Abstract
Collapsin response mediator protein-2 (CRMP-2) plays a crucial role in axonal guidance and neurite outgrowth during neural development and regeneration. We have studied the interaction between calmodulin (CaM) and CRMP-2 and how Ca2+/CaM binding modulates the biological functions of CRMP-2. We have shown that CRMP-2 binds to CaM directly in a Ca2+-dependent manner. The CaM-binding site of CRMP-2 is proposed to reside in the last helix of the folded domain, and in line with this, a synthesized peptide representing this helix bound to CaM. In addition, CaM binding inhibits a homotetrameric assembly of CRMP-2 and attenuates calpain-mediated CRMP-2 proteolysis. Furthermore, a CaM antagonist reduces the number and length of process induced by CRMP-2 overexpression in HEK293 cells. Take together, our data suggest that CRMP-2 is a novel CaM-binding protein and that CaM binding may play an important role in regulating CRMP-2 functions.
Keywords: CRMP-2, proteolysis, calmodulin, calmodulin-binding protein, oligomerization
Introduction
Collapsin response mediator protein-2 (CRMP-2) is a developmentally regulated protein and highly expressed in the nervous system [1, 2]. CRMP-2 has been reported to regulate neuronal polarity and axon elongation. Overexpression of CRMP-2 induces the formation of multiple axons and elongation of the primary axon in hippocampal neurons [3, 4]. CRMP-2 shows 60% identity to the enzyme dihydropyriminidase (DHP) and shares a common structural mode of tetrameric assembly [5]. However, unlike DHP, CRMP-2 lacks catalytic activity of its own, and thus, the molecular basis for its function remains unclear. CRMP-2 is a target for post-translational modifications, including proteolysis, oxidation, glycosylation and phosphorylation, under different neurological disorders, such as epilepsy, stroke, traumatic brain injury and Alzheimer’s disease [6–10]. One common feature of these diseases is the alteration of intra-cellular Ca2+ homeostasis, which appears to play a central role in the mechanisms of the neuronal/axonal injury that underlie these diseases [11–14].
Calmodulin (CaM) is a ubiquitous calcium-sensing protein [10, 11]. Upon elevation of intracellular Ca2+, CaM undergoes conformational change and binds to different CaM-binding proteins (CaMBPs) [15]. The extraordinarily high concentration of CaM (10–100 μM) found in neurons of the central nervous system suggests that CaM binding to enzymes, cytoskeletal proteins, receptors, and ion channels might modulate neuronal response to stimuli. For example, CaM binding to calcineurin (protein phosphatase 2B) and neurofilament (NF) regulates the proteolysis and phosphorylation of NF, which in turn modifies axonal injury after calcium influx [16, 17]. In two independent high-throughput studies, CRMP-2 was identified as a putative CaM ligand in the brain [18, 19]. However, this putative interaction still needs experimental validation.
From a structural point of view, the target sequences of CaM in general are amphipathic helices flanked by basic residues, having a characteristic spacing between large hydrophobic residues. A number of diverging CaM targets have been characterized recently with respect to sequence and binding mode to CaM [20–22]. Given the relevance of the Ca2+/CaM system in the processing of Ca2+ signaling, and taking into account the importance of CRMP-2 in axon formation and elongation [23, 24], we hypothesize that CaM binding may influence CRMP-2 functions, by modulating its involvement in cytoskeletal reorganization upon Ca2+ influx.
Here, we show a direct physical interaction between CRMP-2 and CaM using affinity chromatography, CaM overlay, surface plasmon resonance (SPR), and isothermal titration calorimetry (ITC). We also show that the interaction is calcium dependent and identify a CaM-binding segment in the C-terminal helix of CRMP-2. Furthermore, CaM binding prevents homo-tetramerization of CRMP-2, retards calpain-mediated CRMP-2 proteolysis, and regulates CRMP-2-induced F-actin process formation.
Materials and methods
Protein purification
Recombinant human CaM and CRMP-2 were purified as previously described [22, 23]. Protein purity was determined by SDS-PAGE analysis. Briefly, CaM was expressed using the pETCM vector in E. coli Rosetta (DE3) cells in LB medium, and induced with 0.8 mM IPTG overnight at 37°C. The bacteria were collected by centrifugation and resuspended in ice-cold lysis buffer (10 mM HEPES pH 7.5, 1 mM EDTA, 1 mM NaN3, 10 mM DTT, 10 μg/ml DNase I), and the cells were disrupted by sonication. The sample was centrifuged (20 000 g, 15 min, 4°C), heated at 80°C for 10 min, CaCl2 (5 mM) was added to the supernatant, and the sample was centrifuged again. The supernatant was subjected to calcium-dependent affinity chromatography on phenyl Sepharose followed by gel filtration on a Superdex 75 16/60 column in 50 mM HEPES pH 7.5, 150 mM NaCl, 4 mM CaCl2. The His-tagged human CRMP-2 (residues 1–490) was purified by immobilized metal affinity chromatography and gel filtration as previously described [23, 24]. The peptide (HPLC, >95% purity) YKRIKARSRLAELRGVPR (residues 479–496), corresponding to the last helix of the CRMP-2 crystal structure, and one of the two regions identified as putative CaM binding sites by the CaM target database [21], was purchased from Genscript.
CaM affinity capture and elution
CaM affinity capture and elution were performed as previously described [18]. Rat brain lysates and recombinant proteins were premixed with CaM agarose (Sigma) in TBS (20 mM Tris-HCl pH 7.4 and 150 mM NaCl). A negative control was also prepared using lysis buffer with CaM-agarose. After premixing the lysate with CaM agarose, 10 mM CaCl2 was added, and the reacting mixture was incubated for 90 min at room temperature. The CaM-agarose-bound CaMBPs were collected by centrifugation, washed eight times with TBS with 1 mM CaCl2, and eluted from the resin in TBS containing 15 mM EDTA. Purified CaMBPs were concentrated with a Millipore YM-10 filter unit (Millipore), suspended in sample buffer (Invitrogen), resolved by 10–20% SDS-PAGE, and visualized by Coomassie blue (Bio-Rad R250) staining.
Capillary reversed-phase tandem mass spectrometry-based protein identification
Capillary reversed phase liquid chromatography (RPLC) tandem mass spectrometry (MS/MS) protein identification was performed as described previously [18]. Briefly, Coomassie brilliant blue-stained bands were excised and submitted to in-gel digestion. Sample digests (2 μl) were loaded via an autosampler onto an RP capillary column at 1.5 μl/min. Peptide elution was performed by linear gradient: 5–60% methanol in 0.4% acetic acid over 30 min at 500 nl/min. MS/MS spectra were collected in data-dependent mode (three most intense peaks) on a Thermo Electron LCQ Deca XP plus ion trap mass spectrometer. MS/MS spectra were searched against a National Center for Biotechnology Information (NCBI) rat indexed RefSeq protein database using SEQUEST. Filtering and sorting was performed with DTAselect software by peptide number and SEQUEST cross correlation values. Peptides filtered and sorted by DTAselect were assigned to specific protein accession numbers (NCBI).
