Abstract
The TMC1 channel was identified as a protein essential for hearing in mouse and human, and recognized as one of a family of eight such proteins in mammals. The TMC family is part of a superfamily of seven branches, which includes the TMEM16s. Vertebrate hair cells express both TMC1 and TMC2. They are located at the tips of stereocilia and are required for hair cell mechanotransduction. TMC1 assembles as a dimer and its similarity to the TMEM16s has enabled a predicted tertiary structure with an ion conduction pore in each subunit of the dimer. Cysteine mutagenesis of the pore supports the role of TMC1 and TMC2 as the core channel proteins of a larger mechanotransduction complex that includes PCDH15 and LHFPL5, and perhaps TMIE, CIB2 and others.
Since the first characterization of the hair cell mechanotransduction current in the 1970s, a central quest has been to identify the protein(s) constituting the conductance. Over the years, many proteins have been suggested as transduction channel candidates, often with substantial supporting evidence, but most have been shown to be not essential for transduction when their genes were disrupted. Two proteins, TMC1 and TMC2, are essential, but their role was confused by the emergence of another mechanosensitive conductance in hair cells (mediated by PIEZO2) when TMC1 and TMC2 are deleted. Recent evidence from several laboratories now supports a role for TMC1 and TMC2 as core proteins of a mechanotransduction complex and has suggested a tertiary structure with independent pores in a dimeric channel. Here, we review the function of TMC channels from the early mouse mutants to the first glimpse of a molec-ular structure.
THE DISCOVERY OF TMC1 FROM HEREDITARY DEAFNESS IN MOUSE AND HUMAN
The cloning of genes mutated in hereditary deafness, in both mice and humans, has led to the identification of a great many protein constituents of the hair cell mechanotransduction complex. Similarly, the discovery of TMC1—and by homology, TMC2—came from both deaf mice and deaf humans. In 1958, Deol and Kocher (1958) described a new deaf mouse, the deafness (dn) strain, with recessive inheritance. Steel and Bock (1980; Bock and Steel 1983) showed that it lacked a cochlear microphonic and that its hair cells, present but distorted in neonatal mice, de-generated over the next 2–6 weeks.
Within a few years, human siblings were identified at a school for the deaf in Maharashtra, India, which led to an extended family with recessive deafness linked to 9qll–q21 (DFNB7; Jain et al. 1995). This region of the human genome is syntenic to the mouse dn locus, and Jain et al. suggested that the dn mouse could be a good model for the human DFNB7. The recessive human deafness DFNB11 also maps to the same locus. Kurima et al. (2002) then identified a family with dominant, progressive deafness (DFNA36) in the same interval (9q13–q21) as DFNB7/11. Positional cloning with the DFNA36 and DFNB7/11 families revealed pathogenic variants in a new gene that Kurima et al. named transmembrane channel-like (TMC) 1, for the six or more predicted transmembrane domains. Analysis of similar sequences in the genome showed a closely related gene, TMC2. Kurima et al. (2002) also demonstrated that the deafness mouse carries a 1656-bp deletion in Tmc1. Finally, a companion paper (Vreugde et al. 2002) reported a new ENU-generated mouse mutant with dominantly inherited deaf-ness named Beethoven (Bth), and showed it to have a missense mutation (M412K) in the Tmc1 gene. In these landmark papers, the confluence of both recessive and dominant mutations in both mouse and human TMC1 set the stage for elucidation of TMC1 function, but the path was not to be easy.
THE TMC/TMEM16 SUPERFAMILY
The following year, both Griffith (Kurima et al. 2003) and Heller (Keresztes et al. 2003) used database searches and reverse transcription polymerase chain reaction (RT-PCR) to show that TMC1 and TMC2 belong to a protein family with eight members in mammals and additional homologs in other vertebrates and invertebrates. They predicted up to ten transmembrane domains based on hydrophobicity. A common feature of the family is the TMC domain, corresponding to amino acids 512–627 in mmTMC1 (UniProt ID: Q8R4P5). It begins with very highly conserved amino acids cysteine, tryptophan, glutamic acid, and often threonine—the signature CWET sequence.
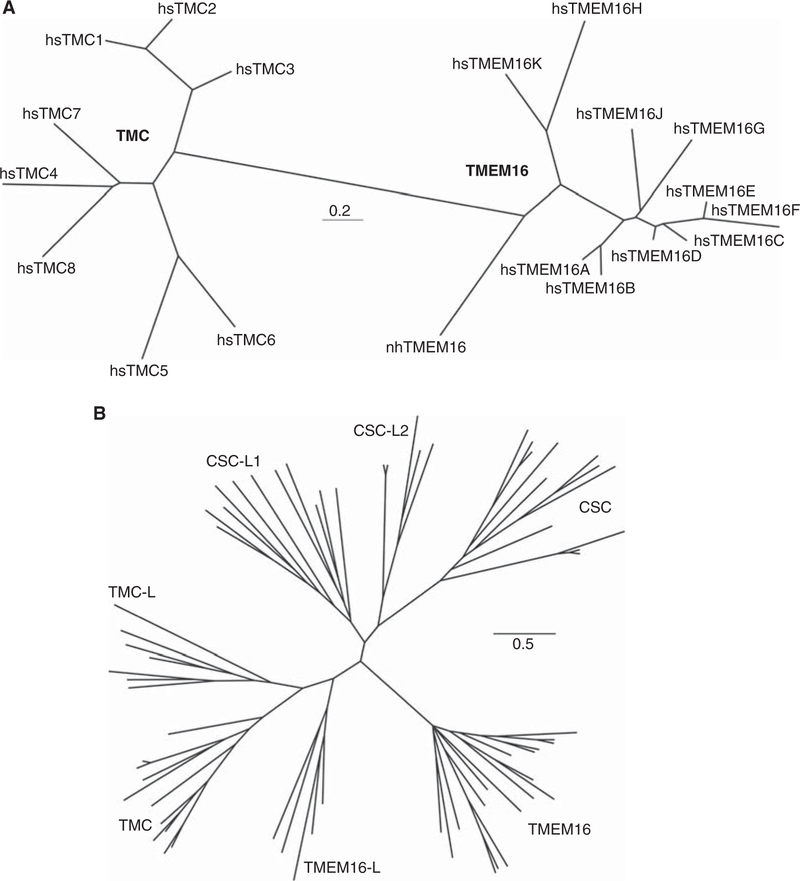
Based on limited sequence similarity, Hahn et al. (2009) suggested that the TMC family is related to the family of ion channels and lipid scramblases called anoctamins or TMEM16s. There are ten TMEM16s in mammals (ANO1–10 or TMEM16A-K). A phylogeny tree derived from an alignment of vertebrate TMC and TMEM16 sequences (Pan et al. 2018) is shown in Figure 1A. Based on the TMC domain, the PFAM database identifies a third group, the osmosensitive calcium-permeable, stress-sensitive cation channel (CSC) family (pfam.xfam.org/family/PF07810#tabview=tab2). Medrano-Soto et al. (2018) then expanded the family with an iterative sequence search, identifying four more branches that they designated ANO-like (ANO-L or TMEM16-L), TMC-like (TMC-L), CSC-like 1 (CSC-L1), and CSC-like 2 (CSC-L2) (Fig. 1B). Three motifs, found in transmembrane do-mains S1, S4–S5, and S7–S8, link the three families. The first, in S1, is of unknown functional significance. A second maps to S4 and S5, which form part of the groove for lipid scrambling in the Nectria lipid scramblase nhTMEM16 and the pore in the mouse chloride channel mmTMEM16A (Whitlock and Hartzell 2016; Jiang et al. 2017). The most conserved motif maps to residues in the S7–S8 region, which includes calcium-binding residues in TMEM16. Because atomic structures for nhTMEM16 and mmTMEM16A have been solved, the homology among families, distant though it is, has provided considerable insight into the structure and function of TMC1 (see below).
