Abstract
Applications of omics technologies in the postgenomics era swiftly expanded from rare monogenic disorders to multifactorial common complex diseases, pharmacogenomics, and personalized medicine. Already, there are signposts indicative of further omics technology investment in nutritional sciences (nutrigenomics), environmental health/ecology (ecogenomics), and agriculture (agrigenomics). Genotype–phenotype association studies are a centerpiece of translational research in omics science. Yet scientific and ethical standards and ways to assess and communicate risk information obtained from association studies have been neglected to date. This is a significant gap because association studies decisively influence which genetic loci become genetic tests in the clinic or products in the genetic test marketplace. A growing challenge concerns the interpretation of large overlap typically observed in distribution of quantitative traits in a genetic association study with a polygenic/multifactorial phenotype. To remedy the shortage of risk assessment and communication tools for association studies, this paper presents the concept of edge effect. That is, the shift in population edges of a multi-factorial quantitative phenotype is a more sensitive measure (than population averages) to gauge the population level impact and by extension, policy significance of an omics marker. Empirical application of the edge effect concept is illustrated using an original analysis of warfarin pharmacogenomics and the VKORC1 genetic variation in a Brazilian population sample. These edge effect analyses are examined in relation to regulatory guidance development for association studies. We explain that omics science transcends the conventional laboratory bench space and includes a highly heterogeneous cast of stakeholders in society who have a plurality of interests that are often in conflict. Hence, communication of risk information in diagnostic medicine also demands attention to processes involved in production of knowledge and human values embedded in scientific practice, for example, why, how, by whom, and to what ends association studies are conducted, and standards are developed (or not). To ensure sustainability of omics innovations and forecast their trajectory, we need interventions to bridge the gap between omics laboratory and society. Appreciation of scholarship in history of omics science is one remedy to responsibly learn from the past to ensure a sustainable future in omics fields, both emerging (nutrigenomics, ecogenomics), and those that are more established (pharmacogenomics). Another measure to build public trust and sustainability of omics fields could be legislative initiatives to create a multidisciplinary oversight body, at arm's length from conflict of interests, to carry out independent, impartial, and transparent innovation analyses and prospective technology assessment.
“Toto, I've a feeling we're not in Kansas anymore . . . It's not a place you can get to by a boat or a train. It's far, far away—behind the moon—beyond the rain. Somewhere, over the rainbow, way up high.”
—Dorothy Gale (Judy Garland) in “The Wizard of Oz” (1939)
Introduction: Omics Science and the New Decade Of Measurements
Mother Nature is not always known for her generosity. Scientists, particularly physicists have painfully accepted the paucity of information available to them in experiments, and that nature imparts information and understanding with stubborn reluctance (Naylor and Cavanagh, 2004). To remedy shortage of experimental data, omics technologies such as proteomics and genomics were introduced over the past 3 decades, starting with the two-dimensional gel electrophoresis for global protein analyses in 1970s and high-throughput DNA sequencing methodologies in 1990s (Anderson and Anderson, 1977, 1996; Kolker et al., 2006; McNally and Glasner, 2006; O'Farrell, 2008; Ozdemir et al., 2006). According to one etymological analysis, the suffix “ome” is derived from the Sanskrit OM (“completeness and fullness”) (Lederberg and McCray, 2001). By combining “gene” and “ome,” Hans Winkler created the term genom(e), referring to “the haploid chromosome set, which, together with the pertinent protoplasm, specifies the material foundations of the species. . . . ” (Lederberg and McCray, 2001; Winkler, 1920). Alternatively, the Oxford English Dictionary suggests that Winkler used genom(e) as a portmanteau of gene and chromosome (Gregory, 2005). Victor McKusick and Frank Ruddle added “genomics” to the scientific lexicon as the title for the new journal they cofounded in 1987, with emphasis on linear gene mapping, DNA sequencing, and comparison of genomes from different species (McKusick and Ruddle, 1987). An account of the events that led to the creation of the word genomics is offered by Thomas H. Roderick (Kuska, 1998).
Omics technologies and various neologisms that define their application contexts, however, are more than a simple play on words. They substantially transformed both the throughput and the design of scientific experiments. The omics technologies allow the generation of copious amounts of data at multiple levels of biology from gene sequence and expression to protein and metabolite patterns underlying variability in cellular networks and function of whole organ systems (Suarez-Kurtz and Cascorbi, 2008; Wilke et al., 2008). In fact, this led to overabundance of data in biomedical experiments recently (Nicholson, 2006). While the 1990s was named as the “decade of the brain,” we are now in the “decade of measurements.” As Judy Garland indicates in her 1939 film, this signals a new era—in how we approach to scientific inquiries. That is, the arrival of “big biology” and a systems (integrative) approach to scientific practice with global measurements of molecular pathways in health and disease (Baccini et al., 2008; Kaput et al., 2006; Naylor et al., 2008; Nicholson and Lindon, 2008; van Ommen, 2004).
In addition to amplified throughput, the process of research is fundamentally altered in “omics science.” Ordinarily, scientists have accustomed to hypothesis-driven research wherein a clearly articulated scientific question/hypothesis would be posed. Subsequently, experiments would be carried out to obtain data in order to test the study hypothesis. With the omics approach, asking an initial research question is not always necessary or a prerequisite. Genome or proteome-wide data can be collected in an omics experiment without an existing hypothesis, followed by generation and testing of biological hypotheses. This reversal from the “first hypothesize-then-experiment” tradition to “first experiment-then-hypothesize” mode of operation offers the promise to discover unprecedented pathophysio-logical mechanisms of disease as well as response and toxicity to drugs and nutrition. The high-throughput omics data obtained in parallel from successive organizational hierarchies of cellular biology also help discern, in real time, the “system level” predictive value of an omics biomarker test, over and above the built-in molecular redundancies preserved in biology during the course of human evolution. Ultimately, it is thought that the omics science and technologies will markedly improve the simplistic and reductionist experimental models that offer merely a temporal snap shot of the much more complex, longitudinal, and dynamic nature of biological networks (and their fluctuations in response to social/environmental exposures) that fundamentally govern human health and disease.
Statement of the Problem: Rapid Transition in Omics Science from Monogenic to Multifactorial Phenotypes
Bearing in mind the promises noted above, can we be confident that the touted omics biotechnologies will bring about major scientific innovations, personalized medicines, knowledge-based economies, and other anticipated benefits for the society (Burgess et al., 2008; Gurwitz and Lunshof, 2008; Knoppers and Joly, 2007; Lunshof et al., 2006; Motulsky, 2002; Nightingale and Martin, 2004; Smart and Martin, 2006; Zika et al., 2006)? In this regard, the very recent confluence of understanding of pharmacogenomics variability in pharmacokinetic and pharmacodynamic pathways for the oral anticoagulant warfarin is a notable positive example (Cooper et al., 2008; Daly and King, 2006; Gage et al., 2008; Nakai et al., 2007; Ozdemir et al., 2008; Perini et al., 2008). Yet the future is still undecided for many of the omics biotechnology applications such as pharmacogenomics, nutrigenomics, and agrigenomics (Brown, 2003; Godard and Ozdemir, 2008; Hedgecoe, 2004; Hopkins et al., 2006; Khoury et al., 2008; McGuire and Burke, 2008; Millstone, 2000; Montpetit et al., 2006; Ozdemir and Godard, 2007). While incremental advances are being achieved, the high expectations from genomics medicine have not been translated to an increase in research products or drug development in a manner that is proportional to the vast R&D investment made in omics biotechnologies thus far (Gurwitz and Motulsky, 2007; Ibarreta, 2008; Nightingale and Martin, 2004; Ozdemir et al., 2007; Wood, 2006). Even after considering the expected lag time of 5 to 10 years from R&D investment to emergence of scientific products, there appears to be a disconnect between what has been promised from genomics technology revolution and its actual impact in the clinic and society (Ibarreta, 2008; Nightingale and Martin, 2004; Ries and Caulfield, 2006). Consistent with this, it was acknowledged in an innovation analysis that “the applied sciences needed for medical product development have not kept pace with the tremendous advances in the basic sciences. The new science is not being used to guide the technology development process in the same way that it is accelerating the technology discovery process” (Food and Drug Administration, 2004).
Why is this gap? Several technical and nontechnical factors have been suggested to explain the difficulties in downstream uptake of omics innovations in the clinic, in particular, the diagnostic tests that predict future disease susceptibility or health intervention outcomes (e.g., toxicity/efficacy) associated with drug treatment and nutrition. These include, for example, lack of cost-effectiveness analysis and inadequate clinical validation of genomics tests, balancing the need for scientific innovation and commercialization with appropriate evidence thresholds for moving technology into health care, criteria for scientific validity that differ between disciplines, education of health professionals, and health technology assessment under a vision that includes both industrialized and developing world (Caulfield, 2003; Compagni et al., 2008; Daar et al., 2007; Frueh and Gurwitz, 2004; Gurwitz et al., 2005; Gurwitz and Motulsky, 2007; Hopkins et al., 2006; Khoury et al., 2008; Morin, 2009; Ozdemir et al., 2007; Pang, 2003; Singh and Daar, 2008).
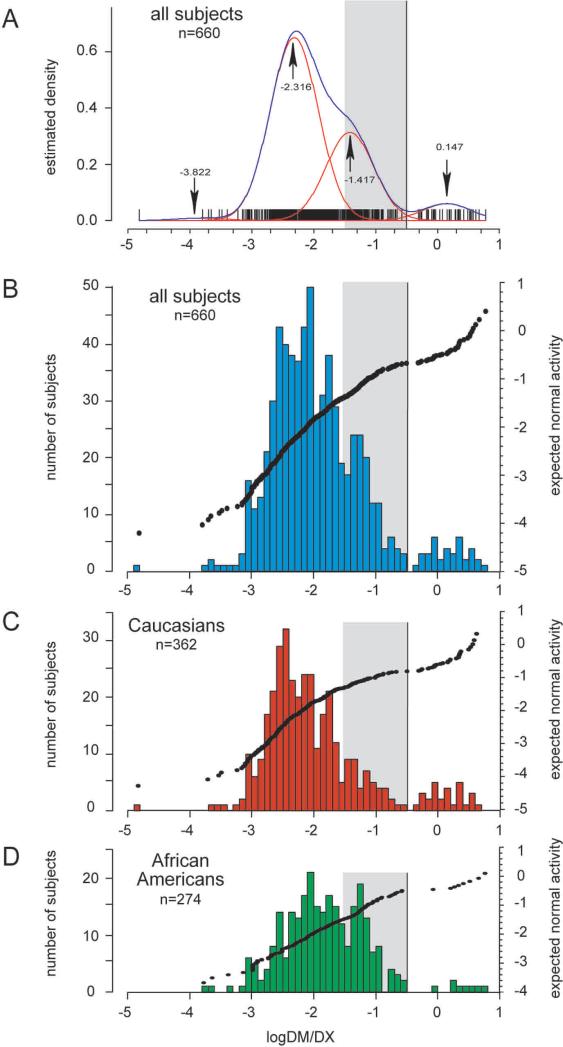
Successful translation of omics innovations from the laboratory bench to the clinic also relies on commensurate development of risk assessment and communication tools to appropriately regulate and engage omics innovations with diverse user groups in society. A specific area of omics science where the need for risk assessment and communication frameworks is most prominent concerns the genotype–phenotype association studies that deal with multifactorial phenotypes such as common human cancers and responses to drugs and nutrition. In terms of the “risk” identified by a genotype–phenotype association study, this paper refers to differences in phenotypic distributions in populations defined by genotypic variants or other types of omics bio-markers (e.g., proteomics, metabolomics, etc.) (Figs. 1 and 2). As omics technologies rapidly transitioned from study of monogenic (Fig. 1) to multifactorial phenotypes (Fig. 2) over the past decade, the existing risk assessment and communication frameworks—originally developed for rare monogenic diseases—became no longer adequate. A current challenge is how to interpret the large overlap typically observed in distribution of quantitative traits in an association study concerning a multifactorial and polygenic phenotype (Fig. 2), as a contrast to monogenic phenotypes that tend to separate into distinctly discernible groups (Fig. 1).
FIG. 1.
Example of a genotype–phenotype association for a monogenic phenotype. Histograms and cluster analysis present the CYP2D6 activity (phenotype), as measured with dextromethorphan as a probe drug. CYP2D6 activity is expressed as the log of the urinary metabolic ratio of dextromethorphan/dextrorphan log(DM/DX). The red lines in A indicate the four population clusters identified by mclust, the blue line represents the summary line. Mean log(DM/DX) are as indicated for each cluster (arrows). Population histograms are shown for all subjects (B), Caucasians (C) and African Americans (D), respectively. Subjects with discordant genotypes were excluded. The mean log(DM/DX) was −1.912 for all subjects, −1.989 for Caucasians and −1.800 for African-Americans. The gray box highlights the activity range for intermediate metabolizers (−1.52288 ≤ log(DM/DX) < −0.52288 or 0.03 ≤ DM/DX < 0.3). Reprinted by permission from Macmillan Publishers Ltd: [Gaedigk et al., Clin Pharmacol Ther 2008; 83, 234−242].
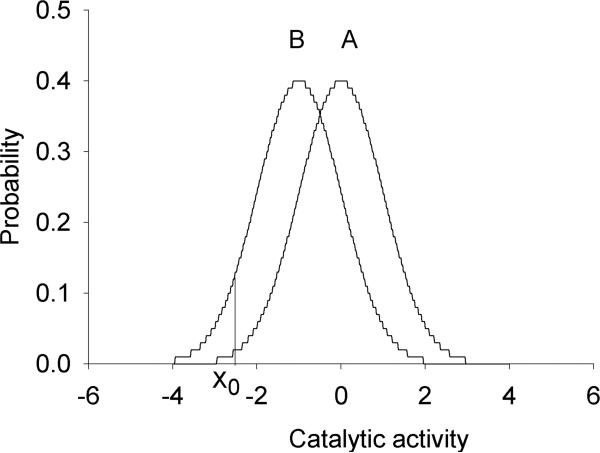
FIG. 2.
