Abstract
Background
Spermatozoa become competent for fertilization during transit through the epididymis. As spermatozoa from the proximal caudal epididymis can fertilize eggs, proteins from the caput and corpus epididymis are required for sperm maturation.
Objectives
Microarray analysis identified that more than 17,000 genes are expressed in the epididymis; however, few of these genes demonstrate expression restricted to the epididymis. To analyze epididymis-enriched gene function in vivo, we generated knockout (KO) mutations in nine genes that are abundantly expressed in the caput and corpus region of the epididymis.
Materials and Methods
KO mice were generated using the CRISPR/Cas9 system. The histology of the epididymis was observed with hematoxylin and eosin staining. KO males were caged with wild-type females for 3 to 6 months to check fertility.
Results
We generated individual mutant mouse lines having indel mutations in Pate1, Pate2, or Pate3. We also deleted the coding regions of Clpsl2, Epp13, and Rnase13, independently. Finally, the 150 kb region encoding Gm1110, Glb1l2, and Glb1l3 was deleted to generate a triple KO mouse line. Histology of the epididymis and sperm morphology of all KO lines were comparable to control males. The females mated with these KO males delivered pups at comparable numbers as control males.
Discussion and Conclusion
We revealed that nine genes abundantly expressed in the caput and corpus epididymis are dispensable for sperm function and male fecundity. CRISPR/Cas9 mediated KO mice generation accelerates the screening of epididymis enriched genes for potential functions in reproduction.
Keywords: genome editing, genetically modified mice, sperm maturation
Introduction
Spermatozoa released from the testis are incapable of fertilizing eggs until they acquire capabilities necessary for fertilization competence (such as motility, capacitation, acrosome reaction and sperm-egg fusion capabilities) by translocating through the epididymis. This step is called “sperm maturation” (Robaire & Hermo 1988; Robaire, Hinton, and Orgebin-Crist 2006). The epididymis is mainly composed of 3 regions: the head (caput), body (corpus), and tail (cauda). In many mammalian species, the sperm journey from the caput region to the cauda region in about 10 days (Robaire & Hermo 1988). The timing of acquisition of fertilization competence by spermatozoa varies among mammalian species; however, almost all spermatozoa isolated from the proximal cauda epididymis have fertilizing ability (Robaire & Hermo 1988), suggesting that factors from the caput and corpus epididymis play important roles in sperm maturation.
Microarray analysis shows that more than 17,000 genes are expressed in the epididymis with gene expression patterns varying greatly among regions of the epididymis (Johnston et al., 2005), suggesting that epididymis-specific or epididymis-enriched genes play important roles during sperm maturation. However, the physiological function of many of these genes remain unknown.
The emergence of the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system opened a new era in mammalian genome editing (Cong et al. 2013; Wang et al. 2013; Yang et al. 2013). The guide RNA (gRNA)/Cas9 complex recognizes 20 nucleotides upstream of the protospacer adjacent motif (PAM) as the target sequences, then the Cas9 nuclease causes a blunt end double-strand break (DSB) between the 3rd and 4th nucleotides upstream of the PAM. After DSB formation, non-homologous end-joining (NHEJ) causes an indel mutation. We previously reported that indel mutations were efficiently obtained by injecting single gRNA and Cas9 expressing plasmids into eggs (Mashiko et al., 2013). In conjunction with conventional ES cell mediated gene-targeting, we also demonstrated that many testis-enriched genes are not essential for male fertility (Miyata et al., 2016). Our previous work demonstrated that CRISPR/Cas9 mediated KO mice generation and phenotype screening is a cost-effective and labor-effective approach to quickly identify essential gene functions in vivo (Mashiko et al., 2014).
In the present study, we have knocked out nine epididymis-enriched genes and discovered these genes are not essential for sperm fertilizing ability and male fecundity.
Materials and Methods
Animals
All mice used in this study were purchased from Japan SLC or CLEA Japan. Mice were acclimated to a 12 hour-light/12 hour-dark cycle. All animal experiments were approved by the Animal Care and Use Committee of the Research Institute for Microbial Diseases, Osaka University, Japan (#Biken-AP-H30–01) and the Institutional Animal Care and Use Committee of Baylor College of Medicine (AN-716).
cDNA and RT-PCR
All tissues [brain, heart, kidney, liver, lung, spleen, thymus, ovary, uterus, testis, epididymis (caput, corpus, and cauda regions), prostate (mixture of dorsal, lateral and ventral regions), coagulating gland, and seminal vesicle] were collected from C57BL/6NCr mice. These samples were homogenized in TRIzol (Ambion). The total RNA was reverse-transcribed to cDNA using SuperScript III First Strand Synthesis System for RT-PCR (Invitrogen). Five ng of cDNA was used for PCR with primer sets (Table S1) and KOD DNA Polymerase (KOD-FX Neo, TOYOBO).
Gene tree
The gene tree was made by GENETYX with the amino acid sequence (method: UPGMA) [for Colipase like 2 (CLPSL2): NP_001030043 (mouse), Q6UWE3 (human), XP_003311311 (chimpanzee), XP_022281476 (dog)] [for Epididymal protein 13 (EPP13): NP_001170887 (mouse), NP_001341587 (human), PNI23622 (chimpanzee), XP_024835189 (cow)] [for GM1110: XP_006510522 (mouse), ENSCAFT00000043177 (dog), ENSBTAT00000016226 (cow)] [for Galactosidase, beta 1-like 2 (GLB1L2): XP_006510360 (mouse), NP_612351 (human), XP_009422813 (chimpanzee), XP_022273772 (dog), XP_010811276 (cow)] [for GLB1L3: AAI32203 (mouse), AAH11001 (human), XP_024203264 (chimpanzee), XP_005620347 (dog), XP_024831720 (cow)] [for Prostate and testis expression 1 (PATE1): NP_001186882 (mouse), NP_612151 (human), XP_024203322 (chimpanzee), XP_022273825 (dog), XP_024843340 (cow)] [for PATE2: XP_011240865 (mouse), NP_997720 (human), XP_001148077 (chimpanzee), XP_005619644 (dog), XP_024843404 (cow)] [for PATE3: NP_001161064 (mouse), NP_001123355 (human), XP_024203289 (chimpanzee), XP_022273771 (dog), XP_024843215 (cow)] [for RNase A family 13 (RNASE13): XP_017171565 (mouse), AAV87186 (human), XP_003804788 (chimpanzee), XP_013974975 (dog), AAI34657 (cow)].
