Abstract
The ubiquitin–proteasome pathway of protein degradation is one of the major mechanisms that are involved in the maintenance of the proper levels of cellular proteins. The regulation of proteasomal degradation thus ensures proper cell functions. The family of proteins containing ubiquitin-like (UbL) and ubiquitin-associated (UBA) domains has been implicated in proteasomal degradation. UbL–UBA domain containing proteins associate with substrates destined for degradation as well as with subunits of the proteasome, thus regulating the proper turnover of proteins.
Keywords: UbL, UBA, Proteasome, Degradation, Ubiquitin
Introduction
Proteasomal degradation is one mechanism by which the cell regulates protein levels. Many cellular proteins are degraded by the proteasome. The proteasome is a large multi-subunit protein complex that maintains cellular homeostasis by contributing to the turnover of short-lived proteins, as well as providing housekeeping functions, such as the degradation of misfolded proteins (reviewed in [1]). The proteasome degrades misfolded or damaged secretory and non-secretory proteins in an ubiquitin- and ATP-dependent manner in the cytosol. In yeast, the proteasome localizes to the nuclear envelope and the endoplasmic reticular (ER) network, while in higher eukaryotes, it is primarily nuclear- and cytoplasmically-localized.
The 26S proteasome holoenzyme consists of two major subunits, the 19S regulatory particle (RP), and the 20S core particle (CP) (reviewed in [1]). The 20S CP is the subunit responsible for proteolysis. It consists of four rings of seven subunits each, stacked on one another. The proteo-lytically active β-type subunits make up the inner two rings with the α-type subunits making up the outer two rings. The 19S RP, or cap complex, consists of a base and a lid, and there is one complete 19S subunit on each end of the CP. The 19S RP is also made up of multiple proteins that are classified as either ATPases (Rpt proteins in S. cerevisiae) or non-ATPases (Rpn proteins in S. cerevisiae) [2]. The cap complex is necessary for the degradation of ubiquitinated target proteins, and it is thought to be involved in the recognition and processing of these proteins before degradation [1]. It has been demonstrated that the proteins in the 19S RP are able to directly bind to ubiquitin and ubiquitinated substrates [3–5].
Protein degradation through the proteasomal pathway must be regulated to prevent indiscriminate degradation. Covalently attached ubiquitin typically marks a protein substrate for degradation by the proteasome. Ubiquitin is a highly conserved 76 amino acid polypeptide that is expressed in all eukaryotes. The covalent attachment of ubiquitin to other cellular proteins occurs through the highly regulated and specific process of ubiquitination (ubiquitylation), which is facilitated by multiple enzymes (reviewed in [6]). The E1 ubiquitin-activating enzyme activates ubiquitin, which is transferred to the E2 ubiquitin-conjugating enzyme. E2s loaded with ubiquitin then associate with E3 ubiquitin ligases, which facilitate the linkage between ubiquitin and the target protein or with another ubiquitin that is already attached to the target protein. Ubiquitin is covalently attached to substrates at lysine (K) residues. In polyubiquitin chains, subsequent ubiquitin moieties can be attached through a variety of linkages, with K29, K48, and K63 linkages being observed in vivo. Polyubiquitin chains consisting of four ubiquitin moieties attached through K48 linkages typically mark a protein for proteasomal degradation. Alterations of the ubiquitination pathway are believed to contribute to the pathogenesis of a range of human diseases, from neurodegenerative diseases to viral infections to cancer. Ubiquitination has been demonstrated to be involved in various processes, although the role of ubiquitin in marking proteins for proteasomal degradation is the best characterized.
Ubiquitin binding proteins
Proteasomal ubiquitin binding proteins
Specific subunits of the proteasome 19S RP have been demonstrated to be ubiquitin receptors, capable of recognizing both free ubiquitin and ubiquitin-conjugated proteins. The most studied proteasome ubiquitin receptor is the S5a/Rpn10/Pus1 (in higher eukaryotes, S. cerevisiae and S. pombe, respectively) subunit. S5a/Rpn10 has been demonstrated to bind to multiubiquitin chains of at least four ubiquitin moieties long [3] through an ubiquitin-interacting motif (UIM) [7]. Rpn10 can bind to both polyubiquitin chains and ubiquitin-conjugated proteins, but interestingly, proteasomes without Rpn10 can still bind them [4]. In addition, yeast Rpn10 mutants are viable and do not have a notable phenotype, and are able to degrade most proteins normally [8]. These observations suggest that other ubiquitin binding proteins may have a role in proteasomal degradation that can compensate for the loss of Rpn10.
Rpn13 is a more recently discovered 19S RP subunit that can also bind ubiquitin [5]. Ubiquitin is bound to Rpn13 through the conserved N-terminally-located Pru (pleckstrin-like receptor for ubiquitin) domain, and appears to have comparable ubiquitin chain binding activity as Rpn10. Rpn13 binds to K48-linked ubiquitin chains, binding to mono- and diubiquitin at 1:1 and tetraubiquitin at 1:2 stoichiometries. Interestingly, proteasomes in rpn10Δrpn13Δ yeast double mutants are unable to bind ubiquitin chains, suggesting that Rpn10 and Rpn13 may be the two major proteasome ubiquitin receptors [5].
Interestingly, extraproteasomal ubiquitin binding proteins have been demonstrated to interact with proteasomal subunits, such as Rpn1 that do not interact with ubiquiti-nated proteins [4, 9, 10]. These ubiquitin binding proteins could serve as adaptors, linking the ubiquitination and proteasomal degradation pathways. The presence of such adapters would be able to compensate for any loss of proteasomal ubiquitin receptors. Thus, ubiquitin binding subunits of the proteasome may not be an absolute requirement for the degradation of ubiquitinated substrates.
The ubiquitin-like (UbL) and ubiquitin-associated (UBA) family of ubiquitin binding proteins
Non-proteasomal ubiquitin binding domain proteins exist that may act to facilitate proteasomal degradation. One class is the UbL–UBA family of proteins (Fig. 1), with the Rad23, Dsk2, and Ddi1 proteins and homologues being most extensively studied. This family of proteins is involved in a variety of additional cell processes, such as nucleotide excision repair (NER), spindle pole body duplication, and cell growth. The properties and functions of these different family members, particularly involving proteasomal degradation, are summarized below.
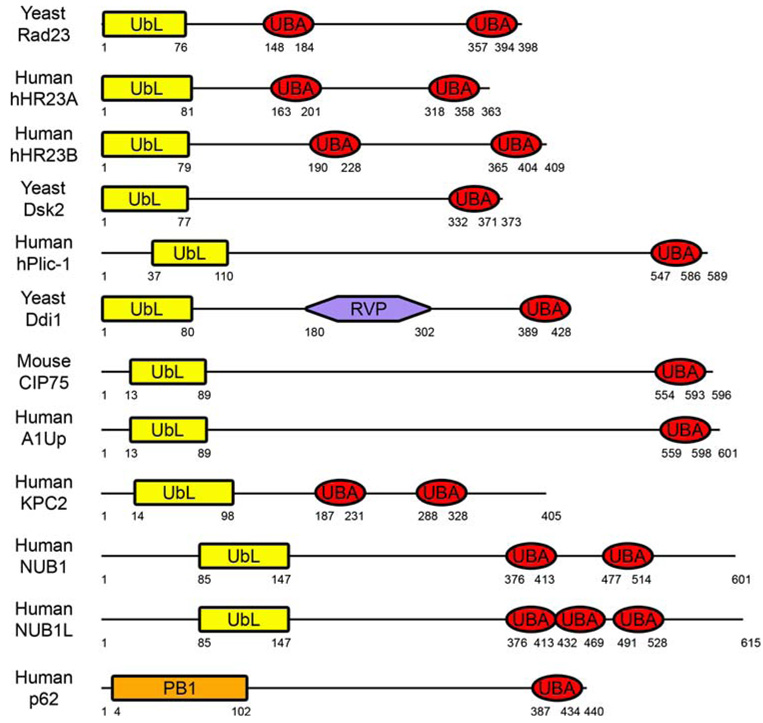
Fig. 1.
Schematic diagram of UbL–UBA domain-containing proteins. The domain structures of yeast Rad23 (accession number NP_010877), human hHR23A (NP_005044) and hHR23B (NP_002865), yeast Dsk2 (P48510), human hPlic-1 (Ubiquilin) (AAD49751), yeast Ddi1 (NP_011070), mouse CIP75 (UBIN) (NP_277068), human A1Up (NP_064516), human KPC2 (NP_057256), human NUB1 (NP_057202) and NUB1L (AAO14547), and human p62 (NP_003891) are shown. The UbL–UBA family of proteins contains an N-terminal UbL domain (or the structurally similar PB1 domain for p62), one or more C-terminal UBA domain(s), and a variable central region (yeast Ddi1 contains a central RVP domain). For family members with multiple UBA domains, the most N-terminal UBA domain is referred to as UBA1 in the text, and additional UBA domains are referred to as UBA2 and UBA3
Functional analyses of the UbL–UBA proteins
Rad23/Rhp23/HR23A/HR23B
Rad23 has been the most extensively studied member of the UbL–UBA family. It was initially identified as having a role in NER by dimerizing with Rad4, a DNA repair protein, and localizing in the nucleus [11, 12]. Rad23 was first identified in S. cerevisiae, followed by the discoveries of homologues in S. pombe (Rhp23) [13] and higher eukaryotes (HR23A and HR23B) [12, 14].
Rad23 UBA domains bind to multiubiquitin
Rad23/ Rhp23 binds tetraubiquitin via the UBA domains, but not monoubiquitin, as well as ubiquitin-conjugated cellular proteins. The more N-terminally-located UBA1 domain may have a larger role in the latter interaction since UBA1 deletion mutants show less binding to ubiquitinated proteins than UBA2 deletion mutants [15, 16]. The UBA domains of the human homologue hHR23A also bind to K48-linked ubiquitin chains and not monoubiquitin [17]. These results suggest that the UBA domain could be a multiubiquitin binding motif. Full-length hHR23A and the isolated UBA1 domain have a more than twofold higher affinity for high molecular weight (HMW) multiubiquitin than hHR23B, suggesting that these two proteins are not functionally redundant [18].
