Clinical Presentation and Family History
A 64-year-old man presented to a dermatologist with a pigmented lesion on his left forearm. The lesion was biopsied to reveal a 1.5-mm-thick malignant melanoma with 2 mitoses/mm2 extending to the radial margin without ulceration. The patient subsequently underwent wide local excision with sentinel lymph node dissection. Pathology of the sample showed skin with melanoma in situ and no residual invasive melanoma identified with margins free of tumor. Two lymph nodes were identified with no tumor seen, as confirmed by Melanoma Antigen Recognized by T Cells 1 (MART1) immunostain. Final pathologic staging was stage IB (pT2aN0) based on the seventh edition of the American Joint Committee on Cancer melanoma staging system. The patient received no additional therapy at that time for his early-stage melanoma.
The patient was in good health for approximately 15 months until he reported bright red blood from his rectum. Colonoscopy revealed a left colon mass. Positron emission tomography/computed tomography revealed an approximately 3.1-cm colonic mass with a standardized uptake value of 26.4; several foci of abnormal [18F]fluorodeoxyglucose uptake in bilateral hepatic lobes and in the retroperitoneum and mesentery; a middle lobe lung nodule measuring 2 × 1.8 cm with a standardized uptake value of 7.5; and a right parietal brain lesion measuring 1.7 × 1.7 cm. Fine-needle aspiration was performed on a left iliac lymph node, which showed malignant cells positive for cytokeratin 20 (CK20) and caudal type homeobox 2 (CDX2) and negative for cytokeratin 7 (CK7), consistent with colorectal carcinoma. Reflex immunostains for mismatch repair (MMR) proteins showed loss of PMS1 homolog 2, mismatch repair system component (PMS2) (Figure 1A). Craniotomy was performed, with pathology showing metastatic melanoma positive for MART1, SRYrelated HMG-box 10 (SOX10), and 100% soluble in ammonium sulfate at neutral pH (S100) in support of the diagnosis (Figure 1B). The patient reported no family history of colon cancer, but he did state that his daughter was diagnosed with ovarian cancer at age 18 years.
Figure 1.
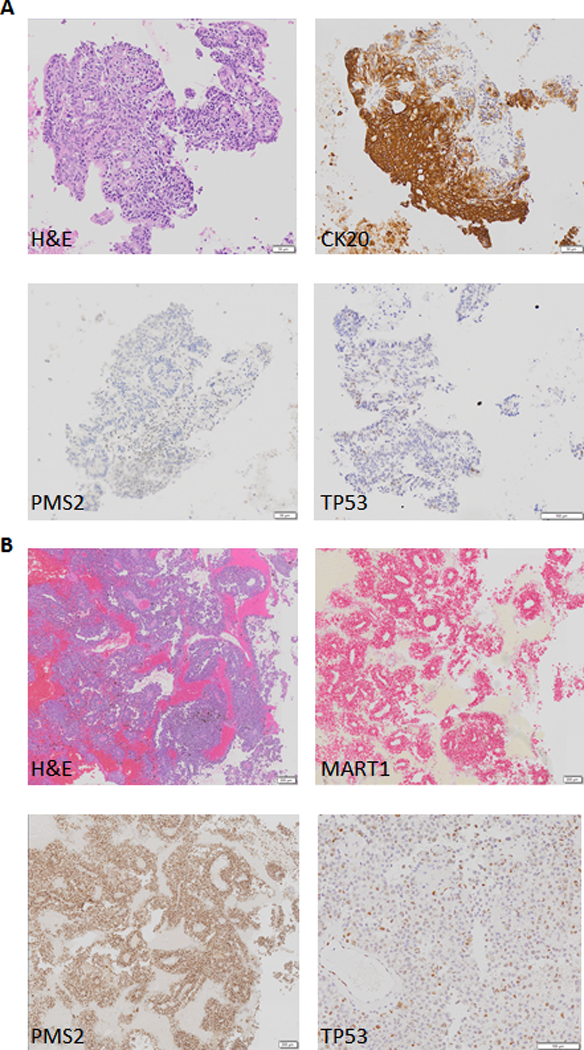
A) Histology of left iliac lymph node involved with colorectal cancer B) Histology of craniotomy specimen with metastatic melanoma. H and E: hematoxylin and eosin.
