Abstract
The guanine-nucleotide exchange factor (GEF) Son-of-Sevenless (SOS) plays a critical role in metazoan signaling by converting Ras•GDP (guanosine diphosphate) to Ras•GTP (guano-sine triphosphate) in response to tyrosine kinase activation. Structural studies have shown that SOS differs from other Ras-specific GEFs in that SOS is itself activated by Ras•GTP binding to an allosteric site, distal to the site of nucleotide exchange. The activation of SOS involves membrane recruitment and conformational changes, triggered by lipid binding, that open the allosteric binding site for Ras•GTP. This is in contrast to other Ras-specific GEFs, which are activated by second messengers that more directly affect the active site. Allosteric Ras•GTP binding stabilizes SOS at the membrane, where it can turn over other Ras molecules processively, leading to an ultrasensitive response that is distinct from that of other Ras-specific GEFs.
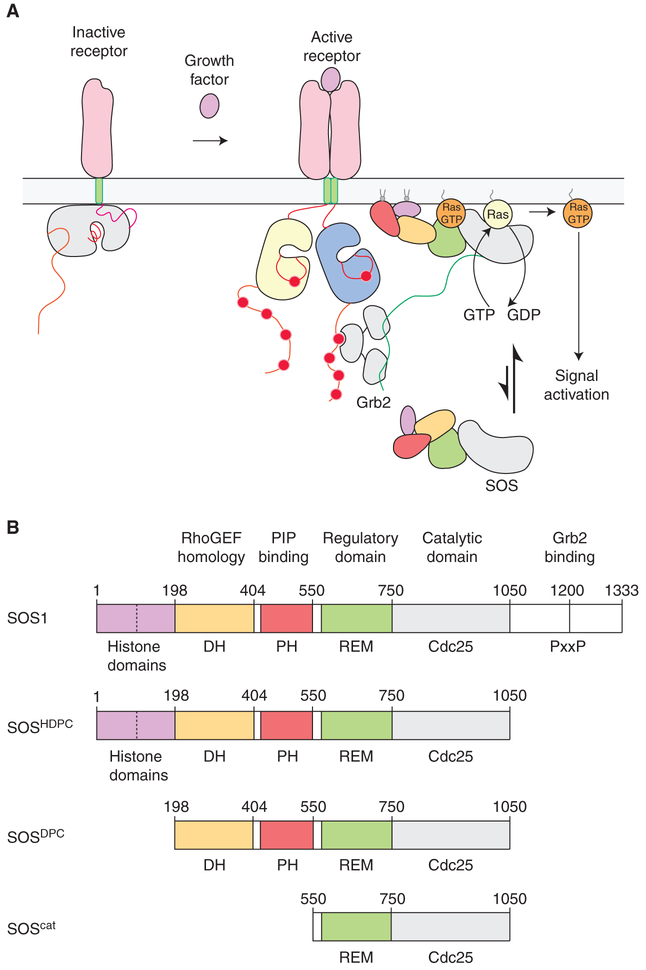
An evolutionary innovation in metazoans is the coupling of Ras activation to the phosphorylation of tyrosine residues on cell-surface receptors. This occurs through the SH2-medi-ated recruitment to these receptors of a Ras-specific guanine nucleotide exchange factor (GEF), Son-of-Sevenless (SOS) (Fig. 1A) (Lowenstein et al. 1992; Buday and Downward 1993; Egan et al. 1993; Gale et al. 1993; Rozakis-Adcock et al. 1993). There are several classes of Ras-specific GEFs in mammalian cells, and a key difference between SOS and other Ras-specific GEFs is that the latter are regulated by soluble second messengers, such as calcium, cyclic AMP (cAMP), and diacylglycerol, whereas SOS is not (Bos et al.2007). Instead, SOS is recruited to the membrane by tyrosine phosphorylation of receptors or scaffold proteins and, in addition, the activation of SOS requires the binding of Ras•TP to an allosteric site (Margarit et al. 2003).
Figure 1.
The activation and domain organization of Son-of-Sevenless (SOS). (A) Ras activation is coupled to the phosphorylation of tyrosine residues on cell-surface receptors. This primarily occurs through the SH2-mediated recruitment of SOS to these receptors by adaptor proteins such as Grb2. Ras activation by SOS subsequently triggers downstream signaling responses in cells. (B) Schematic representation of the various constructs of SOS discussed in this review. DH, Dbl homology; PH, Pleckstrin homology; REM, Ras exchanger motif.
All Ras-specific GEFs share a catalytic module, consisting of a Ras exchanger motif (REM) domain and a Cdc25 domain (Boguski and McCormick 1993). The Cdc25 domain contains the active site, where nucleotide-free Ras is bound during the exchange process. In SOS, allosteric activation occurs by the binding of Ras•TP to a distal site bracketed by the REM and Cdc25 domain (Margarit et al. 2003; Sondermann et al. 2004) but, as far as is known, the principal role of the REM domain in other Ras-specific GEFs appears to be structural. Each class of Ras-specific GEFs has a distinct regulatory mechanism, unrelated to that of SOS. For example, Epac proteins are autoinhibited by a cAMP-binding domain, which prevents Ras binding to the Cdc25 domain in the absence of cAMP (de Rooij et al. 2000; Rehmann et al. 2006, 2008; Bos 2017). Similarly, calcium and diacylglycerol binding to C1 and EF-hand domains, respectively, relieves the autoinhibition of RasGRP proteins at the membrane (Iwig et al. 2013; Jun et al. 2013a; Vercoulen et al. 2017).
In common with other GEFs, SOS activates Ras by causing dissociation of the otherwise tightly bound nucleotide, thereby facilitating the binding of guanosine triphosphate (GTP) (Bos et al. 2007; Cherfils and Zeghouf 2013). SOS is not normally membrane associated, and it was thought that SOS was activated simply by recruitment to the membrane, where it can more efficiently encounter membrane-anchored Ras. SOS is bound to the adaptor protein Grb2, and the SH2 domain of Grb2 binds to activated receptors, either directly or through the bridging action of the adaptor protein Shc (Rozakis-Ad-cock et al. 1992; Buday et al. 1994). In this way, receptor activation results in the membrane localization of SOS. Consistent with this, the constitutive membrane localization of the catalytic module of SOS activates Ras without receptor input (Aronheim et al. 1994). Subsequent studies showed, however, that multiple layers of inhibition are imposed on SOS by its regulatory domains, and these need to be relieved by interaction with lipids in the membrane, such as PIP2 and PA, for SOS to be fully active at the membrane (Corbalan-Garcia et al. 1998; Hall et al. 2002; Sondermann et al. 2004; Gureasko et al. 2008; Yadav and Bar-Sagi 2010). The end point of these actions is the binding of Ras•TP to the allosteric site on SOS, which is blocked prior to activation by the regulatory domains of SOS (Sondermann et al. 2004; Gureasko et al. 2010). Thus, the activation of SOS requires its recruitment to the membrane as well as the release of blockage of the allosteric site of SOS.