Figure 1.
Phylogeny of the TMC and TMEM16 families. (A) An unrooted phylogeny tree of the TMC and TMEM16 families, derived from an alignment of vertebrate TMCs and mammalian TMEM16s. Only Nectria and human orthologs are shown here. Within the TMC branch, TMC1–3 cluster in a distinct subfamily. (B) The TMC/TMEM16 superfamily. (Panel B based on data in Medrano-Soto et al. 2018.)
EXPRESSION AND LOCATION OF MAMMALIAN TMC1 AND TMC2
In vertebrates, Tmc1 and Tmc2 are expressed by hair cells of the inner ear (Kawashima et al. 2011) and of the lateral line in fish (Chou et al. 2017). Tmc1 is expressed in other organs as well such as brain, eye, and colon (Keresztes et al. 2003). In the inner ear, the temporal expression profile differs for the two genes and differs among auditory and vestibular end organs. In the cochlea, Tmc2 messenger RNA (mRNA) expression begins to rise at the base around birth and at the apex around postnatal day 2 (P2). Tmc2 expression peaks during the first postnatal week and declines to near zero by postnatal day 10. As Tmc2 expression declines, Tmc1 expression begins to rise and its expression is maintained into adulthood (Kawashima et al. 2011). In the vestibular organs, Tmc2 mRNA expression also precedes the expression of Tmc1, but both Tmc1 and Tmc2 are expressed in mature vestibular hair cells. In both auditory and vestibular organs, the spatiotemporal expression pattern of Tmc2 is tightly correlated with the spatiotemporal onset of mouse hair cell mecha-nosensitivity (Géléoc and Holt 2003; Lelli et al. 2010; Kawashima et al. 2011).
Appearance of the TMC1 and TMC2 protein parallels that of the mRNA (Kurima et al. 2015). Using fluorescently tagged TMCs in knockin mice, Kurima et al. demonstrated protein localization at the tips of shorter row stereocilia in inner and outer hair cells, the same region where calcium indicators suggested hair cell transduction channels are located (Beurg et al. 2009). TMC2 protein peaked during the first postnatal week, was transiently co-localized with TMC1, and then declined. The rise of TMC1 localization at the tips of hair cell stereocilia paralleled, but preceded, the on-set of auditory function in mice by a few days. The significance of this developmental shift in expression from TMC2 to TMC1 is unclear, but seems necessary for normal auditory function, as forced expression of TMC2 cannot compensate for loss of expression of TMC1 in auditory hair cells (Asai et al. 2018; Nakanishi et al. 2018).
MECHANOTRANSDUCTION CURRENT MEDIATED BY TMC1 AND TMC2
In addition to the difference in the spatiotemporal expression profile, physiological evidence suggests that TMC1 and TMC2 play distinct roles. Mouse hair cells that express just TMC1 or just TMC2 have sensory transduction cur-rents with different calcium selectivity (Kim and Fettiplace 2013; Pan et al. 2013; Corns et al. 2017). With TMC2 alone, selectivity for calcium is approximately sixfold higher than for cesium, whereas with TMC1 alone, selectivity for calcium is only two-to threefold higher. Transduction currents are larger in auditory hair cells that express TMC2. Together with the higher calcium selectivity of TMC2, cells ex-pressing TMC2 may be challenged with a significantly higher calcium influx. In vestibular organs, which do not have a large endocochlear potential, the driving force on calcium is lower, which may permit persistent expression of TMC2 into adulthood, whereas the endocochlear potential in the cochlea may necessitate the developmental switch in TMC expression to channels with lower calcium permeability (Asai et al. 2018; Nakanishi et al. 2018). Whether greater calcium selectivity has any secondary consequences for hair cell function is not known. However, it is clear that cells lacking the proper TMC1 and TMC2 expression fail to acquire a number of physiological properties that typify mature hair cells (Marcotti et al. 2006; Nakanishi et al. 2018), suggesting that disruption of calcium signaling pathways in TMC mutant hair cells may prevent normal development.
A key property of any ion channel is its single-channel conductance. While there is agreement that single-channel conductance for the mechanotransduction current varies along the length of the cochlea and among cochlear hair cells expressing TMC1 or TMC2, the amplitude is controversial. In part, the controversy may be a result of the difficulty of estimating single-channel current levels in the whole-cell configuration, where numerous endogenous ion channels are also active. The Fettiplace group has focused on a technique for destroying all tip links in a hair bundle except one, and then using a fluid jet to stimulate the one remaining tip link and its associated channel. Examination of closed-to-open transitions has yielded estimates of single-channel conductance that range from 50 pS (Beurg et al. 2018) to 260 pS (Beurg et al. 2006). They have argued that multiple channels may be simultaneously gated to appear as a single-channel opening and that a gradient of TMC1 expression in outer hair cells along the cochlea—and a gradient in the number of coupled channels per tip link—may account for the broad range of apparent single-channel conductances (Beurg et al. 2018).
Using two alternate methods in inner hair cells expressing just TMC1, the Holt group re-ported relatively uniform single-channel conductances of ∼140 pS (Pan et al. 2013, 2018). In one case, fine-tipped stimulus probes were used to deflect single inner hair cell stereocilia (Pan et al. 2013), while for the other method, nonstationary noise analysis of whole-cell currents was used to estimate single-channel currents (Pan et al. 2018). That the two independent methods yielded similar results lends support for the reported values. However, reconciliation of the values reported by the Holt and Fettiplace groups may require more conventional methods of recording single-channel events in an excised patch of membrane that includes just one channel, which in turn may depend on membrane expression and gating of TMC1 in heterologous cells.
TMC1/2-BINDING PARTNERS AND THE MECHANOTRANSDUCTION COMPLEX
The proper targeting of TMC1 and TMC2 to the tips of hair cell stereocilia, as well as their functional integrity as pore-forming channel subunits, depends critically on their working partnership with a number of other essential proteins (see also Cunningham and Müller 2018). Some may be essential for targeting, and others for function. Like TMC1, genes encoding these partners are often deafness genes as well.
