Abstract
Background and Purpose:
Accurate automated infarct segmentation is needed for acute ischemic stroke studies relying on infarct volumes as an imaging phenotype or biomarker that require large numbers of subjects. This study investigates whether an ensemble of convolutional neural networks (CNN) trained on multiparametric DWI maps outperforms single networks trained on solo DWI parametric maps.
Materials and Methods:
CNNs were trained on combinations of DWI, ADC, and low b-value-weighted images from 116 subjects. The performances of the networks (measured by Dice score, sensitivity and precision) were compared to one another and to ensembles of 5 networks. To assess the generalizability of the approach, the best performing model was applied to an independent evaluation cohort of 151 subjects. Agreement between manual and automated segmentations for identifying patients with large lesions volumes was calculated across multiple thresholds (21 cm3, 31 cm3, 51 cm3, and 70 cm3).
Results
An ensemble of CNNs trained on DWI, ADC and low b-value-weighted images produced the most accurate acute infarct segmentation over individual networks (p<0.0001). Automated volumes correlated with manually measured volumes (Spearman’s ρ=0.91, p<0.0001) for the independent cohort. For the task of identifying patients with large lesion volumes, agreement between manual outlines and automated outlines was high (Cohen’s κ 0.86 to 0.90, p<0.0001).
Conclusion
Acute infarcts are more accurately segmented using ensembles of CNNs trained with multi-parametric maps than using a single model trained with a solo map. Automated lesion segmentation can perform with high agreement with manual techniques for identifying patients with large lesion volumes.
INTRODUCTION
Accurate acute infarct segmentation on DWI is important for many aspects of ischemic stroke patient management such as deciding whether or not to triage the patient to an intensive care unit, monitoring brain swelling, aiding prognosis, assessing risk for complications, and predicting functional outcome. Robust automated segmentation of acute infarcts also has great potential for use in clinical trials where precise volume measurements are needed to assess differences between groups or to monitor lesion growth. Various automated algorithms for segmenting tissue have been presented.1-3 However, many of these methods focus on only using a solo diffusion parametric map, such as the isotropic high b-value DWI1 or ADC image2. There have been studies that combined DWI and ADC maps,4-6 but these did not include the nondiffusion-weighted low b-value image (b=0 s/mm2, LOWB), which can potentially be used to measure early vasogenic edema. Another study has proposed using multiple b-values, up to 2000 s/mm2 (which is typically not acquired in the acute setting), but whether the data was acquired in the acute or subacute stage was not reported and the effects of using combinations of parameters were not investigated.7
We hypothesize that a multimodal approach can improve the performance of automated segmentation algorithms. Indeed, most radiologists use other sequences in addition to DWI when assessing extent of acute infarction. We tested this hypothesis by comparing the accuracy of fully automated acute infarct segmentation algorithms that utilize solo diffusion parametric maps to the performance of algorithms that combine multiple parametric maps. We also posit that ensemble models which aggregate segmentation results from multiple algorithms will surpass single algorithms. The superior accuracy of ensemble algorithms has been shown for tumor applications 8 but not yet for acute infarct segmentation. Finally, we assessed the generalizability of our approach by evaluating its performance on an independent cohort. We also tested the clinical utility of automated approaches for triaging patients with large infarct volumes who might not benefit from endovascular treatment.9, 10
METHODS
Subjects
All analyses were performed retrospectively under institutional review board approval. MRI from acute ischemic stroke patients admitted at a single academic medical center between 2005-2007, imaged within 12h from when the patient was last known to be well (LKW), whom did not receive either thrombolysis prior to MRI or experimental therapy were used for training the convolutional neural networks (CNN)11. An independent cohort12, 13 consisting of non-overlapping patients admitted to the same center between 1996-2012 for whom imaging was performed within 24 h LKW and for whom follow-up MRI data sets were available were used for the evaluation group. Both cohorts were drawn from separate repositories for which manual outlines were available that had been drawn several years ago for a study of early stage stroke patterns11 or for studies predicting lesion expansion.12, 13
MRI
Diffusion-weighted MRI was acquired on 1.5T scanners (General Electric Medical Systems) with the following parameters for the majority of subjects: b-value of 1000 s/mm2, 5000 ms repetition time, 88.9 ms echo time, 220 mm field-of-view, 23 5 mm thick slices and 1 mm gap, and 6 diffusion directions (see Supplemental Methods for details). MRI were corrected for eddy current distortions prior to calculation of isotropic trace DWI maps (geometric mean of the high b-value acquisitions), and ADC maps (slope of the linear regression fit of the log of the DWI and LOWB images using techniques described previously.14 Manual outlines had been drawn for prior studies11-13 using the program Display (McConnellBrain Imaging Centre, Montreal, Canada) by a neuroscientist with 15 years of experience (Reader 1: OW, training) and a neuroradiology fellow with 4 years of experience (Reader 2: RB, evaluation) interpreting stroke MRI. The readers were blinded to the results of the automated segmentation algorithm. No a priori thresholds were used for manual segmentation, but concomitant ADC and LOWB maps were referenced to avoid inclusion of susceptibility artifacts and chronic lesions with elevated ADC values. Tissue were considered acute infarcts if they exhibited hyperintensity on the DWI, with hypointensity on the ADC or abnormal T2 prolongation on LOWB. To assess for interrater agreement, 10 subjects from the evaluation cohort were randomly selected and outlines drawn by Reader 1 and two-way intraclass correlation (ICC) calculated.
