INTRODUCTION
Activation of the fibroblast growth factor receptor (FGFR) signaling pathway is commonly involved in carcinogenesis,1 and oncogenic molecular alterations in the FGFR pathway occur across various tumor types.2 Agents that target FGFR have been demonstrated to inhibit tumor growth and angiogenesis—and, in some cases, to reverse acquired resistance to anticancer agents—in patients with FGFR alterations in urothelial carcinoma,3,4 cholangiocarcinoma,5 glioblastoma,6 and non–small-cell lung cancer (NSCLC).7 Here, we present the first report, to our knowledge, of a patient with head and neck squamous cell carcinoma (HNSCC) with FGF amplifications who achieved a complete response to an FGFR inhibitor.
CASE PRESENTATION
A 49-year-old man from Kuwait was diagnosed February 2012 with regionally advanced carcinoma of the right anterior tongue and floor of the mouth. A positron emission tomography–computed tomography (PET-CT) scan in April 2012 demonstrated extensive malignancy in the oral cavity with bilateral level IB node metastases and a suggestive fluorodeoxyglucose-avid sub-cutaneous lesion posterior to the sixth cervical spinous process. Surgical biopsy of the tumor in the mouth demonstrated invasive squamous cell carcinoma with basaloid features and a ≥ 1-mm depth of invasion. The subcutaneous lesion was suspicious for metastasis, but no biopsy was performed. Staging was T4aN2cM1. The patient was not tested for human papillomavirus infection. He had a smoking history of 60 pack-years and denied alcohol use, and he had no family history of cancer.
The patient received induction chemotherapy with docetaxel, cisplatin, and fluorouracil. After two cycles, he experienced mild-to-moderate hearing loss and received one cycle of docetaxel, carboplatin, and fluorouracil. He experienced a substantial response, and after multidisciplinary discussion, it was decided to administer radiation and concomitant weekly carboplatin, which was completed August 2012. PET-CT imaging in November 2012 demonstrated a complete metabolic response. The patient was monitored every 3 months with PET-CT imaging. In March 2014, CT of the chest demonstrated bilateral lung parenchymal nodularity. Biopsy of a lung nodule confirmed metastatic squamous cell carcinoma. The patient was enrolled in a randomized, placebo-controlled, phase II study that consisted of docetaxel and cisplatin or carboplatin with or without erlotinib (ClinicalTrials.gov identifier: NCT01064479). He achieved a partial response after six cycles and received maintenance therapy for eight cycles until July 2015, when he developed progressive disease in the lungs. Unblinding of the study revealed that he received erlotinib.
The patient was referred to the Phase I Clinic and participated in the IMPACT2 trial, a randomized trial in precision medicine (ClinicalTrials.gov identifier: NCT02152254). Biopsy of a lung lesion demonstrated multiple alterations, including FGF19, FGF4, FGF23, FGF3, and FGF6 amplifications (Table 1 and Fig 1). He was randomly assigned to receive targeted therapy and was enrolled in a study with a selective pan-FGFR inhibitor that began in September 2015.
Table 1.