The binding of Ras to the allosteric site tethers SOS to the membrane, and stimulates the nucleotide exchange activity of SOS strongly, in a positive feedback loop (Gureasko et al.2008). The fact that activated SOS can be anchored to the membrane by Ras•TP has important consequences. Upon release of autoinhibition, SOS molecules that are anchored to the membrane by Ras•TP can activate large numbers of Ras molecules processively (Iversen et al. 2014). In cells, the pool of Ras•TP that is required to trigger SOS activity is provided, in part, by the activity of the RasGRP family of GEFs (Roose et al. 2007). This interplay between SOS and other Ras-specific GEFs, such as RasGRP1, can result in a switch-like response to the activation of cell-surface receptors (Das et al. 2009). The allosteric activation of SOS by Ras is important for sustained Erk activation (Boykevisch et al. 2006), and for T-cell signaling (Roose et al. 2007; Das et al. 2009; Prasad et al.2009). The allosteric stimulation of SOS by on cogenic Ras can result in the activation of wild-type alleles of Ras, resulting in stronger growth stimulation (Jeng et al. 2012).
In this review, we focus on the structural basis for the interplay between Ras and SOS that results in their mutual activation. We describe the mechanism of nucleotide exchange by SOS, and the nature of the allosteric stimulation of SOS by Ras. We then discuss how SOS is autoinhibited. Finally, we discuss some of the functional consequences of the allosteric activation of SOS at the membrane.
THE DOMAIN ORGANIZATION OF SOS
There are two isoforms of SOS in humans (SOS1 and SOS2) that have the same domain organization (Fig. 1B). SOS1 has 1333 residues and is 74% identical to SOS2, with a high degree of conservation distributed evenly throughout the sequence of both proteins. Most studies of human SOS have focused on SOS1, which we will refer to simply as “SOS.” SOS acts upon all three principal isoforms of Ras in vertebrates (Buday and Downward 1993; Jaumot et al. 2002; Ehr-hardt et al. 2004). The ~500-residue minimal segment of SOS that promotes efficient nucleotide exchange is the catalytic module, denoted SOScat, consisting of the REM domain and the Cdc25 domain. The Cdc25 domain engages the substrate Ras molecule, causing it to release nucleotide (Boriack-Sjodin et al. 1998). A REM domain is usually found associated with the Cdc25 domain in Ras-specific GEFs (Boguski and McCormick 1993). The sequence of the Cdc25 domain is conserved, and is readily identifiable in fungi and animals, whereas the sequence of the REM domain is more variable across organisms (van Dam et al. 2009).
The amino-terminal ~550 residues of SOS consist of three well-defined structural domains, located just before the catalytic module: a histone-fold domain (Sondermann et al. 2003), a Pleckstrin homology (PH) domain, and a Dbl homology (DH) domain (Soisson et al. 1998). Together, these domains cooperate to inhibit SOS activity by blocking Ras•TP binding to the allosteric site (Corbalan-Garcia et al. 1998; Hall et al. 2002; Sondermann et al. 2004). Both the histone domain and the PH domain have been shown to bind to specific lipids in membranes, an important step in the release of autoinhibition (Gureasko et al. 2008; Yadav and Bar-Sagi 2010). DH domains are typically found in GEFs that are specific for the Rac, Rho, and cdc42 family of GTPases (Soisson et al. 1998; Aghazadeh et al. 2000; Worthylake et al. 2000), and SOS has been implicated in coupling Ras activation to the activation of Rac (Nimnual et al. 1998; Jun et al. 2013b), although it is unclear whether this coupling is direct. The structure of autoinhibited SOS, including the amino-terminal regulatory domains and the catalytic module, but excluding the carboxy-terminal tail, has been determined (Sondermann et al. 2004; Gureasko et al. 2010). These structures have revealed multiple ways in which the regulatory domains exert control on SOS activity, highlighting the exceptionally tight control of Ras activation, and its release by membrane interactions (Findlay and Pawson 2008).
The carboxy-terminal ~280 residues of SOS are disordered, and this segment contains the proline-rich motifs that are binding sites for the SH3 domain of Grb2 (Chardin et al. 1993; Li et al. 1993). Mutagenesis and nuclear magnetic resonance (NMR) studies have provided insights into the interaction of Grb2 with SOS (Lim et al. 1994; Terasawa et al. 1994; Wittekind et al. 1994). The carboxy-terminal tail also inhibits SOS (Corbalan-Garcia et al. 1998), and its presence weakens the affinity of Ras•TP for the allosteric site, although the precise structural details of this mechanism are unknown (Lee etal. 2017).
SOS INDUCES A WIDE OPENING OF THE ACTIVE SITE OF Ras, DISPLACING NUCLEOTIDE
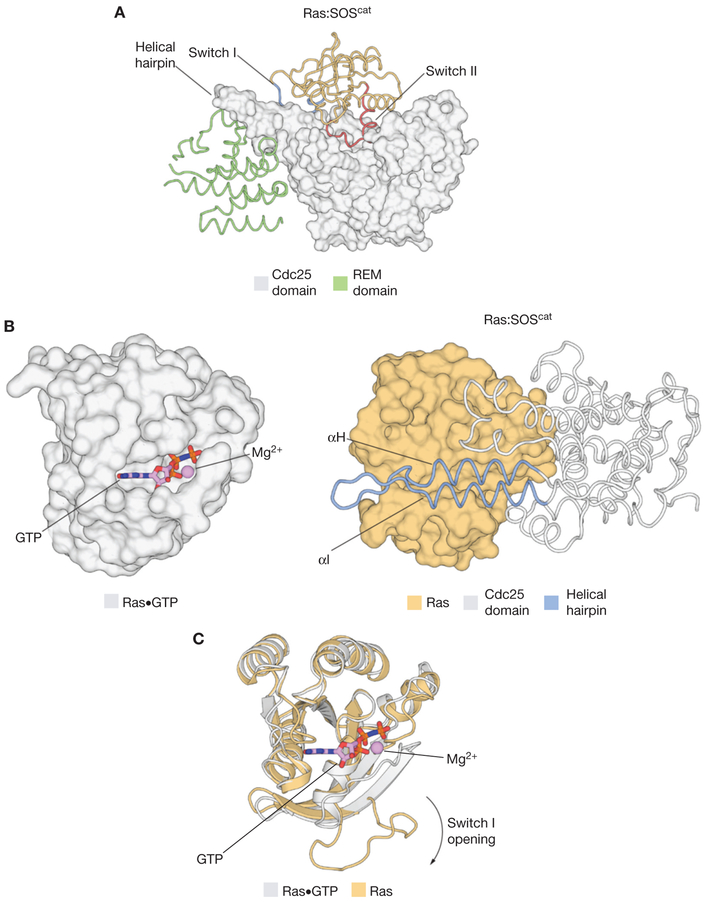
Ras binds with very high affinity to both guano-sine diphosphate (GDP) and GTP, with dissociation constants in the picomolar range (Goody et al. 1991), and Ras is unstable in the absence of nucleotide. The structure of a 1:1 Ras:SOScat complex showed that SOS stabilizes the nucleotide-free form of Ras by forming an extensive interface involving the Switch I and Switch II elements of Ras (Fig. 2A) (Boriack-Sjodin et al. 1998; Hall et al. 2001). Switch II provides the main anchor point for SOS on Ras. A helical hairpin element that protrudes from the main body of the Cdc25 domain of SOS is inserted between the Switch I and Switch II elements of Ras, acting as a molecular “tongue depressor” that causes the active site of Ras to open widely and expel the nucleotide (Fig. 2B). The principal conformational change in Ras is a peeling away of Switch I from the rest of Ras, caused by the insertion of the helical hairpin of SOS (Fig. 2C). The insertion of the helical hairpin into the Switch I region does not occlude the guanine and ribose binding sites in Ras, thereby facilitating the rebinding of nucleotide and the dislodging of SOS. Instead, residues within the helical hairpin, and the associated conformational changes in Ras, prevent the coordination of the phosphate groups of the nucleotide and the associated magnesium ion. Key features of this mechanism are preserved in other Ras-specific GEFs (Rehmann et al. 2008; Vercoulen et al. 2017).