Egg collection
Pregnant mare serum gonadotropin (PMSG) (5 units, ASKA Pharmaceutical) or CARD HyperOva (0.1 mL, Kyudo) was injected into the abdominal cavity of B6D2F1 females, followed by human chorionic gonadotropin (hCG) (5 units, ASKA Pharmaceutical) and natural mating with B6D2F1 males 48 hours after PMSG or CARD HyperOva. After 20 hours, we collected fertilized eggs with 2 pronuclei for genome editing.
Pronuclear injection
Pronuclear injection with 5 ng/μL of gRNA/Cas9 expressing plasmid (for Pate1, Pate2 and Pate3) was performed as previously reported (Mashiko et al., 2013; Noda et al., 2017). The crRNA and tracrRNA (Sigma) were diluted with nuclease free water (non-DEPC treated, Ambion). The mixture was denatured at 95°C for 1 minute and allowed to anneal by cooling gradually to room temperature (~1 hour). Each gRNA was mixed with Cas9 protein solution (Thermo Fisher Scientific) and T10E0.1 buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 7.4), and then incubated at 37°C for 5 minutes to prepare the gRNA/Cas9 RNPs. For multiple gene targeting, we prepared the gRNA/Cas9 RNP solution separately and then combined them [final concentration: 30 ng/μL (≈ 200 nM) Cas9 for 20 ng/μL (≈ 600 nM) of each gRNA (Table S2)]. After centrifugation at 20,000 g at 4°C for 10 minutes, the mixture was used for pronuclear injection (for Clpsl2, Epp13, Rnase13, Gm1110, Glb1l2, Glb1l3).
Electroporation
The gRNA was prepared as described above. The gRNA was mixed with Cas9 protein solution and opti-MEM media (Thermo Fisher Scientific), and then incubated at 37°C for 5 minutes to prepare the gRNA/Cas9 RNPs [final concentration: 50 ng/μL (≈ 330 nM) Cas9 for 20 ng/μL (≈ 600 nM) of each gRNA (Table S2)]. The gRNA/Cas9 RNP solution was placed between the electrodes with a 5 mm gap in the NEPA21 Super Electroporator (Nepagene). Fertilized eggs were arranged between the electrodes, and then the electroporation was done with the following conditions [resistance value: 550~600 Ω, poring pulse: 225 V (voltage), 2 ms (pulse amplitude), 50 ms (pulse interval), 4 (pulse number), 10% (attenuation), + (polarity), transfer pulse: 20 V (voltage), 50 ms (pulse amplitude), 50 ms (pulse interval), ± 5 (pulse number), 40% (attenuation), +/− (polarity)].
Egg transfer
Injected and electroporated embryos were transplanted into the uterus of pseudo-pregnant ICR recipients. After 19 days, offspring were obtained by natural birth or Caesarean section.
Epididymis histology and sperm morphology
Epididymides were fixed in Bouin’s fluid (Polysciences) at 4°C overnight. Fixed epididymides were dehydrated by increasing ethanol concentrations, and then were embedded with paraffin. Paraffin sections (5-μm) were stained with Mayer hematoxylin solution for 3 to 5 minutes, counterstained with eosin Y solution [53% (v/v) ethanol, 0.3% (v/v) eosin, and 0.5% (v/v) acetic acid] for 2 to 5 minutes, dehydrated in increasing ethanol concentrations, and finally mounted in Permount or Entellan new (Merck). The caudal epididymal spermatozoa were observed with phase contrast microscopy.
Mating test
KO male mice were caged with 2 or 3 B6D2F1 females (for Pate1, Pate2, Pate3, Epp13, Rnase13, and Glb1l2-Gm1110 mutant mice), or 1 hybrid female (C57BL/6J × 129S5/SvEvBrd.) (for Clpsl2) for 3 to 6 months. Frozen spermatozoa from Pate1-, Pate2-, Pate3-, Clpsl2-disrupted males (B6D2-Pate1<em1Osb>, RBRC#09830, CARD#2450; B6D2-Pate2<em1Osb>, RBRC#09831, CARD#2451; B6D2-Pate3<em1Osb> Pate3<em2Osb>, RBRC#09832, CARD#2452; B6D2-Clpsl2<em1Osb>, RBRC#10341, CARD#2711) will be available through RIKEN BRC (http://en.brc.riken.jp/index.shtml) and CARD R-BASE (http://cardb.cc.kumamoto-u.ac.jp/transgenic/).
Statistical analysis
All values are shown as the mean ± SD of at least three independent experiments. Statistical analyses were performed using Student’s t-test (Figure 2 to 4 and 6) and Mann-Whitney test (Figure 5).
