Abstract
Photoresponsive materials afford spatiotemporal control over desirable physical, chemical and biological properties. For advanced applications, there is need for molecular phototriggers that are readily incorporated within larger structures, and spatially-sequentially addressable with different wavelengths of visble light, enabling multiplexing. Here we describe spectrally tunable (λmax = 420–530 nm) ruthenium polypyridyl complexes functionalized with two photolabile nitrile ligands that present terminal alkynes for subsequent crosslinking reactions, including hydrogel formation. Two Ru crosslinkers were incorporated within a PEG-hydrogel matrix, and sequentially degraded by irradiation with 592 nm and 410 nm light.
Graphical Abstract
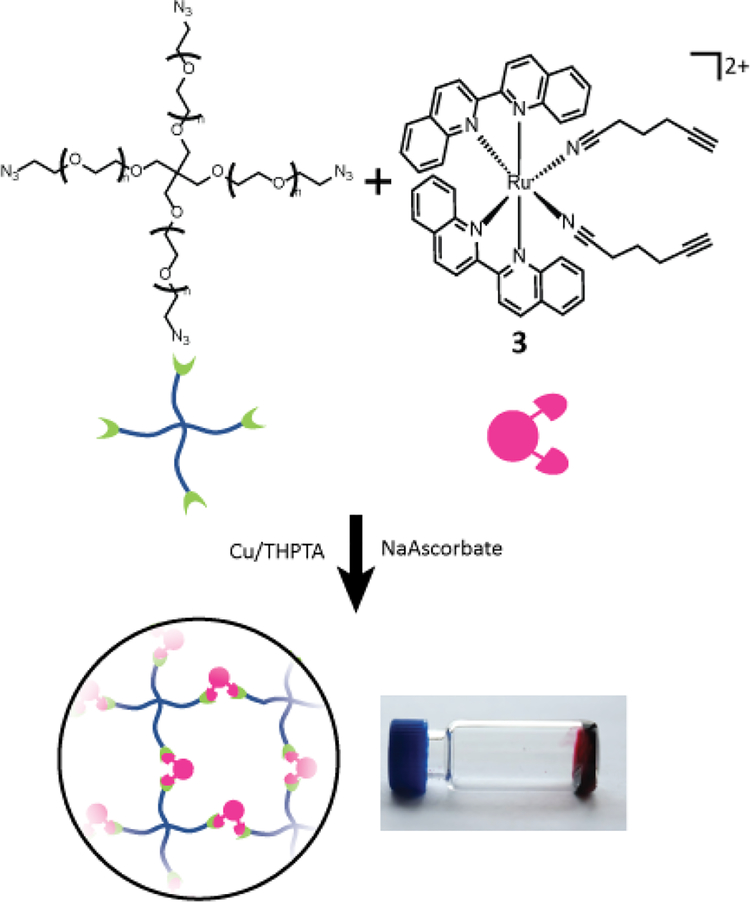
PEG hydrogel incorporating two ruthenium crosslinkers can be photodegraded using two different wavelengths of visible light to tune materials properties.
Introduction
Photoresponsive molecules and materials are transforming multiple areas of research, from drug delivery,[1–6] to materials engineering,[7–15] and biology.[16,17,26,18–25] Many natural biological processes are not photoresponsive, making light a versatile trigger for controlling complex biological systems.[27] The incorporation of photoactive moieties within biomolecules,[24] small-molecule drugs,[28] and materials[7] provides a method for modulating their activity. Likewise, photoactive moieties incorporated within soft materials, e.g., polymers, hydrogels, and elastomers, enable spatiotemporally precise, light-guided modulation of structure-function properties. Photoresponsive hydrogels in particular have long been used as platforms for cell growth and delivery, for small and large molecule drug delivery,[29,30] and for basic materials applications.[31] To expand methods for tuning soft material properties, e.g., shape and viscosity, we developed differentially photoresponsive ruthenium moieties suitable for hydrogel formation and subsequent multiplexed ligand dissociation.
A drawback to most current photoresponsive molecules is the high-energy light required for bond dissociation. Common photoresponsive organic chromophores, e.g., o-nitrobenzyl,[32] azobenzene,[14] and coumarin,[30,33] respond to near-UV and blue light, which barely penetrates most biomaterials or live tissue. Attempts to red-shift the activation wavelength have focused on multiphoton excitation,[10,34–37] coupling with upconverting nanoparticles[36,38] or chemically modified chromophores.[39] Some limiting factors include the small activation volume of multiphoton processes, the potential toxicity of embedded nanoparticles, low quantum yields (leading to sample heating and photodamage during repeated illumination), and synthetic complexity.