Rad23 UbL domain binds to the proteasome
Rad23/ Rhp23 exhibits a strong interaction with the 26S proteasome, which appears to be mediated by the Rad23/Rhp23 UbL domain and the Rpn10 subunit of the 19S RP [16, 19, 20]. The rad23ublΔ yeast mutant fails to rescue the Rad23 mutant DNA repair defect, indicating that the interaction with the proteasome is important for DNA repair and points to a connection between the DNA repair and ubiquitin–proteasome pathways [20]. The Rad23 UbL domain can also bind to another 19S proteasomal subunit, Rpn1, and is able to bind to proteasomes lacking Rpn10 [4], providing a possibility for why Rpn10 does not appear to be an essential gene.
Rad23 function—proteasomal degradation or protein stabilization
Overexpressing Rad23 results in a marked increase in ubiquitinated proteins, with an even higher level observed in the UbL deletion mutant [21]. Loss of Rad23 appears to have the same effect. Yeast strains that are mutant for both Rad23 and Rpn10 (rad23Δrpn10Δ) have increased levels of multiubiquitinated proteins. The rpn10Δ mutant exhibited minor proteolytic defects, which were intensified by the deletion of rad23 [22].
Overexpression of Rad23 also increases the amount of ubiquitinated protein associated with the proteasome but only in the presence of Rpn10, supporting the idea that Rad23 is binding to ubiquitinated substrates and regulating their delivery to Rpn10 in the proteasome [15]. This overexpression also prevents the expansion of multiubiquitin chains on model proteasomal substrates, resulting in protein stabilization [9, 15]. In S. pombe, Rhp23 protects multiubiquitinated conjugates against deubiquitination, preventing the deubiquitinating enzyme UBPY from disassembling tetraubiquitin chains, via the UBA domain. Rhp23 stabilizes ubiquitin conjugates, so accumulation of multiubiquitin conjugates may be caused by the inhibition of deubiquitination [23]. Stabilized ubiquitin conjugates resulting from inhibited deubiquitination would thus reduce degradation of these conjugates, perhaps because increased amounts of ubiquitinated proteins or ubiquitin chains would compete for proteasomal binding to reduce the degradation process. High levels of only the Rad23 UbL domain also stabilizes the model substrates, which suggests that unregulated UbL– proteasome interaction could interfere with the delivery of target substrates to the proteasome [15]. hHR23A also has an inhibitory effect on polyubiquitinated substrate degradation mediated by the UBA domains, with the UbL deletion mutant having a stronger inhibitory effect than the full-length protein. The UbL deletion mutant inhibited proteasomal degradation via an UBA-mediated sequestration of polyubiquitin chains [17].
For example, overexpressed Rad23 reduces Rad4 multiubiquitination, and stabilizes Rad4 levels [24]. Loss of Rad23 in conjunction with a Rpn10 mutant deleted for the UIM domain stabilizes the CDK inhibitor, Sic1. The SCFCdc4 substrate Far1 was also stabilized in rad23Δ mutants [25]. In higher eukaryotes, the hHR23A/B C-terminal UBA2 domain interaction with polyubiquitinated p53 shields it from deubiquitination, which blocks p53 degradation. The inhibition of deubiquitination would thus stabilize p53. Knock-down of hHR23A/B by siRNA results in increased p53 levels [26]. However, a conflicting report demonstrated that hHR23A/B siRNA has the opposite effect of accelerating p53 proteasomal degradation, while overexpression still results in p53 accumulation [27]. Both studies though, concluded that hHR23A/B inhibits p53 degradation.
Thus, Rad23 and its homologues appear to function in stabilizing proteins, which involves the blocking of proteasomal degradation, although Rad23 is required for the degradation of certain substrates such as Sic1 and Far1. Rad23 may have different roles in proteasomal degradation depending on the substrates.
Dsk2/Dph1/XDRP1/Plic-1(Ubiquilin)/Plic-2
Dsk2 was initially identified in S. cerevisiae as having a role in spindle pole body duplication and was classified as an ubiquitin-like protein since it was 36% identical to ubiquitin [28]. Dsk2 has homologues in S. pombe (Dph1) and higher eukaryotes (XDRP1 in Xenopus, and Plic or Ubiquilin in mammals).
Dsk2 UBA domain
Similar to Rad23, Dsk2/Dph1 binds to poly- and not monoubiquitin, specifically K48-linked tetraubiquitin (not K29- or K63-linked) via the UBA domain [16, 29]. Xenopus Dsk2-related protein isoforms, XDRP1L and XDRP1S, and human hPlic-1 also bind to polyubiquitin and polyubiquitinated proteins via the UBA domain [30–32].
Dsk2 UbL domain
The Dsk2 UbL domain interacts weakly with the proteasome, and binds with lower affinity than the interaction of Rad23 with the UIM domain of Rpn10 [4, 16, 18, 29, 32–34]. Interestingly, Dsk2 association with the proteasome was found to only occur in the absence of Rpn10. Extraproteasomal Rpn10 competes for Dsk2 binding with the proteasome [34], explaining the lower affinity that Dsk2 has for proteasomal interaction. In addition, the UbL domain is not absolutely required for human hPlic-2–proteasome interaction. Only simultaneous mutation of the UbL domain and deletion of the UBA domain abrogates binding to the proteasome [35].
Dsk2 function—protection from or promotion of degradation
Overexpression of Dsk2 results in the accumulation of large amounts of K48-linked ubiquitin chains in cells, with the UBA domain required for this effect [29, 34]. Similar to Rad23/Rph23, Dsk2/Dph1 protects multiubiquitinated conjugates against deubiquitination, preventing the disassembly of tetraubiquitin chains, and stabilizing ubiquitin conjugates. So again, accumulation of multiubiquitin conjugates may be caused by the inhibition of deubiquitination [23]. Degradation of a model proteasomal substrate is inhibited in Dsk2 mutants [9, 29], suggesting that Dsk2 participates in K48-linked ubiquitin proteasomal degradation. Dsk2 may be a polyubiquitin binding protein acting as an adaptor that facilitates delivery of polyubiquitin chains/proteins to the proteasome [29].
Xenopus
XDRP1L and XDRP1S UBA domains both bind to polyubiquitinated cyclins [36]. The XDPR1S UbL domain binds to monomeric cyclins, and can prevent degradation of cyclins A and B [32, 36]. This interaction is disrupted by cdc2-mediated phosphorylation of either the cyclins or the XDRP1S UbL domain, which allows cyclin degradation. Thus, XDRP1S may contribute to the regulation of cyclin degradation [36].
hPlic-1 and hPlic-2 are human homologues of yeast Dsk2. Overexpression of both results in increased p53 levels and the p53 half-life, and interferes with IκBα degradation. hPlic proteins could functionally link the ubiquitin machinery to the proteasome, where high levels of hPlic would interfere with a step between ubiquitination and proteasomal degradation [33]. Additionally, an UbL triple point mutant (which prevents the UbL domain from binding to the proteasome) can still stabilize p53 levels in the presence of the UBA domain, indicating that the UBA domain is required for interference of proteolysis [35]. Plic-1 has also been found to interact with neuronal GABAA receptors, through its UBA domain. Co-expression of Plic-1 with GABAA receptors results in stabilization of ubiquitinated, ER-localized GABAA receptors, which causes an increase in the number of receptors at the cell membrane [37, 38]. hPlic-1 also interacts with K7, a small membrane protein that acts as an antiapoptotic factor during KSHV (Kaposi’s sarcoma-associated herpesvirus) lytic replication. This interaction occurs via the hPlic-1 UBA domain. Binding to K7 decreases hPlic-1 binding to polyubiquitinated proteins, and inhibits the hPlic-1 function of facilitating p53 and IκBα proteasomal degradation. K7 may antagonize hPlic-1 to promote degradation of cellular antiviral proteins which would establish an environment that is favorable to viral replication [39].
The Plic proteins have also been observed to interact with proteins that are involved in neurodegenerative diseases, such as Huntington’s disease and Alzheimer’s disease. Huntington’s disease is associated with an abnormal expansion of polyglutamine tracts within the protein that potentially results in protein misfolding. Plic-1 interacts with expanded polyglutamine tracts that are ubiquitinated, with stronger interaction to longer tracts. Overexpression of Plic-1 results in increased turnover of the expanded polyglutamine protein and increased cell survival. Polyubiquitination of expanded polyglutamine proteins may favor interaction with Plic-1, which would result in increased turnover of these mutant proteins compared to normal proteins [40, 41].
Plic-1 and Plic-2 interact and colocalize with presenilins (PS), proteins involved in Alzheimer’s disease. This interaction occurs via the C-terminal region containing the UBA domain [42] and results in a reduction of PS2 polyubiquitination and the accumulation of PS2 [31]. PS2 ubiquitination is not an absolute requirement for Plic-1 interaction, although a reduced affinity was observed when certain lysine residues were mutated. The mutated proteins had higher turnover, indicating that Plic-1 interaction might have acted to stabilize PS2 [43]. Interestingly, inhibition of proteasomal degradation with PS2 expression results in the appearance of PS2 positive aggregates that are also positive for ubiquitin and colocalize with Plic-1, supporting Plic-1 binding to ubiquitinated proteins [31]. Drosophila Plic-1 has been found to interact with another Alzheimer’s related protein, the amyloid precursor protein (APP) via the UBA domain, which also results in the stabilization of APP [44].
Thus, Dsk2 and its homologues appear to have dual and opposite roles in protein stabilization: blocking and promoting proteasomal degradation. These roles could be substrate dependent.
Ddi1
Ddi1 (DNA damage inducible 1 ) in S. cerevisiae is another member of the UbL–UBA family, although it has distinct properties from Rad23 and Dsk2. Ddi1 can bind polyubiquitinated proteins via its UBA domain and the 19S proteasome subunit via the UbL domain, but with lower affinity than Rad23 and Dsk2 [45–47]. Ddi1 is enriched in the nucleus, which is dependent upon both its UbL and UBA domains [48].
Ddi1 in protein stabilization
Unlike Rad23 and Dsk2, Ddi1 mutants cannot stabilize a model proteasomal substrate [9]. However, in vivo loss of Ddi1 results in the stabilization of ubiquitinated Ho, an endonuclease, which accumulates in the cytoplasm. The UBA domain of Ddi1 interacts with ubiquitinated Ho. Ho can only interact with the proteasome (Rpn1) in the presence of Ddi1, and only when Ho is ubiquitinated. This suggests that the initial interaction must be between an ubiquitin chain and the UBA domain present in an UbL–UBA protein. Thus, a potential role for Ddi1 is to release Ho from an ubiquitination complex (i.e., SCFUfo1), making Ho available for deubiquitination and unfolding by the 19S regulatory complex [45, 46]. Ddi1 also has a role in the turnover of Ufo1, a F-box protein, by interacting with the Ufo1 UIM domains. Deletion of the UbL domain of Ddi1 abrogates Ufo1 binding, while loss of the UBA domain reduces interaction. Loss of Ddi1 stabilizes Ufo1 that is not observed with Rad23 or Dsk2 deletions, suggesting that Ddi1 has a role in Ufo1 degradation, and therefore, a role in the turnover of the SCFUfo1 complex [46, 49].