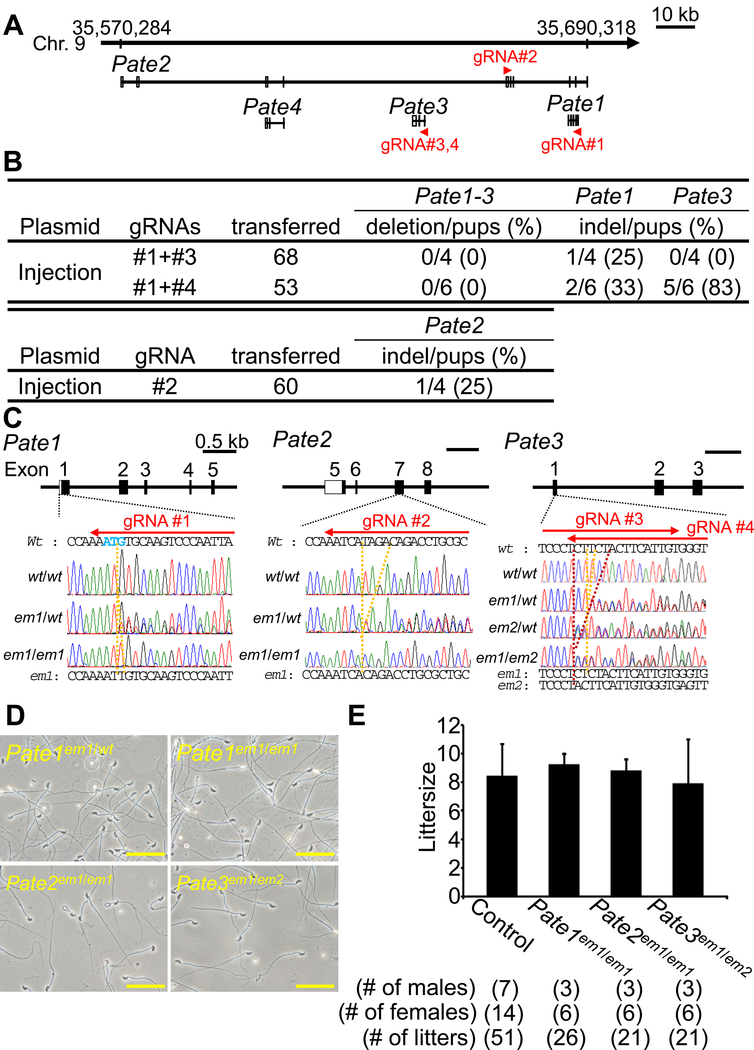
Figure 2. Fecundity of KO males of Pate family genes.
(A) Pate family genes within murine genomic locus |chromosome 9qA4|. (B) Genome editing efficiency of injecting gRNA/Cas9 expressing plasmids into eggs. (C) DNA sequencing of KO mice of Pate family genes. Enzyme mutation (em) 1 for Pate1: 1 base “T” insertion; em1 for Pate2: 4 base “TAGA” deletion; em1 and em2 for Pate3: 1 base “T” and 5 base “CTTCT” deletion. Black boxes: coding region, Blue colored letters: initial methionine. (D) Sperm morphology observed under phase contrast. (E) Male fecundity. There was no difference in average litter size between control and KO males of each gene (Pate1: p = 0.57, Pate2: p = 0.80, Pate3: p = 0.76). Heterozygous males of each gene were used as controls.
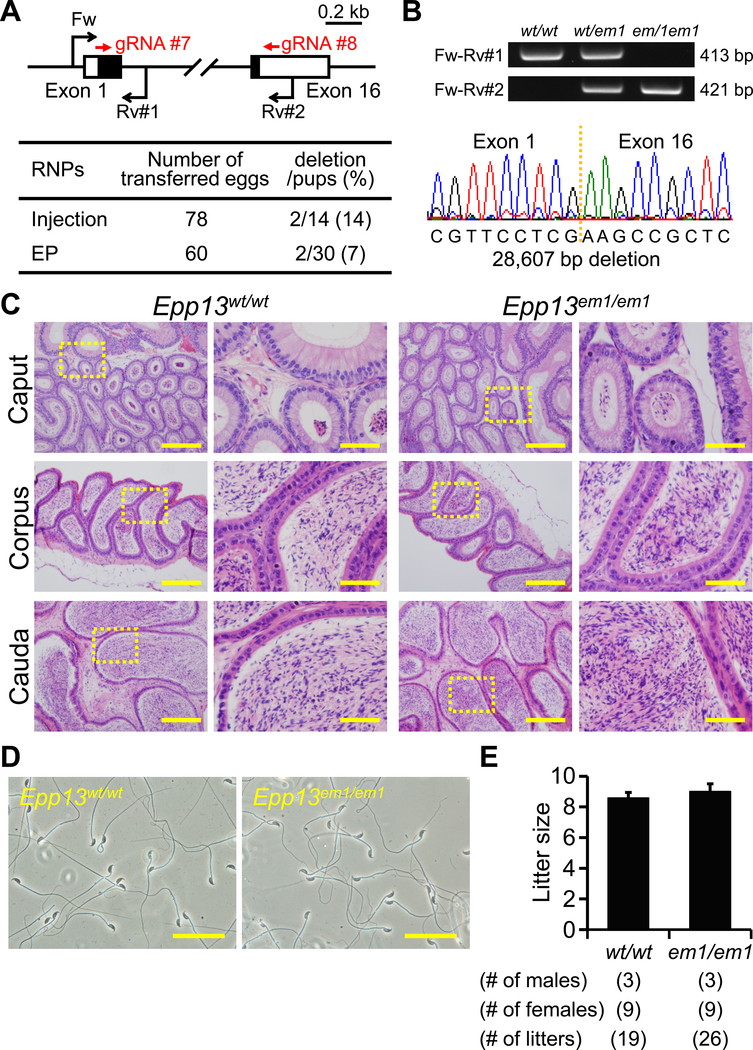
Figure 4. Fecundity of Epp13 KO males.
(A) Genome editing efficiency with gRNA/Cas9 RNPs. Black arrows: primers for genotyping, Black boxes: coding region, (B) Genotyping with PCR and DNA sequencing. Three primers (Fw, Rv#1, and Rv#2) were used for the PCR (also see panel A). em1: 28,607 bp deletion. (C) Histological analysis with H&E staining. Dashed areas in left panels were enlarged (right panels). Scale bars on the left and right panels are 200 μm and 50 μm, respectively. (D) Sperm morphology observed under phase contrast. Scale bars: 50 μm. (E) Male fecundity. There was no difference in average litter size between WT and KO males (p = 0.28).
