Abstract
Electrochemical sensors are major players in the race for improved molecular diagnostics due to their convenience, temporal resolution, manufacturing scalability, and their ability to support real-time measurements. This is evident in the ever-increasing number of health-related electrochemical sensing platforms, ranging from single-measurement point-of-care devices to wearable devices supporting immediate and continuous monitoring. In support of the need for such systems to rapidly process large data volumes, we describe here an open-source, easily customizable, multiplatform compatible program for the real-time control, processing, and visualization of electrochemical data. The software’s architecture is modular and fully documented, allowing the easy customization of the code to support the processing of voltammetric (e.g., square-wave and cyclic) and chronoamperometric data. The program, which we have called Software for the Analysis and Continuous Monitoring of Electrochemical Systems (SACMES), also includes a graphical interface allowing the user to easily change analysis parameters (e.g., signal/noise processing, baseline correction) in real-time. To demonstrate the versatility of SACMES we use it here to analyze the real-time data output by (1) the electrochemical, aptamer-based measurement of a specific small-molecule target, (2) a monoclonal antibody-detecting DNA-scaffold sensor, and (3) the determination of the folding thermodynamics of an electrode-attached, redox-reporter-modified protein.
Graphical Abstract
The advances in electrochemical sensing over the last 2 decades have created a need for software platforms capable of analyzing large, dynamic data sets in real time.1 An example is the continuous glucose sensor,2 which requires software able to process measurements performed several times a minute over the course of weeks.3,4 Following this, other emerging electrochemical technologies, including electrochemical aptamer-based (E-AB) sensors, have been reported that support real-time measurements of, for example, plasma drug levels with seconds- or even subsecond time resolution5–8 over the course of hours, producing exceedingly large data volumes. Unfortunately, however, commercial software for the analysis of such data are expensive9 and cannot be customized without specialized programming skills. This is problematic in academic laboratories where the requisite financial and software development resources are often scarce.10 We believe there is thus a need for the creation and open exchange of efficient software platforms that support the real-time analysis of high-volume electrochemical data.
A software framework for the analysis of electrochemical measurements would ideally be based on (1) an open-source computer language compatible with the three major personal computer operating systems (i.e., Microsoft Windows, Apple macOS, and Linux); (2) a modular design easily customizable to a wide range of electroanalytical applications; and (3) an intuitive graphical user interface (GUI) providing the ability to dynamically control key processing parameters in real time. To ensure broad application, the software should support the analysis of data originating from a diverse set of commercial potentiostats (e.g., CH Instruments, Gamry, Metrohm) employed in single, multichannel, or multiplexed configurations. Finally, to appeal to a broad userbase, the software should have a well-documented standard operating procedure (SOP) containing detailed instructions regarding customization to new applications. Thus, motivated, we describe here an open-source, multiplatform software supporting the real-time analysis of high-volume electrochemical data.
EXPERIMENTAL SECTION
Software.
While designing our software architecture, we incorporated three principle innovations: (1) utilizing a technique called Blitting11 to achieve millisecond temporal resolution and high efficiency over time; (2) the ability to analyze various electrochemical measurements continuously and simultaneously (e.g., multiple electrodes, multiple frequencies); and (3) real-time parameter manipulation to address user needs on the fly.
Blitting (also written Bit Blit or BITBLT) improves our software’s performance. Specifically, most visualization software typically experience CPU bottlenecks when analyzing and animating high-volume data because they consume large amounts of random access memory (RAM) to render images, losing computing efficiency over time as data volumes increase and image rendering demands more memory (which is finite). Instead, Blitting eliminates this problem by only saving a clean “bit-map,” or background canvas, which is then used to only render data originating from new files (it does not save graphical data from previous files). With Blitting, a constant amount of RAM is needed for the program, allowing for larger amounts of data to be visualized without experiencing a decrease in performance over time.
The modular nature of our program allows for easy customization to support the analysis of data originating from many electroanalytical techniques. With minimal programming experience, the user can easily manipulate the script to analyze and visualize not only square-wave voltammetry but other widely used electrochemical methods such as, but not limited to, cyclic voltammetry, linear sweep voltammetry, and chronoamperometry. We provide a fully documented standard operating procedure as Supporting Information to facilitate and promote the adoption of our software.
Reagents and Materials.
Sodium hydroxide, sulfuric acid, tris(hydroxymethyl) aminomethane (Tris), ethylenediaminetetraacetic acid (EDTA), sodium hydrogen phosphate, sodium chloride, potassium chloride, and potassium dihydrogen phosphate were obtained from Fisher Scientific (Waltham, MA). 6-Mercapto-1-hexanol (MCH) and tris(2-carboxyethyl)phosphine (TCEP) were obtained from Sigma-Aldrich (St. Louis, MO). Tobramycin sulfate (USP grade) was obtained from Gold BioTechnology, Inc. (St. Louis, MO). All reagents were used as received. A 1× stock solution of phosphate buffered saline (PBS) was prepared containing 10 mM sodium hydrogen phosphate, 2.7 mM potassium chloride, 137 mM sodium chloride, and 1.76 mM potassium phosphate (pH = 7.4). A 1× stock solution of Tris-EDTA buffer was prepared containing 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. Anti-FLAG monoclonal antibodies were obtained from Sigma-Aldrich (St. Louis, MO). Peptide nucleic acid (PNA) sequences were purchased from PNABio (Newbury Park, CA). Hexanethiol- and methylene blue-modified deoxynucleic acid (DNA) sequences were obtained from Biosearch Technologies (Novato, CA).