PCDH15
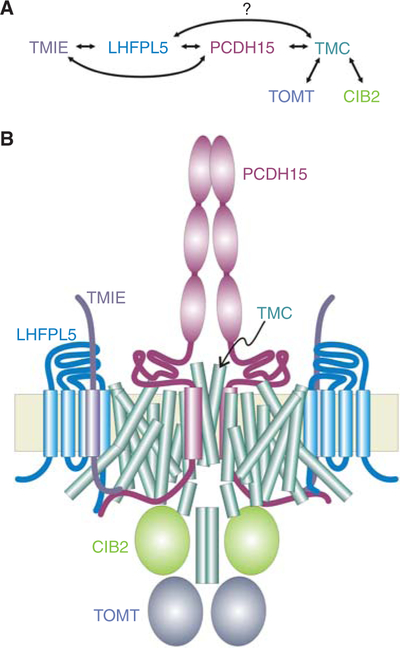
Constituting the tip links between stereocilia, protocadherin-15 (PCDH15) and cadherin-23 (CDH23) proteins feature long extracellular domains with multiple extracellular cadherin (EC) repeats. They each form parallel dimers and bind to each other at their amino-termini to form the tetrameric tip link (Pickles et al. 1984; Kachar et al. 2000; Kazmierczak et al. 2007; Sotomayor et al. 2012; Dionne et al. 2018). PCDH15 and CDH23 are both products of Usher syndrome genes, producing both deafness and blindness when mutated (Ahmed et al. 2006; Alagramam et al. 2001a,b). According to the gating-spring model of mechanotransduction (Corey and Hudspeth 1983; Howard and Hudspeth 1988), the tension in the tip link directly modulates the opening of the mechanotransduction channels. Indeed, at the lower end of the tip link, PCDH15 interacts directly with TMC1 and TMC2 (Maeda et al. 2014), as well as with other channel components (Fig. 2) (Beurg et al. 2015b).
Figure 2.
Possible schematic of the hair cell mechano-transduction apparatus. (A) Reported interactions among six proteins essential for mechanosensory transduction in hair cells. (B) A possible arrangement of proteins within the transduction apparatus. TMC1 and PCDH15 are both dimers, so it is attractive to suppose that there is a one-to-one relationship between them, with one PCDH15 opening the pore in one TMC subunit of the dimeric channel. In addition, PCDH15 and LHFPL5 can form a tetrameric complex (Ge et al. 2018) so LHFPL5 may also be near the dimer interface, rather than as depicted. There is little information about stoichiometries for the other proteins of the complex, nor is it known which are part of a mature complex and which are only necessary for assembly.
Hair cells express several different splice forms of PCDH15, which differ in their cyto-plasmic domains. There are three classes of cytoplasmic isoforms (CD1–3; Ahmed et al. 2006). Mice lacking PCDH15-CD1 or PCDH15-CD3 maintain hearing, whereas mice lacking PCDH15-CD2 are deaf (Webb et al. 2011). On the other hand, in mice lacking any of the three isoforms, tip links are observed at postnatal day 1 (P1), suggesting no isoform is indispensable for tip-link formation (Webb et al. 2011). Using antibodies generated against peptide sequences unique to each isoform, Ahmed et al. (2006) reported that PCDH15-CD2 is intensely labeled in early stages of development but is completely undetectable in mature hair bundles, suggesting that its role may be developmental. In contrast, PCDH15-CD1 is distributed evenly among the length of auditory stereocilia but excluded from the tips, and CD3 is specifically localized to the tips of stereocilia, except for the tallest row (Ahmed et al. 2006). Consistent with the hypothesis that CD3 may be the mature, lower tip-link protein, the distribution of PCDH15-CD3 but not CD1 or CD2 is rapidly affected by calcium chelation, which is known to break tip links. Using a different set of antibodies, on the other hand, Pepermans et al. (2014) demonstrated the presence of PCDH15-CD2 on the tips of stereocilia in mature hair cells and argued that PCDH15-CD2 is the only essential isoform in mature auditory hair cells.
Using a two-hybrid screen, Maeda et al. (2014) reported interactions between zebrafish orthologs of TMCs and PCDH15. Their bait vectors were truncated versions of PCDH15a-CD1 or PCDH15a-CD3, containing the trans-membrane domain and the full cytoplasmic domain, and they identified the TMC2a amino-terminus as interacting. Similarly, Beurg et al. (2015b) found that both mouse TMC1 and TMC2 expressed in HEK cells coimmunopreci-pitated with all three forms of PCDH15.
TMHS/LHFPL5
TMHS (tetraspan membrane protein of hair cell stereocilia), now known as LHFPL5 (lipoma HMGIC fusion partner-like 5, where HMGIC [high mobility group protein I-C], now known as HMGA2), is an essential protein for mechanotransduction (Longo-Guess et al. 2005; Xiong et al. 2012) and is mutated in DFNB67 deafness. The protein encoded by LHFPL5 is a member of a superfamily of tetraspan proteins, which includes the claudin tight junction proteins, gap junction proteins, and peripheral myelin proteins. LHFPL5 is located at stereocilia tips but also near the shaft and ankle links. By the onset of hearing, LHFPL5 localization shows a gradual refinement to the tips of the shorter stereocilia (Mahendrasingam et al. 2017). In the absence of PCDH15, LHFPL5 fails to localize to the tips (Mahendrasingam et al. 2017).
LHFPL5 interacts with the transmembrane and cytoplasmic domain common to all three PCDH15 isoforms (Xiong et al. 2012). It has also been shown to be essential for targeting of TMC1 but not TMC2 (Beurg et al. 2015b). Al-though Beurg et al. (2015b) did not observe interaction between LHFPL5 and TMC1, it would be important to examine this more comprehen-sively. It is interesting that both LHFPL5 and TMC1 are proposed to bind to the same cyto-plasmic domain of PCDH15 and might mediate formation of a ternary complex.
TMIE
The TMIE (transmembrane inner ear) protein is similarly essential for hearing and was also identified through positional cloning of deafness genes in mice (the spinner mouse; Deol and Robins 1962; Mitchem et al. 2002) and humans (DFNB6; Naz et al. 2002). The spinner mouse has malformed stereocilia bundles and no auditory brainstem response; a zebrafish mutant of TMIE also lacks hearing (Gleason et al. 2009). In searching for additional components of the mechanotransduction machinery of hair cells, Zhao et al. (2014a) performed yeast two-hybrid screens with proteins including LHFPL5, PCDH15, and TMC1. TMIE was identified as a binding partner for LHFPL5 and PCDH15. Biochemical data show that TMIE binds to the unique carboxy terminus of PCDH15-CD2. TMIE was also reported to interact with all three PCDH15 isoforms through its interaction with LHFPL5. The relationship of TMIE to TMC1/2 is currently unclear. TMIE does not bind to TMC1/2, and the localization of TMC1/2 in stereocilia does not appear to be affected in mouse hair cells lacking TMIE (Zhao et al. 2014a). Thus, the mechanism by which TMIE affects transduction remains to be established.
CIB2
Calcium and integrin-binding protein 2 (CIB2) is essential for hearing and mutant forms are as-sociated with nonsyndromic deafness (DFNB48) and Usher syndrome type 1J (USH1J) (Riazuddin et al. 2012; but see Booth et al. 2018). Ca2+ binds CIB2 to induce a conformational change. CIB2 was shown to localize to stereocilia but its Ca2+ sensing role there is not clear. CIB2 binds to both TMC1 and TMC2, and these interactions are disrupted by deafness-causing mutations in CIB2 (Giese et al. 2017). The ami-no-terminal domain of TMC1 is essential for this interaction (Giese et al. 2017). Importantly, CIB2 is not essential for the localization of TMC1/2 or PCDH15 (Giese et al. 2017).
TOMT
The transmembrane O-methyltransferase (TOMT/LRTOMT) gene is mutated in nonsyndromic deafness DFNB63 (Ahmed et al. 2008). In HEK293 cells, mouse TOMT and TMC1 can directly interact and, in hair cells, TOMT is required for proper localization of TMC1 and TMC2 (but not LHFPL5, TMIE, or PCDH15) (Cunningham et al. 2017; Erickson et al. 2017). In hair cells, moreover, TOMT is localized in the soma, suggesting it is not part of the transduction complex itself. These suggested that TOMT was an essential accessory protein, mediating TMC transport to the plasma membrane. In HEK293 cells, however, TOMT alone is not sufficient to regulate the transport of TMC1 and TMC2 to the cell membrane.