Figure 2.
The structural mechanism of Ras activation by Son-of-Sevenless (SOS). (A) The structure of the Ras: SOScat complex (Protein Data Bank [PDB] code: 1BKd) showed that SOS stabilizes the nucleotide-free form of Ras by forming an extensive interface involving Switch I and Switch II of Ras. (B)(Left) The structure of Ras•GTP (PDB code: 5P21). (Right) A helical hairpin element that protrudes from the main body of the Cdc25 domain of SOS is inserted between the Switch I and Switch II elements of Ras, and causes the active site of Ras to open widely and expel the nucleotide. (C) After the insertion of the helical hairpin of SOS into Ras, the principal conformational change in Ras that leads to nucleotide release is a peeling away of Switch I from the rest of the protein. REM, Ras exchanger motif; GTP, guanosine triphosphate.
Studies on the Ras-specific GEFs Cdc25 and RCC1 have shown that the GEF does not impose a preference on whether Ras is reloaded with GTP or GDP (Lenzen et al. 1998; Klebe et al. 1995). Once the nucleotide bound to Ras is re-leased by the GEF, Ras is reset by the binding of GTP, driven by the higher cellular concentrations of GTP over GDP (Bennett et al. 2009). Although the data for other GEFs indicate that SOS has no intrinsic preference for loading Ras with either GDP or GTP, it has been suggested recently that SOS is more active when binding and stimulating the exchange of Ras•DP instead of Ras•TP (Vo et al. 2016). This conclusion was not, however, based on a direct comparison of the rates of nucleotide exchange for Ras•DP and Ras•TP by SOS, and so this question needs further investigation.
Peptide-based inhibitors of Ras have been developed that mimic the action of the helical hairpin of SOS (Patgiri et al. 2011). Brefeldin-A is a natural product that inhibits the action of Arf-specific GEFs (Klausner et al. 1992). It does so by binding to the interface between Arf and the GEF, slowing down the release of the complex (Mossessova et al. 2003; Renault et al. 2003). A Brefeldin-A-like molecule has not yet been developed for SOS, but compounds have been found that bind to Ras near the Ras:SOS interface, and enhance the rate of nucleotide exchange by SOS (Burns et al. 2014).
THE ACTIVATION OF SOS BY RAS•GTP
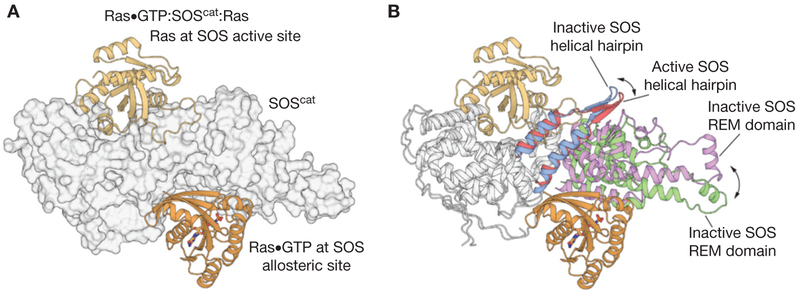
The discovery that SOS is allosterically activated by Ras•TP was unexpected, and it was brought about by the serendipitous crystallization of a 2:1 complex of Ras and SOScat, by Margarit and colleagues (2003). Analysis of the resulting structure revealed that nucleotide-free Ras was bound to the active site in the Cdc25 domain, as seen earlier for the structure of the 1:1 Ras:SOScat complex, but that a second Ras•TP molecule was bound to a distal site, wedged between the REM and Cdc25 domains (Fig. 3A) (Margarit et al. 2003). This site is specific for Ras•TP and, based on comparison to the structures of complexes of Ras•TP bound to effectors such as PI3-kinase (Pacold et al. 2000), the structure suggested that SOS is an “effector” for Ras•GTP. Biochemical studies showed that SOScat and Ras form a positive feedback loop, wherein Ras•GTP produced during the nucleotide exchange process binds to SOScat at the distal binding site, resulting in a dramatic increase in the rate of nucleotide exchange (Margarit et al. 2003; Gu-reasko et al. 2008) and potentiation of Ras activation by the epidermal growth factor receptor (Boykevisch et al. 2006).
Figure 3.
The activation of Son-of-Sevenless (SOS) by Ras•GTP (guanosine triphosphate). (A) The structure of the 2:1 Ras:SOScat complex (Protein Data Bank [PDB] code: 1NVV) revealed that nucleotide-free Ras was bound to the active site of the Cdc25 domain, and that a second Ras•GTP molecule is bound to a distal allosteric site, wedged between the Ras exchanger motif (REM) and Cdc25 domains (B) Comparison of the Ras-free structure of SOScat (PDB code: 2II0) to the 2:1 Ras:SOScat complex shows that the helical hairpin is tilted toward the active site of SOS in the Ras-free state, thereby constricting the site where SOS engages Switch II of nucleotide-free Ras. Allosteric binding of Ras•GTP to SOS leads to the rotation and opening of the helical hairpin, accompanied by the outward rotation of the REM domain, which frees the catalytic site to bind Ras.
The mechanism by which the binding of Ras•GTP to the allosteric site activates SOS was revealed by comparing the structure of isolated SOScat, with no Ras bound to the structures of either the 1:1 Ras:SOS complex or the ternary Ras•GTP:SOScat:Ras complex (Freedman et al. 2006). A prominent difference between the two structures is in the conformation of the helical hairpin of the cdc25 domain of SOS. The Ras-free structure of SOScat shows that the helical hairpin is tilted toward the active site of SOS, thereby constricting the site where SOS engages Switch II of nucleotide-free Ras (Fig. 3B). This suggests that Ras binding to the active site of SOS requires the helical hairpin to be pulled back. Indeed, the allosteric binding of Ras•GTP to SOS facilitates the rotation and opening of the helical hairpin, which frees the catalytic site to bind Ras. This structural observation was validated by mutating residues in the helical hairpin so as to stabilize the catalytically competent open conformation. These mutations led to an increase in the rate of nucleotide exchange (Freedman et al. 2009; Vo et al. 2016).
