To the Editor
Children of Latino ancestry have ~1.6-fold increased risk of acute lymphoblastic leukemia (ALL) relative to non-Latino white children [1], partly explained by the higher frequency of common heritable ALL risk alleles at ARID5B, GATA3, and PIP4K2A in Latinos [2–4]. However, the etiologies of the increased ALL risk in Latinos have not been fully elucidated. We previously performed a large, multi-ethnic genome-wide association study (GWAS) of childhood ALL, including 3,263 cases of which ~60% were of Latino ethnicity [5]. While we identified two novel risk loci, we did not identify Latino-specific risk loci, unlike a recent report from Qian et al. [6]. We have performed whole-genome imputation of our Latino dataset and combined it with GWAS data from two additional, non-overlapping Latino childhood ALL case-control datasets to identify novel and/or Latino-specific risk loci.
The GWAS meta-analysis included the following: (i) 1,949 ALL cases and 2,120 controls from the California Cancer Records Linkage Project (CCRLP-LAT) study, supplemented with 6464 Kaiser GERA study controls [5]; (ii) 38 cases and 49 controls from a Guatemalan ALL case-control study (GTM); and (iii) 312 cases and 454 controls from the California Childhood Leukemia Study (CCLS) [7] (Supplementary Material). Methods for haplotype phasing, whole-genome imputation, and quality-control of imputed genotypes are described in Supplementary Material. Case-control association analyses were performed separately in each study using logistic regression in SNPTEST V2, adjusting for ten ancestry-informative principal components, calculated separately within each dataset. Within-study genomic inflation factors were low (λCCRLP = 1.034, λGTM = 1.01, λCCLS = 1.025). A fixed-effects meta-analysis was performed, and QQ plots indicated adequate control of type I error and minimal population stratification (λMeta = 1.029) (Supplementary Fig. S1).
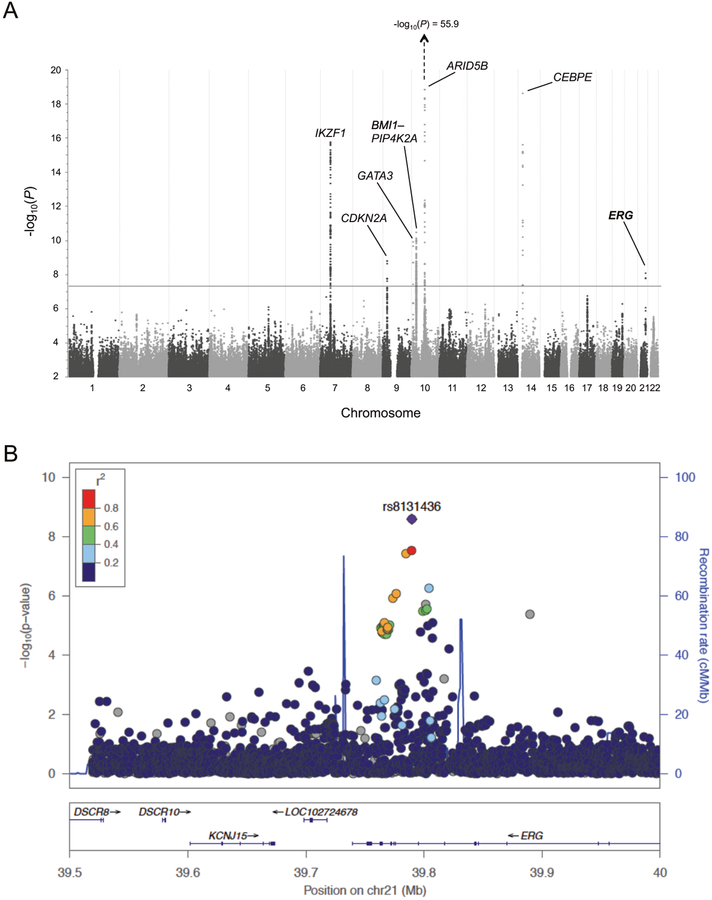
Our GWAS meta-analysis of 2,299 cases and 9,087 controls (Latino only) identified genome-wide significant associations (P < 5.0 × 10−8) at seven well-established risk loci at ARID5B, CEBPE, IKZF1, PIP4K2A, GATA3, CDKN2A, and BMI1 [4, 7–10], plus associations (P < 5.0 × 10−4) at recently identified loci at 17q12/IKZF3, 8q24, LHPP, and ELK3 [5, 11] (Supplementary Table S1). We also identified genome-wide significant association at rs8131436 on chromosome 21q22.2, in an intron of the erythroblast transformation-specific (ETS)-related gene (ERG) (P = 8.76 × 10−9; odds ratio [OR] = 1.23; 95% CI: 1.16–1.31) (Fig. 1a). Targeted re-imputation localized the association to an ~100Kb locus between two recombination peaks (Fig. 1b, Supplementary Table S2).
