Abstract
PARP1/2 inhibitors are effective against BRCA2-deficient tumors. The PARP inhibitor (PARPi) olaparib received FDA breakthrough designation for treatment of metastatic castration-resistant prostate cancers (CRPC) carrying mutations in BRCA1/2 or ATM genes. Emergent resistance to PARPi has been associated with tumor-specific BRCA2 mutations that revert the normal open reading frame rescuing homologous recombination. We describe a case of metastatic CRPC with germline BRCA2 mutation with acquired resistance to olaparib related to biallelic BRCA2 reversion mutations of both the germline and somatic loss of function alleles detected by circulating tumor DNA testing. We also summarize a retrospective analysis of 1,534 prostate cancer cases with ctDNA analysis showing a 1.6% incidence of germline BRCA2 mutations. Within the germline BRCA2-positive cases exposed to platinum chemotherapy or PARP inhibition, the prevalence of reversion mutations was 40%. This report documents the frequency of reversion mutations in a large cohort of prostate cancer patients carrying of BRCA mutations. It also shows the potential utility of ctDNA analyses for early detection of reversion mutation driving tumor resistance.
Introduction
Mutations of BRCA1 or BRCA2 genes BRCA1/2 confer an increased lifetime risk of developing breast, ovarian, pancreatic, and prostate cancers, among others1,2. BRCA1/2-deficient cancer cells from germline BRCA1/2 mutation carriers often lose the second BRCA1/2 allele through deletion of all or part of chromosome 17q or 13q, respectively, or inactivating point mutations or small insertions or deletions3–6. Loss of both alleles leads to impaired homologous recombination of double-strand DNA breaks and increased sensitivity to radiation, platinum-based chemotherapy, and PARP inhibitors7,8.
PARP inhibitors target poly(ADP-ribose) polymerase 1 and 2 (PARP1 and PARP2) enzymes that bind single-strand DNA breaks and catalyze post-translational modification of DNA repair proteins9. In the absence of functional BRCA1 or BRCA2 protein, PARP1/2 inhibition compromises DNA repair and leads to cell cycle arrest and apoptosis10. PARP inhibitors are FDA-approved for the treatment of ovarian and breast cancers with germline BRCA1 and BRCA2 mutations, but have also anti-tumor activity in castration-resistant prostate cancer (CRPC) carrying germline or somatic mutations in genes involved with DNA repair such as BRCA1, BRCA2, ATM, PALB2, FANCA, CHEK2 and CDK1211. In a cohort of 16 patients with metastatic CRPC (mCRPC) carrying mutations in DNA repair genes, the PARP inhibitor olaparib achieved a response rates as high as 88%11. These results fostered ongoing clinical trials of PARP inhibitors in mCRPC and supported breakthrough therapy designation of olaparib by the FDA for the treatment of BRCA1/2- and ATM-mutated mCRPC in January 2016.
In germline BRCA1/2 mutation carriers treated with platinum-based chemotherapy or PARP inhibitors, resistance eventually develops through several mechanisms including acquisition of somatic BRCA1/2 mutations that restore the open reading frame (i.e. BRCA reversion mutations) of the germline allele, which in turn restores production of functional BRCA1/2 protein12–18. BRCA reversion mutations have been reported in BRCA mutated ovarian, breast, and pancreatic cancer cell lines with acquired resistance to platinum compounds or PARP inhibitors13,15–18.
Here we report a case of acquired resistance to the PARP inhibitor olaparib in BRCA2 mutant mCRPC due to multiple acquired reversion mutations detected by circulating tumor DNA (ctDNA) analysis that restored both the BRCA2 germline mutation and the somatic second-hit loss of function mutation on the second allele. We also report the prevalence of BRCA2 reversion mutations among a large cohort of 1,534 patients with mCRPC that had ctDNA testing.
