Abstract
Ascorbic acid (Vitamin C, VC) increases the secretion of mature collagen by promoting the activity of prolyl 4-hydroxylase subunit α 1 (P4HA1). To explore the mechanism involved, we investigated the role of N-linked glycosylation, which can regulate enzyme activity. P4HA1 has two glycosylation sites, Asn (N) 113 and N259. Our studies show that glycosylation of N259, but not N113, by STT3B and magnesium transporter 1 (MAGT1) is augmented by VC. N259 glycosylation on P4HA1 correlates with enhanced pepsin-resistant collagen 1α2 secretion. Downregulation of Stt3b and Magt1 reduces N259 glycans on P4HA1. In collagen 1α2 purified from Stt3b-silenced fibroblasts, decreased hydroxylation is found at 5 specific proline residues, while significantly increased hydroxylation is noted at 2 proline residues. Similarly, in collagen 1α1, reduced proline hydroxylation is detected at 8 sites and increased proline hydroxylation is found at 4 sites. These results suggest that N-linked glycosylation of P4HA1 can direct hydroxylation at specific proline residues and affect collagen maturation.
Keywords: ascorbic acid, type I collagen, prolyl 4-hydroxylase, post-translational modification, STT3B, MAGT1, N-glycosylation
1.1. Introduction
Collagen is the main component of connective tissues in animals, and comprises of 25–35% of the body's protein content. Collagens can be separated into fibrillar and non-fibrillar groups, fibrillar being the most abundant and type I collagen being the main fibrillar collagen. Biosynthesis of type I collagen requires several steps [1]. After preprocollagen is translated on the ribosomes along the rough endoplasmic reticulum (RER), the N-terminal signal is cleaved in the lumen of the RER to form procollagen. In the RER proline and lysine are hydroxylated by prolyl and lysyl hydroxylases, for which ascorbic acid (vitamin C, VC) is a cofactor. The hydroxylation of proline and lysine is essential for collagen maturation [2]. Specific hydroxylysine residues are glycosylated promoting the formation of the mature collagen triple helix that is pepsin-resistant. This triple helix is then transported to Golgi apparatus to be secreted.
Proline is hydroxylated by one or more of three isoforms of prolyl hydroxylase (P4H), P4HA1, P4HA2, and P4HA3[3–6], all of which use VC as a cofactor. Besides P4H, lysyl hydroxylase (Procollagen-Lysine, 2-Oxoglutarate 5-Dioxygenase, PLOD) also uses VC as a cofactor [7,8]. Deficiency of VC compromises P4H and PLOD activity resulting in decreased hydroxylation of collagen leading to the disease condition known as scurvy. Although P4H activity has been reported to be regulated by short-term treatment with VC [11,12], the mechanism of enzymatic activity regulation is not completely understood.
N-linked glycosylation is one of the most common forms of post-translational modification and serves to facilitate protein folding, solubility, and stability, and to regulate the function and activity of multiple enzymes [13].Glycosylation of P4HA1 has been reported [14]; however, the function of the N-glycosylation of P4HA1 remains unclear. In fact, the enzymatic activity of P4HA1 is reported to be unaffected by the lack of glycans[14,15].
Since VC is required for collagen maturation, we sought to better understand the role of VC in collagen production, particularly the mechanism by which VC and P4H accelerate collagen maturation. We report that treatment of fibroblasts for 3–6 hours (hrs) with VC increases the secretion of mature collagen 1α2 (col 1α2) by increasing P4HA1 glycosylation, specifically at N259. N259 glycosylation induces P4HA1 to hydroxylate specific prolines in col 1α2 resulting in more pepsin-resistant collagen. Silencing STT3B or magnesium transporter 1 (MAGT1), the enzymes responsible for N259 glycosylation on P4HA1, significantly decreases hydroxylation of five prolines in col 1α2 and eight prolines in col 1α1. On the other hand, a significant increase in hydroxylation of two prolines in col 1α2 and four prolines in col 1α1 is detected.
1.2. Results
1.2.1. Short term VC treatment increased col1α2 secretion.
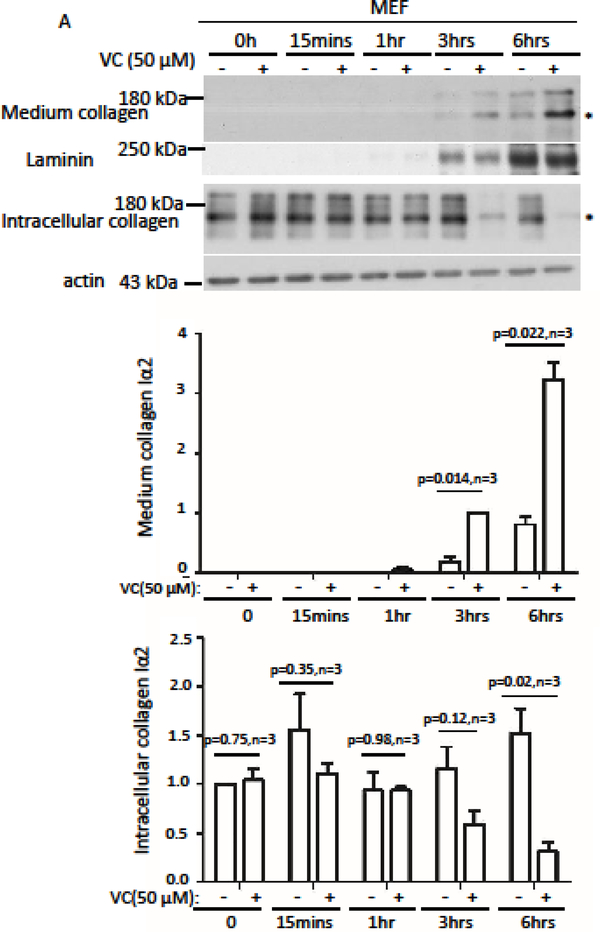
VC treatment stimulated proline or lysine hydroxylation and increased secretion of mature Type I collagen [11,12], which is a triple-stranded helix composed of two α1(I) chains and one α2(I) chain. We verified the effect of VC on Type I collagen secretion on mouse embryonic fibroblast (MEF) cells, human foreskin fibroblast (HFF) cells, and human embryonic lung fibroblast (HEL) cells with 50 μM VC. Two bands were detected by immunoblot analysis 3 hrs after treatment (Figures 1A, 1B, & 1C, top panel; supplemental Figure 1 A). By silencing Col1a2 and producing an antibody specific for col 1α1, we determined that the protein with an apparent molecular weight of 150 kDa was col 1α2 and the 200 kDa protein is col 1α1 (Supplemental Figure 1A,). Since both proteins showed similar pattern after VC treatment, we focused our study on col 1α2 because the antibody is commercially available.
Figure 1.
Short term VC treatment enhanced pepsin-resistant col1α2 secretion via a pathway independent of transcription. Time course of MEF cells (A), HFF cells (B), and HEL cells (C) treated with 50 μM of VC showed that more col 1α2 could be detected in the culture medium compared to non-treated cells. A consistent difference was first detected 3 hrs post treatment of MEF cells (A: middle panel, n=3). By 3 hrs after treatment, less intracellular col 1α2 was detected in VC-treated cells compared with non-treated cells (A: bottom panel, n=3). A consistent difference was detected 6 hrs after treatment of HFF and HEL cells (B&C: middle panel, n=3). Significantly less intracellular col 1α2 could be detected in VC treated cells compared with non-treated cells at 6 hrs after treatment (B&C: bottom panel, n=3). Laminin was used as a loading control for culture medium protein. Actin was used as a loading control for cell lysate protein. * indicates the position of col 1α2. Immunoblot densitometry was quantified using Image J and depicted graphically.
VC treatment enhanced collagen secretion in skin fibroblasts[12], and we verified that 6 hrs of VC treatment significantly increased col 1α2 in the culture medium and reduced col 1α2 in the cell lysate of MEF, HFF, and HEL cells(Figure 1A, 1B, & 1C, middle and bottom panels, n=3). Interestingly, VC treatment did not increase Col1a2 mRNA. Actinomycin D, a transcription inhibitor, did not reduce the short-term effect of VC. (Supplemental Figure 2B, n=3). Thus, a short period of VC treatment did not enhance Col1a2 mRNA transcription, but increased col 1α2 secretion of pepsin-resistant collagen (Supplemental Figure 2 C & 2D).
1.2.2. Prolyl hydroxylase was responsible for VC-stimulated col 1α2 secretion
Proline and lysine hydroxylation by P4H and PLOD, respectively, are important for collagen triple helix formation, which has increased pepsin resistance and secretion rate[16–19]. To determine if VC activates proline or lysine hydroxylases, MEF cells were treated with ethyl-3,4-dihydroxybenzoate (EDBH), a prolyl hydroxylase inhibitor, or minoxidil, an inhibitorof lysyl hydroxylase transcription. EDBH, but not minoxidil, treatment resulted in significant retention of col 1α2 in the cell, reduced col 1α2 secretion, and decreased pepsin-resistant collagen in the culture medium (Supplemental Figure 3A & 3B, n=4). These results suggested that induction of one or more of the P4H isoforms accounted for the increased col 1α2 secretion.