Next-Generation Sequencing Genomic Profile by Foundation One (August 2015)
| Genomic Profile | |
|---|---|
| Genomic alterations | |
| FGF19 amplification | CCND1 amplification |
| FGF4 amplification | CCND2 amplification |
| FGF23 amplification | CDKN2A/B loss |
| FGF3 amplification | CHD2 D213N |
| FGF6 amplification | CREBBP R1392* |
| EMSY amplification | |
| KDM5A amplification | |
| KRAS amplification | |
| MYC duplication exons 2 and 3 | |
| TP53 E204* | |
| Variants of unknown significance | |
| AKT2 E323Q | IGF1R S909C |
| ATM E322D | KLHL6 amplification |
| ATR amplification | NOTCH2 T239S |
| BCL6 amplification | PIK3CA amplification |
| BLM R543H | PIK3CB amplification |
| CARD11 truncation | PIK3R2 R559H |
| CDKN1B amplification | PRDM1 K188E |
| CHD4 amplification | RANBP2 amplification |
| DICER1 A948T | SOX2 amplification |
| EPHB1 amplification | TERC amplification |
| GRM3 amplification | PRKCI amplification |
NOTE. Gene symbols and full gene name as approved by the HUGO Gene Nomenclature Committee. Abbreviations: AKT2, AKT serine/threonine kinase 2; ATM, ATM serine/threonine kinase; ATR, ATR serine/threonine kinase; BCL6, B cell CLL/lymphoma 6; BLM, bloom syndrome RecQ like helicase; CARD11, caspase recruitment domain family member 11; CCND1, cyclin D1; CCND2, cyclin D2; CDKN1B, cyclin-dependent kinase inhibitor 1B; CDKN2A, cyclin-dependent kinase inhibitor 2A; CDKN2B, cyclin-dependent kinase inhibitor 2B; CHD2, chromodomain helicase DNA binding protein 2; CHD4, chromodomain helicase DNA binding protein 4; CREBBP, cAMP-response element binding protein binding protein; DICER1, dicer 1 ribonuclease III; EMSY, BRCA2 (DNA repair associated) interacting transcriptional repressor; EPHB1, EPH receptor B1; FGF3, fibroblast growth factor 3; FGF4, fibroblast growth factor 4; FGF6, fibroblast growth factor 6; FGF19, fibroblast growth factor 19; FGF23, fibroblast growth factor 23; GRM3, glutamate metabotropic receptor 3; IGF1R, insulin-like growth factor 1 receptor; KDM5A, lysine demethylase 5A; KLHL6, kelch-like family member 6; KRAS, KRAS proto-oncogene, GTPase; MYC, MYC proto-oncogene, bHLH transcription factor; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit α; PIK3CB, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit β; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; PRDM1, PR/SET domain 1; PRKCI, protein kinase C ι; RANBP2, RAN binding protein 2; SOX2, SRY-box 2; TERC, telomerase RNA component; TP53, tumor protein p53.
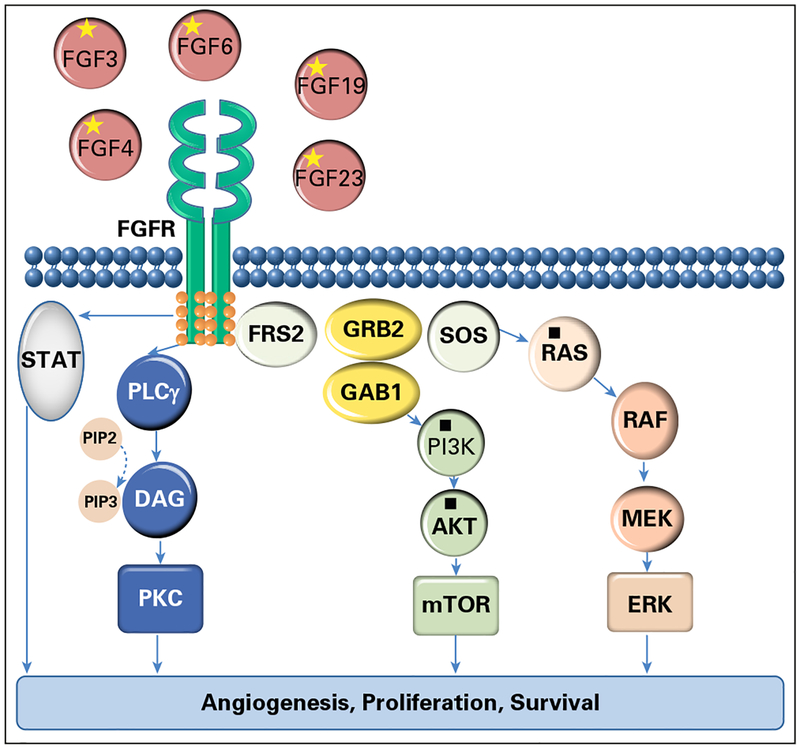
Fig 1.
