SUMMARY
US public health laboratories began reporting Escherichia coli O157 isolates to CDC in 1996. We describe temporal and geographical patterns of isolates reported from 1996 to 2011 and demographics of persons whose specimens yielded isolates. We calculated annual E. coli O157 isolation rates/100000 persons by patient’s state of residence, county of residence, age, and sex using census data. The average annual isolation rate was 0·84. The average isolation rate in northern states (1·52) was higher than in southern states (0·43). Counties with ≥76% rural population had a lower isolation rate (0·67) than counties with ≤ 25%, 26–50%, and 51–75% rural populations (0·81, 0·92, and 0·81, respectively). The highest isolation rate (3·19) was in children aged 1–4 years. Infections were seasonal with 49% of isolates collected during July to September. Research into reasons for higher incidence in northern states and for seasonality could guide strategies to prevent illnesses.
Keywords: Escherichia coli O157, haemolytic uraemic syndrome, HUS, Shiga toxin, surveillance
INTRODUCTION
Escherichia coli O157 causes an estimated 96000 illnesses annually in the USA [1]. Infections typically result in severe abdominal cramping and diarrhoea, which is often bloody [2]. Complications include haemolytic uraemic syndrome (HUS) and death.
E. coli O157 infection became nationally notifiable in the USA in 1994, and passive, national Laboratory-based Enteric Disease Surveillance (LEDS) began collecting information on laboratory-confirmed isolates in 1996. We used these data to describe temporal and geographical patterns of E. coli O157 isolates reported to the US Centers for Disease Control and Prevention (CDC) from 1996 to 2011 and the demographics of the persons whose clinical specimens yielded the isolates.
METHODS
We evaluated data on E. coli O157 isolates reported by state public health laboratories to CDC from 1996 to 2011. Clinical laboratories in all 50 states and territories are requested (and, in some states, required) to forward clinical isolates of E. coli O157 to their state public health department laboratory. State public health laboratories send reports containing information on patient’s sex, age, race, ethnicity, and county and state of residence, as well as specimen source, serotype, and date of collection. Reports are sent electronically to CDC. Isolates with the same state identifiers that also match by age, state, and specimen source and have specimen isolation dates within 30 days of one another are considered duplicates and discarded.
We calculated average annual E.coli O157 isolation rates/100000 persons by patient’s state of residence (including District of Columbia), age, and sex using intercensal population estimates from the US Census Bureau. We used the 37th parallel north to define northern and southern states because it is conveniently the official border of multiple states. We defined a southern state as any state entirely south of the 37th parallel north and the remainder as northern states. In the analysis of northern vs. southern states, we excluded (1) Alaska and Hawaii, because they are not part of the contiguous USA, and (2) California and Nevada, because a large proportion of their population is on both sides of the 37th parallel north.
To explore urban/rural differences in annual E. coli O157 isolation rates, we analysed the data by census-derived categories of counties based on percentage of the population residing in a rural area: 425%, 26–50%, 51–75%, and 576%. We constructed standard and zero-inflated Poisson and negative binomial regression models. The zero-inflated count regression models account for the possibility that zero illness counts reported by some counties are related not to a true lack of illness, but to detection or reporting that is less complete than in other counties [3]. We conducted the analysis on all 3141 counties. To exclude counties that may have been more likely to have incomplete reporting, we also conducted the analysis on a dataset that excluded counties with a population <1000 and those that did not report any Salmonella or E. coli O157 isolates during the 16-year period. We then selected a best model based on quality of fit and epidemiology, and report estimated isolation rates by rural category with their 95% confidence intervals (CIs).
Data were analysed using SAS v. 9.3 (SAS Institute Inc., USA).
