Abstract
Purpose:
Heterogeneity in tumor mutational burden (TMB) quantification across sequencing platforms limits the application and further study of this potential biomarker of response to immune checkpoint inhibitors (ICI). We hypothesized that harmonization of TMB across platforms would enable integration of distinct clinical datasets to better characterize the association between TMB and ICI response.
Methods:
Cohorts of NSCLC patients sequenced by one of three targeted panels or by whole exome sequencing (WES) were compared (total n=7297). TMB was calculated uniformly and compared across cohorts. TMB distributions were harmonized by applying a normal transformation followed by standardization to z-scores. In sub-cohorts of patients treated with ICIs (DFCI n=272; MSKCC n=227), the association between TMB and outcome was assessed. Durable clinical benefit (DCB) was defined as responsive/stable disease lasting ≥6 months.
Results:
TMB values were higher in the panel cohorts than the WES cohort. Average mutation rates per gene were highly concordant across cohorts (Pearson coefficient 0.842-0.866). Subsetting the WES cohort by gene panels only partially reproduced the observed differences in TMB. Standardization of TMB into z-scores harmonized TMB distributions and enabled integration of the ICI-treated sub-cohorts. Simulations indicated that cohorts >900 are necessary for this approach. TMB did not associate with response in never smokers or patients harboring targetable driver alterations, although these analyses were under-powered. Increasing TMB thresholds increased DCB rate, but DCB rates within deciles varied. Receiver operator curves yielded an area under the curve of 0.614 with no natural inflection point.
Conclusion:
Z-score conversion harmonizes TMB values and enables integration of datasets derived from different sequencing panels. Clinical and biologic features may provide context to the clinical application of TMB, and warrant further study.
Introduction
Immune checkpoint inhibitors (ICI) have revolutionized the treatment of multiple advanced cancers2-6. However, only a minority of patients experience clinical benefit, and clinically actionable biomarkers of response are urgently needed.
To date, the only approved biomarkers of ICI response are mismatch repair deficiency and, in NSCLC, programmed death-ligand 1 (PD-L1) expression. However, mounting evidence has demonstrated an association between tumor mutational burden (TMB) and response to ICIs7-17, and there is considerable interest in developing TMB as a clinical biomarker. Importantly, TMB quantification from targeted next generation sequencing (NGS) panels has been shown to correlate with whole exome sequencing-(WES) derived TMB13,18-20 and to associate with ICI response, making the clinical assessment of TMB practically feasible19,21.
However, the proliferation of data related to TMB has also generated confusion, as there are now multiple commercial and academic NGS panels routinely employed, with important differences in gene panel composition, sequencing pipeline, and TMB algorithm22,23. It is unclear how these differences affect TMB quantification, nor is it known how to translate one platform’s TMB values to another for translational discovery or clinical use. Further, the studies describing an association between TMB and response have applied different thresholds to define TMB high vs low groups. It is not known whether this threshold heterogeneity reflects different TMB quantification arising from different platforms, variation across patient cohorts, or unknown clinical or biological effects on the association between TMB and response.
Given these questions, we sought to develop a strategy to harmonize TMB across NGS platforms. We applied this method to integrate multiple clinically annotated cohorts and to more fully characterize the relationship between TMB and ICI response using this larger, pooled dataset, adding nuance and context to our current understanding. We focused on NSCLC due to the early interest in applying TMB to clinical practice in this disease subtype24-26, and to avoid confounding of TMB by tumor type27.
Methods:
Study population
Three cohorts of NSCLC patients whose tumors had been profiled by targeted NGS panel were evaluated. These panel cohorts were compared to a fourth WES cohort from The Cancer Genome Atlas (TCGA).
DFCI Cohort
Patients at the Dana-Farber Cancer Institute (DFCI) whose tumors had undergone OncoPanel NGS were included if they had advanced NSCLC and had consented to institutional review board-approved protocols. The ICI sub-cohort consisted of patients treated with ICIs evaluable for response.
MSKCC Cohort
Molecular profiling from Memorial Sloan Kettering Cancer Center’s (MSKCC) IMPACT NGS panel21 was obtained from the cBioPortal for Cancer Genomics28,29 and limited to NSCLC samples. The ICI sub-cohort consisted of patients treated with ICIs whose tumors had undergone NGS sequencing13.
Foundation Cohort
Patient-level mutation calls for samples sequenced by Foundation Medicine were obtained (study accession phs001179)30 and filtered to include only NSCLC samples.
TCGA Cohort
Somatic WES data from NSCLCs sequenced by TCGA31 were downloaded from the cBioPortal.
Next-generation sequencing
The DFCI cohort was sequenced as previously described32,33. In brief, tumor DNA was extracted and used for custom-designed hybrid capture library preparation. NGS (OncoPanel) was performed, and somatic alterations were identified by custom pipeline. Given the absence of matched normal tissue, common single nucleotide polymorphisms were filtered if present at >0.1% in Exome Variant Server, NHLBI GO Exome Sequencing Project, or gnomAD; variants present ≥2 times in COSMIC were rescued. All variants were reviewed for technical quality34. Finally, to minimize inadvertent inclusion of germline variants, consistent with previous aggregation efforts35, an additional germline filter was applied to exclude events present in the Exome Aggregation Consortium with an allele count >10, after rescuing known somatic events.
The MSKCC, Foundation, and TCGA cohorts were sequenced as described13,30,31,36. The MSK-IMPACT and TCGA WES pipelines use matched normal samples to isolate somatic events. Foundation Medicine uses an internal algorithm to filter putative germline events.