PREDICTED MOLECULAR STRUCTURE OF TMCS
Pan et al. (2018) used a variety of methods, including chemical cross-linking, nonreducing gels, size-exclusion chromatography/multi-angle light scattering, and low-resolution cryoelectron microscopy (cryo-EM), to show that TMC1 assembles as a dimer. Together with sequence similarity (Hahn et al. 2009; Medrano-Soto et al. 2018) and a predicted topology of 10 trans-membrane domains (Pan et al. 2018), this stoichiometry suggested that TMCs are similar to the TMEM16 family of ion channels and lipid scramblases. Similarly, algorithms that identify homologs through iterative sequence similarity searches, such as the Phyre2 and I-TASSER servers (Kelley et al. 2015; Yang and Zhang 2015), consistently identify the TMEM16s as the family of proteins with known structures closest to TMCs. Structures of two TMEM16 family members have been solved, the Nectria hematococca nhTMEM16 scramblase and the mouse mmTMEM16A anion channel (Brunner et al. 2014; Dang et al. 2017; Paulino et al. 2017a,b). Both Phyre2 and I-TASSER then threaded the TMC query sequence onto the known mmTMEM16A structure to create a predicted tertiary structure of a TMC (Fig. 3) (Pan et al. 2018). Ballesteros et al. (2018) also recognized the similarity to the TMEM16 family and, using similar tools, suggested a possible structure.
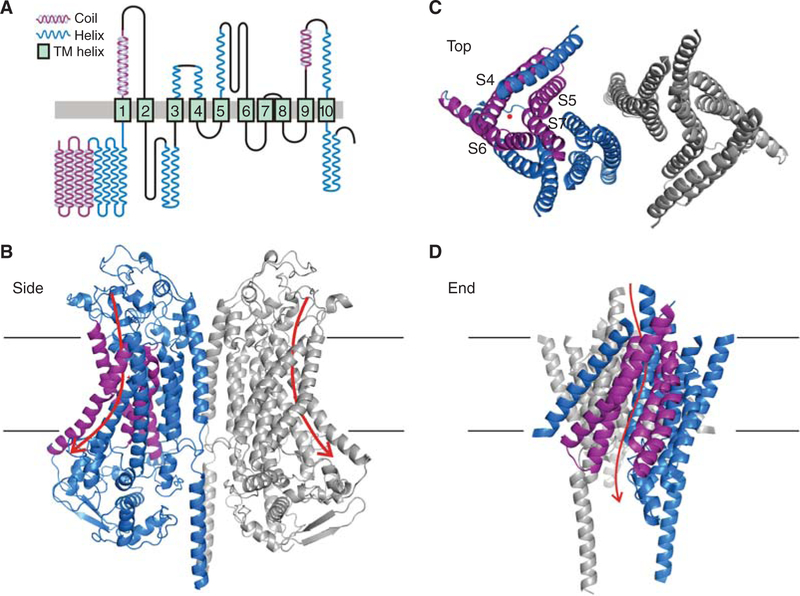
Figure 3.
Predicted secondary and tertiary structure of TMC1. (A) Predicted secondary structure of TMC1. Transmembrane domains assigned by I-TASSER and Phyre2 are based on amino acid similarity to TMEM16A and solved structures of mmTMEM16A and nhTMEM16. Helix and coiled-coil predictions from PSSpred (Yan et al. 2013). (Reprinted from Pan et al. 2018 with permission from the authors.) (B) Tertiary structure of the dimeric TMC1 predicted by I-TASSER. The two subunits are colored blue and gray; the view is from within the plane of the membrane. The S10 transmembrane domains form the dimer interface. Within the blue subunit, the predicted pore-forming helices, S4-S7, are purple. The intracellular and extracellular loops are not well conserved with TMEM16 and are not well modeled by I-TASSER. Arrows show the predicted permeation pathway in each subunit. (C ) Predicted arrangement of transmembrane helices; view from outside the cell. The red dot indicates the general region of the predicted pore. (D) Predicted arrangement of transmembrane helices; view from the plane of the membrane. Transmembrane helices S4–S7 form a groove that lines the pore. Most mutations in pore-facing residues of S4–S7 affect amplitude, single-channel conductance, or selectivity of the transduction current (Pan et al. 2018).
Like the structures of TMEM16s, the predicted structure of TMC1 shows a dimeric channel with 10 transmembrane domains (S1–S10) and intracellular amino-and carboxyl-termini in each monomer. The tenth transmembrane domain, S10, forms the relatively limited dimerization domain. There are four extracellular loops (the S7–S8 connector is a tight hairpin), which have little sequence similarity between TMCs and TMEM16s, and the modeling of loops is not reliable. Four intracellular loops are also not reliably predicted. Two conserved cysteines, one in the CWET sequence in the S5–S6 loop and one in the S9–S10 loop, are close enough to form a disulfide bond and these cysteines show coevolution in analysis of 3000+ TMC sequences, suggesting close apposition (Pan et al. 2018).
Whitlock and Hartzell (2016) proposed that the pathway for lipid head groups in scrambling by TMEM16s is the same as that for ion conduction in the TMEM16s that are channels, providing considerable insight into a possible conduction pathway for TMC1. In striking contrast to better-known channels such as voltage-gated K+ channels or TRP channels, there is not a central pore; instead, each subunit of the TMC dimer has a distinct permeation pathway, bounded by S4, S5, S6, and S7, that faces the lipid (Fig. 3). It is not clear whether the pore is completely bounded by these helices, or bounded in part by other proteins like TMIE, or exposed to the lipid membrane.
Sotomayor and colleagues (Pan et al. 2018) used molecular dynamics simulations of the predicted TMC1 structure embedded in the lipid membrane and surrounded by saline to probe accessibility of the putative pore. In a 100-nsec simulation, water molecules were found in the S4–S7 groove as well as potassium ions, suggesting that this region can be a permeation pathway for cations. The simulation was too short to show complete traversal of the pore by potassium.
But TMEM16A and B are anion channels, whereas TMC1 and TMC2 mediate cation permeability. What differences between them might account for the difference in selectivity? The predicted extracellular S3/4 loops of TMCs 1, 2, and 3 contain a conserved glutamate residue (E399 in mouse TMC1), which bears a negative charge and is not present in TMEM16s. Similarly, the S5/6 loop has two acidic (negative) residues, E514 and E520, that occur in almost all TMCs but not in TMEM16s. TMC1 has three more acidic residues in the S3/4 loop (E386, D391, and D393), although these are less con-served. These might attract cations to a channel vestibule to increase conductivity as do certain acidic residues in the vestibule of TRPA1 (Christensen et al. 2016). We calculated the electrostatic potential near the outer pore of TMEM16A and the putative pore of TMC1 and found that the potential is tens of millivolts more negative for TMC1 than for TMEM16A (N Akyuz, unpubl.), which would tend to attract cations over anions. Finally, TMC1 S6 contains two acidic residues (D528 and D540) that are not in TMEM16s, which might ease cation passage through a pore.
