Abstract
Background:
Pertussis is a highly contagious respiratory disease caused by the bacterium Bordetella pertussis (B. pertussis). The infection is difficult to diagnose especially in underserved or resource-limited areas. We developed a low-cost and instrument-free diagnostic method for rapid and accurate detection of B. pertussis on a point-of-care (POC) testing device.
Methods:
We developed a paper/polymer hybrid microfluidic biochip integrated with loop-mediated isothermal amplification (LAMP) method for the rapid and accurate detection of B. pertussis. This microfluidic approach was validated by testing 100 de-identified remnant clinical nasopharyngeal swabs and aspirates, which were confirmed to be either positive or negative for B. pertussis by a validated real-time PCR assay at the Children’s Hospital Los Angeles.
Findings:
The instrument-free detection results could be successfully read by the naked eye within 45 min with a limit of detection (LOD) of 5 DNA copies per well. Our optimized bacterial lysis protocol allowed the direct testing of clinical samples without any complicated sample processing/preparation (i.e. DNA extraction) or the use of any equipment (e.g. centrifuges). The validation of the microfluidic approach was accomplished by testing 100 clinical samples. High sensitivity (100%) and specificity (96%) with respect to real-time PCR were achieved.
Interpretation:
This microfluidic biochip shows great potential for point-of-care disease diagnosis in various venues including schools and physician’s offices, especially in low-resource settings in developing nations.
Funding:
NIH/NIAID under award number R21AI107415, NIH RCMI Pilot Grant, the Philadelphia Foundation, the Medical Center of the Americas Foundation.
Keywords: Pertussis diagnosis, Point-of-care detection, Microfluidic biochip, Whooping cough, Loop-mediated isothermal amplification (LAMP)
1. Introduction
Pertussis, also known as whooping cough, is a highly contagious respiratory disease that is caused by the bacterium Bordetella pertussis (B. pertussis). Despite high vaccination coverage in many countries for more than 50 years, pertussis remains one of the world’s leading causes of vaccine-preventable deaths [1]. A publication modeling pertussis cases and deaths estimated that about 24·1 million pertussis cases and 160,700 deaths in children younger than 5 years in 2014 worldwide [2]. In the U.S., the most recent peak occurred in 2012 (48,277 cases) but other outbreaks of pertussis occurred in 2013–2014 in all 50 states [3].
Pertussis is commonly underdiagnosed because most of the cases present as mild or subclinical infection [4]. Since pertussis-vaccine-induced protection is not permanent, the immunized host is vulnerable to infection later in life [5]. Although pertussis attack rates remain the highest among infants, the incidence in adults and adolescents has risen significantly. Pertussis can easily be mistaken for other respiratory infections such as Respiratory Syncytial Virus (RSV), rhinovirus, Mycoplasma pneumoniae, and Chlamydophila pneumoniae, especially during the winter season [6]. Similar to these infections, pertussis presents with a runny nose, mild cough, and low-grade fever; and only when a cough becomes persistent or prominent do clinicians tend to suspect pertussis. Furthermore, adults and adolescents may not present with any significant symptoms [7]. As pertussis is highly infectious during the acute phase (10 days) of infection, it is of primary importance to detect and confirm suspected cases as soon as possible and to limit contact with high-risk populations such as infants and the elderly, who are more vulnerable to serious infection and complications.
The current technologies for pertussis diagnosis include culture, serology (ELISA), and real-time PCR. However, these technologies have their limitations. For example, culture takes several days and it is effective in only 80% of cases when the specimen is collected during the acute phase [8]. The sensitivity and specificity of ELISA and other serological tests for pertussis diagnosis are low [1,9]. Another challenge for ELISA-based pertussis diagnosis is collecting both acute and convalescent samples to demonstrate the increase in antibody titer [10]. Most real-time PCR assays target the IS481 sequence for detection of B. pertussis since this insertion element is found in high copy number, providing high sensitivity [11]. However, the IS481 sequence also exists in Bordetella holmesii (B. holmesii) and Bordetella bronchiseptica (B. bronchiseptica) genomes, resulting in low specificity [12]. In contrast, real-time PCR targeting the pertussis toxin (PT) promoter region has high specificity but relatively low sensitivity [13]. As a promising isothermal nucleic acid amplification method, loop-mediated isothermal amplification (LAMP), has been developed to amplify the target DNA at a constant temperature in a range of 60–65 °C within 1 h. The high strand displacement activity and processivity of the Bst polymerase (Bacillus stearothermophilus) and identification of 6 distinct regions from 4 different primers in LAMP results in high specificity. Kamachi et al. [13] initially reported a LAMP method for detection of B. pertussis with high sensitivity and specificity. However, like realtime PCR, LAMP also requires laborious and time-consuming sample preparation such as DNA extraction and purification, and specialized instrumentation such as thermal cyclers, centrifuges, fluorescent microscopes, which limits its broad application, especially in resource-limited settings.
