Abstract
The roles of the thicknesses and grafting densities of the surface biopolymers of four multi-drug resistant (MDR) Escherichia coli bacterial strains that varied in their biofilm formation in controlling cellular elasticities after exposure to ampicillin were investigated using atomic force microscopy. Exposure to ampicillin was carried out at minimum inhibitory concentrations for different duration times. Our results indicated that the four strains resisted ampicillin through variable mechanisms. Strain A5 did not change its cellular properties upon exposure to ampicillin and as such resisted ampicillin through dormancy. Strain H5 increased its biopolymer brush thickness, adhesion and biofilm formation and kept its roughness, surface area and cell elasticity unchanged upon exposure to ampicillin. As such, this strain likely limits the diffusion of ampicillin by forming strong biofilms. At three hours’ exposure to ampicillin, strains D4 and A9 increased their roughness, surface areas, biofilm formation, and brush thicknesses and decreased their elasticities. Therefore, at short exposure times to ampicillin, these strains resisted ampicillin through forming strong biofilms that impede ampicillin diffusion. At eight hours’ exposure to ampicillin, strains D4 and A9 collapsed their biopolymers, increased their apparent grafting densities and increased their cellular elasticities. Therefore, at long exposure times to ampicillin, cells utilized their higher rigidity to reduce the diffusion of ampicillin into the cells. The findings of this study clearly point to the potential of using the nanoscale characterization of MDR bacterial properties as a means to monitor cell modifications that enhance “phenotypic antibiotic resistance”.
Keywords: AFM, ampicillin, biopolymer thickness and grafting density, elasticity, multidrug resistance (MDR)
1. Introduction
Multidrug-resistant (MDR) bacteria are bacterial cells which confer “resistance to one or more antimicrobial agent” (Rzewuska et al., 2015). The increased resistance of these bacteria to nearly all the available antibiotics on the market as well as the emergence of new resistant bacterial strains gained them interest over the years (Spellberg et al., 2011; van Duin and Paterson, 2016). Bacterial infections resulting from these resistant bacterial strains tend to be more difficult to treat and about 1.6-fold as costly compared to infections caused by susceptible bacterial strains (Spellberg et al., 2011). Synthetic and natural antibiotics provide the first line of treatment for MDR bacterial infections (Perry et al., 2009). Among antibiotics, β-lactams have been extensively used against most bacterial infections. Due to their widespread use, Gram-negative bacteria such as Escherichia coli developed complex means to resist these antibiotics (Shaikh et al., 2015). Research has shown that bacterial resistance to antibiotics is encoded in genetics mechanisms; several of which involve the production of hydrolyzing enzymes involved in the hydrolysis of β-lactam ring, modification of the target sites to antibiotics such as changes in the penicillin-binding protein (PBP) resulting in decreased affinity to β-lactams, production of efflux pumps on the cell membrane to expel antibiotics outside the cell, and production of key extracellular polymeric substances (EPS) that affect the formation of biofilms capable of impeding the diffusion of antibiotics into biofilms (Aeschlimann, 2003; Babic, Hujer and Bonomo, 2006; Yang et al., 2006; Mohanty et al., 2012; Read and Woods, 2014; Fruci and Poole, 2016). These complex and interdependent mechanisms employed by resistant bacterial cells to withstand antibiotics are held responsible for the current challenges faced in controlling their infections (Aeschlimann, 2003; Babic, Hujer and Bonomo, 2006; Yang et al., 2006; Mohanty et al., 2012; Read and Woods, 2014; Fruci and Poole, 2016).
For β-lactam antibiotics to be effective against bacterial infections, they must gain entrance into the cell cytoplasm and initiate their actions. Such entrance can be initiated through surface interactions to bind to their target sites such as proteins including PBPs (Holtje, 1998), DNA (Drlica et al., 2008), RNA (Zeng and Xie, 2014) and other cellular organelles (Aeschlimann, 2003). Studying how a bacterial cell and its surface biopolymers interact with antibiotics can be very challenging due to the complex and heterogeneous nature of the bacterial cell surface (van der Mei and Busscher, 2012). The cell wall of Gram-negative bacteria consists of two distinct layers, an inner membrane (IM) and an outer membrane (OM) separated by a periplasmic space with abundant gelatinous substances (Costerton and Cheng, 1974). The IM contains phospholipids and a thin layer of peptidoglycans (PGs) which protects the cell membrane from the possible rupture that may be caused by the high internal osmotic pressure as well as control the integrity and morphology of the cell (Costerton and Cheng, 1974; Vollmer and Seligman, 2010). PGs in the cell wall are the major targets for β-lactams through binding to PBPs (Jacoby and Medeiros, 1991; Holtje, 1998; Bonnet, 2004) and the modification of PBPs contributes to the development of resistance to various β-lactams (Ghuysen, 1994; Vollmer and Seligman, 2010). The OM consists of surface proteins, phospholipids, and lipopolysaccharides (LPS) (Costerton and Cheng, 1974; Vollmer and Seligman, 2010). Numerous strains of Gram-negative bacteria are ornamented with negatively charged LPS, a complex matrix of macromolecules that consist of O-polysaccharides and lipid A (Adams et al., 2014; Miller, 2016). LPS functions as an impermeable barrier against toxic compounds such as antibiotics whose targets are beyond the outer membrane (Miller, 2016). Overcoming this barrier allows for the β-lactams to access PGs and affect the cellular components (Miller, 2016).
Investigations of the antibacterial activity of β-lactams on susceptible bacterial strains using biochemical techniques (Kumarasamy et al., 2010; El Seedy et al., 2017), scanning electron microscope (SEM), (Hartmann et al., 2010; Napiroon et al., 2017), and transmission electron microscope (TEM) (Hartmann et al., 2010; Bæk et al., 2014; Napiroon et al., 2017) is well documented in the literature. For example, biochemical techniques such as polymerase chain reaction (PCR) could assess specific resistance genes that may be expressed in antibiotic-resistant bacteria upon exposure to antibiotics (Fluit and Schmitz, 2001; Kumarasamy et al., 2010). The use of PCR has confirmed many potential antibiotic-resistant genes and their concomitant mechanisms (Kumarasamy et al., 2010; Ali et al., 2014; El Seedy et al., 2017). Despite the huge potentials, genetic methods provide in exploring antibiotic resistance, there have few limitations. These include: (1) the need for large samples of organisms to provide reliable sensitivity; (2) lots of samples are discarded in the medium during the RNA preparation; and (3) each antibiotic to be investigated requires an individual assay (Cockerill, 1999). In comparison, both SEM and TEM could directly depict the bacterial cell envelope and clearly show morphological changes in the single cell and cell colonies in response to antibiotics (Hartmann et al., 2010; Bæk et al., 2014; Napiroon et al., 2017). However, despite the high resolution of these imaging techniques, they require multiple sample preparation steps before and after exposure to antibiotics that can dry the bacterial cell and induce damage to the cell membrane leading to misinterpretation of bacterial surface structure change resulting from exposure to antibiotics (Beniac et al., 2014; Golding et al., 2016). Furthermore, visualizing cells in their physiological environment is almost impossible with the SEM and TEM because wet samples tend to reduce the contrast and the resolution of the microscopes (Golding et al., 2016). Atomic force microscope (AFM), on the other hand, has provided new potentials to probe bacterial cell surface changes in response to antibiotics and their related agents under physiological conditions (Meincken and Rautenbach, 2005), (Uzoechi and Abu-Lail). Through simultaneous imaging and quantification of variable bacterial functions such as adherence, hydrophobicity, elasticity, and biopolymer brush thickness and grafting density, AFM is unique in its abilities to provide structure-function relationships (Braga and Ricci, 1998; Camesano et al., 2000; Abu-Lail, 2003; Perry et al., 2009; Neethirajan and DiCicco, 2014). Investigations of how MDR bacterial cells change their surface biopolymers’ conformational properties including grafting density and thickness, adhesion, membrane permeability, roughness, and biofilm formation to resist β-lactams will be essential to developing effective antibiotics for the clinical treatment of bacterial infections as well as for expanding the scope of our understanding of bacterial resistance to antibiotics.
Many AFM studies in the literature have investigated how β-lactams induce various membrane disruptions to the surfaces of susceptible E. coli strains (Braga and Ricci, 1998; Yang et al., 2006; Perry et al., 2009; Laskowski et al., 2018). For example, Perry et al compared the effects of ampicillin on the membrane ultrastructure and elasticity of a susceptible E. coli strain to that of untreated cells (Perry et al., 2009). This study reported a decrease in Young’s modulus of E. coli from 2.2 ± 0.09 MPa to 1.1 ± 0.13 MPa after exposure to ampicillin at 25 μg/ml (Perry et al., 2009). Yang et al used AFM to study various effects of amoxicillin and penicillin on bacterial cell envelope (Yang et al., 2006). It was found that both amoxicillin and penicillin-induced nanopore damage to the cell membrane of susceptible E. coli, but the level of damage caused by amoxicillin was greater than that of penicillin. Amoxicillin is a semisynthetic β-lactam while penicillin is a pure synthetic antibiotic (Yang et al., 2006). The chemical structure of amoxicillin protects the β-lactam’s ring and broaden its the antimicrobial spectrum (Yang et al., 2006). The studies above, even though they were limited to susceptible and not resistant bacterial strains, demonstrate the potential of AFM to be used as a tool to investigate morphological and mechanical changes to resistant bacterial cells upon exposure to β-lactams. (Braga and Ricci, 1998; Yang et al., 2006; Perry et al., 2009; Neethirajan and DiCicco, 2014; Laskowski et al., 2018).
Here, the effects of a model β-lactam (ampicillin) on the conformational properties of the bacterial surface biopolymers and how such conformations affect the elasticities of resistant E. coli cells was investigated by AFM under water. In our previous study, the effects of ampicillin on the morphology, growth rates, permeability, hydrophobicity, nanoscale adhesion to model surfaces and biofilm formation of the same E. coli strains used here were investigated. The findings from this study combined with those from our previous study were utilized to suggest possible mechanisms through which MDR cells can develop resistance to ampicillin.
2. Materials and Methods
2.1. Chemicals and Materials
Domestic multidrug-resistant Escherichia coli (MDR-E. coli) strains arbitrarily labeled as A5, D4, A9, and H5 were obtained from Prof. Douglas R. Call of the Paul G. Allen School of Global Animal Health, Washington State University. 100 mg/ml ampicillin and gelatin G2500–100G were acquired from Sigma-Aldrich, St. Louis, MO while Luria-Bertani (LB) broth and agar were obtained from RPICorp, IL. The LB broth was prepared by dissolving 10 g of NaCl, 5 g of yeast extract and 10 g of tryptone in 1 L of deionized (DI) water per manufacturer’s instructions.
2.2. Choice of MDR E. coli as a Bacterial Model
E. coli was selected for this study as the model bacterium because of the following four reasons. First, it is well investigated in the literature for its membrane properties and composition, genetics and MDR responses; the fact that will enable us to complement our findings to existing literature (Braga and Ricci, 1998; Yang et al., 2006; Perry et al., 2009; Neethirajan and DiCicco, 2014; Laskowski et al., 2018). Second, MDR E. coli are commonly found in a wide range of infections that range from curable to fatal (Nikaido, 2009). These include urinary tract infection, gastroenteritis, meningitis, septicemia, and peritonitis (Von Baum and Marre, 2005; Tadesse et al., 2012). Third, E. coli possess an impermeable OM to antibiotics that distinguishes them from Gram-positive bacteria (Torres and Kaper, 2005; Nikaido, 2009; Wang et al., 2015). Fourth, these strains vary in their abilities to form biofilms and to resist multiple antibiotics including our model antibiotic choice in this study (ampicillin). Upon exposure to antibiotics, the variations above are expected to result in E. coli surface modifications that can be quantified by AFM (Braga et al., 1998; Meincken and Rautenbach, 2005; Perry et al., 2009).
2.3. Choice of the Model Antibiotic-Ampicillin
Ampicillin was chosen as a model β-lactam antibiotic to use in this study. In this group of antibiotics, the side chain in the structure regulates the antimicrobial spectrum and pharmacokinetic properties of the compound (Filho et al., 2010). The β-lactam ring in the ampicillin core structure is responsible for the antibacterial function via the inhibition of the PBP [Figure S2D]. PBPs synthesize the PGs that form the rigid cell walls of bacteria such as E. coli (Ghuysen, 1994). As a result of exposure to ampicillin at a minimum inhibitory concentration (MIC), we hypothesize that conformational changes to the biopolymers making up the bacterial OM will take place. Such changes can be quantified with AFM in the liquid environment (Abu-Lail, 2003; Ramezanian et al., 2018).
