Abstract
BACKGROUND AND PURPOSE:
Susceptibility-weighted imaging (SWI) is valuable for characterization of intracranial hemorrhage and mineralization, but suffers from long acquisition times. We compared a highly-accelerated Wave-CAIPI SWI sequence (Wave-SWI) with two commonly used alternatives, standard SWI and T2*-weighted Gradient-Echo (T2*w-GRE), for routine clinical brain imaging at 3T.
MATERIALS AND METHODS:
246 consecutive adult patients were prospectively evaluated using a conventional susceptibility-weighted sequence (SWI or T2*w-GRE) and an optimized Wave-SWI sequence that was 3- to 5-times faster than the standard sequence. Two blinded radiologists scored each sequence for the presence of hemorrhage, number of microhemorrhages, and severity of motion artifacts. Wave-SWI was then evaluated in head-to-head comparison to the conventional sequences for visualization of pathology, artifacts, and overall diagnostic quality. Forced choice comparisons were used for all scores. Wave-SWI was tested for superiority relative to T2*w-GRE, and for non-inferiority relative to standard SWI using a 15% non-inferiority margin.
RESULTS:
Compared to T2*w-GRE, Wave-SWI detected hemorrhage in more cases (P<0.001) and detected more microhemorrhages (P<0.001). Wave-SWI was superior to T2*w-GRE for visualization of pathology, artifacts, and overall diagnostic quality (all P<0.001). Compared to standard SWI, Wave-SWI showed no difference in the presence or number of hemorrhages identified. Wave-SWI was non-inferior to standard SWI for visualization of pathology (P<0.001), artifacts (P<0.01), and overall diagnostic quality (P<0.01). Motion was less severe with Wave-SWI than standard SWI (P<0.01).
CONCLUSIONS:
Wave-SWI provided superior visualization of pathology and overall diagnostic quality compared to T2*w-GRE, and was non-inferior to standard SWI with reduced scan time and reduced motion artifact.
Introduction
Susceptibility-weighted imaging (SWI) is widely applied for clinical brain imaging due to its exquisite sensitivity for the detection and characterization of blood products, superior to that of conventional T2*-weighted gradient-echo (T2*w-GRE) imaging.1 SWI is useful in the evaluation of a broad range of pathology including vascular malformations, hemorrhagic or calcified neoplasms, vasculopathies, and neurodegenerative disorders associated with mineralization or brain iron accumulation.2,3 However, conventional SWI is associated with long acquisition times (typically 5 minutes or more),1 which may contribute to motion artifacts4 and patient anxiety.5
Wave-CAIPI (controlled aliasing in parallel imaging) is a rapid acquisition approach that combines a corkscrew gradient trajectory with CAIPI shifts in the ky and kz directions to efficiently encode k-space and uniformly spread the voxel aliasing, taking full advantage of the 3D coil sensitivity information to provide high acceleration factors with negligible artifact and g-factor penalty.6,7 SWI is well-suited for this aggressive acceleration strategy due to the intrinsically high contrast between the pathology of interest (which manifests as a signal void) and background brain tissue. The resulting decrease in acquisition time may facilitate broader clinical application of SWI, especially in motion-prone populations (e.g., children, elderly, and acutely ill patients). Wave-CAIPI has shown the potential to accelerate susceptibility-weighted acquisitions in healthy volunteers,8,9 but has not been systematically evaluated in a clinical setting.
The goal of this study was to compare a highly-accelerated SWI sequence based on Wave-CAIPI (Wave-SWI) to two commonly used alternatives, conventional 3D SWI and 2D T2*w-GRE. We hypothesized that Wave-SWI would be superior to T2*w-GRE for visualization of pathology and overall diagnostic quality in a similar scan time, and non-inferior to the standard SWI sequence with a 3- to 5-fold reduction in scan time.
Materials and Methods
Subjects and Study Design
A prospective comparative study was performed at a single institution. Consecutive adult patients (age > 19 years; N = 246) undergoing clinical brain MRI were prospectively evaluated, including both inpatient and outpatient exams. Imaging was performed on a 3T MAGNETOM Prisma scanner from March to June 2018, and on a 3T MAGNETOM Skyra MRI scanner from May to June 2018 (Siemens Healthcare GmbH, Erlangen, Germany). There were no exclusion criteria beyond those for routine clinical MR imaging. The study was Health Insurance Portability and Accountability Act (HIPAA) compliant and approved by our Institutional Review Board (IRB). Verbal consent was obtained prior to MRI scanning. Written consent was waived by the IRB. Demographics of the study subjects and clinical indications for MRI scanning are shown in Online Table 1.
