Abstract
The Cyclic-AMP Response Element Binding (CREB) proteins comprise a family of transcription factors that stimulate or repress the expression of a wide variety of genes by binding to nucleotide sequences known as cAMP Response Elements (CREs). CREB-mediated transcription has been implicated in a wide variety of important physiological processes, including long-term memory, and enhancement of CREB signaling has been suggested as an attractive therapeutic strategy for human memory disorders. To identify small molecule compounds that enhance CREB pathway signaling, we have optimized and validated a cell-based β-lactamase reporter gene CREB pathway assay in 1536-well plate format. The LOPAC library of 1280 compounds was screened in triplicate in this assay on a quantitative high throughput screening (qHTS) platform. A variety of compounds which affect known members of the CREB pathway were identified as active, including twelve known phosphodiesterase (PDE) inhibitors, and forskolin, a known activator of adenylate cyclase, thus validating the assay’s performance. This qHTS platform assay will facilitate identification of novel small molecule CREB signaling enhancers, which will be useful for chemical genetic dissection of the CREB pathway and as starting points for potentially memory-enhancing therapeutics.
Introduction
The cyclic-AMP Response Element Binding Protein (CREB) is a key effector in coupling neuronal activation with changes in gene expression required for long-term memory (LTM) formation [1–4]. The transcription factors in the CREB family bind to nucleotide sequences (5′-TGACGTCA) termed cAMP Response Elements (CRE) in the promoters of CREB-regulated genes to activate their transcription. In response to activity of a number of cellular receptors, cAMP is generated and stimulates cAMP-dependent protein kinase (PKA) to translocate to the nucleus, where it phosphorylates CREB at Ser133 [5], causing CREB to bind as a dimer to CRE DNA target sequences [6]. Bound phospho-CREB then recruits the transcriptional co-activators CREB-Binding Protein (CBP) and p300 and assembly of additional proteins into a larger transcriptional complex [7]. This complex promotes chromatin remodeling by means of the histone acetyl transferase activity of CBP. Negative regulation of CREB occurs via protein phosphatases 1 (PP1) and 2A (PP2A) catalyze Ser133 dephosphorylation [8–9], and calcium-calmodulin protein kinase II, which catalyzes phosphorylation of CREB at Ser142, thereby promoting dissociation of the CREB dimer and reducing CREB-mediated gene transcription [10–11]. In addition, CRE signaling is negatively regulated by phosphodiesterases (PDEs), which degrade the cAMP that would otherwise stimulate PKA.
CREB signaling plays a critical role in LTM formation. Over-expression of a CREB transcriptional repressor (dCREB2b) blocked the formation of LTM in transgenic flies [12], and over expression of a CREB-activator isoform enhanced memory in the transgenic flies [13]. Mouse behavioral studies have also demonstrated that CREB is necessary for long-term memory formation [14–15]. On the basis of these studies, it has been suggested that potenitation of CREB signaling pathway might represent an attractive target for memory enhancement [16].
There is a growing interest in the use of cellular pathway assays to identify small molecule modulators of important physiological functions, including memory formation [17]. In the present study we have adapted and optimized a cell-based assay in a 1536-well format for potentiators of the CREB signaling pathway that utilizes a β-lactamase reporter under the control of the cAMP response element (CRE). Using this optimized assay, we have screened the LOPAC (Library of Pharmacologically Active Compounds) collection of 1280 compounds in quantitative high-throughput screening (qHTS) format, which provides concentration-dependent pharmacological information on all compounds directly from the primary screen [18]. The assay was highly reliable and reproducible in 1536-well format. The adenylate cyclase activator, forskolin, and twelve known phosphodiesterase (PDE) inhibitors were found to be active, validating this assay as robust and suitable for identifying novel compounds which enhance the CREB signal transduction pathway from larger screening collections.