Preparation of biotinylated CaM
Purified bovine or human brain CaM (Calbiochem) was dialyzed with PBS using Slide-A-Lyzer MINI Dialysis Units (Pierce, 5-kDa MWCO). The concentration of dialyzed CaM was determined by the DC protein assay (Bio-Rad). CaM was then biotinylated using EZ-linkTM, sulfo-NHS-LC-LC-biotin (Pierce) following the manufacturer’s instructions.
CaM-overlay experiments
The recombinant GST-CRMP-2 (Kinasource, UK), GST (Biovision, USA) and calcineurin protein (Upstate) were subjected to SDS-PAGE. Proteins were separated by SDS-PAGE and electrotransferred onto a PVDF membrane. The PVDF membranes were blocked with 5% (w/v) bovine serum albumin in TBST, containing either 2 mM CaCl2 or 2 mM EDTA. Biotinylated bovine or human brain CaM was then added (20 ng/ml) and incubated for another hour. After washing the membranes (four times for 10 min in TBST in the presence of 2 mM CaCl2 or EDTA), the blots were probed for 1 h with avidin-conjugated alkaline phosphatase in the presence or absence of Ca2+. After washing, the positive CaMBP bands were detected using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate.
Surface plasmon resonance
The SPR assays were run on a Biacore 3000 instrument (Biacore AB, Sweden). For SPR, His-CRMP-2 was immobilized onto a CM5 chip (GE Healthcare) using standard amine coupling chemistry. The immobilization buffer was sodium acetate, pH 4.5, and the running buffer was 50 mM HEPES pH 7.5, 150 mM NaCl, 4 mM CaCl2. Serial dilutions of CaM (0–650 μM) were prepared in the running buffer, and duplicate samples at each concentration were injected onto the chip for interaction analysis. The concentration-dependent responses were then used to estimate Kd for the interaction using both the protocols in the BIAevaluation software and non-linear least squares curve fitting and Scatchard plots in GraphPad Prism.
Isothermal titration calorimetry
ITC was performed using the Microcal VP-ITC apparatus. CaM and the CRMP-2 peptide were extensively dialyzed against the assay buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 10 mM CaCl2). The titration was carried out at 30°C, and the used concentrations were 72 μM for CaM and 720 μM for the peptide. Data analysis to derive thermodynamic parameters was carried out in Microcal Origin.
Size exclusion chromatography
The effect of CaM binding to CRMP-2 oligomerization was investigated with gel filtration chromatography, using a Superdex 200 HR 10/30 column (GE Healthcare) connected to an ÄKTApurifier FPLC system (GE Healthcare). The column was equilibrated with 10 mM HEPES pH 7.5, 100 mM NaCl, 200 mM CaCl2. The flow was 0.5 ml/min, the injection volume 500 μl, the concentration of CRMP-2 15 μM, and the concentration of CaM 75 μM. Both proteins were run both separately and as a mixture. Sample elution was followed by UV absorbance testing at 280 nm.
Cell lysate collection and preparation
PC-12 cells were collected and lysed for 90 min at 4°C in lysis buffer (50 mM Tris pH 7.4, 5 mM EDTA, 1% Triton X-100, 1 mM DTT). The lysates were centrifuged at 10 000 g for 5 min at 4°C and then snap-frozen and stored at −80°C until use.
In vitro calpain-1 or -2 digestion of cell lysate or purified CRMP-2
PC-12 cell lysate was prepared as above. In vitro digestion of lysate (20 mg) or purified human CRMP-2 protein was performed with purified proteases, human erythrocyte calpain-1 and rat recombinant calpain-2 (Calbiochem), in a buffer containing 100 mM Tris-HCl (pH 7.4), 20 mM DTT and 1 mM CaCl2, with or without 50 μM W7, at room temperature for 30 min. Protease reactions were stopped by the addition of 30 μM calpain inhibitor (SJA 1670) or a protease inhibitor cocktail solution (Roche Biochemicals).
Plasmid constructs and transfection
The cDNA of human CRMP-2 was amplified by PCR from the IMAGE clone 3870039 using the primers 5′-ACC-AGAATTCAGATGTCTTATCAGGGGAAGAAAA-3′ and 5′-ATCAGTCGACCTAGCCCAGGCTG-GTGATGTT-3′. Primers were purchased from IDT (Coralville). The PCR products were subcloned into pAcGFPV1 (Clontech). HEK293 cells were transfected with GFP-hCRMP-2 or GFP vector using Lipofectamine 2000 (Invitrogen). Stable GFP-hCRMP-2 expression HEK293 cells were selected using media containing 400 μg/ml G418.
Immunocytochemistry
F-actin was fluorescently labeled with 5 U/ml rhodamine-conjugated phalloidin for 30 min at room temperature. The cells were mounted using medium containing 4,6-diamidine-2-phenylindole (Vector Laboratories). Fluorescence images were captured with a 20× objective on Zeiss Axioplan 2 fluorescence microscope with a CCD camera, and combined using SPOT imaging software (Diagnostic Instruments).
Results
Identification of CRMP-2 as a CaMBP
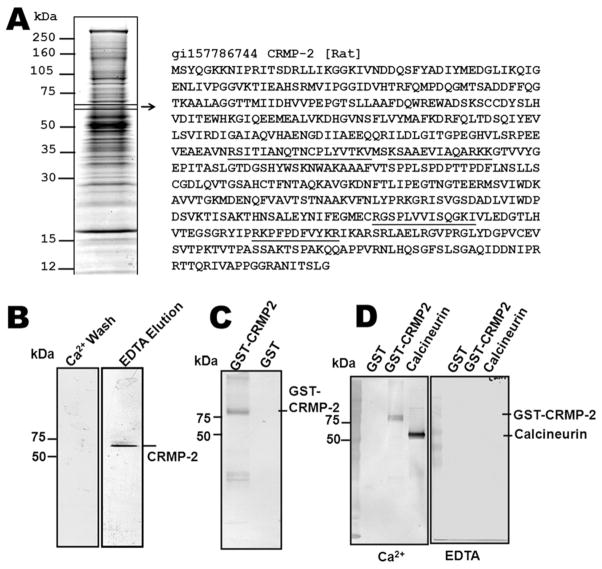
In our previous studies, we developed a novel CaM affinity capture method coupled with RPLC-MS/MS to enrich and identify brain proteins or peptides that have CaM-binding domains (CaMBDs) [18]. These data suggested that CRMP-2 might be a novel CaMBP. Here, the CaM-binding proteome of rat brain was visualized by Coomassie blue staining (Fig. 1A). Mass spectrometry was used to identify four peptides of CRMP-2 that were sequenced by RPLC-MSMS. By comparing these peptides to the full-length rat CRMP-2 sequence, they were found to correspond to residues 238–255, 258–270, 440–452, and 470–481 of the CRMP-2 sequence (underlined in Fig. 1A). The overall sequence coverage of CRMP-2 identified by RPLC-MS/MS was 9.6%. Moreover, the peptides 258–270 and 470–481 were found twice.