Thus, Ddi1 appears to function in facilitating proteasomal degradation.
CIP75/UBIN/A1Up
Mouse UBIN was discovered in a screen for proteins that interact with an ER-resident molecular chaperone, HSP47. UBIN is localized primarily in the cytosol, partially in vesicles, and none in the nucleus. It interacts with the HSP47 signal sequence, and also with the ER signal sequence of other proteins. This is a specific interaction because it does not interact with the mitochondrial targeting signal sequence. This interaction does not occur via either the UbL or UBA domains of UBIN [50]. Mouse CIP75 (connexin43-interacting protein of 75 kDa) was initially discovered in a yeast two hybrid screen using the cytoplasmically located, COOH-terminal region of the gap junction protein, connexin43 (Cx43), as bait. CIP75 also interacts with Cx43 via its UBA domain, and like other UbL–UBA proteins, interacts with 19S RP subunits via the UbL domain. The interaction between CIP75 and Cx43 appears to stimulate the proteasomal degradation of Cx43 [10]. Mouse CIP75 is 100% identical to the UBIN protein [10].
A1Up (ataxin-1 ubiquitin-like interacting protein) is the human homologue of mouse CIP75/UBIN, with 96% identity. A1Up interacts with the S5a/Rpn10 protein of the 19S RP via the UbL domain, an interaction which is disrupted by the binding of ataxin-1 to A1Up. The UBA domain binds to polyubiquitin chains and affects A1Up stability and localization. Overexpressed full-length A1Up and the UbL deletion mutant can stabilize ataxin-1 [51]. A1Up is distributed throughout the cell with large accumulations in both the nucleus and cytoplasm. Colocalization of A1Up with the 20S CP has been observed in both the nucleus and cytoplasm. The UbL deletion mutant is also present in both the nucleus and cytoplasm with predominance in the nucleus, but does not colocalize with the 20S CP [51, 52]. A working model suggests that the distribution of A1Up is dependent upon different conformational states of the protein [51].
In summary, the mouse protein CIP75/UBIN appears to facilitate proteasomal degradation, while the human homologue A1Up stabilizes proteins, suggesting that there may be different effects on proteasomal degradation based on interactions of these UbL–UBA proteins with different substrates. Discovering other substrates for these proteins may help to elucidate their function.
KPC2
KPC2 (Kip1 ubiquitylation-promoting complex) forms an ubiquitin ligase complex with KPC1, the catalytic RING domain-containing protein [53]. Excess amounts of KPC2 inhibit p27 ubiquitination. KPC2 interacts with multiple proteasomal subunits, and both UbL and UBA domains contribute to the interaction. The KPC2 UBA domain is also required for interaction with polyubiquitin. KPC2 stabilizes KPC1, recruits polyubiquitinated proteins, and interacts with the 26S proteasome, which leads to p27 degradation. The studies have led to the proposal that KPC2 functions to deliver ubiquitinated proteins to the proteasome [54]. Thus, KPC2 may normally function to facilitate protein degradation and the level of KPC2 determines whether KPC2 promotes or inhibits degradation.
NUB1/NUB1L
NUB1 (NEDD8 Ultimate Buster-1) and NUB1L (NUB1 long) are unique members of the UbL–UBA family that interact with NEDD8, instead of ubiquitin [55–57]. NUB1L is a splice variant of NUB1 that has a 14 amino acid insert, resulting in an additional C-terminal UBA domain (UBA2) [57, 58]. NUB1 expression is induced by interferon b in a time- and dose-dependent manner. Both NUB1 and NUB1L are localized primarily to the nucleus along with a weak cytoplasmic localization. Coexpression of NUB1/NUB1L and NEDD8 severely reduces the levels of free NEDD8 and NEDD8-conjugated proteins [55–57]. NUB1L appears to have more affinity for NEDD8 than NUB1, possibly due to the additional UBA domain [57].
NUB1/NUB1L can also interact with 19S RP proteasomal proteins, such as S5a, and they have been detected in 19S and 26S proteasomal cell fractions [55, 57, 59, 60]. Interestingly, the interaction between NUB1 and S5a does not appear to be mediated through the NUB1 UbL domain, but rather through a region at its C-terminus [60]. The relevance of this observation has not yet been determined. NEDD8 demonstrates an increased interaction with the proteasome when NUB1 is coexpressed, implicating NUB1 as an adaptor. NUB1 could serve as a direct link between the NEDD8 conjugation system and proteasomal degradation [55].
NUB1L also interacts with the ubiquitin-like modifier FAT10 via its UBA domains, leading to an acceleration of degradation. FAT10 has two UbL domains and causes rapid degradation when it is fused to the N-terminus of a protein. The N-terminal UbL domain of FAT10 interacts with NUB1L UBA domains. Interestingly, deletion of the UBA domains results in the loss of FAT10 binding; however, degradation is still accelerated. Upregulation of NUB1L expression, by interferon γ and TNFα, also results in accelerated FAT10 degradation. FAT10 can also bind the proteasome directly. The NUB1L UbL domain may bind to the 26S proteasome to induce a conformational change in the 19S RP subunit that favors binding and degradation of FAT10 and FAT10-conjugated proteins at a separate binding site [58, 59].
Thus, NUB1L facilitates the proteasomal degradation of NEDD8- and FAT10-conjugated proteins.
Structural analysis of the UbL–UBA domains: intermolecular and intramolecular interactions
The UbL–UBA domain proteins are hypothesized to have a role in proteasomal degradation because of their ability to interact with both components of the proteasome and proteins that are targeted for degradation by ubiquitination [10, 15, 16, 29, 33, 45–47, 54]. Extensive structural analysis primarily through in vitro NMR and surface plasmon resonance studies have demonstrated that the UbL domain is able to interact with the S5a/Rpn10 proteasomal subunit whereas the UBA domain binds ubiquitin, supporting the cellular observations. In addition, intramolecular interaction between the UbL and UBA domains of the same protein and intermolecular interaction between the domains of different family members have also been detected. These interactions could shed some light onto potential mechanisms for how these proteins are regulated.
UbL binding to proteasomal proteins
The UbL domain is able to bind to the UIM domain of the S5a/Rpn10 protein [61]. The hHR23A UbL domain interacts with the second UIM (UIM-2) of S5a in a 1:1 stoichiometry and the conserved hydrophobic residues of both the UbL and UIM-2 domains are required for this interaction [62]. The hHR23A UbL binding surface to S5a is conserved in hPlic-2 (which also binds S5a), where the conserved residues are geometrically similar and also have similar electrostatic potentials. This S5a binding surface is required for the hPlic-2 interaction with the proteasome [61, 63]. NMR studies have demonstrated that the human Rpn13 subunit of the 19S RP is also able to bind to the UbL domains of hHR23A and hPlic-2 [5].
UBA binding to ubiquitin
The structures of the UBA domains of Rad23 and Dsk2 have been solved. These UBA domains form three helix bundles that are of similar lengths [64–66]. These helices are stabilized by a hydrophobic core and have unusually large and conserved hydrophobic surface patches, which are often binding sites for other proteins [64]. Interestingly, while there is a low level of sequence conservation between the Rad23 UBA1 and UBA2 domains, both UBA domains have similar structures [64, 67]. The residues that are most conserved between the two UBA domains form the hydrophobic surface patches, suggesting that these patches could be a common binding interface for all UBA domains [64]. The UBA domains were demonstrated to bind to ubiquitin [65, 68, 69], and the UBA hydrophobic patch is the ubiquitin binding interface and is important for efficient interaction with polyubiquitin [65, 66, 69, 70]. In support of biochemical pulldown results, tetraubiquitin has a higher affinity for both single and double UBA domains than monoubiquitin [65, 69]. Interestingly, the UBA domains interact with the ubiquitin surface containing K48, and can block the assembly of K48-multiubiquitin chains [69, 70], providing a mechanism for the inhibition of ubiquitin chain elongation or blocking deubiquitination that has been seen for certain UbL–UBA proteins [9, 15, 23, 26].
A study was conducted to compare the interaction properties of different UBA domains with polyubiquitin, resulting in a classification of UBA-containing proteins. This in vitro study utilized NMR and surface plasmon resonance with isolated UBA domains and various polyubiquitin chains [71]. For the UbL–UBA family members, the Rad23-UBA2 domains were placed in Class 1 (selectively binds to K48-linked tetraubiquitin) [71, 72], while Rad23-UBA1 was placed in Class 2 (prefers K63-linked tetraubiquitin) [71]. Nub1L UBA domains were placed in Class 3 (no binding to any tested chains) [71]. The UBA domains of hPlic-1, Dsk2, and Ddi1 were placed in Class 4, binding to K48-, K63-, and K29/6-linked tetraubiquitin chains, as well as monoubiquitin [66, 71]. Thus, different UbL–UBA family members have varying affinities for the distinct types of ubiquitin chain linkages, which may contribute to different functions and interactions with assorted proteins.
UbL–UBA intramolecular interactions
Rad23 and Dsk2 UBA and UbL domains display intramolecular interaction, although the binding affinity is low [68, 69, 73]. Initial reports had not detected this intramolecular interaction in Dsk2 [65], most likely because of the low binding affinity. hHR23A UBA binding to ubiquitin results in structural changes to the rest of the hHR23A protein where residues within the UbL domain that reside in the UBA contact surface shift, suggesting that binding of ubiquitin to the UBA domain would prevent the intramolecular interaction with the UbL domain [70].
