Abstract
The outbreak of a novel betacoronavirus (2019-nCoV) represents a pandemic threat that has been declared a public health emergency of international concern. The CoV spike (S) glycoprotein is a key target for vaccines, therapeutic antibodies, and diagnostics. To facilitate medical countermeasure (MCM) development, we determined a 3.5 Å-resolution cryo-EM structure of the 2019-nCoV S trimer in the prefusion conformation. The predominant state of the trimer has one of the three receptor-binding domains (RBDs) rotated up in a receptor-accessible conformation. We also show biophysical and structural evidence that the 2019-nCoV S binds ACE2 with higher affinity than SARS-CoV S. Additionally, we tested several published SARS-CoV RBD-specific monoclonal antibodies and found that they do not have appreciable binding to 2019-nCoV S, suggesting antibody cross-reactivity may be limited between the two RBDs. The structure of 2019-nCoV S should enable rapid development and evaluation of MCMs to address the ongoing public health crisis.
The novel coronavirus 2019-nCoV has recently emerged as a human pathogen in the city of Wuhan in China’s Hubei province, causing fever, severe respiratory illness and pneumonia—a disease recently named COVID-19 (1, 2). According to the World Health Organization (WHO) on February 16th, 2020, there had been over 51,000 confirmed cases globally, leading to at least 1,600 deaths. The emerging pathogen was rapidly characterized as a novel member of the betacoronavirus genus, closely related to several bat coronaviruses as well as severe acute respiratory syndrome coronavirus (SARS-CoV) (3, 4). Compared to SARS-CoV, 2019-nCoV appears to be more readily transmitted from human-to-human, spreading to multiple continents and leading to the WHO declaration of a Public Health Emergency of International Concern (PHEIC) on January 30th, 2020 (1, 5, 6).
2019-nCoV makes use of a densely glycosylated spike (S) protein to gain entry into host cells. The S protein is a trimeric class I fusion protein that exists in a metastable prefusion conformation that undergoes a dramatic structural rearrangement to fuse the viral membrane with the host-cell membrane (7, 8). This process is triggered when the S1 subunit binds to a host-cell receptor. Receptor binding destabilizes the prefusion trimer, resulting in shedding of the S1 subunit and transition of the S2 subunit to a stable postfusion conformation (9). In order to engage a host-cell receptor, the receptor-binding domain (RBD) of S1 undergoes hinge-like conformational movements that transiently hide or expose the determinants of receptor binding. These two states are referred to as the “down” conformation and the “up” conformation, where down corresponds to the receptor-inaccessible state and up corresponds to the receptor-accessible state, which is thought to be less stable (10–13). Due to the indispensable function of the S protein, it represents a target for antibody-mediated neutralization, and characterization of the prefusion S structure would provide atomic-level information to guide vaccine design and development.
Based on the first reported genome sequence of 2019-nCoV (4), we expressed ectodomain residues 1−1208 of 2019-nCoV S, adding two stabilizing proline mutations in the C-terminal S2 fusion machinery based on a previous stabilization strategy that proved effective for other betacoronavirus S proteins (11, 14). Figure 1A shows the domain organization of the expression construct and figure S1 shows the purification process. We obtained roughly 0.5 mg/L of the recombinant prefusion-stabilized S ectodomain from FreeStyle 293 cells, and the protein was purified to homogeneity by affinity chromatography and size-exclusion chromatography (fig. S1). Cryo-EM grids were prepared using this purified, fully glycosylated S protein and preliminary screening revealed a high particle density with little aggregation near the edges of the holes.
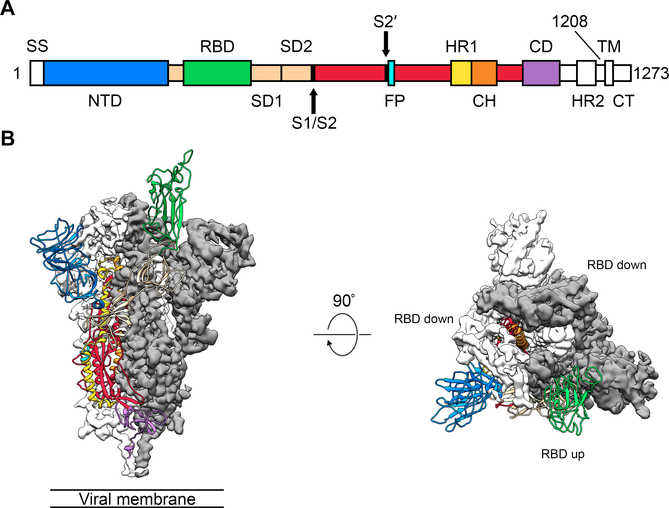
Figure 1. Structure of 2019-nCoV S in the prefusion conformation.