Methods
Blood for cfDNA analysis was drawn during the patients’ regularly schedule clinic visit. The cfDNA NGS analysis was performed at Guardant Health, Inc. (Guardant360; Redwood City, CA), a CLIA-certified, College of American Pathologists (CAP)-accredited, New York State Department of Health-approved laboratory. Barcoded sequencing libraries were generated from five to 30 ng of plasma cfDNA. The exons of 73 cancer genes, were captured using biotinylated custom bait oligonucleotides (Agilent), resulting in a 148,000 base-pair (78 kb) capture footprint. The mean cfDNA loaded into each sequencing reaction was 22ng (range 5ng-30ng). Samples were paired-end sequenced on an Illumina Hi-Seq 2500, followed by algorithmic reconstruction of the digitized sequencing signals. The coverage depth across all coding sequences in all samples averaged approximately 15,000X. Illumina sequencing reads were mapped to the hg19/GRCh37 human reference sequence, and genomic alterations in cfDNA were identified from Illumina sequencing data by proprietary bioinformatics algorithms. These algorithms quantify the absolute number of unique DNA fragments at a given nucleotide position, thereby enabling circulating tumor DNA to be quantitatively measured as a fraction of total cfDNA. The Guardant360 assay detects single-nucleotide variants (SNV), indels, fusions and copy number alterations (CNA) in cfDNA with a reportable range of ≥0.04%, ≥0.02%, ≥0.04%, and ≥2.12 copies, respectively (http://www.guardanthealth.com/).19
This research was approved by Quorum IRB for the generation of de-identified data sets for research purposes (Guardant protocol) and Northwestern University IRB (protocol STU00205723).
Case Report
The patient is a 63-year-old Caucasian male of Ashkenazi Jewish ancestry who underwent a radical prostatectomy revealing Gleason 5+4=9 adenocarcinoma20 with involvement of seminal vesicles, perineural invasion, and negative margins. He received adjuvant androgen deprivation therapy and radiation therapy (7000cGy, 35 fractions) and developed biochemical recurrence one year later when he was treated with bicalutamide. Two years later, prostate specific antigen (PSA) rose to 218 ng/ml. Computed tomography (CT) scan showed retroperitoneal and pelvic lymphadenopathy and a vertebral body metastasis. He received 7 cycles of docetaxel followed by prolonged control of disease with 13 cycles of cabazitaxel before a new liver metastasis was identified on scans. Liver biopsy confirmed prostate adenocarcinoma and NGS (Foundation Medicine, Inc. Cambridge, MA) of the liver biopsy identified two mutations in BRCA2 - c.5946delT (p.Ser1982fs*, also known as 6174delT) and c.5754_5755delTA (p.His1918fs*5). BRCA2 allelic loss was not reported. Tissue NGS also revealed CDKN2a (p16INK4a H83Y; p14ARF A97V), as well as losses of PTEN and FAS and 12 variants of unknown significance. Germline testing confirmed a heterozygous BRCA2 c.5946delT mutation in the patient, which was inherited from his father, who had died of colon cancer at age 81.
After liver biopsy, the patient was treated with olaparib (400mg BID) resulting in rapid reduction of PSA from 821 to 300 ng/ml and improvement of lymphadenopathy and liver lesions. One year after starting olaparib, PSA rose to 779 ng/ml. CT scans showed stable adenopathy and liver lesions, but bone scan demonstrated marked progression of disease. ctDNA analysis was performed at the time of disease progression on olaparib. The patient passed away three months later.
Results
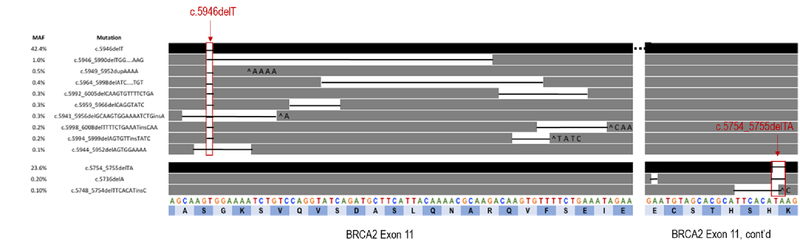
Analysis of ctDNA identified the BRCA2 c.5946delT (p.Ser1982fs*) mutation at a mutant allele fraction (MAF) of 42.4%, consistent with germline origin, and the c.5982_5983delTA (p.His1918fs) mutation at 23.6%, consistent with secondary somatic mutation. ctDNA also detected 11 additional somatic BRCA2 mutations not identified in the pre-PARP liver biopsy specimen (Table 1), all of which occurred at low MAFs (range 0.1%−1.0%), consistent with sub-clonal somatic origin. Nine of these 11 somatic BRCA2 mutations occurred in cis with the germline mutation and 3 overlapped with the original germline mutation (Figure 1). All occurred within 0–52 nucleotides of the c.5946delT germline mutation and restored the BRCA2 open reading frame. Interestingly, the 2 remaining somatic BRCA2 mutations (c.5736delA and c.5749_5754delTCACAT) occurred in close proximity to the putative somatic “second hit”, c.5754_5755delTA. Both were in cis to the c.5754_5755delTA mutation and both were predicted to restore the open reading frame, suggesting that these acquired somatic variants occur on the alternate allele relative to the germline BRCA2 c.5946delT mutation. In addition to multiple BRCA2 mutations, ctDNA analyses revealed the following alterations: TP53 F113fs; GATA3 D336D; ARID1A S1755T; MYC P72A; and amplification of MYC, KRAS, CCND2, and BRAF.
