Abstract
This study aimed to identify biomarkers of major depressive disorder (MDD), by relating neuroimage-derived measures to binary (MDD/control), ordinal (severe MDD/mild MDD/control), or continuous (depression severity) outcomes. To address MDD heterogeneity, factors (severity of psychic depression, motivation, anxiety, psychosis and sleep disturbance) were also used as outcomes. A multi-site, multimodal imaging (diffusion MRI, dMRI, and structural MRI, sMRI) cohort (52 controls and 147 MDD patients) and several modeling techniques- penalized logistic regression (PLR), random forest (RF) and support vector machine (SVM)- were used. An additional cohort (25 controls and 83 MDD patients) was used for validation. The optimally performing classifier (SVM) had a 26.0% misclassification rate (binary), 52.2±1.69% accuracy (ordinal) and r =0.36 correlation coefficient (p-value<0.001, continuous). Using SVM, R2 values for prediction of any MDD factors were <10%. Binary classification in the external dataset resulted in 87.95% sensitivity and 32.00% specificity. Though observed classification rates are too low for clinical utility, four image-based features contributed to accuracy across all models and analyses- two dMRI-based measures (average fractional anisotropy in the right cuneus and left insula) and two sMRI-based measures (asymmetry in the volume of the pars triangularis and the cerebellum) and may serve as a priori regions for future analyses. The poor accuracy of classification and predictive results found here reflects current equivocal findings and sheds light on challenges of using these modalities for MDD biomarker identification. Further, this study suggests a paradigm (e.g. multiple classifier evaluation with external validation) for future studies to avoid non-generalizable results.
Keywords: Major Depressive Disorder (MDD), Magnetic Resonance Imaging (MRI), diffusion MRI, structural MRI, Support Vector Machine (SVM)
Introduction
Major depressive disorder (MDD) is a common and debilitating disease. Characterized by recurrent feelings of sadness, hopelessness and inability to feel pleasure, 16.6% of the US population (Kessler, Berglund, et al., 2005) and 350 million people worldwide (Kessler, Chiu, Demler, Merikangas, & Walters, 2005; World Health Organization, 2012) suffer from MDD, up to 15% of whom will eventually die by suicide (Palucha & Pilc, 2007). Further, MDD is a growing problem. Originally predicted by World Health Organization (WHO) to be the second leading cause of disability worldwide by 2020 (Murray & Lopez, 1996), MDD fulfilled this prediction in 2013 (Global Burden of Disease Study 2013 Collaborators, 2015).
Due to the worldwide impact of MDD, it is important to gain a greater understanding of the illness. Despite decades of inquiry, however, there are currently no objective MDD biomarkers (Mossner et al., 2007). A biomarker is a characteristic that can be objectively measured and used as an indicator of either normal or pathogenic processes (Singh & Rose, 2009). As pointed out by Peterson et al, a biomarker for MDD could aid in diagnosis, the search for genetic and environmental causes, predicting course, identifying those at increased risk and developing the next generation of treatments (Peterson & Weissman, 2011). As such, a biomarker could help reduce the morbidity and mortality of MDD, as it has in other areas of medicine (e.g. breast cancer, macular degeneration and myocardial infarction) (Gonzalez de Castro, Clarke, Al-Lazikani, & Workman, 2013; Mihaly et al., 2013; Newman et al., 2012; Ziegler, Koch, Krockenberger, & Grosshennig, 2012). Neuroimaging techniques, such as structural and diffusion-weighted magnetic resonance imaging (MRI) may be able to provide such a biomarker for MDD, and numerous studies have evaluated this possibility (Aizenstein, Khalaf, Walker, & Andreescu, 2014; M. L. Phillips, 2012).
From structural MRI, both regional volumes and cortical thickness (i.e., the distance between the gray matter/white matter surface and the pial surface) can be estimated. When comparing depressed subjects to healthy volunteers, some studies report widespread volumetric differences in cortical gray matter regions (Grieve, Korgaonkar, Koslow, Gordon, & Williams, 2013; Guo et al., 2014; Takahashi et al., 2010; van Tol et al., 2010) such as smaller gyri of the caudal middle frontal and medial orbitofrontal cortices (Han et al., 2014; Qiu, Huang, et al., 2014), and smaller volume in subcortical regions, such as the amygdala and hippocampus (Amico et al., 2011; Eker & Gonul, 2010; Huang et al., 2013; Jaworska, MacMaster, Yang, et al., 2014; Kupfer, Frank, & Phillips, 2012; Whittle et al., 2014) in MDD patients. Smaller volumes of the hippocampus, basal ganglia, orbitofrontal cortex and prefrontal cortex are also frequently observed in MDD patients (Lorenzetti, Allen, Fornito, & Yucel, 2009). However, findings remain highly variable in terms of which brain regions show abnormalities and the degree to which they are affected across studies (Han et al., 2014; Shizukuishi, Abe, & Aoki, 2013). Similarly, decreased (Mackin et al., 2013; Peterson et al., 2009; Tu et al., 2012), increased (Qiu, Lui, et al., 2014; Reynolds et al., 2014) or bidirectional (Fallucca et al., 2011; Peterson et al., 2009; Tu et al., 2012) differences in cortical thickness have been reported in MDD. Regions found to have cortical thinning in the largest study to date (~1,900 adult MDD subjects), such as the medial orbitofrontal cortex (although with effect sizes likely too small for clinical meaning)(Schmaal et al., 2016), have been previously reported to be thicker (Qiu, Lui, et al., 2014) or the same (Perlman et al., 2017) in other studies of depressed individuals.
Diffusion MRI (dMRI) is used to evaluate orientation and diffusion characteristics of white matter and, by inference, white matter microstructure (Murphy & Frodl, 2011). Fractional anisotropy (FA), is a common measure used in dMRI to determine integrity of white matter fibers by estimating the direction of movement of water molecules (Liao et al., 2013; Murphy & Frodl, 2011). Characteristics of healthy white matter include parallel organization of white matter fibers and myelination, which leads to restricted movement of water lateral to the direction of fiber tracts and more movement along the tract, generally resulting in higher estimates of FA. FA values range from zero (isotropic diffusion) to one (anisotropic diffusion) (Delorenzo et al., 2013).
dMRI and FA measures have been used to study white matter microstructure abnormalities in mood disorders (Henderson et al., 2013; Korgaonkar et al., 2011; Olvet et al., 2014; Peng et al., 2013). A 2009 meta-analysis of dMRI studies reported that, in 21 of the 27 studies examined, subjects with mood disorders had lower FA in frontal and temporal lobes (Sexton, Mackay, & Ebmeier, 2009). Another meta-analysis documented similar findings, in which patients with MDD showed reduced FA values in the white matter of bilateral frontal and right occipital areas (Liao et al., 2013). Similar to the volumetric/thickness analyses, however, although a trend of reduced FA in MDD has been noted in literature (Murphy & Frodl, 2011; Shizukuishi et al., 2013), not all studies detect these differences. Increased and decreased FA values in the corpus callosum, parietal and frontal lobes (Aghajani et al., 2014; Osoba et al., 2013) or no significant differences between groups (Abe et al., 2010; Kieseppa et al., 2010; Olvet et al., 2016; Ugwu, Amico, Carballedo, Fagan, & Frodl, 2015) have been reported.
Beyond the first level analyses of volume, cortical thickness or FA differences in MDD, of even greater uncertainty are the laterality effects of depression which are still not well characterized (Amico et al., 2011; Jaworska, MacMaster, Yang, et al., 2014) as some studies have found more robust structural deficits in the right compared to the left cerebral hemispheres (Mackin et al., 2013; Peterson et al., 2009; Qiu, Huang, et al., 2014) and vice versa (Bijanki, Hodis, Brumm, Harlynn, & McCormick, 2014; Treadway et al., 2015) related to MDD severity (Jaworska, MacMaster, Gaxiola, et al., 2014; Jaworska, MacMaster, Yang, et al., 2014).
Disagreement in MDD-related neurobiological findings within any one modality potentially reflects the variability in depression itself (Joober, 2013) and suggests that multiple modalities of imaging and clinical assessment may be required to uncover disease biology (M. L. Phillips, 2012). Multimodal imaging potentially reveals crucial variations that could only be partially visible in a single modality and therefore could potentially unify conflicting findings (Sui, Huster, Yu, Segall, & Calhoun, 2013). Further, multimodal features used to achieve the most accurate classification (between depressed and control subjects) or prediction of outcome (such as depression severity) can provide biological insight into the differences between diagnostic groups. Therefore, in the present study, we used both structural and diffusion MRI to classify depressed subjects versus controls on an individual level and to predict other outcomes such as overall depression severity or severity of specific depression symptoms/factors.
The challenge in such studies is in analyzing the large volume of data, as each modality can produce hundreds (regional) to hundreds of thousands (voxel) of variables, yet the number of subjects is often limited. To handle these challenges, machine-learning techniques have been applied. A recent systematic review highlighted 19 MRI-based studies of classification in MDD (Arbabshirani, Plis, Sui, & Calhoun, 2016). Though classification accuracies of the 19 studies ranged from 54.6% (Serpa et al., 2014) to 90.3% (Mwangi, Ebmeier, Matthews, & Steele, 2012), none of the selected discriminating features have been replicated or translated into clinical practice. There may be a few reasons for this. One is due to relatively small sample sizes. Only two studies included 40 or more depressed subjects, the maximum number of depressed subjects was 57 and 12 studies included 30 MDD subjects or fewer (Arbabshirani et al., 2016). This is a significant issue, as accuracy decreases with decreasing sample size, and is considered the most critical factor (Arbabshirani et al., 2016). Another issue is feature selection bias. This occurs when the features with the highest discrimination were both extracted from, and used for, classification within the same dataset. This leads to overly inflated accuracy estimates (Arbabshirani et al., 2016). Furthermore, “overfitting” is more likely to occur with complex models, particularly if the process of both training and testing is repeated multiple times, with varying model parameters (Arbabshirani et al., 2016). Cross-validation can compensate for this by providing relatively robust estimate of prediction performance. However, most studies used only leave-one-out cross validation, which may not always lead to consistent model estimates (Shao, 1993). Replication in a separate subject sample is a more robust way of ruling out effects of overfitting.
In addition to the above concerns, only one MDD study from this meta-analysis combined data from multiple modalities (task-based functional MRI [fMRI], resting state fMRI and diffusion MRI). That study involved participants with late life depression (LLD) compared to elderly controls and predicted LLD diagnosis and treatment response with accuracies of 87.27% and 89.47% respectively, suggesting the benefits of multimodal imaging (Patel et al., 2015). Though they did not assess classification accuracy, three additional studies have incorporated multimodal brain imaging techniques to explore depression pathophysiology, focusing on uncovering group-level differences (K. Choi, Craddock, R.C., Holtzheimer, P.E., Yang, Z., Hu, X., Mayberg, H., 2008; Matthews et al., 2011; Sexton et al., 2012). Our study is therefore unique in that it involves the combination of two MR imaging modalities, a focus on MDD (diagnosis and factors) classification and one of the largest cohorts (n = 307) reported to date. Critically, our study uses separate cohorts for training and validation, in which only a single set of parameters (identified as optimal from the training) was applied to the validation set. The advantage of this approach is that it avoids the potential bias of within-sample cross-validation. As the purpose of the classification is to identify components of the structural and diffusion MRI that may serve as biomarkers of MDD, we evaluated two potential classification schemes: (1) MDD vs controls and (2) severe MDD vs mild MDD vs controls. We also examined the ability of structural and diffusion MRI to predict depression severity (continuous measure). Finally, to reduce the heterogeneity within groups, we examined the ability of structural and diffusion MRI to predict the severity of factors of depression derived from a factor analysis of the 24-item Hamilton Depression Scale, which are continuous measures. The analysis predicting factors was performed because clinical management may require deconstructing MDD into multiple dimensions, or symptom clusters (Hamilton, 1960). Individual factors comprise different combinations of partially orthogonal symptoms. These factors may have different risk factors (Fried, Nesse, Zivin, Guille, & Sen, 2014) and may associate with different neurobiological anomalies on structural and diffusion MRI. Therefore, we examined whether we could obtain higher sensitivity for relating neurobiology to components of clinical presentation versus the entire syndrome.
By identifying structural and diffusion MRI-based measures that contribute the most to each classification/predictive model, this study thus aims to bridge the gap between neuroscience and behavior, in order to enhance current understanding of the pathophysiological mechanisms of major depression.
Materials and Methods
Subjects
All participating individuals provided informed consent for the study, following explanation of the experimental procedures of the study. This study was approved by the Institutional Review Board (IRB) of each institution. The study was performed in compliance with the Code of Ethics of the World Medical Association (Declaration of Helsinki) and the standards established by each institution’s IRB and each investigator’s granting agency.
Data for 217 participants (training set: 25 Healthy Controls, 114 MDD; validation set: 12 Healthy Controls, 66 MDD) in this analysis were acquired from the Establishing Moderators and Biosignatures of Antidepressant Response for Clinical Care (EMBARC) study (U01 MH092250, http://embarc.utsouthwestern.edu/). Details on the EMBARC study design and randomization are reported by Trivedi et al (Trivedi et al., 2016). EMBARC dMRI and sMRI samples have been used in previous publications including: dMRI- (Olvet et al., 2016) or sMRI- (Perlman et al., 2017) only examinations of MDD versus controls and a dMRI-only study of anxious depression versus non-anxious depressed groups (Delaparte et al., 2017). These single modality studies showed no group differences, motivating the interest in the present multimodal examination.
To ensure that the MDD sample was representative and as large as possible, data for an additional 90 participants (training set: 27 Healthy Controls, 33 MDD; validation set: 13 Healthy Controls, 17 MDD) were drawn from ten neuroimaging and depression-related studies conducted at the New York State Psychiatric Institute/Columbia University Medical Center, from 10/2007 through 10/2011. The dMRI data from 20 of these subjects was previously reported in an analysis of suicide attempters (Olvet et al., 2014) and impaired attention in MDD (Rizk et al., 2017).
To maximize generalizability, all cohorts were represented in both the training and validation sets.
Across all eleven protocols, subjects were between the ages of 18 and 65 years old and had the capacity to provide informed consent. MDD common inclusion criteria were DSM-IV MDD diagnosis, determined via the Structured Clinical Interview for the DSM (SCID), and in a current depressive episode. Common exclusion criteria for all subjects were current pregnancy, lifetime history of psychosis or bipolar disorder, meeting DSM-IV criteria for substance dependence in the past 6 months or substance abuse in the past 2 months, unstable psychiatric or general medical conditions that may require hospitalization or contraindicate study medication, clinically significant laboratory abnormalities, history of epilepsy or condition requiring an anticonvulsant, protocol excluded medications (including but not limited to antipsychotics, and mood stabilizers), or significant risk of suicide. Common exclusion criteria for controls also included any other Axis I disorders. All subjects were free of antidepressant medication for at least 21 days at the time of scanning.
All image analyses were performed by a single image analysis lab within a standardized processing pipeline. All technicians were blinded to subject diagnoses.
