Abstract
The proteasome, the most complex protease known, degrades proteins that have been conjugated to ubiquitin. It faces the unique challenge of acting enzymatically on hundreds and perhaps thousands of structurally diverse substrates, mechanically unfolding them from their native state and translocating them vectorially from one specialized compartment of the enzyme to another. Moreover, substrates are modified by ubiquitin in myriad configurations of chains. The many unusual design features of the proteasome may have evolved in part to endow this enzyme with a robust ability to process substrates regardless of their identity. The proteasome plays a major role in preserving protein homeostasis in the cell, which requires adaptation to a wide variety of stress conditions. Modulation of proteasome function is achieved through a large network of proteins that interact with it dynamically, modify it enzymatically, or fine-tune its levels. The resulting adaptability of the proteasome, which is unique among proteases, enables cells to control the output of the ubiquitin-proteasome pathway on a global scale.
Introduction
Quality control (QC) of proteins and organelles in eukaryotic cells is mediated by a vast and incompletely charted set of activities. QC pathways can target proteins that are misfolded, aggregated, mutated, chemically modified, mislocalized, mistranslated, or that have failed to assemble into a multisubunit complex. The significance of QC to human disease as well as aging is well recognized and owes to the marked toxicity of many misfolded proteins. Molecular chaperones, autophagy, and the ubiquitin-proteasome system (UPS) are all key players in QC pathways (see review in this issue by Hegde and Zavodszky). While molecular chaperones work in part to prevent and reverse misfolding events, they cannot correct all QC problems by any means, and therefore the activity of molecular chaperones is complemented by autophagy and the UPS, which safeguard proteostasis by destroying misfolded and toxic species. At a mechanistic level, molecular chaperones, autophagy, and the UPS often work hand in hand. For example, molecular chaperones frequently assist in targeting proteins to the UPS, and selective autophagy is often driven by ubiquitination of autophagic cargo.
Here we focus on the UPS, and in particular on the proteasome, a 2.5–3 MDa protease that degrades proteins that have been conjugated to ubiquitin (Hough et al. 1987; Waxman et al. 1987). The proteasome is of interest as the enzyme at which all substrates converge in the UPS, as one of the most complex enzymes in nature, as a regulatory hub of the UPS, and as a major therapeutic target. Excellent recent reviews have covered proteasome structure and function (Collins and Goldberg 2017; Bard et al. 2018), ubiquitin recognition by the proteasome (Saeki 2017), substrate processing by the proteasome (Yu and Matouschek 2017), proteasomal deubiquitinating enzymes (de Poot et al. 2017), and proteasome assembly (Budenholzer et al. 2017; Rousseau and Bertolotti 2018).
Assembly of the proteasome from the regulatory and core particles
All cells carry out selective protein degradation primarily through ATP-dependent proteases whose proteolytic sites are sequestered from the cytoplasmic space to minimize nonspecific proteolytic events. The proteasome is on the same evolutionary lineage as the archaeal protease PAN, although the latter is formed from three distinct gene products and the proteasome 33 gene products. The PAN protease has a proteolytic core particle (CP; also known as the 20S complex) composed of α-type and β-type subunits arranged in rings that are stacked into a barrel-like α7β7β7α7 assembly (Lowe et al. 1995). Thus, the heptameric α rings occupy the ends of the barrel, whereas the inner rings are formed by β subunits, which are proteolytically active. The CP of the eukaryotic proteasome differs mainly in that the α and β rings are heteromeric rather than homomeric (Groll et al. 1997).
Sequestration of the CP’s proteolytic sites, which face the interior of the barrel, restricts their enzymatic activity when the CP is in an isolated state. However, a variety of activating complexes can derepress the CP by opening a gate in the center of the α ring, through which substrates will pass (Groll et al. 2000; Whitby et al. 2000; Stadtmueller and Hill 2011). This gate provides tightly regulated access into the proteolytic chamber of the CP. In the case of PAN, a homohexameric ATPase ring mediates activation. The C-termini of the ATPases insert into intersubunit pockets within the α ring, which control opening of the gate (Smith et al. 2007; Majumder et al. 2018).
Gate opening is required but not sufficient for rapid and specific protein degradation. Folded domains within proteins prevent their passage through the narrow channel into the CP; thus, domains must be unfolded prior to degradation. The ATPase ring uses the energy of ATP hydrolysis to actively unfold the substrate as it is vectorially translocated into CP (Fig. 1). Unfolding is driven mechanically by ATP-dependent translocation, as the substrate is forced to negotiate the narrow channel (Prakash et al. 2004; Yu and Matouschek 2017). The substrate translocation channel of the ATPase ring is lined with substrate-contacting “pore loops” on each ATPase, whose ATP-directed motion drives substrates into the CP (de la Pena et al. 2018; Dong et al. 2018). Favorable proteasome substrates must contain an element that can make initial contact with the proteasome, the target of which is thought to be the pore loops, so as to initiate degradation. This step is known as engagement and requires a flexible segment of at least 20–30 amino acids in the initiator element, though sequence requirements are minimal (Yu et al. 2016; Yu and Matouschek 2017).
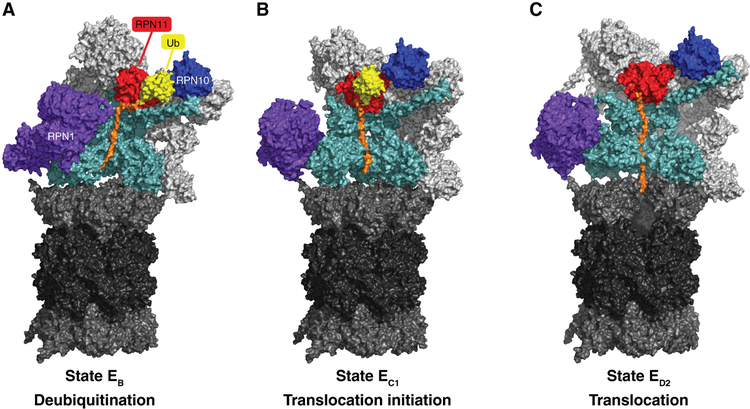
Figure 1. Conformational states of substrate-bound human proteasomes.
Cryo-EM density maps of substrate (orange) bound human proteasomes in three different stages of substrate processing are represented. A shows a conformation that is compatible with the deubiquitylation of the bound substrate. B) and C) represent two consecutive conformations of substrate translocation (EC1 and ED2) into the CP. Rpt1 and Rpt5 density maps were removed from the ATPase rings to expose the substrate translocation channel of the RP. In ED2, subunit α1 (Psma1) was also removed to show the substrate translocation channel of the CP. EB (PDB: 6MSE), EC1 (PDB: 6MSG), and ED2 (PDB: 6MSK) density maps were obtained from (Dong et al. 2018).
Gate opening and substrate unfolding are still not sufficient for protein degradation by the proteasome, as ubiquitin modification is required for most substrates. Ubiquitin docks a substrate on the proteasome, forming an enzyme-substrate complex that is sufficiently stable that substrate can be productively engaged by the pore loops, and translocation can initiate. With the innovation of ubiquitination in the eukaryotic lineage, the PAN-like ancestor protease was greatly elaborated to form the proteasome. As we have seen for the CP, the homomeric ATPase ring evolved into a heteromeric ring (subunits Rpt1-Rpt6), but more importantly many new components were added (Rpn-type subunits) to generate a 19-subunit assembly referred to as the proteasome regulatory particle (RP) or 19S complex. Many of the new subunits and associated factors adapt the proteasome to recognize and process ubiquitin conjugates, functioning either as ubiquitin receptors or deubiquitinating enzymes (Fig. 2). Other CP activators, such as PA28 and PA200, seem to lack both ATPase and ubiquitin-recognizing activities. They will not be considered here but the reader is referred to other reviews (Stadtmueller and Hill 2011; Tanaka et al. 2012). Finally, we note that numerous tissue-specific isoforms of the proteasome have been described, most notably the thymoproteasome and immunoproteasome, which are critical for the ontogeny and selectivity of adaptive immunity, respectively (Qian et al. 2013; Huang et al. 2016a; Murata et al. 2018).
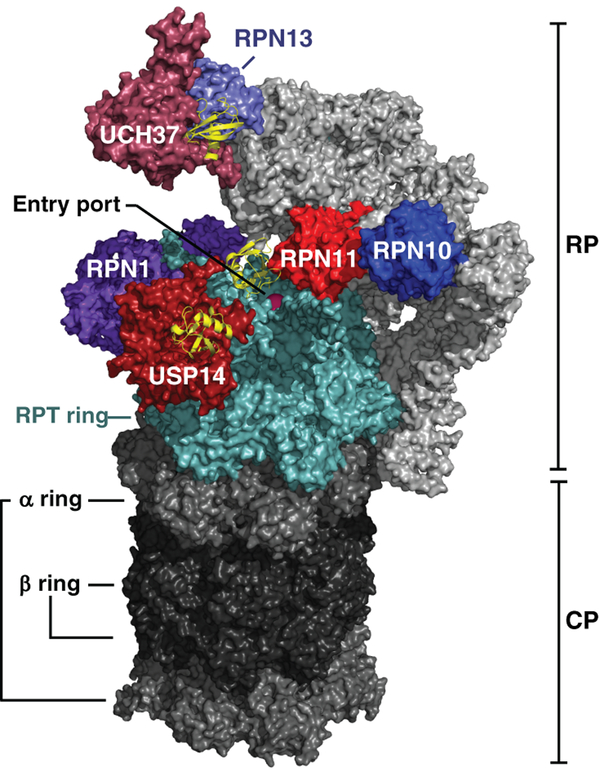
Figure 2. Overview of DUBs and Ub/Ubl receptors of the human proteasome.
A cryo-EM density map of the human proteasome is represented with all of its known ubiquitin receptors (Rpn1, Rpn10 and Rpn13 in purple tones) and DUBs (Rpn11, USP14 and UCH37 in red tones). The cryo-EM structure of the proteasome bound to USP14-ubiquitin-aldehyde was obtained from (Huang et al. 2016b) (PDB: 5GJQ). The location of UCH37 and Rpn13 were modeled as shown in (de Poot et al. 2017) using density maps from (Sahtoe et al. 2015; Vander Linden et al. 2015) (PDB: 4UEL and 4WLQ). All six subunits of the Rpt ring are shown in light blue and ubiquitin monomers bound to the DUBs are shown in yellow ribbon representations.
Ubiquitin recognition and processing
The most critical ubiquitin receptor of the proteasome is Rpn10. This subunit recognizes ubiquitin through an amphipathic α-helix known as the ubiquitin-interacting motif (UIM). In mice, the UIM appears to be essential for embryonic development, and liver-specific conditional mutants lacking the UIM exhibit substantial defects in the breakdown of ubiquitin-protein conjugates in this tissue (Hamazaki et al. 2007). Despite the importance of the UIM, inactivating mutants do not have a striking growth phenotype in yeast (Fu et al. 1998), indicating that other receptors also have significant roles in ubiquitin recognition. The Pru domain of Rpn13 (Husnjak et al. 2008; Schreiner et al. 2008) and the T1 element of Rpn1 (Chen et al. 2016b; Shi et al. 2016; Dong et al. 2018) have also been shown to function in ubiquitin recognition (Fig. 2). However, subunits Rpn13, Rpn1, and Rpn10 do not function only as ubiquitin receptors–they can also recognize ubiquitin indirectly by reversibly binding extrinsic ubiquitin receptors, which themselves carry substrate. The canonical receptors of this class are the UBL-UBA proteins (Fig. 3), which bind ubiquitin via their UBA domains and the proteasome via their ubiquitin-like (UBL) domains. An elegant proteomic study concluded that the UBL-UBA pathway is the major route by which proteins are targeted to the proteasome in yeast (Tsuchiya et al. 2017), though this study tracked biochemical interactions with the proteasome rather than protein degradation per se. The three UBL-UBA proteins in yeast, Rad23, Dsk2, and Ddi1, have each been amplified into a small gene family in mammals (see Fig. 3 and (Saeki 2017)), and mutations in a Dsk2 homolog, Ubiquilin2, underlie a familial form of amyotrophic lateral sclerosis (ALS; (Deng et al. 2011)).
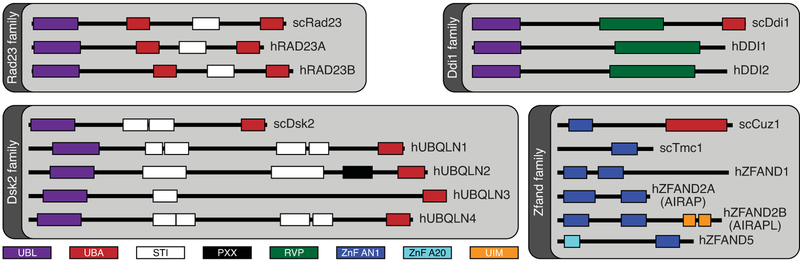
Figure 3. Extrinsic ubiquitin receptors of the proteasome.
