Abstract
The identification of prognostic factors and building of risk assessment prognostic models will continue to play a major role in 21st century medicine in patient management and decision making. Investigators are often interested in examining the relationship between host, tumor-related, and environmental variables in predicting clinical outcomes. We make a distinction between static and dynamic prediction models. In static prediction modelling, typically variables collected at baseline are utilized in building models. On the other hand, dynamic predictive models leverage the longitudinal data of covariates collected during treatment or follow-up, and hence provide accurate predictions of patients prognoses. To date, most risk assessment models in oncology have been based on static models. In this article, we cover topics that are related to the analysis of prognostic factors, centering on factors that are both relevant at the time of diagnosis or initial treatment and during treatment. We describe the types of risk prediction and then provide a brief description of the penalized regression methods. We then review the state-of-the art methods for dynamic prediction and compare the strengths and the limitations of these methods. While static models will continue to play an important role in oncology, developing and validating dynamic models of clinical outcomes need to take a higher priority. It is apparent that a framework for developing and validating dynamic tools in oncology is still needed. One of the limitations in oncology that modelers may be constrained by the lack of access to the longitudinal biomarker data. It is highly recommended that the next generation of risk assessments consider the longitudinal biomarker data and outcomes so that prediction can be continually updated.
Introduction
Identifying prognostic factors and building risk assessment prognostic models will continue to play a major role in 21st century medicine in patient management and decision making [1]. Prognostic factors in oncology associate host and tumor variables to clinical outcomes independent of treatment [2]. Gospodarowicz et al., classify factors as either tumor-related, host or environmental factors [3]. Tumor-related factors are variables that are related to the tumor and reveal the tumor biology and pathology (such as size of tumor, lymph node involvement, presence of metastasis and molecular markers (over expression of PTEN gene, presence of androgen receptor variant AR-V7). Host-related factors are associated to the patients’ characteristics, such as age and comorbities. Lastly, environmental factors are external to the patient such as access to healthcare [3]. Prognostic models are increasingly used in the design, conduct, and analysis of clinical trials. For example, in several trials of prostate cancer, randomization was stratified by the predicted survival probability determined by prognostic model of overall survival [4–7]. In the TAILORx trial, OncotypeDx, a 21-gene score that predicts likelihood of recurrence, is used to classify women with breast cancer by their risk group of recurrence [8]. Prognostic factors have also been used for enriching the patient population in trials with targeted therapies. For example, in the TOGA trial, gastric cancer patients with HER-2 positive were randomized to either trastuzumab plus chemotherapy or chemotherapy alone [9].
In this article, we cover topics that are related to the analysis of prognostic factors, centering on factors that are both relevant at the time of diagnosis or initial treatment and during treatment or follow-up. We use the terms prognostic models, risk models, and risk assessments interchangeably. This article is organized in the following way. We first describe the type of risk prediction and then provide a brief description of the penalized regression methods. We then review the state of the art methods for dynamic prediction and compare the strengths and the limitations of these methods. We next offer a concise discussion of validation and metrics for assessing models. Lastly, we present recommendation for the next generation of risk assessments methods to be built in modern oncology.
I. Types of Risk Predictions
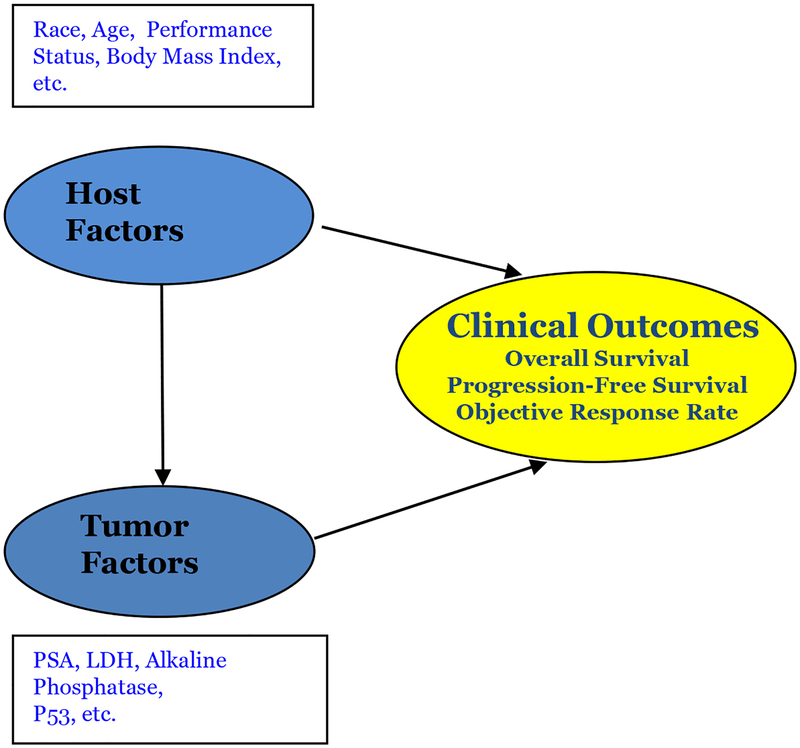
Investigators are interested in examining the relationship between host and tumor-related in predicting clinical outcomes (Figure 1A). We make a distinction between static and dynamic prediction. In static prediction modelling, typically variables collected at baseline are utilized in building models. For example, prostate specific antigen (PSA) measurements at baseline are utilized for prediction of recurrence. On the other hand, dynamic predictive models explicitly leverage the longitudinal data of covariates that are collected during treatment or follow-up. In advanced patients with cancer, the disease has substantially evolved and has a heterogeneous presentation within the patient [10]. The inter and intra-patient variability should be taking into account in statistical modeling [11, 12]. Dynamic prediction incorporates time-dependent covariates so that risk prediction would be continually updated with new observations to reflect the patient’s prognosis.
Figure 1A.
Relationship between host and tumor related factors and clinical outcomes. Modified from Seminars in Oncology[2].
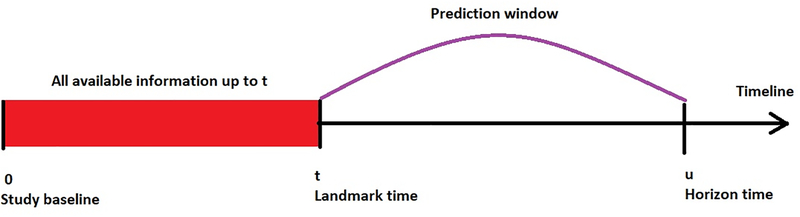
We define terminology that is typically used in dynamic risk prediction. The term landmark time is defined as a current time point at which we have data (host, tumor-related variables and outcomes). The term horizon time is defined as a future time point at which we want to predict a time-to-event outcome, such as overall survival. Dynamic predictive models essentially capture the historical information of the longitudinal measurements from the study baseline (t) to the landmark time (u), such that the risk at horizon time can be accurately predicted (Figure 1B).
Figure 1B.