Materials and Methods
Cell line and culture conditions
CellSensor™ CRE-bla CHO-K1 cell line, obtained from Invitrogen (Madison, WI), stably expresses a β-lactamase reporter gene under the regulation of a cAMP response element (CRE). Cells were cultured in DMEM medium supplemented with 10% dialyzed fetal bovine serum, 2 mM L-glutamine, 0.1 mM non-essential amino acids, 1mM sodium pyruvate, 25 mM HEPES, 50 U/ml penicillin and 50 μg/ml streptomycin, and 5 μg/ml of blasticidin at 37°C under a humidified atmosphere and 5% CO2.
CRE β-lactamase reporter gene assay
CellSensor™ CRE-bla CHO-K1 cells were suspended in assay medium (DMEM with 1% dialyzed fetal bovine serum, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 25 mM HEPES pH 7.3, 100 U/ml penicillin, and 100 μg/ml streptomycin), and were dispensed at 2000 cells/5 μL/well in 1536-well tissue culture treated black/clear bottom plates (Greiner Bio-One North America, NC) using a Flying Reagent Dispenser (Aurora Discovery, Carlsbad, CA). After the cells were incubated at 37°C overnight, 23 nL of compound or DMSO was transferred into the assay plates by a pin tool (Kalypsys, San Diege, CA) resulting in a 261-fold dilution. One μl of 30 nM NKH 477 (final concentration) was dispensed in all the columns except column 3 which one μl medium was added. The positive control plate format was as follows: Column-1 IBMX concentration-response titration from 2 nM to 67 μM, column-2, IBMX 38 μM +30 nM NKH 477, column 3, DMSO only, and columns 4 to 48, 30 nM NKH 477. The plates were incubated 3 hours at 37°C. One μl of LiveBLAzer™ (Invitrogen) detection mix was added, the plates incubated at room temperature for 2 hrs, and fluorescence intensity at 460 and 530 nm emission was measured at 405 nm excitation by an Envision (Perkin Elmer, Boston, MA) detector. Data was expressed as the ratio of 460nm/530nm emissions.
qHTS and data analysis
Approximately 1280 compounds from Library of Pharmacologically Active Compounds (LOPAC), which was purchased from Sigma (St. Louis, MO), were screened in this qHTS. Compound plates were prepared as interplate titrations of fifteen dilutions with the four left-most columns left empty in each plate. Pin tool transfer of compounds to assay plates resulted in a 261-fold dilution. The final compound concentration in the 6 μl assay volume ranged from 0.5 nM to 38 μM. The primary data analysis was performed as previously described [18]. Briefly, raw plate reads for each titration point were first normalized relative to the IBMX control (38 μM, 100%) in the presence of 30 nM NKH 477 and DMSO only wells (basal, 0%), and then corrected by applying a pattern correction algorithm using compound-free control plates (DMSO plates). Concentration-response titration points for each compound were fitted to the Hill equation yielding concentrations of half-maximal activity (EC50) and maximal response (efficacy) values. The concentration response curves of the compounds were classified into four major classes (1–4) based on the completeness of curve, goodness of fit, and efficacy [18]. The compounds with class 1.1, 1.2, and 2.1 curves are statistically the most reliable, while the compounds with class 2.2 and 3 curves are less reliable. Class 4 compounds which show no concentration response were defined as inactive compounds.
Results and Discussion
Assay validation and miniaturization of CRE β-lactamase reporter gene assay
As a start to identifying small molecule potentiators of the CREB signaling pathway, we conducted experiments to validate the CRE β-lactamase reporter gene assay using CRE-bla CHO-K1 cells expressing β-lactamase under the control of CRE. In 384-well plate format, forskolin, an adenylate cyclase activator, and NKH 477, a water soluble forskolin analogue [19], concentration dependently induced CRE coupled β-lactamase activity after 5 hr incubation with the cells. The EC50s of forskolin and NKH 477 were 0.14 μM and 0.05 μM, respectively (Fig. 1). The signal-to-background ratios were 4.5 and 4.3 for forskolin and NKH 477, respectively. NKH 477 was chosen as a control potentiator in the assay due to its water solubility.
