Abstract
Background
Brain tumor clinical trials requiring solid tumor assessment typically rely on the 2-dimensional manual delineation of enhancing tumors (2D-T1) by two or more expert readers, a time-consuming step with poor inter-reader agreement. As a solution, we developed quantitative delta T1 (dT1) maps for the delineation of enhancing lesions. This retrospective analysis compares dT1 to 2D-T1 acquired at two time points during the post-therapeutic surveillance period of the ACRIN 6677/RTOG 0625 clinical trial.
Materials
Patients enrolled in ACRIN 6677/RTOG 0625, a multicenter, randomized phase II trial of bevacizumab in recurrent glioblastoma, underwent standard MRI before and after treatment initiation. For 123 patients from 23 institutions, both 2D-T1 and dT1 datasets were evaluable at weeks 8 (n=74) and 16 (n=57). Using dT1, radiologic response and progression were assessed at each time point. Percent agreement with adjudicated 2D-T1 reads and association between progression status and overall survival (OS) were determined.
Results
For identification of progression, dT1 and adjudicated 2D-T1 reads were in perfect agreement at week 8 with 73.7% agreement at week 16. Both methods showed significant differences in OS at each time point. When non-progressors were further divided into responders versus nonresponders/nonprogressors the agreement decreased to 70.3% and 52.6%, yet dT1 showed a significant difference in OS at week 8 (p=0.01), suggesting that dT1 may provide greater sensitivity for stratifying subpopulations.
Conclusion
This study shows that dT1 can predict early progression comparable to the standard method but offers the potential for substantial time and cost savings for clinical trials.
Keywords: central reader analysis, clinical trial, brain tumor segmentation, T1+C, delta T1 (dT1)
Introduction
Objective, accurate, and reproducible methods to measure glioblastoma volume are important for clinicians to assess treatment response and guide appropriate therapy, both in daily practice and in clinical trials. Contrast-enhanced MRI is the most widely used approach, and the focus of recent consensus brain tumor imaging protocol recommendations.1 However, although contrast-enhanced MRI has excellent spatial resolution, even slight variations in image acquisition parameters or vendor platforms can greatly impact image quality, lesion conspicuity, and measurement of tumor volume.2 These problems are compounded by the fact that glioblastoma is histopathologically and radiographically heterogeneous in appearance, with geographically irregular margins, variable contrast enhancement, and regions of central necrosis or cystic changes.3 Furthermore, assessment of post-treatment tumor volume can be confounded by the presence of blood products that appear bright on post-contrast MRI and that mimic contrast-enhancing tumor4, or in the context of therapies reducing blood brain barrier permeability and contrast agent extravasation. For example, bevacizumab,5 used to treat recurrent glioblastoma, can decrease contrast agent extravasation independent of its effect on tumor biology.3, 6 These challenges contribute to large inter-observer differences (up to 50%–60%) in assessing tumor burden and evaluating treatment response that impact both daily practice and clinical trials.4, 7
As a solution difference maps, created from the subtraction of pre-contrast from post-contrast images, have been used to highlight regions of contrast enhancement.8 However, unlike X-ray angiography or computed tomography, pixel values in magnetic resonance images can vary widely due to multiple factors, even for identical pulse sequences and tissue types, which can result in nonenhancing regions appearing in the subtraction image. In response, we developed quantitative “delta T1” (dT1) images of contrast enhancement, which eliminate much of the normal variability in image contrast due to MRI system instabilities, field strength, slight differences in imaging parameters (TR, TE, etc.), and sources of bright signal apparent on pre-contrast T1w images.9, 10 Because dT1 images are quantitative, delineation of enhancing lesions can be automated by applying the same predetermined threshold across time points and patients.
Consequently, the dT1 tool has the potential to cause a paradigm shift in how brain tumor burden is assessed. This study compares the use of dT1 technology for semiautomatic lesion identification with the accepted standard that relies on expert readers to manually delineate enhancing lesions. The approach was to determine whether the semi-automatic determination of regions of interest (ROIs) using dT1 images would compare similarly to the adjudicated reads from the RTOG 0625/ACRIN 6677 central reader study (as reported in the primary paper11) with regard to association with patient outcome.
Materials and Methods
The Radiation Therapy Oncology Group (RTOG, now NRG Oncology), in collaboration with the American College of Radiology Imaging Network (ACRIN, now Eastern Cooperative Oncology Group [ECOG]-ACRIN), both funded by the National Cancer Institute, conducted a prospective, randomized, phase II multicenter trial (ACRIN 6677/RTOG 0625) of bevacizumab in recurrent glioblastoma multiforme (GBM). Each participating institution obtained institutional review board approval before subject accrual and conducted the trial in compliance with the Health Insurance Portability and Accountability Act. Informed consent was obtained for all subjects.