2.4. Determination of Bacterial MDR and Minimum Inhibitory Concentration (MIC)
The MDR was characterized with standardized known concentrations of ampicillin (32 μg/ml) using Mueller Hinton agar (Jenkins and Schuetz, 2012). MICs were determined in planktonic cultures using a broth dilution test as established in The Clinical & Laboratory Standards Institute (CLSI) guidelines (Turnidge and Paterson, 2007). Briefly, three colonies from each strain were taken from three LB plates, inoculated into 5 ml LB broth and incubated for 24 hours at 37°C with shaking at a 150 rpm. Culture tubes containing two-fold diluted concentrations of ampicillin ranging from 0.2 to 400 μg/ml in LB media were prepared in triplicates. Each tube with a given concentration medium was then inoculated with a 100 μL of MDR-E. coli cells’ suspension grown until the late exponential phase of growth (optical density measured at a wavelength of 600 nm (OD600) = 0.5) (Mazzola et al., 2009). After an overnight incubation, turbidity in the test tubes was used as an evidence of visible cell growth (Turnidge and Paterson, 2007). The lowest concentration of the antibiotic such as ampicillin needed to prevent cell growth was used as the MIC for the subsequent experiments (Turnidge and Paterson, 2007). To verify the MIC obtained, the cultures from the test tubes were inoculated into fresh LB medium without ampicillin (n = 3). MIC was confirmed as the concentration of the test tube from which cells were not growing. MDR-E. coli strains were found resistant to ampicillin at MICs 50 μg/ml for E. coli A5 and H5 and 45 μg/ml of E. coli D4 and A9 (Uzoechi and Abu-Lail).
2.5. Bacterial Growth Kinetics
Four MDR-E. coli strains were cultured overnight in LB broth at 37 °C with continuous shaking at 150 rpm until the cells reached the late exponential phase of growth (Braga and Ricci, 1998; Cui et al., 2012; Longo and Kasas, 2014). Subsequently, the four MDR-E. coli strains were subcultured into a fresh LB medium at 1:100 dilution supplemented with ampicillin at different MICs for 24 hours. Cultures with no ampicillin supplemented with the LB broth served as the negative control (untreated). All experiments were carried in triplicates and were prepared with independent colonies. The OD600 was measured to determine the viability of the cells at variable time points using a UV/Vis spectrophotometer, Thermo Spectronic, USA. Specifically, the two strains treated with 45 μg/ml, D4 and A9 showed two exponential phases followed by a stationary phase that coincided with data collected for the strains in the absence of ampicillin. Conversely, the two strains treated with 50 μg/ml, A5 and H5, showed typical growth curves with no indications of inhibition zones. Because of that, in further investigations, strains were only tested after 3 hours of growth in their stationary phase. Growth curves constructed for all strains were previously published. A summary of the main characteristics of these strains as quantified from these growth curves is provided in our previous publication.
2.6. Preparation of Gelatin Coated Mica Substrates for AFM Studies
E. coli has a negatively charged surface and the mica used here to attach the bacteria to for the AFM studies is also negatively charged when unmodified (Allison et al., 2011). This will render the immobilization of E. coli on mica very challenging due to the expected electrostatic repulsion between the cell surface and the substrate (Doktycz et al., 2003; Allison et al., 2011). To solve this problem, the mica surface was coated with a positively charged gelatin layer capable of electrostatically attracting the bacterial cells for improved adhesion. Note that gelatin is a water-soluble polypeptide made from hydrolysis of collagen (Visai and Bozzini, 1990). Studies have shown that gelatin is non-immunogenic, biocompatible and biodegradable (Lonergan and Sullivan, 2014). Furthermore, gelatin has polar groups, such as carboxyl and amines which facilitate binding to the receptors on the bacterial surface (Rose et al., 2014). A 12 mm diameter mica disc (Ted Pella, Inc, Redding, CA, USA) was cleaved several times and hydrated in deionized (DI) water and dried for 1 hour to obtain a flat surface (Doktycz et al., 2003). A 0.5 % (w/v) gelatin solution was prepared by dissolving 0.5 g of powder gelatin from the porcine skin in a 100 mL of deionized hot water and allowed to cool to 60–70°C (Allison et al., 2011). Afterward, the freshly cleaved mica was gently submerged into the gelatin solution and withdrawn immediately. The gelatin-coated mica was supported on the edge of a paper towel to dry prior to use for bacterial immobilization.
2.7. AFM Experiments
Prior to AFM imaging, a 1 ml bacterial suspension cultured in LB broth to an OD600nm between 0.5 – 0.7 in the presence or absence of ampicillin was pelleted by centrifugation at 5000g for 5 minutes and washed three times with DI water. The pellet was then re-suspended in a 500 μL DI water. A 100 μL of this suspension was dropped onto freshly prepared gelatin-coated mica prepared following previously published protocols (Park and Abu-Lail, 2011). The sample was then incubated at room temperature for 10 minutes in DI water and rinsed with DI water to remove any loosely attached bacterial cells. Per strain, three gelatin-coated mica substrates were prepared, and bacteria attached to them were taken from three different cultures grown with independent colonies. For all strains investigated, 15 cells were probed for each culture and triplicate cultures were investigated per treatment unless otherwise stated. On each bacterium, 25 points that covered the entire bacterial cell surface were mapped for force measurements. Force measurements were recorded using silicon nitride (Si3N4) cantilevers (DNP-10, Bruker Inc., Santa Barbara, CA). Prior to force measurements, the spring constant of each cantilever was determined from the power spectral density of the thermal noise fluctuations in DI water (Hutter and Bechhoefer, 1993; Park and Abu-Lail, 2011). The average spring constant was found to be 0.07 ± 0.01 N/m (n=18), close to the reported manufacturer’s value of 0.06 N/m. The deflection sensitivity was measured on a cleaned unmodified mica surface in water and found to be 45.26 ± 4.61 nm/V (n=12). All images of bacterial cells were captured at an average scan rate of 0.41± 0.03 Hz (n=12) and at a resolution of 256 samples per line. Forces on individual bacterial cells were measured in DI water with a Multimode AFM equipped with a Nanoscope IIIa controller and extender module (Bruker AXS Inc., Camarillo, CA). Since our interest is in probing the differences in physical properties such as elasticities and bacterial surface conformations of the MDR strains biopolymers in response to ampicillin, typical AFM Si3N4 cantilevers were used. Approach and retraction force-distance curves were collected under tapping mode to avoid any bacterial surface damage caused by the lateral force exerted by the AFM cantilever (Camesano et al., 2000). Here, only approach curves where analyzed for quantifying the cell elasticity, biopolymer brush thickness, and grafting density respectively. In our previous publication, retraction curves were analyzed. To confirm that the tip was not contaminated during AFM measurements, force measurements were made on a bacteria-free area of the gelatin-coated mica disk before and after making the measurement on a bacterial cell (Park and Abu-Lail, 2011). Equality of the measurements ensured that the tip properties had not been altered by contact with the bacterial surface biopolymers (Park and Abu-Lail, 2011), [Figure S1].
2.8. Determination of the Length and Grafting Density of the Bacterial Surface Biopolymer Brush using the Steric Model
The repulsive steric interaction forces expected between the negatively charged MDR E. coli cell surface biopolymers and the negatively charged model surface of the AFM Si3N4 cantilever and measured by AFM in the approach data were fitted to a steric model (Alexander, 1977; Butt et al., 1999; Camesano et al., 2000; Abu-Lail, 2003). The steric model describes the conformational properties of the bacterial surface biopolymers in terms of two fitting parameters; these are the brush thickness (Lo/nm) and the grafting density per unit area (Γ/m−2). The steric force, Fst (nN), in this model, is described by:
| (1) |
Where KB is Boltzmann’s constant (1.3801 ×10−23 J/K), T is absolute temperature (298 K), R is the tip radius (40 nm) taken as reported by the manufacturer, L0 and Γ are the thickness (nm) and the grafting density (m−2) of the bacterial surface biopolymer, respectively, and h is the separation distance between the AFM tip and bacterium surface (nm). In equation (1), L0 and the Γ are the two fitting parameters (Camesano et al., 2000; Abu-Lail, 2003). For each MDR E. coli strain investigated, a total of 45 cells (15 cells for each of the triplicate cultures investigated) were investigated unless otherwise stated. Each approach curve was fitted individually to the steric model represented in equation (1), [Figure 1 and Figure S2A–C] due to the observed heterogeneity in the data. Histograms representing how the thickness of the biopolymer brush thickness (nm), and the grafted polymer density (m−2) values varied among strains upon exposure to ampicillin in comparison to the untreated cells (control) were then constructed [Figure 3 and Figure 4]. The histograms were then fitted to either the Gaussian or the log-normal peaks dynamic functions with two parameters as was previously described.
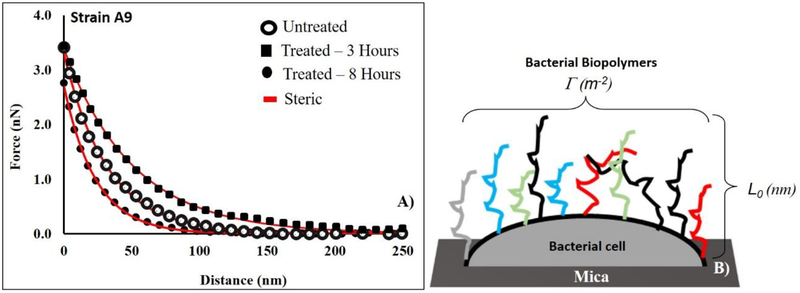
Figure 1:
A comparison between the approach curves collected for untreated E. coli A9 cells and treated ones with ampicillin for 3 and 8 hours at MIC, respectively. Each curve represents the average of five individual curves. Red lines represent the fits of the steric model to the experimental symbolic data. For the untreated data, L0 and Γ were 286 ± 140 nm and 11035 ± 1670 μm−2 respectively, r2= 0.99. For the treated data, L0 and Γ were 403 ± 95 nm and 29496 ± 7880 μm−2 for 3 hours exposure respectively, r2= 0.97. The L0 and Γ for 8 hours exposure was 267 ± 51 nm and 14519 ± 2001 μm−2 respectively, r2= 1.00. (B) A schematic representation of the conformations of the bacterial surface biopolymers showing the biopolymer brush thickness (Γ, μm−2) and the grafting density (Γ, μm−2) respectively. Different colors used for biopolymer chains are meant to represent the heterogeity in these biopolymers. Steric model results for individual cells at different conditions are reported in Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
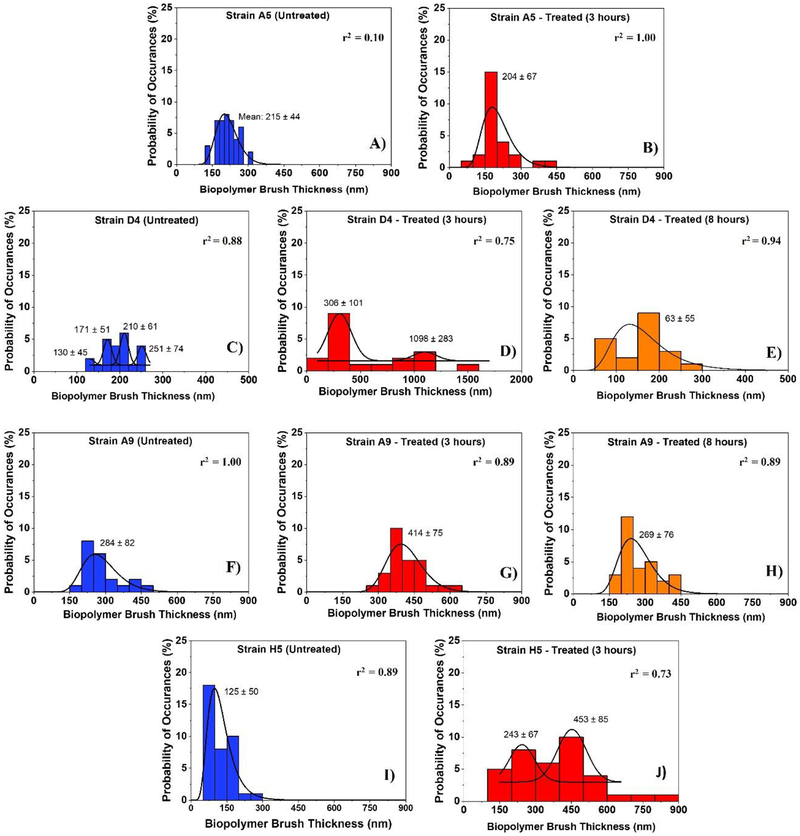
Figure 3:
Histograms showing the distributions of biopolymer brush thicknesses of the MDR-E. coli strains estimated using the steric model for untreated cells (negative control) and for treated ones at different time points (3 and 8 hours). Solid lines show the dynamic peak function model fits the data presented in the histograms (Origin v9.4, OriginLab Corp., Northampton, MA). Lognormal peak function was used for unimodal distributions while the Gaussian peak function was used to fit the multimodal distributions (Arslan et al., 2018). Peaks are reported in nm. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
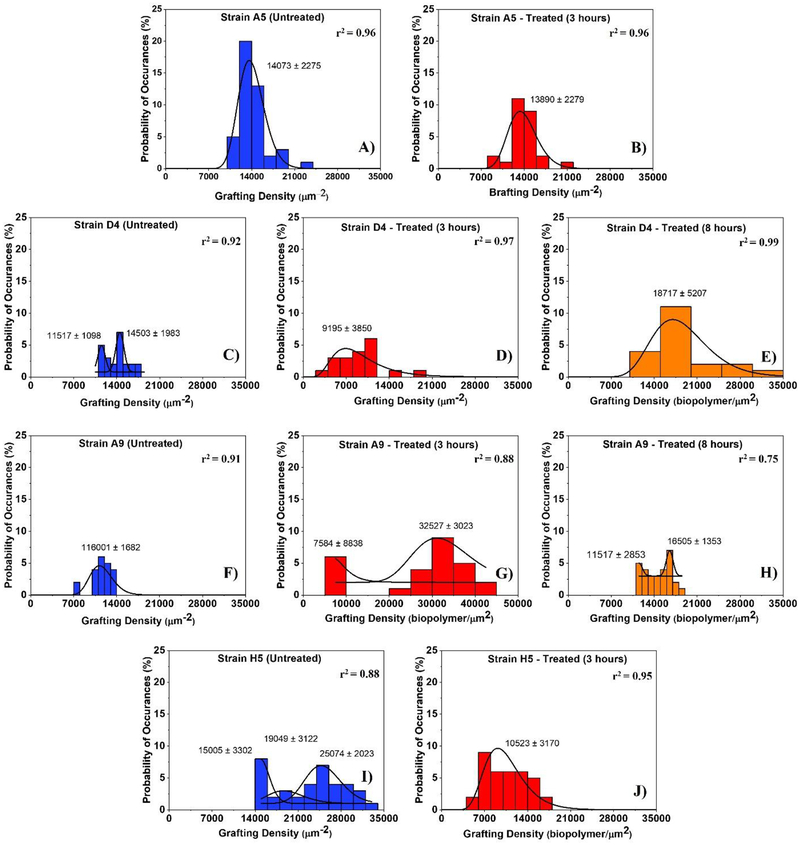
Figure 4:
Histograms showing the distributions of biopolymer grafting densities for MDR-E. coli strains estimated using the steric model for untreated (control) and treated cells at different time points (3 and 8 hours exposure to ampicillin). Solid lines show the dynamic peak function model fits the data presented in the histograms (Origin v9.4, OriginLab Corp., Northampton, MA). Lognormal peak function was used for unimodal distributions while the Gaussian peak function was used to fit the multimodal distributions (Arslan et al., 2018). Peaks are reported in μm−2. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a. Figures like A for all strains and treatments are provided in the supporting material Figure S2 A–C. Figure 1B is adopted from Abu-Lail et al 2003 (Abu-Lail, 2003) with modifications.