(A) Schematic of 2019-nCoV S primary structure, colored by domain. Domains that were excluded from the ectodomain expression construct or could not be visualized in the final map are colored white. SS= signal sequence, NTD= N-terminal domain, RBD= receptor-binding domain, SD1= subdomain 1, SD2= subdomain 2, S1/S2= S1/S2 protease cleavage site, S2′= S2′ protease cleavage site, FP= fusion peptide, HR1= heptad repeat 1, CH= central helix, CD= connector domain, HR2= heptad repeat 2, TM= transmembrane domain, CT= cytoplasmic tail. Arrows denote protease cleavage sites. (B) Side and top views of the prefusion structure of the 2019-nCoV S protein with a single RBD in the up conformation. The two RBD-down protomers are shown as cryo-EM density in either white or gray and the RBD-up protomer is shown in ribbons, colored corresponding to the schematic in Fig 1A.
After collecting and processing 3,207 micrograph movies, we obtained a 3.5 Å-resolution 3D reconstruction of an asymmetrical trimer in which a single RBD was observed in the up conformation. (Fig. 1B, fig. S2, table S1). Due to the small size of the RBD (~21 kDa), the asymmetry of this conformation was not readily apparent until ab initio 3D reconstruction and 3D classification were performed (Fig. 1B, fig. S3). By using the 3D variability feature in cryoSPARC v2 (15), we observed breathing of the S1 subunits as the RBD underwent a hinge-like movement, which likely contributed to the relatively poor local resolution of S1 compared to the more stable S2 subunit (movies S1 and S2). This seemingly stochastic RBD movement has been captured during structural characterization of the closely related betacoronaviruses SARS-CoV and MERS-CoV, as well as the more distantly related alphacoronavirus porcine epidemic diarrhea virus (PEDV) (10, 11, 13, 16). The observation of this phenomenon in 2019-nCoV S suggests that it shares the same mechanism of triggering that is thought to be conserved among the Coronaviridae, wherein receptor-binding to exposed RBDs leads to an unstable 3 RBD-up conformation that results in shedding of S1 and refolding of S2 (11, 12).
Because the S2 subunit appeared to be a symmetric trimer, we performed a 3D refinement imposing C3 symmetry, resulting in a 3.2 Å-resolution map, with excellent density for the S2 subunit. Using both maps, we built most of the 2019-nCoV S ectodomain, including glycans at 44 of the 66 N-linked glycosylation sites per trimer (fig. S4). Our final model spans S residues 27–1146, with several flexible loops omitted. Like all previously reported coronavirus S ectodomain structures, the density for 2019-nCoV S begins to fade after the connector domain (CD), reflecting the flexibility of the heptad repeat 2 (HR2) domain in the prefusion conformation (fig. S4A) (13, 16–18).