Catheters (22 G) and 1 mL syringes were purchased from Becton Dickinson (Franklin Lakes, NJ). PTFE-insulated gold, platinum, and silver wires (75 μm diameter) were purchased from A-M systems. To employ the silver wires as reference electrodes, they were immersed in concentrated sodium hypochlorite (commercial bleach) overnight to form a silver chloride film. Heat-shrink polytetrafluoroethylene insulation (PTFE, HS Sub-Lite-Wall, 0.02, 0.005, 0.003 ± 0.001 in, black-opaque, lot no. 17747112-3), used to electrically insulate gold, silver, and platinum wires, was purchased from ZEUS (Branchburg Township, CA). Commercial 2 mm gold disc electrodes employed in the protein experiments were purchased from CH Instruments (Austin, TX).
In Vitro Sensor Fabrication.
E-AB sensors used for in vitro measurements were fabricated as described in previous reports.6,12 In short, segments of pure gold (5.5 cm) were cut and soldered to gold-plated contacts using 60% tin/40% lead rosin-core solder (0.8 mm diameter) and then insulated with two layers of PTFE heat-shrink insulation. The sensor window (i.e., the region devoid of insulation) of the gold wire was made to be approximately 3 mm in length. To increase the microscopic surface area of the gold working electrodes (to obtain larger peak currents), the gold surface was roughened using a technique previously described.12 Briefly, the electrode surface was electrochemically roughened via immersion in 0.5 M sulfuric acid followed by stepping the potential between Einitial = 0.0 V to Ehigh = 2.0 V vs Ag/AgCl, back and forth, for 16 000 pulses using chronoamperometry. Potential steps were applied with a 20 ms duration with no delay between pulses. Immediately following the completion of the roughening step, the electrodes were removed from sulfuric acid and rinsed with DI water. In parallel, aliquots of aminoglycoside-binding aptamer:
were reduced with a 1000-fold molar excess of TCEP for 30 min at 25 °C. Then, freshly roughened electrodes were immersed in a 200 nM solution of DNA aptamer in PBS for 1 h at 25 °C. Following this, the sensors were immersed in a 10 mM solution of MCH overnight at 4 °C. Finally, the sensors were rinsed with deionized water prior to use.
In Vivo Sensor Fabrication.
The E-AB sensors used for in vivo measurements were fabricated as described in previous reports.5,13 Briefly, segments of insulated gold, platinum, and silver wire were cut to 7.75 cm in length, and 7.5 mm of insulation was removed from each end with a scalpel. One end of the silver wire was incubated in 100% bleach to form a silver chloride film. A length of 4.75 cm of heat shrinkable tubing was then used to cover the gold wire with 3 mm of the gold exposed on one end. Thus, the insulated gold wire and a bare platinum wire were then inserted into 4.5 cm of heat shrinkable tubing and bound together with a heat gun so that 4 mm of the platinum wire lay just behind the bare gold wire. Lastly, the insulated gold and platinum wires were inserted into 4.25 cm of heat shrinkable tubing with 4 mm of the silver chloride end of the silver wire laying behind the platinum wire, then bound to the other wires with heat. The resulting gold electrodes were then electrochemically roughened as described above. Following roughening, the electrodes were cleaned in 0.5 M sulfuric acid using cyclic voltammetry by cycling the electrodes rapidly (4 V s−1) between −0.35 and 1.5 V for 20 scans. After roughening and cleaning, the electrodes were immersed in a solution of 200 nM aptamer in 1× PBS for 30 min. Following this, the electrodes were emplaced into a 22-gauge PTFE catheter and incubated in a 10 mM solution of MCH.
Animal Procedures.
Male and female Sprague–Dawley rats from Charles River, Inc. (Santa Cruz, CA) weighing between 300 and 400 g were pair-housed in a standard light cycle room (0800 ON, 2000 OFF) and given ad-libitum access to food and water. All animal procedures were performed in accordance with the guidelines set forth by the Institutional Animal Care and Use Committee at the University of California Santa Barbara.14
In Vivo E-AB Measurements.
Continuous in vivo E-AB measurements were performed on anesthetized rats implanted with an intravenous drug-delivery catheter made from Silastic tubing and a 22-gauge bent steel cannula in the left jugular vein (as we have previously described5,13). Briefly, rats were induced under 5% isoflurane gas and maintained under 2–3% isoflurane gas for the duration of the surgery. The chest was shaved and cleaned with alternating solutions of betadine and 70% ethanol. A small incision was made above the left jugular vein with surgical scissors, and then the vein was isolated and cut open with spring-loaded microscissors. The catheter was introduced into the vein and tied into place with a 6-0 sterile silk suture. In total, 300 units of heparin were then infused into the catheter followed by 0.5 mL of 0.9% sterile saline solution. After implantation of the catheter into the left jugular vein, the right jugular vein was isolated, cut, and implanted with the E-AB sensor and tied off with a 6-0 sterile silk suture. Prior to implantation, the E-AB sensor was incubated first in 70% CIDEX-in-water solution for 20 min and then in heparin solution (1 000 units) for at least 10 min.
In Vitro E-AB Measurements.
Continuous in vitro measurements were performed in whole undiluted serum from BioIVT, Inc. (Westbury, NY). Prior to performing any electrochemical measurements the sensors were interrogated with 30 preliminary square-wave voltammetry scans to establish a stable peak current baseline. During the course of the experiment, tobramycin was titrated into solution using either a pipet and hand mixed, or a low-pressure neMesys pump (Cetoni, Germany) for the alternating target experiments, to ensure homogeneous distribution of free drug. No sensor incubation time was used in between titrant additions or square-wave voltammetry (SWV) measurements.