To address these challenges, we have worked to develop inorganic photoactive molecules that absorb orange-red light, which has greater penetration depth and is less prone to photodamage in clinical applications.[37] Our laboratory has expanded the use of photolabile ruthenium crosslinkers for applications in biology and soft materials. The first Ru-based crosslinker, (Ru(bipyridine)2(3-ethynylpyridine)2) (Ru-BEP), presented two alkynes for the circularization of antisense bis-azide-modified oligonucleotides for light-activated gene knockdown in zebrafish embryos.[21] A related compound, Ru(bipyridine)2(3-pyridinaldehyde)2 (RuAldehyde), provided a light-responsive crosslinker for hydrogel formation, site-selective degradation and protein release.[29]
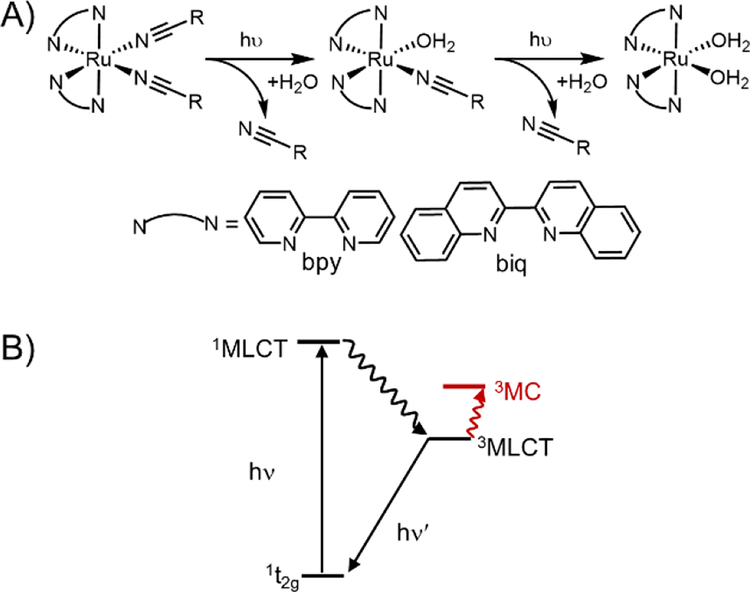
These Ru polypyridyl complexes share the unique ability to exchange a monodentate pyridine ligand with solvent upon irradiation with visible light. Single-site photo-substitution has been observed for other [Ru(polypyridyl)2X2]2+ complexes, where X = pyridine-[2,40–42] or sulphur-containing[43] ligands. Alternatively, two nitrile ligands[44,45] can both undergo rapid photo-substitution (Figure 1A). Excitation into the singlet metal-to-ligand charge transfer (1MLCT) band initiates intersystem crossing to a low-lying triplet state (Figure 1B). In most photo-responsive Ru-polypyridyl complexes this triplet state is primarily 3MLCT in character, with another triplet metal-centred (3MC) state close enough in energy to be thermally populated.
Figure 1.
Photoinitiated ligand exchange in ruthenium polypyridyl complexes. A) Photolysis observed for Ru(II)-nitrile complexes is a two-step process in which both ligands are exchanged with coordinating solvent. B) Jablonski diagram showing excited states responsible for ligand.
In the current study, our goal was to red-shift the absorption of Ru crosslinkers for multiplexing applications, while incorporating two photolabile nitrile-based ligands for maximum photodissociation within a hydrogel.[46] Inspired by previous work from Turro and coworkers, we designed a series of Ru crosslinkers incorporating biquinoline ligands that red-shift the maximum absorption wavelength, λmax.[46] The biquinoline also increased the steric strain around the Ru center, increasing the quantum yield of photorelease, Φpr.[47] This technique has led to several applications of red-light-absorbing, photoresponsive materials incorporating polypyridyl ruthenium compounds.[48,49] Here, we present the first examples of red-shifted Ru compounds that incorporate crosslinking functionality and achieve hydrogel formation, while enabling wavelength-selective degradation with visible light.