Fig. 1.
Novel ALL GWAS association locus at ERG on chromosome 21. a Manhattan plot displaying genome-wide −log10(P) values from a meta-analysis of three Latino ALL GWAS (n = 2,299 cases and 9,087 controls), in the California Cancer Records Linkage Project (CCRLP), the California Childhood Leukemia Study (CCLS), and a Guatemalan ALL study. Grey horizontal line represents the genome-wide significance threshold of P = 5.0 × 10−8. Genome-wide significant signals were observed at 7 known loci at, in chromosomal order, IKZF1, CDKN2A, GATA3, BMI1, PIP4K2A, ARID5B, and CEBPE, as well as a novel locus at ERG on chromosome 21. b Locus Zoom plot of a ~500Kb region at chromosome 21q22.2 encompassing ERG. The −log10(P) values were calculated from meta-analysis of the three Latino case-control studies. The ALL association peak is flanked by two recombination hotspot peaks, represented by vertical blue lines
The effect of this locus on ALL risk was recently reported to increase with increasing global Native American (NA) ancestry [6]. Here we examined local ancestry at the ERG locus (Supplementary Material, Supplementary Fig. S2), and found a larger effect size for rs8131436 in Latinos with ≥1 copy of the NA haplotype (OR = 1.30; 95% CI = 1.15–1.47; P = 2.4 × 10−5) than in Latinos with zero NA haplotypes (OR = 1.15; 95% CI = 0.98–1.34; P = 0.09), further supporting a positive association between NA ancestry and the effect of ERG heritable variation on ALL risk. The frequency of NA haplotypes at rs8131436 was slightly higher in cases (42.7%) than controls (40.9%) (Supplementary Fig. S3); however, taking into account the proportion of global NA ancestry, the case-control difference in local NA ancestry at ERG was not significant (P = 0.44) (Supplementary Table S3).
Next, we investigated whether any ERG SNPs were associated with ALL risk in non-Latino whites (n = 1184 cases, 3551 controls from CCRLP-EUR) [5]. Of the top 10 ERG SNPs in our discovery Latino ALL GWAS meta-analysis, SNP rs2836371 was also associated with ALL in non-Latino whites (P = 8.40 × 10−3), albeit with a smaller effect size (OR = 1.15, 95% CI: 1.05–1.25) (Supplementary Table S2).
ERG is within the Down syndrome (DS) critical region on chromosome 21, and children with trisomy 21 have an ~20-fold increased risk of ALL [12]. Therefore, we explored whether ERG variation may contribute to DS-ALL risk. We genotyped rs2836371 (lead SNP across Latino discovery and non-Latino white replication sets) using a Taqman SNP genotyping assay in a Latino case-control set (DS-ALL cases, n = 103 and DS non-leukemia controls, n = 96) from the International Study of Down Syndrome Acute Leukemia (IS-DSAL, Supplementary Material). Trisomic genotypes were manually clustered to delineate the two heterozygote genotypes (TTC or TCC) (Supplementary Fig. S4). We found that rs2836371 was significantly associated with risk of DS-ALL (P = 0.016) with a per-allele OR of 1.44 (95% CI: 1.08–1.96), which was noticeably but non-significantly higher than that in non-DS Latinos (OR = 1.19, Supplementary Table S2) (Pinteraction = 0.21). Furthermore, subjects with three risk alleles at rs2836371 (CCC genotype) had a 3.7-fold increased risk of ALL compared to DS subjects harboring no risk alleles (TTT), rather than the 2.99-fold increased risk predicted under an allelic additive model (Table 1, Supplementary Fig. S5). In a smaller set of non-Latino white DS-ALL cases (n = 83) and DS controls (n = 78), rs2836371 was not significantly associated with DS-ALL risk (OR = 1.07, 95% CI: 0.77–1.49), reflecting similar inter-ethnic differences in effect size observed in non-DS participants.
Table 1.