1.2.3. P4HA1 enhanced pepsin-resistant col 1α2 secretionin MEF cells treated with VC.
Three isoforms of P4H have been shown to hydroxylate proline in collagen. P4HA1 and P4HA2 were expressed in MEF cells (Supplemental Figure 4A). Expression of P4HA3, which is found primarily in cancer cells [6], was not assessed due to lack of a commercially available antibody for mouse P4HA3. Silencing P4ha1 in MEF cells with shRNAi did not affect protein or mRNA levels of P4ha2 (Figure 2A, left and middle panels), and did not impact the mRNA levels of Col 1a2 (Figure 2A, left panel). The amount of VC-stimulated pepsin-resistant col 1α2 in culture medium, however, was significantly decreased (Figure 2A, right and middle bottom panels). Knocking out P4ha2 in MEF cells did not impact the P4HA1 expression or the amount of pepsin-resistant col 1α2 in the medium (Figure 2B). Thus, silencing P4ha1, but not P4ha2, significantly impacted col 1α2 secretion indicating that P4HA1 is one of the isoforms responsible for VC-induced proline hydroxylation on col 1α2.
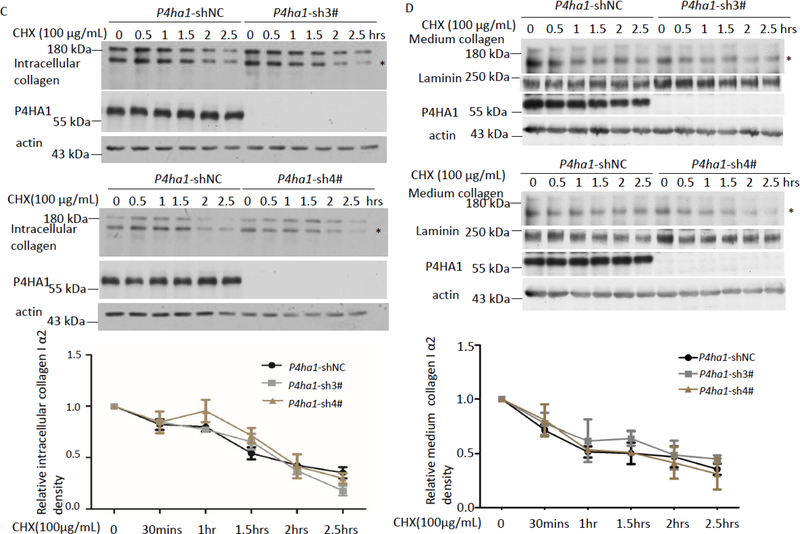
Figure 2.
P4HA1 contributed to VC-enhanced col1α2 secretion. (A) qPCR showed that silencing of P4ha1 did not affect the expression of Col1a2 and P4ha2 mRNA but significantly reduced pepsin-resistant col 1α2 in the culture medium. P4HA1, P4HA2, or Col 1A2 mRNA or protein were detected with quantitative PCR or immunoblotting, respectively. NC: P4ha1-shRNAi-negative control; 3#: P4ha1-shRNAi-3#; 4#: P4ha1-shRNAi-4# (n=3) (left panel). (B) Knockout of P4ha2 did not impact P4HA1 protein level, its glycosylation, or pepsin resistant col 1α2. Downregulation of P4ha1 did not reduce the stability of intracellular col 1α2 (C) and secreted col 1α2 (D). 50 μM of VC treated P4ha1-silenced or control cells were treated with 100 μg/mL CHX for varying times and cell lysates or culture media were immunoblotted for col 1α2 (C: panels 1&4 for intracellular collagen stability; D: panels 1&5 for stability of collagen in culture media). Wherever possible, Laminin was used as a loading control for culture medium protein whereas actin was used as a loading control for cell lysate protein. Densitometry of cell lysate col 1α2 or secreted col 1α2 was quantitated using image J and depicted graphically (n=3, p=NS). * indicates col 1α2.
Silencing P4ha1 could result in reduced col 1α2 protein stability. To determine the effect on protein stability, control and P4ha1 shRNAi transfected MEF cells were incubated with 100 μg/ml cyclohexamide (CHX) and intracellular collagen was measured from 0 to 2.5 hrs. Immunoblot analysis showed no detectable collagen secretion during this time course (data not shown). No significant differences in the rate of intracellular collagen degradation were observed between control and P4ha1 silenced MEF cells (Figure 2C, n=3). These results indicated that silencing P4ha1 did not reduce col 1α2 stability. In addition, no significant difference in col 1α2 stability in culture media was detected with P4ha1-silenced or control MEF cells after VC treatment (Figures 2D, n=3).
1.2.4. Short-term VC treatment stimulated N259 glycosylation on P4HA1.
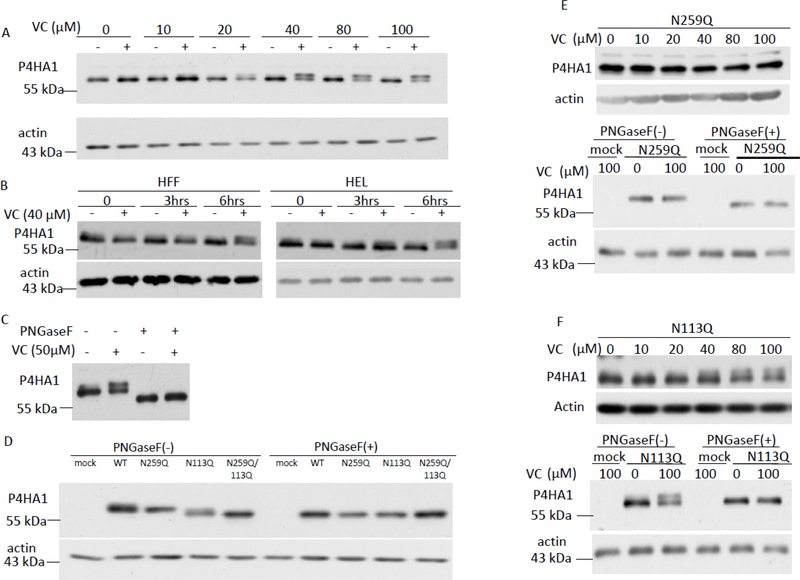
During the preceding studies, it was noted that the P4HA1 antibody recognized an additional protein band in VC-treated MEF cells that was not present in untreated cells (Figure 2A). This suggested that VC treatment caused a post-translational modification of P4HA1. In MEF cells treated with varying concentrations of VC for 6 hrs, a dose-dependent appearance of this additional protein band was noted with obvious effect at 40 μM of VC (Figure 3A). This presumed post-translational modification of P4HA1 was also detected in HEL and HFF cells treated with 40 μM VC (Figure 3B), raising the possibility that the conserved post-translational modification induced by short-term VC treatment might modulate the activity of the enzyme.
Figure 3.
VC induced N259-linked glycosylation on P4HA1. (A) Treatment of MEF cells for 6 hrs with VC concentration of ≥40 μM induced expression of an additional protein band reacting to P4HA1 specific antibody. (B) HFF cells and HEL cells also expressed a second protein reacting to antibody specific to P4HA1 at 6 hrs after VC (40 μM) treatment. (C) PNGaseF treatment of VC-treated and non-treated MEF cell lysate reduced the apparent molecular weight of the proteins reacting to antibody specific for P4HA1. (D) HA-tagged WT, N259Q, N113Q or N113Q/N259Q double mutants were expressed in P4ha1-silenced MEF cells in the absence of VC. The cell lysates were immunoblotted with HA-tag antibody. The N259Q mutant showed the same apparent molecular weight as WT, whereas the apparent molecular weights of N113Q and N113Q/N259Q double mutants were equal but lower than WT (left panel). WT, N113Q, N259Q and N113Q/N259Q double mutants all showed the same motility as N113Q or N113Q/N259Q double mutants after PNGase F treatment (right panel). (E) N259Q mutated P4HA1 did not show a motility shift after VC treatment (top panel) whereas PNGaseF treatment increased motility of N259Q mutated P4HA1 with or without VC stimulation (third panel). (F) 40 μM or higher concentrations of VC induced an additional protein band reacting to HA-tagged antibody for N113Q mutated P4HA1 (top panel), which was sensitive to PNGaseF treatment (third panel).
N-linked glycosylation of P4HA1 has been reported [21], suggesting that the additional protein band induced by VC treatment could represent a glycosylated P4HA1 protein. Therefore, MEF cell lysates were treated with peptide-N-glycosidase F (PNGase F) to remove N-linked oligosaccharides. This abrogated the double band pattern induced by VC treatment and resulted in a single band with enhanced electrophoretic mobility (Figure 3C). These results suggested that P4HA1 undergoes constitutive N-linked glycosylation, and may be further modified by additional N-linked glycans in response to short term VC treatment.