Selected genomic alterations are indicated by an asterisk (*): fibroblast growth factor receptor (FGFR) pathway (FGF19, FGF4, FGF23, FGF3, and FGF6 amplifications). Selected variants of unknown significance are indicated by a filled square (■): KRAS amplification, PIK3CA and PIK3CB amplifications, and PIK3R2 and protein kinase B (AKT) mutations. DAG, dystroglycan; ERK, extracellular signal–regulated kinase; FRS2, fibroblast growth factor receptor substrate 2; GAB1, GRB2 associated binding protein 1; GRB2, growth factor receptor bound protein 2; MEK, mitogen-activated protein kinase; mTOR, mechanistic target of rapamycin; PI3K, phosphatidylinositide 3-kinase; PIP2, phosphatidylinositol 4,5-bisphosphate; PIP3, phosphatidylinositol (3,4,5)-trisphosphate (PtdIns(3,4,5)P3); PKC, protein kinase C; PLCγ, phospholipase C γ; RAF, RAF proto-oncogene serine/threonine-protein kinase; RAS, rat sarcoma viral oncogene homolog; SOS, son of sevenless homolog, Ras/Rac guanine nucleotide exchange factor; STAT, signal transducer and activator of transcription.
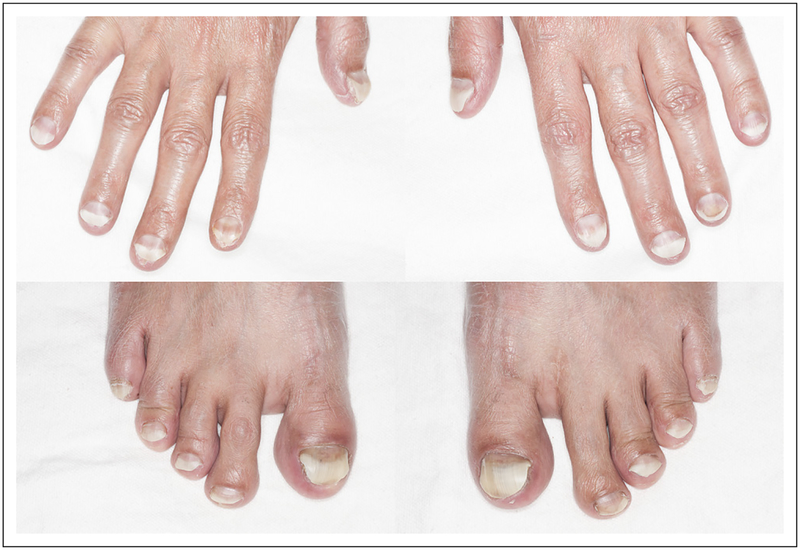
After two cycles, despite a low-phosphorus diet and Sevelamer carbonate treatment, the patient required a dose reduction of the FGFR inhibitor by one dose level for management of hyperphosphatemia. One month after the initiation of treatment, the patient developed nail changes. At the third cycle, his nails had a spoon-like appearance and discoloration (Fig 2). Other adverse events included grade 1 hand-foot syndrome, xerostomia, and mucositis.
Fig 2.
Nail toxicity at 3 months after the initiation of a selective pan–fibroblast growth factor receptor inhibitor.