Wave-CAIPI SWI Pulse Sequence and Reconstruction
Wave-SWI was implemented using a work-in-progress dual-echo 3D gradient-echo pulse sequence6 (WIP1058C, Siemens Healthcare GmbH, Erlangen, Germany). On-line reconstruction was performed using an auto-calibrated procedure for simultaneous estimation of the parallel imaging reconstruction and true k-space trajectory (which accounts for potential gradient hardware imperfections),7 with reconstruction time of approximately 60 seconds. This included phase unwrapping of the reconstructed multi-echo data and a weighted combination that accounted for the TE phase evolution. The standard vendor SWI processing was then performed to produce high-pass filtered phase images and associated SWI images. Pulse sequence parameters could not be exactly matched between the Wave-SWI and standard SWI sequences due to vendor constraints on the available parameter options, but were approximated as closely as possible within the allowable range of parameter values (Online Table 2).
MR Imaging Protocol
Each patient underwent brain MRI on one of two 3T MRI scanners using commercially available 20- and 32-channel receiver coil arrays (Siemens Healthcare GmbH, Erlangen, Germany). Standard institutional brain MRI protocols were selected by the radiologist based on the provided clinical indication. Each scan included a conventional magnetic susceptibility-weighted sequence (either standard SWI or T2*w-GRE, decided at the discretion of the protocoling radiologist based on the clinical indication), and a highly-accelerated Wave-SWI sequence performed immediately before or after the conventional susceptibility-weighted sequence. Acceleration factors of R=3×2 and R=3×3 were chosen on the 20-channel and 32-channel coils respectively to balance scan time with acceptable SNR for each coil configuration.10 The acquisition order alternated on a weekly basis to control for the possible effect of increased patient motion on the later sequence. A summary of the magnetic susceptibility-weighted sequence parameters is provided in Online Table 2.
Image Evaluation
A semi-quantitative grading system based on predetermined criteria was used to compare Wave-SWI with the conventional susceptibility-weighted sequence acquired for each patient (standard SWI or T2*w-GRE). The DICOM datasets were anonymized and transferred to an independent workstation. Blinded to patient information and protocol type, two neuroradiologists (O.R. and S.Y.H.) with 17 and 7 years of experience, respectively, independently reviewed all images in randomized order. To obtain optimal visualization, adjustments of window widths and levels were allowed. Only the magnetic susceptibility-weighted sequences were evaluated.
To provide a comprehensive assessment of the diagnostic performance of the Wave-SWI and conventional susceptibility-weighted sequences, the review sessions were divided into (a) an individual image series analysis (“individual analysis”), where images obtained using each sequence were presented in isolation, and (b) a head-to-head image series analysis (“head-to-head analysis”), where images obtained using the Wave-SWI and standard magnetic susceptibility-weighted sequence were presented side-by-side with randomly selected left and right screen positions. The individual analysis was conducted in two sessions, such that only one sequence per patient was presented to the reviewer in each session. The individual analysis sessions were separated by at least 2 weeks to minimize memory bias.
In the individual analysis, reviewers evaluated: the presence of hemorrhage; the number and location of the cerebral microhemorrhages (if present); the degree of motion; and whether the images were of diagnostic quality (yes or no). Motion was scored on a predefined 4-point scale. To minimize subjectivity, representative images of each motion score were available to reviewers during the evaluation (Supplemental Figure). Microhemorrhages were defined according to the Microbleed Anatomical Rating Scale (MARS)11 and counted by a single reviewer, excluding cases with gross structural lesions (e.g., large parenchymal hematoma, infarct or surgical cavity) that may limit the reliability of the microhemorrhage counts. A subset of 20 randomly selected cases were evaluated by a second reviewer to determine inter-rater reliability of the microhemorrhage counts.
In the head-to-head analysis, all cases in which either reviewer identified an abnormality on either of the two conventional susceptibility sequences in the individual analysis were presented to the reviewers in a separate blinded review, with the Wave-SWI and conventional susceptibility-weighted sequence simultaneously displayed on the left and right half of the screen labeled ‘image A’ and ‘image B’. The order of the cases and the screen position of the sequences were randomized. The raters compared and scored the two sequences for each of the following variables: visualization of pathology (primary outcome); presence of artifacts (including motion, signal dropout, and parallel imaging artifacts); and overall diagnostic quality. A predefined 5-point scale was used, where positive numbers favored the sequence on the right and negative numbers favored the sequence on the left side of the screen (Online Table 3). Disagreements between readers were adjudicated by a third neuroradiologist (P.W.S.) with over 20 years of experience.