Schematic representation of four protein families of S. cerevisiae and H. sapiens that include proteins thought to deliver ubiquitinated targets to the proteasome (Rad23, Ddi1, Dsk2, and Zfand). Some of these proteins do not appear to ubiquitin receptors (see text for details). SMART (Simple Modular Architecture Research Tool (Letunic and Bork 2018)) was used to identify SMART and Pfam protein domains. All proteins and domains are drawn to scale.
Yeast strains in which the RAD23, DSK2, and DDI1 genes have all been deleted, and the ubiquitin-binding elements of Rpn10, Rpn13, and Rpn1 mutationally inactivated, are sensitive to stress but nonetheless viable, suggesting that additional ubiquitin receptors for the proteasome remain to be identified (Shi et al. 2016). Why are there so many proteasomal ubiquitin receptors? One reason may be to promote substrate association with the proteasome in multiple alternative orientations–“orientation sampling” (Shi et al. 2016). According to this model, some substrates docked at the proteasome may initiate degradation inefficiently because their initiation elements are not positioned closely enough to the substrate entry port. However, if this were the case when the ubiquitin chain is bound to one receptor, it might not be when bound to another. Thus, positioning ubiquitin receptors at different locations in the RP could allow the proteasome to function more robustly as a protease. A second, not mutually exclusive model, is that having multiple receptors is advantageous in allowing avid or multipoint recognition of ubiquitin conjugates (Meyer and Rape 2014; Lu et al. 2015; Shi et al. 2016). Multipoint recognition would enhance the ability of the proteasome to recognize substrates modified with branched ubiquitin chains and with chains attached at multiple lysines within the substrate. As with orientation sampling, the most general effect would be a more robust capacity to degrade diverse cellular proteins.
Deubiquitinating enzymes on the proteasome prevent ubiquitin from being degraded together with the substrate to which it is attached (de Poot et al. 2017). Release of ubiquitin can promote substrate degradation, because ubiquitin has an unusually stable folded structure, and thus it can impede translocation kinetically (Verma et al. 2002; Yao and Cohen 2002; Worden et al. 2017). Deubiquitinating enzyme Rpn11 is positioned directly above the substrate entry port of the actively translocating proteasome (Figs. 1 and 2). Due in part to this positioning, it seems to remove ubiquitin primarily from substrates that have already been committed to degradation by virtue of engagement with the pore loops of the ATPase ring. The ATP-dependence of Rpn11 activity is a signature of this coupling to substrate engagement, given that Rpn11 is not itself an ATPase. Proteasomal deubiquitinating enzymes that function independently of ATP, such as USP14/Ubp6 and UCH37/UCHL5 (Fig. 2), can remove ubiquitin prior to substrate engagement and therefore can antagonize degradation by promoting substrate dissociation from the proteasome prior to the “engagement” step (Yao and Cohen 2002; Lee et al. 2010; Lee et al. 2016). The catalytic activities of UCH37/UCHL5, USP14/Ubp6, and Rpn11/PSMD14 are all activated by the proteasome and subject to other types of regulation as reviewed below and elsewhere (de Poot et al. 2017; Bard et al. 2018).
Conformational States of the Proteasome
Cryo-electron microscopy has recently been applied to the proteasome with great success, and many distinct conformational have been resolved (Bard et al. 2018). Although the total number of distinct populated states is high, and remains to be established, studies of the yeast proteasome have to date resolved six conformational states, s1-s6 (Eisele et al. 2018), even in the absence of substrate. The s1 state is the principal species in idling proteasomes, that is, in the presence of ATP and the absence of substrate. (Substrate-free proteasomes continually hydrolyze ATP.) Key features of the proteasome are misaligned in s1; the substrate translocation channel of the ATPase ring is not flush with the CP gate, and importantly the active site of Rpn11 is 25 Ångstroms away from the substrate entry port of the ATPase ring (Fig. 1). Although the CP gate was for many years viewed as being opened as a direct consequence of CP-RP association, this is clearly not the case as the gate is closed in the s1, s2, and s3 states. The increased access of small molecule probes such as LLVY-AMC upon holoenzyme assembly in the absence of substrate may reflect a mixture of holoenzyme states in the sample, although the open-gate states s4, s5, and s6 are not well populated. In any case, the closed CP gate seen in s1 state proteasomes is an additional hallmark of their latency.
The proteasome can be converted into a degradation-competent state even in the absence of substrate, for example by replacing ATP with the slowly hydrolyzed nucleotide ATPγS. Under this condition the major form is s3, which shows productive axial alignment of Rpn11, the RP substrate translocation channel, and the CP gate. But the gate remains closed, being opened only in the s4 state (Wehmer et al. 2017; Eisele et al. 2018). In general, gate opening has been regarded, both from biochemical and initial cryoEM studies, as a consequence of the insertion of “HbYX” type ATPases into their cognate α pockets. The HbYX motif, whose signature is a penultimate tyrosine, was identified in the C-terminus of PAN, and found to be present as well in the three of the six proteasomal ATPases (Rpt2, Rpt3, and Rpt5) (Smith et al. 2007). However, a key role was subsequently found for α pocket contacts to the C-terminus of Rpt6 (Park et al. 2013; Sokolova et al. 2015; Eisele et al. 2018), which does not conform to the HbYX consensus. Optimal gate opening is apparently only observed when the C-termini of both Rpt6 and Rpt1 are engaged with α ring (Eisele et al. 2018), which will depend on the nucleotide state of the proteasome’s ATPase ring, or “Rpt ring,” as discussed below.
Recent work has visualized numerous substrate-engaged states by substrate trapping, using approaches such as stripping Zn++ ion from Rpn11 or adding ATPγS to proteasomes that are in the act of substrate degradation (Matyskiela et al. 2013; de la Pena et al. 2018; Dong et al. 2018). These studies present a detailed model of the proteasome’s ATP hydrolytic cycle and coupling to substrate translocation. The hydrolytic cycle of the engaged proteasome is thought to involve a “rotary” mechanism in which an ATP hydrolytic event in one Rpt protein is followed by one in the adjacent subunit, proceeding continuously around the ring (de la Pena et al. 2018; Dong et al. 2018; Majumder et al. 2018). Substrate-engaged pore loops travel “downwards,” or towards the CP, in conveyor-belt fashion, under the direction of the sequential ATP hydrolytic events. When a pore loop reaches the CP-proximal position, it disengages from the substrate, then returns to a CP-distal position where it will subsequently engage another segment of the translocating substrate. Disengaged subunits are displaced outward from the ring so that their pore loops lose contact with substrate (Fig. 4). The coupling of disengagement to distal movement of the pore loop underlies the vectoriality of substrate movement, in that only engaged pore loops move towards the CP. Contemporaneously with these motions, the four subunits that are engaged with substrate move in concert, with one step of the process translocating the substrate towards the CP by a distance corresponding to approximately two amino acids in the substrate. The dynamics of the Rpt ring and the central role of the pore loops in driving substrate translocation are similar to those generally observed in AAA ATPase complexes (Puchades et al. 2017), which include other proteases, helicases, and additional biochemical activities.
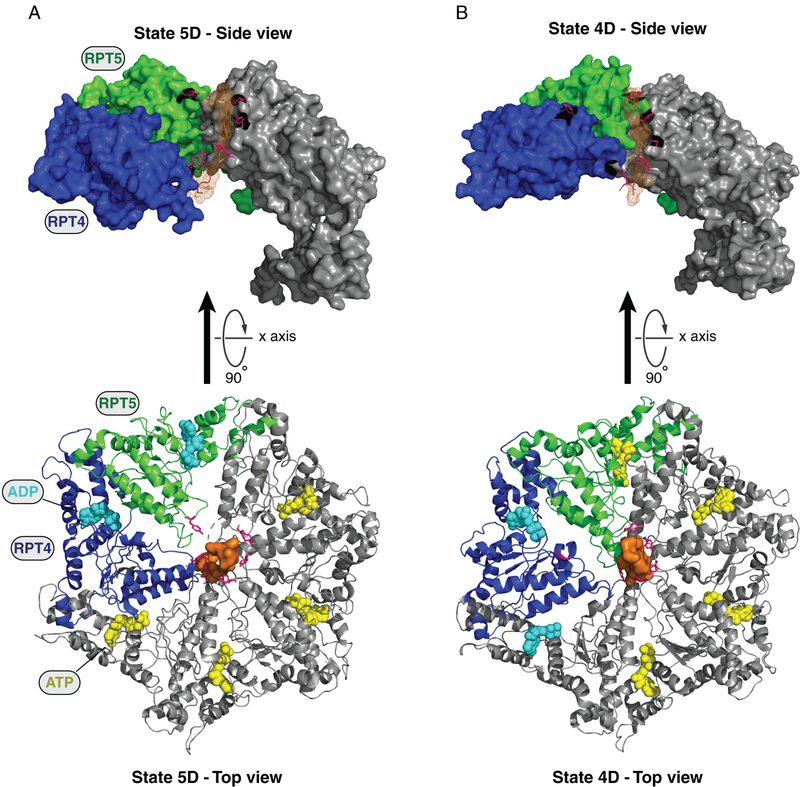
Figure 4. Representation of the ATP hydrolytic cycle and substrate translocation.
In A and B, two different cryo-EM structures of the Rpt ring at different states of substrate (orange mesh) translocation in the yeast proteasome holoenzyme are shown (states 5D and 4D, respectively). Images are modified from de la Pena et al. 2018. Ribbon representations of the top view of the Rpt rings are shown in the bottom panels, where substrate disengagement of Rpt5 (green) and Rpt4 (blue) can be observed (A and B, respectively). A 90° rotation about the x axis was applied to show a lateral view of the Rpt ring (top panels). While the substrate polypeptide is surrounded by a spiral-staircase of pore 1 loop tyrosines (highlighted in pink) projecting from substrate-engaged Rpt subunits, the Tyr of the Rpt subunits that are disengaged [Rpt5 in (A) and Rpt4 in (B)] are displaced from the substrate. In the bottom panels, spherical representations of ATP (yellow) and ADP (blue) are also included. For a better view of the spiral-staircase model in the top panels, two Rpt subunits (Rpt4 and Rpt3) and three amino acids (Arg261, Glu293, and Gly294 of Rpt4) were omitted from the density maps. States 5D and 4D were obtained from (de la Pena et al. 2018) (PDB: 6EF1 and 6EF3, respectively).
Tomographic Resolution of Proteasome States within Cells
Remarkable advances in cryo-electron tomography have recently enabled the visualization of proteasomes under essentially native conditions within cells (Asano et al. 2015; Guo et al. 2018). The level of resolution is sufficient to assign conformational states in reference to cryo-EM data on purified proteasomes. The tomographic approach has been particularly revealing in the study of familial amyotrophic lateral sclerosis (fALS). The C9orf72 gene is often expanded in G4C2 repeats in fALS, and these are aberrantly translated into a variety of toxic dipeptide repeat containing proteins. One of these, poly-Gly-Ala (poly-GA), forms densely packed cytoplasmic fibrils that specifically recruit proteasomes, with poly-GA aggregates concentrating proteasomes ~30-fold over neighboring sites within the cell (Guo et al. 2018). Unlike the general pool of proteasomes, sequestered proteasomes are frequently found in the s4 state and thus are likely to be engaged with substrate. Yet poly-GA proteins are not favorable proteasome substrates. These findings suggest that the sequestered proteasomes may be unproductively engaged with substrate, i.e., stalled. The sequestration and inhibition of proteasome by poly-GA aggregates could potentially play a significant role in their neurodegenerative effect.
Substrate Recognition and the Ubiquitin Code
Unlike many protein modifications, ubiquitination exhibits marked structural variation, arising principally from the formation of ubiquitin-ubiquitin adducts. The seven lysine residues of ubiquitin as well as its N-terminal amino group are all subject to modification (Chau et al. 1989; Spence et al. 1995; Kirisako et al. 2006; Xu et al. 2009; Kwon and Ciechanover 2017). Ligases and ubiquitin conjugating enzymes with high specificity in the formation of ubiquitin-ubiquitin linkages generate homogeneous chains (Chau et al. 1989; VanDemark et al. 2001), while other ligases generate mixed linkages (Kirkpatrick et al. 2006), or in some cases no chains whatsoever (Braten et al. 2016; Nguyen et al. 2017; Yanagitani et al. 2017). Some chains have branched topologies (Meyer and Rape 2014; Yau et al. 2017; Samant et al. 2018), where an acceptor ubiquitin is modified by more than one ubiquitin donor. Phosphorylation and acetylation of ubiquitin provide further important variation (Harper et al. 2018), and another protein modifier, the ubiquitin-like protein SUMO, can form mixed chains with ubiquitin, which also target proteins to the proteasome (Liebelt and Vertegaal 2016). Chains vary in length (Tsuchiya et al. 2018), and with approximately ~100 distinct deubiquitinating enzymes (DUBs) encoded in the mammalian genome (Leznicki and Kulathu 2017), it is not surprising that chains are in addition highly dynamic. Chain editing processes may fundamentally alter chain structures over time (Wertz and Dixit 2014; Chen et al. 2017).