Image Acquisition: Structural MRI (sMRI)
Details on the EMBARC study’s scanning and processing protocols are reported by Iscan et al (Iscan et al., 2015). In brief, T1 anatomical images were acquired with 3T scanners across 5 sites: University of Texas Southwestern Medical Center (TX: Philips Achieva, 8-channel head coil), University of Michigan (UM: Phillips Ingenia, 15-channel), Massachusetts General Hospital (MGH: Siemens TrioTim, 12-channel), Columbia University Medical Center (CU: GE Signa HDx, 8-channel & GE Discovery MR750, 8-channel), and Stony Brook University Medical Center (SBU: Siemens TrioTim, 12-channel). MPRAGE sequences were used for T1 acquisition at TX, UM, MGH, and SBU, while an IR-FSPGR sequence was used at CU. The following MR sequence parameters were maintained across the 4 sites: TR: 5.9–8.2ms, TE: 2.4–4.6ms, Flip Angle: 8–12°, Acquisition Matrix: 256×256 or 256×243, Acceleration Factor: 2, Sagittal Slices: 174–78, and Voxel Dimensions: 1mm3 isotropic. Structural MRIs from the other protocols were all acquired on a 3T GE Signa HDx scanner, using comparable acquisition parameters.
Image Processing: Structural MRI (sMRI)
Region-wise cortical thickness was computed on a Linux-based computing cluster for 68 Desikan-Killiany (DK) atlas regions (Desikan et al., 2006) with FreeSurfer 5.3’s cortical reconstruction pipeline (http://surfer.nmr.mgh.harvard.edu/). The pipeline’s subroutines have been described in previous publications, but in brief, the processing steps include skull-stripping (Segonne et al., 2004), Talairach transformation, subcortical grey/white matter segmentation (Fischl et al., 2002), intensity normalization (Sled, Zijdenbos, & Evans, 1998), grey/white matter tessellation, topology correction (Fischl, Liu, & Dale, 2001; Segonne, Pacheco, & Fischl, 2007) and intensity gradient based surface deformation to generate grey/white and grey/cerebrospinal fluid surface models (Dale, Fischl, & Sereno, 1999; Fischl et al., 2001; Segonne et al., 2007). The resulting surface models were then inflated and registered to a spherical surface atlas, allowing parcellation of cortical regions of interest and estimation of regional volumes (Fischl, Sereno, & Dale, 1999; Fischl, Sereno, Tootell, & Dale, 1999; Fischl et al., 2004). Finally, regional cortical thicknesses were computed by taking the mean of the white-pial distance at all vertices within each parcellated region (Fischl & Dale, 2000). The surface models (used to calculate cortical thickness) then underwent an empirical, systematic inspection process (see (Iscan et al., 2015) for details). In short, a trained technician carefully inspected 2D sections of the pial and white surface models, overlaid on the T1w image, for fidelity to visible tissue class boundaries. Cases where inaccurate tissue delineation persisted for ≥6 consecutive coronal and axial slices were deemed inaccurate and thus disqualified from further analyses.
Image Acquisition: Diffusion Weighted MRI (dMRI)
In the EMBARC sample, diffusion images were acquired using a single-shot EPI (echo planar imaging) sequence. Scan parameters were as follows: TR=8310–9500 ms, TE=95–96.3 ms, flip angle 90°, slice thickness=2.5 mm, FOV=240×240 mm2, voxel dimensions 2.5 mm×2.5 mm×2.5 mm or 1.9 mm×1.9 mm×2.5 mm, acquisition matrix=96 × 96, b value = 1000 s/mm2, and 64 collinear directions with 1 or 5 non-weighted images. Diffusion images in the other protocols were acquired with comparable parameters. However, 25 collinear directions, voxel dimensions of 2.5 mm×2.5 mm×2.5 mm, and an FOV of 256×256 mm2 were used.
Image Processing: Diffusion Weighted MRI
Each dMRI image was run through a series of quality assurance tests for common artifacts, including ghost, ring, slice-wise intensity, venetian blind, and gradient-wise motion artifacts (Liu et al., 2010). Diffusion images were then corrected for distortion induced by gradient coils and simple head motion using the eddy current correction routine within FSL (FMRIB Software Library, http://www.fmrib.ox.ac.uk/fsl/). FSL’s Brain Extraction Tool (BET) removed non-brain tissue from the image. Following this, Camino (http://web4.cs.ucl.ac.uk/research/medic/camino/pmwiki/pmwiki.php) was used to estimate FA by computing the least-squares-fit diffusion tensor with non-linear optimization using a Levenburg-Marquardt algorithm, constrained to be positive by fitting its Cholesky decomposition (Alexander & Barker, 2005; Jones & Basser, 2004).
The dMRI images were coregistered to the cropped T1 images using Advanced Normalization Tools (ANTS; http://www.picsl.upenn.edu/ANTS/) and the inverse transformation was applied to the Freesurfer-derived cortical map in order to place the regions of interest into dMRI space for analysis. Finally, mean FA values in white matter were computed for each region. A trained technician manually inspected each aspect of the dMRI analysis including level of artifact (based on the cutoffs defined in Liu et al), distortion correction, coregistration and FA histogram.
Data Preparation Statistics
In the EMBARC sample, of the 193 MDD subjects in the training set, 178 (92%) unique baseline MRI sessions possessed both dMRI and sMRI acquisitions, 121 of these 178 sessions (68%) passed Freesurfer surface validation, and 114 of the 121 (94%) passed dMRI validation. Of the 93 MDD subjects in EMBARC’s validation set, 82 (88%) unique baseline MRI sessions possessed both dMRI and sMRI acquisitions, 66 of these 82 (80%) passed Freesurfer surface validation, and all remaining 66 passed dMRI validation. Of EMBARC’s 40 healthy control (HC) scans, 93% passed validation. Two-thirds of the all scans were used for the training dataset. Ninety (88%) of the 102 qualifying scans from the other 10 protocols passed sMRI and dMRI validation. Similar to the EMBARC sample, two-thirds of the validated MDD and HC scans from the other protocols were randomized to the training dataset.
Features
Since the biological underpinnings of MDD are unknown, a large number of potential features were examined. For each subject, 225 features were included: age at evaluation, sex, handedness, 145 sMRI-based and 77 dMRI-based features.
sMRI features included:
(1) bilateral gray matter volume of 34 Desikan-Killiany (DK, (Desikan et al., 2006)) and 11 subcortical Center for Morphometric Analysis (CMA, (Fischl et al., 2004)) regions (68 + 22 = 90 features),
(2) volumes of brainstem, CSF and subdivided corpus callosum (7 features),
(3) whole-brain measures: bilateral mean thickness and whole-brain volume (2 + 1= 3 features) to supplement the regional measures in (1), and:
(4) the asymmetry index (), where L is the measure on the left and R is the measure on the right, was designed to gauge the magnitude and direction of morphological asymmetry (Cherbuin, Reglade-Meslin, Kumar, Sachdev, & Anstey, 2010), and was computed for the bilateral CMA and DK regions above (45 features).
dMRI features included: the average FA in white-matter segmentations of the 34 bilateral DK regions (68 features), 5 corpus callosum regions (5 features), and the bilaterally divided cerebrum and cerebellum (4 features).
Outcome Measures
Discrete measure: We evaluated two potential classification schemes: (1) MDD vs controls and (2) severe MDD vs mild MDD vs control. 68 patients had severe depression with a Hamilton Depression Rating Scale (HAMD) 17 item total score >19. Subjects’ characteristics and HAMD scores for training and validation datasets are listed in Tables 1 and 2.
Table 1:
Subject characteristics and Hamilton Depression Rating Scale (HAMD 17-item) scores by severity index for the training samples. p-values were based on ANOVA comparing severe MDD, mild MDD and HC. For those variables that had significant differences among three groups, all pair-wise comparisons were still significant except that there was no significant age difference between mild MDD and HC. MDD: Major Depressive Disorder; HC: Healthy Controls
| Variable | MDD (N=147) | Severe MDD (N=68) | Mild MDD (N=79) | Healthy Control (N=52) | p-values* | p-value for Severe vs Mild | p-value for Severe vs HC | p-value for Mild vs HC |
|---|---|---|---|---|---|---|---|---|
| Male | 53 (36.05%) | 30 (44.12%) | 23 (29.11%) | 21 (40.38%) | 0.1473 | |||
| Age (years) | 36.78±12.94 | 40.83±12.31 | 33.29±12.51 | 32.48±12.15 | 0.0002 | 0.0003 | 0.0003 | 0.7113 |
| Left handedness | 11 (7.48%) | 7 (10.29%) | 4 (5.06%) | 1 (1.92%) | 0.4204 | |||
| Right handedness | 127 (86.39%) | 57 (83.82%) | 70 (88.61%) | 48 (92.31%) | ||||
| HAMD | 18.83±4.63 | 22.84±2.80 | 15.38±2.71 | 1.04±1.45 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 1 Psychic Depression (max=16.04) |
6.63±1.97 | 7.40±1.94 | 5.97±1.75 | 0.15±0.57 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 2 Motivation (max=5.84) |
2.36±1.20 | 3.05±1.12 | 1.76±0.91 | 0.05±0.19 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 3 Psychosis (max=7.2) |
0.44±0.60 | 0.52±0.67 | 0.36±0.53 | 0.00±0.00 | 0.1046 | |||
| Factor 4 Anxiety (max=10.24) |
2.62±1.25 | 3.14±1.18 | 2.17±1.13 | 0.27±0.40 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 5 Sleep (max=4.32) |
2.11±1.41 | 2.93±1.21 | 1.40±1.17 | 0.12±0.32 | <.0001 | <.0001 | <.0001 | <.0001 |
Table 2:
Subject characteristics and Hamilton Depression Rating Scale (HAMD 17-item) scores by severity index for the validation samples. p-values were based on ANOVA comparing severe MDD, mild MDD and HC. For those variables that had significant differences among three groups, all pair-wise comparisons were still significant except that there was no significant age difference between mild MDD and HC. MDD: Major Depressive Disorder; HC: Healthy Controls
| Variable | MDD (N=83) | Severe MDD (N=32) | Mild MDD (N=51) | Healthy Control (N=25) | p-values* | p-value for Severe vs Mild | p-value for Severe vs HC | p-value for Mild vs HC |
|---|---|---|---|---|---|---|---|---|
| Male | 32 (38.55%) | 10 (31.25%) | 28 (54.9%) | 12 (48%) | 0.3944 | - | - | - |
| Age (years) | 35.66±12.44 | 34.94±12.48 | 36.12±12.52 | 33.72±13.43 | 0.7484 | - | - | - |
| Left handedness | 10 (12.05%) | 2 (6.25%) | 8 (15.69%) | 2 (8%) | 0.0624 | - | - | - |
| Right handedness | 69 (83.13%) | 26 (81.25%) | 43 (84.31%) | 22 (88%) | ||||
| HAMD | 18.76±4.50 | 23.59±2.38 | 15.73±2.30 | 1.28±2.01 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 1 Psychic Depression (max=11.05) |
6.70±1.82 | 7.47±1.72 | 6.21±1.73 | 0.18±0.35 | <.0001 | 0.0004 | <.0001 | <.0001 |
| Factor 2 Motivation (max=5.84) |
2.35±1.13 | 3.16±1.08 | 1.85±0.83 | 0.02±0.10 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 3 Psychosis (max=2.18) |
0.30±0.50 | 0.37±0.50 | 0.25±0.50 | 0.00±0.00 | 0.3138 | - | - | - |
| Factor 4 Anxiety (max=6.32) |
2.49±1.21 | 3.35±1.23 | 1.95±0.83 | 0.31±0.45 | <.0001 | <.0001 | <.0001 | <.0001 |
| Factor 5 Sleep (max=4.32) |
2.24±1.28 | 2.99±1.00 | 1.77±1.21 | 0.25±0.63 | <.0001 | <.0001 | <.0001 | 0.0001 |
Continuous measure: This included depression severity (HAMD total score) and factors. Each factor is a sum of the products of the factor’s HAMD items and corresponding loading values. The loading values were obtained from a previous factor analysis of the HAMD, which was optimized for self-report measures with potentially correlated factors by using polychoric correlation (PCC) and a non-orthogonal rotation (Milak et al., 2005). Factor scores for the training and validation datasets are shown in Tables 1 and 2.
Factor 1: Psychic Depression, including HAMD items 1–3, 8, 22–24, signifying depressed mood, guilt, suicidality, retardation, helplessness, hopelessness and worthlessness;
Factor 2: Loss of Motivated Behavior, including HAMD items 7, 12, 14, 16, involves work and activities, somatic and genital symptoms and weight loss;
Factor 3: Psychosis, including HAMD items 17, 19–21, evaluates lack of insight, depersonalization, derealization, paranoia, obsessive and compulsive behavior. (This factor was not evaluated because the majority of subjects had scores of 0.);
Factor 4: Anxiety, including HAMD items 9–11, 15, involving agitation, hypochondrias, psychic or somatic anxiety); and
Factor 5: Sleep Disturbance, including items 4–6, relating to insomnia.
The predictive modeling systems
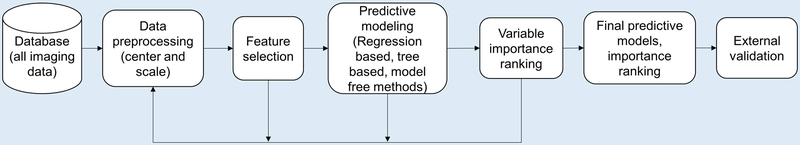
With the separate dataset available to validate model findings, we took an exhaustive approach in applying predictive models to the training data. Figure 1 illustrates the workflow of the predictive modeling system. It starts with data preprocessing, followed by feature selection, predictive modeling and variable importance ranking evaluation blocks, which in turn provide additional information for better feature selection. Validation was performed on the final classifier built on the training dataset.
Figure 1:
The general workflow of the predictive modeling system, which starts with data preprocessing, followed by feature selection, predictive modeling and variable importance ranking evaluation blocks, which in turn provide additional information for better feature selection. Validation was performed on the final classifier built on the training dataset.
Initial feature selection was based on between-feature correlation after applying centering and scaling to all features. Highly correlated image features that had an average correlation coefficient with the rest of the features >0.7 were eliminated here. Fifty-six features were removed in this step: 36 sMRI measures and 20 dMRI measures. Another initial feature selection performed was based on having a well-conditioned matrix of pairwise correlation coefficients among all features. A matrix is considered to be ill-conditioned if the 2-norm condition number (the ratio of its smallest to the largest eigenvalue) is smaller than a tolerance value (2e-15). Jollifee’s method (Jolliffe, 2002) was used to select a subset of features that have a well-conditioned correlation coefficient matrix. Using this method, 27 additional features were removed: 23 sMRI measures, and 4 dMRI measures.