Table 1.
Summary and description of BRCA2 mutations identified by circulating tumor DNA analysis.
| Mutation (HGVS designation) | Protein (HGVS designation) | Indel type | Functional consequence | Indel length (nt) | Net nt loss resulting from | Mutant allele fraction in ctDNA | Cis/trans to germline mutation | |
|---|---|---|---|---|---|---|---|---|
| ALLELE 1 | c.5946delT* | p.SeM982fs | Deletion | Germline loss of function | −1 | NA | 42.4% | N/A |
| c.5946_5990delTGGAAAATCTGTCCAGGTATCAGATGCTTCATTACAAAACGCAAG | p.Ser1982_Ala1996del | Deletion | Somatic reversion | −45 | −46 | 1.0% | cis | |
| C.5949_5952dupAAAA | p.Ser1985fs | Duplication | Somatic reversion | +4 | −3 | 0.5% | cis | |
| c.5964_5998delATCAGATGCTTCATTACAAAACGCAAGACAAGTGT | p.Ser1989fs | Deletion | Somatic reversion | −35 | −36 | 0.4% | cis | |
| c.5959_596SclelCAGGTATC | p.Gln1987fs | Deletion | Somatic reversion | −8 | −9 | 0.3% | cis | |
| c.5992_6005delCAAGTGTTTTCTGA | p.Gln1998fs | Deletion | Somatic reversion | −14 | −15 | 0.3% | cis | |
| c.5941_5956delGCAAGTGGAAAATCTGinsA | pAla1981_Val1986delinslle | Insertion-Deletion | Somatic reversion | −15 | −15 | 0.3% | cis | |
| c.599_5S99delAGTGTTinsTATC | p.Gln1998fs | Insertion-Deletion | Somatic reversion | −3 | −3 | 0.2% | cis | |
| c.5998_6008demTTCTGAAATinsCAA | p.Phe2000fs | Insertion-Deletion | Somatic reversion | −8 | −9 | 0.2% | cis | |
| c.5944_5952delAGTGGAAAA | p.Ser1982_Lys1984del | Deletion | Somatic reversion | −9 | −9 | 0.1% | cis | |
| ALLELE 2 | c.5754_5755dBlAT | p.His1918fs | Deletion | Somatic secondary mutation | −2 | NA | 23.6% | trans |
| c.5736pelA | p.Glu1912fs | Deletion | Somatic reversion | −1 | −3 | 0.2% | tranŝ | |
| c.5748_5754delTTCACATinsC | p.Seri917_His1918del | Insertion-Deletion | Somatic reversion | −6 | −6 | 0.1% | tranŝ |
The c.5946delT mutation corresponds to the 6174delT mutation in BRCA2. c.5946delT uses the Human Genomic Variation Society (HGVS) nomenclature and 6174delT, the Breast Cancer International Consortium (BIC) nomenclature. Indel = Insertion or deletion or compound insertion/deletion. nt = nucleotide.
Figure 1.
Schematic representation of the germline c.5946delT and secondary c.5754_5755delTA loss of function mutations (black bars-top row and 3rd from bottom row) in relation to the acquired somatic reversion mutations (dark gray bars). Black lines between bars represent the nucleotides deleted. ^ indicates an insertion. Letters across the bottom two rows represent the nucleotide (A, C, G, T) and amino acid (dark and light blue bars) sequences of wild type BRCA2 exon 11.
Prevalence of BRCA reversion mutations in a large genomic database of prostate cancer patients
Since the biallelic reversion of both germline and truncal somatic BRCA1/2 alterations contrasted the generally accepted model of monoallelic reversion of germline BRCA1/2 mutations, we attempted to estimate the relative prevalence of germline vs. somatic BRCA1/2 reversion events in mCRPC patients using a large genomic database including comprehensive ctDNA results from over 40,000 patients with a variety of solid tumors. Between October 5, 2015 and April 25, 2017, 1,765 samples from 1,534 unique patients with mCRPC had ctDNA testing (Guardant Health, Inc.), which included complete sequencing of all BRCA1 and BRCA2 exons and exon-intron borders. Of these, 24 patients (1.6%) had a deleterious BRCA2 mutation falling within the germline mutant allele fraction (40–80%). There were no putative germline mutations in the BRCA1 gene in this mCRPC cohort. Five of these 24 patients were on either a PARP inhibitor or platinum-based chemotherapy at the time of the blood draw. Two of the 5 patients , one on olaparib and one on carboplatin, had BRCA2 reversion mutations detected by the ctDNA analysis. Therefore, in this germline mutation-positive, platinum/PARP-exposed cohort, the frequency of BRCA2 reversion was 40% (2 of 5). A third reversion case was identified but had no previous exposure to platinum-based chemotherapy or PARP inhibitors.