Table 1:
A summary of the estimated values for the biopolymer brush thickness (nm), biopolymer grafting density (μm2) and cellular elasticity (MPa) for the untreated cells (NT) and for cells treated with ampicillin for 3 or 8 hours at MICs. The quality of the Hertz or steric models fit the data is provided using the r2 mean values.
| Strain | MIC | TE | a Steric Model Results | b Hertz Model Results | |||||
|---|---|---|---|---|---|---|---|---|---|
| ID | (μg/ml) | (hrs) | n | L0 (nm) | Γ (μm−2) | r2 | E (MPa) | δ (nm) | r2 |
| D4 | 45 | 3 | 37 | 612 ± 290* | 8886 ± 3511* | 0.97 | 0.09 ± 0.03* | 151 ± 32* | 0.97 |
| 45 | 8 | 44 | 169 ± 64* | 18505 ± 5569* | 0.97 | 0.26 ± 0.09* | 127 ± 29* | 0.98 | |
| NT | 5 | 43 | 200 ± 39 | 13840 ± 1726 | 1.00 | 0.13 ± 0.04 | 138 ± 28 | 0.98 | |
| A9 | 45 | 3 | 32 | 403 ± 95* | 29496 ± 7880* | 0.95 | 0.10 ± 0.05* | 221 ± 41* | 0.97 |
| 45 | 8 | 32 | 267 ± 51* | 14519 ± 2001* | 0.91 | 0.14 ± 0.04** | 144 ± 45** | 0.97 | |
| NT | 5 | 26 | 286 ± 140 | 11035 ± 1670 | 1.00 | 0.13 ± 0.05 | 147 ± 48 | 0.98 | |
| A5 | 50 | 3 | 33 | 204 ± 77 | 13890 ± 2274 | 0.99 | 0.19 ± 0.05 | 113 ± 28* | 0.98 |
| NT | 5 | 44 | 215 ± 50 | 14056 ± 2092 | 0.99 | 0.18 ± 0.05 | 134 ± 39 | 0.97 | |
| H5 | 50 | 3 | 40 | 385 ± 14* | 10602 ± 3000* | 1.00 | 0.43 ± 0.13 | 88 ± 20* | 0.98 |
| NT | 5 | 39 | 128 ± 81 | 22637 ± 5487 | 1.00 | 0.37 ± 0.10 | 113 ± 25 | 0.98 | |
Values are statistically significant (99% confidence) from both the untreated (negative control) and the treatment group.
Values are statistically significant (99% confidence) from the treatment group but not from the negative control, p < 0.001. The values are the means ± standard deviations of average measurements performed on 26 to 44 cells obtained from three independent cultures. NT means cells grown in LB without ampicillin supplementation, TE represents the time of exposure (hours), E (MPa) represents the average estimated cellular elasticity, δ(nm) is the indentation distance corresponding to 1 nN indentation force, n is the number of individual cells probed in each condition, L0 (nm) and Γ (μm−2) represent the thickness and the grafting density of the bacterial surface biopolymer brush layer estimated from the steric model, respectively.
2.9. Determination of Young’s Modulus of the bacterial cell
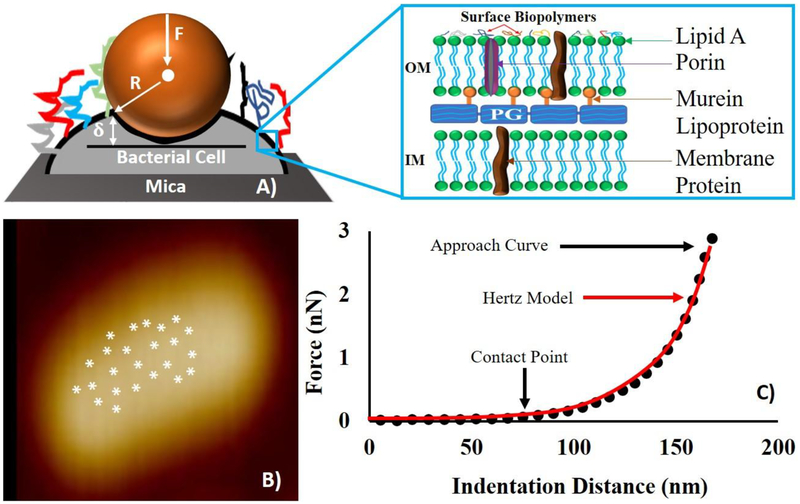
Twenty-five force measurements were performed away from the edges on the bacterial surface [Figure 2B]. After deciding on the location of the contact point (CP) for each force-distance curve, a maximum force of 1 nN was used as the force at which indentation was measured throughout the analysis [Figure 2C]. It is worth mentioning that the CP is the point on the AFM approach curve where the Si3N4 tip first makes a contact with the cell surface before indenting the cell wall [Figure 2C] (Gavara, 2016). The Elastic modulus was estimated from applying the Hertz’s model of contact mechanics to the force-indentation data [Figure 2C]. The Hertz model takes into account the effects of the sample properties and tips shape and dimensions onto the estimated Young’s modulus (Camesano and Logan, 2000; Abu-Lail, 2003; Perry et al., 2009; Vadillo-Rodriguez and Dutcher, 2009). According to the Hertz’s model, the loading force (F) applied by the AFM non-deformable AFM tip required to indent a distance (δ) of a deformable elastic half-space of the bacterium surface is represented by equation 2, Figure 2A.
Figure 2:
(A) Schematic (not drawn to scale) illustrating the nanoindentation of the AFM cantilever through the bacterial surface biopolymers. In the schematic, (δ) represents the indentation distance, F represents the loading force and R is the radius of the AFM tip, and the inset depicts the common structure of a Gram-negative bacterial cell envelope. OM and IM represent the bacterial outer and inner membranes respectively. (B) Shows how the 25 points were distributed onto the surface of a representative cell far from cell edges for force measurements. (C) A representative force-indentation curve depicting the portion of the curve (solid circled line) that was fitted to the Hertz model of contact mechanics (red line).
| (2) |
In equation (3), E represents Young’s modulus of the bacterium cell and is the only fitting parameter in equation 2, R depicts the AFM tip sphere radius which acts as the indenter and provided by the manufacturer as 40 nm and v is the Poisson ratio taken as 0.5 for biological materials (Camesano and Logan, 2000; Abu-Lail, 2003; Perry et al., 2009; Vadillo-Rodriguez and Dutcher, 2009). Both F and δ are measured by AFM. Hertz model was fitted to the experimental data manually using the AFM Nanoscope Analysis 1.5 software (Bruker, Camarillo, CA) (Park and Abu-Lail, 2011; Ramezanian et al., 2018). The results are reported in Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1b. Each curve was considered individually due to the heterogeneity observed among the force-indentation profiles measured. The Young’s modulus values estimated from all approach curves under a given treatment were then averaged for comparison purposes. Histograms representing how Young’s modulus values varied among strains upon exposure to ampicillin in comparison to the control were then constructed [Figure 5]. The histograms were then fitted to log-normal peak dynamic function as was previously described. Peaks are reported in MPa.
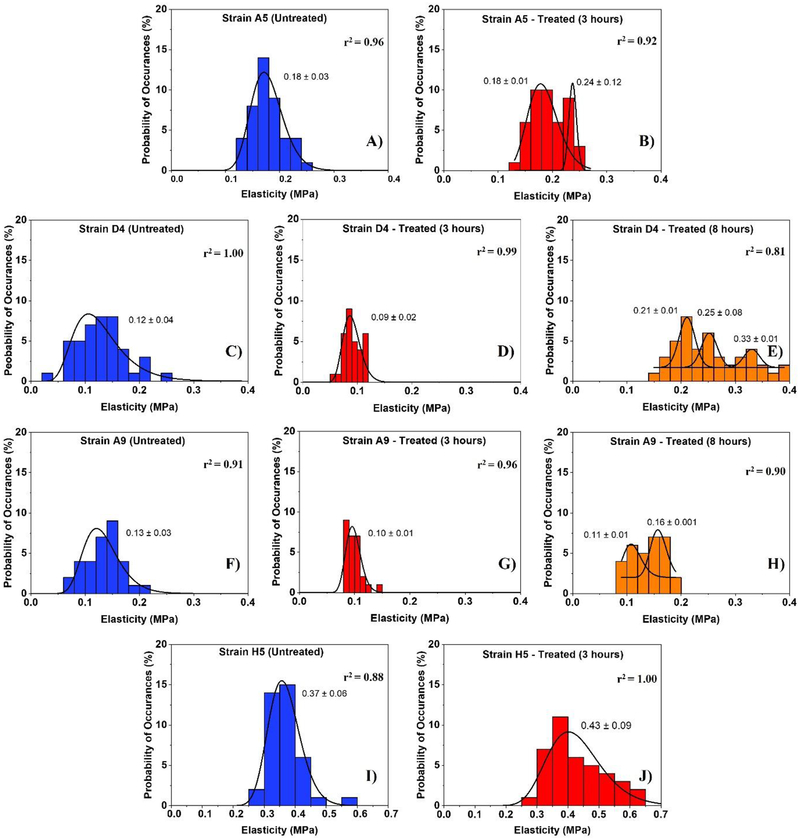
Figure 5:
The distribution of cellular elasticities estimated from applying the Hertz model (Equation 2) to the force-indentation data. Solid lines show the dynamic peak function model fits the data presented in the histograms (Origin v9.4, OriginLab Corp., Northampton, MA). The lognormal dynamic peak function model was used to fit unimodal distributions while the Gaussian dynamic peak function model was used to fit multimodal distributions. Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Hertz model is appropriate to model force-indentation data when the forces between the contact surfaces are insignificant compared to the maximum load (Gilbert et al., 2007). What this implies is that if adhesion is present in the approach curves, the model will not apply to the data. This was not the case in this study. All AFM approach curves measured between the bacterial surface biopolymers and the AFM tip demonstrated mainly repulsive forces, hence adhesion effects on Young’s modulus can be ignored (Ramezanian et al., 2018). Furthermore, the Hertz model does not account for the substrate effects on the estimated Young’s modulus. Here, the average indentation distance (~140 ± 5.00 nm) measured on the bacterial cell is much smaller than the average bacterial thickness (~733 ± 0.01 nm). Due to that, the effect of the beneath subtract on estimated Young’s modulus can be ignored (Ramezanian et al., 2018).