The microfluidic lab-on-a-chip technology offers a unique opportunity for various biomedical applications and development of point-of-care testing devices to improve global health [14–18]. We previously developed a microfluidic approach integrated with LAMP for bacterial meningitis diagnosis [19–21]. However, the microfluidic approach hasn’t been validated by testing clinical samples. Herein, we report the development and validation of another paper/polymer hybrid microfluidic device for the rapid and accurate pertussis diagnosis. The results can be easily read by the naked eye under a handheld UV light pen without using any specialized laboratory instruments. This microfluidic diagnostic approach can be used for rapid detection of pertussis and other pathogens at the point of care and in some other low-resource settings.
2. Methods
2.1. Culture and DNA Preparation
B. pertussis (ATCC 9797) was obtained from American Type Culture Collection (ATCC, Rockville, MD), and B. holmesii and B. parapertussis were clinical isolates obtained from the Wisconsin Department of Health Services, which were de-identified. B. pertussis was grown on Bordet-Gengou agar (BD, Sparks, MD, USA) supplemented with 10% casamino acid (Fisher Scientific, USA). B. holmesii and B. parapertussis were grown in 5% sheep blood agar (BD, Sparks, MD, USA). The microorganisms were incubated in an aerobic environment with sufficient humidity at 35 °C for 3-4 days. Genomic DNA from B. pertussis, B. holmesii and B. parapertussis was extracted and purified by using the Qiagen DNA Mini kit (Catalog No. 51304, Valencia, CA, USA) following the protocol from the manufacturer. For each organism, bacterial colonies were transferred into a centrifuge tube containing 5 mL of sterile saline adjusting the turbidity to 0.5 McFarland standard (ProLab Diagnostics, Round Rock, TX, USA) to achieve a cell density of ~1.5 × 108. Cells were harvested by centrifugation in a Beckman centrifuge, using a C0650 rotor with adapters for 15 mL tubes, at 7500 rpm (5000 ×g) for 10 min. Bacterial cell pellets were collected to proceed with the solid-phase extraction of DNA as indicated by the Qiagen protocol. Eluted DNA samples were measured by using Nanodrop (Nanodrop 1000, Thermo Scientific, MA, USA).
2.2. Hybrid Microfluidic Biochip Design
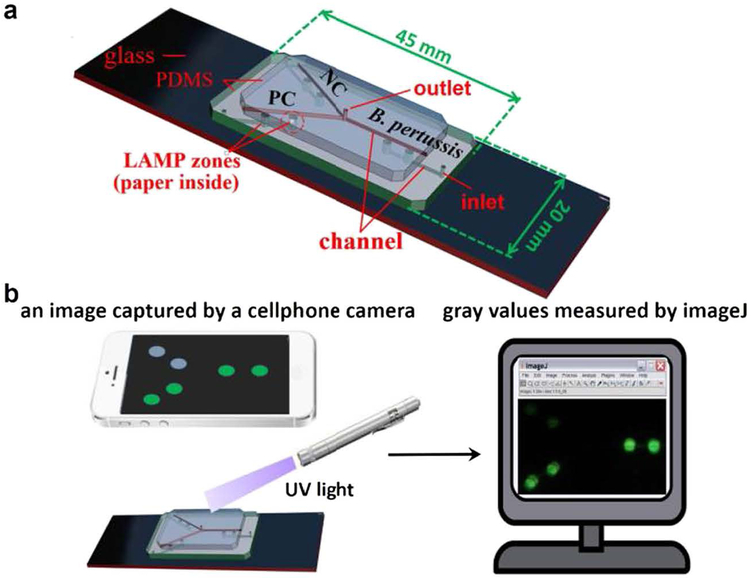
As shown in Fig. 1a, the paper/polymer hybrid microfluidic biochip consists of a top polydimethylsiloxane (PDMS) layer with an outlet reservoir (diameter 1·0 mm, depth 1·5 mm) and microchannels (width 100 μm, depth 100 μm) for reagent delivery; a middle PDMS layer with detection wells (diameter 2·0 mm, depth 1·5 mm) for LAMP reactions and detection of B. pertussis as well as the negative control (NC), 3 inlet reservoirs (diameter 1·0 mm, depth 1·5 mm), and microchannels for reagent delivery as well; and a glass slide for structure support. A paper disk (diameter 2·0 mm) was placed inside each detection wells, serving as a porous 3D substrate for storage and preservation of the pre-loaded specific LAMP primers of B. pertussis.
Fig. 1.
Design of the paper/polymer hybrid microfluidic biochip (a) and schematic of the cellphone-based detection system (b) for POC detection of pertussis. Green spots in (b) indicate positive detection results. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.3. On-chip LAMP Process
Specific LAMP primers of B. pertussis were pre-loaded on a paper disk (diameter 2 · 0 mm) inside each well of the paper/polymer hybrid microfluidic biochip. A 13 μL LAMP reaction buffer with or without samples added was prepared and introduced from each inlet reservoir to their corresponding wells of the biochip for sample test and negative control (NC) respectively. Then the inlet and outlet reservoirs were sealed with Epoxy glue to prevent evaporation. The biochip was heated by using a portable battery-powered heater devised by our laboratory or a water bath at 63 °C for 45 min for LAMP reactions, followed by increasing the temperature to 95 °C for 2 min for the termination of LAMP reactions.