After initial feature selection, different predictive models were built to predict binary outcomes: MDD vs HC and ordinal outcomes: HC, mild MDD and severe MDD. These included commonly used approaches such as the penalized logistic regression (PLR) model with elastic net penalty, random forest (RF) and support vector machine (SVM) with linear or nonlinear kernels such as cubic polynomial and radial basis function kernels. Extensions of these three classifiers for ordinal classification were used for predicting ordinal outcomes: under the PLR framework, cumulative logit model, adjacent category model, backward continuation ratio model and forward continuation ratio model were used; under the SVM framework, results from binary classifiers were aggregated using three different decoding methods – robust tree decoding, maximum vote decoding for the “one-against-all” scheme and most frequent vote decoding using the “one-against-one” scheme. Predictive models for predicting continuous HAMD and factor scores among MDD patients included penalized linear regression with elastic net penalty, random forest and support vector regression.
All predictive models were built using R 3.3.1 (R Core Team, 2015). Prediction performance of binary classifiers was measured by area under the receiver operating curve (AUC), misclassification rate, sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV). Prediction performance of ordinal classifiers was measured by percentage correctly classified (PCC) and the rank correlations between the predicted class and true class such as Spearman’s ρ, Kendall’s τ, Goodman-Kruskal Γ, and Cohen’s κ. Prediction performance of models for continuous scores was measured by root mean squared error (RMSE) and R2. All tuning parameters of these predictive models were chosen based on 10 repeated 5-fold cross validation in the training dataset.
Variable importance ranking was based on the predictive models that had the highest average AUCs, highest average PCC, or smallest average RMSE after 10-repeated 5-fold cross validation. For predictive models using PLR framework, the features in the final model were ranked by their absolute coefficient estimates: the larger the absolute value of the estimated coefficient, the greater the contribution this feature provided to the final prediction. For predictive models using the SVM framework, the contribution of each feature was reflected through its nonzero weights. For predictive models using RF, the variable importance rankings were based on the Gini impurity index (Breiman, 2001). All top ranked features from these predictive models were used in the final predictive models.
Validation
Calibration is an essential aspect of external validation (Steyerberg et al., 2010). Calibration in the large was used to determine whether the mean predicted probability of MDD is equal to the mean observed MDD rate in the validation data set (Van Calster et al., 2016). The ideal value is zero difference between predicted and observed probabilities. The assessment of the overall predictive effect was graphically evaluated in a calibration plot and used for estimation of a calibration slope. A calibration plot displays the relationship between predicted MDD risk (x-axis) and observed true group label (MDD=1, HC=0, y-axis) by fitting a flexible nonlinear calibration curve using a nonparametric regression method called loess, the local regression using polynomials (Austin & Steyerberg, 2014). The estimation of the calibration slope b, is by fitting the following model: , where a is the model intercept, and is the predicted risk. Therefore, the calibration slope summarizes the relationship between the predicted risks and the observed true labels. For example, using this validation, a calibration slope less than 1 reflects an overestimation of MDD risk, and vice versa for a calibration slope greater than 1 (Van Calster et al., 2016).
Results
After an initial round of model building for predicting different outcomes, features were ranked accordingly. The final predictive models for each type of outcome contained 39 features that were top ranked. Table A1 has a complete list of these 39 features: 16 sMRI measures, 21 dMRI measures, sex and age.
Binary classification for predicting MDD vs HC
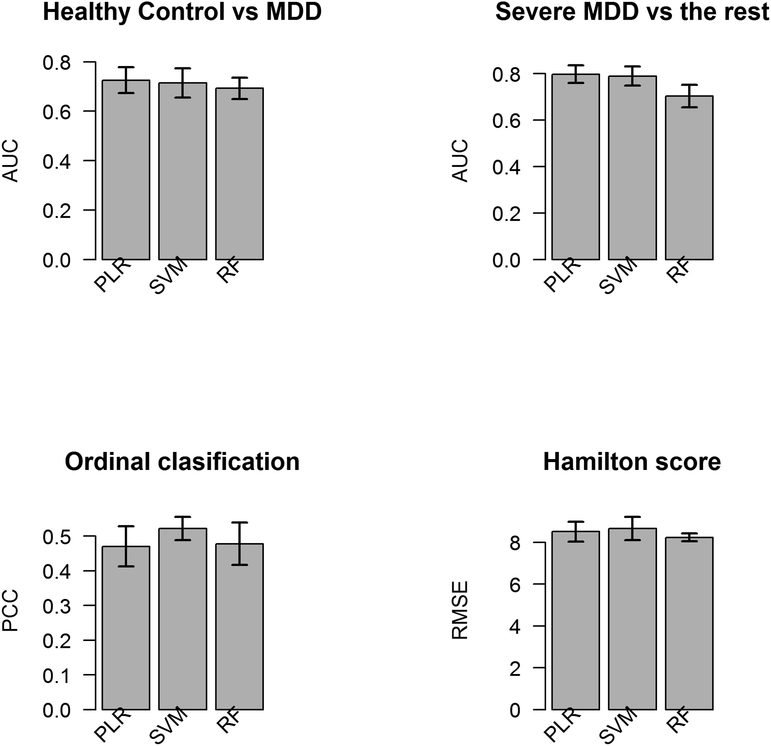
The best binary classifiers in PLR, SVM and RF for predicting MDD had AUC ranges from 0.69 to 0.74 with accuracy rates ranging from 73.45% to 75.05% (Table 3). The classifier using SVM with a radial basis function kernel had the best AUC of 0.74±0.02. Table 4 lists a combination of all of the top 10 ranked features from each of the three binary classifiers. The mean FA in the left medial orbitofrontal cortex and right cuneus contributed highly to predicting MDD in this analysis.
Table 3:
Predictive accuracy of binary classification. Mean±SD were based on 10 sets of average performance measures from repeated 5-fold cross validation using 39 features. MDD: Major Depressive Disorder; HC: Healthy Controls; PLR: penalized logistic regression model with elastic net penalty, SVM: support vector machine, RF: random forest
| Outcomes | Area Under the Curve (AUC) | Accuracy Rate | ||||
|---|---|---|---|---|---|---|
| PLR | SVM | RF | PLR | SVM | RF | |
| MDD vs HC | 0.73 ± 0.03 | 0.74 ± 0.02 | 0.69 ± 0.02 | 73.45% ± 1.57% | 74.00% ± 1.32% | 75.05% ± 1.17% |
Table 4:
Combination of the top 10 important features from each classifier for binary classification using penalized logistic regression (PLR) model with elastic net penalty, random forest (RF) and support vector machine (SVM). Numbers in the parentheses are the average rank from 10 repeated 5-fold cross-validation. Average Rank refers to the mean rank across the three algorithms.
| Measure | Region | Rank in PLR classifier | Rank in SVM classifier | Rank in RF classifier | Average Rank |
|---|---|---|---|---|---|
| Mean FA (dMRI) | Left Medial Orbitofrontal | 1(2.52) | 1(1.30) | 1(1.12) | 1 |
| Mean FA (dMRI) | Right Cuneus | 2(15.82) | 2(2.82) | 2(3.40) | 2 |
| Gray Matter Volume (sMRI) | Inferior Temporal | 6(47.88) | 4(6.16) | 3(6.56) | 4.33 |
| Mean FA (dMRI) | Left Middle Temporal | 8(49.66) | 7(11.74) | 4(7.16) | 6.33 |
| Mean FA (dMRI) | Left Lateral Orbitofrontal | 5(44.62) | 5(6.54) | 10(13.22) | 6.67 |
| Gray Matter Volume (sMRI) | Left Pars Orbitalis | 11(62.82) | 3(4.58) | 8(9.66) | 7.33 |
| Mean FA (dMRI) | Right Entorhinal | 3(31.18) | 6(9.30) | 22(20.78) | 10.33 |
| Gray Matter Volume Asymmetry (sMRI) | Cuneus | 10(61.40) | 8(11.94) | 14(16.54) | 10.67 |
| Mean FA (dMRI) | Left Lateral Occipital | 4(44.62) | 10(12.50) | 20(20.06) | 11.33 |
| Gray Matter Volume Asymmetry (sMRI) | Pars Triangularis | 7(49.54) | 11(13.06) | 19(19.54) | 12.33 |
| Mean FA (dMRI) | Left Banks of the Superior Temporal Sulcus | 9(57.12) | 24(25.66) | 6(8.72) | 13 |
| Mean FA (dMRI) | Right Rostral Anterior Cingulate | 13(68.08) | 19(21.68) | 9(10.36) | 13.67 |
| Mean FA (dMRI) | Right Fusiform | 22(81.58) | 23(25.22) | 5(8.00) | 16.67 |
| Volume Asymmetry (sMRI) | Cerebellum | 18(74.02) | 9(12.08) | 26(25.12) | 17.67 |
| Mean FA (dMRI) | Left Insula | 39(141.30) | 22(24.02) | 7(8.80) | 22.67 |
To evaluate the influence of the feature selection on algorithm output, an independent classification method was also applied to predict MDD vs. healthy control. Half of training data patients were randomly selected to tune the algorithm and the rest were used as to evaluate the results. Due to the imbalance of the classes, the training set was downsampled while all the validation data were used. No feature selection was used. RF and classification trees were built for classification. The fact that splitting variables (the most predictive variables for RF) used in these two tree-based classification were among the features selected for predictive model building confirms the robustness of our feature selection strategies.
Ordinal classification for severity index: severe MDD, mild MDD and HC
Different ordinal classifiers under the SVM and PLR framework in addition to RF were constructed to predict severe MDD, mild MDD and HC. The predictive performance of the best classifiers using SVM, PLR and RF is summarized in Table 5. The highest average PCC from 10 repeated 5-fold cross-validation, 52.2%, was from an SVM classifier assigning subjects to each class using most frequent vote based on pairwise SVM classifiers. A combination of all of the top 10 ranked features from these ordinal classifiers is listed in Table 6. Mean FA in the left medial orbitofrontal cortex and right cuneus again contributed highly in placing subjects into correct subcategories in each of three ordinal classifiers.
Table 5:
Predictive accuracy of ordinal classification. Mean±SD were based on 10 sets of average performance measures from repeated 5-fold cross validation using 39 features.
| Model | Percentage Correctly Classified (PCC) | Spearman’s rho | Kendall’s tau | Goodman-Kruskal’s gamma | Cohen’s Kappa |
|---|---|---|---|---|---|
| SVM with most frequent class based on pairwise classification | 52.20 ± 1.69% | 0.3591 ± 0.0378 | 0.3267 ± 0.0340 | 0.4828 ± 0.0474 | 0.3468 ± 0.0394 |
| PLR with forward continuation | 47.05 ± 2.87% | 0.3736 ± 0.0320 | 0.3359 ± 0.0305 | 0.5062 ± 0.0473 | 0.3674 ± 0.0339 |
| Random forest | 47.80 ± 3.04% | 0.2935 ± 0.0492 | 0.2687 ± 0.0451 | 0.4213 ± 0.0638 | 0.1313 ± 0.0253 |
Table 6:
Combination of the top 10 ranked features from the best ordinal classifiers using SVM, RF and PLR. Numbers in the parentheses are the average rank from 10 repeated 5-fold cross-validation. Average Rank refers to the mean rank across the three algorithms. MDD: Major Depressive Disorder; HC: Healthy Controls; PLR: penalized logistic regression model with elastic net penalty, SVM: support vector machine, RF: random forest
| Measure | Region | Rank in SVM classifier | Rank in RF classifier | Rank in PLR classifier | Average Rank |
|---|---|---|---|---|---|
| Mean FA (dMRI) | Left Medial Orbitofrontal | 1(10.54) | 2(4.08) | 2(5.06) | 1.67 |
| Mean FA (dMRI) | Right Cuneus | 5(13.82) | 3(8.94) | 6(7.74) | 4.67 |
| Mean FA (dMRI) | Left Lateral Orbitofrontal | 3(12.42) | 6(12.05) | 12(14.89) | 7 |
| Gray Matter Volume Asymmetry (sMRI) | Pericalcarine | 4(13.76) | 22(22.61) | 1(4.87) | 9 |
| Gray Matter Volume Asymmetry (sMRI) | Precentral | 8(14.34) | 5(11.94) | 19(19.13) | 10.67 |
| Mean FA (dMRI) | Right Rostral Anterior Cingulate | 7(14.32) | 20(22.42) | 7(8.21) | 11.33 |
| Mean FA (dMRI) | Right Lingual | 9(15.56) | 7(13.00) | 20(19.31) | 12 |
| Gray Matter Volume Asymmetry (sMRI) | Pars Triangularis | 11(16.60) | 21(22.45) | 4(6.90) | 12 |
| Gray Matter Volume (sMRI) | Left Pars Orbitalis | 21(21.02) | 15(21.58) | 3(5.32) | 13 |
| Mean FA (dMRI) | Right Entorhinal | 19(20.50) | 18(21.94) | 5(7.08) | 14 |
| Gray Matter Volume (sMRI) | Inferior Temporal | 10(16.28) | 14(20.59) | 21(21.43) | 15 |
| Gray Matter Volume (sMRI) | Right Pars Triangularis | 6(14.06) | 23(22.85) | 17(18.18) | 15.33 |
| Mean FA (dMRI) | Left Insula | 12(16.60) | 10(17.77) | 28(26.43) | 16.67 |
| Age | Age | 16(17.64) | 1(3.64) | 33(29.42) | 16.67 |
| Mean FA (dMRI) | Right Caudal Anterior Cingulate | 2(10.98) | 36(24.98) | 16(17.18) | 18 |
| Gray Matter Volume Asymmetry (sMRI) | Lingual | 14(17.40) | 33(24.57) | 9(12.00) | 18.67 |
| Mean FA (dMRI) | Left Lateral Occipital | 23(21.24) | 24(22.95) | 10(12.66) | 19 |
| Cortical Thickness (sMRI) | Left Hemisphere (Average) | 15(17.62) | 9(14.11) | 38(32.09) | 20.67 |
| Gray Matter Volume (sMRI) | Right Choroid Plexus | 20(20.92) | 8(13.52) | 35(29.91) | 21 |
| Volume Asymmetry (sMRI) | Cerebellum | 28(23.04) | 28(23.73) | 8(11.77) | 21.33 |
| Cortical Thickness | Right Hemisphere (Average) | 34(24.64) | 4(9.55) | 36(30.32) | 24.67 |
Prediction of Hamilton scores among Patients with MDD
When using PLR, SVM and RF to build predictive models for HAMD total score and its factor scores, SVM using a radial basis function kernel had the best predictive performance for HAMD score, Factor 1 and 4; RF had the best predictive performance for factor 5; PLR had the best performance for Factor 2 (Table 7). Permutation tests applied to these best models for predicting each continuous outcome suggested that the corresponding RMSEs were not significantly below chance levels, except for Factor 2 and Factor 5. Frequently top-ranked variables in predicting all 5 continuous scores can be found in Table 8. Two variables that contribute highly in predicting all five different continuous scores are mean FA in the right cuneus and the volume of the right choroid plexus.