Statistical analysis
In the individual analysis, we used the McNemar test for comparison of dichotomous variables and the Wilcoxon signed-rank test for comparison of ordinal variables between sequences. For motion grading, scores of 0 and 1 (i.e., “no motion” and “mild motion that is not clinically significant”) were combined in a single category, since this distinction was not clinically meaningful. In the head-to-head analysis, we tested for superiority of Wave-SWI compared to T2*w-GRE, and for non-inferiority of Wave-SWI compared to standard SWI. This approach was selected a priori based on our hypothesis that the Wave-SWI images were comparable to the standard SWI but superior to the T2*w-GRE images. For superiority testing, we compared the ordinal radiologist scores using the Wilcoxon signed-rank test, with the null hypothesis (H0) of no difference between sequences. For non-inferiority testing,12 a non-inferiority margin (Δ) of 15% was chosen, with the null hypothesis (H0) that the proportion of cases where standard SWI was preferred over Wave-SWI was > 15%. We used the Z statistic to calculate the probability of the standard sequence being preferred over the Wave-SWI sequence in more than 15% of cases (H0>Δ), with a type 1 error rate (α) of 0.05. We also calculated the upper bound of the 95% confidence interval for the proportion of cases where standard SWI was preferred over Wave-SWI, i.e., the critical value, Pcritical.13 The required sample size was estimated as described by Cohen14 for a single proportion (the proportion of cases where visualization of pathology was preferred on standard SWI over Wave-SWI), for an effect size of 0.15, a type 1 error rate (α) of 0.05, a power (1‒β) of 0.90. According to this calculation, a minimum of 63 abnormal cases was required. For dichotomous variables, inter-rater agreement was evalauted using the unweighted Cohen κ coefficient. For ordinal variables, inter-rater agreement was reported using the quadratically weighted Cohen κ, to disproportionately penalize larger disagreements. For the numerical microhemorrhage counts, inter-rater agreement was reported using the intraclass correlation coefficient (ICC). Agreement was interpreted according to Landis and Koch.15 We applied a Bonferroni correction for 5 comparisons (presence of hemorrhage, motion artifact, visualization of pathology, artifacts, and overall diagnostic quality), with a corrected threshold for statistical significance of 0.05 / 5 = 0.01. We also performed exploratory univariate testing evaluate for the possible effect of age and study indication on motion scores. For age, we calculated the Pearson correlation coefficient between patient age and motion score. For indication, we performed a multinomial logistic regression with the indication as the independent variable and motion score as the dependent variable. All statistical calculations were performed using R version 3.4.3.
Results
Wave-SWI images were successfully acquired, reconstructed and evaluated in all 246 cases, including 139 (56.5%) with comparison to T2*w-GRE (Figure 1) and 107 (43.5%) with comparison to standard SWI (Figure 2). Abnormalities were identified in 85 cases with comparison to T2*w-GRE and 65 cases with comparison to standard SWI, which were included in the head-to-head analysis. In the individual analysis, inter-rater agreement ranged from moderate to almost perfect (κ = 0.85 [0.80 – 0.89] for presence of hemorrhage, 0.52 for motion [0.13 – 0.90], and 0.49 [0.35 – 0.63] for diagnostic quality, where square parentheses indicate the 95% confidence interval). Inter-rater agreement for microhemorrhage counts according to the MARS was almost perfect (ICC = 0.84 [0.64 – 0.93] for infratentorial, 0.95 [0.89 – 0.98] for deep, and 0.98 [0.96 – 0.99] for lobar microhemorrhages). In the head-to-head analysis, inter-rater agreement ranged from moderate to substantial (κ = 0.73 [0.44 – 1.00] for evaluation of pathology, 0.63 [0.37 – 0.93] for artifacts, 0.47 [0.01 – 1.00] for diagnostic quality. There was a weak positive correlation between patient age and motion score (r = 0.289, P < 0.01). There was no significant effect of clinical indication on motion score.
