Abstract
A transcriptional program implemented by transcription factors and epigenetic regulators governs cardiac development and disease. Mutations in these factors are important causes of congenital heart disease. Here we review selected recent advances in our understanding of the transcriptional and epigenetic control of heart development, including determinants of cardiac transcription factor chromatin occupancy, the gene regulatory network that regulates atrial septation, the chromatin landscape and cardiac gene regulation, and the role of BAF, NuRD, and Polycomb epigenetic regulatory complexes in heart development.
The mammalian heart develops through an intricate dance that transforms the linear heart tube into a four-chambered structure while simultaneously maintaining effective embryonic blood flow. The intricate choreography required perhaps explains why among liveborn infants the heart is the organ most frequently affected by major malformations. (Pierpont et al. 2018) Initial studies of rare forms of inherited heart malformations identified mutations of the transcriptional regulators T-Box 5 (TBX5) (Basson et al. 1997), NK2 homeobox 5 (Nkx2–5) (Schott et al. 1998), and GATA binding protein 4 (GATA4) (Garg et al. 2003) as causes of familial congenital heart disease. This initial impression that mutations in regulators of gene transcription are important causes of CHD has been borne out in whole exome sequencing studies of patients with apparently sporadic, non-syndromic congenital heart disease, which showed that CHD probands have an excess burden of de novo mutations that impact transcriptional and epigenetic regulatory pathways (Zaidi et al. 2013; Homsy et al. 2015; Jin et al. 2017).
This review will highlight recent advances in understanding the genetic and epigenetic control of heart development. Given the wealth of emerging information on transcriptional control of cardiac development, a comprehensive review was not possible within the allocated space and instead we focused on selected topics.
Cardiac transcription factor chromatin occupancy
The genome sequence contains the instructions to turn genes on and off. How this information is encoded and the machinery that reads and implements these regulatory instructions is incompletely understood. It is clear that DNA-binding transcription factors (TFs) are central players, since they recognize specific binding sites in DNA and thereby recruit other regulatory proteins, including factors that lack intrinsic sequence specificity. However, the principles that determine transcription factor chromatin occupancy remain obscure. Several studies have provided new insights about the regulation of cardiac transcription factor occupancy during heart development.
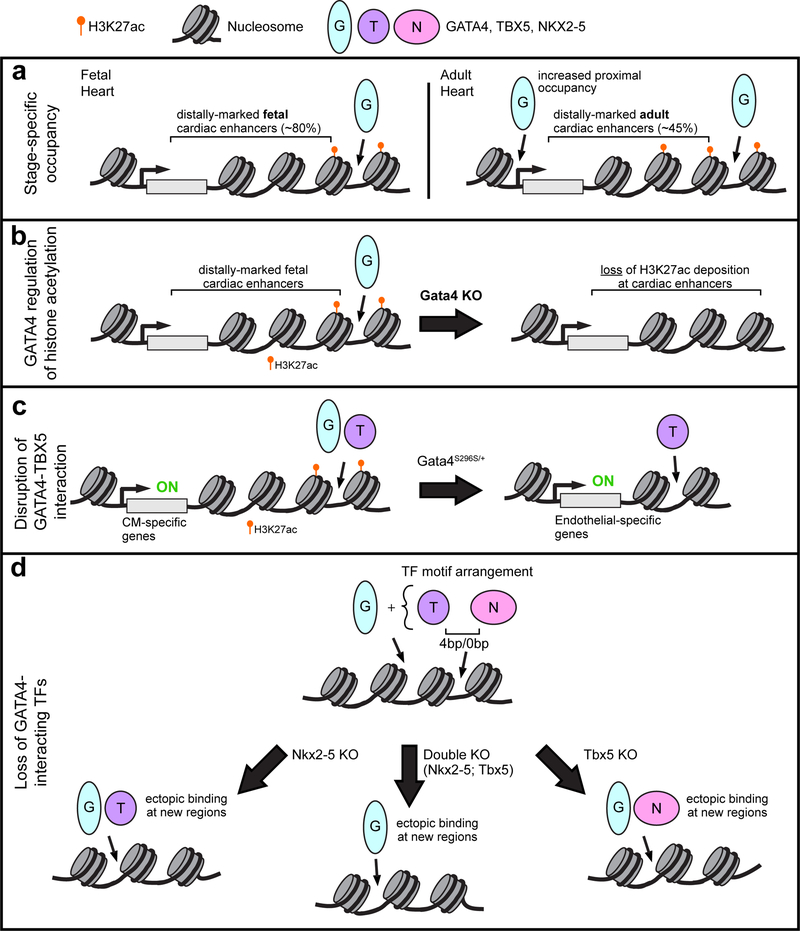
GATA4 is an essential regulator of cardiac development that is mutated in familial as well as sporadic congenital heart disease (Rajagopal et al. 2007; Zeisberg et al. 2005; Pu et al. 2004; Garg et al. 2003; Molkentin et al. 1997). Groups have studied the dynamic change of chromatin occupancy by the key cardiac TF GATA binding protein 4 (GATA4) during heart development (He et al. 2014). A high affinity epitope tag (AviTag, which undergoes in vivo biotinylation by the E. coli enzyme BirA) that was knocked into the C-terminus of GATA4 to allow highly sensitive and reproducible GATA4 bioChIP-seq. The majority of regions occupied by GATA4 in fetal (E12.5) heart were not occupied by GATA4 in adult heart. Interestingly adult GATA4 occupancy occurred closer to promoters (Fig. 1a). GATA4 regions were highly enriched for the active enhancer mark H3K27ac and GATA4 inactivation reduced H3K27ac signal at a subset of GATA4 regions (Fig. 1b). This is consistent with GATA4’s known interaction with EP300 (Dai et al. 2002), which writes H3K27ac. Pathological stress (pressure-overload) also dynamically altered GATA4 occupancy; interestingly, only a minority of pressure-overload-induced GATA4 regions reflected re-acquisition of fetal occupancy, whereas most of these disease-associated GATA4 regions were not observed during normal heart development. Contrary to the adage that pathological stress activates a fetal gene program, this observation suggests that pathological stress predominantly activates a transcriptional program that is distinct from the program that regulates genes during normal development. Ongoing studies of several other central cardiac transcription factors indicates that dynamic transcription factor chromatin occupancy is a common theme, rather than a unique property of GATA4 (Akerberg, Gu, and Pu, unpublished data).
