Abstract
The apicomplexan parasite Cryptosporidium is a leading global cause of severe diarrheal disease and an important contributor to early childhood mortality. Currently there are no fully effective treatments or vaccines available. Parasite transmission occurs through ingestion of oocysts, through either direct contact or contaminated water or food. Oocysts are meiotic spores and the product of parasite sex. Cryptosporidium has a single host lifecycle where both asexual and sexual processes unfold in the intestine of infected hosts. Here we use the ability to genetically engineer Cryptosporidium to make life cycle progression and parasite sex tractable. We derive reporter strains to follow parasite development in culture and infected mice and define the genes that orchestrate sex and oocyst formation through mRNA sequencing of sorted cells. After two days, parasites in cell culture show pronounced sexualization, but productive fertilization does not occur and infection falters. In contrast, in infected mice male gametes successfully fertilize female parasites, leading to meiotic division and sporulation. To rigorously test for fertilization, we devised a two-component genetic crossing assay employing a Cre recombinase activated reporter. Our findings suggest obligate developmental progression towards sex in Cryptosporidium, which has important implications for the treatment and prevention of the infection.
Diarrheal diseases account for 9% of global child mortality1 and infection with Cryptosporidium is a leading cause of severe pediatric diarrhea2. Malnourished children are particularly susceptible to cryptosporidiosis resulting in recurrent or persistent infection and death2–4. Cryptosporidium is also an important cause of malnutrition5, and infection can result in lasting growth defects6. Even in high income countries outbreaks are frequent and more than 50% of U.S. waterborne infection is due to Cryptosporidium7,8. The current treatment of cryptosporidiosis is of limited efficacy for those patients in most urgent need9.
Cryptosporidium is a member of the eukaryotic phylum Apicomplexa and has a lifecycle that alternates asexual and sexual reproduction. However, in contrast to most other apicomplexans, the entire cycle unfolds in a single host. Sex results in the production of oocysts, environmentally hardy meiotic spores. Sex and production of oocysts are thus essential to transmission but may also play a role for continued infection of the host10. Chronic infection could be sustained by asexual replication with facultative sex driving host to host transmission. Alternatively, progression to sexual stages might be obligatory. Cryptosporidium oocysts are unique in that they mature within the host tissue and are auto-infective. Thus, they could reset the developmental cycle and maintain infection. Which of these two models applies is a fundamental yet unanswered question that has important implications for the disease and the development of drugs and vaccines. Here, we develop molecular markers to observe and analyze Cryptosporidium lifecycle progression and use them to demonstrate that a block in fertilization limits parasite growth in culture, supporting a model of obligate sexual developmental progression to maintain infection.
Results
A reporter parasite to track Cryptosporidium lifecycle progression
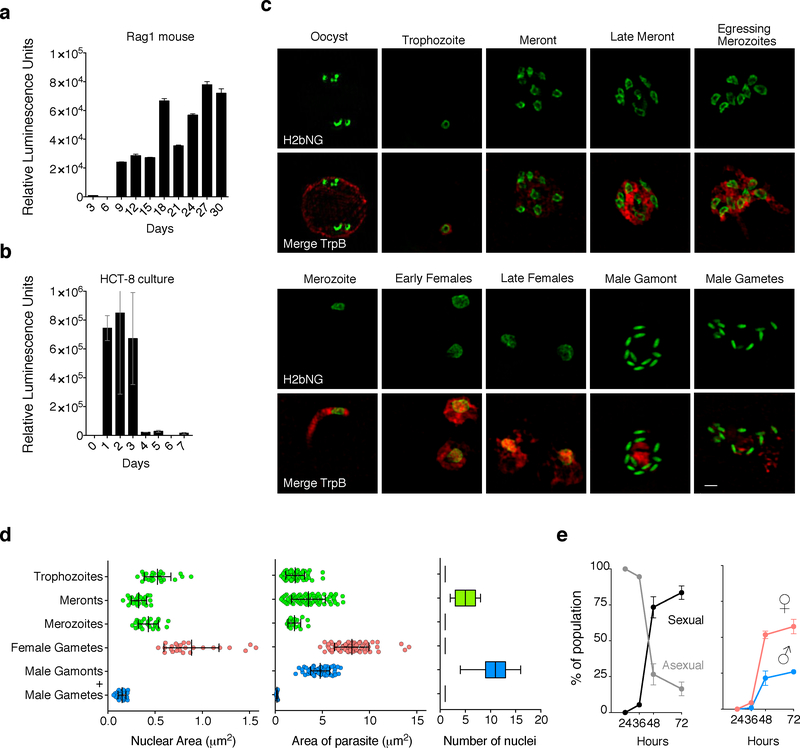
In the absence of adaptive immunity, humans and mice develop long-lasting Cryptosporidium infections and the parasite replicates continuously (Fig. 1a). Immortalized epithelial cell lines such as Caco2, HT-29, and HCT-8 are readily infected, but growth ceases after three days and the infection cannot be maintained by serial passage11 (Fig. 1b). During this time period, morphological stages consistent with asexual and sexual development have been observed and different sets of genes appear to be expressed in succession12–14. However, rigorous stage-specific markers are lacking. We thus sought to engineer transgenic parasites that delineate lifecycle progression and took advantage of well-documented changes in parasite nuclear morphology10,15. We introduced a fusion of C. parvum histone H2B (cgd5_3170) with the fluorescent reporter mNeon16 (see Supplementary Fig. 1 for detail). HCT-8 cells infected with these parasites were fixed after 24 and 48 hours, then imaged by super-resolution structured illumination microscopy. All parasites showed nuclear fluorescence. We recorded morphometric data for each parasite and its nucleus (Fig. 1d) and were able to distinguish multiple stages. At 24 hours, we observed trophozoites, small rounded intracellular stages with a single nucleus and stages of increasing size with unsegmented cytoplasm and two and four nuclei which we interpret as intermediate stages. We also observed mature meronts with eight nuclei, prior and mid egress, as well as free merozoites (Fig. 1c).
Figure 1: Cryptosporidium lifecycle stages revealed by Histone2b-mNeon transgene.
C. parvum infection was monitored by luciferase activity in mice lacking mature T and B cells (a) (feces were measured every 3 days) and HCT-8 cultures (b). Data were derived from three independent biological replicates and represented as mean ± SD. (c) HCT-8 cultures were infected with H2b-mN transgenic parasites and fixed at 24, 36, 48 and 72 h. Note nuclei in green and cytoplasm in red (antibody to tryptophan synthase B, cgd5_4560). This experiment was performed three times with similar results (Scale Bar= 1 μm) (d) Morphometric analyses of size (n= 25) and number (n= 100) of nuclei, and area for each stage (n= 75) based on the markers shown in (c). Nuclear and total area of parasites stages are shown as mean ± SD of individual values represented as dots. The number of nuclei of particular parasite stages are represented as box plots. The box shows median and quartile range and whiskers represent extreme values, respectively. (e) Time course experiment scoring stages using parameters defined in (d) revealed abrupt sexualization of cultures at 48 hours into culture. Data were derived from three independent biological replicates and represented as mean ± SD.
At 48 hours, we observed sexual stages (note that we use the terms male and female gametes following the convention of the extensive literature on sex in the malaria parasite Plasmodium17). Female or macrogametes had a single nucleus significantly larger (0.89 μm2) than the nuclei of asexual stages (0.43 μm2, p<0.0001, unpaired t-test) and male or microgametes, which had dense, bullet-shaped nuclei (0.15 μm2, p<0.0001, unpaired t-test). We found up to 16 of these nuclei in male or microgamonts (the precursor stage of the male gamete). Next, we conducted time course experiments and assigned a stage to all parasites observed using the morphometric characteristics defined above. Initially, all parasites in culture are asexual meronts and trophozoites. After 36 hours the culture rapidly sexualizes with gamonts and gametes representing >80% of all stages after 72 hours (Fig. 1e).