THE AMINO-TERMINAL REGULATORY DOMAINS OF SOS BLOCK THE BINDING OF RAS•GTP AT THE ALLOSTERIC SITE
SOS contains a ~350-residue DH-PH module located immediately before the catalytic module. DH domains, which are usually associated with PH domains, are GEFs for Rho-family GTPases (Cerione and Zheng 1996; Whitehead et al. 1997; Bishop and Hall 2000; Rossman et al. 2005). PH domains bind to specific phospholipids in the membrane (Lemmon and Ferguson 2000). The structure of the DH-PH module of SOS was the first to reveal how the two domains are integrated into a structural module, but, ironically, the DH domain of SOS is not active as a Rho-family GEF (Soisson et al. 1998). Comparison of the crystal structures of the DH-PH modules of SOS with the structure of DH-PH modules that act as Rho-family GEFs indicated that the conformation of the DH-PH module of SOS is not compatible with binding to small GTPases, unless a large conformational change occurs (Soisson et al. 1998; Aghazadeh et al. 2000; Worthylake et al. 2000). Although it is possible that such a conformational change could occur when the PH domain engages the membrane, sequence features of the DH-PH module of SOS also suggest that it does not act directly as a nucleotide exchange factor (Soisson et al. 1998).
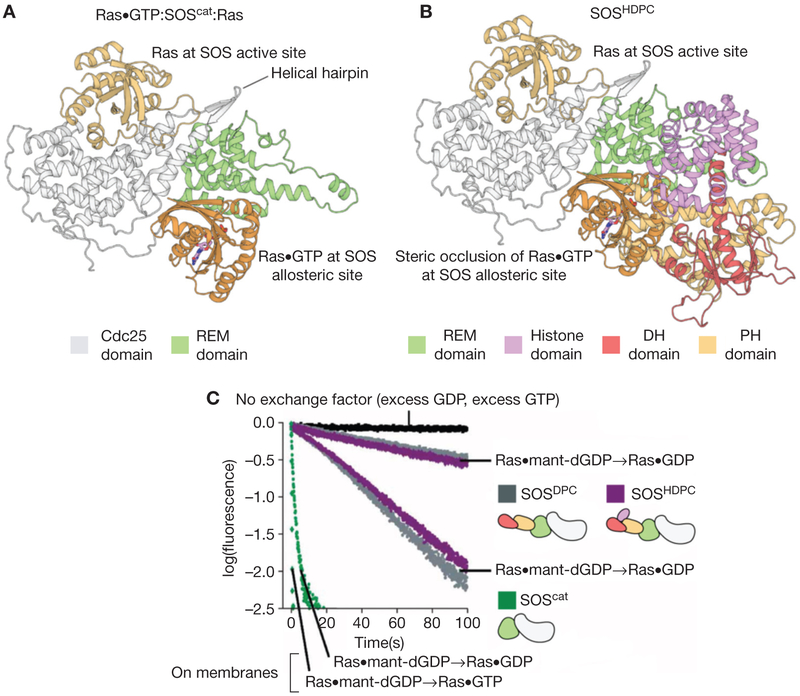
The role of the DH-PH domain in SOS is to regulate binding of Ras•GTP to the allosteric site of SOS (Fig. 4A). The structure of a construct of SOS containing the DH-PH module and the catalytic module (SOSDPC) showed that the location of the DH-PH module is such that it steri-cally occludes the binding of Ras•GTP at the allosteric site (Fig. 4B). Biochemical measurements show that SOSDPC has lower activity than SOScat (Fig. 4C) (Sondermann et al. 2004; Gureasko et al. 2010). Relief of autoinhibition occurs when the PH domain binds to negatively charged membrane phospholipids. This binding event triggers a conformational change in the DH domain that ultimately reveals the allosteric Ras•GTP binding site on SOS (Gureasko et al. 2008).
Figure 4.
The autoinhibition of Son-of-Sevenless (SOS) by regulatory domains. (A) The structure of the 2:1 Ras:SOScat complex, depicting Ras bound to the Cdc25 domain and Ras•GTP (guanosine triphosphate) bound to the allosteric site of SOS. (B) The structure of SOS containing the Dbl homology (DH)-Pleckstrin homology (PH) module and the catalytic module (SOSDPC) (Protein Data Bank [PDB] code: 3KSY) show that the DH-PH module sterically occludes the binding of Ras•GTP at the allosteric site. (C) Biochemical measurements show that SOSDPC has lower activity than SOScat. The fastest traces are for membrane-bound Ras (Gureasko et al. 2008). REM, Ras exchanger motif; GDP, guanosine diphosphate; dGDP, deoxyguanosine diphosphate.
The first ~200 residues of SOS comprise an amino-terminal tandem histone-fold domain, the presence of which in a cytoplasmic domain is unusual (Sondermann et al. 2003). Reflecting the organization of the nucleosome, most nonhistone proteins that contain histone-like domains are present in the nucleus as dimers, and play a role in nucleic acid metabolism (Baxevanis et al. 1995). The two histone domains of SOS are also arranged similarly to two histone proteins in the dimeric units that make up the nucleosome. The DNA-binding face of the “histone dimer” of SOS retains the pattern of positive charge that, in histones, binds DNA. This suggested that the histone domain of SOS may play a role in lipid binding, by using these positively charged residues to interact with the phosphate headgroups of lipids, instead of with the phosphate backbone of DNA. Indeed, mutations in this region affect the affinity of the histone fold for membrane lipids (Yadav and Bar-Sagi 2010; Saliba et al. 2014).
The crystal structure of SOSHDPC shows that the histone domain binds to the linker connecting the DH-PH module to the REM domain, thereby stabilizing the autoinhibited state of SOS (Sondermann et al. 2004; Gureasko et al. 2010; Lee et al. 2017). Notably, the presence of the histone domain suppresses the ability of membrane lipids to release the inhibitory action of the DH-PH module on SOS activity. Arg 552, located in the linker connecting the DH-PH module to the REM domain, stabilizes the auto-inhibited conformation of SOS by interacting with the histone domain (Sondermann et al. 2005), and mutation of this arginine residue activates SOS. Such gain-of-function mutations in SOS have been associated with Noonan syndrome (Roberts et al. 2007; Tartaglia et al. 2007).
ACTIVATION OF SOS AT THE MEMBRANE
Although it was known that the constitutive membrane localization of SOS activates Ras (Aronheim et al. 1994), a series of experiments showed that the activity of SOS is greatly enhanced by allosteric binding of Ras•GTP at the membrane (Gureasko et al. 2008). In these experiments, Ras•GTP was either covalently at tached to supported lipid bilayers or tethered to lipid vesicles, and the rate of nucleotide exchange was measured upon membrane recruitment and allosteric activation of SOScat. On vesicles, the nucleotide exchange rate was enhanced by approximately 500-fold compared to a solution measurement, and the nucleotide exchange rate on lipid bilayers increased as a function of Ras•GTP surface density, in a manner that is dependent on Ras•GTP binding to the allosteric site of SOS. Mutations that weaken the allosteric binding of Ras•GTP fail to recruit SOScat to the membrane, and therefore prevent the stimulation of nucleotide exchange.