Figure 1:
Transcription factor (TF) interactions. a. GATA4 occupies regions in a stage specific manner. GATA4 bioChIP-seq data revealed that GATA4 predominantly binds distal (>2kb from transcription start sites, TSS) enhancers (H3K27ac) in fetal heart. The majority of fetal binding sites were not occupied by GATA4 in adult heart, and overall a greater proportion of GATA4 occupancy was proximal to the TSS. b. Depletion of GATA4 in the embryonic mouse heart resulted in loss of H3K27ac epigenetic marks at candidate cardiac enhancers. c. GATA4 and TBX5 coordinately regulate transcription of cardiac genes through binding enhancers, which are often marked by H3K27ac. A GATA4 point mutation found in a family with septal defects and cardiomyopathy abolishes physical interaction with TBX5 and leads to dysregulated gene expression, in part through ectopic TBX5 occupancy at endothelial-related genes. d. A subset of sides co-occupied by NKX2–5 and TBX5 revealed a distinct binding motif orientation and spacing (0 bp or 4 bp) and frequent co-occupancy by GATA4. Loss of either NKX2–5 or TBX5 resulted in ectopic binding (binding to regions in mutant that were not found in wild-type) of the other TF partner in combination with GATA4. Similarly, in cells with double knock-out of both NKX2–5 and TBX5, GATA4 was found ectopically bound to new genomic regions.
One important determinant of cardiac transcription factor occupancy is the interaction with other TFs. Mutations in GATA4 that perturb its interaction with TBX5 cause both septal and atrioventricular defects (Garg et al. 2003; Maitra et al. 2009; Misra et al. 2012). Interestingly, both mice and humans that lack GATA4 are at risk for developing cardiac dysfunction (Bisping et al. 2006; Ang et al. 2016); in mice this risk varies by genetic background, suggestive of important genetic modifiers (Rajagopal et al. 2007). Srivastava and colleagues used human induced pluripotent stem cell (iPSC)-derived cardiomyocytes (iPSC-CM) to study the pathogenic mechanisms by which a heterozygous GATA4-G296S mutation, causes septal defects and cardiomyopathy (Ang et al. 2016). This mutation both disrupts GATA4-TBX5 interaction and reduces GATA4 DNA binding affinity (Garg et al. 2003). iPSC-CMs harboring this mutation showed reduced contractile function, increased sarcomere disorganization, calcium handling abnormalities, and abnormal metabolism. Transcriptome analysis of the iPSC-derived cardiac progenitors and cardiomyocytes showed that downregulated genes were enriched for functional annotations related to heart development and contraction, whereas upregulated genes were enriched for vascular development. These gene expression changes were linked to reduced activation of cardiac genes and impaired silencing of endothelial genes. In wild-type iPSC-CMs, over 50% of GATA4 sites were co-occupied by TBX5, which were enriched for functional terms related to heart muscle and contraction. The GATA4-G296S mutation resulted in loss of 54% of normal GATA4 binding sites. Interestingly, 15% of the sites occupied by the mutant protein were “ectopic”, i.e. not occupied by the wild-type protein. Furthermore, the GATA4 mutation also altered TBX5 occupancy, with 26% of normal TBX5 sites being lost and 24% of its binding sites being ectopic in the mutant cells. Reduction of both GATA4 and TBX5 occupancy was linked to loss of cardiac super-enhancers, dense clusters of transcriptional enhancers that are implicated in lineage identify (Whyte et al. 2013), and downregulation of cardiac genes, whereas reduction of GATA4 and gain of TBX5 occupancy was associated with endothelial gene upregulation (Fig. 1c). Overall, this study revealed that GATA4-TBX5 interaction promotes proper chromatin occupancy by both TFs, which is required to activate cardiac genes and repress endothelial genes in cardiac cells.
Harvey and colleagues studied the effect of NKX2–5 mutations on NKX2–5 chromatin occupancy at gene promoters using DNA adenine methyltransferase identification (DamID) (Bouveret et al. 2015). Two different NKX2–5 mutations -- NKX2–5-Y191C, in which a human CHD missense mutation interferes with normal DNA binding, and NKX2–5ΔHD, in which the DNA binding domain has been deleted -- were compared to wild-type NKX2–5. Both mutants retains the ability to occupy chromatin at a subset of the wild-type sites, via heterodimerization with the wild-type protein through a tyrosine rich domain. Furthermore, the NKX2–5 mutants occupied ectopic sites, which were not enriched for cardiac TF motifs, specifically the NKX2–5 motif. Rather, the mutant proteins occupied these regions by heterodimerizing with ELK1/4, a subset of widely expressed ETS family transcription factors that normally interact with wild-type NKX2–5 at a subset of binding sites. This study reinforces the conclusion that CHD-causing TF mutations result in not only loss of normal TF occupancy, but also drive ectopic TF occupancy and aberrant gene expression through protein-protein interactions with other TFs.
Bruneau and colleagues studied the interplay of TBX5 and NKX2–5 in cardiac transcriptional regulation using murine embryonic stem cells directed to undergo cardiac differentiation (Luna-Zurita et al. 2016). Knockout of TBX5 or NKX2–5 individually (TKO or NKO) or both factors together (DKO) identified complex individual and combined requirement of these factors for normal expression of cardiac genes, regulation of cell cycle genes, and repression of ectopic, non-cardiac genes. High resolution mapping of TBX5, NKX2–5, and GATA4 chromatin occupancy via chromatin immunoprecipitation combined with exonuclease digestion (ChIP-exo) demonstrated that regions co-occupied by all three factors had the highest chromatin occupancy signal, and neighboring genes were most likely to be differentially expressed in the knockout cells. This analysis also identified a preferred spacing of 4 bp or 0 bp and head-to-head orientation of TBX5 and NKX2–5 motifs, suggesting a preferred composite motif recognized by TBX5-NKX2–5 heterodimers (Fig. 1d). This conclusion was supported by a crystal structure of a TBX5-NKX2–5-DNA ternary complex, which demonstrated a small TBX5-NKX2–5 protein-protein interface that contributes to cooperative binding of the factors to DNA. Knockout of NKX2–5 or TBX5 alone or in combination resulted in loss of a subset of normal binding sites by the remaining factors, as well as gain of ectopic binding sites (Fig. 1d). Both loss and ectopic occupancy changes were functionally significant, as they were associated with differential expression of subsets of genes in the knockout cell lines. This study reinforced the conclusion that the interplay of TFs establishes normal TF chromatin occupancy and prevents ectopic TF binding with associated abnormal gene activation.
Cardiac TF Gene Regulatory Networks in Atrial Septation
Given the interdependence between GATA4, NKX2–5, TBX5, and other cardiac TFs for chromatin occupancy and gene activation, it is not surprising that these factors genetically interact. Patients and mice with heterozygous GATA4, NKX2–5, and TBX5 mutations often have defects in atrial septation (Garg et al. 2003; Schott et al. 1998; Basson et al. 1997; Rajagopal et al. 2007; Biben et al. 2000; Bruneau et al. 2001). Recent studies have defined TBX5- and GATA4- dependent gene regulatory networks that govern atria septation.