Finally, details of how the conformations of biopolymer brushes as well as cell elasticities can be distinguished from the same approach curve can be found in section S.3 in the supporting information along with a Figure S.3 that demonstrates such differences.
3. Statistical Analysis
Variations between the ampicillin treated samples and their corresponding untreated samples (controls), as well as variations among strains, in steric parameters or Young’s moduli, were determined using Dunn’s Test based on One Way Analysis of Variance (ANOVA) available in Sigma Plot 11.0 (Systat Software Inc., San Jose, CA). Because the sizes of the treatment groups were not always equal, Dunn’s method was used to compare the groups of interest. Statistical significance was considered at 99% confidence interval (p<0.001). Log-normal and Gaussian dynamic peak functions were used to fits the data presented in the histograms [Figure 3, Figure 4 and Figure 5] (Origin v9.4, OriginLab Corp., Northampton, MA), (Arslan et al., 2018). The goodness of Hertz and steric models’ fits to the experimental data were determined based on the estimated values of r2 in individual curves.
4. Results
Because E. coli strains D4 and A9 were characterized by an inhibition phase between growth hours 3 and 8, their elasticities and biopolymer brush conformational properties were quantified for the negative control (untreated) and for cells treated for 3 and 8 hours with ampicillin. Strains A5 and H5 showed regular growth patterns with no inhibition zones and as such, their elasticities and biopolymer brush conformational properties where quantified for the untreated (control) and for cells treated with ampicillin for at 3 hours.
4.1. Variations in the Thicknesses and Grafting Densities of Bacterial Surface Biopolymer Brushes in Response to Ampicillin’s Treatment
The ability of the steric model to fit the AFM experimental approach data was determined by an overall average r2 value of 0.99 [Table 1a]. The bacterial surface biopolymer brush thicknesses for all investigated strains ranged from 200 to 612 nm [Table 1a]. The bacterial grafting densities ranged from 8886 to 29,496 μm−2. Because both the brush thickness as well as the grafting density of biopolymer brushes spanned a range each, the distributions of their data were investigated in the forms of histograms [Figure 3 and Figure 4]. To represent all data in the histogram, the average brush thickness and the average grafting density of the bacterial surface biopolymer brushes for each of the four investigated MDR-E. coli strains were calculated (Table 1a).
Among all the strains investigated, strain D4 exhibited multimodal distribution for the thicknesses of the biopolymer brushes of the untreated cells and a bimodal distribution for the thicknesses of cells treated with ampicillin for 3 hours [Figure 3C and Figure 3D]. In comparison, strain H5 demonstrated a bimodal distribution of the brush thicknesses for cells exposed to ampicillin for 3 hours [Figure 3J]. Other strains (A9 and A5) displayed unimodal distributions for their brush thicknesses at all treatment conditions [Figure 3]. When the peaks estimated for all strains were averaged, the biopolymer brush thicknesses ranged from 125 ± 50 to 284 ± 82 nm in the absence of ampicillin [Figure 3A, 3C, 3F, and 3I], 204 ± 67 to 702 ± 192 nm for cells exposed to ampicillin for 3 hours [Figure 3B, 3D, 3G, and 3J] and from 63 ± 55 to 269 ± 76 nm for those exposed to ampicillin for 8 hours [Figure 3E and 3H]. Histograms in Figure 3 indicate that the brush thicknesses are heterogeneous. Exposure to ampicillin for three hours increased the heterogeneity of all brush thicknesses for all strains as evident from the increased widths of histograms. Exposure to ampicillin to 8 hours restored the heterogeneity levels in the brush thicknesses more or less to that observed for the untreated cells.
The summary of average brush thicknesses and grafting densities for all the strains investigated is presented in Table 1a. In E. coli strains D4 and A9, the average biopolymer brush thicknesses increased by 67% and 29% upon exposure to ampicillin for 3 hours compared to untreated cells, respectively [p<0.001, Table 1a]. When E. coli strains D4 and A9 cells exposed to ampicillin for 8 hours were compared to untreated cells, the average brush thicknesses decreased by 16% and 7%, respectively [p<0.001, Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a]. When the effects of the exposure to ampicillin for 3 and 8 hours were compared for strains D4 and A9, the average biopolymer brush thicknesses estimated were 3.6 and 1.5-fold higher at 3 hours compared to those estimated at 8 hours. In general, longer biopolymer brush thicknesses are directly proportional to higher electrosteric repulsions [Figure 1 and Figure S2]. The average biopolymer brush thickness for cells of strain A5 exposed to ampicillin for 3 hours was 1.0-fold (5%) lower than the untreated and did not show any significant change from that estimated for the untreated cells [p>0.001, Table 1a]. The biopolymer brush thickness of strain H5 was higher than the untreated with 3.0-fold change increase after 3 hours of exposure to ampicillin (p<0.001).
The histograms that show the distributions of biopolymer grafting densities for different treatment group are shown in Figure 4. Strains D4 and H5 were characterized by the presence of multiple peaks in their grafting density distributions in the absence of ampicillin (untreated) [Figure 4C and Figure 4I]. In comparison, strain A9 was characterized by a bimodal distribution or their grafting densities in the presence of ampicillin at 3 and 8 hours, respectively [Figure 4G and Figure 4H]. Strain A5 showed a unimodal distribution of the grafting densities with statistically similar peaks in all conditions [Figure 4A and Figure 4B]. When the peaks estimated for all strains were averaged, the grafting densities ranged from 13010 ± 1541 to 116001 ± 1882 μm−2 in the absence of ampicillin [Figure 4C, Figure 4A, Figure 4F and Figure 4I], 9195 ± 3850 to 20056 ± 5931 μm−2 for cells exposed to ampicillin for 3 hours [Figure 4D, Figure 4J, Figure 4B and Figure 4G] and from 14011 ± 2103 to 18717 ± 5207 μm−2 for those exposed to ampicillin for 8 hours [Figure 4H and Figure 4E]. The grafting densities were heterogeneous for all the strains investigated and these heterogeneities reflect the differences in compositions of the MDR-E. coli strains surfaces. The heterogeneity of the grafting densities for 3 hours exposure to ampicillin is consistent with our observations for the brush thicknesses distributions, the biopolymer brushes became more heterogeneous as evident from the wider histograms. Exposure to 8 hours to ampicillin resulted in a reduction in the width of the histograms almost to that observed for untreated cells. In E. coli strains D4 and A9, the average grafting densities decreased by 36% (1.0-fold) in strain D4 and increased by 61% (2.6-fold) for strain A9 upon exposure to ampicillin for 3 hours compared to untreated cells, respectively [p<0.001, Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a]. When E. coli strains D4 and A9 cells exposed to ampicillin for 8 hours were compared to untreated cells, the average grafting densities increased by 1.3-fold (25%) and 1.3-fold (21%), respectively [p<0.001, Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a]. In general, longer biopolymer brush thicknesses are directly proportional to lower grafting densities for strains D4 and A9, respectively [Table 1a]. The average grafting densities quantified for strains A5 and H5 for 3 hours exposure to ampicillin decreased by 1.0-fold (1%, p>0.001) and 2.1-fold (53%, p<0.001), respectively [Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a].
4.2. Bacterial Cellular Elasticity Variations in Response to Exposure to Ampicillin
The cellular elasticities of MDR E. coli cells were estimated from the Hertz model fits the force-indentation data (equation 4). Due to the heterogeneities in the estimated elasticities, histograms of the estimated populations were constructed as a function of ampicillin treatment [Figure 5]. The average values of cellular elasticities reported in each histogram were calculated [Table 1b]. As presented in Figure 5, strain A5 showed more than one Gaussian peak for its elasticity values at 3 hours exposure to ampicillin [Figure 5B]. Using Originlab, two separate populations were quantified. The average for the first and second populations were 0.18 ± 0.002 MPa and 0.24 ± 0.119 MPa respectively [Figure 5B]. Interestingly, the average of the first population was the same as the average of strain A5 population measured in the absence of ampicillin [Figure 5A and Figure 5B]. This suggests that a population of cells remain unaffected by the exposure to ampicillin. The cells in the second population are 1.3-fold stiffer than those in the first population and the untreated. Similar to strain A5 strains D4 and A9 exhibited multiple Gaussian peaks at 8 hours when compared to untreated cells and cells exposed to ampicillin for 3 hours [Figure 5E and Figure 5H]. Our results indicate that cells of strains D4 and A9 are softer at 3 hours exposure by 0.8-fold and 0.7-fold compared to untreated cells, respectively [Figure 5D and Figure 5G]. Treatment of strains D4 and A9 with ampicillin for 8 hours increased the elasticities of their cells by 2.2-fold and 1.0-fold when compared to the elasticity of cells in the absence of ampicillin, respectively [Figure 5E, Figure 5H and Figure 5C, Figure 5F]. When compared, the elasticities of strains D4 and A9 cells at 8 hours’ exposure to ampicillin were 2.6 and 1.5-fold stiffer compared to cells exposed to ampicillin for 3 hours. Finally, strain H5 exhibited a unimodal distribution at 3 hours’ exposure to ampicillin and showed a 1.2-fold increase in cellular elasticity compared to untreated cells [Figure 5I and Figure 5J].
A summary of the average cellular elasticity calculated for all data in each histogram presented in Figure 5 is shown in Table 1b. Strains D4 and A9 responded to ampicillin very similarly. Both strains increased their elasticities after the 3 hours’ exposure to ampicillin and went back to almost the elasticities reported for the untreated cells after exposure to ampicillin at 8 hours’ exposure to ampicillin [Table 1b]. Specifically, the average elasticity of the 3 hours’ treated D4 cells was significantly lower than that of the untreated cells by 0.04 MPa (31%, r2 = 0.98, P<0.001). When the ampicillin resistance persisted until 8 hours of growth, the elasticity of strain D4 significantly increased compared to that measured at 3 hours (66%, r2 = 0.98, p<0.001) as well as compared to that of untreated cells, (50%, r2 = 0.97, p<0.001) [Table 1b]. The average elastic moduli of cells of E. coli strains A5 and H5 treated with ampicillin at MIC were not significantly higher than those estimated for the untreated cells, 0.02 MPa (8%, r2 = 0.98, P>0.001) and 0.06 MPa (14%, r2 = 0.98, P>0.001) respectively [Table 1b]. The average elasticity of E. coli H5 cells was approximately as twice as stiff compared to all strains investigated with an elastic modulus ranging from 0.37 ± 0.10 MPa (r2 = 0.98) when grown in LB alone to 0.43 ± 0.13 MPa (r2 = 0.98) after exposure to ampicillin for 3 hours [Table 1b].
5. Discussion
This study aimed at investigating how MDR-E. coli strains modify their mechanical properties represented in terms of the bacterial surface biopolymer brushes’ thicknesses and grafting densities as well as bacterial cells’ elasticities upon exposure to ampicillin for different time durations at their respective MICs. To our knowledge, very few studies in the literature investigated the effects of antibiotics (Perry et al., 2009; G. Longo et al., 2013) and/or their related agents (Liu et al., 2006; Camesano et al., 2007; Abu-Lail, 2012; Algré et al., 2014) on the properties listed above and they were only performed on susceptible bacterial cells to antibiotics. It is worth noting that these studies did not investigate the mechanical properties of MDR bacterial cells. Irrespective of their shortcomings, these investigations highlighted the possible means through which bacteria respond to antibiotics and explored how various AFM modes and theoretical models can be employed to estimate bacterial mechanical properties in different environmental conditions (Atabek and Camesano, 2007; Perry et al., 2009; Gordesli and Abu-Lail, 2012; G. Longo et al., 2013).
5.1. Effect of Ampicillin on the Conformations of Bacterial Surface Biopolymers
Peaks are reported in MPa. When more than one peak is present in the histogram, the reported r2 value represents the average for all fits.
Table 1a compares the surface biopolymers brush thicknesses (Lo, nm) and their grafting densities (Γ, μm−2) estimated for the different E. coli strains investigated as a function of ampicillin and time of incubation. Our results indicate that at short exposure time to ampicillin (3 hours), the brush thicknesses for all the strains investigated significantly increased compared to control except for strain A5. Cells of strain A5 were similar to untreated cells in their conformational properties. This latter finding suggests that conformations of the A5 surface biopolymers are not affected by exposure to ampicillin. When strains D4 and H5 were considered, the increase in the brush thickness resulted in a reduction of the grafting densities of the strains. This result suggests that the bacterial biopolymers of these strains swell in response to ampicillin and adopt extended and reduced entropic conformations (Janshoff et al., 2000). This conformation may have been associated with the unfolding of some of the chains, the reason that got them extended and decreased the number of possible times a chain meets the surfaces resulting in a decreased apparent grafting density. Exposure to ampicillin to eight hours resulted in strain D4 reduction in brush thickness and increased grafting densities compared to the three hours’ treatment. Values were closer to those untreated indicating that the population of cells exposed to 8 hours ampicillin became similar in characteristics to the untreated cells reflecting some initiation of antibacterial resistance. Contrary to strains D4 and H5, the significant increase in brush thickness of strain A9 was associated with a significant increase in the density of the biopolymers on the strain’s surface. This suggests that the strain expressed more molecules on its surface in response to the short treatment with ampicillin. Upon extended exposure to ampicillin for 8 hours, strain A9 decreased its brush thickness and decreased its length to values nearing those of untreated cells. This again suggests that after 8 hours of exposure to ampicillin, cells started to develop resistance to ampicillin and that is indicative by the similarity the treated cells have with the untreated cells in biopolymer conformational properties. The reduction of the brush thickness after long exposure to ampicillin (8 hours) could be the result of membrane remodeling. Exposure to ampicillin could make the surface biopolymers less negative resulting in decreased steric repulsions to the negatively-charged Si3N4 as evident from the approach curves shown in [Figure 1A], (Atabek and Camesano, 2007).