Dynamic risk prediction framework
II. Identification of Prognostic Factors
Several popular strategies exist for identifying prognostic factors in static risk assessment. Standard variable selection approaches such as forward selection, backward selection, etc. with logistic regression for binary endpoints [13], proportional hazards regression for time-to-event endpoints have been applied [14]. Criticism for the stepwise variable selection has been well documented [15, 16]. It is also worth noting that classification trees, such as recursive partitioning for both binary and time to event endpoints [17–20]have been frequently utilized [2, 21–26].
We concentrate on penalized methods that fit and shrink p predictors and in doing so reduce the variance of the coefficient estimates [27]. Thus, these method would improve the accuracy of the model [28] The least absolute shrinkage and selection operator (LASSO) and adaptive LASSO (ALASSO) have been widely employed to develop prognostic models of clinical outcomes [29, 30]. We will briefly describe ridge and penalized methods. Ridge regression minimizes the residual sum of squares function, but it has a caveat as it does not reduce all the coefficients exactly to zero [28]. Let yi be the response, xij is the jth covariate value (j = 1,2,…,p) corresponding to the ith individual , βj is the regression coefficient jth covariate and λ is a tuning parameter. Similar to ridge regression, the first term in LASSO is the residual sum of squares and LASSO minimizes this function subject to the l1 penalty (Eq 1):
| (1) |
A large tuning parameter causes the coefficients estimates to be equal to zero, then the LASSO will have the sparsity property [28]. LASSO is an improvement over ridge regression, although it has the main limitation of tending to select too many unimportant variables and it also performs poorly in situations when p>n [20, 31–33]. Adaptive LASSO has been proposed as an improvement over LASSO in order to overcome the limitation of LASSO [34]. ALASSO minimizes this function (Eq 2):
| (2) |
ALASSO utilizes a weighted penalty term in the L1 penalty where w = (w1,w2,…,wp) is the weight vector. If is a -consistent estimator, e.g. (OLS), of β (= β1, β2,…, βp), then an appropriate choice of the weight w is . ALASSO is considered to be an improvement over LASSO as it has consistent variable selection as well as lower prediction error. Consequently, ALASSO tends to select fewer non-zero coefficients than the LASSO despite having smaller prediction error. The ALASSO enjoys the oracle property [30, 34].
Elastic net regression utilizes a combination of l1 penalty and ridge l2 penalty and is a compromise between LASSO and ridge regression, and one of its main advantages when p > n is that it retains more than n variables in the model [35]. Hastie et al. provide a thorough comparison of these shrinkage techniques [28].
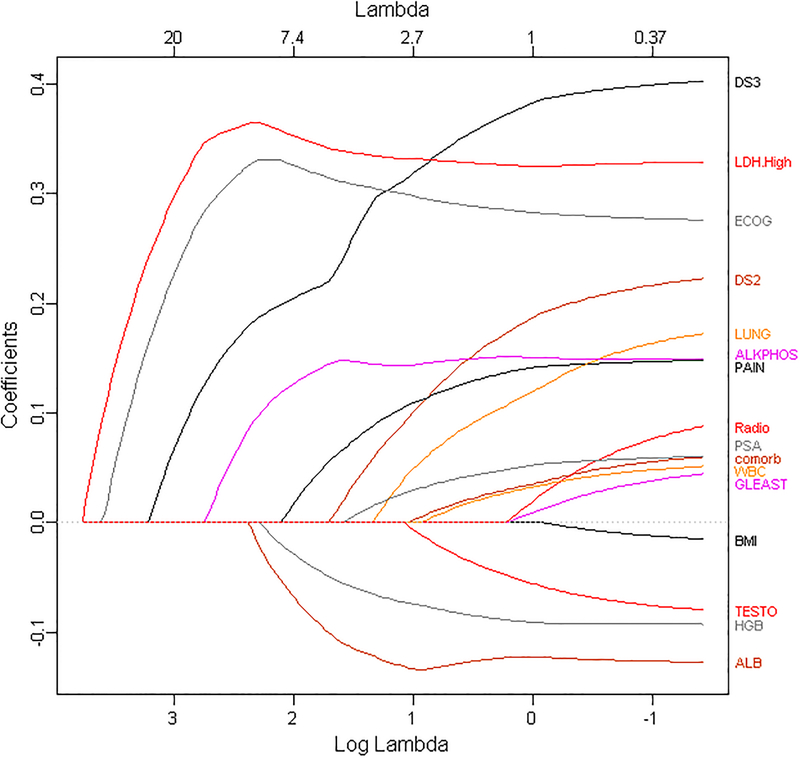
We applied LASSO and ALASSO from CALGB 90401, a phase III clinical trial in advanced prostate cancer with the overall goal of building a model of overall survival [5]. We had 22 variables that were common between CALGB 90401 and the Enthuse trial (external data set) [36]. Because of missing data in the covariates, we utilized methods to impute them as described by [37]. The regression’s estimates from the proportional hazards model, LASSO and ALASSO are presented in Table 1. We considered LASSO and ALASSO and applied both the Akaike information criteria and the Bayesian information criterion to choose the optimal model of overall survival. LASSO and ALASSO selected eight and nine variables, respectively (Table 1). We determined the ALASSO model as the final optimal model since it included the site of metastases for bone disease. Figure 2 presents the solution path for ALASSO and we observe that LDH greater than upper limit of normal and ECOG performance status were selected early in the l1 path compared to the other variables. This is followed by visceral disease, alkaline phosphatase, albumin, hemoglobin, pain, bone metastases and then PSA (the BIC stopped at PSA). The final model selected the following prognostic factors: LDH greater than upper limit of normal, ECOG performance status, metastatic site (presence of visceral disease, presence of bone metastases), PSA, alkaline phosphatase, albumin, hemoglobin and analgesic opioid use.
Table 1.
Identified Prognostic Factors by LASSO and ALASSO in the Training Set
| Description of variable (variable name) | Cox model | LASSO | ALASSO |
|---|---|---|---|
| Site of Metastases: Bone (DS 2) | 0.234 | - | 0.058 |
| Visceral (DS 3) | 0.400 | 0.056 | 0.293 |
| Liver (LIVER) | 0.013 | - | - |
| Lung (LUNG) | 0.199 | - | - |
| Opioid analgesic use (PAIN) | 0.136 | 0.077 | 0.088 |
| Age in years (AGE) | −0.003 | - | - |
| Body Mass Index (BMI) | −0.021 | - | - |
| Race (Caucasian) | 0.034 | - | - |
| ECOG Performance Status (ECOG) | 0.278 | 0.190 | 0.305 |
| Comorbidity (Comorb) | 0.070 | - | - |
| Gleason Score (GLEAST) | 0.052 | - | - |
| Prior treatment with radiotherapy (Radio) | 0.105 | - | - |
| LDH ≥ ULN (LDH.High) | 0.325 | 0.203 | 0.335 |
| Albumin (ALB) | −0.133 | −0.080 | −0.122 |
| Bilirubin (BILI) | −0.017 | - | - |
| Hemoglobin (HGB) | −0.094 | −0.085 | −0.065 |
| Platelets (PLT) | 0.000 | - | - |
| White blood cells (WBC) | 0.055 | - | - |
| Alkaline Phosphatase (ALKPHOS) | 0.145 | 0.138 | 0.145 |
| Aspartate Aminotransferase (AST) | 0.025 | - | - |
| Prostate Specific Antigen(PSA) | 0.063 | 0.026 | 0.015 |
| Testosterone (TESTO) | −0.088 | - | - |
| Training C-index | 0.662 | 0.660 | |
| Integrated Time AUC | 0.742 | 0.740 |
Figure 2.