Discussion
We report a case of acquired resistance to olaparib in BRCA2 germline positive mCRPC due to multiple acquired BRCA2 reversion mutations of both the germline mutation and a “second-hit” somatic mutation on the opposite allele. This case is similar to one recently reported by Goodall et al. in which acquired reversion mutations restore the open reading frame of not only the primary germline mutation, but also the secondary loss of function mutation21. While previous studies in ovarian cancer have established that reversion of the germline allele is necessary and sufficient to restore normal BRCA protein function, this case suggests functional comparability of the variants despite their origin (i.e. somatic or germline). This observation challenges the established model of BRCA1/2 reversion as restricted to germline mutations and suggests that the germline-somatic origin of the allele may not play a critical biological role in this mechanism of resistance.
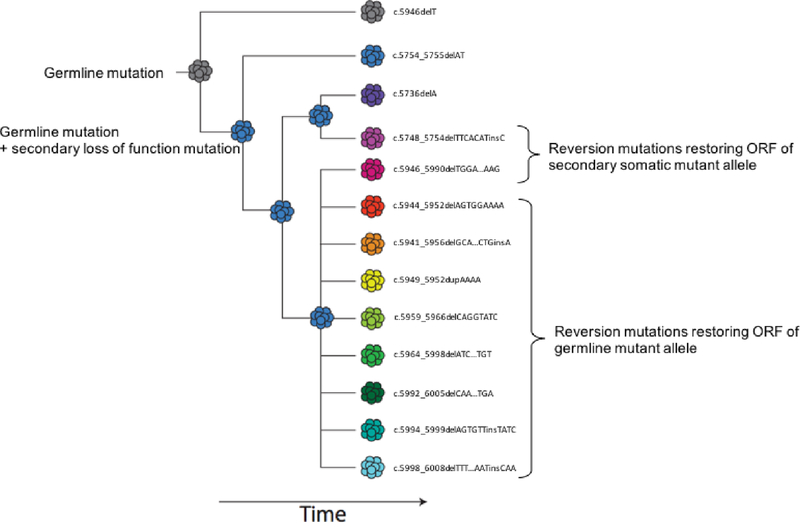
Furthermore, this case is a powerful illustration of convergent evolution of multiple BRCA2 reversion mutations arising in different clones of the metastatic lesion or within multiple metastases (Figure 2), as has been described22. Other studies of acquired resistance have compared ctDNA to tissue-based testing on biopsies from multiple metastatic lesions in the same patient. These studies have shown that a single tissue biopsy often does not capture the full spectrum of acquired resistance mutations, whereas ctDNA may provide a more global summary of tumor heterogeneity, as seen in this case23,24. ctDNA analyses also enable monitoring and early detection of mutations driving treatment resistance to PARPi with meaningful clinical implications.
Figure 2.
ctDNA profiling of a patient progressing on PARPi a known germline frameshift and a somatic “second hit” frameshift as well as 11 additional frame shift mutations. Phasing the mutation using a dollo parsimony model allows a presumptive evolutionary history of the tumor population to be inferred. Both somatic and germline lineage contain multiple independent revertant subclones. ORF= Open reading frame
Once a BRCA1/2 mutation is detected, longitudinal monitoring with ctDNA can be relevant for early detection of reversion BRCA1/2 mutations to predict resistance to PARP inhibitors as illustrated by the case presented on this manuscript. In women with platinum-resistant ovarian cancer, the presence of BRCA reversion mutations was a more accurate predictor of response to subsequent platinum or PARP inhibitor therapy than duration of response to previous lines of platinum therapy25. Another study identified reversion of germline BRCA1/2 mutations in high-grade serous ovarian carcinoma using ctDNA and was able to predict treatment responses26. There is limited data on the prevalence of BRCA reversion mutations and rates of resistance to platinum or PARP inhibitors in mCRPC. Estimates of BRCA1/2 reversion rates in women with platinum-resistant ovarian cancer range from 25%−70%, but are based on very small series18,25. Analysis of genomic data from large databases may be one way to overcome this limitation. Our ctDNA NGS study estimates a frequency of 40% among patients with mCRPC carrying BRCA2 germline mutations exposed to platinum/PARP therapy. However, caution should be used when interpreting the reversion frequency reported here, as it is based on a small series of platinum/PARPi exposed patients. Larger prospective studies are needed to determine he true frequency of reversion mutations in a platinum/PARP exposed cohort.