Table 7:
Performance of predicting Hamilton Depression Rating Scale (HAMD) total score and its factor scores in Major Depressive Disorder. Mean±SD were based on 10 sets of average performance measures from repeated 5-fold cross validation using 39 features. PLR: penalized logistic regression model with elastic net penalty, SVM: support vector machine, RF: random forest; RMSE: root mean squared error
| Variable | RMSE | R2 | ||||
|---|---|---|---|---|---|---|
| PLR | SVM | RF | PLR | SVM | RF | |
| HAMD Score | 4.6019 ± 0.0851 | 4.3408 ± 0.0628 | 4.5138 ± 0.0793 | 0.0516 ± 0.0219 | 0.1301 ± 0.0274 | 0.0632 ± 0.0237 |
| Factor 1: Psychic Depression |
2.0498 ± 0.0381 | 1.9742 ± 0.0140 | 2.0226 ± 0.0393 | 0.0261 ± 0.0096 | 0.0334 ± 0.0100 | 0.0302 ± 0.0173 |
|
Factor 2: Motivation |
1.1802 ± 0.0245 | 1.1904 ± 0.0138 | 1.2130 ± 0.0189 | 0.0662 ± 0.0265 | 0.0349 ± 0.0198 | 0.0224 ± 0.0150 |
| Factor 4: Anxiety |
1.2812 ± 0.0202 | 1.2328 ± 0.0095 | 1.2933 ± 0.0199 | 0.0286 ± 0.0262 | 0.0374 ± 0.0233 | 0.0358 ± 0.0170 |
| Factor 5: Sleep |
1.4344 ± 0.0204 | 1.3926 ± 0.0141 | 1.3649 ± 0.0157 | 0.0299 ± 0.0098 | 0.0502 ± 0.0115 | 0.0856 ± 0.0227 |
Table 8:
Frequently top-ranked variables in predicting HAMD total score and its factor scores (F1-F5) in Major Depressive Disorder; 1 means the feature is top ranked in predicting one type of score while 0 means it is not.
| Measure | Region | HAMD | F1 | F2 | F4 | F5 | Total count of appearance in top ranked features |
|---|---|---|---|---|---|---|---|
| Mean FA (dMRI) | Right Cuneus | 1 | 1 | 1 | 1 | 1 | 5 |
| Gray Matter Volume (sMRI) | Right Choroid Plexus | 1 | 1 | 1 | 1 | 1 | 5 |
| Gray Matter Volume Asymmetry (sMRI) | Lingual | 1 | 1 | 1 | 0 | 1 | 4 |
| Gray Matter Volume Asymmetry (sMRI) | Pericalcarine | 1 | 1 | 1 | 1 | 0 | 4 |
| Gray Matter Volume (sMRI) | Right Frontal Pole | 1 | 1 | 1 | 1 | 0 | 4 |
| Mean FA (dMRI) | Left Inferior Parietal | 1 | 1 | 1 | 1 | 0 | 4 |
| Mean FA (dMRI) | Left Transverse Temporal | 1 | 0 | 1 | 1 | 1 | 4 |
| Volume Asymmetry (sMRI) | Cerebellum | 1 | 1 | 0 | 1 | 1 | 4 |
| Gray Matter Volume Asymmetry (sMRI) | Cuneus | 1 | 1 | 1 | 0 | 0 | 3 |
| Gray Matter Volume Asymmetry (sMRI) | Pars Triangularis | 1 | 0 | 1 | 0 | 1 | 3 |
| Gray Matter Volume Asymmetry (sMRI) | Precentral | 1 | 0 | 1 | 1 | 0 | 3 |
| Cortical Thickness (sMRI) | Left Hemisphere (Average) | 1 | 0 | 0 | 1 | 1 | 3 |
| Mean FA (dMRI) | Left Insula | 1 | 1 | 0 | 1 | 0 | 3 |
| Mean FA (dMRI) | Left Pars Triangularis | 1 | 1 | 0 | 0 | 1 | 3 |
| Mean FA (dMRI) | Left Precuneus | 1 | 1 | 0 | 0 | 1 | 3 |
Comparisons across Models
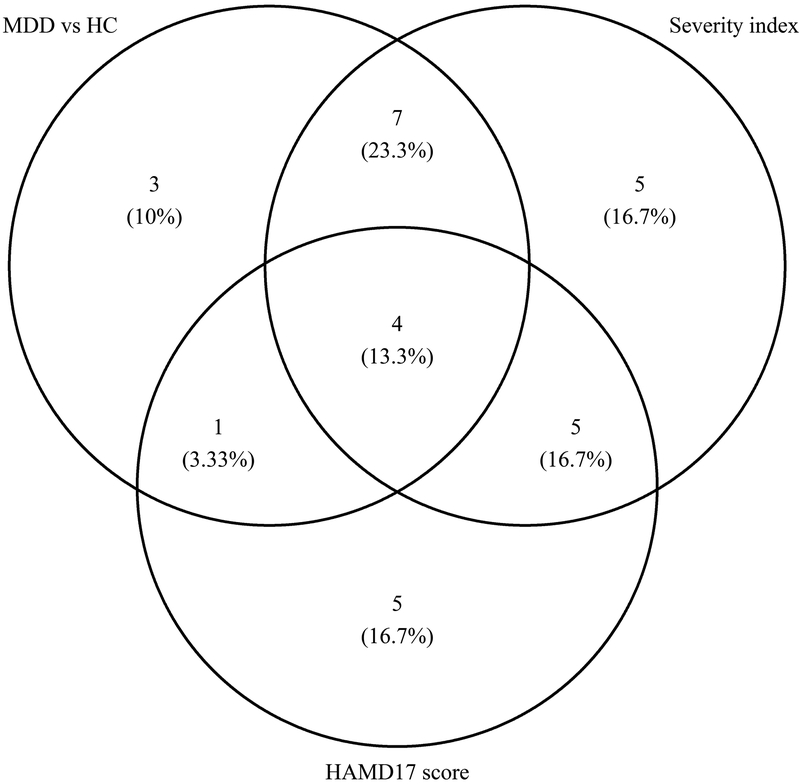
Among all top ranked features from predictive models built for all outcomes here, four common elements contributed to model accuracy, as indicated in the center area of Figure 2. These include mean FA in the right cuneus and left insula and asymmetry in the volume of the pars triangularis and cerebellum.
Figure 2:
Venn diagram for the number of top ranked variables for all predictive models based on training data set (Predictive model for continuous Hamilton Depression Rating Scale [HAMD17] scores was based on MDD patients only). MDD: Major Depressive Disorder; HC: Healthy Controls
As an additional check of the importance of these four features, the above models were re-run with the inclusion of all features (i.e., without doing any feature selection). Without feature selection, these four common features remained highly ranked in one or more of the analyses: mean FA in the right cuneus (binary: rank=1; ordinal: 2; continuous outcomes: 2), mean FA in the left insula (binary: 47; ordinal: 7; continuous: 3); volume asymmetry in the pars triangularis (binary: 38; ordinal: 16; continuous: 4); and volume asymmetry in the cerebellum (binary: 37; ordinal: 81; continuous: 4).
As mentioned in the feature reduction step, highly correlated features were removed. Only one feature removed in this stage was highly correlated to any of the four common features. This was grey matter volume of the left ventral diencephalon (Pearson’s correlation coefficient = 0.8520 with mean FA in the left insula).
External Validation on predicting MDD vs HC
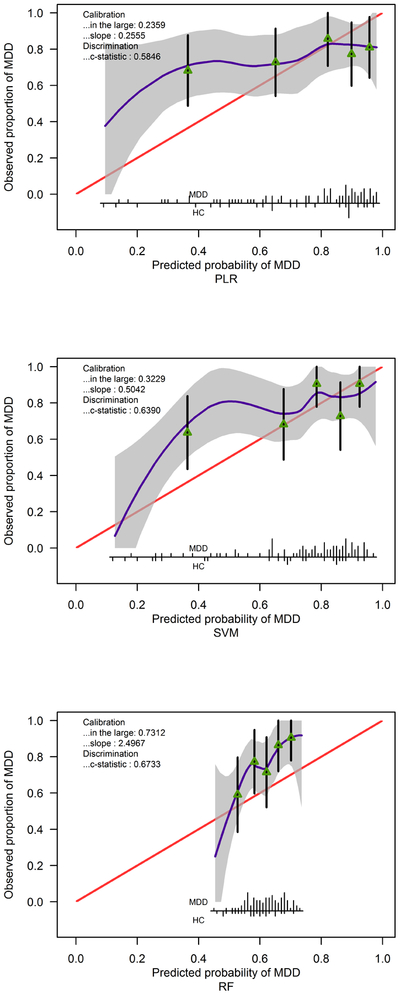
Because of the low performance of both ordinal classification and predictive modeling, external validation analysis was only performed on the binary classifier for MDD vs HC. A patient was classified as having MDD if her/his predicted probability of having MDD was greater than 50%. The prediction performance for three predictive models (as in Table 3) are summarized in Tables 9 and 10. The binary classifier based on RF had the highest accuracy rate of 78.7% and the highest AUC value of 0.6733, but similar to other two classifiers, the specificity was very low. (This result supports the criticism about RF on imbalanced datasets (Dudoit & Fridlyand, 2003).) The calibration plot of this binary classifier actually suggests that it consistently underestimates the probability of MDD and hence even though this method has a better discrimination index (AUC=0.6733, 95% CI: 0.5508 – 0.7957), the calibration in the large is worse than SVM or PLR (Figure 3).
Table 9:
Confusion matrices of binary classifiers on validation dataset.
| Model | Validation outcomes | true Healthy Controls | true Major Depressive Disorder |
|---|---|---|---|
| Penalized Logistic Regression | HC | 6 | 11 |
| MDD | 19 | 72 | |
| Support Vector Machine | HC | 8 | 10 |
| MDD | 17 | 73 | |
| Random Forest | HC | 4 | 2 |
| MDD | 21 | 81 |
Table 10:
Other performance metrics and their 95% confidence intervals for binary classifiers on the validation dataset. PLR: Penalized Logistic Regression; SVM: Support Vector Machine; RF: Random Forest; AUC: Area under the Curve
| Model | AUC | Accuracy (%) | Sensitivity (%) | Specificity (%) | Positive Predictive Value (%) | Negative Predictive Value (%) |
|---|---|---|---|---|---|---|
| PLR | 0.5846(0.4523–0.7169) | 72.22(62.78–80.41) | 86.75(77.52–93.19) | 24.00(9.36–45.13) | 79.12(69.33–86.94) | 35.29(14.21–61.67) |
| SVM | 0.639(0.5085–0.7696) | 75(65.75–0.82.83) | 87.95(78.96–94.07) | 32.00(14.95–53.5) | 81.11(71.49–88.59) | 44.44(21.53–69.24) |
| RF | 0.6733(0.5508–0.7957) | 78.7(69.78–86) | 97.59(91.57–99.71) | 16.00(4.54–36.08) | 79.41(70.27–86.78) | 66.67(22.28–95.67) |
Figure 3:
Calibration plot for predicting diagnosis based on three methods and 39 features. Calibration in the large quantifies the difference between mean predicted probability of having Major Depressive Disorder (MDD) and observed proportion of MDD patients. The closer to 0, the better the calibration is. The calibration slope different from 1 suggests that the overall predictive performance of 39 features was different from that observed in the validation data. A calibration slope less than 1 reflects an overestimation of MDD risk, and vice versa for a calibration slope greater than 1 (Van Calster et al., 2016). The c-statistic is identical to the AUC values and its confidence intervals are in Table 10. Spikes at the bottom of the graph indicate the probability distribution for those with MDD and Healthy Controls (HC). Triangles indicate quintiles of subjects according to predicted probability with 95% confidence intervals for the observed proportions of patients with MDD. For example, the fact that for PLR and SVM, the spikes mostly appear near 0.9, and the triangles are near the right hand side, is consistent with the calibration slope less than 1, i.e., overestimating MDD risk.
Discussion
Major Depressive Disorder is a prevalent disease with a growing global impact. Although numerous imaging studies have uncovered neurobiological differences associated with MDD, clinically translatable markers have yet to be identified. This may be due to limited sample sizes used in previous studies, leading to overfitting of data, and not using a separate replication sample, resulting in inconsistency of results across studies. To overcome these previous limitations, the current study involved an exploration in 199 subjects, using a multi-site design and validation of findings in a separate cohort of 108 subjects.
Modeling/Methodology
To represent a generalizable sample, image-derived data in this study were acquired from 8 sites with 7 different MRI scanners. Because systematic differences in image-derived measures across scanners have been reported (Iscan et al., 2015; Madan, 2017), adjusting for site/scanner differences was considered. Two ways of adjusting for these differences were explored (data not shown): 1) using linear regression to estimate the site/scanner differences after controlling for age, sex and handedness and then normalizing each imaging feature to the reference site/scanner with the most samples; 2) using quantile normalization. In most cases, adjusting for site/scanner within this study did not improve predictive performance, and in a few cases, this adjustment reduced predictive performance (data not shown). Further, top ranked features were similar among models with and without adjusting for these differences. Therefore, with the intention of generalizing our predictive models, no site/scanner adjustment was implemented in the current study.
In this study, multiple classification techniques were applied to a training set of 199 subjects (52 HC, 147 MDD) with two different imaging modalities (dMRI and sMRI). Regional grey matter (volume, asymmetry and thickness from sMRI) and white matter (fractional anisotropy from dMRI) measures comprised 222 features. In addition to these image-based features, sex, age at diagnosis and handedness were used as predictors of clinical status. We did not include clinical factors such as length of illness or number of depressive episodes in our prediction analysis. Though doing so could potentially improve prediction accuracy, there are challenges in accurately assessing these variables (Kruijshaar et al., 2005; Patten, 2003; Takayanagi et al., 2014; Wells & Horwood, 2004), and the focus of this work was to relate objective measures of brain biology to depression outcomes. Further, potential correlations between these variables and depression measures (Kessler et al., 2007) could confound biological interpretations.
We did not restrict the dataset to a priori regions due to a lack of consensus on MDD neurobiology. The large number of initial features, however, required data reduction prior to analysis. Therefore, highly correlated features and those with ill-conditioned pairwise matrices (matrices where one input has a large effect on the outcome) were removed to reduce dimensionality.
An iterative procedure was then used for final feature selection, with 39 out of 225 features chosen for building the final predictive models (see Table A1 in Appendix). This procedure involved applying the three classifiers discussed below to the binary, ordinal and continuous outcomes, ranking the variables in terms of prediction, and then compiling the top 10 features across the three classifiers for each analysis. This resulted in 37 top image-based features in addition to age and sex. To determine whether feature selection is sensitive to choice of data reduction procedure, we also performed the MDD/control and continuous prediction without feature selection and obtained similar results. Further, a recently proposed variable selection algorithm, stability selection, was also applied (data not shown) (Hofner, Boccuto, & Goker, 2015; Hofner & Hothorn, 2017; Meinshausen & Bühlmann, 2010; Shah & Samworth, 2013). All features except one selected by this method for different outcomes fall within 39 features in Table A1. Iterative sure independence screening methods, in which variable selection is integrated into the model building process, were also applied (Fan, Feng, & Song, 2011; Fan & Li, 2001; Fan, Samworth, & Wu, 2009; Tibshirani, 1996; Zhang, 2010). However, the predictive performance did not improve and hence the related results were not reported. Nonetheless, a combination of the top 10 important features ranked by these methods were similar to those reported in Tables 4, 6 and 8. These results provide confidence that the results were not sensitive to the use, or choice of, feature selection technique.
