Abstract
The accumulation of aggregates of amyloidogenic peptides is associated with numerous human diseases. One well studied example is the association between deposition of amyloid β (Aβ) and Alzheimer’s disease. Insulin degrading enzyme and neprilysin are involved in the clearance of Aβ, and presequence peptidase is suggested to play a role in the degradation of mitochondrial Aβ. Recent structural analyses reveal that these three peptidases contain a catalytic chamber (crypt) that selectively encapsulates and cleaves amyloidogenic peptides, hence the name cryptidase. The substrate selectivity of these cryptidases is determined by the size and charge distribution of their crypt as well as the conformational flexibility of substrates. The interaction of Aβ with the catalytic core of these cryptidases is controlled by conformational changes that make the catalytic chambers accessible for Aβ binding. These new structural and biochemical insights into cryptidases provide potential therapeutic strategies for the control of Aβ clearance.
Keywords: Alzheimer's disease, amyloid-β peptide, metalloprotease, anti-amyloidogenic therapy, X-ray crystallography, insulin-degrading enzyme, presequence peptidase, neprilysin
Introduction
In humans, metalloproteases are the most abundant (186 of 553 genes) of the five protease classes (aspartic, metallo, cysteine, serine and threonine) [1]. Similar to other proteases, metalloproteases can selectively modify biological functions of targeted substrates to influence diverse biological processes [2]. Recent structural analyses of insulin degrading enzyme (IDE), presequence peptidase (PreP), and neprilysin, have revealed a new common feature of two metalloprotease family members, M16 (IDE and PreP) and M13 (neprilysin) (Figure 1) [3–8]. These enzymes possess a catalytic chamber that is large enough to encapsulate a variety of peptide substrates that are less than 70 amino acid residues. Access to the catalytic chamber of these enzymes is controlled by protein conformational changes, and the entrapment of substrates and their interaction within this chamber play a key role in substrate recognition and selection. We describe these proteases as cryptidases, from crypt-containing peptidases.
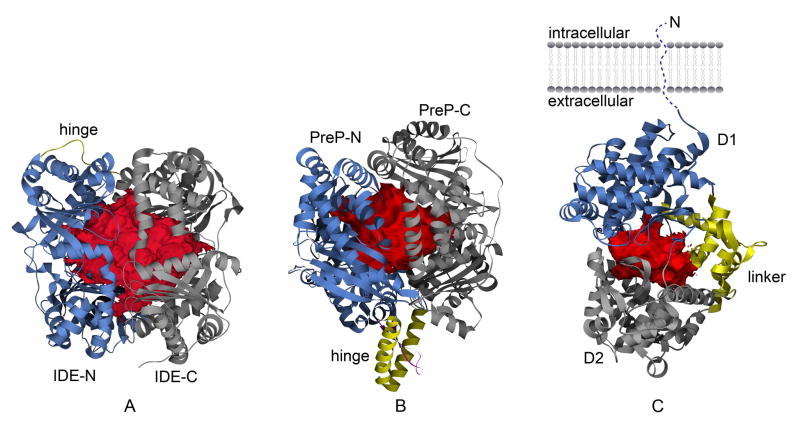
Figure 1.
Catalytic crypts of Aβ-degrading enzymes. Catalytic chambers of IDE (A) PreP (B) and neprilysin (C) are depicted as red surfaces. N-terminal domain, hinge region, and C-terminal domain, are depicted as blue, yellow, and gray ribbon, for IDE and PreP, respectively. Catalytic domain (D1), smaller domain (D2), and the three inter-domain linker fragments (linker) of neprilysin are depicted as blue, gray, and yellow ribbon, respectively, in (C). The insertion between helices making the hinge region of PreP is depicted as magenta in (B). The surface of the catalytic crypts derived from volume data, as generated by VOIDOO [89], were input in Chimera [90] for representation.
Cryptidases target small bioactive peptides. This differs from other crypt-containing proteases such as the 26S proteasome, which targets poly-ubiquitinated proteins [9, 10] and HtrA, an ATP-independent protease-chaperone system that refolds or degrades partially unfolded proteins [11, 12]. Cryptidases are highly relevant in human disease since they have been implicated in the degradation of multiple peptides/hormones including several forms of Aβ [8, 13–19]. Derived from the amyloid precursor protein (APP), Aβ is a processed peptide that is implicated in the development of Alzheimer’s disease. Thus far, therapeutic strategies against Alzheimer’s disease have focused primarily on the family of proteases that are implicated in the generation of Aβ. Yet, increasing either Aβ efflux to the periphery or the rate of Aβ degradation represents another promising and valuable alternative [20–22]. This review focuses on recent advances in structural and functional analyses of the cryptidases, IDE, PreP, and neprilysin and their therapeutic potential toward the control of Aβ levels in humans.
IDE
IDE (M16.002) was discovered based on its ability to rapidly degrade insulin by Mirsky and Broh-Kahn in 1949 [23]. Such activity is biologically relevant since insulin is a key hormone involved in regulating blood glucose levels and possesses a short half life in humans (4–6 minutes). Subsequently, Mirsky and colleagues discovered that inhibitors of this activity could potentiate the action of insulin in rabbits [24]. Biochemical purification and characterization of IDE revealed that it is a relatively large (~100–120 kDa) zinc metalloprotease. Cloning and subsequent bioinformatic analysis of IDE placed it in clan ME of the M16A family of metalloproteases [2, 25, 26]. IDE is evolutionarily conserved and possesses alternatively spliced and initiated variants [27, 28].
A. Substrate specificity of IDE
IDE binds insulin with high affinity (~100 nM) and degrades insulin into several fragments (Figure 2). Mice with the knockout of the IDE gene develop hyperinsulinemia [15]. Furthermore, a single nucleotide polymorphism of the human IDE gene is associated with type 2 diabetes [29]. In addition to degrading insulin, IDE has also been implicated in the degradation of Aβ in vitro (Figure 2, Table 1) [14, 30, 31]. Consistent with in vitro activity, mice with the knockout of IDE gene and the GK rat, with known missense mutations in the IDE gene, result in both elevated levels of insulin in the blood and Aβ in the brain [15, 32, 33]. Additional peptide hormones such as insulin-like growth factor 2, atrial natriuretic peptide, bradykinin, endorphin, and glucagon, have been demonstrated to be degraded by IDE in vitro [14, 34, 35]. However, further study is still required to demonstrate in vivo relevance.
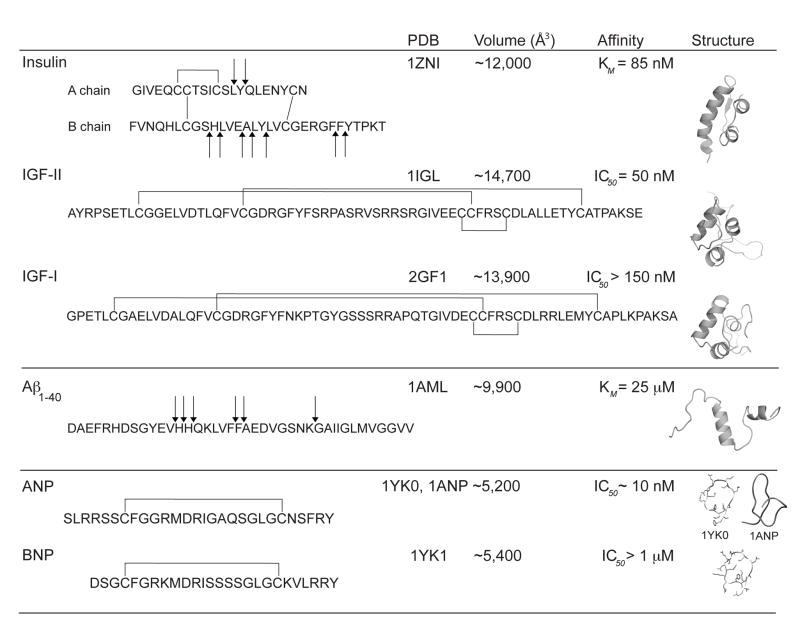
Figure 2.