Figure 1.
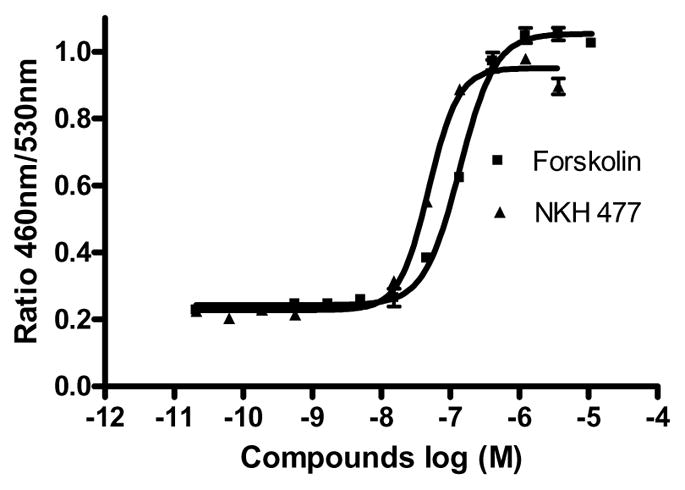
Forskolin and NKH 477 stimulated β-lactamase activity in CRE-bla CHO cells in 384-well format.
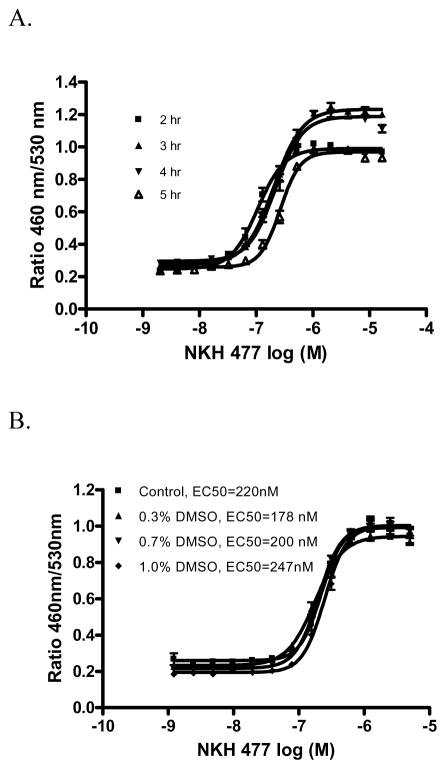
The assay was miniaturized into 1536-well plate format with a final 6 μl assay volume. To optimize the incubation time, the cells were incubated with various concentrations of NKH 477 for 2 to 5 hrs. The EC50s of NKH 477 are 0.11, 0.21, 0.22 and 0.26 μM for 2, 3, 4, and 5 hr incubation, respectively (Fig. 2A). The signal-to-background ratio from 3 hr incubation was 4.7 fold, which was the highest compared to other time points so was chosen for use in the assay. The effect of DMSO on NKH 477 stimulated β-lactamase activity was also evaluated, and showed that DMSO at concentrations up to 1% did not alter NKH 477 stimulated β-lactamase activity (Fig. 2B). Since the final DMSO concentration in the screening conditions chosen was 0.38 %, the assay is clearly tolerant of DMSO in the compound solution.
Figure 2.