Since N113 and N259 (based upon numbering from the N-terminus of the immature protein) are the two known glycosylation sites on P4HA1, we investigated which glycosylation site was affected by VC. N113Q, N259Q, and N113Q/N259Q P4HA1 mutants were generated, and plasmids were separately transfected into P4ha1-silenced MEF cells. Interestingly, while electrophoretic mobility of influenza hemagglutinin epitope (HA) tagged wild type (WT) P4HA1 and HA tagged N259Q P4HA1 was identical in the absence of VC treatment, mobility of the N113Q P4HA1 and N113Q/N259Q P4HA1 was increased equally (Figure 3D). Treatment with PNGase F demonstrated that differences in the electrophoretic mobility of HA tagged P4HA1 were due to differences in the degree of N-linked glycosylation (Figure 3D). These results suggested that N113glycosylation occurs constitutively in the absence of VC treatment. To prove that N113 glycosylation was VC treatment independent, we treated MEF cells bearing N259Q mutated P4HA1 with or without VC for 6 hrs. We found that the migration of N259Q mutated P4HA1 was not affected by VC treatment (Figure 3E, top panels). In addition, when the cell lysates were subjected to PNGaseF treatment, the mobility of the protein was consistently enhanced compared to cell lysate without PNGaseF treatment (Figure 3E, bottom panels). Thus, N113 was glycosylated, but the glycosylation was not induced by VC treatment.
To show that N259 glycosylation depended on VC treatment, P4ha1-silenced MEF cells were transfected with the N113Q P4HA1 mutant and treated with VC for 6 hrs. Enhanced N259 glycosylation of P4HA1 was induced by VC (Figure 3F, top panel), although with apparently lower efficiency than in WT MEF cells (compare Figures 3A & 3F, top panel). When those cell lysates were treated with or without PNGaseF, the higher molecular weight band disappeared but the bottom band remained suggesting that it was resistant to PNGaseF treatment (Figure 3F, bottom panel). Thus, N259 glycosylation of P4HA1 is induced by VC treatment.
1.2.5. N259 glycosylation on P4HA1 was post-translationally modified by STT3B/MAGT1 complex upon VC treatment.
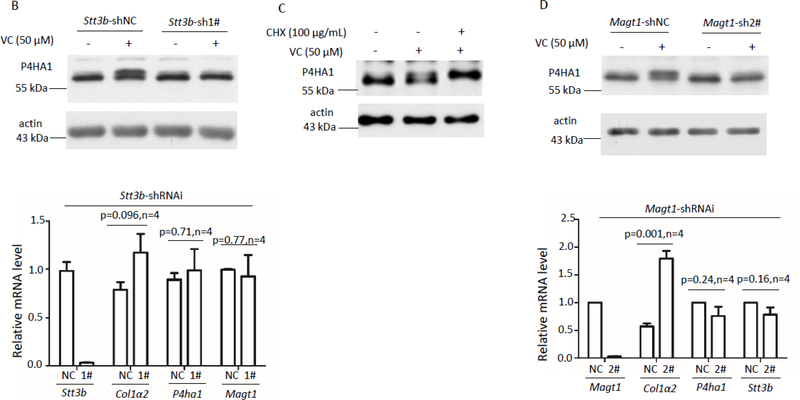
N-linked glycosylation can be co-translational or post-translational. Co-translational N-linked glycosylation depends on STT3A whereas post-translational N-linked modification depends on STT3B/MAGT1 complex[22]. To investigate the nature of N113 and N259 glycosylation, we silenced Stt3a or Stt3b separately in MEF cells (Figures 4A & 4B). Silencing Stt3a or Stt3b did not reduce P4HA1 protein and did not result in glycan-deficient P4HA1 in untreated cells (Figures 4A & 4B). Thus, STT3A or STT3B did not affect N113 glycosylation on P4HA1. In addition, silencing Stt3a neither reduced N259 glycosylation induced by VC nor impacted the mRNA level of Stt3b (Figure 4A). These results suggested that N259 glycosylation induced by VC was independent of STT3A. On the other hand, downregulation of Stt3b significantly impacted N259 glycosylation induced by VC (Figures 4B). These results indicated that VC induced N259 glycosylation was STT3B-dependent and thus post-translational. If so, N259 glycosylation could be derived from the turnover of the pre-existing N113 glycosylated P4HA1. In MEF cells incubated with VC and CHX, the N113 glycosylated band disappeared while the top glycosylated band was enhanced (Figure 4C). Thus, the top glycosylated band was built upon N113 glycosylated P4HA1. Since MAGT1 is in the complex with STT3B as a post-translational modification complex, Magt1 was silenced to investigate the effect on N259 glycosylation. Downregulation of Magt1 also significantly reduced N259 glycosylation without affecting P4HA1 protein level (Figure 4D). These results suggest that N259 is post-translationally modified by STT3B/MAGT1 complex when MEF cells are treated with VC.
Figure 4.
Silencing Stt3b or Magt1 reduced VC stimulated N259 glycosylation on P4HA1. (A) Downregulation of Stt3a (white bars of top panel) did not reduce N113 glycosylation (middle panel) or the mRNA levels of Stt3b (black bars of top panel). NC: Stt3a-shRNAi-negative control; 1#: Stt3a-shRNAi-1#; 2#: Stt3a-shRNAi-2#. Immunoblotting showed that in the absence or presence of VC, downregulation of Stt3a did not affect N113 glycosylation or N259 glycosylation of P4HA1 or P4HA1 protein levels (n=6). Silencing of Stt3b (B) or Magt1 (D) decreased N259 glycosylation on P4HA1 induced by VC without affecting P4HA1 protein levels (left panel). qPCR confirmed efficient downregulation of Stt3b (B) or Magt1 (D). Silencing Stt3b did not affect the mRNA levels of P4ha1, Col1a2, and Magt1 (B). NC: Stt3b-shRNAi-negative control; 1#: Stt3b-shRNAi-1# (n=4). However, silencing Magt1 enhanced Col1a2 but not P4ha1 and Stt3b mRNA levels (D). NC: Magt1-shRNAi-negative control; 2#: Magt1-shRNAi-2# (n=4). (C) Incubation of cells with VC plus CHX at the same time for 6 hrs enhanced N259 glycosylation on P4HA1.
1.2.6. Downregulation of Stt3b and Magt1 reduces pepsin-resistant col 1α2 in culture medium by impacting its entry into the Golgi apparatus.
The apparent correlation between col 1α2 secretion and appearance of N259 glycosylation on P4HA1 after MEF cells were treated with VC for 6 hrs suggested a causal relation between N259 glycosylation and proline hydroxylation. In a time course study, col 1α2 was detected in the medium 3 hrs after VC treatment. This time point correlated with the appearance of N259 glycans on P4HA1, although limited N259 glycosylation could be detected earlier (Figure 5A). In addition, pepsin-resistant col 1α2 could be detected when VC concentration was 20 μM or higher, concentrations that also produced increased N259 glycosylation of P4HA1 (Figure 5B). These results showed a strong correlation between col 1α2 secretion and appearance of N259 glycans on P4HA1.
Figure 5.