Substrate selectivity of IDE. Atomic coordinates are from the PDB with ID: 1ZNI [91], 1IGL [92], 2GF1 [93], 1AML [94], 1ANP [95], 1YK0 and 1YK1 [96]. Volumes were calculated by VOIDOO [89] using a probe radius of 1.5Å. Values of KM and IC50 for insulin, IGF-II, IGF-I, Aβ(1–40), ANP and BNP, are from references [3, 37, 97, 98]. Coordinates of ANP and BNP (1YK0, 1YK1) are from complexes with natriuretic peptide receptor C-type (NPR-C) [96]. For comparison, native conformation of ANP from PDB ID 1ANP [95] is also shown.
Table 1.
Comparison of proteolytic chambers (crypts) of Aβ degrading enzymes.
| Classification (clan/family/subfamily) | PDB ID | Catalytic cavities (Å3)* | KM (μM) | |
|---|---|---|---|---|
| IDE | ME/M16/A | 2JG4 | ~15,700 | 25 (Aβ1–40) [3] |
| PreP | ME/M16/C | 2FGE | ~12,000 | - |
| Neprilysin | MA/M13 | 1DMT | ~5,100 | 11 (Aβ1–40), 7 (Aβ1–42) [66] |
| Aβ1–42 | -- | 1Z0Q | ~9,800 | -- |
Volumes of enzymes’ cavities and Aβ were calculated as solvent-excluded volumes by VOIDOO [89] with a probe-radius of 1.5Å, which is added to the Van der Waals radii of the atoms. Coordinates from the PDB with code 2JG4 [3], 2FGE [4], and 1DMT [5], for IDE, PreP, and Neprilysin, respectively, were used. The solvent-excluded volume of the Aβ1–40 was calculated on the model from PDB code 1AML [94].
One striking feature of IDE is its substrate selectivity (Figure 2). Extensive in vitro enzymatic assays reveal how IDE has high nanomolar affinity for substrates that are diverse in sequence and structure (Figure 2) [34]. At the same time, IDE is unable to exert its proteolytic activity on many other peptides that resemble known substrates such as pro-insulin, glucagon-like peptide 1, nerve growth factor, and somatostatin [34]. For example, IDE prefers to cleave ANP over brain-derived natriuretic peptide as well as to digest IGF-2 over IGF-1 (Figure 2) [36–39]. Interestingly, while IDE peptide substrates share little or no homology of primary amino acid sequence they are amyloidogenic in nature [14, 40].
Another unique feature of IDE is the distribution and stochastic nature of the cleavage sites of its substrates. IDE cuts its substrates multiple times (Figure 2). This makes good physiological sense since the role of IDE is to completely inactivate targeted hormones and bio-active peptides. Thus, multiple cleavages are required to achieve this goal. However, this seems to be in conflict with the observation that IDE has only one catalytic center and has tight binding properties for its substrates (e.g., insulin). This tight binding of substrates to IDE with a sole reaction center should result in specific cleavage at a defined site. To make the matter more complicated, cleavage sites of substrates by IDE are distributed to selected regions with certain preferences (Figure 2).
An interesting link exists between the two physiological relevant substrates of IDE insulin and Aβ [35, 41]. Patients with type 2 diabetes have an increased risk of Alzheimer's disease. Furthermore, boosting insulin levels in human subjects increases the presence of Aβ in the cerebrospinal fluid [42]. Since IDE is more efficient at degrading insulin than Aβ, the concomitant increase in insulin and Aβ levels may lead to a redistribution of available IDE away from its function as an Aβ-degrading enzyme [41]. The link between these two substrates and IDE requires further investigation.
B. Structural basis for the substrate selectivity and regulation of human IDE
The structural basis for substrate recognition and regulation of IDE can be understood based on crystal structures of substrate-bound and substrate-free human IDE (Figure 1A) [3, 8]. X-ray crystallographic analysis reveals that IDE is made of two roughly equal-size 55 kDa domains, IDE-N and IDE-C, that are connected by a 28 residue extended loop (Figure 1A). These structures resemble a closed clam shell with two bowl-shaped halves (IDE-N and IDE-C). Similar to other metalloproteases containing the conserved HXXEH motif such as mitochondrial processing peptidase (MPP), pitrilysin, and PreP, the catalytic center is located in IDE-N and contains a zinc ion coordinated by two conserved histidines (H108 and H112) and one glutamate (E189) [3, 4, 8, 43, 44]. Glutamate 111 located in IDE-N domain serves as the base that activates the catalytic water for the hydrolysis of the peptidyl bond [3]. Interestingly, residues from the IDE-C domain are also required for substrate recognition [8]. Consistent with the domain organization of IDE for substrate recognition, biochemical analysis shows that IDE-N alone is able to bind insulin although it has no or little catalytic activity [45]. IDE-C alone has no detectable proteolytic activity. When IDE-N and IDE-C are mixed together, the catalytic activity of IDE is partially restored.
The structures of substrate bound IDE reveal the molecular basis of selectivity of IDE. IDE-N and IDE-C together form an enclosed chamber with a total volume of approximately 16,000 Å3. This chamber can engulf peptides of different sizes (estimated to be < 70 amino acids in length) and is shown to bind full-length Aβ1–40, whose volume is calculated to be ~10,000 Å3 (Table 1 and Figure 3) [8]. The inner surface of IDE-N is mostly neutral or negatively charged whereas IDE-C is largely positive in charge. This unique feature enables IDE to select and exclude substrates by charge complementarity and charge repulsion, respectively. Therefore, size and charge distribution of peptide substrates are two key factors for the substrate selectivity of IDE [8].
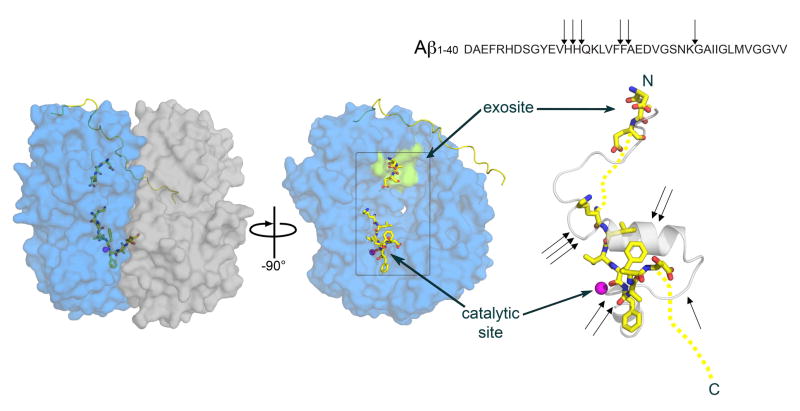
Figure 3.
Interactions between Aβ and IDE. Left panel: surface representation of IDE in complex with Aβ as from PDB ID 2G47 [8]. IDE is depicted as transparent surface to allow visualization of the bound Aβ in the cavity. IDE-N and IDE-C are depicted as transparent blue and gray surfaces, respectively. The hinge region of IDE is depicted as yellow ribbon, and Aβ is depicted as sticks with carbon, nitrogen, and oxygen atoms colored yellow, blue, and red, respectively. Central panel: view of IDE-N after a rotation of -90 degree around the y axis separating IDE-N and IDE-C has been applied. Catalytic site and exosite are indicated by the catalytic zinc ion, depicted as magenta sphere, and by the green surface, respectively. Right panel: details of Aβ conformation as observed in pdb ID 2G47 (depicted as yellow sticks) superimposed to Aβ alone structure from pdb ID 1AML [94] (depicted as transparent gray cartoon). Yellow dash lines show segments of Aβ not revealed in the electron density map of 2G47. Black arrows show the position of the known cleavage sites of Aβ by IDE, according also to the sequence shown above. Figure generated by Pymol [99].
Conformational flexibility of peptide substrates is an additional property for the substrate selectivity of IDE. IDE substrates undergo substantial conformational change upon binding to IDE [8]. For example, a segment of the Aβ1–40 peptide undergoes a conformational switch from an α-helix to a β-strand in order to fit into the catalytic site of IDE. Thus, conformational flexibility of substrates is a pre-requisite for being effectively degraded by IDE. The propensity to undergo this conformational switch also plays a key role in the formation of Aβ plaques. The tendency for substrates to interconvert from α-helix to β-strands could substantiate the observation that substrates for IDE tend to be amyloidogenic in nature [14].