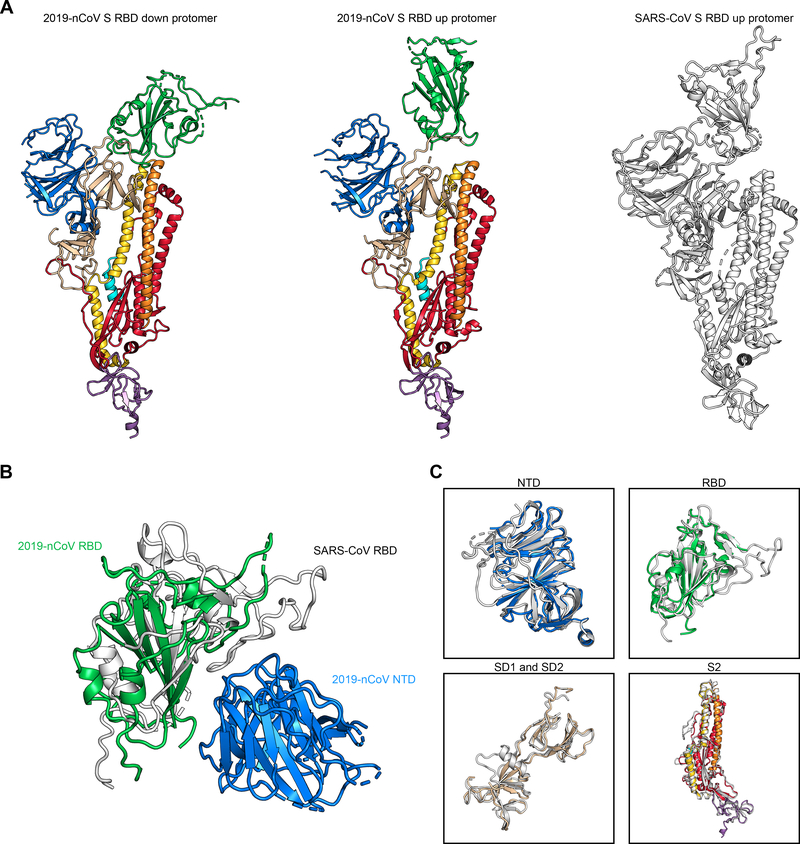
The overall structure of 2019-nCoV S resembles that of SARS-CoV S, with a root mean square deviation (RMSD) of 3.8 Å over 959 Cα atoms (Fig. 2A). One of the larger differences between these two structures (although still relatively minor) is the position of the RBDs in their respective down conformations. Whereas the SARS-CoV RBD in the down conformation packs tightly against the N-terminal domain (NTD) of the neighboring protomer, the 2019-nCoV RBD in the down conformation is angled closer to the central cavity of the trimer (Fig. 2B). Despite this observed conformational difference, when the individual structural domains of 2019-nCoV S are aligned to their counterparts from SARS-CoV S, they reflect the high degree of structural homology between the two proteins, with the NTDs, RBDs, subdomains 1 and 2 (SD1 and SD2) and S2 subunits yielding individual RMSD values of 2.6 Å, 3.0 Å, 2.7 Å and 2.0 Å, respectively (Fig. 2C).
Figure 2. Structural comparison between 2019-nCoV S and SARS-CoV S.
(A) A single protomer of 2019-nCoV S with the RBD in the down conformation (left) is shown in ribbons, colored according to Fig. 1. A protomer of 2019-nCoV S in the RBD-up conformation is shown (center) next to a protomer of SARS-CoV S in the RBD-up conformation (right), displayed as ribbons and colored white (PDB ID: 6CRZ). (B) The RBDs of 2019-nCoV and SARS-CoV have been aligned based on the position of the adjacent NTD from the neighboring protomer. The 2019-nCoV RBD is colored green and the SARS-CoV RBD is colored white. The 2019-nCoV NTD is colored blue. (C) The following structural domains from 2019-nCoV S have been aligned to their counterparts from SARS-CoV S: NTD (top left), RBD (top right), SD1 and SD2 (bottom left), and S2 (bottom right).
2019-nCoV S shares 98% sequence identity with the S protein from the bat coronavirus RaTG13, with the most notable variation arising from an insertion in the S1/S2 protease cleavage site that results in an “RRAR” furin recognition site in 2019-nCoV (19), rather than the single arginine in SARS-CoV (fig. S5) (20–23). Notably, in influenza viruses, amino acid insertions that create a polybasic furin site in a related position in influenza hemagglutinin proteins are often found in highly virulent avian and human influenza viruses (24). In the structure reported here, the S1/S2 junction is in a disordered solvent-exposed loop. In addition to this insertion of residues in the S1/S2 junction, 29 variant residues exist between 2019-nCoV S and RaTG13 S, with 17 of these positions mapping to the RBD (fig. S5 and S6). We also analyzed the 61 available 2019-nCoV S sequences in the Global Initiative on Sharing All Influenza Data (GISAID, gisaid.org) database and found that there were only 9 amino acid substitutions among all deposited sequences. Most of these substitutions are relatively conservative and are not expected to have a dramatic effect on the structure or function of the 2019-nCoV S protein (fig. S6).