2.4. Instrument five Detection and Confirmatoiy Tests
In the reaction buffer, the fluorescence of the detection dye (i.e. calcein) is initially quenched by manganese ions (Mn2+). During the LAMP reactions, the by-product pyrophosphate ions () form complexes (MnP2O7) with Mn2+, As a result, calcein is free to combine with magnesium ions (Mg2+), resulting in the emission of bright fluorescent signals under UV light [22]. The schematic of the cellphone-based detection system is demonstrated in Fig. 1b. After on-chip LAMP reactions, a portable UV light pen was applied to shine LAMP products on the biochips to obtain the visual detection results. The generated fluorescence could be observed by the naked eye, or captured by a cellular phone camera (e.g. iPhone 5). The cellphone camera captured images were processed with the software lmageJ (NIH) (https://imagej.nih.gov/ij/) to obtain the mean gray value of each well to measure the brightness of the selected area (i.e. LAMP zones). The mean gray value is the sum of the gray values of all the pixels in the selection divided by the number of pixels. It is calculated by converting each pixel to grayscale using the formula gray = 0.299 red + 0.587 green + 0.114 blue. The detection results were confirmed by gel electrophoresis analysis in a 1 · 5% agarose gel in Tris-acetic acid ED’l’A buffer (TAE, pH 8.3) at 90 V for 1 h. Electrophoresis was done using a Sub-Cell GT tank (Catalog No. 1704401, BioRad, CA, USA).
2.5. Direct Detection of Nasopharyngeal Swab Samples and Clinical Samples
We first prepared nasopharyngeal swab samples to mimic the patient samples for evaluating a simple centrifuge-free bacterial lysis approach. The nasopharyngeal swabs were sterile twisted applicator rayon tipped from Copan and rinsed in a saline buffer. B. pertussis bacterial was then spiked into the nasopharyngeal swab. The bacterial density was −1.5 × 108 CFU/mL by adjusting to McFarland Standard 0.5 (Key Scientific Products, TX) [23]. In addition, one hundred clinical samples obtained from pediatric patients presenting with signs and symptoms suspicious for whooping cough between February 1 to May 30, 2015, were included in the study. Only remnant samples were enrolled upon completion of clinical testing and per the Children’s Hospital Los Angeles (CHLA) Institutional Review Board they did not meet the definition of human subjects research per 45 CFR 46.102 (CHLA-15-00014). Briefly, 66 nasopharyngeal swab samples collected in universal transport medium (UTM, BD Biosciences, Franklin Lakes, NJ) and 34 nasal aspirates collected in sterile containers were initially tested for B. pertussis and B. parapertussis using the Simplexa Bordetella Universal Direct kit (DiaSorin Molecular, Cypress, CA) within 3 days of collection. This is a real-time PCR assay using scorpion primers that target the well-conserved region of the IS481 (86 bp) (B. pertussis) and the IS1001 (73 bp) (B. parapertussis) genes. An endogenous internal control is also used to monitor the extraction process and to detect PCR inhibition. The performance of the assay was validated at CHLA with a limit of detection of 75 CFU/mL for B. pertussis and 300 CFU/mL for B. parapertussis. As part of the validation, analytical sensitivity, analytical specificity, accuracy, and precision were determined and deemed to be highly acceptable. A past study reported a sensitivity of 97% compared to a laboratory-developed assay [24]. In addition, no cross-reactivity with other genetically similar organisms was identified. Upon completion of testing, samples were stored at — 80 °C in a freezer that is continuously monitored for temperature deviations. The 100 clinical samples enrolled in the study were continuously collected within a 4 month (February 1 to May 30,2015) period, regardless of gender, age or ethnicity at CHLA to avoid bias. Only samples with insufficient volume were excluded from the study. The positive samples had Ct values ranging from 27 to 38. The nasopharyngeal swab samples and clinical samples were directly tested using a simple centrifuge-free bacterial lysis approach: 3 μL of the nasopharyngeal swab samples or clinical samples was mixed with 3 μL bacterial lysis buffer (50 mM Tris buffer, pH 7·5, 4 M Urea and 0·1% Triton), and incubated at room temperature for about 10 min. Then 1 μL of the lysate was directly added to the LAMP reaction mixture for the subsequent on-chip LAMP reaction.
2.6. Statistical Analysis
Sensitivity was calculated as to measure the proportion of actual positives that were correctly identified. Specificity was calculated as to measure the proportion of actual negatives that were correctly identified. Overall agreement was calculated as . We used extract binomial confidence intervals to provide the 95% confidence interval (CI) for sensitivity, specificity and overall agreement.