The binding of IDE catalytic residues to specific sequences of substrates also plays a key role in the selectivity [8]. The interaction of IDE catalytic site with four different substrates provides the structural basis for the distribution and stochastic nature of the cleavage sites of IDE. The hydrophobic nature of S1 and S1’ sites at the catalytic center of IDE allow it to accommodate the bulky, hydrophobic residues of P1 and P1’ substrates. However, the catalytic cavity of IDE is relatively shallow, which allows non-hydrophobic residues to fit without significant energetic penalty. This helps to explain why although IDE has preferred cleavage sites, such sequence preference is degenerative.
Another unique feature of substrate recognition of IDE is the anchoring of the N-terminus of IDE substrates to a highly conserved exosite located in the substrate-binding chamber of IDE (Figure 3) [3, 8]. This interaction requires the N-terminal region of IDE substrates to adapt a β-strand conformation. Such binding does not require a specific N-terminal sequence since the main chains of the N-terminal β-strand of IDE substrates form hydrogen bonds with the β-sheet of IDE substrate-binding chamber. Thus, this anchoring provides a structural basis of how IDE mostly cleaves sequences at least 9–10 amino acid residues away from the N-terminus (Figure 3). In addition, the anchoring of the N-terminus of IDE substrates also allows IDE to cut different regions of IDE substrates in a stochastic manner (Figure 3).
The conformational switch of IDE between open and closed states plays a key role for substrate binding [3, 8, 46] (Figure 4). The crystal structures of substrate-bound and substrate-free IDE revealed closed conformation of IDE and a lack of any opening that may gather substrates into the active site. Moreover, there are extensive interactions between IDE-N and IDE-C, burying a large surface with good surface complementarity and numerous hydrogen bonds. These bonds form a latch, keeping the cryptidase closed. This closed state of IDE prevents entry and exit of substrates. This prompted the hypothesis that mutations to destroy the extended hydrogen bonding tend to speed the protease opening and binding of substrates more rapidly, which is consistent with recent biochemical findings [3, 8].
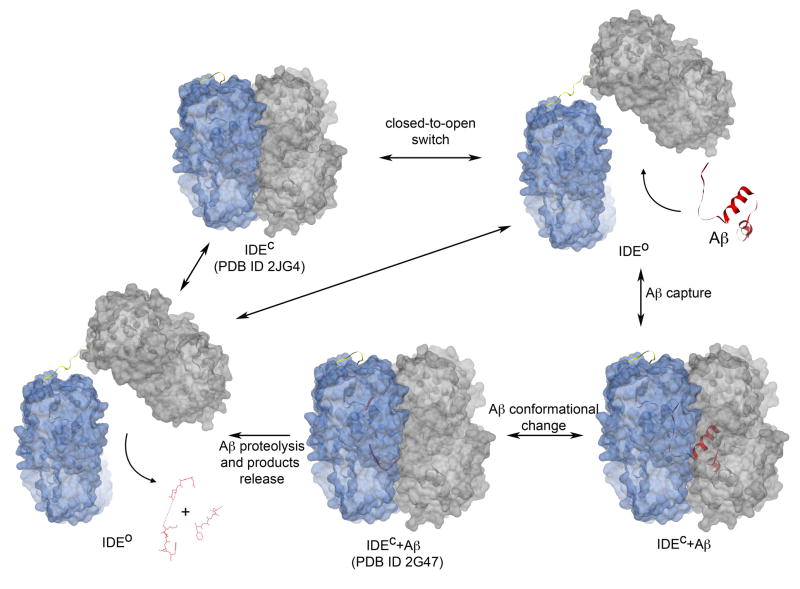
Figure 4.
Model of catalysis for IDE. The first step, the substrate-free closed state (IDEC), corresponds to atomic coordinates deposited in the PDB as 2JG4 [3]. The open state (IDEO) depicted on the top right is a theoretical model, where the C-terminal half of IDE has been arbitrarily rotated as a rigid body to depict the state that allows capture of Aβ into the catalytic cavity. This is modeled based on substrate-free E. coli pitrilysin structure [43]. Once Aβ is entrapped in the catalytic chamber (IDEC+Aβ), it undergoes conformational changes, as revealed by the structure of IDEC+Aβ deposited in the PDB as 2G47 [8]. In this closed stage, IDE performs Aβ proteolysis and subsequently releases the cleavage products by switching back to an open state. The catalytic cycle likewise goes towards either the initial substrate-free closed IDE (the starting IDEC) completing the catalytic cycle, or it incorporates another molecule of Aβ continuing the cycle. Aβ is depicted as red cartoon for PDB ID 1AML and as ribbon and sticks for Aβ1–40 from PDB ID 2G47. IDE-N and IDE-C are depicted as transparent blue and grey surfaces, respectively. The loop hinge of IDE is depicted as yellow ribbon. Figures generated by Chimera [90].
The catalytic cycle of IDE can be envisioned as the following (Figure 4). IDE would normally be in a stable substrate free closed state (IDEC). The rearrangement of IDE-N and IDE-C, triggered by a thermodynamic perturbation or other unknown event(s), yields an open state (IDEO) to allow substrate access to the catalytic site (Figure 4). The IDE-substrate complex then switches back into its closed state, either driven by the binding of substrates or by the attraction between the interface of IDE-N and IDE-C and/or the tension exerted by the loop that joins IDE-N and IDE-C. Once entrapped inside the catalytic chamber, the N-terminus of IDE substrates becomes anchored to the exosite of IDE. In addition, IDE substrates undergo a secondary structure rearrangement to fit into the catalytic site. The conjunction of exosite anchoring and structural rearrangement of substrates would allow IDE to perform successive cleavages in each cycle of entrapment (Figure 3). The final step of the reaction consists of product dissociation that requires IDE to open and release proteolytic fragments (Figure 4). IDE at this stage is either able to incorporate a new substrate to start a new degradation cycle, or to return to the initial substrate-free closed conformation. Therefore, the frequency with which IDE executes the repeated opening and closing is a key to modulate the overall rate of catalysis.
PreP
PreP (M16.012) was initially characterized as a 110 kDa human metalloprotease 1 (hMP1), an evolutionarily conserved metalloprotease sensitive to inhibitors of Zn2+-dependent metalloproteases and ubiquitously expressed with higher abundance in heart and muscle [47].. PreP was subsequently found in mitochondria and chloroplasts and degrades mitochondrial targeting peptides such as the F1B subunit of ATP synthase and the chloroplastic transit peptide SStpNt (54, 55).
A. Substrates of PreP
PreP is postulated to degrade mitochondrial targeting peptides that are cleaved off by mitochondrial processing peptidases following import. Thus, those mitochondrial targeting peptides rich in positively charged, hydroxylated, and hydrophobic residues will not accumulate and cause mitochondrial toxicity. Recently, PreP was shown to degrade several forms of Aβ (Aβ1–40 and Aβ1–42), which are present in mitochondria [13]. This could be highly significant since the Aβ-induced mitochondrial toxicity is associated with Alzheimer's disease [48, 49].
B. Structural analysis of the closed conformation of PreP
The crystal structure of Arabidopsis thaliana PreP (AtPreP) with a co-purified peptide provides structural basis for the substrate recognition and regulation of PreP (Figure 1B) [4]. Similar to human IDE and pitrilysin, AtPreP is made of two homologous domains, PreP-N and PreP-C (Figure 1B). PreP-N also contains the HXXEH motif at the catalytic site, which has a Zn2+ ion coordinated by two histidines (H77 and H81) and a glutamate (E177) [4]. A glutamate residue (E80) serves as the catalytic base that deprotonates a water molecule for hydrolysis of the peptidyl bond. Similar to IDE, residues belonging to PreP-C and located about 800 amino acid residues away from the active site also contribute to substrate binding; thus, both PreP-N and PreP-C are required for substrate binding and catalysis [4]. Enclosed between PreP-N and PreP-C is a catalytic cavity, which in analogy with the closed form of IDE, has no visible opening for the access of substrates. Disulfide bond crosslinking between PreP-N and PreP-C locks PreP in the closed conformation, rendering the enzyme inactive [4]. This indicates that a conformational switch between open and closed state plays a key role in catalysis.