Genomic Analysis
As part of reflex testing performed at our hospital for patients with advanced colon cancer and advanced melanoma, the patient’s left iliac lymph node had KRAS, NRAS, and BRAF sequencing performed, whereas the craniotomy had BRAF, NRAS, and KIT sequencing. The colon cancer sample was found to have a KRAS c.183A>T, p.Gln61His mutation, whereas the melanoma harbored a KIT c.1669T>G, p.Trp557Gly mutation.
The patient subsequently had his blood analyzed for circulating tumor DNA (ctDNA) using the Foundation One Liquid platform (Foundation Medicine, Cambridge, MA), which consisted of hybrid capture sequencing of 70 genes for single nucleotide variants, indels, and copy number alterations with rearrangement detection performed for seven of these genes. This test is also able to identify microsatellite instability. Testing detected the KRAS c.183A>T, p.Gln61His mutation identified previously in the colon cancer specimen at 17.41% allele frequency and the KIT c.1669T>G, p.Trp557Gly mutation identified previously in the melanoma sample at 0.23%. The specimen was identified as having high microsatellite instability, consistent with the loss of PMS2 by immunohistochemistry (IHC) in the colon cancer specimen. Intriguingly, when all of the alterations identified using the ctDNA assay were viewed, there seemed to be two main allelic fractions, one with alterations at an allele frequency of less than 1% and another with allele frequencies from 13.43% to 35.9% (Table 1). The higher allele fraction seemed to correlate with known drivers of colon cancer tumorigenesis, which included the KRAS c.183A>T, p.Gln61His mutation; three APC mutations (c.4666_4667insA, p.T1556fs*3; c.1178C>A, p.S393*; and c.904C>T, p. R302*), and a PTEN c.800delA, p.K267fs*9 frameshift mutation. On the basis of clonal analysis of these variants and assuming ctDNA content of approximately 35% colon cancer cells,1 the KRAS and APC variants had allele frequencies compatible with being heterozygous in the tumor, whereas the PTEN variant was at an allele frequency compatible with being homozygous in the tumor as a result of loss of heterozygosity or gene conversion to inactivate the second wild-type allele. These observations, in addition to the KIT mutation having an allele fraction of 0.23%, raised the possibility that the lower allele fraction alterations may be arising from the patient’s melanoma (Figure 2). Several studies have shown that ctDNA correlates with tumor burden, and it may be that the only melanoma tissue mass in this patient was in his brain.2,3 The reflex testing of the patient’s colon cancer and melanoma was performed using the Illumina Trusight Tumor 15 gene panel (Illumina, San Diego, CA), with results masked to only report out specific genes depending on the indication (i.e., KRAS, NRAS, and BRAF for colon cancer and BRAF, NRAS, and KIT for melanoma). When the original sequencing results were examined for all 15 genes, the KRAS c.183A>T, p.Gln61His mutation was only seen in the patient’s colon cancer at 43.4% allele frequency, and the KIT c.1669T>G, p.Trp557Gly mutation was only seen in the patient’s melanoma at 38.8% allele frequency. Astonishingly, despite the high allele frequencies of the KRAS and KIT mutations, the TP53 c.730G>T, p.G244C mutation identified in ctDNA was not identified in either sample. Studies have shown that clonal mutations in the hematopoietic compartment may be detected on tumor tissue testing.4,5 The results suggest that ctDNA analysis in this patient identified not only clonal DNA from two separate solid tumors, but also an additional third clonal mutation in the hematopoietic compartment, as has been recently described.6 The patient was evaluated at Rutgers Cancer Institute of New Jersey and provided informed consent to participate in a prospective study trial for tumor genomic profiling (ClinicalTrials.gov identifier: NCT02688517). The results of genomic profiling, clinical course, and pathology were reviewed at the Rutgers Cancer Institute of New Jersey Molecular Tumor Board (MTB), which was approved by the Rutgers University New Brunswick Health Sciences Institutional Review Board (Pro2012002075). The patient is now deceased, but his informed consent to participate in the NCT02688517 trial covers approval for publication of these results.