Solution Path for ALASSO. Printed with permission from Chapman & Hall/CRC [96].
We have focused on variable selection when the number of predictors is small relative to the sample size. There are two main challenges in identifying potential prognostic factors in high dimensional space: computational intensity and a high false discover rate [38, 39]. It is important to point that several pre-screening methods are useful in identifying prognostic features in both the large p, small n problem and in ultra-high dimensional space [31, 32, 39–41].
The concept of variable selection is more challenging in building dynamic models as the main goal is to identify important factors that are related to the longitudinal process and the outcomes. In recent years, a few statistical studies extended the penalized method for the joint modeling of longitudinal data and survival outcomes [42, 43]. The general idea is to postulate the joint likelihood linking the two submodels via latent random variables and to add shrinkage operators to select fixed and random effects. He et al. [42] proposed a variable selection method for joint modeling with a univariate longitudinal outcome, Chen and Wang [43] extended the framework to incorporate multiple longitudinal outcomes. While these methods have not been implemented in oncology, the statistical development paves the way for dynamic risk prediction.
Heterogeneity of treatment effect (HTE) is another important area to consider when building prognostic models. It is the nonrandom, explainable variability in the direction and magnitude of treatment effects for individuals within a population [44]. There are different sources for HTE and it may arise from an underlying causal mechanism, artifacts, measurements or methods [45, 46]. One main goal of the HTE analyses is to predict whether a patient might benefit from a treatment. Traditionally, the Cox regression method has been employed to identify subgroups of patients who may benefit from treatment [14]. Recursive partitioning has also been utilized to identify a subgroup of patients [47]. While these classification tress methods have several advantages, they can create complicated structures and produce overfitting [19, 41, 47]. Other methods, such as permutation methods, SIDES, doubly robust augmented inverse probability weighted estimator, and virtual twins have been developed to take HTE into consideration [45, 48–52]. The personalized prediction can be adequately addressed by the dynamic prediction framework, although it is an area for future research.
III. Estimating Patient-Specific Outcome Prediction and Constructing Risk Groups
Once the final model is chosen, the next step is estimating patient-specific outcome prediction. The estimated survival function at time t is (Eq 3):
| (3) |
where R is the estimated linear predictor or risk score for the ith individual , is the baseline survival function, and is the baseline cumulative hazard function. Turning to our prognostic model of overall survival in prostate cancer, we computed a risk score from the estimated regression coefficients and the predicted survival at 24-months using the baseline cumulative hazard. We present the profiles of two patients with different baseline prognostic factors and their predicted overall survival at 24 months (Table 2) [5]. We observe that Patient 2 has a worse predicted survival probability at 24-months than Patient 1.
Table 2.
Profiles of Patient-Specific Predicted Probabilities [5].
| Patient Number | Disease Site | Opiate Use | PS ECOG | LDH>ULN | ALB (g/dL) | HGB (g/dL) | ALK (U/L) | PSA (ng/ml) | Predicted Probability at 24 months (95% CI)* |
|---|---|---|---|---|---|---|---|---|---|
| 1 | Bone | Yes | 1 | No | 4 | 14 | 130 | 90 | 0.47 (0.42–0.52) |
| 2 | Visceral | Yes | 1 | Yes | 3 | 10 | 90 | 75 | 0.28 (0.19–0.38) |
Another important task in static predictive modelling is to construct prognostic risk groups, which can be formed based on their quantiles from the estimated linear predictor. In our overall survival model, we constructed two- and three- prognostic risk groups and determined the cut-points from the training set based on quantiles (33th, 50th and 67th percentiles) [5]. While demonstrating that the overall survival curves differ across the three risk groups are appropriate, it is not a sufficient approach [53]. The optimal strategy would be to compute a measure of discriminative ability of the model.
IV. Methods for Dynamic Modelling
Let Ti denote the true failure time, let fi (t) denote a set of longitudinal measurements at some time points up to landmark time t. We are interested in predicting the probability that a new patient i* is event-free at least up to horizon time u > t given survival up to t. The conditional probability is defined as (Eq 4)
| (4) |
where Dn denotes a sample from the target population and on which the prediction is based. This formulation enables a dynamic updating scheme. Indeed, if a new measurement for patient i* is observed at time t′ > t, we can update the risk prediction by calculating πi*(u|t′).
There are two general dynamic risk prediction frameworks: joint modelling and landmark analysis. Joint modeling is comprised of two linked submodels, one for the longitudinal process, one for the time-to-event data, and both are dependent on a common set of latent random variables [54, 55]. In particular, the longitudinal data are usually modeled by a linear mixed effects model. The time-to-event data are modeled by the proportional hazards model with true longitudinal process as time-varying covariates. The Cox regression coefficient quantifies the association between the latent longitudinal process and the hazard rate at time t. The longitudinal process and the event time process are assumed to be independent given the latent random effects, and the joint likelihood can be derived. The model can be estimated either using a frequentist approach that attains maximum likelihood through EM algorithm [54, 56–58] or a Bayesian approach that uses Markov Chain Monte Carlo (MCMC) to obtain posterior means [59–61]. Assuming that the parameters are readily estimated from the observed data, then the conditional probability πi*(u|t) can be computed. A Monte Carlo estimate of πi*(u|t) can be obtained by sampling the random effects and the parameters from the corresponding distributions [62].
On the other hand, landmarking [63–66] consists of a series of related Cox regression models, each one is defined at a distinct landmark time t [63–66]. For each pair of {u, t}a separate model is fitted to the individuals who remain in the study and have not yet experienced the event of interest. The baseline hazard can be estimated using the Breslow’s estimator [67]. Then πi*(u|t) is computed as the survival probability treating the longitudinal observation at time t as a baseline covariate.
V. Comparison between Joint Modeling and Landmarking
Joint modeling and landmarking approaches differ in three aspects: model assumptions, information utilized, and computational complexity. Joint modeling models the dual distribution of the longitudinal process and the failure times, and hence satisfies the consistency conditions for dynamic prediction [68]. Moreover, it exploits the full information of collected data and takes into account of the measurement error of the longitudinal data. This latter point is critical as this implies that joint modelling is a more efficient than landmarking. However, joint modeling needs to specify a correct model for the longitudinal process, and requires stronger assumptions than landmarking. It also takes a considerable effort to estimate the parameters and the computational cost is high as it involves complicated joint distribution and numerical integration. In contrast, landmarking circumvents the aforementioned model assumptions and computational burden, but it is not a comprehensive probability model of the longitudinal process and the failure times, and as such does not satisfy the consistency conditions. Another major shortcoming is that landmarking only considers the patients at risk at the landmark time and does not fully explore the information.