Figure 1.
Collapsin response-mediator protein-2 (CRMP-2) is a calmodulin-binding protein (CaMBP). (A) Identification of CRMP-2 as a putative CaMBP by tandem mass spectrometry (MS/MS). The rat brain CaM-binding proteome was visualized by Coomassie blue staining. Each visible band was excised, in-gel digested and analyzed by RPLC-MS/MS. The position of the CRMP-2 band, matching the peptide sequences, is boxed. The identified CRMP-2 peptides are underlined. (B) Immunoblot of CRMP-2 as a putative CaMBP from rat brain lysate. CaM-agarose flow through and eluate from a rat brain lysate were separated by SDS-PAGE and analyzed by immunoblot for CRMP-2. (C) Immunoblot of CaM agarose elution of recombinant GST-CRMP-2 and GST. The same amount of recombinant GST-CRMP-2 and GST (2 μg) was run through the CaM-agarose column and eluted by EDTA. The eluates were separated by SDS-PAGE and probed with an anti-GST antibody. (D) CaM-overlay experiments with recombinant GST, GST-CRMP-2 and calcineur-in. Equal amounts of recombinant GST-CRMP-2, GSTand bovine brain calcineurin were analyzed by Western blotting, and probed with 20 ng/ml biotinylated bovine CaM, in the presence of 2 mM CaCl2 or 2 mM EDTA.
To confirm that CRMP-2 is a calcium-dependent CaMBP, a rat brain lysate was subjected to CaM affinity purification in the presence of calcium, followed by EDTA elution. Both the EDTA eluate and the Ca2+flow through were probed with a CRMP-2 antibody (Fig. 1B). The detection of CRMP-2 in the CaM-agarose elution with EDTA, however, does not exclude the possibility that CRMP-2 was isolated because of indirect association with other CaMBPs. Hence, recombinant GST-CRMP-2 and a negative control, GST, were analyzed using a CaM affinity column. GST-CRMP-2 was also found to bind to and elute from the CaM affinity column, demonstrating that the interaction between the two proteins is both direct and calcium dependent (Fig. 1C). We also performed a biotinylated CaM-overlay experiment with recombinant GST-CRMP-2, GSTand calcineurin on a PVDF membrane in either the presence or absence of Ca2+. In the presence of Ca2+, biotinylated CaM binds to calcineurin and GST-CRMP-2, but not to GST. However, in the absence of Ca2+, no CaM binding was observed (Fig. 1D). Therefore, CaM binds to CRMP-2 directly and specifically in a Ca2+-dependent manner.
Analysis of the CRMP-2–CaM interaction by SPR and ITC
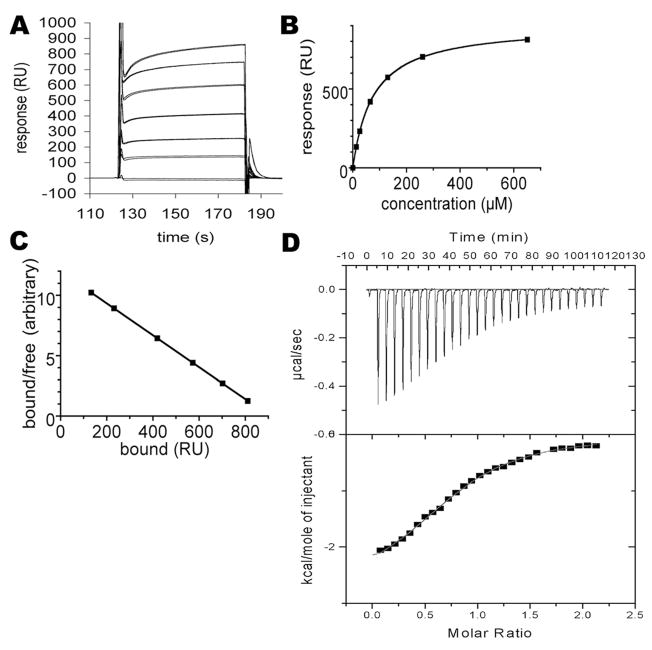
An analysis of the interaction was carried out by SPR, with purified His-tagged CRMP-2 (residues 1–490) immobilized on a CM5 chip. The binding was then analyzed by injecting CaM over the coated surface at different concentrations (Fig. 2A–C), and an apparent Kd value was estimated to be 75 μM. When compared to the Kd values of most other CaM ligands, the relatively low affinity observed may be due to the fact that one of the binding partners, CRMP-2, was covalently immobilized in the experiment. The interaction of CaM with its ligands generally involves large-scale conformational changes and these could have been affected by immobilization. To analyze the putative direct binding of the peptide representing the last helix of CRMP-2 (residues 479–496) to CaM, an ITC experiment was carried out (Fig. 2D). The affinity was found to be approximately five times higher than the outcome from SPR, and binding enthalpy and entropy were both favorable (Table 1). Nevertheless, the affinity is lower than for many other CaM ligand peptides, and in many cases of canonical CaM-peptide recognition involving CaM collapse, binding enthalpy is much more favorable. Our recent study on the interaction between CaM and a peptide from the myelin basic protein (MBP) indicated similar energetics to those observed here. The Kd for the MBP peptide was also in the micro-molar range and both enthalpy and entropy were favorable. This may indicate that CaM does not collapse around the CRMP-2 peptide, but may stay in an open conformation similar to that seen for the CaM-MBP peptide complex [22].
Figure 2.
Analysis of CRMP-2 and CaM interaction by surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC). (A) The sensorgrams for injecting various concentrations of CaM over the immobilized recombinant human CRMP-2 (residues 1–490). Duplicate samples are shown. The concentrations are (from highest to lowest response, respectively): 650, 260, 130, 65, 26, 13, and 0 μM. (B) Plotting of the responses against CaM concentration. Duplicate samples are shown, but the values for duplicates are essentially identical. (C) Scatchard plot of the binding data. The points fit perfectly on a straight line, indicating a single binding event. (D) Calorimetric analysis of the binding of CaM to the putative binding site on CRMP-2. Top: The peptide (residues 479–496) was injected into a solution of CaM. Bottom: After integrating the peak area for each injection, the binding isotherm was used to obtain thermodynamic parameters for the interaction.
Table 1.