Tumor Mutational Burden
TMB was uniformly calculated for each sample as the number of non-synonymous mutations per megabase (Mb) of genome covered. DFCI mutation counts were divided by the number of bases covered in each OncoPanel version; v1: 0.753334 Mb; v2: 0.826167 Mb; v3: 1.315078 Mb. For MSKCC samples, the mutation count was divided by 0.896665, 1.016478, and 1.139322 Mb for the 341-, 410- and 468-gene panels, respectively. For Foundation samples, 1.1 Mb was used as the length of genome covered19. For TCGA samples, 38 Mb was used to approximate exome size, as previously described19.
PD-L1 Testing
A subset of ICI-treated patients had tissue evaluated for PD-L1 expression, reported as the percentage of tumor cells with membranous PD-L1 staining. MSKCC specimens were stained as previously described13; DFCI specimens were stained using clone E1L3N (Cell Signaling Technology, Danvers, MA) at 1:200 dilution with pressure cooker antigen retrieval in citrate buffer.
Immunotherapy outcomes
Patients in the DFCI and the MSKCC ICI sub-cohorts were annotated for treatment response to anti-PD-(L)1 monotherapy or in combination with anti-cytotoxic T-cell lymphocyte-4 (anti-CTLA-4). Scans were reviewed by thoracic radiologists at each institution, and response determined using Response Evaluation Criteria in Solid Tumors (RECIST), version 1.137. Progression-free survival was assessed from the start of ICI treatment until the date of progression/death; patients without progression were censored at last scan. Consistent with prior studies8,13, complete response (CR), partial response (PR), or stable disease (SD) >6 months was defined as durable clinical benefit (DCB); no durable benefit (NDB) was defined as progressive disease (PD) or SD ≤6 months. Patients censored before 6 months of follow-up were considered not evaluable.
Statistical analysis
Cohort-specific gene mutation averages were calculated by summing the number of mutations in each gene within a cohort and dividing the total by the number of patients in the cohort. The means were then transformed to a normal distribution by natural logarithmic transformation. The linear correlation between log average mutations per gene in the panel cohorts vs TCGA was evaluated using Pearson’s correlation coefficient.
Power transformations were used to normalize cohort-specific TMB distributions; Tukey’s Ladder of Powers38 in the “rcompanion” package39 was used to identify the optimal transformation coefficient. The normalized distributions were then standardized into z-scores by subtracting the transformed distribution mean and dividing by the standard deviation. Overlap between normalized distributions was calculated using the “overlapping” package40.
TMB comparisons were made using the Mann-Whitney U test. The Fisher’s exact test was used to test for differences in categorical variables. All p-values are two-sided, taking significance at p<0.05. Receiver operator curve (ROC) analyses were performed using the pROC and OptimalCutpoints packages41,42. Exploratory cutoffs were selected to: maximize the distance to the y=x line (Youden’s index); maximize specificity with sensitivity > 80%; maximize both sensitivity and specificity; maximize the kappa statistic; and maximize the diagnostic odds ratio. All statistical analyses were performed in R (version 3.4.2).
Results:
Comparison of TMB quantification across panel and WES platforms
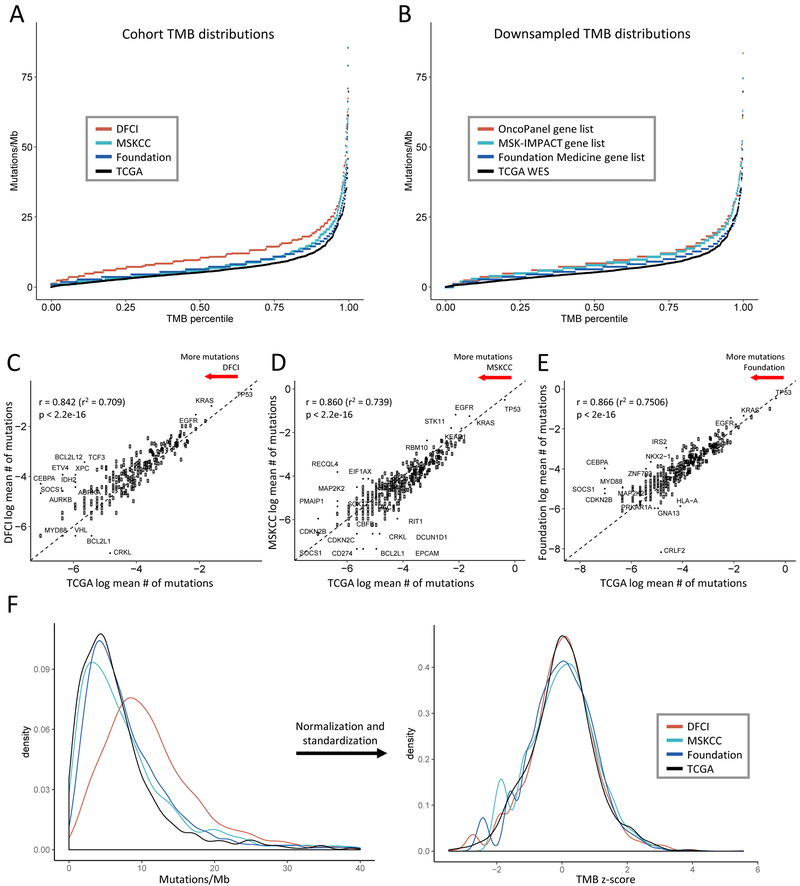
NSCLC patients whose tumors had undergone sequencing via OncoPanel (n=1157), MSK-IMPACT (n=1520), Foundation Medicine (n = 3476), or TCGA (n = 1144) were included (total n=7297, cohort diagram Supplemental Figure S1, clinical characteristics Supplemental Tables S1-4). To determine whether TMB differed between platforms, we plotted the distribution of TMB within each cohort (Figure 1A). TMB distributions differed between cohorts, and targeted panels were associated with higher TMB values than WES. Because targeted panels sequence fewer bases with focused inclusion of mutated cancer genes, we hypothesized that the higher TMB measurements associated with NGS panels were attributable to gene selection. We tested this by subsetting the WES data to include only those genes captured by the targeted panels (“downsampling”, Supplemental Methods) (Figure 1B), and found that downsampled distributions retained greater TMB counts than the unfiltered TCGA distribution, suggesting that gene panel composition contributes to the observed difference in TMB distributions between cohorts. However, the relative differences were less pronounced than in the real-world cohort comparisons, suggesting that assay-specific differences, such as depth of sequencing and the absence of a paired germline sample, might also contribute to inter-test variation.