DNA Scaffold Measurements.
E-DNA scaffold sensors were fabricated following a previously reported procedure.15 First, 2 mm gold electrodes were mechanically polished (first with 1 μm-coarse diamond paper and then with 0.05 μm alumina slurry) and electrochemically cleaned (through successive scans in 0.5 M sulfuric acid and 0.1 M sulfuric acid + 0.01 M potassium chloride). Meanwhile the DNA scaffold molecule:
was reduced with a stock solution of 10 mM TCEP and then diluted to 25 nM final concentration in PBS. Freshly cleaned electrodes were first incubated in the resulting DNA scaffold solution for 1 h at 25 °C and then in 3 mM MCH overnight at 4 °C. Following this, the electrodes were incubated for 2 h in a solution of 100 nM PNA molecule complementary to the DNA scaffold and presenting the FLAG epitope (sequence, DYKDDDDK). The platform was interrogated via SWV using a three-electrode cell consisting of a Ag/AgCl reference electrode and a platinum counter electrode, contained within a glass cell. The platform worked as follows: in the absence of target, the DNA scaffold was free to move generating a high electrochemical signal. However, upon addition of anti-FLAG monoclonal antibody binding to the epitope generates steric hindrance (the antibody is larger than the DNA scaffold) that limits the movement of the DNA scaffold sensor generating a decrease in the electrochemical signal.
Measurements from Protein-Modified Electrodes.
Protein-functionalized electrodes were fabricated following a previously published protocol.16,42 Briefly, commercial 2 mm diameter gold electrodes were cleaned using a series of electrochemical cleaning steps and functionalized with protein by incubating the electrodes for 10 min in a solution 20 mM sodium phosphate and 130 mM sodium chloride (pH 7.0) containing 0.1–1 μM protein. The proteins used for these experiments were modified with a site-specifically conjugated methylene blue redox reporter and an amino-terminal alkyl-thiol anchor that enables site-specific surface-tethering via the proteins’ amino termini. Following protein deposition, the electrode surface was immersed in a solution of 5 mM MCH to form a passivating, hydroxyl-coated, self-assembled monolayer. Prior to use, the electrodes employed in the experiments shown in Figure 2 also underwent four washes in 20 mM sodium phosphate, 0.13 M sodium chloride (pH 7.0) solutions alternating the presence and absence of 7 M guanidinium chloride. The washing solutions contained 1 mM MCH to passivate any bare gold exposed by desorption of nonspecifically adsorbed protein during the washes. Immediately prior to initiation of each experiment, the electrodes were repeatedly interrogated by SWV at 60 or 120 Hz and 25 mV amplitude until producing a stable peak current (typically around 50 interrogations). Automated guanidinium chloride titrations were performed in 20 mM sodium phosphate, 0.5 M sodium chloride (pH 7.0), and 0.45 M trimethylamine N-oxide on Hamilton 500C dual-syringe dispensers connected to a CH Instruments 660D potentiostat (CH Instruments, Austin, TX) and a BASi C3 Cell Stand (Bioanalytical Systems, Inc.). MCH (50 μM) was included in the buffer to minimize baseline drift, and ionic strength was regulated by inclusion of specific concentrations of sodium chloride in the buffer. All electrochemical measurements were performed against a Ag/AgCl reference electrode with saturated potassium chloride and a platinum counter electrode, at 25 mV amplitude and in 5 mL total bulk solution volume.
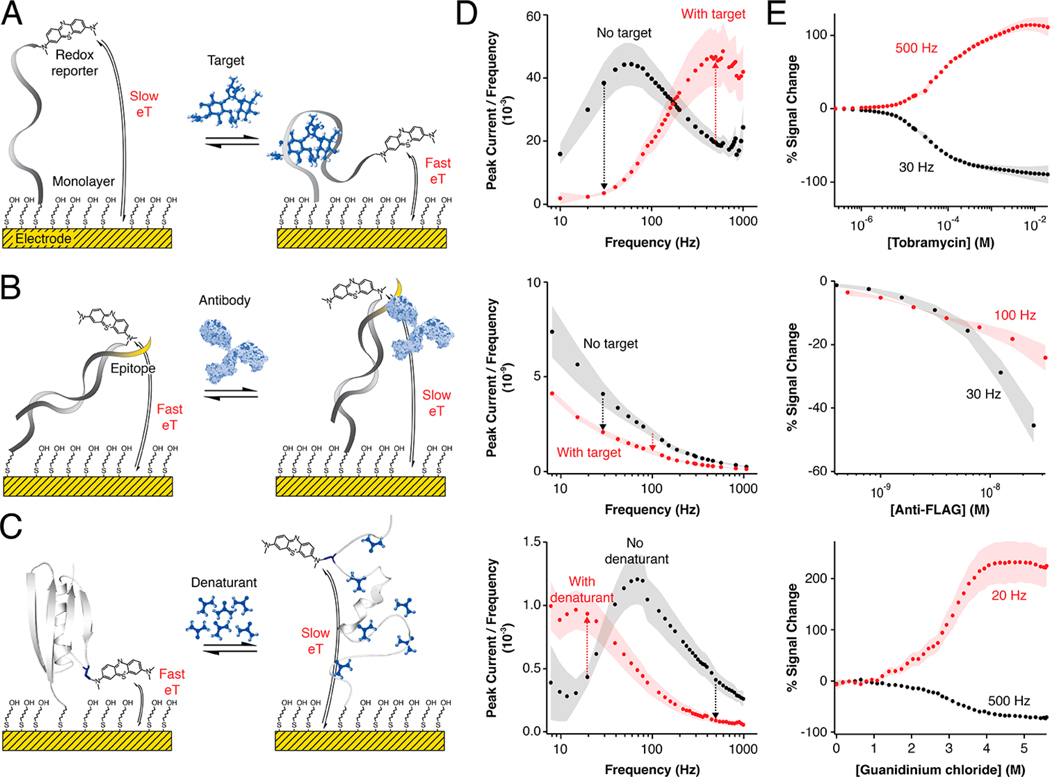
Figure 2.