Characterization of the collapsin response-mediator protein-2 interaction with calmodulin by surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC).
| SPR | |
|---|---|
| Kd (μM) | 75 ± 0.05 |
| ITC | |
| n | 0.81 ± 0.01 |
| Ka (M−1) | 5.8 × 104 ± 2.9 × 103 |
| Kd (μM) | 17 ± 0.9 |
| ΔH (kcal/mol) | −2.8 ± 0.05 |
| −TΔS (kcal/mol) | −3.8 |
| ΔG (kcal/mol) | −6.6 |
CaM binding prevents CRMP-2 homotetramerization
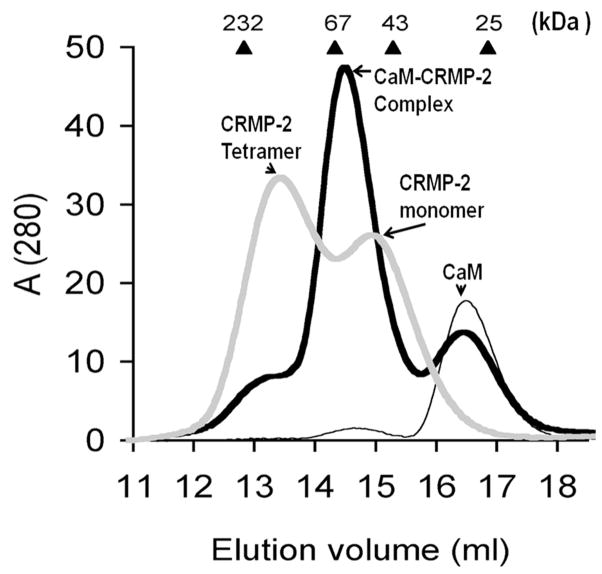
The potential complex between CaM and CRMP-2 was investigated using size exclusion chromatography in the presence of CaCl2. In the absence of CaM, CRMP-2 elutes as two peaks [10, 25] corresponding to the tetrameric and monomeric states (Fig. 3). Due to its elongated form, CaM always elutes at an apparent molecular mass of 30 kDa. In the sample containing both proteins, the tetramer peak of CRMP-2 is nearly completely lost, and the monomer peak shifts towards a higher molecular weight, indicating that CaM binds to monomeric CRMP-2, and that its binding prevents CRMP-2 homotetramer formation.
Figure 3.
Size exclusion chromatography analysis of CRMP-2 and CaM interaction. Gel filtration of recombinant human CRMP-2 in the presence and absence of CaM was performed in the presence of 200 mM CaCl2. The elution profiles of CRMP-2 (thick gray line), CaM (thin black line), and the CRMP-2-CaM complex (thick black line) are shown. The absorbance at 280 nm is shown in milli-absorbance units. The elution volumes for molecular weight standards (in kDa) are shown at the top of the graph.
The location of the CaM-binding site in the 3D structure of CRMP-2
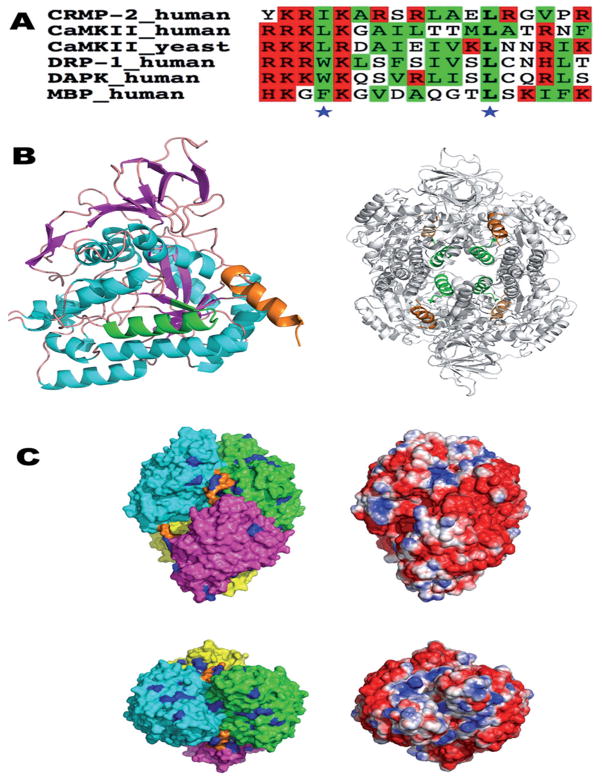
The CaM target database [21] suggests two possible binding sites for CaM; one is located between residues 258 and 275, and the other between residues 475 and 493 (Fig. 4). The first putative CaM-binding site only shows weak homology with other CaM-binding motifs (data not shown) and forms an integral part of the CRMP-2 fold. On the other hand, the C-terminal putative CaM-binding helix is homologous to binding sites in other known CaMBPs, such as CaM-dependent protein kinases and MBP (Fig. 4A). In particular, a ’1–10’ motif of conserved large hydrophobic residues is found, flanked by a cluster of basic residues at the N terminus of the binding site. Within the CRMP protein family, composed of five isoforms in humans, CRMP-1, −3, and −4 share a region homologous to the proposed CaM-binding site of CRMP-2. On the other hand, CRMP-5 and the catalytically active, but structurally related, DHP lack any significant homology at this position (data not shown).
Figure 4.
The putative CaM-binding site of CRMP-2 in the context of the 3D structure of CRMP-2. (A) A sequence alignment of the used CRMP-2 peptide with known CaM ligands. Basic and hydrophobic residues are highlighted in red and green, respectively, and the conserved hydrophobic positions in the 31–103 mode are indicated by asterisks. The sequences are from the CaM-binding domains of CaM-dependent protein kinase II (CaMKII), DAPK-related protein 1 (DRP-1), death-associated protein kinase (DAPK), and myelin basic protein (MBP). (B) The location of the CaM-binding domain (CaMBD) peptide sequence in the 3D structure of CRMP-2 (PDB entry 2GSE). On the left, only a monomer form is shown. The helices are in cyan, sheets in magenta, the first potential CaMBD (residues 258–275) in green, and the C-terminal CaMBD helix in orange. For the tetramer (right), the coloring of the CaMBDs is the same as in the left panel, while the rest of the tetramer is colored gray for clarity. The first CaMBD is located in the central lumen of the tetramer, while the C-terminal helices point outwards from the surface. (C) Surface representation of tetrameric CRMP-2. The top and bottom views are related by a rotation of 90°about the x axis. On the left, the molecular surface of the four monomers is color-coded according to polypeptide chain, and the C-terminal helix is indicated by orange. All arginine residues are colored blue. On the right, the solvent-accessible surface is colored based on electrostatics (red, −2 kT/e – blue, +2 kT/e), calculated using APBS.
Structurally, CRMP-2 can be divided into two distinct regions: the folded domain between residues 1 and 490, with a calculated pI of 5.4, and the C-terminal tail, between residues 491 and 572, with a calculated pI of 10.9 and predicted to be structurally disordered. Thus, the disordered basic C-terminal tail could also be involved in interactions with the highly acidic CaM molecule (Fig. 4B). In this respect, it is interesting to note that CaM target proteins often have disordered regions near or at the CaMBD prior to binding [24] and that many intrinsically unfolded proteins, such as MBP [22, 25], have been characterized as CaMBPs. At the tetramer level, the C-terminal CaMBDs of the CRMP-2 monomers point outwards from a groove at an intermolecular contact (Fig. 4B). An analysis of the electrostatic environment suggests that each CaMBD is surrounded by an overall positive charge potential in the tetramer. Furthermore, nearly all surface arginine residues of CRMP-2 are clustered in 3D space around the CaMBD, possibly playing a role in CaM recognition. Thus, it can be envisaged that upon CaM binding, conformational changes may occur, which in turn dissociate the CRMP-2 homodimers (Fig. 4C).