The bacterial biopolymer brush thicknesses estimated here and how they modulate changes in grafting densities as well as the magnitudes of the grafting densities agree, in part, with what has been previously reported in the literature (Abu-Lail, 2003; Abu-Lail, 2008; Park and Abu-Lail, 2011). For example, investigating the difference in the conformational properties of the surface biopolymers of virulent and avirulent strains of Listeria monocytogenes showed that the virulent strains were characterized by 25% longer and 49% denser brushes compared to the avirulent strains (B J Park and Abu-Lail, 2011). Lengths of brushes ranged from 134 to 178 nm and densities ranged from 13200 to 27100 μm−2, agreeing with ranges reported here. However, in general, caution should be experienced when comparing results obtained from different species and from bacterial strains that are Gram-negative and those that are Gram-positive. Bacterial type, genetic makeup, and solvents in which force measurements were performed are expected to largely affect the biopolymer conformations estimated (Abu-Lail, 2003; Abu-Lail, 2008; Park and Abu-Lail, 2011).
5.2. Effect of Ampicillin on the Heterogeneity of Bacterial Surface Biopolymers
The bacterial surface is decorated with surface biopolymers of lengths that can extend up to hundreds of nanometers into the environment (Camesano and Abu-Lail, 2002). These biopolymers have different structures and functions which include inducement of genes that regulate cell shape, attachment and biofilm formation (van der Mei and Busscher, 2012). Because of the many biopolymeric molecules that cover the bacterial surface, a range of L0 and Γ values are expected (Camesano and Abu-Lail, 2002). This was indeed the case. Approach curves measured between the bacterial surface biopolymers and Si3N4 were heterogeneous and thus were individually fit to the steric model to obtain the conformational properties of biopolymers. The resulting values of L0 and Γ were a plot in forms of histograms to assess and describe the heterogeneity in the populations of the brush thicknesses and grafting densities [Figure 3 and Figure 4], respectively. Our results indicate that MDR-E. coli surface biopolymers are heterogeneous as evident from the large standard deviations obtained in the calculated means of L0 and Γ. The histograms of L0 and Γ span a range each and when fit with the log-normal or Gaussian dynamic peak functions, individual and often times, multiple peaks in each histogram were observed [Figure 3 and Figure 4].
When brush thickness histograms of untreated cells were considered, all strains were characterized by unimodal distributions except for strain D4 that was characterized by 4 subpopulations [Figure 3]. This indicates that even though heterogeneous, the untreated cells all fall within one population with the majority of the cells being characterized by L0 values that are close to the mean of the population. The fact that strain D4 has 4 subpopulations to start with even when untreated indicates that this strain has inherent heterogeneity in its population of cells and on its surfaces. The distributions of the brush thicknesses in the presence of ampicillin at all exposure times (3 or 8 hours) demonstrated single peak distributions except for those of strains D4 and H5 at 3 hours exposure, respectively [Figure 3C and Figure 3J]. The observation for strain D4 is to be expected as the strain was inherently heterogeneous even when untreated. Our data suggest that strain H5 became more heterogeneous as was exposed to ampicillin. Heterogeneity in the bacterial cell surface biopolymers has been suggested to enhance bacterial attachment affinities to surfaces (Camesano and Abu-Lail, 2002). Similarly, histograms of biopolymers’ grafting densities estimated from the steric model for MDR-E. coli strains are shown in [Figure 4]. Interestingly, the histograms of the grafting densities were heterogeneous for all strains investigated. Similar to the histograms of brush thicknesses, the highest heterogeneity was found for 3 hours’ exposure to ampicillin with the standard deviation of 4,232 μm2 when compared to the histograms of cells exposed to ampicillin for 8 hours (3,138 μm2) or for untreated cells (2,212 μm2) [Figure 4]. The heterogeneity in the grafting density in part mirrors the conformation and composition of the bacterial surface biopolymers brushes (Camesano et al., 2000; Atabek and Camesano, 2007; Eskhan and Abu-Lail, 2014).
When the short and long treatments were compared, the short treatment with ampicillin for 3 hours resulted in a higher heterogeneity compared to the longer 8 hours’ ampicillin treatment. This result agrees well with our findings here and our previous findings as well in which we have observed that many of the bacterial traits and functions go back to those of the untreated cells upon longer exposure to antibiotics suggesting that by such long time (8 hours), the cells are likely already adapted to ampicillin and incurred resistance. Whether or not ampicillin induces heterogeneity in bacterial surface biopolymers has not been investigated in the literature before. To the best of our knowledge, this is the first study to estimate bacterial surface biopolymers’ heterogeneity in the presence of ampicillin. We anticipate that the higher heterogeneity observed upon 3 hours’ exposure to ampicillin might be associated with the way cells survive antibiotics. Note that cells may increase their roughness, brush thickness, grafting density, surface area and surface area/volume to resist ampicillin, all of which likely induce heterogeneity in the cellular populations (Costerton et al., 1995; Touhami et al., 2006; Young, 2006; Justice et al., 2007; O’Toole, 2011). Note that the expression of cells of response genes to antibiotics is cell and strain dependent and that may lead to heterogeneities in cell populations with variable abilities and means to resist antibiotics (Wang et al., 2014). We do not have the genetic makeup of the strains investigated. However, the heterogeneities observed here suggest that the surface biopolymers of the MDR-E. coli strains are diverse and play a role in their survival strategies employed against ampicillin, especially at early and short exposure times.
5.3. Effects of Antibiotics on the Bacterial Cell Elasticity
The ability of bacteria to resist antibiotics is in part due to changes made to their cell wall composition, conformations of biopolymers and heterogeneity, other properties such charge and hydrophobicity as well as to how such surface changes affect cellular elasticities and enable cellular interactions with the environment (Ramadan et al., 1995; Vidya, Mallya and Rao, 2005; Longo et al., 2013; Tajkarimi et al., 2016). The bacterial cell wall is known to protect the cell membrane from rupture caused by the high internal turgor pressure as well as controls the integrity and morphology of the cell (Ghuysen, 1994; Vollmer and Seligman, 2010). The bacterial rigidity in terms of Young’s modulus can be directly assessed from AFM force-indentation data by fitting the approach curves to the Hertz model of contact mechanics. This elastic modulus mirrors the resistance of the bacterial cell wall to mechanical compression and the stiffness of the peptidoglycan network and other cell wall constituents such as surface biopolymers (Cerf et al., 2009; Perry et al., 2009; Formosa et al., 2012).
In our previous work, we have observed that some of the E. coli strain investigated here resisted ampicillin by increasing their surface areas (D4 and A9) while others did that by going into dormancy (A5). Our study here adds to our previous findings and points out to the important role played by cellular elasticities in modulating how cells of different strains resist antibiotics. For example, at 3 hours exposure to ampicillin strains D4 and A9 became softer and that directly enhances their potentials to increase their surface areas and as a result increase their biofilm formation. In order for the cells to be softer, they extended their biopolymer brushes and decreased their grafting densities [Table 1b, Table 2]. Despite these changes, the overall topographical morphologies of strains D4 and A9 were unaffected within the AFM resolution. This latter finding is quite different from observations reported in the literature for susceptible strains in which the morphology of cells from these strains are severely disintegrated (Perry et al., 2009; Vollmer and Seligman, 2010; Longo et al., 2013). Even though cells experienced softening upon short exposures to antibiotics, their membranes were still integrated and capable of providing them with protection upon exposure to antibiotics. The latter statement is supported in part by our previous study which investigated cellular morphology and membrane permeability of these strains and showed that MDR-E. coli strains maintained their membrane integrity at their corresponding MICs as revealed by AFM and fluorescence microscopy images. This suggests that MIC could only cause growth delay and elongation in strains D4 and A9 but not cell wall damage or cell lysis. When the longer ampicillin treatment effects on strains D4 and A9 elasticities was explored, our data suggest that D4 cells became as twice rigid as the untreated cells. This is expected due to the increased grafting density and collapsed biopolymer brushes on the bacterial cells as a result of the longer exposure to ampicillin [Table 1]. In comparison, cells for strain A9 went back to the elasticity of that of untreated cells [Table 1]. When strains A5 and H5 were considered, adaptation to ampicillin was not associated with different inhibition stages. Unlike strains D4 and A9, 3 hours exposure to ampicillin did not change the elasticities of cells of strains A5 and H5 [p>0.001, Table 1b, Table 2]. Strain A5 did not change its biopolymer conformational properties in response to ampicillin either [Table 2]. These A5 findings indicate that manipulating the cellular mechanics may not be an important mean through which this strain responds to antibiotics. Strain H5 is interesting in the fact that the increase in the grafting density was accompanied by a reduction in the brush thickness observed in response to ampicillin treatment for three hours. This balance is probably why we have not seen an appreciable difference in the elasticity because of exposure to ampicillin. Another reason comes from the high rigidity of the H5 cells. When compared to all other investigated strains, E. coli H5 cells were on average as twice as stiff as all other strains investigated with an elastic modulus ranging from 0.37 ± 0.10 MPa (r2 = 0.98) when grown in LB alone to 0.43 ± 0.13 MPa (r2 = 0.98) after exposure to ampicillin for 3 hours [Table 1b, Table 2]. This high rigidity is likely why changes in biopolymer conformations may not directly affect the overall cell elasticity.
Table 2:
A summary of the nanoscale adhesion, root mean square (RMS) roughness, surface area (SA), biofilm formation, elasticity, thicknesses and grafting densities of surface biopolymer brushes and elasticity as a function of ampicillin treatment at corresponding MIC for different durations.
| Strain D4 | F (nN) | RMS (nm) | SA (μm2) | Biofilms | L0 (nm) | Γ (μm−2) | E (MPa) |
|---|---|---|---|---|---|---|---|
| NT | 1.1 ± 0.5 | 36 ± 11 | 7.4 ± 1.5 | 0.20 ± 0.03 | 200 ± 39 | 13840±1726 | 0.13 ± 0.04 |
| 3 hours | 1.3 ± 0.3 | 89 ± 14 | 25.6 ± 12.6 | 0.29 ± 0.02 | 612±290 | 8886±3511 | 0.09 ± 0.03 |
| 8 hours | 1.2 ± 0.5 | 63 ± 21 | 10.1 ± 1.2 | 0.26 ± 0.3 | 169 ± 64 | 18505±5569 | 0.26 ± 0.09 |
| Strain A9 | F (nN) | RMS (nm) | SA (μm2) | Biofilms | L0 (nm) | Γ (μm−2) | E (MPa) |
| NT | 0.3 ± 0.1 | 48 ± 10 | 8.6 ± 1.6 | 0.19 ± 0.03 | 286 ±140 | 11035±1670 | 0.13 ± 0.05 |
| 3 hours | 0.6 ± 0.2 | 77 ± 13 | 53.9 ± 22.4 | 0.34 ± 0.04 | 403 ± 95 | 29496±7880 | 0.10 ± 0.05 |
| 8 hours | 0.9 ± 0.5 | 71 ± 13 | 39.1± 20.1 | 0.30 ± 0.04 | 267 ± 51 | 14519±2001 | 0.14 ± 0.04 |
| Strain A5 | F (nN) | RMS (nm) | SA (μm2) | Biofilms | L0 (nm) | Γ (μm−2) | E (MPa) |
| NT | 0.2 ± 0.1 | 68 ± 13 | 10.4 ± 2.0 | 0.16 ± 0.02 | 215 ± 50 | 14056±2092 | 0.18 ± 0.05 |
| 3 hours | 0.2 ± 0.1 | 74 ± 15 | 9.6 ± 1.0 | 0.18 ± 0.02 | 204 ± 77 | 13890±2274 | 0.19 ± 0.05 |
| Strain H5 | F (nN) | RMS (nm) | SA (μm2) | Biofilms | L0 (nm) | Γ (μm−2) | E (MPa) |
| NT | 0.2 ± 0.1 | 48 ± 9 | 9.5 ± 1.4 | 0.11 ± 0.02 | 128 ± 81 | 22637 ± 5487 | 0.37 ± 0.10 |
| 3 hours | 0.3 ± 0.1 | 52 ± 9 | 9.5 ± 1.7 | 0.18 ± 0.03 | 385 ± 14 | 10602±3000 | 0.43 ± 0.13 |
The values reported representing the mea ns ± standard deviations of average measurements performed on 26 to 44 cells obtained from three independent cultures. F (nN) is the adhesion forces measured between the bacterial surface biopolymers and the Si3N4, RMS(nm)is the bacterial root mean square roughness, SA (μm2) represents the surface area of the cell calculated from section analyses of captured AFM images, biofilms’ intensities are reported in terms of arbitrary units that represent the optical densities measured in a crystal violet assay at 580 nm, L0 (nm) and Γ (μm−2) is the average thicknesses and the grafting densities of the bacterial surface biopolymer brush layers estimated from the steric model respectively and E (MPa) represents the average estimated cellular elasticity from the Hertz model. The color scheme used in the Table indicates statistical differences among treatments in comparison to untreated cells. Green indicates that a given measure is statistically higher than the control, red indicates that a given measure is statistically lower than the control and yellow indicates that a given measure is statistically similar to the control. Statistical significance was considered at 99% confidence interval (p<0.001).