ERG SNP rs2836371 genotype frequencies in Latino and non-Latino white Down syndrome (DS) ALL cases and DS non-leukemia controls from the International Study of Down Syndrome Acute Leukemia (IS-DSAL), measured by Taqman SNP genotyping
| rs2836371 genotype | Latinos | Non-Latino whites | ||||||
|---|---|---|---|---|---|---|---|---|
| Cases n = 103 (%) | Controls n = 96 (%) | P-value | OR (95% CI) | Cases n = 83 (%) | Controls n = 78 (%) | P-value | OR (95% CI) | |
| TTT | 26 (25.2) | 34 (35.4) | 0.016 | 1.44 (1.08–1.96) | 24 (28.9) | 26 (33.3) | 0.69 | 1.07 (0.77–1.49) |
| TTC | 33 (32.0) | 35 (36.5) | 36 (43.4) | 30 (38.5) | ||||
| TCC | 30 (29.1) | 22 (22.9) | 14 (16.9) | 15 (19.2) | ||||
| CCC | 14 (13.6) | 5 (5.2) | 9 (10.8) | 7 (9.0) | ||||
| Risk allele (C) frequency | 0.437 | 0.326 | 0.365 | 0.346 | ||||
| CCC vs. TTT* | 0.02 | 3.66 (1.17–11.47) | 0.57 | 1.39 (0.45–4.32) | ||||
P values and odds ratios (OR) are calculated using logistic regression tests, assuming the additive model unless specified
P values and ORs calculated using chi-square tests
Observed inter-ethnic differences in SNP effect size suggest potential interactions with environmental factors, or with additional germline or somatic genetic alterations. Intriguingly, several published GWAS loci for white blood cell (WBC) traits in adults lie ~50Kb downstream of rs2836371 within ERG [13]. These SNPs are in very low linkage disequilibrium (LD) with our ALL-associated SNPs, and are positioned on the other side of a strong recombination peak (Supplementary Fig. S6). Novel analysis of selection signals across ERG in Latinos revealed no evidence of positive selection for ALL risk SNPs, but identified a strongly significant signal (population branch statistic >99th percentile genome-wide; haplotype statistic >97th percentile) at the downstream WBC trait locus (Supplementary Fig. S6). SNP rs2836426 showed the strongest selection signal (P = 2.2 × 10−4) and, though in low LD with ALL risk SNP rs2836371 (D′ = 0.16 in AMR, 1000Genomes), it is in high LD with several WBC trait-associated SNPs (D′ = 1 in AMR). No direct association was detected between the low-frequency WBC trait-associated SNPs and ALL risk; however, we found marginally significant synergistic interaction between ALL-associated SNP rs2836371 and three perfectly linked WBC trait SNPs (rs80109907, rs7275212, and rs58030288) on ALL risk in Latinos (P = 0.079, OR = 2.00) but not in non-Latino whites (P = 0.48, OR = 0.78) (Supplementary Table S4), suggesting Latino-specific cooperation between these two independent trait-associated loci in ALL predisposition.
To explore potential functional effects of ALL-associated SNPs in ERG, we assessed 32 SNPs with P < 5.0 × 10−5 in the Latino meta-analysis, of which 19 replicated in the European data (P < 0.05). ERG protein is expressed at low levels in lymphoblastoid cell lines, which prevented accurate expression quantitative trait locus (eQTL) analysis within Genotype Tissue Expression (GTEx) or GEUVADIS RNASeq datasets. In silico analyses, using Haploreg, RegulomeDB, UCSC Genome Browser, and Epigenome Browser, revealed no protein-coding variants, nor any obvious functional candidates based on overlap with putative regulatory elements and transcription factor binding sites.
A recently identified ALL tumor subtype, “DUX4-rear-ranged ALL”, is characterized by somatic DUX4 rearrangements that result in alternative splicing of ERG using an alternative start site at “exon 6 alt” [14]. ALL-associated SNPs at ERG did not alter known DUX4 binding motifs, and TF-binding motif analysis did not reveal any SNPs creating novel DUX4 binding motifs.