TBX5 is required for atrial septation in a subdomain of the second heart field, the posterior second heart field, that contains cardiopulmonary progenitors (CPPs), which give rise to both atrial and pulmonary structures (Xie et al. 2012). Within CPPs, TBX5 directly activates expression of WNT2/2b, which signals to endoderm to promote pulmonary endoderm specification (Steimle et al. 2018). Pulmonary endoderm expresses sonic hedgehog (Shh), which in turn signals back to CPPs via GLI TFs. GLI and TBX5 collaborate to drive atrial septal progenitor specification and proliferation (Xie et al. 2012; Hoffmann et al. 2014).
The requirement for GATA4 in atrial septation was also localized the posterior second heart field (pSHF) (Zhou et al. 2017a). GATA4 is genetically upstream of Shh signaling, because constitutive activation of Shh in CPPs rescued embryos from atrial septal defects caused by heterozygous GATA4 mutation. Consistent with this observation, GATA4 directly regulated the expression of Shh signaling components within CPPs. Interestingly, the penetrance of atrial septal defects in mice with heterozygous GATA4 mutation is strain dependent (Rajagopal et al. 2007), suggesting genetic modifiers that modulate the sensitivity of CPPs to GATA4 dosage.
Epigenetic regulation of heart development
DNA is tightly wound around histones, which both compacts the genome and regulates its accessibility for transcription. This creates an epigenetic layer of gene regulation that is superimposed upon the primary genome sequence. CHD patients have increased burden of de novo mutations in epigenetic regulatory factors, directly implicating these protein complexes the pathogenesis of structural heart disease (Zaidi et al. 2013).
Dynamic chromatin landscape
Epigenetic chromatin features, including DNA methylation, post-translational covalent histone modifications, and chromatin accessibility, are important regulators of gene expression and development. For example, histone H3 acetylated at lysine 27 (H3K27ac) or monomethylated at lysine 4 (H3K4me1) are associated with active and poised transcriptional enhancers, and H3K27me3 is associated with transcriptional silencing. Extensive efforts have documented the cardiomyocyte epigenome during stem cell to cardiomyocyte differentiation (Wamstad et al. 2012; Paige et al. 2012) and normal heart development (Nord et al. 2013; He et al. 2014; Gilsbach et al. 2014, 2018). These studies show that the epigenome is highly dynamic, with both DNA methylation and histone modifications changing extensively between fetal, adult, and disease states.
Cis-regulatory elements
Availability of these genome-wide datasets has permitted prediction of the function of genomic regions. Attention has primarily been focused on transcriptional enhancers, in part because the best developed experimental assay of cis-regulatory activity, transient transgenesis, measures enhancer activity. Enhancers predicted by specific chromatin features, such as occupancy by H3K27ac (May et al. 2012; Nord et al. 2013), EP300 (Visel et al. 2009; Blow et al. 2010; Zhou et al. 2017b), or cardiac TFs (He et al. 2011, 2014), have been experimentally tested by transient transgenesis, and the results of thousands of tests across multiple developing organs are accessible through the VISTA Enhancer Browser (Visel et al. 2007). Generally, approximately half of the regions decorated by these candidate features have heart activity in transient transgenic assays.
Based largely on this database of regions with validated heart activity, efforts have been made to develop algorithms that integrate information on multiple chromatin features to score a region’s likelihood to be an active cardiac enhancer (Dickel et al. 2016; Erwin et al. 2014; van Duijvenboden et al. 2016). While such predictive algorithms are sorely needed, these efforts were limited by available training data. While the VISTA enhancer database has been invaluable, it is limited (1912 human and 1062 murine enhancers tested; 140 and 160 with cardiac activity, respectively) and inherently biased by the manner in which the regions to be tested were selected. Application of high throughput enhancer assays (Inoue and Ahituv 2015) to cardiac development and disease will create a more extensive and representative database of cardiac enhancer activity. Using these more robust training data and predictive algorithms, it will be possible to develop more extensive and accurate annotations of cardiac enhancer activity within non-coding regions.
Chromatin Remodeling Complexes
ATP-dependent chromatin remodeling complexes catalyze nucleosome eviction or repositioning (Hota and Bruneau 2016). The most studied remodeling complex in the heart is the SWItch/Sucrose Non-Fermentable (SWI/SNF) family, a diverse class of remodeling complexes that share either that has been most studied in the heart. This heterogenous complex contains at least 10 subunits, with the catalytic subunit encoded by either Brg1 or Brm, leading to the alternative moniker of Brg/Brahma-associated factor (BAF) complex. BAF complexes occupy both gene promoters and distal cis-regulatory regions, and both activate and repress genes in a context-dependent manner (Hota and Bruneau 2016; Hota et al. 2019).
BAF complexes have specific subunit compositions in different cell types, and the normal transition between BAF complex subunits has been implicated in precursor differentiation to definitive cell types. The subunit composition of the BAF complex was recently defined at several stages during differentiation of murine ESCs to cardiomyocytes by mass spectrometry analysis of proteins that co-immunoprecipitate with BRG1 (Hota et al. 2019). This revealed the changing composition of the BAF complex during cardiomyocyte differentiation, and identified novel interacting proteins. Among these interacting proteins was WDR5, a member of the MLL complex that deposits H3K4me3. WDR5 is mutated in patients with CHD (Zaidi et al. 2013).
Among the most notable BAF subunit changes was BAF60C (Smarcd3) and BAF170 replacing BAF60A (Smarcd1) and BAF155, respectively, BAF in cardiac progenitors and cardiomyocytes (Hota et al. 2019). This change in BAF60 isoforms is consistent with observations in murine embryos, in which Smarcd3 was selectively expressed in the heart between E7.5-E9.5, and its depletion by RNAi or Cre-loxP conditional knockout (Nkx2–5Cre) disrupted cardiac morphogenesis and impaired proliferation and differentiation of second heart field progenitors and cardiomyocytes (Lickert et al. 2004; Sun et al. 2018). BAF60C interacted with TBX5, NKX2–5, and GATA4 and promoted their activity in luciferase transcriptional assays, suggesting that it acts as an interface between cardiac TFs and the BAF complex (Lickert et al. 2004). In keeping with TBX5 and GATA4 recruitment of the BAF complex, BRG1-occupied regions significantly overlapped with TBX5 and GATA4 in cardiomyocyte progenitors and cardiomyocytes (Hota et al. 2019). In fact, ectopic expression of BAF60C with TBX5 and GATA4 drove non-cardiac mesoderm to differentiate into beating cardiomyocytes (Takeuchi and Bruneau 2009). BAF60C is required for expression of multiple components of the contractile machinery through interactions with myocardin (MYOCD), a transcriptional co-activator of cardiac TFs SRF and MEF2C (Sun et al. 2018). Interestingly, ablation of BAF60C or BAF170 did not abrogate BAF complexes but rather altered their composition, with associated changes in BRG1 occupancy and chromatin accessibility. These data indicate that the subunit composition is developmentally regulated and cell type specific, and critical for BAF complex activation or repression of genes. The cardiac-enriched subunits interface with cardiac transcription factors and co-activators to direct cardiac gene expression.