3. Results
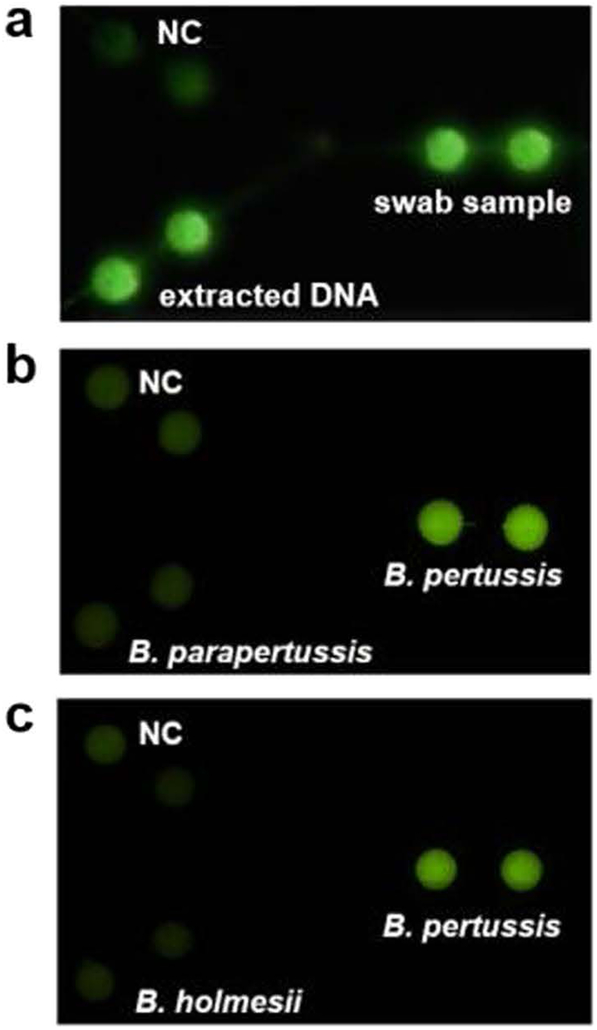
We first tested the feasibility of the paper/polymer hybrid microfluidic biochip for direct detection of B. pertussis bacteria using nasopharyngeal swab samples and our centrifuge-free lysis buffer. Fig. 2a shows the detection results. Strong fluorescence was obtained for both the nasopharyngeal swab sample and the B. pertussis extracted DNA sample (7·5 × 104 copies/μL), without noticeable differences observed. Their signals were much higher than the NC. These results demonstrated that the centrifuge-free lysis procedure was completely compatible with the LAMP reactions, without noticeable inhibition observed. Additionally, the biochip effectively discriminates B. pertussis from two other closely related Bordetella species, B. parapertussis and B. holmesii (Fig. 2b and c), demonstrating that our approach is highly specific for the detection of B. pertussis.
Fig. 2.
Direct detection of nasopharyngeal swab samples and specificity test. (a) Fluorescence image of LAMP products from both the B. pertussis extracted template DNA and nasopharyngeal swab sample spiked with B. pertussis bacteria. (b–c) Fluorescence images for the specificity test among B. pertussis, B. parapertussis (b) and B. holmesii (c). In the specificity test, all the detection wells were preloaded with B. pertussis primers; the reaction mixtures with different DNA samples of B. pertussis, B. parapertussis, and B. holmesii (5 × 106 copies per well), as well as the NC (without template DNA), were introduced into different detection wells separately.
