Abstract
HIV-1 can evolve HLA-specific escape variants in response to HLA-mediated cellular immunity. HLA alleles that are common in the host population may increase the frequency of such escape variants at the population level. When loss of viral fitness is caused by immune escape variation, these variants may revert upon infection of a new host who does not have the corresponding HLA allele. Furthermore, additional escape variants may appear in response to the nonconcordant HLA alleles. Because individuals with rare HLA alleles are less likely to be infected by a partner with concordant HLA alleles, viral populations infecting hosts with rare HLA alleles may undergo a greater amount of evolution than those infecting hosts with common alleles due to the loss of preexisting escape variants followed by new immune escape. This hypothesis was evaluated using maximum likelihood phylogenetic trees of each gene from 272 full-length HIV-1 sequences. Recent viral evolution, as measured by the external branch length, was found to be inversely associated with HLA frequency in nef (p<0.02), env (p<0.03), and pol (p≤0.05), suggesting that rare HLA alleles provide a disproportionate force driving viral evolution compared to common alleles, likely due to the loss of preexisting escape variants during early stages postinfection.
Introduction
HLA-mediated cytotoxic T cell (CTL) responses are thought to be critical for effective control of HIV-1. HLA class I molecules can present peptide antigens on the surface of infected cells, targeting them for removal by the cytotoxic T lymphocytes. However, HIV-1 can escape the HLA-mediated CTL response through the evolution of variants within the targeted epitopes, abolishing recognition by the CTL. Escape from a CTL response can be balanced by detrimental impacts on viral replication. For example, individuals with HLA B*57 or B*5801 will select for the evolution of the escape variant T242N in Gag.1,2 However, this variant has a decreased replication capacity, identified in vitro, that is partially maintained even after the selection of compensatory mutations.3,4 This may explain the reduced viral loads observed in HLA B*57 or B*5801-infected individuals compared to others.2,4-8 T242N will revert when transmitted to an individual without HLA B*57 or B*5801.1,2 Similarly, other CTL escape variants have been identified that cause lower viral fitness than the nonescaped variant through a variety of mechanisms.3,9-12
HLA alleles that are common in a given population may also cause the increased frequency of corresponding CTL escape variants. In particular, mutations with slower reversion times might be most likely to have a greater frequency in a given population, Slow reversion times could be caused by the lack of a strong detrimental impact on viral replication capacity, the presence of compensatory mutations that partially stabilize the escape variant, or amino acid changes that require more than one nucleotide change. Thus, in a transmission chain including several individuals with common HLA alleles, escape mutations that are slow to revert may be maintained at high frequency. Because a large proportion of HIV transmission is thought to occur during the acute stage of infection13 when there is little adaptive immune response in the host, preexisting escape mutations can conceivably be transmitted to an individual without the corresponding HLA alleles, but not have sufficient time to revert before being transmitted again to another individual. In this way, CTL escape variants could be maintained in a transmission chain involving some individuals without the corresponding HLA alleles.
Individuals with rare HLA alleles are more likely to be infected by viral variants with preexisting escape mutations to discordant HLA alleles, compared to individuals with common HLA alleles. After infection, viral variants with optimal fitness will dominate the viral population within the host, resulting in the loss of some preexisting escape mutations. In addition, the virus population is likely to evolve additional escape mutations in response to cellular immunity mediated by the rare HLA allele or alleles. Thus, we hypothesized that a greater amount of viral evolution will be necessary for the virus to achieve an adapted state in an individual with rare HLA alleles compared to those with common alleles, possibly providing additional time for the immune response to be effective during the early stages of infection.