CaM binding attenuates calpain-mediated CRMP-2 proteolysis
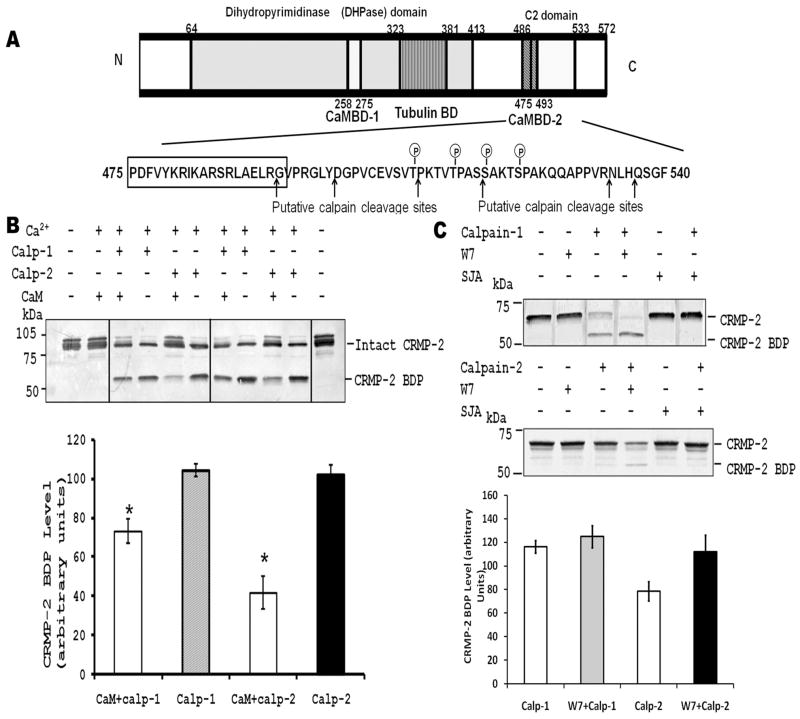
Our previous studies showed that calpain mediates CRMP-2 truncation within the C-terminal residues 499–550 in traumatic brain injury and excitotoxic neural injury [7]. Interestingly, CaMBD-2 overlaps with the C-terminal C2 domain (residues 486–533), which is close to the calpain cleavage sites (Fig. 5A). This suggests that CaM binding might affect proteolysis of CRMP-2.
Figure 5.
CaM retards calpain-mediated CRMP-2 proteolysis in vitro. (A) Schematic diagram of CaMBDs and calpain cleavage sites in CRMP-2. As indicated, the boxed CaMBD (475–493) is close to the phosphorylation and calpain cleavage sites. (B) CaM attenuates calpain-mediated CRMP-2 proteolysis. Recombinant full-length GST-CRMP-2 protein (1 μg) was incubated with calpain-1 or -2 and Ca2+ in the presence or absence of CaM (10 μM). The top panel is a representative Western blot of CRMP-2 and CRMP-2 breakdown product (BDP). The bottom panel shows the densitometric analysis of CRMP-2 BDP on immunoblots under different conditions. Values were mean ± SEM, n=4. (C) Calpain mediated CRMP-2 proteolysis in cell lysates is enhanced by CaM antagonists in vitro. A PC-12 cell lysate (20 μg protein) was incubated with calpain-1 or-2 in the presence of the CaM antagonist W7 or the calpain inhibitor SJA-6017 (30 μM), and analyzed by Western blotting with anti-CRMP-2 (C4G).
To assess the effect of CaM binding on CRMP-2 proteolysis by calpain in vitro, we compared the cleavage efficiency of calpains towards CRMP-2 with and without CaM. Purified GST-CRMP-2 was treated with either calpain-1 or calpain-2 in the presence or absence of CaM. The addition of CaM to GST-CRMP-2 significantly attenuated CRMP-2 proteolysis as noted by the decrease in the level of the CRMP-2 breakdown product (CRMP-2-BDP) in both the calpain-1 and calpain-2 treatments (Fig. 5B).
To further examine the effect of endogenous CaM binding on the proteolysis of CRMP-2, PC-12 cell lysates was used as a model since it contains high levels of endogenous CaM. We treated PC-12 cell lysates with calpain-1 or calpain-2, with or without the CaM antagonist W7 that dissociates endogenous CaM from CaMBPs. The W7-treated samples exhibited higher levels of CRMP-2 BDP when compared to the untreated cells (Fig. 5C). Similar data were obtained with another CaM antagonist, calmidazolium (5 μM, data not shown). This indicated that CaM antagonism enhances CRMP-2 proteolysis by calpain-1 and -2 in vitro. Altogether, these results demonstrate that CaM binding protects CRMP-2 against calpain-mediated proteolysis.
CaM regulates CRMP-2 mediated process formation
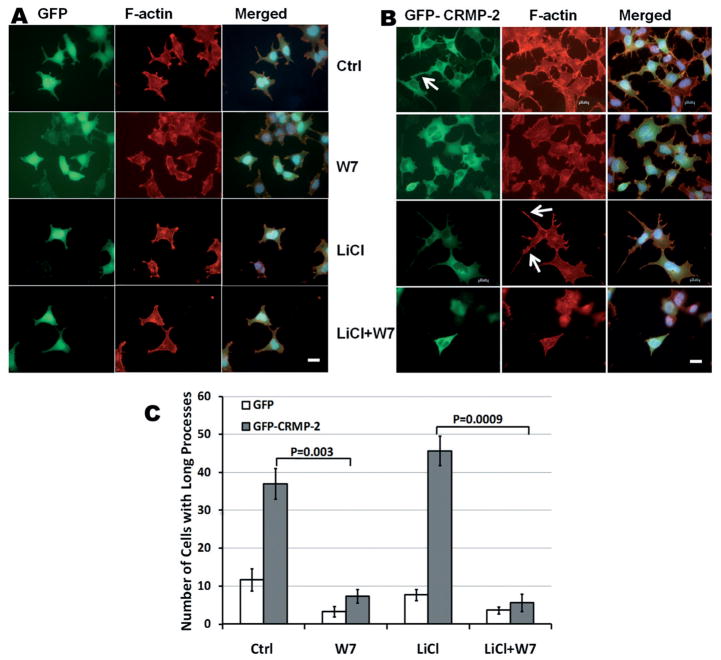
Many known CaMBPs, such as CaMKII, GAP43 and MARCKS, have been found to interact with actin and modulate the formation of processes and microspikes by CaM [26–28]. Previous studies showed that full-length CRMP-2 is co-localized with F-actin [29–31], and one member of the CRMP family, CRMP-4, served as an F-actin-bundling protein [32]. Therefore, we suspected that CaM may modulate CRMP-2-mediated process formation. It has been shown that overexpressing CRMP-2 in hippocampal neurons induces axon formation [33]. To study this process, HEK293 cells overexpressing the full-length GFP-CRMP-2 fusion protein were used as a model (Fig. 6). As a control, GFP was overexpressed in HEK293 cells. The GFP-CRMP-2-overexpressing HEK293 cells were found to have longer rhodamine-phalloidin-stained processes, when compared to GFP-over-expressing cells (Fig. 6). After 30-min incubation with a CaM antagonist, W7 or calmidazolium (not shown), cells with longer processes were dramatically reduced (Fig. 6). In GFP-overexpressing HEK293 cells, W7 also reduced basal processes, possibly via inhibition of other endogenous CaMBPs that interact with F-actin. We also studied the effects of the CaM antagonists in the presence of LiCl, a GSK3β inhibitor. GSK3β has been implicated in axonal/neurite formation. Inhibition of GSK3β affects dephosphorylation of CRMP-2, thus resulting in axon elongation [34]. LiCl treatment alone appears to slightly enhance long filopodial formation. However, the application of LiCl did not prevent the retraction of long processes induced by the CaM antagonists W7 (Fig. 6C) and calmidazolium (data not shown). Taken together, our data suggest that CaM regulates CRMP-2-mediated process formation, independently of the GSK3β pathway.