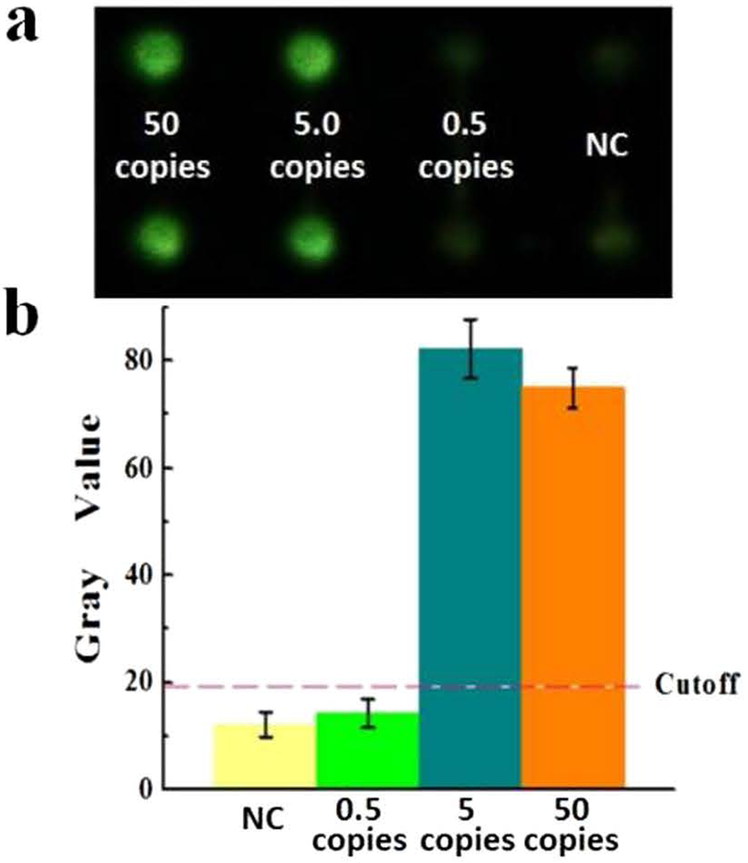
The limit of detection (LOD) was investigated by testing a series of 10-fold dilutions of B. pertussis DNA samples. The initial copy number of DNA template loaded was 50, 5, and 0·5 copies per well, respectively. As shown in Fig. 3a, strong fluorescence of the LAMP products was observed even when the initial DNA template was as low as 5 copies per reaction well. However, when the initial DNA template was less than 5 copies, the fluorescence of the LAMP products was as dim as the NC. The image was further processed by using the software ImageJ to obtain the gray values. The gray values of the LAMP products from 5 copies of initial DNA template were much higher than the cutoff gray value as shown by the dashed line in Fig. 3b, which was calculated on the basis of 3-fold standard deviations of the mean gray value of the NC. Therefore, we concluded that the LOD of the on-chip LAMP approach for detection of B. pertussis was as low as 5 DNA copies per detection well (5 CFU/reaction).
Fig. 3.
LOD investigation. (a) Fluorescence image of LAMP products using B. pertussis template DNA ranging from 50, 5 and 0·5 copies per well, as well as the NC. (b) Corresponding gray values measured by ImageJ. The error bars represent the standard deviation, n = 6.
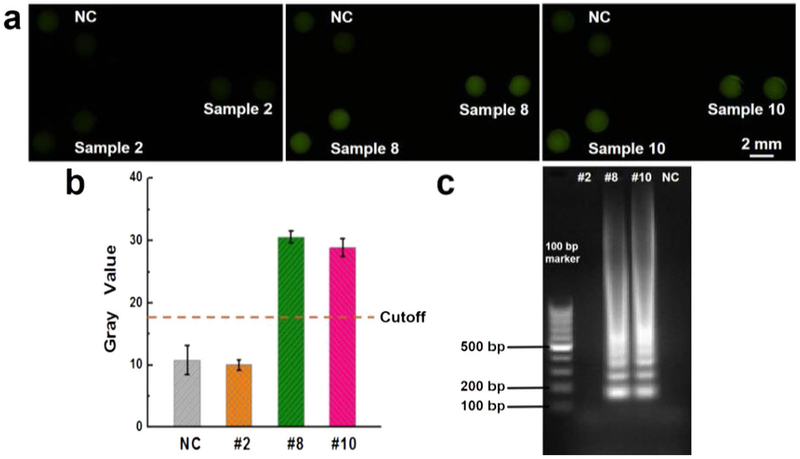
We further investigated the capability of the microfluidic biochip for direct detection of clinical samples and the results of three randomly selected pertussis clinical samples (i.e. #2, #8, and #10) were shown in Fig. 4. According to the real-time PCR test, sample #2 is negative, and samples #8 and #10 are positive. The captured fluorescence images in Fig. 4a shows that samples #8 and #10 exhibited bright green fluorescence while sample #2 and NC only showed very weak fluorescence. As shown in Fig. 4b, the gray values of the samples #8 and 10 were well above the cutoff line and were ~3 folds higher than #2 and NC. In addition, the biochip has versatile functionality, as different on-chip LAMP amplicons can be further separately collected into PCR tubes for other conventional confirmatory tests. Fig. S1 shows similar detection results using those extracted samples in separate PCR tubes placed under the portable UV light pen. The collected LAMP products were also applied for gel electrophoresis analysis to confirm the results, as shown in Fig. 4c. No DNA bands from the LAMP products of sample #2 were shown. The multiple ladder-pattern DNA bands from LAMP products of samples #8 and #10 confirmed the success of the on-chip LAMP reaction.
Fig. 4.
Clinical sample test. (a) Fluorescence images of LAMP products from samples #2, #8 and #10, as well as the NC. (b) Gray values measured by ImageJ; (c) Gel electrophoresis confirmation. The error bars represent the standard deviation, n = 8. (For interpretation of the references to color in this figure, the reader is referred to the web version of this article.)