Intermolecular interactions
Earlier studies indicated that Rad23 can form homodimers through its UBA domains, with both UBA domains participating in dimerization [74]. Dsk2 was also observed to form homodimers through its UBA domain [65]. The Dsk2 human homologue hPlic-1 also displayed self-interaction. hPlic-1 mutants deleted for either the UbL or UBA domain retain the ability to interact with each other, indicating that the central region is required for the interaction. This is in contrast with the Dsk2 homologue; however, Dsk2 and hPlic-1 have a weak overall homology, with only 19% sequence identity [75], possibly explaining the variation in dimerization domains. Ddi1 can form homodimers even with deletion of the UbL or UBA domains, indicating that the dimerization is also mediated by the central region [74]. The X-ray crystal structure of Ddi1 confirmed this result, showing a Ddi1 dimer containing a fold similar to retroviral proteases, with the dimerization occurring in the central region of the protein [76]. This central region was named the RVP (central retroviral aspartyl-protease) domain (Fig. 1) [48]. A1Up self-interaction was observed in yeast that was also detected at a high molecular mass, which suggested the possibility of multimers. Interestingly, the UBA deletion mutant has an even higher molecular mass [51, 52], suggesting that multimerization occurs through the UbL domain and/or the central region of the protein. Thus, it appears that homodimerization occurs through either interaction between UBA domains or through the central domains located between the UbL and UBA domains.
Intermolecular interactions between different members of the UbL–UBA family have also been detected. Rad23 and Ddi1 are reported to form heterodimers via the UBA domains (UBA1 for Rad23) [74]. However, a later conflicting report demonstrated that Rad23 and Ddi1 heterodimers are formed through interaction between UbL and UBA domains, with no detectable UBA–UBA interaction [68]. The UBA domain of hPlic-2 can interact with the hHR23A UbL domain, with the reverse also being true (although only with hHR23A UBA2). This intermolecular binding prevents the hHR23A intramolecular UbL–UBA interaction. The same hHR23A UbL region that interacts with its own UBA domain and S5a also interacts with the hPlic-2 UBA domain. In addition, the UbL domain from both proteins uses the same surface to bind to the other UBA domain [77]. Thus, heterodimerization may occur through UbL–UBA interaction between the different proteins.
p62— a special relation to the UbL–UBA family
The p62 scaffolding protein (aka ZIP/sequestosome 1) was initially found as an interacting partner of atypical protein kinase C (aPKC) enzymes. It was demonstrated to act as a scaffolding protein, facilitating the interaction of aPKCs with substrates of the kinases, and regulating TRAF6 ubiquitination (reviewed in [78, 79]). p62 is not a member of the UbL–UBA family. However, it displays many properties that are similar to the UbL–UBA proteins. p62 is a member of the Phox/Bem1p family, which contain PB1 domains (Fig. 1). PB1 domains are protein–protein interaction domains found in atypical PKC isoenzymes, members of MAPK modules, and in several scaffold proteins. The PB1 domain mediates p62 oligomerization, and also mediates p62 interaction with other family members such as NBR1 (next to breast cancer 1) [80]. NMR studies of the PB1 domain indicate that it forms an ubiquitin-like, β-grasp fold, similar to the UbL domain [81], and thus could have similar binding or interaction properties as the UbL domain.
The p62 UBA domain binds to polyubiquitinated substrates, with a preference for the K63-linkage. p62 expression results in increased amounts of HMW polyubiquitin and small p62 aggregates colocalize with ubiquitin. The UBA deletion mutant blocks the survival-promoting effects of p62. p62 interacts with S5a and Rpt1 via the PB1 domain, similarly to the interactions mediated by UbL domains. Depletion of p62 results in inhibition of ubiquitin-mediated proteasomal degradation and accumulation of polyubiquitinated proteins. p62 has been proposed to function like the UbL–UBA shuttling factors in facilitating proteasomal degradation [82].
p62 also interacts with the K63-polyubiquitinated tau protein via its UBA domain. Overexpression of p62 results in enhanced tau turnover, while reduced p62 causes slower tau turnover. p62 is required for tau interaction with the proteasomal subunit Rpt1 and is necessary to shuttle tau to the proteasome for degradation [83]. p62 also functions as a shuttling protein in the interaction between K63-polyubiquitinated TrkA and Rpt1, with the p62 UBA domain binding to TrkA and the PB1 domain interacting with Rpt1. p62 is required for the interaction of Rpt1 and TrkA. Reduced p62 levels enhance TrkA stability, resulting in polyubiquitinated TrkA accumulation and contributing to an accumulation of K63-linked ubiquitinated proteins. p62 plays a role in the presentation of ubiquitinated TrkA to the proteasome for deubiquitination, regulating TrkA degradation [84, 85].
Thus, p62 appears to function in facilitating proteasomal degradation by acting as a shuttle or linker between K63-polyubiquitinated substrates and the proteasome in a manner similar to that proposed for some members of the UbL–UBA domain family.
Models
The role of the UbL–UBA family of proteins in proteasomal degradation is complex and has been an area of active debate, and is under continued revision. Based on current data, the UBA domain interaction with ubiquitin could serve to: (1) block deubiquitination (which could either facilitate degradation by maintaining existing polyubiquitin chains or stabilize ubiquitin-conjugates, thereby reducing degradation); (2) prevent polyubiquitination (which could reduce degradation by preventing the formation of the necessary polyubiquitin chain required by the proteasome for recognition as a degradation substrate); or (3) sequester ubiquitin (so the proteasome is blocked from recognizing the ubiquitin tag).
The UbL domain interactions could be a mechanism to regulate UbL–UBA protein function, and the domain has been demonstrated to interact with non-proteasomal proteins. For example, the Rad23/Dsk2 UbL domain binds to Pth2 (peptidyl-tRNA hydrolase 2). Overexpression of Pth2 results in the accumulation of polyubiquitinated proteins, preventing ubiquitin-mediated degradation, and leading to growth inhibition. It also inhibits Rad23/Dsk2 interaction with the proteasome by competing for binding and may be involved in the release of Rad23/Dsk2 from the proteasome. It could induce a structural change in the UbL domain or perhaps induce the “closed” conformation of Rad23/Dsk2 [86]. The Rad23 UbL domain can also bind to the yeast E4 ligase, Ufd2 [9]. This suggests a model where the Rad23–Ufd2 interaction facilitates Rad23 recognition of ubiquitinated substrates and enhances the stability of a Rad23-substrate complex, coupling the ubiquitination step to the substrate transport process. Ufd2 would then release Rad23, which would subsequently be able to bind to Rpn1, transporting the substrate to the proteasome [9]. These interactions may serve to regulate or facilitate UbL–UBA protein function.
Shuttle-factor hypothesis
The shuttle-factor hypothesis has been proposed for UbL–UBA protein function [15, 22]. In this model, the UbL–UBA protein binds to ubiquitinated proteins via the UBA domain, and then subsequently interacts with the proteasome via the UbL domain, which permits the transfer of degradation substrates to the proteasome [15, 22]. hHR23A has been shown to simultaneously bind to the 19S RP S5a UIM and ubiquitin [70], thus it is feasible that UbL–UBA proteins can act as shuttling factors. Interestingly, replacing the UbL domain with ubiquitin can act as a substitute for Rad23, but only in the presence of Rpn10, indicating the involvement of a different proteasome targeting mechanism. In the shuttle-factor hypothesis, it appears that there are two structural requirements: the ability to bind to ubiquitinated proteins, and the presence of a targeting signal to promote interaction with the proteasome [87].
Regulation of UbL–UBA protein function: effects of affinity, intramolecular and intermolecular interactions, and conformation changes
The hHR23A UbL domain is structurally similar to ubiquitin. Interestingly, the binding epitopes of ubiquitin and polyubiquitin are similar to the UbL binding epitope on the S5a UIM [62, 69]. All the major S5a UIM contact sites in the hHR23B UbL domain are conserved in ubiquitin, thus mutants that affect UIM–UbL binding also affect tetraubiquitin binding [88]. This suggests that ubiquitin and the UbL domain might compete for the S5a binding sites [62, 69]. However, the S5a UIM-2 domain has a stronger affinity for the hHR23B UbL domain than ubiquitin [69], and therefore the UbL–UBA domain proteins may have a function in enhancing ubiquitin-conjugated protein association with the proteasome.
The intramolecular and intermolecular associations between the UbL and UBA domains have the potential to be major regulating factors in controlling the function of the UbL–UBA proteins. For example, the UbL domain could be shielded by the UBA domain(s) in order to reduce conditions that would be unfavorable for S5a/ Rpn10 interactions until the appropriate conditions occur where ubiquitin binding to the UBA domain would open up the protein conformation, allowing binding to the proteasome [69]. It has also been shown that, when residues are mutated in the UbL domain that affects UBA interaction, polyubiquitin binding increases. This effect is similar to an UbL deletion mutant, suggesting that the UbL–UBA intramolecular interaction reduces the ability of the full-length protein to bind to ubiquitin via its UBA domain. When expressed in yeast, the Rad23 UbL triple point mutant is unable to interact with either the proteasome or the UBA domain, indicating that Rad23 binds to the proteasome through the same surface as it does intramolecularly to the UBA domain. This indicates that disrupting an intramolecular UbL–UBA interaction may be necessary to facilitate the Rad23–polyubiquitin interaction [89]. Similarly, multiple studies have demonstrated that deletion mutants missing one domain can sometimes have a stronger effect than the full-length protein, hinting at the inhibitory potential of the individual domains [15, 17, 21, 35, 51, 52]. A model was proposed where proteasomal or polyubiquitin binding to Rad23 disrupts the intramolecular interaction and results in the full-length protein adopting a more open conformation. This facilitates interaction with ubiquitinated proteins and/or the proteasome, which could result in Rad23 facilitating a more efficient docking of ubiquitinated substrates at the proteasome. Additionally, similar intermolecular UbL–UBA interactions could also have a regulatory role [89], as dimerization was proposed to play an important role in preventing unnecessary ubiquitin chain elongation or premature chain disassembly while substrates are in transit to the proteasome [68].
Interestingly, depletion of ubiquitinated proteins in E1 mutant yeast cells reduces Rad23 interaction with the proteasome. Addition of K48-linked tetraubiquitin can promote the specific interaction of Rad23 and the proteasome. This suggests that ubiquitin chain recognition might precede and activate Rad23 proteasomal targeting, perhaps by disrupting intramolecular or intermolecular interactions. Ubiquitin conjugates may promote association of ubiquitin binding substrate receptor proteins, such as the UbL–UBA proteins, with the proteasome [90].