A previous study compared the CTL responses to peptides targeted by HLA B*1503 from two populations, one in a subtype B-infected population where the HLA allele was rare and the other in a subtype C-infected population where the HLA allele was common.14 Subtype-specific sequence differences in the consensus peptide epitopes were found to be associated with the lack of recognition of several subdominant epitopes, suggesting that these differences were escape variants that had reached fixation in the population where the HLA allele was common.14 In the same study, HLAB*1503 was associated with lower viral load among the subtype B cohort where it was rare and no such association was found among the subtype C cohort where it was common, suggesting a protective effect for those from the population where the allele was rare. Another study found lower viral loads among individuals with rare HLA supertypes, suggesting that HIV had partially adapted to the common alleles, providing a selective advantage to those with rare HLA alleles.15 Furthermore, in Thailand, common HLA alleles were found to be associated with higher viral loads.16 Although these previous studies all suggest that rare HLA alleles may confer a selective advantage, no one has yet evaluated the underlying mechanism at the population level, that is, whether rare HLA alleles drive additional viral evolution. This study investigated the association of viral evolution and HLA frequency within a South African population using near full-length HIV-1 genome sequences.
Materials and Methods
Study population
The study participants were from KwaZulu Natal Province, South Africa and have been described separately.17 Previously reported full-length viral sequences18,19 from 272 members of the cohort were analyzed as part of a study approved by the University of Washington and University of KwaZulu Natal Internal Review Boards for which all participants gave informed consent. Four nonsubtype C and six hypervariable sequences were eliminated from further analysis. HLA class I genotypes were available on a larger subset of the South African participants (n=1119).17 CD4 counts were available for 164 participants with both HIV-1 sequence and HLA genotype information.17
Phylogenetic analysis
Sequences were aligned using ClustalW20 and MacClade version 4.08.21 Maximum likelihood trees were generated for gag, nef, env, pol, vpu, vpr, tat, rev, and vif gene sequences using PhyML.22 Tree parameters are given in Table 1 and the tree files are available at http://mullinslab.microbiol.washington.edu//publications/rousseau_2008_2/. Terminal branch lengthswere tabulated from the Newick tree file using NewickTermBranch (available at http://indra.mullins.microbiol.washington.edu) and the sums of branch lengths to the MRCA were calculated using DistParser (available at http://indra.mullins.microbiol.washington.edu).
Table 1.
Phylogenetic Tree Parameters
|
Nucleotide frequencies |
GTR relative rate parameters |
Gamma shape parameter |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | A | C | G | T | A↔C | A↔G | A↔T | C↔G | C↔T | G↔T | |
| gag | 0.366 | 0.182 | 0.242 | 0.210 | 2.250 | 6.816 | 1.034 | 0.969 | 9.124 | 1 | 0.573 |
| pol | 0.396 | 0.162 | 0.227 | 0.215 | 2.094 | 9.528 | 1.007 | 0.787 | 13.252 | 1 | 0.655 |
| vif | 0.359 | 0.167 | 0.244 | 0.230 | 2.777 | 6.860 | 0.684 | 0.397 | 8.170 | 1 | 0.667 |
| vpr | 0.351 | 0.198 | 0.246 | 0.204 | 2.604 | 7.951 | 1.124 | 0.843 | 15.944 | 1 | 0.719 |
| tat exon 1 | 0.321 | 0.230 | 0.220 | 0.230 | 3.775 | 6.324 | 1.199 | 1.087 | 10.843 | 1 | 0.595 |
| rev exon 2 | 0.288 | 0.199 | 0.283 | 0.230 | 1.773 | 5.553 | 0.515 | 0.460 | 5.006 | 1 | 0.767 |
| vpu | 0.396 | 0.086 | 0.286 | 0.233 | 2.218 | 2.377 | 0.658 | 0.817 | 12.063 | 1 | 0.694 |
| env | 0.345 | 0.171 | 0.238 | 0.247 | 2.250 | 5.738 | 0.845 | 0.660 | 5.895 | 1 | 0.611 |
| nef | 0.312 | 0.217 | 0.257 | 0.214 | 2.527 | 5.883 | 1.797 | 1.425 | 7.582 | 1 | 0.674 |
HLA data analysis
HLA (Class I; A, B, C) genotypes were determined for the 1119 infected individuals from South Africa.17 When only two-digit genotypes were resolved (n=405), the four-digit genotypes were probabilistically inferred using HLA Completion (http://microsoft.com/science), a novel tool that predicts the probabilities of the missing HLA information based on haplotype frequencies in similar populations.23
Linkage disequilibrium at the HLA locus was identified using HLA Linkage Disequilibrium (http://www.hiv.lanl.gov/content/immunology/hla/hla_linkage.html) and two-digit HLA allele designations. Adjustment for multiple comparisons was made using a p-value significance cutoff equal to 0.05/(total number of tests).