Gamete gene expression is controlled by stage specific promoters
To validate our stage assignments, we next defined exclusive molecular markers for gametes. The female gamete produces and stores components of the oocyst wall in wall-forming bodies previously described in related parasites18 and earlier studies identified Cryptosporidium Oocyst Wall Protein-1 (COWP1)19. We tagged the COWP1 protein (cgd6_2090) by C-terminal insertion of either a fluorescent protein or an HA epitope into the native locus (see Supplementary Fig. 2a–b). Transgenic parasites showed strong labeling of the oocyst wall (Fig. 2a). When infected cell cultures were examined, no expression was apparent at 24 hours, but numerous fluorescent parasites were observed at 48 hours. These parasites had a single large nucleus and multiple small foci of COWP1 consistent with wall forming bodies (Fig. 2b). We next observed COWP1 expression in parasites over a detailed in vitro time course (the COWP1-HA tagged strain was used here). Parasites expressing COWP1-HA closely matched the stages we identified as female gametes by H2B-mNeon in morphology and the proportion of the overall parasite population at the observed time points (Fig. 2d and Supplementary Fig. 3). To explore what controls the stage specificity of gene expression, we placed fluorescent protein reporters under the control of the presumptive COWP1 promoter region and ectopically expressed these constructs (Fig. 2c–d and Supplementary Fig. 2). Fluorescence (now cytoplasmic) was exclusively associated with female gametes and temporal expression matched that of the native locus demonstrating that promoters, and thus likely transcription initiation, control stage specificity (Fig. 2d).
Figure 2: Exclusive molecular markers for C. parvum sexual stages.
C. parvum were engineered to express Cowp1-mNeon (a & b, Scale Bar = 1 μm) and Cowp1-HA (Supplementary Figure. 3) out of the native locus or cowp1 promoter driven tdTomato out of the ectopic TK locus (c, Scale Bar = 1 μm). Note mNeon labeling of the wall in oocyst purified from infected mice and punctate labeling in female gametes observed in infected HCT-8 cells (Scale Bar= 1 μm). Labeling becomes apparent 42 h into culture and is never observed in asexual meronts or male gametes (b & d). The Cowp1 promoter alone is sufficient to confer female-specific expression to a reporter protein (c-d). Anti-H3K9Ac was used to label the nuclei of females as they stain poorly with DAPI. (e) Male gametes show a characteristic array of microtubules around the nucleus upon staining with an anti-tubulin antibody (e, Scale Bar = 0.5 μm). (d) For time courses cultures were infected with the indicated transgenic strains and triplicate coverslips were fixed and processed for IFA. Parasite stages were scored for HA staining, the mean percentage ± SD of HA positive stages among all parasites is shown for three independent biological replicates. When parasites were engineered to express HAP2-HA out of the native locus antibody staining revealed exclusive labeling of male gamonts (g, Scale Bar = 1 μm) and free gametes (f, Scale Bar = 1 μm). HAP2 labels a single pole per mature gamete. This staining becomes apparent at 42 hours of culture (d, blue). All microscopy experiments shown in this figure were performed three independent times.
Male gametes express HAP2 and appear at the same time as female gametes
Male gametes in most apicomplexans move with the aid of flagella and their exclusive presence in males provides numerous marker proteins. Cryptosporidium male gametes lack flagella but have a peculiar set of microtubules that are associated with and run along the length of their spindle-shaped nuclei15 which we visualized by super resolution microscopy (Fig. 2e). We also identified a C. parvum homolog of Hapless2 (HAP2, Supplementary Fig. 4) a class II membrane fusion protein that is required for gamete fusion in a range of organisms including Plasmodium falciparum and Chlamydomonas reinhardtii, and is expressed by the male or minus gamete, respectively20. We epitope tagged the C-terminus of HAP2 and infected cell cultures with transgenic parasites. HAP2-HA labeling was exclusively found in male gamonts (Fig. 2g) and gametes and was restricted to one end of the polarized male gamete (Fig. 2f). Time course experiments demonstrated appearance of males 42 h into culture (Fig. 2d). We note that both sexes emerge at the same time and at a male to female ratio of 1:2 for gamonts and 6:1 for gametes. We engineered multiple strains (within the HAP2 locus and ectopically) in an effort to drive fluorescent protein expression with the HAP2 promoter in a male specific fashion but did not observe fluorescence.
Define stage specific Cryptosporidium gene expression
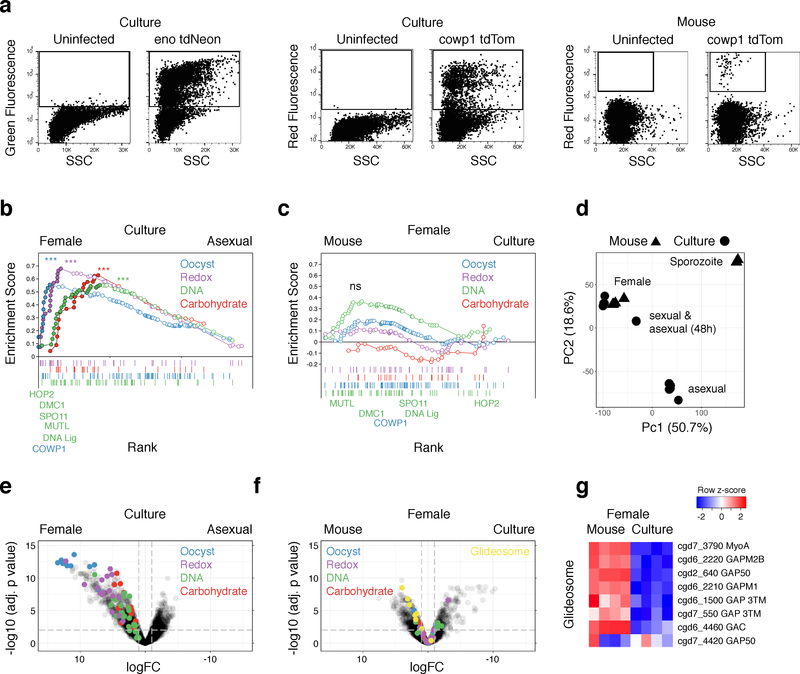
To discover the genes associated with sex in C. parvum, we sought to isolate specific parasite stages. We developed flow cytometry protocols to sort infected cells based on the expression of fluorescent proteins by the parasite (Supplementary Fig. 5). Fig. 3a shows sorts from cell culture and mice, where infected and uninfected cells are readily discernable. Next, we conducted mRNA sequencing experiments using cells sorted for eno tdNeon (see Supplementary Fig. 6) and Cowp1 tdTom from 24 or 48 h cultures to isolate asexual and female stages, respectively, as well as females from infected mice (Supplementary Fig. 7). We obtained between 5 and 35 million reads for each sample, with 50,000 to 7,000,000 mapping to the C. parvum transcriptome, representing 2500–3400 of the 3885 C. parvum genes (see Methods and Supplementary File 7 for detail).
Figure 3: Isolation of parasite stages by cell sorting and RNA sequencing.