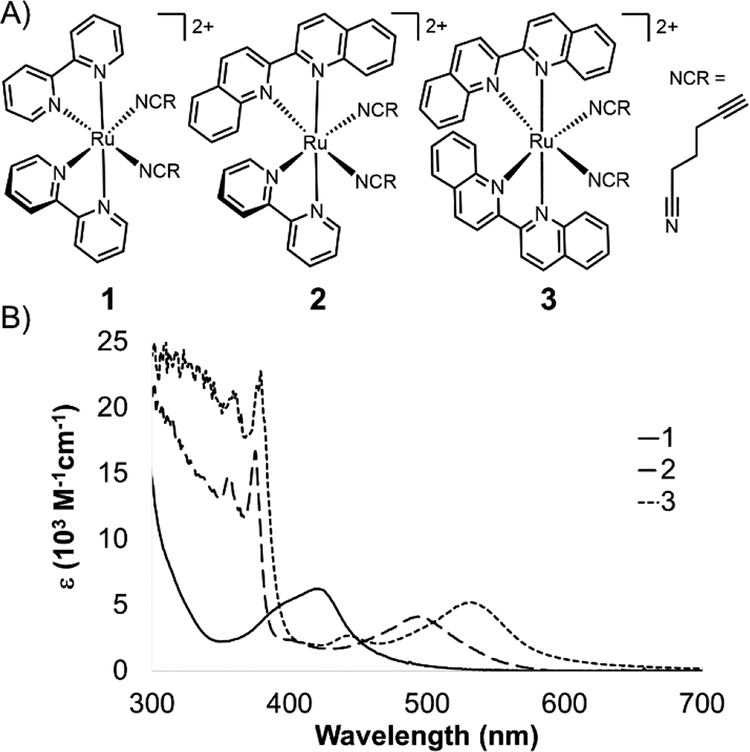
We present a series of alkyne-bearing Ru(II) compounds with nitrile-based photolabile ligands (compounds 1–3, Figure 2). Starting from Ru(bipyridine)2(5-hexynenitrile)2, λmax was sequentially red-shifted by incorporating 1 or 2 biquinoline ligands (Figure 2). A crystallographic analysis confirmed that 5-hexynenitrile appropriately positions the pendant alkyne for subsequent reaction with an azide-modified branched polyethylene glycol (PEG) polymer (10 kDa) via copper(I)-catalyzed alkyne-azide cycloaddition (CuAAC).[50,51] The resulting hydrogels, formed with Ru crosslinkers 1 and 3, allowed spatially selective degradation via two different wavelengths of visible light (592 and 410 nm).
Figure 2.
Ru crosslinkers with two photolabile nitrile ligands. A) Three compounds synthesized in this study. B) Molar absorption spectra for 1–3.
Results and Discussion
Compound 1 was synthesized from commercially available Rubpy2Cl2 and 5-hexynenitrile through the Ru(bpy)2(H2O)2 intermediate generated by the addition of AgPF6 to form AgCl precipitate. 1–3 were purified as the PF6− salt via silica column flash chromatography (1:4 acetonitrile:methylene chloride mobile phase), and isolated as the nitrate salt using an Amberlite© IRA-410 column in good yield (54%) (see SI for synthetic details). The nitrate counterion gave Ru2+ polypyridyl complexes with excellent solubility and stability in water (Figure S1).
To generate Ru(bpy)(biq)Cl2 for 2 we found it necessary to use the benzene ruthenium dimer [(benzene)RuCl2]2 to ensure conversion to the mixed ligand product. Bipyridine was coordinated first to generate Ru(bpy)Cl42-, which was purified by filtration, followed by addition of biquinoline and heating to give Ru(bpy)(biq)Cl2, which was purified by precipitation into diethyl ether, in 55% yield. Subsequent coordination of two 5-hexynenitrile ligands gave 2 in a final overall yield of 13.5%. Compound 3 was synthesized starting with RuCl3; 2.2 equivalents of biquinoline were added with hydroquinone as the reducing agent and excess LiCl to generate the intermediate Rubiq2Cl2, which was isolable by precipitation into ether in 33% yield. Coordination of 5-hexynenitrile proceeded by the same procedure as for 1 and 2, giving 3 as nitrate salt in overall 24% yield. All compounds were characterized by 1H NMR spectroscopy, high-resolution ESI mass spectrometry, and UV-Vis absorption spectroscopy (see SI).
Ruthenium polypyridyl complexes exhibit strong absorbance in the visible region due to low-lying metal-to-ligand charge transfer (MLCT) band. In this state, electrons are excited from the ground state orbital located primarily on the metal center to a low-lying excited orbital located on the polypyridyl ligand, at higher energy for bipyridine than biquinoline.[46] Ligands with more extended pi bonding tend to lower the energy of the 1MLCT band, and red shift the absorbance.
The 1MLCT absorption maxima for 1, 2, and 3 were 419, 491, and 529 nm, respectively (ε reported in Table 1). A shift of over 70 nm was observed with the first substitution of a bipyridine for biquinoline ligand, from 1 to 2 (Figure 2A), followed by a nearly 40 nm red-shift from 2 to 3. This shows good agreement with previously published spectra for Ru(phen)2(MeCN)2 (λmax = 420 nm), Ru(phen)(biq)(MeCN)2 (λmax = 497 nm), and Ru(biq)2(MeCN)2 (λmax = 535 nm).[46]
Table 1.