Recent articles have focused on the comparison of joint modeling and landmarking. Rizopoulos et al. [69] compared the two prediction frameworks and proposed a compromise between joint modeling and landmarking. Suresh et al. [70] contrasted joint modeling and landmarking for dynamic risk prediction in the context of a binary longitudinal marker, and also applied these approaches to a prostate cancer study [70]. Ferrer et al. [71] compared the two approaches in case of model misspecification, and they aimed for predicting competing risks of prostate cancer from the PSA history.
Dynamic risk prediction (joint modelling) relies on model assumptions, and hence its performance suffers from model misspecification. Functional data analysis, which is a nonparametric framework, has received considerable attention recently in medical studies as it is a flexible tool for modeling nonlinear longitudinal process [72–74]. In particular, these methods have been incorporated with joint modeling [75] and are exploited to construct dynamic prediction models [61, 76–78].
VI. Examples
There are a few examples in cancer where longitudinal data were modelled with clinical outcomes. We have previously explored whether PSA decline at different landmark times is prognostic for overall survival in patients with advanced prostate cancer [79]. Fontein et al. developed a dynamic model for predicting overall survival in patients with breast cancer [80]. The authors validated the overall survival model using dynamic cross-validated c-index and reported c-index of 0.72, 0.76 and 0.79 at 1-, 2- and 3- years, respectively. Suresh et al. [70] and Ferrer et al. [71] extended the landmarking approach and used prostate cancer studies as the testing bed. In addition, Proust-Lima and Taylor [81] developed a dynamic prognostic tool based on joint modeling using PSA as a biomarker for prostate cancer recurrence. There are other applications of dynamic models in prostate cancer [82–85] and colorectal cancer [86].
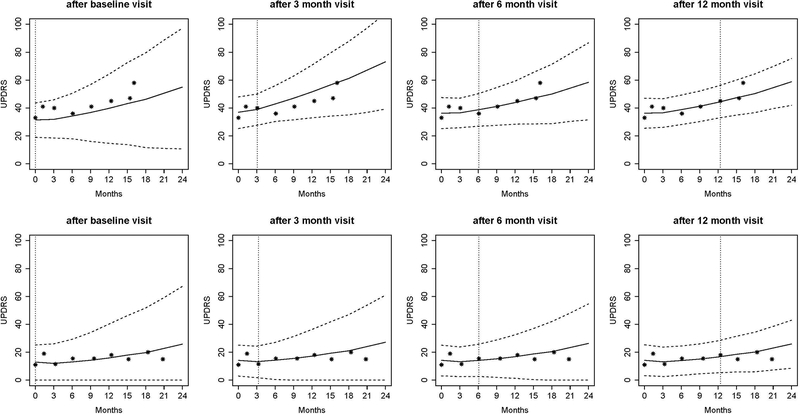
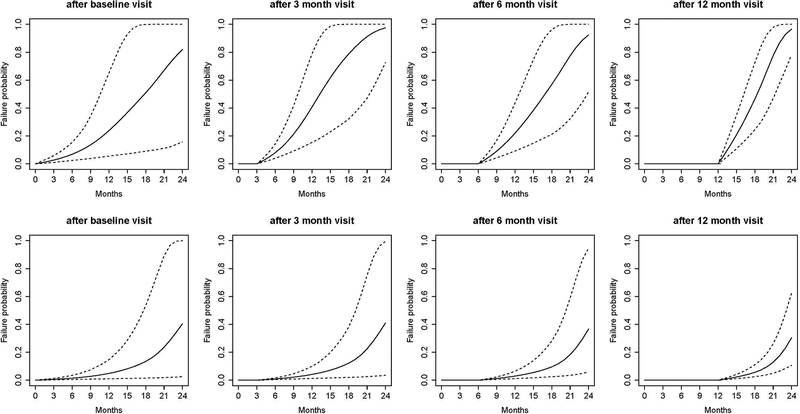
We demonstrate the application of dynamic risk prediction utilizing the DATATOP study [87], a clinical trial that was designed to examine the benefits of deprenyl and alpha-tocopherol in slowing the progression of Parkinson’s disease (PD) [88]. Multiple longitudinal biomarkers were collected in the DATATOP study, including Unified PD Rating Scale (UPDRS) total score, modified Hoehn and Yahr (HY) scale, and Schwab and England activities of daily living (SEADL). The biomarkers measured at baseline, 1-month, and every 3-months show strong correlation between the PD symptoms and the terminal event. We applied a joint modeling framework to account for the informative event times. We developed a Bayesian approach for parameter estimation and predicted patients’ future outcome trajectories (Figure 3A) and risk of functional disability (Figure 3B). A more severe Patient 169 with earlier development of functional disability and a less severe Patient 718 were selected to illustrate the patient-specific predictions at a clinically relevant future time points, conditional on their available longitudinal measurements. The predicted UPDRS trajectories were biased with wide uncertainty bands when only baseline measurements were used. Although dynamic prediction for longitudinal trajectories is an advantage of the joint modeling, our major interest was to predict the probability of functional disability after visits at time t given the patient’s longitudinal profiles and the event-free status up to time t. We found a similar pattern in that the risk predictions with higher accuracy were achieved based on more longitudinal observations. For example, based on the longitudinal profiles of the first 12 months, the predicted probabilities in the next 3, 6, 9, and 12 months were 0.21, 0.46, 0.78, 0.97 and 0.02, 0.06, 0.13, 0.30 for Patient 169 (Figure 3B, last plot first row) and Patient 718 (Figure 3B, last plot second row), respectively. Therefore, Patient 169 with a higher risk of functional disability in the next few months might need attentive medical intervention to control disease progression. Meanwhile, the Brier’s scores were 0.216 and 0.108, respectively, which implied a better prediction in terms of calibration given more follow-up data.
Figure 3A.
Predicted UPDRS trajectories for Patient 169 (first row) and Patient 718 (second row). Solid lines are the means of 2000 MCMC samples. Dashed lines are the 2.5% and 97.5% percentiles of the 2000 MCMC samples. The dotted vertical lines represent the landmark time. Printed with permission from Annals of Applied Statistics [88].
Figure 3B.
Predicted conditional failure probabilities for Patient 169 (first row) and Patient 718 (second row). Solid lines are the means of 2000 MCMC samples. Dashed lines are the 2.5% and 97.5% percentiles of the 2000 MCMC samples. Printed with permission from Annals of Applied Statistics [88].