Figure 6.
CaM antagonist induces process retraction in CRMP-2-overexpressing HEK293 cells. HEK293 cells with overex-pressed GFP (A) or GFP-CRMP-2 (B) were treated with CaM antagonist W7 (10 μM), LiCl (5 mM) or both. GFP fluorescence was imaged (green), and F-actin was stained by rhodamine-phalloidin (red). Yellow color in merged photographs corresponds to the colocalization of CRMP-2 and F-actin. Arrows indicate the CRMP-2-induced F-actin-enriched processes. Scale bar = 20 μM. (C) Quantification of cells with at least one process greater than half the diameter of the cell body following various treatments. For each experiment, 50 cells per slide were randomly measured. The values shown are the mean of three independent experiments. Error bars represent SEM. Significant differences between the different treatments between two groups were analyzed by Student’s t-test.
Discussion
In this study, we have shown that CRMP-2 binds to CaM directly in a Ca2+-dependent manner (Figs 1, 2). There are two predicted CaMBDs in CRMP-2: CaMBD-1 (residues 258–275) and CaMBD-2 (residues 475–493). The C-terminal CRMP-2 is a stronger candidate CaM-binding target, as this region is rich in basic and hydrophobic residues, and since CaMBD-1 is an integral part of the TIM barrel fold of CRMP-2, located on the inside of the tetramer. A helical wheel projection of CaMBD-2 shows that this region can form an appropriate amphiphilic helix with the characteristic hydrophobic amino acid residues (Phe475, Ala 484, Leu488 and Leu491) on one face, and, on the other, a cluster of charged amino acid residues (data not shown). This is in agreement with previous studies showing that the C-terminal CaM-binding sequence forms a basic amphiphilic α-helix [23]. Thus, CaM most likely binds to CRMP-2 through the CaMBD-2 (Fig. 4). In this study, we have also shown that the peptide representing this helix bound directly to CaM with an affinity in the micromolar range.
CRMP-2 has recently been shown to exist in both monomeric and homotetrameric forms [10]. Here, gel filtration experiments showed that CaM binds only to monomeric CRMP-2, and that CaM binding inhibits the formation of the homotetrametric assembly (Fig. 3). It is possible that the tetrameric form of CRMP-2 has different subcellular localizations and/or binding partners; in such a case, CaM could be a regulator of such processes. Clearly, this area needs to be further investigated.
The C-terminal CaMBD of CRMP-2 is very close to the calpain cleavage site, suggesting that CaM binding might affect proteolysis [7, 35]. We have shown here for the first time that CaM protects CRMP-2 against calpain cleavage (Fig. 5). We interpret this effect to be a direct consequence of CaM binding to the corresponding domain of the CRMP-2 CaMBD (residues 475–493), thus preventing access to the adjacent calpain cleavage site. As a Ca2+ sensor, CaM binds to numerous different CaMBPs, resulting in specific intracellular responses to the Ca2+ signal. For example, CaM binding to the plasma membrane Ca2+-ATPase and GAP43 leads to inhibition of calpain-mediated cleavage at several sites located within the CaMBD of these proteins [15]. In other cases, CaM either accelerated the proteolysis by calpain (brain spectrin, calponin and calcineurin) or changed the pattern of cleavage (myosin, myosin light chain kinase) [36–39]. Furthermore, studies have demonstrated that CaM binding to the N-methyl-D-aspartate receptor (NMDAR) subunit 1 facilitates calpain-mediated inactivation of NMDAR, which, in turn, serves as a negative feedback regulator to fine tune Ca2+ influx after injury [28, 40]. Since previous studies have shown that proteolysis of CRMP-2 also occurs in ischemia and TBI [7, 35], it is possible that CaM binding to CRMP-2 also prevents its degradation and stabilizes growth cone structure in a negative feedback loop to fine tune the response to injury in vivo. It has been established that the actual extension of an axon occurs at its distal tip, the growth cone. The peripheral and central domains of the growth cone are composed of two F-actin-based structures, filopodia and lamellipodia [41]. These actin-rich structures contribute to the force necessary for the forward extension of the growth cone. It is, therefore, important to understand how actin dynamics are regulated, and this could provide key insights into how axonal regeneration may be promoted. Several studies have suggested that CRMP-2 interacts with actin, and the C terminus of the protein is essential for the interaction [29]. Our data suggest that CRMP-2 co-localizes with F-actin, and the application of CaM antagonists reduced the number and length of F-actin processes induced by CRMP-2 overexpression (Fig. 6). We used two different CaM inhibitors (W7 and calmidazolium) in this study, and obtained similar results. However, CaM affects many different pathways in the cell, and the effects observed here could be partially mediated by other CaMBPs. Thus, further physiological interaction experiments with CRMP-2 mutants disabling CaM interaction, peptide competition, CRMP-2 mutations in calpain binding sites or CaM overexpression may provide more direct evidence.
Taken together, our data show that CRMP-2 may be regulated by CaM in at least three ways: (i) CaM inhibits the CRMP-2 homotetrameric assembly, (ii) CaM retards the calpain-mediated CRMP-2 proteolysis, and (iii) CaM might facilitate CRMP-2 mediated process growth. Our proposed working model is that, under physiological Ca2+ signaling conditions, CaM may facilitate CRMP-2 and F-actin interaction, promoting growth cone formation and neurite outgrowth. However, if neurons experience initiation of Ca2+ overload (e.g., in neurotoxic challenge), CaM protects CRMP-2 against calpain-mediated proteolysis and neuronal damage. To go a step further, a recent study suggested that CRMP-2 fragments themselves might be neurotoxic [42, 43]. Thus, by reducing CRMP-2 proteolysis, there may be additional neuro-protective effects. Finally, understanding the signaling mechanisms of Ca2+/CaM and CRMP-2 in cytoskeletal dynamics at the molecular level may open up possibilities of developing novel strategies to alleviate cell death and promote axon regeneration in vivo.