Example of a genotype–phenotype association for a multifactorial phenotype. As a phenotype, standardized normal distributions of catalytic activity of a hypothetical metabolic pathway (subject to multifactorial regulation) are presented (Population A: fast metabolism; Population B: slow metabolism). Xo reflects the threshold activity below which drug or food toxicity is observed. Note that an overlapping phenotypic distribution is a mainstay occurrence in association of a multifactorial phenotype with an omics marker. Reprinted by permission from Bentham Science Publishers: [Ozdemir et al., Curr Pharmacogenomics Person Med 2005; 3, 53−71].
Establishing robust genotype–phenotype associations is central to translation of omics science to clinical applications. Yet scientific and ethical standards and ways to assess and communicate the risk information derived from association studies have been neglected to date. This is a significant regulatory vacuum because association studies may decisively influence which genetic loci become a genetic test in the clinic or a product in the genetic test marketplace (McGuire and Burke, 2008).
Aims of the Paper and the Knowledge Gap to be Addressed
To remedy the shortage of risk assessment and communication tools for omics marker associations with multifactorial phenotypes, this paper presents the concept of edge effect. That is, the shift in population tails of a multifactorial quantitative trait is a more sensitive measure (than population averages) to gauge the population level impact and by extension, policy significance of an omics marker. A case study of the empirical application of the edge effect concept is presented using the recent example of warfarin pharmacogenomics and the VKORC1 genetic variation in a Brazilian population sample. These edge effect analyses are examined in relation to future regulatory guidance and policy development for association studies in omics science, and diagnostic (predictive) medicine more generally.
As omics fields rapidly proliferate from pharmacogenomics to nutrigenomics and ecogenomics, it is important to bear in mind that the scientific practice gains an increasingly significant social dimension. Science and technology markedly impact society, culture, and ways we live and relate to others in the community, with both intended and unintended consequences (UNESCO, 2000). Science is inherently value laden and embedded in a social, ethical, and political context (Master and Ozdemir, 2008). Thus, closing the gap between the omics laboratory and society is crucial for prospective assessment and anticipatory governance of omics science and to forecast its diverse downstream applications in society. While the edge effect analyses and the risk assessment/communication frameworks presented in this paper contribute toward these objectives, they will not materialize unless a deeper understanding of the omics history is established, nor will the omics science and its products be sustainable unless certain nontechnical factors are taken into account, in parallel with scientific considerations. Hence, in addition to the concept of edge effect, this paper addresses the following hitherto neglected issues in the literature.
Building on an interdisciplinary analysis of the omics history and diagnostic medicine (see next section), we explain why and how risk assessment/communication frameworks will benefit the association studies that drive translational research and innovations from the omics laboratory to society.
Tertiary care hospitals and formularies are discussed as a potential avenue for proof-of-concept studies of the edge effect analyses.
Nontechnical factors such as human values embedded in scientific practice and the past lessons from omics history are contextualized, as they relate to future sustainability of emerging omics fields (nutrigenomics) and considerations (in addition to risk assessment/communication tools) to effectively bridge the gap between the omics laboratory and society.
Tracing the “Omics Origins” in Diagnostic Medicine from Ancient Times to the Postgenomics Era
While omics technologies forge ahead with yet another application in nutritional sciences, as illustrated under the overarching vision of nutrigenomics in this second special consecutive issue of the OMICS, it is essential to recall the origins of omics science, the individuals who pioneered the key concepts, as well as the human values and sociotechnical factors that decisively shaped the field. As we trace the “omics origins” deeper in the archives of technoscience history, some threads of commonality will thus become apparent. This could shed light on how best to prepare for a sustainable future in nutrigenomics by surfacing the embedded factors—beyond the laboratory bench—that might limit or facilitate the innovations in the postgenomics era across transdisciplinary boundaries (Maienschein et al., 2008; Maienschein and Smith, 2008; Motulsky, 2002).
Diagnostic medicine in ancient times
Omics science has a strong motive to characterize individual and population differences in biology. In this sense, the focus of omics science on biological “variability questions” is rooted in diagnostic medicine. In fact, long before the introduction of omics technologies and the present popularity of diverse omics fields, understanding (and diagnosing) individual differences in health and disease have been a preoccupation since ancient times. Early diagnosticians not only lacked the microarrays that the modern-day genomics scientists now possess, but they were also limited in access to tissue specimens. In ancient times, patients could be observed and touched by physicians, but it was illegal to practice invasive procedures. Access to diagnostic tissue specimens was essentially limited to those that were naturally passed from the body such as urine. The Babylonians and Sumerians thus utilized urine to diagnose the health status of individuals as early as 4000 BCE, whereas Hippocrates (460—ca. 370 BCE, the Island of Cos, Greece) suggested the presence of bubbles in the urine as an indication of chronic kidney disease (White, 1991).
Pythagoras in Croton (Calabria region), Italy noted in 510 BCE that the ingestion of fava beans resulted in a potentially fatal reaction in some, but not all, individuals (Nebert, 1999; Pirmohamed, 2001). Pythagoras considered beans as sacred, and prohibited their consumption, as well as walking through the bean fields (Detienne, 1972; Mahlknecht and Voelter-Mahlknecht, 2005). The scientific basis of this adverse reaction to food, that is, favism, was inherited deficiency of glucose-6-phosphate dehydrogenase (Carson et al., 1956), one of the early documented accounts of metabolic genetic polymorphism.
20th Century diagnostic medicine and Archibald Garrod: The rise of human biochemical genetics and personalized medicine
Biological individuality is a fundamental property of all living organisms. This individuality, and its reciprocal corollary—individual differences—form the very basis of modern day attempts to develop molecular diagnostics with omics technologies for common complex diseases or to forecast response to pharmaceuticals and foodstuff. While these research efforts are now relatively well accepted, this was not always so at the beginning. Despite the early hints of interest in diagnostic medicine in ancient times, until the end of 19th century, “biological variability questions” were not a legitimate component of the mainstream scientific inquiries. In the early 20th century, Archibald Garrod (1857−1936) suggested the chemical individuality of humans as a basis for certain inborn errors of metabolism such as alkaptonuria (Garrod, 1902, 1909). Garrod did not use the term genetics (this was coined by his colleague William Bateson), but he presciently foresaw the role of human chemical individuality and genetic factors in most human diseases. While his initial thoughts centered on rare familial inborn errors of metabolism (Garrod, 1902), he later broadened the concept of human biochemical genetics to include majority of human diseases (Garrod, 1931):
. . . substances contained in particular foods, certain drugs, and exhalations of animals or plants produce in some people affects wholly out of proportion to any which they bring about in average individuals. Some effects vary from a slight and temporary discomfort or morbid syndromes which amount to severe or fatal illness.
Following Garrod's early insights on biological variability questions, the concept of human biochemical genetics was advanced by J.B.S. Haldane (1892−1964), further cementing the roots of contemporary diagnostic medicine.
Study of taste perception is another area that catalyzed research interest in person-to-person differences in human biology (Blankeslee and Salmon, 1931; Snyder, 1931). Taste is a nutritional gatekeeper for the body influencing which foodstuff is ingested or rejected (Tepper, 2008). Strong bitter taste is universally rejected, and often serves as an indicator of contamination of food with toxins. Moderate bitter taste can be appealing and desired in certain food such as wine, beer, and cheese. In 1931, A.L. Fox accidentally discovered that the ability to taste the synthetic compounds phenylthiocarbamide (PTC) and 6-n-propylthiouracil (PROP) varied markedly among humans (Fox, 1932). PTC powder accidentally escaped into the laboratory air while synthesizing nonnutritive sweeteners. Fox did not perceive anything but his colleagues complained from moderately to intensely bitter taste on their lips. Subsequent testing showed that PTC was tasteless to about 30% of Caucasians, with marked population-to-population variability in frequency of the nontaster phenotype (Fox, 1932; Kim and Drayna, 2004; Tepper, 2008; Wooding, 2006). The mode of transmission was initially thought to be simple Mendelian recessive inheritance (Blankeslee and Salmon, 1931; Snyder, 1931;). Further research indicated that multiple alleles or genes are involved (Guo and Reed, 2001; Kim and Drayna, 2004; Tepper, 2008).
These advances in human biochemical genetics in the first half of the 20th century set the stage for the idea (role of genetic factors in drug effects) proposed by Arno G. Motulsky (Seattle), in his seminal article in October 1957, with the programmatic title “Drug Reactions, Enzymes and Biochemical Genetics,” indicating the confluence of biochemistry and genetics within the specific context of pharmacology (Motulsky, 1957). Two years later, the term pharmacogenetics was coined by Friedrich Vogel of Heidelberg, Germany (i.e., long before “pharmacogenomics” became a popular term and research topic) (Vogel, 1959). The first book on pharmacogenetics was published by Werner Kalow (Toronto), which definitively established the field of pharmacogenetics (Kalow, 1962). These events coincided with the report on acetylator polymorphism (Evans et al., 1960).
After the field of pharmacogenetics was defined by the three pioneers Motulsky, Vogel, and Kalow from 1957 to 1962, the progress towards understanding the genetic basis of drug efficacy and toxicity remained sluggish. Skepticism initially prevailed on the extent to which heredity played a role, if any, in determining drug treatment outcomes, given the multitude of environmental factors that can confound drug action in humans. This concern was addressed in part by Elliot S. Vesell in Hershey, Pennsylvania, in studies of monozygotic and dizygotic twins (Vesell and Page, 1968). These series of twin studies firmly indicated a genetic basis for individual differences in drug metabolism and pharmacokinetics. Subsequently, the sparteine/debrisoquine (CYP2D6) monogenic polymorphism was identified by Eichelbaum in Germany (Dengler and Eichelbaum, 1977; Eichelbaum et al., 1975a, 1975b, 1979) and Smith in England (Mahgoub et al., 1977).
Long before the launch of the Human Genome Project and the present popularity of genomics research, Norman G. Anderson and N. Leigh Anderson, a father and son team, attempted to develop an index of all human proteins (the Human Protein Index, or HPI) in 1970s, at a scale that could be considered as one of the very first investigations with an “omics” vision (Anderson and Anderson, 1977, 1996; Anderson et al., 2001). The Andersons were ahead of their time; the proposal for the HPI remained unfunded initially. In hindsight, the Andersons provide the following personal account: “ . . . because DNA sequencing technology is inherently simpler and more scalable than protein analytical technology, and because the finiteness of genomes invited a spirit of rapid conquest, the notion of genome sequencing has displaced that of protein databases in the minds of most molecular biologists” (Anderson et al., 2001). This insight (on the origins of proteomics science) is noteworthy because it provides a grounded historical perspective well beyond the genocentric representations of human biology in omics science. It also underscores the nonlinear, ostensibly chaotic, and often unpredictable ways in which novel ideas may (or may not) turn into innovations and diagnostic products in the clinic and marketplace.
The two decades after the initial proposal for the HPI, the 1980s and the 1990s, were defined and shaped by advances in molecular biology applications to decipher the genetic basis of inter-individual and population-to-population variation in key drug metabolizing enzymes such as CYP2D6 and phase I and phase II drug metabolism pathways (Gonzalez et al., 1988; Masimirembwa et al., 1993; Masimirembwa et al., 1996; for a review see: Flockhart, 2007; Gunes and Dahl, 2008; Ingelman-Sundberg et al., 2008; Owen et al., 2008). Early hints supported the idea that cytochrome P450s also contribute to metabolism of endogenous compounds in humans and possibly, of neurotransmitters, thereby potentially influencing behavior and personality (Bertilsson et al., 1989; for recent findings on the latter subject, see Gonzàlez et al., 2008; LLerena et al., 2009). The first peer-reviewed journal dedicated to the field of pharmacogenetics was established in 1991 (presently, Pharmacogenetics and Genomics) that created a formal publication platform to report on the study of drug–gene interactions. Genomics research on drug transporters and drug targets (e.g., receptors) followed the work on drug metabolizing enzymes, providing a much needed integrative insight to understand individual variability in drug intervention outcomes (Chinn and Kroetz 2007; Giaco-mini et al., 2007). In parallel, the last 2 decades of the 20th century witnessed the rise of population approaches and related mathematical models to describe and forecast inter-and intraindividual variability in pharmacokinetics and pharmacodynamics in clinical pharmacology by the seminal works of Lewis B. Sheiner and others (Blaschke and Giaco-mini, 2004). Regrettably, despite their shared focus on variability questions in biology, the population pharmacokinetic–pharmacodynamic modeling and pharmacogenomics approaches did not intersect appreciably to this date. Nonetheless, the intellectual momentum and the lore established by these advances in the 20th century collectively created a solid foundation for in-depth investigations towards personalized health interventions (drugs, nutrition, vaccines) using omics technologies in the first decade of the 21st century in the postgenomics era (Poland et al., 2008; Ozdemir et al., 2009).