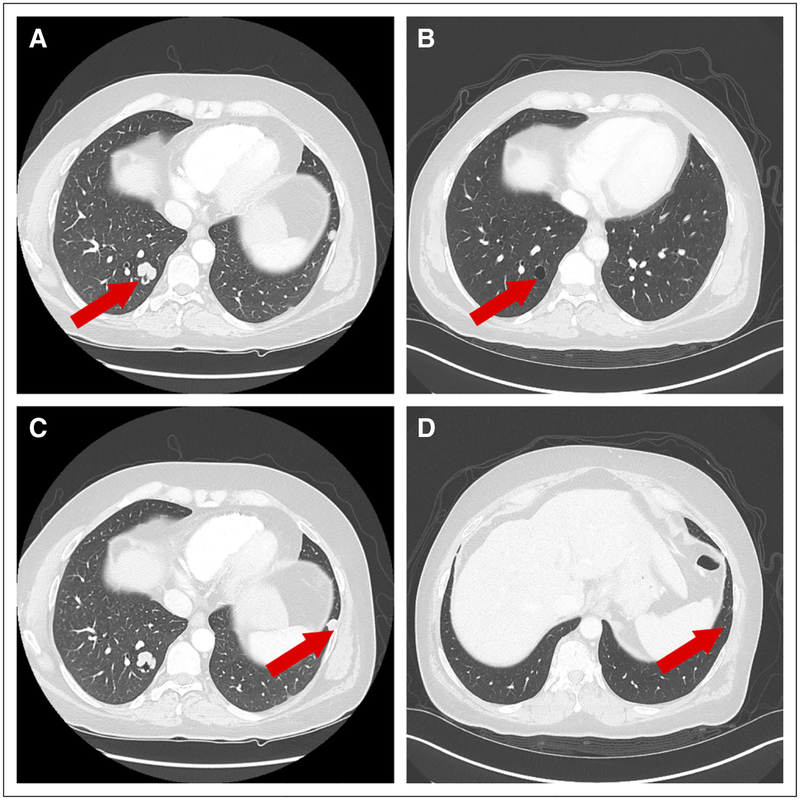
In March 2016, CT imaging after six cycles demonstrated a complete radiologic response (Fig 3). After 15 cycles (December 2016), CT imaging indicated a right infrahilar mass (3.4 × 1.8 cm). Pathologic examination of this mass showed poorly differentiated squamous cell carcinoma consistent with the primary tumor. The patient discontinued treatment. Immunohisto-chemical analysis of the biopsy specimen demonstrated programmed death-ligand 1 expression in 5% of tumor cells. In March 2017, the patient was started on a clinical trial of nivolumab and an investigational agent. In July 2017, he developed progressive disease and was enrolled in a study with another investigational agent. After the initiation of the second cycle, the patient developed brain metastases and treatment was discontinued. He then underwent whole-brain radiation therapy, and 5 weeks after the completion of this treatment, he died of acute myocardial infarction.
Fig 3.
Radiologic response to treatment with fibroblast growth factor receptor inhibitor. (A and B) Baseline scans, September 2015. (C and D) Best response, March 2016.
DISCUSSION
To our knowledge, this is the first case of a complete response to an FGFR inhibitor in a patient with HNSCC harboring several fibro-blast growth factor (FGF) amplifications. Alterations of the FGF/FGFR signaling pathways play important roles in carcinogenesis, stimulating cancer cell proliferation and angiogenesis.8 Deregulation of the FGFR pathway occurs in FGF ligands (FGFs) or receptors via gene mutations, translocations, fusions, or amplifications that lead to protein overexpression that results in the activation of the downstream signaling pathway.9 The FGFR family has four highly conserved transmembrane receptor tyrosine kinases (FGFR1 to FGFR4) that differ in their ligand affinity and tissue distribution. FGFs include more than 20 polypeptides (FGF1 to FGF10 and FGF16 to FGF23) that signal through FGFRs.10 FGFRs can bind FGFs that form complexes with heparan sulfate proteoglycans in an autocrine and paracrine fashion or endocrine FGFs (FGF19, FGF21, and FGF23).11 Each FGFR can be activated by several FGFs, and FGFs can activate more than one receptor. Binding of FGFs to FGFRs induces dimerization of the intracellular domain of the receptor and downstream activation of signaling.10
With the advent of next-generation sequencing, aberrations in FGFRs have been better characterized than FGF alterations. The most common mechanisms of FGF activation are gene amplification that leads to overexpression and mutations that lead to increased affinity for FGFRs.9 FGF amplifications have been observed in several tumor types.12 In HNSCC, FGFR1 overexpression has been reported in > 75% of both human papillomavirus (HPV)–positive and –negative HNSCC, and it is associated with poor survival.13 FGFR1, FGFR2, and FGFR3 amplifications, as well as FGFR3-TACC3 fusions, have been identified in HPV-positive tumors.14 FGF3, FGF4, FGF19, and CCND1, also amplified in this patient, colocalize on the same amplicon of 11q13 and are frequently coamplified. In addition, 11q13 amplification is prevalent in HNSCC—particularly HPV unrelated and tobacco induced—and has been reported to be associated with poor prognosis.15,16 Although the HPV status of our patient was unknown, he was a heavy smoker throughout the course of his disease.