The three classifiers applied were PLR, RF and SVM. Each model has differing strengths. For example, SVM has advantages when dealing with binary class data whereas RF is advantageous for multi-class data with outliers (Hastie, Tibshirani, & Friedman, 2001). Penalized logistic regression has also been shown to handle outliers better than the SVM (Hastie et al., 2001). The No Free Lunch (NFL) theory asserts that there is no one optimal classifier across different data sets (Wolpert & Macready, 1997). Therefore, the optimal modeling strategy may be data dependent. For this reason, three of the most popular and effective modeling techniques were applied.
Model Results
For binary classification (MDD/HC), all three models had similar accuracy, with misclassification rates of ~26%. In the validation set, results were poor, with a mean 87.95% sensitivity but only 32% specificity. Note that, in the validation analysis, a subject was predicted as having MDD if her/his predicted probability of having MDD was greater than 50%. Raising this threshold did not improve results (data not shown). The low specificity results from false classification of the majority of healthy controls as depressed subjects. Due to the limited number of misclassified MDD patients, it becomes challenging to determine patient or site characteristics associated with the misclassification.
The low specificity may have been an effect of the imbalance in subject numbers between the two classes: ~74% of the sample were MDD patients. Literature in the machine learning field has recognized the influence of imbalanced data on the performance of most traditional machine learning methods (Sun, Wong, & Kamel, 2009). The most popular approach to handle class imbalance is the synthetic minority oversampling technique (SMOTE), which oversamples by introducing new, non-replicated minority class examples using the nearest neighbors of these cases (Chawla, Bowyer, Hall, & Kegelmeyer, 2002). The SMOTE resampling technique was used, but did not improve the performance of predicting MDD or severe MDD (data not shown). In the binary classification of MDD/healthy control, the specificity increased to 0.3–0.49 for different predictive models but at the expense of decreasing sensitivity from 0.95–0.99 to 0.51–0.74. The overall accuracy decreased as well as AUC values. Similarly, in a classification of severe MDD (i.e., 131 non-severe vs 68 severe subjects), the sensitivity increased from ~0.3 to ~0.6 but at the expense of a specificity drop from ~0.8 to ~0.6, as well as a decrease in accuracy and AUC values. Therefore, it is unlikely that this finding is a result of the imbalance, and results without using any resampling technique are presented here.
The relatively high misclassification rate in the binary classification analysis may be a result of treating all depressed patients as a single group. Depression is a heterogeneous disease. In fact, there are nearly 1,500 combinations of symptoms that meet DSM criteria for a depression diagnosis and MDD patients may share only a single symptom (Ostergaard, Jensen, & Bech, 2011). Such heterogeneity may arise from differing neurobiological underpinnings (Joober, 2013). To reduce the heterogeneity, therefore, the same models used in the binary classification were also used to determine whether neurobiology can be used to stratify individuals based on levels of depression severity (control vs mild MDD vs severe MDD). However, the best predictor model was SVM with a percentage correctly classified (PCC) close to chance (52.20 ± 1.69%). Despite the lack of predictive success, the top ranked regions overlapped with those of the binary analysis, providing some confidence in the importance of these regions as classifiers. Specifically, 11 of the 15 top ranked binary features (Table 4) are top ranked features in the ordinal (control vs mild MDD vs severe MDD) analysis (Table 6, 21 top features). Further, the top two predictive features across all models were the same as the binary analysis - mean FA in the left medial orbitofrontal cortex and the right cuneus, with average ranks of 1.67 and 4.67, respectively.
To examine whether finer resolution of depression severity is needed in order to relate severity to neurobiology, we also examined prediction of a continuous severity measure (HAMD total score). However, the highest correlation between combinations of high-ranking features and HAMD scores explained only 13% of the variance.
Although the above analyses increase in resolution (from binary classification, to three groups, to a continuous measure), they still rely on aggregate measures of depression severity, which does not overcome the issue of depression heterogeneity. We therefore sought to also examine clusters of correlated symptoms, using our previously published factor analysis of the HAMD. This is in line with NIMH’s Research Domain Criteria (RDoC (Insel & Cuthbert, 2009)), which provides a neuroscience-based approach to classifying psychopathology using an expanding set of domains relating to different functions (e.g., “anxiety” or “arousal”). These factors included psychic depression, motivation, anxiety, and sleep (excluding psychosis). However, the finer resolution of symptoms did not result in improved model accuracy, as the R2 value of the prediction was less than 0.14 in all cases.
Despite the disparate nature of the symptom categories, many of the same features were implicated in predicting each of the factors (Table 8), as well as overall severity (as assessed by the HAMD). This suggests that, despite the low accuracy of any individual model, which would prevent clinical translation, examining aggregate model results provides insight into the neurobiological underpinnings of MDD. 36.6% (11 features) were implicated across binary and severity prediction and 4 features were implicated across all measurements (Figure 2), although rankings for each feature differed across classifiers. These four features consisted of two dMRI-based measures (average FA in the right cuneus and left insula) and two sMRI-based measures (asymmetry in the volume of the pars triangularis and the cerebellum).
The cuneus is a region in the occipital lobe containing the primary visual cortex and is associated with the processing of visual cues (Parker, Zalusky, & Kirbas, 2014). White matter tracts through the cuneus connect the precuneus to the parietal lobe (Parker et al., 2014). The precuneus has been shown to be a critical component of the default mode network (DMN) (Cunningham, Tomasi, & Volkow, 2017; Fransson & Marrelec, 2008; Klaassens et al., 2017; Utevsky, Smith, & Huettel, 2014), the network of brain regions implicated in self-referential thought and activated in the absence of a specific task. MDD has been associated with an inability to downregulate the DMN (Sheline et al., 2009), which might be associated with maladaptive rumination and difficulties disengaging from negative cues. As such, connectivity from the precuneus (through the cuneus) may be altered in MDD. Further, the orbitofrontal cortex receives information regarding visual cues indirectly from the primary visual areas (Rolls, 2004a). The orbitofrontal cortex, which was an important feature in the ordinal analysis, is implicated in both reward processing and the integration of sensory and emotional information (Hare, O’Doherty, Camerer, Schultz, & Rangel, 2008; Kringelbach & Rolls, 2004; Price & Drevets, 2010; Rolls, 2004b). Obitofrontal-cuneus structural connectivity, which may affect cuneus FA, may therefore be altered in MDD. Although the group-wise FA differences in the cuneus were not significant (HC FA: 0.37±0.03, MDD FA: 0.36±0.04, p-value = 0.35), these FA measures contributed to overall classification. Potentially relating to these dMRI-based findings, cuneus volumetric asymmetry was a significant predictor in both the binary and continuous analysis. Examining the data revealed that the right cuneus volume was 3.2±11.9% larger than left cuneus volume in the controls and 5.5±11.9% larger than the left cuneus volume in the MDD cohort. This may reflect right-sided hyperactivity in MDD (Briceno et al., 2013) or right hemisphere selective involvement in processing negative emotion and negative self-referential thinking, in conjunction with left hemisphere hypoactivity and bias for positive stimuli and pleasure (Hecht, 2010).
Unlike the cuneus, group-level differences in FA were observable in the left insula at a trend level (HC FA: 0.49±0.04, MDD FA: 0.48±0.03, p-value = 0.07), although the average difference was too small to be clinically meaningful. The insula is involved in integrating sensory interoception signals, cognition and motivation (Namkung, Kim, & Sawa, 2017). As such, insula dysfunction (including structural and functional abnormalities) has been implicated in MDD (Namkung et al., 2017). The insula also has extensive connections to the DMN, and differences in connectivity between the insula and the DMN network as well as the amygdala may result in pathological inward focus in MDD (Sliz & Hayley, 2012). Further, the right and left anterior insula may respond to differing stimuli, with the left being activated by prominent sensory input and emotional feelings (Sliz & Hayley, 2012).
Consistent with the lateral findings in the cuneus, the right pars triangularis (also referred to as BA45) volume was 12.0±15.2% larger than left pars triangularis volume in the controls and 15.2±16.0% larger than the left pars triangularis volume in the MDD cohort. Reflecting this, the average pars triangularis laterality measure was negative in both cohorts (HC laterality: −7.1±8.2, MDD laterality: −9.0±9.2, p-value = 0.18). The pars triangularis is part of the inferior frontal gyrus and, along with BA44, is considered part of Broca’s area, in which language processing occurs (Ardila, Bernal, & Rosselli, 2016). Interestingly, a recent meta-analysis using activation likelihood estimation (ALE) on 28 studies including 403 participants determined that the functional connectivity network of the inferior temporal gyrus (another critical language area) in healthy controls consists of the left prefrontal cortex (including BA45), the left insula, bilateral precuneus, cerebellum and occipital areas (as well as the left temporal lobe) (Ardila, Bernal, & Rosselli, 2015). As such, this language network includes all four top predictors in this work. Dysfunction in this network may be one reason why depression is associated with slower speech and an increase in pausing (Maser, 1987). Further, changes in verbal fluency appear to be a hallmark of the disease (Lim et al., 2013).
Consistent with the lateral findings in the cuneus and pars triangularis, though to a lesser magnitude, the right cerebellum volume was 1.1±3.1% larger than left cerebellum volume in the controls and 2.3±4.4% larger than the left cerebellum volume in the MDD cohort. Reflecting this, the average cerebellum laterality measure was negative in both cohorts (HC laterality: −0.6±1.6, MDD laterality: −1.2±2.3, p-value = 0.06). The role of the cerebellum in psychiatric disorders continues to be elucidated (Baldacara, Borgio, Lacerda, & Jackowski, 2008; J. R. Phillips, Hewedi, Eissa, & Moustafa, 2015; Shakiba, 2014). In MDD, cerebellar volume may be reduced, activity may be increased and connectivity with cortical brain regions disrupted (J. R. Phillips et al., 2015).
The above suggest that these features (mean FA in the right cuneus and left insula, asymmetry in the volume of the pars triangularis and cerebellum) may play a significant role in MDD, and should be examined in future studies. Also, importantly, the significance of these features is not immediately apparent from examining them in isolation (i.e. examining group differences). This emphasizes the need for techniques examining multiple features in parallel. As the pathophysiology of MDD continues to be studied, more insight can be gained into the specific roles of these features, and importantly, the laterality effect, which is not often addressed, may be uncovered.
Limitations
Although we evaluate one of the largest MDD sMRI and dMRI imaging cohorts, across geographically diverse imaging centers, our inclusion/exclusion criteria may prevent these findings from representing all MDD patients, particularly those with comorbities or on medication, who were excluded from the current study. Further, there are numerous other measures that may be extracted from sMRI and dMRI modalities that were not examined in this study, and may yield more clinically significant results. Additionally, the choice of brain atlas for the regional analysis can impact model results. There are available atlases with finer parcellations than the Desikan-Killiany atlas used in this work. Those would increase the number of model variables (and therefore complexity) but also would allow detection of smaller regional effects that may be subsumed by large regional averages. To balance these concerns, future work could involve finer parcellations of the four regions implicated in this study. Finally, MDD is a heterogeneous disease. Though our analysis of depression factors attempts to account for this, there still may be multiple biological pathways resulting in the same symptom manifestation, which would confound study results. And, as these factors were derived from a previous study, they may not be universally applicable to all populations. For example, using Cronbach’s alpha and Loevinger’s coefficient of homogeneity (Olsen, Jensen, Noerholm, Martiny, & Bech, 2003) confirmed good internal reliability of the HAMD and factors 1 and 5, with safely acceptable reliability of factor 2 and just acceptable reliability of factor 4.
Conclusion
From this study, we can draw several important conclusions. 1) Despite our use of multiple models with differing advantages, a large training dataset, and a separate validation analysis, the final overall model performance was too low for clinical application. 2) Although four features (mean FA in the right cuneus and left insula, asymmetry in the volume of the pars triangularis and cerebellum) were implicated across all analyses, low classification and prediction accuracy using these features indicates that they cannot represent the entire pathophysiology of MDD. However, they may be relevant for future investigations of MDD neurobiology. 3) It has already been suggested that dMRI-based measures cannot be used to distinguish MDD in large samples (K. S. Choi et al., 2014) and this could be one reason for the equivocal results to date. In agreement with lack of previous consensus among sMRI and dMRI findings in MDD, the results of our powerful, comprehensive approach suggest that the sMRI and dMRI features used here may not provide a usable marker for diagnostic classification or prediction of depression severity on their own.
To improve predictive power, future work would involve utilizing these study characteristics (large cohort, multimodality features, robust methods, external validation) to combine the four sMRI and dMRI measures implicated across all analyses with other potential neurobiomarkers such as those derived from PET and/or EEG, or other behavioral measures. Such an approach could bring us closer to the first clinically relevant biomarker of MDD.
Acknowledgements
We would like to thank Rose Boyle and Alexander Hogenhuis for their diligent work performing literature searches and aiding in the development of this manuscript. We acknowledge the biostatistical consultation and support provided by the Biostatistical Consulting Core at School of Medicine, Stony Brook University. This publication was supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant number UL1 TR000040. This work was also supported by U01 MH092250 (EMBARC, PIs: Weissman, Parsey, McGrath), P50 MH62185 and R01 MH040695 (PI: Mann), K08 MH079033 (PI: Sublette), K08 MH076258 (PI: Milak), K08 MH067015 (PI: Sullivan), R01 MH074813 (PI: Parsey) as well as the Stony Brook School of Medicine and the Office of the Vice President for Research through the Targeted Research Opportunity Program – FUSION Award (PIs: DeLorenzo, Ahn, Yang).