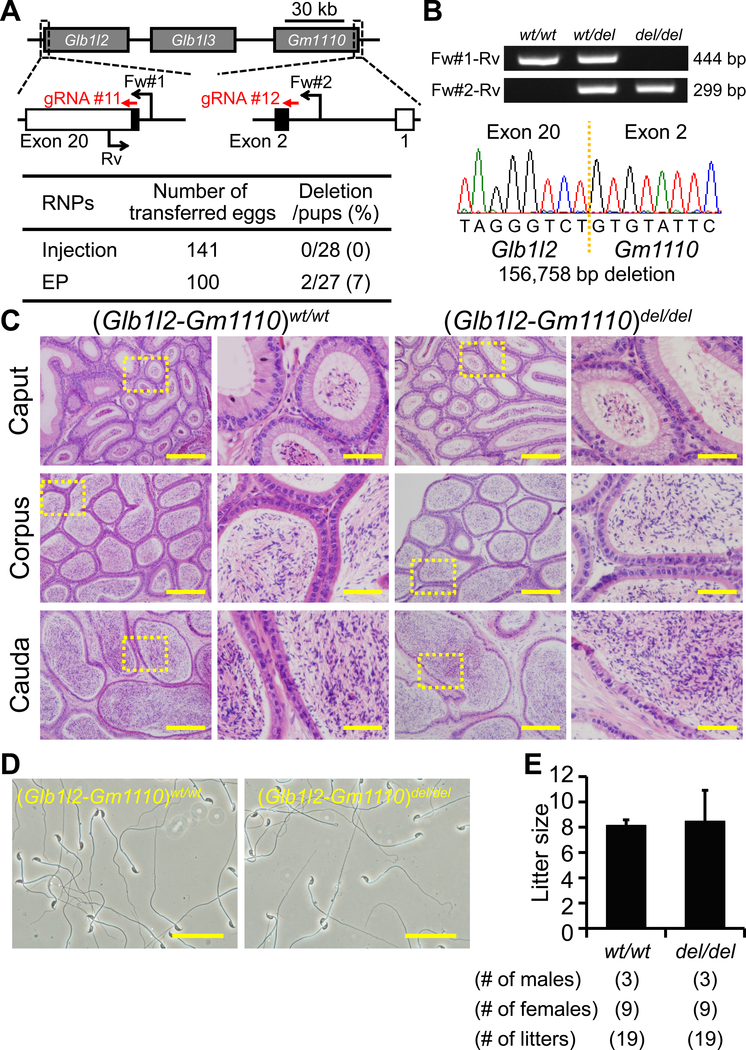
Figure 6. Fecundity of Glb1l2-Gm1110 KO mice.
(A) Genome editing efficiency with gRNA/Cas9 RNPs. Black arrows: primers for genotyping, Black boxes: coding region. (B) Genotyping with PCR and DNA sequencing. Three primers (Fw#1, Fw#2, and Rv) were used for the PCR (also see panel A). em1: 156,758 bp deletion. (C) Histological analysis with H&E staining. Dashed areas in left panels were enlarged (right panels). Scale bars on the left and right panels are 200 μm and 50 μm, respectively. (D) Sperm morphology observed under phase contrast. Scale bars: 50 μm. (E) Male fecundity. There was no difference in average litter size between WT and KO males (p = 0.83).
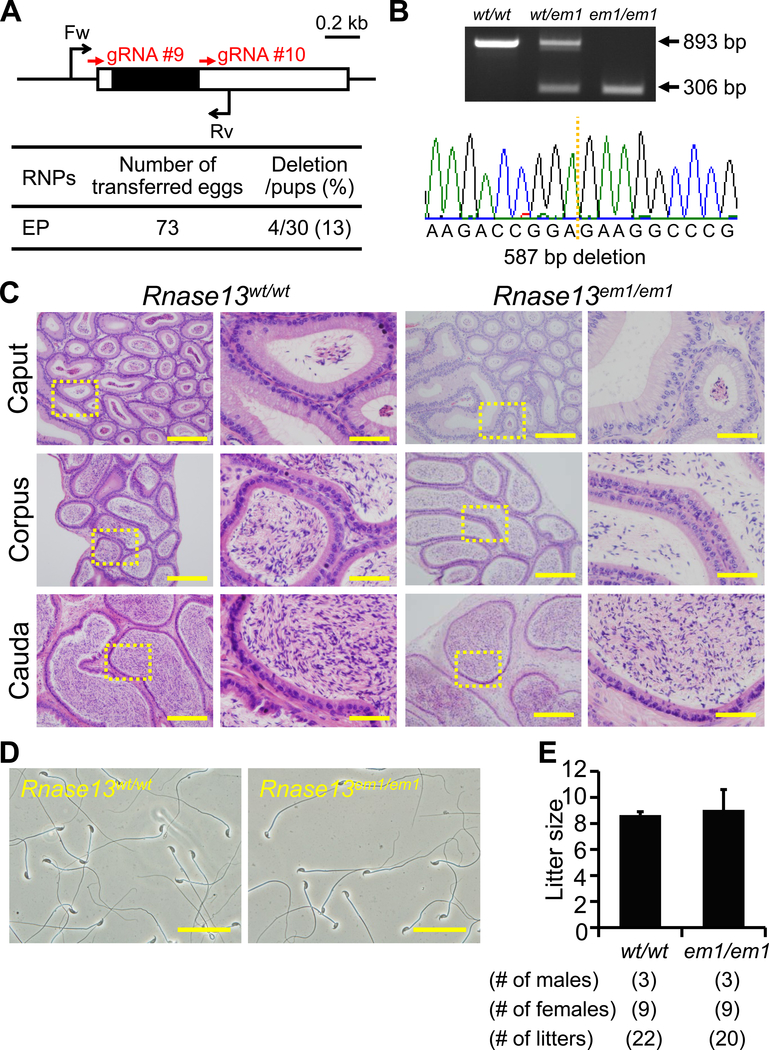
Figure 5. Fecundity of Rnase13 KO males.