Absorptivities and quantum yields for 1–3
| ε (M−1cm−1) | Φpr | |
|---|---|---|
| 1 | 6140 ± 100 | 0.16 ± 0.02 @450 nm |
| 2 | 1900 ± 100 | 0.19 ± 0.005 @532 nm |
| 3 | 7400 ± 400 | 0.07 ± 0.01 @532 nm |
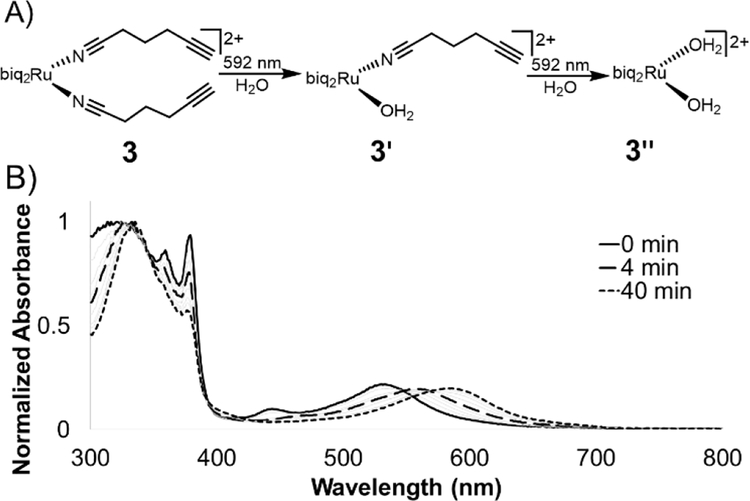
The photolysis of ruthenium polypyridyl compounds can be observed directly using UV-Vis spectroscopy. As the compound undergoes ligand exchange of a coordinated ligand for a solvent molecule, a significant red shift is observed in the MLCT band. Under continuous irradiation, compounds 1-3 sequentially exchanged both nitrile ligands (Figures 3A, S2). UV-Vis photolysis curve for 3 is shown in Figure 3B, where peaks at 560 and 590 nm indicated a stepwise process, with a monoaquated intermediate. The clear isosbestic points at 550 and 570 nm also indicated the stepwise transition from 3 to monoaquated 3’ to bisaquated 3”, although the first transition point at 550 nm included early formation of 3” under continuous irradiation.
Figure 3.
Photolysis of 3 in water. A) Compounds 1–3 undergo a stepwise ligand exchange of both nitrile ligands when irradiated in water. The second step takes much longer than the first. B) Photolysis trace of 3 in water under irradiation from 592 nm LED (25 mW/cm2).
The loss of the second nitrile ligand in 3 was slower, occurring on the order of 40 min (Figure 3B), compared to the first ligand exchange event, which was completed within 4 min of constant irradiation in the bulk sample. This trend was observed for 1 and 2 as well (Figure S2). The Ru MLCT band extends well beyond the λmax, which can be used to induce ligand exchange at longer wavelengths of light; irradiation at 600 – 700 nm (red incandescent light bulb, 5 mW) was less efficient but led to complete photolysis of 3 in 4 h (Figure S3).
Photolysis data were fit to an equation derived from pseudo-first order kinetics process, and the time constants were determined (Figure S4). The value of Φpr was found for the first exchange event from the rate constant coupled with the laser power (Figure S4). As expected, Φpr decreased roughly 2-fold as the MLCT band was shifted further to the red, from 0.16 (in 1) to 0.07 (in 3), Table 1.[52,53] Similarly, the efficiency of photolysis for 3 (ε × Φpr = 520 M−1cm−1) was lower than 1 at 980 M−1cm−1. Although these values were lower than other published ruthenium caging groups, with efficiencies that range from 2,000[46] to 4,000 M−1cm−1,[29] they are significantly improved over other green-light-sensitive caging groups like BODIPY, with Φpr on the order of 100 M−1cm−1.[54,55]
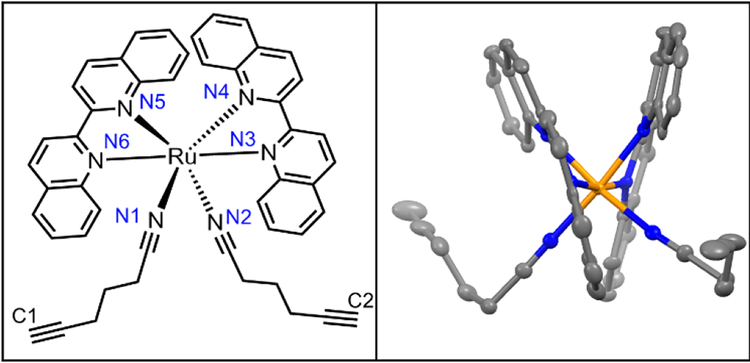
Diffraction-quality crystals of 3 as PF6− salt were grown via vapour diffusion from 3–5 mg dissolved in acetonitrile/methanol/THF (0.1 mL each) with diethyl ether, stored at −20 °C for 2 weeks (Figure 4). Bond lengths between Ru2+ and ligands were within expected ranges, with variations due to the steric strain in the system. The angle between the nitrile ligands is stretched significantly to >95° perhaps due to the strain caused by bulkier biq ligands coordinated to Ru2+. In the less crowded Ru(bpy)(biq)(5-hexynenitrile)2 compound, the nitrile-Ru-nitrile angle is ~90° (Figure S5). Crystal structure of 3 shows alkynes positioned 4.3 Å and 4.9 Å from biquinolines, angled such that they are accessible for cycloaddition with azide and copper catalyst.
Figure 4.
Crystal structure of 3.