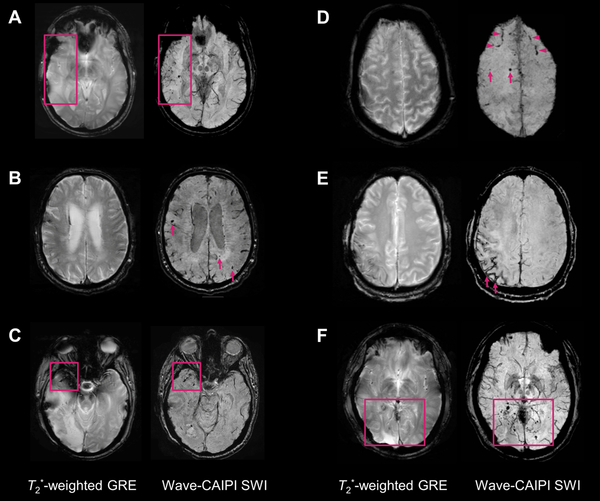
Figure 1.
Representative images comparing T2*-weighted gradient-echo (T2*w-GRE) and Wave-CAIPI susceptibility-weighted imaging (Wave-SWI). A. Small hemorrhagic foci in the right temporal lobe are clearly seen on Wave-SWI but not well visualized on T2*w-GRE. These abnormalities fall within the radiation field of a treated right temporal lobe oligoastrocytoma, and are presumed to reflect sequelae of radiation-induced vasculopathy. B. Multiple cerebral microhemorrhages in a 72 year old man with history of cardiopulmonary bypass for aortic valve replacement and ascending aortic aneurysm repair, some of which are better seen on Wave-SWI and some of which are only seen on Wave-SWI (arrows). C. Hemorrhagic foci within a right anterior temporal lobe glioblastoma are clearly visualized on Wave-SWI, but obscured on T2*w-GRE due to signal dropout artifact on the GRE sequence. D. Scattered foci of parenchymal (arrows) and subarachnoid (arrowheads) hemorrhage in the bilateral frontal lobes are better visualized on Wave-SWI than T2*w-GRE. E. Superficial hemosiderosis in the right frontoparietal region is better visualized on Wave-SWI than T2*wGRE (arrows), in a patient with history of multiple craniotomies for a recurrent anaplastic astrocytoma. F. Scattered foci of posterior predominant susceptibility effect are well visualized on Wave-SWI and not well seen on T2*w-GRE (box), in a 58 year old man with remote radiotherapy to a posterior fossa atypical meningioma. The findings were presumed to reflect post-radiation changes (microhemorrhages and/or small cavernous malformations).
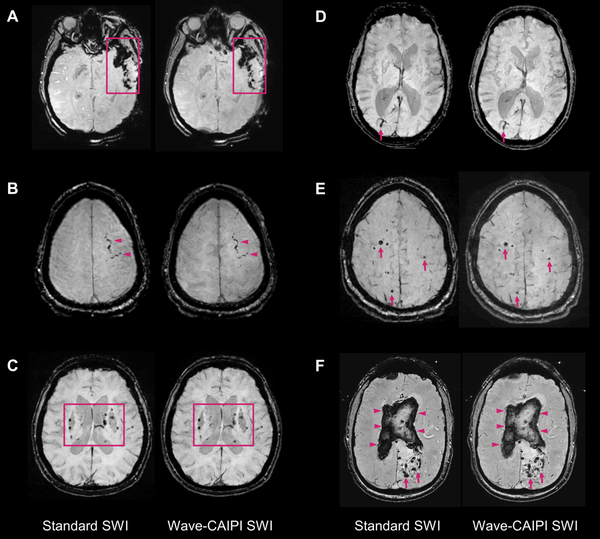
Figure 2.
Representative images comparing standard susceptibility-weighting imaging (SWI) and Wave-CAIPI SWI (Wave-SWI). A. Extensive susceptibility effect in the left temporal region in a 33 year old woman with history of trauma, corresponding to a combination of parenchymal contusion, subdural and subarachnoid hemorrhage. B. Focal subarachnoid hemorrhage in the left superior frontal sulcus (arrowheads). C. Scattered microhemorrhages throughout the bilateral basal ganglia in a patient with chronic poorly controlled hypertension. D. Incidental finding of a right occipital lobe developmental venous anomaly (arrows). E. Scattered foci of susceptibility effect in a patient with familial multiple cavernous malformations (arrows). F. Serpiginous foci of susceptibility effect in the left occipital region corresponding to an arteriovenous malformation (arrows), with associated rupture and extensive diffuse intraventricular hemorrhage (arrowheads). In all cases, visualization of the pathology was rated equivalent (score of 0, see Online Table 3) by both interpreting radiologists.