VII. Validation and Assessment of Prognostic Models
The primary goal of a risk assessment is to provide accurate outcome prediction in new patients [16, 89]. Overfitting remains one of the main challenges in model building. Overfitting occurs when a high predictive accuracy is estimated from a model that has been applied to the training set, but has low accuracy when assessed in an independent dataset [90]. A good example of overfitting is provided in Halabi and Owzar [2]
Validating a prognostic model is considered as a critical step after a risk assessment model has been built. There are two types of validation: external and internal [16, 89, 90]. External validation where the frozen model from the training data is applied to an independent data set, is the most rigorous approach. However, often investigators may not have access to external data set. It is important to emphasize that other types of resampling methods, such as cross-validation, bootstrapping and bootstrapping using 0.632+ are considered appropriate approaches of model validations [32, 53, 91, 92].
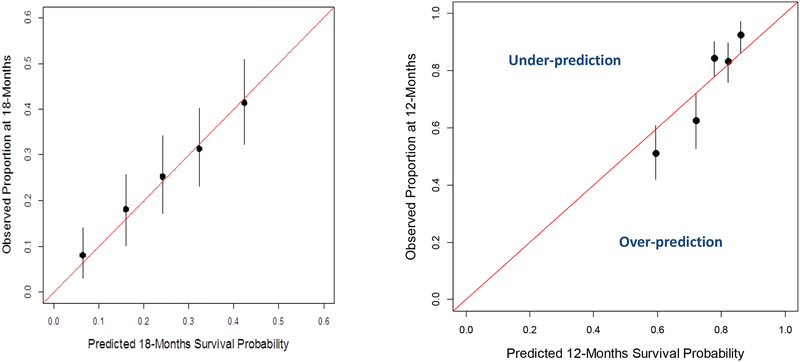
Assessing the performance of the model is usually conducted by examining the calibration and the discriminative ability of the model. Calibration signifies the extent of the match between the predicted and observed outcome [16]. Often investigators plot the predicted versus the observed outcome. The model would be calibrated if the data fall on a 45-degree line. Using data from two phase III clinical trials, we evaluated the overall survival model for calibration at different landmarks [36, 93]. Figure 4A shows the predicted survival probabilities at 18-months were close to the proportion of patients surviving 18-months using the Enthuse 33 trial. On the other hand, Figure 4B demonstrates that the model was not well-gauged as the observed-predicted data points did not fall on the 45-degree line. The first two points (circles) show that the model over-predicted the proportion of patients surviving 12-months, whereas the third and fifth data points demonstrate that the model under-predicted the proportion of patients surviving 12-months.
Figures 4A-4B.
Calibration of the Overall Survival Model on Two Datasets.
Discrimination describes the ability of a prognostic model to distinguish between patients with and without the outcome of interest [16]. Several metrics are employed to report the performance of a model. A widely used measure is the concordance (c-index), which is the agreement between observed outcomes and prediction. Another widely used measure of predictive accuracy is the time-dependent area under the receiver operating characteristic curve (tAUROC) [94], which can be combined to form an integrated area under the receiver operating characteristic curve for the whole range of the study (iAUROC) [95].
Circling back to our prognostic model in prostate cancer, we evaluated the performance of the model by implementing Uno’s integrated measure for the time dependent area under the receiver operating characteristic curve [95], which were 0.73 (95% CI=0.70–0.73) and 0.76 (95% CI=0.72–0.76) in the testing and validation sets, respectively [5].
It is important to emphasize that dynamic models need also to be assessed for their discriminative ability and calibration. These measures can evaluate the performance of the model at various time points of the prediction. The tAUROC and Brier’s score are widely used for dynamic prediction validation [69, 70, 82, 87]. In the DATATOP study [87], we applied 5-fold cross-validation to evaluate the predictive performance of our framework. Conditional on the longitudinal history up to month 3 and month 12, our model yielded 0.744 and 0.766 tAUROC, respectively, for correctly assigning higher risk of functional disability by month 15 to more severe patients.
Criteria for evaluating risk assessments have been published by the Precision Medicine Core of the American Joint Commission on Cancer [53]), the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis Or Diagnosis [92] and the Critical Appraisal and Data Extraction for Systematic Reviews of Prediction Modelling Studies [91]. Investigators are encouraged to follow these guidelines as more rigorous tools of clinical outcomes would be developed in oncology. It is anticipated that these models will be implemented in both the design and conduct of future trials.
While static models will continue to play an important role in oncology, developing and validating dynamic models of clinical outcomes need to take a higher priority. It is apparent that a framework for developing and validating dynamic tools in oncology is needed. One of the limitations is that modelers may be constrained by the lack of access to the longitudinal biomarker data. It is highly recommended that the next generation of risk assessments take into consideration the longitudinal biomarker data and outcomes so that prediction are updated.
In summary, risk assessment will remain an important research task in precision oncology. We advocate for good clinical practice in risk assessment studies and recommend that investigators design these studies prospectively, in order to obtain accurate individual outcome prediction and prognostic risk group classification. Prognostic studies should begin by asking fundamental questions that are pertinent to patient outcomes, define the primary endpoint a priori, justify the sample size, and describe the appropriate methods for variable selection and model assessment. Lastly, they should be validated using external datasets if available.
Understanding the longitudinal relationship between host and tumor related factors and their impact on clinical outcomes is critical. We expect to see an upsurge in dynamic risk assessments in oncology, and as such, the AJCC and the TRIPOD guidelines should be extended to dynamic predictive modelling. Regardless whether static or dynamic modelling is the primary objective, we envision that this review will serve as a catalyst to bridge gaps in knowledge and motivate investigators to take risk assessment as a discipline by itself. Funding opportunities with the primary goal of building and validating high quality prognostic models will be critical for personalized predictions.
Context Summary.
Key objective:
Building prognostic models will continue to play a role in 21st century medicine in patient management. We make a distinction between static and dynamic predictive models and provide a review of the state-of-the art methods for dynamic predictive models to promote them for future use.
Knowledge generated:
To date, most risk assessment models in oncology have been based on static prognostic models. Understanding the longitudinal relationship between host and tumor related factors and their impact on clinical outcomes is critical. Regardless whether static or dynamic modelling is the primary objective, we envision that this review will serve as a catalyst to encourage investigators to take risk assessment as a discipline by itself.
Relevance:
We expect to see an upsurge in dynamic risk assessments and as such it is highly recommended that the next generation of models consider the longitudinal data and outcomes so that prediction are updated.
Acknowledgments
This work was funded in part by the United States Army Medical Research Awards W81XWH-15-1-0467, W81XWH-18-1-0278 and the Prostate Cancer Foundation. This article draws and elaborates upon references [2, 41, 96]. Sheng Luo’s research was partly supported by National Institutes of Health (R01NS091307).