To validate the paper/PDMS hybrid microfluidic biochip for accurate pertussis diagnosis, a total of 100 clinical samples were further tested. NC (no template DNA) was included in each run. The sensitivity and specificity were calculated by using real-time PCR as a reference assay. As shown in Table 1, we found that all the 53 samples that were tested positive by real-time PCR were also positive by our method. Of the 47 samples that were negative by real-time PCR, 45 were confirmed by the biochip; an additional 2 samples were found to be positive by biochip. Within those 53 samples, one was initially negative by real-time PCR results but positive using our biochip. Upon repeat testing by real-time PCR, the sample was confirmed to be positive, verifying the sensitivity of our biochip. Test sensitivity and specificity were calculated by using real-time PCR as the reference method, which were 100·0% and 95·7%, respectively. The overall agreement was found to be 98·0%. The relevant calculation equations are shown in the Supplementary material.
Table 1.
Results of clinical samples from the microfluidic LAMP test and the real-time PCR test.a
| Real-time PCR (+) | Real-time PCR (−) | No. of clinical samples | Sensitivity, 95% CI (%) | Specificity, 95% CI (%) | Overall agreement, 95% CI (%) | |
|---|---|---|---|---|---|---|
| On-chip LAMP (+) | 53 | 2 | 100 | 100 · 0, | 95 · 7, | 98 · 0, |
| On-chip LAMP (−) | 0 | 45 | [93 · 3–100 · 0] | [85 · 5–99 · 5) | [93 · 0–99 · 8] |
Assuming real-time PCR results (not a gold standard method) are correct.
4. Discussion
The low-cost, instrument-free paper/polymer hybrid microfluidic biochip integrated with LAMP was successfully validated as a reliable tool for rapid and accurate diagnosis of pertussis. The total cost per assay was ~3 dollars (Table S3 and S4). The whole assay procedures were less than 1 h, and no special instruments such as thermal cyclers, centrifuges, or detection facilities were required.
The hybrid microfluidic biochip takes advantages of both the polymer and paper substrates. The polymer substrate is transparent, providing easy visual detection. It is also easy to fabricate and has high performance in liquid manipulation. The paper substrate has a high surface-to-volume ratio property, which makes it an ideal 3D porous substrate for storage and preservation of the pre-loaded primers. Our previous study demonstrated that the hybrid biochips could enable a stable diagnostic performance for >3 months at room temperature compared to paper-free non-hybrid biochips [20].
This is the first low-cost POC biochip for rapid and accurate pertussis diagnosis that has been validated using clinical samples. The successful direct detection of clinical samples indicated that our centrifuge-free bacterial and cell lysis approach was very efficient and totally compatible with our on-chip LAMP reactions (Figs. 2a and 4). Thus, it avoided complicated and time-consuming sample preparation process (i.e. DNA extraction), as well as the use of special facilities in a laboratory (e.g. centrifuge, and water bath). The LAMP methodology was also proved less subjected to inhibitors from clinical samples compared to the PCR method (Fig. S2) [25].
LAMP requires identification of 6 distinct regions of a target DNA sequence by using at least 4 different primers, resulting in high specificity. By simultaneous testing of B. pertussis with its similar species B. parapertussis and B. holmesii, we found that our method was very specific for B. pertussis detection (Fig. 2b and c). In contrast, real-time PCR assays, usually target the IS481 sequence, cannot specifically identify and distinguish B. pertussis from B. holmesii since they all have the IS481 sequence [11,26]. Moreover, given that the LOD of 5 DNA copies per well has been achieved within 45 min (Fig. 3), our method overcomes lengthy assay time and low-sensitivity issues in conventional methods for pertussis diagnosis. For instance, the culture usually takes several days and has low detection sensitivity, and requires collecting specimens during the acute phase [8]. The sensitivity and specificity of ELISA and other serological tests require collecting both acute and convalescent samples, and have low sensitivity and specificity [10,27]. Compared with those current diagnostic methods (Table S4), our method can achieve both high specificity and high sensitivity for rapid pertussis diagnosis.
We further compared our results with real-time PCR results (Table 1). The real-time PCR assay developed at CHLA was thoroughly validated on >100 positive and negative clinical specimens and demonstrated sensitivity, specificity, and agreement of 100%. Our findings showed 2 samples with conflicting results, which were negatives according to the real-time PCR assay but positives according to our LAMP-biochip method. LAMP amplification systems have been reported to be more sensitive assays than real-time PCR by producing a higher amount of amplicons (>103 folds) [28,29]. Our study demonstrated the high sensitivity of our approach identifying B. pertussis with a LOD as low as 5 DNA copies per well (Fig. 3). Therefore, we concluded that the 2 positive samples, detected by our biochip, might be weak positives due to the higher sensitivity of our on-chip LAMP method, However, we couldn’t be able to confirm the diagnostic results of these two conflicting samples because the limited amounts of the samples were not enough for the tests by using other methods. The overall agreement in terms of specificity (96%) and sensitivity (100%) indicated that our test has high accuracy. The results are comparable to those obtained by the real-time PCR test, which is however costly and has to be conducted in well-equipped laboratories [30].