The conformational flexibility of the nitrile-alkyl ligands required synthesis and testing of several Ru compounds to identify competent crosslinkers. Initially, Ru compounds employing a shorter 4-pentynenitrile ligand were synthesized and found to be incapable of Cu(I)-mediated PEG gelation (Figure S6). Incorporation of longer 5-hexynenitrile ligands led to functional Ru crosslinkers, but only after mild synthetic conditions for nitrile coordination were employed. Ru-(5-hexynenitrile) coordination performed at elevated temperatures and longer reaction times resulted in Ru compounds found to be incapable of Cu(I)-mediated PEG gelation. X-ray crystal structure analysis of one such example shows alkyne positioned much closer to the biquinoline, only 3.7 Å (Figure S5). The cis-alkane conformation should disfavour Cu(I)-mediated alkyne-azide cycloaddition chemistry. Ru compounds 1-3 were synthesized using the mild conditions detailed in the Synthetic Procedure and confirmed to be excellent crosslinkers in gelation studies.
CuAACs have been widely used for materials design, with several studies showing the generation of hydrogel materials. Hyaluronic acid,[56,57] polyethylene glycol (PEG),[58] dextran,[59] poly(vinyl) alcohol (PVA),[60] along with several other polymers have been modified with azides and terminal alkynes to facilitate hydrogel formation. The need for a Cu(I) catalyst has limited some bio-applications as it can be toxic to cells,[61] but can also provide spatiotemporal control. In one example Bowman and co-workers used a photocatalyst to reduce Cu(II) for the formation of a hydrogel with precise control.[62] Copper can be dialysed away from preformed hydrogels, which is acceptable for many drug delivery platforms.
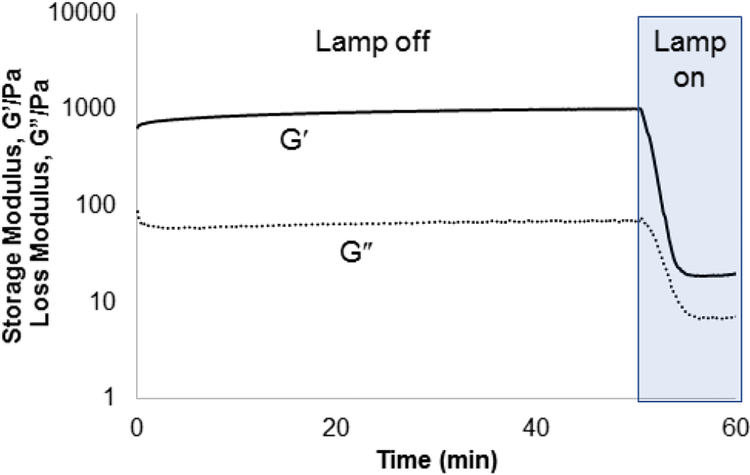
Compounds 1–3 were tested for crosslinking reaction with azido-PEG (MW 10,000 Da) in the presence of CuSO4, THPTA ligand, and sodium ascorbate reducing agent (Scheme 1), forming a strong hydrogel within 30 s (results shown for 3, Figure 5). Hydrogels formed at a final weight percent of 7.5 wt% with stoichiometric ruthenium crosslinker, generating elasticity nearing 1 kPa (Figure 5). As expected, when exposed to visible light (400 – 500 nm) the hydrogel rapidly lost its elastic properties, becoming a viscous liquid within 5 min (Figure 5).
Scheme 1.
Gelation of branched PEG via crosslinking reaction with 3
Figure 5.
Rheometry demonstrating gelation formed from the incorporation of 3 into a PEG hydrogel. The hydrogel was rapidly degraded under irradiation with 400 – 500 nm light (25 mW/cm2).