Patients
A total of 123 patients from 23 institutions, with recurrent histologically proven glioblastoma or gliosarcoma, were enrolled to the ACRIN 6677/RTOG 0625 trial. Detailed inclusion and exclusion criteria are available on the RTOG website https://www.rtog.org/ClinicalTrials/ProtocolTable/StudyDetails.aspx?study=0625. All patients were treated with bevacizumab (10 mg/kg i.v. on days 1 and 15 of a 28-day cycle) in combination with either temozolomide or irinotecan.11
Of the 123 patients enrolled, 107 patients met the inclusion criteria, defined as having imaging beyond baseline and progression data. Of these, 105 datasets could be analyzed by 2D-T1 central reader analysis, which required having an interpretable baseline image and at least one additional interpretable time point. Of the 105 datasets, matched pre- and post-contrast T1 images were available for 83 patients enabling the creation of dT1 images. A matched dataset is defined as one for which the same imaging sequence and the same scanning options (e.g., flow-compensation is either on or off for both) are used for both the pre- and post-contrast T1-weighted images. Slight differences in TR and TE between the pre- and post-contrast images are acceptable. Restricting attention to the week 8 and week 16 time points, a total of 74 cases were evaluable for comparison between the same T1–2D and dT1 image datasets at week 8, and 57 cases were evaluable for comparison at week 16.
Data Acquisition Methods
MRI was performed at both 1.5T (Siemens Espree, Siemens Magnetom Avanto, GE Signa Excite, GE Signa HDx) and 3T (GE Signa HDx, GE Signa Excite). Conventional MRI included pre-contrast T1w, T2-weighted, fluid-attenuated inversion recovery (FLAIR), and diffusion-weighted imaging.11 For both pre- and post-contrast T1w imaging, all sites were required to collect the data using a spin-echo sequence with the following parameter ranges: TE/TR = min (<15 ms)/400–600 ms, FOV=220–240 mm, phase FOV = 75%, slice thickness/gap = 5 mm/1 mm, matrix = 256 × 256, NEX = 1. The imaging protocol remained fixed at each site and across all time points. Following i.v. of 0.1 mmol/kg of a standard gadolinium-based contrast agent (The brand used was dictated by each site’s preference.), axial 2D spin-echo (2D-T1) and 3D volumetric T1w post-contrast images were acquired. Patients participating in the optional advanced component of the trial had dynamic contrast-enhanced, dynamic susceptibility contrast (DSC), and/or spectroscopic MRI at baseline, week 2, and after every two cycles of treatment. Results from these advanced imaging cohorts were previously reported.12–14 A complete listing of all MRI parameters for this protocol can be found on the ACRIN website. (https://www.acrin.org/PROTOCOLSUMMARYTABLE/PROTOCOL6677/6677ImagingMaterials.aspx).
Image Analysis
Central Reader Analysis
As previously described,11 all local imaging was transmitted to ACRIN for central review by two primary readers and one adjudicator, each with neuroradiology Certificates of Added Qualification and 8, 6, and 3 years, respectively, of post-fellowship experience. For each distinct contrast-enhancing target lesion (≥1 cm diameter, ≥1 cm from other enhancing lesions), the largest diameter of contrast enhancement and its maximum perpendicular diameter were measured. A 2D tumor area was computed by summing over all lesions the product of maximum perpendicular diameters. Pre- and post-contrast images were reviewed simultaneously to exclude blood products from 2D measurements.
For all evaluable patients, images at each available time point were presented in random order to both of the primary readers who then independently made 2D-T1 measurements. After completing measurements for all time points, the primary readers were unblinded to the order of exams. Consistent with Macdonald and Response Assessment in Neuro-Oncology (RANO) criteria, each reader determined the time of progression on 2D-T1 when there was a >25% increase with respect to nadir in maximal cross-sectional enhancing areas, or an appearance of any new measurable enhancing tumor. Radiologic response was defined as a ≥50% decrease with respect to baseline, confirmed on the subsequent time point. Steroid dosage and clinical status were unavailable to the readers for this study. The adjudicator settled discordant times to progression between the readers by opining on the most correct times to progression. FLAIR images were not used to determine outcomes for either the 2D-T1 or dT1 analysis.