Similar to IDE, AtPreP’s catalytic chamber is primarily polar [4]. PreP-N and PreP-C are characterized by an overall negative and positive electrostatic potential, respectively. The highly negatively charged surface could explain the substrate selectivity of PreP to degrade presequences, which are rich in positive charges [4]. The catalytic chamber of PreP in its closed conformation has a volume of ~12,000Å3, which is large enough to accommodate Aβ1–42 (volume is ~10,000Å3) (Table 1). Thus, similar to IDE, PreP also uses size and charge distribution of the catalytic chamber for substrate selectivity. Differing from IDE, however, no exosite is present in the catalytic chamber of PreP [3]. To date, the precise interaction of PreP with Aβ remains elusive [13].
In contrast with the model postulated for IDE-mediated catalysis, PreP is hypothesized to normally exist in an unbound open state that, upon substrate binding, switches to a closed state able to perform proteolysis [4]. PreP-N and PreP-C are linked by a hinge of 82 amino acid residues made of two α-helices protruding to form a V shape (Figure 1B). Instead, an extended loop is found to connect N- and C-terminal halves in IDE and pitrilysin. Electrostatic forces are postulated to keep AtPreP open and a hinge-bending motion triggered by peptide binding switches AtPreP to the closed state [4]. The hypothesis that unbound AtPreP stays in the open state is largely based on the open conformation observed in the structure of substrate-free E. coli pitrilysin [43], which shares substantial structural homology with AtPreP and human IDE. Interestingly, PreP-N and PreP-C make substantial contact in the closed state with a large buried surface and good shape complementarity (buried surface =11,150 Å2 with shape complementarity score of 0.60) [3]. Thus, it is possible that AtPreP is in a dynamic equilibrium between the open and closed conformations in the absence of peptide substrates, while the binding of the peptides could favor equilibrium toward the closed state. This would provide an additional layer of regulation to control the proteolytic activity of PreP in mitochondria.
Neprilysin family
Neprilysin (M13.001), also named kidney brush border neutral proteinase, enkephalinase, endopeptidase 24.11, neutral endopeptidase (NEP), common acute lymphoblastic leukemia antigen and CD10 based on repeated independent discovery was officially named by IUBMB in 1992 and has been in depth reviewed elsewhere [2, 20, 22, 50]. This long history reflects the wide range of cell types and processes in which neprilysin is implicated. We will focus on the role and function of neprilysin in the nervous system regarding Aβ degradation. Neprilysin is an 85–93 kDa type II membrane protein with the catalytic domain facing the extracellular/lumen space and is glycosylated at multiple sites (Figure 1C) [2, 5, 51, 52]. A related group of proteases, endothelin-converting enzymes 1 (M13.002) and 2 (M13.003) (ECE-1 and ECE-2) share striking similarities in sequence and domain organization to neprilysin [53, 54]. ECE enzymes were originally implicated in the processing of pro-hormone forms of endothelin, a highly potent vasoconstrictor. All three enzymes are shown to be capable of degrading Aβ in vitro and in vivo [17, 19, 55, 56].
A. Aβ degradation by neprilysin family
Several lines of evidence implicate neprilysin in the degradation of Aβ. Neprilysin was shown to degrade Aβ1–42 in vivo, and selective inhibition of neprilysin blocked Aβ degradation in rat hippocampus [52]. Consistent with the role of Aβ degradation in vivo, long-term administration of thiorphan, which is a selective inhibitor of neprilysin, caused an increase of Aβ1–42 plaques in rat hippocampus and the cortical region outside the hippocampus [52]. Inhibition of the neprilysin family of metalloproteases with metalloprotease inhibitor phosphoramidon was also shown to increase Aβ level in mice models [56]. The deletion of the neprilysin gene in mice further supports the role of neprilysin in Aβ degradation [19]. These neprilysin knockout homozygote mice have an impaired ability to degrade externally administered Aβ1–42 and elevated levels of Aβ1–40 and Aβ1–42. Finally, correlative studies in patients with sporadic Alzheimer’s disease and in normally aged mice and humans implicate a role for neprilysin in Aβ degradation [57–60].
Neprilysin can be used to control Aβ levels in vivo. The use of lentivirus to deliver human neprilysin to primary neural progenitors (HCN) showed an enhanced ability to degrade Aβ1–42 in vitro [61]. In addition, the injection of this virus on one side of the hippocampus of APP transgenic mice decreased plaque burden [61]. Ex-vivo gene delivery of a soluble form of neprilysin, via fibroblasts, into transgenic APP mice also demonstrated a robust clearance of plaques [18]. Interestingly, mRNA and protein levels of neprilysin can be induced by intracranial injection of Aβ1–42, which also reduced the accumulation of amyloid plaques [62].
Similar to neprilysin, ECE-1 has been shown to degrade Aβ in vitro [55]. Homozygous knockouts for ECE-1 are lethal [63] and heterozygous animals show an increased amount of Aβ1–40 and Aβ1–42 in the brain [17]. In the same study, homozygous ECE-2 knockout mice showed a gene dose dependent increase of both forms of Aβ in the brain as well. Furthermore, the exploration of the combinatorial roles of neprilysin, ECE-1 and ECE-2 through the use of knockout mice shows additive effects of this protease family for Aβ degradation [56]. In this study, the crossing of neprilysin homozygote knockout mice with ECE-1 heterozygote or homozygote ECE-2 knockout mice yielded higher Aβ accumulation than any one knockout by itself. This supports the hypothesis that multiple proteases are involved in the degradation of Aβ in the brain.
B. Structural analysis of neprilysin
Neprilysin consists of a short N-terminal cytoplasmic tail, a single transmembrane helix and a large C-terminal extracellular ectodomain (Figure 1C). The N-terminal tail of neprilysin is shown to directly bind the ezrin/radixin/moesin (ERM) protein, which could serve as a scaffold for the organization of membrane-associated cytoskeleton and cell signaling [64]. X-ray structures of the extracellular domain of neprilysin in complex with phosphoramidon and other inhibitors [5–7] and of ERM in complex with the N-terminal tail of neprilysin [65] have been recently solved. The ellipsoid shaped ectodomain of neprilysin is largely made of α-helices with 6 disulfide bridges and at least 3 N-glycosylation sites. This ectodomain can be divided into 2 domains connected by four inter-domain linkers (Figure 1C). Domain 1 is the juxta-transmembrane domain made of three non-consecutive polypeptide segments belonging to both the N- and the C-terminal regions. The catalytic site, with the conserved HExxH motif necessary for zinc coordination is located within domain 1. Domain 2 is smaller than domain 1 and is connected to domain 1 by three non-consecutive α-helical fragments (Figure 1C).
While there is no structural similarity of neprilysin to IDE and PreP, neprilysin also possesses a proteolytic chamber that is enclosed between domains 1 and 2 and flanked by the four inter-domain linkers (Figure 1C). The catalytic chamber of neprilysin has a total volume of ~5,000 Å3, which is only about half of the volume of Aβ (~10,000 Å3) (Table 1). This raises the challenge of explaining how neprilysin degrades Aβ [19, 66]. In addition to the non-consecutive linker fragments joining domain 1 and 2, a second major difference with IDE and PreP is that a small opening, constituted primarily of charged residues, connects the catalytic chamber of neprilysin to the solvent. The side-chains of residues lining this opening are separated by ~7 Å distance at their closest point, and on one side are located on a ~20 aa-long loop (residues 523–544) positioned to occlude the catalytic chamber (Figure 1C). Both the size of the opening and the chamber volume of neprilysin are apparently incompatible with diffusion and fitting of folded Aβ into the cavity.
Neprilysin likely undergoes conformational change upon Aβ binding. The protein motion of neprilysin could be both a close-to-open switch mediated by the linker connecting domain 1 and 2 (similar to the hinge-mediated motion hypothesized to take place in IDE and PreP), as well as a localized shift of the loop in front of the catalytic chamber. Charge-charge interactions between Aβ and neprilysin could be responsible for inducing these conformational changes in neprilysin [67]. Once neprilysin partially accommodates Aβ in the catalytic cavity, it could perform proteolysis in a progressive manner, generating peptides of different amino acids in length (in a proteasome-like manner). To date, the molecular details of the interaction of neprilysin with Aβ remain elusive.