NKH 477 stimulated β-lactamase activity in CRE-bla CHO cells in 1536-well format. A. Time course of NKH 477 stimulated β-lactamase activity in CRE-bla CHO cells in 1536-well format. B. DMSO effect on the NKH 477 stimulated β-lactamase activity assay
Assay optimization in potentiator screening mode
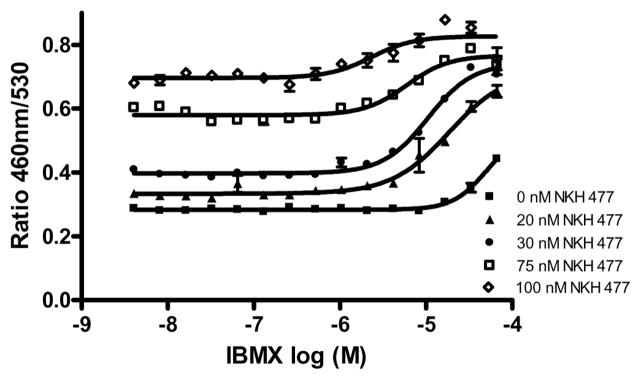
It has been reported that the over expression of CREB alone had no effect on memory enhancement, but the synergetic effect on CREB signaling enhanced LTM [3]. Thus, the potentiator of CREB function might be a better approach to enhance LTM. We have optimized a cell-based assay for compound screening in the presence of small amount of NKH477, an activator of CREB signaling. To find an appropriate concentration of NKH 477 that could be used to capture the response of potentiators without a significant reduction in the signal-to-background ratio, the concentration response of 3-Isobutyl-1-methylxanthine (IBMX) [20], a known PDE inhibitor, was determined in the presence of several different sub-EC50 concentrations of NKH 477. Fig. 3 showed IBMX concentration-dependently increased β-lactamase activity in the presence of NKH 477. The highest signal to background ratio of approximately 2 fold was observed in the presence of 30 nM of NKH 477. The EC50 of IBMX in the present of 30 nM NKH 477 was 10.5 μM. In the screen 30 nM of NKH477 was used to stimulate a low level of cAMP production and IBMX was chosen as a positive control.
Figure 3.
The concentration response curves of IBMX in the presence of indicated concentrations of NKH 477. CRE-bla CHO cells were incubated with various concentrations of IBMX in the presence of indicated concentrations of NKH 477 for 3 hours, and then the β-lactamase activity was measured.
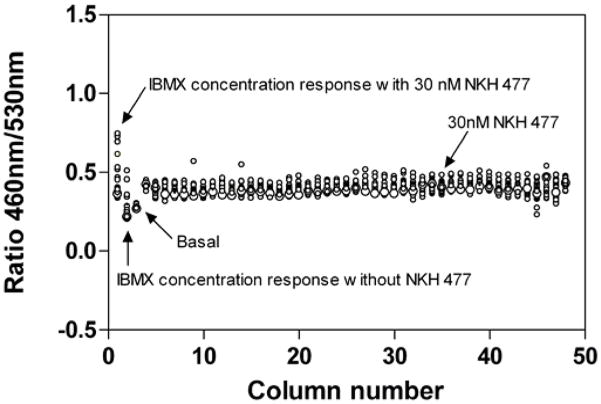
To evaluate the assay performance, DMSO plate without compounds in the presence of 30 nM NKH 477 was tested in 1536-well plate format. The signal to background ratio (S/B) was 2.0 fold, CV (coefficient of variation) was 8.6% and Z factor was 0.5 indicating that this β-lactamase assay in a 1536-well format was robust and ready for high throughput screening (Fig. 4).
Figure 4.
Screening test of β-lactamase reporter gene assay in CRE-bla CHO cells in 1536-well format. Column-1 is a concentration response curve of IBMX, the known potentiator in duplicate, in the presence of 30 nM NKH 477. Column-2 is a concentration response curve of IBMX in the absence of NKH 477. Column-3 was the negative control with assay buffer only. Column-4 to 48 was DMSO with 30 nM NKH 477.
Assay reproducibility and data quality in qHTS
To test the data quality on each plate screened in the qHTS process, the concentration titration of IBMX, the positive control compound, was performed in each plate. The concentration response curves of IBMX from 51 plates seem to reproduce well in each plate, with an average EC50 value of 11 ± 2 μM. The average signal to background ratio from 51 plates was 2.7 ± 0.24 and average CV (%) was 7 ± 4. The average Z′ factor and Z factor were 0.64 ± 0.07 and 0.60 ± 0.07. The high data quality with small inter-plate variation obtained from this screening could be accredited to the use of high quality liquid handlers, plate readers and the robust nature of this β-lactamase reporter gene assay. This ensured the quality of results from the inter-plate compound dilutions used in the qHTS campaign.