Figure 1. Comparison of tumor mutational burden (TMB) distribution and gene mutation rates across targeted panel and whole exome sequencing (WES) cohorts.
(A-B) TMB distributions (A) in the panel cohorts and (B) obtained by subsetting the WES cohort to the panel gene sets (downsampling) are higher than the WES TMB distribution. The x-axis depicts the percentile of each TMB value, the y-axis depicts TMB in mutations/megabase. 8 high outliers in (A) not shown. (C-E) Average mutation rates per gene in each panel cohort are highly correlated with average gene mutation rates in the WES cohort. Natural log average mutations per gene in (C) DFCI, (D) MSKCC, and (E) Foundation cohorts are shown on the y-axis, natural log average mutations per gene in the TCGA cohort are on the x-axis. Each point represents a gene. Dashed line depicts y = x. Pearson’s correlation coefficients are shown. (F) Normalization and standardization of TMB distributions bring the NGS and WES cohort distributions into alignment. The left panel shows the kernel density plot of unadjusted TMB values in each cohort, the right panel shows the transformed density plot of TMB z-scores demonstrating high overlap.
To further examine assay-specific sources of variation in TMB across panels, we compared the average number of mutations per gene in each cohort against the TCGA averages, surmising that this could reflect differences in assay performance or mutation calling (Figure 1C-E). Concordance between the panel cohorts and TCGA was high (Pearson coefficient 0.842–0.866), and only rarely mutated genes emerged as outliers, suggesting minimal gene-specific variability. Comparison of variant classes demonstrated differential enrichment in the panel vs WES cohorts, also consistent with assay-specific differences in mutation filtration (Supplemental Figure S2).
Harmonization of TMB across platforms using Z-score standardization
We first attempted to harmonize TMB values across cohorts by linearly mapping panel TMB distributions onto the TCGA TMB distribution (Supplemental Figure S3A-B). However, inconsistent variation in TMB across distributions prohibited use of a linear constant. As above, this variability was diminished but still present when the downsampled TMB values were analyzed (Supplemental Figure S3C-D). Consequently, we instead pursued the strategy of transforming unadjusted TMB values into standardized z-scores that could be compared across panels. Use of a power transformation converted the right-skewed TMB distributions (Figure 1F) to normal distributions (skewness values ≤0.06, Supplemental Figure S4), and standardization to z-scores brought the TMB distributions into good concordance (Figure 1F), with >85% overlap (Supplemental Figure S5). Cohort size simulation (Supplemental Methods) demonstrated that cohorts of > 900 patients are necessary for this approach (Supplemental Figure S6).
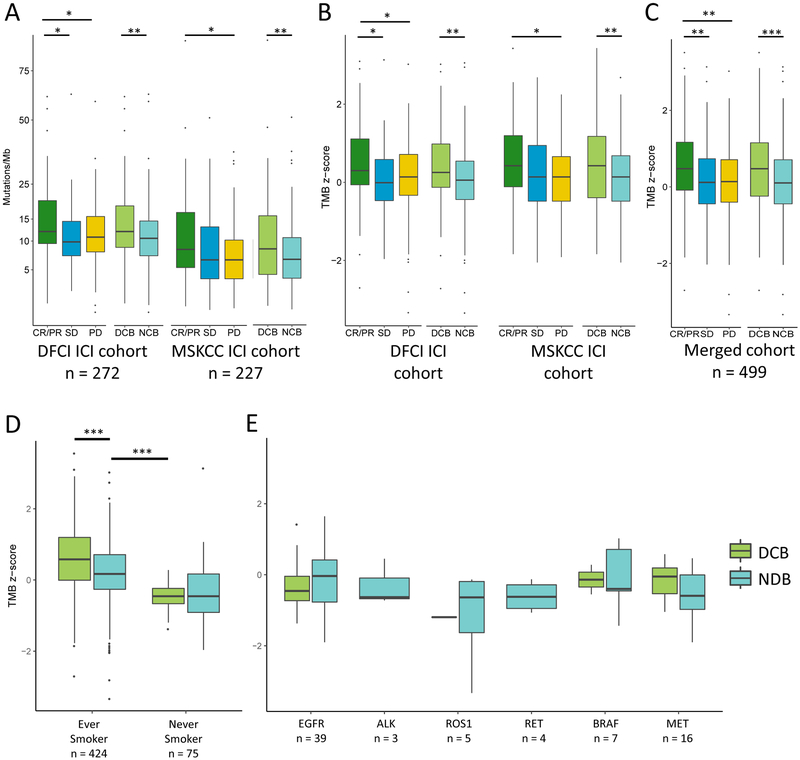
TMB z-scores correlate with ICI outcome and allow for cross cohort comparison
We then applied this transformation to TMB in the DFCI and MSKCC ICI-treated sub-cohorts (n=272 and 227, respectively; total n=499; demographic features Table 1; detailed clinical characteristics Supplemental Table S5) and examined whether the derived TMB z-scores associated with response13,24,26. We confirmed that TMB was higher in patients with CR/PR or DCB than with PD or NDB in both sub-cohorts pre-transformation (Figure 2A), and that this association held when post-transformation z-scores were used (Figure 2B). We noted that while the unadjusted DFCI TMB values were higher than the unadjusted MSKCC TMB values, the z-score standardization produced overlapping distributions (Supplemental Figure S7), allowing us to combine the DFCI and MSKCC ICI cohorts. In the merged cohort, TMB z-scores remained significantly higher in responders (Figure 2C).