Table 1.
Variants detected in circulating tumor DNA.
| Gene | Amino Acid Variant | Variant Allele Frequency (%) | Total Depth | Chromosome & Position | [Variant] & Sequence Context | Molecular Tumor Board Discussion |
|---|---|---|---|---|---|---|
| ABL1 | M318T | 0.22 | 3,594 | chr9:133748292 | GTTCA[T>C]GACCT | Putative melanoma |
| KIT | W557G | 0.23 | 3,069 | chr4:55593603 | TACAG[T>G]GGAAG | Pathogenic / Melanoma |
| CCND1 | P220T | 0.24 | 3,321 | chr11:69462845 | GGAGC[C>A]CCAAC | Putative melanoma |
| SMO | T336A | 0.26 | 4,632 | chr7:128846076 | TCCTC[A>G]CCTAT | Putative melanoma |
| BRCA2 | H910R | 0.27 | 3,640 | chr13:32911221 | ACTTC[A>G]TGAAA | Putative melanoma |
| RAF1 | L546R | 0.27 | 3,673 | chr3:12626652 | AAGGA[A>C]GCTCC | Putative melanoma |
| MTOR | N2219fs*60 | 0.29 | 3,400 | chr1:11184560 | CTGAG[T>–]TTTTC | Pathogenic / Indel in homopolymer region / Confirmed melanoma |
| ATM | S214P | 0.32 | 2,820 | chr11:108114823 | TTTTT[T>C]CCAAG | Putative melanoma / Substitution in homopolymer region |
| ALK | N1095fs*14 | 0.36 | 4,448 | chr2:29446284 | TAGTT[G>–]GGGTT | Indel in homopolymer region |
| MDM2 | Y287H | 0.38 | 3,169 | chr12:69230470 | CTGTG[T>C]ATCAG | Pathogenic / Putative melanoma |
| NF1 | A1224V | 0.41 | 3,163 | chr17:29560194 | TATAG[C>T]GATGG | Pathogenic / Putative melanoma |
| CDK12 | R298* | 0.48 | 3,513 | chr17:37619216 | GTAGG[C>T]GACAG | Pathogenic / Putative melanoma |
| STK11 | F25L | 0.48 | 3,127 | chr19:1206985 | ACACG[T>C]TCATC | Putative melanoma |
| ERRFI1 | F115fs*6 | 0.50 | 2,999 | chr1:8074313 | TTCTT[A>–]AAACC | Pathogenic / Indel in homopolymer region / Putative melanoma |
| TP53 | G244C | 0.54 | 2,795 | chr17:7577551 | GCCGC[G>T]CATGC | Putative CHIP |
| MTOR | L1460P | 0.58 | 3,097 | chr1:11217299 | CCACA[A>C]GGGCA | Pathogenic / Putative melanoma |
| PTPN11 | V243A | 0.70 | 3,122 | chr12:112893839 | TAAAG[T>C]CAAAC | |
| BRCA1 | S1387P | 0.70 | 3,131 | chr17:41242987 | CTGAG[A>G]GGATA | |
| PDCD1LG2 | P186fs*14 | 1.71 | 4,155 | chr9:5549529 | CACCC[C>–]CTGGC | Indel in homopolymer region |
| EGFR | S1104A | 1.90 | 3,527 | chr7:55272987 | CTGGC[T>G]CTGTG | |
| BRCA2 | L929fs*31 | 13.43 | 3,261 | chr13:32911277 | TGGTT[T>–]TATAT | Pathogenic / Indel in homopolymer region |
| KRAS | Q61H | 17.41 | 2,837 | chr12:25380275 | TCCTC[T>A]TGACC | Pathogenic / Confirmed colon |
| APC | R302* | 17.46 | 3,952 | chr5:112151261 | CACCT[C>T]GAAGG | Pathogenic / Putative colon |
| APC | S393* | 18.21 | 4,294 | chr5:112154907 | TCACT[C>A]ACAGC | Pathogenic / Putative colon |
| ATM | K2465E | 18.49 | 3,662 | chr11:108201026 | TATGT[A>G]AAGCA | Putative colon |
| GNAS | R199H | 20.22 | 3,036 | chr20:57484415 | GCTTC[G>A]CTGCC | Putative colon |
| APC | T1556fs*3 | 22.00 | 4,169 | chr5:112175957 | AAAAA[–>A]CTATT | Pathogenic / Indel in homopolymer region / Putative colon |
| PTEN | K267fs*9 | 35.90 | 3,309 | chr10:89717774 | TAAAA[A>–]AGGTT | Pathogenic / Indel in homopolymer region / Putative colon |
Note: Variants described as pathogenic were reported on the front page of the Foundation One Liquid assay, Foundation Medicine (Cambridge, MA) report in genomic findings. Other listed variants were reported by Foundation Medicine as variants of unknown significance.