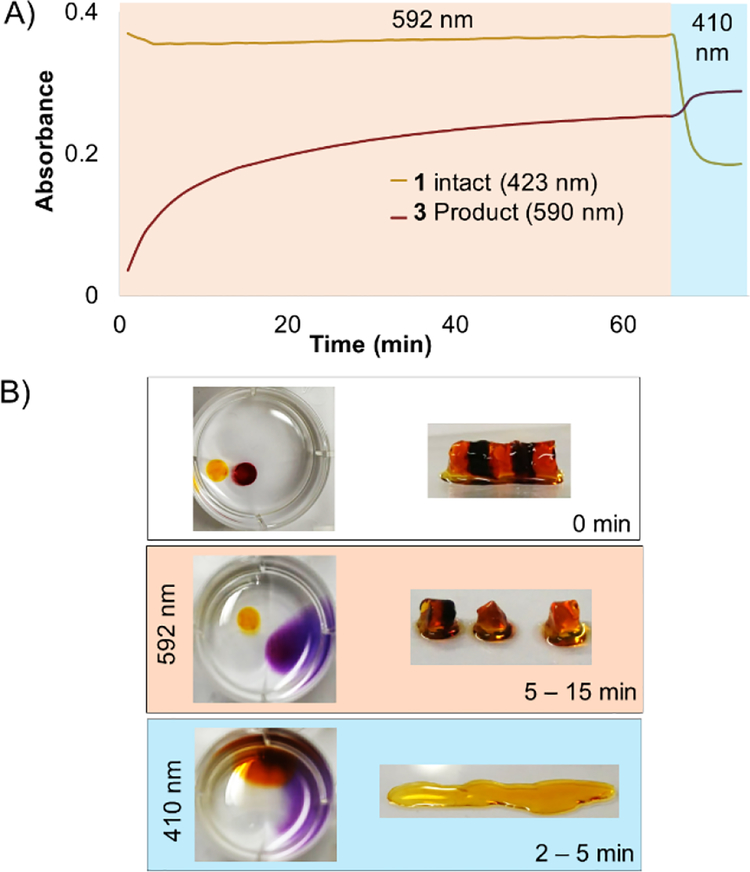
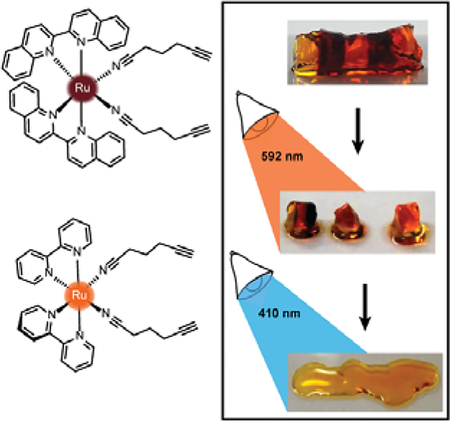
Next, a striped hydrogel was formed for multiplexing experiment via “layer-by-layer” reaction of azido-PEG with crosslinker 1, alternating with crosslinker 3 (Figure 6). Orange light (592 nm) was used to degrade 3 selectively while leaving 1 intact, as demonstrated both in a solution experiment with equal parts 1 and 3 (Figure 6A) and in the gel (Figure 6B). A significant increase in absorbance at 590 nm confirmed the formation of the bisaquated product Ru(biq)2(H2O)2, 3” (Figures 6A, 3B). Finally, irradiation at 410 nm led to a significant decrease in absorbance at 423 nm and small increase at 590 nm due to formation of Ru(bpy)2(H2O)2, 1” (Figures 6A, S1), and rapidly degraded the remaining hydrogel sections crosslinked by 1 (Figure 6B). The sequence of irradiation is important in this case, as compound 3 absorbs both 410 and 592 nm light and will be degraded by both wavelengths.
Figure 6.
Selective degradation of 1 and 3 in solution and hydrogel. A) Irradiation at 592 nm photolyzed 3 in solution while leaving 1 intact, until irradiation at 410 nm. B) Striped hydrogel incorporating alternating sections of 1 and 3 was selectively degraded at 592 nm, leaving the orange gel regions intact. The remaining sections crosslinked by 1 were degraded by 410 nm light.
We developed spectrally tuneable ruthenium polypyridyl crosslinkers with two pendant alkynes for hydrogel formation, and high photolysis efficiency for multiplexed, visible-light gel degradation. Replacing bipyridine with more pi-conjugated biquinoline ligands red-shifted the absorbance, generating a series of sequentially red-shifted compounds. Incorporation of two flexible 5-hexynenitrile ligands at the Ru2+ center enabled CuAAC crosslinking reactions, while also facilitating subsequent photodegradation of gels incorporating these Ru crosslinkers. This represents the first example to our knowledge of a two-color hydrogel system that can be selectively activated by two different visible wavelengths. Such Ru crosslinkers may be applied broadly in materials chemistry, or alternately employed for generating photoactive versions of circular oligos,[21] peptides, or other bis-azide containing molecules.
Supplementary Material
Table 2.
Select Bond Lengths
| Bond | 3 (Å) | |
|---|---|---|
| Ru-biq | Ru-N4 | 2.084(6) |
| Ru-N3 | 2.093(6) | |
| Ru-N≡C | Ru-N1 | 2.025(6) |
| Ru-N2 | 2.024(6) | |
| C≡C to biq | C1-biq | 4.267 |
| C2-biq | 4.926 |
Acknowledgements
Many thanks to Jonathan Gallarga for his assistance with the rheometry experiments, and Pat Carroll for his work in solving the crystal structures. We also thank the NIH (R01-GM083030) and the UPenn Chemistry Department for financial support.
Footnotes
Supporting information for this article is given via a link at the end of the document. CCDC 1568985 contains the supplementary crystallographic data for this paper. These data are provided free of charge by The Cambridge Crystallographic Data Centre.