Table 1. ICI-Treated Sub-Cohort Patient Characteristics.
Abbreviations: CR, complete response; CTLA-4, cytotoxic T-cell lymphocyte associated protein-4; DCB, durable clinical benefit; ICI, immune checkpoint inhibitor; NDB, no durable benefit; PD, progressive disease; PD-(L)1, programmed cell death-1 or programmed death-ligand 1; PR, partial response; SD, stable disease.
| DFCI ICI Cohort | MSKCC ICI Cohort | ||
|---|---|---|---|
| Characteristic | No. (%) | Characteristic | No. (%) |
| No of patients | 272 | No of patients | 227 |
| Med age at diagnosis (range) | 65 (24-90) | Med age at diagnosis (range) | 66 (22-92) |
| Sex | Sex | ||
| Male | 132 (49) | Male | 116 (51) |
| Female | 140 (51) | Female | 111 (49) |
| Histology | Histology | ||
| Squamous | 37 (14) | Squamous | 31 (14) |
| Adenocarcinoma | 213 (78) | Adenocarcinoma | 179 (79) |
| Other | 22 (8) | Other | 17 (7) |
| Smoking status | Smoking status | ||
| Ever | 241 (89) | Ever | 183 (81) |
| Never | 31 (11) | Never | 44 (19) |
| Line of therapy | Line of therapy | ||
| First | 86 (32) | First | 47 (21) |
| Second | 132 (48) | Second | 122 (54) |
| Third or higher | 54 (20) | Third or higher | 58 (26) |
| Treatment | Treatment | ||
| PD-(L)1, monotherapy | 251 (92) | PD-(L)1, monotherapy | 195 (86) |
| PD-(L)1 + CTLA-4 combination | 21 (8) | PD-(L)1 + CTLA-4 combination | 32 (14) |
| Best overall response | Best overall response | ||
| CR/PR | 56 (21) | CR/PR | 39 (17) |
| SD | 72 (26) | SD | 80 (35) |
| PD | 144 (53) | PD | 108 (48) |
| Clinical benefit | Clinical benefit | ||
| DCB | 83 (31) | DCB | 69 (30) |
| NDB | 189 (69) | NDB | 158 (70) |
| Actionable mutations | Actionable mutations | ||
| EGFR | 21 (8) | EGFR | 18 (8) |
| BRAF V600E | 6 (2) | BRAF V600E | 1 (<1) |
| ALK | 2 (1) | ALK | 1 (<1) |
| ROS1 | 1 (<1) | ROS1 | 4 (2) |
| RET | 3 (1) | RET | 1 (<1) |
| MET exon 14Δ | 9 (3) | MET exon 14A | 7 (3) |
Figure 2. Correlation of tumor mutational burden (TMB) and TMB z-scores with outcome.
(A) In both the DFCI and MSKCC ICI cohorts, TMB is greater in patients with complete or partial response (CR/PR) than with progressive disease (PD) (DFCI: median, 12.0 vs 10.8 mut/Mb, P = 0.03; MSKCC: median, 8.9 vs 6.9 mut/Mb, P = 0.02), and in those with durable clinical benefit (DCB) vs no durable benefit (NDB) (DFCI: median, 12.0 vs 10.5 mut/Mb, P = 0.003; MSKCC: median 8.9 vs 6.9 mut/Mb, P = 0.007). (B) DFCI and MSKCC TMB z-score distributions overlap. Z-scores are higher in patients with CR/PR vs PD, and DCB vs NDB in each cohort respectively (DFCI: median, CR/PR 0.30 vs PD 0.14, P = 0.03; median, DCB 0.30 vs NDB 0.10, P = 0.003; MSKCC: median, CR/PR 0.46 vs 0.17 mut/Mb, P = 0.02; median, DCB 0.46 vs NDB 0.17, P = 0.006). (C) TMB z-scores are higher in patients with CR/PR and DCB in the joint cohort (median, CR/PR 0.47 vs PD 0.14, P = 0.002; median, CR/PR 0.47 vs SD 0.14, P = 0.007; median, DCB 0.46 vs NDB 0.14, P = 6.43e-05). (D) TMB z-scores in the joint cohort are higher in ever smokers with DCB vs NDB (median 0.579 vs 0.171, P = 6.046e-05), but were not associated with response in never smokers (median −0.456 vs −0.456, P = 0.749). TMB z-scores were significantly higher in ever smokers with NDB than never smokers with DCB (median 0.171 vs −0.456, P = 0.00097). (E) TMB z-scores in the joint cohort do not associate with response in patients with mutations in targetable oncogenic drivers. *p<0.05; **p<0.01, ***p<0.001. Box plots represent medians, interquartile ranges, and vertical lines extend to the 95th percentiles.