There has been exponential progress in the field of FGFR targeting, owing to the development of novel agents that inhibit FGF ligands and receptors. These agents include nonselective or selective tyrosine kinase inhibitors (TKIs), monoclonal antibodies, and FGF ligand traps. Nonselective TKIs17–20 are effective against FGFRs as a result of the structural similarity of their kinase domains. Because they simultaneously target vascular endothelial growth factor receptor, platelet-derived growth factor receptor, and FGFR signaling pathways, these compounds are being developed primarily as antiangiogenic agents. Selective FGFR TKIs21–23 are also in clinical development. AZD4547 has been investigated in FGFR2-amplified gastric cancers and in FGFR1-amplified breast cancer and NSCLC.24–26 BGJ398 induced a 40% overall response rate in patients with urothelial cancer and FGFR alterations.21 DEBIO 1347 may inhibit the FGFR2 mutation V564F, which causes resistance to other drugs.27 Monoclonal antibodies that target FGF/FGFR signaling by blocking ligand binding or preventing receptor dimerization are in clinical development.28
Adverse effects associated with nonselective FGFR inhibitors include vascular endothelial growth factor receptor–related toxicities, and with selective FGFR inhibitors, hyperphosphatemia as a result of FGF23 signaling.17,29 In our patient, hyperphosphatemia was managed with a low-phosphate diet, phosphate binders, and a dose reduction. Our patient experienced ungual, cutaneous, and mucosal toxicity. Ungual toxicity occurs in 35% of patients—selective inhibitors—and is dose dependent.17,22 He did not experience nausea and vomiting, diarrhea, anorexia, or ocular effects.28
This case report demonstrates the benefits of the precision medicine approach of using tumor molecular profiling to select therapy. The patient achieved a complete response to an FGFR inhibitor that targeted his multiple FGF amplifications. This response is particularly intriguing given his disease progression while on erlotinib, which suggests resistance to epidermal growth factor receptor (EGFR) inhibition. Other investigators suggest that the cross-talk between other signaling pathways and FGFR signaling may explain the development of acquired resistance to anticancer therapies.30 Activation of the FGF2–FGFR1 autocrine pathway may be a mechanism of acquired resistance to gefitinib, an EGFR TKI, in patients with EGFR-mutant NSCLC.30,31 In vitro studies have demonstrated that increasing FGFR2 and FGFR3 mRNA in NSCLC cell lines32 and FGF9 upregulation in colorectal cancer may induce acquired resistance to EGFR inhibitors.33 In our patient, it is plausible that the FGF/FGFR pathway alterations could have been acquired secondary to exposure to erlotinib.
As FGFR signals through both the phosphatidylinositol 3-kinase/AKT/mammalian target of rapamycin and mitogen-activated protein kinase pathways, it is plausible that activating mutations in these pathways might confer resistance to FGFR inhibitors.34 Molecular testing was not performed at disease progression.
In conclusion, this case report exemplifies the value of tumor molecular profiling and the selection of targeted therapy on the basis of tumor molecular alterations. Use of a selective pan-FGFR inhibitor that targets multiple FGF amplifications—FGF3, FGF4, FGF6, FGF19, and FGF23—was associated with a durable complete response in a patient with HNSCC. FGF amplifications could have arisen secondary to the activation of an escape pathway after the use of an EGFR inhibitor. Monitoring of patients via repeated molecular profiling may provide valuable information about disease pathogenesis and guide therapy. Targeting acquired molecular alterations may prevent or delay tumor progression. Future clinical trials should investigate the role of targeting evolving alterations before evidence of clinical progression in patients who receive targeted therapy.
Support
Funded by Foundation Medicine (IMPACT2 trial; A.M.T.).