A neuroradiologist with 12 years of experience (WAC) assigned each patient to one of the following categories, based on lesion location: brainstem, cerebellum, supratentorial/cortical, or supratentorial/subcortical. The “supratentorial/cortical” designation was used if any portion of one or more infarcts involved the cortex. Patients with both supra- and infratentorial lesions, or lesions involving both the brainstem and cerebellum, were assigned to a fifth category: “multiple.”
Image Preprocessing
DWI, ADC and LOWB images were resampled to an isotropic voxel size of 1mm3. The LOWB brain mask was computed using the Brain Extraction Tool (FSL version 5.0.9).15, 16 Mean and standard deviation were calculated from intensities within the brain mask limited to the [1, 99] percentile range to normalize values to mean 0 and standard deviation 1.0.
CNN Training
CNNs were trained to classify voxels as lesion or non-lesion on a NVIDIA Tesla K40 GPU using the DeepMedic (v0.7.0) framework with two pathways (see original publication17 and Supplemental Methods). Supplemental Figure A1 shows the architecture. DeepMedic is a 3D CNN that operates on multi-resolution pathways to allow efficient and accurate supervised segmentation. This framework was chosen over other approaches since it performed best in the Ischemic Stroke Lesion Segmentation Challenge (ISLES) 2015.18 Additional studies have also shown DeepMedic had better or comparable performance compared to other neural network architectures (see Supplemental Methods). Separate CNNs were trained on single or different combinations of diffusion parametric maps (DWI, ADC, and LOWB individually, DWI+ADC, ADC+LOWB, DWI+LOWB, DWI+ADC+LOWB). To generate ensemble segmentations, the class posteriors from the softmax layers of five independent CNNs were averaged voxel-wise. The results of all models were resampled back to the original image resolution, thresholded at 50% and masked with the resampled brain mask created at the normalization step. Performance within the training data was assessed via 5-fold cross validation. For subjects in each fold, lesion segmentations were generated using a CNN that was trained on data from the other four folds. Training a single CNN with DWI+ADC+LOWB maps on the full training cohort of 116 subjects required approximately 16 hours. Applying the trained CNN to an individual subject to segment the lesion took on average 35 seconds. With sequential evaluation of 5 CNNs, merging their output and resampling, we estimate a full segmentation will require less than 5 minutes.
Performance Evaluation
Binarized segmentation performances were assessed with the Dice score (measure of overlap between automated and manual lesion segmentations), precision and sensitivity metrics. Dice score, precision and sensitivity were computed as follows: Dice = 2TP/(2*Tp+FP+FN); Precision = TP/(TP+FP); Sensitivity = TP/(TP+FN) for which TP=true positives, FP=false positive and FN=false negatives. All metrics range from 0-100%, for which higher values indicate better performances.
To evaluate generalizability of the approach, the best performing network was re-trained on the full training cohort and applied to the independent cohort. The evaluation cohort was also segmented with an approach that has been utilized in clinical trials19. In brief, the technique combined thresholding of ADC (<615×10−6mm2/s), DWI and exponential attenuation maps with morphological operations (opening with a 2-voxel structural element). ADC images were first masked with a LOWB brain mask prior to thresholding. We evaluated the algorithm on images that had been resampled to 1 mm resolution for processing and on images that were segmented in their original resolution. Segmented outputs from all algorithms were evaluated at 1 mm resolution to reduce potential confounds from different MRI acquisition resolutions. Effects of lesion volume and location on performance were investigated using univariable and multivariable regression analysis as a function of the manually segmented lesion volumes (MLV). We also compared algorithm accuracy between very small MLV < 1 cm3 (Group I-A) and larger MLV ≥ 1cm3 (Group I-B).
To assess the accuracy of using automatically segmented lesion volumes (ALV) in place of MLV for identifying patients who have lesion volumes that are too large to likely benefit from endovascular treatment, we explored the agreement between ALV and MLV for MLV <21 cm2 (Group II-A) vs ≥ 21 cm3 (Group II-B), MLV <31 cm3 (Group III-A) vs ≥ 31 cm3 (Group III-B), MLV <51 cm3 (Group IV-A) vs ≥ 51 cm3 (Group IV-B) and MLV < 70 cm3 (Group V-A) vs ≥ 70 cm3 (Group V-B) to determine potential misclassification rates of patients with large lesions using automated algorithms compared to manual volumes. The thresholds (21, 31, 51 and 70 cm3) were selected based on values that had been used for enrollment in prospective endovascular clinical trials of expanded-window interventions.9,10 To be eligible for endovascular treatment using the DAWN trial criteria10, patients have to meet inclusion and exclusion criteria of 1 of the following 3 groups: Group A >=80 years of age, NIHSS >=10 and infarct volume <21 cm3, Group B <80 years of age, NIHSS>=10, and infarct volume<31 cm3; Group C <80 years of age, NIHSS>=20, and infarct volume of 31 to < 51 cm3. For the MRI cohort, the infarct volume was measured on DWI. Similarly, to be eligible for late window endovascular treatment using DEFUSE 3 MRI criteria,9 patients have to exhibit an infarct volume on DWI <70 cm3. Although there may be other volume thresholds that might be useful for patient selection,20 we focused on thresholds that were used in positive prospective clinical trials.