This hybrid microfluidic biochip integrated with LAMP provides a POC testing platform for fast and accurate pertussis diagnosis that can be used in physician’s offices, schools, health centers, and it is ideal for limited resource settings in developing nations. Moreover, we are working on developing another POC biochip for simultaneous identification of multiple Bordetella species. This microfluidic diagnostic approach has the potential for broad applications in POC testing of various pathogens in various fields.
Supplementary Material
Research in context.
Evidence Before This Study
We searched PubMed on February 23, 2018, using the terms “loop-mediated isothermal amplification AND pertussis” and found 16 published papers. Another PubMed search with the terms “point-of-care detection AND pertussis” found 6 published papers, 2 of which were related to pertussis diagnostic methods and loop-mediated isothermal amplification method. However, none of those searched papers were related to point-care-of testing devices, microfluidics, or direct detection of clinical samples. Moreover, none of the point-of-care testing devices or microfluidic platforms whether integrated with LAMP methodology or not, have been reported for pertussis diagnosis. Our study is the first point-of-care testing device based on LAMP methods for pertussis diagnosis.
Added Value of This Study
To the best of our knowledge, this is the first study to develop a microfluidic platform integrated with a LAMP method as a point-of-care testing device for pertussis diagnosis. The findings from the study show that the developed hybrid microfluidic biochips were ready for use with robust diagnostic performance. This microfluidic approach laid a solid foundation for point-of-care detection of other bacteria (e.g. foodborne pathogens) as well as its applicability in other fields such as food industry, agriculture, environmental analysis, etc.
Implications of All the Available Evidence
Accurate diagnosis of pertussis requires laboratory testing, which is not accessible to everyone. The LAMP assay on this hybrid microfluidic biochip that we developed provides a low-cost and instrument-free POC testing platform for fast and accurate detection of B. pertussis. The biochip can be used in physician’s offices, health care centers, schools, and daycares. Particularly, the biochip is ideal for limited resource settings.
Acknowledgments
We would like to acknowledge the financial support from the National Institute of Allergy and Infectious Disease of the NIH (R21AI107415), the Philadelphia Foundation, the Medical Center of the Americas Foundation, and the U.S. NSF-PREM program (DMR 1205302 and 1827745). Financial support from the National Institute of General Medical Sciences of the NIH (SC2GM105584), the NIH RCMI Pilot grant, the NIH BUILDing Scholar Summer Sabbatical Award (NIGMS Award Numbers RL5GM118969, TL4GM118971, and UL1GM11897), the University of Texas at El Paso (UTEP) for the IDR Program, and University of Texas System for the STARS award is also greatly acknowledged. They had no influence in study design, data collection, data analysis, interpretation, or writing of the report. We would like to express special thanks to the staff of the Genomic Analysis Core Facility of UTEP. This core is supported by Grant G12MD007592 from the National Institutes on Minority Health and Health Disparities (NIMHD).
Footnotes
Conflict of Interests
XJL and DCD received a grant from the National Institutes of Health (NIH), USA. XJL, MD and DCD filed patent US #15/701,886 related to the pertussis LAMP assays used in this study. NM, FS and JDB declare no competing interests.
Appendix A. Supplementary Data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eclinm.2019.02.008.
References
- [1].Kilgore PE, Salim AM, Zervos MJ, Schmitt H-J. Pertussis: microbiology, disease, treatment, and prevention. Clin Microbiol Rev 2016;29(3):449–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Yeung KHT, Duclos P, Nelson EAS, Hutubessy RCW. An update of the global burden of pertussis in children younger than 5 years: a modelling study. Lancet Infect Dis 2017;17(9):974–80. [DOI] [PubMed] [Google Scholar]
- [3].CDC. Centers for Disease Control and Prevention, about pertussis outbreaks, https://www.cdc.gov/pertussis/outbreaks/abouthtml; 2017.