T2*w-GRE × Wave-SWI Comparison
In the individual analysis, hemorrhage was detected more frequently using Wave-SWI than T2*w-GRE (P<0.001, Online Table 4). In 21 cases (15.1%), hemorrhage was identified only on the Wave-SWI sequence. These cases included 15 cases of microhemorrhage, three cases of parenchymal hemorrhage (>10 mm), two cases of post-operative extra-axial hemorrhage, and one case of intraventricular hemorrhage. More microhemorrhages were detected using Wave-SWI compared to T2*w-GRE (P < 0.01 for infratentorial, P = 0.01 for deep, and P < 0.001 for lobar microhemorrhages, Online Table 4). Motion artifacts were rated less severe on T2*w-GRE compared to Wave-SWI (P<0.001, Online Table 4); however, there was no significant difference in the number of cases that were rated as non-diagnostic (7 cases with T2*w-GRE versus 6 cases with Wave-SWI).
The results of the head-to-head comparison of Wave-SWI and T2*w-GRE are shown in Figure 3. Wave-SWI was rated superior to T2*w-GRE for visualization of pathology, presence of artifacts, overall diagnostic quality, and visualization of normal anatomic structures (all P<0.001).
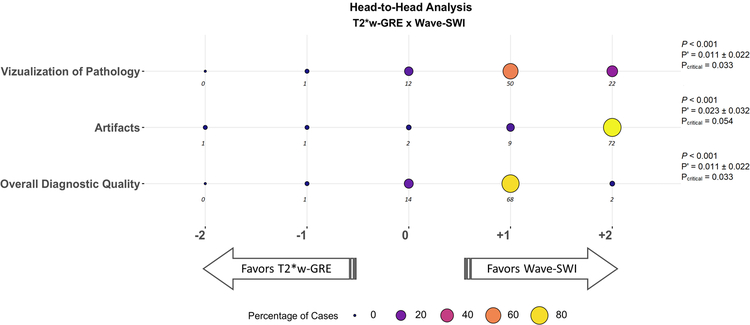
Figure 3.
Balloon plot showing the results of the head-to-head comparison of T2*-weighted gradient-echo (T2*w-GRE) and Wave-CAIPI susceptibility-weighted imaging (Wave-SWI). The size and color of each circle represents the percentage of cases that were assigned a given score, from a total of 85 abnormal cases. The actual number of cases receiving a given score is indicated below each circle. Negative scores (left) favor T2*w-GRE, and positive scores (right) favor Wave-SWI. The proportion of cases where T2*w-GRE was preferred over Wave-SWI (P’) and the 95% confidence interval for this proportion are indicated at the right of the figure. The critical value (Pcritical) is also provided, corresponding to the upper bound of the 95% confidence interval for P’. Superiority testing was performed as described in the methods, and the corresponding P-values are shown in the figure. Wave-SWI was superior to T2*w-GRE for the three variables evaluated.
Standard SWI × Wave-SWI Comparison
In the individual analysis, there was no significant difference in the presence or number of hemorrhages identified on the Wave-SWI and standard SWI sequences (Online Table 4). Motion artifacts were rated less severe with Wave-SWI compared to standard SWI (P<0.01), with twice as many cases receiving a motion score of 3 (severe motion that may obscure major findings) on the standard SWI sequence (21 cases, 19.6%) compared to the Wave-SWI sequence (9 cases, 9.3%). More cases were rated non-diagnostic on standard-SWI (9 cases, 8.4%) than Wave-SWI (3 cases, 2.8%), however the difference was not statistically significance (P=0.08).
The results of the head-to-head comparison and the associated non-inferiority testing are shown in Figure 4. Wave-SWI was non-inferior to standard SWI for visualization of pathology (P<0.001), artifacts (P<0.01), and overall diagnostic quality (P<0.01) with a 15% non-inferiority margin. The critical value for each non-inferiority test is shown in Figure 4, corresponding to the upper bound of a 95% confidence interval on the proportion of cases in which standard-SWI is preferred over Wave-SWI. This upper bound ranged from 4% for visualization of pathology to 11% for artifacts and overall diagnostic quality.
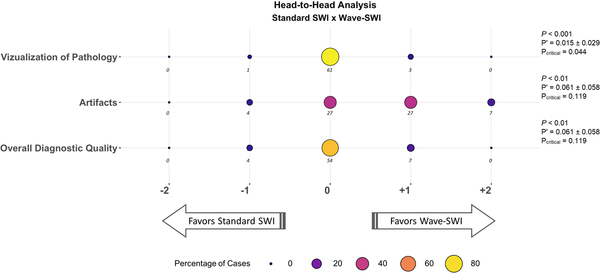
Figure 4.