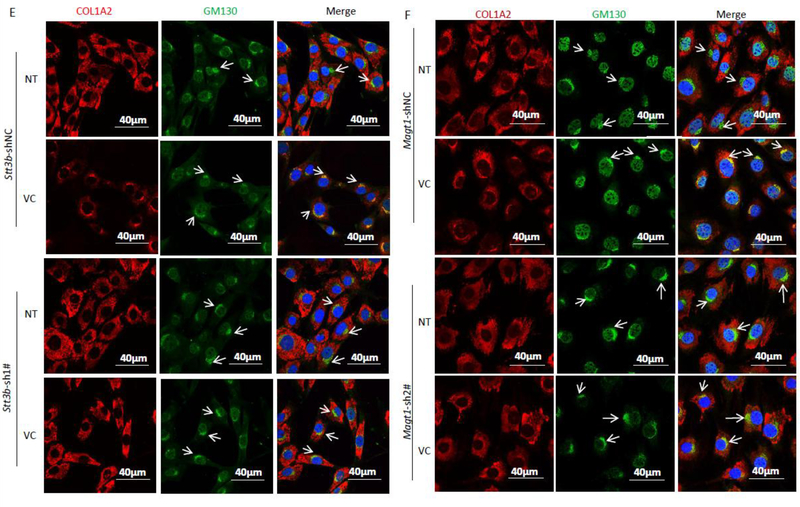
VC-induced N259-linked glycosylation on P4HA1 altered col 1α2 secretion. Enhanced col 1α2 in the culture medium and pepsin-resistant col 1α2 could be detected 3 hrs after VC treatment of MEF cells (top two panels) (A) or when VC concentration was ≥40 μM (top two panels) (B). MEF cells were treated with 50 μM of VC for different times (A) or with different concentrations of VC for 6 hs (B). Col 1α2 in cell lysates or in the culture medium was detected. Quantification of collagen in the medium and N113/N259 and N113glycosylated P4HA1 confirmed a correlation between N259 glycosylation and collagen in the medium ((A) & (B) graphic pictures, n=3). N113/N259 glycosylated P4HA1 (or di-glycosylated P4HA1, Di-) is the top band, N113 glycosylated P4HA1 (or mono-glycosylated P4HA1, Mono-) is the bottom band. Silencing Stt3b (C & E & G) or Magt1 (D & F) reduced VC-induced pepsin-resistant col 1α2 in the culture medium (C & D) and VC activated col 1α2 (red) entry into Golgi apparatus (green, indicated by arrow) (E&F). However, silencing Stt3b did not alter P4HA1 localization in ER (G). Stt3b- or Magt1-silenced or control MEF cells were treated with 50 μM of VC or without VC treatment for 6 hrs, col 1α2 was detected in the cell lysates or in the culture medium (C & D) or was detected by confocal immunofluorescence staining (E & F & G). The ER marker KDEL-EGFP was expressed in Stt3b downregulated MEF cells. P4HA1 was labeled with polyclonal antibody specific to P4HA1. Most P4HA1 co-localized with KDEL-EGFP. Nuclear staining for P4HA1 was non-specific. Quantification of intracellular collagen and pepsin-resistant collagen showed that silencing Stt3b or Magt1 significantly altered VC-induced pepsin-resistant collagen (n=4). Laminin was used as a loading control for culture medium protein whereas actin was used as a loading control for cell lysate. NT: no VC treatment; VC: VC treatment. * indicates col 1α2. Densitometry of immunoblots was quantified using Image J and depicted graphically.
Since STT3B/MAGT1 complex regulated VC-induced N259 glycosylation, Stt3b or Magt1 silencing should impact col 1α2 secretion if N259 glycans play a role for P4HA1 enzymatic activity. When Stt3b was downregulated and MEF cells were treated with VC, col 1α2 in the culture medium was slightly reduced and intracellular col 1α2 was increased (Figures 5C & 5D). Much of the secreted col 1α2 after Stt3b or Magt1 silencing was not pepsin-resistant (Figures 5C & 5D, n=4). Since Stt3b or Magt1 silencing did not reduce P4HA1 protein, Col 1α2 mRNA, or col 1α2 protein, but did decrease N259 glycosylation of P4HA1 (Figures 4B & 4D), these results supported the role of N259 glycosylation in P4HA1 activity and subsequent col 1α2 secretion.
To investigate which step of col 1α2 secretion was affected by decreased N259 glycosylation on P4HA1, intracellular trafficking of col 1α2 was assessed in MEF cells after VC treatment. Stt3b or Magt1 silencing decreased col 1α2 entry into the Golgi apparatus (Figures 5E & 5F). In addition, confocal immunofluorescence microscopy showed that most col 1α2 protein was retained in ER when Stt3b was downregulated (Supplemental Figure 5). To verify that silencing Stt3b did not altered P4HA1 subcellular localization, leading to reduced proline hydroxylation on Type I collagen, P4HA1 location in the Stt3b downregulated cells was assessed. Most P4HA1 co-localized with KDEL-EGFP (Figure 5G), a protein that indicates ER location. The same phenomenon was observed in the control cells. Thus, silencing Stt3b did not alter the subcellular localization of P4HA1. The nuclear staining for P4HA1 was likely to be non-specific because the staining remained in nuclei in P4ha1-silenced cells (Supplemental Figure 6). These results, together with the observation that silencing Stt3b reduced col 1α2 entry into Golgi apparatus, indicated that loss of N259 glycans could alter P4HA1 binding to collagen, thus decreasing collagen hydroxylation and secretion.
1.2.7. Reduced N259 glycosylation on P4HA1 decreased binding between P4HA1 and col 1α2 and attenuated proline hydroxylation on col 1α2.
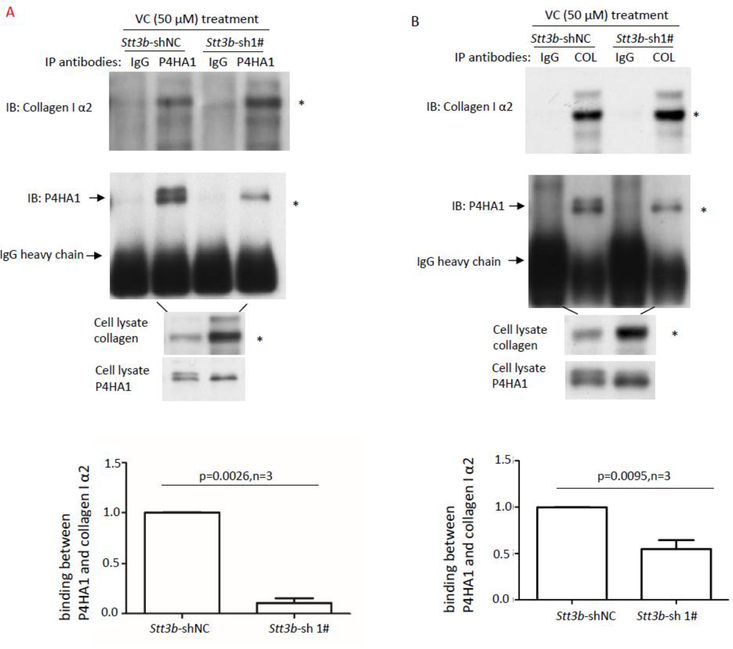
Lack of glycans on P4HA1 was reported to have no effect on its enzymatic activity in vitro [14,15], but N259 glycans on P4HA1 affected col 1α2 secretion in MEF cells. Therefore, the effect of N259 glycosylation on the interactions between P4HA1 and col 1α2 in cells was assessed. Co-immunoprecipitation (Co-IP) of P4HA1 and col 1α2 with anti-P4HA1 or anti-Type I collagen antibody showed significantly less P4HA1 was associated with col 1α2 in Stt3b-silenced cells (Figure 6A & 6B, n=3). The decreased association could impact on subsequent hydroxylation of the substrate (col 1α2).
Figure 6.
Lack of N259 glycosylation on P4HA1 reduces P4HA1 association with col 1α2. Co-IP was performed with anti-P4HA1 (A) or anti-Type I collagen (B) antibody (2μg) using 1 mL of cell lysate. Co-purified col 1α2 and P4HA1 were identified by immunoblot analysis using corresponding antibodies. IgG heavy chain was marked. Immunoblot analysis for col 1α2 was performed with 20 μL of cell lysate. Presence of N259 glycosylation on P4HA1 was indicated by two bands recognized by P4HA1 antibody. Statistical analysis showed that presence of N259 glycans on P4HA1 significantly enhanced the association between col 1α2 and P4HA1 when Co-IP was performed with anti-P4HA1 antibody (n=3) (A) and anti-Type I collagen antibody (n=3) (B). The ratio between P4HA1 and col 1α2 in control MEF cells was defined as 1. The relative ratio between P4HA1 and col 1α2 in Stt3b-silenced cells was calculated based on densitometry on immunoblots assessed by Image J using ratio between P4HA1 and col 1α2 from control cells as standard. (C) Immunoblotting with Type I collagen antibody confirmed that Type I collagen was loaded equivalently from Stt3b-silenced or control MEF cells (indicated by * and #). The slices in a Coomassie brilliant blue stained gel corresponding to the immunoblotting were cut and subjected to mass spectrometry analysis. (D) Significant differences in proline hydroxylation on col 1α2 were observed between Stt3b-silenced and control cells. The number of peptide containing specific proline sites identified by mass spectrometry was included in the bracket. P-value was indicated for each proline site which showed significant differences in hydroxylation between control and Stt3b-silenced cells. NC: Stt3b-shRNAi-negative control (white bars); 1#: Stt3b-shRNAi-1# (black bars). Chi-square analysis was used to analyze differences at each proline site on Type I collagen.
To prove that P4HA1 without N259 glycosylation hydroxylated fewer proline residues on col 1α2, col 1α2 from Stt3b-silenced and control MEF cells was assessed by mass spectrometry. Although similar amounts of col 1α2 were concentrated from Stt3b-silenced and control MEF cells (Figure 6C), significant differences in proline hydroxylation on col 1α2 were noted. When Stt3b was silenced, col 1α2 had reduced proline hydroxylation, especially on proline at sites 96, 294, 777, 795, and 1038 (based upon numbering from the N-terminus of the immature protein) (Figure 6D). However, the reduction of proline hydroxylation was not universal; significantly more hydroxylated proline was detected at sites 564 and 921 on col 1α2 (Figure 6D). Similar results were observed for col 1α1 with reduced proline hydroxylation detected at 8 sites and more hydroxylated proline found at 4 sites (Supplemental Figure 7). Three sets of mass spectrometry results showed similar findings (Supplemental Table 1). These data supported the idea that VC-induced N-linked glycosylation of P4HA1 alters the association between enzyme and substrate and the subsequent proline hydroxylation and collagen maturation.