(A) Genome editing efficiency with gRNA/Cas9 RNPs. Black arrows: primers for genotyping, Black boxes: coding region. (B) Genotyping with PCR and DNA sequencing. Two primers were used for PCR (also see panel A). em1: 587 bp deletion, (C) Histological analysis with H&E staining. Dashed areas in left panels were enlarged (right panels). Scale bars on the left and right panels are 200 μm and 50 μm, respectively. (D) Sperm morphology observed under phase contrast. Scale bars: 50 μm. (E) Male fecundity. There was no difference in average litter size between WT and KO males (p = 0.51).
RESULTS
Selection of the target genes
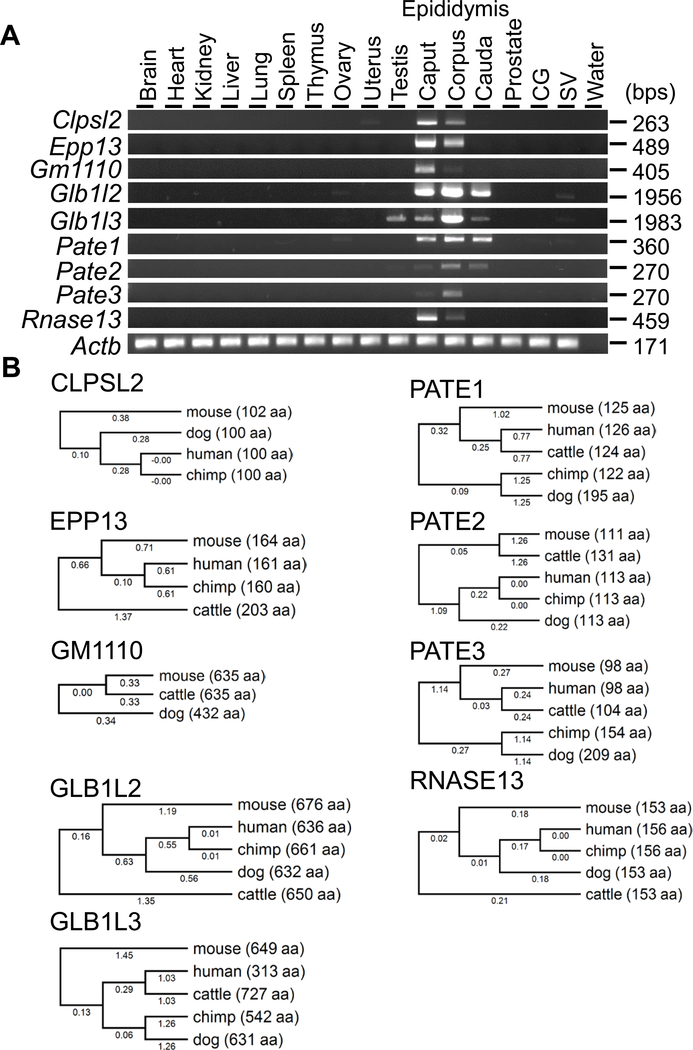
We selected nine genes abundantly expressed in the epididymis and reconfirmed their expression by RT-PCR (Figure 1A). While Glb1l3 showed detectable expression in testis, the remaining genes showed epididymis-enriched expression. Clpsl2, Epp13, Gm1110, and Rnase13 were strongly expressed in the caput region, while Glb1l2, Glb1l3, Pate2, and Pate3 were strongly expressed in the corpus region, and Pate1 was expressed in all regions at comparable levels. It should be noted that all genes examined in the study, except Gm1110, are conserved in human and various species (Figure 1B). Because Gm1110, Glb1l2, and Glb1l3 located closely on the same chromosome and show high similarity in amino acid sequence (Figure S1), we decided to generate a null mice without these three genes.
Figure 1. Tissue expression analysis and conservation of target genes between some mammalian species.
(A) RT-PCR analysis. Actin beta (Actb) was used as the control. (B) Phylogenetic trees. The values under branches and parentheses show the distances and the lengths of the amino acid sequences, respectively.
Fertility of Pate family knockout mice
Four and thirteen Pate family genes form a cluster on human chromosome 11 and mouse chromosome 9 (Levitin et al., 2008), respectively. While Pate4 is abundantly expressed in mouse seminal vesicles and not expressed in the epididymis (Noda et al., 2018), Pate1, 2, and 3 are exclusively expressed in the epididymis (Figure 1A). Because we reported that Pate4 KO mice are subfertile due to an impaired vaginal plug formation and semen leakage (Noda et al., 2018), we knocked out the remaining members to determine their roles in fertility. We attempted to delete the region covering all Pate1, Pate2, and Pate3, by injecting sgRNA/Cas9 co-expressing plasmids into zygotes (Figure 2A). Although we could not delete the region [deletion/pups = 0/4 (sgRNA #1 + #3), 0/6 (sgRNA #1 + #4), Figure 2B], we obtained F0 indel mutant mice in Pate1 and Pate3 independently [Pate1: indel/pups = ¼ (sgRNA #1 + #3), 2/6 (sgRNA #1 + #4), Pate3: indel/pups = 0/4 (sgRNA #1 + #3), indel/pups = 5/6 (sgRNA #1 + #4), Figure 2B]. We additionally generated Pate2 F0 mutant mice [Pate2: indel/pups = ¼ (sgRNA #2), Figure 2B]. Because small indels are difficult to detect by PCR and subsequent agarose gel electrophoresis (1 bp insertion in Pate1, 4 bp deletion in Pate2, 1 and 5 bp deletion in Pate3), we genotyped mutant mice by sequencing PCR amplicons (Figure 2C). KO mice were obtained by heterozygous F1 intercrosses. All the KO mice produced spermatozoa with normal morphology as shown in Figure 2D. When we examined 3 males for each KO lines, all KO males showed normal fecundity [littersize: 9.3 ± 0.7 for Pate1em1/em1, 8.8 ± 0.8 for Pate2em1/em1, and 7.9 ± 3.1 for Pate3em1/em2, while control showed 8.5 ± 2.2 (Figure 2E)]. These results show that Pate1, 2, 3 are dispensable for the male fecundity.