The complexity of chain architectures reflects that ubiquitin serves numerous signaling roles apart from targeting to the proteasome. Ubiquitination of cell surface proteins targets them to the endosome and ultimately the lysosome for degradation (Piper et al. 2014). In addition, selective autophagy is frequently dependent on ubiquitination of cargo proteins (Grumati and Dikic 2018). Aside from these degradative pathways, many ubiquitination events do not target substrate degradation at all, as commonly seen in the signaling pathways of innate immunity and in targets of modification in chromatin, such as histones and PCNA. It is clear that chain linkage specificity underlies much of the selectivity of these targeting events, with chains linked through Lys63 and Met1 being instrumental in nonproteolytic signaling. Yet why the proteasome acts so poorly, if at all, on substrates modified by such chains is still unresolved. Purified proteasomes discriminate only weakly against chains, such as Lys63 chains, that appear to exhibit negligible targeting activity in vivo (Hofmann and Pickart 2001). One explanation may be that the in vivo activity of proteasomes on such targets is limited by kinetic competition with other (Lys63-specific) ubiquitin receptors (Nathan et al. 2013). In other cases, the Lys63 chain selectivity effect may be misconstrued in that it does not owe to the chain itself but instead to the substrate components of these conjugates being inherently poor proteasome substrates, for example by being resistant to unfolding by the proteasome.
A key problem in substrate recognition by the proteasome is how UBL-UBA proteins promote the process. Since the proteasome-binding UBL domain and the ubiquitin-binding UBA domain are joined through long, flexible linkers, a substrate docked by these bridging factors would have greater freedom of orientation with respect to the proteasome than one docked directly onto Rpn1 or Rpn13, which could result in faster and more efficient orientation sampling (Shi et al. 2016). The UBA domains within UBL-UBA proteins suppress deubiquitination of bound conjugates, which would also promote degradation (Hartmann-Petersen et al. 2003). Binding of the proteasome by the UBL domains of these proteins may also play a regulatory role, as purified UBL domains from mammalian homologs of Rad23 and Dsk2 (hHR23B and Ubiquilin1, respectively; Fig. 3) stimulate peptide hydrolysis by the proteasome, albeit partially (Kim and Goldberg 2018). As mentioned above, only a subset of the conformational states of the assembled proteasome holoenzyme have an open CP channel. Therefore, such a stimulation of peptide hydrolysis could suggest that UBL binding somehow redistributes the ensemble of conformational states that are present at steady-state in the proteasome samples tested.
Additional, more specific functions of UBL-UBA proteins have also been reported. For example, ubiquilins (mammalian homologues of Dsk2) specifically promote the proteasomal degradation of mislocalized mitochondrial proteins through recognizing exposed transmembrane (TM) domains (Itakura et al. 2016; Suzuki and Kawahara 2016; Whiteley et al. 2017). This ubiquilin function is particularly evident in activated B cells, which have elevated levels of mitochondrial protein mislocalization (Whiteley et al. 2017). The domain implicated in TM interaction is neither the UBL nor UBA, but one within the flexible linker between them–the STI domain, also known as the M domain (Fig. 3)–that has so far been subject to little study (Itakura et al. 2016). As ubiquilins are hypothesized to recruit an E3 to ubiquitinate such proteins, this model holds that, at least for these substrates, ubiquilin binding actually precedes and helps to direct ubiquitination events.
Studies in yeast point to a critical role for UBL-UBA proteins (Rad23 and Dsk2) in the transfer of ubiquitin conjugates from Cdc48 to the proteasome. Cdc48, known in mammals as p97, is an interesting ATPase-ring complex from the same (AAA+) protein family as the Rpt proteins of the proteasome (van den Boom and Meyer 2018). It appears to unfold proteins, primarily ubiquitinated proteins, by a mechanism involving threading of the substrate through a narrow axial channel, as described for the proteasome above (Blythe et al. 2017; Bodnar and Rapoport 2017). Such substrates are in many (but not all) cases destined to the proteasome for degradation, in that the Cdc48 complex itself has no proteolytic activity. One possible model of Cdc48-proteasome coupling is that the ATPase ring of Cdc48 replaces the RP complex at the cylinder end of the CP, injecting substrate directly into the CP (Barthelme and Sauer 2012; Esaki et al. 2017). A model with more direct support to date is that UBL-UBA proteins ferry the substrate from Cdc48 to a canonical RP-CP proteasome (Kim et al. 2004; Richly et al. 2005; Tsuchiya et al. 2017).
A cofactor of Cdc48 known as Ufd2, an E4 ubiquitin ligase (i.e., it extends pre-existing ubiquitin chains), directly interacts with the UBL domains of Rad23 and Dsk2 (Hanzelmann et al. 2010). In fact Ufd2 has a considerably higher affinity for these UBL domains than does the proteasome. As chains are extended by a Ufd2-Rad23 complex on Cdc48, the UBA domain in Rad23 can bind the growing chain as it reaches approximately six ubiquitins in length, blocking further elongation. The Rad23-substrate complex can then dissociate from Ufd2, freeing its UBL domain to bind proteasomal receptors such as Rpn1 and Rpn13.
Recent studies have reported a crucial role of Ubiquilin2 (Hjerpe et al. 2016) and its yeast homolog Dsk2 (Samant et al. 2018) in the turnover of misfolded or aggregated proteins in the nucleus, including misfolded proteins generated by heat shock. Aggregates of polyQ-expanded Huntingtin also appear to be cleared through Ubiquilin2 (Hjerpe et al. 2016). In these studies the UBL-UBA protein is thought to cooperate with Hsp70 chaperones rather than Cdc48/p97. This pathway to the proteasome may be of special importance in the nucleus because of the absence of autophagy in this compartment.
Several ubiquitin-receptor proteins that are not in the UBL-UBA family also bind proteasomes and appear to target proteins to the proteasome (reviewed in (Saeki 2017)). The ZFAND family of zinc finger proteins is of special interest (Fig. 3). Initial work defined AIRAP (a.k.a. ZFAND2A) as a protein that is induced by the proteotoxic agent arsenic, binds proteasomes, and enhances their activity through an unknown mechanism (Stanhill et al. 2006). A constitutive homolog located in the endoplasmic reticulum, AIRAPL (a.k.a. ZFAND2B), was later identified, and C. elegans lacking this activity were found to have a decreased lifespan and to be hypersensitive to the expression of misfolded proteins (Yun et al. 2008). Although the expression of AIRAPL is not induced by arsenic, the protein is recruited to the proteasome under these conditions. ZFAND1, a.k.a. Cuz1, was originally characterized in S. cerevisiae, where it binds both proteasomes and Cdc48 and confers resistance to arsenic (Sa-Moura et al. 2013; Hanna et al. 2014). It was recently found to bind arsenite-induced cytoplasmic stress granules (SGs) in human (HEK293) cells and in yeast, and to promote binding of both proteasomes and p97 to these structures (Turakhiya et al. 2018). ZFAND1 also mediates the elimination of SGs in a proteasome- and p97-dependent manner. SGs form by phase separation when translation is suppressed, and contain ribonucleoprotein complexes stalled at a pre-initiation step. In general, SGs can be induced by diverse stresses, and curiously only those induced by arsenite stress are ZFAND1-responsive (Turakhiya et al. 2018).
ZFAND5 is unique among these ZFAND family members in having no obvious responsiveness to arsenic. It is instead strongly induced in atrophic muscle after treatments such as fasting (Hishiya et al. 2006), and Zfand5 null mutant mice are substantially deficient in muscle atrophy. These mutants also exhibit elevated ubiquitin conjugate levels under atrophy-inducing conditions, consistent with defective conjugate degradation. ZFAND5 has complex stimulatory effects on the proteasome and notably binds ubiquitin via its A20 domain, which is necessary for stimulation of ubiquitin conjugate degradation (Hishiya et al. 2006; Lee et al. 2018a). The ubiquitin-binding capacity of ZFAND5 is shared with ZFAND2B/AIRAPL, although ZFAND2B binds ubiquitin via tandem UIM elements rather than an A20 domain (Rahighi et al. 2016). In summary, two members of the ZFAND family are ubiquitin-binding proteins and are assumed to deliver conjugates to the proteasome in analogy to UBL-UBA proteins. Others, such as AIRAP, also stimulate proteasome activity, ostensibly by coupling the proteasome to p97, but perhaps also through other still unidentified mechanisms.
Regulation of proteasome levels by Rpn4 and Nrf1
Early in the development of the ubiquitin field, it was assumed that proteasomes were present at levels adequate to handle the incoming flux of ubiquitin conjugates, and that therefore the exact level and activity of this enzyme were not of major biological significance. Today control of the level of the proteasome is understood to be mediated by an elaborate network that contributes to global regulation of protein degradation. Proteasome levels are especially significant when cells are under proteotoxic stress or exposed to proteasome inhibitors in anticancer treatments.
In elegant studies, proteasome levels were shown in yeast to be under negative feedback control by the transcriptional factor Rpn4 (Xie and Varshavsky 2001). All genes encoding proteasome subunits have PACE elements in their promoters, which confer Rpn4 control (Fig. 5). Feedback is mediated by proteasomal degradation of Rpn4, whose basal half-life of ~2 minutes makes it one of the most rapidly turned over proteins in yeast. Rpn4 is ubiquitinated by the ligase Ubr2 but is also degraded through a ubiquitin-independent pathway (Wang et al. 2004; Ha et al. 2012). The latter pathway is very unusual for a high-turnover protein, and ensures that Rpn4 is sensitive principally to fluctuations in proteasome activity rather than ubiquitin levels, which are separately regulated (Hanna et al. 2007) (see also below). In the absence of proteostatic stress, proteasome levels are reduced in rpn4 null mutants (Xie and Varshavsky 2001), indicating that Rpn4 mediates tonic control over proteasome levels under favorable conditions. However, multiple transcriptional sensors of proteostatic stress bind to and regulate the promoter of the RPN4 gene (Wang et al. 2008) including those sensing heat stress (Hsf1) and oxidative stress (Yap1). Mediators of the pleiotropic drug response (Pdr1 and Pdr3) can also induce RPN4 transcription. These findings indicate that the Rpn4-proteasome negative feedback loop is integrated into a broader stress-responsive regulatory network. Indeed, Rpn4 itself has many transcriptional targets other than proteasome subunit genes (Jelinsky et al. 2000).
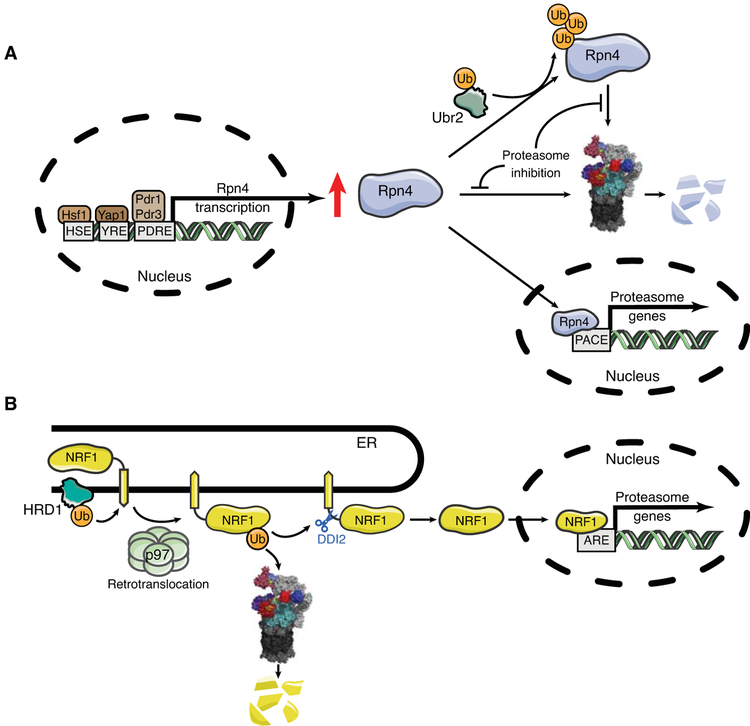
Figure 5. Feedback control of proteasome levels in yeast and mammals.
A) In S. cerevisiae, transcription factor Rpn4 controls the level of expression of proteasome subunits. Rpn4 is stabilized under specific proteotoxic stresses, inducing the transcription of proteasomal subunit genes regulated by PACE elements. B) In mammals, NRF1 is the key transcriptional regulator of proteasome subunit genes. A resident of the ER, NRF1 is ubiquitinated by HRD1 and retrotranslocated by p97. NRF1 protein can then be degraded via the proteasome as an ERAD substrate or cleaved endoproteolytically by the protease DDI2, resulting in its liberation from the ER membrane. The cleaved version of NRF1 is translocated to the nucleus where it binds ARE elements and induces the transcription of genes for proteasome subunits. The nuclear form of NRF1 is also rapidly degraded by the proteasome (see main text).