BAF complexes are also essential in the non-cardiomyocyte components of the heart. Brg1 inactivation in the endothelial (Tie2:Cre), but not myocardial, compartment caused defective myocardial trabeculation (Stankunas et al. 2008). This defect was linked to depletion of cardiac jelly, the extracellular matrix between endocardium and myocardium which is required for signaling pathways essential for trabeculation. Endocardium depleted for BRG1 expressed abnormally high levels ADAMTS1, a metalloproteinase that degrades key cardiac jelly components. Adamts1 transcription was directly repressed by BRG1 in endocardial cells, and its inhibition rescued cardiac jelly and trabeculation. Endocardial BRG1 is also required for normal semilunar valve development, since endocardium-specific (NfatcCre) deletion of Brg1 caused abnormal ECM organization in the developing semilunar valves that impaired endocardial-to-mesenchymal transitions in the proximal outflow tract, resulting in bicuspid aortic valve (Akerberg et al. 2015). Together these studies show that BAF complex repressive activity governs endocardial cell extracellular matrix deposition, which regulates development of both the myocardium and the cardiac valves.
Additionally, the BAF complex is also required in the epicardium, the mesothelial sheet that covers the myocardium (Vieira et al. 2017). During embryonic heart development, BRG1 bound within the promoter and first intron of Wt1, a transcription factor required for normal epicardium development and formation of epicardium-derived cells (EPDC) via epithelial-to-mesenchymal transition. Loss of Brg1 in EPDCs (Wt1:Cre) significantly diminished Wt1 levels, reduced formation of EPDCs and their derivatives, such as coronary vascular smooth muscle cells.
Nucleosome Remodeling and Histone Deacetylation (NuRD)
The Nucleosome Remodeling and Histone Deacetylation (NuRD) complex couples ATP-dependent chromatin remodeling with histone deacetylation. Like BAF complexes, NuRD is a multi-subunit complex with alternative isoforms for several components. The ATP-dependent chromatin remodeling activity is provided by CHD3 and CHD4, also known as Mi-2α and Mi-2β, respectively, and the histone deacetylase activity resides in HDAC1 or HDAC2 (Torchy et al. 2015). The complex contains proteins that recognize multiple chromatin features, including histone tails, methylated lysine, and DNA methylation, suggesting that the complex can integrate information from these epigenetic features to regulate histone deacetylation and nucleosome positioning. In the absence of ATP, the NuRD complex primarily deacetylases histone octamers, not nucleosomes, suggesting that chromatin remodeling is required for HDAC activity on chromatin (Xue et al. 1998).
Chd4 mutations cause congenital heart disease including atrial and ventricular septal defects (Homsy et al. 2015). In mouse models, Chd4 is required for early cardiogenesis, as early inactivation by Nkx2–5Cre resulted in thinning of the compact myocardium and reduced myocardial trabeculation, which was due to decreased cardiomyocyte proliferation. Transcriptome analysis revealed derepression of smooth and skeletal muscle gene expression programs. Both up- and down-regulated genes were associated with enrichment of CHD4 binding, and the most differentially expressed genes (DEGs) correlated with CHD4 enrichment distal to promoter regions, suggesting a role for the NuRD complex to regulate transcriptional enhancer usage (Wilczewski et al. 2018). Chd4 inactivation in skeletal muscle led to the reciprocal observation that cardiac-specific muscle gene isoforms were derepressed (Gómez-Del Arco et al. 2016).
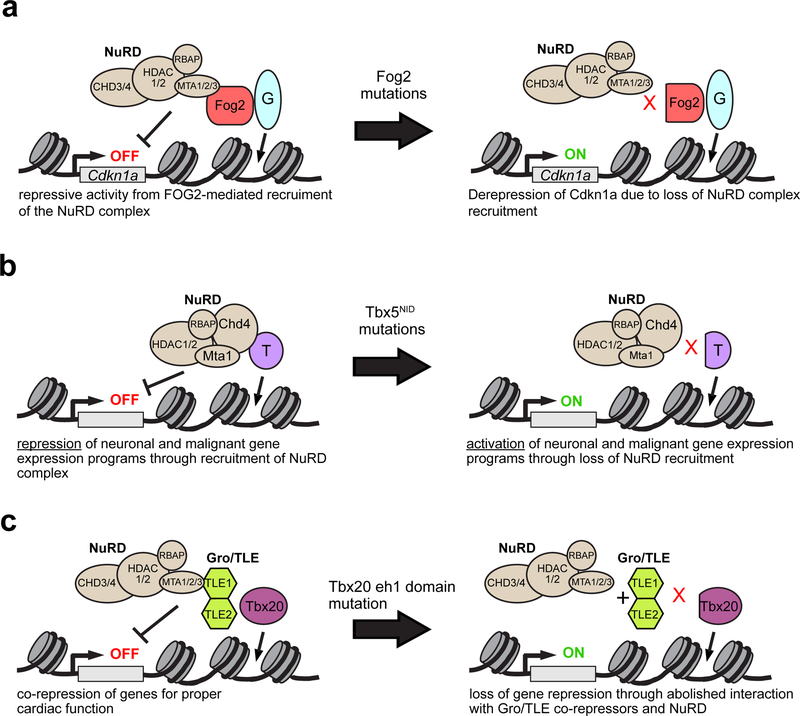
NuRD interacts with at least three key cardiac transcriptional regulators: FOG2 (Roche et al. 2008), TBX5 (Waldron et al. 2016), and TBX20 (Kaltenbrun et al. 2013). FOG2, a protein that binds to GATA4 and often represses genes at a subset of targets, interacts with MTA and RbAp subunits of NuRD via a 12 amino acid sequence at its N-terminus. This interaction was required for FOG2-mediated transcriptional repression in vitro. In vivo, disruption of this interaction via introduction of precise mutations into the FOG2 N-terminal interaction domain causes atrial and ventricular septal defects, thin ventricular myocardium, and reduced cardiomyocyte proliferation (Garnatz et al. 2014) (Fig. 2a).
Figure 2:
Nucleosome Remodeling and Deacetylase (NuRD) complex and cardiac gene regulation.a. The NuRD complex interacts with FOG2, a direct modulator of GATA4 transcriptional activity. One target of GATA4-FOG2-NuRD repression was Cdkn1a. b. TBX5 directly interacts with NuRD complex subunits CHD4, HDAC2 and MTA1 to repress gene expression programs not normally expressed during heart development. TBX5 mutations in its NuRD interaction domain (NID) abolished direct interaction with the NuRD complex, leading to expression of normally repressed, ectopic gene programs. c. Physical interaction between TBX20 and the NuRD complex is mediated through Gro/TLE co-repressors. Specifically, TBX20 eh1 domain is essential for proper transcriptional regulation by the Gro/TLE-NuRD complexes.