There are several limitations to the current study. Although genomic testing on tissue prior to PARP inhibitor therapy was performed for the index case, this information was not available for the additional cases showing evidence of reversion mutation. All patients underwent cfDNA analysis at the time of clinical progression suggesting that they had developed platinum or PARP inhibitor resistance but the duration of their response on therapy or presence of reversion mutations were present prior to exposure is unknown. With regard to the retrospective cohort analysis, our BRCA2 germline mutation rate was lower than that previously described in the literature27. Possible explanations for this include exclusion of putative germline missense and nonsense mutations in the analysis and overly restrictive germline mutant allele fraction thresholds resulting in exclusion of putative germline mutations in patients with more severe allele imbalance. Lastly, one case with evidence of a reversion mutation had no prior exposure to PARPi or platinum. Review of the patient’s treatment history revealed therapy with taxane based chemotherapy, radium-223, as well as mitoxantrone, a DNA intercalating agent used in the treatment of breast cancer, prostate cancer and acute myeloid leukemia. Interestingly, Ikeda et al. reported a patient with Fanconi anemia due to biallelic BRCA2 mutations. The patient developed acute myeloid leukemia and experienced relapse after treatment with chemotherapy and mitoxantrone. At the time of relapse, a bone marrow biopsy was performed and a patient derived cell line showed loss of the FA phenotype due to mono-allelic reversion of the BRCA2 mutation and restoration of wild type BRCA2 function. The authors suggest that DNA intercalating agents other than platinum may have the ability to induce reversion mutations and lead to resistance28.
Compared to biopsy, cfDNA analyses allow easier monitoring and potentially earlier detection of mutations that result in treatment resistance. cfDNA analysis, which allows detection of both somatic and germline mutations in a single test, is well suited to distinguish whether a somatic BRCA mutation represents a second-hit loss of function or a reversion of the germline BRCA mutation. To make the distinction, the exact location of the mutations must be known, since in order to restore the reading frame a revert must be located near the inactivating mutation (i.e. before the end of the same exon)26. Furthermore, cfDNA may provide a more global summary of tumor heterogeneity and the full spectrum of acquired resistance mutations than a single tissue biopsy23,24. The case presented here illustrates convergent evolution of multiple BRCA2 reversion mutations arising in different clones of the metastatic lesion or within multiple metastases (Figure 2), as has been described elsewhere22. Incorporation of routine cfDNA analyses into standard of care of BRCA1/2 mutated cancers treated with PARP inhibitors or platinum-based chemotherapy requires validation of the germline calls from cfDNA, but may allow early detection of treatment resistant and subsequent change in therapy before significant disease progression.