Creation of dT1 Maps
The dT1 method quantitatively compares calibrated pre-(T1) and post-contrast (T1+C) anatomic images, where the calibration rule was machine-learned from input data of a given type (e.g., T1w spin-echo).9, 10 Specifically, learning the calibration rule (historically referred to as the “standardization step”)15, 16 requires the determination of mean intensity values at predefined landmarks, which correspond to percentiles in the distribution of pixel values, using a dataset of training images. This training step is performed only once. Next, each new input image of a given type is transformed to the standardized space (i.e., calibrated) using a piecewise linear intensity mapping function. The result is a constant dynamic range for the calibrated images such that for a given tissue type, it is possible to establish fixed gray-level windows without the need for a per-case window level adjustment.16
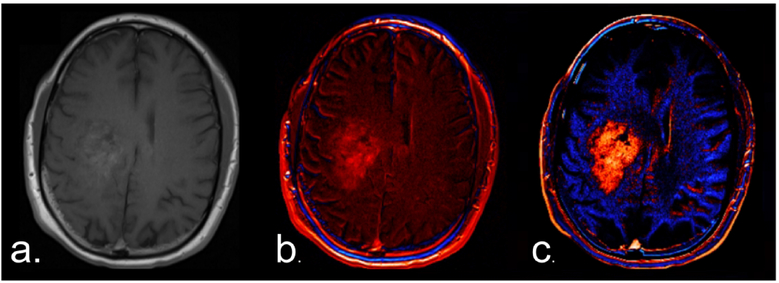
For routine analysis, 2D pre- and post-contrast 2D-T1 images were co-registered using a rigid mutual information cost function, followed by application of the machine-learned calibration rule to each T1w image. The calibrated-registered pre-contrast T1w image was subtracted from the calibrated-registered post-contrast T1w image, resulting in a dT1 image. Figure 1 illustrates the superior conspicuity of a glioblastoma with dT1 compared with a simple difference map constructed from noncalibrated images.
Figure 1.
Standardization is required for the creation of robust deltaT1 (dT1) maps. (a) Post-contrast T1w image obtained in a patient with a glioblastoma. (b) A difference map created without calibration. (c) A dT1 map created from the difference between calibrated post- and pre-contrast T1w images.
Because dT1 maps are quantitative, delineation of enhancing lesions can be semiautomated by choosing a fixed threshold and applying it consistently across time points and patients. The threshold of 3000 (calibrated units) was determined by an experienced (>20 years) neuroradiologist (SDR), as previously described.10 Briefly, dT1 voxels were spatially correlated with raw DSC-MRI data. Voxels with no visually discernable DSC-MRI signal (i.e., a lack of perfused tissue) were used to confirm a lack of contrast-agent-perfused tissue and, thus, a lack of contrast-agent enhancement. A threshold of 3000 was found to reliably make this distinction and is now routinely applied to dT1 images for the semiautomatic determination of contrast-agent enhancing ROI. Note that the perfusion signal was used for the initial determination of a threshold. Its collection and use are not required for the routine use of dT1 maps. Generation of dT1 images was built into the IB Delta Suite™ software (Imaging Biometrics LLC, Elm Grove, WI) used for this study.
A nonexpert reader (i.e., an engineer with <4 years of MRI experience at the time of annotation) blinded to the central reader analyses coarsely defined the bounding region on each image slice using the dT1 maps. Care was taken to exclude the choroid plexus, vessels, and scalp. All pixels within the bounding region that were above 3000 were included as the final enhancing tumor ROI. No manipulation of the tumor ROI was performed beyond identification of the initial bounding region. An experienced neuroradiologist (SDR) blinded to the central reader results reviewed and approved the final ROIs for any difficult cases. This approach mimics the current practice of having technologists preprocess data and radiologists perform a final review.
Statistical Analysis
2D-T1 results were reported previously.11 Using dT1, for each post-baseline time point, the dT1 volume was measured against the nadir value, and progression and response (R) were determined as described above for the central reader analysis. If neither the progression nor R criteria were satisfied, the time point was labeled as nonresponder/nonprogressor (NR-NP). Uninterpretable time points were ignored.
Agreement between the adjudicated 2D-T1 assessments and the derived dT1 assessments at week 8 and week 16 was determined using a simple percent agreement, as well as the Krippendorff alpha statistic. The latter statistic corrects for chance agreement17 such that when the methods agree perfectly, alpha = 1, and when the methods agree as if chance had produced the results, alpha = 0.
For both 2D-T1 and dT1, separate landmark analysis sets were created for progression by week 8 and progression by week 16, and association with overall survival (OS) was reported using Kaplan-Meier curves with the log-rank test.
Statistical computations were performed using SAS v9.4 software (SAS Institute, Cary, NC) or R v3.4.4 software (R project; http://www.r-project.org/), with P values <0.05 considered statistically significant.