Our software enables the multielectrode, multifrequency processing of square-wave voltammograms. Here we illustrate this via the analysis of three systems commonly used in our laboratories: (A) a small molecule drug-detecting E-AB sensor,5,13 (B) an antibody-detecting DNA-scaffold sensor,15,40,41 and (C) a protein site-specifically attached to an electrode in support of biophysical studies of its folding thermodynamics.6,16,42 By supporting the simultaneous analysis of voltammograms recorded on multiple electrodes and at multiple frequencies, our software facilitates the characterization of (column D) the electron transfer behavior and (column E) the frequency-dependent signal gain (relative signal change43) of each system. Simultaneously it performs real-time statistical analysis of electrode-to-electrode variation in the peak current or area under the voltammogram (AUC), illustrated here as the shaded areas in columns D and E. The arrows in panel D indicate the expected signal ON or OFF response of each platform at such frequencies; the arrows also marked the frequencies used for the calibration of each platform in column E. The illustration of the antibody in panel B was modified from ref 44.
RESULTS AND DISCUSSION
We chose the Python programming language17 to develop our software framework. As required to produce sharable code, Python is developed under an Open Source Initiative-approved license, which makes our scripts freely distributable.18 Moreover, as required to ensure easy portability, Python is a cross-platform language, such that a Python program written in one operating system can be run on any other (for Microsoft Windows, a free interpreter19 must be installed first). Finally, Python offers a large standard library of well-documented functions, operators and toolkits,20 as well as a community-supported library containing over 130 000 open-sourced software packages to implement graphical user interfaces (GUIs), automation, text/image processing, and scientific computing.21–24
Our script, which we call Software for the Analysis and Continuous Monitoring of Electrochemical Systems (SACMES), can be freely downloaded at the link https://github.com/netzlab/SACMES.git and uses Python’s de facto GUI, Tkinter, which is included in all standard installs of Python. The GUI controls all aspects of SACMES, running an event-driven loop that handles data analysis, real-time data processing, and visualization (Figure 1A). To start the Data Analysis module, a Tkinter-based recursive loop searches for files in a user-designated Path folder that contains files created by an external device such as, for example, a potentiostat. When new files are found, the loop enters the Data Analysis module to read and analyze the data contained within those files. If no new files are found, however, the program will exit the Data Analysis module and continue on to handle any GUI events (e.g., button clicks or parameter inputs) while it waits for new files to be created. After every new file is analyzed, the resulting processed data is returned to an “Animation Class” module to be visualized. Finally, the program returns to the Tkinter loop to continue searching for new files while it handles any GUI events that occurred while the Data Analysis module was in use.
Figure 1.
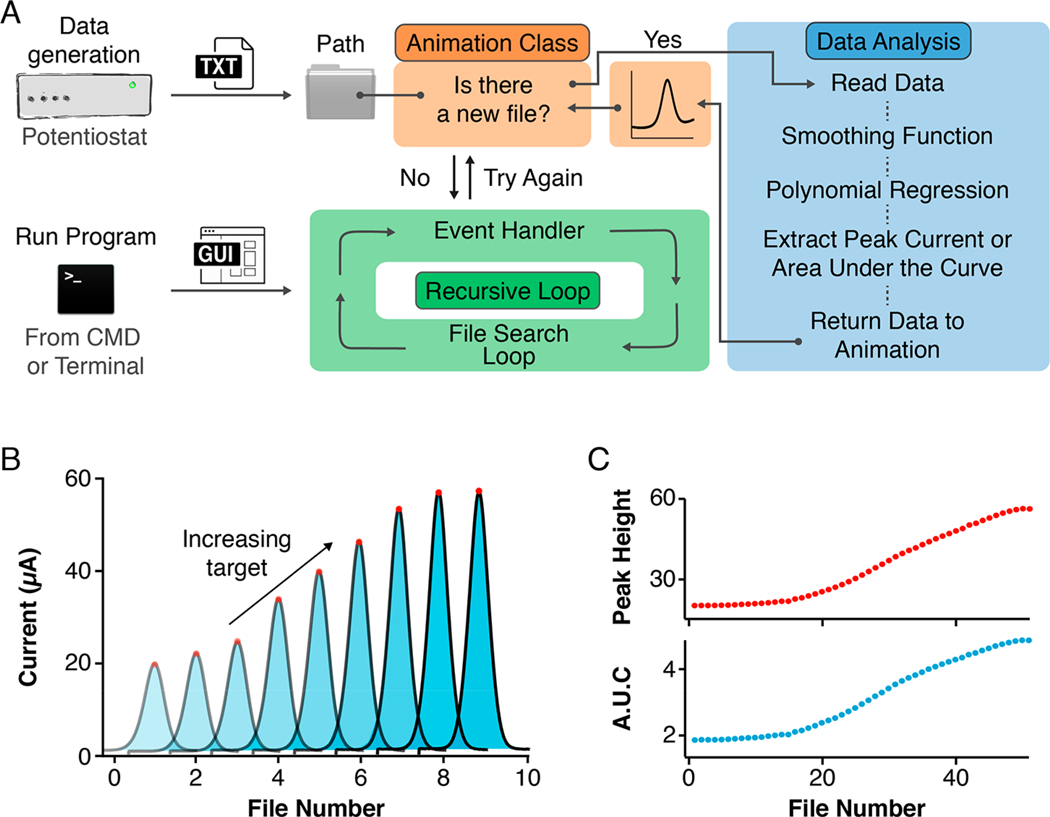
Software framework for the real-time analysis of high-volume electrochemical data. We have developed an open-source, easily customizable script written in the Python programming language to enable the real-time analysis of electrochemical measurements collected with multiple platforms. (A) Our script uses a recursive loop to continuously handle user-interface events while simultaneously looking in a user-defined Path folder for new files generated by, for example, a potentiostat. Once a data file exists, the script activates a Data Analysis module which performs a series of data processing steps including signal averaging and least-squares fitting to a user-defined function and returns the processed data to an Animation Class to be visualized. The terms “CMD” and “Terminal” refer to programs installed by default in Microsoft Windows and Apple macOS, respectively, which provide text-based access to control the operating systems. (B) In this work, we used square-wave voltammograms arising from electrochemical sensors to illustrate the capability of our code to extract peak currents (calculated as the difference between peak maximum and its two minima) and Areas Under the Curve (AUCs). (C) The script outputs and visualizes these values in real-time as the experiment progresses. See Figures S1 and S2 for snapshots of the software’s graphical user interface.