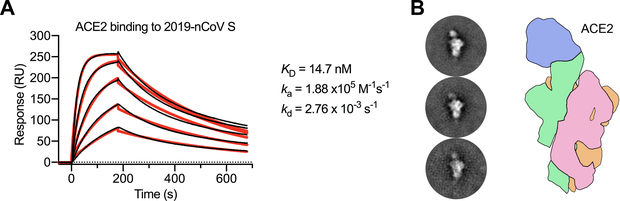
Recent reports demonstrating that 2019-nCoV S and SARS-CoV S share the same functional host-cell receptor—angiotensin-converting enzyme 2 (ACE2) (22, 25–27)—prompted us to quantify the kinetics of this interaction by surface plasmon resonance (SPR). ACE2 bound to 2019-nCoV S ectodomain with ~15 nM affinity, which is approximately 10- to 20-fold higher affinity than ACE2 binding to SARS-CoV S (Fig. 3A, fig. S7) (14). We also formed a complex of ACE2 bound to the 2019-nCoV S ectodomain and observed it by negative-stain EM, where it strongly resembled the complex formed between SARS-CoV S and ACE2, which has been observed at high-resolution by cryo-EM (Fig. 3B) (14, 28). The high affinity of 2019-nCoV S for human ACE2 may contribute to the apparent ease with which 2019-nCoV can spread from human-to-human (1), however, additional studies are needed to investigate this possibility.
Figure 3. 2019-nCoV S binds human ACE2 with high affinity.
(A) SPR sensorgram showing the binding kinetics for human ACE2 and immobilized 2019-nCoV S. Data are shown as black lines and the best fit of the data to a 1:1 binding model is shown in red. (B) Negative-stain EM 2D class averages of 2019-nCoV S bound by ACE2. Averages have been rotated so that ACE2 is positioned above the 2019-nCoV S protein with respect to the viral membrane. A cartoon depicting the ACE2-bound 2019-nCoV S protein is shown (right) with ACE2 in blue and S protein protomers colored tan, pink and green.
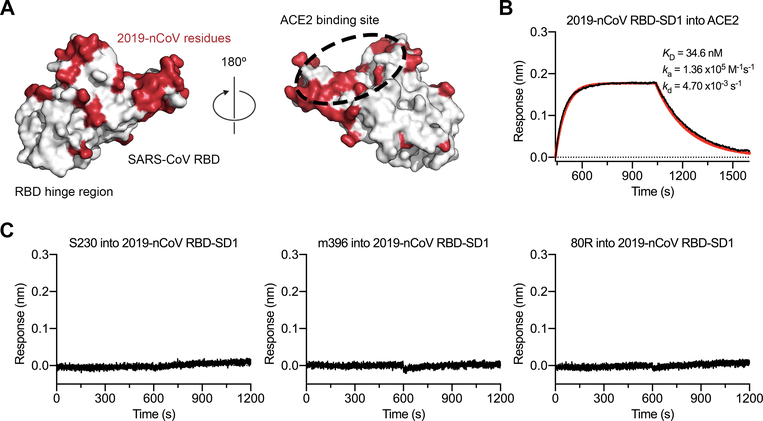
The overall structural homology and shared receptor usage between SARS-CoV S and 2019-nCoV S prompted us to test published SARS-CoV RBD-directed monoclonal antibodies (mAbs) for cross-reactivity to the 2019-nCoV RBD (Fig. 4A). A 2019-nCoV RBD-SD1 fragment (S residues 319–591) was recombinantly expressed, and appropriate folding of this construct was validated by measuring ACE2 binding using biolayer interferometry (BLI) (Fig. 4B). Cross-reactivity of the SARS-CoV RBD-directed mAbs S230, m396 and 80R was then evaluated by BLI (12, 29–31). Despite the relatively high degree of structural homology between the 2019-nCoV RBD and the SARS-CoV RBD, no binding to the 2019-nCoV RBD could be detected for any of the three mAbs at the concentration tested (1 μM) (Fig. 4C), in contrast to the strong binding we observed to the SARS-CoV RBD (fig. S8). Although the epitopes of these three antibodies represent a relatively small percentage of the surface area of the 2019-nCoV RBD, the lack of observed binding suggests that SARS-directed mAbs will not necessarily be cross-reactive and that future antibody isolation and therapeutic design efforts will benefit from using 2019-nCoV S proteins as probes.
Figure 4. Antigenicity of the 2019-nCoV RBD.
