Summary paragraph
Inflammatory bowel diseases (IBD), which include Crohn’s disease (CD) and ulcerative colitis (UC), affect several million individuals worldwide. CD and UC are complex diseases and heterogeneous at the clinical, immunological, molecular, genetic, and microbial levels. Extensive study has focused on individual contributing factors. As part of the Integrative Human Microbiome Project (HMP2), 132 subjects were followed for one year each to generate integrated longitudinal molecular profiles of host and microbial activity during disease (up to 24 time points each; in total 2,965 stool, biopsy, and blood specimens). These provide a comprehensive view of the gut microbiome’s functional dysbiosis during IBD activity, showing a characteristic increase in facultative anaerobes at the expense of obligate anaerobes, as well as molecular disruptions in microbial transcription (e.g. among clostridia), metabolite pools (acylcarnitines, bile acids, and short-chain fatty acids), and host serum antibody levels. Disease was also marked by greater temporal variability, with characteristic taxonomic, functional, and biochemical shifts. Finally, integrative analysis identified microbial, biochemical, and host factors central to the dysregulation. The study’s infrastructure resources, results, and data, available through the Inflammatory Bowel Disease Multi’omics Database (http://ibdmdb.org), provide the most comprehensive description to date of host and microbial activities in IBD.
Introduction
Inflammatory bowel diseases (IBD) affect >3.5 million people, with increasing incidence worldwide1. These diseases, the most prevalent forms of which are Crohn’s disease (CD) and ulcerative colitis (UC), are characterized by debilitating and chronic relapsing and remitting inflammation of the gastrointestinal tract (for CD) or the colon (in UC). Extensive study has demonstrated that these conditions result from a complex interplay between host2, 3, microbial4–6, and environmental7 factors. Drivers in the human genome include over 200 risk variants, many of which are responsible for host-microbial interactions3. Common gut microbiome changes in IBD include an increase in facultative anaerobes, including E. coli8, and a decrease in obligately anaerobic short-chain fatty acid (SCFA) producers4. Here, to support a systems-level understanding of the etiology of the IBD gut microbiome that goes beyond previously reported metagenomic profiles, we introduce the Inflammatory Bowel Disease Multi’omics Database (IBDMDB), as part of the Integrative Human Microbiome Project (HMP2).
Results
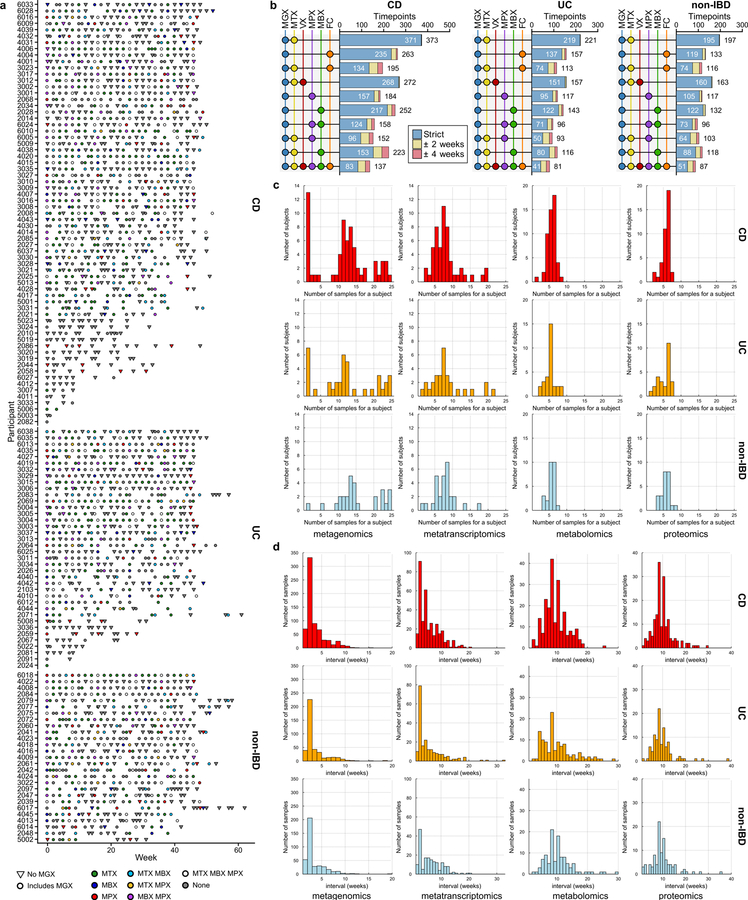
132 participants were recruited from five academic medical centers (three pediatric sub-cohorts: Cincinnati Children’s Hospital, Massachusetts General Hospital [MGH] Pediatrics, and Emory University Hospital, and two adult: MGH and Cedars-Sinai Medical Center, Fig. 1A, Extended Data Table 1, Methods). We analyzed 651 biopsies (baseline) and 529 blood samples (approximately quarterly) which were collected in clinic and 1,785 stool samples which were collected biweekly using a home shipment protocol for one year (Fig. 1B). The latter yielded primarily microbially-focused profiles: metagenomes (MGX), metatranscriptomes (MTX), proteomes (MPX), metabolomes (MBX), and viromes (VX) at several “global” time points across all subjects (Fig. 1B), as well as denser, more intensive sampling for individuals with more variable disease activity (Methods, Extended Data Fig. 1A–D). Multiple measurement types were generated from many individual stool specimens, including 305 samples that contain all stool-derived measurements, and 791 MGX-MTX pairs (Fig. 1C, Extended Data Fig. 1B). Biopsies yielded host-and microbially-targeted human RNA-seq (HTX), epigenetic Reduced Representation Bisulfite Sequencing (RRBS), and 16S rRNA gene amplicon sequencing (16S), which are matched with human exome sequencing, serological profiles, and RRBS from blood. All data are available at https://ibdmdb.org/.
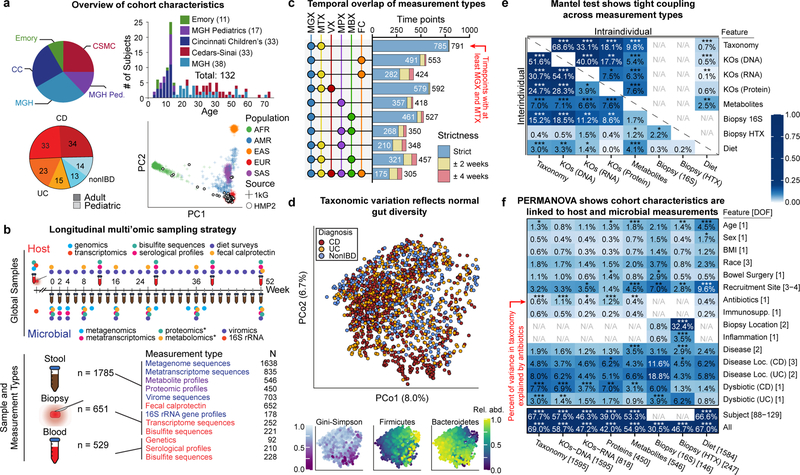
Figure 1. Multi’omics of the IBD microbiome in the IBDMDB study.
A) 132 CD, UC, and non-IBD control subjects were followed for one year each across a range of ages from five clinical centers. Principal component analysis (PCA) of SNP profiles shows the resulting IBDMDB cohort is mostly of European ancestry as compared to the 1000 Genomes reference (Methods). B) The study yielded host and microbial data from colon biopsy (baseline), blood (~quarterly), and stool (biweekly), assessing global time points for all subjects and dense time courses for a subset. Raw, non-quality-controlled sample counts are shown. C) Overlap of multi’omic measurements from the same sample (strict) or from near-concordant time points (with up to 2 or 4 weeks’ time difference; Methods). D) Principal coordinates analysis (PCoA) based on species-level Bray-Curtis dissimilarity; most variation is driven by a tradeoff between phylum Bacteroidetes vs. Firmicutes. IBD samples (CD in particular) had weakly lower Gini-Simpson alpha diversity (Wald test p-values 0.26 and 0.014 for UC and CD vs non-IBD, respectively). E) Mantel tests quantifying variance explained (square of Mantel statistic) between measurement type pairs, with differences across subjects (inter-individual) or within-subject over time (intra-individual; Methods). Sample sizes in (F). F) PERMANOVA shows that inter-individual variation is largest for all measurement types, with even relatively large effects (e.g. antibiotics or IBD phenotype) capturing less variation (Methods). Stratified tests (CD/UC) consider only samples within the indicated phenotype (note that sample counts decrease for these, resulting in larger expected covariation by chance). Stars show FDR-corrected statistical significance (* FDR p≤0.05, ** ≤0.01, *** ≤0.001). Variance is estimated for each feature independently (Methods). “All” refers to a model with all metadata. Total n for each measurement type is shown in [], distributed across up to 132 subjects (Extended Data Fig. 1A, Methods).
Multi’omic IBD gut microbiome perturbations
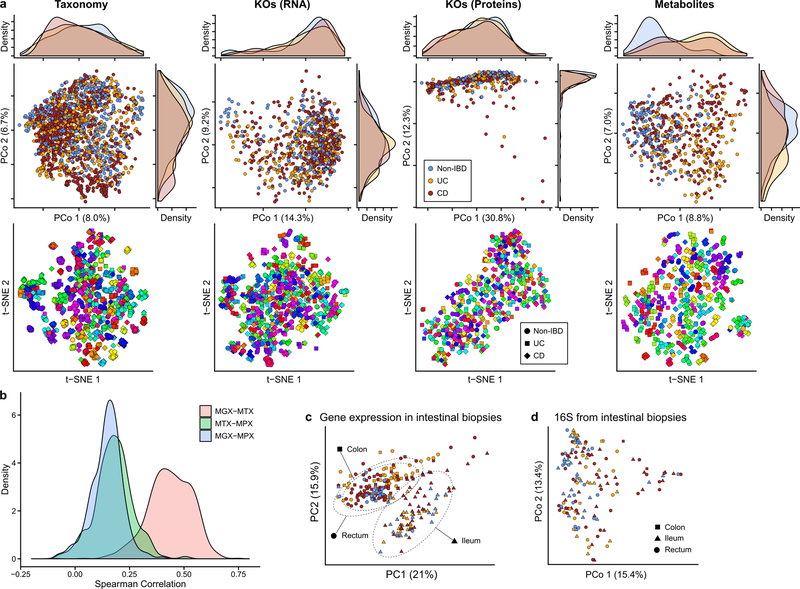
Consistent with prior studies4, 5, although IBD subsets (CD in particular) contributed to the second axis of taxonomy-based principal coordinates (Fig. 1D, Extended Data Fig. 2A), inter-individual variation accounted for the majority of variance for all measurement types5, 9, 10 (Fig. 1E, F, Extended Data Fig. 2A). Even relatively large effects, such as disease status, physiological, and technical factors, explained a smaller proportion of variation (Fig. 1F), which as consistent across measurement types although these captured distinct aspects of IBD dysbiosis (below).
Most measurement types captured correlated changes among and within subjects, cross-sectionally and longitudinally (Fig. 1E). Functional profiles, measured from MGX, MTX, and MPX, were the most tightly coupled (Fig. 1E), although some individual feature-wise correlations were weak (Spearman MGX-MTX 0.44±0.10 [mean ± standard deviation], MGX-MPX 0.14±0.083, and MTX-MPX 0.18±0.096, Extended Data Fig. 2B). Surprisingly, characterized enzymes tended to be only weakly correlated with their known substrates or products (Supplementary Fig. S1). Our dietary characterization, though from a very broad-level food frequency questionnaire, provides a first characterization of longitudinal diet-microbiome coupling in a substantial population over many months, and accounted for a small but significant 3% (FDR p = 7.4 × 10−4) of taxonomic variation between subjects, and 0.7% (FDR p = 4.3 × 10−4) of variation longitudinally.
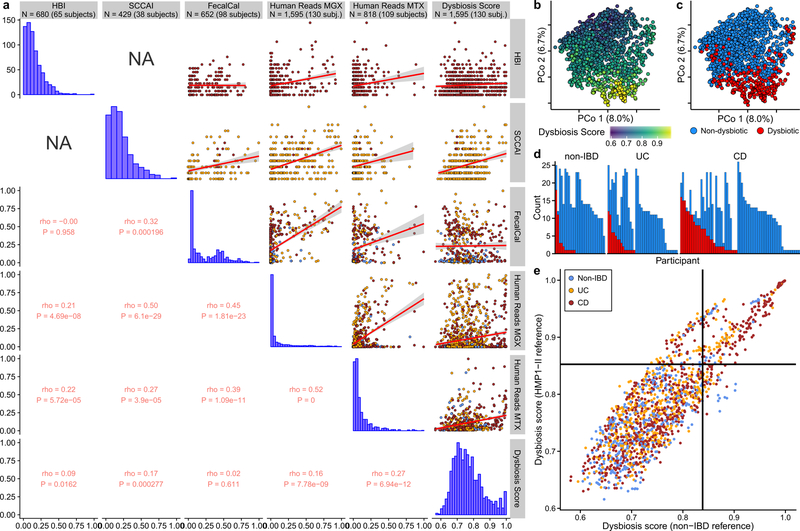
Simple cross-sectional differences among IBD and non-IBD phenotypes (Supplementary Tables S1–14) were most apparent in the metabolome (Fig. 1F, Fig. 2A, Extended Data 2A, 2C–D, Methods). Overall, metabolite pools were less diverse in IBD, paralleling previous observations for microbial diversity (Supplementary Table S2), possibly caused by poor absorption, greater water/blood content, and shorter transit times in IBD11. The smaller number of more-abundant compounds included polyunsaturated fatty acids such as adrenate and arachidonate. Pantothenate and nicotinate (vitamin B5 and B3, respectively) were particularly depleted in the IBD gut, notable since these are not typically part of the vitamin B deficiencies observed in the serum of IBD patients12, although low nicotinate levels have been detected during active CD13. Both vitamins are required to produce cofactors used in lipid metabolism14, and nicotinate plays anti-inflammatory and anti-apoptotic roles in the gut15. Interestingly, nicotinuric acid, a metabolite of nicotinate16, was found almost exclusively in the stool of IBD cases. Fecal calprotectin (FC) and the Harvey-Bradshaw Index (HBI), two measures of disease severity in CD, did not show significant agreement, while the Simple Clinical Colitis Activity Index17 (SCCAI) in UC did weakly correlate with FC (Fig. 2B).
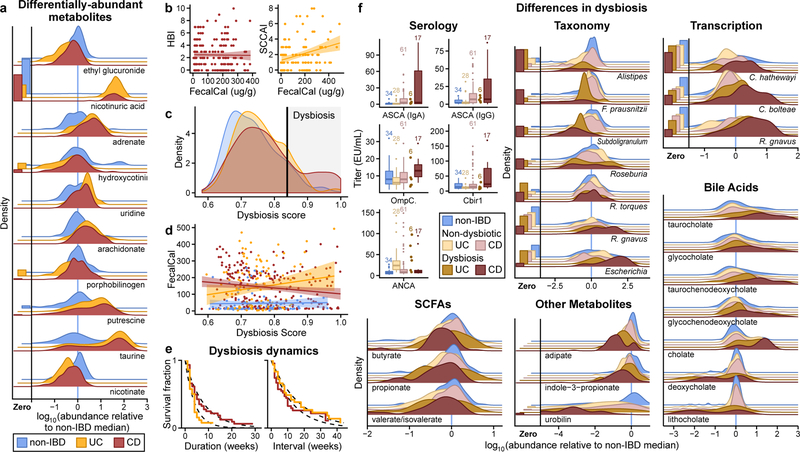
Figure 2. Metagenomic, metatranscriptomic, and stool metabolomic profiles are disrupted during IBD activity.
A) Relative abundance distributions for 10 of the most cross-sectionally significantly differentially abundant metabolites in IBD, as a ratio to the median relative abundance in non-IBD (Wald test; all FDR p < 0.003; Methods; full results in Supplementary Tables S1–14). Fraction of samples below detection limit on left (Methods). N=546 samples from 106 subjects. B) Relationships between two different measures of disease activity: patient-reported (Harvey-Bradshaw Index, HBI, in CD N=680 samples from 65 subjects; Simple Clinical Colitis Activity Index, SCCAI, in UC N=429 samples from 38 subjects) and host molecular (fecal calprotectin43 N=652 samples from 98 subjects). Linear regression shown with 95% confidence bound. C) Distribution of microbial dysbiosis scores as a measure of disease activity (median Bray-Curtis dissimilarity between a sample and non-IBD samples; Methods) and D) its relationship with calprotectin (N=652 samples from 98 subjects). Linear regression with 95% confidence. E) Kaplan-Meier curves for the distributions of the durations of (left) and intervals between (right) dysbiotic episodes for UC and CD subjects. Both are approximately exponential (fits in dashed lines), with a means of 4.1 and 17.2 weeks for UC, and 7.8 and 12.8 weeks for CD (Methods). F) Relative abundance distributions of significantly different metagenomic species (N=1,595 samples from 130 subjects), metabolites (N=546 samples from 106 subjects), and microbial transcribers (N=818 samples from 106 subjects) in dysbiotic samples compared to non-dysbiotic samples from the same disease group (Wald test; all FDR p < 0.05; full results in Supplementary Tables S15–28). Also shown are antibody titers for anti-neutrophil cytoplasmic antibodies (ANCA), anti-Saccharomyces cerevisiae mannan antibodies (ASCA, IgG or IgA), anti-OmpC, and anti-CBir1 antibodies (N=146 from 61 subjects). Boxplots show median and lower/upper quartiles; whiskers show inner fences; sample sizes above boxes.