References
- 1.Osullivan B, Brierley J, Gospodarowicz M: Prognosis and classification of cancer. 2015:23–33. [Google Scholar]
- 2.Halabi S, Owzar K: The importance of identifying and validating prognostic factors in oncology. Semin Oncol 2010, 37(2):e9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gospodarowicz MK, O’Sullivan B, Sobin LH, International Union against C: Prognostic factors in cancer. Hoboken: Wiley-Liss; 2006. [Google Scholar]
- 4.Halabi S, Small EJ, Kantoff PW, Kattan MW, Kaplan EB, Dawson NA, Levine EG, Blumenstein BA, Vogelzang NJ: Prognostic model for predicting survival in men with hormone-refractory metastatic prostate cancer. J Clin Oncol 2003, 21(7):1232–1237. [DOI] [PubMed] [Google Scholar]
- 5.Halabi S, Lin CY, Kelly WK, Fizazi KS, Moul JW, Kaplan EB, Morris MJ, Small EJ: Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2014, 32(7):671–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly WK, Halabi S, Carducci M, George D, Mahoney JF, Stadler WM, Morris M, Kantoff P, Monk JP, Kaplan E et al. : Randomized, double-blind, placebo-controlled phase III trial comparing docetaxel and prednisone with or without bevacizumab in men with metastatic castration-resistant prostate cancer: CALGB 90401. J Clin Oncol 2012, 30(13):1534–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris MJ, Heller G, Bryce AH, Armstrong AJ, Beltran H, Hahn OM, McGary EC, Mehan PT, Goldkorn A, Roth BJ et al. : Alliance A031201: A phase III trial of enzalutamide (ENZ) versus enzalutamide, abiraterone, and prednisone (ENZ/AAP) for metastatic castration resistant prostate cancer (mCRPC). JCO Journal of Clinical Oncology 2019, 37(15_suppl):5008. [Google Scholar]
- 8.Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, Geyer CE Jr., Dees EC, Goetz MP, Olson JA Jr. et al. : Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med 2018, 379(2):111–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bang YJ, Van Cutsem E, Feyereislova A, Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T et al. : Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2-positive advanced gastric or gastro-oesophageal junction cancer (ToGA): a phase 3, open-label, randomised controlled trial. Lancet 2010, 376(9742):687–697. [DOI] [PubMed] [Google Scholar]
- 10.Nowell PC: The clonal evolution of tumor cell populations. Science (New York, NY) 1976, 194(4260):23–28. [DOI] [PubMed] [Google Scholar]
- 11.Bedard PL, Hansen AR, Ratain MJ, Siu LL: Tumour heterogeneity in the clinic. Nature Nature 2013, 501(7467):355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Junttila MR, de Sauvage FJ: Influence of tumour micro-environment heterogeneity on therapeutic response. Nature 2013, 501(7467):346–354. [DOI] [PubMed] [Google Scholar]
- 13.Hosmer DW, Lemeshow S, Sturdivant RX: Applied logistic regression. Oxford: Wiley-Blackwell; 2013. [Google Scholar]
- 14.Cox DR: Regression Models and Life-Tables. J R Stat Soc B 1972, 34(2):187.-+. [Google Scholar]
- 15.Altman DG: Studies investigating prognostic factors: conduct and evaluation In: Prognostic Factors in Cancer Edited by MK GB OS HS, 3rd edn. Hoboken, New Jersey: Wiley-Liss; 2006: 39–54. [Google Scholar]
- 16.Harrell FE: Regression Modeling Strategies With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis Introduction. Springer Ser Stat 2015:1–11. [Google Scholar]
- 17.Breiman L, Friedman JH, Olshen RA, Stone CJ: Classification and regression trees. Boca Raton, Fla.: CRC Press; 2017. [Google Scholar]
- 18.LeBlanc M, Crowley J: Survival Trees by Goodness of Split. jamerstatasso Journal of the American Statistical Association 1993, 88(422):457–467. [Google Scholar]
- 19.Hothorn T, Hornik K, Zeileis A: Unbiased Recursive Partitioning: A Conditional Inference Framework. JOURNAL OF COMPUTATIONAL AND GRAPHICAL STATISTICS 2006, 15(3):651–674. [Google Scholar]
- 20.Banerjee M, George J, Song EY, Roy A, Hryniuk W: Tree-based model for breast cancer prognostication. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2004, 22(13):2567–2575. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Liu K-Y, Wong STC: Cancer classification and prediction using logistic regression with Bayesian gene selection. Journal of Biomedical Informatics Journal of Biomedical Informatics 2004, 37(4):249–259. [DOI] [PubMed] [Google Scholar]
- 22.Sparano JA: Prognostic gene expression assays in breast cancer: are two better than one? NPJ Breast Cancer 2018, 4:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller KD, O’Neill A, Gradishar W, Hobday TJ, Goldstein LJ, Mayer IA, Bloom S, Brufsky AM, Tevaarwerk AJ, Sparano JA et al. : Double-Blind Phase III Trial of Adjuvant Chemotherapy With and Without Bevacizumab in Patients With Lymph Node-Positive and High-Risk Lymph Node-Negative Breast Cancer (E5103). J Clin Oncol 2018, 36(25):2621–2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart GC, Azzato EM, Greenberg DC, Rashbass J, Kearins O, Lawrence G, Caldas C, Pharoah PD: PREDICT: a new UK prognostic model that predicts survival following surgery for invasive breast cancer. Breast Cancer Research 2010, 12(R1):3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rudloff U, Jacks LM, Goldberg JI, Wynveen CA, Brogi E, Patil S, Van Zee KJ: Nomogram for Predicting the Risk of Local Recurrence After Breast-Conserving Surgery for Ductal Carcinoma In Situ. JCO Journal of Clinical Oncology 2010, 28(23):3762–3769. [DOI] [PubMed] [Google Scholar]
- 26.Halbesma N, Jansen DF, Heymans MW, Stolk RP, de Jong PE, Gansevoort RT, Group PS: Development and validation of a general population renal risk score. Clin J Am Soc Nephrol 2011, 6(7):1731–1738. [DOI] [PubMed] [Google Scholar]
- 27.Hastie T, Tibshirani R, Friedman JH: The elements of statistical learning : data mining, inference, and prediction; 2017. [Google Scholar]
- 28.Hastie T, Friedman J, Tibshirani R: The Elements of Statistical Learning Data Mining, Inference, and Prediction; 2001. [Google Scholar]
- 29.Tibshirani R: The lasso method for variable selection in the Cox model. Stat Med 1997, 16(4):385–395. [DOI] [PubMed] [Google Scholar]
- 30.Zhang HH, Lu WB: Adaptive lasso for Cox’s proportional hazards model. Biometrika 2007, 94(3):691–703. [Google Scholar]
- 31.Fan J, Feng Y, Wu Y: High-dimensional variable selection for Cox’s proportional hazards model. 2010. [Google Scholar]
- 32.Fan J, Lv J: Sure Independence Screening for Ultrahigh Dimensional Feature Space. jroyastatsocise4 Journal of the Royal Statistical Society Series B (Statistical Methodology) 2008, 70(5):849–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meinshausen N, Bühlmann P: High-dimensional graphs and variable selection with the Lasso. The Annals of Statistics, 2006, 34(3):1436–1462. [Google Scholar]
- 34.Zou H: Theory and Methods - The Adaptive Lasso and Its Oracle Properties. Journal of the American Statistical Association 2006, 101(476):1418–1429. [Google Scholar]