TMB is lower in never smokers and may not associate with response
We performed subgroup analyses using the transformed, pooled cohort (DFCI and MSKCC ICI-treated patients, n=499) to determine whether specific clinical and biological features impact the association between TMB and response. TMB z-scores in ever smokers were higher than never smokers (median 0.312 vs −0.456, P<0.0001); notably, TMB in ever smokers with NDB was higher than TMB in never smokers with DCB (median 0.171 vs −0.456, P=0.00097) (Figure 2D). Among never smokers, TMB did not differ between DCB versus NDB (median −0.456 vs −0.456, P=0.749). Sampling simulations (Supplemental Methods) suggest this negative finding may be due to decreased power in this subset, although lower TMB values and distinct biology may also contribute (Supplemental Figure S8). Similar exploratory analyses of patients harboring targetable oncogenic drivers did not demonstrate an association between TMB z-score and DCB (total n=74), although power in these small driver sub-groups was also limited (Figure 2E). Power simulations suggest that cohort sizes >300 may be necessary to detect a difference in TMB between patients with DCB vs NDB in groups with lower response rates or effect sizes (Supplemental Figure S9).
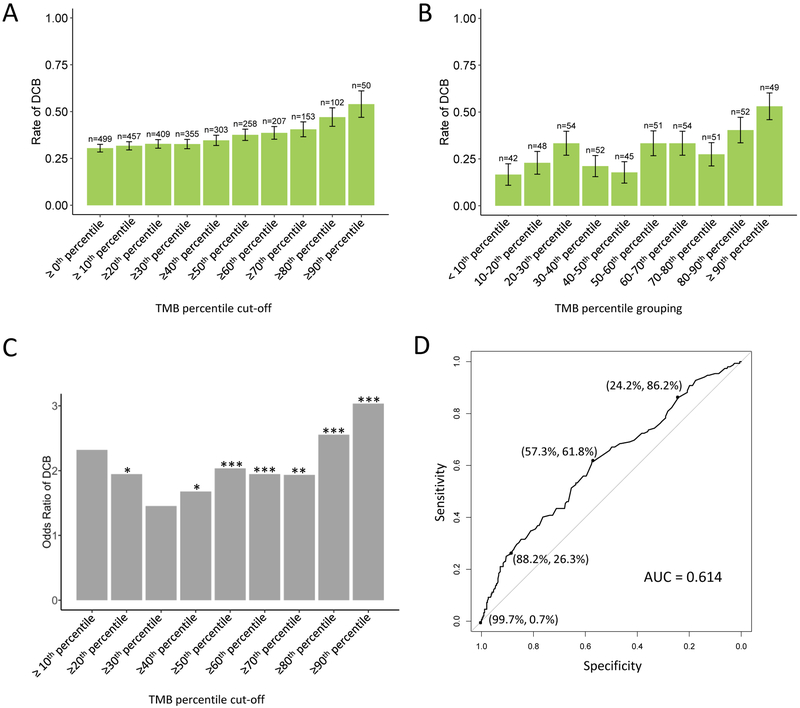
TMB thresholds and response
Given the heterogeneity in previously identified thresholds and the percentile cut-points used to identify such thresholds, we used our pooled cohort to systematically explore the relationship between TMB and response to ICI across the TMB distribution. We calculated the rate of DCB and CR/PR with increasing TMB thresholds in the pooled and separate ICI cohorts (Figure 3A, Supplemental Figure S10). Table 2 illustrates the TMB z-scores and values associated with each threshold. We observed a gradual increase in rate of DCB with increasing TMB thresholds. We noted, however, that this could arise from enriching for high TMB outliers, and therefore calculated the rate of DCB within each TMB decile (joint cohort Figure 3B, separate cohorts Supplemental Figure S11). In this analysis, we noted high DCB rates in the highest deciles (DCB rate 40.4% in patients with TMB z-scores between the 80th and 90th percentiles, DCB rate 53.1% in patients with TMB z-scores ≥90th percentile), and low rates in the lowest deciles (DCB rate 16.7% in patients with TMB z-score <10th percentile). However, the middle deciles exhibited greater heterogeneity in DCB rate. Accordingly, the odds ratio of DCB with increasing TMB thresholds was highest with TMB cutoffs ≥80th percentile, and more heterogeneous at lower thresholds (Figure 3C). Similar trends were observed in smokers; there was no increase in DCB rates with increasing TMB threshold in never smokers (Supplemental Figure S12). The pattern of association between PD-L1 and DCB was similar to TMB, with increasing rates of DCB with higher PD-L1 thresholds, but more variability within PD-L1 score groupings (Supplemental Figure S13).
Figure 3. Rate of response to immune-checkpoint inhibitors by tumor mutational burden (TMB) threshold.
(A) Rate of durable clinical benefit (DCB) increases with increasing TMB cut-off thresholds. TMB z-score deciles were selected as cut-points, and rate of DCB was calculated for patients in the joint cohorts whose TMB z-scores were ≥ the cut-point. Bars depict rate of DCB. Left-most bar depicts rate of DCB in the unselected cohort (TMB ≥ 0th percentile). (B) Rate of DCB within TMB z-score deciles is more variable. Bars depict rate of DCB among patients whose TMB z-scores are ≥ the lower bound and < the upper bound. (C) Odds ratio of DCB varies with increasing TMB z-score cut-points. Bars depict odds ratio, significant p-values are indicated above bars. (D) Receiver operator characteristic (ROC) curve demonstrates a trade-off in sensitivity vs specificity of durable clinical benefit at varying TMB z-score values (area under the curve [AUC], 0.614). Exploratory cut-points, with their associated specificity and sensitivity, are indicated. *p<0.05; **p<0.01, ***p<0.001. Error bars represent standard error. Numbers over bar graphs indicated number of patients in each group.