An unbiased proteomics screen of TBX5-interacting proteins identified NuRD components CHD4 and MTA1 (Waldron et al. 2016). This interaction is likely functionally significant, as Tbx5+/−;Mta1+/− doubly heterozygous mice developed more severe cardiac septation defects than Tbx5+/− mice. The study identified a region of TBX5 necessary and sufficient for NuRD interaction. This NuRD Interaction Domain (NID) is affected by Tbx5 mutations that cause Holt Oram Syndrome and its associated cardiac defects, and these mutations impaired TBX5-NURD interaction (Waldron et al. 2016) (Fig. 2b). A similar proteomics screen of TBX20-interacting proteins also identified NuRD components including MTA1 and HDAC2, and mutation of the TBX20 interaction domain disrupted TBX20-Gro/TLE-NuRD-mediated transcriptional repression in Xenopus embryos (Kaltenbrun et al. 2013) (Fig. 2c).
Polycomb Repressor Complexes
The polycomb genes were initially discovered in Drosophila by their requirement for silencing Hox gene expression (Lewis 1978). Many of these genes encode factors active in epigenetic repression. Interestingly, subsequent screens for polycomb antagonists uncovered the trithorax genes, many of which encode BAF complex subunits (Schuettengruber et al. 2017).
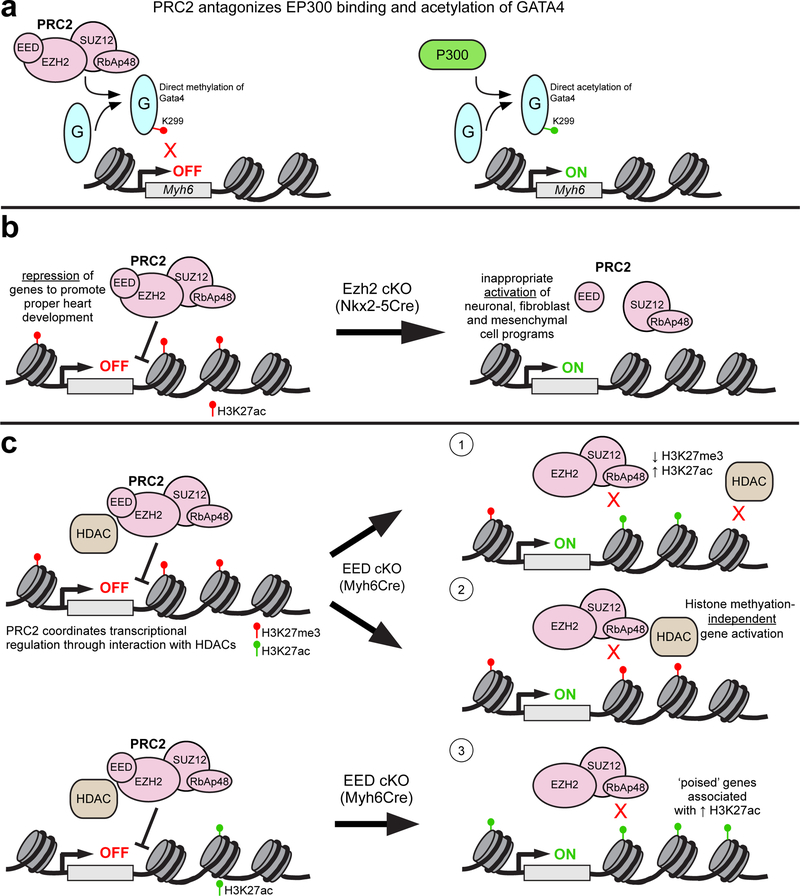
Based on biochemical purification experiments, polycomb proteins have been grouped into two main complexes: polycomb repressive complex 1 (PRC1) and polycomb repressive complex 2 (PRC2). The core subunits of PRC2 contains enhancer of zeste (either EZH1 or EZH2), which catalyzes trimethylation of histone H3 on lysine 27 (H3K27me3), embryonic ectoderm development (EED), and suppressor of zeste (SUZ12). H3K27me3 is a major repressive epigenetic mark, and writing of this mark has been thought to be a major mechanism of PRC-mediated gene repression. Surprisingly, EZH2 directly interacted with and monomethylated GATA4 (He et al. 2012b). GATA4 methylation limited its recruitment of the transcriptional co-activator EP300, which activates cardiac gene transcription by acetylating GATA4 (Trivedi et al. 2010) as well as histone H3 at lysine 27 (Fig. 3a). This study identified PRC2 methylation of non-histone targets as additional mechanisms of gene repression.
Figure 3:
Polycomb Repressive Complex 2 (PRC2). a. PRC2 binds to directly methylate K299 residue on GATA4 to attenuate transcriptional activation of downstream targets such as Myh6. This repressive activity of PRC2 is balanced by the EP300, which competes with PRC2 for GATA4 binding and directly acetylates GATA4 at the same residue, resulting in target gene activation. b. EZH2, the catalytic subunit of the PRC2 complex, is required for proper heart development. Conditional knock-out (cKO) of EZH2 by Nkx2–5Cre in cardiac progenitors globally decreased H3K27me3 levels and inappropriately de-repressed neuronal, fibroblast and mesenchymal gene programs in embryonic mouse hearts. c. Myh6-specific deletion of EED, a core PRC2 component, leads to derepression of many genes. These de-repressed genes fall into three categories based on their associated epigenetic marks: 1) increased H3K27me3 and increased H3K27ac, consistent with loss of EED-dependent HDAC activity; 2) retained H3K27me3, consistent with histone methylation-independent gene regulation; and 3) little H3K27me3 in control and increased H3K27ac in EEDKO, suggestive of “poised” genes that are activated by relief of PRC2 and HDAC repression.
Through gene inactivation studies in mice, the function of PRC2 in cardiac development and postnatal cardiac function has been carefully dissected. Inactivation of EZH2 in cardiac progenitors (Nkx2–5Cre or Mef2-SHF-Cre) severely disrupted heart morphogenesis by de-repressing ectopic gene expression programs and cell cycle inhibitors, which reduced cardiomyocyte proliferation (He et al. 2012a; Delgado-Olguin et al. 2012) (Fig. 3b). Although selective EZH2 inactivation in the second heart field by Mef2-SHF-Cre disrupted normal cardiac development, it was compatible with survival to adulthood, and affected adult cardiomyocytes were significantly enlarged (Delgado-Olguin et al. 2012). However, EZH2 inactivation in differentiated cardiomyocytes did not affect heart development or postnatal heart function, suggesting that these defects in gene expression, proliferation, and postnatal cardiomyocyte hypertrophy reflect epigenetic scars induced by EZH2 loss in cardiac progenitors (He et al. 2012a; He and Pu 2012). Likely EZH2 is dispensable in differentiated cardiomyocytes due to functional redundancy with EZH1; indeed, embryos with cardiomyocyte deficiency of either EZH1 or EZH2 developed normally, but ablation of both factors caused lethal heart malformations (Ai et al. 2017b). Intriguingly, EZH1 but not EZH2 was required for neonatal heart regeneration. These studies demonstrate that EZH1 and EZH2 have partially overlapping functions, but each also possesses unique activity that is unmasked in specific developmental stages or disease contexts.