Proposed model
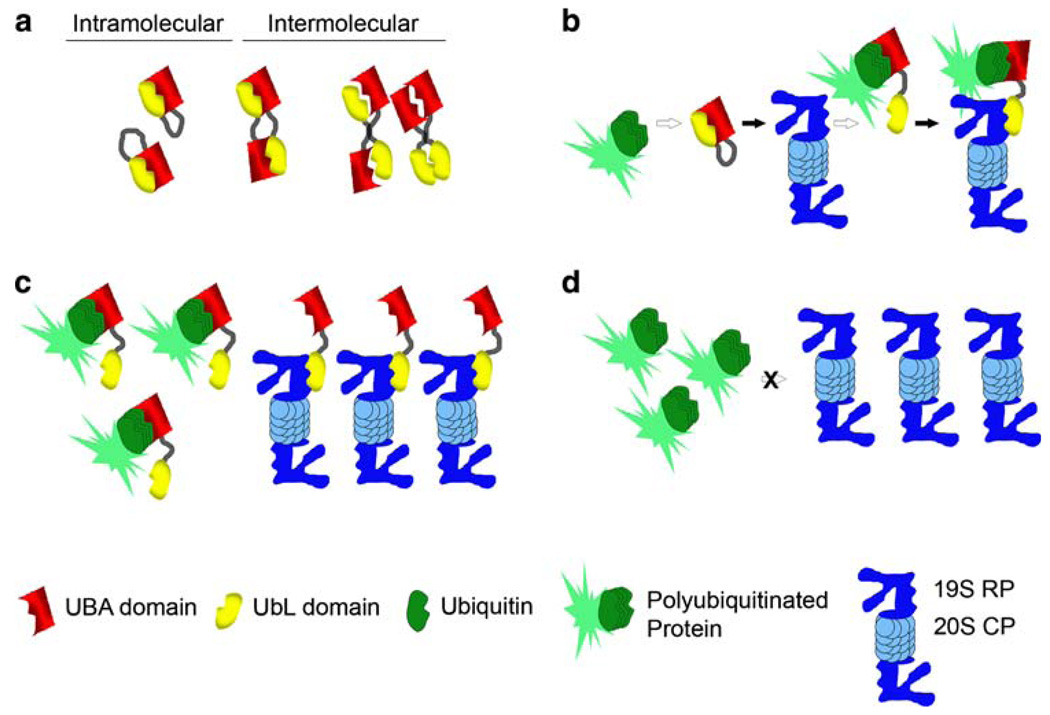
Based on the available data, we would like to propose a model for the mechanism of UbL–UBA function in proteasomal degradation (Fig. 2) that is similar to one proposed for hHR23A [89]. Under “resting” or uninduced conditions, the UbL–UBA protein resides in a conformation that is not amenable to binding to the proteasomal proteins or ubiquitinated substrates, due to UbL–UBA interactions (either intramolecular or intermolecular homoor heterodimers) (Fig. 2a). Upon induction of more favorable conditions (perhaps through an increase in the levels of ubiquitinated proteins), the UbL or UBA domains bind to the proteasome or the ubiquitinated target substrates, respectively, which induces a conformational change in the UbL–UBA protein, allowing the other domain to interact with its binding partner (Fig. 2b). The resultant multimeric complex containing the UbL–UBA shuttle protein, the ubiquitinated target substrate, and components of the proteasome, such as the S5a/Rpn10 protein, may lead to the proteasomal degradation of the target protein. We also propose that this effect could be concentration-dependent, where excessively high levels of UbL–UBA proteins could have a dominant negative effect (similar to expressing only one domain): either ubiquitinated substrates are bound by the UBA domain or proteasomal receptors are bound by the UbL domain of different proteins, depending upon the relative affinities (Fig. 2c). This would result in the sequestering of the substrate or proteasomal protein and the inability of the UbL–UBA protein to simultaneously bind both the target substrate and the proteasome. Conversely, loss of UbL– UBA proteins would represent the lack of the necessary shuttle factor to carry the target protein to the proteasome (Fig. 2d).
Fig. 2.
Model for the mechanism of UbL–UBA function in proteasomal degradation. a Intramolecular and intermolecular interactions between UbL and UBA domains. The UbL and UBA domains within the same protein can interact. They can also interact in either homoor heterodimeric fashion when present in separate proteins. b Under “resting”’ conditions, the UbL–UBA protein may reside in a conformation that is not amenable to binding to the proteasomal proteins or ubiquitinated substrates through UbL–UBA interactions (either intramolecular or intermolecular homo- or heterodimers). Upon induction of more favorable conditions (such as increased levels of ubiquitinated proteins), the UbL or UBA domains bind to the proteasome or the ubiquitinated target substrates, which causes a conformational change in the UbL–UBA protein and disruption of the intramolecular UbL–UBA interaction, for example. This allows the other domain to be able to bind as well, thereby creating a complex containing the UbL–UBA shuttle protein, the ubiquitinated target substrate, and the proteasome, which may facilitate the proteasomal degradation of the target protein. This sequence of events may occur for misfolded or damaged ER-localized proteins that are subject to ERAD. c Excessively high levels of the UbL–UBA proteins may exert a dominant negative effect (similar to expressing only one domain), where ubiquitinated substrates are bound by the UBA domain and proteasomal receptors are bound by the UbL domain of different proteins, depending on relative affinities. This would result in a sequestering effect that diminishes the ability of a single UbL– UBA protein to bind the target substrate and the proteasome simultaneously, as occurs in the model described in (b). d Conversely, the loss of UbL–UBA proteins would remove the necessary shuttle factor required to carry the target protein to the proteasome
ER-associated degradation: a role for the UbL–UBA proteins?
Proteasomal degradation functions to regulate proteins throughout the cell. In ER-associated degradation (ERAD), ER-localized proteins are translocated across the ER membrane into the cytoplasm and transported to the proteasome for degradation. This process requires the collaboration of multiple proteins and protein complexes. Proteins which are able to shuttle substrates from the ER after substrate translocation to the proteasome would be useful in facilitating efficient degradation. ERAD targets misfolded proteins in the ER as well as resident ER proteins for degradation. Upon ER stress induction, which can be induced by exposure to conditions that interfere with proper folding, misfolded proteins accumulate in the ER. Efficient ERAD enables the cell to purge itself of these damaged proteins.
Dsk2 and Rad23 appear to be involved in ERAD. rad23Δdsk2Δ yeast double mutants stabilize ER degradation substrates, which accumulate in the cytosol due to the inability to deliver the substrates to the proteasome. Additionally, a larger amount of ubiquitinated protein is detected in soluble cytosolic fractions. The degradation phenotype is specific to ER proteins as cytosolic substrates are not affected [91]. Rad23 also interacts with the de-N-glycosylating enzyme Png1p through its C-terminal region and is required for the interaction of Png1p with the proteasome, suggesting that Rad23 facilitates Png1p– proteasomal interaction [92, 93]. Png1p deglycosylates dislocated substrates in preparation for ERAD [94, 95]. Thus, an interaction between Rad23 and a protein known to be involved in ERAD is suggestive of a potential role for Rad23 in ERAD as well.
The Plic proteins have also been implicated in ERAD. Plic-1 interacts with PDI (protein-disulfide isomerase), a resident ER protein that is upregulated in ER stress, induced by hypoxia, for example. Interestingly, Plic-1 is also upregulated under hypoxic conditions [96]. Overex-pression of Plic-1 inhibits apoptosis induced by prolonged ER stress, specifically by delaying the induction of CHOP (a C/EBP homologue), which normally occurs from prolonged ER stress [96, 97]. The Plic proteins also interact with an ER-resident protein believed to have an important role in ERAD, Herp [98, 99]. The Plic proteins interact with the cytosolic region of Herp. Loss of Plic-1 and Plic-2 by siRNA knockdown results in the stabilization of a known ERAD substrate, and this effect was found specifically with ERAD substrates. Thus, the Plic proteins may also function as shuttle factors in ERAD, transporting substrates to the proteasome for degradation [98].
Additional evidence suggests that the CIP75/UBIN protein may also be involved in ERAD. CIP75/UBIN colocalizes with Cx43 to the region of the ER and facilitates the proteasomal degradation of Cx43, through the direct interaction with the proteasome [10]. Connexins have previously been shown to be degraded through ERAD [100–102]. Since Cx43 has not been shown to directly interact with the proteasome, an adaptor protein is thought to be necessary for Cx43 proteasomal degradation [102, 103], and CIP75/UBIN may fill this role.
Non-proteasomal degradation roles of the UbL–UBA proteins
The different members of the UbL–UBA family have also been reported to interact with proteins where it is unclear whether the UbL–UBA proteins are acting as agents in proteasomal degradation. The hHR23A UBA2 domain binds to Vpr, a protein that mediates various HIV functions during the viral life cycle. Overexpression of hHR23A alleviates the Vpr-mediated G2 cell cycle arrest, which requires the UBA2 domain [104]. hHR23A/B has also been reported to interact with ataxin-3 via the UbL domain [105]; however, the functional importance of this interaction is unclear.
hPlic-1 also binds to the UIM domains of proteins involved in endocytosis, including Eps15 and Hrs, through its UbL domain. Eps15 and Hrs recruit hPlic-1 to ubiquitinrich cytoplasmic aggregates [106]. Further study revealed that Plic-1 interaction with Eps15 is required for aggresome (cytoplasmic inclusion bodies) formation, and that Plic-1 contributes to transport of aggregates to the perinuclear aggresome, especially under conditions of stress that result in misfolded proteins [107]. These data suggest that Plic-1 may have an additional role in response to the presence of misfolded proteins that does not directly involve the proteasome. Interestingly, a role for the Plic proteins in autophagy has recently been reported [108]. Aggregates are autophagic substrates so perhaps the role of Plic proteins in aggresome formation is part of the autophagic pathway. Loss of Plic-1 or Plic-2 causes cells to be more susceptible to starvation. The protective effect of Plic-1/Plic-2 requires autophagy proteins, such as ATG5 and ATG7. In addition, Plic-2 colocalizes with the autophagosome marker LC3, and Plic-2 vesicles could be found close to lysosomal compartments, which is significant because autophagosomes eventually fuse to lysosomes. The UBA domain of Plic-2 is required for starvation protection and for association with LC3. The Plic interaction with the autophagosome appears to be required for autophagosome fusion with the lysosome, as loss of Plic proteins prevents autophagosome lysosomal degradation [108]. Plic-1 also interacts with polyubiquitinated TDP-43 (TAR DNA-binding domain protein) which localizes to aggregates in the neurodegenerative disease, amyotrophic lateral sclerosis (ALS). This interaction occurs via the UBA domain, and Plic-1 overexpression causes an increase in TDP-43 aggregates. These aggregates also contain the autophagosome LC3 marker, suggesting that Plic-1 is involved in sequestering TDP-43 in autophagosomes [109]. In addition, the role of p62 in autophagy has been extensively studied since the first observation that p62 interacted and colocalized with LC3 and that both proteins formed a “shell” around mutant Huntingtin aggregates, conferring a protective effect on cells [110]. Thus, UbL–UBA proteins could be involved in both proteasome degradation and autophagy.