Although proteasome levels in mammals are also controlled by a homeostatic negative feedback loop, the regulators are not homologous to Rpn4. The design of the mammalian circuit is extremely interesting, and is conserved to C. elegans (Lehrbach and Ruvkun 2016). Transcription factor NRF1 is, like Rpn4, constitutively degraded (with a basal half-life of ~30 min), and like Rpn4 it controls the genes for all proteasome subunits in response to proteasome inhibitors (Radhakrishnan et al. 2010; Steffen et al. 2010). NRF1 recognizes well-characterized anti-oxidant response elements (AREs) in target genes, which encode a wide variety of activities (Fig. 5).
NRF1 differs from Rpn4 in being localized to the endoplasmic reticulum (ER), and its constitutive degradation proceeds in part through the ERAD pathway, in which proteins from the ER are retrotranslocated into the cytoplasm (see reviews in this issue: Karagöz et al.; Needham et al.). Retrotranslocation is coupled to ubiquitination, in this case by the HRD1 ligase, which resides in the ER membrane and also appears to be the transmembrane channel for retrotranslocation (Schoebel et al. 2017). The ubiquitinated substrate is extracted from the membrane by p97 and subsequently degraded by the proteasome. When retrotranslocated but not immediately degraded, NRF1 gains access to the nucleus, where it is active, though still rapidly degraded through the action of the ß-TRCP and FBXW7 ligases, and thus significantly accumulating only when its degradation through the proteasome is inhibited. However, translocation into the nucleus, and thus activation of Nrf1 in the cytosol, additionally requires an unusual endoproteolytic cleavage event that separates the transcriptional activation (TAD) and DNA binding domains of NRF1 from its transmembrane domain. Unexpectedly, the pathway does not simply consist in the release of a cytoplasmic TADs from a membrane anchor by endoproteolytic cleavage, because the TAD domains of NRF1 are situated inside the ER lumen prior to retrotranslocation (Fig. 5).
The source of endoproteolytic activity is remarkably enough a UBL-UBA domain protein, DDI2. It has been known for some time that Ddi1, the yeast homolog of mammalian DDI2, contains a functional aspartyl protease domain between its UBL and UBA domains (Krylov and Koonin 2001; Sirkis et al. 2006), but no substrate had been identified prior to NRF1. Possibly DDI2 uses its ubiquitin-binding capacity to selectively cleave ubiquitinated NRF1, working like the proteasome as a ubiquitin-dependent protease and indeed in competition with the proteasome for retrotranslocated NRF1. Such a competition would help to explain why transcription by NRF1 mediates feedback control of proteasome levels in response to proteasome inhibition, although cleaved and nuclearly localized NRF1 is also stabilized by proteasome inhibitors.
Although interest in NRF1 has focused mainly on its role in mediating adaptation to proteasome inhibitors during chemotherapy, there are other interesting aspects to the biology of this circuit. When mammals are exposed to ambient temperatures that are below body temperature, they induce thermogenic pathways to sustain their temperature. Enhanced metabolic activity in brown adipose tissue, or BAT, plays a major role in cold adaptation. Metabolic adaptation is accompanied by transcriptional induction of the Nrf1 gene (Nfe2l1), giving rise to significant elevation of proteasome levels (Bartelt et al. 2018). Under thermogenic conditions, but not at thermoneutrality, proteasome inhibitors delivered systemically compromise the ability of mice to sustain their body temperature. In BAT-specific Nrf1 mutants, numerous BAT proteins were found by global proteomics to be hyperubiquitinated under thermogenic conditions, an accumulation that points to overload of the proteasome in the absence of induction. Interestingly, ~1/3 of the hyperubiquitinated proteins were from mitochondria, which correlates with impaired mitochondrial function in Nrf1 mutants. This is a striking example of how proteasome levels are finely adjusted under physiologically relevant stress conditions.
Subunit Rpn6 promotes proteasome assembly and organismal longevity
Whereas Rpn4 and NRF1 control proteasome levels through coordinate regulation of all genes encoding proteasome subunits, other regulatory pathways are far more targeted and yet surprisingly achieve a similar effect. For example, transcriptional induction of the gene encoding Rpn6/PSMD11, a subunit of the RP, increases proteasome activity with dramatic physiological consequences, as described below (Vilchez et al. 2012a; Vilchez et al. 2012b; Vilchez et al. 2014). Interesting studies of C. elegans revealed the role of Rpn6 in proteasome regulation and set this in a critical physiological context. It has long been recognized that C. elegans mutants that lack a germline have an extended lifespan, which is dependent on the forkhead-class transcription factor DAF-16. The rpn-6.1 gene has an apparent DAF-16 binding site in its promoter and is induced by DAF-16. Overexpression of rpn-6.1 conferred longevity on worms as well as a variety of proteostasis-related phenotypes such as resistance to oxidative and heat stress. The Rpn6 effect is conserved between C. elegans and mammals, and the mechanism underlying this effect has been examined in mammalian cells (Vilchez et al. 2012a). Higher levels of Rpn6 were found to promote more efficient formation of holoenzyme from RP and CP (Vilchez et al. 2012a). Elevated Rpn6 expression was found in human embryonic stem cells, apparently accounting for their relatively high proteasome activities.
Although Rpn6 is to our knowledge the only proteasome subunit used physiologically to fine-tune proteasome holoenzyme levels, several other subunits have been found to drive proteasome assembly when artificially overexpressed. For example, overexpression of Rpn11 in otherwise wild-type D. melanogaster resulted in a substantial increase in mean lifespan from ~27 to ~37 days (Tonoki et al. 2009). Rpn11 overexpression increased proteasome levels and suppressed the accumulation of ubiquitin-protein conjugates that is observed in wild-type flies as they age. Similar effects on longevity have been reported for overexpression of CP subunit ß5 in C. elegans as well (Chondrogianni et al. 2015).
Post-translational regulation of proteasome levels
It has long been recognized that cells subjected to nutritional limitation induce autophagy, which serves to ensure stable amino acid supply from endogenous resources. More recently, notable studies have shown that a comparable response is exhibited by in the UPS and can be elicited by inhibition of the growth-regulatory kinase mTOR (Zhang et al. 2014; Zhao et al. 2015; Rousseau and Bertolotti 2016), which may likewise serve in stabilizing amino acid pools (Suraweera et al. 2012). These studies, however, reach different conclusions on the nature of mTOR regulation of the UPS, which can perhaps be traced back to the differing experimental systems used. Another study suggested that mTOR inhibition suppresses rather than activates the UPS (Zhang et al. 2014). The molecular mechanisms underlying these effects, including an acute elevation of proteasome activity upon mTOR inhibition, have been debated, and the reader is referred to recent reviews that discuss this in detail (Zhao and Goldberg 2016; Rousseau and Bertolotti 2018).
The response of the UPS is surprisingly sensitive to the nature and duration of the nutritional deprivation. In particular, longer deprivation results in down-regulation of proteasome activity (in contrast to that noted above), as cells progressively adopt a quiescent state. Protein synthesis rates fall markedly, and, as protein synthesis and degradation must be held in balance, downregulation of the proteasome should be adaptive. Genetic studies support this view (Marshall and Vierstra 2018). Multiple mechanisms have been shown to underlie this downregulation. It was first noted that as cells proceed into quiescent phase their proteasomes are disassembled into free RP and CP (Bajorek et al. 2003). Upon refeeding of these cultures, RP and CP rapidly reassemble, and thus a level of proteasome activity characteristic of growing culture is quickly re-established (Bajorek et al. 2003; Laporte et al. 2008). Open-channel mutants in CP α subunits lose viability after prolonged stationery phase incubation, indicating that these cells are indeed vulnerable to hyperactive protein degradation (Bajorek et al. 2003). The CP gate may thus play a key role in suppressing protein degradation by free CP, especially in stationary phase cells.
Interestingly, the free RP and CP in quiescent cells collect into structures known as proteasome storage granules, or PSGs (Laporte et al. 2008). PSGs contain both RP and CP but they do not assemble into holoenzyme within the granule. PSG formation also involves translocation of RP and CP from the nucleus to the cytoplasm (Laporte et al. 2008). Over 40 genes have been found to be required for PSG formation (Gu et al. 2017). Although a general understanding of the pathway has not yet been achieved, it is clear that Blm10/PA200 plays a key role in targeting CP into PSGs, while Spg5 does so for RP (Marshall and Vierstra 2018). Blm10 contains a functional HbYX motif at its C-terminus, which inserts into the α5/α6 pocket of the CP. Through this and other interactions, Blm10 occupies the cylinder end of the complex (Sadre-Bazzaz et al. 2010), explaining, at least in part, the failure of CP-RP interaction in the PSG.
PSGs are formed under carbon starvation though surprisingly not under nitrogen starvation. Under the latter condition, with amino acids limiting, proteasomes seem to be sacrificed for their amino acids, and eliminated through selective autophagy, with the CP and RP being independently targeted (Waite et al. 2016). This process, known as proteaphagy, is directed by Cue5 (Marshall et al. 2016), the only autophagy receptor in yeast that is known to recognize ubiquitinated cargo (Lu et al. 2014). The involvement of Cue5 suggests that ubiquitin modification of the proteasome may be required for proteaphagy, and indeed various proteasome components are subject to ubiquitination, particularly upon proteasome inhibition (Crosas et al. 2006; Besche et al. 2014; Marshall et al. 2015). What is the function of PSGs? They are not formed simply as an alternative to proteaphagy, but rather PSGs function to protect proteasome components from a default pathway of autophagy under carbon starvation. Thus, when PSG formation is prevented in carbon-starved cells, their proteasomes are degraded via autophagy, essentially reverting to the program of nitrogen starvation. This has been elegantly shown using null mutants in the above-mentioned PSG recruitment factors Blm10 and Spg5 (Marshall and Vierstra 2018).
Regulation of proteasome activity by USP14
As discussed above, during substrate degradation, attached ubiquitin groups constitute a kinetic impediment to translocation, a problem solved by the incorporation of deubiquitinating enzymes into the proteasome. In yeast, deletion of the UBP6 gene results in accelerated ubiquitin degradation via the proteasome (Hanna et al. 2003). Therefore Rpn11 cannot be fully competent to rescue substrate-attached ubiquitin from degradation. Perhaps there is a distinct class of ubiquitin conjugates that are weak Rpn11 substrates and preferred Ubp6 substrates but there is no indication of what features may underlie this. The specificity of Ubp6 and its mammalian ortholog USP14 is highly unusual in that these enzymes rapidly disassemble ubiquitin-protein conjugates that carry more than one ubiquitin chain (Lee et al. 2016). Conjugates of this type can be deubiquitinated by proteasome-associated, or “activated” Ubp6 on a time scale of milliseconds to seconds, whereas the deubiquitination of single-chain conjugates and disassembly of free chains by activated Ubp6 is so slow as to be challenging to measure. If a conjugate carrying multiple chains docks at the proteasome, Ubp6 removes every chain but one and then stops (Lee et al. 2016). The preservation of a single chain ensures that the substrate will still have a chance of being degraded, depending on the length of the remaining chain, which governs the dissociation rate of the substrate from the proteasome.
Ubp6 and Usp14 have long been viewed as suppressive of substrate degradation by the proteasome (Hanna et al. 2006; Hanna et al. 2007; Lee et al. 2010; Bashore et al. 2015; Lee et al. 2016; Kim and Goldberg 2017). This suppression has two distinct components, one catalytic and one noncatalytic. For brevity, the latter will not be discussed here, as it was recently reviewed (de Poot et al. 2017). The multichain specificity of deubiquitination by USP14 is in agreement with the model that USP14 suppresses substrate degradation, assuming that more than one chain on a substrate can contribute to the strength of binding to the proteasome. Recent work supports this assumption (Lu et al. 2015), and the concept of multivalent recognition of ubiquitin conjugates is in agreement with the multiplicity of ubiquitin receptors on the proteasome (see above).
The model that deubiquitination by Ubp6 or USP14 can suppress degradation requires that deubiquitination is very fast, so as to outrace substrate degradation. This has been validated by quench-flow and single-molecule analysis (Lee et al. 2016). The multichain model has not been validated with endogenous substrates, but many groups have identified endogenous clients of Usp14 that are more rapidly degraded when this activity is inhibited, or when USP14 expression is knocked down (Lee et al. 2010; Homma et al. 2015; Chen et al. 2016a; McKinnon et al. 2016; Nakashima et al. 2016; Sareen-Khanna et al. 2016; Xu et al. 2016; Boselli et al. 2017; Liao et al. 2017; Min et al. 2017b; Song et al. 2017; Wei et al. 2017; Zhu et al. 2017; Li et al. 2018; Liao et al. 2018; Liu et al. 2018; Massa et al. 2018). These experiments have relied mainly on the specific small-molecule inhibitors of USP14, IU1 and IU1–47 (Lee et al. 2010; Boselli et al. 2017). IU1 has recently been co-crystallized with USP14 and shown to inhibit enzymatic activity by blocking access of the C-terminus of ubiquitin to the active site of USP14, as shown in Fig. 6 (Wang et al. 2018).