Statistical analysis
Ten datasets were imputed using the HLA allele probabilities. Because each study participant had two HLA alleles (with corresponding population frequencies) at each locus yet only a single measure of viral evolution, generalized estimating equations were used to evaluate the relationship between viral evolution (either terminal branch lengths or distance to MRCA) and HLA frequency, adjusting for year, cohort, CD4 counts, HLA homozygosity, and the remaining five HLA allele frequencies. The generalized estimating equations involved the assumption of Gaussian distribution of branch length, an independent correlation structure, and the Huber/White/sandwich estimator of variance. Linear regression was used to evaluate the association of HLA supertype frequency and viral evolution, adjusting for year, cohort, CD4 counts, HLA homozygosity, and the remaining five HLA allele frequencies. Linear regression was also used to evaluate the association of the presence of each individual HLA-A allele and terminal branch length adjusting for year, cohort, CD4 counts, HLA homozygosity, and the remaining five HLA allele frequencies. No correction for multiple comparisons was made in the analyses including branch lengths.
Results
The length of the terminal branch in a maximum likelihood phylogeny indicates the amount of evolution that has taken place since the most recent ancestor, including the evolution that took place within the present host. The appearance of escape variants and reversions will increase the length of the terminal branches in a phylogenetic tree. In this study, the average terminal branch length ranged from 0.05 to 0.09 corrected substitutions=site (Table 2). The average estimated HLA-A genotype frequency was 0.01 (0.00001–0.12), HLA-B 0.01 (0.00001–0.12), and HLA-C 0.03 (0.00001–0.15). The HLAA allele frequency was inversely associated with the length of the terminal branch from the maximum likelihood phylogenetic trees of env, nef, and pol (Table 3). Interestingly, all of the significant associations involved HLA-A. However, it is important to note that because HLA loci are clustered on the same chromosome, the alleles at these loci are often coin-herited. Thus, the recovered associations may reflect linkage rather than the effect of individual HLA genes.
Table 2.
Terminal Branch Lengths
| Gene | Average length | Range | Standard error |
|---|---|---|---|
| env | 0.09 | 0.03–0.24 | 0.0016 |
| gag | 0.06 | 0.01–0.11 | 0.0012 |
| nef | 0.06 | 0.01–0.13 | 0.0014 |
| pol | 0.05 | 0.02–0.10 | 0.0008 |
| rev | 0.06 | 0.01–0.17 | 0.0021 |
| tat | 0.05 | 0–0.18 | 0.0019 |
| vif | 0.06 | 0–0.15 | 0.0014 |
| vpr | 0.06 | 0–0.17 | 0.0017 |
| vpu | 0.07 | 0–0.19 | 0.0021 |
Table 3.
HLA Frequency Associated with Terminal Branch Length
| Gene | HLA | Coefficienta | p-value |
|---|---|---|---|
| env | HLA-A | −0.008 | 0.03 |
| nef | HLA-A | −0.008 | 0.02 |
| pol | HLA-A | −0.003 | 0.05 |
Reflecting the change for each 1.0% increase in HLA frequency with adjustment for year, cohort, CD4 T cell counts, HLA homozygocity, and remaining HLA allele frequencies.
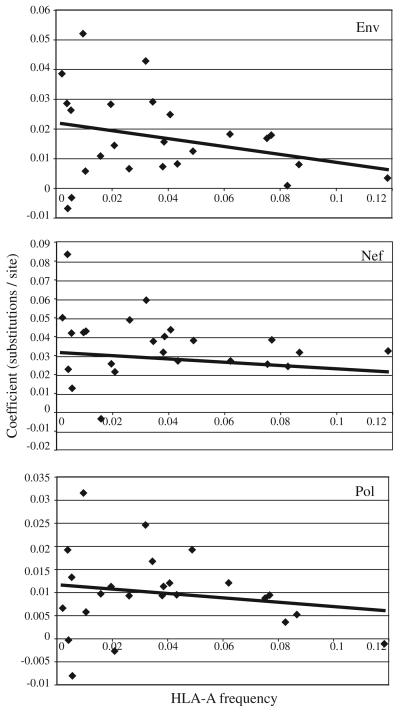
To determine if single alleles are driving these associations, the individual HLA-A alleles were evaluated for their relationship with external branch lengths in env, nef, and pol (Table 4). The impact of each allele on external branch length was evaluated in relation to allele frequency (Figure 1). Again, an inverse relationship was observed between HLA allele frequency and external branch length. Furthermore, two HLA-A alleles were found to be in linkage disequilibrium with HLA-B and HLA-C alleles (Table 4).