We assessed whether any SNPs overlapped ERG exon 6 alt and found that SNP rs2836361, in tight LD with rs2836371 (R2 = 0.93 and D′ = 0.97 in 1000 Genomes individuals of Mexican ancestry; R2 = 0.99 and D′ = 0.99 in Europeans), was located 3 bp upstream of the first exon 6 alt codon (Supplementary Fig. S7). SNP rs2836361 disrupts a strong exonic splicing silencer (ESS), with the risk allele reducing the score of a silencer motif “TCTCCCAA” [15] from 88.1 (TCTGCCAA containing the rs2836361 protective allele) to 70.9 (TCTGTCAA containing the risk allele). This ESS had the highest predicted score within a region encompassing exon 6 alt + /−100bp. Moreover, we found that the rs2836361 risk allele may increase exonic splicing enhancer activity by elevating the RNA recognition motif score for serine/arginine-rich pre-mRNA splicing factor (SRp40). Hence, the rs2836361 risk allele may increase splicing of the non-canonical ERG exon 6 alt, conferring dominant negative effects on wildtype ERG and increased risk of ALL. Further analysis is needed to confirm the causal variant at this locus and its functional effects.
In sum, we report the largest GWAS of childhood ALL among Latinos to date, identifying a risk locus at chromosome 21q22.2, encompassing the hematopoietic transcription factor ERG. This gene is frequently somatically mutated in ALL, adding to a growing list of genes that both predispose to ALL and drive tumorigenesis following somatic mutations. Insufficient patient data were available to investigate the relationship between ERG SNPs and somatic alterations; however, during preparation of this manuscript, Qian et al. reported that the ERG risk genotype was negatively correlated with somatic ERG deletions [6], supporting that the SNP may somewhat mimic effects of somatic loss of ERG.
Novel to our study, we replicated the ERG association in a case-control study of Down syndrome-ALL; this is the first reported heritable risk factor for DS-ALL, and may inform future risk stratification in this vulnerable population. Current methods to accurately assess trisomic geno-types using SNP arrays are sub-optimal; next-generation sequencing strategies are warranted to elucidate the contribution of heritable variation across chromosome 21 to DS-ALL risk.
Our study highlights the importance of Latino subjects in elucidating the germline genetic architecture of childhood ALL, and suggests that larger sample sizes may reveal additional important susceptibility loci that inform the biology of leukemogenesis.
Supplementary Material
Acknowledgements
For recruitment of subjects enrolled in the California Childhood Leukemia Study (CCLS), the authors gratefully acknowledge the clinical investigators at the following collaborating hospitals: University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. Mignon Loh and Katherine Matthay), Children’s Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children’s Hospital (Dr. Gary Dahl), Children’s Hospital Oakland (Drs. James Feusner and Carla Golden), Kaiser Permanente Roseville (formerly Sacramento) (Drs. Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo, Alan Wong, and Denah Taggart), Kaiser Permanente San Francisco (Dr. Kenneth Leung), Kaiser Permanente Oakland (Drs. Daniel Kronish and Stacy Month), California Pacific Medical Center (Dr. Louise Lo), Cedars-Sinai Medical Center (Dr. Fataneh Majlessipour), Children’s Hospital Los Angeles (Dr. Cecilia Fu), Children’s Hospital Orange County (Dr. Leonard Sender), Kaiser Permanente Los Angeles (Dr. Robert Cooper), Miller Children’s Hospital Long Beach (Dr. Amanda Termuhlen), University of California, San Diego Rady Children’s Hospital (Dr. William Roberts), and University of California, Los Angeles Mattel Children’s Hospital (Dr. Theodore Moore). The authors additionally thank the families for their participation in the CCLS (formerly known as the Northern California Childhood Leukemia Study). The IS-DSAL study included biospecimens and/or data obtained from the California Bio-bank Program, (SIS requests # 26 and 572), Section 6555(b), 17 CCR. The California Department of Public Health is not responsible for the results or conclusions drawn by the authors of this publication. The authors would like to thank Robin Cooley and Steve Graham (Genetic Disease Screening Program, California Department of Public Health) for their assistance and expertise in the procurement and management of DBS specimens. We are grateful to the Washington State Department of Health for additional specimen/data access and to William O’Brien of the University of Washington for programming/data management. We are also grateful to the New York State Department of Health Newborn Screening Program, the New York State Cancer Registry, and the New York State Congenital Malformations Registry for additional specimen/data access, and to Drs. Maria Schymura of the NYS Cancer Registry, Marilyn Browne of the NYS Congenital Malformations Registry and Denise Kay of the NYS Newborn Screening Program for case identification, linkage and assistance in the procurement of de-identified DBS specimens and data. This study used biospecimens from the California Biobank Program. Any uploading of genomic data and/or sharing of these biospecimens or individual data derived from these biospecimens has been determined to violate the statutory scheme of the California Health and Safety Code Sections 124980(j), 124991(b), (g), (h), and 103850 (a) and (d), which protect the confidential nature of biospecimens and individual data derived from biospecimens. Certain aggregate results may be available from the authors by request.