(a) Flow cytometry of infected cells with the indicated markers and origins. Gates used for sorting are shown as boxes. This experiment was performed twice. (b & c) Gene set enrichment analysis with multiple testing correction comparing cultured asexual and female parasites (b) or females sorted from mice or culture (c). Custom gene signatures were generated using gene ontology or community datasets available on CryptoDB.org (b, carbohydrate (GO:0005975, NES=1.67, FDR=0.004), DNA (GO:0006259, NES=1.68, FDR=0.005), redox (GO:0055114, NES=1.99, FDR=0) and oocyst wall proteome23 dataset (NES=1.76, FDR=0.001)). Enrichment analysis comparing between females was not significant (n= 4 biological replicates per group). (d) Principal component analysis of all RNAseq datasets generated in this study (see Supplementary Fig. 7 for detail). (e & f) Volcano plots showing C. parvum genes differentially expressed between asexual and female parasites from culture (e) or between in vitro and in vivo female parasites (f, n=4 biological replicates per group). Each symbol represents a C. parvum gene, those genes representing the leading edge from (b) are filled in color according to the pathway they act in. The horizontal dashed line shows an FDR of 0.01, the vertical dashed lines indicate log2 fold change of −1 and +1. (g) Heat map of glideosome components are highlighted in yellow in (f, n= 4 biological replicates per group).
Analysis revealed robust transcriptional differences between asexual and female parasites. Transition to female gametes is accompanied by a 2-fold or greater increase (FDR<0.01) in the expression of 673 genes including COWP1 (Figs. 3e and Supplementary File 2; 451 genes are downregulated). We compared these genes with those associated with female gametogenesis in P. berghei21 (this particular dataset was most comparable to our experiment) and note 72 shared ortholog groups encompassing 73 C. parvum genes and 81 P. berghei genes (~31% of the female C. parvum genes with an identifiable P. berghei homolog) in addition to 595 C. parvum specific genes. We also compared female-specific genes from C. parvum and P. berghei to gametocyte genes in Eimeria tenella22 and identified a set of 41 ortholog groups containing 42 C. parvum genes, 49 P. berghei genes, and 55 E. tenella genes (see Supplementary File 4 for complete gene lists).
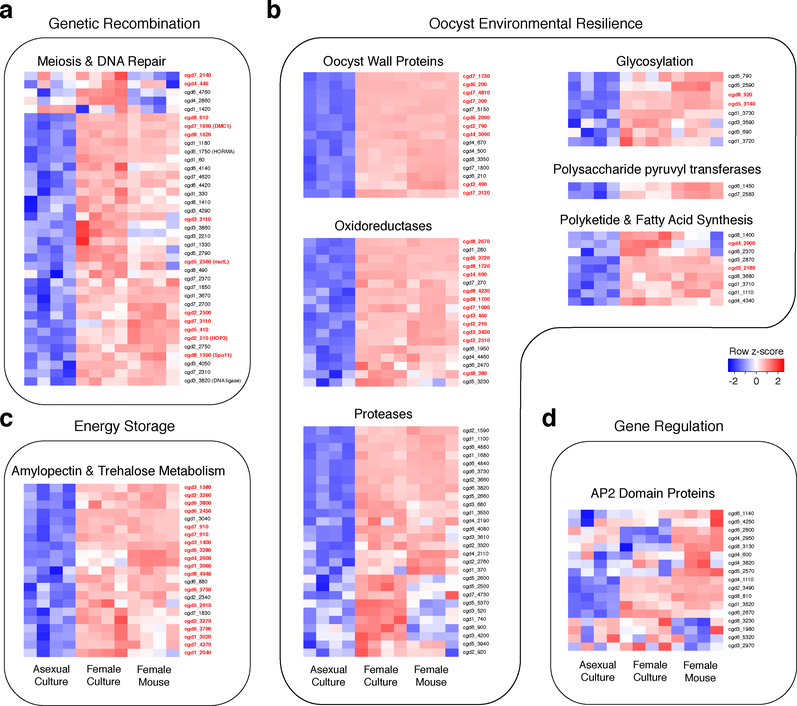
Functional annotation of the C. parvum genome using gene ontology is very limited. We thus assembled pathways using the GO terms for DNA, carbohydrate, and oxidase/reductase metabolism as well as a candidate oocyst wall proteome 23 to conduct enrichment analysis (Fig. 3b). We note significant enrichment when comparing female with asexual parasites (FDR<0.005). Fig. 4 shows additional clustering based on literature-based pathway annotation, those genes also found in the leading edge of the enrichment analysis are highlighted in red. Cryptosporidium is a haplont and meiosis is presumed to follow fertilization. Consistent with this view, we find conserved eukaryotic factors of meiotic recombination including DMC1, Spo11, HORMA and HOP2 to be preferentially expressed in females in addition to proteins with a likely role in meiosis-associated DNA repair including the mismatch repair protein MutL and DNA ligase (Fig. 3b and 4a). We also note chromosome segregation and cell division factors including condensins, cohesins, stage specific-cyclins, cyclin-dependent and NIMA kinases, and cytoskeletal proteins. Overall, we identified 37 genes in this meiosis category and many of these are shared among apicomplexan females (Supplementary Fig. 8 and Supplementary File 4).
Figure 4: Female gametes express genes required for genetic recombination and oocyst formation.
(a-c) Heat maps illustrating expression of genes associated with specific molecular functions that are upregulated in females (by-and-large in vivo and in vitro females concur with some exceptions, n= 4 biological replicates per group). (d) Expression heat maps for all C. parvum AP2 DNA binding proteins. Note pronounced difference identifying four genes upregulated in all females and two only in in vivo females. As we have been unable to sequence males, we cannot formally exclude that some genes that show high female expression may be up regulated in all sexual stages. Expression values are given as row z-score and annotated genes list are available for download as Supplementary files. Genes from the leading edge in Fig. 3b are highlighted in red font.
Cryptosporidium oocysts remain infectious for months24 and female transcription provides candidate mechanisms of this resilience. 22 enzymes required for the metabolism of amylopectin and trehalose are upregulated in females (Fig. 4c). Amylopectin is consumed24 by the sporozoites and the disaccharide trehalose may play a role in energy storage as well as serving to moderate osmotic stress25. Cryptosporidium oocysts are also remarkably resistant to chemical assault, including water chlorination26, due to a complex multilayered wall made of proteins, carbohydrates and lipids27. We identified 69 genes that are preferentially expressed in females and encode proteins with a likely role in oocyst wall synthesis, most of which have a predicted signal peptide. These include previously identified oocyst wall proteins and their homologs, numerous proteases (aspartic peptidases, serine proteases, and subtilases) and protein modifiers like amine oxidases22 that serve to build or modify the proteinaceous components of the wall (Fig. 4b). Many protozoans feature chitin and glucan cyst walls and the Cryptosporidium wall is labeled by various lectins, but no wall polysaccharide has been identified27,28. We similarly do not find a stereotypical chitin or glucan synthase, however, among female transcripts there are numerous glycosyl-transferases. Interestingly this set contains two polysaccharide pyruvyl transferases (cgd7_2583, cgd6_1450) and a UDP-glucose dehydrogenase (cgd8_920) all recently linked to capsule synthesis in pathogenic Acinetobacter29, as well as proteins with lectin domains including chitin binding proteins which suggests a proteoglycan structure (Fig. 4b). Lastly, we find that the two giant lipid synthases: polyketide synthase30 (cgd4_2900) and type I fatty acid synthase (cgd3_2180) that were acquired by horizontal transfer from bacteria are female expressed. In Mycobacterium tuberculosis these enzymes work in series to produce mycolic acids, key components of the mycobacterial wall. We chose two previously uncharacterized genes identified here as likely female (cgd7_4810 and cgd7_5140) for experimental validation and used CRISPR/Cas9 to append a c-myc-epitope tag. Transgenic oocysts reacted strongly with the c-myc antibody, and in culture we noted exclusive staining of female gametes (see Supplementary Fig. 9).