Human variability in gene–environment interactions: The conceptual thread shared by pharmacogenomics, nutrigenomics, and ecogenomics
An event overlooked in the archives of technoscience history, diagnostic medicine, and nutrigenomics is the early hint provided by Roger J. Williams (1893−1988, the discover of pantothenic acid) on differences between healthy individuals, particularly in the context of nutrition and metabolism (Davis et al., 2008; Motulsky, 2002; Williams, 1956). A nutritional biochemist in Austin, Texas, Williams suggested in the 1950s the role of heredity in person-to-person variability in response to external milieu or the environment (e.g., food, drugs), although he did not engage directly with the technology of biochemical genetics (Motulsky, 2002; Williams, 1956). Notably, Williams considered variability among living organisms as a “given,” and that all members of the population are biological deviants (e.g., from the population mean value of a given phenotype), and thus unique. In this sense, Willliams’ thoughts offered wisdom to understand individual differences in response to nutrition and pharmaceuticals, but his ideas on human biological variability remained in relative obscurity initially.
Roger J. Williams’ ideas (and of other pioneers noted above), however, had fundamental implications beyond personalized medicine. Human variation, conferred in part by genetics, environmental, and sociocultural factors, is of relevance as a “population insurance mechanism” for survival of the populations or the species in the event of environmental catastrophes. While a genetic polymorphism may be neutral or insignificant in terms of the general reproductive advantage of a given person, upon exposure to environmental hazards (e.g., consider population level exposure to radiation in cases such as the Chernobyl nuclear disaster), those with “variant” biology and resistance to radiation, toxins, or devastating infections may ensure survival of the population. That is, beyond its relevance to individualize nutrition and drug treatment, human variation can determine the survival and thus evolution of populations of living organisms (Kalow, 2002). This broader vision of gene–environment interactions was first articulated by Haldane, a polymath biologist with astute mathematical insights, by the suggestion that “potter's bronchitis” might be caused due to innate individual differences (Haldane, 1938). Later, Brewer coined the term “ecogenetics” to broaden the concept of gene–environment interactions from drugs and nutrition to include environmental agents in general (Brewer, 1971). Nutrigenomics and pharmacogenomics are thus subspecialties of the broader field of ecogenetics/ecogenomics. The historical origins and the conceptual thread running across pharmacogenomics—nutrigenomics—ecogenomics were endorsed and sealed definitively in a key gathering of the early contributors (Wendell W. Weber, Werner Kalow, Robert L. Smith, Arno G. Motulsky, Bert N. La Du, and others) of these fields in October 3−6, 1999 in Ann Arbor, Michigan, under the auspices of the ASPET, the American Society for Pharmacology and Experimental Therapeutics (Kalow, 2001; Weber and Smith, 2001).
Genotype–phenotype association studies are significant drivers of innovations by answering key biological variability questions across the omics fields ranging from pharmacogenomics, nutrigenomics and ecogenomics and looking further, in agrigenomics research. They serve as a centerpiece of the scientific activity in diagnostic medicine that moves the omics data from the laboratory to society in the 21st century. On the other hand, given the very real challenges in omics science expressed by scientists who are cognizant of the early origins of research on human biological variability (De Leon, 2008; Nebert et al., 2003, 2008) as summarized above, the nascent field of nutrigenomics (being addressed in this second special issue of the OMICS) cannot be left to random drifts, if we are to meaningfully bridge the nutrigenomics laboratory with society. This calls for development of risk assessment and communication tools on genotype–phenotype associations for multifactorial traits, for example, the host response to nutritional interventions.
Genotype–Phenotype Associations: A Centerpiece of Translational Research in 21st Century Diagnostic Medicine
Genotype–phenotype association studies are an essential component of the innovation path from gene discovery on the bench-side to commercially available genetic test in the clinic. Association studies have a dual impact on prioritizing basic genomics discoveries (e.g., focusing bench research on certain candidate genes) as well as translational medicine. There are three essential components of an association study. These include:
the molecular genetic afnalysis of person-to-person differences in the human genome,
the ascertainment of phenotypes (e.g., measurement of beneficial and toxic response to drugs or food) and,
the bioinformatics analysis to evaluate the relationship between (1) and (2); that is, genome–phenotype statistical association analysis.
The genotype–phenotype association studies are rapidly proliferating in the bioscience literature (McGuire and Burke, 2008). With the decreasing costs of genotyping, it is anticipated that this trend will accelerate further in the coming years. The exponential increase in representation of association studies is not limited to the scientific literature; we often encounter in the popular media and news on the discovery of genes that predict response or toxicity to a pharmaceutical agent or foodstuff (Edelson, 2008).
An objective and balanced interpretation of omics marker associations with phenotypes is important—not only because of their proliferating numbers—but also because they determine future public and private investments in omics fields such as pharmacogenomics and nutrigenomics and importantly, which “genetic locus” may become a “genetic test” in the clinic. An incorrect evaluation of the association findings may thus adversely impact translational omics research and the development of downstream innovations and appropriate policy on genetic tests. More worrisome would be the meta-analyses that rely on biased or substandard association studies that may carry forward these inaccuracies to the realm of policy and commercialization of diagnostic tests based on omics technologies. The association studies thus influence which omics test may be included in hospital formularies, or reimbursed by insurers.
At present, there are approximately 1,700 genetic tests: nearly 1,400 available clinically and another 300 available on a research basis (Gene Tests, 2008). The latter tabulation of genetic tests concerns primarily disease susceptibility related phenotypes (Gene Tests, 2008). Conceivably, the actual number of genetics tests including pharmacogenomics and nutrigenomics tests is substantially larger. While some genetic associations may involve apparently innocuous phenotypes and human traits, for example, the type of earwax (Yoshiura et al., 2006), others deal with serious and life-threatening conditions such as susceptibility to common human cancers and effects of drugs with a narrow therapeutic window. In the absence of a widely adopted regulatory framework to interpret and communicate data, genetic association findings could mislead patients as well as researchers by diversion of their valuable time, future research efforts, and limited health care resources.
Omics technologies are increasingly applied to phenotypes subject to regulation by polygenic and multifactorial mechanisms. This is a sharp contrast compared to a decade ago when the phenotypes of interest concerned mostly monogenic traits. A topical example is the genetic studies of the normal variation in human height. Nearly 80% of the phenotypic variation in height can be explained by genetic factors (Preece, 1996; Weedon and Frayling, 2008). Height is therefore a highly heritable classic polygenic trait. Recent large-scale genome-wide association studies (GWAS) identified more than 40 independent genetic loci that strongly influence the normal variation in height (for an in-depth review, see Weedon and Freyling, 2008). However, many of the common genetic variants identified in GWAS confer only a small effect. Although height has 80% heritability, these common variants explain about 5% of this variation, with many more independent loci estimated to be discovered in the very near future (Weedon and Freyling, 2008). A natural consequence of the statistically significant but small genetic effects conferred by individual variants is that the phenotypic subpopulations defined by each genetic marker will overlap substantially (Fig. 2). This creates a challenge—how do we make sense from the overlapping phenotypic distributions of multifactorial quantitative phenotypes in an association study, beyond statistical significance? What are the additional criteria that should be considered to make an informed decision on which genetic locus and association finding are ready for “prime time” applications in the clinic, or deserve the financial investment for further large scale prospective trials? This is particularly relevant given that the existing risk assessment and communication frameworks from genetic association studies still rely, to a large extent, on distinct bimodal phenotypic separations observed in monogenic phenotypes. Regulatory frameworks from the genetics age are ill-suited for genomics and the whole genome based approaches to gene discoveries for polygenic/multifactorial phenotypes (Prainsack et al., 2008). There is a need for innovation in socio-ethical analysis (Lunshof, 2008; Lunshof et al., 2008), risk assessment/communication and policy-making processes in the post-genomics era. Hence, the next section outlines the concept of edge effect and suggests ways in which it can serve as a risk assessment and communication tool in relation to genotype associations with multifactorial phenotypes.
Edge Effect: A Risk Assessment and Communication Tool Based on Shifts in Population Edges in Association Studies
Genotype associations with multifactorial quantitative phenotypes have two notable aspects:
There is marked overlap in phenotype distributions between genotypic groups in an association study.
There is greater range of phenotypic variation within each genotypic group, compared to the difference in mean values between subpopulations (Fig. 2).
As a general rule, population tails (edges) represent the individuals who are either most responsive or refractory to health interventions. Hence, insofar as the nutrigenomics risk assessment is concerned, it makes sense to evaluate the impact of genetic factors on the tails of a phenotypic distribution. Any change in the proportion of individuals who reside in population tails is more likely to translate into clinically meaningful events in the form of food toxicity or lack of efficacy of nutritional interventions.
Figure 2 shows two populations with an overlapping quantitative trait, catalytic function of a metabolic pathway that plays a role in clearance of foodstuff or pharmaceuticals. Each population is represented by a standardized normal distribution with a mean of 0.0 and a standard deviation (SD) of 1.0. It is assumed that clinically manifested drug (or food) toxicity is observed when the catalytic function decreases below a threshold value of Xo. It is notable that even though the mean enzyme activity is modestly different between the two populations with considerable overlap in their distributions, the proportion of patients with toxicity is markedly higher in population B than population A (Fig. 2).
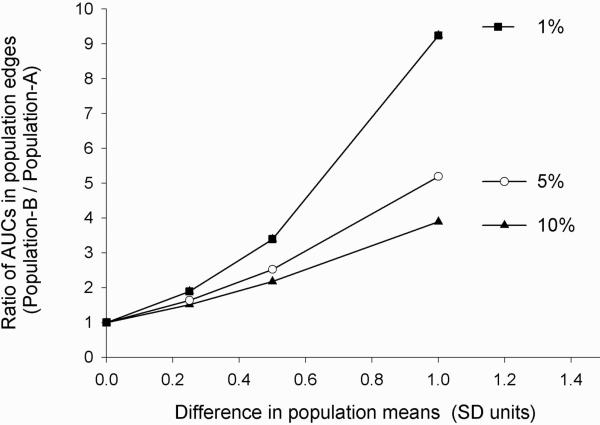
A graphical comparison of the relative changes in the area under the frequency distribution curve (AUC) at population tails exemplifies this point further (Fig. 3). Evidently, the degree of changes in population means underestimates the relative increase in toxicity in population B that has a slower catalytic activity. Importantly, the lower the toxicity threshold Xo (i.e., the rarer an adverse drug reaction) is, the more it will be affected by changes in population mean values (Fig. 3).
FIG. 3.
Ratio of AUCs at two population edges (B/A) as a function of differences in population means when the toxicity threshold is positioned at the 10th, 5th, or the 1st percentile in the reference population A. Note the upward concavity and the nonlinear increase in the ratio of edge AUCs (population B/population A) (ordinate) with linear increments in the difference between population means (abscissa).
The idea of using changes in population tails to gauge the impact of population differences in quantitative phenotypes was earlier named as the “edge effect” (Kalow, 1992). We herein develop this concept further quantitatively and in the new context of innovation analysis in omics science. We reason that this idea has fundamental implications for interpretation of the clinical significance of multifactorial phenotype assocations with omics markers. It can usefully serve as a guidance to omics scientists, research sponsors, diagnosticians, policy makers, and the biotechnology/pharmaceutical industry in making informed decisions and setting priorities on which association finding is worthy of future translational research and policy intervention. Social scientists and bioethicists, too, will be better equipped in their analyses to guide the development and implementation of genetics tests in a manner that reflect scientific nuances.
As an example of forward applications, subpopulations identified by a nutrigenomics biomarker test may presumably be catalogued into different risk categories by considerations of the degree of change in edge AUCs. Risk categories may reflect, for instance, a small (less than twofold), moderate (two- to fivefold) or high/marked (more than fivefold) increase in edge AUC in the variant population (e.g., Population B) relative to the index reference population (e.g., Population A) (Fig. 2). To this end, there is regulatory precedence set by the FDA favoring a graded risk assessment for pharmaceuticals. For instance, using the increments in plasma drug AUC values for the CYP3A substrates, new therapeutic candidates are classified as a strong CYP3A inhibitor when there is “ . . . a ≥ 5-fold increase in the plasma AUC values or more than 80% decrease in clearance of CYP3A substrates (not limited to midazolam, a sensitive CYP3A substrate) in clinical evaluations” (CDER, 2006). A moderate CYP3A inhibitor is one that caused “ . . . a ≥ 2-but < 5-fold increase in the AUC values or 50−80% decrease in clearance of sensitive CYP3A substrates when the inhibitor was given at the highest approved dose and the shortest dosing interval in clinical evaluations” (CDER, 2006). A weak inhibitor is one that caused a ≥1.25 - but less than twofold increase in the AUC values or 20−50% decrease in clearance of sensitive CYP3A substrates (CDER, 2006). This regulatory guidance benefits the drug labels such that varying levels of risk for drug interactions can be presented on the label for a drug found to inhibit CYP3A sub-family. Conceivably, a graded risk assessment approach might also benefit scientists in communication of significance of overlapping subpopulations identified by omics markers to regulators, research sponsors, policy makers, and various user groups for genetic tests.
Table 1 illustrates the data presented in Figure 3 in a numerical form and with the additional assignment of various risk levels to changes in edge AUCs. For example, note that a small 25% SD difference in mean catalytic activity between population A and population B may be reflected by an 89% increase in toxicity when the Xo is set at 1% (Table 1). The threshold values for assignment of the risk into different categories (low, moderate, high) shown in Table 1 may, of course, differ depending on the severity and the nature of clinical endpoint (e.g., food or drug toxicity) in the clinic. In contrast to monogenic traits with bimodal distributions (Fig. 1), it is thus clear that risk assessment for polygenic and multifactorial phenotypes requires population-based considerations to complement decision making on clinical relevance of omics diagnostic tests. The edge effect concept offers additional guidance to expand the regulatory tools available for translation of omics research findings from the laboratory bench and the clinic to the realm of omics policy and society.