Prior literature studies have investigated the elasticities of susceptible cells in response to antibiotics using AFM force-indentation mode and using fits to the Hertz model of contact mechanics to back up the elasticities. To the best of our knowledge, none of these studies were performed on resistant bacterial cells and this study is the first to explore changes in the elasticities of resistant bacterial cells in response to antibiotics. Most of the studies reported a reduction in cell elasticity and a noticeable structural damage upon exposure of susceptible cells to antibiotics (Braga et al., 1998; Perry et al., 2009; Longo et al., 2013). For example, Young’s moduli of E. coli (ATCC 9637) cells were decreased from 2.2 ± 0.09 MPa to 1.1 ± 0.13 MPa upon exposure to ampicillin at 25 μg/ml (Perry et al., 2009). In another experiment, Longo et al measured the mechanical stiffness of susceptible E. coli DH5α upon exposure to ampicillin (Longo et al., 2013). Growing E. coli in LB supplemented with ampicillin below the MIC at 0.18 μg/ml caused dramatic changes in the bacterial morphology ranging from membrane deflation to lysis (Longo et al., 2013). The elasticity of the cells was reduced from 300 ± 70 kPa to 100 ± 20 kPa after exposure to ampicillin, indicating softening in the cell membrane (Longo et al., 2013). In explaining the results obtained on the susceptible strains, the authors suggested that the antibiotics might have interfered with the mechanism of action of the penicillin-binding protein (PBP), leading to inhibiting it from synthesizing peptidoglycans and eventually causing membrane lysis. Upon lysis, cells release their internal content resulting in a reduced turgor pressure and cell elasticity (Perry et al., 2009; Vu et al., 2009; Longo et al., 2013). The magnitudes reported for the elasticities in the studies above are mostly similar to ours except for the elasticity of the native E. coli cells of 2.2. MPa which is several orders of magnitude higher than we have estimated of our wild-type E. coli cells. However, note that here, some of the MDR strains decreased their elasticities in response to ampicillin while others increased them.
5.4. Effect of Ampicillin on the Heterogeneity of Cell Elasticities
Since the elasticity of cells and the conformational properties of biopolymers represented by the biopolymer brush thickness and grafting density are closely associated and since the latter two are heterogeneous [Figure 3 and Figure 4], the elasticities of cells are expected to be heterogeneous as well. To explore this heterogeneity, force-indentation profiles were individually fit to the Hertz model of contact mechanics. Histograms that represent all the estimated Young’s moduli values for each strain and at different ampicillin treatments for different times were generated [Figure 5].
Our data indicate that cells exposed to ampicillin for 3 hours were slightly more heterogeneous than cells exposed to ampicillin for 8 hours or than cells of the control [Figure 5]. This heterogeneity agrees well with our earlier findings which also indicated that the biopolymer conformational thicknesses and grafting densities for strains were the most heterogeneous when cells were exposed to ampicillin for short time (3 hours) compared to long exposure to ampicillin (8 hours) or untreated control [Figure 3 and Figure 4]. A look at all the histograms investigated shows that with the exceptions of few histograms, most histograms were unimodal. Unimodal histograms indicate more or less than the majority of cells in the population are distributed with a ± standard deviation from the mean. The exceptions to the above are the histograms for the treated strain A5 for 3 hours with ampicillin and the treated D4 and A9 strains for 8 hours with ampicillin. When the former is explored, it is interesting to note that the first subpopulation of the two presented in the treated sample (0.18 ± 0.01 MPa) matches statistically the peak of the population of the untreated cells (0.18 ± 0.03 MPa) (P<0.001) [Figure 5A and Figure 5B]. This is interesting and suggests that some of the A5 cells were not affected by ampicillin and retained their elastic properties while a fraction developed resistance through going into dormancy and increased the elasticity of other cells (0.24 ± 0.12 MPa) [Figure 5A]. The same discussion above applies to strain A9. The first subpopulation of the two presented in the treated sample for 8 hours with ampicillin (0.11 ± 0.01 MPa) matches statistically the peak of the population of the untreated cells (0.13 ± 0.03 MPa) (P<0.001) [Figure 5F and Figure 5H]. This is interesting and suggests that some of the A9 cells were able to regain their native cellular elasticities after exposure to ampicillin for 8 hours. The remaining fraction of cells represented by the second peak of the population [Figure 5H] was still experiencing the influence of ampicillin as indicated by the more rigid cells (0.16 ± 0.01 MPa) [Figure 5B]. Strain D4 was slightly different than the above two discussed strains. In all subpopulations reported for the strain as a result of exposure to ampicillin for 8 hours, the strain was always characterized by higher elasticities that the untreated cellular population or those treated for short times [Figure 5C, D and E]. the higher elasticity can be one of the means through which these cells resist antibiotics. When all unimodal histograms were compared, the population of cells of strain H5 was not only characterized by the highest elasticities but was also characterized by the most heterogeneity at 3-hours’ exposure to ampicillin.
5.5. Possible Mechanisms through which Investigated MDR-E. coli Develop Resistance to Ampicillin.
Here, the effects of treating cells of MDR E. coli strains with ampicillin at the MIC for different durations on the conformational properties of the bacterial surface biopolymers as well as the elasticities of these cells were explored. Previously, the effects of ampicillin treatment at MIC’s and for different durations on the nanoscale interaction forces measured between the bacterial surface biopolymers of these same MDR E. coli strains and a model surface of silicon nitride, biofilm formation, cellular roughness, bacterial cell membrane permeability, and cellular morphology and dimensions were investigated. By combining all the results above, possible mechanisms through which MDR-E. coli cells can develop resistance to common antibiotics such as ampicillin are discussed [Table 2].
When all data were combined, the following four possible mechanisms of resistance to ampicillin as employed by the different bacterial strains emerged. First, strain A5 was unique in displaying no differences in all the characterized cellular properties upon exposure to ampicillin at MIC [Table 2]. In our prior work, this strain exhibited a reduction in its cell length upon exposure to ampicillin. This was suggested to associate the cells with possible dormancy behavior. The current study does not really tell us how strain A5 develop resistance to ampicillin. One hypothesis we are testing through which they are possibly developing such resistance is the likelihood of having many persister cells in their population exposed to ampicillin. The characteristics of these persister cells are also being investigated. We anticipate that the population of strain A5 may contain a higher percentage of persister cells or slow-growing cells that are in a state of dormancy to conserve their energies and resist antibiotics (Spoering and Lewis, 2001; Mokrozub et al., 2015; Luby et al., 2016; Sultana et al., 2016).
Second, strain H5 increased its brush thickness, adhesion and biofilm formation as a result of exposure to ampicillin at MIC. The increase in the biopolymer brush thickness can be linked to the increase in adhesion. In the literature, as the brush thickness of the biopolymers of Listeria monocytogenes (Park and Abu-Lail, 2010), those of Pseudomonas putida (Abu-Lail and Camesano, 2003) and those of Escherichia coli (Abu-Lail and Camesano, 2003) increased, their adhesion to surfaces increased as well. This strain did not change its roughness, area or elasticity upon exposure to ampicillin [Table 2]. This strain likely resisted ampicillin by forming stronger biofilms on surfaces which in turn are expected to limit the diffusion of ampicillin within the biofilm to kill the cells. When all strains investigated were compared, it is worth to note that the H5 strain was the most rigid. The rigidity of the strain was associated with outer bacterial membranes that are not as permeable upon exposure to ampicillin as membranes of other strains. Previously, we reported lower frequencies at which the membranes of cells of strain H5 were damaged when cells were treated with ampicillin at a concentration higher than MIC (65 μg/ml) as indicated by less uptake of crystal violet strain compared to all other strains investigated. This indicates that this strain may also resist ampicillin by having a rigid and less permeable outer membrane.
Third, strains D4 and A9 adopted more or less similar resistance mechanisms upon exposure to ampicillin. At short exposure periods of three hours, the cells of both strains increased their roughness, brush thickness, and biofilm formation elongated themselves and that resulted in increased surface areas and decreased their elasticities. The two strains were different in two properties. Upon short period exposure to ampicillin, strain D4 did not change its adhesion and decreased its grafting density while strain A9 increased its adhesion and increased its grafting density [Table 2]. Prior literature studies supported that increased biofilm formation is proportional to increased adhesion (Busscher and Van der Mei, 1997). Increased adhesion has been documented to be proportional to decreased elasticity (Johnson and Greenwood, 1997), increased roughness (Aykent et al., 2010), increased surface area (Aykent et al., 2010) and as indicated above, increased brush thickness. As such, strains D4 and A9 likely resisted ampicillin through stronger biofilm formation that is expected to result in an impediment to ampicillin diffusion. The A9 cells can also utilize the higher grafting density as a means to induce further diffusion limitations to cells.
Finally, when D4 and A9 strains were exposed to ampicillin for a longer period of time (8 hours), both strains collapsed their biopolymers on their surfaces, increased their apparent grafting densities and as a result, increased their cellular elasticities [Table 2]. This indicates that, at long exposure to ampicillin, these strains utilized their mechanical properties to resist ampicillin. Upon the collapse of biopolymers on the bacterial surface, the likelihood of ampicillin meeting the PBPs gets reduced. Note that the β-lactam ring in the ampicillin core structure is responsible for the antibacterial function via the inhibition of the PBP which synthesizes the PGs that form the rigid cell walls of bacteria such as E. coli and thus provide them with integrity. Furthermore, as was described for the H5 strain, higher rigidity is associated with a lower outer membrane permeability which as such reduces the diffusion of ampicillin into the cells. Note that at longer exposures to ampicillin, strain D4 increased its roughness and surface area but did not change their adhesion or biofilm formation. Compared to all strains investigated, strain A9 can claim the most resistant strain title as it utilized every mean to resist antibiotics [Table 2].
6. Conclusions
The effects of introducing a model β-lactam antibiotic, ampicillin, at MIC and for the different duration on the mechanical properties of MDR E. coli cells and their surface biopolymers were investigated. Results from this study along with our prior findings on how ampicillin treatment affected the nanoscale interaction forces measured between the bacterial surface biopolymers of these same MDR E. coli strains and a model surface of silicon nitride, biofilm formation, cellular roughness, bacterial cell membrane permeability and cellular morphology and dimensions were utilized to suggest possible mechanisms through which these bacterial cells resisted ampicillin. Our results indicated that all strains resisted ampicillin through variable mechanisms. Strain A5 was unique in displaying no differences in all its characterized cellular properties upon exposure to ampicillin at MIC. We anticipate that this strain resists ampicillin through dormancy. Strain H5 increased its biopolymer brush thickness, adhesion and biofilm formation and kept its roughness, surface area and cell elasticity unchanged upon exposure to ampicillin at MIC. When all strains investigated were compared, strain H5 was the most rigid. The rigidity was associated with a low outer membrane (OM) permeability. As such, this strain likely limits the diffusion of ampicillin into cells by having rigid cells with low OM permeability and through the biofilms by forming stronger biofilms. At short exposure periods to ampicillin (three hours), the cells of strains D4 and A9 increased their roughness, surface areas, biofilm formation, and brush thicknesses and decreased their elasticities. In addition, strain D4 did not change its adhesion and decreased its grafting density while strain A9 increased its adhesion and increased its grafting density. As such and at short ampicillin treatment duration, strains D4 and A9 likely resisted ampicillin through stronger biofilm formation that is expected to result in an impediment to ampicillin diffusion. Cell-cell communication within the biofilm network can induce the expression of genes that regulate survival in the presence of antibiotics and nutrient deficiency (Staffan Kjelleberg and Soeren Molin, 2002). The A9 cells also utilized the higher grafting density as a means to induce further diffusion limitations to cells. Finally, when strains D4 and A9 were exposed to ampicillin for a longer period of time (8 hours), both strains collapsed their biopolymers, increased their apparent grafting densities and as a result, increased their cellular elasticities. This indicates that, at long exposure to ampicillin, cells utilize their higher rigidity and lower outer membrane permeability to reduce the diffusion of ampicillin into the cells. The findings of this study clearly point to the potential of using the nanoscale characterization of MDR bacterial properties as a means to monitor cell modifications that enhance “phenotypic antibiotic resistance”. As such, our results underscore the importance of characterizing the roles of bacterial variable properties in mediating how antibiotic-resistant bacteria behave during exposure to antibiotics. Such better fundamental understanding is essential for the development of new effective antibiotics capable of invading cell membranes and biofilms.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest.