Appendix
Table A1:
List of the 39 features used in predictive modeling
| Category | Variable |
|---|---|
| Demographics | Sex |
| Age | |
| Gray Matter Volume Asymmetry of Various Cortical Regions (sMRI) | Pars Triangularis |
| Pericalcarine | |
| Precentral | |
| Transverse Temporal | |
| Cuneus | |
| Lingual | |
| Paracentral | |
| Cerebellum | |
| Gray Matter Volume of Various Cortical Regions (sMRI) | Left Inferior Temporal |
| Pars Orbitalis | |
| Frontal Pole | |
| Pars Triangularis | |
| Gray Matter Volume of Various Subcortical Regions (sMRI) | Right Choroid Plexus |
| Volume (sMRI) | Brainstem |
| Mean Thickness of the Entire Hemispheres (sMRI) | Left Mean Thickness |
| Right Mean Thickness | |
| Mean Fractional Anisotropy of Various Regions (dMRI) | Anterior Corpus Callosum |
| Left Banks of the Superior Temporal Sulcus | |
| Left Inferior Parietal | |
| Left Insula | |
| Left Lateral Occipital | |
| Left Lateral Orbitofrontal | |
| Left Medial Orbitofrontal | |
| Left Middle Temporal | |
| Left Pars Triangularis | |
| Left Precuneus | |
| Left Transverse Temporal | |
| Left Cerebellum | |
| Right Banks of the Superior Temporal Sulcus | |
| Right Caudal Anterior Cingulate | |
| Right Cuneus | |
| Left Entorhinal | |
| Right Fusiform | |
| Right Lateral Orbitofrontal | |
| Right Lingual | |
| Right Parahippocampal | |
| Right Rostral Anterior Cingulate |
Figure A1:
Bar plot for the 5-fold cross-validated performances for each task. MDD: Major Depressive Disorder; PLR: penalized logistic regression model with elastic net penalty, SVM: support vector machine, RF: random forest; AUC: area under the curve; PCC: percentage correctly classified; RMSE: root mean squared error
Footnotes
Disclosures
Dr. Miller reports receiving financial compensation for psychiatric evaluations of patients enrolled in medication studies sponsored by Pfizer and Orexigen Therapeutics and family ownership of stock in Johnson & Johnson.
Dr. Trivedi is or has been an advisor/consultant and received fees from (lifetime disclosure): Abbott Laboratories, Inc., Abdi Ibrahim, Akzo (Organon Pharmaceuticals Inc.), Alkermes, AstraZeneca, Axon Advisors, Bristol-Myers Squibb Company, Cephalon, Inc., Cerecor, CME Institute of Physicians, Concert Pharmaceuticals, Inc., Eli Lilly & Company, Evotec, Fabre Kramer Pharmaceuticals, Inc., Forest Pharmaceuticals, GlaxoSmithKline, Janssen Global Services, LLC, Janssen Pharmaceutica Products, LP, Johnson & Johnson PRD, Libby, Lundbeck, Meade Johnson, MedAvante, Medtronic, Merck, Mitsubishi Tanabe Pharma Development America, Inc., Naurex, Neuronetics, Otsuka Pharmaceuticals, Pamlab, Parke-Davis Pharmaceuticals, Inc., Pfizer Inc., PgxHealth, Phoenix Marketing Solutions, Rexahn Pharmaceuticals, Ridge Diagnostics, Roche Products Ltd., Sepracor, SHIRE Development, Sierra, SK Life and Science, Sunovion, Takeda, Tal Medical/Puretech Venture, Targacept, Transcept, VantagePoint, Vivus, and Wyeth-Ayerst Laboratories. In addition, he has received grants/research support from: Agency for Healthcare Research and Quality (AHRQ), Cyberonics, Inc., National Alliance for Research in Schizophrenia and Depression, National Institute of Mental Health and National Institute on Drug Abuse.
In the past two years, Dr. Weissman received funding from the National Institute of Mental Health (NIMH), the National Alliance for Research on Schizophrenia and Depression (NARSAD), the Sackler Foundation, the Templeton Foundation; and receives royalties from the Oxford University Press, Perseus Press, the American Psychiatric Association Press, and MultiHealth Systems.
Dr. McGrath has received funding from the National Institute of Mental Health, New York State Department of Mental Hygiene, Research Foundation for Mental Hygiene (New York State), Forest Research Laboratories, Sunovion Pharmaceuticals, and Naurex Pharmaceuticals (now Allergan).
Dr. Fava has received research support from Abbot Laboratories; Alkermes, Inc.; American Cyanamid; Aspect Medical Systems; AstraZeneca; Avanir Pharmaceuticals; BioResearch; BrainCells Inc.; Bristol-Myers Squibb; CeNeRx BioPharma; Cephalon; Clintara, LLC; Cerecor; Covance; Covidien; Eli Lilly and Company; EnVivo Pharmaceuticals, Inc.; Euthymics Bioscience, Inc.; Forest Pharmaceuticals, Inc.; Ganeden Biotech, Inc.; GlaxoSmithKline; Harvard Clinical Research Institute; Hoffman-LaRoche; Icon Clinical Research; i3 Innovus/Ingenix; Janssen R&D, LLC; Jed Foundation; Johnson & Johnson Pharmaceutical Research & Development; Lichtwer Pharma GmbH; Lorex Pharmaceuticals; Lundbeck Inc.; MedAvante; Methylation Sciences Inc.; National Alliance for Research on Schizophrenia & Depression (NARSAD); National Center for Complementary and Alternative Medicine (NCCAM); National Institute of Drug Abuse (NIDA); National Institute of Mental Health (NIMH); Neuralstem, Inc.; Novartis AG; Organon Pharmaceuticals; PamLab, LLC.; Pfizer Inc.; Pharmacia-Upjohn; Pharmaceutical Research Associates., Inc.; Pharmavite® LLC; PharmoRx Therapeutics; Photothera; Reckitt Benckiser; Roche Pharmaceuticals; RCT Logic, LLC (formerly Clinical Trials Solutions, LLC); Sanofi-Aventis US LLC; Shire; Solvay Pharmaceuticals, Inc.; Stanley Medical Research Institute (SMRI); Synthelabo; Tal Medical; Wyeth-Ayerst Laboratories; he has served as advisor or consultant to Abbott Laboratories; Acadia; Affectis Pharmaceuticals AG; Alkermes, Inc.; Amarin Pharma Inc.; Aspect Medical Systems; AstraZeneca; Auspex Pharmaceuticals; Avanir Pharmaceuticals; AXSOME Therapeutics; Bayer AG; Best Practice Project Management, Inc.; Biogen; BioMarin Pharmaceuticals, Inc.; Biovail Corporation; BrainCells Inc; Bristol-Myers Squibb; CeNeRx BioPharma; Cephalon, Inc.; Cerecor; CNS Response, Inc.; Compellis Pharmaceuticals; Cypress Pharmaceutical, Inc.; DiagnoSearch Life Sciences (P) Ltd.; Dinippon Sumitomo Pharma Co. Inc.; Dov Pharmaceuticals, Inc.; Edgemont Pharmaceuticals, Inc.; Eisai Inc.; Eli Lilly and Company; EnVivo Pharmaceuticals, Inc.; ePharmaSolutions; EPIX Pharmaceuticals, Inc.; Euthymics Bioscience, Inc.; Fabre-Kramer Pharmaceuticals, Inc.; Forest Pharmaceuticals, Inc.; Forum Pharmaceuticals; GenOmind, LLC; GlaxoSmithKline; Grunenthal GmbH; i3 Innovus/Ingenis; Intracellular; Janssen Pharmaceutica; Jazz Pharmaceuticals, Inc.; Johnson & Johnson Pharmaceutical Research & Development, LLC; Knoll Pharmaceuticals Corp.; Labopharm Inc.; Lorex Pharmaceuticals; Lundbeck Inc.; MedAvante, Inc.; Merck & Co., Inc.; MSI Methylation Sciences, Inc.; Naurex, Inc.; Nestle Health Sciences; Neuralstem, Inc.; Neuronetics, Inc.; NextWave Pharmaceuticals; Novartis AG; Nutrition 21; Orexigen Therapeutics, Inc.; Organon Pharmaceuticals; Osmotica; Otsuka Pharmaceuticals; Pamlab, LLC.; Pfizer Inc.; PharmaStar; Pharmavite® LLC.; PharmoRx Therapeutics; Precision Human Biolaboratory; Prexa Pharmaceuticals, Inc.; Puretech Ventures; PsychoGenics; Psylin Neurosciences, Inc.; RCT Logic, LLC Formerly Clinical Trials Solutions, LLC; Rexahn Pharmaceuticals, Inc.; Ridge Diagnostics, Inc.; Roche; Sanofi-Aventis US LLC.; Sepracor Inc.; Servier Laboratories; Schering-Plough Corporation; Solvay Pharmaceuticals, Inc.; Somaxon Pharmaceuticals, Inc.; Somerset Pharmaceuticals, Inc.; Sunovion Pharmaceuticals; Supernus Pharmaceuticals, Inc.; Synthelabo; Taisho Pharmaceutical; Takeda Pharmaceutical Company Limited; Tal Medical, Inc.; Tetragenex Pharmaceuticals, Inc.; TransForm Pharmaceuticals, Inc.; Transcept Pharmaceuticals, Inc.; Vanda Pharmaceuticals, Inc.; VistaGen; he has received speaking or publishing fees from Adamed, Co; Advanced Meeting Partners; American Psychiatric Association; American Society of Clinical Psychopharmacology; AstraZeneca; Belvoir Media Group; Boehringer Ingelheim GmbH; Bristol-Myers Squibb; Cephalon, Inc.; CME Institute/Physicians Postgraduate Press, Inc.; Eli Lilly and Company; Forest Pharmaceuticals, Inc.; GlaxoSmithKline; Imedex, LLC; MGH Psychiatry Academy/Primedia; MGH Psychiatry Academy/Reed Elsevier; Novartis AG; Organon Pharmaceuticals; Pfizer Inc.; PharmaStar; United BioSource, Corp.; Wyeth-Ayerst Laboratories; he has equity holdings in Compellis and PsyBrain, Inc.; he has a patent for Sequential Parallel Comparison Design (SPCD), which are licensed by MGH to Pharmaceutical Product Development, LLC (PPD); and patent application for a combination of Ketamine plus Scopolamine in Major Depressive Disorder (MDD), licensed by MGH to Biohaven; and he receives copyright royalties for the MGH Cognitive & Physical Functioning Questionnaire (CPFQ), Sexual Functioning Inventory (SFI), Antidepressant Treatment Response Questionnaire (ATRQ), Discontinuation-Emergent Signs & Symptoms (DESS), Symptoms of Depression Questionnaire (SDQ), and SAFER; Lippincott, Williams & Wilkins; Wolkers Kluwer; World Scientific Publishing Co. Pte. Ltd.
Dr. Kurian has received research grant support from the following organizations: Targacept, Inc., Pfizer, Inc., Johnson & Johnson, Evotec, Rexahn, Naurex, Forest Pharmaceuticals and the National Institute of Mental Health (NIMH). Mary L. Phillips has received funding from NIMH and the Emmerling-Pittsburgh Foundation.
Over the past three years, Dr. Pizzagalli has received honoraria/consulting fees from Akili Interactive Labs, BlackThorn Therapeutics, Pfizer, and Posit Science.
Dr. Oquendo receives royalties for use of the Columbia Suicide Severity Rating Scale. Her family owns stock in Bristol Myers Squibb.
Dr. Mann receives royalties for commercial use of the Columbia Suicide Severity Rating Scales from the Research Foundation for Mental Hygiene.
No other disclosures are reported.