Abbreviations: CHIP, clonal hematopoiesis of indeterminate potential; MTB, molecular tumor board; Total Depth, total depth of sequencing reads.
Figure 2.
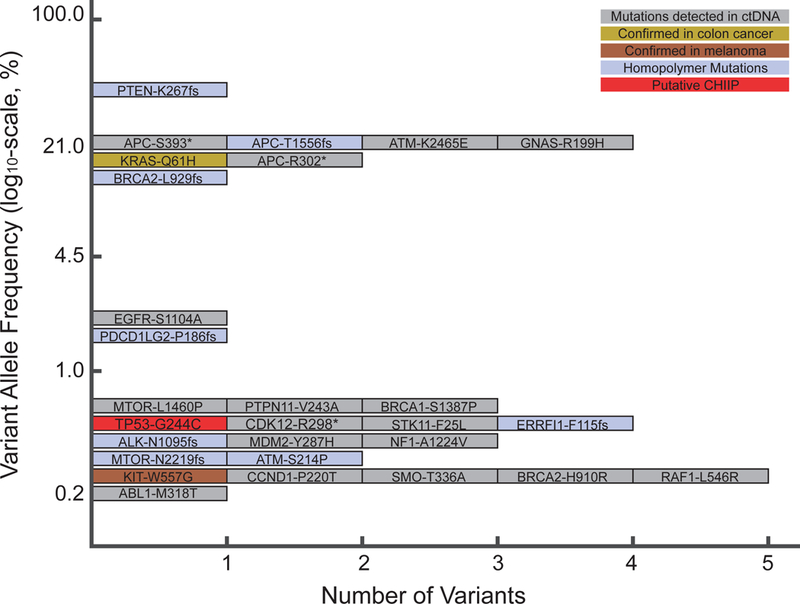
Allele frequencies of variants detected in circulating tumor DNA (ctDNA). CHIP: clonal hematopoiesis of indeterminate potential.