Both in vivo and in vitro females show upregulation of genes required for meiosis, wall formation, and oocyst persistence without significant difference (Fig. 3 c and f); suggesting an overall developmental competence of in vitro females (Fig. 4 a–c). We also transcriptionally profiled sporozoites released from oocysts and infected bulk culture after 24 and 48 hours for comparison and sporozoites are moderately more similar to in vivo than in vitro females (Supplementary Fig. 10 and Fig. 3 d). Overall, we conclude that a fraction of in vivo sorted cells has moved beyond fertilization to the production of sporozoites. This is consistent with the expression of the protein components of the gliding machinery required for the motility of invasive stages only observed in vivo females (Fig. 3f (highlighted in yellow) and g). Among the genes unique to in vivo females are also cgd5_2570 and cgd8_3130, which encode Apetala2 (AP2) domain proteins (Fig. 4d). AP2-type transcriptional regulators have been demonstrated in other apicomplexans to bind to specific genome features31,32 including promoters and emerged as master regulators of lifecycle progression. Cryptosporidium encodes a comparably small set of AP2s33 and many of these genes appear to be developmentally regulated, with expression patterns that differ greatly between asexual from sexual parasites and in vitro and in vivo (Fig. 4d).
Fertilization and meiosis occur within the host cell but only in vivo
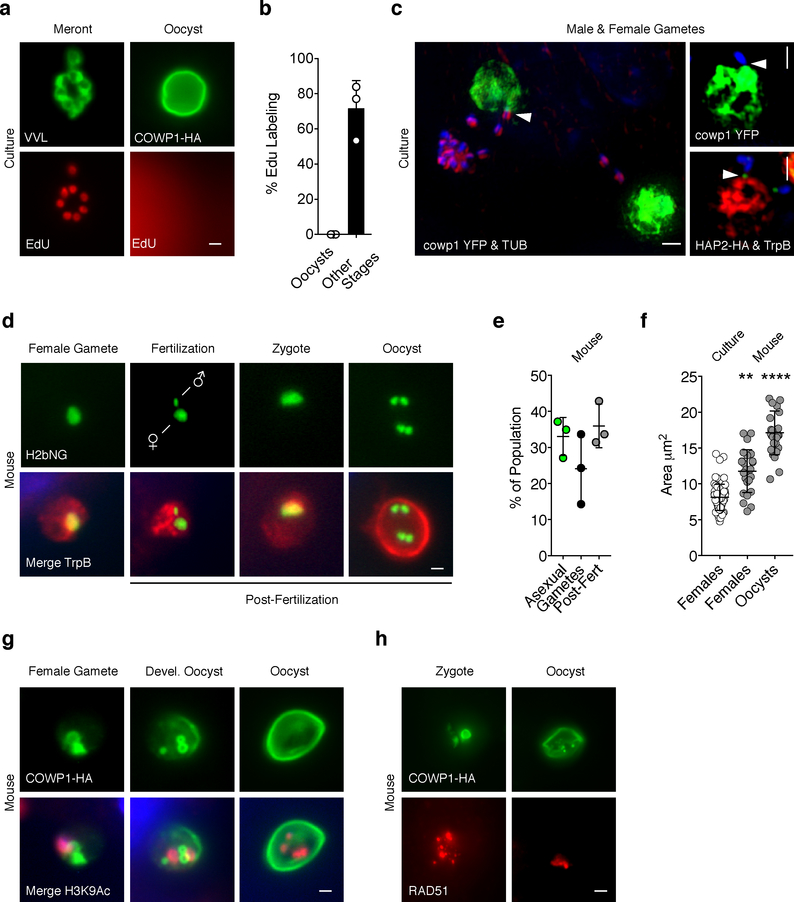
Gametes are plentiful in infected cell monolayers, but oocysts are rare (<0.1% of all stages) which is consistent with our sequencing data. To determine whether these oocysts were produced de novo, we labelled newly synthesized DNA using the thymidine kinase analog, EdU. As reported previously, C. parvum readily incorporates thymidine kinase activated tracers34. In vitro, none of the oocysts observed at 48 hours post infection were labeled (Figs. 5a & b), indicating that these were from the inoculum and not formed in culture. Next we wondered whether male gametes mature and go on to fertilize females in culture. At 48 hours, we find that 10.6±0.8% of all male gametes are released from gamonts and we frequently observed them attached to female gametes (Fig. 5c). At this time point, 16.1±2.2% of all females identified by H2B-mNeon featured an ‘attached’ male. Note that attachment was polar with the HAP2 marked end oriented towards the female. However, we did not observe female gametes with an ‘internalized’ male gamete. To ask how this compares with in vivo infection, where we know fertilization occurs, segments of the small intestine were recovered from infected mice, cryosectioned and processed for immunofluorescence. When using the H2b-mNeon line we rarely spotted males attached to females, yet frequently observed parasites that contained both an identifiable female and male nucleus (~5% of all stages, Fig. 5d). In vivo, zygotes and various intermediates of meiosis with one, two and four nuclei were readily observable and these post-fertilization stages accounted for 35% of all parasites (Fig. 5e). As these stages mature, they grow and their size significantly exceeds that of female gametes observed in culture (p= 0.0023, Fig. 5f). We made very similar observations when studying the COWP1-HA strain in vivo. Meiotic divisions precede and partially overlap with the deposition of the oocyst wall (Fig. 5g). We also observed strong labeling of these stages with RAD51, a DNA repair protein with an important role in homologous cross-over during meiosis (Fig 5h).
Figure 5: Cryptosporidium males locate females in culture, but fertilization and meiosis only occur in vivo.
(a) HCT-8 cells were infected with COWP1-HA C. parvum and after 36 h, the nucleotide analog EdU was added to the medium and 12 h later cells were click labeled and counter stained with anti-HA or VVL (Scale Bar = 1 μm) and scored for nuclear EdU labeling (b). 100 stages were quantified for three biological replicates, and the experiment was performed twice. Data is represented as mean ± SD. (c) Representative images of encounters of male and female gametes in culture, gametes were identified using the indicated transgenes or antibodies and attached males are highlighted by arrowheads (Scale Bar = 1 μm). (d) IFNγ−/− mice were infected with H2b Neon (d, Scale Bar = 1 μm) or COWP1-HA (g & h, Scale Bar = 1 μm) expressing parasites and intestines were sectioned and prepared for IFA and counterstained with TrpB, H3K9Ac, or RAD51. Representative micrographs show progression of events following fertilization. Post fertilization stages are abundant in vivo (e, mean and SD derived from three independent mice) and these stages were significantly larger than those found in vitro (f, each symbol represents a parasite, n=25, mean and SD are shown along with two–sided t-test p values comparing cultured females with in vivo females (0.0023) or vivo oocysts (0.0001). All microscopy experiments shown in (d-h) were performed twice with similar results.