Compartmentalization of Aβ-degrading cryptidases for Aβ degradation
Aβ has a very high clearance rate in human central nervous system (CNS), approximately 8% per hour [68]. The Aβ clearance in CNS is controlled by Aβ degradation in brain and Aβ efflux from CNS to the peripheral circulation [69, 70]. The Aβ high clearance rate is in part achieved by the presence of cryptidases in multiple cellular compartments. The extracellular space is a primary site for Aβ accumulation to form hallmark amyloid plaques observed in Alzheimer's disease and extracellular Aβ can be degraded by cell surface and secreted form of cryptidases including IDE and neprilysin [71–73]. As expected, the neprilysin and ECE family are primarily associated with the cell surface [22]. While IDE is originally described as a cytosolic enzyme and suggested to play a critical role in degradation of intracellular Aβ [74], it is also found in endosomes, on the cell surface, and in the extracellular milieu [34, 71, 72]. The recent finding that IDE serves as the receptor of Varicella-zoster virus further demonstrates the functional role of cell surface localization of IDE [75]. Interestingly, the type of cell in the nervous system may dictate whether IDE is secreted or associated with the cell surface. Primary mouse microglia and the BV-2 cell line are found to secrete IDE, yet primary hippocampal neurons and differentiated PC12 cells only possess membrane associated IDE [71, 73]. Aβ is also found in mitochondria, which represent a large fraction of the volume of a cell (22%) and exist in high density in the dendrites of neurons. Mitochondrial dysfunction in Alzheimer's disease emphasizes the potential role of Aβ in mitochondria [48, 49]. Both PreP and IDE can be found in mitochondria [28, 76] so they are likely involved in mitochondrial Aβ clearance. IDE, ECE-1, and ECE-2 can also be found in other intracellular organelles such as ER, Golgi, and endosomes where Aβ may be present [22, 77, 78]. The subcellular distributions of several cryptidases support the notion that Aβ degradation can be controlled at multiple subcellular compartments.
Aβ-degrading cryptidase-based therapeutic for Alzheimer's disease
Aβ and APP serve key physiological roles in neuronal functions including the control of synaptic activity [79] or neuronal migration [80]. Thus, control of physiological Aβ levels, instead of the complete inhibition of Aβ production, is a key to slow down the progression of Alzheimer's disease [21]. Recent evidence also suggests that plaque formation may occur rapidly [81], positing a challenge to the dynamic control of Aβ levels in the brain. Targeting Aβ clearance represents a potential therapeutic avenue for patients with Alzheimer's disease [21, 70]. A series of studies in Aβ-based immunotherapy reveal that targeting Aβ directly can reduce the Aβ load and the clearance of amyloid plaques [82–86].
Aβ-degrading cryptidase-based therapeutic represents an alternative approach in controlling Aβ clearance. Therapeutic intervention could occur by re-compartmentalization of cryptidases with the gene targeted ex-vivo gene therapy [18]. As described above, the soluble form of neprilysin that was expressed in fibroblasts and injected into the brain of transgenic APP mice led to a reduction of amyloid plaques. Alternatively, instead of using immunotherapy, it could be possible to use engineered Aβ-specific cryptidases that have enhanced Aβ-degrading activity to reduce the Aβ in peripheral circulation, which could in turn reduce the Aβ load in the brain [3, 69]. Small molecular weight drugs to boost the expression and/or activity of cryptidases will likely provide therapeutic benefits as well.
While Aβ-based immunotherapy shows some promise, the first analysis of human neuropathology with Aβ immunization had unusual side effects that may be caused by antibody-mediated inflammation [87, 88]. Although further improvement of this approach is ongoing, future testing using combinations of Aβ-protease based therapeutic with Aβ-based immunotherapy may improve the effectiveness of therapy and minimize unwanted side effects.
Conclusion remarks
Numerous questions regarding the structure and function of Aβ-degrading cryptidases remain to be addressed. Except for IDE in complex with Aβ(1–40) [8], no structural studies exist describing the molecular basis for the recognition of Aβ by cryptidases. While our current structural knowledge suggests conformational changes of both cryptidase and Aβ upon their encounter, little is known about these dynamic processes. In addition, our knowledge of other physiologically relevant substrates of these cryptidases is incomplete, as well as our understanding of the regulation of catalytic activity and the control of expression and subcellular compartmentization of cryptidases is rather limited. However, the rate at which knowledge of cryptidases’ role in Aβ catabolism has expanded during recent years promises exciting further development that will likely lead to new therapeutic avenues for Alzheimer's disease and other human diseases.
Acknowledgments
This work was supported by NIH R01-GM81539 for W-J T, the American Health Assistance Foundation for EM and NIH T32-GM07839 for REH.
References
- 1.Puente XS, Sanchez LM, Overall CM, Lopez-Otin C. Human and mouse proteases: a comparative genomic approach. Nat Rev Genet. 2003;4:544–58. doi: 10.1038/nrg1111. [DOI] [PubMed] [Google Scholar]
- 2.Barrett AJ. Handbook of proteolytic enzymes. Elsevier Academic Press; London ; San Diego: 2004. [Google Scholar]
- 3.Im H, Manolopoulou M, Malito E, Shen Y, Zhao J, Neant-Fery M, Sun CY, Meredith SC, Sisodia SS, Leissring MA, Tang WJ. Structure of substrate-free human insulin-degrading enzyme (IDE) and biophysical analysis of ATP-induced conformational switch of IDE. J Biol Chem. 2007;282:25453–63. doi: 10.1074/jbc.M701590200. [DOI] [PubMed] [Google Scholar]
- 4.Johnson KA, Bhushan S, Stahl A, Hallberg BM, Frohn A, Glaser E, Eneqvist T. The closed structure of presequence protease PreP forms a unique 10,000 Angstroms3 chamber for proteolysis. Embo J. 2006;25:1977–86. doi: 10.1038/sj.emboj.7601080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oefner C, D'Arcy A, Hennig M, Winkler FK, Dale GE. Structure of human neutral endopeptidase (Neprilysin) complexed with phosphoramidon. J Mol Biol. 2000;296:341–9. doi: 10.1006/jmbi.1999.3492. [DOI] [PubMed] [Google Scholar]
- 6.Oefner C, Pierau S, Schulz H, Dale GE. Structural studies of a bifunctional inhibitor of neprilysin and DPP-IV. Acta Crystallogr D Biol Crystallogr. 2007;63:975–81. doi: 10.1107/S0907444907036281. [DOI] [PubMed] [Google Scholar]
- 7.Oefner C, Roques BP, Fournie-Zaluski MC, Dale GE. Structural analysis of neprilysin with various specific and potent inhibitors. Acta Crystallogr D Biol Crystallogr. 2004;60:392–6. doi: 10.1107/S0907444903027410. [DOI] [PubMed] [Google Scholar]
- 8.Shen Y, Joachimiak A, Rosner MR, Tang WJ. Structures of human insulin-degrading enzyme reveal a new substrate recognition mechanism. Nature. 2006;443:870–874. doi: 10.1038/nature05143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann-Petersen R, Gordon C. Proteins interacting with the 26S proteasome. Cell Mol Life Sci. 2004;61:1589–95. doi: 10.1007/s00018-004-4132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55:555–90. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 11.Clausen T, Southan C, Ehrmann M. The HtrA family of proteases: implications for protein composition and cell fate. Mol Cell. 2002;10:443–55. doi: 10.1016/s1097-2765(02)00658-5. [DOI] [PubMed] [Google Scholar]