- 35.Zou H, Hastie T: Regularization and Variable Selection via the Elastic Net. jroyastatsocise4 Journal of the Royal Statistical Society Series B (Statistical Methodology) 2005, 67(2):301–320. [Google Scholar]
- 36.Fizazi K, Higano CS, Nelson JB, Gleave M, Miller K, Morris T, Nathan FE, McIntosh S, Pemberton K, Moul JW: Phase III, randomized, placebo-controlled study of docetaxel in combination with zibotentan in patients with metastatic castration-resistant prostate cancer. J Clin Oncol 2013, 31(14):1740–1747. [DOI] [PubMed] [Google Scholar]
- 37.White IR, Royston P: Imputing missing covariate values for the Cox model. Stat Med 2009, 28(15):1982–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim S, Halabi S: High Dimensional Variable Selection with Error Control. Biomed Res Int 2016, 2016:8209453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pi L, Halabi S: Combined Performance of Screening and Variable Selection Methods in Ultra-High Dimensional Data in Predicting Time-To-Event Outcomes. Diagn Progn Res 2018, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao SD, Li Y: Principled sure independence screening for Cox models with ultra-high-dimensional covariates. J Multivar Anal 2012, 105(1):397–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Halabi S, Pi L: Statistical Considerations for Developing and Validating Prognostic Models of Clinical Outcomes In: Oncology Clinical Trials : Successful Design, Conduct, and Analysis. Edited by Kelly WKDO Halabi SP, 2nd edn. New York: Springer Publishing Company; 2018: 313–322. [Google Scholar]
- 42.He Z, Tu W, Wang S, Fu H, Yu Z: Simultaneous variable selection for joint models of longitudinal and survival outcomes. Biometrics 2015, 71(1):178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y, Wang Y: Variable selection for joint models of multivariate longitudinal measurements and event time data. Statistics in medicine 2017, 36(24):3820–3829. [DOI] [PubMed] [Google Scholar]
- 44.Varadhan R, Segal JB, Boyd CM, Wu AW, Weiss CO: A framework for the analysis of heterogeneity of treatment effect in patient-centered outcomes research. JCE Journal of Clinical Epidemiology 2013, 66(8):818–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabler NB, Duan N, Liao D, Elmore JG, Ganiats TG, Kravitz RL: Dealing with heterogeneity of treatment effects: is the literature up to the challenge? Trials Trials 2009, 10(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kravitz RL, Duan N, Braslow J: Evidence-Based Medicine, Heterogeneity of Treatment Effects, and the Trouble with Averages. The Milbank Quarterly 2004, 82(4):661–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruberg SJ, Lei C, Yanping W: The mean does not mean as much anymore: finding sub-groups for tailored therapeutics. Clinical Trials Clinical Trials: Journal of the Society for Clinical Trials 2010, 7(5):574–583. [DOI] [PubMed] [Google Scholar]
- 48.Wang R, Schoenfeld DA, Hoeppner B, Evins AE: Detecting treatment-covariate interactions using permutation methods. Statistics in medicine 2015, 34(12):2035–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Foster JC, Taylor JM, Ruberg SJ: Subgroup identification from randomized clinical trial data. Stat Med Statistics in Medicine 2011, 30(24):2867–2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dane A, Spencer A, Rosenkranz G, Lipkovich I, Parke T, Analysis PEWGoS: Subgroup analysis and interpretation for phase 3 confirmatory trials: White paper of the EFSPI/PSI working group on subgroup analysis. Pharm Stat 2018. [DOI] [PubMed] [Google Scholar]
- 51.Lipkovich I, Dmitrienko A: Strategies for identifying predictive biomarkers and subgroups with enhanced treatment effect in clinical trials using SIDES. J Biopharm Stat 2014, 24(1):130–153. [DOI] [PubMed] [Google Scholar]
- 52.Lipkovich I, Dmitrienko ABR: Tutorial in biostatistics: data-driven subgroup identification and analysis in clinical trials. Statistics in medicine 2017, 36(1):136–196. [DOI] [PubMed] [Google Scholar]
- 53.Kattan MW, Hess KR, Amin MB, Lu Y, Moons KG, Gershenwald JE, Gimotty PA, Guinney JH, Halabi S, Lazar AJ et al. : American Joint Committee on Cancer acceptance criteria for inclusion of risk models for individualized prognosis in the practice of precision medicine. CA Cancer J Clin 2016, 66(5):370–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wulfsohn MS, Tsiatis AA: A joint model for survival and longitudinal data measured with error. Biometrics 1997:330–339. [PubMed] [Google Scholar]
- 55.Tsiatis AA, Davidian M: Joint modeling of longitudinal and time-to-event data: an overview. Statistica Sinica 2004:809–834. [Google Scholar]
- 56.Henderson R, Diggle P, Dobson A: Joint modelling of longitudinal measurements and event time data. Biostatistics 2000, 1(4):465–480. [DOI] [PubMed] [Google Scholar]
- 57.Rizopoulos D: JM: An R package for the joint modelling of longitudinal and time-to-event data. Journal of Statistical Software 2010, 35(9):1–33.21603108 [Google Scholar]
- 58.Rizopoulos D: Joint models for longitudinal and time-to-event data: With applications in R: Chapman and Hall/CRC; 2012. [Google Scholar]
- 59.Rizopoulos D, Hatfield LA, Carlin BP, Takkenberg JJ: Combining dynamic predictions from joint models for longitudinal and time-to-event data using Bayesian model averaging. Journal of the American Statistical Association 2014, 109(508):1385–1397. [Google Scholar]
- 60.Rizopoulos D: The R Package JMbayes for Fitting Joint Models for Longitudinal and Time-to-Event Data Using MCMC. Journal of Statistical Software 2016, 72(7):1–46 [Google Scholar]
- 61.Li K, Luo S: Dynamic predictions in Bayesian functional joint models for longitudinal and time-to-event data: An application to Alzheimer’s disease. Statistical methods in medical research 2019, 28(2):327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rizopoulos D: Dynamic predictions and prospective accuracy in joint models for longitudinal and time-to-event data. Biometrics 2011, 67(3):819–829. [DOI] [PubMed] [Google Scholar]
- 63.Anderson JR, Cain KC, Gelber RD: Analysis of survival by tumor response. J Clin Oncol 1983, 1(11):710–719. [DOI] [PubMed] [Google Scholar]
- 64.Van Houwelingen H, Putter H: Dynamic prediction in clinical survival analysis: CRC Press; 2011. [Google Scholar]
- 65.Van Houwelingen HC: Dynamic prediction by landmarking in event history analysis. Scandinavian Journal of Statistics 2007, 34(1):70–85. [Google Scholar]
- 66.Zheng Y, Heagerty PJ: Partly conditional survival models for longitudinal data. Biometrics 2005, 61(2):379–391. [DOI] [PubMed] [Google Scholar]
- 67.Breslow NE: Discussion of Professor Cox’s paper. J Royal Stat Soc B 1972, 34:216–217. [Google Scholar]