Results
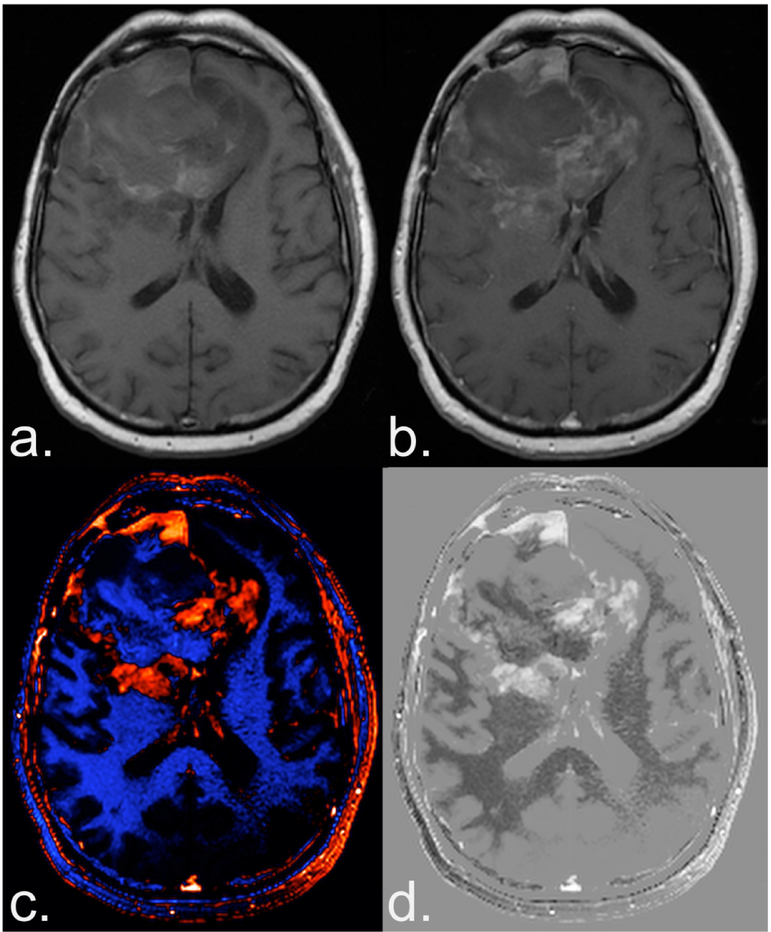
An example of a dT1 image created from a study patient is shown in Figure 2. The dT1 map, shown in color and gray-scale (Fig. 2c,d) clearly highlights the enhancing tumor without being confounded by the bright signal on pre-contrast T1w images (Fig. 2a) or subtle enhancement on the post-contrast T1w images (Fig. 2b).
Figure 2.
Benefit of creating a deltaT1 (dT1) map. Shown are the pre (a) and post-contrast (b) T1-weighted images from a patient with recurrent glioblastoma treated with bevacizumab enrolled in the ACRIN 6677 trial. The bright signal on the pre-contrast image and the subtle enhancement on the post-contrast image, makes it difficult to determine the extent of enhancing tumor. Alternatively, the dT1 map created from the difference between calibrated pre- and calibrated post-contrast T1-weighted images clearly delineates enhancing tumor as displayed with either color (c) or in gray scale (d).
Using dT1, a total of 7/74 (9%) cases progressed by week 8, and 26/57 (46%) cases progressed by week 16. Of the cases that did not progress by week 8, 28/67 (42%) were R and 39/67 (58%) were NR-NP; the counts at week 16 were 23/31 (74%) and 8/31 (26%), respectively.
Cross-tabulation of week 8 status revealed perfect agreement for progression between dT1 and the adjudicated 2D-T1 reads. When nonprogressors were further subdivided into R versus NR-NP, the percent agreement was 70.3% (95% CI: 59.1%−79.5%) with Krippendorf alpha of 0.52 (95% CI: 0.35, 0.70). Agreement for progression by week 16 decreased, with percent agreement of 73.7% (95% CI: 61.0%−83.4%) and Krippendorf alpha of 0.46 (95% CI: 0.21, 0.68). The percent agreement further decreased when nonprogressors were subdivided into R versus NR-NP (percent agreement = 52.6%; 95% CI: 39.9%−65.0%; Krippendorf alpha=0.45; 95% CI: 0.22, 0.65).
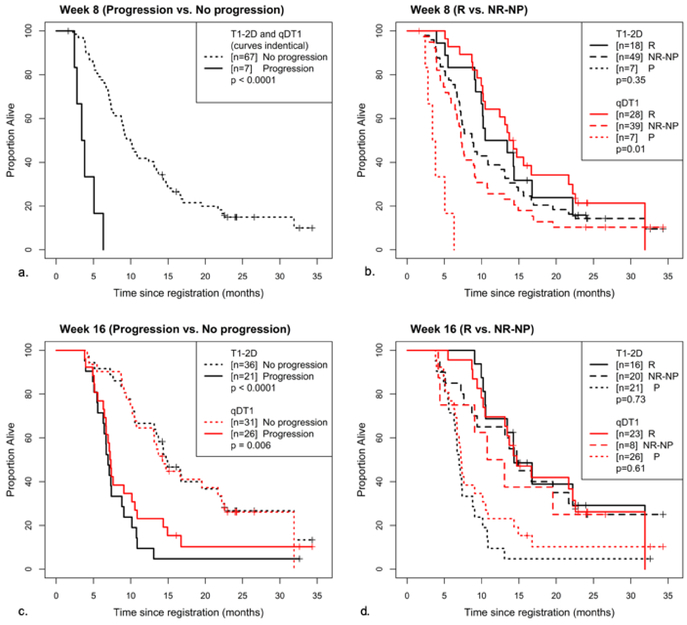
Given the perfect agreement for progression at week 8 between dT1 and the adjudicated 2D-T1 reads, the Kaplan-Meier curves for both methods were identical (Table 1, Figure 3a), with a significant difference in OS (p<0.0001). While 2D-T1 did not further distinguish between R and NR-NP (p=0.35), there was a significant difference in OS between R and NR-NP for dT1 (p=0.01; Table 1, Figure 3b).