Table 1.
Edge Effects in Overlapping Phenotypes Identified by Omics Markers
| Decrease in mean enzyme activity (SD units) ↓ | Ratio of subjects with toxicity in population B versus population A |
|||||
|---|---|---|---|---|---|---|
| Xo < 10% | Risk category | Xo < 5% | Risk category | Xo < 1% | Risk category | |
| 0.25 | 1.51 | Low | 1.63 | Low | 1.89 | Low |
| 0.5 | 2.17 | Moderate | 2.52 | Moderate | 3.39 | Moderate |
| 1 | 3.89 | Moderate | 5.19 | High | 9.24 | High |
Xo represents the threshold catalytic activity below which clinical toxicity is experienced. Relative toxicity risk is calculated by the ratio of the number of individuals who display a catalytic activity below Xo in population B (slower activity) versus population A (faster activity). Risk categories were assigned based on the relative increase in AUC at the edges of population B versus population A. Low risk = less than twofold increase in edge AUC, moderate risk = two-to fivefold increase; high risk = greater than fivefold increase. Splus 4.5 (MathSoft,WA) was used to evaluate the normal and inverse normal cumulative distribution functions and the AUCs.
An Empirical Case Study of Edge Effects in Warfarin Pharmacogenomics
In this section we present a real-life example of the concept of edge effect, using data from a recent study of warfarin pharmacogenomics in 390 Brazilian cardiovascular patients under chronic anticoagulation therapy (Perini et al., 2008). The pharmacological phenotype used in the present new analysis is the stable warfarin dose, defined as the prescribed warfarin weekly dose associated with three consecutive readings of the international normalized ratio of prothrombin time (INR) within the optimal target range (2−3.5) (Perini et al., 2008).
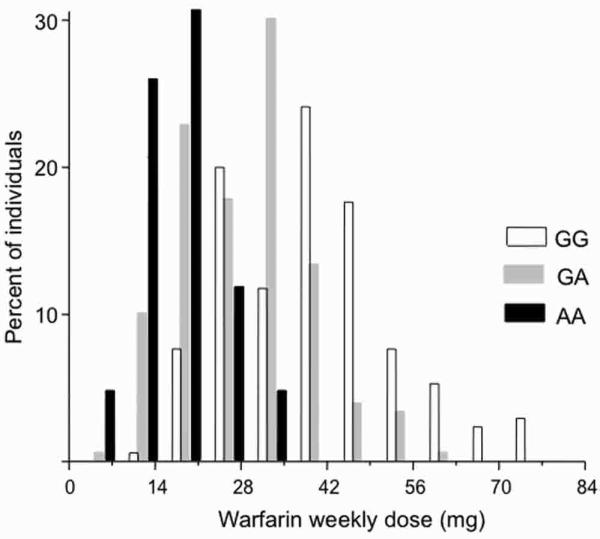
For the analysis of edge effects, the study sample was first catalogued into three groups according to the VKORC1 genotype at 3673G>A (rs9923231) (Fig. 4). The VKORC1 3673G>A genetic variation proved to be the most important predictor of warfarin dose requirement in the multiple regression modeling of the original data (Perini et al., 2008). Accordingly, the warfarin weekly dose decreased significantly (p < 0.00001, ANOVA) from the wild-type VKORC1 3673GG genotype (mean warfarin dose 37.9 mg/week; SD 13.6 mg/week; N 170 subjects), to the GA (28.8, 10.3, 180) and the AA (18.0, 6.2, 40) genotypes. As pointed out in the original publication (Perini et al., 2008), despite these statistically significant group differences in the mean warfarin dose required to achieve the target INR, there was considerable overlap in the distribution of warfarin dose among the three VKORC1 genotypic groups. This is evident in the histograms shown in Figure 4. Hence, while large-scale prospective clinical trials are being planned to assess the significance of genetic factors in routine warfarin therapy, it is essential to have additional criterion and decision aids. This would allow an informed investment on the substantial technical, financial, and human resources that will be necessary to carry out the anticipated clinical pharmacogenomics studies of warfarin in the near future.
FIG. 4.
Frequency histograms of the stable warfarin weekly dose required to achieve an international normalized ratio of prothrombin time (INR) within the optimal target range (2−3.5) in Brazilian patients with cardiovascular disease. The VKORC1 3673G>A polymorphism (rs9923231) was used to stratify the study sample into three genotypic groups. The histogram presents the variability in warfarin dose in the three VKORC1 genotypes: GG (N 170), GA (N 180), and AA (N 40). Data are expressed as percent of the individuals with different warfarin dose requirements in each genotypic group. Note the marked overlap in distribution of warfarin dose among the three VKORC1 genotypes.
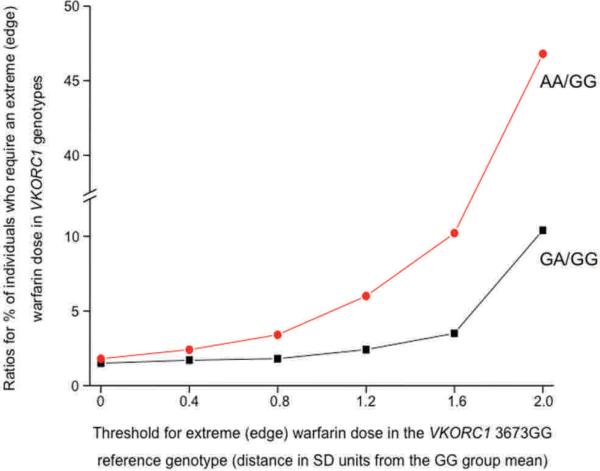
Our conceptual analysis presented in Figure 3 showed markedly larger increases in the ratio of edge AUCs compared to differences between population means, when the threshold (Xo) defining the edge AUC was placed at 10th, 5th, or 1st percentiles of the phenotype distribution in the reference population A. The present empirical case study of warfarin pharmacogenomics (Fig. 5), using real-life clinical pharmacogenomics data from a Brazilian population sample (Perini et al., 2008), lends further experimental support to the edge effect concept. We characterized the changes in the ratio of edge AUCs (i.e., the relative percentage of individuals with an “extreme/edge warfarin phenotype” in a variant and reference population) as a function of the threshold defining the edge AUC in the wild-type VKORC1 3673GG reference genotypic group. Figure 5 displays these edge effect analyses in two population pairs: (1) VKORC1 GA versus GG and, (2) AA versus GG genotypic groups. Accordingly, the ratio of individuals with an edge warfarin phenotype in the VKORC1 GA/GG and AA/GG groups increased as the threshold defining the edge phenotype moved farther away from the reference population VKORC1 3673GG mean phenotypic value (for calculations, see the legend for Fig. 5). For example, the edge sub-population that is located two standard deviations from the mean phenotypic value in the reference VKORC1 GG genotype was “enriched” substantially in the VKORC1 AA genotype—by almost 47-fold (Figs. 4 and 5).
FIG. 5.
The ratio of edge AUCs for the warfarin phenotype (dose required to reach an optimal INR in Brazilian cardiovascular patients) in VKORC1 3673GA versus GG and, (2) AA versus GG genotypes. The change in the ratio of edge AUCs (ordinate) in these paired genotypic groups is presented as a function of the threshold (abscissa) defining the edge for the warfarin phenotype (expressed as the distance in SD units from the mean phenotypic value of the VKORC1 3673GG reference genotype: 37.9 mg/week). The thresholds for the edge AUCs were located 0.4 SD to 2.0 SD distance from the GG group mean. For the edge effect analysis in paired contrasts (GA vs. GG; and AA vs. GG), we used the pooled the SD for each genotype pair (11.9 and 12.5, respectively). The nonlinear increase (with upward concavity) in the ratio of edge AUCs indicates that the edge effect is more pronounced with more extreme phenotypic edges. The following exercise exemplifies the edge effect calculations further: For the GA and GG genotypic contrast, the threshold defining an edge AUC that is 0.8 SD distance from the reference VKORC1 3673GG group mean (37.9 mg/week warfarin dose) = 37.9 − (0.8 pooled SD for the GA and GG pair) = 37.9 − (0.8*11.9) = 28.4 mg/week. Accordingly, at this threshold defining the phenotype edge for the GG reference genotype, the edge effect = (percent of individuals within the GA group requiring warfarin doses equal to, or smaller than 28.4 mg/week)/(percent of individuals within the reference GG group requiring warfarin doses equal to, or smaller than 28.4 mg/week). The edge effect calculations are subsequently repeated for threshold values defining different edge AUCs by thresholds located from 0.4 SD to 2.0 SD distance from the reference VKORC1 3673GG group mean (37.9 mg/week).
Taken together, our edge effect analyses based on conceptual (Fig. 3, Table 1) and empirical data (Figs. 4 and 5) underscore two salient points:
In an association study of polygenic/multifactorial trait, the influence of an omics marker on the population edges of a quantitative phenotypic distribution is more profound than that on the population mean values (Figs. 3 and 5). Policy makers, ethicists, and those engaged in omics diagnostic test evaluation would be advised to consider the individuals at population edges who may be impacted substantially by an otherwise (apparently) modest shift in the mean value for a phenotype.
The edge effect will increase with phenotype edges that are located farther away from the population mean phenotypic value (Figs. 3 and 5).
Because individuals who reside at population edges are the ones with extreme phenotypes, the edge effect analysis can be a useful guidance to set priority levels for a targeted implementation of omics diagnostic tests, particularly in the context of limited healthcare resources. For example, in the case of the warfarin pharmacogenomics, individuals with the VKORC1 3673AA genotype may warrant more intensive clinical monitoring and interventions to adjust (decrease) warfarin dose as this subgroup includes a greater percentage of the patients who require markedly lower warfarin doses.
Edge Effect and Hospital Formulary Decisions
Hospital formularies have particular advantages and disadvantages compared with community settings to carry out field tests for omics diagnostics. It is here that the “proof of concept” for omics technologies and tests can be evaluated prior to implementation in the community where inherent logistical problems may hinder introduction of diagnostic services. In addition, diagnostic services tend to be close at hand and less expensive in a tertiary teaching hospital setting than at private commercial services. The interpretation of an omics diagnostic test can be more readily performed in a tertiary care hospital, because the infrastructure and expertise usually reside in such centers. In many technologically advanced countries’ wards, clinical pharmacists and applied pharmacologists are in a position to play a key role in not only patient identification but also ensuring all pertinent phenotypic data, pathology reports, and environmental factors are recorded, thereby allowing for almost complete data assessment, and hence, making the omics diagnostic test results more easy to interpret. These considerations position tertiary care hospitals as a model setting, and place the hospital formulary decision makers and those involved in diagnostic test reimbursement in a responsible leadership position to assess, recommend or reject omics diagnostic testing. However, most hospital formularies contain a restricted list of medicines that would not completely reflect community practice. As a result, clinical evaluation of omics testing in hospitals would not truly reflect the outside community omics testing which is the ultimate place for such testing. Also, when cost–benefit analyses are conducted, the result can only reflect that of the hospital and its formulary and cannot be used by regulatory bodies and health service providers to make a valued judgment. On the other hand, patients in tertiary care hospitals usually have more severe illness, are treated with multiple medications with inherent drug–drug interactions and tend to represent a broader range of ages. These factors may all result in a larger spread of the phenotypic distributions, and by extension, a greater percentage of individuals with extreme or edge phenotypes. Thus, despite the limitations to generalize results, university tertiary care teaching hospitals may serve as an ideal context to evaluate the proof of concept for omics diagnostics using edge effect analyses.
Future Outlook: 21st Century Diagnostic (Predictive) Medicine
Poised at the threshold of a new application of omics technologies in nutritional sciences (i.e., nutrigenomics) in the postgenomics era, what lessons can we learn—now—from the early observations of the Babylonians, Sumerians and Pythagoras, and the works in the 20th century by Garrod, Haldane, Motulsky, Vogel, Kalow, Williams, Vesell, Weber, Brewer, and other pioneers of predictive medicine, if we are to achieve a measured, mature and balanced entry into the field of nutrigenomics with realistic expectations?
First, nutrigenomics does not represent the end of omics applications. It is merely a passage from pharmacogenomics to broader omics-es such as agrigenomics and ecogenomics (Dunwell, 2009; Ozdemir et al., 2009; Select Biosciences, 2008).
Second, despite the promissory and often positively hyped representations in the media, the “nutrigenomics future” is yet undecided, awaiting multiple future trajectories (Ozdemir and Godard, 2007). Thus, success toward personalized nutritional interventions cannot be taken for granted. Looking back over the past 100 years, it is interesting to note that Garrod's vision of biochemical genetics attracted little attention from his peers, despite holding a Chair at the Oxford University (for a review, see: Motulsky, 2002). Pharmacogenetics remained initially as a fringe intellectual academic curiosity for nearly 40 years (until the 1990s) since the first introduction of the idea by Motulsky in October 1957. Applied social science research and prospective technology assessment will be essential to understand the technical and nontechnical factors that can influence the uptake of nutrigenomics and other omics innovations in diagnostic (predictive) medicine.
Third, perceptions on the value of new ideas and innovations tend to be fluid. Perceptions on the promise of pharmacogenetics, too, shifted with the sands of time. The availability of genomics and other omics technologies, together with the perceived promise of knowledge-based national economies endorsed by governments, brought pharmacogenetics into the mainstream scientific practice in late 1990s. Yet there are risks associated with both premature rejection of an innovative idea as well as its overendorsement to the point of hype with unsustainable claims—the latter is occasionally observed in the present day genomics literature and its representations by different user groups and stakeholders. Importantly, the genomics research in polygenic/multi-factorial complex diseases and pharmacogenomics did not bring about the same degree of success achieved formerly in monogenic diseases. In the meantime, social scientists aptly cautioned for the “hype fatigue” experienced overtly or implicitly by investors and various stakeholders in genomics (Nightingale and Martin, 2004). At this new intersection of genomics and nutritional sciences in 2009 (the focus for the present issue of the OMICS), it is therefore all the more important to reflect and learn constructively from the past history of diagnostic medicine, in order to discern what is realistically achievable in use of omics technologies toward the goal of nutrigenomics and personalized nutritional interventions.