Plic-2 has also been demonstrated to be involved in the negative regulation of G protein-coupled receptor (GPCR) endocytosis. Here, the Plic-2 UbL domain was required to prevent GPCR clustering into clathrin-coated pits. The mechanism of this negative regulation is unclear, although interaction with endocytic proteins such as Eps15 may play a role [111]. Plic-1 also interacts with mTOR, a protein kinase involved in cell cycle progression and cell growth, via its UBA domain. This interaction does not seem to have an effect on mTOR turnover and the function of this interaction is unknown [112].
Ddi1 expression can rescue the pds1ts-induced growth defect, which requires all the major domains (UbL, UBA, and RVP) [48, 113]. The mechanism of this rescue is unclear. Ddi1 also interacts with the v-snares Snc1 and Snc2, and the t-snare protein, Sso1, via its C-terminus; although the UBA domain is not required (interaction occurs with the region between the RVP and UBA domains). Ddi1 may help to regulate the exocytic machinery and cell cycle control [48], although it is unclear whether this would occur through ubiquitin-mediated degradation pathways. Ddi1 also interacts with Pho81p, part of the PHO pathway which regulates phosphate responsive gene expression. However, the ddi1Δ yeast mutant, or rad23Δddi1Δ double mutant, has similar levels of Pho81p as wild-type, indicating that they are not directly responsible for Pho81p degradation [114].
Conclusions
The UbL–UBA family of proteasome/ubiquitin binding proteins is a unique and diverse group of proteins. It is clear from the current body of work that this group of proteins cannot be placed in one simple category of function. There are reported differences in binding partners, in proteasomal function (promotion or inhibition of proteasomal degradation), and in interactions between the domains of the family members. While much work has already been done studying several of these proteins, especially Rad23 and Dsk2, and to a lesser degree Ddi1, elucidating a clearer role that the UbL–UBA family of proteins plays in proteasomal degradation will require more extensive study. In addition, it is becoming increasingly evident that facilitating proteasomal degradation is not the only role for these proteins. Thus, identifying additional interacting partners and studying the effects that the UbL–UBA proteins have on them will likely help to define these additional roles.
Acknowledgments
We would like to thank Dr. Mervyn Monteiro for helpful comments on the manuscript. Work by V.S. and A.F.L. is supported by grant CA052098 from the National Cancer Institute, National Institutes of Health.
Contributor Information
Vivian Su, Natural Products and Cancer Biology Program, Cancer Research Center, University of Hawaii at Manoa, 651 Ilalo Street, BSB 222, Honolulu, HI 96813, USA e-mail: vsu@crch.hawaii.edu.
Alan F. Lau, Natural Products and Cancer Biology Program, Cancer Research Center, University of Hawaii at Manoa, 651 Ilalo Street, BSB 222, Honolulu, HI 96813, USA e-mail: vsu@crch.hawaii.edu Department of Cell and Molecular Biology, John A. Burns School of Medicine, University of Hawaii at Manoa, 651 Ilalo Street, BSB 222L, Honolulu, HI 96813, USA.
References
- 1.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 2.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269:7059–7061. [PubMed] [Google Scholar]
- 4.Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Muller B, Feng MT, Tubing F, Dittmar GA, Finley D. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 5.Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, Hofmann K, Walters KJ, Finley D, Dikic I. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fang S, Weissman AM. A field guide to ubiquitylation. Cell Mol Life Sci. 2004;61:1546–1561. doi: 10.1007/s00018-004-4129-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M. Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 8.van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, Wefes I, Finley D, Vierstra RD. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16:6020–6028. doi: 10.1128/mcb.16.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15:3357–3365. doi: 10.1091/mbc.E03-11-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li X, Su V, Kurata WE, Jin C, Lau AF. A novel connexin43-interacting protein, CIP75, which belongs to the UbL-UBA protein family, regulates the turnover of connexin43. J Biol Chem. 2008;283:5748–5759. doi: 10.1074/jbc.M709288200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van der Spek PJ, Eker A, Rademakers S, Visser C, Sugasawa K, Masutani C, Hanaoka F, Bootsma D, Hoeijmakers JH. XPC and human homologs of RAD23: intracellular localization and relationship to other nucleotide excision repair complexes. Nucleic Acids Res. 1996;24:2551–2559. doi: 10.1093/nar/24.13.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watkins JF, Sung P, Prakash L, Prakash S. The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol. 1993;13:7757–7765. doi: 10.1128/mcb.13.12.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lombaerts M, Goeloe JI, den DulkH, Brandsma JA, Brouwer J. Identification and characterization of the rhp23(+) DNA repair gene in Schizosaccharomyces pombe. Biochem Biophys Res Commun. 2000;268:210–215. doi: 10.1006/bbrc.2000.2100. [DOI] [PubMed] [Google Scholar]
- 14.Masutani C, Sugasawa K, Yanagisawa J, Sonoyama T, Ui M, Enomoto T, Takio K, Tanaka K, van der Spek PJ, Bootsma D, Hoeijmakers JH, Hanaoka F. Purification and cloning of a nucleotide excision repair complex involving the xeroderma pigmentosum group C protein and a human homologue of yeast RAD23. EMBO J. 1994;13:1831–1843. doi: 10.1002/j.1460-2075.1994.tb06452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Madura K. Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol. 2002;22:4902–4913. doi: 10.1128/MCB.22.13.4902-4913.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilkinson CR, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 17.Raasi S, Pickart CM. Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem. 2003;278:8951–8959. doi: 10.1074/jbc.m212841200. [DOI] [PubMed] [Google Scholar]
- 18.Chen L, Madura K. Evidence for distinct functions for human DNA repair factors hHR23A and hHR23B. FEBS Lett. 2006;580:3401–3408. doi: 10.1016/j.febslet.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 19.Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, Hoeijmakers JH, Hanaoka F. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274:28019–28025. doi: 10.1074/jbc.274.39.28019. [DOI] [PubMed] [Google Scholar]
- 20.Schauber C, Chen L, Tongaonkar P, Vega I, Lambertson D, Potts W, Madura K. Rad23 links DNA repair to the ubiquitin/proteasome pathway. Nature. 1998;391:715–718. doi: 10.1038/35661. [DOI] [PubMed] [Google Scholar]
- 21.Hwang GW, Sasaki D, Naganuma A. Overexpression of Rad23 confers resistance to methylmercury in Saccharomyces cerevisiae via inhibition of the degradation of ubiquitinated proteins. Mol Pharmacol. 2005;68:1074–1078. doi: 10.1124/mol.105.013516. [DOI] [PubMed] [Google Scholar]
- 22.Lambertson D, Chen L, Madura K. Pleiotropic defects caused by loss of the proteasome-interacting factors Rad23 and Rpn10 of Saccharomyces cerevisiae. Genetics. 1999;153:69–79. doi: 10.1093/genetics/153.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartmann-Petersen R, Hendil KB, Gordon C. Ubiquitin binding proteins protect ubiquitin conjugates from disassembly. FEBS Lett. 2003;535:77–81. doi: 10.1016/s0014-5793(02)03874-7. [DOI] [PubMed] [Google Scholar]
- 24.Ortolan TG, Chen L, Tongaonkar P, Madura K. Rad23 stabilizes Rad4 from degradation by the Ub/proteasome pathway. Nucleic Acids Res. 2004;32:6490–6500. doi: 10.1093/nar/gkh987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Glockzin S, Ogi FX, Hengstermann A, Scheffner M, Blattner C. Involvement of the DNA repair protein hHR23 in p53 degradation. Mol Cell Biol. 2003;23:8960–8969. doi: 10.1128/MCB.23.24.8960-8969.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brignone C, Bradley KE, Kisselev AF, Grossman SR. A post-ubiquitination role for MDM2 and hHR23A in the p53 degradation pathway. Oncogene. 2004;23:4121–4129. doi: 10.1038/sj.onc.1207540. [DOI] [PubMed] [Google Scholar]
- 28.Biggins S, Ivanovska I, Rose MD. Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol. 1996;133:1331–1346. doi: 10.1083/jcb.133.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ko HS, Uehara T, Tsuruma K, Nomura Y. Ubiquilin interacts with ubiquitylated proteins and proteasome through its ubiquitin-associated and ubiquitin-like domains. FEBS Lett. 2004;566:110–114. doi: 10.1016/j.febslet.2004.04.031. [DOI] [PubMed] [Google Scholar]
- 31.Massey LK, Mah AL, Ford DL, Miller J, Liang J, Doong H, Monteiro MJ. Overexpression of ubiquilin decreases ubiquitination and degradation of presenilin proteins. J Alzhei-mers Dis. 2004;6:79–92. doi: 10.3233/jad-2004-6109. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka K, Funakoshi M, Inoue K, Kobayashi H. Identification of two isoforms of Dsk2-related protein XDRP1 in Xenopus eggs. Biochem Biophys Res Commun. 2006;350:768–773. doi: 10.1016/j.bbrc.2006.09.123. [DOI] [PubMed] [Google Scholar]