References:
- Abe O, Yamasue H, Kasai K, Yamada H, Aoki S, Inoue H, … Ohtomo K (2010). Voxel-based analyses of gray/white matter volume and diffusion tensor data in major depression. Psychiatry Res, 181(1), 64–70. doi: 10.1016/j.pscychresns.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Aghajani M, Veer IM, van Lang ND, Meens PH, van den Bulk BG, Rombouts SA, … van der Wee NJ (2014). Altered white-matter architecture in treatment-naive adolescents with clinical depression. Psychol Med, 44(11), 2287–2298. doi: 10.1017/S0033291713003000 [DOI] [PubMed] [Google Scholar]
- Aizenstein HJ, Khalaf A, Walker SE, & Andreescu C (2014). Magnetic resonance imaging predictors of treatment response in late-life depression. J Geriatr Psychiatry Neurol, 27(1), 24–32. doi: 10.1177/0891988713516541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DC, & Barker GJ (2005). Optimal imaging parameters for fiber-orientation estimation in diffusion MRI. Neuroimage, 27(2), 357–367. doi: 10.1016/j.neuroimage.2005.04.008 [DOI] [PubMed] [Google Scholar]
- Amico F, Meisenzahl E, Koutsouleris N, Reiser M, Moller HJ, & Frodl T (2011). Structural MRI correlates for vulnerability and resilience to major depressive disorder. J Psychiatry Neurosci, 36(1), 15–22. doi: 10.1503/jpn.090186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbabshirani MR, Plis S, Sui J, & Calhoun VD (2016). Single subject prediction of brain disorders in neuroimaging: Promises and pitfalls. Neuroimage. doi: 10.1016/j.neuroimage.2016.02.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila A, Bernal B, & Rosselli M (2015). Language and visual perception associations: meta-analytic connectivity modeling of Brodmann area 37. Behav Neurol, 2015, 565871. doi: 10.1155/2015/565871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila A, Bernal B, & Rosselli M (2016). How Localized are Language Brain Areas? A Review of Brodmann Areas Involvement in Oral Language. Arch Clin Neuropsychol, 31(1), 112–122. doi: 10.1093/arclin/acv081 [DOI] [PubMed] [Google Scholar]
- Austin PC, & Steyerberg EW (2014). Graphical assessment of internal and external calibration of logistic regression models by using loess smoothers. Stat Med, 33(3), 517–535. doi: 10.1002/sim.5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldacara L, Borgio JG, Lacerda AL, & Jackowski AP (2008). Cerebellum and psychiatric disorders. Rev Bras Psiquiatr, 30(3), 281–289. [DOI] [PubMed] [Google Scholar]
- Bijanki KR, Hodis B, Brumm MC, Harlynn EL, & McCormick LM (2014). Hippocampal and left subcallosal anterior cingulate atrophy in psychotic depression. PLoS One, 9(10), e110770. doi: 10.1371/journal.pone.0110770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breiman L (2001). Random forests. Machine Learning, 45(1), 5–32. doi:Doi 10.1023/A:1010933404324 [DOI] [Google Scholar]
- Briceno EM, Weisenbach SL, Rapport LJ, Hazlett KE, Bieliauskas LA, Haase BD, … Langenecker SA (2013). Shifted inferior frontal laterality in women with major depressive disorder is related to emotion-processing deficits. Psychol Med, 43(7), 1433–1445. doi: 10.1017/S0033291712002176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chawla NV, Bowyer KW, Hall LO, & Kegelmeyer WP (2002). SMOTE: synthetic minority over-sampling technique. Journal of Artificial Intelligence Research, 16, 321–357. [Google Scholar]
- Cherbuin N, Reglade-Meslin C, Kumar R, Sachdev P, & Anstey KJ (2010). Mild Cognitive Disorders are Associated with Different Patterns of Brain asymmetry than Normal Aging: The PATH through Life Study. Front Psychiatry, 1, 11. doi: 10.3389/fpsyt.2010.00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Craddock RC, Holtzheimer PE, Yang Z, Hu X, Mayberg H (2008). A Combined Functional–Structural Connectivity Analysis of Major Depression Using Joint Independent Components Analysis. Psychiatric MRI/MRS, 16, 3–9. [Google Scholar]
- Choi KS, Holtzheimer PE, Franco AR, Kelley ME, Dunlop BW, Hu XP, & Mayberg HS (2014). Reconciling variable findings of white matter integrity in major depressive disorder. Neuropsychopharmacology, 39(6), 1332–1339. doi: 10.1038/npp.2013.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham SI, Tomasi D, & Volkow ND (2017). Structural and functional connectivity of the precuneus and thalamus to the default mode network. Hum Brain Mapp, 38(2), 938–956. doi: 10.1002/hbm.23429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale AM, Fischl B, & Sereno MI (1999). Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage, 9(2), 179–194. doi: 10.1006/nimg.1998.0395 [DOI] [PubMed] [Google Scholar]
- Delaparte L, Yeh FC, Adams P, Malchow A, Trivedi MH, Oquendo MA, … DeLorenzo C (2017). A comparison of structural connectivity in anxious depression versus non-anxious depression. J Psychiatr Res, 89, 38–47. doi: 10.1016/j.jpsychires.2017.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorenzo C, Delaparte L, Thapa-Chhetry B, Miller JM, Mann JJ, & Parsey RV (2013). Prediction of selective serotonin reuptake inhibitor response using diffusion-weighted MRI. Front Psychiatry, 4, 5. doi: 10.3389/fpsyt.2013.00005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desikan RS, Segonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, … Killiany RJ (2006). An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 31(3), 968–980. doi: 10.1016/j.neuroimage.2006.01.021 [DOI] [PubMed] [Google Scholar]
- Dudoit S, & Fridlyand J (2003). Classification in microarray experiments In Speed TP (Ed.), Statistical analysis of gene expression microarray data (pp. 93–158). Boca Raton, FL: Chapman & Hall/CRC. [Google Scholar]
- Eker C, & Gonul AS (2010). Volumetric MRI studies of the hippocampus in major depressive disorder: Meanings of inconsistency and directions for future research. World J Biol Psychiatry, 11(1), 19–35. doi: 10.1080/15622970902737998 [DOI] [PubMed] [Google Scholar]
- Fallucca E, MacMaster FP, Haddad J, Easter P, Dick R, May G, … Rosenberg DR (2011). Distinguishing between major depressive disorder and obsessive-compulsive disorder in children by measuring regional cortical thickness. Arch Gen Psychiatry, 68(5), 527–533. doi: 10.1001/archgenpsychiatry.2011.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Feng Y, & Song R (2011). Nonparametric independence screening in sparse ultra-high-dimensional additive models. Journal of the American Statistical Association, 106(494), 544–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, & Li R (2001). Variable selection via nonconcave penalized likelihood and its oracle properties. Journal of the American Statistical Association, 96(456), 1348–1360. [Google Scholar]
- Fan J, Samworth RJ, & Wu Y (2009). Ultrahigh dimensional feature selection: beyond the linear model. Journal of Machine Learning Research, 10, 2013–2038. [PMC free article] [PubMed] [Google Scholar]
- Fischl B, & Dale AM (2000). Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci U S A, 97(20), 11050–11055. doi: 10.1073/pnas.200033797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Liu A, & Dale AM (2001). Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging, 20(1), 70–80. doi: 10.1109/42.906426 [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, … Dale AM (2002). Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron, 33(3), 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, & Dale AM (1999). Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage, 9(2), 195–207. doi: 10.1006/nimg.1998.0396 [DOI] [PubMed] [Google Scholar]
- Fischl B, Sereno MI, Tootell RB, & Dale AM (1999). High-resolution intersubject averaging and a coordinate system for the cortical surface. Hum Brain Mapp, 8(4), 272–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, … Dale AM (2004). Automatically parcellating the human cerebral cortex. Cereb Cortex, 14(1), 11–22. [DOI] [PubMed] [Google Scholar]
- Fransson P, & Marrelec G (2008). The precuneus/posterior cingulate cortex plays a pivotal role in the default mode network: Evidence from a partial correlation network analysis. Neuroimage, 42(3), 1178–1184. doi: 10.1016/j.neuroimage.2008.05.059 [DOI] [PubMed] [Google Scholar]
- Fried EI, Nesse RM, Zivin K, Guille C, & Sen S (2014). Depression is more than the sum score of its parts: individual DSM symptoms have different risk factors. Psychol Med, 44(10), 2067–2076. doi: 10.1017/S0033291713002900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Burden of Disease Study 2013 Collaborators. (2015). Global, regional, and national incidence, prevalence, and years lived with disability for 301 acute and chronic diseases and injuries in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013. Lancet, 386(9995), 743–800. doi: 10.1016/S0140-6736(15)60692-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez de Castro D, Clarke PA, Al-Lazikani B, & Workman P (2013). Personalized cancer medicine: molecular diagnostics, predictive biomarkers, and drug resistance. Clin Pharmacol Ther, 93(3), 252–259. doi: 10.1038/clpt.2012.237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieve SM, Korgaonkar MS, Koslow SH, Gordon E, & Williams LM (2013). Widespread reductions in gray matter volume in depression. Neuroimage Clin, 3, 332–339. doi: 10.1016/j.nicl.2013.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Liu F, Yu M, Zhang J, Zhang Z, Liu J, … Zhao J (2014). Functional and anatomical brain deficits in drug-naive major depressive disorder. Prog Neuropsychopharmacol Biol Psychiatry, 54, 1–6. doi: 10.1016/j.pnpbp.2014.05.008 [DOI] [PubMed] [Google Scholar]
- Hamilton M (1960). A rating scale for depression. J Neurol Neurosurg Psychiatry, 23, 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han KM, Choi S, Jung J, Na KS, Yoon HK, Lee MS, & Ham BJ (2014). Cortical thickness, cortical and subcortical volume, and white matter integrity in patients with their first episode of major depression. J Affect Disord, 155, 42–48. doi: 10.1016/j.jad.2013.10.021 [DOI] [PubMed] [Google Scholar]
- Hare TA, O’Doherty J, Camerer CF, Schultz W, & Rangel A (2008). Dissociating the role of the orbitofrontal cortex and the striatum in the computation of goal values and prediction errors. J Neurosci, 28(22), 5623–5630. doi: 10.1523/JNEUROSCI.1309-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie T, Tibshirani R, & Friedman JH (2001). The elements of statistical learning : data mining, inference, and prediction : with 200 full-color illustrations. New York: Springer. [Google Scholar]
- Hecht D (2010). Depression and the hyperactive right-hemisphere. Neurosci Res, 68(2), 77–87. doi: 10.1016/j.neures.2010.06.013 [DOI] [PubMed] [Google Scholar]
- Henderson SE, Johnson AR, Vallejo AI, Katz L, Wong E, & Gabbay V (2013). A preliminary study of white matter in adolescent depression: relationships with illness severity, anhedonia, and irritability. Front Psychiatry, 4, 152. doi: 10.3389/fpsyt.2013.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofner B, Boccuto L, & Goker M (2015). Controlling false discoveries in high-dimensional situations: boosting with stability selection. BMC Bioinformatics, 16, 144. doi: 10.1186/s12859-015-0575-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofner B, & Hothorn T (2017). stabs: Stability Selection with Error Control, R package version R package version 0.6–2. Retrieved from https://CRAN.R-project.org/package=stabs
- Huang Y, Coupland NJ, Lebel RM, Carter R, Seres P, Wilman AH, & Malykhin NV (2013). Structural changes in hippocampal subfields in major depressive disorder: a high-field magnetic resonance imaging study. Biol Psychiatry, 74(1), 62–68. doi: 10.1016/j.biopsych.2013.01.005 [DOI] [PubMed] [Google Scholar]
- Insel TR, & Cuthbert BN (2009). Endophenotypes: bridging genomic complexity and disorder heterogeneity. Biol Psychiatry, 66(11), 988–989. doi: 10.1016/j.biopsych.2009.10.008 [DOI] [PubMed] [Google Scholar]
- Iscan Z, Jin TB, Kendrick A, Szeglin B, Lu H, Trivedi M, … DeLorenzo C (2015). Test-retest reliability of freesurfer measurements within and between sites: Effects of visual approval process. Hum Brain Mapp, 36(9), 3472–3485. doi: 10.1002/hbm.22856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska N, MacMaster FP, Gaxiola I, Cortese F, Goodyear B, & Ramasubbu R (2014). A preliminary study of the influence of age of onset and childhood trauma on cortical thickness in major depressive disorder. Biomed Res Int, 2014, 410472. doi: 10.1155/2014/410472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworska N, MacMaster FP, Yang XR, Courtright A, Pradhan S, Gaxiola I, … Ramasubbu R (2014). Influence of age of onset on limbic and paralimbic structures in depression. Psychiatry Clin Neurosci, 68(12), 812–820. doi: 10.1111/pcn.12197 [DOI] [PubMed] [Google Scholar]
- Jolliffe IT (2002). Principal component analysis (2nd ed.). New York: Springer. [Google Scholar]
- Jones DK, & Basser PJ (2004). “Squashing peanuts and smashing pumpkins”: how noise distorts diffusion-weighted MR data. Magn Reson Med, 52(5), 979–993. doi: 10.1002/mrm.20283 [DOI] [PubMed] [Google Scholar]
- Joober R (2013). On the simple and the complex in psychiatry, with reference to DSM 5 and research domain criteria. J Psychiatry Neurosci, 38(3), 148–151. doi: 10.1503/jpn.130051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Amminger GP, Aguilar-Gaxiola S, Alonso J, Lee S, & Ustun TB (2007). Age of onset of mental disorders: a review of recent literature. Curr Opin Psychiatry, 20(4), 359–364. doi: 10.1097/YCO.0b013e32816ebc8c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, & Walters EE (2005). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry, 62(6), 593–602. doi: 10.1001/archpsyc.62.6.593 [DOI] [PubMed] [Google Scholar]
- Kessler RC, Chiu WT, Demler O, Merikangas KR, & Walters EE (2005). Prevalence, severity, and comorbidity of 12-month DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry, 62(6), 617–627. doi: 10.1001/archpsyc.62.6.617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieseppa T, Eerola M, Mantyla R, Neuvonen T, Poutanen VP, Luoma K, … Isometsa E (2010). Major depressive disorder and white matter abnormalities: a diffusion tensor imaging study with tract-based spatial statistics. J Affect Disord, 120(1–3), 240–244. doi: 10.1016/j.jad.2009.04.023 [DOI] [PubMed] [Google Scholar]
- Klaassens BL, van Gerven JMA, van der Grond J, de Vos F, Moller C, & Rombouts S (2017). Diminished Posterior Precuneus Connectivity with the Default Mode Network Differentiates Normal Aging from Alzheimer’s Disease. Front Aging Neurosci, 9, 97. doi: 10.3389/fnagi.2017.00097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korgaonkar MS, Grieve SM, Koslow SH, Gabrieli JD, Gordon E, & Williams LM (2011). Loss of white matter integrity in major depressive disorder: evidence using tract-based spatial statistical analysis of diffusion tensor imaging. Hum Brain Mapp, 32(12), 2161–2171. doi: 10.1002/hbm.21178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kringelbach ML, & Rolls ET (2004). The functional neuroanatomy of the human orbitofrontal cortex: evidence from neuroimaging and neuropsychology. Prog Neurobiol, 72(5), 341–372. doi: 10.1016/j.pneurobio.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Kruijshaar ME, Barendregt J, Vos T, de Graaf R, Spijker J, & Andrews G (2005). Lifetime prevalence estimates of major depression: an indirect estimation method and a quantification of recall bias. Eur J Epidemiol, 20(1), 103–111. [DOI] [PubMed] [Google Scholar]
- Kupfer DJ, Frank E, & Phillips ML (2012). Major depressive disorder: new clinical, neurobiological, and treatment perspectives. Lancet, 379(9820), 1045–1055. doi: 10.1016/S0140-6736(11)60602-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Huang X, Wu Q, Yang C, Kuang W, Du M, … Gong Q (2013). Is depression a disconnection syndrome? Meta-analysis of diffusion tensor imaging studies in patients with MDD. J Psychiatry Neurosci, 38(1), 49–56. doi: 10.1503/jpn.110180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Oh IK, Han C, Huh YJ, Jung IK, Patkar AA, … Jang BH (2013). Sensitivity of cognitive tests in four cognitive domains in discriminating MDD patients from healthy controls: a meta-analysis. Int Psychogeriatr, 25(9), 1543–1557. doi: 10.1017/S1041610213000689 [DOI] [PubMed] [Google Scholar]