1.3. Discussion
Type I collagen is the most abundant collagen in the body and collagen production is essential for many physiological processes including tissue repair. Understanding how collagen matures and is secreted will aid in understanding disease conditions due to deficiency or over-deposition of collagen. VC is a cofactor for collagen homeostasis in haplorrhini (including humans, guinea pigs, and some bats), which lack L-gulonolactone oxidase. Although the chemical reactions have been largely solved for proline and lysine hydroxylation on collagen[23–27], it remains unclear how specific proline and lysine sites on collagen are selected by these hydroxylases. Our studies show that prolyl hydroxylation is modulated by VC-induced enhancement of N259 glycosylation on P4HA1 (Figure 6). However, N259 glycosylation does not seem to enhance affinity between P4HA1 and Type I collagen because no enrichment of N259 glycosylated P4HA1 was detected in the Co-IP studies (Figure 6B). The mechanism by which this occurs warrants further investigation.
Mammalian cells possess two oligosaccharyltransferase complexes (STT3A and STT3B) to catalyze N-linked glycosylation, which may facilitate protein folding, enhance protein stability, modify immune recognition, and regulate protein interactions [29,30]. Glycosylation of many NXT/S sites in glycoproteins is not reduced when Stt3a or Stt3b is silenced by siRNA-based method [31,32]. Downregulation of Stt3a or Stt3b does not affect N113 glycosylation on P4HA1. This could be due to low efficiency of downregulation or to the existence of other oligosaccharyltransferase complexes. Work is in progress to understand this. In addition, the effect of N113 glycosylation on P4HA1 activity and collagen maturation remains unclear.
Downregulation of Stt3b or Magt1 does not eliminate VC-stimulated secretion of pepsin-resistant col 1α2 into the culture medium (Figure 5C & 5D). Thus, there is an additional mechanism for VC-activated Type I collagen secretion. Interestingly, it has been reported that VC can stimulate procollagen biosynthesis by enhancing the association between endoplasmic reticulum and ribosome [33]. Whether this is the mechanism responsible for the observed col 1α2 in the culture medium of Stt3b- or Magt1-silenced cells after incubation with VC remains to be investigated.
When Magt1 is silenced, the level of Col1a2 mRNA is significantly elevated (Figure 4D, right panel), without a concomitant increase in intracellular protein level of col 1α2 (Figure 5D). The explanation for this remains unclear. Col1a2 translation efficacy or protein stability could be reduced.
It is reported that glycans on P4HA1 do not change its total enzymatic activity when purified enzymes are used to test enzymatic activity in vitro [14,15]. In Stt3b-silenced cells, reduced N259 glycosylation decreases P4HA1 association with col 1α2 and altered hydroxylation of specific proline residues. The mechanism by which P4HA1 N259 glycosylation alters P4HA1 association with col 1α2 or selection of proline residues for hydroxylation remains unknown. Silencing Stt3b and subsequent decrease in N259 glycosylation of P4HA1 appear to reduce P4HA1 binding to its antibody when the antibody was used in Co-IP studies (Figure 6A).
Although N259 glycosylation on P4HA1 modulates proline hydroxylation, it does not have a universal effect on col 1α2. In Stt3b-silenced cells, five proline sites of col 1α2 had decreased hydroxylation and two had increased hydroxylation (Figure 6). This suggests that N259 glycans fine tune the interaction between col 1α2 and P4HA1. Similar effects were detected for col 1α1. Identification of the mechanism by which the N259 glycans regulate proline hydroxylation at specific sites of collagen will require detailed structural studies.
1.4. Materials and Methods
1.4.1. Antibodies and reagents
Ascorbic acid (A7506) and ethyl-3,4-dihydroxybenzoate (EDHB) (E24859) were purchased from Sigma-Aldrich (Darmstadt, Germany). GM130 rabbit polyclonal antibody (A5344), P4HA1 rabbit polyclonal antibody (A3999), Rabbit polyclonal antibody anti-Laminin (LAMC1) (A16020), Rabbit anti-HA-Tag polyclonal antibody (AE036), HRP-conjugated goat anti-mouse IgG (AS003), goat anti-rabbit IgG (AS014), and HRP conjugated rabbit anti-goat IgG (AS029) were purchased from ABclonal (Cambridge, MA, USA). Goat anti-human Type I collagen (1310–01s) antibody, which was shown to react to mouse Type I collagen, was purchased from Southern Biotech (Birmingham, AL, USA),. Proteinase inhibitor cocktail (11697498001) and 4', 6-diamidino-2-phenylindole (DAPI) (10236276001) were purchased from Roche Diagnostics (Mannheim, Germany). Alexa Fluor 555 conjugated donkey anti-goat IgG (ANT082) was purchased from AntGene Biotech (Wuhan, China). Alexa Fluor 488 conjugated donkey anti-rabbit IgG (A21206)waspurchased from Invitrogen (Eugene, OR, USA). Protein G Agarose Fast Flow beads (16–266) were purchased from Merck Millipore (Darmstadt, Germany). PNGase F (P0704S) was purchased from New England BioLabs (Beverly, MA, USA).
1.4.2. Fibroblast Cell Culture
MEF cells were purchased from ATCC and were grown in Dulbecco’s modified Eagle’s medium (DMEM; high glucose without ascorbic acid), supplemented with 1% antibiotic Penicillin-Streptomycin solution (PS) (03–031-1, Biological Industries, Kibbutz Beit-Haemek, Israel) plus 10% fetal bovine serum (FBS) (04–001-1, Biological Industrie,Kibbutz Beit-Haemek, Israel). HFF cells and HEL cells were isolated and provided by Dr. Min-hua Luo at Wuhan Institute of Virology, Chinese Academy of Sciences and used between cell passages six and ten.All cells were cultured in a 37°, 5% CO2, 95% humidity incubator.
To evaluate the effect of VC on collagen production, all fibroblasts were seeded in 6-well plates and cultured for 24 hrs in 10% FBS DMEM until reaching 85% confluence. Cells were then maintained in 1% FBS DMEM overnight and stimulated for 0, 1, 3, and 6 hrs in serum-free DMEM containing 50 μM VC or as indicated in the manuscript. At the end of treatment, conditioned media were collected and spun at 1000×g for 30 minutes (mins). Collected media containing Type Icollagen were precipitated with 176 mg/mL ammonium sulfate for 24 hrs at 4°. Samples were then centrifuged at 10,000×g for 1 hr, and the pellet was solubilized in a solution containing 0.4 M NaCl, 0.1 M Tris, pH 7.4.
The cells were rinsed twice with ice cold phosphate buffered saline (PBS) and scraped into cell lysis buffer (20 mMTris-HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 2.5 mM sodium pyrophosphate, 1 mM b-glycerophosphate, 1 mM Na3VO4, 1% Triton, 1 mM PMSF and 1 × proteinase inhibitor cocktail) and sonicated at 1J for six seconds on ice. The cell debris was removed by centrifugation and protein concentration was quantified using a Bio-Rad Protein Assay Kit II.
1.4.3. SDS-Page and Western blotting analysis
To determine the level of type I collagen secreted from control and VC-treated fibroblasts, western blots were performed on the cell lysate and conditioned media prepared as above. The samples were mixed with 4 × sodium dodecyl sulfate (SDS) reducing sample buffer (40% glycerol (V/V), 250 mM Tris-HCl pH 6.8 (V/V), 8% sodium dodecyl sulfate (W/V), 0.04% bromophenol blue (W/V), 20% 2-mercaptoethanol (ME) (V/V)). The samples were then heated at 100 °C for 10 mins and loaded in a 7.5% SDS-PAGE. After electrophoresis, proteins were transferred to nitrocellulose membranes (HATF00010, Merck Millipore), which were further blocked with bovine serum albumin (BSA) in tris-buffered saline (TBS) with 0.1% Tween-20 (TBST) at room temperature (RT) for 3 hrs and then incubated with anti-Type I collagen antibody (0.4 μg/mL), anti-Laminin antibody (1 μg/mL), anti-P4HA1 (1 μg/mL), anti-P4HA2 (1 μg/mL), or anti-actin antibody (0.2 μg/mL) overnight at 4 °C. Bound primary antibodies were detected with rabbit anti-goat, goat anti-rabbit or mouse HRP-conjugated secondary antibody (0.12μg/mL), respectively. The amount of type I collagen was quantified by densitometry using the Image J software (NIH) and were normalized to actin loading controls. The data were presented as the mean ±SEM.