Statistical Analysis:
Differences between model performance metrics were tested by two-way ANOVA followed by post-hoc paired Wilcoxon signed rank test. Correlations were assessed via Spearman correlation coefficient (ρ). Univariate analysis was performed with Wilcoxon two-sample rank sum test for continuous variables, or two-sided Fisher’s Exact Test for categorical variables. Cohen’s Kappa (κ) assessed agreement between MLV and ALV. Statistical tests were conducted with JMP Pro 14.0 (SAS Institute, Cary, NC). P-values less than 0.05 were considered significant. Figures of MRI data were generated using FSLeyes (version 0.27).21
RESULTS
Subject demographics for training (N=116) and evaluation cohorts (N=151) are shown in Table 1. Although there were imbalances in sex and time-to-MRI likely due to different inclusion and exclusion criteria of the two cohorts (i.e. patients for whom follow-up MRI are ordered clinically which made up the Evaluation Cohort tend to be more severe), there was no statistical difference in the distribution of MLVs. The median volume of the 10 subjects randomly selected from the evaluation cohort for ICC analysis was 9.7 [2.7-32.6] cm3, ranging from 1.2 to 94.4 cm3. The ICC for the two readers was excellent (ICC=0.997, p<0.0001).
Table 1:
Demographics for Training and Evaluation Subjects. Differences as a factor of training cohort are shown.
| Characteristic | Training (N=116) |
Evaluation (N=151) |
P-value |
|---|---|---|---|
| Age, years | 67.9±17.2 | 65.2±15.5 | 0.11 |
| Sex, male | 57 (49.1%) | 104 (68.9%) | 0.0016 |
| NIHSS Score | 7 [3-15.75]a | 6 [3-13]b | 0.53 |
| Time-to-MRI, h | 5.0 [2.9-6.8] | 6.2 [3.8-8.3] | 0.002 |
| Manual Lesion Volumes, cm3 | 9.0 [1.5-28.4] | 10.6 [2.0-32.4] | 0.60 |
Data are shown as median [Interquartile Range] or mean±Standard Deviation or N (%).
N=112
N=115
Effect of selection of diffusion parametric maps on CNN performance
Significant differences (p<0.0001) were found between all performance metrics (Dice, precision, sensitivity) across all models (Table 2). Precision could not be calculated for cases for which models could not detect a lesion.
Table 2:
Comparison of Performance Metrics of Segmentations for Different CNN Models. Of the non-ensemble models, significant differences in Dice, precision and sensitivity were found (p<0.0001). The ensemble models, E2 and E3, were superior to all other models (p<0.0001).
| Model | Dice | Precision | Sensitivity |
|---|---|---|---|
| LOWB | 6.5 [0.3-20.9] | 5.7 [0.3-32.7] | 8.5 [0.3-28.5] |
| ADC† | 56.4 [27.1-75.4] | 59.4 [22.3-78.4] | 58.2 [32.7-78.9] |
| DWI | 72.3 [46.2-82.5] | 73.0 [38.3-88.1] | 84.0 [62.4-90.8] |
| ADC+LOWB | 76.5 [51.9-86.1] | 78.1 [47.2-88.8] | 79.2 [66.6-89.7] |
| DWI+LOWB | 76.7 [58.4-85.4] | 79.4 [52.0-89.8] | 83.0 [64.8-90.6] |
| DWI+ADC | 79.0 [57.1-86.4] | 79.0 [62.1-90.5] | 82.6 [68.4-91.4] |
| DWI+ADC+LOWB | 78.9 [56.2-86.2] | 77.4 [55.0-89.8] | 83.4 [71.3-91.8] |
| E2 (DWI+ADC) | 82.0 [62.9-88.1] | 82.0 [65.1-92.6] † | 84.1 [71.0-92.6] |
| E3 (DWI+ADC+LOWB) | 82.2 [64.9-88.9] | 83.2 [67.7-93.3] | 83.9 [71.9-92.4] |
All metrics denoted in % as median [Interquartile Range]. CNN=Convolutional Neural Network, E2=Ensemble of 5 CNNs trained on DWI+ADC, E3=Ensemble of 5 CNNs trained on DWI+ADC+LOWB, LOWB=Low b-value diffusion-weighted image.
Excludes one subject with automatically segmented lesion volume of zero since precision is undefined in this circumstance.
Individual Diffusion Maps
The CNN trained on DWI yielded significantly higher Dice scores, compared to the CNN trained on ADC (p<0.0001) or LOWB (p<0.0001) maps (see Supplemental Figure A2, Table 2). Findings for the CNNs’ precision (DWI vs ADC, p<0.0001; vs LOWB, p<0.0001) and sensitivity (DWI vs ADC, p<0.0001; vs LOWB, p<0.0001) were analogous to those for the Dice score. Of the networks trained with a single parametric map, the CNN models that use the DWI parametric map performed best, followed by the model based on the ADC map, with the LOWB-based model having the worst scores.