A genetic two component assay demonstrates gamete fusion in vivo but not in vitro
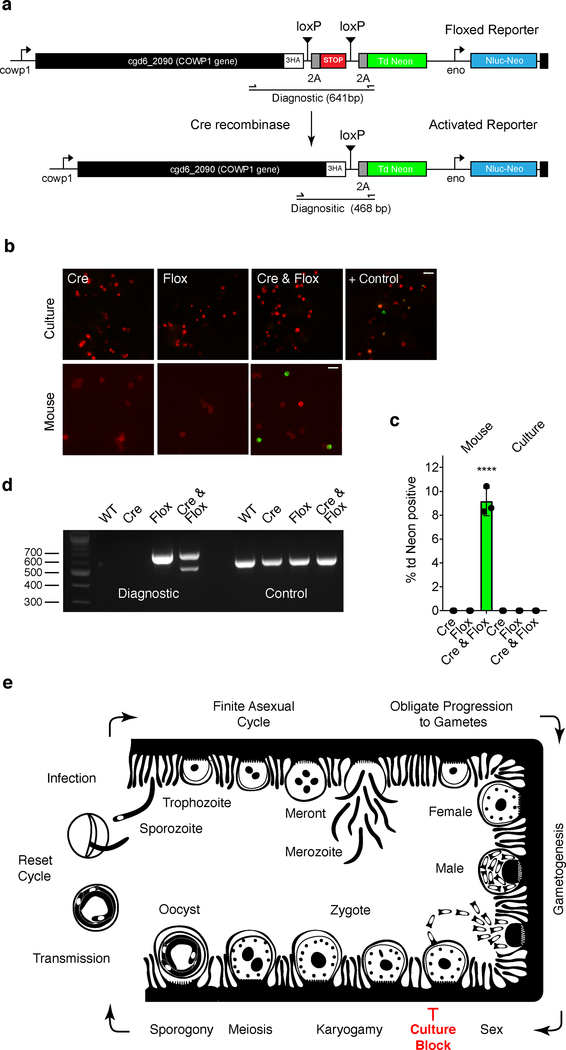
Our experiments suggested lack of fertilization in vitro. To test this rigorously we devised a genetic assay for Cryptosporidium gamete fusion. We engineered a two-component system that produces a reporter signal only upon cytoplasmic fusion of two strains. The first component is a driver strain that expresses Cre recombinase, an enzyme that excises DNA segments flanked by loxP recognition sequences (the 34 bp loxP sequence is absent from the C. parvum genome). Cre is driven by a constitutive promoter and detected in transgenics using a specific antibody (Supplementary Fig. 11). The second component is a strain carrying a tdNeon reporter in the COWP1 locus linked by a 2A skip peptide. A terminator sequence flanked by loxP sites blocks expression, Cre-mediated excision will release the block (see Fig. 6a for detail). Mice were infected with each parasite strain individually or with both in equal proportion. Only infection with both strains resulted in shedding of green fluorescent oocysts (~10% of total oocyst from days 3–10, see Figs. 6b–d). We next performed this assay in vitro and tested for tdNeon expression at 48 and 72 h post infection. In contrast to mice, we did not detect expression of tdNeon in HCT-8 cells coinfected with Cre and floxed strains (Figs 6b–c, p= 0.0002). Fluorescence could be readily detected in our positive control, HCT-8 cells infected with oocysts obtained by Cre/flox coinfection in mice. We conclude that gamete fusion occurs in vivo, but not in vitro, and this block in fertilization prevents the formation of new oocysts and continued growth in culture.
Figure 6: A genetic fusion assay demonstrates fertilization in vivo but not in vitro.
To detect gamete fusion, we engineered two C. parvum strains, one that constitutively expresses Cre recombinase (Supplementary Fig. 11) and a second that carries a floxed tdNeon reporter in the COWP1 locus (a). Cre-mediated excision of a terminator results in reporter expression. HCT-8 cultures and IFNγ−/− mice were infected with each strain individually or in combination (b). This experiment was performed twice. Cultured parasites were counterstained with Anti-TrpB, oocysts with Macula pomifera agglutinin (both red) and scored for tdNeon expression (Scale bar= 10 μm). (c) Three replicates were quantified for green fluorescence and 1000 cells were counted for each replicate, means and SD are shown. Green fluorescence is only observed upon in vivo infection and only when both strains are present (P= 0.0002, two-sided t-test). As a positive control, cells were infected with parasites crossed in vivo (b). (d) PCR mapping of the floxed (diagnostic) and α-tubulin (control) loci using the primer pair shown in (a). Genomic DNA was isolated from wild type parasites as well as oocysts from the mouse infection experiments. Crossing resulted in a new amplicon consistent with precise Cre excision. This experiment was performed twice with similar results. (e) Schematic model of the C. parvum lifecycle highlighting the model of obligate progression to sex and the fertilization block in HCT-8 culture. We do not show Type II meronts here, which are often depicted as an obligate step towards gametes. While we observe meronts with 4 and 8 nuclei we did not find a quantitative link between the meronts with 4 nuclei and gametes (see Supplementary Fig. 12 for detail).
Discussion
The complex lifecycles of parasites are among the most fascinating aspects of their biology. Cryptosporidium is a minute protist with a highly reduced genome, and yet it continuously transforms itself into a menagerie of specialized stages that amplify asexually, transform into male and female gametes, undergo fertilization, and build a resilient spore. Here we trace and analyze this lifecycle, label and isolate specific stages, and discover the genes that define them to provide a road map for the molecular dissection of parasite sex. We rigorously demonstrate that in HCT-8 culture Cryptosporidium undergoes sexual differentiation, but a block in gamete fusion prevents the development of new oocysts and parasite cultures ultimately arrest (Fig. 6e). The cause of this block remains to be elucidated but appears to be linked to host rather than parasite physiology. This may be overcome partially by culture modalities that provide structured environments to transformed cells35,36 or by using stem cell derived models that self-organize into more complex organoids37. It is unclear whether this is due to differences in the infected host cells themselves or to factors secreted by more complex assemblages. In all cases improved growth is linked to appearance of oocysts. Overall, this is consistent with a model of obligate developmental progression and suggests that interventions targeting sex could potentially not only block transmission but cure ongoing infection. Antibodies to HAP2 block fertilization in Plasmodium20,38, ‘contraceptive’ vaccination may thus offer protection against cryptosporidiosis. This idea could be further tested by genetic ablation of the sex genes identified here, including HAP2. We define the stage specificity of numerous AP2 transcription factors, and the ablation or forced expression of these regulators32,39 opens the door to engineer the Cryptosporidium lifecycle.
Fertilization, the fusion of two gametes, typically requires direct contact guided by interaction of surface displayed fusion proteins and their ligands40,41. Cryptosporidium male gametes lack flagella and yet they are capable of locating females (Fig. 5c). Importantly, male gametes find females while they are inside of an infected cell and fertilization and subsequent meiosis are intracellular events (Figure 6e). This could be guided by pheromones42, or alternatively by a ligand dispatched to the host cell surface by the female, translocons capable of such export are known in other Apicomplexa43,44. How the male overcomes the three membranes that separate it from its female counterpart is unknown, but appears to be the very step blocked in vitro. In Chlamydomonas, membrane fusion and HAP2 are restricted to fertilization tubules that form in proximity to the basal body45. HAP2 in the Cryptosporidium male is restricted to the pole that faces the host cell carrying the female. Intriguingly, the male gamete also carries a polar basal body15,46 and adjacent membrane structures. These structures attract speculation, and with genetic and cell biological experiments now feasible, invite further mechanistic studies.
Methods
Plasmid construction
Guide oligonucleotides (Sigma-Aldrich, St. Louis. MO) were introduced into the C. parvum Cas9/U6 plasmid16 by restriction cloning. See Pawlowic et al.47 for a detailed discussion of guide design for C. parvum. Transfection plasmids were constructed by Gibson assembly using NEB Gibson Assembly® Master Mix (New England Biolabs, Ipswich. MA.).