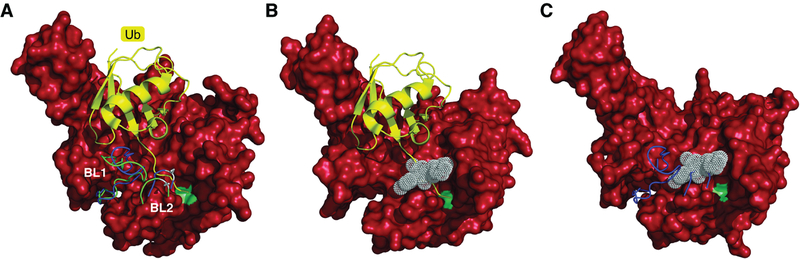
Figure 6. Activation and inhibition of Usp14.
A) Structure of substrate-engaged USP14 (red) bound to ubiquitin aldehyde (Ub-Al, in yellow), a model of the enzyme’s activated state. The principal difference from the ubiquitin-free (inactive) state of USP14 is in the positioning of the two blocking (BL) loops (Hu et al. 2005). The BL loops of USP14-Ub-Al are in green, those of the free USP14 in blue. To the right of these loops, the active-site Cys is represented as a light green patch. In the free form of USP14, the BL loops block the access of the C-terminus of ubiquitin to the active site. This occlusion is relieved in the substrate-engaged form. Ser432 (grey), located within the BL2 loop, is subject to phosphorylation by AKT, providing partial activation (Xu et al. 2015). B) Structure of USP14 bound to IU1–47 (white) shows that this inhibitor occludes access of the C-terminus of ubiquitin to the catalytic site (Wang et al. 2018). To better visualize the IU1–47 binding pocket, both BL loops were removed from the density maps. C) Structure of free USP14 with IU1–47 superimposed. This model shows how the BL2 loop (blue) occludes access of IU1–47, consistent with previous evidence that IU1 series compounds do not inhibit the free form of USP14 (Lee et al. 2016). Density maps of the USP14 catalytic domain in its free form (PDB: 2AYN), USP14 bound to Ub-Al (PDB: 2AYO), and the USP14-IU1–47 complex (PDB: 6IIL) were obtained from (Hu et al. 2005; Wang et al. 2018). To better visualize the catalytic site of USP14, Gln197 was removed from all USP14 structures in this figure.
In addition to suppressing the degradation of numerous proteins, USP14 has remarkably been found to also suppress the degradation of an organelle (Chakraborty et al. 2018). In both mammalian cells and D. melanogaster, USP14 controls the rate of basal mitophagy. Although specific USP14 substrates that mediate this effect have not been identified, it may reflect that basal mitophagy can be signaled by ubiquitination of proteins of the mitochondrial membrane (Villa et al. 2018), as described for the depolarization-induced mitophagy pathway that is mediated by the ubiquitin ligase Parkin (Harper et al. 2018). In D. melanogaster mutants that lack Parkin-dependent mitophagy, reduction of USP14 activity confers substantial lifespan extension and enhances motor function. These effects apparently result from improved mitochondrial function, as increased basal mitophagy provides for more efficient clearance of dysfunctional mitochondria that would otherwise be cleared by the Parkin pathway. Suppression of Parkin mutant phenotypes was evident both with Usp14 knockdown mutations and with IU1 provided in the flies’ food. Phenotypic suppression by loss of USP14 activity was also seen with mutants in Pink1, the protein kinase that activates Parkin (Chakraborty et al. 2018).
Ubp6/USP14 is activated 300–800 fold by proteasome association when assayed with a model substrate having a small leaving group, such as ubiquitin-AMC (Leggett et al. 2002). Repression of the free form of Ubp6/USP14 is thought to be mediated, at least in part, by two loops, BL1 and BL2 (Fig. 6), that block access of the C-terminus of ubiquitin to the active site of the enzyme (Hu et al. 2005; Huang et al. 2016b). Free Ubp6/USP14 is often assayed using diubiquitin, but interestingly no activation is evident with this substrate (Lee et al. 2016). A likely resolution to this conundrum is that complex formation with the proteasome, as seen by cryoEM (Aufderheide et al. 2015; Huang et al. 2016b), brings the active site of Ubp6/USP14 so close to the proteasome that the folded structure of the proximal ubiquitin is occluded (Lee et al. 2016). Accordingly, Ubp6/USP14 release chains en bloc rather than whittling them down from their tips. Interestingly, because proximal ubiquitin in a diubiquitin adduct occupies the position of substrate in a ubiquitin-substrate adduct, the idea that the body of the proteasome blocks access of bulky leaving groups to Ubp6/USP14 also carries the prediction that Ubp6/USP14 discriminates against folded domains in the substrate protein itself. Ubiquitin groups or chains attached to domains that are folded when the substrate docks at the proteasome may therefore be reserved for Rpn11, because substrate translocation draws these domains towards Rpn11 while their accessibility to Rpn11’s active site is ensured by ATP-dependent unfolding.
As discussed above, Ubp6 is needed in yeast to prevent excessive ubiquitin degradation. In yeast engineered to express low levels of ubiquitin, Ubp6 is dramatically induced by a homeostatic feedback circuit (Hanna et al. 2007). USP14 activity is subject to multiple controls though the mechanisms differ from that of Ubp6 in yeast. Perhaps surprisingly, four distinct microRNAs control the expression of the protein, exerting a variety of physiological effects (Wu et al. 2014; Sun et al. 2016; Zhu et al. 2017; Lee et al. 2018b). At the post-translational level, USP14 is phosphorylated on Ser432 by the growth-regulatory kinase AKT, which results in partial activation of its deubiquitinating activity (Xu et al. 2015). As Ser432 lies within the BL2 loop (Fig. 6), its modification presumably antagonizes the above-described autoinhibitory mechanism. In summary, growth signals such as insulin activate AKT, which among its many activities will potentiate USP14, likely leading to reduced protein breakdown by the proteasome.
The most dramatic regulation of USP14 described to date is post-translational and involves the protein TRIM11, which binds the N-terminal ubiquitin-like domain of USP14 in competition with the proteasome (Chen et al. 2018). Thus, TRIM11 functions as an endogenous USP14 inhibitor. Heat shock induces TRIM11, promoting cell survival apparently by releasing the proteasome from the inhibitory effect of USP14 (Chen et al. 2018). The intricate regulation of USP14 on multiple levels in mammals seems to imply that USP14 is used not simply to promote ubiquitin recovery from proteasome substrates but also as a vehicle for fine tuning of proteasome activity.
Proteasome-associated ubiquitin-conjugating activity
The proteasome is associated with not only deubiquitinating activity but also ubiquitin ligase activity. A number of ligases have been shown to interact with the proteasome, if in some cases weakly (Verma et al. 2000; Xie and Varshavsky 2000; Leggett et al. 2002; Xie and Varshavsky 2002; Crosas et al. 2006; Besche et al. 2009; Martinez-Noel et al. 2012; Kuhnle et al. 2018). One view of this phenomenon is that the potency of a ligase will be enhanced if it ubiquitinates its substrates at the proteasome, since the temporal lag between ubiquitination and proteasome docking, during which the conjugate is vulnerable to deubiquitination, is reduced (Xie and Varshavsky 2000). Another view, proposed for the yeast Hul5 ligase (Crosas et al. 2006), is that conjugating activity could be directed preferentially towards proteasome-bound proteins–typically proteins that are already ubiquitinated by another ligase–and function as a general regulator of the proteasome. In vitro, Hul5 efficiently adds ubiquitin to model target proteins such as tetraubiquitin and ubiquitinated cyclin B, while not taking unmodified cyclin B as a substrate (Crosas et al. 2006). These reactions are dependent on the presence of the proteasome. Because Hul5 modified these proteins only if they have already been ubiquitinated, Hul5 appeared to act in these cases as an “E4” or (essentially) a ubiquitin chain extending enzyme (Crosas et al. 2006).
Hul5 and its mammalian homolog UBE3C have repeatedly been identified as proteasome processivity factors in studies carried out with artificial fusion proteins that are ubiquitinated by an E3 other than Hul5 and degraded by the proteasome (Kohlmann et al. 2008; Aviram and Kornitzer 2010; Martinez-Noel et al. 2012; Chu et al. 2013). In hul5 and Ube3C loss of function mutants, the proteasome fails to fully degrade these proteins, releasing a truncated product. The activity seems to be unique to Hul5/UBE3C. These observations are consistent with a generalized E4 activity in Hul5, which serves to promote the degradation of stalled substrates that may otherwise fail to complete translocation from the RP to the CP. Seemingly in agreement with these findings, Hul5 and UBE3C are rapidly recruited to proteasomes whose activity has been compromised by mutation, inhibitors of the proteasome, or inhibitors of USP14 (Park et al. 2011; Kuo and Goldberg 2017). This recruitment effect is seen with USP14 as well (Kuo and Goldberg 2017), and the presence of USP14 enhances UBE3C recruitment just as Ubp6 enhances Hul5 recruitment (Crosas et al. 2006; Kuo and Goldberg 2017). Such inducible recruitment of USP14 to the proteasome may underlie the cytoprotective effects of USP14 inhibitors observed under some stress conditions (Sareen-Khanna et al. 2016; Min et al. 2017a; Chen et al. 2018; VerPlank et al. 2018). Defective proteasomes also recruit Ecm29, a large, HEAT-repeat-containing protein (Park et al. 2011; De La Mota-Peynado et al. 2013). Like USP14, Ecm29 exerts a negative influence on the proteasome. Binding in proximity to Rpt5, it suppresses ATPase activity, gate opening, and the degradation of ubiquitin-protein conjugates (De La Mota-Peynado et al. 2013).
An interesting inducer of Hul5-dependent ubiquitination is heat stress, pointing to a role of Hul5 in quality control degradation (Fang et al. 2011). hul5Δ mutants show a strong delay in the resumption of cell division after a heat shock. Moreover, as much as 50% of the ubiquitination that is acutely induced by heat shock is Hul5-dependent, with assays of the degradation of newly synthesized proteins giving comparable results. Most Hul5 substrates identified in this study were still monoubiquitinated in the absence of Hul5, in agreement with the model that Hul5 functions as an E4 in heat-shocked cells (Fang et al. 2011).
The conjugating activity of Hul5 and UBE3C is not strictly limited to proteasome substrates; they can also modify proteasome subunits as well, such as ubiquitin receptors Rpn10 and Rpn13 (Crosas et al. 2006; Besche et al. 2014). Ubiquitin modification of ubiquitin receptors typically results in their functional inactivation (Hoeller and Dikic 2010). Therefore, ubiquitination of Rpn10 and Rpn13 could mediate negative regulation of the proteasome under certain, perhaps extreme, stress conditions, in contrast to the positive regulation seen in many studies as described above.
Control of proteasome activity by phosphorylation
Protein modification by phosphorylation is widely used to integrate signaling, metabolic, and gene expression networks. Hundreds of sites on the proteasome are subject to phosphorylation but to date only a few have been characterized (reviewed by (Guo et al. 2017; VerPlank and Goldberg 2018)). Modification of Rpt3 at Thr25 by the Dyrk2 kinase places proteasome activity under cell cycle control, with a Thr25Ala knockin mutant being defective in total protein degradation to a remarkable 66% (Guo et al. 2016). Among other things this suggests minimal substrate specificity in the effect of phosphorylation. The mutation impairs cellular proliferation as well as the in vivo growth of triple-negative breast cancer cells, perhaps to be expected given the extent to which these proteasomes are inhibited. Why the knockin proteasomes function so poorly remains mysterious. The mutation does not affect proteasome levels or ostensibly their assembly (Guo et al. 2016). It has not been possible to visualize Rpt3-Thr25 in the proteasome to date but it would seem to be located far from the core elements of the RP such as the translocation channel and Rpn11.
Another critical phosphorylation site is Ser14 of Rpn6, a target of the cAMP-dependent protein kinase (PKA) (Lokireddy et al. 2015). Overexpression of a nonphosphorylatable Ser14Ala mutant of Rpn6 is suppressive of protein degradation, whereas that of the phosphomimetic Ser14Asp mutant stimulates degradation. As cAMP is used for signaling in diverse physiological and cellular contexts, including fasting and exercise, this pathway may be widely utilized. cAMP levels can be elevated experimentally using rolipram, a specific inhibitor of phosphodiesterase-4, which acts on cAMP. Rolipram increases the levels of assembled holoenzyme in cells, particularly the double-capped RP2CP form of holoenzyme, possibly accounting for the stimulation of protein degradation in treated cells (Lokireddy et al. 2015).
Rolipram accentuates proteasome activity in the mouse brain, presumably by inducing the phosphorylation of Rpn6-Ser14 (Myeku et al. 2016). In mice expressing a pathogenic form of the tau protein (tau-Pro301Leu) that induces neurodegeneration, rolipram significantly reduced the levels of insoluble tau species. These findings suggest that the stimulation of proteasome phosphorylation and activity in the brain attenuates tauopathies and may have implications for other neurodegenerative disorders.