Combinations of Two Diffusion Maps
Including additional diffusion parametric maps as training data improved segmentation results. When training CNNs on two parametric maps (Supplemental Figure A2b, Table 2), all three of the CNNs that utilized combinations of 2 maps yielded higher Dice scores than all single-map CNNs (DWI+ADC vs LOWB, vs ADC, vs DWI, p<0.0001; DWI+LOWB vs LOWB, vs ADC, vs DWI, p<0.0001; ADC+LOWB vs LOWB, vs ADC, vs DWI, p<0.001). DWI+ADC had the highest Dice score compared to the other combinations (ADC+LOWB, p=0.0005; DWI+LOWB, p=0.03). Similarly, all CNNs trained with combinations of 2 parametric maps had higher precision than CNNs trained with one map (DWI+ADC vs LOWB, vs ADC, vs DWI, p<0.0001; DWI+LOWB vs LOWB, vs ADC, vs DWI, p<0.0001; ADC+LOWB vs LOWB, vs ADC, vs DWI, p<0.0001). However, there was no significant difference in precision between the combinations (DWI+ADC vs DWI+LOWB, p=0.67; DWI+LOWB vs ADC+LOWB, p=0.28), except for DWI+ADC vs ADC+LOWB, p=0.03. DWI+ADC similarly outperformed the individual parametric maps (LOWB, p<0.0001; ADC, p<0.0001) except for DWI (p=0.28) in terms of sensitivity. ADC+LOWB outperformed the individual LOWB (p<0.0001) and ADC (p<0.0001) models but not DWI (p=0.24). Similar results were found for the DWI+LOWB model as compared to the individual parametric maps (LOWB, p<0.0001; ADC, p<0.0001; DWI, p=0.83). DWI+ADC had comparable sensitivity to DWI+LOWB (p=0.11) and improved sensitivity with respect to ADC+LOWB (p=0.0007). DWI+LOWB and ADC+LOWB were equally sensitive (p=0.06).
Combination of 3 diffusion maps
The CNN model that combined all 3 parametric maps had a significantly greater Dice score (vs LOWB, ADC, DWI, p<0.0001; DWI+LOWB, p=0.01; ADC+LOWB, p=0.0003) compared to all other combinations with the exception of DWI+ADC (p=0.49). Precision results showed improvement against models using individual maps (vs LOWB, ADC, DWI, p<0.0001) but not against the other combinations (DWI+ADC, p=0.47; DWI+LOWB, p=0.69; ADC+LOWB, p=0.19). Similar results were found for sensitivity (vs LOWB, ADC, p<0.0001; DWI, p=0.008; DWI+ADC, p=0.10; DWI+LOWB, p=0.007; ADC+LOWB, p<0.0001).
Ensemble of CNNs
Five CNNs were trained each using either DWI+ADC or DWI+ADC+LOWB, the two best performing models. The Dice performances of each of the 5 individual CNNs were slightly different using DWI+ADC (Supplemental Table A2, Figure A3, ANOVA p=0.02, with differences between CNN #2 and CNN #3, p=0.04; and CNN #4 and CNN #5 p=0.04 ) but were similar to one another using DWI+ADC+LOWB (Supplemental Table A3, Figure A4, ANOVA p=0.60). Aggregating results of the individual CNNs to create ensembles (E2: DWI+ADC CNNs, E3: DWI+ADC+LOWB CNNs) significantly improved the Dice performance over individual CNNs (p<0.0001). Both ensembles yielded similar results to one another in terms of Dice (p=0.66) and precision (p=0.62), but both surpassed the other CNNs (Table 2, p<0.0001). E3 and E2 had similar sensitivity to one another (p=0.46), and to the DWI+ADC+LOWB model (vs E2 p=0.58; vs E3 p=0.12), but outperformed the others (p<0.01, Table 2).