If negatively charged residues facilitate cat-ion flux through TMC1 channels, positively charged residues should inhibit cation flux. Indeed, the mutant Beethoven mouse line (Bth) (Vreugde et al. 2002) shows dominant progressive hearing loss; it carries a single missense mutation in mmTMC1 (M412K) that is situated near the middle of putative S4. Bth homozygous mice show lower single-channel conductance and lower selectivity for Ca2+ than do wild-type (Pan et al. 2013). M412 faces the putative pore, and placement there of a large, positive lysine side chain would be expected to impede passage of cations, especially divalent cations. Similarly, the same mutation at the orthologous position in human (M418K) (Zhao et al. 2014b) causes the dominantly inherited deafness DFNA36.
EVIDENCE THAT TMCs ARE PORE-FORMING SUBUNITS
TMC1 meets a number of criteria required for molecular components of the hair cell transduction channel. Expression of TMC1 or TMC2 is an absolute requirement for hair cell sensory transduction (Kawashima et al. 2011; Pan et al. 2013). Both the mRNA and the protein are ex-pressed at the right time and in the right place to mediate hair cell transduction (Kawashima et al. 2011; Kurima et al. 2015). A single-point mutation (Bth: M412K) in TMC1 alters ionic selectivity (Pan et al. 2013: Beurg et al. 2015a; Corns et al. 2016). Two different TMC1 mutations (M412K, D569C) alter binding affinity for the pore blocker dihydrostreptomycin (Corns et al. 2016; Pan et al. 2018).
Despite the growing body of evidence that supports TMC1 as a component of the hair cell transduction channel, an important criterion remains unfulfilled: reconstitution of TMC1 channels and mechanosensitivity in a heterologous cell. Because TMC1 does not traffic to the mem-brane in heterologous cells, Pan et al. (2018) adopted an alternate strategy. They used native hair cells of Tmc1/Tmc2-null mice, which lack transduction current, and reintroduced Tmc1 bearing individual cysteine substitutions at 17 different sites predicted to be in or near the channel pore. Sixteen of the substitutions yielded viable transduction currents. The mutations themselves or exposure to cysteine modification reagents reduced calcium selectivity (11 sites), whole-cell transduction current (five sites), and single-channel current (three sites tested). Be-cause these are core permeation properties intimately associated with an ion channel pore, the affected sites must be part of the pore of TMC1 channels. Furthermore, because the effects of cysteine modification were protected by channel closure using negative hair bundle deflections or open-channel blockers, the alternate explanation that these sites are on the outside of the channel and cause an allosteric or conformational change in the channel complex that indirectly alters the channel pore can be dismissed. Given the large body of evidence, it is hard to support alternate interpretations that do not include TMC1 as part of the ion channel pore in hair cell sensory transduction channels.
The location of the cysteine substitutions examined by Pan et al. (2018) are also consistent with the proposed TMEM16-based structure for TMC1. Sixteen cysteine mutations are at sites within S4, S5, S6, and S7, supporting the prediction that these transmembrane domains form the pore. Whether helices S4–S7 form the entire pore is not yet clear. If reconstitution of hair cell–like mechanosensitivity in a heterologous system eventually proves successful, it will likely require expression of TMC1 in the correct lipid environment, coexpression with the correct binding partners, scaffold proteins, and chaperones.
IMPLICATIONS FOR A MECHANOTRANSDUCTION COMPLEX
Arrangement within a Complex
The lower end of the tip link is composed of a homodimer of PCDH15 (Kachar et al. 2000; Ahmed et al. 2006; Kazmierczak et al. 2007; Dionne et al. 2018), and graphical representations of the transduction apparatus often depict the two PCDH15 strands each binding to distinct channel complexes (Kazmierczak and Muller 2012; Fettiplace 2016). The dimeric structure of TMC1 instead suggests the attractive possibility that a dimer of PCDH15 associates with a dimer of TMC1 or TMC2. In this arrangement, each monomer of PCDH15 would bind to each TMC monomer (Fig. 2B) to regulate the opening of one pore (Pan et al. 2018).
Gating
A central question for any force-gated ion channel is to understand the conformational change associated with gating. The hair-cell transduction channel has a particularly large gating movement of 4 nm or more (Howard and Hud-speth 1988; Cheung and Corey 2006). Although this increases sensitivity (with a gating movement of 4 nm, a force of just 1 pN would lower the relative energy of the channel open state by ∼1 kT), it is large compared to the size of the protein. However, for TMEM16s, comparison of different structures suggests a possible gating movement of ∼1 nm at the outer end of S4 and a large shift in the intracellular end of S6 by 3–4 nm (Brunner et al. 2014; Jiang et al. 2017; Paulino et al. 2017a; Peters et al. 2018). It may be that a similar shift underlies gating in TMC1. Moreover, a shift in the outer end of S4 is consistent with a gate for the hair-cell transduction channel that is outside the binding site for charged blockers (Marcotti et al. 2005).
Are the two pores gated simultaneously or independently? Opening of one TMC1 monomer would relieve tension in its associated PCDH15 filament, which would transfer tension to the partner PCDH15, increasing the like-lihood that the partner TMC1 subunit would open. Thus, gating might be highly cooperative. Most published single-channel current records, obtained by cutting all but one tip link, do not show two-step openings, suggesting that if pores are gated separately, a second opening must follow in a very short time.
Conductance
How do these gating scenarios affect electro-physiological estimates of single-channel conductance? If TMC channel dimers include two separate permeation pathways, it may be necessary to define a single-pore conductance of the monomer that is half the single-channel conductance of the dimer. Pan et al. (2018) used noise analysis to measure TMC1 single-channel currents of ∼13 pA, corresponding to single-channel conductances of ∼150 pS, consistent with prior estimates (Beurg et al. 2006; Pan et al. 2013). Interestingly, Beurg et al. (2014, 2015a) reported values for hair-cell sensory transduction channels of ∼6 pA, approximately half those reported by Pan et al. If the Beurg et al. values represent single-pore estimates and the Pan et al. values represent single-channel values for a TMC1 dimer, it would reconcile the apparent discrepancy. Last, because FRET experiments showed that TMC1 can formheteromeric dimers with TMC2 (Panet al. 2018), itsuggests at least three different dimeric configurations in hair cells: TMC1 homodimers, TMC1–TMC2 heterodimers and TMC2 homodimers. If each monomer has a distinct single-pore conductance, wild-type hair cells expressing both TMC1 and TMC2 may have at least three single-channel conductance levels, consistent with the Pan et al. (2013) data for early postnatal cochlear inner hair cells.
A remarkable feature of the mechanotransduction channels studied in cochlear hair cells is that their single-channel conductances increase by almost threefold from the low-frequency to high-frequency end of the sensory epithelium. This persists in mice lacking TMC2 (Beurg et al. 2014) so it cannot be accounted for by hetero-or homo-multimerization with TMC2. RNA editing or posttranslational modification may modify channel properties. It is interesting that mass spectroscopy of purified hsTMC1 identified six phosphorylation sites (Pan et al. 2018); these might play a role in tuning single-channel conductance. Gradients in TMC1 expression level may also account for the gradient of apparent single-channel conductance along the length of the cochlea if groups of channels open together (Beurg et al. 2018).