RESULTS
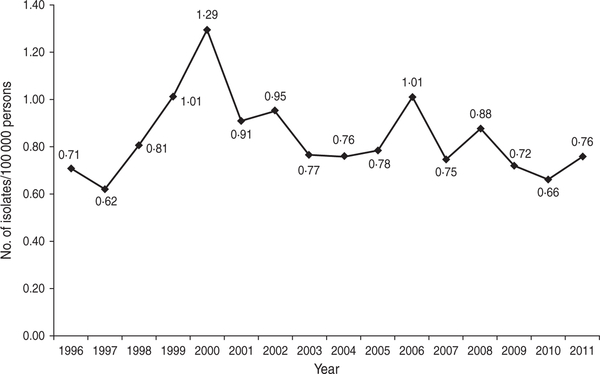
During 1996–2011, CDC received reports of 38895 laboratory-confirmed E. coli O157 isolates from all 50 states and the District of Columbia. Of these, 36841 (95%) were identified as O157:H7. Of the 35095 isolates with known sources, 33886 (97%) were from stool specimens and the remainder were from other sources including urine (163, 0·5%), blood (91, 0·3%), and wounds or abscesses (40, 0·1%). The average annual isolation rate nationally was 0·84/100000 persons. In 1996, the isolation rate was 0·71/100 000 persons, which decreased to 0·62/100000 persons in 1997 (Fig. 1). Isolation rates then steadily increased to a peak of 1·29/100000 persons in 2000, followed by a decline to 0·76/100000 persons in 2004. From 2004–2011, isolation rates were relatively stable with the exception of increases in 2006 and 2008.
Fig.1.
Annual isolation of E. coli O157, USA, Laboratory-based Enteric Disease Surveillance, 1996–2011.
Of the isolates with patient data available, 53% were from female patients and the median age in patients was 15 (range 0–108) years. Comparatively, the median age of the general population was 37 years in 2010 [4]. Based on the 33937 (87%) isolates with known patient’s age and sex data, the highest isolation rate (3·19/100000 persons) was in children aged 1–4 years (Table 1). The rate declined with increasing age to a nadir of 0·30/100 000 persons in those aged 30–39 years, then steadily increased to 0·62/100 000 persons in those aged 70–79 years and was similar (0·58) in patients aged ≥80 years.
Table 1.
Average annual isolation rate (isolates/100000 persons) of E. coli O157 by age group and sex, USA, Laboratory-based Enteric Disease Surveillance, 1996–2011
| Age group (years) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| <1 | 1–4 | 5–9 | 10–19 | 20–29 | 30–39 | 40–49 | 50–59 | 60–69 | 70–79 | ≥80 | |
| Female | 1.08 | 2.93 | 1.47 | 0.92 | 0.64 | 0.35 | 0.37 | 0.52 | 0.64 | 0.63 | 0.58 |
| Male | 1.01 | 3.06 | 1.60 | 1.12 | 0.49 | 0.21 | 0.22 | 0.30 | 0.37 | 0.52 | 0.49 |
| Total | 1.14 | 3.19 | 1.61 | 1.07 | 0.59 | 0.30 | 0.31 | 0.43 | 0.54 | 0.62 | 0.58 |
For children other than infants (aged <12 months), laboratory-confirmed isolation rates were higher in boys than girls (Table 1). For infants and those in age groups 520–29 years, the rates were higher in females than males.
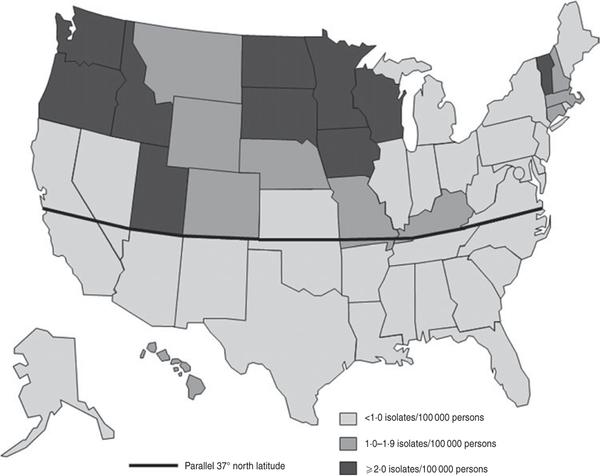
In the contiguous USA, the average annual isolation rate was 1·52/100000 persons in states in the north and 0·43 in states in the south. All states with an average annual isolation rate >1·0 were in the north (Fig. 2).