Footnotes
Disclaimers: Nagy RJ and Lanman RB are shareholders of Guardant Health
References
- 1.Steichen-Gersdorf E, Gallion HH, Ford D, et al. Familial site-specific ovarian cancer is linked to BRCA1 on 17q12–21. Am J Hum Genet. 1994;55(5):870–875. [PMC free article] [PubMed] [Google Scholar]
- 2.Wooster R, Bignell G, Lancaster J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378(6559):789–792. [DOI] [PubMed] [Google Scholar]
- 3.Cleton-Jansen AM, Collins N, Lakhani SR, et al. Loss of heterozygosity in sporadic breast tumours at the BRCA2 locus on chromosome 13q12-q13. Br J Cancer. 1995;72(5):1241–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gudmundsson J, Johannesdottir G, Bergthorsson JT, et al. Different tumor types from BRCA2 carriers show wild-type chromosome deletions on 13q12-q13. Cancer Res. 1995;55(21):4830–4832. [PubMed] [Google Scholar]
- 5.Neuhausen SL, Marshall CJ. Loss of heterozygosity in familial tumors from three BRCA1-linked kindreds. Cancer Res. 1994;54(23):6069–6072. [PubMed] [Google Scholar]
- 6.Collins N, McManus R, Wooster R, et al. Consistent loss of the wild type allele in breast cancers from a family linked to the BRCA2 gene on chromosome 13q12-13. Oncogene. 1995;10(8):1673–1675. [PubMed] [Google Scholar]
- 7.Farmer H, McCabe N, Lord CJ, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434(7035):917–921. [DOI] [PubMed] [Google Scholar]
- 8.Tutt AN, Lord CJ, McCabe N, et al. Exploiting the DNA repair defect in BRCA mutant cells in the design of new therapeutic strategies for cancer. Cold Spring Harb Symp Quant Biol. 2005;70:139–148. [DOI] [PubMed] [Google Scholar]
- 9.Durkacz BW, Omidiji O, Gray DA, Shall S. (ADP-ribose)n participates in DNA excision repair. Nature. 1980;283(5747):593–596. [DOI] [PubMed] [Google Scholar]
- 10.Bryant HE, Schultz N, Thomas HD, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434(7035):913–917. [DOI] [PubMed] [Google Scholar]
- 11.Mateo J, Carreira S, Sandhu S, et al. DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med. 2015;373(18):1697–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bouwman P, Jonkers J. Molecular pathways: how can BRCA-mutated tumors become resistant to PARP inhibitors? Clin Cancer Res. 2014;20(3):540–547. [DOI] [PubMed] [Google Scholar]
- 13.Dhillon KK, Swisher EM, Taniguchi T. Secondary mutations of BRCA1/2 and drug resistance. Cancer Sci. 2011;102(4):663–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lord CJ, Ashworth A. PARP inhibitors: Synthetic lethality in the clinic. Science. 2017;355(6330):1152–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barber LJ, Sandhu S, Chen L, et al. Secondary mutations in BRCA2 associated with clinical resistance to a PARP inhibitor. J Pathol. 2013;229(3):422–429. [DOI] [PubMed] [Google Scholar]
- 16.Edwards SL, Brough R, Lord CJ, et al. Resistance to therapy caused by intragenic deletion in BRCA2. Nature. 2008;451(7182):1111–1115. [DOI] [PubMed] [Google Scholar]
- 17.Sakai W, Swisher EM, Jacquemont C, et al. Functional restoration of BRCA2 protein by secondary BRCA2 mutations in BRCA2-mutated ovarian carcinoma. Cancer Res. 2009;69(16):6381–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451(7182):1116–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanman RB, Mortimer SA, Zill OA, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10(10):e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Epstein JI, Egevad L, Amin MB, et al. The 2014 International Society of Urological Pathology (ISUP) Consensus Conference on Gleason Grading of Prostatic Carcinoma: Definition of Grading Patterns and Proposal for a New Grading System. Am J Surg Pathol. 2016;40(2):244–252. [DOI] [PubMed] [Google Scholar]
- 21.Goodall J, Mateo J, Yuan W, et al. Circulating Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov. 2017;7(9):1006–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of Circulating Cell-free DNA Identifies Multi-clonal Heterogeneity of BRCA2 Reversion Mutations Associated with Resistance to PARP Inhibitors. Cancer Discov. 2017;7(9):999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Mattos-Arruda L, Weigelt B, Cortes J, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014;25(9):1729–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Russo M, Siravegna G, Blaszkowsky LS, et al. Tumor Heterogeneity and Lesion-Specific Response to Targeted Therapy in Colorectal Cancer. Cancer Discov. 2016;6(2):147–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Norquist B, Wurz KA, Pennil CC, et al. Secondary somatic mutations restoring BRCA1/2 predict chemotherapy resistance in hereditary ovarian carcinomas. J Clin Oncol. 2011;29(22):3008–3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christie EL, Fereday S, Doig K, Pattnaik S, Dawson SJ, Bowtell DDL. Reversion of BRCA1/2 Germline Mutations Detected in Circulating Tumor DNA From Patients With High-Grade Serous Ovarian Cancer. J Clin Oncol. 2017;35(12):1274–1280. [DOI] [PubMed] [Google Scholar]
- 27.Pritchard CC, Offit K, Nelson PS. DNA-Repair Gene Mutations in Metastatic Prostate Cancer. N Engl J Med. 2016;375(18):1804–1805. [DOI] [PubMed] [Google Scholar]
- 28.Ikeda H, Matsushita M, Waisfisz Q, et al. Genetic reversion in an acute myelogenous leukemia cell line from a Fanconi anemia patient with biallelic mutations in BRCA2. Cancer Res. 2003;63(10):2688–2694. [PubMed] [Google Scholar]