Table 1.
Overall survival stratified by progression status at week 8.
| Sequence | Progression by week 8? | Progression by week 8? (R vs NR-NP) | |||
|---|---|---|---|---|---|
| No | Yes | R | NR-NP | P | |
| N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | |
| 2D | 67 (91%) 303 (232, 400) |
7 (9%) 110 (74, 192) |
18 (24%) 364 (280, 510) |
49 (66%) 268 (217, 378) |
7 (9%) 110 (74, 192) |
| P<0.0001 (N=74) | P=035: R vs NR-NP | ||||
| dT1 | 67 (91%) 303 (232, 400) |
7 (9%) 110 (74, 192) |
28 (38%) 425.5 (311, 660) |
39 (53%) 223 (180, 276) |
7 (9%) 110 (74, 192) |
| P<0.0001 (N=74) | P=0.01: R vs NR-NP | ||||
Figure 3.
Kaplan-Meier curves for 2D-T1 and dT1-determined enhancing tumor. Shown are overall survival curves stratified by whether the participant (a) progressed at or prior to week 8 (b) progressed at or prior to week 8, broken into progressing (P) responding (R) and non-responding/non-progressing (NR-NP), (c) progressed at or prior to week 16 and (d) had progressed at or prior to week 16, broken into P, R and NR-NP.
At week 16, a highly significant difference in OS was observed between progressors and nonprogressors for both T1–2D and dT1 (p<0.0001, p=0.006; Table 2, Figure 3c). No difference in OS was observed between R and NR-NP for either method (p=0.73 and p=0.61; Table 2, Figure 3d).
Table 2.
Overall survival stratified by progression status at week 16.
| Sequence | Progression by week 16? | Progression by week 16? (R vs NR-NP) | |||
|---|---|---|---|---|---|
| No | Yes | R | NR-NP | P | |
| N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | N (%) Median OS (days) (95% CI) | |
| 2D | 36 (63%) 448 (321, 660) |
21 (37%) 212 (170, 268) |
16 (28%) 437 (319, 971) |
20 (35%) 451 (232, 687) |
21 (37%) 212 (170, 268) |
| P<0.0001 (N=57) | P=0.73: R vs NR-NP | ||||
| dT1 | 31 (54%) 434 (319, 676) |
26 (46%) 221.5 (196, 309) |
23 (40%) 448 (319, 687) |
8 (14%) 362.5 (127, -) |
26 (46%) 221.5 (196, 309) |
| P=0.006 (N=57) | P=0.61: R vs NR-NP | ||||
Discussion
The results of this study support the integration of dT1 into central reader analysis for the delineation of contrast-enhancing brain tumor. The dT1 method was comparable to expert reads for determination of early tumor progression and proved superior for further distinguishing R vs. NR-NP at the week 8 time point. While the agreement between the methods decreased at week 16, both methods showed a significant difference in OS based on progression status.