1.4.4. Intracellular and medium collagen stability in P4ha1 silencing MEF cells
To determine intracellular collagen stability after P4ha1 silencing, P4ha1-silenced or control MEF cells were incubated with 100 μg/mL of CHX, the protein translation inhibitor, for various times. Cell lysates were collected and assayed by immunoblot analysis as indicated.
To determine medium collagen stability after P4ha1 silencing, P4ha1-silenced or control MEF cells were treated with 50μM VC for 3 hrs, then 100 μg/mL of CHX added for varying times (time 0 was defined as time of CHX addition).After treatment, cell lysates and media were collected and assayed by immunoblot analysis as indicated.
1.4.5. Immunofluorescence (IF) Staining
To investigate the effect of silencing Stt3b or Magt1 on Type I collagen’s entry into Golgi apparatus after VC treatment, cells were seeded on confocal petri dishes (801002, NEST, China) and cultured for 24 hrs in 10% FBS DMEM until reaching 85% confluence. Cells were then maintained in DMEM containing 1% FBS overnight and stimulated for 6 hrs in DMEM containing 1% FBS with 50 μM VC. After treatment, IF staining was performed to detect the col 1α2 localization. The cells were rinsed twice with ice cold PBS and then fixed with 4% (W/V) paraformaldehyde in PBS for 10 mins at RT. After washing three times, fixed cells were permeabilized with 0.1% (V/V) Triton X-100 in PBST for 10 mins. Permeabilized cells were blocked with 5% BSA in PBST for 2 hrs at RT. To detect the co-localization of col1α2 and Golgi apparatus, goat anti-type I collagen or rabbit anti-GM130 antibody were applied for 2 hrs at RT. Bound primary antibodies were then detected with Alexa Fluor-conjugated donkey anti-rabbit or goat IgG. Nuclei were counterstained with DAPI (500 ng/ml) for 5 mins.
To investigate whether silencing Stt3b affects P4HA1 sub-cellular localization, Stt3b-silenced and control MEF cells were seeded on confocal petri dishes and cultured for 12hrs in 10 % FBS DMEM until 50 % confluent. The ER marker EGFP-KDEL was transiently transfected in above cells with Lipofectamine 2000 reagent according to the manufacturer's protocol. Briefly, the Lipofectamine 2000 reagent (6 μL)or vector (2 μg) was diluted in Opti-MEM Medium (150 μL ). The diluted vector was added to diluted Lipofectamine 2000 Reagent (1:1 ratio). After 20 mins incubation, the mixture was added to the cells, and they were cultured for 24 hrs.Then cells were maintained in DMEM containing 1% FBS overnight and stimulated for 6 hrs in DMEM containing 1% FBS, 50 μM VC. IF staining was performed as above.
1.4.6. Establishment of stable knockdown and knockout cell lines.
To generate stable P4ha1, Stt3a, Stt3b and Magt1-silenced cells, shRNA against P4ha1, Stt3a, Stt3b, and Magt1 were designed using online BLOCK-iT™ RNAi Designer software.
shRNAi against the above genes were sub-cloned into pLKO.1 Vector (VEC-PRT-0002,Oligoengine, US) according to the manufacturer’s instructions. Briefly, the oligonucleotide sequences were annealed and ligated into pLKO.1 Vector at 22 °for 2 hrs. The ligation product was transformed into E. coli (DH5a). Positive clones were selected after EcoRI and NcoI digestion showing two fragments, a 2 kb fragment and a 5 kb fragment.
Lentivirus particles were generated. Briefly, HEK293T cells were co-transfected with pLKO.1 recombinant construct and the packaging plasmids pMD2.G and psPAX2 (a gift supplied by Professor Cunyi Zheng, Wuhan University, China) for 72 hrs. Media was then collected and filtered with a 0.45 μm filter. The shRNA retroviral stocks were used to infect MEF cells in the presence of 7.5 μg/ml Polybrene (107689, Sigma, USA). The stably silenced cells were screened with 2 μg/ml puromycin (ant-pr-1, InvivoGen, USA) for at least two weeks. The interference effect was assessed with qPCR and immunoblotting. The target sequences by shRNAi were listed in Table 1.
Table 1.
The P4ha1, P4ha2, Stt3a, Stt3b and Magt1 sequences targeted by shRNAi or sgRNA.
| Name | Sequence |
|---|---|
| P4ha1-shRNAi-sh3# | 5’-GGAGTGCAACATAAGTCTTTC-3’ |
| P4ha1-shRNAi-sh4# | 5’-GCTGTCTACTGTAGATAAAGT-3’ |
| Stt3b-shRNAi-sh1# | 5’-GCTGCTTGCTTCATTGCTATC -3’ |
| Magt1-shRNAi-sh2# | 5’-GGACTTGTGTATCTGCGAAGA-3’ |
| Stt3a-shRNAi-sh1# | 5’-GCATGCTGCTTACTTACTACA-3’ |
| Stt3a-shRNAi-sh2# | 5’-GCTGTAATGGTGCGTCTAATG-3’ |
| P4ha2-sgRNA-1# | 5’-CGATCTGATTTACGCAGAGA-3’ |
| P4ha2-sgRNA-4# | 5’-AGATCAGCTGCCGACCCCGA-3’ |
To generate P4ha2 null MEF cells, CRISPR/Cas systems were used. The primers for P4ha2 knockout in MEF cells were listed in Table 1. To knockout P4ha2 in MEF cells, the primers (100μM) were annealed and ligated to pX459. The recombinant plasmids were sequenced and transfected into MEF cells with Lipofectamine 2000 reagent according to the manufacturer's protocol. Three days post-transfection, cells were screened with 2 μg/ml puromycin. P4ha2 knockout MEF cells were verified by DNA sequencing and western blotting.
1.4.7. RNA Isolation and Quantitative Real-Time PCR (qPCR)
To evaluate whether silencing P4ha1, Stt3b, Magt1, or Stt3a in MEF cells influenced other genes expression, RNA was isolated from control and corresponding silenced cells using the total RNA purification kit (GeneMark, number TR01–150, Taichung, Taiwan). RNA (1 μg) was reverse transcribed using a PrimeScript TM RT reagent kit (TaKaRa, number RR047A, Shiga, Japan). qPCR was carried out using SYBR Green Supermix (Bio-Rad, number170–8882AP, Hercules, CA, USA) on a Bio-Rad Connect TM real-time PCR instrument (CFX Connect TM Optics Module). Each reaction volume of 10 μL contained cDNA templates, primer pairs, and SYBR Green Supermix. Amplification occurred after initial denaturation at 95° for 3 mins, followed by 40 cycles at 95° for 10 seconds, 53° for 15 seconds, and 72° for 15 seconds. β-Actin was used as a reference gene. Gene-specific primers were listed in Table 2.
Table 2.
Primers used to amplify Col1α2, β-Actin, P4ha1, Stt3b, Magt1, and Stt3a from MEF cells.
| Gene name | Sequence |
|---|---|
| Col1α2 | |
| Sense: | 5’-GCGGTGAAGAAGGAAAGAGA-3’ |
| Antisense: | 5’- CCAGGAGACCCAGGAAGAC-3’ |
| P4ha1 | |
| Sense: | 5’-CAAGCAGGAGGACGAGTGG-3’ |
| Antisense: | 5’-TGGGTTTGAAATGGTGGC-3’ |
| P4ha2 | |
| Sense: | 5’- GTGTGGACGACTGCTTTGG-3’ |
| Antisense: | 5’-TTGACAGTGATTTCCCTCTTTCT-3’ |
| Stt3b | |
| Sense: | 5’-TGGTCCTTGCCTCCTACAAT-3’ |
| Antisense: | 5’-GCCAATCACTCCTCCGAAA-3’ |
| Magt1 | |
| Sense: | 5’-TGTTATTGGTGGACTTGTGTATC-3’ |
| Antisense: | 5’-GAAGAACGATGTGGGTTTCA-3’ |
| Stt3a | |
| Sense: | 5’-CCTGTCTGATGCTCGGATTT-3’ |
| Antisense: | 5’-CCCACTCGCCACCTCATT-3’ |
| β-Actin | |
| Sense: | 5’-CGTGCGTGACATCAAAGAGAAGC-3’ |
| Antisense: | 5’-TGGATGCCACAGGATTCCATACC-3’ |
1.4.8. Construction of plasmids
pHAGE-P4HA1 was constructed as below. Briefly,P4HA1 was amplified with primers listed in Table 3 using the cDNA from MEF cells. The PCR amplified fragment was gel purified and digested with NotI and NheI at 37° for 2 hrs. The digested sample was further gel purified and ligated into pHAGE-CMV-MCS-IZsGreen vector by standard molecular biology techniques.
Table 3.