- 33.Kleijnen MF, Shih AH, Zhou P, Kumar S, Soccio RE, Kedersha NL, Gill G, Howley PM. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- 34.Matiuhin Y, Kirkpatrick DS, Ziv I, Kim W, Dakshinamurthy A, Kleifeld O, Gygi SP, Reis N, Glickman MH. Extrapro-teasomal Rpn10 restricts access of the polyubiquitin-binding protein Dsk2 to proteasome. Mol Cell. 2008;32:415–425. doi: 10.1016/j.molcel.2008.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleijnen MF, Alarcon RM, Howley PM. The ubiquitin-associated domain of hPLIC-2 interacts with the proteasome. Mol Biol Cell. 2003;14:3868–3875. doi: 10.1091/mbc.E02-11-0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka K, Funakoshi M, Kobayashi H. A Cdc2-sensitive interaction of the UbL domain of XDRP1S with cyclin B mediates the degradation of cyclin B in Xenopus egg extracts. Biochem Biophys Res Commun. 2006;350:774–782. doi: 10.1016/j.bbrc.2006.09.122. [DOI] [PubMed] [Google Scholar]
- 37.Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat Neurosci. 2001;4:908–916. doi: 10.1038/nn0901-908. [DOI] [PubMed] [Google Scholar]
- 38.Saliba RS, Pangalos M, Moss SJ. The ubiquitin-like protein Plic-1 enhances the membrane insertion of GABAA receptors by increasing their stability within the endoplasmic reticulum. J Biol Chem. 2008;283:18538–18544. doi: 10.1074/jbc.M802077200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng P, Scott CW, Cho NH, Nakamura H, Chung YH, Monteiro MJ, Jung JU. Kaposi’s sarcoma-associated herpesvirus K7 protein targets a ubiquitin-like/ubiquitin-associated domaincontaining protein to promote protein degradation. Mol Cell Biol. 2004;24:3938–3948. doi: 10.1128/MCB.24.9.3938-3948.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington’s disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- 41.Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mah AL, Perry G, Smith MA, Monteiro MJ. Identification of ubiquilin, a novel presenilin interactor that increases presenilin protein accumulation. J Cell Biol. 2000;151:847–862. doi: 10.1083/jcb.151.4.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ford DL, Monteiro MJ. Studies of the role of ubiquitination in the interaction of ubiquilin with the loop and carboxyl terminal regions of presenilin-2. Biochemistry. 2007;46:8827–8837. doi: 10.1021/bi700604q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gross GG, Feldman RM, Ganguly A, Wang J, Yu H, Guo M. Role of X11 and ubiquilin as in vivo regulators of the amyloid precursor protein in Drosophila. PLoS ONE. 2008;3:e2495. doi: 10.1371/journal.pone.0002495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaplun L, Tzirkin R, Bakhrat A, Shabek N, Ivantsiv Y, Raveh D. The DNA damage-inducible UbL-UbA protein Ddi1 participates in Mec1-mediated degradation of Ho endonuclease. Mol Cell Biol. 2005;25:5355–5362. doi: 10.1128/MCB.25.13.5355-5362.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplun L, Ivantsiv Y, Bakhrat A, Tzirkin R, Baranes K, Shabek N, Raveh D. The F-box protein, Ufo1, maintains genome stability by recruiting the yeast mating switch endonuclease, Ho, for rapid proteasome degradation. Isr Med Assoc J. 2006;8:246–248. [PubMed] [Google Scholar]
- 47.Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitinlike proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293:986–992. doi: 10.1016/S0006-291X(02)00340-6. [DOI] [PubMed] [Google Scholar]
- 48.Gabriely G, Kama R, Gelin-Licht R, Gerst JE. Different domains of the UBL-UBA ubiquitin receptor, Ddi1/Vsm1, are involved in its multiple cellular roles. Mol Biol Cell. 2008;19:3625–3637. doi: 10.1091/mbc.E07-05-0462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ivantsiv Y, Kaplun L, Tzirkin-Goldin R, Shabek N, Raveh D. Unique role for the UbL-UbA protein Ddi1 in turnover of SCFUfo1 complexes. Mol Cell Biol. 2006;26:1579–1588. doi: 10.1128/MCB.26.5.1579-1588.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuda M, Koide T, Yorihuzi T, Hosokawa N, Nagata K. Molecular cloning of a novel ubiquitin-like protein, UBIN, that binds to ER targeting signal sequences. Biochem Biophys Res Commun. 2001;280:535–540. doi: 10.1006/bbrc.2000.4149. [DOI] [PubMed] [Google Scholar]
- 51.Riley BE, Xu Y, Zoghbi HY, Orr HT. The effects of the polyglutamine repeat protein ataxin-1 on the UbL-UBA protein A1Up. J Biol Chem. 2004;279:42290–42301. doi: 10.1074/jbc.M406284200. [DOI] [PubMed] [Google Scholar]
- 52.Davidson JD, Riley B, Burright EN, Duvick LA, Zoghbi HY, Orr HT. Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum Mol Genet. 2000;9:2305–2312. doi: 10.1093/oxfordjournals.hmg.a018922. [DOI] [PubMed] [Google Scholar]
- 53.Kamura T, Hara T, Matsumoto M, Ishida N, Okumura F, Hatakeyama S, Yoshida M, Nakayama K, Nakayama KI. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol. 2004;6:1229–1235. doi: 10.1038/ncb1194. [DOI] [PubMed] [Google Scholar]
- 54.Hara T, Kamura T, Kotoshiba S, Takahashi H, Fujiwara K, Onoyama I, Shirakawa M, Mizushima N, Nakayama KI. Role of the UBL-UBA protein KPC2 in degradation of p27 at G1 phase of the cell cycle. Mol Cell Biol. 2005;25:9292–9303. doi: 10.1128/MCB.25.21.9292-9303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamitani T, Kito K, Fukuda-Kamitani T, Yeh ET. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J Biol Chem. 2001;276:46655–46660. doi: 10.1074/jbc.M108636200. [DOI] [PubMed] [Google Scholar]
- 56.Kito K, Yeh ET, Kamitani T. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J Biol Chem. 2001;276:20603–20609. doi: 10.1074/jbc.M100920200. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka T, Kawashima H, Yeh ET, Kamitani T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J Biol Chem. 2003;278:32905–32913. doi: 10.1074/jbc.M212057200. [DOI] [PubMed] [Google Scholar]
- 58.Hipp MS, Raasi S, Groettrup M, Schmidtke G. NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J Biol Chem. 2004;279:16503–16510. doi: 10.1074/jbc.M310114200. [DOI] [PubMed] [Google Scholar]
- 59.Schmidtke G, Kalveram B, Weber E, Bochtler P, Lukasiak S, Hipp MS, Groettrup M. The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. J Biol Chem. 2006;281:20045–20054. doi: 10.1074/jbc.M603063200. [DOI] [PubMed] [Google Scholar]
- 60.Tanji K, Tanaka T, Kamitani T. Interaction of NUB1 with the proteasome subunit S5a. Biochem Biophys Res Commun. 2005;337:116–120. doi: 10.1016/j.bbrc.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 61.Walters KJ, Kleijnen MF, Goh AM, Wagner G, Howley PM. Structural studies of the interaction between ubiquitin family proteins and proteasome subunit S5a. Biochemistry. 2002;41:1767–1777. doi: 10.1021/bi011892y. [DOI] [PubMed] [Google Scholar]
- 62.Mueller TD, Feigon J. Structural determinants for the binding of ubiquitin-like domains to the proteasome. EMBO J. 2003;22:4634–4645. doi: 10.1093/emboj/cdg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walters KJ, Goh AM, Wang Q, Wagner G, Howley PM. Ubiquitin family proteins and their relationship to the proteasome: a structural perspective. Biochim Biophys Acta. 2004;1695:73–87. doi: 10.1016/j.bbamcr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 64.Mueller TD, Feigon J. Solution structures of UBA domains reveal a conserved hydrophobic surface for protein-protein interactions. J Mol Biol. 2002;319:1243–1255. doi: 10.1016/S0022-2836(02)00302-9. [DOI] [PubMed] [Google Scholar]
- 65.Sasaki T, Funakoshi M, Endicott JA, Kobayashi H. Budding yeast Dsk2 protein forms a homodimer via its C-terminal UBA domain. Biochem Biophys Res Commun. 2005;336:530–535. doi: 10.1016/j.bbrc.2005.08.126. [DOI] [PubMed] [Google Scholar]
- 66.Zhang D, Raasi S, Fushman D. Affinity makes the difference: nonselective interaction of the UBA domain of Ubiquilin-1 with monomeric ubiquitin and polyubiquitin chains. J Mol Biol. 2008;377:162–180. doi: 10.1016/j.jmb.2007.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mueller TD, Kamionka M, Feigon J. Specificity of the interaction between ubiquitin-associated domains and ubiquitin. J Biol Chem. 2004;279:11926–11936. doi: 10.1074/jbc.M312865200. [DOI] [PubMed] [Google Scholar]
- 68.Kang Y, Vossler RA, Diaz-Martinez LA, Winter NS, Clarke DJ, Walters KJ. UBL/UBA ubiquitin receptor proteins bind a common tetraubiquitin chain. J Mol Biol. 2006;356:1027–1035. doi: 10.1016/j.jmb.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Ryu KS, Lee KJ, Bae SH, Kim BK, Kim KA, Choi BS. Binding surface mapping of intra- and interdomain interactions among hHR23B, ubiquitin, and polyubiquitin binding site 2 of S5a. J Biol Chem. 2003;278:36621–36627. doi: 10.1074/jbc.M304628200. [DOI] [PubMed] [Google Scholar]
- 70.Wang Q, Goh AM, Howley PM, Walters KJ. Ubiquitin recognition by the DNA repair protein hHR23a. Biochemistry. 2003;42:13529–13535. doi: 10.1021/bi035391j. [DOI] [PubMed] [Google Scholar]
- 71.Raasi S, Varadan R, Fushman D, Pickart CM. Diverse polyubiquitin interaction properties of ubiquitin-associated domains. Nat Struct Mol Biol. 2005;12:708–714. doi: 10.1038/nsmb962. [DOI] [PubMed] [Google Scholar]
- 72.Raasi S, Orlov I, Fleming KG, Pickart CM. Binding of polyubiquitin chains to ubiquitin-associated (UBA) domains of HHR23A. J Mol Biol. 2004;341:1367–1379. doi: 10.1016/j.jmb.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 73.Lowe ED, Hasan N, Trempe JF, Fonso L, Noble ME, Endicott JA, Johnson LN, Brown NR. Structures of the Dsk2 UBL and UBA domains and their complex. Acta Crystallogr D Biol Crystallogr. 2006;62:177–188. doi: 10.1107/S0907444905037777. [DOI] [PubMed] [Google Scholar]
- 74.Bertolaet BL, Clarke DJ, Wolff M, Watson MH, Henze M, Divita G, Reed SI. UBA domains mediate protein-protein interactions between two DNA damage-inducible proteins. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 75.Ford DL, Monteiro MJ. Dimerization of ubiquilin is dependent upon the central region of the protein: evidence that the monomer, but not the dimer, is involved in binding presenilins. Biochem J. 2006;399:397–404. doi: 10.1042/BJ20060441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sirkis R, Gerst JE, Fass D. Ddi1, a eukaryotic protein with the retroviral protease fold. J Mol Biol. 2006;364:376–387. doi: 10.1016/j.jmb.2006.08.086. [DOI] [PubMed] [Google Scholar]