SACMES can analyze and visualize a range of electrochemical outputs. As our first example we customized its Data Analysis module to process SWV, an electroanalytical technique that measures currents at specific time intervals following a square-shaped potential program to enable the effective discrimination of faradaic currents from charging currents.25 Because of this, SWV is becoming one of the preferred methods in the field of electrochemical sensors, where removing double-layer charging effects is fundamental to achieving improved detection limits.26 For example, SWV has been used for the interrogation of electrochemical biosensors27 employing different molecular recognition elements such as single- and double-stranded nucleic acids,28–32 non-natural nucleic acids,4 and peptides.33–35 SWV has also been used in the detection of environmental pollutants like heavy metals via electrodeposition and stripping analysis.36,37 More recently, SWV has been used to interrogate electrochemical aptamer-based (E-AB) sensors38 supporting the high-frequency, real-time measurement of plasma drug levels in situ in the living body.5,13,39
We designed SACMES with the ability to continuously process square-wave voltammograms via two methods: (1) peak current extraction, calculated as the difference between the voltammogram’s maximum current and its baseline, and (2) area under the curve (AUC), calculated using a Riemann sum of the area under each voltammogram and above its baseline (Figure 1B and Figures S1 and S2). Moreover, these methods support the simultaneous processing of multiple sensors and multiple square-wave frequencies. The results of these analyses are then plotted over time (or over file number) to produce data series. For example, a titration experiment in which an E-AB sensor is challenged with its molecular target produces curves where, as expected, peak currents and AUCs vary monotonically with increasing target concentration (Figure 1C). These curves are exported in real time into text files that, if needed, can later be analyzed to determine physicochemical parameters such as binding affinity (from a titration with the target), folding thermodynamics (from titration with a denaturant), or analyte concentrations (e.g., from a sensor monitoring drug pharmacokinetics in vivo).
The ability of SACMES to analyze square-wave voltammograms is independent of the analytical system being interrogated. We illustrate this by presenting the analysis of data collected from three systems commonly employed in our groups: (1) E-AB sensors that measure specific small-molecule targets5,45 (Figure 2A); (2) DNA-scaffold sensors detecting monoclonal antibodies15,40,41 (Figure 2B); and (3) redox-reporter-modified proteins site-specifically attached to an electrode to study surface effects on protein stability16,42 (Figure 2C). Through these data, we demonstrate the ability of SACMES to analyze measurements recorded on multiple electrodes at multiple square-wave frequencies. For example, SACMES can be used to estimate electron transfer kinetics from the analysis of peak currents extracted from voltammograms measured at over 40 frequencies using the Lovrić approach25,43,46 (Figure 2D). SACMES also supports the determination of peak currents and AUCs from voltammograms recorded on multiple electrodes simultaneously (Figure 2E) and can perform the statistical analysis of electrode-to-electrode variation in these parameters. Finally, since the shape of square-wave voltammograms can be dependent on changing experimental conditions (e.g., changes in the baseline of an E-AB sensor deployed in vivo), SACMES is equipped with baseline control that allows the user to define, in real time, the range of data points to be considered during the analysis of voltammograms (Figure 3). Thus, we have created a software that is truly adaptable to the analysis of both low and high signal-to-noise electrochemical measurements.
Figure 3.