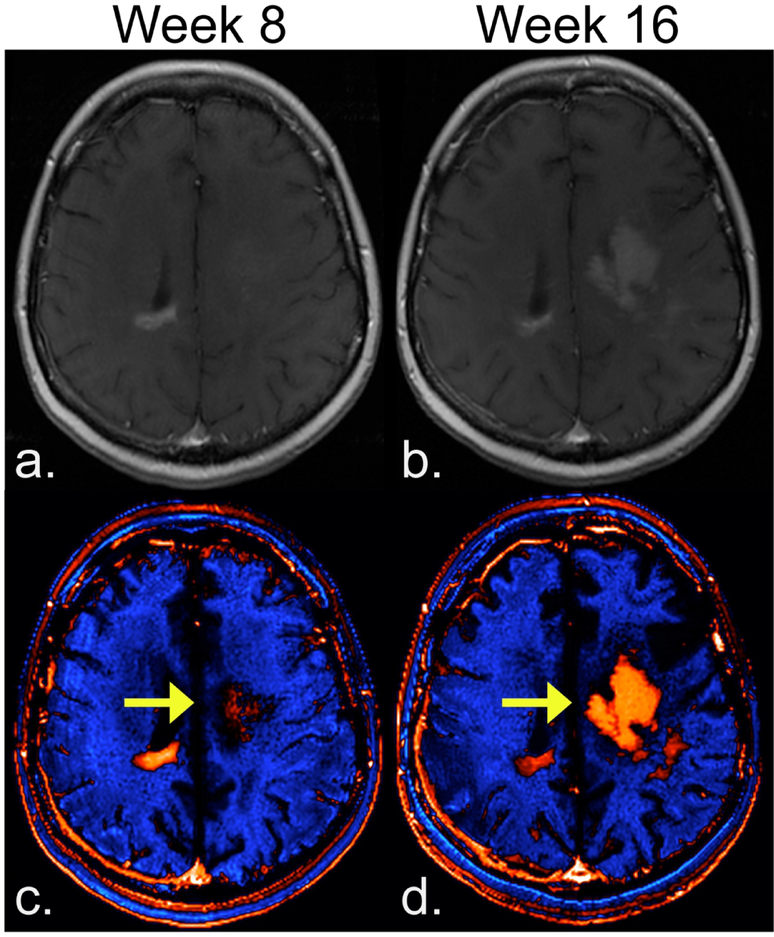
The result that dT1 proved better for stratifying subpopulations may be explained in several possible ways. First, due to the process of standardization (calibration) followed by subtraction, dT1 provides a consistent and objective delineation of enhancing lesion. It is less confounded by both systematic differences (vendor platforms, slight variations in imaging settings) and the subjectivity (inter-reader differences) that influences current approaches. This in turn can result in an improved sensitivity to enhancing lesion, which is not apparent on T1+C images, as illustrated in Figure 4. In addition, the superiority of using dT1 may be explained by the demonstrated benefit of volumetric measurements over standard bi-dimensional approaches for measuring tumor size18 and the application of a fixed physiology-based threshold to dT1 images to determine enhancing tumor burden.
Figure 4.
Potential of dT1 for improved sensitivity to enhancing lesion. For T1+C images (a,b) progression of enhancing lesion, which is clearly visible at week 16 (b) was not discernable at week 8 (a). With dT1 the enhancing visible earlier at week 8 (c), becoming even more apparent at week 16 (d).
Prior to 2010, the MacDonald criteria were widely used to assess treatment response of high-grade glioma,19 and included the 2D measurement of enhancing tumor in conjunction with a clinical assessment and corticosteroid dose. Tumor progression on FLAIR and the recognition that contrast enhancement is nonspecific prompted the development of the updated response assessment criteria (RANO) that added FLAIR to the MacDonald criteria.20 which also has important limitations and remains controversial.21 In fact, the parent study did not find a statistically significant survival time reduction among the isolated FLAIR progressors compared with nonprogressors.11 Even so, we are not suggesting that dT1 replace RANO as the standard assessment criteria. Rather, we are suggesting that dT1 has the potential to replace the current approach for delineating enhancing lesion volumes, which is one aspect of the RANO assessment.
Measurement of the contrast-enhancing lesion remains central to the assessment of treatment response and was the focus of a recent effort to standardize imaging protocols for tumor volume assessment.1 Even as new imaging biomarkers, derived from perfusion- or diffusion-weighted MRI, for example, are proving useful for the biologic assessment of tumor response, the analysis of such biomarkers depends on the accurate delineation of enhancing tumor. Therefore, it is necessary to be able to process these pre- and post-contrast T1w data in a robust manner for both routine care and clinical trials.
The standard approach for lesion segmentation is the labor-intensive and time-consuming manual delineation of contrast-enhancing lesion by expert readers. Due to the subjective nature of this approach, clinical trials rely on multiple expert readers and involve additional readers to adjudicate cases for which there is disagreement. In a study that enlisted eight board-certified radiologists to measure high-grade tumor diameters, substantial inter-reader disagreement was demonstrated with a rate of consensus regarding tumor progression of only 45% and only moderate reproducibility.22 This lack of agreement necessitates frequent adjudication. The primary study, from which this secondary analysis obtained its data, reported a 43% adjudication rate when using 2D-T1 and 42% for 3D-T1.11. Even more detrimental is that the turnaround time for central analysis may preclude certain study designs that require assessment of progression within 48 hours, for example.
By comparison, the dT1 technology can be used by nonradiologists, as demonstrated in the present study, and requires only seconds to identify enhancing lesion. Since it is a semi-automated, machine-learned approach, dT1 overcomes the subjectivity that confounds current methods and, therefore, has the potential to provide greater consistency in lesion identification across time points and patients. These capabilities derive from the unique standardization (calibration) algorithm incorporated into the process of creating dT1 difference maps.10, 15, 23 The standardization algorithm serves to diminish slight differences in TE and TR settings for a given sequence type16 and thus lessens the importance of such variations that can result in differences in lesion conspicuity. Finally, the standardization algorithm, which has also been trained for use in the creation of rCBV (relative cerebral blood volume) maps, resulted in substantial improvements in repeatability24 as well as consistency across time.25 Therefore, it is expected that dT1 also should result in greater repeatability; this hypothesis should be tested in prospective studies.