To minimize false positives and false negatives that are common with single-concentration screening paradigms, and obtain immediate information on compound potencies and efficacies, we screened the compounds in 7 different concentrations ranging from 0.5 nM to 38 μM in quantitative high-throughput screening (qHTS) format [18]. The qHTS analysis and concentration-response curve classification schema has been published previously [18]. In this study, compounds producing curve classes 1.1, 1.2, or 2.1 were defined as active, those producing class 2.2 or 3 curves were defined as inconclusive, and compounds producing class 4 curves defined as inactive.
To evaluate assay robustness, traditional Z′- and Z factors [21], coefficients of variation (CV), and signal-to-background ratio were used, but were interpreted somewhat differently given the nature of the concentration-response data generated by qHTS. In traditional HTS in which compounds are tested at a single concentration, Z factors less than 0.5 or CV (over 10%) are frequently used as cut-offs for acceptable assays, since they indicate assay performance below which compound activities cannot be reliably determined. However, we have found that the Z factor and CV are less important in qHTS comparing to the single concentration screening because the hits are selected based on the significance of concentration-response of the compounds. In this qHTS, robust and reproducible results were obtained though the Z factor was 0.41 at the 38 μM compound concentration, and CVs were 12% and 18% at the 7.6 and 38 μM compound concentrations – both outside the usually acceptable HTS parameters. This illustrates a general feature of qHTS, which is that it allows screening of assays that would be unscreenable in traditional HTS paradigms.
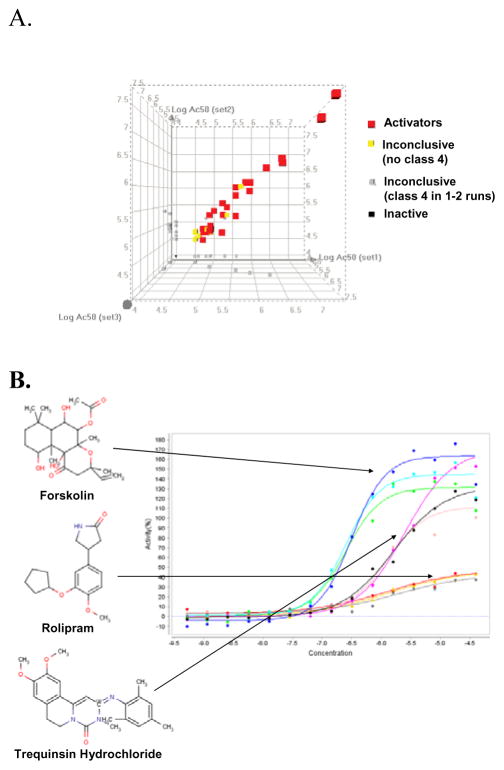
To evaluate data reproducibility, the LOPAC library was screened independently in CRE-bla CHO cells on three separate days. Of these 1280 compounds, 44 compounds (3.4%) showed a concentration dependent response (curve classes 1–3) in all three independent runs. The EC50 values from the three runs correlated well with an average R2 of 0.94 (Fig 5A). Example replicate concentration-response curves for some of these compounds are shown in figure 5B. 1185 compounds were inactive in all three runs. Twenty-five compounds showed activity in only one or two of the three runs, but the curve classes of these compounds were mostly class 2.2 or 3, indicating a lower degree of data reliability.
Figure 5.
qHTS reproducibility of β-lactamase reporter gene assay. (A) LOPAC library was screened independently in CRE-bla CHO cells on three separate times. Linear correlations of EC50s from 48 compounds with curve classes 1–3 in three independent screening yielded an average R2 = 0.94 ± 0.02 (0.95, run 1 vs run 2; 0.94, run 1 vs run 3; and 0.92, run 2 vs run 3). 47 compounds were inactive in one or two runs and are colored gray in the figure. 1185 compounds were inactive in all three runs. (B) Examples of triplicate concentration-response curves for some of known PDE inhibitors, forskolin, rolipram and trequinsin hydrochloride, identified from the LOPAC library.