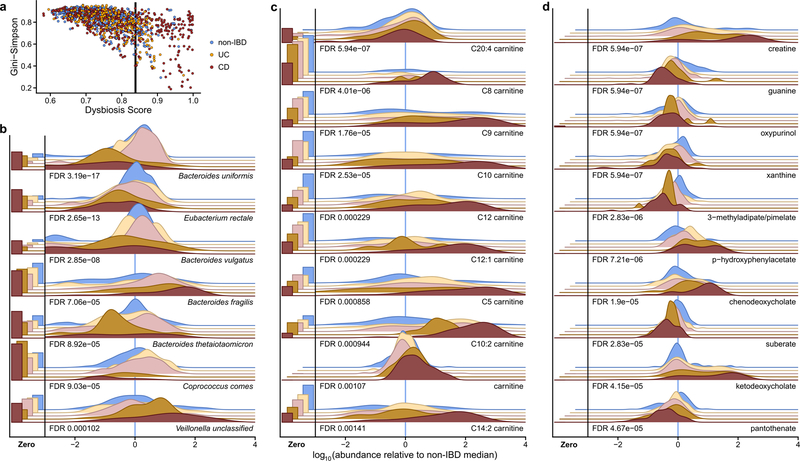
Notably, no metagenomic species were significantly different between IBD and non-IBD phenotypes after correction for multiple hypothesis testing (Supplementary Table S1), in contrast with prior work4, 5, 18. We hypothesized this was due to the differentiation of IBD patients into two subsets, one with relatively inactive disease (either due to remission or recent onset) and the other with greater activity. This differentiation has been observed in several IBD cohorts5, 18, though is more pronounced here since samples are not specifically from subjects selected for active disease. We thus classified samples with taxonomic compositions highly unlike those from non-IBD samples as “dysbiotic” (Fig. 2C, see definition and discussion in Methods, Extended Data Fig. 3A–E). Dysbiotic excursions in this cohort did not correspond with disease location (e.g. ileal CD; F-test p=0.11, Methods), and occurred longitudinally within subjects; they were weakly correlated with patient-reported and molecular measures of disease activity (Fig. 2D, Extended Data Fig. 3A). In total, 272 dysbiotic samples occurred in 78 full periods of dysbiosis and 9 censored periods (i.e. subjects which were dysbiotic at the end of the time series, Methods), or 17.1% of all samples (n=178 [24.3%] in CD and n=51 [11.6%] in UC). The durations of and times between dysbiotic periods were both approximately exponential, suggesting that transitions are triggered, at least in part, by events with constant probability over time (i.e. potentially stochastic, Fig. 2E).
Using the resulting definition, dysbiosis corresponded with a larger fraction of variation in all measurement types than did overall IBD phenotype (see Fig. 1F; Supplementary Tables S15–28), likely reflecting a clearer delineation between active and less active disease states within extremely heterogeneous subjects over time. Though it is unclear which aspects of dysbiosis are cause or consequence in IBD, characterizing these changes will lead to a greater understanding of microbial dynamics in disease. As in previous cross-sectional studies of established disease4, differences between dysbiotic and non-dysbiotic samples from CD subjects were more pronounced than in UC (see Fig. 1F). Notably, dysbiosis also distinguished between independent host measures, such as individuals with high and low ASCA, ANCA, OmpC (outer membrane protein C), and Cbir1 (anti-flagellin) antibody titers in serological profiles (Fig. 2F; Fisher’s combined probability test p-value 0.00044 from Wilcoxon tests between dysbiotic and non-dysbiotic CD). Dysbiosis was not significantly associated with demographics or medication (logistic regression with subject as random effect, all FDR p > 0.05). Dysbiosis recapitulated a known decrease in alpha diversity in active disease, but also identified numerous communities with normal complexity as dysbiotic (Extended Data Fig. 4A). Taxonomic perturbations during dysbiosis, strikingly, mirrored those previously observed cross-sectionally in IBD6, such as the depletion of obligate anaerobes including Faecalibacterium prausnitzii and Roseburia hominis in CD and the enrichment of facultative anaerobes such as Escherichia coli (Fig. 2F; Extended Data Fig. 4B). Ruminococcus torques and R. gnavus, two prominent species in IBD19, were also differentially-abundant in dysbiotic CD and UC, respectively (FDR p=0.041 and 0.0087). A smaller subset of species also increased significantly in transcriptional activity (mean total transcript relative abundance relative to genomic abundance; Methods) in addition to abundance differences, including Clostridium hathewayi, C. bolteae, and R. gnavus (Fig. 2F). All had significantly increased expression during dysbiosis (all FDR p<0.07), and thus their roles in IBD may be more pronounced than suggested solely by their genomic abundance differences.
In the metabolome, short-chain fatty acids (SCFAs) were generally reduced in dysbiosis (Fig. 2F). The reduction in butyrate in particular is consistent with the previously-observed depletion of butyrate producers6 such as F. prausnitzii and R. hominis, also observed here (Fig. 2F). We also detected enrichments in the primary bile acids cholate and its glycine and taurine conjugates (glycocholate q=5.2 × 10−5, taurocholate q=1.3 × 10−5) in dysbiotic CD. Similarly, glycochenodeoxycholate q=1.1 × 10−4 was also enriched. In contrast, secondary bile acids lithocholate and deoxycholate (q=5 × 10−7 and 1.8 × 10−4, respectively) were reduced in dysbiosis, suggesting that secondary bile-acid producing bacteria are depleted in IBD-related dysbiosis, or that transit time through the colon is too short for them to be metabolized20, 21. These highly significant metabolomic differences during microbial dysbioses, which were in turn concordant with changes expected during disease, provide further evidence that the dysbiosis measure is specifically relevant in IBD. We also observed several novel biochemical differences during dysbiosis, such as large changes in acylcarnitines. Many acylcarnitines were significantly enriched in dysbiosis (Extended Data Fig. 4C), while base metabolites were typically reduced (Fig. 2F, Extended Data Fig. 4D). Of note, however, arachidonoyl carnitine (C20:4 carnitine) was reduced while free arachidonate, a precursor of prostaglandins involved in inflammation, was increased (see Fig. 2A). Like bile acids, carnitines are microbially-modified compounds can have competing phenotypic effects depending on the precise modifications: L-carnitine, for example, tends to be anti-inflammatory, while fatty acid-conjugated carnitine does not act uniformly on gut inflammation22. These opposing changes in biochemically related metabolites further suggest that these differences do not stem simply from the wholesale dilution of stool. Numerous other metabolites were also significantly altered in dysbiotic IBD (117 of 548 tested known metabolites with FDR p<0.05; Extended Data Fig. 4D, Supplementary Table S16), showing a large-scale dysregulation of metabolite pools in tandem with host-and microbiome-specific taxonomic and molecular features (Fig. 2F). Finally, though we found only a single poorly-characterized bacteriophage to be differentially-prevalent in both IBD and in dysbiosis (interestingly with reduced prevalence in IBD, Supplementary Tables S3, S17), we note that several subjects were observed to have a spike in viral load prior to a dysbiotic period (Supplementary Fig. S2).
Decreased IBD gut microbiome stability
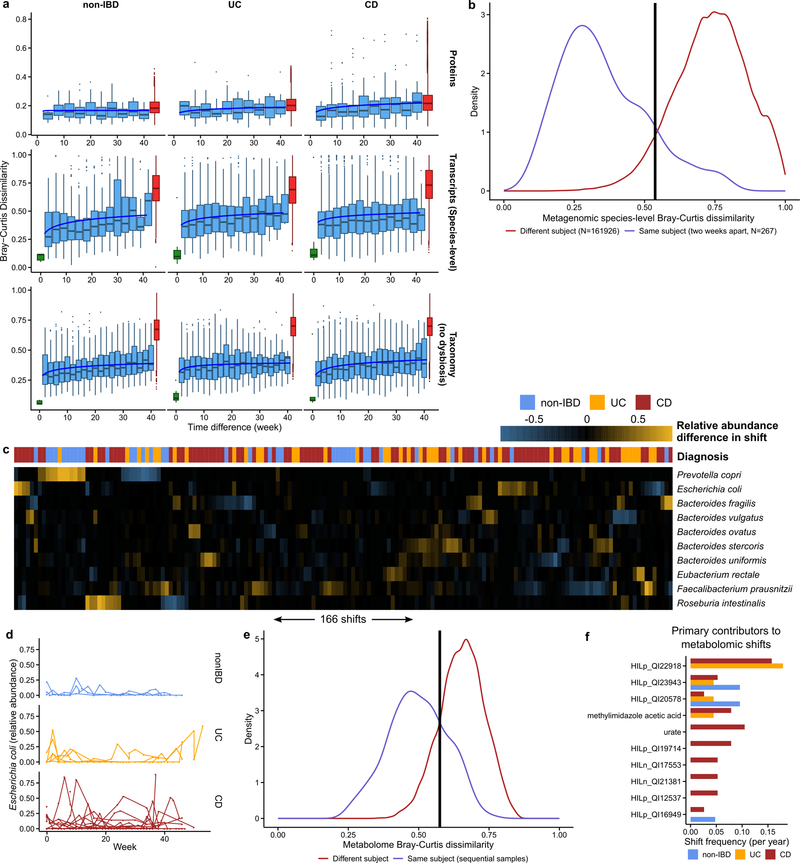
The IBDMDB’s dense time series for stool-derived multi’omics from many subjects enabled in-depth longitudinal analysis integrating multiple measurements of the microbiome. Each subject’s microbiome tended to change more over time for metagenomic, metatranscriptomic, and metabolomic profiles (Fig. 3A; F-test power law fit p-values < 10−24; Methods). These changes were most pronounced for CD and UC subjects’ taxonomic profiles (F-test difference in power law fits p-values < 10−9), where a single subject’s microbiome may have almost no species in common with itself at an earlier time point (dissimilarity of 1, Fig. 3A), consistent with previous observations9. Transcripts summarized within species (Extended Data Fig. 5A) showed similar trends (all F-test p-values < 8 × 10−4) to metagenomic species abundances. Meanwhile, gene family transcripts (KOs), metabolites (Fig. 3A), and proteins (KOs, Extended Data Fig. 5A) varied much more rapidly, with essentially as much change after ~two weeks as over longer time periods (increasing trends less-or non-significant: non-IBD, UC, CD F-test p-values 0.0006, 0.001, and 0.04, respectively for transcripts; 0.02, 0.06, and 0.003 for metabolites; and 0.5, 0.15, and 0.06 for proteomics). This indicates that these features all vary rapidly in both IBD and control subjects’ guts and lack additional, more extreme excursions during disease.
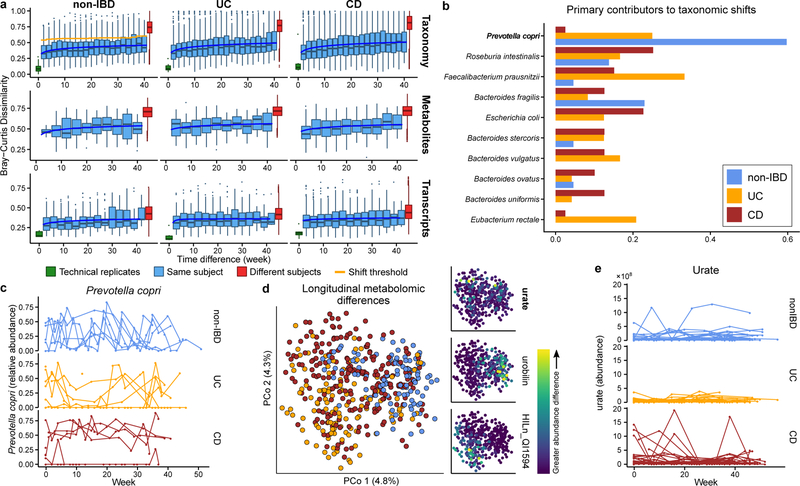
Figure 3. Temporal shifts in the microbiome are more frequent and more extreme in IBD.
A) Bray-Curtis dissimilarities within subject as a function of intervening time difference, as compared to different people or technical replicates; calculated for metagenomic taxonomic profiles (species; N=1,595 samples from 130 subjects), metabolomics (N=546 samples from 106 subjects), and functional profiles (KO30 gene families; N=818 samples from 106 subjects). Boxplots show median and lower/upper quartiles; whiskers show inner fences. Least-squares power-law fits in blue, thresholds for microbiome shifts in orange (Methods). Proteomics and species-level transcripts in Extended Data Fig. 5A. Within-subject changes are significantly more extreme in UC (F-test p-value 3.9 × 10−10) and CD (F-test 1.2 × 10−18) for taxonomic profiles and transcripts (latter F-test p-values 0.00016 and 1.7 × 10−5), with mixed differences for metabolites (F-test p-values 0.012 and 0.23). Technical replicates shown (when possible) at 0 weeks. B) Shift frequencies for the top 10 species with greatest change during shifts, ranked by number of shifts as primary contributor, stratified by disease phenotype(s) (full table Supplementary Table S29). C) Prevotella copri is of interest in arthritis23 and international populations44, and it uniquely retained stable abundances in CD but bloom-relaxation dynamics in controls (Two-tail Wilcoxon test of absolute differences between consecutive time points p=4.2 × 10−6 between non-IBD and UC, and 1.1 × 10−4 between non-IBD and CD). Plot shows 22 subjects with at least one time point with >10% differential abundance (N=267 samples). D) Ordination of temporally adjacent samples within individual, based on metabolomics (Bray-Curtis principal coordinates on normalized absolute abundance differences). Disease groups separate significantly (N=440 sample pairs from 106 subjects; PERMANOVA R2 = 2.8%, p<10−4). Urobilin, urate, and an unidentified untargeted feature that segregate with disease groups in the PCoA are shown (right); HILn_QI1594 (HILIC-neg method m/z 152.0354, RT: 4.16 min). E) As in (C), but for urate (Two-tail Wilcoxon test p=0.0012 non-IBD-UC, and 0.044 non-IBD-CD; N=546 samples from 106 subjects).
We further characterized large-scale temporal differences by searching for “shifts” in the microbiome between consecutive time points, defined as Bray-Curtis dissimilarities more similar to those between different people than within person (Fig. 3A, Extended Data Fig. 5B, Methods). First considering only metagenomic taxonomic profiles, 166 such shifts were observed, with 39 in non-IBD (of 382 total possible), 44 in UC (of 381), and 83 in CD (of 650) (Supplementary Table S29). Due to differences in total observation times, the rate of shifts was only marginally higher in CD and UC compared to non-IBD (2.09 and 1.83 shifts/year versus 1.79 in non-IBD), and these were generally confined to a subpopulation of dysbiotic individuals (Fig. 3A). However, the species with the greatest changes in relative abundance differed dramatically (Fig. 3B). Shifts in non-IBD subjects primarily occurred in individuals with high abundances of Prevotella copri, which underwent repeated expansion and relaxation cycles over the course of weeks to months (Fig. 3C). This organism is of particular interest due to its behavior as a population-scale outgroup and enrichment during new-onset rheumatoid arthritis23. The lack of shifts due to P. copri in IBD subjects was not due to an absence of P. copri in these individuals or an overabundance in non-IBD subjects (6 of 27 non-IBD subjects had at least one time point with >10% P. copri, consistent with healthy populations10, 24). Instead, the relative abundances that were present remained more stable in the IBD population (Fig. 3C). Taxonomic shifts in IBD subjects mirrored earlier observations of relative reductions of obligate anaerobes and overgrowths of facultative anaerobes (Fig. 3B, Extended Data Fig. 5C), and frequently corresponded with entry into/exit from dysbiosis (28 and 23 shifts marked entries and exits in IBD, respectively, accounting for 40% of shifts in IBD). E. coli in particular contributed to a large number of shifts in IBD, though there was no clear pattern in which species it trades abundance with (Extended Data Fig. 5C–D).
Defining shifts in a similar manner for metabolomics profiles (Extended Data Fig. 5E), we find approximately half the overall rate of shifts as in the metagenome (1.05 shifts/year in non-IBD subjects, 0.99 shifts/year in UC and 1.36 shifts/year in CD), though this is strongly affected by the availability of fewer metabolomics samples (Extended Data Fig. 5E). Examining differences in metabolite profiles between adjacent samples for the same subject, we find a significant separation by diagnosis (Fig. 3D; PERMANOVA p<10−4). These differences were largely driven by unknown compounds, emphasizing the need for further compound annotation efforts and follow up to determine their significance in IBD. Features with the greatest differences included urobilin (with larger differences in non-IBD), urate largely in CD, and a feature with m/z of 152.0354 and retention time (RT) of 4.16 min (potentially the formic acid adduct of pyridinaldehyde), accounting for differences largely specific to UC. The primary contributors to shifts were largely unidentified compounds (Extended Data Fig. 5F, Supplementary Table S30). HILp_QI22918, an unknown feature with m/z of 648.43067 and RT of 5.03 min, contributed the most (10) shifts exclusively in IBD subjects. Among known compounds, methylimidazole acetic acid and urate were the primary contributors to the most shifts (4 shifts each; Fig. 3E).
Microbiome-associated host factors
When incorporating host molecular measurements, primarily from intestinal biopsies taken colonoscopically at baseline, into analysis of the IBD microbiome, the main influences on population variability were strikingly different from those affecting the microbiota alone. In particular, tissue location is a major driver of intestinal epithelial gene expression (Extended Data Fig. 2C) even in the face of microbial variation25 (Extended Data Fig. 2D). Microbiome and phenotypic association analyses were thus performed independently for each standardized biopsy location (Methods).