(A) The SARS-CoV RBD is shown as a white molecular surface (PDB ID: 2AJF), with residues that vary in the 2019-nCoV RBD colored red. The ACE2 binding site is outlined with a black dotted line. (B) A biolayer interferometry sensorgram that shows binding to ACE2 by the 2019-nCoV RBD-SD1. Binding data are shown as a black line and the best fit of the data to a 1:1 binding model is shown in red. (C) Biolayer interferometry to measure cross-reactivity of the SARS-CoV RBD-directed antibodies S230, m396 and 80R. Sensortips with immobilized antibodies were dipped into wells containing 2019-nCoV RBD-SD1 and the resulting data are shown as a black line.
The rapid global spread of 2019-nCoV, prompting the PHEIC declaration by WHO signals the urgent need for coronavirus vaccines and therapeutics. Knowing the atomic-level structure of the 2019-nCoV spike will allow for additional protein engineering efforts that could improve antigenicity and protein expression for vaccine development. The structural data also facilitates the evaluation of 2019-nCoV spike mutations that will occur as the virus undergoes genetic drift and help define whether those residues have surface exposure and map to sites of known antibody epitopes for other coronavirus spike proteins. In addition, the structure provides assurance that the protein produced by this construct is homogeneous and in the prefusion conformation, which should maintain the most neutralization-sensitive epitopes when used as candidate vaccine antigens or B-cell probes for isolating neutralizing human monoclonal antibodies. Furthermore, the atomic-level detail will enable the design and screening of small molecules with fusion-inhibiting potential. This information will support precision vaccine design and discovery of anti-viral therapeutics, accelerating medical countermeasure development.
Supplementary Material
Acknowledgments
We thank Dr. John Ludes-Meyers for assistance with cell transfection and the rest of the members of the McLellan laboratory for critical reading of the manuscript. We would also like to thank Dr. Aguang Dai from the Sauer Structural Biology Laboratory at the University of Texas at Austin for his assistance with microscope alignment.
Funding: This work was supported in part by an NIH/NIAID grant awarded to J.S.M. (R01-AI127521) and by intramural funding from the National Institute of Allergy and Infectious Diseases (B.S.G). The Sauer Structural Biology Laboratory is supported by the University of Texas College of Natural Sciences and by award RR160023 of the Cancer Prevention and Research Institute of Texas (CPRIT).
Footnotes
Competing interests: N.W., K.S.C., B.S.G, and J.S.M are inventors on US patent application No. 62/412,703 (Prefusion Coronavirus Spike Proteins and Their Use) and D.W., N.W., K.S.C., O.A., B.S.G, and J.S.M are inventors on US patent application No. 62/972,886 (2019-nCoV Vaccine).
Data and materials availability: Atomic coordinates and cryo-EM maps of the reported structure have been deposited in the Protein Data Bank under accession code 6VSB and in the Electron Microscopy Data Bank under accession codes EMD-21374 and EMD-21375. Plasmids are available from B.S.G. under a material transfer agreement with the National Institutes of Health or J.S.M. under a material transfer agreement with The University of Texas at Austin.
References and Notes
- 1.Chan JF et al. , A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet, (2020). doi: 10.1016/S0140-6736(20)30154-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang C et al. , Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet, (2020). doi: 10.1016/s0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu R et al. , Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet, (2020). doi: 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu F et al. , A new coronavirus associated with human respiratory disease in China. Nature, (2020). doi: 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen N et al. , Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet, (2020). doi: 10.1016/S0140-6736(20)30211-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q et al. , Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N Engl J Med, (2020). doi: 10.1056/NEJMoa2001316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu Rev Virol 3, 237–261 (2016). doi: 10.1146/annurev-virology-110615-042301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bosch BJ, van der Zee R, de Haan CA, Rottier PJ, The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77, 8801–8811 (2003). doi: 10.1128/jvi.77.16.8801-8811.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walls AC et al. , Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci U S A 114, 11157–11162 (2017). doi: 10.1073/pnas.1708727114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gui M et al. , Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res 27, 119–129 (2017). doi: 10.1038/cr.2016.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pallesen J et al. , Immunogenicity and structures of a rationally designed prefusion MERS-CoV spike antigen. Proc Natl Acad Sci U S A 114, E7348–E7357 (2017). doi: 10.1073/pnas.1707304114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls AC et al. , Unexpected Receptor Functional Mimicry Elucidates Activation of Coronavirus Fusion. Cell 176, 1026–1039 e1015 (2019). doi: 10.1016/j.cell.2018.12.