Table 2.
TMB z-score associated with each decile cut-off in the joint ICI cohort. Equivalent TMB values in the DFCI, MSKCC, Foundation and TCGA cohorts are shown.
| Percentile | TMB z-score | DFCI TMB | MSKCC TMB | Foundation TMB |
TCGA TMB (mutation count) |
|---|---|---|---|---|---|
| 10th | −1.04 | 4.81 | 2.27 | 2.83 | 1.84 (55) |
| 20th | −0.47 | 7.22 | 3.89 | 4.45 | 3.35 (101) |
| 30th | −0.24 | 8.42 | 4.78 | 5.30 | 4.18 (125) |
| 40th | 0.00 | 9.87 | 5.90 | 6.36 | 5.25 (158) |
| 50th | 0.17 | 11.07 | 6.89 | 7.27 | 6.10 (183) |
| 60th | 0.45 | 13.24 | 8.76 | 8.97 | 7.58 (228) |
| 70th | 0.70 | 15.47 | 10.82 | 10.80 | 9.41 (282) |
| 80th | 0.95 | 18.05 | 13.34 | 13.00 | 11.31 (339) |
| 90th | 1.38 | 23.49 | 19.10 | 17.90 | 15.43 (463) |
Given the heterogeneity of response rates over the mid TMB distribution, we plotted a receiver operator characteristic (ROC) curve to formally quantify how well TMB z-scores discriminate between DCB and NDB. ROC analysis yielded an area under the curve (AUC) of 0.614. The Youden Index cutoff was associated with a sensitivity of 61.8% and a specificity of 57.3%, resulting in undertreatment of 12% of patients, and overtreatment of 30% (Figure 3D, Table 3). Application of other thresholds demonstrated better specificity at higher TMB z-score thresholds, but at the expense of missing patients who would have responded. Cutoffs and their associated sensitivity/specificity were similar in the cohorts considered separately (Supplemental Figure S14). Application of the clinically-used PD-L1 threshold ≥50% was associated with undertreatment of 13% and overtreatment of 19% of patients. TMB z-score did not discriminate between DCB and NDB in never smokers or targetable driver mutated patients (AUC 0.493, data not shown). Analysis of TMB thresholds with respect to PFS, rather than response, demonstrated similar results (Supplemental Figure S15).
Table 3.
Test characteristics and performance of TMB cut-off values in the joint ICI cohort (n = 499). Right-most columns describe number of patients with DCB who would not have been treated at that threshold (false negative), and the number of patients who would have been treated with NDB (false positive).
Abbreviations: DCB, durable clinical benefit; NDB, no durable benefit; OR, odds ratio; Sens, sensitivity; Spec, specificity; TMB, tumor mutational burden.
| Z-score cut-off |
DFCI TMB |
MSKCC TMB |
% Sens. | % Spec. | % DCB | OR (p-value) |
# ≥ cut- off (%) |
# with DCB not treated (%) |
# treated w/ NDB (%) |
|---|---|---|---|---|---|---|---|---|---|
| −0.46 | 7.34 | 3.77 | 86.2 | 24.2 | 33 | 1.99 (0.008) | 394 (79) | 21 (4) | 263 (53) |
| 0.28 | 11.9 | 7.18 | 61.8 | 57.3 | 39 | 2.10 (< 0.001) | 245 (49) | 58 (12) | 151 (30) |
| 1.08 | 20.6 | 15.1 | 26.3 | 88.2 | 54 | 2.99 (< 0.01) | 80 (16) | 111 (22) | 39 (8) |
| 3.10 | 61.4 | 68.2 | 0.7 | 99.7 | 67 | 4.60 (0.22) | 3 (1) | 150 (30) | 1 (0.2) |
Discussion:
We present a pragmatic comparison of TMB calculated from targeted panels and WES, and apply TMB z-score conversion to enable harmonized analyses. We demonstrate that this approach can translate TMB values across tests and can be used to integrate distinct datasets for discovery and further analyses. Additionally, our use of real-world datasets allows us to incorporate and account for sources of variation not captured by in silico downsampling analyses, such as differences in mutation/indel calling pipelines, depth of coverage, and germline filtration. This approach is distinct from other parallel harmonization efforts43,44, which focus on standardization of TMB definitions and reporting, and eventually aim to generate ‘gold standard’ cell lines for benchmarking. We anticipate that our approach will be of immediate use to both clinicians and researchers, and further anticipate that this approach can be easily applied to other platforms and relevant tumor types.
Although the association between TMB and response to ICIs in NSCLC has been demonstrated, less is understood about how clinical and biologic features affect this association. Here we found that TMB did not associate with DCB in never smokers and in patients harboring targetable oncogenic mutations. Importantly, these analyses were underpowered to detect a difference, and our power simulations indicated that larger cohorts are needed, cautioning against definitive conclusions in these small subgroup analyses. However, we also observed that never smokers who benefitted from ICIs had markedly lower TMB values than ever smokers who did not, suggesting that further study to identify TMB-independent predictors of response in never smokers may be warranted, and raising the important possibility that the clinical application of TMB as a biomarker will need to take clinical and biologic features into account.