Fig. 2.
Average annual isolation rate of E. coli O157 by state, USA, Laboratory-based Enteric Disease Surveillance, 1996–2011 (n=38895).
For the rurality analysis, we found significant over-dispersion of illness counts relative to simple Poisson regression. In 3141 counties, 35% never reported anE. coli O157 isolate over the 16-year time span; 56% of the non-reporting counties had 576% rural populations. We examined the occurrence of many zero illness counts in zero-inflated and other models, and found no evidence of zero inflation. A negative binomial regression model had the best fit. The model applied to the complete dataset estimated isolation rates in counties with 425%, 26–50%, 51–75%, and 576% rural populations as 0·81 (95% CI 0·73–0·89), 0·92 (95% CI 0·84–1·0), 0·81 (95% CI 0·74–0·88), and 0·67 (95% CI 0·61–0·73)/100000 persons, respectively. We found similar results when this model was applied to the dataset that excluded the 105 (3%) counties that may have been more likely to have incomplete reporting.
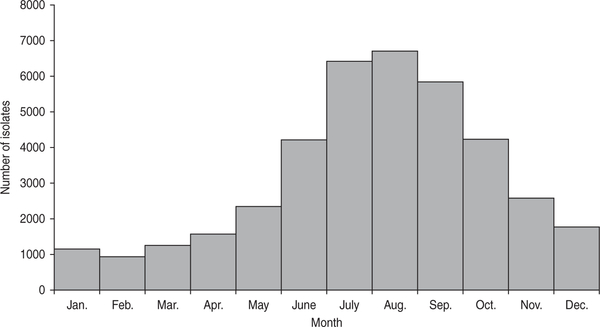
Infections were sharply seasonal, with 49% of isolates collected during July–September and only 9% during January–March (Fig. 3). This summer predominance was similarly present in both northern and southern regions.
Fig. 3.
Number of E. coli O157 isolates by month, USA, Laboratory-based Enteric Disease Surveillance, 1996–2011 (n=38895).
DISCUSSION
This study is the first to evaluate longitudinal US national data on E. coli O157 infections and highlights the value of national surveillance programmes to evaluate geographically disperse data. Isolation rates were highest in northern states, during summer months, and in children aged 1–4 years.
The reason that states in the northernmost latitudes have higher incidence rates is not known, but this finding is consistent with other studies and reports [2, 5]. A multicentre hospital-based study during 1990–1992 reported a northern predominance [2], and the northernmost Foodborne Diseases Active Surveillance System (Food Net) sites have reported a higher incidence than other sites [6], even after adjusting for outbreaks and testing practices [7]. In addition, analysis of outbreak data indicates a higher rate of outbreaks in northern than in southern states (CDC, unpublished data). This increasing incidence with increasing proximity to the poles may be a global phenomenon. Within continental Europe, reported rates tend to be higher in northern European countries, such as Germany and The Netherlands, than in southern European countries, such as Spain and Italy [8]. In addition, in the UK and North America, incidence is highest in the northernmost countries, Scotland and Canada, respectively [9]. Conversely, in the southern hemisphere, higher rates of illness tend to be reported from the more southerly nations. For example, Argentina, which has a high cattle density, has the highest reported incidence rates of HUS globally; most of Argentina’s HUS cases are caused by Shiga toxin-producing E. coli infection, and the predominant sero group is E. coli O157 [10, 11]. The southern Africa region was among the first areas to report HUS cases [12] and reported the largest E. coli. coli.E.coli O157 outbreak in Africa [13]. A latitude effect suggests a climatic role in transmission of E. coli O157 or shedding in cattle, the primary reservoir [14]. Number of daylight hours has been reported to correlate positively with increased shedding of E. coli O157 by cattle. In an experimental study, cattle exposed daily for 60 days to ~ 12 h of natural light followed by an additional 5 h of artificial light were found to shed more E. coli O157 organisms than cattle exposed only to the 12 h of natural light [15]. These authors suggested a possible role of melatonin, a hormone with seasonal fluctuations and possible effects on the immune system. Such a mechanism may explain our findings, given the longer daylight hours in northern than southern states in the USA during the summer.