Fecundity of colipase like 2 (Clpsl2) knockout mice
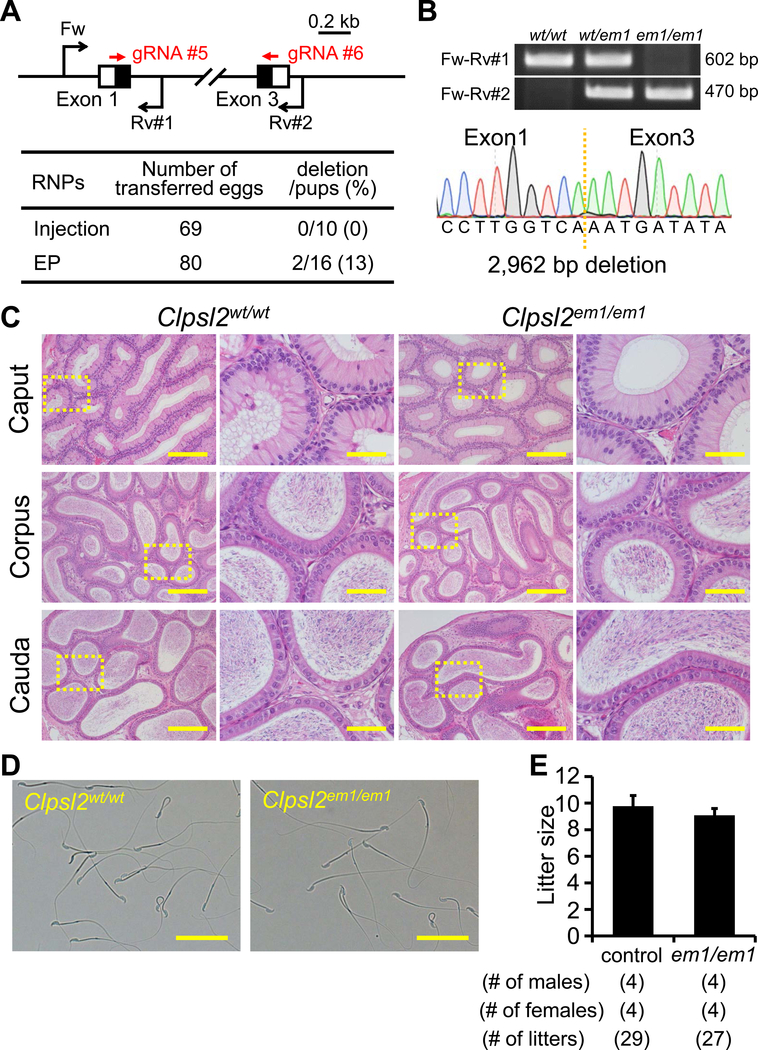
CLPSL2 is conserved between mice and humans. Using RNAi methods, a previous paper suggested that Clpsl2 is required for sperm maturation (Lu et al., 2018). To evaluate these findings and determine the essential roles of CLPSL2 in the complete absence of the protein, we generated KO mice lacking the gene. Hereafter, we applied both sgRNA/Cas9 co-expressing plasmids and gRNA/Cas9 ribonucleoprotein (RNP) because the genome editing efficiency is comparable in various cell lines (Liang et al., 2015). We also applied a zygote electroporation system because many eggs can be treated in a shorter time. When we injected 2 gRNA/Cas9 RNPs into zygotes, we could not obtain any F0 mutant mice (deletion/pups = 0/10); however, by electroporation, we could obtain F0 deletion mutants (deletion/pups = 2/16) (Figure 3A). The deletion of the coding region was confirmed by sequencing the PCR amplicons (Figure 3B). KO mice were obtained by heterozygous F1 intercrosses. Histological analysis of the epididymis and analysis of sperm morphology failed to reveal any apparent differences between Clpsl2em1/em1 and control males (Figure 3C and D). The Clpsl2em1/em1 males showed normal fecundity (litter size: 9.8 ± 0.8 for controls, 9.1 ± 0.5 for Clpsl2em1/em1, Figure 3E). Thus, Clpsl2 is not required for male fecundity.
Figure 3. Fecundity of Clpsl2 KO males.
(A) Genome editing efficiency with gRNA/Cas9 RNPs. Black arrows: primers for genotyping, Black boxes: coding region, EP: electroporation. (B) Genotyping with PCR and DNA sequencing. Three primers (Fw, Rv#1, and Rv#2) were used for the PCR (also see panel A). em1: 2,962 bp deletion. (C) Histological analysis with H&E staining. Dashed areas in left panels were enlarged (right panels). Scale bars on the left and right panels are 200 μm and 50 μm, respectively. (D) Sperm morphology observed under phase contrast. Scale bars: 50 μm. (E) Male fecundity. There was no difference in average litter size between WT and KO males (p = 0.20).
Fertility of Epp13 knockout mice
Epp13 is conserved between mice and humans (also known as EDDM13). We generated Epp13 F0 mutant mice with pronuclear injection and electroporation of gRNA/Cas9 RNPs (injection: deletion/pups = 2/14, electroporation: deletion/pups = 2/30) (Figure 4A). The complete deletion of the coding region was confirmed by PCR and direct sequencing (Figure 4A and B). KO mice were obtained by heterozygous F1 intercrosses. There were no apparent differences between Epp13em1/em1 and control males in histology of epididymis and sperm morphology (Figure 4C and D). The Epp13em1/em1 males showed normal fecundity (litter size: 8.6 ± 0.3 for controls, 9.0 ± 0.5 for Epp13em1/em1, Figure 4E). Thus, Epp13 is not required for male fecundity.