The importance of context is further emphasized by our analysis of TMB thresholds. Prior analyses have generally focused on identifying a single threshold to define ‘TMB high’ and ‘TMB low’ subgroups, with variation in selected thresholds across studies13,15,26,45. Our systematic analysis of TMB thresholds illustrates additional nuances in the relationship between TMB and response. We observed enrichment in DCB with higher TMB thresholds as expected, but weaker discrimination in the mid-range of TMB values without a single, natural biological inflection point. These findings may account for some of the observed heterogeneity among previously proposed thresholds, as there may be a range of values that discriminate similarly between responders and non-responders. Additionally, our data suggest that the choice of a given threshold must be decided within a goal-specific context that considers the relative efficacy of the alternative treatment; a TMB threshold selected to enrich for response to first-line therapy may be different than a threshold selected for second-line therapy. Notably, TMB is independent of PD-L1 expression12,13, with similar biomarker performance: increasing expression is associated with improved efficacy without a natural cut-point; there is variability in DCB enrichment within deciles of expression; and distinct thresholds are appropriately applied based on the specific treatment scenario (i.e. >/=50%, >/=1%, or none)3,5,46. Ultimately, these data do not answer whether and how TMB should be applied to clinical practice, as this must be examined through prospective clinical trials, but add nuance to our understanding of how TMB associates with response.
One limitation of this study is that our comparison of TMB assumes that the observed distinctions reflect differences in platform rather than patient/samples. We were not able to account for clinical and tumor features due to inconsistent sample annotation, but note that our large cohorts help mitigate any sampling bias, and the overall consistencies in shape of distribution are reassuring. It is an open question as to whether TMB distributions should be more narrowly defined by sample features such as histology or stage, and the normalization we describe here can be adjusted as more is learned. At present, however, TMB is compared across patients and biopsy specimens without reference to these sample characteristics, making this aggregated approach consistent with current clinical practice.
In conclusion, we provide a practical approach to the challenge of standardizing TMB across platforms, and we apply this approach to integrate distinct datasets to better understand how TMB associates with response. Much remains to be learned about how and why TMB associates with response to ICI, and how best to apply TMB in the clinic for precision immunotherapy.
Supplementary Material
Context Summary:
Key Objective: It is not known how to account for differences in tumor mutational burden (TMB) generated by different sequencing assays. We sought to address assay heterogeneity in TMB quantification by developing a technique to harmonize TMB across assays, and applied this technique to pool distinct clinical cohorts to better characterize the association between TMB and response to immune checkpoint inhibitors.
Knowledge generated: TMB differs across sequencing assays due to differences in gene panels and sequencing pipelines. Standardization of TMB into z-scores enabled inter-assay comparison and pooling of distinct clinical cohorts. From this pooled analysis, we observed that TMB may not associate with response in patients who are never smokers or harbor targetable oncogenes, and TMB thresholds yield significant trade-offs in sensitivity and specificity.
Relevance: Z-score standardization harmonizes TMB values across assays for pooled analysis. Clinical and biologic features may modulate the association between TMB and response.
Acknowledgement of Research Support
Supported by the Damon Runyon Foundation (E.M.V.) and by NIH R01CA227388 (E.M.V.); by Damon Runyon Foundation (DL), NIH K08CA234458 (DL), Conquer Cancer Foundation YIA (DL), and Society for Immunotherapy of Cancer-BMS Translational Fellowship (DL); by Memorial Sloan Kettering Cancer Center Support Grant/Core Grant (P30 CA008748) and the Druckenmiller Center for Lung Cancer Research at MSKCC; MDH is a Damon Runyon Clinical Investigator supported (in part) by the Damon Runyon Cancer Research Foundation (CI-98-18) and is a member of the Parker Institute for Cancer Immunotherapy. M.N. is supported by R01CA203636 and U01CA209414 (NCI).
Footnotes
Prior Presentations: This study has been presented in part at the International Association for the Study of Lung Cancer (IASLC) Targeted Therapies 2019 conference, at the 2019 ASCO-SITC Clinical Immuno-Oncology Symposium, and at the AACR Annual Meeting 2019.