Table 4.
Individual HLA Alleles Associated with Terminal Branch Lengths
| Gene | HLA allele | Coefficienta | Standard error | p-value | Upper 95% CI | Lower 95% CI | HLA frequency |
|---|---|---|---|---|---|---|---|
| env | A4301 | 0.0428 | 0.0215 | 0.0470 | 0.0006 | 0.0850 | 0.0321 |
| nef | A3009 | 0.0435 | 0.0206 | 0.0360 | 0.0029 | 0.0840 | 0.0022 |
| A0214 | 0.0770 | 0.0204 | <0.001 | 0.0368 | 0.1172 | 0.0039 | |
| A2901 | 0.0353 | 0.0179 | 0.0490 | 0.0002 | 0.0705 | 0.0054 | |
| A2601 | 0.0364 | 0.0165 | 0.0280 | 0.0039 | 0.0688 | 0.0106 | |
| A2402b | 0.0423 | 0.0162 | 0.0090 | 0.0105 | 0.0741 | 0.0262 | |
| A4301 | 0.0528 | 0.0165 | 0.0020 | 0.0204 | 0.0852 | 0.0321 | |
| A0205 | 0.0310 | 0.0155 | 0.0460 | 0.0006 | 0.0614 | 0.0346 | |
| pol | A6602 | 0.0315 | 0.0111 | 0.0050 | 0.0096 | 0.0534 | 0.0097 |
| A4301 | 0.0245 | 0.0088 | 0.0060 | 0.0072 | 0.0419 | 0.0321 | |
| A0205 | 0.0167 | 0.0083 | 0.0440 | 0.0005 | 0.0330 | 0.0346 | |
| A3402c | 0.0192 | 0.0089 | 0.0320 | 0.0016 | 0.0368 | 0.0490 |
Representing the average branch length among those without the allele compared to those with the allele.
Linked to both HLA*B07 and HLA*C07, p-value=2.69×10−7.
Linked to both HLA*B44 and HLA*C04, p-value=6.50×10−8.
FIG. 1.
The HLA-A allele frequency is plotted along the x-axis and the linear regression coefficient along the y-axis. The coefficient can be interpreted as the change in terminal branch length (corrected substitutions per site) in the presence compared to the absence of each given allele. The line represents the linear regression line (p-value=n.s. for all) and is included for clarity.
The distance to the most recent common ancestor of the entire population, likely encompassing evolution throughout the duration of the subtype C epidemic, was not predicted to be influenced by the HLA from the most recently infected individual. And as expected, no significant association was found with HLA frequency and distance to the MRCA for any of the trees generated from each HIV gene.
HLA alleles can be grouped into supertypes24,25 based on the recognition of common motifs. HLA supertype frequency was not found to be associated with terminal branch length for any HIV gene, suggesting that this grouping, although increasing the power of the analysis by increasing sample size for each association evaluated, may have introduced misclassification bias by grouping HLA alleles with different impacts on HIV evolution, driving the association toward the null hypothesis.
Under the hypothesis of heterozygote advantage,26 individuals with heterozygous HLA alleles would select for a greater amount of viral evolution than homozygous individuals. Indeed, homozygosity at HLA-B was associated with shorter branch lengths in gag (two-sided t-test p < 0.0006), and homozygosity at HLA–A with shorter branch lengths in tat (two-sided t-test p < 0.012).