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y et al. , Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun 8, 15092 (2017). doi: 10.1038/ncomms15092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirchdoerfer RN et al. , Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci Rep 8, 15701 (2018). doi: 10.1038/s41598-018-34171-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 16.Wrapp D, McLellan JS, The 3.1-Angstrom Cryo-electron Microscopy Structure of the Porcine Epidemic Diarrhea Virus Spike Protein in the Prefusion Conformation. J Virol 93, (2019). doi: 10.1128/JVI.00923-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls AC et al. , Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol 23, 899–905 (2016). doi: 10.1038/nsmb.3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kirchdoerfer RN et al. , Pre-fusion structure of a human coronavirus spike protein. Nature 531, 118–121 (2016). doi: 10.1038/nature17200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coutard B et al. , The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res, 104742 (2020). doi: 10.1016/j.antiviral.2020.104742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch BJ, Bartelink W, Rottier PJ, Cathepsin L functionally cleaves the severe acute respiratory syndrome coronavirus class I fusion protein upstream of rather than adjacent to the fusion peptide. J Virol 82, 8887–8890 (2008). doi: 10.1128/JVI.00415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glowacka I et al. , Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85, 4122–4134 (2011). doi: 10.1128/JVI.02232-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W et al. , Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426, 450–454 (2003). doi: 10.1038/nature02145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Belouzard S, Chu VC, Whittaker GR, Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106, 5871–5876 (2009). doi: 10.1073/pnas.0809524106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen J et al. , Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95, 409–417 (1998). doi: 10.1016/s0092-8674(00)81771-7 [DOI] [PubMed] [Google Scholar]
- 25.M. K-W, Hoffmann H; Krüger N; Müller M; Drosten C; Pöhlmann S, The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRXiv, (2020). doi: 10.11101/2020.01.31.929042 [DOI] [Google Scholar]
- 26.Wan Y, Shang J, Graham R, Baric RS, Li F, Receptor recognition by novel coronavirus from Wuhan: An analysis based on decade-long structural studies of SARS. J Virol, (2020). doi: 10.1128/JVI.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou P et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature, (2020). doi: 10.1038/s41586-020-2012-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song W, Gui M, Wang X, Xiang Y, Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog 14, e1007236 (2018). doi: 10.1371/journal.ppat.1007236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang WC et al. , Structural basis of neutralization by a human anti-severe acute respiratory syndrome spike protein antibody, 80R. J Biol Chem 281, 34610–34616 (2006). doi: 10.1074/jbc.M603275200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prabakaran P et al. , Structure of severe acute respiratory syndrome coronavirus receptor-binding domain complexed with neutralizing antibody. J Biol Chem 281, 15829–15836 (2006). doi: 10.1074/jbc.M600697200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian X et al. , Potent binding of 2019 novel coronavirus spike protein by a SARS coronavirus-specific human monoclonal antibody. BioRXiv, (2020). doi: 10.1101/2020.01.28.923011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carragher B et al. , Leginon: an automated system for acquisition of images from vitreous ice specimens. J Struct Biol 132, 33–45 (2000). doi: 10.1006/jsbi.2000.4314 [DOI] [PubMed] [Google Scholar]
- 33.Tegunov D, Cramer P, Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods 16, 1146–1152 (2019). doi: 10.1038/s41592-019-0580-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramirez-Aportela E et al. , Automatic local resolution-based sharpening of cryo-EM maps. Bioinformatics 36, 765–772 (2020). doi: 10.1093/bioinformatics/btz671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sali A, Blundell TL, Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234, 779–815 (1993). doi: 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- 36.Pettersen EF et al. , UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612 (2004). doi: 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 37.Adams PD et al. , PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58, 1948–1954 (2002). doi: 10.1107/s0907444902016657 [DOI] [PubMed] [Google Scholar]
- 38.Croll TI, ISOLDE: a physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr D Struct Biol 74, 519–530 (2018). doi: 10.1107/S2059798318002425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). doi: 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 40.Morin A et al. , Collaboration gets the most out of software. eLife 2, e01456 (2013). doi: 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grant T, Rohou A, Grigorieff N, cisTEM, user-friendly software for single-particle image processing. eLife 7, (2018). doi: 10.7554/eLife.35383 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.