Another phosphorylation event observed with brain proteasomes is the modification of Rpt6-Ser120 by the calcium/calmodulin-dependent kinase CaMKIIα, a critical and very abundant activity in the brain. Phosphorylation at Ser120 stimulates proteasome activity and interestingly it is induced in particular by cocaine (Gonzales et al. 2018). For reasons that are unclear, phosphomimetic Rtp6-Ser120Asp mouse mutants fail to develop cocaine sensitization, an effect closely linked to addiction.
Concluding remarks
Although derived from a common type of ATP-dependent protease expressed in archaea, the proteasome has evolved within the eukaryotic lineage into a unique molecular machine. Presumably the initial driving force behind the evolution of the complexity of the proteasome was the inherent complications involved in degrading ubiquitin-protein conjugates, as reflected in the unanticipated multiplicity of ubiquitin receptors and deubiquitinating enzymes associated with the proteasome. The complexity of the proteasome does not stop at its traditional borders; numerous cofactors, regulators, and post-translational modifications of the proteasome are integral to its function, and this broader network provides for striking adaptability in response to growth stimuli, nutritional limitation, and proteostatic stress. Thus, contrary to longstanding perceptions of the proteasome, extensive variation of the magnitude and nature of its activity are tolerated, and indeed these variations are promoted through conserved regulatory mechanisms. Because of the importance of the proteasome in proteostasis, these insights may have significant implications for our understanding of disease and aging. Future studies should define more clearly how proteasome regulators impact the increasingly well-defined core mechanisms of the enzyme and its ensemble of conformational states. In addition, proteomic studies will play a central role in defining how specific modes of proteasome regulation perturb its output on a global scale.
Acknowledgments and Disclosure statement
We gratefully acknowledge members of the Finley laboratory for helpful comments on the manuscript. Patents 8933087 and 9201073 are held on IU1, IU1–47, and USP14 inhibition, filed by Harvard University on behalf of D.F. and others. These patents have been licensed to Proteostasis Therapeutics.
References
- Asano S, Fukuda Y, Beck F, Aufderheide A, Forster F, Danev R, Baumeister W. 2015. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science 347: 439–442. [DOI] [PubMed] [Google Scholar]
- Aufderheide A, Beck F, Stengel F, Hartwig M, Schweitzer A, Pfeifer G, Goldberg AL, Sakata E, Baumeister W, Forster F. 2015. Structural characterization of the interaction of Ubp6 with the 26S proteasome. Proc Natl Acad Sci U S A 112: 8626–8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aviram S, Kornitzer D. 2010. The ubiquitin ligase Hul5 promotes proteasomal processivity. Mol Cell Biol 30: 985–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajorek M, Finley D, Glickman MH. 2003. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr Biol 13: 1140–1144. [DOI] [PubMed] [Google Scholar]
- Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A. 2018. Structure and Function of the 26S Proteasome. Annu Rev Biochem 87: 697–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A, Widenmaier SB, Schlein C, Johann K, Goncalves RLS, Eguchi K, Fischer AW, Parlakgul G, Snyder NA, Nguyen TB et al. 2018. Brown adipose tissue thermogenic adaptation requires Nrf1-mediated proteasomal activity. Nat Med 24: 292–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. 2012. Identification of the Cdc48·20S proteasome as an ancient AAA+ proteolytic machine. Science 337: 843–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashore C, Dambacher CM, Goodall EA, Matyskiela ME, Lander GC, Martin A. 2015. Ubp6 deubiquitinase controls conformational dynamics and substrate degradation of the 26S proteasome. Nat Struct Mol Biol 22: 712–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Haas W, Gygi SP, Goldberg AL. 2009. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48: 2538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besche HC, Sha Z, Kukushkin NV, Peth A, Hock EM, Kim W, Gygi S, Gutierrez JA, Liao H, Dick L et al. 2014. Autoubiquitination of the 26S proteasome on Rpn13 regulates breakdown of ubiquitin conjugates. EMBO J 33: 1159–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe EE, Olson KC, Chau V, Deshaies RJ. 2017. Ubiquitin- and ATP-dependent unfoldase activity of P97/VCP·NPLOC4·UFD1L is enhanced by a mutation that causes multisystem proteinopathy. Proc Natl Acad Sci U S A 114: E4380–E4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar NO, Rapoport TA. 2017. Molecular Mechanism of Substrate Processing by the Cdc48 ATPase Complex. Cell 169: 722–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boselli M, Lee BH, Robert J, Prado MA, Min SW, Cheng C, Silva MC, Seong C, Elsasser S, Hatle KM et al. 2017. An inhibitor of the proteasomal deubiquitinating enzyme USP14 induces tau elimination in cultured neurons. J Biol Chem 292: 19209–19225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braten O, Livneh I, Ziv T, Admon A, Kehat I, Caspi LH, Gonen H, Bercovich B, Godzik A, Jahandideh S et al. 2016. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc Natl Acad Sci U S A 113: E4639–4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budenholzer L, Cheng CL, Li Y, Hochstrasser M. 2017. Proteasome Structure and Assembly. J Mol Biol 429: 3500–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty J, von Stockum S, Marchesan E, Caicci F, Ferrari V, Rakovic A, Klein C, Antonini A, Bubacco L, Ziviani E. 2018. USP14 inhibition corrects an in vivo model of impaired mitophagy. EMBO Mol Med 10 10.15252/emmm.201809014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chau V, Tobias JW, Bachmair A, Marriott D, Ecker DJ, Gonda DK, Varshavsky A. 1989. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243: 1576–1583. [DOI] [PubMed] [Google Scholar]
- Chen L, Paquette N, Mamoor S, Rus F, Nandy A, Leszyk J, Shaffer SA, Silverman N. 2017. Innate immune signaling in Drosophila is regulated by transforming growth factor beta (TGFbeta)-activated kinase (Tak1)-triggered ubiquitin editing. J Biol Chem 292: 8738–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Zhu G, Johns EM, Yang X. 2018. TRIM11 activates the proteasome and promotes overall protein degradation by regulating USP14. Nat Commun 9 10.1038/s41467-018-03499-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Meng Q, Qin Y, Liang P, Tan P, He L, Zhou Y, Chen Y, Huang J, Wang RF et al. 2016a. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol Cell 64: 105–119. [DOI] [PubMed] [Google Scholar]
- Chen X, Randles L, Shi K, Tarasov SG, Aihara H, Walters KJ. 2016b. Structures of Rpn1 T1:Rad23 and hRpn13:hPLIC2 Reveal Distinct Binding Mechanisms between Substrate Receptors and Shuttle Factors of the Proteasome. Structure 24: 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrogianni N, Georgila K, Kourtis N, Tavernarakis N, Gonos ES. 2015. 20S proteasome activation promotes life span extension and resistance to proteotoxicity in Caenorhabditis elegans. FASEB J 29: 611–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu BW, Kovary KM, Guillaume J, Chen LC, Teruel MN, Wandless TJ. 2013. The E3 ubiquitin ligase UBE3C enhances proteasome processivity by ubiquitinating partially proteolyzed substrates. J Biol Chem 288: 34575–34587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins GA, Goldberg AL. 2017. The Logic of the 26S Proteasome. Cell 169: 792–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosas B, Hanna J, Kirkpatrick DS, Zhang DP, Tone Y, Hathaway NA, Buecker C, Leggett DS, Schmidt M, King RW et al. 2006. Ubiquitin chains are remodeled at the proteasome by opposing ubiquitin ligase and deubiquitinating activities. Cell 127: 1401–1413. [DOI] [PubMed] [Google Scholar]
- De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, Roelofs J. 2013. The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J Biol Chem 288: 29467–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Pena AH, Goodall EA, Gates SN, Lander GC, Martin A. 2018. Substrate-engaged 26S proteasome structures reveal mechanisms for ATP-hydrolysis-driven translocation. Science 362 10.1126/science.aav0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Poot SAH, Tian G, Finley D. 2017. Meddling with Fate: The Proteasomal Deubiquitinating Enzymes. J Mol Biol 429: 3525–3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, Yang Y, Fecto F, Shi Y, Zhai H et al. 2011. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature 477: 211–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Zhang S, Wu Z, Li X, Wang WL, Zhu Y, Stoilova-McPhie S, Lu Y, Finley D, Mao Y. 2018. Cryo-EM structures and dynamics of substrate-engaged human 26S proteasome. Nature 565: 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele MR, Reed RG, Rudack T, Schweitzer A, Beck F, Nagy I, Pfeifer G, Plitzko JM, Baumeister W, Tomko RJ Jr. et al. 2018. Expanded Coverage of the 26S Proteasome Conformational Landscape Reveals Mechanisms of Peptidase Gating. Cell Rep 24: 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esaki M, Islam MT, Tani N, Ogura T. 2017. Deviation of the typical AAA substrate-threading pore prevents fatal protein degradation in yeast Cdc48. Sci Rep 7 10.1038/s41598-017-05806-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang NN, Ng AH, Measday V, Mayor T. 2011. Hul5 HECT ubiquitin ligase plays a major role in the ubiquitylation and turnover of cytosolic misfolded proteins. Nat Cell Biol 13: 1344–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Sadis S, Rubin DM, Glickman M, van Nocker S, Finley D, Vierstra RD. 1998. Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26 S proteasome subunit Mcb1. J Biol Chem 273: 1970–1981. [DOI] [PubMed] [Google Scholar]
- Gonzales FR, Howell KK, Dozier LE, Anagnostaras SG, Patrick GN. 2018. Proteasome phosphorylation regulates cocaine-induced sensitization. Mol Cell Neurosci 88: 62–69. [DOI] [PubMed] [Google Scholar]
- Groll M, Bajorek M, Kohler A, Moroder L, Rubin DM, Huber R, Glickman MH, Finley D. 2000. A gated channel into the proteasome core particle. Nat Struct Biol 7: 1062–1067. [DOI] [PubMed] [Google Scholar]
- Groll M, Ditzel L, Lowe J, Stock D, Bochtler M, Bartunik HD, Huber R. 1997. Structure of 20S proteasome from yeast at 2.4 A resolution. Nature 386: 463–471. [DOI] [PubMed] [Google Scholar]
- Grumati P, Dikic I. 2018. Ubiquitin signaling and autophagy. J Biol Chem 293: 5404–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu ZC, Wu E, Sailer C, Jando J, Styles E, Eisenkolb I, Kuschel M, Bitschar K, Wang X, Huang L et al. 2017. Ubiquitin orchestrates proteasome dynamics between proliferation and quiescence in yeast. Mol Biol Cell 28: 2479–2491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Lehmer C, Martinez-Sanchez A, Rudack T, Beck F, Hartmann H, Perez-Berlanga M, Frottin F, Hipp MS, Hartl FU et al. 2018. In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment. Cell 172: 696–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Huang X, Chen MJ. 2017. Reversible phosphorylation of the 26S proteasome. Protein Cell 8: 255–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Wang X, Wang Z, Banerjee S, Yang J, Huang L, Dixon JE. 2016. Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat Cell Biol 18: 202–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha SW, Ju D, Xie Y. 2012. The N-terminal domain of Rpn4 serves as a portable ubiquitin-independent degron and is recognized by specific 19S RP subunits. Biochem Biophys Res Commun 419: 226–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamazaki J, Sasaki K, Kawahara H, Hisanaga S, Tanaka K, Murata S. 2007. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol 27: 6629–6638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D. 2006. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: 99–111. [DOI] [PubMed] [Google Scholar]
- Hanna J, Leggett DS, Finley D. 2003. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol 23: 9251–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna J, Meides A, Zhang DP, Finley D. 2007. A ubiquitin stress response induces altered proteasome composition. Cell 129: 747–759. [DOI] [PubMed] [Google Scholar]
- Hanna J, Waterman D, Isasa M, Elsasser S, Shi Y, Gygi S, Finley D. 2014. Cuz1/Ynl155w, a zinc-dependent ubiquitin-binding protein, protects cells from metalloid-induced proteotoxicity. J Biol Chem 289: 1876–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P, Stingele J, Hofmann K, Schindelin H, Raasi S. 2010. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J Biol Chem 285: 20390–20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper JW, Ordureau A, Heo JM. 2018. Building and decoding ubiquitin chains for mitophagy. Nat Rev Mol Cell Biol 19: 93–108. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R, Hendil KB, Gordon C. 2003. Ubiquitin binding proteins protect ubiquitin conjugates from disassembly. FEBS Lett 535: 77–81. [DOI] [PubMed] [Google Scholar]