References
- [1].Smith NA, Zhang P, Greenough SE, Horbury MD, Clarkson GJ, McFeely D, Habtemariam A, Salassa L, Stavros VG, Dowson CG, et al. , Chem. Sci 2017, 8, 395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Li A, Turro C, Kodanko JJ, Chem. Commun 2018, 54, 1280–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Reeßing F, Szymanski W, Curr. Med. Chem 2018, 24, 4905–4950. [DOI] [PubMed] [Google Scholar]
- [4].Döbber A, Phoa AF, Abbassi RH, Stringer BW, Day BW, Johns TG, Abadleh M, Peifer C, Munoz L, ACS Med. Chem. Lett 2017, 8, 395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Dcona MM, Mitra D, Goehe RW, Gewirtz DA, Lebman DA, Hartman MCT, Chem. Commun 2012, 48, 4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ki Choi S, Thomas T, Li M-H, Kotlyar A, Desai A, Baker JR Jr., Chem. Commun 2010, 46, 2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kloxin AM, Kasko AM, Salinas CN, Anseth KS, Science 2009, 324, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ovsianikov A, Deiwick A, Van Vlierberghe S, Dubruel P, Möller L, Dräger G, Chichkov B, Biomacromolecules 2011, 12, 851–858. [DOI] [PubMed] [Google Scholar]
- [9].Shin D-S, You J, Rahimian A, Vu T, Siltanen C, Ehsanipour A, Stybayeva G, Sutcliffe J, Revzin A, Angew. Chem. Int. Ed 2014, 53, 8221–8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peng K, Tomatsu I, van den Broek B, Cui C, Korobko AV, van Noort J, Meijer AH, Spaink HP, Kros A, Soft Matter 2011, 7, 4881. [Google Scholar]
- [11].Kirschner CM, Alge DL, Gould ST, Anseth KS, Adv. Healthc. Mater 2014, 3, 649–657. [DOI] [PubMed] [Google Scholar]
- [12].Arakawa CK, Badeau BA, Zheng Y, DeForest CA, Adv. Mater 2017, 29, 1703156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Griffin DR, Kasko AM, J. Am. Chem. Soc 2012, 134, 13103–13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Rosales AM, Mabry KM, Nehls EM, Anseth KS, Biomacromolecules 2015, 16, 798–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Brown TE, Marozas IA, Anseth KS, Adv. Mater 2017, 29, 1605001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu L, Wang Y, Wu J, Lv C, Wang J, Tang X, Nucleic Acids Res. 2013, 41, 677–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kröck L, Heckel A, Angew. Chem. Int. Ed 2005, 44, 471–473. [DOI] [PubMed] [Google Scholar]
- [18].Young DD, Lively MO, Deiters A, J. Am. Chem. Soc 2010, 132, 6183–6193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yamazoe S, Liu Q, McQuade LE, Deiters A, Chen JK, Angew. Chem. Int. Ed 2014, 53, 10114–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yamazoe S, Shestopalov IA, Provost E, Leach SD, Chen JK, Angew. Chem. Int. Ed 2012, 51, 6908–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Griepenburg JC, Rapp TL, Carroll PJ, Eberwine J, Dmochowski IJ, Chem. Sci 2015, 6, 2342–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Griepenburg JC, Ruble BK, Dmochowski IJ, Bioorg. Med. Chem 2013, 21, 6198–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Goguen BN, Hoffman BD, Sellers JR, Schwartz MA, Imperiali B, Angew. Chem. Int. Ed 2011, 123, 5785–5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Riggsbee CW, Deiters A, Trends Biotechnol. 2010, 28, 468–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Ankenbruck N, Courtney T, Naro Y, Deiters A, Angew. Chem. Int. Ed 2018, 57, 2768–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hemphill J, Borchardt EK, Brown K, Asokan A, Deiters A, J. Am. Chem. Soc 2015, 137, 5642–5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mura S, Nicolas J, Couvreur P, Nat. Mater 2013, 12, 991–1003. [DOI] [PubMed] [Google Scholar]
- [28].Huisman M, White JK, Lewalski VG, Podgorski I, Turro C, Kodanko JJ, Chem. Commun 2016, 52, 12590–12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Rapp TL, Highley CB, Manor BC, Burdick JA, Dmochowski IJ, Chem. Eur. J 2018, 24, 2328–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Azagarsamy MA, Anseth KS, Angew. Chem. Int. Ed 2013, 52, 13803–13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wong DY, Griffin DR, Reed J, Kasko AM, Macromolecules 2010, 43, 2824–2831. [Google Scholar]