Unlike EZH1/2, the essential PRC2 component EED is not genetically redundant. Inactivation of EED in developing cardiomyocytes caused lethal cardiac morphogenesis defects including impaired cardiomyocyte proliferation (He et al. 2012a), consistent with similar findings in embryos lacking EZH1 and EZH2 (Ai et al. 2017b). Interestingly, postnatal deletion of EED caused lethal dilated cardiomyopathy associated with de-repression of ectopic gene programs, such as expression of skeletal muscle gene isoforms (Ai et al. 2017a). Surprisingly, de-repressed genes were not enriched for loss of H3K27me3, suggesting that PRC2 represses these genes through alternative, non-canonical mechanisms (Fig. 3c). There was a global increase of H3K27ac in EED knockout cardiomyocytes and de-repressed genes gained this activating mark. AAV-mediated EED re-expression rescued the cardiomyopathy phenotype and restored H3K27ac but not H3K27me3. Mechanistically, EED was found to interact with multiple HDACs and to increase HDAC activity. In support of this model, AAV-mediated HDAC1/2 over-expression partially rescued the EED knockout cardiomyopathy phenotype. Collectively these data indicate that EED regulation of gene expression in postnatal cardiomyocytes occurs through non-canonical mechanisms. Most studies of PRC2 activity have been performed in actively cycling cells, which may emphasize repressive mechanisms that rely on PRC2-mediated H3K27me3 deposition. However, in non-cycling cells such as postnatal cardiomyocytes, EED regulates gene expression through alternative, HDAC-dependent mechanisms.
PRC1 is the second major complex formed by polycomb group proteins. The core of PRC1 is composed of RING1 proteins (RING1A (RNF1) or RING1B (RNF2)), which catalyze H2A lysine 119 ubiquitination, and one of six polycomb group ring-finger domain proteins (PCGF1-PCGF6). The canonical PRC1 complex contains a chromobox protein (CBX2, CBX4, or CBX6–CBX8) and a polyhomeotic (Ph) homologous protein (PHC1–3), whereas non-canonical PRC1 complexes contain RING1 And YY1 Binding Protein (RYBP) (Schuettengruber et al. 2017). The roles of these PRC1 complexes in heart development have not been studied as extensively as PRC2, although both canonical and non-canonical complexes have been implicated. Loss of RNF2, common to both canonical and non-canonical PRC1, in zebrafish de-repressed Tbx2/3 in working myocardium, resulting in downregulation of chamber specific genes (Chrispijn et al. 2019). Non-canonical PRC1 subunit RYBP was required for formation of beating cardiomyocytes in murine ESC differentiation (Ujhelly et al. 2015), and inactivation of canonical PRC1 subunit PHC1 (also known as Rae28) caused aberrant cardiac morphogenesis and impaired expression of Nkx2–5 (Shirai et al. 2002). The molecular mechanism by which repressor inactivation leads to Nkx2–5 downregulation remains to be determined.
Although polycomb-mediated gene repression is clearly essential for cardiac development and adult heart function, the specific mechanisms that govern its chromatin occupancy are poorly defined. In general, polycomb complexes are recruited to chromatin by several potential mechanisms. In mammals, a subset of polycomb proteins occupy hypomethylated CG islands (Boyer et al. 2006), suggesting that DNA sequence composition and methylation influence recruitment. Polycomb complexes also directly interact with some transcription factors, including GATA4 (He et al. 2012b) and YY1 (Basu et al. 2014), which are active in cardiac progenitors and cardiomyocytes. Long non-coding RNAs regulate cardiac development and function (Salamon et al. 2018), and some have been shown to recruit PRC2 to specific repressed loci (Wang et al. 2016; Plath et al. 2003; Kanhere et al. 2010). Polycomb complexes can also be recruited by binding to epigenetic marks. For example, EED binds to H3K27me3 and H3K9me3, and this interaction contributes to H3K27me3 maintenance (Margueron et al. 2009). PRC1 chromobox proteins also bind H3K27me3, suggesting a model in which PRC2 hierarchically recruits PRC1 (Wang et al. 2004). PRC1 chromatin occupancy is also determined by controlled chromatin expulsion, as BRG1 has been shown to catalyze ATP-dependent PRC1 eviction (Stanton et al. 2017). It will be important to determine how these general polycomb recruitment strategies are deployed in heart development and disease to regulate cardiac gene expression.
Concluding comments
Over the past decade, application of exome and genome sequencing to congenital heart disease has highlighted the important pathogenic contribution of mutations in genes that regulate transcription during cardiac development (Zaidi et al. 2013; Homsy et al. 2015; Jin et al. 2017). Mechanistic studies in model organisms has yielded insights into the intricate process by which transcription factors and epigenetic readers and writers interact to shape the chromatin landscape and activate and repress gene expression in a precise, spatiotemporally regulated manner. Continuing basic and translational efforts will reveal answers to many important questions, including: the systematic annotation of non-coding transcriptional regulatory regions in heart development, homeostasis, and disease; the contribution of sequence variants in non-coding transcriptional regulatory regions to congenital heart disease; the role of three dimensional chromatin structure in cardiac development and disease; the influence of mutations in transcriptional and epigenetic regulators on long term function of cardiac structures, in addition to their impact on initial cardiac morphogenesis; and the effect of oligogenic mutations on heart development.
Acknowledgements
BNA was supported by T32HL07572. WTP was supported by HL098166 and HL146634.