- Hishiya A, Iemura S, Natsume T, Takayama S, Ikeda K, Watanabe K. 2006. A novel ubiquitin-binding protein ZNF216 functioning in muscle atrophy. EMBO J 25: 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjerpe R, Bett JS, Keuss MJ, Solovyova A, McWilliams TG, Johnson C, Sahu I, Varghese J, Wood N, Wightman M et al. 2016. UBQLN2 Mediates Autophagy-Independent Protein Aggregate Clearance by the Proteasome. Cell 166: 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeller D, Dikic I. 2010. Regulation of ubiquitin receptors by coupled monoubiquitination. Subcell Biochem 54: 31–40. [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM. 2001. In vitro assembly and recognition of Lys-63 polyubiquitin chains. J Biol Chem 276: 27936–27943. [DOI] [PubMed] [Google Scholar]
- Homma T, Ishibashi D, Nakagaki T, Fuse T, Mori T, Satoh K, Atarashi R, Nishida N. 2015. Ubiquitin-specific protease 14 modulates degradation of cellular prion protein. Sci Rep 5 10.1038/srep11028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hough R, Pratt G, Rechsteiner M. 1987. Purification of two high molecular weight proteases from rabbit reticulocyte lysate. J Biol Chem 262: 8303–8313. [PubMed] [Google Scholar]
- Hu M, Li P, Song L, Jeffrey PD, Chenova TA, Wilkinson KD, Cohen RE, Shi Y. 2005. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J 24: 3747–3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Haratake K, Miyahara H, Chiba T. 2016a. Proteasome activators, PA28gamma and PA200, play indispensable roles in male fertility. Sci Rep 6 10.1038/srep23171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Luan B, Wu J, Shi Y. 2016b. An atomic structure of the human 26S proteasome. Nat Struct Mol Biol 23: 778–785. [DOI] [PubMed] [Google Scholar]
- Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. 2008. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature 453: 481–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Zavodszky E, Shao S, Wohlever ML, Keenan RJ, Hegde RS. 2016. Ubiquilins Chaperone and Triage Mitochondrial Membrane Proteins for Degradation. Mol Cell 63: 21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Estep P, Church GM, Samson LD. 2000. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol 20: 8157–8167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HT, Goldberg AL. 2017. The deubiquitinating enzyme Usp14 allosterically inhibits multiple proteasomal activities and ubiquitin-independent proteolysis. J Biol Chem 292: 9830–9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2018. UBL domain of Usp14 and other proteins stimulates proteasome activities and protein degradation in cells. Proc Natl Acad Sci U S A 115: E11642–E11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Mi K, Rao H. 2004. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell 15: 3357–3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K. 2006. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25: 4877–4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick DS, Hathaway NA, Hanna J, Elsasser S, Rush J, Finley D, King RW, Gygi SP. 2006. Quantitative analysis of in vitro ubiquitinated cyclin B1 reveals complex chain topology. Nat Cell Biol 8: 700–710. [DOI] [PubMed] [Google Scholar]
- Kohlmann S, Schafer A, Wolf DH. 2008. Ubiquitin ligase Hul5 is required for fragment-specific substrate degradation in endoplasmic reticulum-associated degradation. J Biol Chem 283: 16374–16383. [DOI] [PubMed] [Google Scholar]
- Krylov DM, Koonin EV. 2001. A novel family of predicted retroviral-like aspartyl proteases with a possible key role in eukaryotic cell cycle control. Curr Biol 11: R584–587. [DOI] [PubMed] [Google Scholar]
- Kuhnle S, Martinez-Noel G, Leclere F, Hayes SD, Harper JW, Howley PM. 2018. Angelman syndrome-associated point mutations in the Zn(2+)-binding N-terminal (AZUL) domain of UBE3A ubiquitin ligase inhibit binding to the proteasome. J Biol Chem 293: 18387–18399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CL, Goldberg AL. 2017. Ubiquitinated proteins promote the association of proteasomes with the deubiquitinating enzyme Usp14 and the ubiquitin ligase Ube3c. Proc Natl Acad Sci U S A 114: E3404–E3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YT, Ciechanover A. 2017. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem Sci 42: 873–886. [DOI] [PubMed] [Google Scholar]
- Laporte D, Salin B, Daignan-Fornier B, Sagot I. 2008. Reversible cytoplasmic localization of the proteasome in quiescent yeast cells. J Cell Biol 181: 737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, Gartner C, Dimova N, Hanna J, Gygi SP et al. 2010. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 467: 179–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Lu Y, Prado MA, Shi Y, Tian G, Sun S, Elsasser S, Gygi SP, King RW, Finley D. 2016. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature 532: 398–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D, Takayama S, Goldberg AL. 2018a. ZFAND5/ZNF216 is an activator of the 26S proteasome that stimulates overall protein degradation. Proc Natl Acad Sci U S A 115: E9550–E9559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SW, Oh YM, Lu YL, Kim WK, Yoo AS. 2018b. MicroRNAs Overcome Cell Fate Barrier by Reducing EZH2-Controlled REST Stability during Neuronal Conversion of Human Adult Fibroblasts. Dev Cell 46: 73–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, Walz T, Ploegh H, Finley D. 2002. Multiple associated proteins regulate proteasome structure and function. Mol Cell 10: 495–507. [DOI] [PubMed] [Google Scholar]
- Lehrbach NJ, Ruvkun G. 2016. Proteasome dysfunction triggers activation of SKN-1A/Nrf1 by the aspartic protease DDI-1. Elife 5 10.7554/eLife.17721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leznicki P, Kulathu Y. 2017. Mechanisms of regulation and diversification of deubiquitylating enzyme function. J Cell Sci 130: 1997–2006. [DOI] [PubMed] [Google Scholar]
- Li H, Zhao Z, Ling J, Pan L, Zhao X, Zhu H, Yu J, Xie B, Shen J, Chen W. 2018. USP14 promotes K63-linked RIG-I deubiquitination and suppresses antiviral immune responses. Eur J Immunol. 10.1002/eji.201847603. [DOI] [PubMed] [Google Scholar]
- Liao Y, Liu N, Hua X, Cai J, Xia X, Wang X, Huang H, Liu J. 2017. Proteasome-associated deubiquitinase ubiquitin-specific protease 14 regulates prostate cancer proliferation by deubiquitinating and stabilizing androgen receptor. Cell Death Dis 8 10.1038/cddis.2016.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Xia X, Liu N, Cai J, Guo Z, Li Y, Jiang L, Dou QP, Tang D, Huang H et al. 2018. Growth arrest and apoptosis induction in androgen receptor-positive human breast cancer cells by inhibition of USP14-mediated androgen receptor deubiquitination. Oncogene 37: 1896–1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebelt F, Vertegaal AC. 2016. Ubiquitin-dependent and independent roles of SUMO in proteostasis. Am J Physiol Cell Physiol 311: C284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Jiang S, Li M, Xiong X, Zhu M, Li D, Zhao L, Qian L, Zhai L, Li J et al. 2018. Proteome-wide analysis of USP14 substrates revealed its role in hepatosteatosis via stabilization of FASN. Nat Commun 9 10.1038/s41467-018-07185-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokireddy S, Kukushkin NV, Goldberg AL. 2015. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A 112: E7176–7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe J, Stock D, Jap B, Zwickl P, Baumeister W, Huber R. 1995. Crystal structure of the 20S proteasome from the archaeon T. acidophilum at 3.4 A resolution. Science 268: 533–539. [DOI] [PubMed] [Google Scholar]
- Lu K, Psakhye I, Jentsch S. 2014. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 158: 549–563. [DOI] [PubMed] [Google Scholar]
- Lu Y, Lee BH, King RW, Finley D, Kirschner MW. 2015. Substrate degradation by the proteasome: a single-molecule kinetic analysis. Science 348 10.1126/science.1250834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder P, Rudack T, Beck F, Danev R, Pfeifer G, Nagy I, Baumeister W. 2018. Cryo-EM structures of the archaeal PAN-proteasome reveal an around-the-ring ATPase cycle. Proc Natl Acad Sci U S A. 10.1073/pnas.1817752116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, Li F, Gemperline DC, Book AJ, Vierstra RD. 2015. Autophagic Degradation of the 26S Proteasome Is Mediated by the Dual ATG8/Ubiquitin Receptor RPN10 in Arabidopsis. Mol Cell 58: 1053–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall RS, McLoughlin F, Vierstra RD. 2016. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Rep 16: 1717–1732. [DOI] [PubMed] [Google Scholar]
- Marshall RS, Vierstra RD. 2018. Proteasome storage granules protect proteasomes from autophagic degradation upon carbon starvation. Elife 7 10.7554/eLife.34532. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Martinez-Noel G, Galligan JT, Sowa ME, Arndt V, Overton TM, Harper JW, Howley PM. 2012. Identification and proteomic analysis of distinct UBE3A/E6AP protein complexes. Mol Cell Biol 32: 3095–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa F, Tammaro R, Prado MA, Cesana M, Lee BH, Finley D, Franco B, Morleo M. 2018. The deubiquitinating enzyme USP14 controls ciliogenesis and hedgehog signalling. Hum Mol Genet. 10.1093/hmg/ddy380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matyskiela ME, Lander GC, Martin A. 2013. Conformational switching of the 26S proteasome enables substrate degradation. Nat Struct Mol Biol 20: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon C, Goold R, Andre R, Devoy A, Ortega Z, Moonga J, Linehan JM, Brandner S, Lucas JJ, Collinge J et al. 2016. Prion-mediated neurodegeneration is associated with early impairment of the ubiquitin-proteasome system. Acta Neuropathol 131: 411–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HJ, Rape M. 2014. Enhanced protein degradation by branched ubiquitin chains. Cell 157: 910–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min JW, Lu L, Freeling JL, Martin DS, Wang H. 2017a. USP14 inhibitor attenuates cerebral ischemia/reperfusion-induced neuronal injury in mice. J Neurochem 140: 826–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min Y, Lee S, Kim MJ, Chun E, Lee KY. 2017b. Ubiquitin-Specific Protease 14 Negatively Regulates Toll-Like Receptor 4-Mediated Signaling and Autophagy Induction by Inhibiting Ubiquitination of TAK1-Binding Protein 2 and Beclin 1. Front Immunol 8 10.3389/fimmu.2017.01827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata S, Takahama Y, Kasahara M, Tanaka K. 2018. The immunoproteasome and thymoproteasome: functions, evolution and human disease. Nat Immunol 19: 923–931. [DOI] [PubMed] [Google Scholar]
- Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, Duff KE. 2016. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med 22: 46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima A, Ohnuma S, Kodani Y, Kaneko YS, Nagasaki H, Nagatsu T, Ota A. 2016. Inhibition of deubiquitinating activity of USP14 decreases tyrosine hydroxylase phosphorylated at Ser19 in PC12D cells. Biochem Biophys Res Commun 472: 598–602. [DOI] [PubMed] [Google Scholar]
- Nathan JA, Kim HT, Ting L, Gygi SP, Goldberg AL. 2013. Why do cellular proteins linked to K63-polyubiquitin chains not associate with proteasomes? EMBO J 32: 552–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Prado MA, Schmidt PJ, Sendamarai AK, Wilson-Grady JT, Min M, Campagna DR, Tian G, Shi Y, Dederer V et al. 2017. UBE2O remodels the proteome during terminal erythroid differentiation. Science 357 10.1126/science.aan0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Kim W, Tian G, Gygi SP, Finley D. 2011. Structural defects in the regulatory particle-core particle interface of the proteasome induce a novel proteasome stress response. J Biol Chem 286: 36652–36666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Li X, Kim HM, Singh CR, Tian G, Hoyt MA, Lovell S, Battaile KP, Zolkiewski M, Coffino P et al. 2013. Reconfiguration of the proteasome during chaperone-mediated assembly. Nature 497: 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper RC, Dikic I, Lukacs GL. 2014. Ubiquitin-dependent sorting in endocytosis. Cold Spring Harb Perspect Biol 6 10.1101/cshperspect.a016808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. 2004. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol 11: 830–837. [DOI] [PubMed] [Google Scholar]
- Puchades C, Rampello AJ, Shin M, Giuliano CJ, Wiseman RL, Glynn SE, Lander GC. 2017. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science 358 10.1126/science.aao0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian MX, Pang Y, Liu CH, Haratake K, Du BY, Ji DY, Wang GF, Zhu QQ, Song W, Yu Y et al. 2013. Acetylation-mediated proteasomal degradation of core histones during DNA repair and spermatogenesis. Cell 153: 1012–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. 2010. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell 38: 17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahighi S, Braunstein I, Ternette N, Kessler B, Kawasaki M, Kato R, Matsui T, Weiss TM, Stanhill A, Wakatsuki S. 2016. Selective Binding of AIRAPL Tandem UIMs to Lys48-Linked Tri-Ubiquitin Chains. Structure 24: 412–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly H, Rape M, Braun S, Rumpf S, Hoege C, Jentsch S. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120: 73–84. [DOI] [PubMed] [Google Scholar]
- Rousseau A, Bertolotti A. 2016. An evolutionarily conserved pathway controls proteasome homeostasis. Nature 536: 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2018. Regulation of proteasome assembly and activity in health and disease. Nat Rev Mol Cell Biol 19: 697–712. [DOI] [PubMed] [Google Scholar]