There are no conflicts to declare by authors.
Note to reviewers:
Our published reference that has the data we refer to in this manuscript was removed for the purposes of the double blind review such as our identities are not known.
7. References
- Abu-Lail and and Camesano (2003) ‘Role of lipopolysaccharides in the adhesion, retention, and transport of Escherichia coli JM109’, Environmental Science and Technology, 37(10), pp. 2173–2183. doi: 10.1021/es026159o. [DOI] [PubMed] [Google Scholar]
- Abu-Lail L (2012) ‘Using Atomic Force Microscopy to Measure Anti-Adhesion Effects on Uropathogenic Bacteria, Observed in Urine after Cranberry Juice Consumption’, Journal of Biomaterials and Nanobiotechnology, 03(04), pp. 533–540. doi: 10.4236/jbnb.2012.324055. [DOI] [Google Scholar]
- Abu-Lail NI (2003) ‘The Effect of Biopolymer Properties on Bacterial Adhesion: an Atomic Force Microscopy (AFM) Study’, p. 320. [Google Scholar]
- Abu-Lail NI and Camesano TA (2003) ‘Role of ionic strength on the relationship of biopolymer conformation, DLVO contributions, and steric interactions to bioadhesion of pseudomonas putida KT2442’, Biomacromolecules, 4(4), pp. 1000–1012. doi: 10.1021/bm034055f. [DOI] [PubMed] [Google Scholar]
- Adams PG et al. (2014) ‘Lipopolysaccharide-induced dynamic lipid membrane reorganization: Tubules, perforations, and stacks’, Biophysical Journal, 106(11), pp. 2395–2407. doi: 10.1016/j.bpj.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschlimann JR (2003) ‘The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria’, Pharmacotherapy, 23(7), pp. 916–924. doi: 10.1592/phco.23.7.916.32722. [DOI] [PubMed] [Google Scholar]
- Alexander S (1977) ‘Adsorption of chain molecules with a polar head a scaling description’, Journal de Physique, 38(8), pp. 983–987. doi: 10.1051/jphys:01977003808098300. [DOI] [Google Scholar]
- Algré E et al. (2014) ‘Design and construction of the motion mechanism of an XY micro-stage for precision positioning’, Applied Physics Letters, 11(1), pp. 1–10. doi: 10.1103/PhysRevLett.56.930. [DOI] [Google Scholar]
- Ali R et al. (2014) ‘Role of Polymerase Chain Reaction (PCR) in the detection of antibiotic-resistant Staphylococcus aureus’, Egyptian Journal of Medical Human Genetics. Production and hosting by Elsevier B.V., 15(3), pp. 293–298. doi: 10.1016/j.ejmhg.2014.05.003. [DOI] [Google Scholar]
- Allison DP et al. (2011) ‘Bacterial Immobilization for Imaging by Atomic Force Microscopy’, Journal of Visualized Experiments, (54), pp. 5–7. doi: 10.3791/2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arslan B et al. (2018) ‘Characterizing interaction forces between actin and proteins of the tropomodulin family reveals the presence of the N-terminal actin-binding site in leiomodin’, Archives of Biochemistry and Biophysics. Elsevier, 638(November 2017), pp. 18–26. doi: 10.1016/j.abb.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atabek and Camesano (2007) ‘Atomic force microscopy study of the effect of lipopolysaccharides and extracellular polymers on adhesion of Pseudomonas aeruginosa’, Journal of Bacteriology, 189(23), pp. 8503–8509. doi: 10.1128/JB.00769-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aykent et al. (2010) ‘Effect of different finishing techniques for restorative materials on surface roughness and bacterial adhesion’, Journal of Prosthetic Dentistry. The Editorial Council of the Journal of Prosthetic Dentistry, 103(4), pp. 221–227. doi: 10.1016/S0022-3913(10)60034-0. [DOI] [PubMed] [Google Scholar]
- Babic M, Hujer AM and Bonomo RA (2006) ‘What’s new in antibiotic resistance? Focus on beta-lactamases’, Drug Resistance Updates, 9(3), pp. 142–156. doi: 10.1016/j.drup.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Bæk KT et al. (2014) ‘β-lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease’, Antimicrobial Agents and Chemotherapy, 58(8), pp. 4593–4603. doi: 10.1128/AAC.02802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Baum H and Marre R (2005) ‘Antimicrobial resistance of Escherichia coli and therapeutic implications’, International Journal of Medical Microbiology, 295(6–7), pp. 503–511. doi: 10.1016/j.ijmm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Beniac DR et al. (2014) ‘A Filtration Based Technique for Simultaneous SEM and TEM Sample Preparation for the Rapid Detection of Pathogens’, Viruses, pp. 3458–3471. doi: 10.3390/v6093458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet R (2004) ‘MINIREVIEW Growing Group of Extended-Spectrum β-Lactamases: the CTX-M Enzymes’, Antimicrobial Agents and Chemotherapy, 48(1), Antimicrobial Agents and Chemotherapy, pp. 1–14. doi: 10.1128/AAC.48.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braga and Ricci (1998) ‘Atomic Force Microscopy: Application to Investigation of Escherichia coli Morphology before and after Exposure to Cefodizime Atomic Force Microscopy: Application to Investigation of Escherichia coli Morphology before and after Exposure to Cefodizime Dow’, Antimicrobial Agents and Chemotherapy, 42(1), pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busscher and Van der Mei (1997) ‘Advances in Dental Research’, Adv Dent Res, 11(1), pp. 24–32. doi: 10.1177/08959374970110011301. [DOI] [PubMed] [Google Scholar]
- Butt HJ et al. (1999) ‘Steric forces measured with the atomic force microscope at various temperatures’, Langmuir, 15(7), pp. 2559–2565. doi: 10.1021/la981503+. [DOI] [Google Scholar]
- Camesano et al. (2007) ‘Cranberry prevents the adhesion of bacteria: overview of relevant health benefits’, Agro Food Industry Hi-Tech, 18(1, S), pp. 24–27. [Google Scholar]
- Camesano TA et al. (2000) ‘Observation of changes in bacterial cell morphology using tapping mode atomic force microscopy’, Langmuir, 16(18), pp. 4563–4572. [Google Scholar]
- Camesano TA and Abu-Lail NI (2002) ‘Heterogeneity in bacterial surface polysaccharides, probed on a single-molecule basis’, Biomacromolecules, 3(4), pp. 661–667. doi: 10.1021/bm015648y. [DOI] [PubMed] [Google Scholar]
- Camesano T. a. and Logan BE (2000) ‘Probing Bacterial Electrosteric Interactions Using Atomic Force Microscopy’, Environmental Science & Technology, 34(16), pp. 3354–3362. doi: 10.1021/es9913176. [DOI] [Google Scholar]
- Cerf A et al. (2009) ‘Nanomechanical properties of dead or alive single-patterned bacteria’, Langmuir, 25(10), pp. 5731–5736. doi: 10.1021/la9004642. [DOI] [PubMed] [Google Scholar]
- Cockerill FR (1999) ‘Genetic methods for assessing antimicrobial resistance’, Antimicrobial Agents and Chemotherapy, 43(2), pp. 199–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton et al. (1974) ‘Structure and function of the cell envelope of gram-negative bacteria.’, Bacteriological reviews, 38(1), pp. 87–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton J et al. (1995) ‘Microbial biofilms’, Annu Rev Microbiol, 49, pp. 711–745. [DOI] [PubMed] [Google Scholar]
- Cui Y et al. (2012) ‘AFM Probing the Mechanism of Synergistic Effects of the Green Tea Polyphenol (−)-Epigallocatechin-3-Gallate (EGCG) with Cefotaxime against Extended-Spectrum Beta-Lactamase (ESBL)-Producing Escherichia coli’, PLoS ONE, 7(11). doi: 10.1371/journal.pone.0048880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doktycz MJ et al. (2003) ‘AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces’, Ultramicroscopy, 97(1–4), pp. 209–216. doi: 10.1016/S0304-3991(03)00045-7. [DOI] [PubMed] [Google Scholar]
- Drlica K et al. (2008) ‘Quinolone-mediated bacterial death’, Antimicrobial Agents and Chemotherapy, 52(2), pp. 385–392. doi: 10.1128/AAC.01617-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Duin D and Paterson DL (2016) ‘Multidrug-Resistant Bacteria in the Community: Trends and Lessons Learned’, Infectious Disease Clinics of North America. Elsevier Inc, 30(2), pp. 377–390. doi: 10.1016/j.idc.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskhan and Abu-Lail (2014) ‘A new approach to decoupling of bacterial adhesion energies measured by AFM into specific and nonspecific components’, Colloid Polym Sci, 292(2), pp. 343–353. doi: 10.1021/nl061786n.Core-Shell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filho RP et al. (2010) ‘Prodrugs available on the Brazilian pharmaceutical market and their corresponding bioactivation pathways’, Brazilian Journal of Pharmaceutical Sciences, 46(3), pp. 393–420. doi: 10.1590/S1984-82502010000300003. [DOI] [Google Scholar]
- Fluit ADC, Visser MR and Schmitz F (2001) ‘Molecular Detection of Antimicrobial Resistance’, Clinical microbiology reviews, 14(4), pp. 836–871. doi: 10.1128/CMR.14.4.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa C et al. (2012) ‘Nanoscale analysis of the effects of antibiotics and CX1 on a Pseudomonas aeruginosa multidrug-resistant strain’, Scientific Reports, 2(1), p. 575. doi: 10.1038/srep00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruci M and Poole K (2016) ‘Bacterial Stress Responses as Determinants of Antimicrobial Resistance’, Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria, 1(April), pp. 115–136. doi: 10.1002/9781119004813.ch10. [DOI] [Google Scholar]
- Gavara N (2016) ‘Combined strategies for optimal detection of the contact point in AFM force-indentation curves obtained on thin samples and adherent cells’, Scientific Reports. Nature Publishing Group, 6(February), pp. 1–13. doi: 10.1038/srep21267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghuysen J-M (1994) ‘Molecular structures of penicillin-binding proteins and P-lactamases’. [DOI] [PubMed] [Google Scholar]
- Gilbert et al. (2007) ‘Single-molecule force spectroscopy and imaging of the vancomycin/D-Ala-D-Ala interaction.’, Nano Lett, 7(3), pp. 796–801. doi: 10.1021/nl0700853. [DOI] [PubMed] [Google Scholar]
- Golding CG et al. (2016) ‘The scanning electron microscope in microbiology and diagnosis of infectious disease’, Scientific Reports. Nature Publishing Group, 6(February), pp. 1–8. doi: 10.1038/srep26516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordesli FP and Abu-Lail NI (2012) ‘Impact of ionic strength of growth on the physiochemical properties, structure, and adhesion of Listeria monocytogenes polyelectrolyte brushes to a silicon nitride surface in water.’, Journal of colloid and interface science, pp. 1–11. doi: 10.1016/j.jcis.2012.08.048. [DOI] [PubMed] [Google Scholar]
- Hartmann M et al. (2010) ‘Damage of the bacterial cell envelope by antimicrobial peptides gramicidin S and PGLa as revealed by transmission and scanning electron microscopy’, Antimicrobial Agents and Chemotherapy, 54(8), pp. 3132–3142. doi: 10.1128/AAC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV (1998) ‘Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli’, Microbiology and molecular biology reviews: MMBR, 62(1), pp. 181–203. doi: S0104-11692007000700004 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong W, Zeng J and Xie J (2014) ‘Antibiotic drugs targeting bacterial RNAs’, Acta Pharmaceutica Sinica B. Elsevier, 4(4), pp. 258–265. doi: 10.1016/j.apsb.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter JL and Bechhoefer J (1993) ‘Calibration of atomic-force microscope tips’, Review of Scientific Instruments, 64(7), pp. 1868–1873. doi: 10.1063/1.1143970. [DOI] [Google Scholar]
- Jacoby GA and Medeiros AA (1991) ‘More extended-spectrum beta-lactamases.’, Antimicrobial Agents and Chemotherapy, 35(9), pp. 1697–1704. doi: 10.1128/AAC.35.9.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janshoff A et al. (2000) ‘Force Spectroscopy of Molecular Systems—Single Molecule Spectroscopy of Polymers and Biomolecules’, Angewandte Chemie, 39(18), pp. 3212–3237. doi: . [DOI] [PubMed] [Google Scholar]
- Jenkins SG and Schuetz AN (2012) ‘Current concepts in laboratory testing to guide antimicrobial therapy’, Mayo Clinic Proceedings. Elsevier Inc., 87(3), pp. 290–308. doi: 10.1016/j.mayocp.2012.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson and Greenwood (1997) ‘An Adhesion Map for the Contact of Elastic Spheres’, Journal of Colloid and Interface Science, 192(2), pp. 326–333. doi: 10.1006/jcis.1997.4984. [DOI] [PubMed] [Google Scholar]