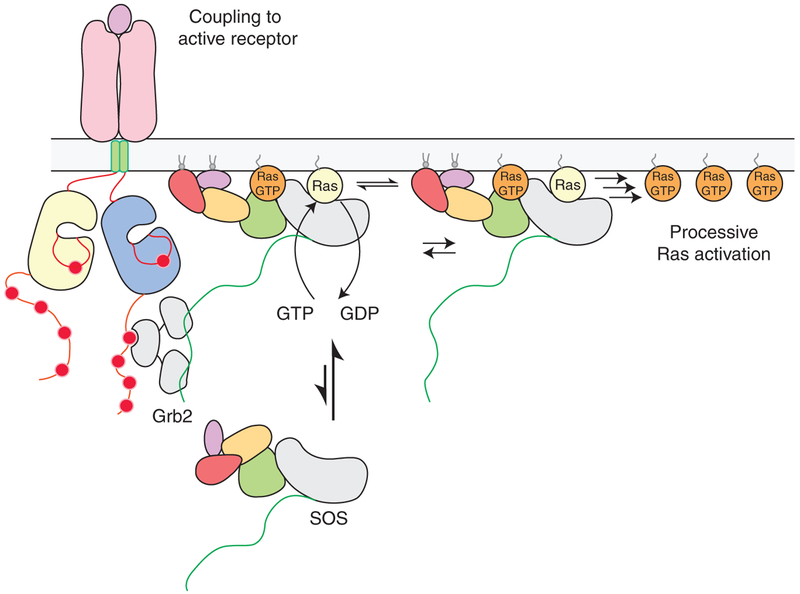
Once a single molecule of SOS is activated allosterically by Ras•GTP at the membrane, hundreds of Ras molecules can then be activated processively by that SOS molecule (Fig. 5). This was shown by integrating structural and bulk biochemical observations with single-molecule measurements of SOS on membranes (Iversen et al. 2014). Solution experiments have shown that Ras•GDP binds about 10-fold less tightly to the allosteric site than Ras•GTP, and a crystal structure revealed that Ras with hydrolyzed GTP (GDP+Pi) can be bound to the allosteric site (Sondermann et al. 2004). The feedback activation of SOS by Ras occurs at two levels: first, Ras•GTP is more efficient at recruiting SOS to the membrane, by virtue of having a higher binding affinity to the allosteric site than Ras•GDP. Second, on the membrane, Ras•GTP is more effective at enabling SOS to transition from states of lower GEF activity to states of much higher activity, as demonstrated by single-molecule measurements of the nucleotide exchange rates of SOScat corralled within micrometer-size partitions on a supported lipid bilayer (Iversen et al. 2014). In these experiments, a spectrum of SOS activity was observed, and a minority of highly active states of SOS dominated the nucleotide exchange reaction. Once SOS is anchored to the membrane by either Ras•GDP or Ras•GTP, the difference between the nucleotide-loading state of Ras bound at the allosteric site is only manifested in the efficiency of accessing the most highly active states. The structural basis for the interconversion of highly and lowly active states of SOS is not yet understood.
Figure 5.
The activation of Son-of-Sevenless (SOS) at the membrane. SOS is initially recruited to the membrane by adaptor proteins, such as Grb2, that bind to activated cell-surface receptors. Once a single molecule of SOS is activated allosterically by Ras•GTP (guanosine triphosphate) at the membrane, many more Ras molecules can then be activated processively by that SOS molecule, even if it were to disengage from the receptor. GDP, Guanosine diphosphate.
The slow off-rate of Ras•GTP at the allosteric site contributes to SOScat retention on the membrane in the absence of regulatory domains (Iversen et al. 2014). However, once SOSHDPC is recruited to the membrane, it is retained there much longer than SOScat (Christensen et al. 2016), largely as a result of interactions between lipid-binding regulatory domains with membrane lipids. In cells, SOS is also tethered to transmembrane receptors via interactions with Grb2, and SOS activation is balanced by the reversible binding kinetics of the SOS:Grb2 interaction (Christensen et al. 2016). Furthermore, SOS remains at the membrane until actively being removed by membrane endocytosis, thus preventing new Ras molecules from being activated (Christensen et al. 2016). Recent work has pointed to the existence of phase-separated signaling clusters on membranes, where multivalent interactions between the T-cell receptor, costimulatory receptors, SOS, Grb2, and other proteins lead to downstream signal transduction in T cells (Huang et al. 2016; Su et al. 2016). The ability of SOS to make multiple interactions to scaffold proteins via Grb2 and Shc, as well as its ability to stay engaged to Ras•GTP at the membrane, are critical properties that are likely to facilitate these phase transitions.
BISTABILITY IN RAS ACTIVATION BY SOS IN CELLS
In lymphocytes, RasGRP exchange factors produce Ras•GTP that subsequently allosterically activates SOS, potentiating the direct activation of SOS via Grb2 (Roose et al. 2007). The subsequent production of Ras•GTP by SOS leads to sharp responses in signaling pathways that control the generation of self-tolerant immune cells (Das et al. 2009). RasGRP proteins are activated by binding to membrane-localized diacylgly-cerol or phosphorylation by protein kinase C (Ebinu et al. 1998, 2000), and the strength of the external stimulus can further tune the activity of SOS and RasGRP proteins. Therefore, the interplay between SOS and RasGRP proteins, and their ability to be activated by distinct stimuli, leads to dynamic downstream signaling responses in cells.
In the case of SOS, low levels of receptor stimulation, and consequently low levels of SOS activation, lead to a predominant population of cells containing a low amount of Ras•GTP. Conversely, high levels of receptor stimulation led to a predominant population of cells containing a high amount of Ras•GTP (Das et al. 2009). Intermediate levels of receptor stimulation lead to two distinct cell populations, containing low and high amounts of Ras•GTP, respectively. Therefore, an intermediate level of SOS activity introduces a bistability in the amount of Ras•GTP produced in response to receptor stimulation (Das et al. 2009). This bistability disappears past a threshold amount of SOS activity, suggesting that T lymphocytes occupy either one of two binary “on” or “off” states with regard to Ras activation, in a “digital” manner (Das et al. 2009). For RasGRP proteins, increasing levels of receptor stimulation lead to an increasing number of cells containing Ras•GTP, and bistability in Ras•GTP levels is not observed. This suggests a graded, or “analog,” response of Ras•GTP production to receptor stimulation.
Once the allosteric site of SOS is primed and engaged by Ras•GTP, the bistable nature of SOS activation emerges, and might give rise to a molecular memory of past signals. This “hysteresis” in the Ras response was predicted by computational simulations, and was validated in T cells subject to repeated stimulation over the course of tens of minutes (Das et al. 2009). Hysteresis in Ras activation allows T cells to produce sustained levels of Ras•GTP in response to potentially weaker receptor stimulation, in a manner that is dependent on the feedback interaction between Ras and SOS.
SOS MAY HAVE CONSTRAINED Ras EVOLUTION IN METAZOANS
SOS is the only Ras-specific GEF that is known to be allosterically activated by Ras•GTP, pointing to an evolutionary divergence between SOS and other Ras-specific GEFs. Insight into the unique dependence of SOS on Ras may be gained from analyzing the sequence conservation of Ras in metazoans, relative to other Ras family GTPases. The three principal isoforms of Ras (H-Ras, K-Ras, and N-Ras) are generally highly conserved in metazoans (Rojas et al. 2012; Johnson et al. 2017), though less so in invertebrates compared to vertebrates (Bandaru et al. 2017). The sequence of Ras from the choa-noflagellate Salpingoeca rosetta, thought to be the closest living relative of metazoans, is 72% identical to human Ras, whereas Ras is nearly invariant (>90% identical to human) in vertebrates. In comparison, the sequence of the Ras-family GTPase, Rap, in S. rosetta is 92% identical to that of human Rap, indicating that the selective pressures on Rap did not change as much in the transition into the vertebrate lineage. It is intriguing to note that the high conservation of Ras in vertebrates can possibly be traced to interactions with SOS, which may have been a factor that constrained the evolution of Ras (Bandaru et al. 2017).