Funding This study was supported by Alex’s Lemonade Stand Foundation “A” Awards (A.J.D., K.M.W.), the Emerging Investigator Fellowship Grant from the Pediatric Cancer Research Foundation (A.J.D.), The Children’s Health and Discovery Initiative of Translating Duke Health (K.M.W.), and research grants from the National Institutes of Health (R01 CA155461 to J.L.W. and X.M., R01 ES009137 to C.M., P24ES004705 to C.M., R24ES028524 to C.M. and L.M., and R21 ES021819 to L.M.). The 2016–2019 CLIC Scientific Annual Meetings were supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number U13ES026496. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The collection of cancer incidence data used in the CCRLP study was supported by the California Department of Public Health pursuant to California Health and Safety Code Section 103885; Centers for Disease Control and Prevention’s (CDC) National Program of Cancer Registries, under cooperative agreement 5NU58DP003862–04/DP003862; the National Cancer Institute’s Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute.
Footnotes
Supplementary information The online version of this article (https://doi.org/10.1038/s41375-019-0514-9) contains supplementary material, which is available to authorized users.
Publisher's Disclaimer: Disclaimer
Publisher's Disclaimer: The ideas and opinions expressed herein are those of the author(s) and do not necessarily reflect the opinions of the State of California, Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors.
Conflict of interest The authors declare that they have no conflict of interest.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dores GM, Devesa SS, Curtis RE, Linet MS, Morton LM. Acute leukemia incidence and patient survival among children and adults in the United States, 2001–2007. Blood. 2012;119:34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu H, Cheng C, Devidas M, Pei D, Fan Y, Yang W, et al. ARID5B genetic polymorphisms contribute to racial disparities in the incidence and treatment outcome of childhood acute lymphoblastic leukemia. J Clin Oncol. 2012;30:751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh KM, Chokkalingam AP, Hsu LI, Metayer C, de Smith AJ, Jacobs DI, et al. Associations between genome-wide Native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013;27:2416–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walsh KM, de Smith AJ, Chokkalingam AP, Metayer C, Roberts W, Barcellos LF, et al. GATA3 risk alleles are associated with ancestral components in Hispanic children with ALL. Blood. 2013;122:3385–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wiemels JL, Walsh KM, de Smith AJ, Metayer C, Gonseth S, Hansen HM, et al. GWAS in childhood acute lymphoblastic leukemia reveals novel genetic associations at chromosomes 17q12 and 8q24.21. Nat Commun. 2018;9:286–017-02596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qian M, Xu H, Perez-Andreu V, Roberts KG, Zhang H, Yang W, et al. Novel susceptibility variants at the ERG locus for childhood acute lymphoblastic leukemia in Hispanics. Blood. 2019;133:724–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh KM, de Smith AJ, Hansen HM, Smirnov IV, Gonseth S, Endicott AA, et al. A heritable missense polymorphism in CDKN2A confers strong risk of childhood acute lymphoblastic leukemia and is preferentially selected during clonal evolution. Cancer Res. 2015;75:4884–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Hosking FJ, Vijayakrishnan J, Price A, Olver B, Sheridan E, et al. Loci on 7p12.2, 10q21.2 and 14q11.2 are associated with risk of childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1006–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31–12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105:733–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trevino LR, Yang W, French D, Hunger SP, Carroll WL, Devidas M, et al. Germline genomic variants associated with childhood acute lymphoblastic leukemia. Nat Genet. 2009;41:1001–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vijayakrishnan J, Kumar R, Henrion MY, Moorman AV, Rachakonda PS, Hosen I, et al. A genome-wide association study identifies risk loci for childhood acute lymphoblastic leukemia at 10q26.13 and 12q23.1. Leukemia. 2017;31:573–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hasle H, Clemmensen IH, Mikkelsen M. Risks of leukaemia and solid tumours in individuals with Down’s syndrome. Lancet. 2000;355:165–9. [DOI] [PubMed] [Google Scholar]
- 13.Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016; 167:1415–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, McCastlain K, Yoshihara H, Xu B, Chang Y, Churchman ML, et al. Deregulation of DUX4 and ERG in acute lymphoblastic leukemia. Nat Genet. 2016;48:1481–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sironi M, Menozzi G, Riva L, Cagliani R, Comi GP, Bresolin N, et al. Silencer elements as possible inhibitors of pseudoexon splicing. Nucleic Acids Res. 2004;32:1783–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.