Fourth, omics science, including nutrigenomics, is not only about high-throughput measurements of complex biological systems in response to environmental perturbations such as food, pharmaceuticals or toxins. The concepts and embedded professional values in “scientific laboratory” were fundamentally transformed with omics research, and societies that increasingly rely on knowledge-based national economies. The metaphor of a scientist as a person wearing a white coat at the laboratory bench is no longer adequate to capture the complex realities of research with omics technologies, and their promiscuous application contexts in human diseases, pharmacology, nutriscience, ecology, and agriculture. As evident in the primary focus of the journal OMICS, “integration” will be the key ingredient of success in the postgenomics era to move from a “parts list” offered by the Human Genome Project to the complex biological, environmental, economic, and social realities of the living organisms that are inseparable. A transdisciplinary approach to omics science and attendant innovations is important for their future sustainability and to understand downstream and lateral impacts on society and various user groups.
Metaphors are tenacious and powerful human constructs that resist change. The actual practice of omics research in the first decade of the 21st Century increasingly takes place in hitherto unprecedented social, economic and political spaces and consortiums (Gottweis, 2005; McNally and Glasner, 2006; Pohlhaus and Cook-Deegan, 2008; see also: Harding, 1998), replacing what was once described by the previous scientific metaphors of the 20th century—that scientific practice is (or ought to be) a value-free intellectual activity often limited to the physical confines of the laboratory space (Merton, 1942a; Merton, 1942b). Fueled by the rapid transition of scientific practice with the introduction of omics technologies, and their applications shifting from monogenic to multifactorial phenotypes, a growing gap emerged between the actual practice of science in the 21st century, and the ways in which it continues to be perceived by the society drawing on the metaphors of the past. This creates de facto a ‘social double-vision’ (i.e., social diplopia) conflicted with the actual realities and motivations of omics scientific practice, and perceptions endorsed by enduring metaphors. To understand the lived experience in practice of the 21st century diagnostic medicine and the attendant values of scientists and society, this gap has to be bridged.
There is a growing need for “recalibration” of human values, expectations, and public understanding of omics science. That is, omics research extends well beyond the laboratory space, and includes numerous user groups and stake-holders such as governments, private consortiums, charities, insurers, and nongovernmental organizations (NGOs). The multiplicity of human (e.g., “big science research groups”) and nonhuman actors (e.g., technologies, microarrays) engaged in omics innovations, and the complex ways in which they cooperate and compete for representation in the future (Brown et al., 2000), demand transparency and standardization efforts in omics fields, and the evidence base supporting the public health value of products emerging from omics applications (e.g., nutrigenomics tests in the marketplace).
Fifth, to the extent that nutri-genomics aims to predict nutri-gene interactions and associated health risks (e.g., benefit and harm from bioactive food constituents), we need to acknowledge that foodstuff is much more complex than well-defined and measurable units of pharmaceuticals. Discerning the “exposure–gene–health outcome” relationships will be substantially more difficult in nutrigenomics than pharmacogenomics. This predicament is further compounded by the present lack of frameworks for interpretation and communication of results from genotype associations discovered in the “omics laboratory” to the larger society.
Sixth, one could suggest that a deeper appreciation of human variability (as illustrated by the history of diagnostic medicine and omics science and its broad ramifications for evolution—reviewed earlier) can offer lateral gains toward participatory democracy in society. It could serve as an essential starting point to earnestly work toward a civil society that is not only tolerant of differences but keenly appreciative of human diversities, whether genetic, environmental, social, and/or cultural. Facing the deep fault lines that separate and fracture the 21st century human civilization and communities that stand on an increasingly unsustainable precarious pedestal, remembrance of the origins of omics technologies in “human variability questions” posed by pharmacogenomics, nutrigenomics, and ecogenomics (and their importance for population survival and sustainability) may thus offer wisdom beyond personalized medicine: toward reconciliation among human populations, cultures, ways of living and knowing, to build a more hopeful future for the global civil society (de Bernières, 2004; Havel, 1999; Pamuk, 2005; Power, 2008).
Reconnecting Omics Science and Society
Innovations resemble teenagers and the culinary art of wine making; they need to be tended. When left unattended, they may (potentially) turn sour or stop thriving and undermine those who conceived and invested in them. Nutrigenomics, the confluence of genomics and nutritional science, promises innovations in our understanding of the mechanisms of nutri-gene interactions, if not the elusive goal of personalized diets. Nutrigenomics future, however, is still uncertain. It demands careful anticipatory governance to cultivate a conceptual framework on how nutrigenomics data are interpreted and communicated from the nutrigenomics laboratory to society. This makes risk assessment and communication tools on laboratory findings an essential prerequisite, for nutrigenomics science to meaningfully and transparently engage with its complex set of actors and user groups in society. Because the genotype-phenotype association study design is the translational bedrock on which nutrigenomics and many other omics innovations rest, risk assessment and communication frameworks need to be developed specifically for the association studies.
In the final days of the 20th century, the United Nations Educational, Scientific, and Cultural Organization (UNESCO) in cooperation with the International Council for Science (ICSU), and the Government of Hungary, organized the World Conference on Science from 26 June to 1 July 1999, in Budapest. The conference was attended by over 1,800 delegates from 155 countries, including 28 intergovernmental organizations, and more than 60 international NGOs. The aims of this conference were “to help strengthen the commitment of ICSU and UNESCO member states and other major stakeholders to science education, research and development, and to define a strategy that would ensure that science responds better to society's needs and aspirations in the twenty-first century” (UNESCO, 2000). While this commendable conference recognized the important role science played in society, there remains a gap between omics science, particularly genotype–phenotype association studies, and society. With the very rapid transition from monogenic to multifactorial phenotypes in genomics over the past decade, risk assessment and communication concepts are unfortunately still driven, to a large extent, by what was available from previous studies of monogenic phenotypes. This “carryover effect” creates an expectation for distinctly separated phenotypic subpopulations in omics marker associations with multifactorial phenotypes. This is an unrealistic expectation in the context of polygenic/multifactorial phenotypes (Weedon and Frayling, 2008), and indeed, could be misleading, in interpretation of the merit of new nutrigenomics tests.
There appears to be an “asymmetrical progress” in the recent history of omics innovations: high-throughput technologies are extremely well developed and increasingly available in many parts of the world, but ways to interpret and communicate the research output from omics technology applications severely lagged behind, particularly in a policy context. Edge effect analyses presented in this paper may benefit the development of future risk assessment and communication strategies on association studies in nutrigenomics science and other omics fields. Undoubtedly, edge effect analysis is only one of the many other conceivable approaches that will be necessary to interpret and communicate the risk information obtained from association studies to society. This could permit moving from hindsight to foresight in biotechnology evaluation (Einsiedel, 2009), prevent the creation of a “nutrigenomics straw-man,” and ultimately, facilitate prospective policy development for future applications of omics technologies as well (e.g., ecogenomics and agrigenomics).
Empiricism and the scientific method transformed human curiosity into technological breakthroughs that enormously benefited the society over the past four centuries. This also contributed to framing of science as an invariably and uniformly ‘useful’ and objective activity (e.g., captured in the enthusiastic aphorism ‘knowledge is power’) (Harrison, 2001). The legitimacy of a body of knowledge is now considered to rely upon following a strict set of experimental procedures dictated by the standards of a given discipline (e.g., genomics, pharmacology). While the importance of experimental integrity cannot be denied, the process of acquisition of knowledge starts much earlier, including which research question is being asked and later, how data are interpreted and communicated. In addition to experimental accuracy, integrity of scientific knowledge thus relies heavily on awareness of the ethical issues that may arise before an experiment is conducted (e.g., why and how a certain research question is asked or not asked at all) as well as after the empirical experiments are completed (e.g., how the primary data and covariates in an association study are analyzed, reported or suppressed). Fault lines in this entire process of scientific inquiry and acquisition, interpretation or communication of new knowledge can have ethical significance (Birmingham, 2001; De Vries and Lemmens, 2006; Fish-man, 2004; Guston et al., 2009; Healy, 2007; Landefeld and Steinman, 2009; Lemmens and Miller, 2006; Lexchin, 2008; Mintzes et al., 2005; Rhodes and Strain, 2004; Turner et al., 2008). Development of risk assessment and communication standards for data generated in genotype-phenotype association studies would contribute to integrity in omics science. For this vision to materialize, a broad awareness of the following issues is needed within the omics research community:
The knowledge (e.g., generated in the omics laboratory) and the knower (e.g., a scientist) are not separable (Harrison, 2001). Knowledge is embedded in the values of its producer. That is, the human values and social responsibilities of scientists are of paramount importance and could impact which data are generated, reported or suppressed, in much the same way as being an innovative and curious scientist. Interpretation of the data, too, can be impacted by the values and set of prejudices of each person who reviews a body of evidence, whether they are scientists, policy-makers or the public (Choi, 2005; Marrot, 2004). This recognition predicts that the relationship between evidence and policy is not linear and that governments and policy-makers often balance multiple competing interests—beyond scientific evidence—in forming policies (Marmot, 2004). This also highlights that science and scientific practice cannot be neatly separated from their political underpinnings.
Experimental data do not speak on their own in a uniform manner unless accompanied by scientific and ethical standards.
These are essential prerequisite insights before the need for risk assessment and communication tools and standards can be appreciated. A sole reliance on technical and experimental integrity of science may not suffice to ensure long term public trust in emerging omics fields such as nutrigenomics, and could presumably create invisible roadblocks in uptake of future omics innovations. With this in mind, the OMICS Nutrigenomics Special Issues also highlighted some of the human values that can influence interpretation and communication of data between omics laboratory and user groups in society (for a review, see: Ozdemir et al., 2009).
Interpreting emerging scientific information that is incomplete, uncertain, and contested is a formidable challenge (Guston, 2007; Guston, 2008; Saltelli and Funtowicz, 2004). The multiplicity of actors engaged in omics science, the complex ways in which the omics stakeholders (hyper)-compete, and the strong focus on product development (e.g., genetic tests) in translational research in applied genomics (with the attendant economic and political stakes) call for attention to genealogy of scientific knowledge and the processes by which knowledge is produced (Kaye, 2008). A genealogical approach to innovation analysis would take into account views that are both dominant and dissenting (or silenced), and ways in which the latter did not become established and recognized because of the institutionalization of knowledge by those in power (Foucault, 1994; Dreyfus and Rabinow, 1982). As noted by the Editors of the Science, Technology & Human Values “. . . great research may be published anywhere and in any language. Truly groundbreaking work may be more likely to appear from marginal, dissident, or unexpected sources, rather than from a well-established and entrenched mainstream” (ST&HV Editors, 2009). Such concerns apply equally well to emergence of new unprecedented forms of art and public knowledge outside the traditional gallery and museum system (e.g., consider the original graffiti art and the chalk figures drawn by Keith Haring on New York City subway posters in the 1980s) (Aubert, 1989). As with arts, discourses on omics innovations cannot be complete, nor balanced, unless due attention is given to a broad range of knowledge sources in society, and how knowledge is processed and communicated from the laboratory to society (and vice versa). Genealogical approach to analysis of scientific knowledge would also consider notions of discontinuity, and precisely what determines just what makes it into the ostensible evidence base that supports or rejects new ideas, clinical guidelines (George, 2003; Stevenson and Cutcliffe, 2006), genetic tests and innovations more generally. Hence, the future nutrigenomics tests need to be developed in the presence of appropriate regulatory guidance and risk assessment/communication frameworks informed by, and firmly grounded in the genealogy of omics knowledge and diagnostic medicine. Additionally, legislative initiatives to create a publicly funded multidisciplinary oversight body could be timely to proactively build public trust in new omics fields, and to carry out independent, impartial and integrated innovation analyses across the omics fields from pharmacogenomics to nutrigenomics and ecogenomics (Ozdemir et al., 2009).
An unintended consequence of rapid technological advances in omics science, in the absence of commensurate developments “outside the laboratory space,” is the deep laceration (discontinuity) in omics history that formed between the past and the present-day practice of omics science, as well as between the omics laboratory and society. For human populations who are forcibly amputated from their past history, it is difficult to meaningfully forecast and engage with their future (de Bernières, 2004; Pamuk, 2005). This is no less applicable in the case of sustainability of scientific innovations. The modern-day representations of omics innovations are detached from the history of diagnostic medicine, with displays of unrestricted optimism that portray scientific advances as a linear process from the laboratory bench to society. The history of diagnostic (predictive) medicine and omics science firmly indicates that new ideas follow a nonlinear trajectory and may remain in obscurity for some time. Innovators in many fields of human inquiry (and their ideas) endure rejection, ridicule, and at times, persecution. In this regard, it may suffice to recall that many poems and novels that are now considered as classics were considered unpublishable initially or banned entirely (e.g., consider the Ulysses by James Joyce, or the Human Landscapes from My Country by Nazim Hikmet) (Hikmet, 2009; Joyce, 1998). For a deeper understanding of the present-day omics fields and potential future trajectories, they need to be situated within the past history of diagnostic medicine and omics science.