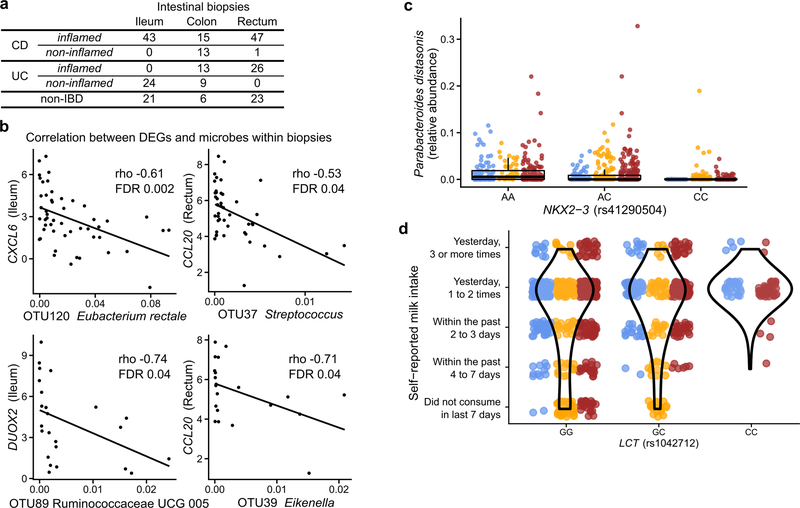
We identified genes that were significantly differentially expressed (DEGs) in patient biopsies taken in inflamed locations of the ileum (CD subjects) and rectum (CD and UC subjects) compared to non-IBD controls (Extended Data Fig. 6A). This targeted 305 ileal and 920 rectal genes, primarily overexpressed, for further analysis (together representing 1,008 unique genes, negative binomial model FDR p<0.05 and fold-change > 1.5. Fig. 4A, Supplementary Table S31). These included genes that can affect commensal microbes directly, such as the antimicrobial CXCL6 (a cell membrane disruptor26) and SAA2 (a Gram-negative growth inhibitor27), as well as indirect microbial modulators such as DUOX2 (reactive oxygen species production28) and LCN2 (iron starvation through sequestration29, Fig. 4B). Enrichment analysis testing for overrepresentation of KEGG30 pathways among DEGs also confirmed strong representation of immune related pathways (one-sided hypergeometric test, FDR p<0.05). Particularly, the IL-17 signaling pathway, whose components have been previously reported in gene expression studies of ileal biopsies of CD patients31, 32, was enriched in up-regulated DEGs in both ileum and rectum (FDR p=2.8 × 10−14, Fig. 4A, Supplementary Table S32). Among up-regulated DEGs in rectal biopsies of UC patients, we found further enrichment of the complement cascade (FDR p=4.4 × 10−10), a component of innate immunity33 which has been implicated in IBD in early studies25, 34, 35.
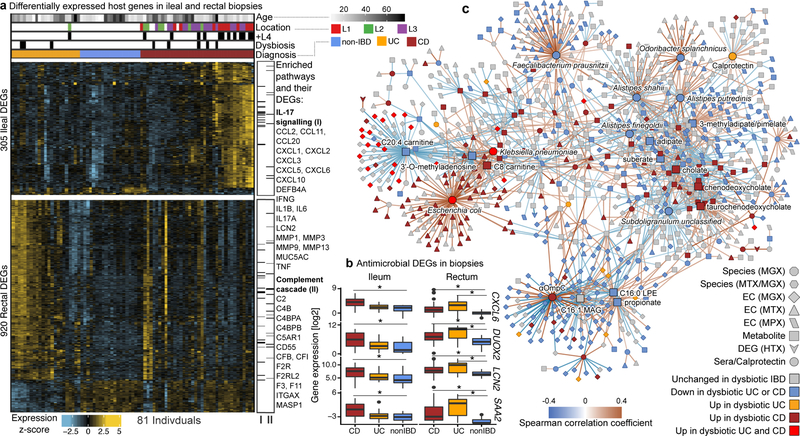
Figure 4. Colonic epithelial molecular processes perturbed during IBD and in tandem with multi’omic host-microbial interactions.
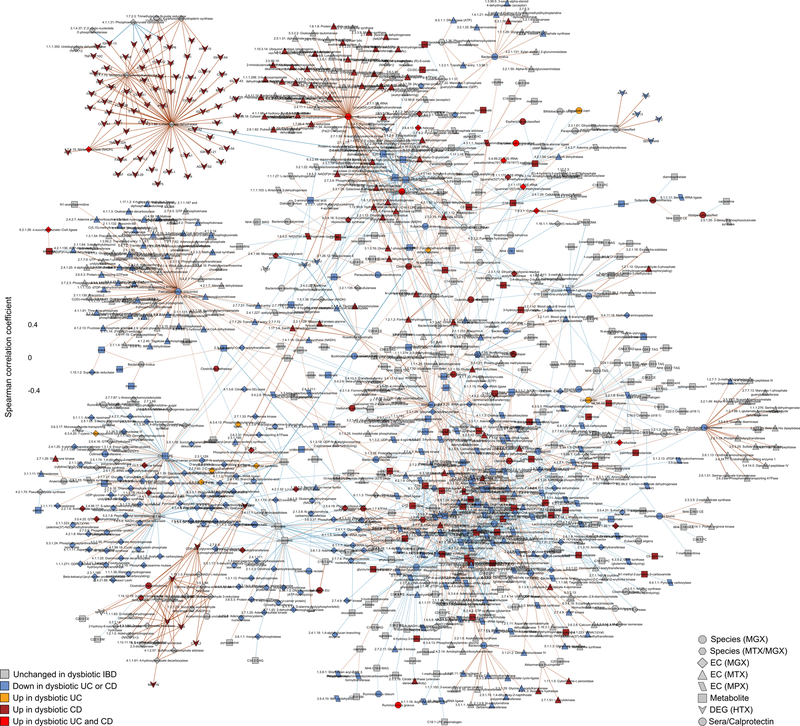
A) Differentially expressed human genes (DEGs) (negative binomial FDR p<0.05, min. fold change 1.5; Supplementary Table S31) from 81 subjects with paired ileal and rectal biopsies. Ordering by diagnosis, clustering within diagnosis. IL-17 signaling (I) showed strongest enrichment in ileal DEGs (FDR p=8.2 × 10−12)31, while the complement cascade (II)45 was enriched in rectal DEGs of UC patients (FDR p=5.2 × 10−8; KEGG30 gene sets, Supplementary Table S32). Example DEGs shown with I and II. B) Expression of four genes involved in host-microbial interactions26–29. Inflamed biopsy samples are shown for CD in ileum (left column, N=20, 23, 39 independent samples for non-IBD, UC, CD respectively); for CD and UC in rectum (right column; N=22, 25, 41 independent samples for non-IBD, UC, CD); non-IBD samples were non-inflamed. Stars indicate significant differential expression compared to non-IBD (Fisher’s exact, FDR p<0.05; p-values in Supplementary Table S31). Boxplots show median and lower/upper quartiles; whiskers inner fences. C) Significant associations among 10 different aspects of host-microbiome interactions: metagenomic species, species-level transcription ratios, functional profiles captured as EC gene families (MGX, MTX and MPX), metabolites, host transcription (rectum and ileum), serology, and calprotectin (sample counts in Fig. 1B–C). Network shows top 300 significant correlations (FDR p<0.05) between each pair of measurement types (for serology, FDR p<0.25). Nodes colored by the disease group they are “high” in, edges by sign and strength of association. Spearman correlations use residuals of a mixed-effects model with subjects as random effects (or a simple linear model when only baseline samples were used, i.e. biopsies) after covariate adjustment (Methods). Time points approximately matched with maximum separation 4 weeks (Methods). Singletons pruned for visualization (Extended Data Fig. 8). Hubs (nodes with ≥20 connections) emphasized.
To identify components of the microbiome most associated with these changes, we tested for transcripts covarying with microbe relative abundance measured directly from the same specimens using 16S amplicon sequencing. We identified 31 and 106 significant gene-OTU pairs in the ileum and rectum, respectively, with no overlap between the two sites, consistent with the different overall gene expression patterns that separate them (partial Spearman correlation FDR p<0.05, Methods, Extended Data Fig. 6B, Supplementary Table S33). Genes involved included known IBD-associated host-microbial interaction factors such as DUOX2 and its maturation factor DUOXA2 that produce reactive oxygen species31, 36, both negatively associated with abundance of Ruminococcaceae UCG 005 (OTU 89) in the ileum. Expression of several chemokines, some of which have reported antimicrobial properties37 (CXCL6, CCL20), were negatively correlated with the relative abundance of Eubacterium rectale (OTU 120) in the ileum, and Streptococcus (OTU 37) and Eikenella (OTU 39) in the rectum, suggesting that these species are the most susceptible to the activity of these chemokines. Finally, while this cohort was not designed for genetic association discovery (Supplementary Discussion, Extended Data Fig. 6C–D, Supplementary Table S34), we do also provide exome sequencing of 92 subjects such that these results may be integrated with larger populations in the future.
Dynamic, multi’omic microbiome interactions
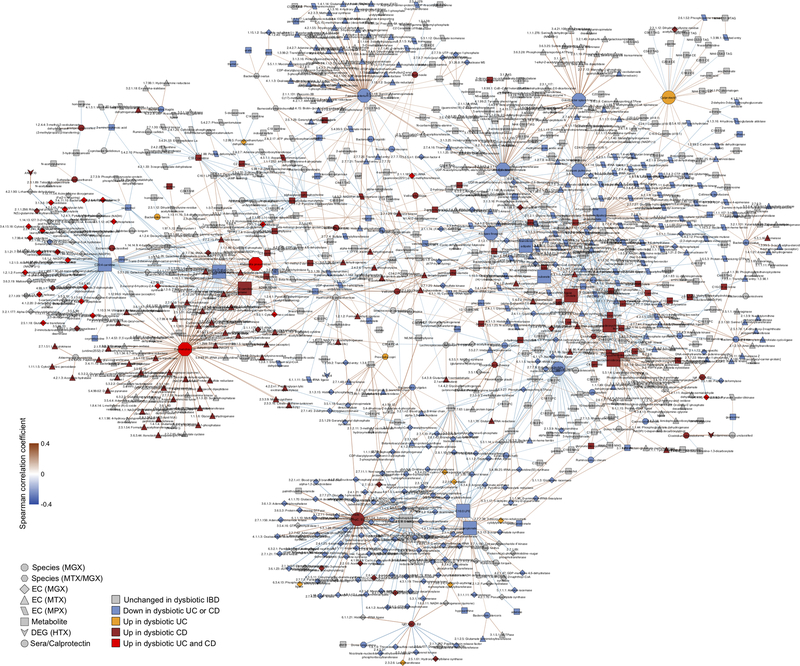
We next searched for putative host and microbial molecular interactions underlying disease activity in IBD by constructing a large-scale cross-measurement type association network incorporating 10 different microbiome measurements: metagenomic species, species-level transcription ratios, functional profiles captured as Enzyme Commission (EC) gene families (MGX, MTX and MPX), metabolites, host transcription (rectum and ileum separately), serology, and fecal calprotectin. To identify co-variation between components of the microbiome above and beyond those linked strictly to inflammation and disease state, each measurement type was first residualized using the same mixed-effects model (or linear model when appropriate) used to determine differential abundance (“adjusted” network; Methods). This residualization utilizes the longitudinal measurements to minimize any inter-individual variation (including IBD status), as well as dysbiotic excursions as drivers of the detected associations, and thus highlights associations that occur within-person over time. The resulting network contained 53,161 total significant edges (FDR p<0.05) and 2,916 nodes spanning features from all measurement types (Supplementary Table S35). A filtered subnetwork was then constructed for visualization from the top 300 edges (by p-value) per measurement type in which at least one connected node was dysbiosis-associated (Fig. 4C).
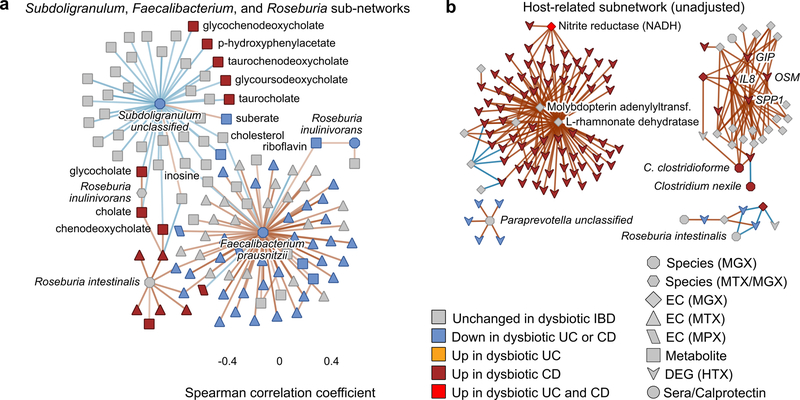
Representatives from the five stool-derived measurements occurred as hubs (defined as nodes with ≥20 connections) in this network, all of which were identified as differentially abundant in dysbiosis. Particularly connected taxonomic features (from metagenomes and metatranscriptomes) included the abundances of F. prasunitzii and unclassified clades related to Subdoligranulum38, which are closely phylogenetically related, although the only molecular features common to both organisms were covariation with the abundances of cholesterol and inosine (Extended Data Fig. 7A). F. prausnitzii accounted for some of the strongest associations overall, including the expression of numerous down-regulated ECs in dysbiosis. On the other hand, E. coli (and to lesser extent Haemophilus parainfluenzae) accounted for a large fraction of up-regulated ECs. Members of the Roseburia genus were also associated, metatranscriptionally as well as metagenomically, with both the bile acids and a number of acylcarnitines, suggesting that Roseburia (together with Subdoligranulum) play an integral role in the carnitine and bile acid dysregulation observed in IBD.
Acylcarnitines and bile acids as overall chemical classes featured prominently in the network, related in part to their changes during dysbiosis. Acylcarnitines were associated with numerous dysbiosis-associated species including R. hominis (9 acylcarnitines, FDR p < 0.05; Supplementary Table S35), Klebsiella pneumoniae (3), and H. parainfluenzae (3), as well as expression of C. boltae (3), suggesting that multiple scales of regulation are involved—long-term growth-based and short-term transcriptional. Particularly notable biochemical hubs in the network included C8 carnitine, another acylcarnitine significantly increased in dysbiotic CD, cholate, chenodeoxycholate, and taurochenodeoxycholate, together accounting for 107 edges (6%; Fig. 4C). Other prominent metabolite associations included several long-chain lipid hubs and the SCFA propionate; antibodies against OmpC were strongly associated with these, as well as the metagenomic abundances of the numerous ECs involved in the system’s biosynthesis or as interactors. Calprotectin, as the sole feature in its own measurement type, was weakly associated with a number of metabolites that were not differentially abundant in dysbiosis, as well as with the metagenomic abundance of several dysbiosis-associated ECs. Three host genes appear in this high-significance subnetwork: the ileal expression of GIP, NXPE4, and ANXA10. Expression of RNA polymerase was also a prominent node in the network, though not a hub, up-regulated in dysbiosis (Extended Data Fig. 8). This essential enzyme class’s regulation is growth rate-dependent39, suggesting that microbial communities as a whole are more often in higher growth conditions in dysbiotic IBD.
Finally, we also identified associations among features in the microbiome that did take dysbiosis into account, resulting in a second network using the same methodology but without adjusting for dysbiosis (“unadjusted”; Supplementary Discussion, Extended Data Figs. 7B and 9, Supplementary Table S36). Together, these networks contextualize the multiple different types of microbiome disruptions observed in IBD, with associations among many different molecular feature types representing potential targets for follow-up studies on the mechanisms underlying IBD and gastrointestinal inflammation.
Conclusions
As part of the HMP2, we have developed the IBDMDB, one of the first integrated studies of multiple molecular features of the gut microbiome implicated in IBD dynamics. While overall population structure was comparable among measurements of the microbiome—metagenomic, metatranscriptomic, metabolomic, and others—each measurement identified complementary molecular components of longitudinal dysbioses in CD and UC. Some, such as taxonomic shifts in favor of aerotolerant, pro-inflammatory clades, have been captured by previous studies; others, such as greater gene expression by clostridia during disease, were only able to be newly discovered using other measurements (i.e. metatranscriptomes). Temporal stability of multiple microbiome measurements likewise differed across IBD phenotypes and disease activity, with distinct impacts on molecular components of the microbiome (including an unexpected stability of the relative abundance of P. copri in diseased individuals). The study provides a complete catalog of new relationships between multi’omic features identified as potentially central during IBD, in addition to data, protocols, and relevant bioinformatic approaches to enable future research.
By leveraging a multi’omic view on the microbiome, our results single out a number of new host and microbial features for follow-up characterization. An unclassified Subdoligranulum species, recently shown to form a complex of new species-level clades38, was both drastically reduced in IBD and central to the functional network, associating with a wide range of IBD-linked metabolites both identifiable (e.g. bile acids and polyunsaturated fatty acids) and unidentifiable. The clade likely contains at least seven species closely related to the Subdoligranulum, Gemmiger, and Faecalibacterium genera, typically butyrate-producers considered to be beneficial particularly in IBD40. Therefore, the isolation and characterization of additional species—particularly in tandem with these associated metabolites—is likely to reveal its physiological and immunological interactions and the consequences of its depletion in IBD. More generally, strain-level profiling of implicated microbes remains to be carried out, particularly in direct association with host epithelium and corresponding molecular changes. This is feasible with existing data from this study, and will serve to pinpoint the specific organisms responsible for IBD-associated accumulation of primary unconjugated bile acids and depletion of secondary bile acids41. Only very few, low-abundance species are currently known to be capable of secondary bile acid metabolism42, and thus expanding the range of strains known to carry appropriate metabolic cassettes will indicate potential new targets for therapeutic restoration. Beyond short-chain fatty acids and bile acids, the large-scale acylcarnitine dysbiosis observed here may also provide a promising new target for IBD, particularly after determining whether this large-scale shift in metabolite pools is host or microbially-driven.
We stress that it has not yet been determined whether these multi’omic features of the microbiome are predictive of disease events before their occurrence, nor have the disease-relevant time scales of distinct molecular events been identified (e.g. static host genetics, relatively slow epigenetics or microbial growth, rapid host and microbial transcriptional changes). It may also be fruitful to seek out the earliest departures from a subject-specific baseline state that, while themselves still “eubiotic,” may predict the subsequent onset of dysbiosis or disease symptoms. Some such characterization may be possible in data from this study, although other causal analysis may be better carried out at finer-grained time scales or using interventional study designs. It will be most important to take these molecular results back to the clinic, in the form of better predictive biomarkers of IBD progression and outcome, and as a set of new host-microbial interaction targets for which treatments to ameliorate the disease may be developed.