- 12.Kim DY, Kim KK. Structure and function of HtrA family proteins, the key players in protein quality control. J Biochem Mol Biol. 2005;38:266–74. doi: 10.5483/bmbrep.2005.38.3.266. [DOI] [PubMed] [Google Scholar]
- 13.Falkevall A, Alikhani N, Bhushan S, Pavlov PF, Busch K, Johnson KA, Eneqvist T, Tjernberg L, Ankarcrona M, Glaser E. Degradation of the amyloid beta-protein by the novel mitochondrial peptidasome, PreP. J Biol Chem. 2006;281:29096–104. doi: 10.1074/jbc.M602532200. [DOI] [PubMed] [Google Scholar]
- 14.Kurochkin IV. Insulin-degrading enzyme: embarking on amyloid destruction. Trends Biochem Sci. 2001;26:421–5. doi: 10.1016/s0968-0004(01)01876-x. [DOI] [PubMed] [Google Scholar]
- 15.Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, Frosch MP, Eckman CB, Tanzi RE, Selkoe DJ, Guenette S. Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A. 2003;100:4162–7. doi: 10.1073/pnas.0230450100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leissring MA, Farris W, Chang AY, Walsh DM, Wu X, Sun X, Frosch MP, Selkoe DJ. Enhanced proteolysis of beta-amyloid in APP transgenic mice prevents plaque formation, secondary pathology, and premature death. Neuron. 2003;40:1087–93. doi: 10.1016/s0896-6273(03)00787-6. [DOI] [PubMed] [Google Scholar]
- 17.Eckman EA, Watson M, Marlow L, Sambamurti K, Eckman CB. Alzheimer's disease beta-amyloid peptide is increased in mice deficient in endothelin-converting enzyme. J Biol Chem. 2003;278:2081–4. doi: 10.1074/jbc.C200642200. [DOI] [PubMed] [Google Scholar]
- 18.Hemming ML, Patterson M, Reske-Nielsen C, Lin L, Isacson O, Selkoe DJ. Reducing amyloid plaque burden via ex vivo gene delivery of an Abeta-degrading protease: a novel therapeutic approach to Alzheimer disease. PLoS Med. 2007;4:e262. doi: 10.1371/journal.pmed.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Iwata N, Tsubuki S, Takaki Y, Shirotani K, Lu B, Gerard NP, Gerard C, Hama E, Lee HJ, Saido TC. Metabolic regulation of brain Abeta by neprilysin. Science. 2001;292:1550–2. doi: 10.1126/science.1059946. [DOI] [PubMed] [Google Scholar]
- 20.Hamaguchi T, Ono K, Yamada M. Anti-amyloidogenic therapies: strategies for prevention and treatment of Alzheimer's disease. Cell Mol Life Sci. 2006;63:1538–52. doi: 10.1007/s00018-005-5599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanzi RE, Moir RD, Wagner SL. Clearance of Alzheimer's Abeta peptide: the many roads to perdition. Neuron. 2004;43:605–8. doi: 10.1016/j.neuron.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 22.Turner AJ, Nalivaeva NN. New insights into the roles of metalloproteinases in neurodegeneration and neuroprotection. Int Rev Neurobiol. 2007;82:113–35. doi: 10.1016/S0074-7742(07)82006-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Broh-Kahn RH, Mirsky IA. The inactivation of insulin by tissue extracts; the effect of fasting on the insulinase content of rat liver. Arch Biochem. 1949;20:10–4. [PubMed] [Google Scholar]
- 24.Mirsky IA, Perisutti G, Diengott D. Effect of insulinase-inhibitor on destruction of insulin by intact mouse. Proc Soc Exp Biol Med. 1955;88:76–8. doi: 10.3181/00379727-88-21497. [DOI] [PubMed] [Google Scholar]
- 25.Affholter JA, Fried VA, Roth RA. Human insulin-degrading enzyme shares structural and functional homologies with E. coli protease III. Science. 1988;242:1415–8. doi: 10.1126/science.3059494. [DOI] [PubMed] [Google Scholar]
- 26.Rawlings ND, Morton FR, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2006;34:D270–2. doi: 10.1093/nar/gkj089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farris W, Leissring MA, Hemming ML, Chang AY, Selkoe DJ. Alternative splicing of human insulin-degrading enzyme yields a novel isoform with a decreased ability to degrade insulin and amyloid beta-protein. Biochemistry. 2005;44:6513–25. doi: 10.1021/bi0476578. [DOI] [PubMed] [Google Scholar]
- 28.Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, Guarente L, Selkoe DJ. Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J. 2004;383:439–46. doi: 10.1042/BJ20041081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–5. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 30.Kurochkin IV, Goto S. Alzheimer's beta-amyloid peptide specifically interacts with and is degraded by insulin degrading enzyme. FEBS Lett. 1994;345:33–7. doi: 10.1016/0014-5793(94)00387-4. [DOI] [PubMed] [Google Scholar]
- 31.Selkoe DJ. Clearing the brain's amyloid cobwebs. Neuron. 2001;32:177–80. doi: 10.1016/s0896-6273(01)00475-5. [DOI] [PubMed] [Google Scholar]
- 32.Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, Eckman CB, Tanzi RE, Selkoe DJ. Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol. 2004;164:1425–34. doi: 10.1016/s0002-9440(10)63229-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller BC, Eckman EA, Sambamurti K, Dobbs N, Chow KM, Eckman CB, Hersh LB, Thiele DL. Amyloid-beta peptide levels in brain are inversely correlated with insulysin activity levels in vivo. Proc Natl Acad Sci U S A. 2003;100:6221–6. doi: 10.1073/pnas.1031520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duckworth WC, Bennett RG, Hamel FG. Insulin degradation: progress and potential. Endocr Rev. 1998;19:608–24. doi: 10.1210/edrv.19.5.0349. [DOI] [PubMed] [Google Scholar]
- 35.Qiu WQ, Folstein MF. Insulin, insulin-degrading enzyme and amyloid-beta peptide in Alzheimer's disease: review and hypothesis. Neurobiol Aging. 2006;27:190–8. doi: 10.1016/j.neurobiolaging.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 36.Misbin RI, Almira EC, Duckworth WC, Mehl TD. Inhibition of insulin degradation by insulin-like growth factors. Endocrinology. 1983;113:1525–7. doi: 10.1210/endo-113-4-1525. [DOI] [PubMed] [Google Scholar]
- 37.Muller D, Baumeister H, Buck F, Richter D. Atrial natriuretic peptide (ANP) is a high-affinity substrate for rat insulin-degrading enzyme. Eur J Biochem. 1991;202:285–92. doi: 10.1111/j.1432-1033.1991.tb16374.x. [DOI] [PubMed] [Google Scholar]
- 38.Muller D, Schulze C, Baumeister H, Buck F, Richter D. Rat insulin-degrading enzyme: cleavage pattern of the natriuretic peptide hormones ANP, BNP, and CNP revealed by HPLC and mass spectrometry. Biochemistry. 1992;31:11138–43. doi: 10.1021/bi00160a026. [DOI] [PubMed] [Google Scholar]
- 39.Roth RA, Mesirow ML, Yokono K, Baba S. Degradation of insulin-like growth factors I and II by a human insulin degrading enzyme. Endocr Res. 1984;10:101–12. doi: 10.3109/07435808409035411. [DOI] [PubMed] [Google Scholar]
- 40.Bennett RG, Duckworth WC, Hamel FG. Degradation of amylin by insulin-degrading enzyme. J Biol Chem. 2000;275:36621–5. doi: 10.1074/jbc.M006170200. [DOI] [PubMed] [Google Scholar]
- 41.Taubes G. Neuroscience. Insulin insults may spur Alzheimer's disease. Science. 2003;301:40–1. doi: 10.1126/science.301.5629.40. [DOI] [PubMed] [Google Scholar]