Generation of transgenic parasites
5X107 C. parvum oocysts Iowa II strain (procured from Bunchgrass Farms or the University of Arizona) were incubated at 37°C for 1hr in 0.8% sodium taurocholate to induce excystation. Excysted sporozoites were then transfected using an Amaxa 4D Electropotrater (Lonza, Basel, Switzerland) with parasites suspended in SF buffer using program EH100 and 50μg of each Cas9/U6 plasmid, and PCR repair construct. The repair encodes the neomycin phosphotransferase drug selection marker fused to a nanoluciferase reporter flanked by 50 bp homologous regions to guide insertion into the parasite genome. IFNγ−/− mice were infected with transfected parasites by oral gavage. Stomach acid was neutralized with 100μl of 8% NaHCO3 solution by gavage prior to infection. Note that this modification replaces surgery-based infection and significantly streamlines the protocol. Stable transformants were selected with paromomycin, given to mice ad libitum in their drinking water (16 mg/mL) and parasite shedding was monitored by measuring nanoluciferase activity in the feces of infected mice.
To purify transgenic parasites from collected feces we used sucrose flotation followed by a cesium chloride gradient47. Briefly, collected mouse feces were homogenized in tap water using a LabGen 125 homogenizer (Cole-Parmer, Vernon Hills, IL, USA) and filtered through a 250-um mesh filter. This filtrate was diluted 1:1 with a saturated sucrose solution (specific gravity 1.33) and centrifuged at 1,000g for 5 min. The supernatant was collected, resuspended in 0.85% saline solution and overlaid onto CsCl solution (1.15 specific gravity) and centrifuged at 16,000g for 3 min. Purified oocysts were collected from the saline-CsCl interface and resuspended in cold PBS.
Immunofluorescence Assay
HCT-8 cells were infected with bleached and washed oocysts. Infected cells were maintained in RPMI-1640 medium (Sigma-Aldrich, St. Louis. MO) containing 1% fetal bovine serum. Infected cells were fixed with 4% paraformaldehyde (Electron Microscopy Science, Hatfield. PA) in PBS and then permeabilized with PBS containing 0.25% Triton X-100. Cells were blocked with 3% bovine serum albumin (BSA) solution, followed by an incubation with primary antibodies. Cells were washed with PBS and then incubated with appropriate fluorophore conjugated secondary antibodies and counterstained with 4,6-diamidino-2-phenylindole (DAPI). Coverslips were then mounted on glass slides with fluorogel (Electron Microscopy Science, Hatfield. PA) mounting medium.
For in vivo staining, infected mice were killed, and the small intestine was resected and flushed with 10% neutral buffered formalin (Sigma-Aldrich, St. Louis. MO), then ‘swiss-rolled’ and fixed overnight in 4% paraformaldehyde followed by an overnight incubation in 30% sucrose solution. Samples were embedded in OCT medium (Tissue-Tek, Sakura Finetek. Japan) and cryo-sectioned. Tissue sections were blocked with 10% BSA and 0.1% Triton X-100 in PBS. Sections were stained with antibodies in PBS 0.1% Triton X-100 as described above counterstained with DAPI and mounted.
Super resolution structured illumination microscopy (SR-SIM) was conducted on Carl Zeiss Elyra (UGA Biomedical Microscopy Core) or a GE OMX (PennVet Imaging Core) microscopes. Widefield microscopy was performed using a Leica LAS X microscope (PennVet Imaging Core) and images were processed and analyzed using Carl Zeiss ZEN 2.3 SP1, GE Softworxs and NIH ImageJ software.
Cre-lox based fertilization assay
To measure sex between two different strains, IFNγ−/− mice were infected with 50,000 oocysts of either the Cre or COWP1 HA flox tdNeon strain or coinfected with both the strains. Oocysts were purified from fecal samples collected from days 3–10 post infection. Oocysts were fixed with 4% PFA, stained with biotinylated Macula pomifera agglutinin48 (Vector Laboratories, Inc, Burlingame. CA), washed and incubated with Streptavidin-594, and settled onto poly-L-lysine (Sigma-Aldrich, St. Louis. MO) coated coverslips prior to mounting with fluorogel. The same strains were used to infect HCT-8 cells. HCT-8 cells infected with oocysts obtained from Cre X COWP1 HA flox tdNeon coinfection were used as positive controls. Cells were fixed 48 and 72 hours post infection and parasites were stained by using rabbit anti tryptophan synthase B antibody.
EdU labelling to detect DNA synthesis
HCT-8 cells were infected with 100,000 oocysts of the COWP1-HA strain. EdU was added to cultures 36 hours post infection to a final concentration of 10μM and cells were fixed 12 hours later. The Click-iT Edu Alexa-Fluor 594 kit (Thermo Fischer Scientific, Waltham. MA) was used to label incorporated EdU. Parasites were stained with anti-HA antibody or fluorescein conjugated Vicia villosa lectin (Vector Laboratories, Inc, Burlingame, CA).
Flow sorting of intracellular stages and RNA extraction
HCT-8 cells grown in 6 well plates were infected with 300,000 oocysts of eno tdNeon (constitutive reporter strain) or COWP1 tDTomato (female reporter strain). Infected cultures were trypsinized with TrypLE Express Enzyme (Thermo Fischer Scientific, Waltham. MA), extensively washed with PBS, passed through a 40μm filter (BD Biosciences, San Jose, CA) and pelleted. Cells were resuspended in 400μl of buffer and sorted on a BD FACSJazz sorter (BD Biosciences, San Jose, CA). Uninfected HCT-8 control cells were used to gate on the singlet host cell population. 10,000 positive cells were directly sorted into the RLT lysis buffer of the Qiagen micro RNA extraction kit (Qiagen, Hilden, Germany).
4 IFNγ−/− mice were infected with 200,000 oocysts of COWP1 tDTomato reporter strain. Mice were killed 2 days post infection, the small intestine was resected, cut into small pieces and incubated in RPMI-1640 medium containing 10% FBS, 25mM HEPES, 5mM EDTA, 50μM β-mercaptoethanol and 0.145mg/ml dithiothrietol for 20 mins. The cell suspension was filtered through kitchen mesh, 70μ, and 40μ filters (BD Biosciences, San Jose, CA). Cells were pelleted, resuspended in buffer, stained with anti-CD45.2 antibody and sorted. Intestinal cells isolated from uninfected mice were used as controls. 1000 tdTomato-positive cells from each replicate were sorted directly into 350μl of RLT lysis buffer.
The Qiagen micro RNA extraction kit was used to extract RNA from sorted cells. RNA was finally eluted in RNAse free water and samples were then stored at −80°C.
RNA sequencing of sorted cells
cDNA was generated using the SMART-Seq v4 Ultra Low Input RNA Kit (Takara Bio USA Inc., Mountain View. CA), and barcoded, sequence-ready libraries were prepared using the Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA). Total RNA and libraries were quality checked and quantified on an Agilent Tapestation 4200 (Agilent Technologies, Santa Clara, CA) and Qubit 3 (Thermo Fischer Scientific, Waltham. MA), respectively. All samples were pooled and single-end reads were generated on an Illumina NextSeq 500 sequencer.