As a result of the colon cancer being deficient for PMS2, the ctDNA showing high microsatellite instability, and the benefit in a subset of patients with melanoma, the primary MTB recommendation was to use immune checkpoint inhibitors to treat this patient.7 Interestingly, when the genomic context of alterations identified in the ctDNA analysis was analyzed, three alterations in homopolymer regions were identified at higher allele frequency, whereas five homopolymer alterations at low allele frequency were found, raising the possibility that the melanoma was also MMR deficient (Table 1 and Figure 2). For the patient’s melanoma, the MTB discussed data showing responses to imatinib in patients with activating KIT mutations.8 These data suggested acquired NRAS mutations or KIT amplification as a resistance mechanism. A published report of a patient with rectal melanoma harboring an identical KIT c.1669T>G, p.Trp557Gly mutation with a 7-month response to sunitinib was presented. When this patient ultimately experienced relapse, a new NRAS p.Q61K mutation was detected.9 The MTB noted that this patient had a previously described activating mutation in MTOR (c. 4379T>C, p. L1460P) at allele frequencies compatible with being present in the patient’s melanoma.10
Other alterations that the MTB discussed as alternative targets of treatment included a BRCA2 c.2786delT, p.L929fs*31 truncating mutation identified in ctDNA at 13.43% allele frequency. It was likely that only one allele was inactivated in BRCA2 in the colon cancer based on the allele frequency being compatible with it being heterozygous. Because the alteration occurred in a homopolymer region and BRCA2 is a large gene, this may have been a passenger alteration as a result of the tumor being deficient in MMR. Nonetheless, because ctDNA could not determine whether both alleles were inactivated, the MTB suggested that platinum drugs or poly (ADP-ribose) polymerase inhibitors could be effective. In addition, a CDK12 c.892C>T, p. R298* inactivating mutation was identified as possibly being in the patient’s melanoma on the basis of its allele frequency. Recent work has shown that CDK12 regulates DNA damage response genes, including BRCA1, and is sensitive to DNA-damaging agents and poly (ADPribose) polymerase inhibitors.11,12 The MTB suggested that these agents may be an option with the potential to target both the BRCA2 truncating mutation and the CDK12 inactivating mutation. In addition, during the MTB meeting, accumulating evidence suggesting that loss of genes involved in DNA damage repair, such as BRCA2 or CDK12, may have a role in sensitivity to immune checkpoint inhibitors was presented.13,14
It is unusual that three truncating APC mutations that likely arose from the patient’s colon cancer at heterozygous levels were identified. APC mutations are typically correlated with microsatellite-stable colon cancer, whereas one mutation of each allele would be sufficient to inactivate the protein. It may be that the colon cancer initially developed along the microsatellite-stable pathway with biallelic inactivation of APC before acquiring an additional frameshift mutation as a result of loss of MMR function, with APC being particularly susceptible to being damaged by a loss of MMR because it is a large gene. One PTEN allele seems to have been inactivated as a result of the defect in MMR with loss of heterozygosity or gene conversion of the second allele, whereas BRCA2 likely only had one allele inactivated by MMR deficiency in a similar manner to the third APC mutation.
Additional recommendations from the MTB included genetic counseling with the patient harboring a PMS2-deficient colon cancer and having a daughter with a questionable history of ovarian cancer at a young age, PMS2 IHC to be performed on the patient’s craniotomy melanoma specimen, and consideration for sequencing washed peripheral-blood mononuclear cells to determine whether the TP53 c.730G>T, p.G244C mutation was present in the hematopoietic compartment.
Treatment Outcome
The patient was started on nivolumab. Shortly after, the patient became septic, developed endocarditis, and ultimately died during an operation to remove the aortic valve. After the MTB meeting, PMS2 IHC was performed on the melanoma from the craniotomy, which showed intact PMS2 (Figure 1B). Therefore, the five lower allele frequency variants showing alterations in homopolymer regions most likely represent subclonal alterations in the colon cancer as a result of the defect in MMR. Also after the MTB meeting, sequencing of a lymph node negative for tumor from the patient’s original sentinel lymph node dissection on the Illumina Trusight Tumor 15 gene panel was performed. This was also found to be negative for TP53 at c.730G>T, p.G244C. Although finding this mutation in the lymph node would have provided strong evidence that it arose in the hematopoietic compartment, not finding it does not disprove this hypothesis. The TP53 p.G244C mutation was present at an allele fraction of 0.54% in the blood, and although it would be expected to be detected in the melanoma or colon cancer tissue samples if it was present, it could be present at this relatively low fraction in the blood and not be present in a lymph node. In addition, because the lymph node was removed 18 months before the patient’s blood was sequenced, the TP53 mutation may not have even been present at that time or present at even lower levels. As ancillary confirmation, TP53 IHC was performed on both the craniotomy with melanoma and the iliac lymph node fine-needle aspiration with colon cancer. In both cases, TP53 was negative in tumor cells, although melanin and occasional TP53-positive cells were observed in the melanoma (Figures 1A and 1B). TP53 somatic missense mutations in cancer are generally reported to be positive by IHC, and p.G244C was reported to be positive in 17 of 18 samples.15 In total, the negative sequencing and IHC results strongly suggest the possibility of the TP53 p.G244C mutation having arisen in the hematopoietic compartment.