Balloon plot showing the results of the head-to-head comparison of standard susceptibility-weighted imaging (SWI) and Wave-CAIPI SWI (Wave-SWI). The size and the color of each circle represents the percentage of cases that were assigned a given score, from a total of 65 abnormal cases. The actual number of cases receiving a given score is indicated below each circle. Negative scores (left) favor standard SWI, and positive scores (right) favor Wave-SWI. The proportion of cases where T2*w-GRE was preferred over Wave-SWI (P’) and the 95% confidence interval for this proportion are indicated at the right of the figure. The critical value (Pcritical) is also provided, corresponding to the upper bound of the 95% confidence interval for P’. Non-inferiority testing was performed as described in the methods, and the corresponding P-values are shown in the figure. Wave-SWI was non-inferior to standard SWI for the three variables evaluated.
Discussion
This study compared a highly-accelerated Wave-SWI sequence to two commonly used alternative sequences in terms of technical feasibility, visualization of pathology including hemorrhage, and overall diagnostic quality. We included consecutive MRI exams obtained for a wide range of indications in order to understand the implications of adopting this sequence for routine clinical brain imaging.
Compared to standard T2*w-GRE, Wave-SWI provided superior visualization of pathology and overall diagnostic quality, with acquisition times that were approximately 1.5 times faster than standard T2*w-GRE when the 20-channel coil was used and 2 times faster when the 32-channel coil was used. The differences in diagnostic quality were clinically significant, including 21 cases (15.1%) in which hemorrhage was seen only on Wave-SWI. With increasing utilization of MRI in the emergency setting,16 the ability to rapidly detect acute pathology including hemorrhage is particularly desirable. Detection of even a small number of microhemorrhages is often clinically important. For example, in anticoagulated patients with a history of atrial fibrillation and recent stroke, the presence of microhemorrhages is associated with intracranial hemorrhage risk and may inform anticoagulation decisions.17 Artifacts were rated more severe on T2*w-GRE, likely due to thicker slices resulting in greater through-plane dephasing and signal dropout near the skull base (Figure 1C). Motion was rated less severe on T2*w-GRE, likely due to the intrinsically lower motion sensitivity of multi-slice 2D versus 3D sequences.18 For this reason, the reduced motion sensitivity of T2*w-GRE (and the even faster echo planar imaging based GRE sequences) may provide advantages over Wave-SWI in the setting of extreme motion. Our findings suggest that Wave-SWI could replace T2*w-GRE for most indications, providing superior visualization of pathology with a 1.5- to 2-fold decrease in scan time, encouraging greater adoption of SWI across clinical brain imaging protocols. Wave-SWI also provides additional information over T2*w-GRE in the form of filtered phase maps, which can be useful in distinguishing blood products from mineralization.19
Wave-SWI was non-inferior to standard SWI for visualization of pathology, artifacts, and overall diagnostic quality, with reduced scan time (approximately 3 times faster than standard SWI on the 20-channel coil and 5 times faster on the 32-channel coil), and reduced motion artifacts. Twice as many scans received a motion score of 3 (severe motion that may obscure major findings) using standard SWI, likely a direct consequence of the longer acquisition time. Motion is a common source of image degradation in MRI, especially in emergency and inpatient settings, and is associated with substantial institutional costs.20 Our findings suggest that Wave-SWI could replace standard SWI for most indications, improving utilization of MRI resources while reducing motion artifacts and costs associated with repeat sequence attempts or repeat exams. The decreased scan time of Wave-CAIPI may provide synergistic benefits with other prospective21 and retrospective22 motion correction strategies. For example, the recently proposed Targeted Motion Estimation and Reduction (TAMER) algorithm employs a joint-optimization similar to the auto-calibrated Wave-CAIPI reconstruction,22 and future work incorporating TAMER directly into the joint reconstruction may provide even further reduction in the motion sensitivity of Wave-SWI. Comparison of Wave-CAIPI to other acceleration strategies based on compressed sensing23 and exploiting possible synergies between these approaches would be a valuable area for future study. Further evaluation of multi-echo approaches, including the optimal number and spacing of echoes for R2* mapping and quantitative susceptibility mapping, would also be a valuable extension of this work.