Acknowledgments
The authors thank Jingxiang Mo for the CRMP-2 construct and Dr. Nobuhiro Hayashi for the pETCM vector. We wish to thank Drs Andrew K. Ottens, Michael Bubb, Juan A. Martinez and Stephen F. Larner for insightful discussions and editorial assistance. This work was supported by the Department of Defense grant DAMD-03-1-0066, NIH grants R01 NS049175-01-A1 and R01 NS051431, and by the Academy of Finland. K. K. W. Wang and R. L. Hayes own stock of Banyan Biomarkers Inc., and may benefit financially as a result of the outcome of this research or the work reported in this publication.
References
- 1.Bretin S, Reibel S, Charrier E, Maus-Moatti M, Auvergnon N, Thevenoux A, Glowinski J, Rogemond V, Premont J, Honnorat J, Gauchy C. Differential expression of CRMP1, CRMP2A, CRMP2B, and CRMP5 in axons or dendrites of distinct neurons in the mouse brain. J Comp Neurol. 2005;486:1–17. doi: 10.1002/cne.20465. [DOI] [PubMed] [Google Scholar]
- 2.Wang LH, Strittmatter SM. A family of rat CRMP genes is differentially expressed in the nervous system. J Neurosci. 1996;16:6197–6207. doi: 10.1523/JNEUROSCI.16-19-06197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fukata Y, Itoh TJ, Kimura T, Menager C, Nishimura T, Shiromizu T, Watanabe H, Inagaki N, Iwamatsu A, Hotani H, Kaibuchi K. CRMP-2 binds to tubulin heterodimers to promote microtubule assembly. Nat Cell Biol. 2002;4:583–591. doi: 10.1038/ncb825. [DOI] [PubMed] [Google Scholar]
- 4.Fukata Y, Kimura T, Kaibuchi K. Axon specification in hippocampal neurons. Neurosci Res. 2002;43:305–15. doi: 10.1016/s0168-0102(02)00062-7. [DOI] [PubMed] [Google Scholar]
- 5.Wang LH, Strittmatter SM. Brain CRMP forms heterotetramers similar to liver dihydropyrimidinase. J Neurochem. 1997;69:2261–2269. doi: 10.1046/j.1471-4159.1997.69062261.x. [DOI] [PubMed] [Google Scholar]
- 6.Jiang SX, Kappler J, Zurakowski B, Desbois A, Aylsworth A, Hou ST. Calpain cleavage of collapsin response mediator proteins in ischemic mouse brain. Eur J Neurosci. 2007;26:801–809. doi: 10.1111/j.1460-9568.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Z, Ottens AK, Sadasivan S, Kobeissy FH, Fang T, Hayes RL, Wang KK. Calpain-mediated collapsin response mediator protein-1, -2, and -4 proteolysis after neurotoxic and traumatic brain injury. J Neurotrauma. 2007;24:460–472. doi: 10.1089/neu.2006.0078. [DOI] [PubMed] [Google Scholar]
- 8.Gu Y, Hamajima N, Ihara Y. Neurofibrillary tangle-associated collapsin response mediator protein-2 (CRMP-2) is highly phosphorylated on Thr-509, Ser-518, and Ser-522. Biochemistry (Mosc) 2000;39:4267–4275. doi: 10.1021/bi992323h. [DOI] [PubMed] [Google Scholar]
- 9.Kobeissy FH, Ottens AK, Zhang Z, Liu MC, Denslow ND, Dave JR, Tortella FC, Hayes RL, Wang KK. Novel differential neuroproteomics analysis of traumatic brain injury in rats. Mol Cell Proteomics. 2006;5:1887–1898. doi: 10.1074/mcp.M600157-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Majava V, Loytynoja N, Chen WQ, Lubec G, Kursula P. Crystal and solution structure, stability and post-translational modifications of collapsin response mediator protein 2. FEBS J. 2008;275:4583–4596. doi: 10.1111/j.1742-4658.2008.06601.x. [DOI] [PubMed] [Google Scholar]
- 11.Sola C, Barron S, Tusell JM, Serratosa J. The Ca2+/calmodulin signaling system in the neural response to excitability. Involvement of neuronal and glial cells. Prog Neurobiol. 1999;58:207–232. doi: 10.1016/s0301-0082(98)00082-3. [DOI] [PubMed] [Google Scholar]
- 12.Czogalla A, Sikorski AF. Spectrin and calpain: a ’target’ and a ’sniper’ in the pathology of neuronal cells. Cell Mol Life Sci. 2005;62:1913–1924. doi: 10.1007/s00018-005-5097-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haeseleer F, Imanishi Y, Sokal I, Filipek S, Palczewski K. Calcium-binding proteins: intracellular sensors from the calmodulin superfamily. Biochem Biophys Res Commun. 2002;290:615–623. doi: 10.1006/bbrc.2001.6228. [DOI] [PubMed] [Google Scholar]
- 14.O’Day DH, Myre MA. Calmodulin-binding domains in Alzheimer’s disease proteins: extending the calcium hypothesis. Biochem Biophys Res Commun. 2004;320:1051–1054. doi: 10.1016/j.bbrc.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 15.Wang KKW, Villalobo A, Roufogalis BD. Calmodulin-binding proteins as calpain substrates. Biochem J. 1989;262:693–706. doi: 10.1042/bj2620693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia Z, Storm DR. The role of calmodulin as a signal integrator for synaptic plasticity. Nat Rev Neurosci. 2005;6:267–276. doi: 10.1038/nrn1647. [DOI] [PubMed] [Google Scholar]
- 17.Johnson GV, Greenwood JA, Costello AC, Troncoso JC. The regulatory role of calmodulin in the proteolysis of individual neurofilament proteins by calpain. Neurochem Res. 1991;16:869–873. doi: 10.1007/BF00965535. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Ottens AK, Golden EC, Hayes RL, Wang KK. Using calmodulin-affinity capture to study the rat brain calmodulin binding proteome and its vulnerability to calpain and caspase proteolysis. Calcium Binding Proteins. 2006;1:125–134. [Google Scholar]
- 19.Berggard T, Arrigoni G, Olsson O, Fex M, Linse S, James P. 140 mouse brain proteins identified by Ca2+-calmodulin affinity chromatography and tandem mass spectrometry. J Proteome Res. 2006;5:669–687. doi: 10.1021/pr050421l. [DOI] [PubMed] [Google Scholar]
- 20.Ikura M, Ames JB. Genetic polymorphism and protein conformational plasticity in the calmodulin superfamily: two ways to promote multifunctionality. Proc Natl Acad Sci USA. 2006;103:1159–1164. doi: 10.1073/pnas.0508640103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yap KL, Kim J, Truong K, Sherman M, Yuan T, Ikura M. Calmodulin target database. J Struct Funct Genomics. 2000;1:8–14. doi: 10.1023/a:1011320027914. [DOI] [PubMed] [Google Scholar]