- Liu Z, Wang Y, Gerig G, Gouttard S, Tao R, Fletcher T, & Styner M (2010). Quality control of diffusion weighted images. Paper presented at the SPIE Medical Imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzetti V, Allen NB, Fornito A, & Yucel M (2009). Structural brain abnormalities in major depressive disorder: a selective review of recent MRI studies. J Affect Disord, 117(1–2), 1–17. doi: 10.1016/j.jad.2008.11.021 [DOI] [PubMed] [Google Scholar]
- Mackin RS, Tosun D, Mueller SG, Lee JY, Insel P, Schuff N, … Weiner MW (2013). Patterns of reduced cortical thickness in late-life depression and relationship to psychotherapeutic response. Am J Geriatr Psychiatry, 21(8), 794–802. doi:10.1016/j.jagp.2013.01.01310.1016/j.jagp.2013.01.01310.1097/JGP.0b013e31825485a110.1097/JGP.0b013e31825485a1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan CR (2017). Advances in Studying Brain Morphology: The Benefits of Open-Access Data. Front Hum Neurosci, 11, 405. doi: 10.3389/fnhum.2017.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser JD (1987). Depression and expressive behavior. Hillsdale, N.J.: L. Erlbaum Associates. [Google Scholar]
- Matthews SC, Strigo IA, Simmons AN, O’Connell RM, Reinhardt LE, & Moseley SA (2011). A multimodal imaging study in U.S. veterans of Operations Iraqi and Enduring Freedom with and without major depression after blast-related concussion. Neuroimage, 54 Suppl 1, S69–75. doi: 10.1016/j.neuroimage.2010.04.269 [DOI] [PubMed] [Google Scholar]
- Meinshausen N, & Bühlmann P (2010). Stability selection. Journal of the Royal Statistical Society: Series B (Statistical Methodology), 72(4), 417–473. [Google Scholar]
- Mihaly Z, Kormos M, Lanczky A, Dank M, Budczies J, Szasz MA, & Gyorffy B (2013). A meta-analysis of gene expression-based biomarkers predicting outcome after tamoxifen treatment in breast cancer. Breast Cancer Res Treat, 140(2), 219–232. doi: 10.1007/s10549-013-2622-y [DOI] [PubMed] [Google Scholar]
- Milak MS, Parsey RV, Keilp J, Oquendo MA, Malone KM, & Mann JJ (2005). Neuroanatomic correlates of psychopathologic components of major depressive disorder. Archives of General Psychiatry, 62(4), 397–408. doi:Doi 10.1001/Archpsyc.62.4.397 [DOI] [PubMed] [Google Scholar]
- Mossner R, Mikova O, Koutsilieri E, Saoud M, Ehlis AC, Muller N, … Riederer P (2007). Consensus paper of the WFSBP Task Force on Biological Markers: biological markers in depression. World J Biol Psychiatry, 8(3), 141–174. doi: 10.1080/15622970701263303 [DOI] [PubMed] [Google Scholar]
- Murphy ML, & Frodl T (2011). Meta-analysis of diffusion tensor imaging studies shows altered fractional anisotropy occurring in distinct brain areas in association with depression. Biol Mood Anxiety Disord, 1(1), 3. doi: 10.1186/2045-5380-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJ, & Lopez AD (1996). Evidence-based health policy--lessons from the Global Burden of Disease Study. Science, 274(5288), 740–743. [DOI] [PubMed] [Google Scholar]
- Mwangi B, Ebmeier K, Matthews K, & Steele J (2012). Multi-center diagnostic classification of individual structural neuroimaging scans from patients with major depressive disorder. Brain, 135(5), 1508–1521. [DOI] [PubMed] [Google Scholar]
- Namkung H, Kim SH, & Sawa A (2017). The Insula: An Underestimated Brain Area in Clinical Neuroscience, Psychiatry, and Neurology. Trends Neurosci, 40(4), 200–207. doi: 10.1016/j.tins.2017.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA, … Radeke MJ (2012). Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med, 4(2), 16. doi: 10.1186/gm315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen LR, Jensen DV, Noerholm V, Martiny K, & Bech P (2003). The internal and external validity of the Major Depression Inventory in measuring severity of depressive states. Psychol Med, 33(2), 351–356. [DOI] [PubMed] [Google Scholar]
- Olvet DM, Delaparte L, Yeh FC, DeLorenzo C, McGrath PJ, Weissman MM, … Parsey RV (2016). A Comprehensive Examination of White Matter Tracts and Connectometry in Major Depressive Disorder. Depress Anxiety, 33(1), 56–65. doi: 10.1002/da.22445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvet DM, Peruzzo D, Thapa-Chhetry B, Sublette ME, Sullivan GM, Oquendo MA, … Parsey RV (2014). A diffusion tensor imaging study of suicide attempters. J Psychiatr Res, 51, 60–67. doi: 10.1016/j.jpsychires.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osoba A, Hanggi J, Li M, Horn DI, Metzger C, Eckert U, … Walter M (2013). Disease severity is correlated to tract specific changes of fractional anisotropy in MD and CM thalamus--a DTI study in major depressive disorder. J Affect Disord, 149(1–3), 116–128. doi: 10.1016/j.jad.2012.12.026 [DOI] [PubMed] [Google Scholar]
- Ostergaard SD, Jensen SO, & Bech P (2011). The heterogeneity of the depressive syndrome: when numbers get serious. Acta Psychiatr Scand, 124(6), 495–496. doi: 10.1111/j.1600-0447.2011.01744.x [DOI] [PubMed] [Google Scholar]
- Palucha A, & Pilc A (2007). Metabotropic glutamate receptor ligands as possible anxiolytic and antidepressant drugs. Pharmacol Ther, 115(1), 116–147. doi: 10.1016/j.pharmthera.2007.04.007 [DOI] [PubMed] [Google Scholar]
- Parker JG, Zalusky EJ, & Kirbas C (2014). Functional MRI mapping of visual function and selective attention for performance assessment and presurgical planning using conjunctive visual search. Brain Behav, 4(2), 227–237. doi: 10.1002/brb3.213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel MJ, Andreescu C, Price JC, Edelman KL, Reynolds CF, & Aizenstein HJ (2015). Machine learning approaches for integrating clinical and imaging features in latelife depression classification and response prediction. Int. J. Geriatr. Psychiatry, 30, 1056–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten SB (2003). Recall bias and major depression lifetime prevalence. Soc Psychiatry Psychiatr Epidemiol, 38(6), 290–296. doi: 10.1007/s00127-003-0649-9 [DOI] [PubMed] [Google Scholar]
- Peng HJ, Zheng HR, Ning YP, Zhang Y, Shan BC, Zhang L, … Li LJ (2013). Abnormalities of cortical-limbic-cerebellar white matter networks may contribute to treatment-resistant depression: a diffusion tensor imaging study. BMC Psychiatry, 13, 72. doi: 10.1186/1471-244x-13-72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman G, Bartlett E, DeLorenzo C, Weissman M, McGrath P, Ogden T, … Parsey R (2017). Cortical thickness is not associated with current depression in a clinical treatment study. Hum Brain Mapp, 38(9), 4370–4385. doi: 10.1002/hbm.23664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BS, Warner V, Bansal R, Zhu H, Hao X, Liu J, … Weissman MM (2009). Cortical thinning in persons at increased familial risk for major depression. Proc Natl Acad Sci U S A, 106(15), 6273–6278. doi: 10.1073/pnas.0805311106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BS, & Weissman MM (2011). A brain-based endophenotype for major depressive disorder. Annual review of medicine, 62, 461–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JR, Hewedi DH, Eissa AM, & Moustafa AA (2015). The cerebellum and psychiatric disorders. Front Public Health, 3, 66. doi: 10.3389/fpubh.2015.00066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips ML (2012). Neuroimaging in psychiatry: bringing neuroscience into clinical practice. Br J Psychiatry, 201(1), 1–3. doi: 10.1192/bjp.bp.112.109587 [DOI] [PubMed] [Google Scholar]
- Price JL, & Drevets WC (2010). Neurocircuitry of mood disorders. Neuropsychopharmacology, 35(1), 192–216. doi: 10.1038/npp.2009.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Huang X, Zhang J, Wang Y, Kuang W, Li J, … Gong Q (2014). Characterization of major depressive disorder using a multiparametric classification approach based on high resolution structural images. J Psychiatry Neurosci, 39(2), 78–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L, Lui S, Kuang W, Huang X, Li J, Li J, … Gong Q (2014). Regional increases of cortical thickness in untreated, first-episode major depressive disorder. Transl Psychiatry, 4, e378. doi: 10.1038/tp.2014.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2015). A language and environment for statistical computing. Retrieved from https://www.R-project.org/.
- Reynolds S, Carrey N, Jaworska N, Langevin LM, Yang XR, & Macmaster FP (2014). Cortical thickness in youth with major depressive disorder. BMC Psychiatry, 14, 83. doi: 10.1186/1471-244X-14-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk MM, Rubin-Falcone H, Keilp J, Miller JM, Sublette ME, Burke A, … Mann JJ (2017). White matter correlates of impaired attention control in major depressive disorder and healthy volunteers. J Affect Disord, 222, 103–111. doi: 10.1016/j.jad.2017.06.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolls ET (2004a). Convergence of sensory systems in the orbitofrontal cortex in primates and brain design for emotion. Anat Rec A Discov Mol Cell Evol Biol, 281(1), 1212–1225. doi: 10.1002/ar.a.20126 [DOI] [PubMed] [Google Scholar]
- Rolls ET (2004b). The functions of the orbitofrontal cortex. Brain Cogn, 55(1), 11–29. doi: 10.1016/S0278-2626(03)00277-X [DOI] [PubMed] [Google Scholar]
- Schmaal L, Hibar DP, Samann PG, Hall GB, Baune BT, Jahanshad N, … Veltman DJ (2016). Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol Psychiatry. doi: 10.1038/mp.2016.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segonne F, Dale AM, Busa E, Glessner M, Salat D, Hahn HK, & Fischl B (2004). A hybrid approach to the skull stripping problem in MRI. Neuroimage, 22(3), 1060–1075. doi: 10.1016/j.neuroimage.2004.03.032 [DOI] [PubMed] [Google Scholar]
- Segonne F, Pacheco J, & Fischl B (2007). Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans Med Imaging, 26(4), 518–529. doi: 10.1109/TMI.2006.887364 [DOI] [PubMed] [Google Scholar]
- Serpa M, Ou Y, Schaufelberger M, Doshi J, Menezes P, Scazufca M, … Zanetti M (2014). Neuroanatomical classification in a population-based sample of psychotic major depression and bipolar I disorder with 1 year of diagnostic stability. Biomed. Res. Int, 706157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton CE, Allan CL, Le Masurier M, McDermott LM, Kalu UG, Herrmann LL, … Ebmeier KP (2012). Magnetic resonance imaging in late-life depression: multimodal examination of network disruption. Arch Gen Psychiatry, 69(7), 680–689. doi: 10.1001/archgenpsychiatry.2011.1862 [DOI] [PubMed] [Google Scholar]
- Sexton CE, Mackay CE, & Ebmeier KP (2009). A systematic review of diffusion tensor imaging studies in affective disorders. Biol Psychiatry, 66(9), 814–823. doi: 10.1016/j.biopsych.2009.05.024 [DOI] [PubMed] [Google Scholar]
- Shah RD, & Samworth RJ (2013). Variable selection with error control: another look at stability selection. Journal of the Royal Statisical Society, 75(1), 55–80. [Google Scholar]
- Shakiba A (2014). The role of the cerebellum in neurobiology of psychiatric disorders. Neurol Clin, 32(4), 1105–1115. doi: 10.1016/j.ncl.2014.07.008 [DOI] [PubMed] [Google Scholar]
- Shao J (1993). Linear Model Selection by Cross-Validation Journal of the American Statistical Association, 88(422), 486–494. [Google Scholar]
- Sheline YI, Barch DM, Price JL, Rundle MM, Vaishnavi SN, Snyder AZ, … Raichle ME (2009). The default mode network and self-referential processes in depression. Proc Natl Acad Sci U S A, 106(6), 1942–1947. doi: 10.1073/pnas.0812686106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizukuishi T, Abe O, & Aoki S (2013). Diffusion tensor imaging analysis for psychiatric disorders. Magn Reson Med Sci, 12(3), 153–159. [DOI] [PubMed] [Google Scholar]
- Singh I, & Rose N (2009). Biomarkers in psychiatry. Nature, 460(7252), 202–207. doi: 10.1038/460202a [DOI] [PubMed] [Google Scholar]
- Sled JG, Zijdenbos AP, & Evans AC (1998). A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging, 17(1), 87–97. doi: 10.1109/42.668698 [DOI] [PubMed] [Google Scholar]
- Sliz D, & Hayley S (2012). Major depressive disorder and alterations in insular cortical activity: a review of current functional magnetic imaging research. Front Hum Neurosci, 6, 323. doi: 10.3389/fnhum.2012.00323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, … Kattan MW (2010). Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology, 21(1), 128–138. doi: 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Huster R, Yu Q, Segall JM, & Calhoun VD (2013). Function-structure associations of the brain: Evidence from multimodal connectivity and covariance studies. Neuroimage. doi: 10.1016/j.neuroimage.2013.09.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wong AKC, & Kamel MS (2009). Classification of imbalanced data: a review. International Journal of Pattern Recognition and Artificial Intelligence, 23(4), 687–719. [Google Scholar]
- Takahashi T, Yucel M, Lorenzetti V, Walterfang M, Kawasaki Y, Whittle S, … Allen NB (2010). An MRI study of the superior temporal subregions in patients with current and past major depression. Prog Neuropsychopharmacol Biol Psychiatry, 34(1), 98–103. doi: 10.1016/j.pnpbp.2009.10.005 [DOI] [PubMed] [Google Scholar]
- Takayanagi Y, Spira AP, Roth KB, Gallo JJ, Eaton WW, & Mojtabai R (2014). Accuracy of reports of lifetime mental and physical disorders: results from the Baltimore Epidemiological Catchment Area study. JAMA Psychiatry, 71(3), 273–280. doi: 10.1001/jamapsychiatry.2013.3579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R (1996). Regression shrinkage and selection via the lasso. Journal of the Royal Statistical Society, Series B (Methodological), 58(1), 267–288. [Google Scholar]
- Treadway MT, Waskom ML, Dillon DG, Holmes AJ, Park MTM, Chakravarty MM, … Pizzagalli DA (2015). Illness progression, recent stress, and morphometry of hippocampal subfields and medial prefrontal cortex in major depression. Biol Psychiatry, 77(3), 285–294. doi: 10.1016/j.biopsych.2014.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi MH, McGrath PJ, Fava M, Parsey RV, Kurian BT, Phillips ML, … Weissman MM (2016). Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): Rationale and design. J Psychiatr Res, 78, 11–23. doi: 10.1016/j.jpsychires.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu PC, Chen LF, Hsieh JC, Bai YM, Li CT, & Su TP (2012). Regional cortical thinning in patients with major depressive disorder: a surface-based morphometry study. Psychiatry Res, 202(3), 206–213. doi: 10.1016/j.pscychresns.2011.07.011 [DOI] [PubMed] [Google Scholar]
- Ugwu ID, Amico F, Carballedo A, Fagan AJ, & Frodl T (2015). Childhood adversity, depression, age and gender effects on white matter microstructure: a DTI study. Brain Struct Funct, 220(4), 1997–2009. doi: 10.1007/s00429-014-0769-x [DOI] [PubMed] [Google Scholar]
- Utevsky AV, Smith DV, & Huettel SA (2014). Precuneus is a functional core of the default-mode network. J Neurosci, 34(3), 932–940. doi: 10.1523/JNEUROSCI.4227-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Calster B, Nieboer D, Vergouwe Y, De Cock B, Pencina MJ, & Steyerberg EW (2016). A calibration hierarchy for risk models was defined: from utopia to empirical data. J Clin Epidemiol, 74, 167–176. doi: 10.1016/j.jclinepi.2015.12.005 [DOI] [PubMed] [Google Scholar]
- van Tol MJ, van der Wee NJ, van den Heuvel OA, Nielen MM, Demenescu LR, Aleman A, … Veltman DJ (2010). Regional brain volume in depression and anxiety disorders. Arch Gen Psychiatry, 67(10), 1002–1011. doi: 10.1001/archgenpsychiatry.2010.121 [DOI] [PubMed] [Google Scholar]
- Wells JE, & Horwood LJ (2004). How accurate is recall of key symptoms of depression? A comparison of recall and longitudinal reports. Psychol Med, 34(6), 1001–1011. [DOI] [PubMed] [Google Scholar]
- Whittle S, Lichter R, Dennison M, Vijayakumar N, Schwartz O, Byrne ML, … Allen NB (2014). Structural brain development and depression onset during adolescence: a prospective longitudinal study. Am J Psychiatry, 171(5), 564–571. doi: 10.1176/appi.ajp.2013.13070920 [DOI] [PubMed] [Google Scholar]
- Wolpert DH, & Macready WG (1997). No free lunch theorems for optimization. IEEE Transactions on Evolutionary Computation, 1(1), 67–87. [Google Scholar]
- World Health Organization. (2012). Depression. Fact sheet N 369. Retrieved from http://www.who.int/mediacentre/factsheets/fs369/en/
- Zhang C-H (2010). Nearly unbiased variable selection under minimax concave penalty. The Annals of Statistics, 894–942. [Google Scholar]
- Ziegler A, Koch A, Krockenberger K, & Grosshennig A (2012). Personalized medicine using DNA biomarkers: a review. Hum Genet, 131(10), 1627–1638. doi: 10.1007/s00439-012-1188-9 [DOI] [PMC free article] [PubMed] [Google Scholar]