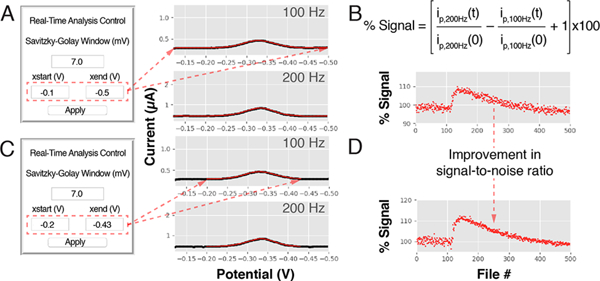
Real-time control of software parameters improves the analysis of electrochemical data. We have implemented the ability to control parameters in real time in our software, a feature that can be useful for the analysis of electrochemical measurements recorded under fluctuating conditions or for repetitive measurements performed over long experimental times in harsh conditions. (A) Here, for example, we show square-wave voltammograms recorded with a tobramycin-detecting E-AB sensor emplaced in the jugular vein of a live rat. Using our software we extracted peak currents from the voltammograms by performing (1) a five point rolling average to remove high frequency noise, (2) a polynomial regression of the data, and (3) calculating the difference between the peak current and the lowest of the two current minima (the “valleys” in the voltammograms) from the best-fit curve obtained from the regression. (B) We serially interrogated the sensor every 10 s over 80 min at square-wave frequencies of 100 and 200 Hz. The visualization module produced graphs of the relative signal change. (C) By adjusting the data range analyzed from each voltammogram considered in our analysis (red line limited by the red arrows in panel C), we corrected for poor voltammogram baselines and (D) improved the signal-to-noise ratio of our measurements 2-fold.
While the ability to process multielectrode, multifrequency data should be of significant value for the interrogation of many electrochemical systems, SACMES is uniquely suited for the processing of rapid, time-dependent measurements. In its current form, the program’s Tkinter loop searches for files at speeds as fast as 3 ms per cycle, enabling the continuous, real-time processing of data files even for applications demanding a high temporal resolution. To achieve these speeds, we implemented Python’s “High-Level Threading Interface20” in our code, which allows SACMES to run multiple tasks in parallel (as opposed to serially) to dramatically improve processing speed and GUI responsiveness without relying on expensive computers. For example, the program supports the real-time analysis of chronoamperometrically interrogated E-AB sensors sampled as frequently as every few hundred milliseconds6 (Figure 4). To support the analysis of chronoamperometric measurements, we modified the Data Analysis module to (1) read chronoamperograms, as opposed to voltammograms, and (2) perform regression analysis between the chronoamperograms and a monoexponential function6,47 to determine, from the best fit, the current decay time constants from every measurement. Finally, we modified the Animation Class to display each chronoamperogram in a log–log format, which facilitates the analysis of current and time data covering many orders of magnitude. The program for the analysis of the chronoamperometric measurements can be downloaded from the link: https://github.com/netzlab/SACMES.git. In the same link we have also incorporated an example program for the continuous processing of cyclic voltammograms.
Figure 4.
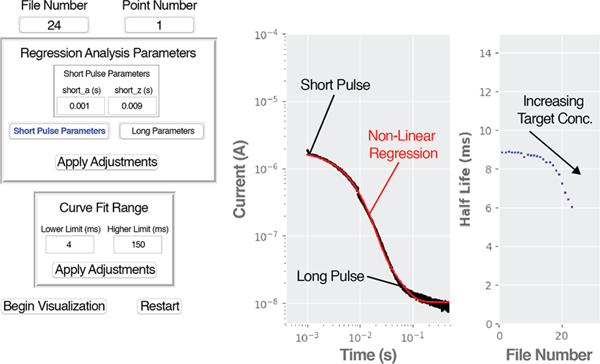
With a processing speed of a few milliseconds per cycle, our program enables the real-time analysis of data acquired at fast rates. To illustrate this, we modified our code so it would read chronoamperograms, as opposed to voltammograms. For each measurement, we merged two chronoamperograms (one lasting 10 ms and a second one lasting 500 ms) which we acquired with the same sampling frequency (one point every 10 μs) but different “current sensitivity” so we could resolve with high signal-to-noise ratio decays spanning 4 orders of magnitude in current. We continuously recorded these two-pulse, merged chronoamperograms every 500 ms from a tobramycin-binding E-AB sensor. The program acquired these measurements, merged them according to the limits set by the user in real time in the “Regression Analysis Parameters” box, visualized them as a single trace, and fit them to a monoexponential decay with range limits set in the “Curve Fit Range” box. The right-hand panel shows how the half-life of the current decay decreases monotonically with increasing tobramycin concentrations. This decay occurs because the sensor’s aptamer folds into a more compact conformation upon drug binding, which increases the efficiency of electron transfer from the redox reporter.6
Besides achieving high processing speeds, SACMES supports constant-speed analysis and animation of large data volumes collected over extended periods of time without building temporary (RAM) memory. To achieve this, we integrated a computer graphics technique called “Blitting”.11 Whereas most image-rendering techniques require the rendering of an entirely new image for each data file, Blitting saves a clean background canvas, or “bit map”, in which only new data is rendered, thus keeping the time needed to visualize each file constant. Whether SACMES analyzes a single file or thousands, the speed of analysis under a specific set of technique parameters is thus constant, a feature that enables the visualization of measurements taken over extended periods without the need for high-performance computers. To illustrate this, we ran two experiments: (1) monitoring the output of a tobramycin-detecting E-AB sensor (measurement frequency of 20 s) alternatively challenged every 30 min with either tobramycin-containing serum or drug-free serum over 14 h (Figure 5A) and (2) the continuous monitoring of signaling current, every 2 min, from an electrode functionalized with redox-reporter-modified protein and immersed in a solution that unfolds the protein (10 M urea) for 24 h (Figure 5B). These experiments illustrate the ability of SACMES to uninterruptedly and continuously analyze data every few seconds over the course of hours or days.