Identification of potentiators of the CREB pathway
The triple LOPAC library screen revealed 44 potentiators of the CREB pathway. The relatively high active rate (3.4%) is likely due to both the nature of the LOPAC library comprised of pharmacologically active compounds, and the nature of the assay, which detects compounds active against any molecular target in the CREB pathway. Among the 44 compounds, twelve were known PDE inhibitors, including papverine [22], trequinsin [23], Ro20-1724 [23–24], rolipram [23–24], zardaverine [24], diprydamole [23], IBMX, methoxymethyl-IBMX, etazolate [25], ibudilast [26], T-1032 [27], and propentofylline [28]. Inhibition of PDE leads to a decrease in the cAMP degradation, which enhances CREB signaling. A direct activator of adenylate cyclase, forskolin, and two cAMP analogues, 8-bromo-cAMP and 8-(4-chlorophenylthio)-cAMP were also found active. The EC50 values of these known compounds from the screen are listed in Table 1. These data suggest that this CRE qHTS assay is robust and can be used to identify small molecule compounds that potentiate CREB signaling from large compound libraries.
Table 1.
Summary of compound potencies (μM) with known activity in the CREB pathway identified from screening of the LOPAC library in qHTS
| Name | First Run | Second Run | Third Run | Mean ± SD |
|---|---|---|---|---|
| Forskolin | 0.32 | 0.25 | 0.25 | 0.27±0.04 |
| 8-(4-Chlorophenylthio)-cAMP | 25.12 | 25.12 | 15.85 | 22.03±5.35 |
| 8-Bromo-cAMP | 25.12 | 15.85 | 25.12 | 22.03±5.35 |
| Papaverine hydrochloride | 1.58 | 0.79 | 0.79 | 1.06±0.46 |
| Trequinsin (HL-725) | 2.51 | 1.58 | 1.26 | 1.79±0.65 |
| Rolipram | 1.26 | 1.26 | 2.00 | 1.50±0.43 |
| Ro 20-1724 | 5.01 | 12.59 | 2.00 | 6.53±5.46 |
| Etazolate hydrochloride | 12.59 | 15.85 | 15.85 | 14.76±1.88 |
| Ibudilast | 10.00 | 12.59 | 12.59 | 11.73±1.49 |
| Tracazolate | 5.01 | 5.01 | 5.01 | 5.01±0.00 |
| T-1032 | 12.59 | 12.59 | 12.59 | 12.59±0.00 |
| 3-Isobutyl-1-methylxanthine | 15.85 | 15.85 | 15.85 | 15.85±0.00 |
| 8-Methoxymethyl-3-isobutyl-1-methylxanthine | 25.12 | 15.85 | 15.85 | 18.94±5.35 |
| Propentofylline | 25.12 | 25.12 | 15.85 | 22.03±5.35 |
| Zardaverine | 10.00 | 15.85 | 12.59 | 12.81±2.93 |
Conclusion
We describe the development and validation of cell-based CRE β-lactamase reporter gene assay for the screening of potentiators of the CREB signaling pathway in a qHTS format. This cell-based assay has been miniaturized into a 1536-well format and is suitable for large scale library screening [29]. The data from the qHTS allowed us to quickly and efficiently evaluate compound potencies and efficacies. From the LOPAC library, we identified compounds known to act on multiple different targets within the CREB pathway, including PDE inhibitors, activators of adenylate cyclase, and cAMP analogues. These data suggest that a larger compound screen may identify compounds that enhance CREB signaling via a variety of mechanisms of action, including action on previously unidentified members of this important pathway.
Acknowledgments
We thank Sam Michael, Carleen Klumpp, and Paul Shinn for assistance with automated screening and compound management. This research was supported by the Molecular Libraries Initiative of the NIH Roadmap for Medical Research and the Intramural Research Program of the National Human Genome Research Institute and National Heart, Lung, and Blood Institute, National Institutes of Health.