References:
- 1.Garon EB, Rizvi NA, Hui R, et al. : Pembrolizumab for the treatment of non-small-cell lung cancer. N Engl J Med 372:2018–28, 2015 [DOI] [PubMed] [Google Scholar]
- 2.Borghaei H, Paz-Ares L, Horn L, et al. : Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med 373:1627–39, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herbst RS, Baas P, Kim DW, et al. : Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 387:1540–50, 2016 [DOI] [PubMed] [Google Scholar]
- 4.Fehrenbacher L, Spira A, Ballinger M, et al. : Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387:1837–46, 2016 [DOI] [PubMed] [Google Scholar]
- 5.Reck M, Rodriguez-Abreu D, Robinson AG, et al. : Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 375:1823–1833, 2016 [DOI] [PubMed] [Google Scholar]
- 6.Brahmer J, Reckamp KL, Baas P, et al. : Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med 373:123–35, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snyder A, Makarov V, Merghoub T, et al. : Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rizvi NA, Hellmann MD, Snyder A, et al. : Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124–8, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Van Allen EM, Miao D, Schilling B, et al. : Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350:207–211, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le DT, Uram JN, Wang H, et al. : PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med 372:2509–20, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson DB, Frampton GM, Rioth MJ, et al. : Targeted Next Generation Sequencing Identifies Markers of Response to PD-1 Blockade. Cancer Immunol Res 4:959–967, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carbone DP, Reck M, Paz-Ares L, et al. : First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med 376:2415–2426, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi H, Sanchez-Vega F, La K, et al. : Molecular Determinants of Response to Anti-Programmed Cell Death (PD)-1 and Anti-Programmed Death-Ligand 1 (PD-L1) Blockade in Patients With Non-Small-Cell Lung Cancer Profiled With Targeted Next-Generation Sequencing. J Clin Oncol 36:633–641, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miao D, Margolis CA, Vokes NI, et al. : Genomic correlates of response to immune checkpoint blockade in microsatellite-stable solid tumors. Nat Genet 50:1271–1281, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gandara DR, Paul SM, Kowanetz M, et al. : Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med 24:1441–1448, 2018 [DOI] [PubMed] [Google Scholar]
- 16.Samstein RM, Lee CH, Shoushtari AN, et al. : Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cristescu R, Mogg R, Ayers M, et al. : Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garofalo A, Sholl L, Reardon B, et al. : The impact of tumor profiling approaches and genomic data strategies for cancer precision medicine. Genome Med 8:79, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chalmers ZR, Connelly CF, Fabrizio D, et al. : Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med 9:34, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szustakowski JD, Green G, Geese WJ, et al. : Abstract 5528: Evaluation of tumor mutation burden as a biomarker for immune checkpoint inhibitor efficacy: A calibration study of whole exome sequencing with FoundationOne<sup>®</sup>. Cancer Research 78:5528, 2018 [Google Scholar]
- 21.Zehir A, Benayed R, Shah RH, et al. : Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703–713, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.33rd Annual Meeting & Pre-Conference Programs of the Society for Immunotherapy of Cancer (SITC 2018): Late-Breaking Abstracts: Washington, D.C., USA 7–11 November 2018 Journal for Immunotherapy of Cancer 6:1–13, 2018. 29298730 [Google Scholar]
- 23.Hendriks LE, Rouleau E, Besse BJTLCR: Clinical utility of tumor mutational burden in patients with nonsmall cell lung cancer treated with immunotherapy. 2018 7:647–660, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hellmann MD, Nathanson T, Rizvi H, et al. : Genomic Features of Response to Combination Immunotherapy in Patients with Advanced Non-Small-Cell Lung Cancer. Cancer Cell 33:843–852 e4, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hellmann MD, Ciuleanu TE, Pluzanski A, et al. : Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N Engl J Med 378:2093–2104, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ready N, Hellmann MD, Awad MM, et al. : First-Line Nivolumab Plus Ipilimumab in Advanced Non-Small-Cell Lung Cancer (CheckMate 568): Outcomes by Programmed Death Ligand 1 and Tumor Mutational Burden as Biomarkers. J Clin Oncol:JCO1801042, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lawrence MS, Stojanov P, Polak P, et al. : Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499:214–218, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, et al. : Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6:pl1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, et al. : The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2:401–4, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartmaier RJ, Albacker LA, Chmielecki J, et al. : High-Throughput Genomic Profiling of Adult Solid Tumors Reveals Novel Insights into Cancer Pathogenesis. Cancer Res 77:2464–2475, 2017 [DOI] [PubMed] [Google Scholar]
- 31.Campbell JD, Alexandrov A, Kim J, et al. : Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet 48:607–16, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia EP, Minkovsky A, Jia Y, et al. : Validation of OncoPanel: A Targeted Next-Generation Sequencing Assay for the Detection of Somatic Variants in Cancer. Arch Pathol Lab Med 141:751–758, 2017 [DOI] [PubMed] [Google Scholar]
- 33.Sholl LM, Do K, Shivdasani P, et al. : Institutional implementation of clinical tumor profiling on an unselected cancer population. JCI Insight 1:e87062, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li MM, Datto M, Duncavage EJ, et al. : Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn 19:4–23, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Consortium APG: AACR Project GENIE: Powering Precision Medicine through an International Consortium. Cancer Discov 7:818–831, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frampton GM, Fichtenholtz A, Otto GA, et al. : Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–31, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eisenhauer EA, Therasse P, Bogaerts J, et al. : New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 45:228–47, 2009 [DOI] [PubMed] [Google Scholar]
- 38.Tukey JW: Exploratory data analysis Reading, Mass, Addison-Wesley Pub. Co., 1977 [Google Scholar]
- 39.Mangiafico SS: Summary and Analysis of Extension Program Evaluation in R, version 1.15.0. rcompanion.org/handbook/, 2016
- 40.Pastore: Overlapping: a R package for Estimating Overlapping in Empirical Distributions. Journal of Open Source Software 3:1023, 2018 [Google Scholar]
- 41.Robin X, Turck N, Hainard A, et al. : pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12:77, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.López-Ratón M, Rodríguez-Álvarez MX, Cadarso-Suárez C, et al. : OptimalCutpoints: An R Package for Selecting Optimal Cutpoints in Diagnostic Tests. Journal of Statistical Software; Vol 1, Issue 8 (2014), 2014 [Google Scholar]
- 43.Stenzinger A, Allen JD, Maas J, et al. : Tumor mutational burden standardization initiatives: Recommendations for consistent tumor mutational burden assessment in clinical samples to guide immunotherapy treatment decisions. Genes, Chromosomes and Cancer 0, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.33rd Annual Meeting & Pre-Conference Programs of the Society for Immunotherapy of Cancer (SITC 2018): Late-Breaking Abstracts Journal for ImmunoTherapy of Cancer 6:1–13, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Heeke S, Hofman P: Tumor mutational burden assessment as a predictive biomarker for immunotherapy in lung cancer patients: getting ready for prime-time or not? Translational Lung Cancer Research 7:631–638, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Antonia SJ, Villegas A, Daniel D, et al. : Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med 377:1919–1929, 2017 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.