- 42.Watson GS, Peskind ER, Asthana S, Purganan K, Wait C, Chapman D, Schwartz MW, Plymate S, Craft S. Insulin increases CSF Abeta42 levels in normal older adults. Neurology. 2003;60:1899–903. doi: 10.1212/01.wnl.0000065916.25128.25. [DOI] [PubMed] [Google Scholar]
- 43.Maskos K. In: Handbook of metalloproteins. Messerschmidt A, Dode W, Cygler M, editors. Vol. 3. John Wiley &Sons; 2004. pp. 190–198. [Google Scholar]
- 44.Taylor AB, Smith BS, Kitada S, Kojima K, Miyaura H, Otwinowski Z, Ito A, Deisenhofer J. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure. 2001;9:615–25. doi: 10.1016/s0969-2126(01)00621-9. [DOI] [PubMed] [Google Scholar]
- 45.Li P, Kuo WL, Yousef M, Rosner MR, Tang WJ. The C-terminal domain of human insulin degrading enzyme is required for dimerization and substrate recognition. Biochem Biophys Res Commun. 2006;343:1032–7. doi: 10.1016/j.bbrc.2006.03.083. [DOI] [PubMed] [Google Scholar]
- 46.Leissring MA, Selkoe DJ. Structural biology: enzyme target to latch on to. Nature. 2006;443:761–2. doi: 10.1038/nature05210. [DOI] [PubMed] [Google Scholar]
- 47.Mzhavia N, Berman YL, Qian Y, Yan L, Devi LA. Cloning, expression, and characterization of human metalloprotease 1: a novel member of the pitrilysin family of metalloendoproteases. DNA Cell Biol. 1999;18:369–80. doi: 10.1089/104454999315268. [DOI] [PubMed] [Google Scholar]
- 48.Caspersen C, Wang N, Yao J, Sosunov A, Chen X, Lustbader JW, Xu HW, Stern D, McKhann G, Yan SD. Mitochondrial Abeta: a potential focal point for neuronal metabolic dysfunction in Alzheimer's disease. Faseb J. 2005;19:2040–1. doi: 10.1096/fj.05-3735fje. [DOI] [PubMed] [Google Scholar]
- 49.Lustbader JW, Cirilli M, Lin C, Xu HW, Takuma K, Wang N, Caspersen C, Chen X, Pollak S, Chaney M, Trinchese F, Liu S, Gunn-Moore F, Lue LF, Walker DG, Kuppusamy P, Zewier ZL, Arancio O, Stern D, Yan SS, Wu H. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer's disease. Science. 2004;304:448–52. doi: 10.1126/science.1091230. [DOI] [PubMed] [Google Scholar]
- 50.Roques BP, Noble F, Dauge V, Fournie-Zaluski MC, Beaumont A. Neutral endopeptidase 24.11: structure, inhibition, and experimental and clinical pharmacology. Pharmacol Rev. 1993;45:87–146. [PubMed] [Google Scholar]
- 51.Hooper NM. Families of zinc metalloproteases. FEBS Lett. 1994;354:1–6. doi: 10.1016/0014-5793(94)01079-x. [DOI] [PubMed] [Google Scholar]
- 52.Iwata N, Tsubuki S, Takaki Y, Watanabe K, Sekiguchi M, Hosoki E, Kawashima-Morishima M, Lee HJ, Hama E, Sekine-Aizawa Y, Saido TC. Identification of the major Abeta1–42-degrading catabolic pathway in brain parenchyma: suppression leads to biochemical and pathological deposition. Nat Med. 2000;6:143–50. doi: 10.1038/72237. [DOI] [PubMed] [Google Scholar]
- 53.Turner AJ, Tanzawa K. Mammalian membrane metallopeptidases: NEP, ECE, KELL, and PEX. Faseb J. 1997;11:355–64. doi: 10.1096/fasebj.11.5.9141502. [DOI] [PubMed] [Google Scholar]
- 54.Bur D, Dale GE, Oefner C. A three-dimensional model of endothelin-converting enzyme (ECE) based on the X-ray structure of neutral endopeptidase 24.11 (NEP) Protein Eng. 2001;14:337–41. doi: 10.1093/protein/14.5.337. [DOI] [PubMed] [Google Scholar]
- 55.Eckman EA, Reed DK, Eckman CB. Degradation of the Alzheimer's amyloid beta peptide by endothelin-converting enzyme. J Biol Chem. 2001;276:24540–8. doi: 10.1074/jbc.M007579200. [DOI] [PubMed] [Google Scholar]
- 56.Eckman EA, Adams SK, Troendle FJ, Stodola BA, Kahn MA, Fauq AH, Xiao HD, Bernstein KE, Eckman CB. Regulation of steady-state beta-amyloid levels in the brain by neprilysin and endothelin-converting enzyme but not angiotensin-converting enzyme. J Biol Chem. 2006;281:30471–8. doi: 10.1074/jbc.M605827200. [DOI] [PubMed] [Google Scholar]
- 57.Iwata N, Takaki Y, Fukami S, Tsubuki S, Saido TC. Region-specific reduction of A beta-degrading endopeptidase, neprilysin, in mouse hippocampus upon aging. J Neurosci Res. 2002;70:493–500. doi: 10.1002/jnr.10390. [DOI] [PubMed] [Google Scholar]
- 58.Russo R, Borghi R, Markesbery W, Tabaton M, Piccini A. Neprilysin decreases uniformly in Alzheimer's disease and in normal aging. FEBS Lett. 2005;579:6027–30. doi: 10.1016/j.febslet.2005.09.054. [DOI] [PubMed] [Google Scholar]
- 59.Yasojima K, Akiyama H, McGeer EG, McGeer PL. Reduced neprilysin in high plaque areas of Alzheimer brain: a possible relationship to deficient degradation of beta-amyloid peptide. Neurosci Lett. 2001;297:97–100. doi: 10.1016/s0304-3940(00)01675-x. [DOI] [PubMed] [Google Scholar]
- 60.Yasojima K, McGeer EG, McGeer PL. Relationship between beta amyloid peptide generating molecules and neprilysin in Alzheimer disease and normal brain. Brain Res. 2001;919:115–21. doi: 10.1016/s0006-8993(01)03008-6. [DOI] [PubMed] [Google Scholar]
- 61.Marr RA, Rockenstein E, Mukherjee A, Kindy MS, Hersh LB, Gage FH, Verma IM, Masliah E. Neprilysin gene transfer reduces human amyloid pathology in transgenic mice. J Neurosci. 2003;23:1992–6. doi: 10.1523/JNEUROSCI.23-06-01992.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mohajeri MH, Wollmer MA, Nitsch RM. Abeta 42-induced increase in neprilysin is associated with prevention of amyloid plaque formation in vivo. J Biol Chem. 2002;277:35460–5. doi: 10.1074/jbc.M202899200. [DOI] [PubMed] [Google Scholar]
- 63.Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998;125:825–36. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- 64.Iwase A, Shen R, Navarro D, Nanus DM. Direct binding of neutral endopeptidase 24.11 to ezrin/radixin/moesin (ERM) proteins competes with the interaction of CD44 with ERM proteins. J Biol Chem. 2004;279:11898–905. doi: 10.1074/jbc.M212737200. [DOI] [PubMed] [Google Scholar]
- 65.Terawaki S, Kitano K, Hakoshima T. Structural basis for type II membrane protein binding by ERM proteins revealed by the radixin-neutral endopeptidase 24.11 (NEP) complex. J Biol Chem. 2007;282:19854–62. doi: 10.1074/jbc.M609232200. [DOI] [PubMed] [Google Scholar]
- 66.Shirotani K, Tsubuki S, Iwata N, Takaki Y, Harigaya W, Maruyama K, Kiryu-Seo S, Kiyama H, Iwata H, Tomita T, Iwatsubo T, Saido TC. Neprilysin degrades both amyloid beta peptides 1–40 and 1–42 most rapidly and efficiently among thiorphan- and phosphoramidon-sensitive endopeptidases. J Biol Chem. 2001;276:21895–901. doi: 10.1074/jbc.M008511200. [DOI] [PubMed] [Google Scholar]
- 67.Wade RC, Gabdoulline RR, Ludemann SK, Lounnas V. Electrostatic steering and ionic tethering in enzyme-ligand binding: insights from simulations. Proc Natl Acad Sci U S A. 1998;95:5942–9. doi: 10.1073/pnas.95.11.5942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bateman RJ, Munsell LY, Morris JC, Swarm R, Yarasheski KE, Holtzman DM. Human amyloid-beta synthesis and clearance rates as measured in cerebrospinal fluid in vivo. Nat Med. 2006;12:856–61. doi: 10.1038/nm1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DeMattos RB, Bales KR, Cummins DJ, Paul SM, Holtzman DM. Brain to plasma amyloid-beta efflux: a measure of brain amyloid burden in a mouse model of Alzheimer's disease. Science. 2002;295:2264–7. doi: 10.1126/science.1067568. [DOI] [PubMed] [Google Scholar]
- 70.Tanzi RE, Bertram L. Twenty years of the Alzheimer's disease amyloid hypothesis: a genetic perspective. Cell. 2005;120:545–55. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 71.Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang J, Podlisny MB, Rosner MR, Safavi A, Hersh LB, Selkoe DJ. Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J Biol Chem. 1998;273:32730–8. doi: 10.1074/jbc.273.49.32730. [DOI] [PubMed] [Google Scholar]