Other possible reasons for our geographical findings include geographical differences in animal carriage, meat processing, or urbanization. In addition, environmental factors including water sources and cattle density may play a role. Studies in Canada and Germany found cattle density to be positively associated with incidence of E. coli O157 infections in humans [16–18]. For this reason, we expected to find higher isolation rates in rural counties but instead found the lowest rate in the group of counties with the highest rural population. A large proportion of rural counties did not report a single case over the 16-year period. It is possible that persons in rural counties are less likely to have a stool cultured, or that clinical laboratories that serve rural areas test fewer samples for E. coli O157 or report cases less consistently; if so, our finding may be an artifact of diagnosis or surveillance. However, we found no evidence that non-reporting counties influenced our findings. Our finding is plausible if persons in rural counties have many opportunities for low-level exposure that results in immunological protection without clinical illness. An estimated 68% of E. coli O157 infections nationally are transmitted by food, so rural residence may not have a large role in determining variation in rates of illness [1].
Consistent with other reports [2, 19], we found that infections were most common in the summer months. While a study based on FoodNet Population Survey data found no seasonal variation in ground beef consumption patterns [20], a higher proportion of cattle shed E. coli O157 during the summer months [21], which probably relates to the increased prevalence of E. coli O157 contamination of beef during those months [22]. In addition, other modes of transmission such as waterborne and direct animal contact may also be affected by this increased shedding.
Incidence was highest in children aged 1–4 years. Person-to-person transmission has been well documented, accounting for 14% of US E. coli O157 outbreaks from 1982 to 2002, mostly in child daycare centres [23]. Hygienic factors may result in more frequent exposure in children, and immunological susceptibility may also play an important role.
Sources of E. coli O157 infection include food, particularly ground beef, water, contact with animals or their environment, and direct contact with another person or fomite [23]. Most prevention efforts have focused on decreasing the contamination of ground beef. The decline in incidence from 2000 to 2003 mirrored the 76% decline in contamination of ground beef samples with E. coli O157 from 2000 to 2004 [24].
The pattern of isolation rates observed in our data, including the decrease in incidence from 2000 to 2004, correlates well with trends in FoodNet sites, which included 15% of the US population in 2009 [5]. The increase in incidence from 1996 to 2000 may be due to increased testing by clinical laboratories [25]. In 2002, in response to continued outbreaks and ground beef recalls, USDA tightened regulations and the beef grinding industry made changes that were followed by marked declines in 2003 and 2004 in the proportion of ground beef samples that yielded E. coli O157 which possibly contributed to decreasing incidence [26, 27]. The spikes in incidence in 2006 and 2008 may reflect actual increases in illnesses but could have been related to increased testing related to several large, multistate outbreaks. In 2006, an outbreak of E. coli O157 infections from contaminated spinach resulted in 205 illnesses in 26 states [28, 29]. In 2008, a widespread outbreak of Salmonella serotype Saintpaul infections associated mostly with contaminated jalapeño and Serrano peppers caused 1500 illnesses in 43 states [30]. The increased awareness and publicity caused by these outbreaks may have led to increased patient visits and increased stool testing by physicians.
We did not calculate confidence intervals in our analysis because the surveillance system is designed to capture all isolates reported at the national level and does not represent a subset of isolates (e.g. National Antimicrobial Resistance Monitoring System) or of the population (e.g. FoodNet). Therefore, with no sampling within the surveillance system, there are no appropriate statistical methods to assess the uncertainty within it.