- 77.Kang Y, Zhang N, Koepp DM, Walters KJ. Ubiquitin receptor proteins hHR23a and hPLIC2 interact. J Mol Biol. 2007;365:1093–1101. doi: 10.1016/j.jmb.2006.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Moscat J, Diaz-Meco MT, Wooten MW. Signal integration and diversification through the p62 scaffold protein. Trends Biochem Sci. 2007;32:95–100. doi: 10.1016/j.tibs.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 79.Seibenhener ML, Geetha T, Wooten MW. Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 2007;581:175–179. doi: 10.1016/j.febslet.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lamark T, Perander M, Outzen H, Kristiansen K, Overvatn A, Michaelsen E, Bjorkoy G, Johansen T. Interaction codes within the family of mammalian Phox and Bem1p domain-containing proteins. J Biol Chem. 2003;278:34568–34581. doi: 10.1074/jbc.M303221200. [DOI] [PubMed] [Google Scholar]
- 81.Hirano Y, Yoshinaga S, Ogura K, Yokochi M, Noda Y, Sumimoto H, Inagaki F. Solution structure of atypical protein kinase C PB1 domain and its mode of interaction with ZIP/p62 and MEK5. J Biol Chem. 2004;279:31883–31890. doi: 10.1074/jbc.M403092200. [DOI] [PubMed] [Google Scholar]
- 82.Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW. Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol. 2004;24:8055–8068. doi: 10.1128/MCB.24.18.8055-8068.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Babu JR, Geetha T, Wooten MW. Sequestosome 1/p62 shuttles polyubiquitinated tau for proteasomal degradation. J Neurochem. 2005;94:192–203. doi: 10.1111/j.1471-4159.2005.03181.x. [DOI] [PubMed] [Google Scholar]
- 84.Geetha T, Seibenhener ML, Chen L, Madura K, Wooten MW. p62 serves as a shuttling factor for TrkA interaction with the proteasome. Biochem Biophys Res Commun. 2008;374:33–37. doi: 10.1016/j.bbrc.2008.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wooten MW, Geetha T, Babu JR, Seibenhener ML, Peng J, Cox N, Diaz-Meco MT, Moscat J. Essential role of sequestosome 1/p62 in regulating accumulation of Lys63-ubiquitinated proteins. J Biol Chem. 2008;283:6783–6789. doi: 10.1074/jbc.M709496200. [DOI] [PubMed] [Google Scholar]
- 86.Ishii T, Funakoshi M, Kobayashi H. Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin-proteasome pathway. EMBO J. 2006;25:5492–5503. doi: 10.1038/sj.emboj.7601418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lambertson D, Chen L, Madura K. Investigating the importance of proteasome-interaction for Rad23 function. Curr Genet. 2003;42:199–208. doi: 10.1007/s00294-002-0350-7. [DOI] [PubMed] [Google Scholar]
- 88.Fujiwara K, Tenno T, Sugasawa K, Jee JG, Ohki I, Kojima C, Tochio H, Hiroaki H, Hanaoka F, Shirakawa M. Structure of the ubiquitin-interacting motif of S5a bound to the ubiquitinlike domain of HR23B. J Biol Chem. 2004;279:4760–4767. doi: 10.1074/jbc.M309448200. [DOI] [PubMed] [Google Scholar]
- 89.Goh AM, Walters KJ, Elsasser S, Verma R, Deshaies RJ, Finley D, Howley PM. Components of the ubiquitin-proteasome pathway compete for surfaces on Rad23 family proteins. BMC Biochem. 2008;9:4. doi: 10.1186/1471-2091-9-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ghaboosi N, Deshaies RJ. A conditional yeast E1 mutant blocks the ubiquitin-proteasome pathway and reveals a role for ubiquitin conjugates in targeting Rad23 to the proteasome. Mol Biol Cell. 2007;18:1953–1963. doi: 10.1091/mbc.E06-10-0965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Medicherla B, Kostova Z, Schaefer A, Wolf DH. A genomic screen identifies Dsk2p and Rad23p as essential components of ER-associated degradation. EMBO Rep. 2004;5:692–697. doi: 10.1038/sj.embor.7400164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Park H, Suzuki T, Lennarz WJ. Identification of proteins that interact with mammalian peptide:N-glycanase and implicate this hydrolase in the proteasome-dependent pathway for protein degradation. Proc Natl Acad Sci USA. 2001;98:11163–11168. doi: 10.1073/pnas.201393498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki T, Park H, Kwofie MA, Lennarz WJ. Rad23 provides a link between the Png1 deglycosylating enzyme and the 26 S proteasome in yeast. J Biol Chem. 2001;276:21601–21607. doi: 10.1074/jbc.M100826200. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki T, Park H, Hollingsworth NM, Sternglanz R, Lennarz WJ. PNG1, a yeast gene encoding a highly conserved peptide:N-glycanase. J Cell Biol. 2000;149:1039–1052. doi: 10.1083/jcb.149.5.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Suzuki T, Park H, Kitajima K, Lennarz WJ. Peptides glycosylated in the endoplasmic reticulum of yeast are subsequently deglycosylated by a soluble peptide: N-glycanase activity. J Biol Chem. 1998;273:21526–21530. doi: 10.1074/jbc.273.34.21526. [DOI] [PubMed] [Google Scholar]
- 96.Ko HS, Uehara T, Nomura Y. Role of ubiquilin associated with protein-disulfide isomerase in the endoplasmic reticulum in stress-induced apoptotic cell death. J Biol Chem. 2002;277:35386–35392. doi: 10.1074/jbc.M203412200. [DOI] [PubMed] [Google Scholar]
- 97.Lu A, Hiltunen M, Romano DM, Soininen H, Hyman BT, Bertram L, Tanzi RE. Effects of ubiquilin 1 on the unfolded protein response. J Mol Neurosci. 2009;38:19–30. doi: 10.1007/s12031-008-9155-6. [DOI] [PubMed] [Google Scholar]
- 98.Kim TY, Kim E, Yoon SK, Yoon JB. Herp enhances ER-associated protein degradation by recruiting ubiquilins. Biochem Biophys Res Commun. 2008;369:741–746. doi: 10.1016/j.bbrc.2008.02.086. [DOI] [PubMed] [Google Scholar]
- 99.Kokame K, Agarwala KL, Kato H, Miyata T. Herp, a new ubiquitin-like membrane protein induced by endoplasmic reticulum stress. J Biol Chem. 2000;275:32846–32853. doi: 10.1074/jbc.M002063200. [DOI] [PubMed] [Google Scholar]
- 100.Kelly SM, Vanslyke JK, Musil LS. Regulation of ubiquitin-proteasome system mediated degradation by cytosolic stress. Mol Biol Cell. 2007;18:4279–4291. doi: 10.1091/mbc.E07-05-0487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.VanSlyke JK, Deschenes SM, Musil LS. Intracellular transport, assembly, and degradation of wild-type and diseaselinked mutant gap junction proteins. Mol Biol Cell. 2000;11:1933–1946. doi: 10.1091/mbc.11.6.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.VanSlyke JK, Musil LS. Dislocation and degradation from the ER are regulated by cytosolic stress. J Cell Biol. 2002;157:381–394. doi: 10.1083/jcb.200111045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Musil LS, Le AC, VanSlyke JK, Roberts LM. Regulation of connexin degradation as a mechanism to increase gap junction assembly and function. J Biol Chem. 2000;275:25207–25215. doi: 10.1074/jbc.275.33.25207. [DOI] [PubMed] [Google Scholar]
- 104.Dieckmann T, Withers-Ward ES, Jarosinski MA, Liu CF, Chen IS, Feigon J. Structure of a human DNA repair protein UBA domain that interacts with HIV-1 Vpr. Nat Struct Biol. 1998;5:1042–1047. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 105.Wang G, Sawai N, Kotliarova S, Kanazawa I, Nukina N. Ataxin-3, the MJD1 gene product, interacts with the two human homologs of yeast DNA repair protein RAD23, HHR23A and HHR23B. Hum Mol Genet. 2000;9:1795–1803. doi: 10.1093/hmg/9.12.1795. [DOI] [PubMed] [Google Scholar]
- 106.Regan-Klapisz E, Sorokina I, Voortman J, de Keizer P, Roovers RC, Verheesen P, Urbe S, Fallon L, Fon EA, Verkleij A, Benmerah A, van Bergen en Henegouwen PM. Ubiquilin recruits Eps15 into ubiquitin-rich cytoplasmic aggregates via a UIM-UBL interaction. J Cell Sci. 2005;118:4437–4450. doi: 10.1242/jcs.02571. [DOI] [PubMed] [Google Scholar]
- 107.Heir R, Ablasou C, Dumontier E, Elliott M, Fagotto-Kaufmann C, Bedford FK. The UBL domain of PLIC-1 regulates aggresome formation. EMBO Rep. 2006;7:1252–1258. doi: 10.1038/sj.embor.7400823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.N’Diaye EN, Kajihara KK, Hsieh I, Morisaki H, Debnath J, Brown EJ. PLIC proteins or ubiquilins regulate autophagy-dependent cell survival during nutrient starvation. EMBO Rep. 2009;10:173–179. doi: 10.1038/embor.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim SH, Shi Y, Hanson KA, Williams LM, Sakasai R, Bowler MJ, Tibbetts RS. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, Stenmark H, Johansen T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005;171:603–614. doi: 10.1083/jcb.200507002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.N’Diaye EN, Hanyaloglu AC, Kajihara KK, Puthenveedu MA, Wu P, von Zastrow M, Brown EJ. The ubiquitin-like protein PLIC-2 is a negative regulator of G protein-coupled receptor endocytosis. Mol Biol Cell. 2008;19:1252–1260. doi: 10.1091/mbc.E07-08-0775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wu S, Mikhailov A, Kallo-Hosein H, Hara K, Yonezawa K, Avruch J. Characterization of ubiquilin 1, an mTOR-interacting protein. Biochim Biophys Acta. 2002;1542:41–56. doi: 10.1016/s0167-4889(01)00164-1. [DOI] [PubMed] [Google Scholar]
- 113.Diaz-Martinez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1:28. doi: 10.1186/1747-1028-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Auesukaree C, Fuchigami I, Homma T, Kaneko Y, Harashima S. Ddi1p and Rad23p play a cooperative role as negative regulators in the PHO pathway in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2008;365:821–825. doi: 10.1016/j.bbrc.2007.11.044. [DOI] [PubMed] [Google Scholar]