In general, CD4 counts decrease as the disease progresses. Thus, it would be expected that branch lengths would be inversely associated with CD4 counts, because the virus continues to evolve throughout the course of infection. Indeed, we observed that CD4 counts were inversely associated with vif (p < 0.008) and rev (p < 0.031) external branch lengths. Similarly, CD4 disease stage categories were associated with vif and rev external branch lengths (Table 5). However, viral load demonstrated no association with viral evolution in any HIV gene.
Table 5.
Associations with Terminal Branch Length
| Variable | HIV-1 gene | Coefficient | p-value |
|---|---|---|---|
| HLA homozygocitya | gag | — | 0.0006 |
| tat | — | 0.012 | |
| CD4 countsb | vif | −0.000018 | 0.008 |
| rev | −0.000021 | 0.031 | |
| CDC disease categoriesc | |||
| <200 cells/μl | vif | 0.010 | 0.005 |
| <200 cells/μl | rev | 0.013 | 0.002 |
Student's t-test (two-sided).
Bivariate linear regression with CD4 count as a linear variable.
Bivariate linear regression with >500 cell/μl as the reference group.
Discussion
In our study, HLA allele frequency was found to be inversely associated with HIV-1 terminal branch lengths for env, nef, and pol, suggesting that rare HLA alleles may be associated with additional viral evolution in these HIV-1 genes. This supports previous reports suggesting that HIV escape variants can be transmitted1,27-31 and may become common in the interhost viral population.14 Furthermore, some escape variants can impart a replication cost to the virus3,9-12 and these variants may be lost during early stages of infection.1,32-34 In fact, the proportion of such events has been approximated in previous studies and ranges from 8 to 42%.32,34 Together these investigations suggest that the frequency of the HLA allele in the host population impacts the evolution of HIV by influencing the amount of escape and reversion during early stages of infection. Genetic drift also affects viral evolution but, by definition, it is not impacted by the immune response. The passage of time is associated with genetic drift and for this reason we adjusted for time in our multivariate analysis, reducing the potential for confounding due to this variable. Furthermore, the duration of infection will also impact the external branch lengths, due to genetic bottlenecking that takes place at the time of transmission. For this reason, we adjusted for CD4 count as a measure of disease progression. Neither genetic drift nor duration of infection was likely to be associated with HLA allele frequency, reducing the likelihood that these variables confounded our results.
One of the most daunting challenges in the design of an effective vaccine against HIV-1 is the enormous amount of genetic diversity observed among sequences identified worldwide. Developing a vaccine that targets elements common to all HIV-1 infections is a challenging task. One approach has been to identify sequences present in early infection with the hope that the factors selecting for the transmitted variants will result in common sequences or sequence patterns that can be incorporated into effective immunogens. An encouraging idea is that viruses undergo a procession toward an ancestral state of higher fitness prior to the evolution of escape mutations.35 A vaccine targeting this ancestral state, theoretically common to all infections, could prevent the virus from achieving high replication capacity very early after transmission, possibly resulting in eradication of the virus or reduced disease progression. Our study suggests that individuals with rare HLA alleles will have a greater number of evolutionary events, possibly due to increased reversions toward this ancestral state. Thus, sequences from individuals with rare HLA alleles may optimally assist in the identification of this ancestral state.
Viruses infecting individuals with rare HLA alleles would likely have both reduced replication capacity due to preexisting escape mutations as well as susceptibility to the immune response because the preexisting escape mutations would not protect against the immune response mediated by the rare HLA allele. This could lead to reduced viral load among those with rare HLA alleles. Supporting this hypothesis, two recent studies have identified an indirect association of viral load with the presence of transmitted escape variants selected for by discordant HLA earlier in the transmission chain.8,36 While we were able to detect an association between HLA frequency and viral evolution, no significant association was found in our analysis when HLA frequency was evaluated in relation to viral load. We may not have observed an association in this study because the difference in viral load among those with and without the escape variants may be most striking during the initial stages of infection. Because our study was cross-sectional, the majority of the study participants were likely to be in the asymptomatic stage of the infection. In addition, different HLA alleles have been shown to be associated both with increased and decreased disease progression,37 suggesting that the HLA-mediated immune response may have a complex set of effects, preventing the observation of an overall association of HLA-driven evolution with disease outcome. These and other unidentified misclassifications could explain why the clinical impact of a putative advantage of rare HLA alleles, possibly detectable in prospective analyses, may have been obscured in this cross-sectional study.