Fertility of Rnase13 knockout mice
Rnase13, conserved between mice and humans, is a member of the ribonuclease A superfamily members (Cho et al., 2005), of which some members, Rnase9 and Rnase10, have been shown to be required for sperm maturation (Krutskikh et al., 2012; Westmuckett et al., 2014). We generated Rnase13 F0 mutant mice with 2 gRNAs (deletion/pups = 4/30) (Figure 5A). The deletion of the coding region was confirmed by PCR and direct sequencing (Figure 5B). KO mice were obtained by heterozygous F1 intercrosses. There were no apparent differences between Rnase13em1/em1 and control males in histology of epididymis and sperm morphology (Figure 5C and D). The Rnase13em1/em1 males showed normal fecundity (litter size: 8.6 ± 0.2 for controls, 9.0 ± 1.6 for Rnase13em1/em1, Figure 5E). Thus, Rnase13 is not required for male fecundity.
Fertility of mice lacking Glb1l2, Glb1l3, and Gm1110
Glb1l2 and Glb1l3 are categorized as galactosidases. Gm1110 is located next to Glb1l3 on mouse chromosome 9 (Figure 6A) (Zhen et al., 2009). The homology rates between GLB1L2, GLB1L3, and GM1110 are high at the amino acid level (Figure S1, Gm1110 vs GLB1L2: 47.4%, Gm1110 vs GLB1L3: 44.9%), suggesting redundant functions of these genes on sperm maturation. Gm1110 is identified as Glb1l4 in rats and a previous report suggested that GLB1L4 is required for the sperm maturation (Zhen et al., 2009). Thus, we generated 2 gRNAs against Glb1l2 and Gm1110 and deleted all 3 genes (injection: deletion/pups = 0/28, electroporation: deletion/pups = 2/27) (Figure 6A). The deletion was confirmed by PCR and direct sequencing (Figure 6B). KO mice were obtained by heterozygous F1 intercrosses. There were no apparent differences between triple KO (Glb1l2-Gm1110)del/del and control males in histology of epididymis and sperm morphology (Figure 6C and D). The triple KO males showed normal fecundity [litter size: 8.2 ± 0.4 for controls, 8.5 ± 2.4 for (Glb1l2-Gm1110)del/del, Figure 6E]. Thus, Glb1l2, Glb1l3, and Gm1110 are not required for male fecundity.
Discussion
It is known that 4 and 13 genes of the Pate family members exist in human and mouse, respectively, and that Pate1 to Pate4 are conserved in both species (Levitin et al., 2008). In human, PATE1 was localized to the sperm equatorial segment, and the anti-PATE1 antibody treatment decreased sperm motility (Liu et al., 2015). Further, previous studies showed asthenozoospermia patients having reduced levels of PATE1 (Liu et al., 2015), and that single nucleotide polymorphisms were found in idiopathic asthenozoospermia (Zhang et al., 2016). These results suggest that human PATE1 is required for sperm function, but we revealed that mouse Pate1 KO males are fertile (Figure 2D and E). In addition, we showed that mouse Pate2 and Pate3 are dispensable for male fecundity. Because more Pate family genes exist in mouse, we cannot rule out the possibility of gene complementation. Further studies will be required to fully uncover the role of PATE family protein in sperm functions.
Clpsl2 is conserved in mouse and human, and predominantly expressed in the caput epididymis (Figure 1). CLPSL2 from the caput epididymis binds to the sperm acrosome and principal piece of the sperm tail (Lu et al., 2018). The previous study showed that knockdown of mouse Clpsl2 by in vivo lentivirus-based RNAi resulted in decreased sperm motility, acrosome reaction, and sperm count in the cauda epididymis, leading to the male subfertility (Lu et al., 2018). However, we could not observe any defects in Clpsl2 KO (null) males. The discrepancy may be explained by experimental approaches; while KO mice specifically suppressed the target gene, RNAi interferes not only with the target gene, but also genes carrying similar sequences. While one can claim that the gRNA may also have off-target effects, our data support that Clpsl2 is not essential for male fertility.
RNASE9, 10, and 13 in the ribonuclease A superfamily were detected in the epididymis (Penttinen et al., 2003; Westmuckett et al., 2014) and show high homology rates at the amino acid level (Cho et al., 2005). Rnase9 KO males were fertile, but KO spermatozoa showed impaired sperm maturation (Westmuckett et al., 2014). Rnase10 KO males were severely subfertile due to the disruption of ADAM3 processing (Krutskikh et al., 2012). However, we revealed that Rnase13 is dispensable for male fecundity. We cannot rule out the possibility that other members complement RNASE13 function in vivo, so the additional KO approaches will be necessary to address this question. Because RNase A family genes are located on Chromosome 14, two gRNAs mediated deletion approaches as shown in Figure 6 are feasible.
Here, we showed that nine genes, which are abundantly expressed in epididymis and conserved among several mammalian species, are dispensable for sperm maturation and male fecundity. The CRISPR/Cas9 mediated KO approach is a powerful tool to determine whether genes of interests are essential for fertility. Screening for genes through a CRISPR/Cas9 mediated KO approach has accelerated the study of severed genes implicated in reproduction, especially of genes within gene families.
Supplementary Material
Supplemental Figure S1. Multiple sequence alignment with GM1110, GLB1L2, and GLB1L3. The amino acid sequences of mouse GM1110 (XP_006510522), GLB1L2 (XP_006510360), and GLB1L3 (AAI32203) were used for the alignment analysis using Clustal omega.