RNA sequencing of sporozoites & 24/48hr infected bulk cultures
Oocysts from Sterling farms (University of Arizona, AZ) were first induced to excyst through resuspension in 0.8% sodium deoxytaurocholate and incubation at 37°C for 2 hours. RNA from the released sporozoites was then isolated using an RNeasy Mini Kit (Qiagen, Hilden, Germany) following manufacturer’s instructions. Each biological replicate was isolated from 40mil sporozoites. For the 24/48hr in vitro time-points, oocysts from Sterling farms (University of Arizona, AZ) were first treated with dilute household bleach (1:4 in dH2O) for 10 minutes on ice to sterilize them. Oocysts were then washed twice with cold PBS and resuspended in 0.8% sodium deoxytaurocholate for a 10-minute incubation on ice. Oocysts were washed once more with cold PBS then used to infect HCT-8 cell monolayers grown to 80% confluency in a 24 well plate. For 24hr timepoints, 1mil oocysts were infected into each well with 3 biological replicates. For 48hr timepoints, 100,000 oocysts were infected into each well with 3 biological replicates. During infection standard RPMI growth media was used with 1% fetal bovine serum. RNA from infected cells was isolated using an RNAeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocols. Sequencing libraries for sporozoites and in vitro timepoints were prepared using a Nextera XT DNA Library Preparation Kit (Illumina Inc., San Diego, CA) and 150bp paired end reads were collected with an Illumina MiSeq (Illumina Inc., San Diego. CA).
RNA sequence analysis
Raw reads were mapped to the C. parvum Iowa II reference (Ensembl, ASM16534v1) using Kallisto v0.45.049. All subsequent analyses were carried out using the statistical computing environment, R version 3.6 in RStudio version 1.1.463 and Bioconductor. Briefly, transcript level quantification data was summarized to genes using the tximport package and data were normalized using the TMM method (implemented in EdgeR). Only genes with >10 counts per million in ≥ 3 or 4 samples (depending on the analysis) were carried forward for analysis. Precision weights were applied to each gene based on the mean-variance relationship using the VOOM function in Limma. Linear modeling and Bayesian statistics carried out in Limma were used to identify differentially expressed genes with a false discovery rate (FDR) ≤ 0.01 and an absolute log2 fold change ≥ 1, after correcting for multiple testing using Benjamini-Hochberg. When necessary, batch correction was carried out using the empirical Bayes-moderated adjustment for unwanted covariates function (empiricalBayesLM) in the WGCNA package. All code used in these analyses is available in the supplementary code file, and on GitHub (https://github.com/dpbisme/CryptoSex_manuscript). For Plasmodium berghei, files were downloaded from the NCBI Sequence Read Archive BioProject ID: PRJNA37491821 and forward reads were mapped as described above for the sorted C. parvum samples to the P. berghei reference transcriptome (Ensembl, PBANKA01). For Eimeria tenella, differentially expressed gametocyte genes were obtained from Walker, et al.22. Cross species comparisons and ortholog identifications were done on EuPathDB.org. See Supplementary File 7 for full detail including a link to all code used for RNA analyses performed here.
Functional enrichment analysis
Gene Set Enrichment Analysis was carried out using GSEA software50. Four custom gene signatures for C. parvum were generated using gene ontology or community datasets available on CryptoDB.org. A 28 gene signature for ‘carbohydrate metabolism’ was generated using the Gene Ontology term GO:0005975. A 63 gene signature for ‘DNA metabolic process’ was generated using GO:0006259. A 48 gene signature for ‘oxidation-reduction’ was generated using GO:0055114. An 85 gene oocyst signature was generated by using CryptoDB to mine a published oocyst wall proteome dataset from Truong and Ferrari23 to retrieve only genes that had ≥ 20 unique peptide sequences per sample. All four signatures were used for GSEA analysis with 1000 permutations of gene sets to generate P values, and multiple testing correction was applied to generate FDRs. GSEA results were used to create enrichment plots in DataGraph v4.4 (Visual Data Tools, Inc).
Statistical methods
GraphPad PRISM was used for all statistical analyses. When measuring the difference between two populations, we used a standard T-test. No statistical tests were used to predetermine sample size and no animals were excluded from results.
Animal Ethics Statement
All protocols for animal experimentation were approved by the Institutional Animal Care and Use Committee of the University of Georgia (protocol A2016 01–028-Y1-A4) and/or the Institutional Animal Care and Use Committee of the University of Pennsylvania (protocol #806292). 4-week old IFNγ−/− & Rag1 KO female mice strains of Mus musculus were used for all the experiments. No statistical tests were used to predetermine the sample size of mice used for experiments. Mice were not randomized or blinded prior to any experiments.
Data availability
RNA-seq data generated in this study are available through the GEO database repository under accession number GSE129267. Additional RNA-seq data that support the findings of this study are available through the NCBI Sequence Read Archive BioProject ID: PRJNA374918.
Supplementary Material
Acknowledgements
We thank Carrie Brooks, Emily Myers, Gillian Herbert, Stephanie Cave, Muthugapatti Kandasamy and Paul Hallberg for help and technical support. This work was funded in part by the National Institutes of Health through grant R01AI127798 to BS and fellowships and career awards to AS (F32AS124518, K99AI137442) and JG (T32AI055400) and by grant OPP1161001 from the Bill & Melinda Gates Foundation to BS.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Liu L et al. Global, regional, and national causes of under-5 mortality in 2000–15: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet 388, 3027–3035, doi: 10.1016/S0140-6736(16)31593-8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kotloff KL et al. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382, 209–222, doi: 10.1016/S0140-6736(13)60844-2 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Checkley W et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for cryptosporidium. Lancet Infect Dis 15, 85–94, doi: 10.1016/S1473-3099(14)70772-8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Platts-Mills JA et al. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3, E564–E575, doi: 10.1016/S2214-109x(15)00151-5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korpe PS & Petri WA Jr. Environmental enteropathy: critical implications of a poorly understood condition. Trends Mol Med 18, 328–336, doi: 10.1016/j.molmed.2012.04.007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khalil IA et al. Morbidity, mortality, and long-term consequences associated with diarrhoea from Cryptosporidium infection in children younger than 5 years: a meta-analyses study. Lancet Glob Health 6, e758–e768, doi: 10.1016/S2214-109X(18)30283-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scallan E et al. Foodborne illness acquired in the United States--major pathogens. Emerg Infect Dis 17, 7–15, doi: 10.3201/eid1701.P1110110.3201/eid1701.091101p1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Painter JE et al. Cryptosporidiosis surveillance -- United States, 2011–2012. MMWR Suppl 64, 1–14 (2015). [PubMed] [Google Scholar]
- 9.Amadi B et al. High dose prolonged treatment with nitazoxanide is not effective for cryptosporidiosis in HIV positive Zambian children: a randomised controlled trial. BMC infectious diseases 9, 195, doi: 10.1186/1471-2334-9-195 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Current WL & Reese NC A comparison of endogenous development of three isolates of Cryptosporidium in suckling mice. J Protozool 33, 98–108 (1986). [DOI] [PubMed] [Google Scholar]
- 11.Upton SJ, Tilley M & Brillhart DB Comparative development of Cryptosporidium parvum (Apicomplexa) in 11 continuous host cell lines. FEMS Microbiol Lett 118, 233–236 (1994). [DOI] [PubMed] [Google Scholar]
- 12.Current WL & Haynes TB Complete development of Cryptosporidium in cell culture. Science 224, 603–605 (1984). [DOI] [PubMed] [Google Scholar]