Our study has several limitations. First, it is not possible to compare the diagnostic accuracy of Wave-SWI and standard SWI in vivo due to the absence of a reference standard. Because of the similar contrast and image quality of these sequences, and the variable nature of patient motion and other image artifacts, detection of a small hemorrhage on Wave-SWI but not standard SWI does not necessarily imply a false positive, and could alternatively reflect motion artifact obscuring the hemorrhage on the standard sequence. To validate Wave-SWI as an index test in the absence of a reference standard, we followed the approach described by Reitsma et al24 by relating the result of the index test to other clinically relevant characteristics ‒ in our case, the impact on the final clinical diagnosis determined by the interpreting radiologist (Online Table 3). Under this framework, we demonstrated non-inferiority of the Wave-SWI sequence with respect to the major factors relevant to the interpreting radiologist in rendering a clinical diagnosis. Although we used a standardized image-based definition of microhemorrhages,11 we did not have CT correlation to confirm the presence of calcification or follow-up imaging to confirm persistence or expansion of hemorrhage, and it is possible that some of the microhemorrhages we identified were in fact calcifications or other mimics. Second, the selection of a suitable non-inferiority margin for imaging studies is often challenging. Our selection was informed by a review of similar imaging-based non-inferiority studies,25,26 and consensus among our group of neuroradiologists that the new sequence could be considered non-inferior if the standard sequence was preferred in fewer than 15% of cases. Because this threshold is inherently subjective, we also reported the critical value (Pcritical), equivalent to the upper bound on a 95% confidence interval for the proportion of cases in which the standard sequence was preferred. The critical value allows us to understand how selection of a different non-inferiority margin would impact the results. For example, with a 25% non-inferiority margin, Wave-SWI would be deemed non-inferior to standard SWI for all of the variables considered (Pcritical < 25% in all cases, Figure 4). With a 5% non-inferiority margin, Wave-SWI would be deemed non-inferior for visualization of pathology (the primary outcome), but not the remaining variables. Third, although readers were blinded to the acquisition protocol, some features of the images may allow the readers to identify the pulse sequence being evaluated. In the case of Wave-SWI versus T2*w-GRE, the difference is visibly apparent. In the case of Wave-SWI versus standard SWI, we minimized this possibility by matching the most important parameters that determine image quality and image contrast (including nominal voxel dimensions, slice thickness, and effective echo time) as closely as possible within the range of allowable parameter values.
In summary, Wave-SWI provided superior visualization of pathology and overall diagnostic quality compared to T2*w-GRE, and was non-inferior to standard SWI with reduced scan time and reduced motion artifact. Broader clinical application of the Wave-CAIPI approach may result in more efficient utilization of MRI resources without loss of clinically important information.
Supplementary Material
Supplemental Figure. Reference images available to readers during the image review, illustrating representative images for the grading of motion artifact. Motion was graded according to the following scale: 0 - none; 1 - mild (motion is perceptible but not clinically relevant); 2 - moderate (motion is present and may obscure subtle findings); and 3 - severe (motion is present and may obscure major findings).
Funding information:
Supported by the National Institutes of Health (P41 EB015896, R01 EB020613, UL 1TR002541), an RSNA Research Resident Grant (S.Y.H.), and Siemens Healthineers. This work was conducted with support from Harvard Catalyst | The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health.
ABBREVIATIONS:
- SWI
Susceptibility-Weighted Imaging
- CAIPI
Controlled Aliasing In Parallel Imaging
- T2*w-GRE
T2*-Weighted Gradient-Echo
References
- 1.Nandigam RNK, Viswanathan A, Delgado P, et al. MR imaging detection of cerebral microbleeds: Effect of susceptibility-weighted imaging, section thickness, and field strength. Am J Neuroradiol 2009;30:338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haacke EM, Mittal S, Wu Z, et al. Susceptibility-Weighted Imaging: Technical Aspects and Clinical Applications, Part 1. Am J Neuroradiol 2009;30:19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mittal S, Wu Z, Neelavalli J, et al. Susceptibility-weighted imaging: Technical aspects and clinical applications, part 2. Am J Neuroradiol 2009;30:232–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havsteen I, Ohlhues A, Madsen KH, et al. Are movement artifacts in magnetic resonance imaging a real problem?-a narrative review. Front Neurol 2017;8:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munn Z, Pearson A, Jordan Z, et al. Patient anxiety and satisfaction in a magnetic resonance imaging department: Initial results from an action research study. J Med Imaging Radiat Sci 2015;46:23–9. [DOI] [PubMed] [Google Scholar]