The mutational sensitivity of human H-Ras was determined by screening a library consisting of all possible point mutations to the protein sequence, in the presence of a GTPase-activating protein (GAP) and a GEF (Bandaru et al. 2017). It was found that the overall conservation of Ras was a consequence of most residues being involved in binding the nucleotide, maintaining protein stability, interacting with the GAP and the GEF, as well as suppressing spontaneous activation. Interestingly, four regions in the sequence of Ras are more tolerant of mutation, and these contain residues that differed between invertebrate and human Ras, yet are nearly invariant in vertebrates. Residues in two of the four variable regions of Ras are directly involved in allosteric binding to SOS, and residues in the remaining two variable regions are involved in a two-state conformational transition that might affect the affinity of Ras for the allosteric site of SOS. This conformational transition may be triggered by the binding of an allosteric effector between the α3 and α4 helices of Ras (Buhrman et al. 2010), leading to the outward rotation of the Tyr 71 residue, which appears inconsistent with Ras binding to the allosteric site of SOS. The structure of S. rosetta Ras adopts the outward conformation of Tyr 71, whereas the structure human Ras does not, and S. rosetta Ras also fails to allosterically activate SOS (Bandaru et al. 2017). These observations suggest that SOS may have coevolved with Ras in vertebrates.
CONCLUDING REMARKS
Ras is a highly conserved and central molecular switch in metazoan cell signaling. In vertebrates, Ras displays the unique property of being able to activate its own activator, SOS. Structural and functional analysis has revealed the remarkable complexity underlying the interdependent activation of Ras and SOS. The culmination of the release of autoinhibition of SOS by regulatory domains is the allosteric binding of Ras•GTP to SOS. This binding event sets up a positive feedback loop, which allows receptor tyrosine kinases to generate highly nonlinear responses in response to extracellular stimuli. This has profound consequences on cell signaling, most importantly in generating a sharp response in pathways that ultimately control cellular differentiation and growth. It is intriguing that the structural element in SOS that promotes the allosteric activation by Ras is the REM domain, which is also present in other Ras-specific GEFs. An intriguing question for further investigation is whether the REM domains of other Ras-specific GEFs modulate activity by interacting with as-of-yet unidentified allosteric regulators.
REFERENCES
* Reference is also in this collection.
- Aghazadeh B, Lowry WE, Huang XY, Rosen MK. 2000. Structural basis for relief of autoinhibition of the Dbl homology domain of proto-oncogene Vav by tyrosine phosphorylation. Cell 102: 625–633. [DOI] [PubMed] [Google Scholar]
- Aronheim A, Engelberg D, Li N, al-Alawi N, Schlessinger J, Karin M. 1994. Membrane targeting of the nucleotide exchange factor Sos is sufficient for activating the Ras signaling pathway. Cell 78: 949–961. [DOI] [PubMed] [Google Scholar]
- Bandaru P, Shah NH, Bhattacharyya M, Barton JP, Kondo Y, Cofsky JC, Gee CL, Chakraborty AK, Kortemme T, Ran-ganathan R, Kuriyan J. 2017. Deconstruction of the Ras switching cycle through saturation mutagenesis. eLife 6: e27810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxevanis AD, Arents G, Moudrianakis EN, Landsman D. 1995. A variety of DNA-binding and multimeric proteins contain the histone fold motif. Nucleic Acids Res 23: 2685–2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. 2009. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol 5: 593–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop AL, Hall A. 2000. Rho GTPases and their effector proteins. Biochem J 348: 241–255. [PMC free article] [PubMed] [Google Scholar]
- Boguski MS, McCormick F. 1993. Proteins regulating Ras and its relatives. Nature 366: 643–654. [DOI] [PubMed] [Google Scholar]
- Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. 1998. The structural basis of the activation of Ras by Sos. Nature 394: 337–343. [DOI] [PubMed] [Google Scholar]
- * Bos JL. 2017. From ras to Rap and back, a journey of 35 years. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a031468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. 2007. GEFs and GAPs: Critical elements in the control of small G proteins. Cell 129:865–877. [DOI] [PubMed] [Google Scholar]
- Boykevisch S, Zhao C, Sondermann H, Philippidou P, Halegoua S, Kuriyan J, Bar-Sagi D. 2006. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr Biol 16: 2173–2179. [DOI] [PubMed] [Google Scholar]
- Buday L, Downward J. 1993. Epidermal growth factor regulates p21ras through the formation of a complex of receptor, Grb2 adapter protein, and Sos nucleotide exchange factor. Cell 73: 611–620. [DOI] [PubMed] [Google Scholar]
- Buday L, Egan SE, Rodriguez Viciana P, Cantrell DA, Downward J. 1994. A complex of Grb2 adaptor protein, Sos exchange factor, and a 36-kDa membrane-bound tyrosine phosphoprotein is implicated in ras activation in T cells. J Biol Chem 269: 9019–9023. [PubMed] [Google Scholar]
- Buhrman G, Holzapfel G, Fetics S, Mattos C. 2010. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc Natl Acad Sci 107: 4931–4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns MC, Sun Q, Daniels RN, Camper D, Kennedy JP, Phan J, Olejniczak ET, Lee T, Waterson AG, Rossanese OW, et al. 2014. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc Natl Acad Sci 111: 3401–3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerione RA, Zheng Y. 1996. The Dbl family of oncogenes. Curr Opin Cell Biol 8: 216–222. [DOI] [PubMed] [Google Scholar]
- Chardin P, Camonis JH, Gale NW, van Aelst L, Schlessinger J, Wigler MH, Bar-Sagi D. 1993. Human Sos1: A guanine nucleotide exchange factor for Ras that binds to GRB2. Science 260: 1338–1343. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. 2013. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol Rev 93: 269–309. [DOI] [PubMed] [Google Scholar]
- Christensen SM, Tu HL, Jun JE, Alvarez S, Triplet MG, Iwig JS, Yadav KK, Bar-Sagi D, Roose JP, Groves JT. 2016. Oneway membrane trafficking of SOS in receptor-triggered Ras activation. Nat Struct Mol Biol 23: 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbalan-Garcia S, Margarit SM, Galron D, Yang SS, Bar-Sagi D. 1998. Regulation of Sos activity by intramolecular interactions. Mol Cell Biol 18: 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das J, Ho M, Zikherman J, Govern C, Yang M, Weiss A, Chakraborty AK, Roose JP. 2009. Digital signaling and hysteresis characterize ras activation in lymphoid cells. Cell 136: 337–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij J, Rehmann H, van Triest M, Cool RH, Wittinghofer A, Bos JL. 2000. Mechanism of regulation of the Epac family of cAMP-dependent RapGEFs. J Biol Chem 275: 20829–20836. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Bottorff DA, Chan EY, Stang SL, Dunn RJ, Stone JC. 1998. RasGRP, a Ras guanyl nucleotide- releasing protein with calcium- and diacylglycerol-binding motifs. Science 280: 1082–1086. [DOI] [PubMed] [Google Scholar]