Methods
Recruitment and specimen collection
Recruitment
Five medical centers participated in the IBDMDB: Cincinnati Children’s Hospital, Emory University Hospital, Massachusetts General Hospital, Massachusetts General Hospital for Children, and Cedars-Sinai Medical Center. Patients were approached for potential recruitment upon presentation for routine age-related colorectal cancer screening, work up of other GI symptoms, or suspected IBD, either with positive imaging (e.g. colonic wall thickening or ileal inflammation) or symptoms of chronic diarrhea or rectal bleeding. Subjects could not have had a prior screening or diagnostic colonoscopy. Potential subjects were excluded if they were unable or did not consent to provide tissue, blood, or stool, were pregnant, had a known bleeding disorder or an acute gastrointestinal infection, were actively being treated for a malignancy with chemotherapy, were diagnosed with indeterminate colitis, or had a prior, major gastrointestinal surgery such as an ileal/colonic diversion or j-pouch. Upon enrollment, an initial colonoscopy was performed to determine study strata. Subjects not diagnosed with IBD based on endoscopic and histopathologic findings were classified as “non-IBD” controls, including the aforementioned healthy individuals presenting for routine screening, and those with more benign or non-specific symptoms. This creates a control group that, while not completely “healthy”, differs from the IBD cohorts specifically by clinical IBD status. Differences observed between these groups are therefore more likely to constitute differences specific to IBD, and not differences attributable to general GI distress. In total, 132 subjects took part in the study (Extended Data Table 1).
Regulatory Compliance
The study was reviewed by the Institutional Review Boards at each sampling site: overall Partners Data Coordination (IRB #2013P002215); MGH Adult cohort (IRB #2004P001067); MGH Pediatrics (IRB #2014P001115); Emory (IRB #IRB00071468); Cincinnati Children’s Hospital Medical Center (2013–7586); and Cedars-Sinai Medical Center (3358/CR00011696). All study participants gave their written informed consent before sampling. Each IRB has a federal wide assurance and follows the regulations established at 45 CFR Part 46. The study was conducted in accordance with the ethical principles expressed in the Declaration of Helsinki and the requirements of applicable federal regulations.
Specimen collection and storage
Specimens for research (biopsies, blood draws, and stool samples) were collected during the screening colonoscopy, at up to 5 quarterly follow-up visits at the clinic (termed ‘baseline’, visit 2, etc, occurring at months 0, 3, 6, 9, and 12), and biweekly by mail.
Biopsies
Biopsies were primarily gathered during the initial screening colonoscopy, where approximately four to fourteen biopsies were collected for all subjects. For each location sampled (at least ileum and 10cm from rectum, plus discretionary sites of inflammation), one biopsy was collected for standard histopathology at the individual institution, two biopsies were collected and stored in RNAlater for molecular data generation (host and microbial, stored at −20 °C), and one biopsy was collected and placed in a sterile tube with 5% glycerol (stored at −80 °C). If possible, additional biopsies from inflamed tissue, as well as nearby non-inflamed tissue were taken from CD/UC subjects. For adult subjects, a second set of biopsies were also collected from each location (rectum and ileum) for epithelial cell culture (detailed protocols: http://ibdmdb.org/protocols). All biopsies were stored for up to two months at the collection site, and shipped overnight on dry ice to Washington University for epithelial cell culture or to the Broad Institute for molecular profiling.
Blood draws
Blood draws (whole blood and serum) were taken at the quarterly clinical visits. For whole blood, 1 mL of blood was collected and stored at −80 °C. For serum, blood was drawn into a 5mL SST tube, and left at room temperature for 40 minutes. This was centrifuged for 15 minutes at 3,000 rpm and 0.5 mL portions were immediately aliquoted into 2 mL microtubes. Tubes were stored at −80 °C.
Stool samples
Stool specimens were collected both at the clinical visits as well as biweekly by mail using a home collection kit developed for the project (http://ibdmdb.org/protocols) and previously validated46. Subjects first deposited stool into a collection bowl suspended over a commode. Subject then collected two aliquots using a scoop to transfer stool into two Sarstedt 80.623 tubes: one with approximately 5 mL molecular biology grade 100% ethanol, and one with no preservative. Stool samples were then sent from each participant by FedEx to the Broad Institute where they were processed immediately prior to storage at −80 °C. The ethanol tube was centrifuged to pellet stool, which was subaliquotted, and the supernatant was transferred to a new tube for metabolomic analysis. Stool from ethanol was aliquoted into 2 ml cryovials in ~100–200 mg aliquots, prioritizing specimens for meta’omic sequencing, metabolomics, and viromics in that order. Any remaining stool was stored in additional aliquot tubes. 100mg of the non-ethanol stool was stored for assaying fecal calprotectin and the remainder was saved in a second tube. All samples were stored at −80 °C after receipt prior to processing. This home-collection method was shown previously to produce reproducible results compared to flash-frozen samples46, consistent with previous observations across data types47–49. Note that an accurate estimate of the stool water content could not be obtained, since samples were collected by subjects and preserved in ethanol at room temperature until aliquots were generated for the different data generation platforms.
Participant and sample metadata
Descriptions of each participant and specimen were captured at baseline and accompanying each specimen collection, respectively. At baseline (i.e. during or prior to the screening colonoscopy), subjects completed a Reported Symptoms Questionnaire, the Short Inflammatory Bowel Disease Questionnaire50, a Food Frequency Questionnaire, and an Environmental Questionnaire, and the Simple Endoscopic Score51 for CD subjects or Baron’s Score52 for UC subjects was assessed.
During both follow-up visits and paired with mailed stool samples, subjects completed an Activity Index and Dietary Recall Questionnaire to assess their disease activity index (HBI for CD or SCCAI for UC) and provided a retrospective recall of their recent diet. All questionnaires, as well as detailed protocols (including product numbers) can be found on the IBDMDB data portal at http://ibdmdb.org/protocols. Responses and metadata are available at http://ibdmdb.org/results, and summaries of phenotypes for samples and subjects are available (Supplementary Fig. S3) along with summaries of the final time series for each subject (Supplementary Fig. S2).
Stool specimen processing
Sample selection
Sample selection proceeded in two phases, with an initial round of data generation producing a pilot metagenomics and metatranscriptomics dataset, analyzed separately53. This pilot sample selection included at least one sample per participant that was enrolled in the study at that time, two long time courses per disease group (CD, UC, non-IBD), and multiple shorter time courses, resulting in 300 samples. For a subset of 78 samples, metatranscriptomic data was generated. Samples were chosen based on sample mass, preferentially selecting samples that could be re-sequenced if needed during the later data generation.
For the second, larger phase of data generation, stool samples were selected for different assays with the goal of generating data tiling as many aspects of the cohort as possible, including per-subject time courses, cross-subject global time points, and samples from all patients, phenotypes, age ranges, clinical centers, and so forth (Fig. 1B). The subset of measurements performed for each sample was determined in large part by aliquot requirements (in particular, mass requirements for the assay relative to how much the patient provided) and cost.
For proteomics and metabolomics, 6 global time points were equally distributed over the year-long time series for as many subjects as possible. Restrictions such as available sample mass and missing samples were incorporated by selecting the nearest suitable sample in time, resulting in slight irregularities in the sampling pattern. In total, 546 metabolite profiles and 450 proteomics profiles were generated. From among these samples, a total of 768 were selected for metagenomics, metatranscriptomics, and viromics, corresponding to 8 plates of 96 samples each. Samples already selected for proteomics/metabolomics were prioritised to facilitate integrated data analysis (316 samples had sufficient mass), resulting in 6 global time points for all subjects. In cases where the respective sample was not available for a subject, the nearest suitable sample in time was selected. Subjects with greater fluctuations in their HBI/SCCAI scores were then prioritised for a denser sampling, resulting in 12 long time courses for 5 CD, 4 UC, and 3 non-IBD subjects. The selection also includes 23 technical replicates for metagenomics, metatranscriptomics and viromics.
Finally, 576 additional samples were selected specifically for metagenomic sequencing (6 plates) resulting in a total of 1,344 metagenomic samples. Samples at previously-selected global time points and long time courses which had been restricted by available mass for other measurement types were prioritized. An additional 4 global time points were added by this process, as well as 15 long time courses (representing 10 CD, 10 UC, and 7 non-IBD subjects), and 22 samples which were previously sequenced for the pilot data and represent additional technical replicates. Lastly, 522 samples were selected for fecal calprotectin measurements, prioritizing samples that were selected for any other multi’omics data generation and representing a broad overview of the cohort. Of a total of 2,653 collected stool samples 1,785 generated at least one measurement type (Fig. 1B).
Sample selection for RNA-seq and 16S sequencing from biopsies, and host genotyping from blood draws, aimed to cover the 95 subjects who contributed at least 14 stool samples, as permitted by the availability of biopsies/blood draws for each assay. Sample selection from biopsies additionally aimed to cover biopsies from inflamed and non-inflamed sites. In total, 254 biopsies were selected for RNA-seq, covering 43 CD, 25 UC, and 22 non-IBD subjects, and distributed across biopsy sites and inflammation statuses (Extended Data Fig. 6A); and 161 biopsies were selected for 16S sequencing, covering 36 CD, 21 UC, and 22 non-IBD subjects. Exome sequencing was performed for 46 CD, 24 UC, and 22 non-IBD subjects.
Sample selection for remaining sample types (RRBS, blood serology) included all samples with a suitable sample available.
DNA and RNA isolation for metagenomics and metatranscriptomics
Total nucleic acid was extracted from one aliquot of each assayed stool sample via the Chemagic MSM I with the Chemagic DNA Blood Kit-96 from Perkin Elmer. This kit combines a chemical and mechanical lysis with magnetic bead-based purification. Prior to extraction on the MSM-I, TE buffer, Lysozyme, Proteinase K, and RLT Buffer with beta-mercaptoethanol were added to each stool sample. The stool lysate solution was vortexed to mix.
Samples were then placed on the MSM I unit to automate the following steps: M-PVA Magnetic Beads were added to the stool lysate solution and vortexed to mix. The bead-bound total nucleic acid was then removed from solution via a 96-rod magnetic head and washed in three ethanol-based wash buffers. The beads were then washed in a final water wash buffer. Finally, the beads were dipped in elution buffer to resuspend the DNA sample in solution. The beads were then removed from solution, leaving purified total nucleic acid eluate. The eluate was then split into two equal volumes, one for DNA the other for RNA. SUPERase-IN solution was added to the DNA samples, the reaction was cleaned up using AMPure XP SPRI beads. DNase was added to the RNA samples, and the reaction was cleaned up using AMPure XP SPRI beads.
DNA samples were quantified using a fluorescence-based PicoGreen assay. RNA samples were quantified using a fluorescence-based RiboGreen assay. RNA quality was assessed via smear analysis on the Caliper LabChip GX.
Metagenome sequencing
Metagenomes were generated from the resulting DNA for 1,638 stool samples, selected to obtain both a broad overview of targeted, aligned time points for all subjects (Fig. 1B), complemented by a dense sampling of subjects which tended to have greater disease activity, as determined by their HBI or SCCAI scores.
Whole genome fragment libraries were prepared as follows. Metagenomic DNA samples were quantified by Quant-iT PicoGreen dsDNA Assay (Life Technologies) and normalized to a concentration of 50 pg/uL. Illumina sequencing libraries were prepared from 100–250pg of DNA using the Nextera XT DNA Library Preparation kit (Illumina) according to the manufacturer’s recommended protocol, with reaction volumes scaled accordingly. Prior to sequencing, libraries were pooled by collecting equal volumes (200 nl) of each library from batches of 96 samples. Insert sizes and concentrations for each pooled library were determined using an Agilent Bioanalyzer DNA 1000 kit (Agilent Technologies). Libraries were sequenced on HiSeq2000 or 2500 2×101 to yield ~10 million paired end reads. Post-sequencing de-multiplexing and generation of BAM and FASTQ files were generated using the Picard suite (https://broadinstitute.github.io/picard).
Metatranscriptome sequencing
Metatranscriptomes were generated for 855 stool samples, subsampled from metagenomic selections as above. Illumina cDNA libraries were generated using a modified version of the RNAtag-seq protocol54. Briefly, 500 ng −1 μg of total RNA was fragmented, depleted of genomic DNA, dephosphorylated, and ligated to DNA adapters carrying 5’-AN8–3’ barcodes of known sequence with a 5’ phosphate and a 3’ blocking group. Barcoded RNAs were pooled and depleted of rRNA using the RiboZero rRNA depletion kit (Epicentre). Pools of barcoded RNAs were converted to Illumina cDNA libraries in 2 main steps: (i) reverse transcription of the RNA using a primer designed to the constant region of the barcoded adaptor with addition of an adapter to the 3’ end of the cDNA by template switching using SMARTScribe (Clontech) as described55; (ii) PCR amplification using primers whose 5’ ends target the constant regions of the 3’ or 5’ adaptors and whose 3’ ends contain the full Illumina P5 or P7 sequences. cDNA libraries were sequenced on the Illumina HiSeq2500 platform to generate ~13 million paired end reads.
Viromics
703 stool samples were selected for viral profiling, following the sample selection used for metatranscriptomics, adjusted slightly only when aliquots were unavailable (Fig. 1C). Viral nucleic acids were extracted using the MagMax Viral RNA Isolation Kit (Cat # AM1939, Thermo Fisher Scientific, Waltham, MA). Viral RNA was reverse transcribed using SuperScript II RT (Cat # 18064014, Thermo Fisher, Waltham, MA) and random hexamers. After short molecule and random hexamer removal with ChargeSwitch (Cat # CS12000, Thermo Fisher, Waltham, MA), molecules were amplified and tagged with a BC12-V8A2 construct56 using AccuPrimeTM Taq polymerase and cleaned with ChargeSwitch kit.
The resulting viral amplicons were normalized, pooled, and made into an Illumina library without shearing. The library (150–600 bp) was loaded in an Illumina HiSeq 2000 (Illumina, Carlsbad, CA) and sequenced using the 2×100 bp chemistry. Reads were demultiplexed into a sample bin using the barcode prefixing read-1 and read-2, allowing zero mismatches. Demultiplexed reads were further processed by trimming off barcodes, semi-random primer sequences, and Illumina adapters. This process utilized a custom demultiplexer and the BBDuk algorithm included in BBMap (http://sourceforge.net/projects/bbmap). The resulting trimmed dataset was analyzed using a pipeline created at the Alkek Center for Metagenomics and Microbiome Research at Baylor College of Medicine57. Briefly, the viral analysis pipeline employs a clustering algorithm creates putative viral genomes using a mapping assembly strategy that leverages nucleotide and translated nucleotide alignment information. Viral taxonomies were assigned using a scoring system that incorporates nucleotide and translated nucleotide alignment results in a per-base fashion and optimizes for the highest resolution taxonomic rank.
Metabolomics
Sample selection, receipt, and storage
Sample selection for metabolomics aimed to obtain only a broad sampling of many subjects. In total 546 stool samples were selected for profiling (Fig. 1B). A portion of each selected stool sample (40–100 mg) and the entire volume of originating ethanol preservative were stored in 15 mL centrifuge tubes at −80 °C until all samples were collected.
Sample processing
Samples were thawed on ice and then centrifuged (4 °C, 5,000 x g) for 5 minutes. Ethanol was evaporated using a gentle stream of nitrogen gas using a nitrogen evaporator (TurboVap LV; Biotage, Charlotte, NC) and stored at −80 °C until all samples in the study had been dried. Aqueous homogenates were generated by sonicating each sample in 900 μl of H2O using an ultrasonic probe homogenizer (Branson Sonifier 250) set to a duty cycle of 25% and output control of 2 for 3 minutes. Samples were kept on ice during the homogenization process. The homogenate for each sample was aliquoted into two 10 µL and two 30 µL in 1.5 mL centrifuge tubes for LC-MS sample preparation and 30 µL of homogenate from each sample were transferred into a 50 mL conical tube on ice to create a pooled reference sample. The pooled reference mixture was mixed by vortexing and then aliquoted (100 µL per aliquot) into 1.5 mL centrifuge tubes. Aliquots and reference sample aliquots were stored at −80 °C until LC-MS analyses were conducted.
LC-MS analyses
A combination of four LC-MS methods were used to profile metabolites in the fecal homogenates, as previously published58; two methods that measure polar metabolites, a method that measures metabolites of intermediate polarity (e.g. fatty acids and bile acids), and a lipid profiling method. For the analysis queue in each method, subjects were randomized and longitudinal samples from each subject were randomized and analyzed as a group. Additionally, pairs of pooled reference samples were inserted into the queue at intervals of approximately 20 samples for QC and data standardization. Samples were prepared for each method using extraction procedures that are matched for use with the chromatography conditions. Data were acquired using LC-MS systems comprised of Nexera X2 U-HPLC systems (Shimadzu Scientific Instruments; Marlborough, MA) coupled to Q Exactive/Exactive Plus orbitrap mass spectrometers (Thermo Fisher Scientific; Waltham, MA). The method details are summarized below:
LC-MS Method 1 – HILIC-pos: positive ion mode MS analyses of polar metabolites. LC-MS samples were prepared from stool homogenates (10 µL) via protein precipitation with the addition of nine volumes of 74.9:24.9:0.2 v/v/v acetonitrile/methanol/formic acid containing stable isotope-labeled internal standards (valine-d8, Isotec; and phenylalanine-d8, Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9,000 x g, 4°C), and the supernatants injected directly onto a 150 × 2 mm Atlantis HILIC column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 250 µL/min with 5% mobile phase A (10 mM ammonium formate and 0.1% formic acid in water) for 1 minute followed by a linear gradient to 40% mobile phase B (acetonitrile with 0.1% formic acid) over 10 minutes. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over m/z 70–800 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings are: ion spray voltage, 3.5 kV; capillary temperature, 350°C; probe heater temperature, 300 °C; sheath gas, 40; auxiliary gas, 15; and S-lens RF level 40.