Although we evaluated the full HIV proteome, including all nine genes, no correction for multiple comparisons was performed because these genes were not independent, complicating the identification of an appropriate test. Given a p-value cutoff of 0.05 and the conservative Bonferroni method of correction for multiple comparisons, one would expect ≤1 association due to chance. Because our results identified three of nine associations, we conclude that our analysis provides significant evidence that population HLA class I allelic frequency has a substantial impact on viral evolution. Furthermore, this analysis evaluated a linear relationship between HLA frequency and HIV evolution as a logical first-order approximation and not necessarily the best approximation of the true relationship between these variables.
The three genes identified in this study are likely to reflect those with the strongest associations, allowing their identification in this analysis. It is not surprising to find Env evolution associated with HLA frequency, because this highly variable protein may allow substantial epitope escape in response to HLA-mediated immunity without substantial loss of viral fitness. Because Nef may be most targeted by the CTL response38 and may have the most amino acid variation driven by HLA selection pressure,19 it is also not surprising to find that Nef is likely to demonstrate differential evolution in response to varying CTL-based selection pressures. Finally, Pol, neither highly variable nor the most targeted protein, is an unanticipated result of this analysis. Pol is highly conserved and may not tolerate variation without loss of function, allowing variation only in the presence of strong CTL selection. In this case, a large proportion of the most recent evolution in Pol should be due to evolution toward a more fit form.
It remains unclear why we found associations with HLA-A only. However, this does not rule out the possibility that CTL responses mediated by HLA-B and HLA-C are not involved. Because the HLA genes are located in the same genomic region, they cosegregate, particularly HLA-A and HLA-B due to their proximity. In fact, two HLA alleles found to be associated with phylogenetic branch length also had signifi-cant linkage disequilibrium with HLA-B and HLA-C alleles (Table 4). Thus the associations with HLA-A may simply reflect linked associations with the other HLA genes.
There are several limitations to this study that may have prevented the identification of associations with some HIV-1 proteins. As mentioned earlier, this was a cross-sectional study and the precise time of infection was unknown. Long-itudinal analysis including extensive viral sequencing during the acute stage of infection may be required to thoroughly evaluate viral evolution in response to HLA-mediated immunity. Because this ideal dataset is currently unavailable, our phylogenetic analysis was the best method to approximate those sequence changes. In addition, the external branch lengths we observed likely included evolutionary events that occurred in prior hosts, diluting our ability to identify an association between divergence and HLA frequency. Furthermore, HIV-1 evolves due to other factors, including other adaptive immune responses and protein function, possibly reducing associations evaluated with the CTL response. Finally, the sample size, although large for the extensive and high quality sequence data generated, may be too small to distinguish the impact of the CTL response on viral evolution in every HIV-1 gene when multiple factors are involved. These potential limitations are likely to bias our results toward the null hypothesis, making the identification of significant associations even less likely. Thus, our findings may be limited to only the strongest associations.
We have shown differential evolution of HIV-1 based on HLA frequency, suggesting additional HIV evolutionary events among individuals with rare HLA alleles. These findings support the idea that viral evolution undergoes a regressive path, reverting to states of greater fitness, prior to forward evolution driven by escape from the CTL response and other factors.35 These findings also suggest that CTL vaccine designs based on consensus sequences may include multiple escape variants to common HLA alleles, potentially reducing their effectiveness. Finally, focusing on infected individuals with rare HLA alleles may allow the identification of reversion variants that can be used as candidates in HIV-1 vaccine designs.
Acknowledgments
The authors would like to thank all of the participants who contributed time, information, and blood samples for the study. Funding for this research was provided by University of Washington Center For AIDS Research (AI27757), including a New Investigator Award (AI047734) to C.M.R., by Puget Sound Partners in Global Health (to C.M.R.), and by Microsoft Research (to J.L., C.K., and D.E.H.). This work was previously presented in part at the 14th Conference on Retroviruses and Opportunistic Infections, 2007.