Dominant Deafness in DFNA36
How would the mouse M412K and human M418K mutations cause a dominant phenotype? Dominant mutations block function in other ion channels in several ways. For instance, G285S is a dominant mutation of the KCNQ4 potassium channel (Holt et al. 2007), but in that case dominant suppression is easy to understand; mutation in a single subunit could disrupt permeation through a single pore formed by all subunits— including wild-type—of a tetrameric channel. For TMC1, we argue that each monomer has an independent pore, so it is not clear how a mixture of mutant and wild-type subunits would impair function. It is not simple haploinsufficiency, as hair cells from TMC1wt/KO heterozy-gotes are functional (Kawashima et al. 2011; Pan et al. 2013). One possibility is a loss of function. The Bth mutation causes a reduction in both calcium selectivity (Pan et al. 2013; Beurg et al. 2015b; Corns et al. 2016) and single-channel conductance (Pan et al. 2013), reducing net Ca2+ influx through Bth subunits to perhaps 10%–15% of normal. Heterozygotes would have a little more than half the normal Ca2+ in-flux. Because calcium entry provides a signal for stereocilia growth and maintenance (Vélez-Ortega et al. 2017), the reduction of Ca2+ influx by almost half might not be sufficient to sustain hair cells over the long term. Another is a gain of function. It might be that hair cells with TMC1Bth channels have a higher resting open probability because they have lower Ca2+ entry. We previously found that point mutations in the DEG/ ENaC channels UNC105 and ASIC2acause constitutive opening, which leads to ion influx that is eventually lethal to cells expressing those channels (García-Añoveros et al. 1998; Tannous et al. 2009). Similarly, a mutation in the TRPML3 ion channel causes constitutive opening and death of hair cells, causing a semi-dominant deafness in the varitint-waddler (Va) mouse line (Grimm et al. 2007; Nagata et al. 2008). It is possible that TMC1M412K channels have higher resting current that is eventually toxic to hair cells.
CONCLUDING REMARKS
It has been a long road from the discovery of the deafness mouse in 1958, through the cloning of the TMC1 gene in 2002 and the demonstration that TMC1 and TMC2 are essential for mechanotransduction in 2011, to a proposed structure of a dimeric channel in 2018. Together with new findings on the structures of associated proteins such as PCDH15 and LHFPL5, we are entering a new era of structure and function studies of the hair cell transduction complex at an atomic level. This will involve the challenging task of understanding how half a dozen different proteins bind to each other to form a transduction complex and how force is transmitted through the complex to open a pore. Nonetheless, it will be extraordinarily exciting to finally understand how this exquisite molecular machine actually works.
ACKNOWLEDGMENTS
Research on TMCs in the Corey laboratory is supported by NIH Grant R01-DC000304, and in the Holt laboratory by R01-DC013521. We appreciate helpful discussions with laboratory members and Dr. H. Criss Hartzell.
Footnotes
* Reference is also in this collection.
REFERENCES
- Ahmed ZM, Goodyear R, Riazuddin S, Lagziel A, Legan PK, Behra M, Burgess SM, Lilley KS, Wilcox ER, Riazuddin S, et al. 2006. The tip-link antigen, a protein associated with the transduction complex of sensory hair cells, is proto-cadherin-15. J Neurosci 26: 7022–7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Masmoudi S, Kalay E, Belyantseva IA, Mosrati MA, Collin RW, Riazuddin S, Hmani-Aifa M, Venselaar H, Kawar MN, et al. 2008. Mutations of LRTOMT, a fusion gene with alternative reading frames, cause non-syndromic deafness in humans. Nat Genet 40: 1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagramam KN, Murcia CL, Kwon HY, Pawlowski KS, Wright CG, Woychik RP. 2001a. The mouse Ames waltzer hearing-loss mutant is caused by mutation of Pcdh15, a novel protocadherin gene. Nat Genet 27: 99–102. [DOI] [PubMed] [Google Scholar]
- Alagramam KN, Yuan H, Kuehn MH, Murcia CL, Wayne S, Srisailpathy CR, Lowry RB, Knaus R, Van Laer L, Bernier FP, et al. 2001b. Mutations in the novel protocadherin PCDH15 cause Usher syndrome type 1F. Hum Mol Genet 10: 1709–1718. [DOI] [PubMed] [Google Scholar]
- Asai Y, Pan B, Nist-Lund C, Galvin A, Lukashkin AN, Lu-kashkina VA, Chen T, Zhou W, Zhu H, Russell IJ, et al. 2018. Transgenic Tmc2 expression preserves inner ear hair cells and vestibular function in mice lacking Tmc1. Sci Rep 8: 12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros A, Fenollar-Ferrer C, Swartz KJ. 2018. Structural relationship between the putative hair cell mechanotrans-duction channel TMC1 and TMEM16 proteins. eLife doi: 10.7554/elife.38433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Evans MG, Hackney CM, Fettiplace R. 2006. A large-conductance calcium-selective mechanotransducer channel in mammalian cochlear hair cells. J Neurosci 26: 10992–11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Fettiplace R, Nam JH, Ricci AJ. 2009. Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat Neurosci 12: 553–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Kim KX, Fettiplace R. 2014. Conductance and block of hair-cell mechanotransducer channels in trans-membrane channel-like protein mutants. J Gen Physiol 144: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Goldring AC, Fettiplace R. 2015a. The effects of Tmc1 Beethoven mutation on mechanotransducer chan-nel function in cochlear hair cells. J Gen Physiol 146: 233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Xiong W, Zhao B, Muller U, Fettiplace R. 2015b. Subunit determination of the conductance of hair-cell mechanotransducer channels. Proc Natl Acad Sci 112: 1589–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Cui R, Goldring AC, Ebrahim S, Fettiplace R, Ka-char B. 2018. Variable number of TMC1-dependent me-chanotransducer channels underlie tonotopic conductance gradients in the cochlea. Nat Commun 9: 2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock GR, Steel KP. 1983. Inner ear pathology in the deafness mutant mouse. Acta Otolaryngol 96: 39–47. [DOI] [PubMed] [Google Scholar]
- Booth KT, Kahrizi K, Babanejad M, Daghagh H, Bademci G, Arzhangi S, Zareabdollahi D, Duman D, El-Amraoui A, Tekin M, et al. 2018. Variants in CIB2 cause DFNB48 and not USH1J. Clin Genet 93: 812–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner JD, Lim NK, Schenck S, Duerst A, Dutzler R. 2014. X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516: 207–212. [DOI] [PubMed] [Google Scholar]