- 22.Majava V, Petoukhov MV, Hayashi N, Pirila P, Svergun DI, Kursula P. Interaction between the C-terminal region of human myelin basic protein and calmodulin: Analysis of complex formation and solution structure. BMC Struct Biol. 2008;8:10. doi: 10.1186/1472-6807-8-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenmark P, Ogg D, Flodin S, Flores A, Kotenyova T, Nyman T, Nordlund P, Kursula P. The structure of human collapsin response mediator protein 2, a regulator of axonal growth. J Neurochem. 2007;101:906–917. doi: 10.1111/j.1471-4159.2006.04401.x. [DOI] [PubMed] [Google Scholar]
- 24.Radivojac P, Vucetic S, O’Connor TR, Uversky VN, Obradovic Z, Dunker AK. Calmodulin signaling: analysis and prediction of a disorder-dependent molecular recognition. Proteins. 2006;63:398–410. doi: 10.1002/prot.20873. [DOI] [PubMed] [Google Scholar]
- 25.Libich DS, Hill CM, Haines JD, Harauz G. Myelin basic protein has multiple calmodulin-binding sites. Biochem Biophys Res Commun. 2003;308:313–319. doi: 10.1016/s0006-291x(03)01380-9. [DOI] [PubMed] [Google Scholar]
- 26.Gerendasy D. Homeostatic tuning of Ca2+ signal transduction by members of the calpacitin protein family. J Neurosci Res. 1999;58:107–119. [PubMed] [Google Scholar]
- 27.Krucker T, Siggins GR, McNamara RK, Lindsley KA, Dao A, Allison DW, De Lecea L, Lovenberg TW, Sutcliffe JG, Gerendasy DD. Targeted disruption of RC3 reveals a calmodulin-based mechanism for regulating metaplasticity in the hippocampus. J Neurosci. 2002;22:5525–5535. doi: 10.1523/JNEUROSCI.22-13-05525.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Dalen JJ, Gerendasy DD, de Graan PN, Schrama LH, Gruol DL. Calcium dynamics are altered in cortical neurons lacking the calmodulin-binding protein RC3. Eur J Neurosci. 2003;18:13–22. doi: 10.1046/j.1460-9568.2003.02720.x. [DOI] [PubMed] [Google Scholar]
- 29.Rogemond V, Auger C, Giraudon P, Becchi M, Auvergnon N, Belin MF, Honnorat J, Moradi-Ameli M. Processing and nuclear localization of CRMP2 during brain development induce neurite outgrowth inhibition. J Biol Chem. 2008;283:14751–14761. doi: 10.1074/jbc.M708480200. [DOI] [PubMed] [Google Scholar]
- 30.Kawano Y, Yoshimura T, Tsuboi D, Kawabata S, Kaneko-Kawano T, Shirataki H, Takenawa T, Kaibuchi K. CRMP-2 is involved in kinesin-1-dependent transport of the Sra-1/WAVE1 complex and axon formation. Mol Cell Biol. 2005;25:9920–9935. doi: 10.1128/MCB.25.22.9920-9935.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gungabissoon RA, Bamburg JR. Regulation of growth cone actin dynamics by ADF/cofilin. J Histochem Cytochem. 2003;51:411–420. doi: 10.1177/002215540305100402. [DOI] [PubMed] [Google Scholar]
- 32.Rosslenbroich V, Dai L, Baader SL, Noegel AA, Gieselmann V, Kappler J. Collapsin response mediator protein-4 regulates F-actin bundling. Exp Cell Res. 2005;310:434–444. doi: 10.1016/j.yexcr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 33.Arimura N, Menager C, Fukata Y, Kaibuchi K. Role of CRMP-2 in neuronal polarity. J Neurobiol. 2004;58:34–47. doi: 10.1002/neu.10269. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura T, Kawano Y, Arimura N, Kawabata S, Kikuchi A, Kaibuchi K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell. 2005;120:137–149. doi: 10.1016/j.cell.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 35.Chung MA, Lee JE, Lee JY, Ko MJ, Lee ST, Kim HJ. Alteration of collapsin response mediator protein-2 expression in focal ischemic rat brain. Neuroreport. 2005;16:1647–1653. doi: 10.1097/01.wnr.0000176520.49841.e6. [DOI] [PubMed] [Google Scholar]
- 36.Harris AS, Croall DE, Morrow JS. Calmodulin regulates fodrin susceptibility to cleavage by calcium-dependent protease I. J Biol Chem. 1989;264:17401–17408. [PubMed] [Google Scholar]
- 37.Kosaki G, Tsujinaka T, Kambayashi J, Morimoto K, Yamamoto K, Yamagami K, Sobue K, Kakiuchi S. Specific cleavage of calmodulin-binding proteins by low Ca2+-requiring form of Ca2+-activated neutral protease in human platelets. Biochem Int. 1983;6:767–775. [PubMed] [Google Scholar]
- 38.Tsunekawa S, Takahashi K, Abe M, Hiwada K, Ozawa K, Murachi T. Calpain proteolysis of free and bound forms of calponin, a troponin T-like protein in smooth muscle. FEBS Lett. 1989;250:493–496. doi: 10.1016/0014-5793(89)80783-5. [DOI] [PubMed] [Google Scholar]
- 39.Croall DE, Chacko S, Wang Z. Cleavage of caldesmon and calponin by calpain: Substrate recognition is not dependent on calmodulin binding domains. Biochim Biophys Acta. 1996;1298:276–284. doi: 10.1016/s0167-4838(96)00138-0. [DOI] [PubMed] [Google Scholar]
- 40.Zhang S, Ehlers MD, Bernhardt JP, Su CT, Huganir RL. Calmodulin mediates calcium-dependent inactivation of N-methyl-D-aspartate receptors. Neuron. 1998;21:443–453. doi: 10.1016/s0896-6273(00)80553-x. [DOI] [PubMed] [Google Scholar]
- 41.Dent EW, Gertler FB. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron. 2003;40:209–227. doi: 10.1016/s0896-6273(03)00633-0. [DOI] [PubMed] [Google Scholar]
- 42.Tahimic CG, Tomimatsu N, Nishigaki R, Fukuhara A, Toda T, Kaibuchi K, Shiota G, Oshimura M, Kurimasa A. Evidence for a role of collapsin response mediator protein-2 in signaling pathways that regulate the proliferation of non-neuronal cells. Biochem Biophys Res Commun. 2006;340:1244–1250. doi: 10.1016/j.bbrc.2005.12.132. [DOI] [PubMed] [Google Scholar]
- 43.Touma E, Kato S, Fukui K, Koike T. Calpain-mediated cleavage of collapsin response mediator protein (CRMP)-2 during neurite degeneration in mice. Eur J Neurosci. 2007;26:3368–3381. doi: 10.1111/j.1460-9568.2007.05943.x. [DOI] [PubMed] [Google Scholar]