6):3620–3626 [DOI] [PubMed] [Google Scholar]
- 95.Roy S, Ly D, Castro CD, Li NS, Hawk AJ, Altman JD, Meredith SC, Piccirilli JA, Moody DB, Adams EJ (2016) Molecular Analysis of Lipid-Reactive Vdelta1 gammadelta T Cells Identified by CD1c Tetramers. Journal of immunology 196 (4):1933–1942. doi: 10.4049/jimmunol.1502202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kasmar A, Van Rhijn I, Magalhaes KG, Young D, Cheng TY, Turner M, Schiefner A, Kalathur RC, Wilson A, Bhati M, Gras S, Rossjohn J, Shires J, Jakobsen S, Altman JD, Moody DB (2013) CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. J Immunol 191:4499–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Van Rhijn I, Iwany SK, Fodran P, Cheng TY, Gapin L, Minnaard AJ, Moody DB (2017) CD1b-mycolic acid tetramers demonstrate T-cell fine specificity for mycobacterial lipid tails. European journal of immunology. doi: 10.1002/eji.201747062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Seshadri C, Lin L, Scriba TJ, Peterson G, Freidrich D, Frahm N, DeRosa SC, Moody DB, Prandi J, Gilleron M, Mahomed H, Jiang W, Finak G, Hanekom WA, Gottardo R, McElrath MJ, Hawn TR (2015) T Cell Responses against Mycobacterial Lipids and Proteins Are Poorly Correlated in South African Adolescents. J Immunol 195 (10):4595–4603. doi: 10.4049/jimmunol.1501285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Felio K, Nguyen H, Dascher CC, Choi HJ, Li S, Zimmer MI, Colmone A, Moody DB, Brenner MB, Wang CR (2009) CD1-restricted adaptive immune responses to Mycobacteria in human group 1 CD1 transgenic mice. The Journal of experimental medicine 206 (11):2497–2509. doi: 10.1084/jem.20090898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dascher CC, Hiromatsu K, Xiong X, Morehouse C, Watts G, Liu G, McMurray DN, LeClair KP, Porcelli SA, Brenner MB (2003) Immunization with a mycobacterial lipid vaccine improves pulmonary pathology in the guinea pig model of tuberculosis. International immunology 15 (8):915–925 [DOI] [PubMed] [Google Scholar]
- 101.Porcelli S, Morita CT, Brenner MB (1992) CD1b restricts the response of human CD4–8- T lymphocytes to a microbial antigen. Nature 360 (6404):593–597. doi: 10.1038/360593a0 [DOI] [PubMed] [Google Scholar]
- 102.Moody DB, Ulrichs T, Muhlecker W, Young DC, Gurcha SS, Grant E, Rosat JP, Brenner MB, Costello CE, Besra GS, Porcelli SA (2000) CD1c-mediated T-cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 404 (6780):884–888. doi: 10.1038/35009119 [DOI] [PubMed] [Google Scholar]
- 103.Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T, Bloom BR, Brenner MB, Kronenberg M, Brennan PJ, et al. (1995) CD1-restricted T cell recognition of microbial lipoglycan antigens. Science 269 (5221):227–230 [DOI] [PubMed] [Google Scholar]
- 104.Ernst WA, Maher J, Cho S, Niazi KR, Chatterjee D, Moody DB, Besra GS, Watanabe Y, Jensen PE, Porcelli SA, Kronenberg M, Modlin RL (1998) Molecular interaction of CD1b with lipoglycan antigens. Immunity 8 (3):331–340 [DOI] [PubMed] [Google Scholar]
- 105.Takayama K, Wang C, Besra GS (2005) Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clinical microbiology reviews 18 (1):81–101. doi: 10.1128/CMR.18.1.81-101.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grant EP, Beckman EM, Behar SM, Degano M, Frederique D, Besra GS, Wilson IA, Porcelli SA, Furlong ST, Brenner MB (2002) Fine specificity of TCR complementarity-determining region residues and lipid antigen hydrophilic moieties in the recognition of a CD1-lipid complex. Journal of immunology 168 (8):3933–3940 [DOI] [PubMed] [Google Scholar]
- 107.Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST, Ye S, Reinhold VN, Sieling PA, Modlin RL, Besra GS, Porcelli SA (1997) Structural requirements for glycolipid antigen recognition by CD1b-restricted T cells. Science 278 (5336):283–286 [DOI] [PubMed] [Google Scholar]
- 108.Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME, de Vries N, Godfrey DI, Altman JD, de Jager W, Rossjohn J, Moody DB (2013) A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nature immunology 14 (7):706–713. doi: 10.1038/ni.2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Van Rhijn I, Moody DB (2015) CD1 and mycobacterial lipids activate human T cells. Immunological reviews 264 (1):138–153. doi: 10.1111/imr.12253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J, Mori L, Stenger S, De Libero G, Puzo G, Gilleron M (2009) Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1-restricted T cells. Chemistry & biology 16 (1):82–92. doi: 10.1016/j.chembiol.2008.11.008 [DOI] [PubMed] [Google Scholar]