- Cheung EL, Corey DP. 2006. Ca2+ changes the force sensi-tivity of the hair-cell transduction channel. Biophys J 90: 124–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou SW, Chen Z, Zhu S, Davis RW, Hu J, Liu L, Fernando CA, Kindig K, Brown WC, Stepanyan R, et al. 2017. A molecular basis for water motion detection by the mecha-nosensory lateral line of zebrafish. Nat Commun 8: 2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen AP, Akyuz N, Corey DP. 2016. The outer pore and selectivity filter of TRPA1. PloS ONE 11: e0166167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corey DP, Hudspeth AJ. 1983. Kinetics of the receptor current in bullfrog saccular hair cells. J Neurosci 3: 962–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corns LF, Johnson SL, Kros CJ, Marcotti W. 2016. Tmc1 point mutation affects Ca2+ sensitivity and block by dihydrostreptomycin of the mechanoelectrical transducer current of mouse outer hair cells. J Neurosci 36: 336–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corns LF, Jeng JY, Richardson GP, Kros CJ, Marcotti W. 2017. TMC2 modifies permeation properties of the me-chanoelectrical transducer channel in early postnatal mouse cochlear outer hair cells. Front Mol Neurosci 10: 326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * Cunningham CL, Müller U. 2018. Molecular structure of the hair cell mechanoelectrical transduction complex. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a033167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CL, Wu Z, Jafari A, Zhao B, Schrode K, Harkins-Perry S, Lauer A, Muller U. 2017. The murine cate-cholamine methyltransferase mTOMT is essential for mechanotransduction by cochlear hair cells. eLife 6: e24318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang S, Feng S, Tien J, Peters CJ, Bulkley D, Lolicato M, Zhao J, Zuberbuhler K, Ye W, Qi L, et al. 2017. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature 552: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deol MS, Kocher W. 1958. A new gene for deafness in the mouse. Heredity 12: 463–466. [Google Scholar]
- Deol MS, Robins MW. 1962. The spinner mouse. J Hered 53: 133–136. [DOI] [PubMed] [Google Scholar]
- Dionne G, Qiu X, Rapp M, Liang X, Zhao B, Peng G, Kat-samba PS, Ahlsen G, Rubinstein R, Potter CS, et al. 2018. Mechanotransduction by PCDH15 relies on a novel cis-dimeric architecture. Neuron 99: 480–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson T, Morgan CP, Olt J, Hardy K, Busch-Nentwich E, Maeda R, Clemens R, Krey JF, Nechiporuk A, Barr-Gillespie PG, et al. 2017. Integration of Tmc1/2 into the mechanotransduction complex in zebrafish hair cells is regulated by transmembrane O-methyltransferase (Tomt). eLife 6: e28474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fettiplace R 2016. Is TMC1 the hair cell mechanotransducer channel? Biophys J 111: 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Añoveros J, García JA, Liu JD, Corey DP. 1998. The nematode degenerin UNC-105 forms ion channels that are activated by degeneration-or hypercontraction-causing mutations. Neuron 20: 1231–1241. [DOI] [PubMed] [Google Scholar]
- Ge J, Elferich J, Goehring A, Zhao H, Schuck P, Gouaux E. 2018. Structure of mouse protocadherin 15 of the stereo-cilia tip link in complex with LHFPL5. eLife 7: e38770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Géléoc GS, Holt JR. 2003. Developmental acquisition of sensory transduction in hair cells of the mouse inner ear. Nat Neurosci 6: 1019–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese APJ, Tang YQ, Sinha GP, Bowl MR, Goldring AC, Parker A, Freeman MJ, Brown SDM, Riazuddin S, Fetti-place R, et al. 2017. CIB2 interacts with TMC1 and TMC2 and is essential for mechanotransduction in auditory hair cells. Nat Commun 8: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleason MR, Nagiel A, Jamet S, Vologodskaia M, Lopez-Schier H, Hudspeth AJ. 2009. The transmembrane inner ear (Tmie) protein is essential for normal hearing and balance in the zebrafish. Proc Natl Acad Sci 106: 21347–21352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C, Cuajungco MP, van Aken AF, Schnee M, Jors S, Kros CJ, Ricci AJ, Heller S. 2007. A helix-breaking mutation in TRPML3 leads to constitutive activity underlying deafness in the varitint–waddler mouse. Proc Natl Acad Sci 104: 19583–19588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y, Kim DS, Pastan IH, Lee B. 2009. Anoctamin and transmembrane channel-like proteins are evolutionarily related. Int J Mol Med 24: 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt JR, Stauffer EA, Abraham D, Geleoc GS. 2007. Dominant-negative inhibition of M-like potassium conductances in hair cells of the mouse inner ear. J Neurosci 27: 8940–8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J, Hudspeth AJ. 1988. Compliance of the hair bun-dle associated with gating of mechanoelectrical transduction channels in the bullfrog’s saccular hair cell. Neuron 1: 189–199. [DOI] [PubMed] [Google Scholar]
- Jain PK, Fukushima K, Deshmukh D, Ramesh A, Thomas E, Lalwani AK, Kumar S, Plopis B, Skarka H, Srisailapathy CR, et al. 1995. A human recessive neurosensory non-syndromic hearing impairment locus is potential homo-logue of murine deafness (dn) locus. Hum Mol Genet 4: 2391–2394. [DOI] [PubMed] [Google Scholar]
- Jiang T, Yu K, Hartzell HC, Tajkhorshid E. 2017. Lipids and ions traverse the membrane by the same physical pathway in the nhTMEM16 scramblase. eLife 6: e28671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachar B, Parakkal M, Kurc M, Zhao Y, Gillespie PG. 2000. High-resolution structure of hair-cell tip links. Proc Natl Acad Sci 97: 13336–13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima Y, Geleoc GS, Kurima K, Labay V, Lelli A, Asai Y, Makishima T, Wu DK, Della Santina CC, Holt JR, et al. 2011. Mechanotransduction in mouse inner ear hair cells requires transmembrane channel-like genes. J Clin Invest 121: 4796–4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, Muller U. 2012. Sensing sound: Molecules that orchestrate mechanotransduction by hair cells. Trends Neurosci 35: 220–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak P, Sakaguchi H, Tokita J, Wilson-Kubalek EM, Milligan RA, Muller U, Kachar B. 2007. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature 449: 87–91. [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 web portal for protein modeling, pre-diction and analysis. Nat Protoc 10: 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keresztes G, Mutai H, Heller S. 2003. TMC and EVER genes belong to a larger novel family, the TMC gene family encoding transmembrane proteins. BMC Genomics 4: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KX, Fettiplace R. 2013. Developmental changes in the cochlear hair cell mechanotransducer channel and their regulation by transmembrane channel-like proteins. J Gen Physiol 141: 141–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurima K, Peters LM, Yang Y, Riazuddin S, Ahmed ZM, Naz S, Arnaud D, Drury S, Mo J, Makishima T, et al. 2002. Dominant and recessive deafness caused by mutations of a novel gene, TMC1, required for cochlear hair-cell function. Nat Genet 30: 277–284. [DOI] [PubMed] [Google Scholar]
- Kurima K, Yang Y, Sorber K, Griffith AJ. 2003. Characterization of the transmembrane channel-like (TMC) gene family: Functional clues from hearing loss and epidermodysplasia verruciformis. Genomics 82: 300–308. [DOI] [PubMed] [Google Scholar]