- 68.Jewell NP, Nielsen JP: A framework for consistent prediction rules based on markers. Biometrika 1993, 80(1):153–164. [Google Scholar]
- 69.Rizopoulos D, Molenberghs G, Lesaffre EM: Dynamic predictions with time-dependent covariates in survival analysis using joint modeling and landmarking. Biometrical Journal 2017, 59(6):1261–1276. [DOI] [PubMed] [Google Scholar]
- 70.Suresh K, Taylor JM, Spratt DE, Daignault S, Tsodikov A: Comparison of joint modeling and landmarking for dynamic prediction under an illness-death model. Biometrical Journal 2017, 59(6):1277–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferrer L, Putter H, Proust-Lima C: Individual dynamic predictions using landmarking and joint modelling: validation of estimators and robustness assessment. Statistical methods in medical research 2017:0962280218811837. [DOI] [PubMed] [Google Scholar]
- 72.Yao F, Müller H-G, Wang J-L: Functional data analysis for sparse longitudinal data. Journal of the American Statistical Association 2005, 100(470):577–590. [Google Scholar]
- 73.Yao F, Müller HG, Clifford AJ, Dueker SR, Follett J, Lin Y, Buchholz BA, Vogel JS: Shrinkage estimation for functional principal component scores with application to the population kinetics of plasma folate. Biometrics 2003, 59(3):676–685. [DOI] [PubMed] [Google Scholar]
- 74.Yao F, Müller H-G, Wang J-L: Functional linear regression analysis for longitudinal data. The Annals of Statistics 2005, 33(6):2873–2903. [Google Scholar]
- 75.Yao F: Functional principal component analysis for longitudinal and survival data. Statistica Sinica 2007:965–983. [Google Scholar]
- 76.Yan F, Lin X, Huang X: Dynamic prediction of disease progression for leukemia patients by functional principal component analysis of longitudinal expression levels of an oncogene. The Annals of Applied Statistics 2017, 11(3):1649–1670. [Google Scholar]
- 77.Yan F, Lin X, Li R, Huang X: Functional principal components analysis on moving time windows of longitudinal data: dynamic prediction of times to event. Journal of the Royal Statistical Society: Series C 2018, 67(4):961–978. [Google Scholar]
- 78.Li K, Luo S: Bayesian functional joint models for multivariate longitudinal and time-to-event data. Computational statistics data analysis 2019, 129:14–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Halabi S, Conaway MR, Small EJ, Vogelzang NJ, Dawson NA: Serum Prostate Specific Antigen as a Predictor of Survival in Prostate Cancer Patients Treated with Second-Line Hormonal Therapy (CALGB 9181). TPR The Prostate Journal 2001, 3(1):18–25. [Google Scholar]
- 80.Fontein D, Klinten Grand M, Nortier JW, Seynaeve C, Meershoek-Klein Kranenbarg E, Dirix L, van de Velde C, Putter H: Dynamic prediction in breast cancer: proving feasibility in clinical practice using the TEAM trial. Annals of Oncology 2015, 26(6):1254–1262. [DOI] [PubMed] [Google Scholar]
- 81.Proust-Lima C, Taylor JM: Development and validation of a dynamic prognostic tool for prostate cancer recurrence using repeated measures of posttreatment PSA: a joint modeling approach. Biostatistics 2009, 10(3):535–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sène M, Taylor JM, Dignam JJ, Jacqmin-Gadda H, Proust-Lima C: Individualized dynamic prediction of prostate cancer recurrence with and without the initiation of a second treatment: Development and validation. Statistical methods in medical research 2016, 25(6):2972–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor JM, Park Y, Ankerst DP, Proust-Lima C, Williams S, Kestin L, Bae K, Pickles T, Sandler HJB: Real-time individual predictions of prostate cancer recurrence using joint models. Biometrics 2013, 69(1):206–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yu M, Law NJ, Taylor JM, Sandler HM: Joint longitudinal-survival-cure models and their application to prostate cancer. Statistica Sinica 2004:835–862. [Google Scholar]
- 85.Yu M, Taylor JMG, Sandler HM: Individual prediction in prostate cancer studies using a joint longitudinal survival–cure model. Journal of the American Statistical Association 2008, 103(481):178–187. [Google Scholar]
- 86.Król A, Ferrer L, Pignon JP, Proust-Lima C, Ducreux M, Bouché O, Michiels S, Rondeau V: Joint model for left-censored longitudinal data, recurrent events and terminal event: Predictive abilities of tumor burden for cancer evolution with application to the FFCD 2000–05 trial. Biometrics 2016, 72(3):907–916. [DOI] [PubMed] [Google Scholar]
- 87.Wang J, Luo S, Li L: Dynamic prediction for multiple repeated measures and event time data: An application to Parkinson’s disease. The annals of applied statistics 2017, 11(3):1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shoulson I, Group PS: DATATOP: a decade of neuroprotective inquiry. Annals of neurology 1998, 44(S1 1):S160–S166. [PubMed] [Google Scholar]
- 89.Steyerberg EW: Clinical prediction models : a practical approach to development, validation, and updating; 2010. [Google Scholar]
- 90.Steyerberg EW, Harrell FE: Prediction models need appropriate internal, internal-external, and external validation. JCE Journal of Clinical Epidemiology 2016, 69:245–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Moons KG, de Groot JA, Bouwmeester W, Vergouwe Y, Mallett S, Altman DG, Reitsma JB, Collins GS: Critical appraisal and data extraction for systematic reviews of prediction modelling studies: the CHARMS checklist. PLoS Med 2014, 11(10):e1001744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moons KG, Altman DG, Reitsma JB, Ioannidis JP, Macaskill P, Steyerberg EW, Vickers AJ, Ransohoff DF, Collins GS: Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015, 162(1):W1–73. [DOI] [PubMed] [Google Scholar]
- 93.Petrylak DP, Vogelzang NJ, Budnik N, Wiechno PJ, Sternberg CN, Doner K, Bellmunt J, Burke JM, de Olza MO, Choudhury A et al. : Docetaxel and prednisone with or without lenalidomide in chemotherapy-naive patients with metastatic castration-resistant prostate cancer (MAINSAIL): a randomised, double-blind, placebo-controlled phase 3 trial. Lancet Oncol 2015, 16(4):417–425. [DOI] [PubMed] [Google Scholar]
- 94.Heagerty PJ, Zheng Y: Survival Model Predictive Accuracy and ROC Curves. Biometrics Biometrics 2005, 61(1):92–105. [DOI] [PubMed] [Google Scholar]
- 95.Uno H, Cai TX, Tian L, Wei LJ: Evaluating prediction rules for t-year survivors with censored regression models. Journal of the American Statistical Association 2007, 102(478):527–537. [Google Scholar]
- 96.Halabi S, Pi L, Lin C: Developing and Validating Prognostic Models of Clinical Outcomes In: Textbook of Clinical Oncology: A Statitical Perspective. Edited by Halabi S, Michiels S: Chapman & Hall/CRC; 2019. [Google Scholar]
- 97.First line Metastatic Castrate-Resistant Prostate Cancer Patients. [https://www.cancer.duke.edu/Nomogram/firstlinechemotherapy.html]