The dT1 images are different from the subsequently developed, yet possibly better-known, Gaussian-normalized difference maps.8 Gaussian-normalized maps require the determination of a new normalization for each patient and image, both pre- and post-contrast, which raises questions about consistency across time points and patients. dT1 uses the same calibration and threshold for each patient and image, enabling consistent quantification and automation across time points, patients, and sites.
Simple difference images, which do eliminate some confounding bright signal on pre-contrast T1w images, are limited by variations in sequence parameter settings (e.g., Figure 1). Furthermore, simple difference maps are not quantitative, thereby precluding the ability to automate lesion identification and resulting in little improvement over current methods. Consequently, the dT1 technology has the greatest likelihood of offering a substantial improvement over similar tools, with a greater potential for automation and clinical use.
Also of particular interest is that, in this study, only dT1 could predict differences in outcome for the NP-NR tumors at week 8. This greater sensitivity may result from dT1’s more accurate and possibly more sensitive delineation of enhancing tumor (Figure 4), free of pre-contrast bright signal, or it may be attributed to the physiologically motivated threshold used with dT1. Thus, a qDT1 (quantitative delta T1)-determined lesion may more accurately reflect active brain tumor. However, whether this same threshold should be used for other contrast-enhancing tumor types is unknown and will be the topic of future studies.
A limitation of this study is that only one non-radiologist determined the enhancing ROIs using dT1. A separate study characterizing the inter-reader agreement using the dT1 technology is warranted. Also, it is likely that all cases, particularly those with more complicated lesions, will still require expert review. Yet expert sign-offs are routine, and improving the initial step of tumor delineation with dT1 should result in improved time efficiencies to the radiologists’ workflow.
Another perceived limitation is that many fewer datasets could be analyzed with dT1 compared with 2D-T1. However, this is not intrinsic to the dT1 method. Rather, it is because this is a retrospective analysis of data that were not collected for the purpose of creating dT1 images. Though dT1 is amenable to slight variations in parameter settings such as TE and TR and works well across vendor platforms and field strengths, it requires that the same sequence be used to collect both the pre- and post-contrast T1w images. Consequently, the issue of limited application is not of concern for prospective clinical trials.
A final limitation is that the response assessment using dT1 did not explicitly include the appearance of new lesions as an indicator of progression. However, for all cases included in this sub-study no new measurable lesions > 1cc were present at the time of review. Thus, all statements of progression were made based on the findings at the primary tumor site only. Future studies will explicitly include the presence of new lesions as an additional criterion to determine progression.
Overall, the potential impact of dT1 technology is far-reaching given the approximately 117,000 new diagnoses of primary brain tumor per year,26 the more than 300,000 patients living with brain tumors who undergo repeated imaging follow-ups as part of their standard of care, and the 475 active clinical studies for GBM (clinicaltrials.gov). Therefore, the potential impact of the dT1 technology for daily practice as well as clinical trials is immense.
Conclusions
This study shows that dT1 can predict early progression comparable to the standard method, may be superior for sub-stratification and offers the potential for substantial time and cost savings for clinical trials.
Acknowledgements
Imaging Biometrics LLC (IQ-AI Ltd) for technical support, software development and assistance with data analysis. Funding support through National Institutes of Health / National Cancer Institute U01-CA079778 and U01-CA080098 and the Robert C. Olson MD Endowment.
Funding: Funding support through National Institutes of Health / National Cancer Institute U01-CA079778 and U01-CA080098 and the Robert C. Olson MD Endowment.
Abbreviations:
- 2D-T1
2-dimensional manual delineation of enhancing tumor
- ACRIN
American College of Radiology Imaging Network
- dT1
delta T1
- ECOG
Eastern Cooperative Oncology Group
- FLAIR
Fluid-attenuated inversion recovery
- GBM
Glioblastoma multiforme
- RANO
Response assessment in neuro-oncology
- ROI
Region of interest
- RTOG
Radiation Therapy Oncology Group
Footnotes
Conflict of Interest: Ownership interest in Imaging Biometrics LLC (KMS), IQ-AI Ltd (KMS), Consultation fees from Imaging Biometrics LLC (TRJ), IQ-AI (TRJ), ADMdx Diagnostics (TRJ), Change Healthcare (TRJ).