- Ebinu JO, Stang SL, Teixeira C, Bottorff DA, Hooton J, Blumberg PM, Barry M, Bleakley RC, Ostergaard HL, Stone JC. 2000. RasGRP links T-cell receptor signaling to Ras. Blood 95: 3199–3203. [PubMed] [Google Scholar]
- Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA. 1993. Association of Sos Ras exchange protein with Grb2 is implicated in tyrosine kinase signal transduction and transformation. Nature 363: 45–51. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, David MD, Ehrhardt GRA, Schrader JW. 2004. Distinct mechanisms determine the patterns of differential activation of H-Ras, N-Ras, K-Ras 4B, and M-Ras by receptors for growth factors or antigen. Mol Cell Biol 24: 6311–6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay GM, Pawson T. 2008. How is SOS activated? Let us count the ways. Nat Struct Mol Biol 15: 538–540. [DOI] [PubMed] [Google Scholar]
- Freedman TS, Sondermann H, Friedland GD, Kortemme T, Bar-Sagi D, Marqusee S, Kuriyan J. 2006. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl Acad Sci 103: 16692–16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman TS, Sondermann H, Kuchment O, Friedland GD, Kortemme T, Kuriyan J. 2009. Differences in flexibility underlie functional differences in the Ras activators son of sevenless and Ras guanine nucleotide releasing factor 1. Structure 17: 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale NW, Kaplan S, Lowenstein EJ, Schlessinger J, Bar-Sagi D. 1993. Grb2 mediates the EGF-dependent activation of guanine nucleotide exchange on Ras. Nature 363: 88–92. [DOI] [PubMed] [Google Scholar]
- Goody RS, Frech M, Wittinghofer A. 1991. Affinity of guanine nucleotide binding proteins for their ligands: Facts and artefacts. Trends Biochem Sci 16: 327–328. [DOI] [PubMed] [Google Scholar]
- Gureasko J, Galush WJ, Boykevisch S, Sondermann H, Bar-Sagi D, Groves JT, Kuriyan J. 2008. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat Struct Mol Biol 15: 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gureasko J, Kuchment O, Makino DL, Sondermann H, Bar-Sagi D, Kuriyan J. 2010. Role of the histone domain in the autoinhibition and activation of the Ras activator Son of Sevenless. Proc Natl Acad Sci 107: 3430–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BE, Yang SS, Boriack-Sjodin PA, Kuriyan J, Bar-Sagi D. 2001. Structure-based mutagenesis reveals distinct functions for Ras switch 1 and switch 2 in Sos-catalyzed guanine nucleotide exchange. J Biol Chem 276: 27629–27637. [DOI] [PubMed] [Google Scholar]
- Hall BE, Yang SS, Bar-Sagi D. 2002. Autoinhibition of Sos by intramolecular interactions. Front Biosci 7: d288–d294. [DOI] [PubMed] [Google Scholar]
- Huang WYC, Yan Q, Lin WC, Chung JK, Hansen SD, Christensen SM, Tu HL, Kuriyan J, Groves JT. 2016. Phosphotyrosine-mediated LAT assembly on membranes drives kinetic bifurcation in recruitment dynamics of the Ras activator SOS. Proc Natl Acad Sci 113: 8218–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen L, Tu HL, Lin WC, Christensen SM, Abel SM, Iwig J, Wu HJ, Gureasko J, Rhodes C, Petit RS, et al. 2014. Molecular kinetics. Ras activation by SOS: Allosteric regulation by altered fluctuation dynamics. Science 345: 50–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwig JS, Vercoulen Y, Das R, Barros T, Limnander A, Che Y, Pelton JG, Wemmer DE, Roose JP, Kuriyan J. 2013. Structural analysis of autoinhibition in the Ras-specific exchange factor RasGRP1. eLife 2: e00813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaumot M, Yan J, Clyde-Smith J, Sluimer J, Hancock JF. 2002. The linker domain of the Ha-Ras hypervariable region regulates interactions with exchange factors, Raf-1 and phosphoinositide 3-kinase. J Biol Chem 277: 272–278. [DOI] [PubMed] [Google Scholar]
- Jeng HH, Taylor LJ, Bar-Sagi D. 2012. Sos-mediated cross-activation of wild-type Ras by oncogenic Ras is essential for tumorigenesis. Nat Commun 3: 1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CW, Reid D, Parker JA, Salter S, Knihtila R, Kuzmic P, Mattos C. 2017. The small GTPases K-Ras, N-Ras and H-Ras have distinct biochemical properties determined by allosteric effects. J Biol Chem 292: 12981–12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JE, Rubio I, Roose JP. 2013a. Regulation of ras exchange factors and cellular localization of ras activation by lipid messengers in T cells. Front Immunol 4: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jun JE, Yang M, Chen H, Chakraborty AK, Roose JP. 2013b. Activation of extracellular signal-regulated kinase but not of p38 mitogen-activated protein kinase pathways in lymphocytes requires allosteric activation of SOS. Mol Cell Biol 33: 2470–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausner RD, Donaldson JG, Lippincott-Schwartz J. 1992. Brefeldin A: Insights into the control of membrane traffic and organelle structure. J Cell Biol 116: 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebe C, Prinz H, Wittinghofer A, Goody RS. 1995. The kinetic mechanism of Ran–nucleotide exchange catalyzed by RCC1. Biochemistry 34: 12543–12552. [DOI] [PubMed] [Google Scholar]
- Lee YK, Low-Nam ST, Chung JK, Hansen SD, Lam HYM, Alvarez S, Groves JT. 2017. Mechanism of SOS PR-do-main autoinhibition revealed by single-molecule assays on native protein from lysate. Nat Commun 8: 15061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA, Ferguson KM. 2000. Signal-dependent membrane targeting by Pleckstrin homology (PH) domains. Biochem J 350: 1–18. [PMC free article] [PubMed] [Google Scholar]
- Lenzen C, Cool RH, Prinz H, Kuhlmann J, Wittinghofer A. 1998. Kinetic analysis by fluorescence of the interaction between Ras and the catalytic domain of the guanine nucleotide exchange factor Cdc25Mm. Biochemistry 37: 7420–7430. [DOI] [PubMed] [Google Scholar]
- Li N, Batzer A, Daly R, Yajnik V, Skolnik E, Chardin P, Bar-Sagi D, Margolis B, Schlessinger J. 1993. Guanine-nucleotide-releasing factor hSos1 binds to Grb2 and links receptor tyrosine kinases to Ras signalling. Nature 363: 85–88. [DOI] [PubMed] [Google Scholar]
- Lim WA, Richards FM, Fox RO. 1994. Structural determinants of peptide-binding orientation and of sequence specificity in SH3 domains. Nature 372: 375–379. [DOI] [PubMed] [Google Scholar]