In summary, ctDNA profiling identified alterations at different allele frequencies that seemed to correspond with mutations present in his colon cancer and melanoma. Remarkably, there was a TP53 mutation identified in the liquid biopsy, with subsequent sequencing and IHC results indicating it was unlikely to be present in either of the solid tumors and suggesting a third clonal mutation in the hematopoietic compartment. When ctDNA analysis is performed, knowledge of genomic alterations in the primary cancer may not be available, and occasionally, there may be a second unknown primary cancer. This patient’s case demonstrates potential confounding factors in genomic profiling of ctDNA and the value in presentation at MTB, where members with different expertise can help in properly interpreting the results.
Acknowledgments
Funding Support
This study was supported by the National Institutes of Health Grant No. P30CA072720, an anonymous gift to the Precision Medicine Program at the Rutgers Cancer Institute of New Jersey, and Grant No. R01CA233662 from the National Cancer Institute (H.K.). N.J. is a postdoctoral fellow of the New Jersey Commission on Cancer Research (DCHS19PPC016).
References
- 1.Khiabanian H, Hirshfield KM, Goldfinger M, et al. : Inference of Germline Mutational Status and Evaluation of Loss of Heterozygosity in High-Depth, Tumor-Only Sequencing Data. JCO Precis Oncol 10.1200/PO.17.00148, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diaz LA Jr, Bardelli A: Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 32(6):579–86, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson SJ, Tsui DW, Murtaza M, et al. : Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 368(13):1199–209, 2013 [DOI] [PubMed] [Google Scholar]
- 4.Severson EA, Riedlinger GM, Connelly CF, et al. : Detection of clonal hematopoiesis of indeterminate potential in clinical sequencing of solid tumor specimens. Blood 131(22):2501–2505, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riedlinger G, Hadigol M, Khiabanian H, et al. : Association of JAK2-V617F Mutations Detected by Solid Tumor Sequencing with Coexistent Myeloproliferative Neoplasms. JAMA Oncol 1;5(2):265–267, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu Y, Ulrich BC, Supplee J, et al. : False-Positive Plasma Genotyping Due to Clonal Hematopoiesis. Clin Cancer Res 24(18):4437–4443, 2018 [DOI] [PubMed] [Google Scholar]
- 7.Overman MJ, Lonardi S, Wong KYM, et al. : Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair-Deficient/Microsatellite Instability-High Metastatic Colorectal Cancer. J Clin Oncol 36(8):773–779, 2018 [DOI] [PubMed] [Google Scholar]
- 8.Hodi FS, Corless CL, Giobbie-Hurder A, et al. : Imatinib for melanomas harboring mutationally activated or amplified KIT arising on mucosal, acral, and chronically sun-damaged skin. J Clin Oncol 31(26):3182–90, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minor DR, Kashani-Sabet M, Garrido M, et al. : Sunitinib therapy for melanoma patients with KIT mutations. Clin Cancer Res 18(5):1457–63, 2012 [DOI] [PubMed] [Google Scholar]
- 10.Grabiner BC, Nardi V, Birsoy K, et al. : A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 4(5):554–63, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blazek D1, Kohoutek J, Bartholomeeusen K, et al. : The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev 25(20):2158–72, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joshi PM, Sutor SL, Huntoon CJ, et al. : Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J Biol Chem 289(13):9247–53, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nolan E, Savas P, Policheni AN, et al. : Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med 9(393): pii: eaal4922, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu YM, Cieślik M, Lonigro RJ, et al. : Inactivation of CDK12 Delineates a Distinct Immunogenic Class of Advanced Prostate Cancer. Cell 173(7):1770–1782, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Murnyák B, Hortobágyi T: Immunohistochemical correlates of TP53 somatic mutations in cancer. Oncotarget 7(40):64910–64920, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]