Validation on Independent Cohort
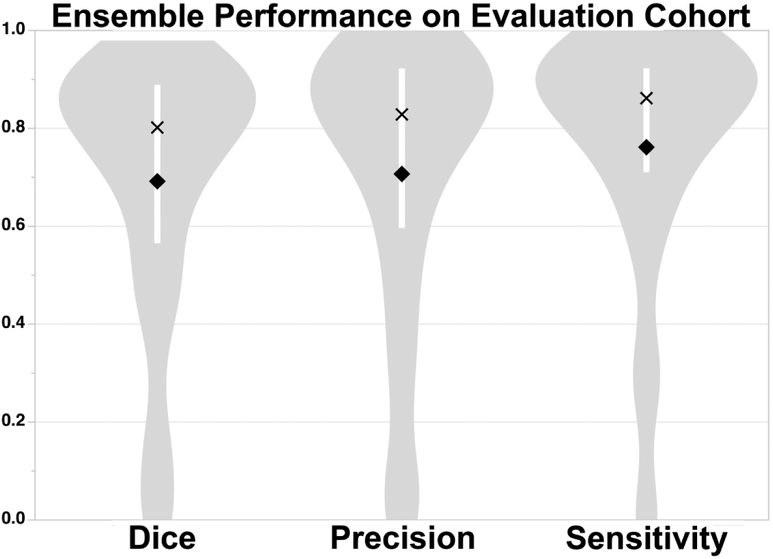
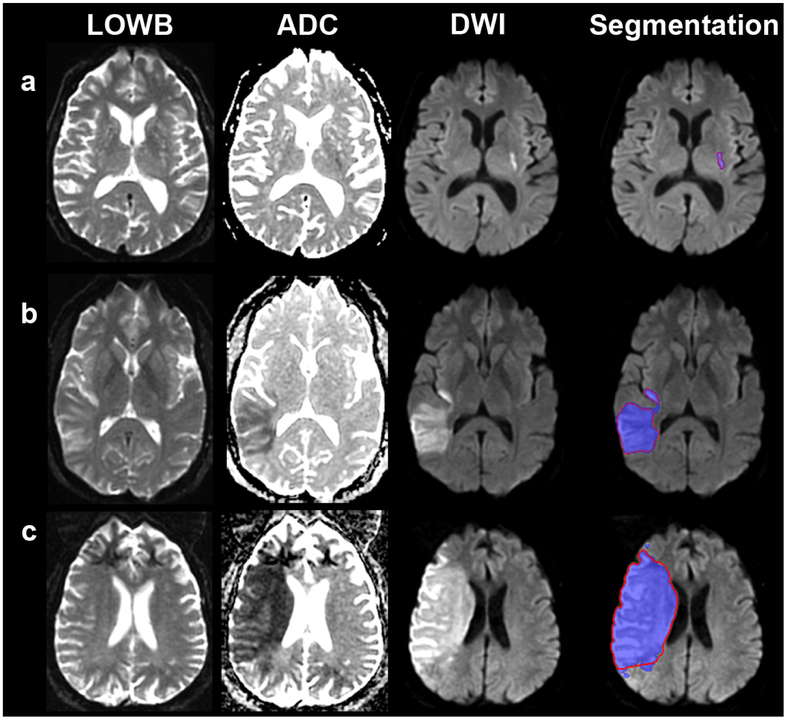
E3 was used for the evaluation studies to assess generalizability of the approach since E3 tended to perform better than E2. Figure 1 shows the results of applying the ensemble E3 to the evaluation cohort. Dice (p=0.59), precision (p=0.35) and sensitivity (p=0.66) were not significantly different from the results for the training cohort. In contrast, the thresholding approach performed significantly worse compared to E3 across all measures (p<0.0001), achieving only a Dice score of 13.3 [2.3-41.6], precision of 7.5 [1.2-34.2] and sensitivity of 60.7 [39.5-72.2] for data analyzed in the original resolution and for data analyzed at 1 mm isotropic resolution (Dice: 11.6 [2.2-34.7], precision: 6.5 [1.1-28.0], sensitivity: 59.4 [37.6-72.2]. We therefore focus the remainder of our analyses on E3 results. Examples of segmentation on subjects from the evaluation cohort using E3 are provided in Figure 2 (see Supplemental Figure A5 for probability maps). Regression analysis showed that Dice scores (p<0.0001), precision (p<0.0001) and sensitivity (p=0.01) improved with larger lesion volumes. The independent evaluation cohort consisted of strokes involving primarily supratentorial/cortical strokes (N=104, 69%) locations and supratentorial/subcortical (N=30, 20%) regions. There were significant differences in MLV, PLV, Dice, precision and sensitivity as a function of location (see Supplemental Table A4). Univariable regression showed lesion location affected Dice scores (p=0.0019), precision (p=0.013) and sensitivity (p<0.0001). However, multivariable analysis taking lesion volume into account (p<=0.0001), lesion location was no longer significantly associated with Dice score (p=0.06) or precision (p=0.17). Interestingly, sensitivity was still associated with location (p=0.0004) but not volume (p=0.085) in multivariable analysis.
Figure 1.
Median [interquartile range] Dice (80.2% [56.6%-88.9%]), Precision (82.9% [59.7%-92.2%]) and Sensitivity (86.2% [71.1%-92.3%]) Scores of DWI+ADC+LOWB Ensemble on Evaluation Cohort. The white bar within the violin plot shows the IQR, mean is represented as diamond, median is marked as cross. LOWB=Low b-value diffusion-weighted image.