Acknowledgements
We thank the Biotechnology Research and Development (NPO) for support to generate the mutant mice. And, the authors would like to thank Dr. Julio Castaneda for critical reading of the manuscript.
Funding information
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology (MEXT)/Japan Society for the Promotion of Science (JSPS) KAKENHI grants (JP18K14612 to T.N., JP17H01394 and JP25112007 to M.I.), Japan Agency for Medical Research and Development (AMED) grant (JP18gm5010001), Takeda Science Foundation grants to M.I., the Eunice Kennedy Shriver National Institute of Child Health and Human Development (P01HD087157 and R01HD088412 to M.M.M. and M.I.), the Bill & Melinda Gates Foundation (Grand Challenges Explorations grant OPP1160866 to M.M.M. and M.I.), a Japan Society for the Promotion of Science Overseas Research Fellowships (20170633) to K.N., and a Training Interdisciplinary Pharmacology Scientists (TIPS) Program (T32 GM120011) predoctoral fellowship position to D.J.D.
Footnotes
COI statement:
The authors declare no conflict of interest.
References
- Cho S, Beintema JJ & Zhang J. (2005) The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics 85, 208–220. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA & Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, & Turner TT. (2005). The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod 73, 404–413. [DOI] [PubMed] [Google Scholar]
- Krutskikh AA, Poliandri A, Cabrera-Sharp V, Dacheux JL, Poutanen M & Huhtaniemi I. (2012). Epididymal protein Rnase10 is required for post-testicular sperm maturation and male fertility. FASEB J 26, 4198–4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin F, Weiss M, Hahn Y, Stern O, Papke RL, Matusik R, Nandana SR, Ziv R, Pichinuk E, Salame S, Bera T, Vincent J, Lee B, Pastan I & Wreschner DH. (2008) PATE gene clusters code for multiple, secreted TFP/Ly-6/uPAR proteins that are expressed in reproductive and neuron-rich tissues and possess neuromodulatory activity. J Biol Chem 283, 16928–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang XJ, Potter J. Kumar S, Zou Y, Quintanilla R, Sridharan M, Carte J, Chen W, Roark N, Ranganathan S, Ravinder N & Chesnut JD. (2015) Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208, 44–53. [DOI] [PubMed] [Google Scholar]
- Liu FJ, Liu X, Han JL, Wang YW, Jin SH, Liu XX, Liu J, Wang WT & Wang WJ. (2015) Aged men share the sperm protein PATE1 defect with young asthenozoospermia patients. Hum Reprod 30, 861–869. [DOI] [PubMed] [Google Scholar]
- Lu X, Ding F, Lian Z, Chen L, Cao Z, Guan Y, Chen R, Cai D & Yu Y. (2018) An epididymis-specific secretory protein Clpsl2 critically regulates sperm motility, acrosomal integrity, and male fertility. J Cell Biochem 119, 4760–4774. [DOI] [PubMed] [Google Scholar]
- Mashiko D, Fujihara Y, Satouh Y, Miyata H, Isotani A & Ikawa M. (2013) Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci Rep 3, 3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiko D, Young SA, Muto M, Kato H, Nozawa K, Ogawa M, Noda T, Kim YJ, Satouh Y, Fujihara Y & Ikawa M. (2014) Feasibility for a large scale mouse mutagenesis by injecting CRISPR/Cas plasmid into zygotes. Dev Growth Differ 56, 122–129. [DOI] [PubMed] [Google Scholar]
- Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A et al. (2016). Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc Natl Acad Sci U S A 113, 7704–7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Fujihara Y, Matsumura T, Oura S, Kobayashi S & Ikawa M. (2018) Seminal vesicle secretory protein 7, PATE4, is not required for sperm function but for copulatory plug formation to ensure fecundity. Biol Reprod, [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T, Oji A & Ikawa M. (2017) Genome Editing in Mouse Zygotes and Embryonic Stem Cells by Introducing SgRNA/Cas9 Expressing Plasmids. Methods Mol Biol 1630, 67–80. [DOI] [PubMed] [Google Scholar]
- Penttinen J, Pujianto DA, Sipila P, Huhtaniemi I & Poutanen M. (2003) Discovery in silico and characterization in vitro of novel genes exclusively expressed in the mouse epididymis. Mol Endocrinol 17, 2138–2151. [DOI] [PubMed] [Google Scholar]
- Robaire B & Hermo L. (1988) Efferent Ducts, Epididymis, and Vas Deferens: Structure, Functions, and Their Regulation In: The Physiology of Reproduction (eds Knobil E & Neill JD, pp. 999–1080. Raven Press. [Google Scholar]
- Robaire B, Hinton BT & Orgebin-Crist MC (2006). The Epididymis In: Knobil and Neill’s Physiology of Reproduction. (eds Knobil E & Neill JD, pp. 1071–1148. Elsevier. [Google Scholar]
- Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F & Jaenisch R. (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153, 910–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmuckett AD, Nguyen EB, Herlea-Pana OM, Alvau A, Salicioni AM & Moore KL. (2014) Impaired sperm maturation in RNASE9 knockout mice. Biol Reprod 90, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila CS, Cheng AW, Shi L & Jaenisch R. (2013) One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 154, 1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang QM, Ding XP, Wang T, Mu XM & Chen ZY. (2016) Association of polymorphisms in PATE1 gene with idiopathic asthenozoospermia in Sichuan, China. J Reprod Immunol 118, 54–60. [DOI] [PubMed] [Google Scholar]
- Zhen W, Li P, He B, Guo J & Zhang YL. (2009) The novel epididymis-specific beta-galactosidase-like gene Glb1l4 is essential in epididymal development and sperm maturation in rats. Biol Reprod 80, 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Multiple sequence alignment with GM1110, GLB1L2, and GLB1L3. The amino acid sequences of mouse GM1110 (XP_006510522), GLB1L2 (XP_006510360), and GLB1L3 (AAI32203) were used for the alignment analysis using Clustal omega.