Primers used to amplify P4ha1 and EGFP-KDEL and mutate P4ha1.
| Gene name | Sequence |
|---|---|
| P4ha1 | |
| Sense: | 5’ATTTGCGGCCGCCACCATGATCTGGGTTGTTTTAATGA-3’ |
| Antisense: | 5’-CTAGCTAGCTTCCAATTCTGACAGGGTG-3’ |
| HA-tagged-P4ha1 | |
| Sense: | 5’-CGTAAAAAACCCTGGATGAGCGTAATCTGGAACATCGTATGGGTAGGCTAAAGACTGAGGAAG-3’ |
| Antisense: | 5’-CTTCCTCAGTCTTTAGCCTACCCATACGATGTTCCAGATTACGCTCATCCAGGGTTTTTTACG-3’ |
| P4ha1-N113Q | |
| Sense: | 5’-ACTGCCTCTGAATGGTTAGTTGAGAGATGAAGCCATCCGAC-3’ |
| Antisense: | 5’-GTCGGATGGCTTCATCTCTCAACTAACCATTCAGAGGCAGT-3’ |
| P4ha1-N259Q | |
| Sense: | 5’-GGTCGCCCGACGCTGACTTTTGGGCATCTTTTTCTTTACTCA-3’ |
| Antisense: | 5’-TGAGTAAAGAAAAAGATGCCCAAAAGTCAGCGTCGGGCGACC-3’ |
| Signal peptide of Calreticulin | |
| Sense: | 5’-ATTTGCGGCCGCCACCATGCTCCTTTCGGTGCCG-3’ |
| Antisense: | 5’-CAGCTCCTCGCCCTTGCTCACTGCGGCGGCCAGGCCGAGGA-3’ |
| Egfp | |
| Sense: | 5’- TCCTCGGCCTGGCCGCCGCAGTGAGCAAGGGCGAGGAGCTG-3’ |
| Antisense: | 5’- GCTCTAGATTACAGCTCATCCTTGTACAGCTCGTCCATGCC-3’ |
Constructs of pHAGE-HA-tagged-P4HA1 were amplified through insertion mutation with mutagenic primers listed in Table 3. Briefly, pHAGE-P4HA1 was amplified with mutagenic primers containing influenza hemagglutinin epitope (HA) sequence. The amplified sample was digested with DpnI at 37° for 2 hrs. The digested sample was directly transformed intoE. Coli (DH5a). pHAGE-HA-tagged-P4HA1-N113Q, P4HA1-N259Q, and P4HA1-N113Q/N259Q (double mutations) were mutated with corresponding primers listed in Table 3.
The lentiviral packaging system consisted of one expression plasmid: pHAGE-CMV-MCS-IZsGreen and two packaging plasmids: pMD2.G and psPAX2. Generation of lentivirus particles containing WT and mutant P4HA1 was similar to shRNA retrovirus described above. The lentivirus stocks were used to infect P4ha1-silenced MEF cells.
Constructions of pcDNA3.1-EGFP-KDEL were amplified through Overlap PCR with the corresponding primers listed in Table 3. The EGFP containing the amino acids KDEL was amplified from the pEGFP-N1 vector (the reverse primer containing the nucleotide sequence of KDEL motif). Signal peptide of calreticulin was amplified using the cDNA from MEF cells. These two DNA fragments were then mixed and annealed to obtain hybrid duplexes. The resulting hybrids were then extended and amplified to yield recombinant PCR products. The product was digested with NotI and XbaI and ligated into pcDNA3.1.
1.4.9. Glycosidase assay
PNGase F treatment was performed according to manufacturer’s instructions. Briefly, MEF cells were stimulated with 50 μM VC for 6 hrs. Cell lysates were extracted and incubated with glycoprotein denaturing buffer. The samples were boiled for 10 mins and then treated with PNGase F for 1 hr at 37°. PNGase F treated samples were then assayed by immunoblotting as above.
N113/N259 glycosylated P4HA1 and N113 glycosylated P4HA1 bands were quantitated by densitometry using Image J. Densitometry of collagen bands and glycosylated P4HA1 bands were depicted graphically.
To investigate which glycosylation site was VC treatment dependent, stably transfected HA-tagged N113Q and N259Q P4HA1 cells were stimulated with 100μM VC for 6 hrs. The cell lysates were treated with PNGase F as above and used for immunoblot analysis to assess glycosylation status of N113Q and N259Q mutated P4HA1.
To investigate if the STT3B/MAGT1 complex glycosylated newly made P4HA1 or pre-existing N113-glycan form of P4HA1, MEF cells were maintained in DMEM containing 1% FBS overnight and treated for 6 hrs in serum-free DMEM containing 50 μM VC alone, or 50 μM VC plus 100 μg/mL CHX. After treatment, cell lysates were immunoblotted to visualize the glycosylation status of P4HA1.
1.4.10. Co-IP
To detect the interaction between P4HA1 and col 1α2 in silenced or control MEF cells, Stt3b-silenced or control cells were grown in 10% FBS DMEM until reaching 85% confluence. The cells were then maintained in DMEM containing 1% FBS overnight and treated for 6 hrs in serum-free DMEM containing 50 μM VC. Cell lysates were collected as above. Co-IP was performed with 2 μg of anti-P4HA1 antibody or anti-Type I collagen antibody and their corresponding isotype control antibodies. Briefly, 1 ml of cell lysate was incubated with P4HA1 or Type I collagen antibody and 40 μL of 50% slurry of protein G Sepharose beads(16–266, Merck Millipore) at 4° overnight. After washing 6 times with cell lysis buffer, the beads were boiled in 40 μl 2×SDS reducing sample buffer. 20 μL cell lysates was used for Co-IP and proteins were separated on a 7.5% SDS-PAGE.
1.4.11. Liquid Chromatograph Mass Spectrometer (LC-MS)
To enrich media Type I collagen for LC-MS analysis,eight 15 cm Petri dishes of Stt3b-silenced and twelve 15 cm Petri dishes of control cells were treated with VC as above. Conditioned media were collected and spun at 1,000×g for 30 mins. Media containing Type I collagen was concentrated with Amicon Ultra-15, 30K (UFC 903001, Millipore). The concentrated media was mixed with 4× SDS reducing sample buffer and separated on a 7.5% SDS-PAGE. The gel was stained by Coomassie brilliant blue. The slices corresponding to immunoblot containing Type I collagen were cut off for LC-MS. The peptide solutions were digested with trypsin and analyzed by Orbitrap Elite LC-MS/MS (Thermo, USA). Peptides were separated on the C18 Reversed Phase HPLC Columns (1.8 μm,0.15×1,00 mm ) at a flow rate of 600 nL/min with a binary gradient. Identification of peptides containing Pro residues and their hydroxylated forms was performed by searching the acquired MS/MS spectra against the Uniprot-Mus musculus.fasta using Proteome Discoverer 1.4 software (Thermo). Quantitation of Pro 4-hydroxylation (4-Hyp) at a specific Pro site was calculated using the ratio of the number of peptides containing corresponding specific 4-Hyp site to the number of total peptides containing corresponding 4-Hyp and Pro site. The experiments were repeated three times.
1.4.12. Statistical analyses
Two major statistical tests were applied in this manuscript. When comparing the measurements under two treatment groups, a two-sample t-test was performed to determine if the two measurements were significantly different. When comparing the proportions of two groups, a Chi-squared test was performed to determine if the two proportions were significantly different. All the experiments were repeated at least 3 times and analyzed by two tailed t-test. Densitometry of immunoblots was quantified by Image J. Chi-square test was used to assess the hydroxylated proline data obtained from Mass Spectrometry. Quantitative data were expressed as the mean ± standard error of the mean. p<0.05 was considered statistically significant.
Supplementary Material
Acknowledgements:
We thank the Core Facility and Technical Support, Wuhan Institute of Virology for assistance with confocal microscope (Dr. Ding Gao) and flow cytometry (Ms. Juan Min).
Funding: This work was supported in part by National Institutes of Health, National Heart, Lung, and Blood Institute (Grant R01-HL41178 to LMG), by MOST (2018YFA0507201 to CL), by the Strategic Priority Research Program A of the Chinese Academy of Sciences (XDA12010309 to CL), by the National Basic Research Priorities Program of China (2013CB911102 to CL), by National Science Foundation of China (31670170 to CL).
Footnotes
Conflict of Interest: None.