Figure 2:
Example Segmentation Results of Ensemble of DWI+ADC+LOWB (blue regions) on Sample Subjects along with manual outlines (red outlines). (a) Small lesion example from a 70-year-old man, with admission NIHSS score of 1, imaged approximately 9 h from LKW: MLV = 0.96 cm3, ALV=1.07 cm3, Dice = 89.4%. (b) Medium lesion example from a 38-year-old woman with admission NIHSS score of 4, imaged approximately 10 h from LKW: MLV = 54.3 cm3, ALV=57.9 cm3, Dice = 95.7%. (c) Large lesion from a 62-year-old man, undocumented admission NIHSS score, imaged approximately 10 h from LKW: MLV = 229.0 cm3, ALV=208.7 cm3, Dice = 94.0%. ALV=Automated Lesion Volume, LKW=Last known to be well, LOWB=Low b-value diffusion-weighted image, MLV=Manual lesion volume
ALV correlated significantly with MLV (ρ =0.91, p<0.0001) and NIHSS (p=0.55, p<0.0001), which was comparable to MLV correlation with NIHSS (ρ=0.46, p<0.0001). Subgroup analysis based on MLV (Table 3) showed that the automated method performed significantly worse on small volumes (< 1 cm3) compared to large volumes for all metrics (Group I-A vs Group I-B, p<0.01). Misclassification rates across all thresholds were low: 21 cm3: 9/151 (6.0%), κ=0.87, p<0.0001; 31 cm3: 6/151 (4.0%), κ=0.90, p<0.0001; 51 cm3: 6/151 (4.0%), κ=0.86, p<0.0001; and 70 cm3: 4/151 (2.6%), κ=0.89, p<0.0001. There were 3 subjects for which the differences in ALV and MLV were greater than 50 cm3-these cases had poor skull stripping as a result of scanner inhomogeneities (see Supplemental Figure A6). Excluding these 3 subjects, the median differences in the misclassified cases were: 21 cm3: 18.7 [8.9-25.2] cm3; 31 cm3: 7.4 [1.7-16.8] cm3; 51 cm3: 8.8 [8.0-14.6] cm3; 70 cm3: 5.3 [3.7-6.9] cm3.
Table 3:
Dependency of Automated Segmentation Performance on Measured Lesion Volume (MLV). Results of E3 (Ensemble of 5 CNNs trained on DWI+LOWB+ADC) applied to the evaluation cohort are shown as a function of different volume thresholds.
| Group | Thresholds | Dice | Precision† | Sensitivity | Correlation |
|---|---|---|---|---|---|
| I-A | MLV < 1 cm3 | 31.0 | 29.6 | 57.5 | ρ=0.09, |
| (n=22) | [0-50.0] | [3.4-54.9] | [0-90.0] | p=0.68 | |
| I-B | MLV≥1 cm3 | 83.5* | 84.9* | 87.6** | ρ=0.90, |
| (n=129) | [71.2-89.3] | [70.3-92.9] | [75.8-92.9] | p<0.0001 | |
| II-A | MLV≥21 cm3 | 71.2 | 71.6 | 81.3 | ρ=0.79, |
| (n=100) | [45.8-84.8] | [38.7-84.9] | [59.8-92.5] | p<0.0001 | |
| II-B | MLV≥21 cm3 | 89.4* | 92.3* | 89.3*** | ρ=0.97, |
| (n=51) | [85.4-92.5] | [85.6-96.1] | [83.0-92.2] | p<0.0001 | |
| III-A | MLV <31 cm3 | 73.6 | 77.2 | 82.5 | ρ=0.83, |
| (n=113) | [48.0-85.8] | [46.3-85.7] | [62.8-92.1] | p<0.0001 | |
| III-B | MLV >31 cm3 | 90.6* | 94.7* | 89.4*** | ρ=0.96, |
| (n=38) | [87.3-93.2] | [88.4-96.8] | [82.8-93.6] | p<0.0001 | |
| IV-A | MLV < 51 cm3 | 75.0 | 78.1 | 83.3 | チ=0.87, |
| (n=124) | [48.9-86.8] | [49.2-86.5] | [65.2-92.5] | p<0.0001 | |
| IV-B | MLV ≥ 51cm3 | 91.5* | 95.9* | 89.2 | ρ=0.92, |
| (n=27) | [89.1-93.6] | [92.2-97.5] | [83.5-92.2] | p<0.0001 | |
| V-A | MLV < 70 cm3 | 77.2 | 79.9 | 84.0 | ρ=0.88, |
| (n=131) | [51.5-87.0] | [54.2-87.0] | [67.8-92.6] | p<0.0001 | |
| V-B | MLV ≥ 70 cm3 | 91.8* | 96.0* | 89.6 | ρ=0.83, |
| (n=20) | [89.4-93.9] | [93.0-96.9] | [85.0-92.0] | p<0.0001 |
Performance metrics in median [Interquartile Range]%. CNN=Convolutional Neural Network, E3=Ensemble of 5 CNNs trained on DWI+ADC+LOWB; LOWB=Low b-value diffusion-weighted image. MLV=Measured Lesion Volume.
p<0.0001
p<0.01
p<0.05 Group A versus Group B
Excludes two subjects in Group A with automatically segmented lesion volumes of zero since precision is undefined in this circumstance.