References
- Ai S, Peng Y, Li C, Gu F, Yu X, Yue Y, Ma Q, Chen J, Lin Z, Zhou P, et al. 2017a. EED orchestration of heart maturation through interaction with HDACs is H3K27me3-independent. Elife 6 10.7554/eLife.24570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai S, Yu X, Li Y, Peng Y, Li C, Yue Y, Tao G, Li C, Pu WT, He A. 2017b. Divergent Requirements for EZH1 in Heart Development Versus Regeneration. Circ Res 121: 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerberg BN, Sarangam ML, Stankunas K. 2015. Endocardial Brg1 disruption illustrates the developmental origins of semilunar valve disease. Dev Biol 407: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Y-S, Rivas RN, Ribeiro AJS, Srivas R, Rivera J, Stone NR, Pratt K, Mohamed TMA, Fu J-D, Spencer CI, et al. 2016. Disease Model of GATA4 Mutation Reveals Transcription Factor Cooperativity in Human Cardiogenesis. Cell 167: 1734–1749.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson CT, Bachinsky DR, Lin RC, Levi T, Elkins JA, Soults J, Grayzel D, Kroumpouzou E, Traill TA, Leblanc-Straceski J, et al. 1997. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15: 30–35. [DOI] [PubMed] [Google Scholar]
- Basu A, Wilkinson FH, Colavita K, Fennelly C, Atchison ML. 2014. YY1 DNA binding and interaction with YAF2 is essential for Polycomb recruitment. Nucleic Acids Res 42: 2208–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biben C, Weber R, Kesteven S, Stanley E, McDonald L, Elliott DA, Barnett L, Koentgen F, Robb L, Feneley M, et al. 2000. Cardiac septal and valvular dysmorphogenesis in mice heterozygous for mutations in the homeobox gene Nkx2–5. Circ Res 87: 888–895. [DOI] [PubMed] [Google Scholar]
- Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, Rajagopal S, Son JK, Ma Q, Springer Z, et al. 2006. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci U S A 103: 14471–14476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, McCulley DJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat Genet 42: 806–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouveret R, Waardenberg AJ, Schonrock N, Ramialison M, Doan T, de Jong D, Bondue A, Kaur G, Mohamed S, Fonoudi H, et al. 2015. NKX2–5 mutations causative for congenital heart disease retain functionality and are directed to hundreds of targets. Elife 4 10.7554/eLife.06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Bruneau BG, Nemer G, Schmitt JP, Charron F, Robitaille L, Caron S, Conner DA, Gessler M, Nemer M, Seidman CE, et al. 2001. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106: 709–721. [DOI] [PubMed] [Google Scholar]
- Chrispijn ND, Elurbe DM, Mickoleit M, Aben M, de Bakker DEM, Andralojc KM, Huisken J, Bakkers J, Kamminga LM. 2019. Loss of the Polycomb group protein Rnf2 results in derepression of tbx-transcription factors and defects in embryonic and cardiac development. Sci Rep 9: 4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y-S, Cserjesi P, Markham BE, Molkentin JD. 2002. The transcription factors GATA4 and dHAND physically interact to synergistically activate cardiac gene expression through a p300-dependent mechanism. J Biol Chem 277: 24390–24398. [DOI] [PubMed] [Google Scholar]
- Delgado-Olguin P, Huang Y, Li X, Christodoulou D, Seidman CE, Seidman JG, Tarakhovsky A, Bruneau BG. 2012. Epigenetic repression of cardiac progenitor gene expression by Ezh2 is required for postnatal cardiac homeostasis. Nat Genet 44: 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickel DE, Barozzi I, Zhu Y, Fukuda-Yuzawa Y, Osterwalder M, Mannion BJ, May D, Spurrell CH, Plajzer-Frick I, Pickle CS, et al. 2016. Genome-wide compendium and functional assessment of in vivo heart enhancers. Nat Commun 7: 12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erwin GD, Oksenberg N, Truty RM, Kostka D, Murphy KK, Ahituv N, Pollard KS, Capra JA. 2014. Integrating diverse datasets improves developmental enhancer prediction. PLoS Comput Biol 10: e1003677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, Schluterman MK, King IN, Butler CA, Rothrock CR, Eapen RS, Hirayama-Yamada K, Joo K, et al. 2003. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424: 443–447. [DOI] [PubMed] [Google Scholar]
- Garnatz AS, Gao Z, Broman M, Martens S, Earley JU, Svensson EC. 2014. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev Biol 395: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R, Preissl S, Grüning BA, Schnick T, Burger L, Benes V, Würch A, Bönisch U, Günther S, Backofen R, et al. 2014. Dynamic DNA methylation orchestrates cardiomyocyte development, maturation and disease. Nat Commun 5: 5288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilsbach R, Schwaderer M, Preissl S, Grüning BA, Kranzhöfer D, Schneider P, Nührenberg TG, Mulero-Navarro S, Weichenhan D, Braun C, et al. 2018. Distinct epigenetic programs regulate cardiac myocyte development and disease in the human heart in vivo. Nat Commun 9: 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-Del Arco P, Perdiguero E, Yunes-Leites PS, Acín-Pérez R, Zeini M, Garcia-Gomez A, Sreenivasan K, Jiménez-Alcázar M, Segalés J, López-Maderuelo D, et al. 2016. The Chromatin Remodeling Complex Chd4/NuRD Controls Striated Muscle Identity and Metabolic Homeostasis. Cell Metab 23: 881–892. [DOI] [PubMed] [Google Scholar]
- He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, Pu WT. 2014. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun 5: 4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. 2011. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci 108: 5632–5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Ma Q, Cao J, von Gise A, Zhou P, Xie H, Zhang B, Hsing M, Christodoulou DC, Cahan P, et al. 2012a. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ Res 110: 406–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Pu WT. 2012. Mature cardiomyocytes recall their progenitor experience via polycomb repressive complex 2. Circ Res 111: 162–164. [DOI] [PubMed] [Google Scholar]
- He A, Shen X, Ma Q, Cao J, von Gise A, Zhou P, Wang G, Marquez VE, Orkin SH, Pu WT. 2012b. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev 26: 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP. 2014. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet 10: e1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. 2015. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science 350: 1262–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Bruneau BG. 2016. ATP-dependent chromatin remodeling during mammalian development. Development 143: 2882–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hota SK, Johnson JR, Verschueren E, Thomas R, Blotnick AM, Zhu Y, Sun X, Pennacchio LA, Krogan NJ, Bruneau BG. 2019. Dynamic BAF chromatin remodeling complex subunit inclusion promotes temporally distinct gene expression programs in cardiogenesis. Development. 10.1242/dev.174086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue F, Ahituv N. 2015. Decoding enhancers using massively parallel reporter assays. Genomics 106: 159–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al. 2017. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nat Genet 49: 1593–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL. 2013. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res 12: 5395–5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell 38: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature 432: 107–112. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MAH, He D, Small EM, Mileikovsky M, et al. 2016. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164: 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maitra M, Schluterman MK, Nichols HA, Richardson JA, Lo CW, Srivastava D, Garg V. 2009. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol 326: 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May D, Blow MJ, Kaplan T, McCulley DJ, Jensen BC, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, et al. 2012. Large-scale discovery of enhancers from human heart tissue. Nat Genet 44: 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra C, Sachan N, McNally CR, Koenig SN, Nichols HA, Guggilam A, Lucchesi PA, Pu WT, Srivastava D, Garg V. 2012. Congenital heart disease-causing Gata4 mutation displays functional deficits in vivo. PLoS Genet 8: e1002690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Lin Q, Duncan SA, Olson EN. 1997. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev 11: 1061–1072. [DOI] [PubMed] [Google Scholar]