There are still limitations to the interpretation of these data. First, only persons with illness severe enough for the person to seek medical care and for which the provider ordered cultures are represented. Second, the completeness of reporting varies between states because public health requirements and infrastructure vary at the state and local level. Because our results correlate with trends found in FoodNet, this variation in reporting appears to have had a minimal effect on the evaluation of national patterns but does dictate caution in the interpretation of state and regional data. This is particularly an issue with Maine, Nebraska, Texas, and Wyoming; after 2006, these states began using a reporting system from which serogroup information could not be retrieved. There is also variability in information provided to the surveillance system. For example, nearly 13% of reports lacked age and gender information. However, there is no evidence on the extent that this may have biased the patterns related to age or gender.
Prevention of E. coli O157 infections depends on understanding mechanisms of transmission. Further research into decreasing carriage and shedding by cattle, preventing contamination of beef and other foods such as produce, and better understanding of the reasons for the higher incidence in northern states and for the summer seasonality could help in developing strategies to prevent illnesses. Analyses that estimate the proportion of illnesses attributable to specific foodborne (e.g. ground beef, leafy vegetables) and non-foodborne transmission routes could help in targeting prevention efforts.
ACKNOWLEDGEMENTS
The authors thank Robert M. Hoekstra (Centers for Disease Control and Prevention, Atlanta, GA), for his statistical support and guidance.
Footnotes
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Scallan E, et al. Foodborne illness acquired in the United States - major pathogens. Emerging Infectious Diseases 2011;17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slutsker L, et al. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiologic features. Annals of Internal Medicine 1997; 126: 505–513. [DOI] [PubMed] [Google Scholar]
- 3.Hilbe J Negative Binomial Regression, 2nd edn New York: Cambridge University Press, 2011. [Google Scholar]
- 4.US Census 2010. (http://www.census.gov/2010census/data/).
- 5.Boyce TG, Swerdlow DL, Griffin PM. Escherichia coli O157:H7 and the hemolytic-uremic syndrome. New England Journal of Medicine 1995; 333: 364–368. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. Preliminary FoodNet data on the incidence of infection with pathogens transmitted commonly through food – 10 states, 2009.Morbidity and Mortalilty Weekly Report 2010; 59: 418–422. [PubMed] [Google Scholar]
- 7.Bender J, et al. Factors affecting surveillance data on Escherichia coli O157 infections collected from Food-Net sites, 1996–1999. Clinical Infectious Diseases 2004; 38: S157–S164. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli A, Tozzi A. Epidemiology of shiga toxin-producing Escherichia coli infections in continental Europe In: Kaper JB, O’Brien AD, eds. Escherichia coli O157:H7 and Other Shiga Toxin-producing E. coli Strains. Washington, D.C.: American Society for Microbiology Press, 1998, pp. 38–48. [Google Scholar]
- 9.Waters JR, Sharp JC, Dev VJ. Infection caused by Escherichia coli O157:H7 in Alberta, Canada, and in Scotland: a five-year review, 1987–1991. Clinical Infectious Diseases 1994;19: 834–43. [DOI] [PubMed] [Google Scholar]
- 10.Rivas M, et al. Characterization and epidemiologic sub-typing of Shiga toxin-producing Escherichia coli strains isolated from hemolytic uremic syndrome and diarrhea cases in Argentina. Foodborne Pathogens and Disease 2006; 3:88–96. [DOI] [PubMed] [Google Scholar]
- 11.Rivas M, et al. Risk factors for sporadic Shiga toxin-producing Escherichia coli infections in children, Argentina. Emerging Infectious Diseases 2008; 14: 763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kibel MA, Barnard PJ. The haemolytic-uraemic syndrome: a survey in Southern Africa. South African Medical Journal 1968; 42: 692–698. [PubMed] [Google Scholar]