LC-MS Method 2 – HILIC-neg: negative ion mode MS analysis of polar metabolites. LC-MS samples were prepared from stool homogenates (30 µL) via protein precipitation with the addition of four volumes of 80% methanol containing inosine-15N4, thymine-d4 and glycocholate-d4 internal standards (Cambridge Isotope Laboratories; Andover, MA). The samples were centrifuged (10 min, 9,000 x g, 4°C) and the supernatants were injected directly onto a 150 × 2.0 mm Luna NH2 column (Phenomenex; Torrance, CA). The column was eluted at a flow rate of 400 µL/min with initial conditions of 10% mobile phase A (20 mM ammonium acetate and 20 mM ammonium hydroxide in water) and 90% mobile phase B (10 mM ammonium hydroxide in 75:25 v/v acetonitrile/methanol) followed by a 10 min linear gradient to 100% mobile phase A. MS analyses were carried out using electrospray ionization in the negative ion mode using full scan analysis over m/z 60–750 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings are: ion spray voltage, −3.0 kV; capillary temperature, 350°C; probe heater temperature, 325 °C; sheath gas, 55; auxiliary gas, 10; and S-lens RF level 40.
LC-MS Method 3 – C18-neg: negative ion mode analysis of metabolites of intermediate polarity (e.g. bile acids and free fatty acids). Stool homogenates (30 µL) were extracted using 90 µL of methanol containing PGE2-d4 as an internal standard (Cayman Chemical Co.; Ann Arbor, MI) and centrifuged (10 min, 9,000 x g, 4 °C). The supernatants (10 µL) were injected onto a 150 × 2.1 mm ACQUITY BEH C18 column (Waters; Milford, MA). The column was eluted isocratically at a flow rate of 450 µL/min with 20% mobile phase A (0.01% formic acid in water) for 3 minutes followed by a linear gradient to 100% mobile phase B (0.01% acetic acid in acetonitrile) over 12 minutes. MS analyses were carried out using electrospray ionization in the negative ion mode using full scan analysis over m/z 70–850 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings are: ion spray voltage, −3.5 kV; capillary temperature, 320°C; probe heater temperature, 300 °C; sheath gas, 45; auxiliary gas, 10; and S-lens RF level 60.
LC-MS Method 4 – C8-pos: Lipids (polar and nonpolar) were extracted from stool homogenates (10 µL) using 190 µL of isopropanol containing 1-dodecanoyl-2-tridecanoyl-sn-glycero-3-phosphocholine as an internal standard (Avanti Polar Lipids; Alabaster, AL). After centrifugation (10 min, 9,000 x g, ambient temperature), supernatants (10 µL) were injected directly onto a 100 × 2.1 mm ACQUITY BEH C8 column (1.7 µm; Waters; Milford, MA). The column was eluted at a flow rate of 450 µL/min isocratically for 1 minute at 80% mobile phase A (95:5:0.1 vol/vol/vol 10 mM ammonium acetate/methanol/acetic acid), followed by a linear gradient to 80% mobile-phase B (99.9:0.1 vol/vol methanol/acetic acid) over 2 minutes, a linear gradient to 100% mobile phase B over 7 minutes, and then 3 minutes at 100% mobile-phase B. MS analyses were carried out using electrospray ionization in the positive ion mode using full scan analysis over m/z 200–1100 at 70,000 resolution and 3 Hz data acquisition rate. Additional MS settings are: ion spray voltage, 3.0 kV; capillary temperature, 300 °C; probe heater temperature, 300 °C; sheath gas, 50; auxiliary gas, 15; and S-lens RF level 60.
Metabolomics data processing
Raw LC-MS data were acquired to the data acquisition computer interfaced to each LC-MS system and then stored on a robust and redundant file storage system (Isilon Systems) accessed via the internal network at the Broad Institute. Nontargeted data were processed using Progenesis QIsoftware (v 2.0, Nonlinear Dynamics) to detect and de-isotope peaks, perform chromatographic retention time alignment, and integrate peak areas. Peaks of unknown ID were tracked by method, m/z and retention time. Identification of nontargeted metabolite LC-MS peaks were conducted by i) matching measured retention times and masses to mixtures of reference metabolites analyzed in each batch and ii) matching an internal database of >600 compounds that have been characterized using the Broad Institute methods. Temporal drift was monitored and normalized with the intensities of features measured in the pooled reference samples.
Proteomics
Sample selection and LC-MS/MS
Sample selection for proteomics largely followed sample selection for metabolomics (Fig. 1B–C), with slight adjustments when aliquots were unavailable. In total, 447 stool samples were targeted for profiling. From the selected samples, proteins were proteolytically digested using trypsin, and each digest subjected to automated offline high pH reversed-phase fractionation with fraction concatenation. LC-MS/MS analysis for each fraction was performed using a Thermo Scientific Q-Exactive Orbitrap mass spectrometer resident at UCLA, outfitted with a custom-built nano-ESI interface. Samples were loaded onto an in-house packed capillary LC column (70 cm x 75 μm, 3 μm particle size), and data were acquired for 120 min. Precursor MS spectra were collected over 400–2000 m/z, followed by data-dependent MS/MS spectra of the twelve most abundant ions, using a collision energy of 30%. A dynamic exclusion time of 30 s was used to discriminate against previously analyzed ions.
Peptide identification and protein data roll-up
Mass spectra from the resulting analyzes were evaluated using the MSGF+ software59 v10072 using the HMP 1 gut reference genomes (HMP_Refgenome-gut_2015–06-18). Briefly, after conversion of the metagenomic assemblies into predicted open reading frames (e.g., predicted proteins), libraries were created using the forward and reverse direction to allow determination of False Discovery Rate (FDR). The reverse decoy database allows measurement of the rate of detection of false hits, which in turn allows for FDR calculation and appropriate filtering of the data to maximize the real peptide identifications while minimizing spurious ones. MSGF+ was then used to search the experimental mass spectra data against both the forward/reverse decoy databases. Cut-offs for data included: MSGF+ spectra probability (>1 × 1010, equivalent to a BLAST e-value), mass accuracy (±20 ppm), protein level FDR of 1% and one unique peptides per protein identification.
Fecal calprotectin
Fecal calprotectin was quantified for 652 stool samples total, which were stored at −80C without preservative prior to processing. Sample selection focused on obtaining a broad survey of all subjects rather than detailed time series (Fig. 1B). Calprotectin was quantified using QUANTA Lite® Calprotectin ELISA (Inova Diagnostics 704770) following manufacturer protocol. Between 80–120 mg of stool was used for input. Incubation time prior to stopping reaction was adjusted to obtain OD 405nm values in the suggested range for assay.
Biopsy specimen processing
Co-isolation of DNA and RNA from frozen tissue
DNA and RNA were extracted from RNA-later-preserved biopsies via the AllPrep DNA/RNA Universal Kit from Qiagen. Biological samples were cut in 20–25mg pieces on a dry ice batch, then placed in tubes with a steel bead for mechanical homogenization and a highly denaturing guanidine isothiocyanate-containing buffer, which immediately inactivates DNases and RNases to ensure isolation of intact DNA and RNA. After homogenization, the lysate was passed through an AllPrep DNA Mini spin column. This column, in combination with the high-salt buffer, allows selective and efficient binding of genomic DNA. On-column Proteinase K digestion in optimized buffer conditions allows purification of high DNA yields from all sample types. The column was then washed and DNA was eluted in TE buffer. Flow-through from the AllPrep DNA Mini spin column was digested by Proteinase K in the presence of ethanol. This optimized digestion, together with the subsequent addition of further ethanol, allowed for appropriate binding of total RNA, including miRNA, to the RNeasy Mini spin column. Samples were then digested with DNase I to ensure high-yields of DNA-free RNA. Contaminants were efficiently washed away and RNA was eluted in water.
16S rRNA gene profiling
178 biopsies were selected for 16S amplicon-based taxonomic profiling. The 16S rRNA gene sequencing protocol was adapted from the Earth Microbiome Project60 and the Human Microbiome Project61–63. Briefly, bacterial genomic DNA was extracted from the total mass of the biopsied specimens using the MoBIO PowerLyzer Tissue and Cells DNA isolation kit and sterile spatulas for tissue transfer. The 16S rDNA V4 region was amplified from the extracted DNA by PCR and sequenced in the MiSeq platform (Illumina) using the 2×250 bp paired-end protocol yielding pair-end reads that overlap almost completely. The primers used contained adapters for MiSeq sequencing and single-index barcodes such that PCR products may be pooled and sequenced directly61, targeting at least 10,000 reads per sample.
Read pairs were demultiplexed and merged using USEARCH v7.0.109064. Sequences were clustered into operational taxonomic units (OTUs) at a similarity threshold of 97% using the UPARSE algorithm65. OTUs were subsequently mapped to a subset of the SILVA database66 containing only sequences from the V4 region of the 16S rRNA gene to determine taxonomies. Abundances were then recovered by mapping the demultiplexed reads to the UPARSE OTUs, producing the final taxonomic profiles. The 150 samples with ≥1000 mapped reads were used in downstream analyses.
Host RNA-Seq
cDNA library construction
In total, 252 biopsies selected for transcriptional profiling. Total RNA was quantified using the Quant-iT™ RiboGreen® RNA Assay Kit and normalized to 5 ng/ul. Following plating, 2 uL of ERCC controls (using a 1:1000 dilution) were spiked into each sample. An aliquot of 200ng for each sample was transferred into library preparation which was an automated variant of the Illumina TruSeq™ Stranded mRNA Sample Preparation Kit. This method preserves strand orientation of the RNA transcript. It uses oligo dT beads to select mRNA from the total RNA sample. It is followed by heat fragmentation and cDNA synthesis from the RNA template. The resultant 500bp cDNA then goes through library preparation (end repair, base ‘A’ addition, adapter ligation, and enrichment) using Broad Institute designed indexed adapters substituted in for multiplexing. After enrichment the libraries were quantified using Quant-iT PicoGreen (1:200 dilution). After normalizing samples to 5 ng/uL, the set was pooled and quantified using the KAPA Library Quantification Kit for Illumina Sequencing Platforms. The entire process is in 96-well format and all pipetting is done by either Agilent Bravo or Hamilton Starlet.
Illumina sequencing
Pooled libraries were normalized to 2nM and denatured using 0.1 N NaOH prior to sequencing. Flowcell cluster amplification and sequencing were performed according to the manufacturer’s protocols using either the HiSeq 2000 or HiSeq 2500. Each run was a 101bp paired-end with an eight-base index barcode read. Data was organized using the Broad Institute Picard Pipeline which includes de-multiplexing and lane aggregation.
Blood specimen processing
Serological analysis
210 sera were analyzed for expression of anti-neutrophil cytoplasmic antibodies (ANCA), anti-Saccharomyces cerevisiae mannan antibodies (ASCA), anti-OmpC, and anti-CBir1 by ELISA as previously described67, 68. Antibody levels were determined and the results expressed as ELISA units (EU/mL), which are relative to laboratory standards consisting of pooled, antigen reactive sera from of patients with well-characterized disease.
DNA isolation from whole blood
DNA was extracted via the Chemagic MSM I with the Chemagic DNA Blood Kit-96 from Perkin Elmer. The kit combines a chemical and mechanical lysis with magnetic bead-based purification. Whole blood samples were incubated at 37 °C for 5–10 minutes to thaw. The blood was then transferred to a deep well plate with protease and placed on the Chemagic MSM I. The following steps were automated on the MSM I.
M-PVA Magnetic Beads were added to the blood, protease solution. Lysis buffer was added to the solution and vortexed to mix. The bead-bound DNA was then removed from solution via a 96-rod magnetic head and washed in three Ethanol-based wash buffers to eliminate cell debris and protein residue. The beads were then washed in a final water wash buffer. Finally, the beads were dipped in elution buffer to resuspend the DNA. The beads were then removed from solution, leaving purified DNA eluate. The resulting DNA samples were quantified using a fluorescence-based PicoGreen assay.
Host exome sequencing
92 host exomes were sequenced from DNA extracts using previously published methods69. Whole exome libraries were constructed and sequenced on an Illumina HiSeq 4000 sequencer with 151 bp paired-end reads. Output from Illumina software was processed by the Picard pipeline to yield BAM files containing calibrated, aligned reads.
Library construction
Library construction was performed as described69 with some slight modifications. Initial genomic DNA input into shearing was reduced from 3 µg to 50 ng in 10 µL of solution and enzymatically sheared. In addition, for adapter ligation, dual-indexed Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter and added to each end.
In-solution hybrid selection for exome enrichment
In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries are pooled into groups of up to 96 samples prior to hybridization. The liquid handling is automated on a Hamilton Starlet. The enriched library pools are quantified via PicoGreen after elution from streptavadin beads and then normalized to a range compatible with sequencing template denature protocols.
Preparation of libraries for cluster amplification and sequencing
Following sample preparation, the libraries prepared using forked, indexed adapters were quantified using quantitative PCR (purchased from KAPA biosystems), normalized to 2 nM using the Hamilton Starlet Liquid Handling system, and pooled by equal volume using the Hamilton Starlet Liquid Handling system. Pools were then denatured using 0.1 N NaOH. Denatured samples were diluted into strip tubes using the Hamilton Starlet Liquid Handling system.
Cluster amplification and sequencing
Cluster amplification of the templates was performed according to the manufacturer’s protocol (Illumina) using the Illumina cBot. Flow cells were sequenced on HiSeq 4000 Sequencing-by-Synthesis Kits, then analyzed using RTA2.7.3
Host genetic data processing
Host genetic exome sequence data were processed using the Broad Institute sequencing pipeline by the Data Sciences Platform (Broad Institute). This was done in three steps -pre-processing (including reads mapping, alignment to a reference genome and data cleanup), variant discovery (including per-sample variant calling and joint genotyping), and variant filtering to produce callset ready for downstream genetic analysis -using Genome Analysis Toolkit (GATK) (detailed documentation at https://software.broadinstitute.org/gatk/documentation/).
Reduced representation bisulfite sequencing
Reduced Representation Bisulfite Sequencing (RRBS) libraries were prepared for 221 biopsies and 228 blood samples as described previously70 with modifications detailed below. Briefly, genomic DNA samples were quantified using a Quant-It dsDNA high sensitivity kit (ThermoFisher, cat# Q33120) and normalized to a concentration of 10 ng/ul. A total of 100 ng of normalized genomic DNA was digested with MspI in a 20 ul reaction containing 1 ul MspI (20 U/ul) (NEB, cat# R0106L) and 2 ul of 10X CutSmart Buffer (NEB, cat# B7204S). MspI digestion reactions were then incubated at 37 °C for 2 hours followed by a 15 min. incubation at 65 °C.
Next, A-tailing reactions were performed by adding 1 ul dNTP mix (containing 10 mM dATP, 1 mM dCTP and 1 mM dGTP) (NEB, cat# N0446S), 1 ul Klenow 3’−5’ exo− (NEB, cat# M0212L) and 1 ul 10X CutSmart Buffer in a total reaction volume of 30ul. A-tailing reactions were then incubated at 30 °C for 20 min, followed by 37 °C for 20 min, followed by 65 °C for 15 min.
Methylated Illumina sequencing adapters70 were then ligated to the A-tailed material (30 ul) by adding 1 ul 10X CutSmart Buffer, 5 ul 10 mM ATP (NEB, cat# P0756S), 1 ul T4 DNA Ligase (2,000,000U/ml) (NEB, cat# M0202M) and 2ul methylated adapters in a total reaction volume of 40 ul. Adapter ligation reactions were then incubated at 16 °C overnight (16–20 hours) followed by incubation at 65 °C for 15 min. Adapter ligated material was purified using 1.2X volumes of Ampure XP according to the manufacturer’s recommended protocol (Beckman Coulter, cat# A63881).
Following adapter ligation, bisulfite conversion and subsequent sample purification was performed using the QIAGEN EpiTect kit according to the manufacturer’s recommended protocol designated for DNA extracted from FFPE tissues (QIAGEN, cat# 59104). Two rounds of bisulfite conversion were performed yielding a total of 40 ul bisulfite converted DNA.
In order to determine the minimum number of PCR cycles required for final library amplification, 50 ul PCR reactions containing 3 ul bisulfite converted DNA, 5 ul 10X PfuTurbo Cx hotstart DNA polymerase buffer, 0.5 ul 100 mM dNTP (25 mM each dNTP) (Agilent, cat# 200415), 0.5 ul Illumina TruSeq PCR primers (25 uM each primer)70 and 1 ul PfuTurbo Cx hotstart DNA polymerase (Agilent, cat# 600412) were prepared. Reactions where then split equally into four separate tubes and thermocycled using the following conditions: denature at 95 °C for 2 min followed by ‘X’ cycles of 95 °C for 30 s, 65 °C for 30 s, 72 °C for 45 s (where ‘X’ number of cycles = 11, 13, 15 and 17), followed by a final extension at 72 °C for 7 min. PCR products were purified using 1.2X volumes of Ampure XP and analyzed on an Agilent Bioanalyzer using a High Sensitivity DNA kit (Agilent, cat# 5067–4626). Once the optimal number of PCR cycles was determined, 200 ul PCR reactions were prepared using 24 ul bisulfite converted DNA, 20 ul 10X PfuTurbo Cx hotstart DNA polymerase buffer, 2 ul 100 mM dNTPs (25 mM each), 2 ul Illumina TruSeq PCR primers (25 uM each) and 4 ul PfuTurbo Cx hotstart DNA polymerase with the thermal cycling conditions listed above. PCR reactions were purified using 1.2X volumes of Ampure XP according to the manufacturer’s recommended protocol and analyzed on an Agilent Bioanalyzer using a High Sensitivity DNA kit.
RRBS sequencing produced an average of 15.0M reads (sd 4.0M reads) over all 504 samples, with 448 (88.9%) samples exceeding 10M reads. Samples were analyzed with Picard 2.9.4 using default parameters, resulting in a mean alignment rate to the human genome hg19 of 95.1%. Mean CpG coverage was 8.9X (sd. 2.1%). As expected, 99.9% (sd. 0.02%) of non-CpG bases and 49.8% (sd 2.8%) of CpG bases were converted.
Data handling
Informatics for microbial community sequencing data
For metagenomes and metatranscriptomes, sequencing reads from each sample in a pool were demultiplexed based on their associated barcode sequence using custom scripts. Up to 1 mismatch in the barcode was allowed provided it did not make assignment of the read to a different barcode possible. Barcode sequences were removed from the first read as were terminal G’s from the second read that may have been added by SMARTScribe during template switching.