Figure 5.
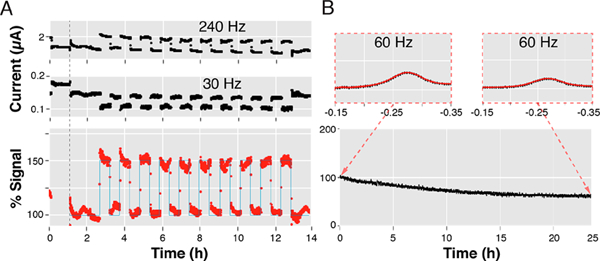
Continuous analysis and visualization of long-duration electrochemical data without building temporary memory. By incorporating into our code high-efficiency visualization techniques such as “blitting” we have created a program that performs data analysis at a constant-speed. (A) To illustrate this, we have followed, in real time, the signaling of a tobramycin-detecting E-AB sensor interrogated every 20 s over 14 h as it was alternatively challenged every 30 min with either tobramycin-containing serum or drug-free serum. The % Signal was calculated via the equation shown in Figure 3B, except using square-wave frequencies 240 and 30 Hz. (B) This ability is ideally suited to evaluate sensor stability, as shown for an electrode functionalized with redox-reporter-modified protein and immersed in a solution containing 10 M denaturant (urea) that we interrogated every 2 min over the course of 25 h.
CONCLUSIONS
Here we describe an open-source program that supports the efficient, customizable, real-time analysis of high-volume electrochemical data. This software, nicknamed SACMES, can easily be used to conduct and control data processing and visualization in real-time, giving the user the ability to dynamically adapt processing parameters “on the fly.” To make SACMES widely applicable and easy to operate we provide an SOP (Manual), sample data, and tutorial videos as Supporting Information. We recognize that even after extensive debugging, the program may need improvement; thus, we also include online access to a GitHub folder (https://github.com/netzlab/SACMES.git) where we will continue to post code fixtures and updates. This folder also contains an open-source license for free software distribution. We encourage the scientific community to actively use and freely distribute our code and to reach out to us with script improvements. We hope this work will further promote the sharing of data processing software in the field of electrochemical biosensors.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health (Grant R01GM125789). N.A.C. thanks Oak Ridge Associated Universities for granting a 2019 Ralph E. Powe Junior Faculty Enhancement Award to support this work.
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.9b02553.
Fully documented SOP explaining how to use, customize, and debug the program (PDF)
Video tutorial on how to use SACMES (MP4)
Video tutorial on installing the Anaconda Distribution (MP4)
Video tutorial on using the Anaconda Prompt (MP4)
Video tutorial on downloading SACMES from GitHub (MP4)
Video tutorial on using Atom Text Editor to edit SACMES (MP4)
Square-wave voltammogram sample data (ZIP
Cyclic voltammogram sample data (ZIP)
The authors declare no competing financial interest.
We recognize that even after extensive debugging, the program may need improvement; thus, we also include online access to a GitHub folder (https://github.com/netzlab/SACMES.git) where we will continue to post code fixtures and updates. This folder also contains an open-source license for free software distribution. We encourage the scientific community to actively use and freely distribute our code and to reach out to us with script improvements.
REFERENCES
- (1).Redmond SJ; Lovell NH; Yang GZ; Horsch A; Lukowicz P; Murrugarra L; Marschollek M. Yearb Med. Inform 2014, 23, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Olczuk D; Priefer R. Diabetes Metab Syndr 2018, 12, 181–187. [DOI] [PubMed] [Google Scholar]
- (3).Liebl A; Henrichs HR; Heinemann L; Freckmann G; Biermann E; Thomas AJ Diabetes Sci. Technol 2013, 7, 500–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Medtronic. Continuous Glucose Monitoring, https://www.medtronicdiabetes.com/treatments/continuous-glucose-monitoring (accessed April 12, 2019).
- (5).Arroyo-Curras N; Somerson J; Vieira PA; Ploense KL; Kippin TE; Plaxco KW Proc. Natl. Acad. Sci. U. S. A 2017, 114, 645–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Arroyo-Curras N; Dauphin-Ducharme P; Ortega G; Ploense KL; Kippin TE; Plaxco KW ACS Sens 2018, 3, 360–366. [DOI] [PubMed] [Google Scholar]
- (7).Santos-Cancel M; Lazenby RA; White RJ ACS Sens 2018, 3, 1203–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Vieira PA; Shin CB; Arroyo-Currás N; Ortega G; Li W; Keller AA; Plaxco KW; Kippin TE Ultra-High-Precision, in-vivo Pharmacokinetic Measurements Highlight the Need for and a Route Toward More Highly Personalized Medicine. Front. Mol. Biosci 2019, DOI: 10.3389/fmolb.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).The MathWorks Inc. MATLAB, R2018b; Natick, MA, 2019. [Google Scholar]
- (10).Weiss CJ Chem. Eng. News 2017, 95, 30–31. [Google Scholar]
- (11).Julio Sanchez MPC, Displaying Bit-Mapped images In Software Solutions for Engineers and Scientists; CRC Press, 2007. [Google Scholar]
- (12).Arroyo-Curras N; Scida K; Ploense KL; Kippin TE; Plaxco KW Anal. Chem 2017, 89, 12185–12191. [DOI] [PubMed] [Google Scholar]
- (13).Arroyo-Currás N; Ortega G; Copp DA; Ploense KL; Plaxco ZA; Kippin TE; Hespanha JP; Plaxco KW ACS Pharmacol. Transl. Sci 2018, 1, 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).IACUC. Guide for the Care and Use of Laboratory Animals,8th ed.; The National Academies Press, 2011. [PubMed] [Google Scholar]
- (15).Cash KJ; Ricci F; Plaxco KW J. Am. Chem. Soc 2009, 131, 6955–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Kurnik M; Ortega G; Dauphin-Ducharme P; Li H; Caceres A; Plaxco KW Proc. Natl. Acad. Sci. U. S. A 2018, 115, 8352–8357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Van Rossum G. Python, https://www.python.org/ (accessed March 28, 2019).