- Justice SS et al. (2007) ‘Morphological plasticity as a bacterial survival strategy’, Nat Rev Micro, advanced o. Available at: file:///c:/pdf/rfd6662.pdf%5Cnfile:///home/ford/Documents/PDF/rfd6662.pdf. [DOI] [PubMed] [Google Scholar]
- Kumarasamy KK et al. (2010) ‘Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: A molecular, biological, and epidemiological study’, The Lancet Infectious Diseases, 10(9), pp. 597–602. doi: 10.1016/S1473-3099(10)70143-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski D et al. (2018) ‘Effect of ampicillin on adhesive properties of bacteria examined by atomic force microscopy’, Micron. Elsevier, 112(May), pp. 84–90. doi: 10.1016/j.micron.2018.05.005. [DOI] [PubMed] [Google Scholar]
- Liu Y et al. (2006) ‘Role of cranberry juice on molecular-scale surface characteristics and adhesion behavior of Escherichia coli’, Biotechnology and Bioengineering, 93(2), pp. 297–305. doi: 10.1002/bit.20675. [DOI] [PubMed] [Google Scholar]
- Lonergan NE, Britt LD and Sullivan CJ (2014) ‘Immobilizing live Escherichia coli for AFM studies of surface dynamics’, Ultramicroscopy. Elsevier, 137, pp. 30–39. doi: 10.1016/j.ultramic.2013.10.017. [DOI] [PubMed] [Google Scholar]
- Longo G et al. (2013) ‘Antibiotic-induced modifications of the stiffness of bacterial membranes’, Journal of Microbiological Methods. Elsevier B.V., 93(2), pp. 80–84. doi: 10.1016/j.mimet.2013.01.022. [DOI] [PubMed] [Google Scholar]
- Longo G et al. (2013) ‘Rapid detection of bacterial resistance to antibiotics using AFM cantilevers as nanomechanical sensors’, Nature Nanotechnology, 8(7), pp. 522–526. doi: 10.1038/nnano.2013.120. [DOI] [PubMed] [Google Scholar]
- Longo G and Kasas S (2014) ‘Effects of antibacterial agents and drugs monitored by atomic force microscopy’, Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology, 6(3), pp. 230–244. doi: 10.1002/wnan.1258. [DOI] [PubMed] [Google Scholar]
- Luby E et al. (2016) ‘Molecular Methods for Assessment of Antibiotic Resistance in Agricultural Ecosystems: Prospects and Challenges’, Journal of Environment Quality, 45(2), p. 441. doi: 10.2134/jeq2015.07.0367. [DOI] [PubMed] [Google Scholar]
- Mazzola PG et al. (2009) ‘Minimal inhibitory concentration (MIC) determination of disinfectant and/or sterilizing agents’, Brazilian Journal of Pharmaceutical Sciences, 45(2), pp. 241–248. doi: 10.1590/S1984-82502009000200008. [DOI] [Google Scholar]
- van der Mei HC and Busscher HJ (2012) ‘Bacterial Cell Surface Heterogeneity: A Pathogen’s Disguise’, PLoS Pathogens, 8(8), pp. 8–11. doi: 10.1371/journal.ppat.1002821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meincken et al. (2005) ‘Atomic Force Microscopy Study of the Effect of Antimicrobial Peptides on the Cell Envelope of Escherichia coli’, Society, 49(10), pp. 4085–4092. doi: 10.1128/AAC.49.10.4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SI (2016) ‘Antibiotic resistance and regulation of the gram-negative bacterial outer membrane barrier by host innate immune molecules’, mBio, 7(5), pp. e01541–16. doi: 10.1128/mBio.01541-16.Copyright. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty S et al. (2012) ‘An investigation on the antibacterial, cytotoxic, and antibiofilm efficacy of starch-stabilized silver nanoparticles’, Nanomedicine: Nanotechnology, Biology, and Medicine. Elsevier Inc., 8(6), pp. 916–924. doi: 10.1016/j.nano.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Mokrozub VV et al. (2015) ‘The role of beneficial bacteria wall elasticity in regulating innate immune response’, EPMA Journal, 6(1), pp. 1–15. doi: 10.1186/s13167-015-0035-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napiroon T et al. (2017) ‘Antibacterial Activity of Three Medicinal Lasianthus (Rubiaceae) Extracts on Human Resistant Pathogenic Bacteria’, European Journal of Experimental Biology, 07(06), pp. 1–7. doi: 10.21767/2248-9215.100037. [DOI] [Google Scholar]
- Neethirajan S and DiCicco M (2014) ‘Atomic force microscopy study of the antibacterial effect of fosfomycin on methicillin-resistant Staphylococcus pseudintermedius’, Applied Nanoscience, 4(6), pp. 703–709. doi: 10.1007/s13204-013-0256-3. [DOI] [Google Scholar]
- Nikaido H (2009) ‘Multidrug Resistance in Bacteria’, Annu Rev Biochem, (2), pp. 119–146. doi: 10.1146/annurev.biochem.78.082907.145923.Multidrug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole GA (2011) ‘Microtiter Dish Biofilm Formation Assay’, Journal of Visualized Experiments, (47), pp. 10–11. doi: 10.3791/2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B-J and Abu-Lail NI (2010) ‘Variations in the Nanomechanical Properties of Virulent and Avirulent Listeria monocytogenes.’, Soft matter, 6(16), pp. 3898–3909. doi: 10.1039/b927260g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ and Abu-Lail NI (2011) ‘The role of the pH conditions of growth on the bioadhesion of individual and lawns of pathogenic Listeria monocytogenes cells’, Journal of Colloid and Interface Science. Elsevier Inc., 358(2), pp. 611–620. doi: 10.1016/j.jcis.2011.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park BJ and Abu-Lail NI (2011) ‘Variations in the nanomechanical propertis of virulent and avirulent Listeria monocytogenes’, Soft Matter, 6(16), pp. 3898–3909. doi: 10.1039/b927260g.Variations. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry CC et al. (2009) ‘Atomic force microscopy study of the antimicrobial activity of aqueous garlic versus ampicillin against Escherichia coli and Staphylococcus aureus’, Journal of the Science of Food and Agriculture, 89(6), pp. 958–964. doi: 10.1002/jsfa.3538. [DOI] [Google Scholar]
- Ramadan MA et al. (1995) ‘Post-antibiotic effect of azithromycin and erythromycin on streptococcal susceptibility to phagocytosis’, Journal of Medical Microbiology, 42(5), pp. 362–366. [DOI] [PubMed] [Google Scholar]
- Ramezanian et al. (2018) ‘Role of ionic strength in the thicknesses of the biopolymer fringes, spring constants, and Young’s moduli of Pseudomonas putida’, Journal of vacuum science and technology. B, Nanotechnology & microelectronics, 36(2), p. 021801-. doi: 10.1520/D0850-11.1. [DOI] [Google Scholar]
- Read AF and Woods RJ (2014) ‘Antibiotic resistance management’, Evolution, Medicine, and Public Health, 2014(1), pp. 147–147. doi: 10.1093/emph/eou024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose JB et al. (2014) ‘Gelatin-based materials in ocular tissue engineering’, Materials, 7(4), pp. 3106–3135. doi: 10.3390/ma7043106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzewuska M et al. (2015) ‘Multidrug Resistance in Escherichia coli Strains Isolated from Infections in Dogs and Cats in Poland (2007–2013)’, The Scientific World Journal, 2015, pp. 1–8. doi: 10.1155/2015/408205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Seedy FR et al. (2017) ‘Polymerase chain reaction detection of genes responsible for multiple antibiotic resistance Staphylococcus aureus isolated from food of animal origin in Egypt’, Veterinary World, 10(10), pp. 1205–1211. doi: 10.14202/vetworld.2017.1205-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh S et al. (2015) ‘Antibiotic resistance and extended spectrum beta-lactamases: Types, epidemiology and treatment’, Saudi Journal of Biological Sciences, 22(1), pp. 90–101. doi: 10.1016/j.sjbs.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellberg B et al. (2011) ‘Combating antimicrobial resistance: Policy recommendations to save lives’, Clinical Infectious Diseases, 52(SUPPL. 5). doi: 10.1093/cid/cir153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoering AMYL and Lewis KIM (2001) ‘Biofilms and Planktonic Cells of Pseudomonas aeruginosa Have Similar Resistance to Killing by Antimicrobials’, Journal of Bacteriology, 183(23), pp. 6746–6751. doi: 10.1128/JB.183.23.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelleberg Staffan and Molin Soeren (2002) ‘Is there a role for quorum sensing signals in bacterial biofilms? Staffan Kjelleberg * and Soeren Molin †’, Current Opinion in Microbiology, pp. 254–258. [DOI] [PubMed] [Google Scholar]
- Sultana et al. (2016) ‘Eradication of Pseudomonas aeruginosa biofilms and persister cells using an electrochemical scaffold and enhanced antibiotic susceptibility’, npj Biofilms and Microbiomes. Springer US, 2(1), p. 2. doi: 10.1038/s41522-016-0003-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadesse DA et al. (2012) ‘Antimicrobial drug resistance in Escherichia coli from humans and food animals, United States, 1950–2002.’, Emerging Infectious Diseases, pp. 741–749. doi: 10.3201/eid1805.111153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajkarimi M et al. (2016) ‘Mechanobiology of antimicrobial resistant Escherichia coli and Listeria innocua’, PLoS ONE, 11(2), pp. 1–13. doi: 10.1371/journal.pone.0149769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Zhou X and Kaper JB (2005) ‘Adherence of Diarrheagenic Escherichia coli Strains to Epithelial Cells MINIREVIEW Adherence of Diarrheagenic Escherichia coli Strains to Epithelial Cells’, Infection and Immunity, 73(1), pp. 18–29. doi: 10.1128/IAI.73.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhami A et al. (2006) ‘Nanoscale Characterization and Determination of Adhesion Forces of Pseudomonas aeruginosa Pili by Using Atomic Force Microscopy Nanoscale Characterization and Determination of Adhesion Forces of Pseudomonas aeruginosa Pili by Using Atomic Force Microscopy’, Journal of …, 188(2), pp. 370–377. doi: 10.1128/JB.188.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnidge J and Paterson DL (2007) ‘Setting and revising antibacterial susceptibility breakpoints’, Clinical Microbiology Reviews, 20(3), pp. 391–408. doi: 10.1128/CMR.00047-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vadillo-Rodriguez et al. (2009) ‘In situ characterization of differences in the viscoelastic response of individual gram-negative and gram-positive bacterial cells’, Journal of Bacteriology, 191(17), pp. 5518–5525. doi: 10.1128/JB.00528-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidya K, Mallya P and Rao P (2005) ‘Inhibition of Bacterial Adhesion Concentrations of Antibiotics By’, 23, pp. 102–105. [DOI] [PubMed] [Google Scholar]
- Visai L, Speziale P and Bozzini S (1990) ‘Binding of collagens to an enterotoxigenic strain of Escherichia coli’, Infection and Immunity, 58(2), pp. 449–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W and Seligman SJ (2010) ‘Architecture of peptidoglycan: more data and more models’, Trends in Microbiology, 18(2), pp. 59–66. doi: 10.1016/j.tim.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Vu B et al. (2009) ‘Bacterial extracellular polysaccharides involved in biofilm formation’, Molecules, 14(7), pp. 2535–2554. doi: 10.3390/molecules14072535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X et al. (2014) ‘Heteroresistance at the Single-Cell Level: Adapting to Antibiotic Stress through a Population-Based Strategy and Growth-Controlled’, 5(1), pp. 1–9. doi: 10.1128/mBio.00942-13.Editor. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z et al. (2015) ‘Influence of core oligosaccharide of lipopolysaccharide to outer membrane behavior of Escherichia coli’, Marine Drugs, 13(6), pp. 3325–3339. doi: 10.3390/md13063325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L et al. (2006) ‘Atomic force microscopy study of different effects of natural and semisynthetic β-lactam on the cell envelope of Escherichia coli’, Analytical Chemistry, 78(20), pp. 7341–7345. doi: 10.1021/ac0604890. [DOI] [PubMed] [Google Scholar]
- Young KD (2006) ‘The Selective Value of Bacterial Shape’, Microbiology and Molecular Biology Reviews, 70(3), pp. 660–703. doi: 10.1128/MMBR.00001-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y (2014) ‘Persisters, persistent infections and the Yin-Yang model.’, Emerging microbes & infections, 3(1), p. e3. doi: 10.1038/emi.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.