- Kurima K, Ebrahim S, Pan B, Sedlacek M, Sengupta P, Millis BA, Cui R, Nakanishi H, Fujikawa T, Kawashima Y, et al. 2015. TMC1 and TMC2 localize at the site of mechano-transduction in mammalian inner ear hair cell stereocilia. Cell Rep 12: 1606–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lelli A, Kazmierczak P, Kawashima Y, Muller U, Holt JR. 2010. Development and regeneration of sensory transduction in auditory hair cells requires functional interaction between cadherin-23 and protocadherin-15. J Neuro-sci 30: 11259–11269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo-Guess CM, Gagnon LH, Cook SA, Wu J, Zheng QY, Johnson KR. 2005. A missense mutation in the previously undescribed gene Tmhs underlies deafness in hurry-scurry (hscy) mice. Proc Natl Acad Sci 102: 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda R, Kindt KS, Mo W, Morgan CP, Erickson T, Zhao H, Clemens-Grisham R, Barr-Gillespie PG, Nicolson T. 2014. Tip-link protein protocadherin 15 interacts with transmembrane channel-like proteins TMC1 and TMC2. Proc Natl Acad Sci 111: 12907–12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahendrasingam S, Fettiplace R, Alagramam KN, Cross E, Furness DN. 2017. Spatiotemporal changes in the distribution of LHFPL5 in mice cochlear hair bundles during development and in the absence of PCDH15. PloS ONE 12: e0185285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W, van Netten SM, Kros CJ. 2005. The aminoglycoside antibiotic dihydrostreptomycin rapidly enters mouse outer hair cells through the mechano-electrical transducer channels. J Physiol 567: 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotti W, Erven A, Johnson SL, Steel KP, Kros CJ. 2006. Tmc1 is necessary for normal functional maturation and survival of inner and outer hair cells in the mouse cochlea. J Physiol 574: 677–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano-Soto A, Moreno-Hagelsieb G, McLaughlin D, Ye ZS, Hendargo KJ, Saier MH Jr. 2018. Bioinformatic characterization of the Anoctamin superfamily of Ca2+-activated ion channels and lipid scramblases. PloS ONE 13: e0192851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchem KL, Hibbard E, Beyer LA, Bosom K, Dootz GA, Dolan DF, Johnson KR, Raphael Y, Kohrman DC. 2002. Mutation of the novel gene Tmie results in sensory cell defects in the inner ear of spinner, a mouse model of human hearing loss DFNB6. Hum Mol Genet 11: 1887–1898. [DOI] [PubMed] [Google Scholar]
- Nagata K, Zheng L, Madathany T, Castiglioni AJ, Bartles JR, Garcia-Anoveros J. 2008. The varitint–waddler (Va) deaf-ness mutation in TRPML3 generates constitutive, inward rectifying currents and causes cell degeneration. Proc Natl Acad Sci 105: 353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Kurima K, Pan B, Wangemann P, Fitzgerald TS, Geleoc GS, Holt JR, Griffith AJ. 2018. Tmc2 expression partially restores auditory function in a mouse model of DFNB7/B11 deafness caused by loss of Tmc1 function. Sci Rep 8: 12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naz S, Giguere CM, Kohrman DC, Mitchem KL, Riazuddin S, Morell RJ, Ramesh A, Srisailpathy S, Deshmukh D, Riazuddin S, et al. 2002. Mutations in a novel gene, TMIE, are associated with hearing loss linked to the DFNB6 locus. Am J Hum Genet 71: 632–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Geleoc GS, Asai Y, Horwitz GC, Kurima K, Ishikawa K, Kawashima Y, Griffith AJ, Holt JR. 2013. TMC1 and TMC2 are components of the mechanotransduction channel in hair cells of the mammalian inner ear. Neuron 79: 504–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan B, Akyuz N, Liu XP, Asai Y, Nist-Lund C, Kurima K, Derfler BH, György B, Limaphichat W, Walujkar S, et al. 2018. TMC1 forms the pore of mechanosensory transduction channels in mammalian inner ear hair cells. Neuron 99: 736–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulino C, Kalienkova V, Lam AKM, Neldner Y, Dutzler R. 2017a. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature 552: 421–425. [DOI] [PubMed] [Google Scholar]
- Paulino C, Neldner Y, Lam AK, Kalienkova V, Brunner JD, Schenck S, Dutzler R. 2017b. Structural basis for anion conduction in the calcium-activated chloride channel TMEM16A. eLife 6: e26232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepermans E, Michel V, Goodyear R, Bonnet C, Abdi S, Dupont T, Gherbi S, Holder M, Makrelouf M, Hardelin JP, et al. 2014. The CD2 isoform of protocadherin-15 is an essential component of the tip-link complex in mature auditory hair cells. EMBO Mol Med 6: 984–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters CJ, Gilchrist JM, Tien J, Bethel NP, Qi L, Chen T, Wang L, Jan YN, Grabe M, Jan LY. 2018. The sixth trans-membrane segment is a major gating component of the TMEM16A calcium-activated chloride channel. Neuron 97: 1063–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickles JO, Comis SD, Osborne MP. 1984. Cross-links be-tween stereocilia in the guinea pig organ of Corti, and their possible relation to sensory transduction. Hear Res 15: 103–112. [DOI] [PubMed] [Google Scholar]
- Riazuddin S, Belyantseva IA, Giese AP, Lee K, Indzhykulian AA, Nandamuri SP, Yousaf R, Sinha GP, Lee S, Terrell D, et al. 2012. Alterations of the CIB2 calcium-and integrin-binding protein cause Usher syndrome type 1J and non-syndromic deafness DFNB48. Nat Genet 44: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotomayor M, Weihofen WA, Gaudet R, Corey DP. 2012. Structure of a force-conveying cadherin bond essential for inner-ear mechanotransduction. Nature 492: 128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel KP, Bock GR. 1980. The nature of inherited deafness in deafness mice. Nature 288: 159–161. [DOI] [PubMed] [Google Scholar]
- Tannous BA, Christensen AP, Pike L, Wurdinger T, Perry KF, Saydam O, Jacobs AH, Garcia-Anoveros J, Weissleder R, Sena-Esteves M, et al. 2009. Mutant sodium channel for tumor therapy. Mol Ther 17: 810–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vélez-Ortega AC, Freeman MJ, Indzhykulian AA, Grossheim JM, Frolenkov GI. 2017. Mechanotransduction current is essential for stability of the transducing stereocilia in mammalian auditory hair cells. eLife 6: e24661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreugde S, Erven A, Kros CJ, Marcotti W, Fuchs H, Kurima K, Wilcox ER, Friedman TB, Griffith AJ, Balling R, et al. 2002. Beethoven, a mouse model for dominant, progressive hearing loss DFNA36. Nat Genet 30: 257–258. [DOI] [PubMed] [Google Scholar]
- Webb SW, Grillet N, Andrade LR, Xiong W, Swarthout L, Della Santina CC, Kachar B, Muller U. 2011. Regulation of PCDH15 function in mechanosensory hair cells by alternative splicing of the cytoplasmic domain. Development 138: 1607–1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock JM, Hartzell HC. 2016. A Pore idea: The ion conduction pathway of TMEM16/ANO proteins is composed partly of lipid. Pflugers Arch 468: 455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong W, Grillet N, Elledge HM, Wagner TF, Zhao B, John-son KR, Kazmierczak P, Muller U. 2012. TMHS is an integral component of the mechanotransduction machinery of cochlear hair cells. Cell 151: 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Xu D, Yang J, Walker S, Zhang Y. 2013. A comparative assessment and analysis of 20 representative sequence alignment methods for protein structure prediction. Sci Rep 3: 2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhang Y. 2015. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res 43: W174–W181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wu Z, Grillet N, Yan L, Xiong W, Harkins-Perry S, Muller U. 2014a. TMIE is an essential component of the mechanotransduction machinery of cochlear hair cells. Neuron 84: 954–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Wang D, Zong L, Zhao F, Guan L, Zhang P, Shi W, Lan L, Wang H, Li Q, et al. 2014b. A novel DFNA36 mutation in TMC1 orthologous to the Beethoven (Bth) mouse associated with autosomal dominant hearing loss in a Chinese family. PloS ONE 9: e97064. [DOI] [PMC free article] [PubMed] [Google Scholar]