- (18).Open Source Initiative. Python License (Python-2.0), https://opensource.org/licenses/Python-2.0 (accessed March 26, 2019).
- (19).For example: Anaconda Environment, https://www.anaconda.com/distribution/.
- (20).Van Rossum G. The Python Standard Library, https://docs.python.org/3/library/ (accessed March 20, 2019).
- (21).Van Rossum G. Python Package Index, https://pypi.org/ (accessed March 20, 2019).
- (22).Van Rossum G. Python: Useful Modules, https://wiki.python.org/moin/UsefulModules (accessed March 20, 2019).
- (23).Jones EOE; Peterson P. et al. SciPy: Open Source Scientific Tools for Python, http://www.scipy.org/ (accessed March 21, 2019).
- (24).Oliphant TE A Guide to NumPy; Trelgol Publishing, 2006. [Google Scholar]
- (25).Mircčeski V, Komorsky-Lovrić S, Lovrić M. Square-Wave Voltammetry: Theory and Application; Springer: Berlin, Germany, 2007. [Google Scholar]
- (26).Chen AC; Shah B. Anal. Methods 2013, 5, 2158–2173. [Google Scholar]
- (27).Liu X; Duckworth PA; Wong DK Biosens. Bioelectron 2010, 25, 1467–73. [DOI] [PubMed] [Google Scholar]
- (28).Ikeda R; Kobayashi S; Chiba J; Inouye M. Chem. - Eur. J 2009, 15, 4822–8. [DOI] [PubMed] [Google Scholar]
- (29).Lubin AA; Plaxco KW Acc. Chem. Res 2010, 43, 496–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Ricci F; Lai RY; Plaxco KW Chem. Commun. (Cambridge, U. K.) 2007, 0, 3768–70. [DOI] [PubMed] [Google Scholar]
- (31).Jolly P; Batistuti MR; Miodek A; Zhurauski P; Mulato M; Lindsay MA; Estrela P. Sci. Rep 2016, 6, 36719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Das J; Ivanov I; Safaei TS; Sargent EH; Kelley SO Angew. Chem., Int. Ed 2018, 57, 3711–3716. [DOI] [PubMed] [Google Scholar]
- (33).Thompson CC; Lai RY ECS Meeting Abstracts 2017, 1938. [Google Scholar]
- (34).González-Fernández E; Staderini M; Avlonitis N; Murray AF; Mount AR; Bradley M. Sens. Actuators, B 2018, 255, 3040–3046. [Google Scholar]
- (35).Puiu M; Idili A; Moscone D; Ricci F; Bala C. Chem. Commun. (Cambridge, U. K.) 2014, 50, 8962–5. [DOI] [PubMed] [Google Scholar]
- (36).Wojclechowski M; Go W; Osteryoung J. Anal. Chem 1985, 57, 155–158. [Google Scholar]
- (37).Feldman BJ; Osterloh JD; Hata BH; D’Alessandro A. Anal. Chem 1994, 66, 1983–7. [DOI] [PubMed] [Google Scholar]
- (38).Xiao Y; Lubin AA; Heeger AJ; Plaxco KW Angew. Chem., Int. Ed 2005, 44, 5456–9. [DOI] [PubMed] [Google Scholar]
- (39).Ferguson BS; Hoggarth DA; Maliniak D; Ploense K; White RJ; Woodward N; Hsieh K; Bonham AJ; Eisenstein M; Kippin TE; Plaxco KW; Soh HT Sci. Transl. Med 2013, 5, 213ra165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Fetter L; Richards J; Daniel J; Roon L; Rowland TJ; Bonham AJ Chem. Commun. (Cambridge, U. K.) 2015, 51, 15137–40. [DOI] [PubMed] [Google Scholar]
- (41).Kang D; Parolo C; Sun S; Ogden NE; Dahlquist FW; Plaxco KW ACS Sens 2018, 3, 1271–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Ortega G; Kurnik M; Dauphin-Ducharme P; Li H; Arroyo-Curras N; Caceres A; Plaxco KW Angew. Chem., Int. Ed 2019, 58, 1714–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dauphin-Ducharme P; Plaxco KW Anal. Chem 2016, 88, 11654–11662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Goodsell DS Oncologist 2001, 6, 547–8. [DOI] [PubMed] [Google Scholar]
- (45).Somerson J; Plaxco KW Molecules 2018, 23, 912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Mirčeski V; Lovrić M. Electroanalysis 1997, 9, 1283–1287. [Google Scholar]
- (47).Dauphin-Ducharme P; Arroyo-Curras N; Kurnik M; Ortega G; Li H; Plaxco KW Langmuir 2017, 33, 4407–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.