DISCUSSION
We have shown that an ensemble of CNNs trained with multi-parametric diffusion maps improves automated segmentation of acute infarcts over methods that use solo maps. Among the individual parameter models, CNNs trained on DWI performed best. However, a model trained on only DWI may incorrectly classify regions with susceptibility artifacts that appear as DWI hyperintensities, or wrongly include subacute “T2-shine through” regions 22. Networks trained only on ADC images provided a fair performance since reduced ADC values represent cytotoxic edema that manifests in hyperacute stroke,23 but may under segment later stage strokes when ADC pseudonormalizes22. CNNs exclusively trained on LOWB performed poorly, likely because our data consisted of mainly early phase stroke patients (median 6 h from LKW), before vasogenic edema is evident on LOWB.24
Combining DWI and ADC improved segmentation, consistent with “standard practice” by expert outliners who typically refer to the ADC image to confirm that the DWI hyperintensity coincides with reduced diffusivity to minimize inclusion of artifacts. Combining LOWB with either ADC or DWI increased Dice, suggesting that LOWB provides complementary information. Although inclusion of LOWB with DWI+ADC did not result in statistically significant improved performance, a tendency towards more accurate segmentation was observed in the ensemble models.
We have also shown that our model performs comparably to humans as reflected by both high Dice scores and correlation between ALV and MLV. Indeed, the Dice score of the E3 algorithm results were comparable with the Dice scores between human readers in our subcohort of 10 patients with outlines from both readers. The time for automated segmentation currently is approximately 5 minutes, which may be similar to times required by an experienced human reader, but we expect with optimization and faster GPUs, the time for segmentation can be further reduced. Furthermore, the primary benefits of our automated approach are that the results will be reproducible, unbiased, and scalable (eg clinical trials that compare lesion volumes for thousands of subjects).
ALV and MLV were closely correlated, but segmentation of small lesion volumes was overestimated. Accurate estimation of small lesion volumes (<1 cm3) is more difficult as they are harder to detect22 and small variation from the ground truth lead to greater aberrations of performance metrics. Small lesion segmentation could possibly be improved by customizing specific CNNs tailored towards detecting lesions by volume size. Nevertheless, we have shown that our automated approach performed comparably to manual lesions delineated by our human experts with regards to patient selection tasks. The cases of disagreement typically occurred when there were image artifacts that led to poor brain extraction which in turn might have led to poor normalization, resulting in over segmentation. A second reason for this failure might be that the networks have not previously seen context outside the brain during training, since it is excluded in most cases where the masks are correctly computed. We did not manually fix the brain masks since we wanted to evaluate a fully automated approach. Refining the automated brain extraction step will likely further improve our algorithms.
There were several limitations to this study. One is the retrospective nature of our analysis that resulted in variable MRI acquisition protocols that changed over the years with clinical practice. However, this is also a strength since our approach will likely be more generalizable to real-world clinical situations and not dependent on a specific MRI protocol, which is often used in clinical trials. This may also explain why the thresholding approach performed poorly on our data as compared to other studies for which MRI acquisition was harmonized as part of a trial.19 Another potential limitation is that a different reader created the manual outlines used for the evaluation cohort from the training cohort. However, the accurate segmentation results on both cohorts suggest that the model is not over-fitted to one particular reader. Another benefit of an automated approach is that it is reproducible and not dependent on the expertise of the reader.
To evaluate the impact of different diffusion maps on segmentation performance, we kept the CNN architecture constant throughout all experiments. In addition to changing the combinations of inputs, we chose to build an ensemble from several CNNs, as ensemble learning is known to boost performances of single classifier algorithms.8, 25 DeepMedic samples randomly from the training cohort, i.e. both the selected subjects and extracted samples differ in each training epoch. Although DeepMedic is very robust in its performance, the variation in sampling inherently results in slightly different models, even when trained with the same architecture. Merging the segmentations of several models reduces false positives, thus improving overall performance. Although strong single networks are desired and necessary to create a high performing ensemble, our CNNs may come with bias specific to DeepMedic. Building an ensemble of different CNN architectures might further enhance the performance. Future investigation will need to analyze the benefits of merging more diverse networks to cancel out each other’s inherent biases8. This diversity of models could be achieved by changing the hyperparameters of DeepMedic, using completely different architectures, or training on a different data set.
In summary, ensembles of CNNs trained on multi-parametric diffusion MRI improved automated segmentation of acute infarcts in comparison to individual CNNs trained on solo diffusion maps, producing results that are comparable with manual lesions drawn by experts.
Supplementary Material
Acknowledgments:
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Tesla K40 GPU used for this research.
Funding: NIH-NINDS R01NS059775, R01NS063925, R01NS082285, P50NS051343, R01NS086905, R01NS051412, R21NS077442, R21NS085574, U01 NS069208, U10NS086729, 1U01NS095869, NIH NIBIB P41EB015896, 1S10RR023401, 1S10RR019307, and 1S10RR023043.