References
- 1.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–45. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 2.Josselyn SA, Nguyen PV. CREB, synapses and memory disorders: past progress and future challenges. Curr Drug Targets CNS Neurol Disord. 2005;4:481–97. doi: 10.2174/156800705774322058. [DOI] [PubMed] [Google Scholar]
- 3.Tully T, Bourtchouladze R, Scott R, Tallman J. Targeting the CREB pathway for memory enhancers. Nat Rev Drug Discov. 2003;2:267–77. doi: 10.1038/nrd1061. [DOI] [PubMed] [Google Scholar]
- 4.Cooke SF, Bliss TV. Long-term potentiation and cognitive drug discovery. Curr Opin Investig Drugs. 2005;6:25–34. [PubMed] [Google Scholar]
- 5.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–80. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 6.Yamamoto KK, Gonzalez GA, Biggs WH, 3rd, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–8. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- 7.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–9. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 8.Hagiwara M, Alberts A, Brindle P, Meinkoth J, Feramisco J, Deng T, Karin M, Shenolikar S, Montminy M. Transcriptional attenuation following cAMP induction requires PP-1-mediated dephosphorylation of CREB. Cell. 1992;70:105–13. doi: 10.1016/0092-8674(92)90537-m. [DOI] [PubMed] [Google Scholar]
- 9.Wadzinski BE, Wheat WH, Jaspers S, Peruski LF, Jr, Lickteig RL, Johnson GL, Klemm DJ. Nuclear protein phosphatase 2A dephosphorylates protein kinase A-phosphorylated CREB and regulates CREB transcriptional stimulation. Mol Cell Biol. 1993;13:2822–34. doi: 10.1128/mcb.13.5.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthews RP, Guthrie CR, Wailes LM, Zhao X, Means AR, McKnight GS. Calcium/calmodulin-dependent protein kinase types II and IV differentially regulate CREB-dependent gene expression. Mol Cell Biol. 1994;14:6107–16. doi: 10.1128/mcb.14.9.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, McMurray CT. Calmodulin kinase II attenuation of gene transcription by preventing cAMP response element-binding protein (CREB) dimerization and binding of the CREB-binding protein. J Biol Chem. 2001;276:1735–41. doi: 10.1074/jbc.M006727200. [DOI] [PubMed] [Google Scholar]
- 12.Yin JC, Wallach JS, Del Vecchio M, Wilder EL, Zhou H, Quinn WG, Tully T. Induction of a dominant negative CREB transgene specifically blocks long-term memory in Drosophila. Cell. 1994;79:49–58. doi: 10.1016/0092-8674(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 13.Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: Induced expression of dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 14.Pittenger C, Huang YY, Paletzki RF, Bourtchouladze R, Scanlin H, Vronskaya S, Kandel ER. Reversible inhibition of CREB/ATF transcription factors in region CA1 of the dorsal hippocampus disrupts hippocampus-dependent spatial memory. Neuron. 2002;34:447–462. doi: 10.1016/s0896-6273(02)00684-0. [DOI] [PubMed] [Google Scholar]
- 15.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 16.Jackson T, Ramaswami M. Prospects of memory-modifying drugs that target the CREB pathway. Curr Opin Drug Discov Devel. 2003;6:712–719. [PubMed] [Google Scholar]
- 17.Scott R, Bourtchuladze R, Gossweiler S, Dubnau J, Tully T. CREB and the discovery of cognitive enhancers. J Mol Neurosci. 2002;19:171–7. doi: 10.1007/s12031-002-0029-z. [DOI] [PubMed] [Google Scholar]