- [4].Singh M, Lingappan K. Whooping cough: the current scene. Chest 2006; 130(5):1547–53. [DOI] [PubMed] [Google Scholar]
- [5].Dominguez D The reemergence of pertussis in immunized populations: a case study. Clin Lab Sci 2005;18(4):233–7. [PubMed] [Google Scholar]
- [6].Mahon CR, Donald CL, Manuselis G. Textbook of diagnostic microbiology.. 4th ed. Maryaland Height MO: W.B. Saunders Co.; 2011. [Google Scholar]
- [7].Munoz FM. Pertussis in infants, children, and adolescents: diagnosis, treatment and prevention. Semin Pediatr Infect Dis 2006;17(1):14–9. [DOI] [PubMed] [Google Scholar]
- [8].Dragsted DM, Dohn B, Madsen J, Jensen JS. Comparison of culture and PCR for detection of Bordetella pertussis and Bordetella parapertussis under routine laboratory conditions. J Med Microbiol 2004;53(8):749–54. [DOI] [PubMed] [Google Scholar]
- [9].Weinberger R, Riffelmann M, Kennerknecht N, et al. Long-lasting cough in an adult German population: incidence, symptoms, and related pathogens. Eur J Clin Microbiol Infect Dis 2018;37(4):665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cherry JD, Grimprel E, Guiso N, Heininger U, Mertsola J. Defining pertussis epidemiology: clinical, microbiologic and serologic perspectives. Pediatr Infect Dis J 2005;24 (5):S25–34. [DOI] [PubMed] [Google Scholar]
- [11].Anderson TP, Beynon KA, Murdoch DR. Comparison of real-time PCR and conventional hemi-nested PCR for the detection of Bordetella pertussis in nasopharyngeal samples. Clin Microbiol Infect 2003;9(7):746–9. [DOI] [PubMed] [Google Scholar]
- [12].Roorda L, Buitenwerf J, Ossewaarde JM, van der Zee A. A real-time PCR assay with improved specificity for detection and discrimination of all clinically relevant Bordetella species by the presence and distribution of three insertion sequence elements. BMC Res Notes 2011;4(1):11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kamachi K, Toyoizumi-Ajisaka H, Toda K, et al. Development and evaluation of a loop-mediated isothermal amplification method for rapid diagnosis of Bordetella pertussis infection. J Clin Microbiol 2006;44(5):1899–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sanjay ST, Fu G, Dou M, et al. Biomarker detection for disease diagnosis using cost-effective microfluidic platforms. Analyst 2015;140(21):7062–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Dou M, Sanjay ST, Benhabib M, Xu F, Li X. Low-cost bioanalysis on paper-based and its hybrid microfluidic platforms. Talanta 2015;145:43–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Dou M, Lopez J, Rios M, et al. A fully battery-powered inexpensive spectrophotometric system for high-sensitivity point-of-care analysis on a microfluidic chip. Analyst 2016;141(12):3898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sanjay ST, Dou M, Sun J, Li X. A paper/polymer hybrid microfluidic microplate for rapid quantitative detection of multiple disease biomarkers. Sci Rep 2016;6:30474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Sanjay ST, Dou M, Fu G, Xu F, Li X. Controlled drug delivery using microdevices. Curr Pharm Biotechnol 2016; 17(9):772–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Dou M, Dominguez DC, Li X, Sanchez J, Scott G. A versatile PDMS/paper hybrid microfluidic platform for sensitive infectious disease diagnosis. Anal Chem 2014; 86(15):7978–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dou M, Sanjay ST, Dominguez DC, Liu P, Xu F, Li X. Multiplexed instrument-free meningitis diagnosis on a polymer/paper hybrid microfluidic biochip. Biosens Bioelectron 2017;87:865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dou M, Sanjay ST, Dominguez DC, Zhan S, Li X. A paper/polymer hybrid CD-like microfluidic SpinChip integrated with DNA-functionalized graphene oxide nanosensors for multiplex qLAMP detection. Chem Commun 2017;53(79):10886–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat Protoc 2008;3(5):877. [DOI] [PubMed] [Google Scholar]
- [23].de Almeida Gomes BPF, Vianna ME, Sena NT, Zaia AA, Ferraz CCR, de Souza Filho FJ. In vitro evaluation of the antimicrobial activity of calcium hydroxide combined with chlorhexidine gel used as intracanal medicament. Oral Surg Oral Med Oral Pathol Oral Radiol Endod 2006; 102(4):544–50. [DOI] [PubMed] [Google Scholar]
- [24].Lanotte P, Plouzeau C, Burucoa C, et al. Evaluation of four commercial real-time PCR assays for detection of Bordetella spp. in nasopharyngeal aspirates. J Clin Microbiol 2011;49(11):3943–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Notomi T, Mori Y, Tomita N, Kanda H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. J Microbiol 2015;53(1):1–5. [DOI] [PubMed] [Google Scholar]
- [26].Williams MM, Taylor TH, Warshauer DM, Martin MD, Valley AM, Tondella ML. Harmonization of Bordetella pertussis real-time PCR diagnostics in the United States in 2012. J Clin Microbiol 2015;53(1):118–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Orenstein WA. Pertussis in adults: epidemiology, signs, symptoms, and implications for vaccination. Clin Infect Dis 1999;28(Supplement 2):S147–50. [DOI] [PubMed] [Google Scholar]
- [28].Meng S, Xu J, Xiong Y, Ye C. Rapid and sensitive detection of Plesiomonas shigelloides by loop-mediated isothermal amplification of the hugA gene. PLoS One 2012;7(10):e41978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Zhao X, Lin C-W, Wang J, Oh DR Advances in rapid detection methods for foodborne pathogens. J Microbiol Biotechnol 2014;24(3):297–312. [DOI] [PubMed] [Google Scholar]
- [30].Suntarattiwong P, Kanjanabura K, Laopipattana T, Kerdsin A, Paveenkittiporn W, Chotpitayasunondh T. Pertussis surveillance in a children hospital in Bangkok, Thailand. Int J Infect Dis 2019;81:43–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.