Taxonomic and functional profiles were generated with the bioBakery meta’omics workflow71 v0.9.0 (http://huttenhower.sph.harvard.edu/biobakery_workflows). Briefly, reads mapping to the human genome were first filtered out using KneadData 0.7.0. Taxonomic profiles of shotgun metagenomes were generated using MetaPhlAn272 v2.6.0, which uses a library of clade-specific markers to provide pan-microbial (bacterial, archaeal, viral, and eukaryotic) profiling (http://huttenhower.sph.harvard.edu/metaphlan2). Functional profiling was performed by HUMAnN273 v0.11.0 (http://huttenhower.sph.harvard.edu/humann2). HUMAnN2 constructs a sample-specific reference database from the pangenomes of the subset of species detected in the sample by MetaPhlAn2 (pangenomes are precomputed representations of the open reading frames of a given species74). Sample reads are mapped against this database to quantify gene presence and abundance on a per-species basis. A translated search is then performed against a UniRef-based protein sequence catalog75 (UniRef release 2014_07) for all reads that fail to map at the nucleotide level. The result are abundance profiles of gene families (UniRef90’s), for both metagenomics and metatranscriptomics, stratified by each species contributing that genes, and which can then be summarized to higher-level gene groupings such as Enzyme Classes (ECs) or KEGG Orthologs (KOs).
Sample counts in Fig. 1B represent the numbers of raw products available. To ensure a reasonable read depth in each sample, only samples (metagenomes and metatranscriptomes) with at least 1 million reads (after human filtering) and at least one non-zero microbial abundance detected by MetaPhlAn2 were used in downstream analyses (Fig. 1C and later). In total, this was 1,595 metagenomic and 818 metatranscriptomic samples. Principal coordinates plots were generated with the cmdscale function in the R package stats. Visualizations were principally generated using ggplot276.
Species-level meta’omic functional profile summaries
Functional profiles per clade (typically species) were further quantified by summing the total sum-normalized stratified abundance attributed to each organism in the HUMAnN2 functional profiles from both metatranscriptomics and metagenomics. For metatranscriptomic analyses, the expression ratio for the species was then also defined as the ratio between these sums.
Metaproteomic gene family functional profiles
Gene family profiles were generated from metaproteomic peptides using UniRef90 identifiers by mapping peptide sequences to the Diamond-annotated reference in HUMAnN2 v0.11.0 with v0.8.22.8477. Each peptide was mapped to the UniRef90 with the highest percent identity (minimum 90% match).
Statistical methods and association testing
PERMANOVA and Mantel tests
Omnibus testing was performed on Bray-Curtis dissimilarity matrices from MGX, MTX, MPX, and biopsy measurements. Functional profiles (MGX, MTX, and MPX) were first summarized to the KO level using HUMAnN2. Profiles were first normalized before calculation of dissimilarities. Dietary distance matrices were calculated by ordering the dietary intake frequencies from less to more frequent, assigning integers to these levels, and calculating the Manhattan distance.
Quantifications of covariation between measurement types in Fig. 1E were done using Mantel tests. To quantify cross-sectional (“inter-individual”) covariation, we first produced an average profile for each subject by taking the feature-wise mean over all samples from the subject. Subject-subject dissimilarity matrices were then generated and compared using the mantel.rtest function in the R package ape4. To quantify longitudinal covariation, we first generated the complete sample-sample dissimilarity matrix, but only calculate the Mantel test statistic (the pearson correlation between distances) from distances between samples from the same subject. Significance in this case was assessed using a permutation test with permutations limited within-subject.
Quantifications of variance explained in Fig. 1F were calculated using PERMANOVA with the adonis function in the R package Vegan78. Apart from the All row in Fig. 1F, the total variance explained by each variable was calculated independently of other variables (i.e. as the sole variable in the model) to avoid issues related to variable ordering, and should thus be considered the total variance explainable by that variable. To account for the repeated measures present for all measurement types tested, relevant permutations were performed blocked within subject for variables that change over time (medication, biopsy location, inflammation status, and dysbiosis). Meanwhile, variables that were constant (or change slowly enough to be considered constant) across samples from the same subject (age, sex, BMI, race, recruitment site, diagnosis, and disease location) were first permuted across subjects and samples were relabeled with the variable from their permuted subject. To determine the significance of models including inter-individual variance (the Subject and All rows), permutations were performed freely. For subjects with incomplete records for BMI, we imputed the mean BMI of the remaining population. The All row is the total variance explained when including all other variables in the model.
Differential microbiome feature abundance
Differential abundance (DA) analysis of all microbial measurement types (except for viruses, which were modeled as presence/absence binary features) were tested as follows. First, an appropriate transformation/normalization method was applied: arcsine square-root transformation for microbial taxonomic and functional relative abundances, log transformation (with pseudo count 1 for zero values) for metabolite profiles and protein abundances, and log transform with no pseudocount for expression ratios (non-finite values removed). Transformed abundances were then fit with the following per-feature linear mixed-effects model:
That is, in each per-feature multivariable model, recruitment sites and subjects were included as random effects to account for the correlations in the repeated measures (denoted by (1 | recruitment site) and (1 | subject) respectively) and the transformed abundance of each feature was modeled as a function of diagnosis (a categorical variable with non-IBD as the reference group) and dysbiosis state as a nested binary variable (with non-dysbiotic as reference) within each IBD phenotype (UC, CD, and non-IBD), while adjusting for consent age as a continuous covariate, and antibiotics as as binary covariate. Pearson’s residual values from the above linear mixed effects models were retained for use in subsequent analyses (Cross-measurement type interaction testing).
Fitting was performed with the nlme package in R79 (using the lme function), where significance of the association was assessed using Wald’s test (except for viruses where a logistic random effects model was considered with the glmer function from the lmer R package). Nominal p-values were adjusted for multiple hypothesis testing with a target false discovery rate of 0.25. In order to reduce the effect of zero-inflation in microbiome data, features with no variance or with >90% zeros were removed before fitting linear models. In addition, a variance filtering step was applied to remove features with very low variance. To further remove the effect of redundancy in KO gene family abundances (explainable by at most a single taxon), only features (both DNA and RNA) with low correlation with individual microbial abundances (Spearman correlation coefficient <0.6) were retained.
Differential host gene expression
Differentially expressed human genes between disease groups were quantified using a quasi-likelihood negative binomial generalized log-linear model (glmQLFit), implemented in the edgeR package in R80, 81. Analysis was performed separately for each section of the intestine on genes with at least 2 CPM (counts per million) in 10 or more samples, with significance threshold FDR p<0.05 and >1.5 log-fold change. Gene enrichment analysis was performed on differentially expressed genes against the KEGG database82 using a one-sided hypergeometric test in the package limma83.
Associations with host gene expression
Associations between host gene expression and biopsy taxonomic profiles were assessed using partial Spearman correlation, accounting for BMI, age at consent, sex and diagnosis. Association testing was performed for each biopsy location independently, as biopsy location was shown to heavily influence expression profiles (Fig. 1F, Fig. 4A–B). This simpler method was used rather than the more complex procedure outlined above for microbial measurement types since host gene expression, once filtered by biopsy location, do not have the same repeat measures problem as the microbial measurement types, allowing a simpler test.
Genetic associations
Genetic principal components for IBDMDB subjects as well as 1,000 Genomes subjects84 were calculated for a set of independent SNPs overlapping between the two datasets and pruned based on linkage disequilibrium (LD). Pruning was first performed in HMP2 using the --indep-pairwise 1500 150 0.1 command in PLINK85 by calculating LD (r2) for each pair of SNPs within a window of 1,500 SNPs, removing one of a pair of SNPs if r2>0.1 and repeating this procedure by shifting the window 150 SNPs forward. We then used the 1,000 Genomes reference phase 3 version 5a data for 2504 participants (http://bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a) to merge with the HMP2 pruned data, resulting in 7,227 overlapping independent SNPs. Using these, we performed genome-wide estimation of identity-by-descent allele sharing on the combined dataset using the --genome function in PLINK, followed by calculation of genetic principal components using the --cluster --mds-plot function for the first two principal components (Fig. 1A).
For association analyses, we used first 20 genetic principal components as covariates, obtained from the same identity-by-descent sharing matrix using --cluster --pca 20 function. We targeted associations in five loci with strong previously-reported associations with IBD and/or implicated in microbial interactions86–88. To avoid confounding by ancestry, we restricted the analysis to subjects of European ancestry, excluding 8 subjects with exomes available from other ancestral backgrounds. When available, we utilized reported SNPs that had minor allele frequency of at least 5% and Hardy-Weinberg equilibrium P<5×10−5. If not, we utilized close proxies (LD r2<0.8 in CEU population using 1,000 genomes phase 3 version 5 reference via http://analysistools.nci.nih.gov/LDlink (MST1, FUT2, IRGM, NKX2–3) or SNPs from the gnomAD browser at http://gnomad.broadinstitute.org (PTGER4)).
We used the following linear mixed effect model with the SNP as a predictor variable, coded with an additive genetic model with the outcome as the arcsine-square root transformed microbial relative abundance measured from stool metagenomes. Age, sex, antibiotic and immunosuppressant use, and the first 20 genetic principal components (PCs) were fitted as covariates with subjects as the random effect:
Optimization was performed using the lme function (from the nlme R package), with p-values calculated using the Wald test.
Associations between the rs1042712 SNP of the LCT locus89 and self-reported milk intake from dietary recall forms were tested using the same mixed effect model. Reported dairy intake options were assigned the following numeric values for regression: 1 (“yesterday, 3 or more times”), 2 (“yesterday, 1 to 2 times”); 3 (“within the past 2 to 3 days”), 4 (“within the past 4 to 7 days”), and 5 (“did not consume in last 7 days”).
Density ridgeline plots of differentially abundant features
To visualize the abundances of features significantly differentially abundant by one of the tests above (Fig. 2 and Extended Data Fig. 4), the bandwidth for kernel density estimation was selected independently for the portion of each feature above the detection limit (non-”Zero”) using the Sheather & Jones method90. Density estimates were scaled such that the maximum density for the plot spanned the distance between baselines for a given disease group. Samples below the detection limit are represented as barplots on the left, where a bar that spans the distance between baselines for a disease group represents 100% zeros. Density estimates were then additionally scaled by the fraction of non-zero samples such that relative differences in densities between groups with differing fractions of zeros are accurately represented. For both density estimates and fraction of zeros, samples were weighted by the inverse of the number of samples obtained from that subject, to avoid biasing estimates towards subjects with more densely sampled time series.
Dysbiosis analyses
Dysbiosis score
To identify samples with highly divergent (dysbiotic) metagenomic microbial compositions, as a complement to baseline disease diagnosis, we defined a dysbiosis score based on Bray-Curtis dissimilarities to non-IBD metagenomes. First, a “reference set” of samples was constructed from non-IBD subjects by taking all samples after the 20th week after the subject’s first stool sample. This was chosen since a subset of the non-IBD subjects at the start of their respective time series may not yet have overcome any gastrointestinal symptoms that triggered the initial visit to a doctor, though ultimately not caused by IBD. The dysbiosis score of a given sample was then defined as the median Bray-Curtis dissimilarity to this reference sample set, excluding samples that came from the same subject (Fig. 2C).
To identify samples which were highly divergent from the reference set, we thresholded the dysbiosis score at the 90th percentile of this score for non-IBD samples. This therefore identifies samples with a feature configuration that has a <10% probability of occurring in a non-IBD subject. By this measure, 272 metagenomes were classified as dysbiotic. Samples from CD and UC subjects are overrepresented in the dysbiotic set, with 24.3% and 11.6% of their samples classified as dysbiotic, respectively. As expected, these samples also tended to locate in the extremes of the taxonomic ordination based on metagenomes (Extended Data Figs. 3B, 3C). Dysbiosis was unevenly distributed among subjects (Extended Data Fig. 3D), with some subjects remaining dysbiotic for all or most of their time series, while others remained non-dysbiotic for their entire time series.
To lend additional support to the dysbiosis definition (i.e. as outliers by one type of microbiome profile), we tested the concordance between dysbiosis classifications made using the same statistical definition, but applied to metabolomic rather than taxonomic profiles. That is, we defined a metabolomic dysbiosis score as the median Bray-Curtis dissimilarity of one metabolomic profile to the non-IBD metabolomic profiles (after the 20th week), and defined the dysbiosis threshold as the 90th percentile of this distribution among non-IBD metabolomic profiles. We then compared these dysbiosis classifications with those from the nearest metagenomic sample (up to two weeks, see the “Cross-measurement type temporal matching” section) and found that dysbiotic samples identified by metagenomics were 4.6 times more likely to be dysbiotic by metabolomics (Fisher’s exact p = 5.9 × 10−9), showing that dysbiosis measurements are highly consistent across measurement types.
To test the sensitivity of the dysbiosis classification to the choice of reference dataset, we also performed the dysbiosis classification using the HMP1-II stool samples10 as the reference sample set instead of the non-IBD samples. The resulting dysbiosis scores (Extended Data Fig. 3E) were highly concordant (Spearman rho = 0.86; p < 2.2 × 10−16), as were the dysbiosis classifications (odds ratio of 56; Fisher’s exact p < 2.2 × 10−16). This shows that, despite the inclusion of subjects with other conditions in the non-IBD group here, as well as large differences in measurement technologies between the datasets, the dysbiosis classification is highly robust. Further, 43/426 (10.1%) of non-IBD samples were classified as dysbiotic using the HMP1-II samples as reference, falling remarkably close to the 10% expected by the definition and showing that the enrichment of IBD samples in the dysbiotic set is not simply a consequence of the definition.
Dysbiosis durations/intervals
Samples of the dysbiosis durations and intervals were obtained by taking the difference in time between stool metagenomes where the dysbiosis state changes, i.e. the time from the first dysbiotic sample in an excursion into dysbiosis until the next non-dysbiotic sample was taken as one sample of the dysbiosis duration distribution. If the subject’s time series ended before this transition occurred, this resulted in a “censored” duration/interval (Fig. 2E). Estimates of the durations of and time between dysbioses were then obtained from a censored maximum likelihood estimator for the mean of an exponential distribution. This incorporates the censored durations and intervals into the estimate to avoid underestimating these durations due to limited observation times.
Association of dysbiosis with disease location
We tested for a relationship between the Montreal disease location classification in CD and periods of dysbiosis to ensure that dysbiosis was not simply detecting different disease locations. For this, an F-test of no significance was used with a Kenward-Roger approximation of degrees of freedom91 in a logistic random effects regression that models dysbiosis as the binary outcome with subjects as a random effect and disease location as covariate, as implemented in the function glmer in the R package lme4. Only subjects with CD were considered.
Temporal analyses
Power-law fits to Bray-Curtis dissimilarities
Power-law fits to species-level metagenomic, metatranscriptomic, and metabolite Bray-Curtis dissimilarities (Fig. 3A; Extended Data Fig. 5A) were performed by fitting a power-law curve with free intercept by least-squares using the neldermead function from the R package nloptr. Significance was assessed using an F-test to compare the fit model with a flat line. Significance of the difference of the fit between disease groups was also assessed using an F-test, comparing a model jointly fit to both disease groups with separate fits to each group.
Microbiome shifts
A microbiome “shift” was defined as having occurred between two consecutive time points from the same person if the Bray-Curtis dissimilarity between their profiles was more likely to have come from a comparison between samples from different people rather than from the same person. (Extended Data Fig. 5B). Since an individual’s microbial profile naturally changes over time10, 92, the Bray-Curtis threshold at which this occurs will increase with the time difference between samples. To determine these thresholds, kernel density estimates were generated for the distribution of Bray-Curtis dissimilarities between profiles from different non-IBD subjects and between samples from the same non-IBD subjects at a range of time differences, using the density function in R. The point where the inter-individual density estimate exceeded the intra-individual density estimate was then taken as the threshold to define a “shift,” with the additional constraint that this must be a monotonically increasing function of the time difference between samples (Fig. 3A). Metabolomic shifts were defined similarly, though due to the more sparse temporal sampling of the metabolomics data (Extended Data Fig. 1) and lack of a strong upward trend in Bray-Curtis dissimilarities with time difference (Fig. 3A), only a single threshold was used based on the distribution of Bray-Curtis dissimilarities from comparisons within-subject over time in non-IBD subjects. Heatmaps of shift differences were generated using the R package pheatmap 1.0.1093.
Longitudinal multi’omic study design
Due to the large variation in microbial profiles between people (Supplementary Figure S2), with relatively smaller variation within-subject over time (Fig. 3A), longitudinal study designs have the potential to be higher-powered than purely cross-sectional studies, particularly in their ability to self-control individuals and to capture transitions between phenotypes (or after interventions) of interest94. Here, while some subjects remained in a dysbiotic state far longer than others (Extended Data Fig. 3D), the heterogeneity observed was enough to discover differences in measurement types other than where dysbiosis was defined. For example, after noting which subjects had unusual, disease-associated microbiome taxonomic profiles, they also proved to have generally shifted serological and/or metabolomic profiles at corresponding time points. Noting these dysbiotic time points offered a complementary set of differences to what was visible cross-sectionally (Fig. 2).
Among microbially-related measurements, metabolomics provided the most robust separation between disease and dysbiosis groups, possibly due to integrating a combination of host, microbial, and dietary differences (Fig. 1E–F). Thus, despite the challenges presented by untargeted metabolomics, such as unknown compound identification, the presence of redundant and background signals, and the complexity of the stool matrix, this measurement type often provides a robust characterization of subjects, their disease state, and of individual small molecules that interface between host and microbiome. Conversely, the current state of viromics assays and reference databases makes this more challenging to work with, though the importance of the virome in microbial community dynamics95 will make this an extremely interesting feature space going forward.