Footnotes
Disclosure Statement
No competing financial interests exist.
References
- 1.Leslie AJ, Pfafferott KJ, Chetty P, et al. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282–289. doi: 10.1038/nm992. [DOI] [PubMed] [Google Scholar]
- 2.Frater AJ, Brown H, Oxenius A, et al. Effective T-cell responses select human immunodeficiency virus mutants and slow disease progression. J Virol. 2007;81:6742–6751. doi: 10.1128/JVI.00022-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Martinez-Picado J, Prado JG, Fry EE, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brockman MA, Schneidewind A, Lahaie M, et al. Escape and compensation from early HLA-B57-mediated cytotoxic T-lymphocyte pressure on human immunodeficiency virus type 1 gag alter capsid interactions with cyclophilin A. J Virol. 2007;81:12608–12618. doi: 10.1128/JVI.01369-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci USA. 2000;97:2709–2714. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaslow RA, Carrington M, Apple R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 7.Gao X, Bashirova A, Iversen AK, et al. AIDS restriction HLA allotypes target distinct intervals of HIV-1 pathogenesis. Nat Med. 2005;11:1290–1292. doi: 10.1038/nm1333. [DOI] [PubMed] [Google Scholar]
- 8.Chopera DR, Woodman Z, Mlisana K, et al. Transmission of HIV-1 CTL escape variants provides HLA-mismatched recipients with a survival advantage. PLoS Pathog. 2008;4:e1000033. doi: 10.1371/journal.ppat.1000033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schneidewind A, Brockman MA, Yang R, et al. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J Virol. 2007;81:12382–12393. doi: 10.1128/JVI.01543-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crawford H, Prado JG, Leslie A, et al. Compensatory mutation partially restores fitness and delays reversion of escape mutation within the immunodominant Hla-B*5703-restricted gag epitope in chronic HIV-1 infection. J Virol. 2007;81:8346–8351. doi: 10.1128/JVI.00465-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanchez-Merino V, Farrow MA, Brewster F, Somasundaran M, Luzuriaga K. Identification and characterization of HIV-1 CD8+ T cell escape variants with impaired fitness. J Infect Dis. 2008;197:300–308. doi: 10.1086/524845. [DOI] [PubMed] [Google Scholar]
- 12.Ueno T, Motozono C, Dohki S, et al. CTL-mediated selective pressure influences dynamic evolution and pathogenic functions of HIV-1 Nef. J Immunol. 2008;180:1107–1116. doi: 10.4049/jimmunol.180.2.1107. [DOI] [PubMed] [Google Scholar]
- 13.Lewis F, Hughes GJ, Rambaut A, Pozniak A, Leigh Brown AJ. Episodic sexual transmission of HIV revealed by molecular phylodynamics. PLoS Med. 2008;5:e50. doi: 10.1371/journal.pmed.0050050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frahm N, Kiepiela P, Adams S, et al. Control of human immunodeficiency virus replication by cytotoxic T lymphocytes targeting subdominant epitopes. Nat Immunol. 2006;7:173–178. doi: 10.1038/ni1281. [DOI] [PubMed] [Google Scholar]
- 15.Trachtenberg E, Korber B, Sollars C, et al. Advantage of rare HLA supertype in HIV disease progression. Nat Med. 2003;9:928–935. doi: 10.1038/nm893. [DOI] [PubMed] [Google Scholar]
- 16.Nguyen L, Chaowanachan T, Vanichseni S, et al. Frequent human leukocyte antigen class I alleles are associated with higher viral load among HIV type 1 seroconverters in Thailand. J Acquir Immune Defic Syndr. 2004;37:1318–1323. doi: 10.1097/01.qai.0000127059.98621.55. [DOI] [PubMed] [Google Scholar]
- 17.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 18.Rousseau C, Birditt BA, McKay AR, et al. Large-scale amplification, cloning and sequencing of near full-length HIV-1 subtype C genomes. J Virol Methods. 2006;136:118–125. doi: 10.1016/j.jviromet.2006.04.009. [DOI] [PubMed] [Google Scholar]