- Sa-Moura B, Funakoshi M, Tomko RJ Jr., Dohmen RJ, Wu Z, Peng J, Hochstrasser M. 2013. A conserved protein with AN1 zinc finger and ubiquitin-like domains modulates Cdc48 (p97) function in the ubiquitin-proteasome pathway. J Biol Chem 288: 33682–33696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadre-Bazzaz K, Whitby FG, Robinson H, Formosa T, Hill CP. 2010. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol Cell 37: 728–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y. 2017. Ubiquitin recognition by the proteasome. J Biochem 161: 113–124. [DOI] [PubMed] [Google Scholar]
- Sahtoe DD, van Dijk WJ, El Oualid F, Ekkebus R, Ovaa H, Sixma TK. 2015. Mechanism of UCH-L5 activation and inhibition by DEUBAD domains in RPN13 and INO80G. Mol Cell 57: 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samant RS, Livingston CM, Sontag EM, Frydman J. 2018. Distinct proteostasis circuits cooperate in nuclear and cytoplasmic protein quality control. Nature 563: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sareen-Khanna K, Papillon J, Wing SS, Cybulsky AV. 2016. Role of the deubiquitinating enzyme ubiquitin-specific protease-14 in proteostasis in renal cells. Am J Physiol Renal Physiol 311: F1035–F1046. [DOI] [PubMed] [Google Scholar]
- Schoebel S, Mi W, Stein A, Ovchinnikov S, Pavlovicz R, DiMaio F, Baker D, Chambers MG, Su H, Li D et al. 2017. Cryo-EM structure of the protein-conducting ERAD channel Hrd1 in complex with Hrd3. Nature 548: 352–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, Finley D, Dikic I, Walters KJ, Groll M. 2008. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature 453: 548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Chen X, Elsasser S, Stocks BB, Tian G, Lee BH, Shi Y, Zhang N, de Poot SA, Tuebing F et al. 2016. Rpn1 provides adjacent receptor sites for substrate binding and deubiquitination by the proteasome. Science 351 10.1126/science.aad9421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirkis R, Gerst JE, Fass D. 2006. Ddi1, a eukaryotic protein with the retroviral protease fold. J Mol Biol 364: 376–387. [DOI] [PubMed] [Google Scholar]
- Smith DM, Chang SC, Park S, Finley D, Cheng Y, Goldberg AL. 2007. Docking of the proteasomal ATPases’ carboxyl termini in the 20S proteasome’s alpha ring opens the gate for substrate entry. Mol Cell 27: 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolova V, Li F, Polovin G, Park S. 2015. Proteasome Activation is Mediated via a Functional Switch of the Rpt6 C-terminal Tail Following Chaperone-dependent Assembly. Sci Rep 5 10.1038/srep14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Ma R, Yang X, Pang S. 2017. The Deubiquitinating Enzyme USP14 Regulates Leukemic Chemotherapy Drugs-Induced Cell Apoptosis by Suppressing Ubiquitination of Aurora Kinase B. Cell Physiol Biochem 42: 965–973. [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15: 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller BM, Hill CP. 2011. Proteasome activators. Mol Cell 41: 8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanhill A, Haynes CM, Zhang Y, Min G, Steele MC, Kalinina J, Martinez E, Pickart CM, Kong XP, Ron D. 2006. An arsenite-inducible 19S regulatory particle-associated protein adapts proteasomes to proteotoxicity. Mol Cell 23: 875–885. [DOI] [PubMed] [Google Scholar]
- Steffen J, Seeger M, Koch A, Kruger E. 2010. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell 40: 147–158. [DOI] [PubMed] [Google Scholar]
- Sun Y, Qin Z, Li Q, Wan JJ, Cheng MH, Wang PY, Su DF, Yu JG, Liu X. 2016. MicroRNA-124 negatively regulates LPS-induced TNF-alpha production in mouse macrophages by decreasing protein stability. Acta Pharmacol Sin 37: 889–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suraweera A, Munch C, Hanssum A, Bertolotti A. 2012. Failure of amino acid homeostasis causes cell death following proteasome inhibition. Mol Cell 48: 242–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, Kawahara H. 2016. UBQLN4 recognizes mislocalized transmembrane domain proteins and targets these to proteasomal degradation. EMBO Rep 17: 842–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mizushima T, Saeki Y. 2012. The proteasome: molecular machinery and pathophysiological roles. Biol Chem 393: 217–234. [DOI] [PubMed] [Google Scholar]
- Tonoki A, Kuranaga E, Tomioka T, Hamazaki J, Murata S, Tanaka K, Miura M. 2009. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol Cell Biol 29: 1095–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Burana D, Ohtake F, Arai N, Kaiho A, Komada M, Tanaka K, Saeki Y. 2018. Ub-ProT reveals global length and composition of protein ubiquitylation in cells. Nat Commun 9 10.1038/s41467-018-02869-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya H, Ohtake F, Arai N, Kaiho A, Yasuda S, Tanaka K, Saeki Y. 2017. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol Cell 66: 488–502. [DOI] [PubMed] [Google Scholar]
- Turakhiya A, Meyer SR, Marincola G, Bohm S, Vanselow JT, Schlosser A, Hofmann K, Buchberger A. 2018. ZFAND1 Recruits p97 and the 26S Proteasome to Promote the Clearance of Arsenite-Induced Stress Granules. Mol Cell 70: 906–919. [DOI] [PubMed] [Google Scholar]
- van den Boom J, Meyer H. 2018. VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 69: 182–194. [DOI] [PubMed] [Google Scholar]
- VanDemark AP, Hofmann RM, Tsui C, Pickart CM, Wolberger C. 2001. Molecular insights into polyubiquitin chain assembly: crystal structure of the Mms2/Ubc13 heterodimer. Cell 105: 711–720. [DOI] [PubMed] [Google Scholar]
- Vander Linden RT, Hemmis CW, Schmitt B, Ndoja A, Whitby FG, Robinson H, Cohen RE, Yao T, Hill CP. 2015. Structural basis for the activation and inhibition of the UCH37 deubiquitylase. Mol Cell 57: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Aravind L, Oania R, McDonald WH, Yates JR 3rd, Koonin EV, Deshaies RJ. 2002. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science 298: 611–615. [DOI] [PubMed] [Google Scholar]
- Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, Deshaies RJ. 2000. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell 11: 3425–3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank JJS, Goldberg AL. 2018. Exploring the Regulation of Proteasome Function by Subunit Phosphorylation. Methods Mol Biol 1844: 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VerPlank JJS, Lokireddy S, Feltri ML, Goldberg AL, Wrabetz L. 2018. Impairment of protein degradation and proteasome function in hereditary neuropathies. Glia 66: 379–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Boyer L, Morantte I, Lutz M, Merkwirth C, Joyce D, Spencer B, Page L, Masliah E, Berggren WT et al. 2012a. Increased proteasome activity in human embryonic stem cells is regulated by PSMD11. Nature 489: 304–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilchez D, Morantte I, Liu Z, Douglas PM, Merkwirth C, Rodrigues AP, Manning G, Dillin A. 2012b. RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489: 263–268. [DOI] [PubMed] [Google Scholar]
- Vilchez D, Saez I, Dillin A. 2014. The role of protein clearance mechanisms in organismal ageing and age-related diseases. Nat Commun 5 10.1038/ncomms6659. [DOI] [PubMed] [Google Scholar]
- Villa E, Marchetti S, Ricci JE. 2018. No Parkin Zone: Mitophagy without Parkin. Trends Cell Biol 28: 882–895. [DOI] [PubMed] [Google Scholar]
- Waite KA, De-La Mota-Peynado A, Vontz G, Roelofs J. 2016. Starvation Induces Proteasome Autophagy with Different Pathways for Core and Regulatory Particles. J Biol Chem 291: 3239–3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Mao X, Ju D, Xie Y. 2004. Rpn4 is a physiological substrate of the Ubr2 ubiquitin ligase. J Biol Chem 279: 55218–55223. [DOI] [PubMed] [Google Scholar]
- Wang X, Xu H, Ju D, Xie Y. 2008. Disruption of Rpn4-induced proteasome expression in Saccharomyces cerevisiae reduces cell viability under stressed conditions. Genetics 180: 1945–1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang Y, Ding S, Li J, Song N, Ren Y, Hong D, Wu C, Li B, Wang F et al. 2018. Small molecule inhibitors reveal allosteric regulation of USP14 via steric blockade. Cell Res 28: 1186–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman L, Fagan JM, Goldberg AL. 1987. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J Biol Chem 262: 2451–2457. [PubMed] [Google Scholar]
- Wehmer M, Rudack T, Beck F, Aufderheide A, Pfeifer G, Plitzko JM, Forster F, Schulten K, Baumeister W, Sakata E. 2017. Structural insights into the functional cycle of the ATPase module of the 26S proteasome. Proc Natl Acad Sci U S A 114: 1305–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Dong S, Bowser RK, Khoo A, Zhang L, Jacko AM, Zhao Y, Zhao J. 2017. Regulation of the ubiquitylation and deubiquitylation of CREB-binding protein modulates histone acetylation and lung inflammation. Sci Signal 10 10.1126/scisignal.aak9660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I, Dixit V. 2014. A20--a bipartite ubiquitin editing enzyme with immunoregulatory potential. Adv Exp Med Biol 809: 1–12. [DOI] [PubMed] [Google Scholar]
- Whitby FG, Masters EI, Kramer L, Knowlton JR, Yao Y, Wang CC, Hill CP. 2000. Structural basis for the activation of 20S proteasomes by 11S regulators. Nature 408: 115–120. [DOI] [PubMed] [Google Scholar]
- Whiteley AM, Prado MA, Peng I, Abbas AR, Haley B, Paulo JA, Reichelt M, Katakam A, Sagolla M, Modrusan Z et al. 2017. Ubiquilin1 promotes antigen-receptor mediated proliferation by eliminating mislocalized mitochondrial proteins. Elife 6 10.7554/eLife.26435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden EJ, Dong KC, Martin A. 2017. An AAA Motor-Driven Mechanical Switch in Rpn11 Controls Deubiquitination at the 26S Proteasome. Mol Cell 67: 799–811. [DOI] [PubMed] [Google Scholar]
- Wu N, Zhang C, Bai C, Han YP, Li Q. 2014. MiR-4782–3p inhibited non-small cell lung cancer growth via USP14. Cell Physiol Biochem 33: 457–467. [DOI] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. 2000. Physical association of ubiquitin ligases and the 26S proteasome. Proc Natl Acad Sci U S A 97: 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2001. RPN4 is a ligand, substrate, and transcriptional regulator of the 26S proteasome: a negative feedback circuit. Proc Natl Acad Sci U S A 98: 3056–3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- -. 2002. UFD4 lacking the proteasome-binding region catalyses ubiquitination but is impaired in proteolysis. Nat Cell Biol 4: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Xu D, Shan B, Lee BH, Zhu K, Zhang T, Sun H, Liu M, Shi L, Liang W, Qian L et al. 2015. Phosphorylation and activation of ubiquitin-specific protease-14 by Akt regulates the ubiquitin-proteasome system. Elife 4 10.7554/eLife.10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Shan B, Sun H, Xiao J, Zhu K, Xie X, Li X, Liang W, Lu X, Qian L et al. 2016. USP14 regulates autophagy by suppressing K63 ubiquitination of Beclin 1. Genes Dev 30: 1718–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, Rush J, Hochstrasser M, Finley D, Peng J. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell 137: 133–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Juszkiewicz S, Hegde RS. 2017. UBE2O is a quality control factor for orphans of multiprotein complexes. Science 357: 472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao T, Cohen RE. 2002. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature 419: 403–407. [DOI] [PubMed] [Google Scholar]
- Yau RG, Doerner K, Castellanos ER, Haakonsen DL, Werner A, Wang N, Yang XW, Martinez-Martin N, Matsumoto ML, Dixit VM et al. 2017. Assembly and Function of Heterotypic Ubiquitin Chains in Cell-Cycle and Protein Quality Control. Cell 171: 918–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Matouschek A. 2017. Recognition of Client Proteins by the Proteasome. Annu Rev Biophys 46: 149–173. [DOI] [PubMed] [Google Scholar]
- Yu H, Singh Gautam AK, Wilmington SR, Wylie D, Martinez-Fonts K, Kago G, Warburton M, Chavali S, Inobe T, Finkelstein IJ et al. 2016. Conserved Sequence Preferences Contribute to Substrate Recognition by the Proteasome. J Biol Chem 291: 14526–14539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun C, Stanhill A, Yang Y, Zhang Y, Haynes CM, Xu CF, Neubert TA, Mor A, Philips MR, Ron D. 2008. Proteasomal adaptation to environmental stress links resistance to proteotoxicity with longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 105: 7094–7099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. 2014. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513: 440–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Goldberg AL. 2016. Coordinate regulation of autophagy and the ubiquitin proteasome system by MTOR. Autophagy 12: 1967–1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhai B, Gygi SP, Goldberg AL. 2015. mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc Natl Acad Sci U S A 112: 15790–15797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Zhang Y, Sui Z, Zhang Y, Liu M, Tang H. 2017. USP14 de-ubiquitinates vimentin and miR-320a modulates USP14 and vimentin to contribute to malignancy in gastric cancer cells. Oncotarget 8: 48725–48736. [DOI] [PMC free article] [PubMed] [Google Scholar]