- [32].Walker JW, Mccray JA, Hess GP, Biochemistry 1986, 25, 1799–1805. [DOI] [PubMed] [Google Scholar]
- [33].Azagarsamy MA, McKinnon DD, Alge DL, Anseth KS, ACS Macro Lett. 2014, 3, 515–519. [DOI] [PubMed] [Google Scholar]
- [34].Aujard I, Benbrahim C, Gouget M, Ruel O, Baudin J-B, Neveu P, Jullien L, Chem. Eur. J 2006, 12, 6865–6879. [DOI] [PubMed] [Google Scholar]
- [35].Wylie RG, Shoichet MS, J. Mater. Chem 2008, 18, 2716. [Google Scholar]
- [36].Yan B, Boyer J-C, Branda NR, Zhao Y, J. Am. Chem. Soc 2011, 133, 19714–19717. [DOI] [PubMed] [Google Scholar]
- [37].Zeng X, Zhou X, Wu S, Macromol. Rapid Commun 2018, 1800034. [DOI] [PubMed] [Google Scholar]
- [38].Yan B, Boyer J-C, Habault D, Branda NR, Zhao Y, J. Am. Chem. Soc 2012, 134, 16558–16561. [DOI] [PubMed] [Google Scholar]
- [39].Wang D, Wagner M, Butt H-J, Wu S, Soft Matter 2015, 11, 7656–62. [DOI] [PubMed] [Google Scholar]
- [40].Leonardo Z, Cecilia C, Pablo A, Baraldo L, Etchenique R, J. Am. Chem. Soc 2003, 125, 882–883. [DOI] [PubMed] [Google Scholar]
- [41].Huisman M, White JK, Lewalski VG, Podgorski I, Turro C, Kodanko JJ, Chem. Commun 2016, 52, 12590–12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lameijer LN, Ernst D, Hopkins SL, Meijer MS, Askes SHC, Le Dévédec SE, Bonnet S, Angew. Chem. Int. Ed 2017, 56, 11549–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Garner RN, Joyce LE, Turro C, Inorg. Chem 2011, 50, 4384–4391. [DOI] [PubMed] [Google Scholar]
- [44].Sharma R, Knoll JD, Martin PD, Podgorski I, Turro C, Kodanko JJ, Inorg. Chem 2014, 53, 3272–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Respondek T, Sharma R, Herroon MK, Garner RN, Knoll JD, Cueny E, Turro C, Podgorski I, Kodanko JJ, ChemMedChem 2014, 9, 1306–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Albani BA, Durr CB, Turro C, J. Phys. Chem. A 2013, 117, 13885–13892. [DOI] [PubMed] [Google Scholar]
- [47].Knoll JD, Albani BA, Durr CB, Turro C, J. Phys. Chem. A 2014, 118, 10603–10610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Zeng X, Zhou X, Wu S, Macromol. Rapid Commun 2018, 1800034. [DOI] [PubMed] [Google Scholar]
- [49].Xie C, Sun W, Lu H, Kretzschmann A, Liu J, Wagner M, Butt H-J, Deng X, Wu S, Nat. Commun 2018, 9, 3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Rostovtsev VV, Green LG, Fokin VV, Sharpless KB, Angew. Chem. Int. Ed 2002, 41, 2596–2599. [DOI] [PubMed] [Google Scholar]
- [51].Tornoe C, Christensen C, Meldal M, J. Org. Chem 2002, 67, 3057–3064. [DOI] [PubMed] [Google Scholar]
- [52].Lameijer LN, Ernst D, Hopkins SL, Meijer MS, Askes SHC, Le Dévédec SE, Bonnet S, Angew. Chem. Int. Ed 2017, 56, 11549–11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Loftus LM, Li A, Fillman KL, Martin PD, Kodanko JJ, Turro C, J. Am. Chem. Soc 2017, 139, 18295–18306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Goswami PP, Syed A, Beck CL, Albright TR, Mahoney KM, Unash R, Smith EA, Winter AH, J. Am. Chem. Soc 2015, 137, 3783–3786. [DOI] [PubMed] [Google Scholar]
- [55].Umeda N, Takahashi H, Kamiya M, Ueno T, Komatsu T, Terai T, Hanaoka K, Nagano T, Urano Y, ACS Chem. Biol 2014, 9, 2242–2246. [DOI] [PubMed] [Google Scholar]
- [56].Crescenzi V, Cornelio L, Di Meo C, Nardecchia S, Lamanna R, Biomacromolecules 2007, 8, 1844–1850. [DOI] [PubMed] [Google Scholar]
- [57].Hu X, Li D, Zhou F, Gao C, Acta Biomater. 2011, 7, 1618–1626. [DOI] [PubMed] [Google Scholar]
- [58].Liu SQ, Rachel Ee PL, Ke CY, Hedrick JL, Yang YY, Biomaterials 2009, 30, 1453–1461. [DOI] [PubMed] [Google Scholar]
- [59].Heller DA, Levi Y, Pelet JM, Doloff JC, Wallas J, Pratt GW, Jiang S, Sahay G, Schroeder A, Schroeder JE, et al. , Adv. Mater 2013, 25, 1449–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ossipov DA, Hilborn J, Macromolecules 2006, 39, 1709–1718. [Google Scholar]
- [61].Jiang Y, Chen J, Deng C, Suuronen EJ, Zhong Z, Biomaterials 2014, 35, 4969–4985. [DOI] [PubMed] [Google Scholar]
- [62].Adzima BJ, Tao Y, Kloxin CJ, DeForest CA, Anseth KS, Bowman CN, Nat. Chem 2011, 3, 256–259. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.