Truly novel innovations (not all innovations are innovative) are often cultivated by crystallization of intellectual entropy in the middle of chaos: that is, at the intersection of new ideas that struggle for survival and future representation on the one hand, and antagonism by existing ideas, institutionalized forms of old knowledge, and human and nonhuman actors in science and society whose power structures may be disrupted by innovations and novel ways of human understanding. Conversely, a new idea may be adopted too soon without proactively building an appropriate evidence base and analytical framework to evaluate their multiple intended and unintended impacts on science and society. If left unchecked, such diagonally opposing influences may result in a wide range of public perceptions and responses, including technophilia and molecular myopia on the one hand, and technophobia on the other extreme. In a recent editorial, Gurwitz and Motulsky make the apt observation that October 1957—the date when the seminal article by Motulsky on the idea of pharmacogenetics appeared in print—coincided with the launch of the Sputnik 1. In their account, “ . . . the contrast between the slow evolution of pharmacogenetics over the last 50 years is striking when compared with the amazing advance of the space program which also started in October 1957 with the launch of the Sputnik 1” (Gurwitz and Motulsky, 2007). The thoughts of Roger J. Williams (1893−1988) on individual differences in response to nutrition and pharmaceuticals were not recognized by his peers immediately. While one may debate whether Pythagoras is a founder of the field of pharmacogenetics (recalling his observations on favism), he nonetheless had to leave his native land Samos (in east Aegean Sea) for Croton, Italy, to escape (flee) the tyrannical governance of Polycrates (Ozdemir, 2007). Innovations thus tend to follow a chaotic, ostensibly anarchic, and unpredictable course (Smil, 2001). These lessons from the history of diagnostic medicine and omics science need to be reconnected with the present-day practice of omics science in order to set realistic expectations from new biotechnology applications such as nutrigenomics. This is significant not only for sustainability of new omics fields and balanced representation of scientific advances. It also has relevance for training of new generation of omics scientists under a vision that acknowledges and appreciates the discontinuous and nonlinear nature of scientific innovations.
We caution the reader that the edge effect analysis does not represent the only approach to risk assessment to interpret the omics marker associations with multifactorial phenotypes. It is used herein as an example to illustrate the need for developing standards over risk assessment and communication tools to support nutrigenomics and other emerging omics innovations. The choice of phenotypic edges should not be arbitrary. This could be guided by biological plausibility and clinical relevance as well as the extent to which different public user groups for omics diagnostic tests understand and value a phenotypic value as being ‘edge’. Definition of edge phenotypes by standard deviation distances from a mean phenotypic value is another solution to express the edges in a quantitative phenotype distribution.
Concluding Remarks
Sustainability of omics fields, both emerging (nutrigenomics, agrigenomics and ecogenomics) and those that are more established (pharmacogenomics), depends in part on connecting these scientific inquiries with society, through timely development of risk assessment and communication tools and standards (both scientific and ethical) for association studies. The history of diagnostic medicine and omics science can enable scientists to attain a richer interdisciplinary understanding and critical scholarship of the present day omics fields, and factors that can limit and facilitate omics innovations in the future. Importantly, recognition of the nontechnical factors that can influence the processes by which omics knowledge is produced, for example, social responsibilities of scientists and human values embedded in scientific practice, can reduce the uncertainty in the omics future. These considerations would collectively render the anticipated journey of omics science from laboratory to society more predictable by cultivating a sustainable and responsible conduit between them.
Acknowledgements
This article is dedicated to Laszlo Endrenyi (Toronto). The work in this manuscript is supported by an ethics operating catalyst grant from the Canadian Institutes of Health Research (CIHR) and a career investigator salary for ethics, science, and society research from the Fonds de la recherche en santé du Québec (FRSQ) to Ozdemir. Research in the Suarez-Kurtz laboratory is supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Coordenação de Aperfeiçoamento de Pessoal do Ensino Superior (CAPES), Financiadora de Estudos e Projetos (Finep), and Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro (Faperj). Raphaëlle Stenne is the recipient of a doctoral bursary for bioethics research in personalized medicine from the Département de Sciences Biomédicales, Université de Montréal.
Helpful discussions with the following individuals on the concept of edge effect, and science and society bridging in genomics are gratefully acknowledged: Drs. Edmund J.D. Lee (Yong Loo Lin School of Medicine, National University of Singapore, Singapore), Laszlo Tothfalusi (Semmelweis University, Budapest), Orhan Arikan (Bilkent University, Ankara, Turkey), Yeşim Yetimalar (Atatürk Eğitim ve Araştirma Hastanesi, Izmir, Turkey), and Collen Masimirembwa (African Institute of Biomedical Science and Technology, Zimbabwe). Professor Edmund Lee has taught pharmacogenomics and personalized medicine for many years (and continues to do so) in Singapore using the concept and examples of edge effects.
We thank Drs. Andrea Gaedigk and Steven Leeder (Children's Mercy Hospital and Clinics, Kansas City) for providing a high resolution copy of the Figure 1 from their published work, and Drs. Serge Dubé and Vincent Castellucci for encouraging transdisciplinary boundary crossing research in bioscience, medicine and humanities.
The history of omics science and predictive medicine in this paper does not necessarily include the entire list of investigators and authors who made important contributions to the field in the 20th Century. However, we trust that the analyses presented herein and the accompanying references will provide the reader with an immediate grasp of the past history and processes of innovations in omics science and diagnostic medicine as well as the enormous challenges and discontinuities faced by the pioneers in this field. Additional information on the development of omics science and its applications is available elsewhere (Nebert et al., 2008). The ideas and analyses presented in this paper reflect entirely the personal views of the authors.
Footnotes
Author Disclosure Statement
No competing financial interests exist. No medical writer or writing assistance was employed in preparation of this work.
References
- Anderson L, Anderson NG. High resolution two-dimensional electrophoresis of human plasma proteins. Proc Natl Acad Sci USA. 1977;74:5421–5425. doi: 10.1073/pnas.74.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson NG, Anderson NL. Twenty years of two-dimensional electrophoresis: past, present and future. Electrophoresis. 1996;17:443–453. doi: 10.1002/elps.1150170303. [DOI] [PubMed] [Google Scholar]
- Anderson NG, Matheson A, Anderson NL. Back to the future: the human protein index (HPI) and the agenda for post-proteomic biology. Proteomics. 2001;1:3–12. doi: 10.1002/1615-9861(200101)1:1<3::AID-PROT3>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Aubert E. Drawing the line [video recording]: a portrait of Keith Haring. Biografilm Associates; W. Long Branch, N.J., USA. Kultur: 1989. [distributor] [Google Scholar]
- Baccini M, Bachmaier EM, Biggeri A, Boekschoten MV, Bouwman FG, Brennan L, et al. The NuGO proof of principle study package: a collaborative research effort of the European Nutrigenomics Organisation. Genes Nutr. 2008;3:147–151. doi: 10.1007/s12263-008-0102-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertilsson L, Alm C, De Las Carreras C, Widen J, Edman G, Schalling D. Debrisoquine hydroxylation polymorphism and personality. Lancet. 1989;1:555. doi: 10.1016/s0140-6736(89)90094-9. [DOI] [PubMed] [Google Scholar]
- Birmingham K. Dark clouds over Toronto psychiatry research. Nat Med. 2001;7(6):643. doi: 10.1038/88975. [DOI] [PubMed] [Google Scholar]
- Blankeslee AF, Salmon MR. Odor and taste-blindness. Eugen News. 1931;16:105–110. [Google Scholar]
- Blaschke TF, Giacomini KM. Obituary. Lewis B. Sheiner, 1940−2004. Clin Pharmacol Ther. 2004;76:650–651. [Google Scholar]
- Brewer GJ. Annotation: human ecology, an expanding role for the human geneticist. Am J Hum Genet. 1971;23:92–94. [PMC free article] [PubMed] [Google Scholar]
- Brown N. Hope against hype—Accountability in biopasts, presents and futures. Sci Stud. 2003;16:3–21. [Google Scholar]
- Brown N, Rappert B, Webster A. Contested futures: a sociology of prospective techno-science. Ashgate; Aldershot, UK: 2000. [Google Scholar]
- Burgess M, O'Doherty K, Secko D. Biobanking in British Columbia: discussions of the future of personalized medicine through deliberative public engagement. Personal Med. 2008;5:285–296. doi: 10.2217/17410541.5.3.285. [DOI] [PubMed] [Google Scholar]
- Carson PE, Flanagan CL, Ickes CE, Alvong AS. Enzymatic deficiency in primaquine sensitive erythrocytes. Science. 1956;124:484–485. doi: 10.1126/science.124.3220.484-a. [DOI] [PubMed] [Google Scholar]
- Caulfield T. Sustainability and the balancing of the health care and innovation agendas: commercialization of genetic research. Saskatchewan Law Rev. 2003;66:629–645. [PubMed] [Google Scholar]
- CDER, Food and Drug Administration, U.S. Department of Health and Human Services [January 2, 2009];Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers. 2006 Available at: http://www.fda.gov/cder/drug/drugInteractions/tableSubstrates.htm#4.
- Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clin Pharmacol Ther. 2007;81:265–269. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- Choi BC, Pang T, Lin V, Puska P, Sherman G, Goddard M, et al. Can scientists and policy makers work together? J Epidemiol Community Health. 2005;59:632–637. doi: 10.1136/jech.2004.031765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagni A, Bartoli S, Buehrlen B, Fattore G, Ibarreta D, de Mesa EG. Avoiding adverse drug reactions by pharmacogenetic testing: a systematic review of the economic evidence in the case of TPMT and AZA-induced side effects. Int J Technol Assess Health Care. 2008;24:294–302. doi: 10.1017/S0266462308080392. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Johnson JA, Langaee TY, Feng H, Stanaway IB, Schwarz UI, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daar AS, Berndtson K, Persad DL, Singer PA. How can developing countries harness biotechnology to improve health? BMC Public Health. 2007;7:346. doi: 10.1186/1471-2458-7-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly AK, King BP. Contribution of CYP2C9 to variability in vitamin K antagonist metabolism. Expert Opin Drug Metab Toxicol. 2006;2:3–15. doi: 10.1517/17425255.2.1.3. [DOI] [PubMed] [Google Scholar]
- Davis DR, Hackert ML, Reed LJ. Roger J. Williams. 1893−1988. A Biographical Memoir. National Academy of Sciences; Washington, DC: 2008. [January 2, 2009]. Available at: http://books.nap.edu/html/biomems/rwilliams.pdf. [Google Scholar]
- de Bernières L. Birds Without Wings. Vintage Books, Random House; London: 2004. pp. 1–625. [Google Scholar]
- de Leon J. The future (or lack of future) of personalized prescription in psychiatry. Pharmacol Res. 2008 Oct 17; doi: 10.1016/j.phrs.2008.10.002. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Dengler HJ, Eichelbaum M. Polymorphismen und Defekte des Arzneimittelstoffwechsels als Ursache toxischer Reaktionen. Arzneim. Forsch. (Drug Res.) 1977;27:1836–1844. [PubMed] [Google Scholar]
- Detienne M. La Mythologie des Aromates en Grèce. Les Jardins d'Adonis. Gallimard; Paris: 1972. pp. 76–113. [Google Scholar]
- De Vries R, Lemmens T. The social and cultural shaping of medical evidence: case studies from pharmaceutical research and obstetric science. Soc Sci Med. 2006;62:2694–2706. doi: 10.1016/j.socscimed.2005.11.026. [DOI] [PubMed] [Google Scholar]
- Dreyfus HL, Rabinow P. Michel Foucault: Beyond Structuralism and Hermeneutics. The University of Chicago Press; Chicago, IL: 1982. [Google Scholar]
- Dunwell JM. Transgenic wheat, barley and oats: future prospects. Methods Mol Biol. 2009;478:333–345. doi: 10.1007/978-1-59745-379-0_20. [DOI] [PubMed] [Google Scholar]
- Edelson E. Genetic test predicts response to warfarin. U.S. News and World Report. 2008 August 14; [Google Scholar]
- Eichelbaum M, Spannbrucker N, Dengler HJ. N-oxidation of sparteine in man and its interindividual differences. Arch. Pharmacol. 1975a;287:R94. [PubMed] [Google Scholar]
- Eichelbaum M, Spannbrucker N, Dengler HJ. Lack of N-oxidation of sparteine in certain healthy subjects.. Sixth International Congress of Pharmacology; Helsinki. July 20−25.1975b. p. 1071. [Google Scholar]
- Eichelbaum M, Spannbrucker N, Steincke B, Dengler HJ. Defective N-oxidation of sparteine in man: a new pharmacogenetic defect. Eur J Clin Pharmacol. 1979;16:183–187. doi: 10.1007/BF00562059. [DOI] [PubMed] [Google Scholar]
- Einsiedel EF. Emerging Technologies: From Hindsight to Foresight. UBC Press; Vancouver: 2009. pp. 1–328. [Google Scholar]
- Evans DA, Manley KA, McKusick VA. Genetic control of isoniazid metabolism in man. Br Med J. 1960;2:4484–4491. doi: 10.1136/bmj.2.5197.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman JR. Manufacturing desire: the commodification of female sexual dysfunction. Soc Stud Sci. 2004;34:187–218. doi: 10.1177/0306312704043028. [DOI] [PubMed] [Google Scholar]
- Flockhart DA. Drug Interactions: Cytochrome P450 Drug Interaction Table. Indiana University School of Medicine; Bloomington, IN: 2007. [January 2, 2009]. http://medicine.iupui.edu/flockhart/table.htm. [Google Scholar]
- Food and Drug Administration . Innovation/stagnation. Challenge and Opportunity on the Critical Path to New Medical Products. Food and Drug Administration; Washington, DC: 2004. US Department of Health and Human Services. [Google Scholar]
- Foucault M. Genealogy and Social Criticism. In: Seidman S, editor. The Post-modern Turn: New Perspectives on Social Theory. Cambridge University Press; Cambridge, MA: 1994. (reprinted from Foucault, M. Power/knowledge. Pantheon Books, 1977) [Google Scholar]
- Fox AL. The relationship between chemical constitution and taste. Proc Natl Acad Sci USA. 1932;18:115–120. doi: 10.1073/pnas.18.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frueh FW, Gurwitz D. From pharmacogenetics to personalized medicine: a vital need for educating health professionals and the community. Pharmacogenomics. 2004;5:571–579. doi: 10.1517/14622416.5.5.571. [DOI] [PubMed] [Google Scholar]
- Gaedigk A, Simon SD, Pearce RE, Bradford LD, Kennedy MJ, Leeder JS. The CYP2D6 activity score: translating genotype information into a qualitative measure of phenotype. Clin Pharmacol Ther. 2008;83:234–242. doi: 10.1038/sj.clpt.6100406. [DOI] [PubMed] [Google Scholar]
- Gage B, Eby C, Johnson J, Deych E, Rieder M, Ridker P, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod AE. Incidence of Alkaptonuria: A study in Chemical Individuality. Lancet. 1902;2:653–656. [PMC free article] [PubMed] [Google Scholar]
- Garrod AE. Inborn Errors of Metabolism. Oxford University Press; London: 1909. [Google Scholar]
- Garrod AE. The Inborn Factors in Disease: An Essay. Clare-don Press; Oxford: 1931. [Google Scholar]
- Gene Tests [January 2, 2009];Funded by the National Institutes of Health. 2008 Available at: www.genetests.org.