- 18.Inglese J, Auld DS, Jadhav A, Johnson RL, Simeonov A, Yasgar A, Zheng W, Austin CP. Quantitative high-throughput screening: a titration-based approach that efficiently identifies biological activities in large chemical libraries. Proc Natl Acad Sci USA. 2006;103:11473–11478. doi: 10.1073/pnas.0604348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morinobu S, Fujimaki K, Okuyama N, Takahashi M, Duman RS. Stimulation of adenylyl cyclase and induction of brain-derived neurotrophic factor and TrkB mRNA by NKH477, a novel and potent forskolin derivative. J Neurochem. 1999;72:2198–205. doi: 10.1046/j.1471-4159.1999.0722198.x. [DOI] [PubMed] [Google Scholar]
- 20.Wagner B, Jakobs S, Habermeyer M, Hippe F, Cho-Chung YS, Eisenbrand G, Marko D. 7-Benzylamino-6-chloro-2-piperazino-4-pyrrolidino-pteridine, a potent inhibitor of cAMP-specific phosphodiesterase, enhancing nuclear protein binding to the CRE consensus sequence in human tumour cells. Biochemical Pharmacology. 2002;63:659–668. doi: 10.1016/s0006-2952(01)00893-0. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomolecular Screening. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Menniti FS, Chappie TA, Humphrey JM, Schmidt CJ. Phosphodiesterase 10A inhibitors: a novel approach to the treatment of the symptoms of schizophrenia. Current Opinion in Investigational Drugs. 2007;8:54–59. [PubMed] [Google Scholar]
- 23.Lugnier C. Cyclic nucleotide phosphodiesterase (PDE) superfamily. A new target for the development of specific therapeutic agents. Pharmacology & Therapeutics. 2006;109:366–98. doi: 10.1016/j.pharmthera.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Teixeira Mauro M, Gristwood Robert W, Cooper Nicola, Hellewell Paul G. Phosphodiesterase (PDE)4 inhibitors: anti-inflammatory drugs of the future? Trends in Pharmacological Sciences. 1997;18:164–170. doi: 10.1016/s0165-6147(97)01049-3. [DOI] [PubMed] [Google Scholar]
- 25.Chasin M, Harris DN, Phillips MS, Hess SM. 1-Ethyl-4-(isopropylidenehydrazinol-H-pyrazolo-(3,4-b)-pyridin-5-carboxylic acid, ethyl ester, hydrochloride (SQ 20009) - potent new inhibitor of cyclic 3′5-nucleotide phosphodiesterase. Biochem Pharmacol. 1972;21:2443–50. doi: 10.1016/0006-2952(72)90414-5. [DOI] [PubMed] [Google Scholar]
- 26.Kishi Y, Ohta S, Kasuya N, Sakita S, Ashikaga T, Isobe M. Ibudilast: a non-selective PDE inhibitor with multiple actions on blood cells and the vascular wall. Cardiovasc Drug Rev. 2001;19:215–25. doi: 10.1111/j.1527-3466.2001.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 27.Inoue H, Koji Yano K, Noto T, Takagi M, Ikeo T, Kikkawa K. Acute and Chronic Effects of T-1032, a Novel Selective Phosphodiesterase Type 5 Inhibitor, on Monocrotaline-Induced Pulmonary Hypertension in Rats. Biol Pharm Bull. 2002;25:1422–6. doi: 10.1248/bpb.25.1422. [DOI] [PubMed] [Google Scholar]
- 28.Meskini N, Némoz G, Okyayuz-Baklouti I, Lagarde M, Prigent AF. Phosphodiesterase inhibitory profile of some related xanthine derivatives pharmacologically active on the peripheral microcirculation. Biochem Pharmacol. 1994;47:781–8. doi: 10.1016/0006-2952(94)90477-4. [DOI] [PubMed] [Google Scholar]
- 29.Xia M, Ruili Huang R, Guo V, Southall N, Cho M, Inglese J, Austin CP, Marshall N. Identification of compounds that potentiate CREB signaling as possible enhancers of long-term memory. Proc Natl Acad Sci USA. 2009;106:2412–17. doi: 10.1073/pnas.0813020106. [DOI] [PMC free article] [PubMed] [Google Scholar]