- 13.Mauzy MJ, Enomoto S, Lancto CA, Abrahamsen MS & Rutherford MS The Cryptosporidium parvum transcriptome during in vitro development. PLoS One 7, e31715, doi: 10.1371/journal.pone.0031715 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilke G et al. Monoclonal Antibodies to Intracellular Stages of Cryptosporidium parvum Define Life Cycle Progression In Vitro. mSphere 3, doi: 10.1128/mSphere.00124-18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostrovska K & Paperna I Cryptosporidium sp. of the starred lizard Agame stellio:ultrastructure and life cycle. Parasitology Research 76, 712–720 (1990). [Google Scholar]
- 16.Vinayak S et al. Genetic modification of the diarrhoeal pathogen Cryptosporidium parvum. Nature 523, 477–480, doi: 10.1038/nature14651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Josling GA & Llinas M Sexual development in Plasmodium parasites: knowing when it’s time to commit. Nature Reviews Microbiology 13, 573–587, doi: 10.1038/nrmicro3519 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Ferguson DJ, Belli SI, Smith NC & Wallach MG The development of the macrogamete and oocyst wall in Eimeria maxima: immuno-light and electron microscopy. Int J Parasitol 33, 1329–1340 (2003). [DOI] [PubMed] [Google Scholar]
- 19.Spano F, Puri C, Ranucci L, Putignani L & Crisanti A Cloning of the entire COWP gene of Cryptosporidium parvum and ultrastructural localization of the protein during sexual parasite development. Parasitology 114 ( Pt 5), 427–437 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Liu YJ et al. The conserved plant sterility gene HAP2 functions after attachment of fusogenic membranes in Chlamydomonas and Plasmodium gametes. Gene Dev 22, 1051–1068, doi: 10.1101/gad.1656508 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeoh LM, Goodman CD, Mollard V, McFadden GI & Ralph SA Comparative transcriptomics of female and male gametocytes in Plasmodium berghei and the evolution of sex in alveolates. BMC Genomics 18, 734, doi: 10.1186/s12864-017-4100-0 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker RA et al. RNA Seq analysis of the Eimeria tenella gametocyte transcriptome reveals clues about the molecular basis for sexual reproduction and oocyst biogenesis. Bmc Genomics 16, doi:ARTN 94, 10.1186/s12864-015-1298-6 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Truong Q & Ferrari BC Quantitative and qualitative comparisons of Cryptosporidium faecal purification procedures for the isolation of oocysts suitable for proteomic analysis. International Journal for Parasitology 36, 811–819, doi: 10.1016/j.ijpara.2006.02.023 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Fayer R, Trout JM & Jenkins MC Infectivity of Cryptosporidium parvum oocysts stored in water at environmental temperatures. Journal of Parasitology 84, 1165–1169, doi:Doi 10.2307/3284666 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Elbein AD, Pan YT, Pastuszak I & Carroll D New insights on trehalose: a multifunctional molecule. Glycobiology 13, 17r–27r, doi: 10.1093/glycob/cwg047 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Korich DG, Mead JR, Madore MS, Sinclair NA & Sterling CR Effects of ozone, chlorine dioxide, chlorine, and monochloramine on Cryptosporidium parvum oocyst viability. Appl Environ Microbiol 56, 1423–1428 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samuelson J, Bushkin GG, Chatterjee A & Robbins PW Strategies To Discover the Structural Components of Cyst and Oocyst Walls. Eukaryotic Cell 12, 1578–1587, doi: 10.1128/Ec.00213-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bushkin GG et al. beta-1,3-glucan, which can be targeted by drugs, forms a trabecular scaffold in the oocyst walls of Toxoplasma and Eimeria. mBio 3, doi: 10.1128/mBio.00258-12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kasimova AA et al. Acinetobacter baumannii K20 and K21 capsular polysaccharide structures establish roles for UDP-glucose dehydrogenase Ugd2, pyruvyl transferase Ptr2 and two glycosyltransferases. Glycobiology 28, 876–884, doi: 10.1093/glycob/cwy074 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Zhu G et al. Cryptosporidium parvum: the first protist known to encode a putative polyketide synthase. Gene 298, 79–89 (2002). [DOI] [PubMed] [Google Scholar]
- 31.De Silva EK et al. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci U S A 105, 8393–8398, doi:0801993105 [pii], 10.1073/pnas.0801993105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kafsack BF et al. A transcriptional switch underlies commitment to sexual development in malaria parasites. Nature 507, 248–252, doi: 10.1038/nature12920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oberstaller J, Pumpalova Y, Schieler A, Llinas M & Kissinger JC The Cryptosporidium parvum ApiAP2 gene family: insights into the evolution of apicomplexan AP2 regulatory systems. Nucleic Acids Res 42, 8271–8284, doi: 10.1093/nar/gku500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Striepen B et al. Gene transfer in the evolution of parasite nucleotide biosynthesis. Proc Natl Acad Sci U S A 101, 3154–3159 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morada M et al. Continuous culture of Cryptosporidium parvum using hollow fiber technology. Int J Parasitol, doi: 10.1016/j.ijpara.2015.07.006 (2015). [DOI] [PubMed] [Google Scholar]
- 36.RePass MAD et al. Novel Bioengineered Three- Dimensional Human Intestinal Model for Long-Term Infection of Cryptosporidium parvum. Infection and Immunity 85, doi:UNSP e00731–16, 10.1128/IAI.00731-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heo I et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat Microbiol 3, 814–+, doi: 10.1038/s41564-018-0177-8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angrisano F et al. Targeting the Conserved Fusion Loop of HAP2 Inhibits the Transmission of Plasmodium berghei and falciparum. Cell Rep 21, 2868–2878, doi: 10.1016/j.celrep.2017.11.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent RS et al. Inducible developmental reprogramming redefines commitment to sexual development in the malaria parasite Plasmodium berghei. Nat Microbiol 3, 1206–1213, doi: 10.1038/s41564-018-0223-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchi E, Doe B, Goulding D & Wright GJ Juno is the egg Izumo receptor and is essential for mammalian fertilization. Nature 508, 483–+, doi: 10.1038/nature13203 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fedry J et al. The Ancient Gamete Fusogen HAP2 Is a Eukaryotic Class II Fusion Protein. Cell 168, 904–915 e910, doi: 10.1016/j.cell.2017.01.024 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Frenkel J, Vyverman W & Pohnert G Pheromone signaling during sexual reproduction in algae. Plant J 79, 632–644, doi: 10.1111/tpj.12496 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Hakimi MA, Olias P & Sibley LD Toxoplasma Effectors Targeting Host Signaling and Transcription. Clinical Microbiology Reviews 30, 615–645, doi: 10.1128/Cmr.00005-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ho CM et al. Malaria parasite translocon structure and mechanism of effector export. Nature 561, 70–75, doi: 10.1038/s41586-018-0469-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Pei J, Grishin N & Snell WJ The cytoplasmic domain of the gamete membrane fusion protein HAP2 targets the protein to the fusion site in Chlamydomonas and regulates the fusion reaction. Development 142, 962–971, doi: 10.1242/dev.118844 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melicherova J et al. Life cycle of Cryptosporidium muris in two rodents with different responses to parasitization. Parasitology 141, 287–303, doi: 10.1017/S0031182013001637 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Pawlowic MC, Vinayak S, Sateriale A, Brooks CF & Striepen B Generating and Maintaining Transgenic Cryptosporidium parvum Parasites. Curr Protoc Microbiol 46, 20B 22 21–20B 22 32, doi: 10.1002/cpmc.33 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chatterjee A et al. Evidence for Mucin-Like Glycoproteins That Tether Sporozoites of Cryptosporidium parvum to the Inner Surface of the Oocyst Wall. Eukaryotic Cell 9, 84–96, doi: 10.1128/Ec.00288-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bray NL, Pimentel H, Melsted P & Pachter L Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34, 525–527, doi: 10.1038/nbt.3519 (2016). [DOI] [PubMed] [Google Scholar]
- 50.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data generated in this study are available through the GEO database repository under accession number GSE129267. Additional RNA-seq data that support the findings of this study are available through the NCBI Sequence Read Archive BioProject ID: PRJNA374918.