Footnotes
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Ecaterina Ileana Dumbrava
No relationship to dislcose
Rasha Alfattal
No relationship to dislcose
Vincent A. Miller Employment: Foundation Medicine
Leadership: Foundation Medicine
Stock and Other Ownership Interests: Foundation Medicine
Patents, Royalties, Other Intellectual Property: Royalties related to T790M patent awarded to Memorial Sloan Kettering Cancer Center
Apostolia Maria Tsimberidou
Consulting or Advisory Role: Roche
Research Funding: EMD Serono (Inst), Baxter (Inst), Foundation Medicine (Inst), Onyx Pharmaceuticals (Inst), Bayer (Inst), Boston Biomedical (Inst), Placon (Inst), Immatics (Inst), Karus Therapeutics (Inst), Stem Cells (Inst), OBI Pharma (Inst)
REFERENCES
- 1.Haugsten EM, Wiedlocha A, Olsnes S, et al. : Roles of fibroblast growth factor receptors in carcinogenesis. Mol Cancer Res 8:1439–1452, 2010 [DOI] [PubMed] [Google Scholar]
- 2.Touat M, Ileana E, Postel-Vinay S, et al. : Targeting FGFR signaling in cancer. Clin Cancer Res 21:2684–2694, 2015 [DOI] [PubMed] [Google Scholar]
- 3.Lamont FR, Tomlinson DC, Cooper PA, et al. : Small molecule FGF receptor inhibitors block FGFR-dependent urothelial carcinoma growth in vitro and in vivo. Br J Cancer 104:75–82, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williams SV, Hurst CD, Knowles MA: Oncogenic FGFR3 gene fusions in bladder cancer. Hum Mol Genet 22:795–803, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borad MJ, Champion MD, Egan JB, et al. : Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet 10:e1004135, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh D, Chan JM, Zoppoli P, et al. : Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 337:1231–1235, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weeden CE, Solomon B, Asselin-Labat M-L: FGFR1 inhibition in lung squamous cell carcinoma: Questions and controversies. Cell Death Discov 1:15049, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itoh N, Ornitz DM: Fibroblast growth factors: From molecular evolution to roles in development, metabolism and disease. J Biochem 149:121–130, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ornitz DM, Itoh N: The fibroblast growth factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4:215–266, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katoh M, Nakagama H: FGF receptors: Cancer biology and therapeutics. Med Res Rev 34:280–300, 2014 [DOI] [PubMed] [Google Scholar]
- 11.Mundhenke C, Meyer K, Drew S, et al. : Heparan sulfate proteoglycans as regulators of fibroblast growth factor-2 receptor binding in breast carcinomas. Am J Pathol 160:185–194, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parish A, Schwaederle M, Daniels G, et al. : Fibroblast growth factor family aberrations in cancers: Clinical and molecular characteristics. Cell Cycle 14:2121–2128, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koole K, Brunen D, van Kempen PMW, et al. : FGFR1 is a potential prognostic biomarker and therapeutic target in head and neck squamous cell carcinoma. Clin Cancer Res 22:3884–3893, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network: Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 517:576–582, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smeets SJ, Braakhuis BJM, Abbas S, et al. : Genome-wide DNA copy number alterations in head and neck squamous cell carcinomas with or without oncogene-expressing human papillomavirus. Oncogene 25:2558–2564, 2006 [DOI] [PubMed] [Google Scholar]
- 16.Katoh M, Katoh M: Comparative genomics on mammalian Fgf3-Fgf4 locus. Int J Oncol 27:281–285, 2005 [PubMed] [Google Scholar]
- 17.Chae YK, Ranganath K, Hammerman PS, et al. : Inhibition of the fibroblast growth factor receptor (FGFR) pathway: The current landscape and barriers to clinical application. Oncotarget 8:16052–16074, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gozgit JM, Wong MJ, Moran L, et al. : Ponatinib (AP24534), a multitargeted pan-FGFR inhibitor with activity in multiple FGFR-amplified or mutated cancer models. Mol Cancer Ther 11:690–699, 2012 [DOI] [PubMed] [Google Scholar]