- Nord AS, Blow MJ, Attanasio C, Akiyama JA, Holt A, Hosseini R, Phouanenavong S, Plajzer-Frick I, Shoukry M, Afzal V, et al. 2013. Rapid and pervasive changes in genome-wide enhancer usage during mammalian development. Cell 155: 1521–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paige SL, Thomas S, Stoick-Cooper CL, Wang H, Maves L, Sandstrom R, Pabon L, Reinecke H, Pratt G, Keller G, et al. 2012. A temporal chromatin signature in human embryonic stem cells identifies regulators of cardiac development. Cell 151: 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont ME, Brueckner M, Chung WK, Garg V, Lacro RV, McGuire AL, Mital S, Priest JR, Pu WT, Roberts A, et al. 2018. Genetic Basis for Congenital Heart Disease: Revisited: A Scientific Statement From the American Heart Association. Circulation 138: e653–e711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300: 131–135. [DOI] [PubMed] [Google Scholar]
- Pu WT, Ishiwata T, Juraszek AL, Ma Q, Izumo S. 2004. GATA4 is a dosage-sensitive regulator of cardiac morphogenesis. Dev Biol 275: 235–244. [DOI] [PubMed] [Google Scholar]
- Rajagopal SK, Ma Q, Obler D, Shen J, Manichaikul A, Tomita-Mitchell A, Boardman K, Briggs C, Garg V, Srivastava D, et al. 2007. Spectrum of heart disease associated with murine and human GATA4 mutation. J Mol Cell Cardiol 43: 677–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche AE, Bassett BJ, Samant SA, Hong W, Blobel GA, Svensson EC. 2008. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J Mol Cell Cardiol 44: 352–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamon I, Saccani Jotti G, Condorelli G. 2018. The long noncoding RNA landscape in cardiovascular disease: a brief update. Curr Opin Cardiol 33: 282–289. [DOI] [PubMed] [Google Scholar]
- Schott JJ, Benson DW, Basson CT, Pease W, Silberbach GM, Moak JP, Maron BJ, Seidman CE, Seidman JG. 1998. Congenital heart disease caused by mutations in the transcription factor NKX2–5. Science 281: 108–111. [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Bourbon H-M, Di Croce L, Cavalli G. 2017. Genome regulation by Polycomb and Trithorax: 70 years and counting. Cell 171: 34–57. [DOI] [PubMed] [Google Scholar]
- Shirai M, Osugi T, Koga H, Kaji Y, Takimoto E, Komuro I, Hara J, Miwa T, Yamauchi-Takihara K, Takihara Y. 2002. The Polycomb-group gene Rae28 sustains Nkx2.5/Csx expression and is essential for cardiac morphogenesis. J Clin Invest 110: 177–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankunas K, Hang CT, Tsun ZY, Chen H, Lee NV, Wu JI, Shang C, Bayle JH, Shou W, Iruela-Arispe ML, et al. 2008. Endocardial Brg1 represses ADAMTS1 to maintain the microenvironment for myocardial morphogenesis. Dev Cell 14: 298–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton BZ, Hodges C, Calarco JP, Braun SMG, Ku WL, Kadoch C, Zhao K, Crabtree GR. 2017. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 49: 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steimle JD, Rankin SA, Slagle CE, Bekeny J, Rydeen AB, Chan SS-K, Kweon J, Yang XH, Ikegami K, Nadadur RD, et al. 2018. Evolutionarily conserved Tbx5-Wnt2/2b pathway orchestrates cardiopulmonary development. Proc Natl Acad Sci 115: E10615–E10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Hota SK, Zhou Y-Q, Novak S, Miguel-Perez D, Christodoulou D, Seidman CE, Seidman JG, Gregorio CC, Henkelman RM, et al. 2018. Cardiac-enriched BAF chromatin-remodeling complex subunit Baf60c regulates gene expression programs essential for heart development and function. Biol Open 7 10.1242/bio.029512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Bruneau BG. 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchy MP, Hamiche A, Klaholz BP. 2015. Structure and function insights into the NuRD chromatin remodeling complex. Cell Mol Life Sci 72: 2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Zhu W, Wang Q, Jia C, Kee HJ, Li L, Hannenhalli S, Epstein JA. 2010. Hopx and Hdac2 interact to modulate Gata4 acetylation and embryonic cardiac myocyte proliferation. Dev Cell 19: 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujhelly O, Szabo V, Kovacs G, Vajda F, Mallok S, Prorok J, Acsai K, Hegedus Z, Krebs S, Dinnyes A, et al. 2015. Lack of Rybp in Mouse Embryonic Stem Cells Impairs Cardiac Differentiation. Stem Cells Dev 24: 2193–2205. [DOI] [PubMed] [Google Scholar]
- van Duijvenboden K, de Boer BA, Capon N, Ruijter JM, Christoffels VM. 2016. EMERGE: A flexible modelling framework to predict genomic regulatory elements from genomic signatures. Nucleic Acids Res 44: e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira JM, Howard S, Villa Del Campo C, Bollini S, Dubé KN, Masters M, Barnette DN, Rohling M, Sun X, Hankins LE, et al. 2017. BRG1-SWI/SNF-dependent regulation of the Wt1 transcriptional landscape mediates epicardial activity during heart development and disease. Nat Commun 8: 16034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA. 2007. VISTA Enhancer Browser--a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, et al. 2016. The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation. Dev Cell 36: 262–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamstad JA, Alexander JM, Truty RM, Shrikumar A, Li F, Eilertson KE, Ding H, Wylie JN, Pico AR, Capra JA, et al. 2012. Dynamic and coordinated epigenetic regulation of developmental transitions in the cardiac lineage. Cell 151: 206–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS. 2004. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell 14: 637–646. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zhang XJ, Ji YX, Zhang P, Deng KQ, Gong J, Ren S, Wang X, Chen I, Wang H, et al. 2016. The long noncoding RNA Chaer defines an epigenetic checkpoint in cardiac hypertrophy. Nature Medicine 22: 1131–1139. 10.1038/nm.4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. 2013. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153: 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilczewski CM, Hepperla AJ, Shimbo T, Wasson L, Robbe ZL, Davis IJ, Wade PA, Conlon FL. 2018. CHD4 and the NuRD complex directly control cardiac sarcomere formation. Proc Natl Acad Sci 115: 6727–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP. 2012. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell 23: 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W. 1998. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2: 851–861. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. 2013. De novo mutations in histone-modifying genes in congenital heart disease. Nature 498: 220–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisberg EM, Ma Q, Juraszek AL, Moses K, Schwartz RJ, Izumo S, Pu WT. 2005. Morphogenesis of the right ventricle requires myocardial expression of Gata4. J Clin Invest 115: 1522–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Liu J, Xiang M, Olson P, Guzzetta A, Zhang K, Moskowitz IP, Xie L. 2017a. Gata4 potentiates second heart field proliferation and Hedgehog signaling for cardiac septation. Proceedings of the National Academy of Sciences 114: E1422–E1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Gu F, Zhang L, Akerberg BN, Ma Q, Li K, He A, Lin Z, Stevens SM, Zhou B, et al. 2017b. Mapping cell type-specific transcriptional enhancers using high affinity, lineage-specific Ep300 bioChIP-seq. Elife 6 10.7554/eLife.22039. [DOI] [PMC free article] [PubMed] [Google Scholar]