References
- 1.Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized Brain Tumor Imaging Protocol in clinical trials. Neuro Oncol 2015;17:1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). European Journal of Cancer 2009;45:228–247 [DOI] [PubMed] [Google Scholar]
- 3.Henson JW, Ulmer S, Harris GJ. Brain tumor imaging in clinical trials. American Journal of Radiology 2008;29:419–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kubben PL, Postma AA, Kessels AGH, et al. Intraobserver and interobserver agreement in volumetric assessment of glioblastoma mulitforme resection. Neurosurgery 2010;67:1329–1334 [DOI] [PubMed] [Google Scholar]
- 5.Hurwitz H, Fehrenbacher L, Novotny W, et al. Bevacizumab plus Irinotecan, Fluorouracil, and Leucovorin for metastic colorectal cancer. The New England Journal of Medicine 2004;350:2335–2342 [DOI] [PubMed] [Google Scholar]
- 6.Norden AD, Young GS, Setayesh K, et al. Bevacizumab for recurrent malignant gliomas: Efficacy, toxicity, and patterns of recurrence. Neurology 2008;70:779–787 [DOI] [PubMed] [Google Scholar]
- 7.Provenzale JM, Ison C, DeLong D. Bidimensional measurements in brain tumors: Assessment of interobserver variability. Am J Roentgenology 2009;193:W515–W522 [DOI] [PubMed] [Google Scholar]
- 8.Ellingson BM, Kim HJ, Woodworth DC, et al. Recurrent glioblastoma treated with bevacizumab: contrast-enhanced T1-weighted subtraction maps improve tumor delineation and aid prediction of survival in a multicenter clinical trial. Radiology 2014;271:200–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen TR, Schmainda KM. Computer-aided detection of brain tumor invasion using multiparametric MRI. J Magn Reson Imaging 2009;30:481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedekar D, Jensen T, Rand S, et al. Delta T1 Method: An automatic postl-contrast RO1 selection technique for brain tumors. Proc Intl Soc Mag Reson Med, 18th Annual Meeting Stockholm, Sweden; 2010 [Google Scholar]
- 11.Boxerman JL, Zhang Z, Safriel Y, et al. Early post-bevacizumab progression on contrast-enhanced MRI as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 Central Reader Study. Neuro Oncol 2013;15:945–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmainda KM, Zhang Z, Prah M, et al. Dynamic susceptibility contrast MRI measures of relative cerebral blood volume as a prognostic marker for overall survival in recurrent glioblastoma: results from the ACRIN 6677/RTOG 0625 multicenter trial. Neuro Oncol 2015;17:1148–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellingson BM, Kim E, Woodworth DC, et al. Diffusion MRI quality control and functional diffusion map results in ACRIN 6677/RTOG 0625: a multicenter, randomized, phase II trial of bevacizumab and chemotherapy in recurrent glioblastoma. Int J Oncol 2015;46:1883–1892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ratai EM, Zhang Z, Snyder BS, et al. Magnetic resonance spectroscopy as an early indicator of response to anti-angiogenic therapy in patients with recurrent glioblastoma: RTOG 0625/ACRIN 6677. Neuro Oncol 2013;15:936–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyul LG, Udupa JK. On standardizing the MR image intensity scale. Magnetic Resonance in Medicine 1999;42:1072–1081 [DOI] [PubMed] [Google Scholar]
- 16.Nyul LG, Udupa JK, Zhang X. New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 2000;19:143–150 [DOI] [PubMed] [Google Scholar]
- 17.Agreement Krippendorf K. and information in the reliability of coding. Communication Methods and Measures 2011;5:93–112 [Google Scholar]
- 18.Dempsey MF, Condon BR, Hadley DM. Measurement of tumor “size” in recurrent malignant glioma: 1D, 2D, or 3D? AJNR Am J Neuroradiol 2005;26:770–776 [PMC free article] [PubMed] [Google Scholar]
- 19.MacDonald DR, Cascino TL, Schold SC, et al. Response criteria for phase II studies of malignant glioma. J Clin Oncol 1990;8:1277–1280 [DOI] [PubMed] [Google Scholar]
- 20.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol 2010;28:1963–1972 [DOI] [PubMed] [Google Scholar]
- 21.Wen PY, Norden AD, Drappatz J, et al. Response assessment challenges in clinical trials of gliomas. Curr Oncol Rep 2010;12:68–75 [DOI] [PubMed] [Google Scholar]
- 22.Provenzale JM, Ison C, Delong D. Bidimensional measurements in brain tumors: assessment of interobserver variability. AJR Am J Roentgenol 2009;193:W515–522 [DOI] [PubMed] [Google Scholar]
- 23.Bedekar D, Jensen TR, Schmainda KM. Standardization of relative cerebral blood volume (rCBV) image maps for ease of both inter and intra-patient comparisons. Magn Reson Med 2010;64:907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prah MA, Stufflebeam SM, Paulson ES, et al. Repeatability of Standardized and Normalized Relative CBV in Patients with Newly Diagnosed Glioblastoma. AJNR Am J Neuroradiol 2015;36:1654–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bedekar D, Jensen T, Schmainda KM. Standardization of relative cerebral blood volume (rCBV) image maps for ease of both inter- and intrapatient comparisons. Magn Reson Med 2010;64:907–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ostrom QT, Gittleman H, Fulop J, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro Oncol 2015;17 Suppl 4:iv1–iv62 [DOI] [PMC free article] [PubMed] [Google Scholar]