All longitudinal microbial measurement types showed significant variation within two weeks (Fig. 3A, Extended Data Fig. 5A). This suggests that even higher sampling rates may be needed to catch relevant microbial variation, particularly prior to onset of more severe clinical symptoms. Our sampling protocol also did not account for other potential sources of within-subject variation, such as transit time or the precise portion of each whole stool that was sampled. In this dataset, we thus cannot distinguish between temporal vs. technical variation within subject. To mitigate the higher costs associated with processing additional samples, a future study aiming to achieve higher temporal resolution might proceed in two phases, thanks to the coupling between data types (Fig. 1E): first, collect samples at a higher frequency, and process these with metagenomic or 16S sequencing. Then only proceed with more expensive and detailed data generation for samples specifically around periods of interest, such as periods of dysbiosis identified during the first stage. To this end, the sampling rate can also be tuned to target a particular probability of missing a dysbiosis period.
Integrative analyses
Lenient cross-measurement type temporal matching
For comparison between multiple measurement types, we first constructed sets of samples corresponding to the same biosample across datasets. However, exact matches were not always possible, e.g. due to specimen limitations during sample selection (see Sample selection section) or due to samples that failed QC. In these cases, matching sample sets across datasets were created using nearby samples. During this process, a degree of leniency was allowed in the matching, allowing samples up to a given time difference (two or four weeks) apart to be “matched”. To perform this matching (sample numbers in Fig. 1C and Extended Data Fig. 1B), we used the following algorithm.
For a given set of measurement types to be matched for a particular subject, find the first time window in which all measurement types have at least one produced sample. Next, within this window, find the time point with the most measurement types produced; in the event of a tie, select earlier time points. Finally, for each dataset, select the nearest sample to this target time point that is within the time window, breaking ties towards earlier time points. This set of selected samples comprises one “matched” sample. For each dataset, all samples up to and including the later of the selected sample or the target time point are then disregarded from future consideration (and thus any sample will only be included in at most one matched time point). This process is repeated for each subject until no such window exists.
Cross-measurement type interaction testing
Significant associations between features from multiple measurement types were identified using two different models: an “unadjusted” model of associations that occur mainly due to dysbiosis, and an “adjusted” model that emphasizes associations in addition to those that are dysbiosis-linked. Associations in both cases incorporated features from 10 datasets: metagenomic species, species-level transcription ratios, functional profiles at the EC level (MGX, MTX and MPX), metabolites, host transcription (rectal and ileal separately), serology and fecal calprotectin.
To detect “adjusted” associations, we first obtained residuals of features from the above data types fit to a mixed-effects model including subjects as random effects as above for the differential abundance testing (or a simple linear model without the random effects when only baseline samples were used) and adjusting for age, sex, diagnosis, dysbiosis status, antibiotic and immunosuppressant use, and bowel surgery status. Residuals from subjects with less than 4 samples in their time series for the measurement type were ignored, and for measurements with no longitudinal samples (e.g. serology and host transcriptomics measurements), the residualization was repeated with the first available samples using a simple linear model without random effects. This allowed the identification of significant (FDR p<0.05 for most measurement types, p<0.25 for serology) Spearman associations using HAllA 0.8.17 (Hierarchical All-against-all Association testing, http://huttenhower.sph.harvard.edu/halla, Supplementary Table S35). Since subject-specific random effects and covariate effects were removed from these residuals, the resulting correlations are likely to be independent of all sources of inter-individual variation as well as confounding effects due to the covariates. See Fig. 4C and Extended Data Figs. 7, 8, 9 for summary visualizations of these results. Similarly, “unadjusted” associations were identified using the same procedure, but without including dysbiosis as a covariate (Supplementary Table S36). Network visualization was done with Cytoscape96 3.6.0.
Extended Data
Extended Data Figure 1. Distribution of sample and measurement types and timing.
A) Measurements available over time for each IBDMDB participant. B) Number of identical or closely-aligned time points available for which each measurement type has been generated (Methods). C) Distributions of the number of processed samples per subject, stratified by disease and measurement type. D) Distributions of time intervals between consecutive samples for each measurement type.
Extended Data Figure 2. Within-individual stability is a major driver of microbiome differences across measurement types.
A) Principal Coordinate Analyses (PCoA) and t-SNE embeddings based on Bray-Curtis dissimilarity matrices from stool species abundances, transcripts, proteins, and metabolites. Marginal densities are shown for the PCoAs showing disease separation for some measurements. In the t-SNEs, each subject has been assigned a different hue, showing that small clusters generally represent individuals’ time courses, since inter-individual differences are the greatest driver of microbiome variation across measurement types (Fig. 1F). Sample counts are shown in Fig. 1B. B) Distributions of correlations between functional profiles, captured as UniRef90100 gene family abundances73, measured from paired metagenomes, metatranscriptomes, and metaproteomes (Methods). C) Human transcriptional expression was mostly determined by biopsy location rather than IBD phenotype and inflammation (N=249 samples from 91 subjects). Ordination shows principal component analysis on gene expression levels normalized by library sizes and represented as counts per million (CPM). Ellipses indicate 95% confidence regions for the indicated sample types. D) Principal Coordinates Plot (Bray-Curtis on OTU profiles) of community profiles from biopsy samples shows that mucosal microbial communities do not significantly differ by biopsy location (shape), unlike human gene expression in epithelial tissue (Fig. 4B).
Extended Data Figure 3. Patient-reported, molecular, and microbial disease activity measures.
A) Relationships between six measures of disease activity: the patient-reported Harvey Bradshaw Index (HBI) in CD, patient-reported Simple Clinical Colitis Activity Index (SCCAI) in UC, fecal calprotectin, the fractions of human reads from stool metagenomes (MGX) and metatranscriptomes (MTX), and a dysbiosis score defined here as departures from control population microbiome configurations (Fig. 2C). Rho values are Spearman correlation with ties broken randomly. Linear regression is shown (red line) with 95% confidence bound (shaded). Sample counts are presented in the title bar, though sample counts for a particular correlation may be less as samples must be paired. B-C) PCoA based on metagenomic species-level Bray-Curtis dissimilarities (N=1,595 samples from 130 subjects), indicating dysbiosis score (B) and whether the sample was defined as dysbiotic (C). D) Number of dysbiotic samples per participant. Color scheme is as in (C). E) Relationship between the dysbiosis score, when using the HMP1-II gut dataset as reference (N=553 from 249 subjects), compared to the non-IBD dataset. The threshold for the dysbiosis classification is also shown (black lines). The two scores are highly correlated (Pearson rho = 0.86; two-sided P < 2.2 × 10−16), as are the resulting dysbiosis classifications (odds ratio of 56; Fisher’s exact test P < 2.2 × 10−16). N=1,595 samples from 130 subjects.
Extended Data Figure 4. Significant microbial and metabolic perturbations during taxonomic dysbioses.
A) Alpha diversity (Gini-Simpson) as a function of the dysbiosis score for the sample (Pearson correlation −0.60; P < 2.2 × 10−16). N=1,595 samples from 130 subjects. B) Seven (chosen for space constraints) most differentially-abundant species not shown elsewhere in this manuscript (N=1,595 samples from 130 subjects; Wald test; Methods; full results in Supplementary Table S15). C) Top 10 differentially-abundant acylcarnitines in dysbiosis (N=546 samples from 106 subjects; Wald test). Carnitine and acylcarnitines are more abundant in dysbiotic CD, while C20:4 carnitine is significantly depleted (Supplementary Table S16). D) Top 10 metabolites differentially abundant during dysbiosis not shown elsewhere in this manuscript (N=546 samples from 106 subjects; Wald test; full results in Supplementary Table S16).
Extended Data Figure 5. Detecting shifts in longitudinal microbiome multi’omics.
A) Distributions of Bray-Curtis dissimilarities as a function of against the time difference between samples for protein profiles, species-level transcriptional activity (Methods), and species-level taxonomy (though excluding subjects with dysbiosis at any time point), otherwise as in Fig. 3A. Removing subjects with dysbiotic samples removes the extreme dissimilarities (near 1) observed in IBD subjects. Boxplots show median and lower/upper quartiles; whiskers show inner fences. B) Distribution of Bray-Curtis dissimilarities between samples from the same subject, two weeks apart vs. those from different individuals, allowing us to define a “shift” in the microbiome as a change more likely to have been drawn from the between-subject distribution than within-subject distances (corresponding to Bray-Curtis > 0.54). C) Relative abundance differences of the top 10 microbes contributing to each of the 183 detected taxonomic shifts among any two within-subject subsequent time points. Shifts are typically reciprocal (i.e. losing a microbe and regaining it later, or vice versa), and microbes with frequent high-abundance shifts generally correspond to frequent contributors in Fig. 3B. Sample ordering is from a hierarchical clustering using average linkage followed by Optimal Leaf Ordering101. D) As in Fig. 3C, but for Escherichia coli (N=322 samples from 24 subjects; Two-tail Wilcoxon test of the absolute differences in relative abundances between consecutive time points p=2.2 × 10−4 for non-IBD to UC, and p=0.029 for non-IBD to CD), frequently implicated in gut inflammation. E) As in (B), but showing Bray-Curtis dissimilarities of metabolomic profiles. Here, 22% (96/440) of sample pairs exceed the shift threshold, whereas 13% (183/1,413) exceed the threshold in (A). If metagenomic profiles are sub-sampled to match the metabolomics samples, this increases to 14% (57/398) of sample pairs, showing that if we increased the sampling rate, this measurement type would likely shift more than the metagenomes. F) As in Fig. 3B, but showing the primary contributors to metabolomic shifts, i.e. the metabolite with the largest change in relative abundance during a shift. Note that other metabolites may still experience large changes in abundance (e.g. for this reason, urate was not a primary contributor to any non-IBD shifts, though large changes are visible for one non-IBD subject in Fig. 3E). Full table of detected metabolomic shifts in Supplementary Table S30. Violin plot shows the density of points around that intake frequency; bandwidth chosen automatically by Silverman’s method102.
Extended Data Figure 6. Mucosal communities and human genetics.
A) Number of biopsy samples available for each biopsy location and inflammation status. B) DEGs (Fig. 4A) with newly identified significant correlations with OTU abundances in biopsies (partial Spearman correlation conditioned on disease status, BMI, age of consent and sex, FDR p<0.05; N=54 in ileum and N=52 independent 16S/RNA-seq pairs; full table in Supplementary Table S33). C) A limited subset of the microbiome trended with genetic variants in targeted testing, including the strongest trend shown here of Parabacteroides distasonis with genotypes of NKX2–3 (a known IBD-associated locus103; boxplots show median and lower/upper quartiles; whiskers show inner fences). This is the most significant association by p-value among all tested associations between metagenomic taxa and five known IBD loci (nominal significance p=0.006; no associations passed FDR p<0.05, mixed effect model with age, sex, antibiotic and immunosuppressant use and first 20 genetic principal components as covariates while specifying subjects as random effects; Wald test; N=84 subjects of European ancestry with exomes and 960 metagenomes; full results in Supplementary Table S34). D) Association between rs1042712 SNP in the LCT locus and self-reported milk intake from dietary recall. Self-reported short-term milk intake (from dietary recalls accompanying stool samples) was significantly associated with the count of C alleles (29.8% allele frequency) at rs1042712 in the LCT gene locus using linear mixed effect model accounting for age, sex, first 20 genetic principal components and with subjects as random effects (p=0.028, linear mixed effect regression with Wald test, Methods). All available data is plotted for unique subjects of European ancestry with exome data (per-genotype subject count (GG/GC/CC):50/26/8). Differences between IBD and non-IBD groups are not statistically significant (odds ratio 0.27; 95% CI 0.05 −1.33; p = 0.10; N=84 subjects of European ancestry with exomes and 960 dietary surveys; model: IBD (yes|no) ~ intercept + SNP + sex + age + PC1-PC20).
Extended Data Figure 7. Microbial and host-related subsets of the multi’omic association networks.
A) Subset of the network in Fig. 4C showing metagenomic abundances (octagons) and expression levels (hexagons) of Subdoligranulum, Roseburia spp and Faecalibacterium prausnitzii and their neighbors (three functionally associated microbial hubs selected for further investigation based on anti-inflammatory associations in the literature, see text). B) The host expression-related subnetwork of the “unadjusted” association network (Extended Data Fig. 9, Supplementary Discussion). Sample counts in Fig. 1B–C.
Extended Data Figure 8. Significant covariation among multi’omic components of the gut microbiome and host interactors in IBD (adjusted).
Detailed labelling of the association network in Fig. 4C (intended for magnification). The network was constructed from 10 datasets: metagenomic species, species-level transcription ratios, functional profiles at the EC levels (MGX, MTX and MPX), metabolites, host transcription (rectal and ileal separately), serology and fecal calprotectin. As in Fig. 4C, measurement types were approximately matched in time with a maximum separation between paired samples of 4 weeks. The top 300 significant correlations (FDR p<0.05) among correlations between features which were differentially abundant in dysbiosis were used to construct the network visualized here (for serology, a threshold of FDR p<0.25 was used). Nodes are colored by the disease group they are “high” in, and edges are colored by the sign and strength of the correlation. For this “adjusted” network, Spearman correlations were calculated using HAllA from the residuals of a mixed-effects model with subjects as random effects (or a simple linear model without the random effects when only baseline samples were used) after adjusting for age, sex, diagnosis, dysbiosis status, recruitment site, and antibiotics (Methods). Appropriate normalization and/or transformation for each measurement type was performed independently before the model fitting (Methods). Singleton node-pairs were pruned from the network. Source associations are in Supplementary Table S35, sample counts in Fig. 1B–C.
Extended Data Figure 9. Significant covariation among multi’omic components of the gut microbiome and host interactors in IBD (unadjusted).
The network was constructed from 10 datasets: metagenomic species, species-level transcription ratios, functional profiles at the EC levels (MGX, MTX and MPX), metabolites, host transcription (rectal and ileal separately), serology and fecal calprotectin. As in Fig. 4C, measurement types were approximately matched in time with a maximum separation between paired samples of 4 weeks. The top 300 significant correlations (FDR p<0.05) among correlations between features which were differentially abundant in dysbiosis were used to construct the network visualized here (for serology, a threshold of FDR p<0.25 was used). Nodes are colored by the disease group they are “high” in, and edges are colored by the sign and strength of the correlation. For this “unadjusted” network, Spearman correlations were calculated using HAllA from the residuals of the same model as in Extended Data Fig. 8, though without adjusting for dysbiosis (Methods). Appropriate normalization and/or transformation for each measurement type was performed independently before the model fitting (Methods). Singleton node-pairs were pruned from the network. Source associations are in Supplementary Table S36, sample counts in Fig. 1B–C.
Extended Data Table 1: IBDMDB cohort characteristics.
Breakdown of the cohort by diagnosis, sex, ethnicity, physical characteristics, and disease location for each clinical site. Numeric data are summarized with mean ± standard deviation, with the range given by [min, max], and with N/A’s removed. Subject counts for disease locations are subset to the respective diseases.
| Cedars-Sinai | Emory | Cincinnati | MGH | MGH Ped. | Total | ||
|---|---|---|---|---|---|---|---|
| Subjects | 33 | 11 | 33 | 38 | 17 | 132 | |
| Diagnosis |
non-IBD | 1 | 1 | 9 | 13 | 3 | 27 |
| UC | 12 | 3 | 7 | 11 | 5 | 38 | |
| CD | 20 | 7 | 17 | 14 | 9 | 67 | |
| Sex |
Female | 24 | 6 | 16 | 15 | 3 | 64 |
| Male | 9 | 5 | 17 | 23 | 14 | 68 | |
| Ethnicity |
American Indian/Alaska Native | 1 | 1 | ||||
| Black or African American | 6 | 2 | 2 | 10 | |||
| More than one race | 3 | 1 | 1 | 5 | |||
| Other | 1 | 3 | 4 | ||||
| White | 32 | 2 | 30 | 32 | 16 | 112 | |
| Age (years) | 45.0 ± 13.0 [26 – 76] | 12.6 ± 3.8[7 – 17] | 12.9 ± 2.9 [6 – 17] | 35.9 ± 14.5 [18 – 74] | 13.9 ± 3.2 [6 – 17] | 27.5 ± 17.3 [6 – 76] | |
| Height (cm) | 169.7 ± 9.5 [152 – 188] | 148.7 ± 10.7 [137 – 158] | 157.9 ± 15.8 [112 – 182] | 169.4 ± 11.4 [142 – 188] | 164. 1 ± 20.8 [114.3 – 193] | 163.5 ± 15.5 [112 – 193] | |
| Weight (kg) | 66.4 ± 13.0 [54.9 – 87.1] | 43.9 ± 4.1 [41.0 – 46.8] | 65.0 ± 23.5 [19.3 – 115.7] | 71.8 ± 16.6 [37 – 100] | 53.9 ± 18.6 [19.6 – 75.0] | 65.1 ± 20. 1 [19.3 – 115.7] | |
| BMI | 28.4 ± 10.2 [19.5 – 50.2] | 18.8 ± 3.2 [16.3 – 22.4] | 21. 1 ± 6.1 [13.5 – 35.6] | 24.4 ± 5.1 [19. 1 – 40.6] | 19.1 ± 4.4 [13.5 – 28.4] | 22.9 ± 7. 1 [13.5 – 50.2] | |
| CD Montreal Location |
L1 | 1 | 2 | 8 | 11 | ||
| L1+L4 | 4 | 1 | 5 | ||||
| L2 | 2 | 2 | 1 | 5 | |||
| L2+L4 | 1 | 1 | |||||
| L3 | 9 | 1 | 5 | 2 | 2 | 19 | |
| L3+L4 | 1 | 7 | 4 | 12 | |||
| N/A | 8 | 3 | 2 | 1 | 14 | ||
| UC Extent |
Ulcerative proctitis | 1 | 1 | 2 | |||
| Left-sided UC | 2 | 2 | 1 | 5 | |||
| Extensive UC I Pancolitis | 2 | 1 | 3 | 2 | 1 | 9 | |
| N/A | 8 | 2 | 1 | 8 | 3 | 22 | |
Supplementary Material
Acknowledgements
The IBDMDB Investigators are: Jason Bishai, Kevin Bullock, Amy Deik, Courtney Dennis, Jess L. Kaplan, Hamed Khalili, Lauren J. McIver, Christopher J. Moran, Long Nguyen, Kerry A. Pierce, Randall Schwager, Alexandra Sirota-Madi, Betsy W. Stevens, William Tan, Johanna J. ten Hoeve, George Weingart, Robin G. Wilson, Vijay Yajnik
We wish to acknowledge Gail Ackermann, Emma X. Li, Jonathan Livny, Daniel McDonald, Chris Moran, Adam Robbins-Pianka, and Serene Sun for their contributions during this project, as well as data generation and management by the Broad Institute of MIT and Harvard Microbial ‘Omics Core, Genomics Platform, and the Harvard T. H. Chan School of Public Health Microbiome Analysis Core. We also thank Theresa Reimels for editorial assistance. This work was supported by National Institutes of Health grants P01DK046763 (DPBM), U01DK062413 (DPBM), U54DK102557 (DPBM), UL1TR001881 (JB), P30DK043351 (RJX), R24DK110499 (CH), R01HG005969 (CH), U54DE023798 (CH and RJX), National Science Foundation grant DBI-1053486 (CH), and Army Research Office grant W911NF-11-1-0473 (CH). Dmitry Shungin was supported by the Swedish Research Council International Career Fellowship (4.1-2016-00416). A. Brantley Hall is a Merck Fellow of the Helen Hay Whitney Foundation. Ashwin N. Ananthakrishnan was supported by the Crohn’s and Colitis Foundation. Dermot P. B. McGovern was supported by the Leona M. and Harry B. Helmsley Charitable Trust. Pacific Northwest National Laboratory is a multi-program laboratory operated by Battelle for the U.S. Department of Energy under Contract DE-AC05-76RL01830.