- George JM. An emerging discourse toward epistemic diversity in nursing. Adv Nurs Sci. 2003;26:44–52. doi: 10.1097/00012272-200301000-00007. [DOI] [PubMed] [Google Scholar]
- Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin Pharmacol Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godard B, Ozdemir V. Nutrigenomics and personalized diet: from molecule to intervention and nutri-ethics. OMICS. 2008;12:227–228. doi: 10.1089/omi.2008.0069. [DOI] [PubMed] [Google Scholar]
- Gonzalez FJ, Skoda RC, Kimura S, Umeno M, Zanger UM, Nebert DW, et al. Characterization of the common genetic-defect in humans deficient in debrisoquine metabolism. Nature. 1988;331:442–446. doi: 10.1038/331442a0. [DOI] [PubMed] [Google Scholar]
- González I, Peñas-Lledó EM, Pérez B, Dorado P, Alvarez M, Lerena A. Relation between CYP2D6 pheno-type and genotype and personality in healthy volunteers. Pharmacogenomics. 2008;29:833–840. doi: 10.2217/14622416.9.7.833. [DOI] [PubMed] [Google Scholar]
- Gottweis H. Governing genomics in the 21st century: between risk and uncertainty. New Genet Soc. 2005;24:175–193. doi: 10.1080/14636770500184818. [DOI] [PubMed] [Google Scholar]
- Gregory TR. Genome size evolution in animals. In: Gregory TR, editor. The Evolution of the Genome. Elsevier; San Diego, CA: 2005. pp. 3–87. [Google Scholar]
- Gunes A, Dahl ML. Variation in CYP1A2 activity and its clinical implications: influence of environmental factors and genetic polymorphisms. Pharmacogenomics. 2008;9:625–637. doi: 10.2217/14622416.9.5.625. [DOI] [PubMed] [Google Scholar]
- Guo SW, Reed DR. The genetics of phenylthiocarbamide perception. Ann Hum Biol. 2001;28:111–142. doi: 10.1080/03014460151056310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurwitz D, Lunshof JE, Dedoussis G, Flordellis CS, Fuhr U, Kirchheiner J, et al. Pharmacogenomics education: International Society of Pharmacogenomics recommendations for medical, pharmaceutical, and health schools deans of education. Pharmacogenomics J. 2005;5:221–225. doi: 10.1038/sj.tpj.6500312. [DOI] [PubMed] [Google Scholar]
- Gurwitz D, Lunshof J. Personalized pharmacotherapy: genotypes, biomarkers, and beyond. Clin Pharmacol Ther. 2008 Nov 12; doi: 10.1038/clpt.2008.228. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Gurwitz D, Motulsky AG. “Drug reactions, enzymes, and biochemical genetics”: 50 years later. Pharmacogenomics. 2007;8:1479–1484. doi: 10.2217/14622416.8.11.1479. [DOI] [PubMed] [Google Scholar]
- Guston D. [February 6, 2009];Toward Anticipatory Governance. 2007 http://nanohub.org/resources/3270.
- Guston DH. Innovation policy: not just a jumbo shrimp. Nature. 2008;454(7207):940–941. doi: 10.1038/454940a. [DOI] [PubMed] [Google Scholar]
- Guston DH, Sarewitz D, Miller C. Scientists not immune to partisanship. Science. 2009;323(5914):582. doi: 10.1126/science.323.5914.582b. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. Heredity and Politics. W.W. Norton; New York: 1938. [Google Scholar]
- Harding S. Is Science Multicultural? Postcolonialisms, Feminisms, and Epistemologies. Indiana University Press; Bloomington, IN: 1998. pp. 1–242. [Google Scholar]
- Harrison P. Curiosity, forbidden knowledge, and the reformation of natural philosophy in early modern England. Isis. 2001;92:265–290. doi: 10.1086/385182. [DOI] [PubMed] [Google Scholar]
- Havel V. Address by Vaclav Havel President of the Czech Republic in acceptance of “Open Society” Prize by the Central European University. Budapest: Jun 24, 1999. [January 2, 2009]. Available online at: http://old.hrad.cz/president/Havel/speeches/1999/2406_uk.html. [Google Scholar]
- Healy D. One flew over the conflict of interest nest. World Psychiat. 2007;6:26–27. [PMC free article] [PubMed] [Google Scholar]
- Hedgecoe A. The Politics of Personalised Medicine—Pharmacogenetics in the Clinic. Cambridge University Press; Cambridge: 2004. Cambridge Studies in Society and the Life Sciences; pp. 1–208. [Google Scholar]
- Hikmet N. Human Landscapes from My Country: An Epic Novel in Verse. Persea Books; New York: 2009. Hirsch, E. (foreword), Blasing, R. (translator), Konuk, M. (translator) [Google Scholar]
- Hopkins MM, Ibarreta D, Gaisser S, Enzing CM, Ryan J, Martin PA, et al. Putting pharmacogenetics into practice. Nat Biotechnol. 2006;24:403–410. doi: 10.1038/nbt0406-403. [DOI] [PubMed] [Google Scholar]
- Ibarreta D. Interview. Personal Med. 2008;5:331–334. doi: 10.2217/17410541.5.4.331. [DOI] [PubMed] [Google Scholar]
- Ingelman-Sundberg M, Daly AK, Nebert DM, editors. [February 6, 2009];Human Cytochrome P450 (CYP) Allele Nomenclature Committee. 2008 http://www.cypalleles.ki.se/index.htm.
- Joyce J. Orchises Press; Washington, DC: 1998. Ulysses: A Facsimile of the First Edition Published in Paris in 1922. [Google Scholar]
- Kalow W. Pharmacogenetics. Heredity and the Response to Drugs. W.B. Saunders Co.; Philadelphia, PA: 1962. [Google Scholar]
- Kalow W. Pharmacoanthropology and the genetics of drug metabolism. In: Kalow W, editor. Pharmacogenetics of Drug Metabolism. Pergamon Press Inc; New York: 1992. pp. 865–877. [Google Scholar]
- Kalow W. Pharmacogenetics in perspective. Drug Metab Dispos. 2001;29(4 Pt 2):468–470. [PubMed] [Google Scholar]
- Kalow W. Both populations and individuals are evolutionary targets: pharmacogenomic and cultural indicators. Pharmacogenomics J. 2002;2:12–14. doi: 10.1038/sj.tpj.6500083. [DOI] [PubMed] [Google Scholar]
- Kaput J, Astley S, Renkema M, Ordovas J, van Ommen B. Harnessing nutrigenomics: development of web-based communication, databases, resources, and tools. Genes Nutr. 2006;1:5–11. doi: 10.1007/BF02829931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaye J. The regulation of direct-to-consumer genetic tests. Hum Mol Genet. 2008;17(R2):R180–183. doi: 10.1093/hmg/ddn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoury MJ, Berg A, Coates R, Evans J, Teutsch SM, Bradley LA. The evidence dilemma in genomic medicine. Health Aff (Millwood) 2008;27:1600–1611. doi: 10.1377/hlthaff.27.6.1600. [DOI] [PubMed] [Google Scholar]
- Kim UK, Drayna D. Genetics of individual differences in bitter taste perception: lessons from the PTC gene. Clin Genet. 2004;67:275–280. doi: 10.1111/j.1399-0004.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- Knoppers BM, Joly Y. Our social genome? Trends Biotechnol. 2007;25:284–288. doi: 10.1016/j.tibtech.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Kolker E, Higdon R, Hogan JM. Protein identification and expression analysis using mass spectrometry. Trends Microbiol. 2006;14:229–235. doi: 10.1016/j.tim.2006.03.005. [DOI] [PubMed] [Google Scholar]
- Kuska B. Beer, Bethesda, and biology: how “genomics” came into being. J Natl Cancer Inst. 1998;90:93. doi: 10.1093/jnci/90.2.93. [DOI] [PubMed] [Google Scholar]
- Landefeld CS, Steinman MA. The Neurontin legacy—marketing through misinformation and manipulation. N Engl J Med. 2009;360:103–106. doi: 10.1056/NEJMp0808659. [DOI] [PubMed] [Google Scholar]
- Lederberg J, McCray AT. “Ome sweet omics”:—a genealogical treasury of words. Scientist. 2001;15:2. [Google Scholar]
- Lemmens T, Miller PB. Regulating the market in human research participants. PLoS Med. 2006;3:e330. doi: 10.1371/journal.pmed.0030330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexchin J. Doctors and industry funding of continuing medical education: guilty as charged. Israeli J Emergency Med. 2008;8(2):23–25. [Google Scholar]
- Llerena A, Dorado P, Peñas-Lledó EM. Pharmacogenetics of debrisoquine and its use as a marker for CYP2D6 hydroxylation capacity. Pharmacogenomics. 2009;10:17–28. doi: 10.2217/14622416.10.1.17. [DOI] [PubMed] [Google Scholar]
- Lunshof JE, Pirmohamed M, Gurwitz D. Personalized medicine: decades away? Pharmacogenomics. 2006;7:237–241. doi: 10.2217/14622416.7.2.237. [DOI] [PubMed] [Google Scholar]
- Lunshof J. PhD Dissertation. Vrije Universiteit; Amsterdam, NL: 2008. [February 6, 2009]. The new genomics: challenges for ethics. Available at: http://hdl.handle.net/1871/13070. [Google Scholar]
- Lunshof JE, Chadwick R, Vorhaus DB, Church GM. From genetic privacy to open consent. Nat Rev Genet. 2008;9:406–411. doi: 10.1038/nrg2360. [DOI] [PubMed] [Google Scholar]
- Mahlknecht U, Voelter-Mahlknecht S. Pharmacogenomics: questions and concerns. Curr Med Res Opin. 2005;21:1041–1047. doi: 10.1185/030079905X50633. [DOI] [PubMed] [Google Scholar]
- Maienschein J, Smith G. What difference does history of science make, anyway? Isis. 2008;99:318–321. doi: 10.1086/588689. [DOI] [PubMed] [Google Scholar]
- Maienschein J, Laubichler M, Loettgers A. How can history of science matter to scientists? Isis. 2008;99:341–349. doi: 10.1086/588692. [DOI] [PubMed] [Google Scholar]
- Mahgoub A, Idle JR, Dring LG, Lancaster R, Smith R. Polymorphic hydroxylation of debrisoquine in man. Lancet. 1977;2:584–586. doi: 10.1016/s0140-6736(77)91430-1. [DOI] [PubMed] [Google Scholar]
- Marmot MG. Evidence based policy or policy based evidence? BMJ. 2004;328:906–907. doi: 10.1136/bmj.328.7445.906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masimirembwa CM, Johansson I, Hasler JA, Ingelman-Sundberg M. Genetic polymorphism of cytochrome P450 CYP2D6 in Zimbabwean population. Pharmacogenetics. 1993;3:275–280. doi: 10.1097/00008571-199312000-00001. [DOI] [PubMed] [Google Scholar]
- Masimirembwa C, Persson I, Bertilsson L, Hasler J, Ingelman-Sundberg M. A novel mutant variant of the CYP2D6 gene (CYP2D6*17) common in a black African population: association with diminished debrisoquine hydroxylase activity. Br J Clin Pharmacol. 1996;42:713–719. doi: 10.1046/j.1365-2125.1996.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Master Z, Ozdemir V. Selling translational research—is science a value-neutral autonomous enterprise? Am J Bioeth. 2008;8:52–54. doi: 10.1080/15265160802109413. [DOI] [PubMed] [Google Scholar]
- McGuire A, Burke W. An unwelcome side effect of direct-to-consumer personal genome testing: raiding the medical commons. JAMA. 2008;300:2669–2671. doi: 10.1001/jama.2008.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKusick VA, Ruddle FH. Toward a complete map of the human genome. Genomics. 1987;1:103–106. doi: 10.1016/0888-7543(87)90001-2. [DOI] [PubMed] [Google Scholar]
- McNally R, Glasner P. Survival of the gene? 21st century visions from genomics, proteomics and the new biology. In: Glasner P, Atkinson P, Greenslade H, editors. New Genetics, New Social Formations. Routledge; London: 2006. pp. 253–278. [Google Scholar]
- Merton RK. Science and technology in a democratic order. J Leg Polit Sociol. 1942a;1:115–126. [Google Scholar]
- Merton RK. The normative structure of science. In: Storer NW, editor. The Sociology of Science. University of Chicago Press; Chicago, IL: 1942b. pp. 267–278. [Google Scholar]
- Millstone E. Analysing biotechnology's traumas. New Genet Soc. 2000;19:117–132. [Google Scholar]
- Mintzes B, Barer M, Lexchin J, Bassett KL. Introduction of direct-to-consumer advertising of prescription drugs in Canada: an opinion survey on regulatory policy. Res Social Adm Pharm. 2005;1:310–330. doi: 10.1016/j.sapharm.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Montpetit E, Rothmayr C, Varone F, editors. The Politics of Biotechnology in North America and Europe: Policy Networks, Institutions and Internationalization. Lexington Books; Lanham, MD: 2006. [Google Scholar]