- 13.Effler E, et al. Factors contributing to the emergence of Escherichia coli O157 in Africa. Emerging Infectious Diseases 2001; 7: 812–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wells JG, et al. Isolation of Escherichia coli serotype O157:H7 and other Shiga-like-toxin-producing E. coli from dairy cattle. Journal of Clinical Microbiology 1991; 29: 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edrington T, et al. Seasonal shedding of Escherichia coli O157:H7 in ruminants: a new hypothesis. Foodborne Pathogens and Disease 2006; 3: 413–421. [DOI] [PubMed] [Google Scholar]
- 16.Frank C, et al. Cattle density and Shiga toxin-producing Escherichia coli infection in Germany: increased risk for most but not all serogroups. Vector-Borne and Zoonotic Diseases 2008; 8: 635–643. [DOI] [PubMed] [Google Scholar]
- 17.Michel P, et al. Temporal and geographical distributions of reported cases of Escherichia coli O157:H7 infection in Ontario. Epidemiology and Infection 1999; 122: 193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Innocent GT, Mellor DJ, McEwen SA, et al. Spatial and temporal epidemiology of sporadic human cases of Escherichia coli O157 in Scotland, 1996–1999. Epidemiology and Infection 2005; 133: 1033–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ostroff SM, et al. Infections with Escherichia coli O157: H7 in Washington state. The first year of statewide disease surveillance. Journal of the American Medical Association 1989; 262: 355–359. [PubMed] [Google Scholar]
- 20.Taylor E, et al. Ground beef consumption patterns in the United States, FoodNet, 2006 through 2007. Journal of Food Protection 2012; 75: 341–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hancock DD, et al. A longitudinal study of Escherichia coli O157 in fourteen cattle herds. Epidemiology and Infection 1997; 118: 193–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williams M, et al. Determining relationships between the seasonal occurrence of Escherichia coli O157:H7 in live cattle, ground beef, and humans. Foodborne Pathogens and Disease 2010; 7: 1247–1254. [DOI] [PubMed] [Google Scholar]
- 23.Rangel JM, et al. Epidemiology of Escherichia coli O157:H7 outbreaks, United States, 1982–2002. Emerging Infectious Diseases 2005; 11: 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naugle AL, et al. Sustained decrease in the rate of Escherichia coli O157:H7-positive raw ground beef samples tested by the food safety and inspection service. Journal of Food Protection 2006; 69: 480–481. [DOI] [PubMed] [Google Scholar]
- 25.Voetsch A, et al. Laboratory practices for stool-specimen culture for bacterial pathogens, including Escherichia coli O157:H7, in the FoodNet sites, 1995–2000. Clinical Infectious Diseases 2004; 38: S190–S197. [DOI] [PubMed] [Google Scholar]
- 26.Naugle AL, et al. Food safety and inspection service regulatory testing program for Escherichia coli O157: H7 in raw ground beef. Journal of Food Protection 2005; 68: 462–468. [DOI] [PubMed] [Google Scholar]
- 27.Naugle AL, et al. Sustained decrease in the rate of Escherichia coli O157:H7-positive raw ground beef samples tested by the food safety and inspection service. Journal of Food Protection 2005; 68: 2504–2505. [DOI] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention. Ongoing multistate outbreak of Escherichia coli serotype O157:H7 infections associated with consumption of fresh spinach – United States, September 2006. Morbidity and Mortalilty Weekly Report 2006; 55: 1045–1046. [PubMed] [Google Scholar]
- 29.Wendel AM, et al. Multistate outbreak of Escherichia coli O157:H7 infection associated with consumption of packaged spinach, August–September 2006: the Wisconsin investigation. Clinical Infectious Diseases 2009; 48: 1079–1086. [DOI] [PubMed] [Google Scholar]
- 30.Barton Behravesh C, et al. 2008 outbreak of Salmonella Saintpaul infections associated with raw produce. New England Journal of Medicine 2011; 364: 918–927. [DOI] [PubMed] [Google Scholar]