Footnotes
Supplementary Information
is linked to the online version of the paper at www.nature.com/nature.
Competing financial and non-financial interests
Jonathan Braun is on the Scientific Advisory Board for Janssen Research & Development, LLC. Curtis Huttenhower is on the Scientific Advisory Board for Seres Therapeutics. Joseph Petrosino and Nadim Ajami own shares at Diversigen Inc. Ramnik Xavier is a consultant to Novartis and Nestle.
Data Availability Statement
Protocols and data (both raw and summarized to data type-dependent profiles) are available at the IBDMDB web site (https://ibdmdb.org), the HMP DACC web portal (https://www.hmpdacc.org/ihmp/), and Qiita97 (https://qiita.ucsd.edu/). Sequence data are available from SRA BioProject PRJNA398089. Expression data has been deposited in NCBI’s Gene Expression Omnibus98 and is accessible through GEO Series accession number GSE111889. Metabolomics data is available at the NIH Common Fund’s Metabolomics Data Repository and Coordinating Center (supported by NIH grant, U01-DK097430) website, the Metabolomics Workbench, http://www.metabolomicsworkbench.org, where it has been assigned Project ID PR000639. Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE99 partner repository with the dataset identifiers PXD008675 and 10.6019/PXD008675. Reprints and permissions information is available at www.nature.com/reprints.
Code Availability Statement
Bioinformatics workflows for metagenomics and metatranscriptomics data are available at https://bitbucket.org/biobakery/hmp2_workflows. Analysis scripts are available at https://bitbucket.org/biobakery/hmp2_analysis.
References
- 1.Kaplan GG The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol 12, 720–727, doi: 10.1038/nrgastro.2015.150 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Hugot JP et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature 411, 599–603, doi: 10.1038/35079107 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Huang H et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature 547, 173–178, doi: 10.1038/nature22969 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morgan XC et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol 13, R79, doi: 10.1186/gb-2012-13-9-r79 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gevers D et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392, doi: 10.1016/j.chom.2014.02.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kostic AD, Xavier RJ & Gevers D The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499, doi: 10.1053/j.gastro.2014.02.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ananthakrishnan AN Environmental risk factors for inflammatory bowel diseases: a review. Dig Dis Sci 60, 290–298, doi: 10.1007/s10620-014-3350-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knights D et al. Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med 6, 107, doi: 10.1186/s13073-014-0107-1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Halfvarson J et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2, 17004, doi: 10.1038/nmicrobiol.2017.4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lloyd-Price J et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature 550, 61–66, doi: 10.1038/nature23889 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vandeputte D et al. Stool consistency is strongly associated with gut microbiota richness and composition, enterotypes and bacterial growth rates. Gut 65, 57–62, doi: 10.1136/gutjnl-2015-309618 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisshof R & Chermesh I Micronutrient deficiencies in inflammatory bowel disease. Curr Opin Clin Nutr Metab Care 18, 576–581, doi: 10.1097/MCO.0000000000000226 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Kuroki F et al. Multiple vitamin status in Crohn’s disease. Correlation with disease activity. Dig Dis Sci 38, 1614–1618 (1993). [DOI] [PubMed] [Google Scholar]
- 14.Depeint F, Bruce WR, Shangari N, Mehta R & O’Brien PJ Mitochondrial function and toxicity: role of the B vitamin family on mitochondrial energy metabolism. Chem Biol Interact 163, 94–112, doi: 10.1016/j.cbi.2006.04.014 (2006). [DOI] [PubMed] [Google Scholar]
- 15.Li J et al. Niacin ameliorates ulcerative colitis via prostaglandin D2-mediated D prostanoid receptor 1 activation. EMBO Mol Med 9, 571–588, doi: 10.15252/emmm.201606987 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Figge HL et al. Comparison of excretion of nicotinuric acid after ingestion of two controlled release nicotinic acid preparations in man. J Clin Pharmacol 28, 1136–1140 (1988). [DOI] [PubMed] [Google Scholar]
- 17.Walmsley RS, Ayres RC, Pounder RE & Allan RN A simple clinical colitis activity index. Gut 43, 29–32 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank DN et al. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci U S A 104, 13780–13785, doi: 10.1073/pnas.0706625104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall AB et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 9, 103, doi: 10.1186/s13073-017-0490-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kruis W, Kalek HD, Stellaard F & Paumgartner G Altered fecal bile acid pattern in patients with inflammatory bowel disease. Digestion 35, 189–198 (1986). [DOI] [PubMed] [Google Scholar]
- 21.Duboc H et al. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 62, 531–539, doi: 10.1136/gutjnl-2012-302578 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Meadows JA & Wargo MJ Carnitine in bacterial physiology and metabolism. Microbiology 161, 1161–1174, doi: 10.1099/mic.0.000080 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scher JU et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2, e01202, doi: 10.7554/eLife.01202 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abu-Ali GS et al. Metatranscriptome of human faecal microbial communities in a cohort of adult men. Nat Microbiol 3, 356–366, doi: 10.1038/s41564-017-0084-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan XC et al. Associations between host gene expression, the mucosal microbiome, and clinical outcome in the pelvic pouch of patients with inflammatory bowel disease. Genome Biol 16, 67, doi: 10.1186/s13059-015-0637-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Linge HM et al. The human CXC chemokine granulocyte chemotactic protein 2 (GCP-2)/CXCL6 possesses membrane-disrupting properties and is antibacterial. Antimicrob Agents Chemother 52, 2599–2607, doi: 10.1128/AAC.00028-08 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckhardt ER et al. Intestinal epithelial serum amyloid A modulates bacterial growth in vitro and pro-inflammatory responses in mouse experimental colitis. BMC Gastroenterol 10, 133, doi: 10.1186/1471-230X-10-133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Hassani RA et al. Dual oxidase2 is expressed all along the digestive tract. Am J Physiol Gastrointest Liver Physiol 288, G933–942, doi: 10.1152/ajpgi.00198.2004 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Bachman MA, Miller VL & Weiser JN Mucosal lipocalin 2 has pro-inflammatory and iron-sequestering effects in response to bacterial enterobactin. PLoS Pathog 5, e1000622, doi: 10.1371/journal.ppat.1000622 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kanehisa M et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42, D199–205, doi: 10.1093/nar/gkt1076 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haberman Y et al. Pediatric Crohn disease patients exhibit specific ileal transcriptome and microbiome signature. J Clin Invest 124, 3617–3633, doi: 10.1172/JCI75436 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kugathasan S et al. Prediction of complicated disease course for children newly diagnosed with Crohn’s disease: a multicentre inception cohort study. Lancet 389, 1710–1718, doi: 10.1016/S0140-6736(17)30317-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hajishengallis G, Reis ES, Mastellos DC, Ricklin D & Lambris JD Novel mechanisms and functions of complement. Nat Immunol 18, 1288–1298, doi: 10.1038/ni.3858 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross IN, Thompson RA, Montgomery RD & Asquith P Significance of serum complement levels in patients with gastrointestinal disease. J Clin Pathol 32, 798–801, doi: 10.1136/jcp.32.8.798 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jain U, Otley AR, Van Limbergen J & Stadnyk AW The complement system in inflammatory bowel disease. Inflamm Bowel Dis 20, 1628–1637, doi: 10.1097/MIB.0000000000000056 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Lipinski S et al. DUOX2-derived reactive oxygen species are effectors of NOD2-mediated antibacterial responses. J Cell Sci 122, 3522–3530, doi: 10.1242/jcs.050690 (2009). [DOI] [PubMed] [Google Scholar]
- 37.Yung SC & Murphy PM Antimicrobial chemokines. Front Immunol 3, 276, doi: 10.3389/fimmu.2012.00276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pasolli E et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 176, 649–662 e620, doi: 10.1016/j.cell.2019.01.001 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ralling G, Bodrug S & Linn T Growth rate-dependent regulation of RNA polymerase synthesis in Escherichia coli. Mol Gen Genet 201, 379–386 (1985). [DOI] [PubMed] [Google Scholar]
- 40.Sokol H et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105, 16731–16736, doi: 10.1073/pnas.0804812105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vitek L Bile acid malabsorption in inflammatory bowel disease. Inflamm Bowel Dis 21, 476–483, doi: 10.1097/MIB.0000000000000193 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Dawson PA & Karpen SJ Intestinal transport and metabolism of bile acids. J Lipid Res 56, 1085–1099, doi: 10.1194/jlr.R054114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walsham NE & Sherwood RA Fecal calprotectin in inflammatory bowel disease. Clin Exp Gastroenterol 9, 21–29, doi: 10.2147/CEG.S51902 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Truong DT, Tett A, Pasolli E, Huttenhower C & Segata N Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res 27, 626–638, doi: 10.1101/gr.216242.116 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ricklin D, Reis ES & Lambris JD Complement in disease: a defence system turning offensive. Nat Rev Nephrol 12, 383–401, doi: 10.1038/nrneph.2016.70 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Franzosa EA et al. Relating the metatranscriptome and metagenome of the human gut. Proc Natl Acad Sci U S A 111, E2329–2338, doi: 10.1073/pnas.1319284111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogtmann E et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. Am J Epidemiol 185, 115–123, doi: 10.1093/aje/kww177 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loftfield E et al. Comparison of Collection Methods for Fecal Samples for Discovery Metabolomics in Epidemiologic Studies. Cancer Epidemiol Biomarkers Prev 25, 1483–1490, doi: 10.1158/1055-9965.EPI-16-0409 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voigt AY et al. Temporal and technical variability of human gut metagenomes. Genome Biol 16, 73, doi: 10.1186/s13059-015-0639-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jowett SL, Seal CJ, Barton JR & Welfare MR The short inflammatory bowel disease questionnaire is reliable and responsive to clinically important change in ulcerative colitis. Am J Gastroenterol 96, 2921–2928, doi: 10.1111/j.1572-0241.2001.04682.x (2001). [DOI] [PubMed] [Google Scholar]
- 51.Daperno M et al. Development and validation of a new, simplified endoscopic activity score for Crohn’s disease: the SES-CD. Gastrointest Endosc 60, 505–512 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Baron JH, Connell AM & Lennard-Jones JE Variation Between Observers in Describing Mucosal Appearances in Proctocolitis. British Med J 1, 89–92 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schirmer M et al. Dynamics of metatranscription in the inflammatory bowel disease gut microbiome. Nat Microbiol 3, 337–346, doi: 10.1038/s41564-017-0089-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shishkin AA et al. Simultaneous generation of many RNA-seq libraries in a single reaction. Nat Methods 12, 323–325, doi: 10.1038/nmeth.3313 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhu YY, Machleder EM, Chenchik A, Li R & Siebert PD Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30, 892–897 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Clem AL, Sims J, Telang S, Eaton JW & Chesney J Virus detection and identification using random multiplex (RT)-PCR with 3’-locked random primers. Virol J 4, 65, doi: 10.1186/1743-422X-4-65 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ajami NJ, Wong MC, Ross MC, Lloyd RE & Petrosino JF Maximal viral information recovery from sequence data using VirMAP. Nat Commun 9, 3205, doi: 10.1038/s41467-018-05658-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kostic AD et al. The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 17, 260–273, doi: 10.1016/j.chom.2015.01.001 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim S & Pevzner PA MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun 5, 5277, doi: 10.1038/ncomms6277 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thompson LR et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 551, 457–463, doi: 10.1038/nature24621 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Caporaso JG et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6, 1621–1624, doi: 10.1038/ismej.2012.8 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214, doi: 10.1038/nature11234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.The Human Microbiome Project Consortium. A framework for human microbiome research. Nature 486, 215–221, doi: 10.1038/nature11209 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461, doi: 10.1093/bioinformatics/btq461 (2010). [DOI] [PubMed] [Google Scholar]
- 65.Edgar RC UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10, 996–998, doi: 10.1038/nmeth.2604 (2013). [DOI] [PubMed] [Google Scholar]
- 66.Pruesse E et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35, 7188–7196, doi: 10.1093/nar/gkm864 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landers CJ et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto-and microbial antigens. Gastroenterology 123, 689–699 (2002). [DOI] [PubMed] [Google Scholar]
- 68.Targan SR et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn’s disease. Gastroenterology 128, 2020–2028 (2005). [DOI] [PubMed] [Google Scholar]
- 69.Fisher S et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol 12, R1, doi: 10.1186/gb-2011-12-1-r1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gu H et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc 6, 468–481, doi: 10.1038/nprot.2010.190 (2011). [DOI] [PubMed] [Google Scholar]
- 71.McIver LJ et al. bioBakery: A meta’omic analysis environment. Bioinformatics, doi: 10.1093/bioinformatics/btx754 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Truong DT et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12, 902–903, doi: 10.1038/nmeth.3589 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Franzosa EA et al. Species-level functional profiling of metagenomes and metatranscriptomes. Nat Methods 15, 962–968, doi: 10.1038/s41592-018-0176-y (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang K et al. MetaRef: a pan-genomic database for comparative and community microbial genomics. Nucleic Acids Res 42, D617–624, doi: 10.1093/nar/gkt1078 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Suzek BE et al. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932, doi: 10.1093/bioinformatics/btu739 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wickham H ggplot2: Elegant Graphics for Data Analysis Springer-Verlag New York; (2016). [Google Scholar]
- 77.Buchfink B, Xie C & Huson DH Fast and sensitive protein alignment using DIAMOND. Nat Methods 12, 59–60, doi: 10.1038/nmeth.3176 (2015). [DOI] [PubMed] [Google Scholar]
- 78.Oksanen J, F. G. B., Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E and Wagner H vegan: Community Ecology Package. R package version 2.5–3 https://CRAN.R-project.org/package=vegan (2018).
- 79.Pinheiro J, D. B., DebRoy S, Sarkar D and the R Development Core Team. nlme: Linear and Nonlinear Mixed Effects Models. R package version 3.1–108 (2013).
- 80.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McCarthy DJ, Chen Y & Smyth GK Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res 40, 4288–4297, doi: 10.1093/nar/gks042 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kanehisa M, Furumichi M, Tanabe M, Sato Y & Morishima K KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45, D353–D361, doi: 10.1093/nar/gkw1092 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ritchie ME et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47, doi: 10.1093/nar/gkv007 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74, doi: 10.1038/nature15393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575, doi: 10.1086/519795 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jostins L et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491, 119–124, doi: 10.1038/nature11582 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu JZ et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 47, 979–986, doi: 10.1038/ng.3359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hall AB, Tolonen AC & Xavier RJ Human genetic variation and the gut microbiome in disease. Nat Rev Genet, doi: 10.1038/nrg.2017.63 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Enattah NS et al. Identification of a variant associated with adult-type hypolactasia. Nat Genet 30, 233–237, doi: 10.1038/ng826 (2002). [DOI] [PubMed] [Google Scholar]
- 90.Sheather SJ & Jones MC A reliable data-based bandwidth selection method for kernel density estimation. J Royal Stat Soc 53, 683–690, doi: 10.2307/2345597 (1991). [DOI] [Google Scholar]
- 91.Kenward MG & Roger JH An improved approximation to the precision of fixed effects from restricted maximum likelihood. Comput Stat Data Anal 53, 2583–2595, doi: 10.1016/j.csda.2008.12.013 (2009). [DOI] [Google Scholar]
- 92.Faith JJ et al. The long-term stability of the human gut microbiota. Science 341, 1237439, doi: 10.1126/science.1237439 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kolde R Pheatmap: pretty heatmaps. R Package Version 1.0.10 (2012).
- 94.Gibbons RD, Hedeker D & DuToit S Advances in analysis of longitudinal data. Annu Rev Clin Psychol 6, 79–107, doi: 10.1146/annurev.clinpsy.032408.153550 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Minot S et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res 21, 1616–1625, doi: 10.1101/gr.122705.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gonzalez A et al. Qiita: rapid, web-enabled microbiome meta-analysis. Nat Methods 15, 796–798, doi: 10.1038/s41592-018-0141-9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Edgar R, Domrachev M & Lash AE Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30, 207–210 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Vizcaino JA et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44, D447–456, doi: 10.1093/nar/gkv1145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Suzek BE, Huang H, McGarvey P, Mazumder R & Wu CH UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288, doi: 10.1093/bioinformatics/btm098 (2007). [DOI] [PubMed] [Google Scholar]
- 101.Bar-Joseph Z, Gifford DK & Jaakkola TS Fast optimal leaf ordering for hierarchical clustering. Bioinformatics 17 Suppl 1, S22–29 (2001). [DOI] [PubMed] [Google Scholar]
- 102.Silverman. 48, eqn (43.31) (1986).
- 103.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678, doi: 10.1038/nature05911 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.