- 72.Vekrellis K, Ye Z, Qiu WQ, Walsh D, Hartley D, Chesneau V, Rosner MR, Selkoe DJ. Neurons regulate extracellular levels of amyloid beta-protein via proteolysis by insulin-degrading enzyme. J Neurosci. 2000;20:1657–65. doi: 10.1523/JNEUROSCI.20-05-01657.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mentlein R, Ludwig R, Martensen I. Proteolytic degradation of Alzheimer's disease amyloid beta-peptide by a metalloproteinase from microglia cells. J Neurochem. 1998;70:721–6. doi: 10.1046/j.1471-4159.1998.70020721.x. [DOI] [PubMed] [Google Scholar]
- 74.Sudoh S, Frosch MP, Wolf BA. Differential effects of proteases involved in intracellular degradation of amyloid beta-protein between detergent-soluble and -insoluble pools in CHO-695 cells. Biochemistry. 2002;41:1091–9. doi: 10.1021/bi011193l. [DOI] [PubMed] [Google Scholar]
- 75.Li Q, Ali MA, Cohen JI. Insulin degrading enzyme is a cellular receptor mediating varicella-zoster virus infection and cell-to-cell spread. Cell. 2006;127:305–16. doi: 10.1016/j.cell.2006.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Stahl A, Nilsson S, Lundberg P, Bhushan S, Biverstahl H, Moberg P, Morisset M, Vener A, Maler L, Langel U, Glaser E. Two novel targeting peptide degrading proteases, PrePs, in mitochondria and chloroplasts, so similar and still different. J Mol Biol. 2005;349:847–60. doi: 10.1016/j.jmb.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 77.Emoto N, Nurhantari Y, Alimsardjono H, Xie J, Yamada T, Yanagisawa M, Matsuo M. Constitutive lysosomal targeting and degradation of bovine endothelin-converting enzyme-1a mediated by novel signals in its alternatively spliced cytoplasmic tail. J Biol Chem. 1999;274:1509–18. doi: 10.1074/jbc.274.3.1509. [DOI] [PubMed] [Google Scholar]
- 78.Mzhavia N, Pan H, Che FY, Fricker LD, Devi LA. Characterization of endothelin-converting enzyme-2. Implication for a role in the nonclassical processing of regulatory peptides. J Biol Chem. 2003;278:14704–11. doi: 10.1074/jbc.M211242200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pearson HA, Peers C. Physiological roles for amyloid beta peptides. J Physiol. 2006;575:5–10. doi: 10.1113/jphysiol.2006.111203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ. A critical function for beta-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci. 2007;27:14459–69. doi: 10.1523/JNEUROSCI.4701-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meyer-Luehmann M, Spires-Jones TL, Prada C, Garcia-Alloza M, de Calignon A, Rozkalne A, Koenigsknecht-Talboo J, Holtzman DM, Bacskai BJ, Hyman BT. Rapid appearance and local toxicity of amyloid-beta plaques in a mouse model of Alzheimer's disease. Nature. 2008;451:720–4. doi: 10.1038/nature06616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bacskai BJ, Kajdasz ST, Christie RH, Carter C, Games D, Seubert P, Schenk D, Hyman BT. Imaging of amyloid-beta deposits in brains of living mice permits direct observation of clearance of plaques with immunotherapy. Nat Med. 2001;7:369–72. doi: 10.1038/85525. [DOI] [PubMed] [Google Scholar]
- 83.Bacskai BJ, Kajdasz ST, McLellan ME, Games D, Seubert P, Schenk D, Hyman BT. Non-Fc-mediated mechanisms are involved in clearance of amyloid-beta in vivo by immunotherapy. J Neurosci. 2002;22:7873–8. doi: 10.1523/JNEUROSCI.22-18-07873.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Janus C, Pearson J, McLaurin J, Mathews PM, Jiang Y, Schmidt SD, Chishti MA, Horne P, Heslin D, French J, Mount HT, Nixon RA, Mercken M, Bergeron C, Fraser PE, St George-Hyslop P, Westaway D. A beta peptide immunization reduces behavioural impairment and plaques in a model of Alzheimer's disease. Nature. 2000;408:979–82. doi: 10.1038/35050110. [DOI] [PubMed] [Google Scholar]
- 85.Morgan D, Diamond DM, Gottschall PE, Ugen KE, Dickey C, Hardy J, Duff K, Jantzen P, DiCarlo G, Wilcock D, Connor K, Hatcher J, Hope C, Gordon M, Arendash GW. A beta peptide vaccination prevents memory loss in an animal model of Alzheimer's disease. Nature. 2000;408:982–5. doi: 10.1038/35050116. [DOI] [PubMed] [Google Scholar]
- 86.Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T, Hu K, Huang J, Johnson-Wood K, Khan K, Kholodenko D, Lee M, Liao Z, Lieberburg I, Motter R, Mutter L, Soriano F, Shopp G, Vasquez N, Vandevert C, Walker S, Wogulis M, Yednock T, Games D, Seubert P. Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature. 1999;400:173–7. doi: 10.1038/22124. [DOI] [PubMed] [Google Scholar]
- 87.Akiyama H, McGeer PL. Specificity of mechanisms for plaque removal after A beta immunotherapy for Alzheimer disease. Nat Med. 2004;10:117–8. doi: 10.1038/nm0204-117. author reply 118–9. [DOI] [PubMed] [Google Scholar]
- 88.Nicoll JA, Wilkinson D, Holmes C, Steart P, Markham H, Weller RO. Neuropathology of human Alzheimer disease after immunization with amyloid-beta peptide: a case report. Nat Med. 2003;9:448–52. doi: 10.1038/nm840. [DOI] [PubMed] [Google Scholar]
- 89.Kleywegt GJ, Jones TA. Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr D Biol Crystallogr. 1994;50:178–85. doi: 10.1107/S0907444993011333. [DOI] [PubMed] [Google Scholar]
- 90.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 91.Bentley G, Dodson E, Dodson G, Hodgkin D, Mercola D. Structure of insulin in 4-zinc insulin. Nature. 1976;261:166–8. doi: 10.1038/261166a0. [DOI] [PubMed] [Google Scholar]
- 92.Torres AM, Forbes BE, Aplin SE, Wallace JC, Francis GL, Norton RS. Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions. J Mol Biol. 1995;248:385–401. doi: 10.1016/s0022-2836(95)80058-1. [DOI] [PubMed] [Google Scholar]
- 93.Cooke RM, Harvey TS, Campbell ID. Solution structure of human insulin-like growth factor 1: a nuclear magnetic resonance and restrained molecular dynamics study. Biochemistry. 1991;30:5484–91. doi: 10.1021/bi00236a022. [DOI] [PubMed] [Google Scholar]
- 94.Sticht H, Bayer P, Willbold D, Dames S, Hilbich C, Beyreuther K, Frank RW, Rosch P. Structure of amyloid A4-(1–40)-peptide of Alzheimer's disease. Eur J Biochem. 1995;233:293–8. doi: 10.1111/j.1432-1033.1995.293_1.x. [DOI] [PubMed] [Google Scholar]
- 95.Fairbrother WJ, McDowell RS, Cunningham BC. Solution conformation of an atrial natriuretic peptide variant selective for the type A receptor. Biochemistry. 1994;33:8897–904. doi: 10.1021/bi00196a006. [DOI] [PubMed] [Google Scholar]
- 96.He XL, Dukkipati A, Garcia KC. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J Mol Biol. 2006;361:698–714. doi: 10.1016/j.jmb.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 97.Chesneau V, Rosner MR. Functional human insulin-degrading enzyme can be expressed in bacteria. Protein Expr Purif. 2000;19:91–8. doi: 10.1006/prep.2000.1217. [DOI] [PubMed] [Google Scholar]
- 98.Misbin RI, Almira EC. Degradation of insulin and insulin-like growth factors by enzyme purified from human erythrocytes. Comparison of degradation products observed with A14- and B26-[125I]monoiodoinsulin. Diabetes. 1989;38:152–8. doi: 10.2337/diab.38.2.152. [DOI] [PubMed] [Google Scholar]
- 99.DeLano W. The PyMOL Molecular Graphics System. DeLano Scientific; Palo Alto, CA, USA: 2002. [Google Scholar]