References:
- 1.Prockop DJ, Kivirikko KI (1995) Collagens: molecular biology, diseases, and potentials for therapy. Annual review of biochemistry 64:403–434. doi: 10.1146/annurev.bi.64.070195.002155 [DOI] [PubMed] [Google Scholar]
- 2.Cardinale GJ, Udenfriend S (1974) Prolyl hydroxylase. Advances in enzymology and related areas of molecular biology 41 (0):245–300 [DOI] [PubMed] [Google Scholar]
- 3.Helaakoski T, Vuori K, Myllyla R, Kivirikko KI, Pihlajaniemi T (1989) Molecular cloning of the alpha-subunit of human prolyl 4-hydroxylase: the complete cDNA-derived amino acid sequence and evidence for alternative splicing of RNA transcripts. Proceedings of the National Academy of Sciences of the United States of America 86 (12):4392–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helaakoski T, Veijola J, Vuori K, Rehn M, Chow LT, Taillon-Miller P, Kivirikko KI, Pihlajaniemi T (1994) Structure and expression of the human gene for the alpha subunit of prolyl 4-hydroxylase. The two alternatively spliced types of mRNA correspond to two homologous exons the sequences of which are expressed in a variety of tissues. The Journal of biological chemistry 269 (45):27847–27854 [PubMed] [Google Scholar]
- 5.Annunen P, Helaakoski T, Myllyharju J, Veijola J, Pihlajaniemi T, Kivirikko KI (1997) Cloning of the human prolyl 4-hydroxylase alpha subunit isoform alpha(II) and characterization of the type II enzyme tetramer. The alpha(I) and alpha(II) subunits do not form a mixed alpha(I)alpha(II)beta2 tetramer. The Journal of biological chemistry 272 (28):17342–17348 [DOI] [PubMed] [Google Scholar]
- 6.Kukkola L, Hieta R, Kivirikko KI, Myllyharju J (2003) Identification and characterization of a third human, rat, and mouse collagen prolyl 4-hydroxylase isoenzyme. The Journal of biological chemistry 278 (48):47685–47693. doi: 10.1074/jbc.M306806200 [DOI] [PubMed] [Google Scholar]
- 7.Gilkes DM, Bajpai S, Chaturvedi P, Wirtz D, Semenza GL (2013) Hypoxia-inducible factor 1 (HIF-1) promotes extracellular matrix remodeling under hypoxic conditions by inducing P4HA1, P4HA2, and PLOD2 expression in fibroblasts. The Journal of biological chemistry 288 (15):10819–10829. doi: 10.1074/jbc.M112.442939 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Ruotsalainen H, Vanhatupa S, Tampio M, Sipila L, Valtavaara M, Myllyla R (2001) Complete genomic structure of mouse lysyl hydroxylase 2 and lysyl hydroxylase 3/collagen glucosyltransferase. Matrix biology : journal of the International Society for Matrix Biology 20 (2):137–146 [DOI] [PubMed] [Google Scholar]
- 9.Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL (2005) Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood 105 (2):659–669. doi: 10.1182/blood-2004-07-2958 [DOI] [PubMed] [Google Scholar]
- 10.Hofbauer KH, Gess B, Lohaus C, Meyer HE, Katschinski D, Kurtz A (2003) Oxygen tension regulates the expression of a group of procollagen hydroxylases. European journal of biochemistry 270 (22):4515–4522 [DOI] [PubMed] [Google Scholar]
- 11.Murad S, Sivarajah A, Pinnell SR (1980) Prolyl and lysyl hydroxylase activities of human skin fibroblasts: effect of donor age and ascorbate. The Journal of investigative dermatology 75 (5):404–407 [DOI] [PubMed] [Google Scholar]
- 12.Murad S, Sivarajah A, Pinnell SR (1981) Regulation of prolyl and lysyl hydroxylase activities in cultured human skin fibroblasts by ascorbic acid. Biochemical and biophysical research communications 101 (3):868–875 [DOI] [PubMed] [Google Scholar]
- 13.Skropeta D (2009) The effect of individual N-glycans on enzyme activity. Bioorganic & medicinal chemistry 17 (7):2645–2653. doi: 10.1016/j.bmc.2009.02.037 [DOI] [PubMed] [Google Scholar]
- 14.Kedersha NL, Tkacz JS, Berg RA (1985) Characterization of the oligosaccharides of prolyl hydroxylase, a microsomal glycoprotein. Biochemistry 24 (21):5952–5960 [DOI] [PubMed] [Google Scholar]
- 15.Lamberg A, Pihlajaniemi T, Kivirikko KI (1995) Site-directed mutagenesis of the alpha subunit of human prolyl 4-hydroxylase. Identification of three histidine residues critical for catalytic activity. The Journal of biological chemistry 270 (17):9926–9931 [DOI] [PubMed] [Google Scholar]
- 16.Uitto J, Dehm P, Prockop DJ (1972) Incorporation of cis-hydroxyproline into collagen by tendon cells. Failure of the intracellular collagen to assume a triple-helical conformation. Biochimica et biophysica acta 278 (3):601–605 [DOI] [PubMed] [Google Scholar]
- 17.Sasaki T, Majamaa K, Uitto J (1987) Reduction of collagen production in keloid fibroblast cultures by ethyl-3,4-dihydroxybenzoate. Inhibition of prolyl hydroxylase activity as a mechanism of action. The Journal of biological chemistry 262 (19):9397–9403 [PubMed] [Google Scholar]
- 18.Myllyharju J, Kivirikko KI (2001) Collagens and collagen-related diseases. Annals of medicine 33 (1):7–21 [DOI] [PubMed] [Google Scholar]
- 19.Peterkofsky B (1991) Ascorbate requirement for hydroxylation and secretion of procollagen: relationship to inhibition of collagen synthesis in scurvy. The American journal of clinical nutrition 54 (6 Suppl):1135s–1140s. doi: 10.1093/ajcn/54.6.1135s [DOI] [PubMed] [Google Scholar]
- 20.Clark CC, Richards CF (1988) Underhydroxylated minor cartilage collagen precursors cannot form stable triple helices. The Biochemical journal 250 (1):65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kedersha NL, Tkacz JS, Berg RA (1985) Biosynthesis of prolyl hydroxylase: evidence for two separate dolichol-media pathways of glycosylation. Biochemistry 24 (21):5960–5967 [DOI] [PubMed] [Google Scholar]
- 22.Kelleher DJ, Karaoglu D, Mandon EC, Gilmore R (2003) Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Molecular cell 12 (1):101–111 [DOI] [PubMed] [Google Scholar]
- 23.Tuderman L, Myllyla R, Kivirikko KI (1977) Mechanism of the prolyl hydroxylase reaction. 1. Role of co-substrates. European journal of biochemistry 80 (2):341–348 [DOI] [PubMed] [Google Scholar]
- 24.Myllyla R, Tuderman L, Kivirikko KI (1977) Mechanism of the prolyl hydroxylase reaction. 2. Kinetic analysis of the reaction sequence. European journal of biochemistry 80 (2):349–357 [DOI] [PubMed] [Google Scholar]
- 25.Myllyla R, Kuutti-Savolainen ER, Kivirikko KI (1978) The role of ascorbate in the prolyl hydroxylase reaction. Biochemical and biophysical research communications 83 (2):441–448 [DOI] [PubMed] [Google Scholar]
- 26.Puistola U, Turpeenniemi-Hujanen TM, Myllyla R, Kivirikko KI (1980) Studies on the lysyl hydroxylase reaction. II. Inhibition kinetics and the reaction mechanism. Biochimica et biophysica acta 611 (1):51–60 [DOI] [PubMed] [Google Scholar]
- 27.Puistola U, Turpeenniemi-Hujanen TM, Myllyla R, Kivirikko KI (1980) Studies on the lysyl hydroxylase reaction. I. Initial velocity kinetics and related aspects. Biochimica et biophysica acta 611 (1):40–50 [DOI] [PubMed] [Google Scholar]
- 28.De Jong L, Kemp A (1984) Stoicheiometry and kinetics of the prolyl 4-hydroxylase partial reaction. Biochimica et biophysica acta 787 (1):105–111 [DOI] [PubMed] [Google Scholar]
- 29.Dalziel M, Crispin M, Scanlan CN, Zitzmann N, Dwek RA (2014) Emerging principles for the therapeutic exploitation of glycosylation. Science (New York, NY) 343 (6166):1235681. doi: 10.1126/science.1235681 [DOI] [PubMed] [Google Scholar]
- 30.Gao Z, Zhang H, Hu F, Yang L, Yang X, Zhu Y, Sy MS, Li C (2016) Glycan-deficient PrP stimulates VEGFR2 signaling via glycosaminoglycan. Cellular signalling 28 (6):652–662. doi: 10.1016/j.cellsig.2016.03.010 [DOI] [PubMed] [Google Scholar]
- 31.Cherepanova NA, Gilmore R (2016) Mammalian cells lacking either the cotranslational or posttranslocational oligosaccharyltransferase complex display substrate-dependent defects in asparagine linked glycosylation. Scientific reports 6:20946. doi: 10.1038/srep20946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Canada C, Kelleher DJ, Gilmore R (2009) Cotranslational and posttranslational N-glycosylation of polypeptides by distinct mammalian OST isoforms. Cell 136 (2):272–283. doi: 10.1016/j.cell.2008.11.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ueno T, Tanaka K, Kaneko K, Taga Y, Sata T, Irie S, Hattori S, Ogawa-Goto K (2010) Enhancement of procollagen biosynthesis by p180 through augmented ribosome association on the endoplasmic reticulum in response to stimulated secretion. The Journal of biological chemistry 285 (39):29941–29950. doi: 10.1074/jbc.M109.094607 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.