- Morin K. Knowledge and attitudes of Canadian consumers and health care professionals regarding nutritional genomics. OMICS. 2009:13. doi: 10.1089/omi.2008.0047. (in press) [DOI] [PubMed] [Google Scholar]
- Motulsky AG. Drug reactions, enzymes and biochemical genetics. JAMA. 1957;165:835–837. doi: 10.1001/jama.1957.72980250010016. [DOI] [PubMed] [Google Scholar]
- Motulsky AG. From pharmacogenetics and ecogenetics to pharmacogenomics. Med Secoli. 2002;14:683–705. [PubMed] [Google Scholar]
- Nakai K, Tsuboi J, Okabayashi H, Fukuhiro Y, Oka T, Habano W, et al. Ethnic differences in the VKORC1 gene polymorphism and an association with warfarin dosage requirements in cardiovascular surgery patients. Pharmacogenomics. 2007;8:713–719. doi: 10.2217/14622416.8.7.713. [DOI] [PubMed] [Google Scholar]
- Naylor S, Cavanagh J. Status of systems biology—does it have a future? Biosilico. 2004;2:171–174. [Google Scholar]
- Naylor S, Culbertson AW, Valentine SJ. Towards a systems level analysis of health and nutrition. Curr Opin Biotechnol. 2008;19:100–109. doi: 10.1016/j.copbio.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Nebert DW. Pharmacogenetics and pharmacogenomics: why is this relevant to the clinical geneticist? Clin Genet. 1999;56:247–258. doi: 10.1034/j.1399-0004.1999.560401.x. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Jorge-Nebert L, Vesell ES. Pharmacogenomics and “individualized drug therapy”: high expectations and disappointing achievements. Am J Pharmacogenomics. 2003;3:361–370. doi: 10.2165/00129785-200303060-00002. [DOI] [PubMed] [Google Scholar]
- Nebert DW, Zhang G, Vesell ES. From human genetics and genomics to pharmacogenetics and pharmacogenomics: past lessons, future directions. Drug Metab Rev. 2008;40:187–224. doi: 10.1080/03602530801952864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson JK. Reviewers peering from under a pile of “omics” data. Nature. 2006;440:992. doi: 10.1038/440992d. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Lindon JC. Systems biology: metabonomics. Nature. 2008;455:1054–1056. doi: 10.1038/4551054a. [DOI] [PubMed] [Google Scholar]
- Nightingale P, Martin P. The myth of the biotech revolution. Trends Biotechnol. 2004;22:564–569. doi: 10.1016/j.tibtech.2004.09.010. [DOI] [PubMed] [Google Scholar]
- O'Farrell PH. The pre-omics era: the early days of two-dimensional gels. Proteomics. 2008 Nov 10; doi: 10.1002/pmic.200800719. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RP, Altman RB, Klein TE. PharmGKB and the International Warfarin Pharmacogenetics Consortium: the changing role for pharmacogenomic databases and single-drug pharmacogenetics. Hum Mutat. 2008;29:456–460. doi: 10.1002/humu.20731. [DOI] [PubMed] [Google Scholar]
- Ozdemir V. What is theragnostics and how will it change drug development, medical innovation and commercialization?. German Sepsis Society Annual Meeting—Symposium on “Theragnostics in Severe Infection.”; Weimar, Germany. September 3−7.2007. [Google Scholar]
- Ozdemir V, Godard B. Evidence-based management of nutrigenomics expectations and ELSIs. Pharmacogenomics. 2007;8:1051–1062. doi: 10.2217/14622416.8.8.1051. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Kalow W, Tothfalusi L, Bertilsson L, Endrenyi L, Graham JE. Multigenic control of drug response and regulatory decision-making in pharmaco genomics: the need for an upper-bound estimate of genetic contributions. Curr Pharmacogenomics Person Med. 2005;3:53–71. [Google Scholar]
- Ozdemir V, Motulsky AG, Kolker E, Godard B. Genome-environment interactions and prospective technology assessment: Evolution from pharmacogenomics to nutrigenomics and ecogenomics. OMICS: A Journal of Integrative Biology. 2009;13(1) doi: 10.1089/omi.2009.0013. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Williams-Jones B, Glatt SJ, Tsuang M, Lohr JB, Reist C. Shifting emphasis from pharmacogenomics to theragnostics. Nat Biotechnol. 2006;28:942–946. doi: 10.1038/nbt0806-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdemir V, Williams-Jones B, Graham JE, Preskorn SH, Gripeos D, Glatt SJ, et al. Asymmetry in scientific method and limits to cross-disciplinary dialogue: towards a shared language and science policy in pharmacogenomics and human disease genetics. J Invest Med. 2007;55:130–141. doi: 10.2310/6650.2007.06042. [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Dubé MP, Tardif JC, de Denus S, Phillips M, Stenne R, et al. A survey of the personalized medicine landscape. Pharmacogenomics. 2008;9:819–823. [Google Scholar]
- Pamuk O. Istanbul: Memories and the City (Paperback) Vintage Books, Random House; London: 2005. [Google Scholar]
- Pang T. Impact of pharmacogenomics on neglected diseases of the developing world. Am J Pharmacogenomics. 2003;3:393–398. doi: 10.2165/00129785-200303060-00006. [DOI] [PubMed] [Google Scholar]
- Perini J, Struchiner C, Silva-Assunção E, Santana I, Rangel F, Ojopi E, et al. Pharmacogenetics of warfarin: development of a dosing algorithm for Brazilian patients. Clin Pharmacol Ther. 2008;84:722–728. doi: 10.1038/clpt.2008.166. [DOI] [PubMed] [Google Scholar]
- Pirmohamed M. Pharmacogenetics and pharmacogenomics. Br J Clin Pharmacol. 2001;52:345–347. doi: 10.1046/j.0306-5251.2001.01498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlhaus JR, Cook-Deegan RM. Genomics research: world survey of public funding. BMC Genomics. 2008;9:472. doi: 10.1186/1471-2164-9-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM. Personalized vaccines: the emerging field of vaccinomics. Expert Opin Biol Ther. 2008;8:1659–1667. doi: 10.1517/14712598.8.11.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power S. Chasing the Flame: Sergio Vieira de Mello and the Fight to Save the World. The Penguin Press; New York: 2008. pp. 1–622. [Google Scholar]
- Prainsack B, Reardon J, Hindmarsh R, Gottweis H, Naue U, Lunshof JE. Personal genomes: Misdirected precaution. Nature. 2008;456(7218):34–35. doi: 10.1038/456034a. [DOI] [PubMed] [Google Scholar]
- Preece MA. The genetic contribution to stature. Horm Res. 1996;45:56–58. doi: 10.1159/000184849. [DOI] [PubMed] [Google Scholar]
- Rhodes R, Strain JJ. Whistleblowing in academic medicine. J Med Ethics. 2004;30:35–39. doi: 10.1136/jme.2003.005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries NM, Caulfield T. First pharmacogenomics, next nutrigenomics: Genohype or genohealthy? Jurimetrics J. 2006;46:281–308. [Google Scholar]
- Saltelli A, Funtowicz S. The precautionary principle: implications for risk management strategies. Int J Occup Med Environ Health. 2004;17:47–57. [PubMed] [Google Scholar]
- Select Biosciences The Inaugural AgriGenomics World Congress.; Amsterdam, The Netherlands. September 29−30.2008. [Google Scholar]
- Singh JA, Daar AS. The 20-year African biotech plan. Nat Biotechnol. 2008;26:272–274. doi: 10.1038/nbt0308-272. [DOI] [PubMed] [Google Scholar]
- Smart A, Martin P. The promise of pharmacogenetics: assessing the prospects for disease and patient stratification. Stud Hist Philos Biol Biomed Sci. 2006;37:583–601. doi: 10.1016/j.shpsc.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Smil V. Millennium essay. Genius loci. The twentieth century was made in Budapest. Nature. 2001;409:21. doi: 10.1038/35051162. [DOI] [PubMed] [Google Scholar]
- Snyder LH. Inherited taste deficiency. Science. 1931;74:151–152. doi: 10.1126/science.74.1910.151. [DOI] [PubMed] [Google Scholar]
- ST&HV, editor. Journals Under Threat: A Joint Response From History of Science, Technology & Medicine. Science, Technology & Human Values. 2009;34:6–8. [Google Scholar]
- Stevenson C, Cutcliffe J. Problematizing special observation in psychiatry: Foucault, archaeology, genealogy, discourse and power/knowledge. J Psychiatr Ment Health Nurs. 2006;13:713–721. doi: 10.1111/j.1365-2850.2006.01023.x. [DOI] [PubMed] [Google Scholar]
- Suarez-Kurtz G, Cascorbi I. São Paulo Research Conference on Molecular Medicine and Pharmacogenetisc, A Joint Meeting with the IUPHAR Sub-Committee on Pharmacogenetics and the Brazilian Pharmacogenetics Network, São Paulo, Brazi, September 18−20, 2008. Curr Pharmacogenomics Personal Med. 2008;6:234–238. [Google Scholar]
- Tepper BJ. Nutritional implications of genetic taste variation: the role of PROP sensitivity and other taste phenotypes. Annu Rev Nutr. 2008;28:14.1–14.22. doi: 10.1146/annurev.nutr.28.061807.155458. [DOI] [PubMed] [Google Scholar]
- Turner EH, Matthews AM, Linardatos E, Tell RA, Rosenthal R. Selective publication of antidepressant trials and its influence on apparent efficacy. N Engl J Med. 2008;358:252–260. doi: 10.1056/NEJMsa065779. [DOI] [PubMed] [Google Scholar]
- UNESCO (Paris) World Conference on Science. “Science for the Twenty-first Century: A New Commitment” (Declaration on Science and the Use of Scientific Knowledge). Sci Technol Soc. 2000;5:81–92. [Google Scholar]
- van Ommen B. Nutrigenomics: exploiting systems biology in the nutrition and health arenas. Nutrition. 2004;20:4–8. [PubMed] [Google Scholar]
- Vesell ES, Page JG. Genetic control of drug levels in man: phenylbutazone. Science. 1968;159:1479–1480. doi: 10.1126/science.159.3822.1479. [DOI] [PubMed] [Google Scholar]
- Vogel F. Moderne problem der humangenetik. Ergeb. Inn. Med. U. Kinderheilk. 1959;12:52–125. [Google Scholar]
- Weber WW, Smith RL. ASPET/AACC Conference. New directions in pharmacogenetics and ecogenetics. Genetic defenses against environmental impacts: responses to infections, foods, and environmental toxicants. Drug Met Disposition. 2001;29:467. [PubMed] [Google Scholar]
- Weedon MN, Frayling TM. Reaching new heights: insights into the genetics of human stature. Trends Genet. 2008;24:595–603. doi: 10.1016/j.tig.2008.09.006. [DOI] [PubMed] [Google Scholar]
- White WI. A new look at the role of urinalysis in the history of diagnostic medicine. Clin Chem. 1991;37:119–125. [PubMed] [Google Scholar]
- Wilke RA, Mareedu RK, Moore JH. The pathway less traveled: moving from candidate genes to candidate pathways in the analysis of genome-wide data from large scale pharmacogenetic association studies. Curr Pharmacogenomics Personal Med. 2008;6:150–159. doi: 10.2174/1875692110806030150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RJ. Biochemical individuality. John Wiley & Sons; New York: 1956. [Google Scholar]
- Winkler H. Verbreitung und Ursache der Parthenogenesis im Pflanzen-und Tierreiche. Verlag von Gustav Fischer; Jena: 1920. [Google Scholar]
- Wood AJJ. A proposal for radical changes in the drug-approval process. N Engl J Med. 2006;355:618–623. doi: 10.1056/NEJMsb055203. [DOI] [PubMed] [Google Scholar]
- Wooding S. Phenylthiocarbamide: a 75-year adventure in genetics and natural selection. Genetics. 2006;172:2015–2023. doi: 10.1093/genetics/172.4.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiura K, Kinoshita A, Ishida T, Ninokata A, Ishikawa T, Kaname T, et al. A SNP in the ABCC11 gene is the determinant of human earwax type. Nat Genet. 2006;38:324–330. doi: 10.1038/ng1733. [DOI] [PubMed] [Google Scholar]
- Zika E, Gurwitz D, Ibarreta D. Pharmacogenetics and Pharmacogenomics: State-of-the-art and Potential Socioeconomic Impacts in the EU. EUR 22214 EN. 2006 ISBN: 92−79−01901−5. [Google Scholar]