- 111.Kasmar AG, van Rhijn I, Cheng TY, Turner M, Seshadri C, Schiefner A, Kalathur RC, Annand JW, de Jong A, Shires J, Leon L, Brenner M, Wilson IA, Altman JD, Moody DB (2011) CD1b tetramers bind alphabeta T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. The Journal of experimental medicine 208 (9):1741–1747. doi: 10.1084/jem.20110665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gras S, Van Rhijn I, Shahine A, Cheng TY, Bhati M, Tan LL, Halim H, Tuttle KD, Gapin L, Le Nours J, Moody DB, Rossjohn J (2016) T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nature communications 7:13257. doi: 10.1038/ncomms13257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR, Besra GS, Cerundolo V, Jones EY (2004) The crystal structure of human CD1b with a bound bacterial glycolipid. Journal of immunology 172 (4):2382–2388 [DOI] [PubMed] [Google Scholar]
- 114.Garcia-Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L, Huc E, Im JS, Sansano S, Brando T, Julien S, Prandi J, Gilleron M, Porcelli SA, de la Salle H, Heck AJ, Mori L, Puzo G, Mourey L, De Libero G (2011) Structural reorganization of the antigen-binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proceedings of the National Academy of Sciences of the United States of America 108 (43):17755–17760. doi: 10.1073/pnas.1110118108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Bohmer G, Prandi J, Mori L, Puzo G, De Libero G (2004) Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. The Journal of experimental medicine 199 (5):649–659. doi: 10.1084/jem.20031097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moody DB, Young DC, Cheng TY, Rosat JP, Roura-Mir C, O’Connor PB, Zajonc DM, Walz A, Miller MJ, Levery SB, Wilson IA, Costello CE, Brenner MB (2004) T cell activation by lipopeptide antigens. Science 303 (5657):527–531. doi: 10.1126/science.1089353 [DOI] [PubMed] [Google Scholar]
- 117.Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J, Costello CE, Rudd PM, Dwek RA, Miller MJ, Brenner MB, Moody DB, Wilson IA (2005) Molecular mechanism of lipopeptide presentation by CD1a. Immunity 22 (2):209–219. doi: 10.1016/j.immuni.2004.12.009 [DOI] [PubMed] [Google Scholar]
- 118.Snow GA, White AJ (1969) Chemical and biological properties of mycobactins isolated from various mycobacteria. The Biochemical journal 115 (5):1031–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Layre E, Paepe DC, Larrouy-Maumus G, Vaubourgeix J, Mundayoor S, Lindner B, Puzo G, Gilleron M (2011) Deciphering sulfoglycolipids of Mycobacterium tuberculosis. Journal of lipid research 52 (6):1098–1110. doi: 10.1194/jlr.M013482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Scharf L, Li NS, Hawk AJ, Garzon D, Zhang T, Fox LM, Kazen AR, Shah S, Haddadian EJ, Gumperz JE, Saghatelian A, Faraldo-Gomez JD, Meredith SC, Piccirilli JA, Adams EJ (2010) The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 33 (6):853–862. doi: 10.1016/j.immuni.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB, Adams EJ (2014) Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by alphabeta T cells. Proceedings of the National Academy of Sciences of the United States of America 111 (43):E4648–4657. doi: 10.1073/pnas.1408549111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Van Rhijn I, van Berlo T, Hilmenyuk T, Cheng TY, Wolf BJ, Tatituri RV, Uldrich AP, Napolitani G, Cerundolo V, Altman JD, Willemsen P, Huang S, Rossjohn J, Besra GS, Brenner MB, Godfrey DI, Moody DB (2016) Human autoreactive T cells recognize CD1b and phospholipids. Proceedings of the National Academy of Sciences of the United States of America 113 (2):380–385. doi: 10.1073/pnas.1520947112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro-Palomino JC, Ritter G, Schmidt RR, Jones EY, Cerundolo V (2002) Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nature immunology 3 (8):721–726. doi: 10.1038/ni821 [DOI] [PubMed] [Google Scholar]
- 124.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic acids research 34 (Web Server issue):W116–118. doi: 10.1093/nar/gkl282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schrodinger LLC (2015) The PyMOL Molecular Graphics System, Version 1.7.6.4