- Lowenstein EJ, Daly RJ, Batzer AG, Li W, Margolis B, Lammers R, Ullrich A, Skolnik EY, Bar-Sagi D, Schlessinger J. 1992. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell 70: 431–442. [DOI] [PubMed] [Google Scholar]
- Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. 2003. Structural evidence for feedback activation by Ras•TP of the Ras-specific nucleotide exchange factor SOS. Cell 112: 685–695. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Corpina RA, Goldberg J. 2003. Crystal structure of ARF1*Sec7 complexed with Brefeldin A and its implications for the guanine nucleotide exchange mechanism. Mol Cell 12: 1403–1411. [DOI] [PubMed] [Google Scholar]
- Nimnual AS, Yatsula BA, Bar-Sagi D. 1998. Coupling of Ras and Rac guanosine triphosphatases through the Ras exchanger Sos. Science 279: 560–563. [DOI] [PubMed] [Google Scholar]
- Pacold ME, Suire S, Perisic O, Lara-Gonzalez S, Davis CT, Walker EH, Hawkins PT, Stephens L, Eccleston JF, Williams RL. 2000. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell 103: 931–943. [DOI] [PubMed] [Google Scholar]
- Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. 2011. An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol 7: 585–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad A, Zikherman J, Das J, Roose JP, Weiss A, Chakraborty AK. 2009. Origin of the sharp boundary that discriminates positive and negative selection of thymocytes. Proc Natl Acad Sci 106: 528–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmann H, Das J, Knipscheer P, Wittinghofer A, Bos JL. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439: 625–628. [DOI] [PubMed] [Google Scholar]
- Rehmann H, Arias-Palomo E, Hadders MA, Schwede F, Llorca O, Bos JL. 2008. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455: 124–127. [DOI] [PubMed] [Google Scholar]
- Renault L, Guibert B, Cherfils J. 2003. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature 426: 525–530. [DOI] [PubMed] [Google Scholar]
- Roberts AE, Araki T, Swanson KD, Montgomery KT, Schiripo TA, Joshi VA, Li L, Yassin Y, Tamburino AM, Neel BG, et al. 2007. Germline gain-of-function mutations in SOS1 cause Noonan syndrome. Nat Genet 39: 70–74. [DOI] [PubMed] [Google Scholar]
- Rojas AM, Fuentes G, Rausell A, Valencia A. 2012. The Ras protein superfamily: Evolutionary tree and role of conserved amino acids. J Cell Biol 196: 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roose JP, Mollenauer M, Ho M, Kurosaki T, Weiss A. 2007. Unusual interplay of two types of Ras activators, RasGRP and SOS, establishes sensitive and robust Ras activation in lymphocytes. Mol Cell Biol 27: 2732–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. 2005. GEF means go: Turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol 6: 167–180. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, McGlade J, Mbamalu G, Pelicci G, Daly R, Li W, Batzer A, Thomas S, Brugge J, Pelicci PG, et al. 1992. Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360: 689–692. [DOI] [PubMed] [Google Scholar]
- Rozakis-Adcock M, Fernley R, Wade J, Pawson T, Bowtell D. 1993. The SH2 and SH3 domains of mammalian Grb2 couple the EGF receptor to the Ras activator mSos1. Nature 363: 83–85. [DOI] [PubMed] [Google Scholar]
- Saliba A-E, Vonkova I, Ceschia S, Findlay GM, Maeda K, Tischer C, Deghou S, van Noort V, Bork P, Pawson T, et al. 2014. A quantitative liposome microarray to systematically characterize protein-lipid interactions. Nat Methods 11: 47–50. [DOI] [PubMed] [Google Scholar]
- Soisson SM, Nimnual AS, Uy M, Bar-Sagi D, Kuriyan J. 1998. Crystal structure of the Dbl and Pleckstrin homology domains from the human Son of sevenless protein. Cell 95: 259–268. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Soisson SM, Bar-Sagi D, Kuriyan J. 2003. Tandem histone folds in the structure of the N-terminal segment of the ras activator Son of Sevenless. Structure 11: 1583–1593. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Soisson SM, Boykevisch S, Yang S-S, Bar-Sagi D, Kuriyan J. 2004. Structural analysis of autoinhibition in the Ras activator Son of sevenless. Cell 119: 393–405. [DOI] [PubMed] [Google Scholar]
- Sondermann H, Nagar B, Bar-Sagi D, Kuriyan J. 2005. Computational docking and solution x-ray scattering predict a membrane-interacting role for the histone domain of the Ras activator son of sevenless. Proc Natl Acad Sci 102: 16632–16637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Ditlev JA, Hui E, Xing W, Banjade S, Okrut J, King DS, Taunton J, Rosen MK, Vale RD. 2016. Phase separation of signaling molecules promotes T cell receptor signal transduction. Science 352: 595–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Pennacchio LA, Zhao C, Yadav KK, Fodale V, Sarkozy A, Pandit B, Oishi K, Martinelli S, Schackwitz W, et al. 2007. Gain-of-function SOS1 mutations cause a distinctive form of Noonan syndrome. Nat Genet 39: 75–79. [DOI] [PubMed] [Google Scholar]
- Terasawa H, Kohda D, Hatanaka H, Tsuchiya S, Ogura K, Nagata K, Ishii S, Mandiyan V, Ullrich A, Schlessinger J, et al. 1994. Structure of the N-terminal SH3 domain of GRB2 complexed with a peptide from the guanine nucleotide releasing factor Sos. Nat Struct Mol Biol 1: 891–897. [DOI] [PubMed] [Google Scholar]
- van Dam TJP, Rehmann H, Bos JL, Snel B. 2009. Phylogeny of the CDC25 homology domain reveals rapid differentiation of Ras pathways between early animals and fungi. Cell Signal 21: 1579–1585. [DOI] [PubMed] [Google Scholar]
- Vercoulen Y, Kondo Y, Iwig JS, Janssen AB, White KA, Amini M, Barber DL, Kuriyan J, Roose JP. 2017. A histidine pH sensor regulates activation of the Ras-specific guanine nucleotide exchange factor RasGRP1. eLife doi: 10.7554/eLife.209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo U, Vajpai N, Flavell L, Bobby R, Breeze AL, Embrey KJ, Golovanov AP. 2016. Monitoring Ras interactions with the nucleotide exchange factor Son of Sevenless (Sos) using site-specific NMR reporter signals and intrinsic fluorescence. J Biol Chem 291: 1703–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead IP, Campbell S, Rossman KL, Der CJ. 1997. Dbl family proteins. Biochim Biophys Acta 1332: F1–F23. [DOI] [PubMed] [Google Scholar]
- Wittekind M, Mapelli C, Farmer BT, Suen KL, Goldfarb V, Tsao J, Lavoie T, Barbacid M, Meyers CA, Mueller L. 1994. Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy. Biochemistry 33: 13531–13539. [DOI] [PubMed] [Google Scholar]
- Worthylake DK, Rossman KL, Sondek J. 2000. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature 408: 682–688. [DOI] [PubMed] [Google Scholar]
- Yadav KK, Bar-Sagi D. 2010. Allosteric gating of Son of sevenless activity by the histone domain. Proc Natl Acad Sci 107: 3436–3440. [DOI] [PMC free article] [PubMed] [Google Scholar]