- 19.Soria J-C, DeBraud F, Bahleda R, et al. : Phase I/IIa study evaluating the safety, efficacy, pharmacokinetics, and pharmacodynamics of lucitanib in advanced solid tumors. Ann Oncol 25:2244–2251, 2014. [Erratum: Ann Oncol 26:445, 2015] [DOI] [PubMed] [Google Scholar]
- 20.Milowsky MI, Dittrich C, Durán I, et al. : Phase 2 trial of dovitinib in patients with progressive FGFR3-mutated or FGFR3 wild-type advanced urothelial carcinoma. Eur J Cancer 50:3145–3152, 2014 [DOI] [PubMed] [Google Scholar]
- 21.Nogova L, Sequist LV, Perez Garcia JM, et al. : Evaluation of BGJ398, a fibroblast growth factor receptor 1–3 kinase inhibitor, in patients with advanced solid tumors harboring genetic alterations in fibroblast growth factor receptors: Results of a global phase I, dose-escalation and dose-expansion study. J Clin Oncol 35:157–165, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabernero J, Bahleda R, Dienstmann R, et al. : Phase I dose-escalation study of JNJ-42756493, an oral pan-fibroblast growth factor receptor inhibitor, in patients with advanced solid tumors. J Clin Oncol 33:3401–3408, 2015 [DOI] [PubMed] [Google Scholar]
- 23.Saka H, Kitagawa C, Kogure Y, et al. : Safety, tolerability and pharmacokinetics of the fibroblast growth factor receptor inhibitor AZD4547 in Japanese patients with advanced solid tumours: A phase I study. Invest New Drugs 35:451–462, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paik PK, Shen R, Berger MF, et al. : A phase Ib open-label multicenter study of AZD4547 in patients with advanced squamous cell lung cancers. Clin Cancer Res 23:5366–5373, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Cutsem E, Bang Y-J, Mansoor W, et al. : A randomized, open-label study of the efficacy and safety of AZD4547 monotherapy versus paclitaxel for the treatment of advanced gastric adenocarcinoma with FGFR2 polysomy or gene amplification. Ann Oncol 28:1316–1324, 2017 [DOI] [PubMed] [Google Scholar]
- 26.Seckl M, Badman PD, Liu X, et al. : RADICAL trial: A phase Ib/IIa study to assess the safety and efficacy of AZD4547 in combination with either anastrozole or letrozole in ER positive breast cancer patients progressing on these aromatase inhibitors (AIs). J Clin Oncol 35:1059, 2017. (suppl) [Google Scholar]
- 27.Henner Voss M, Hierro C, Heist RS, et al. : Debio 1347, an oral FGFR inhibitor: Results from a first-in-human, phase I dose-escalation study in patients with FGFR genomically activated advanced solid tumors. J Clin Oncol 35:2500, 2017. (suppl) [Google Scholar]
- 28.Porta R, Borea R, Coelho A, et al. : FGFR a promising druggable target in cancer: Molecular biology and new drugs. Crit Rev Oncol Hematol 113:256–267, 2017 [DOI] [PubMed] [Google Scholar]
- 29.Hierro C, Rodon J, Tabernero J: Fibroblast growth factor (FGF) receptor/FGF inhibitors: Novel targets and strategies for optimization of response of solid tumors. Semin Oncol 42:801–819, 2015 [DOI] [PubMed] [Google Scholar]
- 30.Kono SA, Marshall ME, Ware KE, et al. : The fibroblast growth factor receptor signaling pathway as a mediator of intrinsic resistance to EGFR-specific tyrosine kinase inhibitors in non-small cell lung cancer. Drug Resist Updat 12:95–102, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Terai H, Soejima K, Yasuda H, et al. : Activation of the FGF2-FGFR1 autocrine pathway: A novel mechanism of acquired resistance to gefitinib in NSCLC. Mol Cancer Res 11:759–767, 2013 [DOI] [PubMed] [Google Scholar]
- 32.Ware KE, Marshall ME, Heasley LR, et al. : Rapidly acquired resistance to EGFR tyrosine kinase inhibitors in NSCLC cell lines through de-repression of FGFR2 and FGFR3 expression. PLoS One 5:e14117, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizukami T, Togashi Y, Naruki S, et al. : Significance of FGF9 gene in resistance to anti-EGFR therapies targeting colorectal cancer: A subset of colorectal cancer patients with FGF9 upregulation may be resistant to anti-EGFR therapies. Mol Carcinog 56:106–117, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Guagnano V, Kauffmann A, Wohrle S, et al. : FGFR genetic alterations predict for sensitivity to NVP-BGJ398, a selective pan-FGFR inhibitor. Cancer Discov 2:1118–1133, 2012 [DOI] [PubMed] [Google Scholar]