Abbreviations:
- ALV
automatically segmented lesion volume
- CNN
convolutional neural network
- E2
ensemble of CNN using DWI and ADC
- E3
ensemble of CNN using DWI, ADC and LOWB
- FN
false negatives
- FP
false positive
- LKW
last known to be well
- LOWB
low b-value diffusion-weighted image (b0)
- MLV
manually segmented lesion volume
- TP
true positives
REFERENCES
- 1.Chen L, Bentley P, Rueckert D. Fully automatic acute ischemic lesion segmentation in DWI using convolutional neural networks. Neuroimage Clin 2017;15:633–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Straka M, Albers GW, Bammer R. Real-time diffusion-perfusion mismatch analysis in acute stroke. J Magn Reson Imaging 2010;32:1024–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacobs MA, Mitsias P, Soltanian-Zadeh H, et al. Multiparametric MRI tissue characterization in clinical stroke with correlation to clinical outcome: Part 2. Stroke 2001;32:950–957 [DOI] [PubMed] [Google Scholar]
- 4.Hevia-Montiel N, Jimenez-Alaniz JR, Medina-Banuelos V, et al. Robust nonparametric segmentation of infarct lesion from diffusion-weighted MR images. Conf Proc IEEE Eng Med Biol Soc 2007;2007:2102–2105 [DOI] [PubMed] [Google Scholar]
- 5.Tsai JZ, Peng SJ, Chen YW, et al. Automatic detection and quantification of acute cerebral infarct by fuzzy clustering and histographic characterization on diffusion weighted MR imaging and apparent diffusion coefficient map. Biomed Res Int 2014;2014:963032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang R, Zhao L, Lou W, et al. Automatic Segmentation of Acute Ischemic Stroke From DWI Using 3-D Fully Convolutional DenseNets. IEEE Transactions on Medical Imaging 2018:1–1 [DOI] [PubMed] [Google Scholar]
- 7.Mujumdar S, Varma R, Kishore LT. A novel framework for segmentation of stroke lesions in Diffusion Weighted MRI using multiple b-value data. Proceedings - International Conference on Pattern Recognition 2012:3762–3765 [Google Scholar]
- 8.Kamnitsas K, Bai W, Ferrante E, et al. Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation In: Crimini A, Bakas S, Kuijf H, Menze B, Reyes M, eds. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: Springer International Publishing; 2018;450–462 [Google Scholar]
- 9.Albers GW, Marks MP, Kemp S, et al. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. N Engl J Med 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nogueira RG, Jadhav AP, Haussen DC, et al. Thrombectomy 6 to 24 Hours after Stroke with a Mismatch between Deficit and Infarct. N Engl J Med 2017 [DOI] [PubMed] [Google Scholar]
- 11.Wu O, Schwamm LH, Garg P, et al. Using MRI as the Witness: Multimodal MRI-based Determination of Acute Stroke Onset. Stroke 2010;41:E273–E273 [Google Scholar]
- 12.Wu O, McIntosh E, Bezerra R, et al. Prediction of lesion expansion in patients using acute MRI. Stroke 2012;43:A3319 [Google Scholar]
- 13.Wu O, Koroshetz WJ, Østergaard L, et al. Predicting tissue outcome in acute human cerebral ischemia using combined diffusion- and perfusion-weighted MR imaging. Stroke 2001;32:933–942 [DOI] [PubMed] [Google Scholar]
- 14.Sorensen AG, Wu O, Copen WA, et al. Human acute cerebral ischemia: detection of changes in water diffusion anisotropy by using MR imaging. Radiology 1999;212:785–792 [DOI] [PubMed] [Google Scholar]
- 15.Smith SM. Fast robust automated brain extraction. Hum BrainMapp 2002;17:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jenkinson M, Beckmann CF, Behrens TE, Woolrich MW, Smith SM. FSL. Neuroimage 2012;62:782–790 [DOI] [PubMed] [Google Scholar]
- 17.Kamnitsas K, Ledig C, Newcombe VFJ, et al. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 2017;36:61–78 [DOI] [PubMed] [Google Scholar]
- 18.Maier O, Menze BH, von der Gablentz J, et al. ISLES 2015 - A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med Image Anal 2017;35:250–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lansberg MG, Lee J, Christensen S, et al. RAPID automated patient selection for reperfusion therapy: a pooled analysis of the Echoplanar Imaging Thrombolytic Evaluation Trial (EPITHET) and the Diffusion and Perfusion Imaging Evaluation for Understanding Stroke Evolution (DEFUSE) Study. Stroke 2011;42:1608–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie-Mazwi TM, Hirsch JA, Falcone GJ, et al. Endovascular Stroke Treatment Outcomes After Patient Selection Based on Magnetic Resonance Imaging and Clinical Criteria. JAMA Neurol 2016;73:43–49 [DOI] [PubMed] [Google Scholar]
- 21.McCarthy P FSLeyes. 2018 [Google Scholar]
- 22.Dijkhuizen RM, Knollema S, van der Worp HB, et al. Dynamics of cerebral tissue injury and perfusion after temporary hypoxia-ischemia in the rat: evidence for region-specific sensitivity and delayed damage. Stroke 1998;29:695–704 [DOI] [PubMed] [Google Scholar]
- 23.Jiang Q, Chopp M, Zhang ZG, et al. The temporal evolution of MRI tissue signatures after transient middle cerebral artery occlusion in rat. JNeurol Sci 1997;145:15–23 [DOI] [PubMed] [Google Scholar]
- 24.Schwamm LH, Wu O, Song SS, et al. Intravenous thrombolysis in unwitnessed stroke onset: MR WITNESS trial results. Ann Neurol 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Breiman L Random Forests. Machine Learning; 2001;1–32 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.