- 19.Rousseau CM, Daniels MG, Carlson JM, et al. HLA class-I driven evolution of human immunodeficiency virus type 1 subtype C proteome: Immune escape and viral load. J Virol. 2008;82(13):6434–6446. doi: 10.1128/JVI.02455-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson JD, Higgins DG, Gibson TJ, CLUSTAL W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maddison WP, Maddison DR. MacClade–Analysis of Phylogeny and Character Evolution. Sinauer Associates, Inc.; Sunderland, MA: 2001. Version 4. [Google Scholar]
- 22.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 23.Listgarten J, Brumme Z, Kadie C, et al. Statistical resolution of ambiguous HLA typing data. PLoS Comput Biol. 2008;4:e1000016. doi: 10.1371/journal.pcbi.1000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sette A, Sidney J. Nine major HLA class I supertypes account for the vast preponderance of HLA-A and -B polymorphism. Immunogenetics. 1999;50:201–212. doi: 10.1007/s002510050594. [DOI] [PubMed] [Google Scholar]
- 25.Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: A revised and updated classification. BMC Immunol. 2008;9:1. doi: 10.1186/1471-2172-9-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doherty PC, Zinkernagel RM. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature. 1975;256:50–52. doi: 10.1038/256050a0. [DOI] [PubMed] [Google Scholar]
- 27.Price DA, Goulder PJ, Klenerman P, et al. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc Natl Acad Sci USA. 1997;94:1890–1895. doi: 10.1073/pnas.94.5.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goulder PJ, Pasquier C, Holmes EC, et al. Mother-to-child transmission of HIV infection and CTL escape through HLA-A2-SLYNTVATL epitope sequence variation. Immunol Lett. 2001;79:109–116. doi: 10.1016/s0165-2478(01)00272-3. [DOI] [PubMed] [Google Scholar]
- 29.Goulder PJ, Brander C, Tang Y, et al. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature. 2001;412:334–338. doi: 10.1038/35085576. [DOI] [PubMed] [Google Scholar]
- 30.Milicic A, Edwards CT, Hue S, et al. Sexual transmission of single human immunodeficiency virus type 1 virions encoding highly polymorphic multisite cytotoxic T-lymphocyte escape variants. J Virol. 2005;79:13953–13962. doi: 10.1128/JVI.79.22.13953-13962.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pillay T, Zhang HT, Drijfhout JW, et al. Unique acquisition of cytotoxic T-lymphocyte escape mutants in infant human immunodeficiency virus type 1 infection. J Virol. 2005;79:12100–12105. doi: 10.1128/JVI.79.18.12100-12105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navis M, Matas DE, Rachinger A, et al. Molecular evolution of human immunodeficiency virus type 1 upon transmission between human leukocyte antigen disparate donor-recipient pairs. PLoS ONE. 2008;3:e2422. doi: 10.1371/journal.pone.0002422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen TM, Altfeld M, Yu XG, et al. Selection, transmission, and reversion of an antigen-processing cytotoxic T-lymphocyte escape mutation in human immunodeficiency virus type 1 infection. J Virol. 2004;78:7069–7078. doi: 10.1128/JVI.78.13.7069-7078.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li B, Gladden AD, Altfeld M, et al. Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J Virol. 2007;81:193–201. doi: 10.1128/JVI.01231-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herbeck JT, Nickle DC, Learn GH, et al. Human immunodeficiency virus type 1 env evolves toward ancestral states upon transmission to a new host. J Virol. 2006;80:1637–1644. doi: 10.1128/JVI.80.4.1637-1644.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goepfert PA, Lumm W, Farmer P, et al. Transmission of HIV-1 Gag immune escape mutations is associated with reduced viral load in linked recipients. J Exp Med. 2008;205:1009–1017. doi: 10.1084/jem.20072457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 38.Frahm N, Korber BT, Adams CM, et al. Consistent cytotoxic-T-lymphocyte targeting of immunodominant regions in human immunodeficiency virus across multiple ethnicities. J Virol. 2004;78:2187–2200. doi: 10.1128/JVI.78.5.2187-2200.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]