- 6.Bilgic B, Gagoski BA, Cauley SF, et al. Wave-CAIPI for highly accelerated 3D imaging. Magn Reson Med 2015;73:2152–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cauley SF, Setsompop K, Bilgic B, et al. Autocalibrated wave-CAIPI reconstruction; Joint optimization of k-space trajectory and parallel imaging reconstruction. Magn Reson Med 2017;78:1093–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bilgic B, Xie L, Dibb R, et al. Rapid multi-orientation quantitative susceptibility mapping. Neuroimage 2016;125:1131–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilgic B, Ye H, Wald LL, et al. Simultaneous Time Interleaved MultiSlice (STIMS) for Rapid Susceptibility Weighted acquisition. Neuroimage 2017;155:577–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conklin J, Cauley S, Setsompop K, et al. Optimization and Clinical Evaluation of Wave-CAIPI Susceptibility-Weighted Imaging (SWI) for Detection of Intracranial Hemorrhage In: Radiological Society of North America. Chicago, IL; 2018:SSE24–04. [Google Scholar]
- 11.Gregoire SM, Chaudhary UJ, Brown MM, et al. The Microbleed Anatomical Rating Scale (MARS): reliability of a tool to map brain microbleeds. Neurology 2009;73:1759–66. [DOI] [PubMed] [Google Scholar]
- 12.Ahn S, Park SH, Lee KH. How to Demonstrate Similarity by Using Noninferiority and. Radiology 2013;267:328–38. [DOI] [PubMed] [Google Scholar]
- 13.Lakens D, Scheel AM, Isager PM. Equivalence Testing for Psychological Research : A Tutorial. Adv Methods Pract Psychol Sci 2018;1:259–69. [Google Scholar]
- 14.Cohen J Statistical Power Analysis for the Behavioral Sciences (Second Edition). 2nd ed New York, NY: Lawrence Erlbaum; 1988. [Google Scholar]
- 15.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics 1977;33:159–74. [PubMed] [Google Scholar]
- 16.Redd V, Levin S, Toerper M, et al. Effects of Fully Accessible Magnetic Resonance Imaging in the Emergency Department Griffey RT, ed. Acad Emerg Med 2015;22:741–9. [DOI] [PubMed] [Google Scholar]
- 17.Wilson D, Ambler G, Shakeshaft C, et al. Cerebral microbleeds and intracranial haemorrhage risk in patients anticoagulated for atrial fibrillation after acute ischaemic stroke or transient ischaemic attack (CROMIS-2): a multicentre observational cohort study. Lancet Neurol 2018;17:539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Runge VM, Wood ML, Kaufman DM, et al. FLASH: clinical three-dimensional magnetic resonance imaging. RadioGraphics 1988;8:947–65. [DOI] [PubMed] [Google Scholar]
- 19.Yamada N, Imakita S, Sakuma T, et al. Intracranial calcification on gradient-echo phase image: depiction of diamagnetic susceptibility. Radiology 1996;198:171–8. [DOI] [PubMed] [Google Scholar]
- 20.Andre JB, Bresnahan BW, Mossa-Basha M, et al. Toward quantifying the prevalence, severity, and cost associated with patient motion during clinical MR examinations. J Am Coll Radiol 2015;12:689–95. [DOI] [PubMed] [Google Scholar]
- 21.Callaghan MF, Josephs O, Herbst M, et al. An evaluation of prospective motion correction (PMC) for high resolution quantitative MRI. Front Neurosci 2015;9:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haskell MW, Cauley SF, Wald LL. TArgeted Motion Estimation and Reduction (TAMER): Data consistency based motion mitigation for mri using a reduced model joint optimization. IEEE Trans Med Imaging 2018;37:1253–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu B, Li W, Guidon A, et al. Whole brain susceptibility mapping using compressed sensing. Magn Reson Med 2012;67:137–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reitsma JB, Rutjes AWS, Khan KS, et al. A review of solutions for diagnostic accuracy studies with an imperfect or missing reference standard. J Clin Epidemiol 2009;62:797–806. [DOI] [PubMed] [Google Scholar]
- 25.Lee SJ, Park SH, Kim AY, et al. A prospective comparison of standard-dose CT enterography and 50% reduced-dose CT enterography with and without noise reduction for evaluating Crohn disease. AJR Am J Roentgenol 2011;197:50–7. [DOI] [PubMed] [Google Scholar]
- 26.Schaefer PJ, Boudghene FP, Brambs HJ, et al. Abdominal and Iliac Arterial Stenoses: Comparative Double-blinded Randomized Study of Diagnostic Accuracy of 3D MR Angiography with Gadodiamide or Gadopentetate Dimeglumine. Radiology 2006;238:827–40. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure. Reference images available to readers during the image review, illustrating representative images for the grading of motion artifact. Motion was graded according to the following scale: 0 - none; 1 - mild (motion is perceptible but not clinically relevant); 2 - moderate (motion is present and may obscure subtle findings); and 3 - severe (motion is present and may obscure major findings).